Abstract
The prevailing method to assess HIV-1 replication and infectivity is to measure the production of p24 Gag protein by enzyme-linked immunosorbent assay (ELISA). Since fluorescent bead-based technologies offer a broader dynamic range and higher sensitivity, this study describes a p24 capture Luminex assay capable of detecting HIV-1 subtypes A, B, C, D, circulating recombinant forms (CRF) CRF01_AE and CRF02_AG, which together are responsible for over 90% of HIV-1 infections worldwide. The success of the assay lies in the identification and selection of a cross-reactive capture antibody (clone 183-H12-5C). Fifty-six isolates that belonged to six HIV-1 subtypes and CRFs were successfully detected with p-values below 0.021; limits of detection ranging from 3.7 to 3×104 pg/ml. The intra- and inter-assay variation gave coefficient of variations below 6 and 14%, respectively. The 183-bead Luminex assay also displayed higher sensitivity of 91% and 98% compared to commercial p24 ELISA and a previously described Luminex assay. The p24 concentrations measured by the 183-bead Luminex assay showed a significant correlation (R = 0.92, p<0.0001) with the data obtained from quantitative real time PCR.
This newly developed p24 assay leverages the advantages of the Luminex platform, which include smaller sample volume and simultaneous detection of up to 500 analytes in a single sample, and delivers a valuable tool for the field.
Keywords: HIV-1, p24, detection, multi-subtype, multiplex assay
1. Introduction
HIV p24 Gag protein is a commonly used marker of HIV replication clinically and scientifically. Although monitoring HIV-1 replication in vitro can be carried out by quantification of viral nucleic acid, the most common method to assess replication and infectivity is to measure p24 production by Enzyme-Linked Immunosorbent Assay (ELISA) (Patton et al., 2006; Schupbach, 2003). p24 is the structural protein of the viral capsid whose highly conserved amino acid sequence (Coplan et al., 2005) and abundance (Summers et al., 1992; Vogt and Simon, 1999) make it an ideal candidate for HIV detection.
Commercially available p24 ELISAs are reliable, specific, sensitive and widely used in the field; however, drawbacks include a narrow dynamic range and high cost. Currently, fluorescent bead-based technologies, like Luminex, offer a broader dynamic range, higher sensitivity and lower cost (Biancotto et al., 2009). Additionally, the Luminex platform requires a smaller sample volume than the ELISA and permits the simultaneous detection of up to 500 analytes in a single sample. The ability to screen up to 1000 samples per day confers an added advantage to this technique. The core technology is based on microspheres internally dyed with fluorophores that are pre-coated with antibodies to capture analytes of interest (C-Ab; capture antibody). Detection of the bound antibody– protein complex is based on the photometric reading of the laser-excited fluorescent detection antibody that sandwiches the captured antigen. As for all immunological assays, the success relies on the avidity and specificity of the C-Abs selected. Biancotto et al. (Biancotto et al., 2009) developed a sensitive assay for the detection of p24 using the Luminex technology that has been used in many HIV-1 subtype B studies (Balzarini et al., 2013; Introini et al., 2013; Merbah et al., 2012; Parrish et al., 2013; Saba et al., 2010; Vanpouille et al., 2012). However, the C-Ab described by Biancotto et al. lacks sensitivity for some HIV-1 non-B subtypes which account for almost 90% of HIV-1 infections worldwide (Hemelaar et al., 2011). Hence, having an assay that allows the assessment of virus production regardless of the subtype is important.
Here we present the development of a Luminex based assay using a p24 capture monoclonal antibody (mAb), that detects B and non-B subtypes. Assay performance was evaluated on a panel of 56 HIV-1 isolates representing subtypes A, B, C, D, CRF01_AE and CRF02_AG, by comparing p24 concentrations measured with the Luminex assay developed by Biancotto et al. (Biancotto et al., 2009) and a commercially available p24 ELISA kit (Tang et al., 2010). Additionally, the performance of the assay was compared with real-time quantitative Polymerase Chain Reaction (qPCR) (Schupbach, 2003; Stefan et al., 2003).
