Abstract
Tumor necrosis factor (TNF) superfamily cytokines play major roles in the regulation of both adaptive and innate immunity. The TNF superfamily cytokine TL1A (TNFSF15) through its cognate receptor DR3 (TNFRSF25) promotes T cell immunity to pathogens and directly costimulates group 2 and 3 innate lymphoid cells. Polymorphisms in the TNFSF15 gene are associated with risk for various human diseases including inflammatory bowel disease. Like other cytokines in the TNF superfamily, TL1A is synthesized as a type II transmembrane protein and cleaved from the plasma membrane by metalloproteinases. Membrane cleavage has been shown to alter or abrogate certain activities of other TNF-family cytokines, but for TL1A, the functional capabilities of membrane bound and soluble forms of this cytokine are not known. Constitutive expression of TL1A in transgenic mice results in expansion of activated T cells and promotes intestinal hyperplasia and inflammation through stimulation of group 2 innate lymphoid cells. Through the generation of membrane restricted TL1A transgenic mice, we demonstrate that membrane TL1A promotes expression of inflammatory cytokines in the lung, dependent on DR3 expression on T cells. Soluble TL1A alone was unable to produce this phenotype but was still able to induce intestinal type 2 inflammation independently of T cells. These data suggest differential roles of membrane and soluble TL1A on adaptive and innate immune cells, and have implications for the consequences of blocking these two forms of TL1A.
Introduction
Interactions between the TNF superfamily cytokine TL1A and its cognate receptor DR3 play a role in multiple murine models of autoimmune and autoinflammatory diseases. Genetic polymorphisms in the TNFSF15 locus encoding TL1A have also been associated with risk for development of inflammatory bowel disease and elevated levels of TL1A have been found in both the serum and synovial fluid of patients with rheumatoid arthritis [1, 2].
TL1A, like other TNF superfamily cytokines, is initially produced as a type 2 transmembrane protein which is subsequently cleaved to yield a soluble product [3]. Membrane cleavage alters the function of other cytokines in the TNF superfamily. For example, membrane restriction of murine TNF results in an attenuated response to Listeria monocytogenes, LPS immunization, and experimental allergic encephalomyelitis, demonstrating a unique role of soluble TNF in host defense and autoimmunity [4, 5]. However, membrane restricted TNF can still support the formation of lymphoid follicle structures that are defective in TNF-deficient mice, suggesting distinct functions are mediated by membrane bound and soluble forms of TNF [5]. The TNF superfamily member Fas ligand (FasL) induces apoptosis more efficiently when restricted to the membrane. However, soluble FasL may have its own unique function, as mice with FasL that can only be produced in a soluble form show increased mortality compared to FasL-deficient mice due to increased autoimmunity and myeloid malignancies [6].
Expression of full-length human TL1A in 293T cells results in its cleavage at leu72 [3]. In addition, soluble endogenous TL1A can be detected in cell culture supernatants of ex vivo stimulated DCs and monocytes [7] and the serum of patients with rheumatoid arthritis [2]. Work by Hedl et al. has suggested that a potential protease responsible for this cleavage is ADAM17 [8]. Further work has also demonstrated the ability of both membrane restricted [9] and soluble [10, 11] forms of TL1A to be able to signal in some capacity in vitro. Which forms of TL1A are biologically active in vivo has not been addressed.
We and others have previously shown that transgenic mice constitutively expressing full-length TL1A either on T cells or dendritic cells all show a similar phenotype with only severity varying between the strains, commensurate with TL1A expression levels [12–14]. TL1A transgenic mice spontaneously develop IL-13-dependent small bowel pathology, systemic T cell activation and an increase in activated T cells and regulatory Foxp3+ CD4+ T cells. The small-intestinal pathology in these mice has been shown to result from IL-13 produced by group 2 innate lymphoid cells (ILCs) upon TL1A stimulation [15–17]. Whether the T cell subset expansions are due to direct or indirect actions of TL1A on T cells is not known, nor is it understood which of these effects of TL1A are due to membrane or soluble forms of the cytokine.
Here we show that TL1A transgenic mice produce both soluble and membrane bound forms of TL1A. We produce a membrane restricted TL1A transgenic mouse, and show that constitutive expression of this form of TL1A only mildly attenuates small bowel pathology. However we find that these mice have increased inflammatory cytokine levels in the lungs. We further show that chronic administration of soluble TL1A is able to induce Il-13 driven pathology in the ileum, but not T cell hyperactivation or lung pathology. Finally, using mice specifically lacking DR3 on T cells, we demonstrate that the lung and T cell activation phenotypes, but not small intestinal pathology, are dependent upon T cell-specific expression of the TL1A receptor DR3. These findings define distinct roles of membrane bound vs. soluble TL1A in the immune system and have implications for the design of therapeutics that block the effects of TL1A.
Methods
Molecular biology
Murine TL1A was cloned from the vector TL1A pCDNA3 (Kind gift from Dr J Mongkolsapaya, MRC, Oxford, UK). The base pairs corresponding to amino acids 69 – 93 were removed (TL1A Δ69-93) using a sewing PCR and the resulting product was cloned, along with an unaltered TL1A, into pEYFP-C1 using Kpn1 and BamH1. For production of the Mem TL1A transgenic vector, TL1A Δ69-93 was subcloned into the CD2 expression vector [18] using EcoR1 after dephosphorylation with TSAP.
Cell culture and transfections
293T cells were transfected using Effectene (Qiagen) with the indicated plasmids and grown in complete DMEM composed of DMEM (Life Technologies) supplemented with 10% FCS, 1 x sodium pyruvate, 1 x GlutaMAX, and 1 x penicillin-streptomycin solution (Sigma). All other cells were cultured in complete RPMI composed of RPMI (Life Technologies) supplemented as complete DMEM with the addition of 52μM 2-ME. Total T cells were purified from the spleens of indicated mice after red cell lysis using the EasySep Mouse T cell isolation Kit (Stemcell Technologies). For activation assays, cells were either cultured with 10ng/ml IL-7 (Peprotech) or 2μg/ml anti-CD3 (145-2c11, eBioscience) and 10μg/ml anti-CD28 (37-51, eBioscience) for up to 48hrs. BMDCs were generated by culturing the bone marrow from indicated mice at 2x105 cells/ml in complete RPMI with 20ng/ml GM-CSF (Peprotech). After 4 days the media was topped up with half the initial volume of the same media and the cells in suspension harvested after 7-8 days. Expression of CD11c was confirmed by flow cytometry (anti-CD11c, N418, eBioscience). For activation assays, 0.5x106 cells were cultured in 1 ml in a 48-well plate with either 20ng/ml GM-CSF alone or with the addition of 100ng LPS (Enzo) for 48hrs.
Custom TL1A Luminex
Anti-TL1A (5G4.6 [12], purified at BioXcell) was conjugated to Bio-plex Xmap luminex beads and 2.5x103 beads were used per sample. Anti-TL1A (Tan 2-2 [19], University of Southampton) was biotinylated (EZ-Link Sulpho NHS biotin, Thermo Fisher Scientific) and used as detection at 2μg/ml. Recombinant TL1A (R&D Systems) was used as the standard. Assays were carried out on a Bioplex 200 or Bioplex Magpix using the manufactures buffers (Biorad).