2. Materials and Methods
2.1. HIV-1 isolates
The 56 virus isolates (Table 1) belong to the International Panel of HIV-1 Isolates (Brown et al., 2005) representing the six major globally prevalent strains A, B, C, D, CRF01_AE and CRF02_AG. Available through the NIH AIDS Reagent Program (Cat #11412), they constitute a resource for standardization for studies using different HIV-1 subtypes and are wildly used (Jones et al., 2012; Rosa Borges et al., 2010; Stoddart et al., 2015; Zhai et al., 2013). To ensure consistency of our measurement over time, a 500 μl or 1 ml vial of virus stock was thawed, divided into 100μl aliquots and stored at -80°C.
Table 1. List of the 56 isolates belonging to the 6 different subtypes.
| A | 00KE_KER2008 |
| 00KE_KER2018 | |
| 00KE_KNH1144 | |
| 00KE_KNH1207 | |
| 00KE KNH1209 | |
| 00KE KSM4030 | |
| 92UG_029 | |
| 99KE_KNH1088 | |
| 99KE_KNH1135 | |
| B | 84US_MNp |
| 85US_Ba-L | |
| 89BZ_167 | |
| 90TH_BK132 | |
| 90US_873 | |
| 91US-1 | |
| 91US-4 | |
| 92FR_BX08 | |
| 94US_33931N | |
| 96TH_NP1538 | |
| C | 00TZ_A125 |
| 00TZ_A246 | |
| 02ET_14 | |
| 02ET 288 | |
| 89SM 145 | |
| 90SE_364 | |
| 93MW 965 | |
| 94IN_20635-4 | |
| 98US_MSC5016 | |
| D | 00KE_NKU3006 |
| 00UG_E08364M4 | |
| 00UG E13613M4 | |
| 93UG_065 | |
| 98UG 57128 | |
| 99UG_A03349M1 | |
| 99UG_A07412M1 | |
| 99UG_A08483M1 | |
| CRF01_AE | 90TH_CM235 |
| 90TH_CM240 | |
| 90TH CM244 | |
| 96TH_M02138 | |
| 96TH_NI1046 | |
| 96TH_NI1149 | |
| 97TH NP1525 | |
| 97TH_NP1695 | |
| 98TH_NP1251 | |
| 99TH_NI1052 | |
| CRF02_AG | 01CM_0005BBY |
| 01CM_0008BBY | |
| 01CM_1475MV | |
| 01CM_CAM0002BBY | |
| 02CM_0013BBY | |
| 02CM_0014BBY | |
| 02CM_0015BBY | |
| 02CM_1970LE | |
| 91DJ_263 | |
| 98US_MSC5007 |
2.2. p24 quantification by ELISA
To measure the p24 concentration of the isolates, a commercial p24 capture ELISA kit (Advance Bioscience Laboratories, Rockville, MD, USA) was used per the manufacturers' instruction. A modified version of this ELISA was also used to test the binding abilities of the C-Ab candidates (Table 2) to the p24 of the different HIV-1 subtypes. The wells of a flat-bottom polystyrene 96-well plate (Thermo Scientific, Boston, MA, USA) were coated with 100μl of bicarbonate buffer (15mM Na2CO3 and 35mM NaHCO3, pH 9.6) containing 4μg/ml of capture-antibody, and incubated overnight at 4°C. The coating solution was then removed and other protein binding sites were blocked by adding 250 μl per well of a blocking solution (1X PBS, 0.5% skim milk powder and 0.1% Tween20, pH 7.4), and incubated at room temperature (RT) for 2 hours. After the blocking solution was removed, the assay was completed by using the commercial reagents from the p24 capture ELISA cited above and following the instruction of the manufacturer.