Mice
WT C57NL/6-Jax mice were bought from Taconic and used for assays involving only WT cells, as breeders for transgenic lines, and as recipients of osmotic pumps. For all other experiments, WT littermate mice were used. Mem+Sol TL1A transgenic mice were from the existing colony, line R6, housed at the National Institutes of Health [12]. Mem TL1A transgenic mice were produced by injection of the CD2-TL1A Δ69-93 plasmid described above at Transgenic Mouse Model Laboratory (SAIC-Frederick). Samples were taken from the transgenic mice for phenotyping at 12 weeks of age (+/- 3 days). Prior to extraction of organs, mice were perfused with PBS under terminal anesthesia. For chronic soluble TL1A experiments, mice were implanted with osmotic pumps (model 2006, Alzet) set to deliver 500ng of soluble TL1A per day via continuous infusion. Osmotic pumps were implanted using sterile technique subcutaneously on the back of the animal. Osmotic pumps remained in the animals for 6 weeks, at which point they were euthanized and samples taken. Soluble TL1A used in these experiments was produced as previously described [11]. DR3 FL/FL mice were produced by Ozgene. LoxP sites were inserted into intron 1 and 6 of the Tnfrsf25 locus. Mice were subsequently crossed to the Mem TL1A transgenic and CD4-cre (Jackson Laboratory, stock number 017336) mouse lines. All animals were used under protocols approved by the National Institute of Arthritis and Musculoskeletal and Skin Diseases animal care and use committee.
Flow cytometry
Flow cytometric analysis was carried out on the spleens of the indicated animals. Spleens were passed through a 100μm cell strainer (Corning) and the red cells lysed. Samples were resuspended, FC receptors blocked (2.4G2, eBioscience) and surface antigens stained in HBSS (Life Technologies) alone or PBS (Life technologies) plus 0.5% (w/v) BSA (Sigma). The Foxp3 Transcription Factor Staining Buffer Set (eBioscience) was used for intracellular staining. The following stains and antibodies were used: Live/dead (Life Technologies), anti-CD4 (GK1.5, Tonbo Biosciences), anti-CD8 (53-67, Tonbo Bioscience), anti-CD11c (N418, eBioscience), anti-CD19 (6D5, eBioscience), anti-CD25 (PC61.5, eBioscience), anti-CD40L (MR1, eBioscience), anti-CD44 (IM7, eBioscience), anti-CD62L (MEL14, eBioscience), anti-CXCR5 (2G8, BD Bioscience), anti-FoxP3 (FJK-16s, eBioscience), PNA (Vector Laboratories), anti-GL7 (GL-7, eBioscience), anti-ICOS (7E.17G9, eBioscience), anti-IgD (11-26c, eBioscience), anti-IgM (eb121-15F9), anti-PD1 (J43, eBioscience), anti-DR3 (BAF2437, R&D) and anti-TL1A (Tan2-2, University of Southampton [19]). Samples were run for analysis on a BD FACSVerse and sorted on a BD FACS Aria II. Flow cytometrey data was analyzed with Flow Jo V9.
Histological processing and assessment
Organs for histological processing were taken after perfusion and immersed in 10% neutral buffered formalin. Samples were processed for paraffin sections and stained with H and E or PAS stain at Histoserv (MD, USA). For histological scoring, samples were blinded and the number of goblet cells were counted on at least 3 in-plane villi per section.
RNA and DNA extraction
For RNA extraction from tissues, the postcaval lobe of the lung or a small section of the terminal ileum was collected into RNALater (Ambion). Tissue was retrieved from the storage solution, placed in Purelink RNA mini kit lysis buffer (Ambion) and homogenized using a Precellys homogenizer (Bertin). RNA was extracted using a Purelink RNA mini kit (Ambion). To extract genomic DNA, cells were lysed and the RNA extracted using a Purelink RNA mini kit (Ambion), following elution of RNA from the column, 50μl of 8mM NaOH was added and incubated at 55°C for 7min. DNA was eluted by spinning the column at 5000xg for 3 min at room temperature. 260:280 ratio was assessed using a Nano Drop spectrophotometer.
Quantitative PCR
250ng-500ng of RNA per reaction was assessed by qPCR using iTaq universal probes one step kit (Bio-Rad). Relative expression of transcripts was calculated by comparison to HPRT using the ΔΔCT method [20]. The following probes (Applied Biosytems) were used; HPRT (Mm00446968_m1), IL17a (Mm00439619_m1), IFNg (Mm00801778_m1), IL-13 (Mm00487160_m1), IL-4 (Mm00445259_m1), IL-9 (Mm00434305_m1), IL-5 (Mm00439646_m1). Reactions were run on a CFX384 (Bio-Rad) instrument for 45-50 cycles.
ELISA
ANA ELISAs were carried out as per manufactures instructions (Alpha Diagnostics). For other ELISAs Maxisorp (Nunc) plates were coted with either anti-IgG (1037-01, Southern Biotech), anti-IgA (1040-01, Southern Biotech) or anti-IgE (1110-01, Southern Biotech) at 0.2μg/ml in bicarbonate buffer overnight. Plates were blocked in PBS (Life Technologies) with 1% (w/v) BSA (Sigma) for 2 hours. PBS/BSA was used for all subsequent steps. Samples were diluted and added for 1 hour and secondary antibody for a further hour. ELISAs were developed with TMB substrate (Biolegend) and reactions stopped with 2N H2SO4 (Sigma). Between steps, plates were washed 3 times using a Synergy H1M (BioTek). Absorbance at 450nm was read using an Epoch Spectrophotometer (BioTek). HRP-conjugated secondary antibodies (used at 1:12000) were as follows: anti-IgG1 (1070-05, Southern Biotech), anti-IgG2b (1090-05, Southern Biotech), anti-IgG2c (1079-05, Southern Biotech), anti-IgA (1040-05, Southern Biotech) and anti-IgE (1110-05, Southern Biotech). For dilution factor calculation, mouse serum was diluted 1/10 and added to the top well. Samples were serial diluted 1/10 and a curved fitted to the absorbance against the dilution of the sample where a 1/100 absolute dilution is equal to 0.01. The dilution factor was calculated as the inverse of the dilution where the absorbance is equal to twice the background of a blank well (PBS/BSA in place of serum). For samples which never reached this (eg IgE in WT mice) a dilution factor of 1 was assigned.
RNA-Seq
mRNA was purified from 1μg of total RNA extracted as above using a NEBNext Poly(A) mRNA Magnetic isolation module (New England Biolabs). First and second strand cDNA was synthesized using NEBNext synthesis modules (New England Biolabs), purified using Agencourt AMPure XP beads (Beckman Coulter), and quantified using Quant-iT PicoGreen dsDNA assay (Life Technologies). Libraries were prepared from 10ng ds-cDNA using an Ovation SP ultralow library system (Nugen) in combination with a Mondrian SP work station (Nugen). Libraries were amplified by PCR for 15 cycles, quantified, denatured and 10pmol of each library were pooled and sequenced on a 1x50bp program on a Hiseq 2000 or 2500 (Illumina). After sequencing, the BCL file was demulitiplexed and converted to a FASTQ file using CASAVA (Illumina). Files were aligned to the mm10 UCSC genome using TopHat splice junction mapper with Bowtie2 [21]. Features were counted and a count table produced using Rsubread[22]. Raw and processed data including batch information and output from differential expression analysis with DESeq2 is available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo) under series record GSE98492.