Table 2. Capture antibodies references and description.
| Capture antibodies | Manufacturer and catalog number | Description |
|---|---|---|
| mAb anti-p24 clone 4F6 | ImmunoDX, Woburn, MA, USA Catalog #1103 | Mouse IgG monoclonal antibody to a linear epitope (manufacturer's communication) of HIV-1 p24; cross-reactivity with HIV-1 subtypes B, C and E (manufacturer's communication). |
| mAb anti-p24 clone 183-H12-5C | NIH AIDS Reagent Program, Bethesda, MD, USA Catalog #3537 | Mouse IgG1κ monoclonal antibody to an unmapped epitope of HIV p24; cross-reactivity with HIV-1, HIV-2 (Chauveau et al., 2015) and SIV p27. Selection criteria: able to detect CRF07_BC (Huang et al., 2014). |
| mAb anti-p24 clone 39/5.4A | Abcam, Cambridge, MA, USA Catalog #ab9071 | Mouse IgG1 monoclonal antibody to HIV p24 -mapping information non available. Selection criteria: known to react with HIV-1 subtypes A to H (manufacturer's communication). |
| HIV-IG | NIH AIDS Reagent Program, Bethesda, MD, USA Catalog #3957 | Human immunoglobulin. Derived from pooled plasma of asymptomatic HIV-positive donors selected for high titer p24 antibody. |
2.3. Beads coupling reaction and validation
The C-Ab yielding the best result was selected for coupling to beads. The content of a vial of carboxylated magnetic beads region 43 (1.25 × 107 beads, 1ml, region 43, 6.5μm in diameter, Luminex Corp. Austin, TX, USA) was transferred into a low binding microcentrifuge tube (Eppendorf, Hamburg, Germany) and centrifuged at 8000 × g for 2 minutes. After a wash in 200μl of H2O, the beads were pelleted, resuspended in 160μl of 100mM NaH2PO4 (Sigma-Aldrich, St. Louis, MO, USA) pH 6.2 and activated for 20 minutes under agitation by adding 20μl of 50mg/ml of Sulfo-NHS (sulfo-N-hydroxysuccinimide) (Pierce Biotechnoloy Inc., Rockford, IL, USA) and 20μl of 50mg/ml of EDC (1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride) (Pierece Biotechnoloy Inc). The solution of Sulfo-NHS and EDC were freshly prepared and checked for the correct pH before each coupling procedure. The activated beads were washed twice in 250μl of 100M of MES (2[N-Morpholino] ethanesulfonic acid) (Fisher Scientific, Pittsburgh, PA, USA) solution pH 6, centrifuged and resuspended in 250μl of 100M of MES. 110μg of C-Ab diluted in PBS were added and the total volume brought to 1ml with 100mM MES pH6. The coupling reaction proceeded for 2 hours at RT with a gently stirring at 4°C overnight. The coupled beads were then centrifuged, resuspended and incubated under constant stirring at RT for 30 minutes in 500μl of PBS-TBN (1X PBS, 0.1% BSA, 0.02% Tween 20 and 0.05% Sodium Azide). After two washes with 500μl of PBS-TBN, the coupled beads were finally resuspended in 1ml of PBS-TBN. The bead concentration was determined with a hemocytometer, and the C-Ab-beads were stored at 4°C in the dark. During the coupling procedure, the beads were protected from the light as much as possible. All centrifugation steps were performed at ≥ 8000 × g for 2 minutes. After each wash the beads were vortexed at maximum speed for 30 seconds and sonicated for 15 seconds.
The coupling procedure was then validated by incubating the protein-coupled beads with serially diluted PE-labeled antibody solutions, ranging from 6.25μg/ml to 200 μg/ml for 1 hour. The beads were then washed and resuspended in 200μl of BD FACSFlow™ (BD Biosciences, San Diego, CA, USA) and acquired on a LSRII flow cytometer (BD Biosciences, San Diego, CA, USA) using DIVA software version 8.0.1 (BD Biosciences, San Diego, CA, USA), and analyzed with FlowJo version 9.8.2 (Tree Star, Ashland, OR).