Statistics
For non RNA-Seq data, statistics were calculated using GraphPad Prism with the test indicated in the figure legend. For RNA-Seq data, differential expression analysis was carried out using DESeq2 including sex and sequencing batch as covariates in the liner model [23]. GSEA was performed by ranking genes in each comparison by the inverse of the unadjusted p value with the sign of the log fold change calculated by DESeq2. The generated ranked gene lists were tested for enrichment of gene sets defined in the Hallmarks database from MSigDB using GSEA [24].
Results
Cell type-specific and molecular requirements for TL1A cleavage
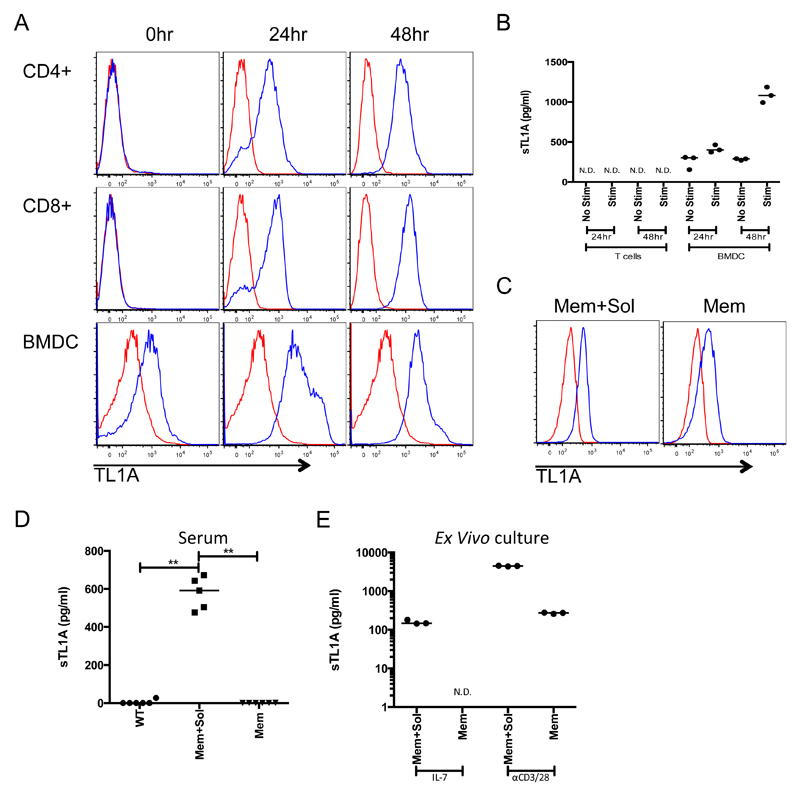
Given previous reports of TL1A cleavage [3] we investigated the production of membrane and soluble forms of TL1A from murine splenic T cells and bone marrow derived dendritic cells (BMDCs). Using flow cytometry, we found expression of surface TL1A on CD4+ and CD8+ T cells by 24 hours after activation through CD3 and CD28. BMDCs spontaneously expressed surface TL1A, which increased upon treatment with LPS (Figure 1A). In the same cultures, soluble TL1A was detectable only in BMDCs, with increased amounts upon stimulation (Figure 1B). These data show that soluble TL1A is preferentially produced by DCs, whereas both T cells and DCs can express membrane bound TL1A, particularly after activation.
Figure 1. TL1A is differentially cleaved dependent upon the cell of origin.
A Total T cells were purified from C57BL/6 mouse spleens and activated with anti CD3 and anti CD28 for 48 hours. BMDC were activated with LPS. Surface expression of TL1A was measured by flow cytometry at the indicated time points with anti TL1A (blue line) and compared to an isotype control (red line). B Supernatants were taken from cells stimulated as in A (Stim) or cultured for the same amount of time (No Stim) without activation (IL-7 for T cells or GM-CSF for BMDCs) and levels of soluble TL1A measured. C Splenic CD4+ naive T cells from Mem+Sol and Mem TL1A transgenic mice. TL1A surface expression was compared as for A on naive (CD62LhighCD44lowCD25-) CD4+ T cells. D Levels of TL1A in the serum from 12 week-old TL1A transgenic mice and WT controls. E CD4+ transgenic T cells were activated as in A for 48 hours and soluble TL1A was measured in the supernatant. Bar indicates median. ** 0.01>P>0.001 using Mann Whitney test. N.D. not detected.
We have previously described transgenic mice that constitutively express TL1A in T cells under the CD2 promoter [12]. Using line R6 from our original publication we found these mice had high levels of soluble TL1A detectable in their serum and on the surface of T cells (Figure 1C and D, group Mem+Sol). Because of these findings, we will refer to this line as Mem+Sol. To prevent TL1A cleavage, we deleted the mouse TL1A region homologous to the region of human TL1A previously identified as the site of cleavage [9] (TL1AΔ69-93, Supplemental Figure 1A). When expressed by 293T cells, TL1AΔ69-93 showed increased surface expression and greater than 250-fold reduced production of soluble TL1A compared to full length murine TL1A (Supplemental Figure 1B-C).
Soluble as well as membrane bound TL1A is required for maximal small bowel pathology
To study the effects of membrane restricted TL1A in vivo, we produced a transgenic mouse line expressing the TL1AΔ69-93 TL1A mutant under the control of the CD2 promoter. From multiple founder lines, we identified one strain in which surface TL1A levels matched those found on naive T cells from Mem+Sol TL1A transgenic mice (Figure 1C); we designated this line Mem. After 48 hours of culture with IL-7 alone, only Mem+Sol CD4+ T cells produced soluble TL1A in the culture supernatant (Figure 1E). The level of soluble TL1A was greatly increased in Mem+Sol transgenic T cells following T cell activation. However, only after activation was a small amount of soluble TL1A detectable in the supernatant from Mem cells, though it was reduced by more than 10-fold compared to the cells from Mem+Sol mice (Figure 1E). Importantly, levels of soluble TL1A in the serum of Mem transgenic mice was no higher than those of wild type (WT) mice, whereas high levels of soluble TL1A were present in the serum from Mem+Sol transgenic mice (Figure 1D).