2.4. p24 Luminex capture assay
The p24 Luminex capture assay was performed in a flat bottom 96-well plate. The washes were performed using an automatic magnetic washer; and the plates were kept from the light during incubation. The magnetic beads were diluted in the assay buffer (20mM Tris Hcl pH6, 0.1% BSA and 0.05% Tween 20) to a concentration of 60,000 beads/ml and 50μl (3000 beads) were added in each well. The samples and the p24 standards were lysed by adding a 10% volume of a 10X lysis solution (10X Triton X-100 and 0.05% Tween 20) for 1 hour at 37°C, and then diluted in assay buffer. 50μl of the lysed samples or standard were added to the magnetic beads and incubated for 1 hour at RT under agitation. The plates were then washed twice with 200μl of a wash solution (20mM Tris Hcl pH6 and 0.05% Tween 20) using a magnetic microplate washer (ELx405, BioTek, Winooski, VT, USA). 100μl of the detection antibody anti-p24 KC57 (Beckman Coulter, Fullerton, CA, USA) was added to each well, and the plates were incubated for 1 hour at RT under agitation. The plates were washed twice with 200μl of a wash solution. The magnetic beads were resuspended in 100μl of assay buffer and agitated for 10 minutes before the analysis. The analysis of the magnetic beads was done on a Luminex 100 using the Bioplex manager software version 6.0 (Bio-Rad, Hercules, CA) and a minimum of 300 beads per well were analyzed.
2.5. Quantitative PCR for HIV-1 Gag RNA
The quantification of HIV-1 Gag by quantitative PCR was performed by extracting HIV-1 RNA from the viral stocks using the QIAamp viral RNA mini kit (QIAGEN Gmbh, Hilden, Germany) per the manufacturer's instruction. The viral RNA was reverse transcribed to complementary DNA synthesis (cDNA) using the SuperScript III RT kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions and 5′Primer (5′-AGT TCT TCT GAT CCT GT-3′). Number of gag cDNA copies was quantified against dilutions of cells engineered to express only 1 copy of HIV gag DNA (8E5 cell line) per cell using 7500 Real Time PCR System (Applied Biosystems, Foster City, CA, USA). Briefly, duplicate PCR reactions at a final volume of 12.5 μl containing 1.25 μl of 10X PCR buffer, 200 μM dNTP, 3.5mM MgCl2, 800 nM of primers (Forward primer 5′-TGA CTA GCG GAG GCT AGA A-3′, Reverse primer 5′-CTC YCT GCT TGCCCA TA-3′), 1.25 U of Platinum Taq DNA Polymerase (Invitrogen Carlsbad, CA, USA), and 2 μl of cDNA was subjected to pre-amplification using an MJ Research PTC-225 thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA). The cycling condition was 94° for 2 min followed by 12 cycles of 95° for 30 sec and 60° for 1 min. A 2 μl aliquot of each pre-amplification products was subjected to a Taqman real-time PCR and a reaction mixture contained 1.25 μl of 10X PCR buffer, 200 μM dNTP, 3.5 mM, MgCl2, 800 nM of primers, 2.5 U of Platinum taq (Invitrogen Carlsbad, CA), 0.375μl of ROX reference dye (Invitrogen Carlsbad, CA, USA) and 0.2 pM of probe (5′-[6FAM] AAA ATT CGG TTA AGS CCA GGG GGA AAG AA[BHQ1]-3′). The cycling condition was 94° for 2 min followed by 45 cycles of 95° for 30 sec and 60° for 1 min and performed using a 7900HT Fast Real Time PCR System (Applied Biosystems, Foster City, CA, USA).
2.6. Statistical analysis
The statistical analyses were done using GraphPad Prism 6. Statistical significance was evaluated with a one sample t-test (with theoretical mean = 0) or multiple t-tests. The correlation coefficient between HIV-1 RNA copy number and p24 antigen concentration was evaluated using the non-parametric Pearson correlation.
3. Results
3.1. Identification and validation of cross-reactive p24 C-Abs
Two mAb directed towards HIV-1 p24, 183-H12-5C and 39/5.4A were selected based on their reported ability to react with non-B HIV-1 subtypes (Table 2) (Chauveau et al., 2015; Huang et al., 2014); a third candidate, a mix of human HIV immunoglobulins (HIV-IG) (NIH AIDS Reagent Program) was also added. In preliminary ELISA tests, all three C-Ab were able to detect the protein of six subtypes (A, B, C, D and CRF01_AE and CRF02_AG) isolates (data not shown). The three candidates were further evaluated on the Luminex platform. Capture beads were generated using the mAb 183-H12-5C (183-beads) and 39/5.4A (39/5.4A-beads), and the HIV-IG (IG-beads). The clone 4F6 (4F6-beads), previously used by Biancotto et al. (Biancotto et al., 2009), served as a control. The efficiency of the coupling reactions was validated by mixing the coupled microspheres with dilutions of a phycoerythrin (PE)-labeled anti-species IgG detection antibody (ranges from 6.25-200 μg/ml) and assessed by flow cytometry. The intensity of the fluorescent signal was proportional to the amount of detection antibody on the surface of the beads (Figure 1), indicating that all coupling procedures were successful. The functionality of the capture beads was further assessed by mixing them with serial dilutions of purified p24 protein. The fluorescence read by the Luminex is shown in Figure 2. The fluorescence intensity (FI) of the 183-beads showed a dose-dependent response comparable to the 4F6-beads. The 39/5.4A- and IG-beads failure to respond in a dose dependent manner indicated that they did not bind to the protein, or that their binding prevented the binding of the detection antibody. The clone 183-H12-5C was the only C-Ab that met the criteria for further development.
Figure 1.
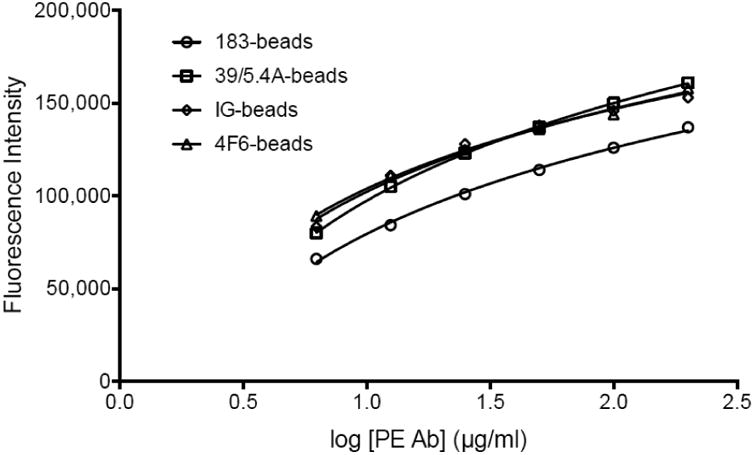
Validation of the coupling reaction by flow cytometry. The different coupled microspheres (183-beads, empty circles; 39/5.4A-beads, empty squares; IG-beads, empty diamonds and 4F6-beads, empty triangles) were mixed with serial dilutions of PE-labeled detection antibody (6.25-200μg/ml) and assessed by flow cytometry. The graph represents the fluorescence intensity depending on the amount of detection antibody.
Figure 2.