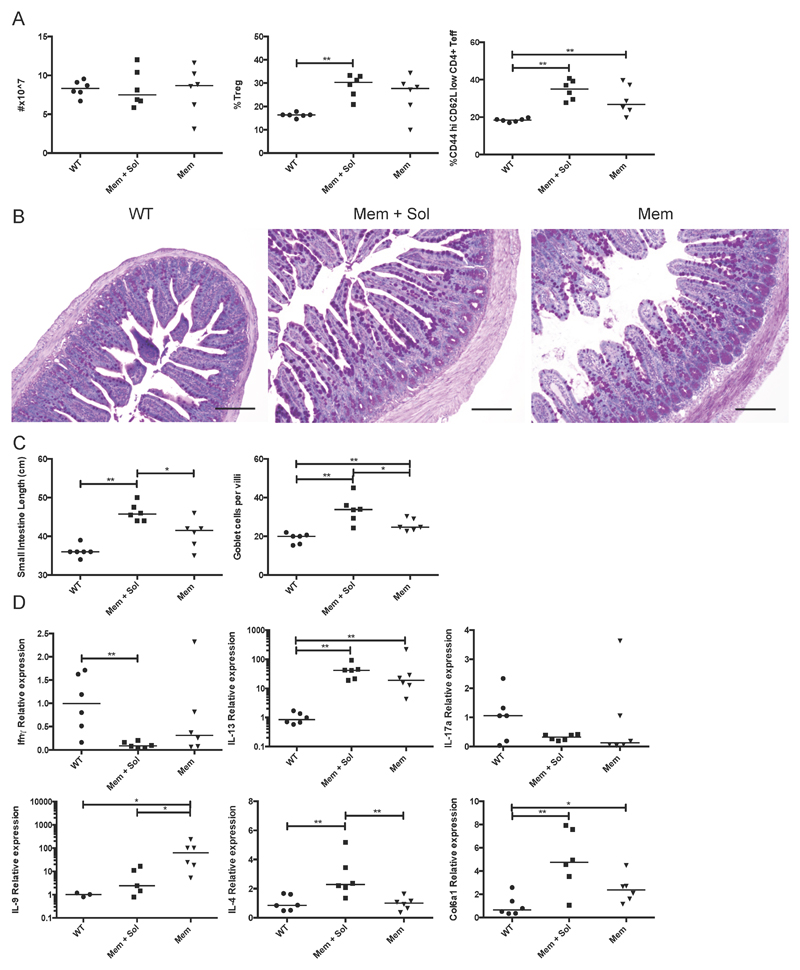
To determine the in vivo effects of membrane restricted TL1A, we examined the phenotype of the Mem and Mem+Sol TL1A transgenic mice compared to WT littermate controls at 12 weeks of age. There was a small but significant increase in the proportion of CD4+ T cells and a reduction in CD19+ B cells in the spleen of Mem+Sol but not Mem TL1A transgenic mice (Supplemental Figure 2A), with total numbers of splenocytes unaffected (Figure 2A). Recent studies have reported negative effects of TL1A on in vitro proliferation of B cells [25]. We therefore examined the B cell and T follicular helper cell compartments and found an increase in the percentage of class switched B cells that was statistically significant only in Mem mice (Supplemental Figure 2A). However, there was a significant increase in germinal center B cells in both strains. We also found significant elevation of total IgG1, IgA and IgE in Mem+Sol mice but only IgG2b, and to a lesser extent, IgA in Mem mice (Supplemental Figure 2B). Mem+Sol but not Mem TL1A transgenic mice also had mildly elevated anti-nuclear antibody titers (ANA). Similar to previous descriptions of transgenic TL1A overexpression [12–14] we found an elevation of both regulatory and activated T cells in the spleens of Mem+Sol transgenic mice (Figure 2A), which was similar in Mem mice.
Figure 2. Systemic and small bowel immune cell alterations and pathology induced by Mem vs. Mem+Sol TL1A transgenic mice.
TL1A transgenic mice from the indicated strain and WT littermates were taken at 12 weeks. A Total splenocytes were counted and the fraction of Tregs (CD4+ FoxP3+ of the CD4+ population) and effector T cells (CD44 high CD62L low CD4+ FoxP3- of the CD4+ population) were measured by flow cytometry. B Sections of the terminal ileum were stained with PAS. Scale bars are equal to 150µm. C The length of the small bowel was measured and the mean number of goblet cells calculated for at least 3 full in-plane villi per section. D Cytokine levels as assessed by qPCR in the terminal ileum. Bar indicates median. * 0.05>P>0.01 ** 0.01>P>0.001 using Mann Whitney test.
In the small intestine, Mem TL1A transgenic mice developed goblet cell and paneth cell hyperplasia reminiscent of the previous findings in Mem+Sol mice (Figure 2B). However, Mem mice had reduced lengthening of the small intestine and an fewer number of goblet cells per villus compared with Mem+Sol TL1A transgenic animals (Figure 2C). The transcriptional profile of the terminal ileum was assessed by quantitative PCR (Figure 2D). IL-13 mRNA was highly elevated in both Mem+Sol and Mem transgenic strains. However, only the Mem+Sol ileum exhibited significantly reduced levels of IFN-γ and elevated IL-4, which are cytokines primarily produced by T cells. Mem+Sol transgenic mice also had the highest expression of collagen Col6a1, indicative of increased fibrosis in these mice. Conversely, membrane restricted TL1A transgenic mice showed the greatest elevation in IL-9, a cytokine recently shown to be induced by TL1A in T cells [26]. Taken together, these data show that membrane bound TL1A is less efficient at inducing small bowel pathology than is soluble TL1A. As IL-13 levels were comparable between the two lines, soluble TL1A may promote increased severity of ileal pathology and cytokine production through other pathways or cell types besides IL-13-producing ILC2.
Membrane restricted TL1A transgenic mice have increased pro-inflammatory cytokine production in the lung
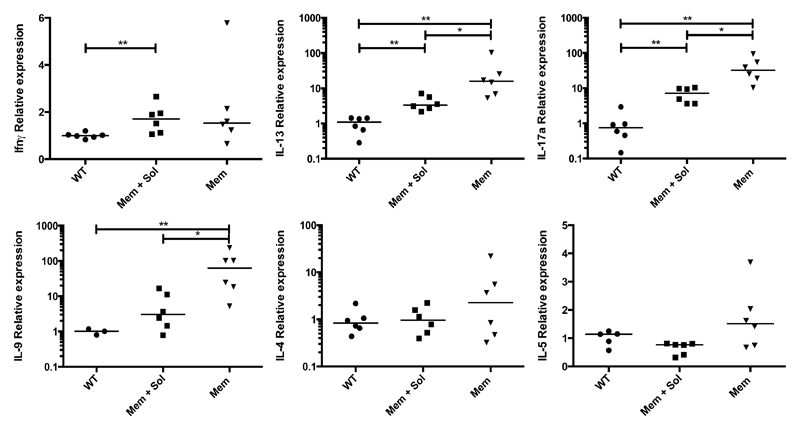
Given the role for TL1A and DR3 in mouse models of asthma [15, 26], we assessed cytokine production by Mem and Mem+Sol transgenic mice in the lungs (Figure 3). Unlike in the ileum, Mem TL1A transgenic mice were found to have higher levels of multiple inflammatory cytokines, including IL-13, IL-17a, and IL-9. We also assessed the lungs for histological changes but no consistent alterations were observed (data not shown).
Figure 3. Altered cytokine profiles in the lungs of TL1A transgenic mice.
qPCR was performed for the indicated cytokines on RNA isolated from the postcaval lobe of the lung. Bar indicates median. * 0.05>P>0.01 ** 0.01>P>0.001 using Mann Whitney test.