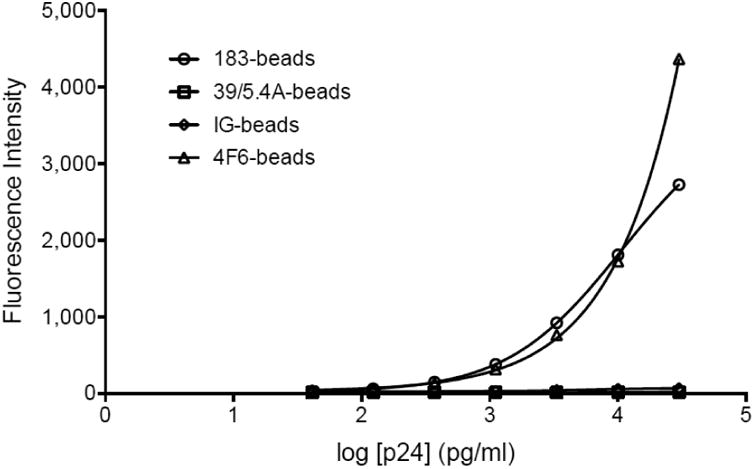
Validation of the functionality of the capture antibodies post coupling. The capture microspheres (183-beads, empty circles; 39/5.4A-beads, empty squares; IG-beads, empty diamonds and 4F6-beads, empty triangles) were mixed with serial dilutions of purified p24 protein, the binding was revealed by PE-labeled anti-p24 and read by Luminex. The graph represents standard curves fitted by a logistic 5P regression using Prism6.
3.2. Performances of the HIV-1 p24 capture Luminex assay - using an internationalpanel of HIV-1 isolates
The p24 capture Luminex assay using the 183-beads (183-beads Luminex) was performed on a panel of HIV-1 isolates (Brown et al., 2005) representing 6 major subtypes and recombinant forms (A, B, C, D, CRF01_AE and CRF02_AG). Out of 60 isolates from the panel, 56 were available for quantification (Table 1). To determine the p24 concentrations, a p24 standard curve (Figure 3) was run in quadruplicate and fitted by a logistic 5-parameter regression (R2 = 0.9993, EC= 17,528 pg/ml, Hill slope = 1.14). The broad dynamic range of the assay and its reproducibility are shown in Figure 3; the intra-assay variation of the quadruplicates and inter- assay of four different experiments gave coefficients of variation (CVs) below 6 and 14% respectively. The range of detection ran from 3.7 pg/ml to 30,000 pg/ml. The quantification of the HIV-1 isolates presented in Figure 4 demonstrates that, with no exception, the 56 isolates were detected by the assay (p-value range: <0.0001 -0.0208, one sample t-test) with p24 concentrations ranging from 3.2× 102 to 5.1×106 pg/ml. The CVs of the samples, run in quadruplicates, were between 0.92 and 29.57%.
Figure 3.
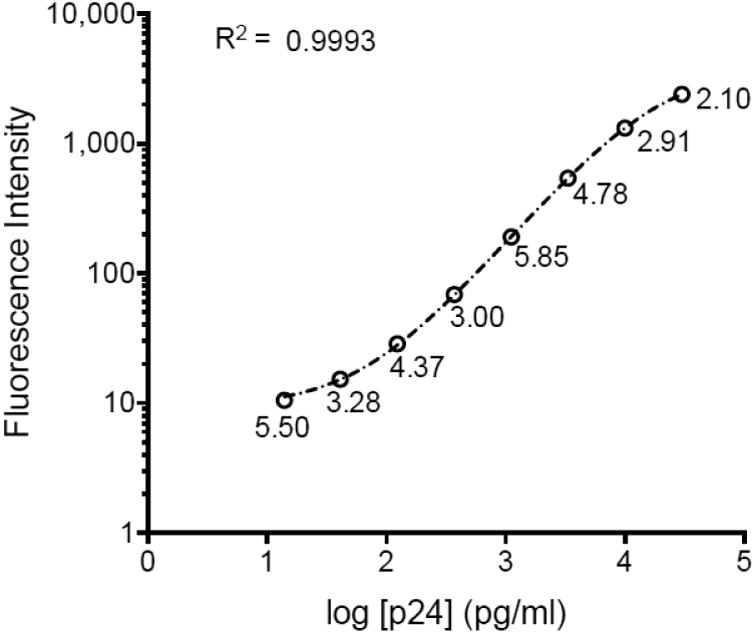
p24 standard curve using the clone 183-H12-5C. The graph represents the mean fluorescence intensity of a standard curve run in quadruplicate fitted by a logistic 5P regression using Prism6, with R2=0.9993. The CV of the data is indicated below each data point..