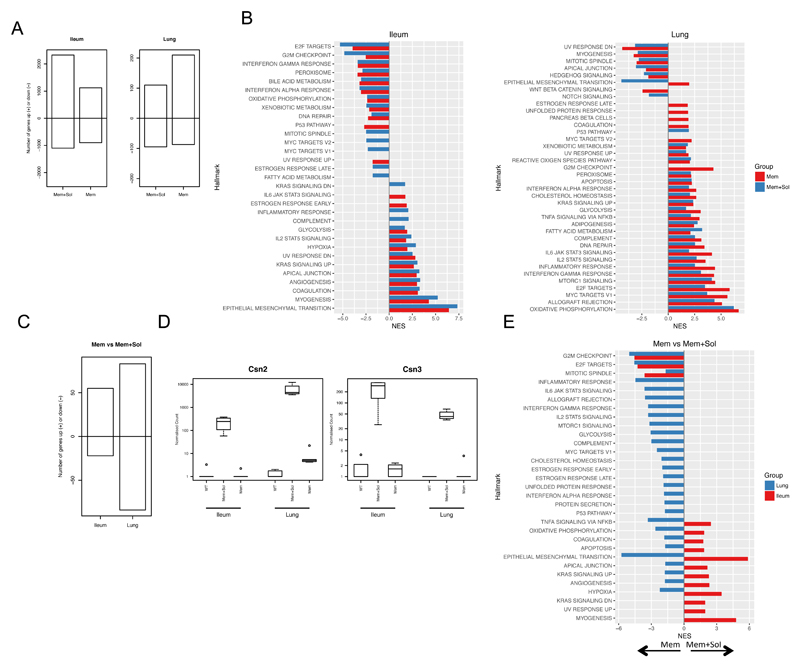
To compare the effects of an excess of either membrane restricted TL1A alone or both membrane and soluble TL1A in an unbiased manner, we performed RNA Sequencing (RNA-Seq) on the lung and the ileum from Mem and Mem+Sol mice. These were compared to non-transgenic (WT) control animals (Full results of differential gene expression analysis deposited as supplementary file in GEO series record GSE98492). In the ileum, we found that the Mem+Sol mouse had more genes with altered expression compared to WT mice than did the Mem mouse. However in the lungs the Mem strain showed greater differences (Figure 4A). Gene set enrichment analysis (GSEA) [24] indicated enrichment of different pathways in the lung compared to the ileum, with a few unique pathways upregulated by only one of the two forms of TL1A (Figure 4B). For example, genes in the interferon pathways were negatively enriched in the ileum but positively enriched in the lungs. However, there was some concordance between the organs as both lung and small intestine showed a positive enrichment for the IL-2 Stat5 signaling pathway, indicating that TL1A drives a mixture of organ dependent and independent effects (Figure 4B).
Figure 4. Transcriptional analysis of TL1A transgenic mice.
RNA-Seq was carried out on the ileum and lung from WT, Mem or Mem+Sol mice. A RNA-Seq results from Mem and Mem+Sol were compared to WT mice using DESeq2, and the number of genes with an absolute log2 fold-change in expression greater than 1 between each TL1A transgenic strain and WT were enumerated for both ileum and lung. The positive extreme of the bar indicates the number of genes upregulated and the negative downregulated in the indicated strain and tissue with respect to WT. B Genes resulting from the differential comparisons in A were ranked according to the inverse of the unadjusted p value with the sign of the log2 fold-change and a gene set enrichment analysis was run against the hallmarks data set of MSigDB. Only pathways with FDR less than 0.05 are plotted. The normalized enrichment score (NES) is plotted for each pathway relative to WT. Bars are colored according to the strain examined. C A differential expression analysis was carried out using DESeq2 for Mem+Sol relative to Mem. Numbers of genes filtered as for A were plotted in the same manner. D The normalized counts of Csn2 and Csn3 from the complete dataset. E GSEA for the differential expression analysis from C was carried out as for B. Color of the bar indicates the organ of origin. A positive NES indicates enrichment in Mem+Sol mice and a negative NES indicates enrichment in Mem mice.
As the predominant effect within the TL1A transgenic mice was based on the tissue type, we subsequently carried out differential expression analysis between the Mem and Mem+Sol groups within each organ. We found that the greatest difference between the transcriptomes induced by the two forms of TL1A was in the lung, where there was a greater number of genes with an absolute log fold change greater than 1 in the Mem+Sol TL1A compared to Mem TL1A mice (Figure 4C). However, soluble TL1A induced a number of genes independent of organ. An example of this are the Casein genes Csn2 and Csn3. Expression of these genes was found in both Ileum and Lung, but only in the Mem+Sol TL1A transgenic mice and not in WT or Mem TL1A lines (Figure 4D). To the best of our knowledge, this is the first report of induction of these genes outside of processes involved in milk production. Further, these transcripts were expressed at a similar levels by both male and female mice. To assess differential pathway enrichment in each organ between the two transgenic strains, we performed GSEA as before, relative to the Mem TL1A transgenic line (Figure 4E). Genes with a positive normalized enrichment score indicate a relative enrichment of that pathway in Mem+Sol mice and a negative score indicates relative enrichment within Mem mice. Strikingly pathway enrichment differed substantially by organ. For example both TNF signaling via NFκB and oxidative phosphorylation pathways were preferentially promoted by membrane restricted TL1A in the lung. However, in the ileum these same pathways were promoted in Mem+Sol TL1A transgenic mice (Figure 4E).
Taken together, these data indicate that although there is overlap between the transcriptional effects of soluble and membrane bound TL1A, there are some transcripts differentially induced in the ileum and lung by the same forms of TL1A, and some unique effects of each form of TL1A that can span these two target tissues.
Chronic administration of soluble TL1A promotes small bowel but not lung pathology or T cell activation
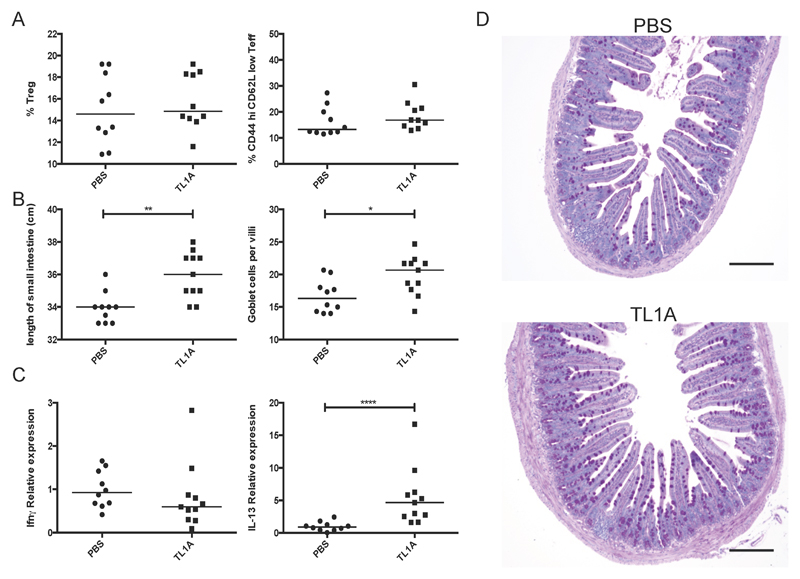
To study of the effects of excess soluble TL1A without the contribution of the membrane-bound form, 8-10 week old mice were implanted with subcutaneous osmotic pumps calibrated to deliver 500pg soluble recombinant TL1A per day (or an equivalent volume of PBS alone) at a constant rate for 6 weeks. At the end of the treatment, the mice were sacrificed and assessed for pathology. Unlike either of the transgenic strains, we found no effect of soluble TL1A on Treg numbers or the activation status of CD4+ T cells in the spleen (Figure 5A). We did however observe a mild effect on total IgA levels in the serum (Supplemental Figure 3A) and elevated IL-13 as well as IL-13-associated small bowel pathology similar to that observed in TL1A transgenic mice (Figure 5 B-D). Importantly, soluble TL1A did not induce IL-13 or IL-17 in the lung (Supplemental Figure 3B), although these were elevated in the lungs of Mem and to a lesser extent Mem+Sol TL1A transgenic mice (Figure 3). These data suggest that soluble TL1A at the concentration utilized is able to co-stimulate the production of ILC2-associated cytokines and induce small bowel pathology, but not co-stimulate T cells or replicate the lung phenotype observed in the Mem and Mem+sol TL1A transgenic mice.
Figure 5. Chronic soluble TL1A promotes small bowel but not systemic pathology.
Mice were continually infused with soluble recombinant TL1A for 6 weeks at 500 ng/day. A The Treg percentage of total CD4+ T cells and the CD44 high CD62L low percentage of CD4+ FoxP3- cells in the spleen. B The length of the small intestine was measured and sections made from the terminal ileum. These were subsequently scored for goblet cells per villi and the mean of at least 3 in-plane villi per section depicted. C qPCR analysis of cytokines from the terminal ileum. D Sections of the terminal ileum were stained with PAS; scale bars are equal to 150µm. Bar indicates median. * 0.05>P>0.01 ** 0.01>P>0.001 **** P>0.0001 using Mann Whitney test. Data combined from 2 independent experiments.
T cell specific DR3 expression is required for abnormal elevation of lung cytokines and development of a TL1A-associated splenic phenotype
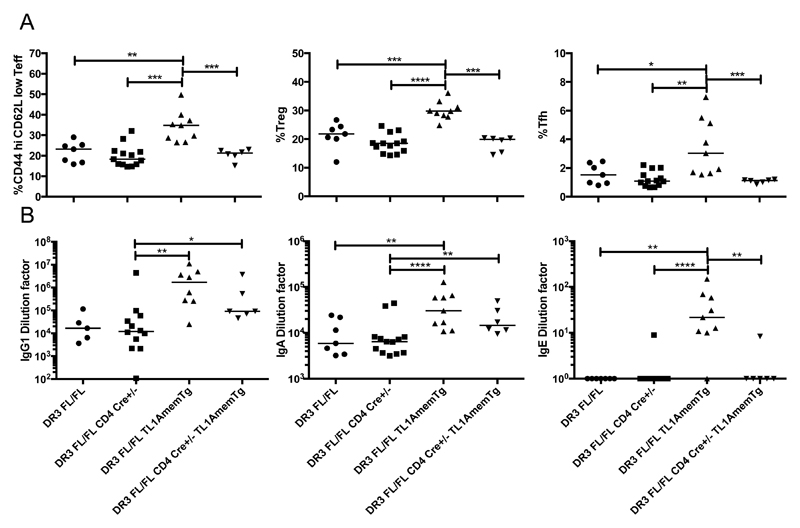
To further clarify which cell types mediate the effects of TL1A, we generated a mouse with T cell specific DR3 deficiency through insertion of loxP sites into introns 1 and 6 of the Tnfrsf25 locus and subsequent expression of Cre under the CD4 promoter. DR3 deletion could be demonstrated in the genomic DNA and also by flow cytometry of DR3 expression on CD4+ cells, but not CD19+ cells isolated from the spleen in these mice (Supplemental Figure 4). Due to Cre expression at the CD4+CD8+ double positive stage of thymocyte development, CD8+ T cells will also lack DR3. We subsequently crossed this mouse with the Mem TL1A transgenic mouse to enable us to dissect the role of membrane bound TL1A signaling via DR3 expressed on T cells (designated DR3FL/FLCD4 Cre+/- TL1AmemTg). We found that T cell activation, Treg expansion, increased numbers of T follicular helper cells, and GC B cell expansion observed in Mem TL1A transgenic mice were directly dependent upon DR3 expression in T cells (Figure 6A and Supplemental Figure 4E). T cell specific deletion of DR3 reduced serum IgE back to baseline levels, but despite the decrease in T follicular helper cells, IgG1 and IgA were only slightly reduced (Figure 6B).
Figure 6. Systemic effects conferred by direct T cell DR3 signaling in TL1A transgenic mice.
A membrane restricted TL1A transgenic strain was crossed with a T cell conditional DR3-deficient strain. Phenotypes of the indicated strains were examined at 12 weeks of age. A T cell populations were examined within the spleen. The proportion of CD44 high CD62L low cells within CD4+ FoxP3- cells was examined. Proportion of Tregs (CD4+ FoxP3-) and Tfh cells (CD4+ ICOS+ CD40L+ PD1+ CXCR5+) within total CD4+ splenocytes was examined. B Serum was collected at 12 weeks of age, and total levels of the indicated antibody isotypes examined.
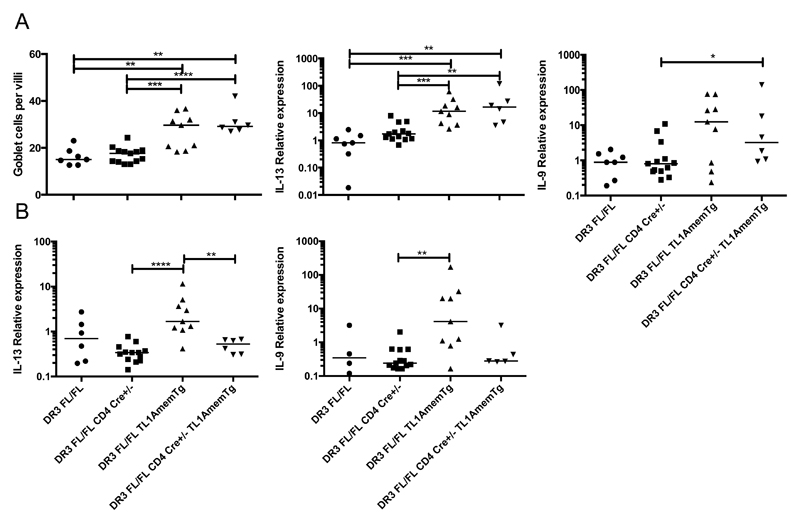
Similar to previous findings in TL1A transgenic mice crossed onto an OT-II Rag1-deficient background, which prevents TCR signaling [12], intestinal goblet cell hyperplasia and elevated IL-13 mRNA in the ileum were not affected by the lack of DR3 on T cells (Figure 7A and Supplemental Figure 4C-D). However, elevation of inflammatory cytokines in the lungs observed in both the Mem+Sol and, to a greater extent, Mem TL1A transgenic mice was lost (Figure 7B), reinforcing the role of DR3 on T cells in this aspect of TL1A-driven pathology.
Figure 7. T cell DR3 is required for pathology in the lung but not the ileum in TL1A transgenic mice.
qPCR analysis for the indicated cytokines was performed at 12 weeks of age on RNA isolated from the terminal ileum (A) and postcaval lobe of the lung (B). Bar indicates median. ** 0.01>P>0.001, *** 0.001>P>0.0001, **** P<0.0001 using Mann Whitney test.
Discussion
Here we have demonstrated that TL1A has distinct functions when expressed as a membrane-bound and soluble cytokine. TL1A cleavage is also cell-type dependent, adding another layer of regulation to the differential expression kinetics of TL1A in T cells and DCs that we have previously shown [27]. We found both cell-type and organ-specific differences in the effects of TL1A in vivo.
In the small intestine, soluble TL1A can efficiently induce type 2 inflammatory cytokines, goblet cell hyperplasia, and transcriptional responses. The effects of TL1A on small intestinal IL-13 expression and goblet cell hyperplasia are independent of DR3 expression on T cells. TL1A has previously been shown to be a potent costimulator of group 2 innate lymphoid cells leading to IL-13 production [15, 16]. Our results suggest that soluble TL1A may costimulate ILC2 more efficiently than the membrane restricted form.
In contrast to the small intestine, we report here for the first time that membrane bound TL1A can promote production of pro-inflammatory cytokines IL-13, IL-17 and IL-9 in the lung, dependent on expression of DR3 by T cells. Infusion of soluble TL1A did not cause detectable elevation of these inflammatory T-cell-dependent cytokines, indicating that increased expression of membrane bound TL1A in the absence of the soluble form of this cytokine triggers these T-cell-dependent effects. This conclusion is further supported by the greater number of differentially expressed genes found by transcriptomic analysis in lung tissue from Mem TL1A transgenic mice compared to the Mem+Sol line.
Differences in the signaling of membrane bound and soluble forms of TL1A between organs was further demonstrated by chronic delivery of soluble TL1A via an osmotic pump which induced IL-13 and goblet cell hyperplasia in the ileum but had no effect in the lung or on splenic T cells. The effect observed in the ileum was considerably less than that seen in the transgenic mice, but still present. This is most likely due to their shorter exposure to chronic soluble TL1A. Whilst this does not rule out the possibility of soluble TL1A being able to stimulate T cells, it does indicate that the phenotype observed in the ileum requires a much lower dose of soluble TL1A and suggests ILC2s have a heightened ability to respond to TL1A compared to T cells.
Interestingly, we were unable to see any evidence of the recently reported anti-proliferative effect of TL1A on B cells in either transgenic mouse strain [25]. By contrast, we saw an elevated number of germinal center B cells in the spleen as well as an elevation in the levels of multiple immunoglobulins in the serum. This increase was most apparent in the Mem+Sol TL1A transgenic line, but of those B cell phenotypes that were also elevated in the Mem TL1A transgenic line, many were reduced or ablated in the absence of DR3 on T cells. The strong increase in IgE and IgA levels in Mem+Sol TL1A transgenic mice may suggest that TL1A is able to promote Th2 skewing in vivo, though this would likely be indirect, as TL1A does not promote Th2 differentiation in vitro [26] Of note, elevated IgA was not fully resolved when T cells lacked DR3 expression. This is particularly interesting because we also found elevated IgA in mice treated with soluble TL1A, where no T cell-dependent effects were observed. Taken together, these data suggest that TL1A can promote GC B cell formation and immunoglobulin production in a predominantly T cell dependent manner, but there is some effect of TL1A acting independently of T cells on specific immunoglobulin levels. Further, this effect appears to be enhanced by the presence of soluble TL1A. At this stage, it is not possible to determine whether T cell-independent effects of TL1A on B cell phenotypes are a consequence of TL1A acting on the ileum, on another responding cell type, or directly on B cells.
There are multiple potential mechanisms by which membrane bound and soluble forms of TL1A may signal differently. A simple explanation could be spatial separation of the membrane restricted form of the ligand from responder cells. This spatial separation would be bypassed by soluble TL1A. A further reason for differential signaling may be due to differential expression of DR3 isoforms. Previous work has demonstrated that alternative splicing of DR3 occurs in response to T cell activation [28]. We have observed that the shorter isoform of DR3 that lacks exon 6 shows a reduced affinity for soluble TL1A compared to the full length DR3 isoform (unpublished observations JRF and AAS). If different cell types at rest express alternative isoforms of DR3, this may explain the observed bias for soluble TL1A to preferentially affect ILC2s. Another possible explanation for differences in the effect of stimulation could be different orders of clustering between TL1A in a cleaved soluble form and a membrane bound form, but we were unable to corroborate this possibility (data not shown). Finally, attenuation of the cytokine response in the lungs of Mem+Sol TL1A transgenic mice compared to Mem TL1A transgenic mice suggests that, at least in this setting, soluble TL1A may be able to act as a competitive inhibitor of membrane-bound TL1A signaling through DR3 on T cells. Alternatively soluble TL1A may trigger an anti-inflammatory profile through specific stimulation of Tregs or production of anti-inflammatory cytokines, although there is no difference in the proportion of splenic Tregs between the Mem and Mem+Sol TL1A transgenic mouse lines.
Overall, our results suggest that although there is overlap between the functions of membrane bound vs. soluble TL1A, membrane TL1A appears to act on T cells more efficiently in vivo, whereas soluble TL1A exerts its effects primarily through innate lymphoid cells. As therapeutic agents can be designed to differentially affect soluble and membrane bound forms of TNF and other TNF superfamily cytokines [29], a better understanding of the differential roles of membrane versus soluble TL1A in physiological and pathological immune responses may help guide development of therapeutic agents against TL1A, which have been considered in multiple diseases [1, 2].
Supplementary Material
Acknowledgements
This work utilized the computational resources of the NIH HPC Biowulf cluster. We would also like to thank the NIH veterinary staff, the flow cytometry group and Next Gen sequencing facility of the Office of Science and Technology in NIAMS. We would also like to thank Erika Hayes, Sarah Geiger, Anthony Cruz, Jane Willoughby, Sarah Buchan, Anne Rogel and Vadim Taraban for helpful discussion and technical assistance, and Amanda Bell for critical reading of this manuscript.
Funding Information
This work was jointly funded by a Wellcome-NIH PhD studentship (Awarded to JRF) and the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health. ACR was funded by the NIH-Oxford-Cambridge Scholars Program.
References
- 1.Richard AC, Ferdinand JR, Meylan F, Hayes ET, Gabay O, Siegel RM. The TNF-family cytokine TL1A: from lymphocyte costimulator to disease co-conspirator. J Leukoc Biol. 2015 doi: 10.1189/jlb.3RI0315-095R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siakavellas SI, Sfikakis PP, Bamias G. The TL1A/DR3/DcR3 pathway in autoimmune rheumatic diseases. Semin Arthritis Rheum. 2015;45(1):1–8. doi: 10.1016/j.semarthrit.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Migone TS, Zhang J, Luo X, Zhuang L, Chen C, Hu B, Hong JS, Perry JW, Chen SF, Zhou JX, Cho YH, et al. TL1A is a TNF-like ligand for DR3 and TR6/DcR3 and functions as a T cell costimulator. Immunity. 2002;16(3):479–92. doi: 10.1016/s1074-7613(02)00283-2. [DOI] [PubMed] [Google Scholar]
- 4.Alexopoulou L, Kranidioti K, Xanthoulea S, Denis M, Kotanidou A, Douni E, Blackshear PJ, Kontoyiannis DL, Kollias G. Transmembrane TNF protects mutant mice against intracellular bacterial infections, chronic inflammation and autoimmunity. Eur J Immunol. 2006;36(10):2768–80. doi: 10.1002/eji.200635921. [DOI] [PubMed] [Google Scholar]
- 5.Ruuls SR, Hoek RM, Ngo VN, McNeil T, Lucian LA, Janatpour MJ, Korner H, Scheerens H, Hessel EM, Cyster JG, McEvoy LM, et al. Membrane-bound TNF supports secondary lymphoid organ structure but is subservient to secreted TNF in driving autoimmune inflammation. Immunity. 2001;15(4):533–43. doi: 10.1016/s1074-7613(01)00215-1. [DOI] [PubMed] [Google Scholar]
- 6.O'Reilly LA, Tai L, Lee L, Kruse EA, Grabow S, Fairlie WD, Haynes NM, Tarlinton DM, Zhang JG, Belz GT, Smyth MJ, et al. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature. 2009;461(7264):659–63. doi: 10.1038/nature08402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prehn JL, Thomas LS, Landers CJ, Yu QT, Michelsen KS, Targan SR. The T cell costimulator TL1A is induced by FcgammaR signaling in human monocytes and dendritic cells. J Immunol. 2007;178(7):4033–8. doi: 10.4049/jimmunol.178.7.4033. [DOI] [PubMed] [Google Scholar]
- 8.Hedl M, Abraham C. A TNFSF15 disease-risk polymorphism increases pattern-recognition receptor-induced signaling through caspase-8-induced IL-1. Proc Natl Acad Sci U S A. 2014;111(37):13451–6. doi: 10.1073/pnas.1404178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biener-Ramanujan E, Gonsky R, Ko B, Targan SR. Functional signaling of membrane-bound TL1A induces IFN-gamma expression. FEBS Lett. 2010;584(11):2376–80. doi: 10.1016/j.febslet.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 10.Bittner S, Knoll G, Fullsack S, Kurz M, Wajant H, Ehrenschwender M. Soluble TL1A is sufficient for activation of death receptor 3. FEBS J. 2016;283(2):323–36. doi: 10.1111/febs.13576. [DOI] [PubMed] [Google Scholar]
- 11.Slebioda TJ, Rowley TF, Ferdinand JR, Willoughby JE, Buchan SL, Taraban VY, Al-Shamkhani A. Triggering of TNFRSF25 promotes CD8(+) T-cell responses and anti-tumor immunity. Eur J Immunol. 2011 doi: 10.1002/eji.201141477. [DOI] [PubMed] [Google Scholar]
- 12.Meylan F, Song YJ, Fuss I, Villarreal S, Kahle E, Malm IJ, Acharya K, Ramos HL, Lo L, Mentink-Kane MM, Wynn TA, et al. The TNF-family cytokine TL1A drives IL-13-dependent small intestinal inflammation. Mucosal Immunol. 2011;4(2):172–85. doi: 10.1038/mi.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taraban VY, Slebioda TJ, Willoughby JE, Buchan SL, James S, Sheth B, Smyth NR, Thomas GJ, Wang EC, Al-Shamkhani A. Sustained TL1A expression modulates effector and regulatory T-cell responses and drives intestinal goblet cell hyperplasia. Mucosal Immunol. 2011;4(2):186–96. doi: 10.1038/mi.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shih DQ, Barrett R, Zhang X, Yeager N, Koon HW, Phaosawasdi P, Song Y, Ko B, Wong MH, Michelsen KS, Martins G, et al. Constitutive TL1A (TNFSF15) expression on lymphoid or myeloid cells leads to mild intestinal inflammation and fibrosis. PLoS ONE. 2011;6(1):e16090. doi: 10.1371/journal.pone.0016090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meylan F, Hawley ET, Barron L, Barlow JL, Penumetcha P, Pelletier M, Sciume G, Richard AC, Hayes ET, Gomez-Rodriguez J, Chen X, et al. The TNF-family cytokine TL1A promotes allergic immunopathology through group 2 innate lymphoid cells. Mucosal Immunol. 2014;7(4):958–68. doi: 10.1038/mi.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu X, Pappu R, Ramirez-Carrozzi V, Ota N, Caplazi P, Zhang J, Yan D, Xu M, Lee WP, Grogan JL. TNF superfamily member TL1A elicits type 2 innate lymphoid cells at mucosal barriers. Mucosal Immunol. 2014;7(3):730–40. doi: 10.1038/mi.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Longman RS, Diehl GE, Victorio DA, Huh JR, Galan C, Miraldi ER, Swaminath A, Bonneau R, Scherl EJ, Littman DR. CX(3)CR1(+) mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J Exp Med. 2014;211(8):1571–83. doi: 10.1084/jem.20140678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhumabekov T, Corbella P, Tolaini M, Kioussis D. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J Immunol Methods. 1995;185(1):133–40. doi: 10.1016/0022-1759(95)00124-s. [DOI] [PubMed] [Google Scholar]
- 19.Bull MJ, Williams AS, Mecklenburgh Z, Calder CJ, Twohig JP, Elford C, Evans BA, Rowley TF, Slebioda TJ, Taraban VY, Al-Shamkhani A, et al. The Death Receptor 3-TNF-like protein 1A pathway drives adverse bone pathology in inflammatory arthritis. J Exp Med. 2008;205(11):2457–64. doi: 10.1084/jem.20072378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 23.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavallini C, Lovato O, Bertolaso A, Pacelli L, Zoratti E, Zanolin E, Krampera M, Zamo A, Tecchio C, Cassatella MA, Pizzolo G, et al. The TNF-family cytokine TL1A inhibits proliferation of human activated B cells. PLoS One. 2013;8(4):e60136. doi: 10.1371/journal.pone.0060136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richard AC, Tan C, Hawley ET, Gomez-Rodriguez J, Goswami R, Yang XP, Cruz AC, Penumetcha P, Hayes ET, Pelletier M, Gabay O, et al. The TNF-family ligand TL1A and its receptor DR3 promote T cell-mediated allergic immunopathology by enhancing differentiation and pathogenicity of IL-9-producing T cells. J Immunol. 2015;194(8):3567–82. doi: 10.4049/jimmunol.1401220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meylan F, Davidson TS, Kahle E, Kinder M, Acharya K, Jankovic D, Bundoc V, Hodges M, Shevach EM, Keane-Myers A, Wang EC, et al. The TNF-family receptor DR3 is essential for diverse T cell-mediated inflammatory diseases. Immunity. 2008;29(1):79–89. doi: 10.1016/j.immuni.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Twohig JP, Marsden M, Cuff SM, Ferdinand JR, Gallimore AM, Perks WV, Al-Shamkhani A, Humphreys IR, Wang EC. The death receptor 3/TL1A pathway is essential for efficient development of antiviral CD4(+) and CD8(+) T-cell immunity. FASEB J. 2012;26(8):3575–86. doi: 10.1096/fj.11-200618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horiuchi T, Mitoma H, Harashima S, Tsukamoto H, Shimoda T. Transmembrane TNF-alpha: structure, function and interaction with anti-TNF agents. Rheumatology (Oxford) 2010;49(7):1215–28. doi: 10.1093/rheumatology/keq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.