Figure 4.
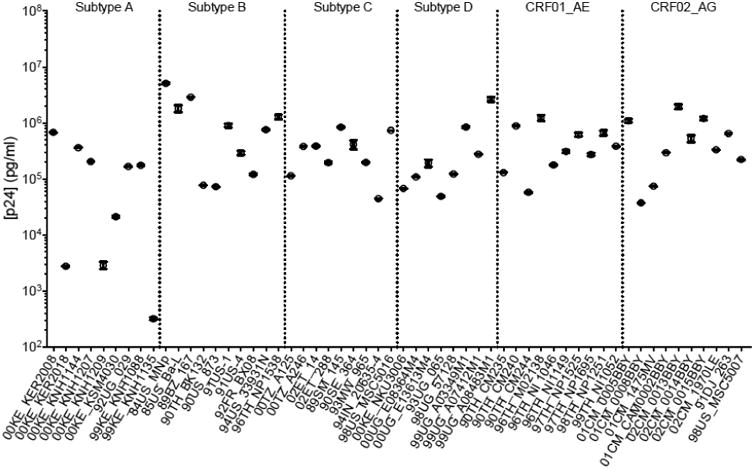
p24 capture Luminex assay of the 56 isolates representing 6 HIV-1 subtypes. The graph represents the mean concentration (in pg/ml) of each sample ran in quadruplicate. The error bars represent the SEM (Standard Error of the Mean).
To assess the inter-assay variation, three additional independent experiments were carried out in duplicate on 30 isolates over a period of six months. The four experiments are represented in Figure 5; the CVs measured were between 14.31 and 34.46% (n=10).
Figure 5.
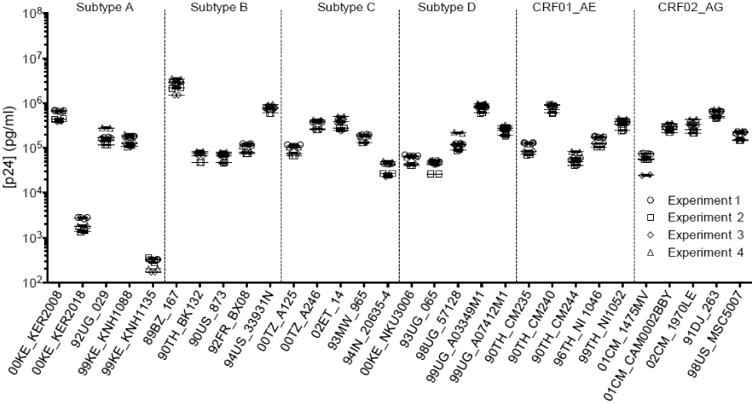
Inter-assay variation. p24 capture Luminex assay of the 30 isolates representing 6 HIV-1 subtypes. The graph represents the mean concentrations (in pg/ml) of 30 isolates run in duplicates in four idependant experiments carried out over a period of time of several months. The error bars represent the SEM.
3.3. Validation against established HIV-1 quantification methods
The newly developed 183-bead Luminex assay was further compared to three different HIV-1 quantification methods: a commercial p24 ELISA kit, broadly used in the field (Humeau et al., 2004; Kuroishi et al., 2009); the p24 capture assay described by Biancotto et al. (Biancotto et al., 2009) (using the 4F6-beads Luminex); and quantification of viral nucleic acid by qPCR.
The p24 ELISA kit was used to evaluate the p24 concentration of the 56 isolates. As shown in Figure 6 by the shift observed along the x-axis, the 183-bead Luminex was more sensitive for 51 isolates: the concentrations ranged from 1.3- to 11.9-fold higher than the concentrations measured with the ELISA kit. With the caveat that the ELISA assay was more sensitive with subtype A viruses, exceptions were found for 5 isolates (Figure 6, empty circle): subtype A (A-) 00KE_KER2018, A-00KE_KNH1209, A-00KE_KSM4030, A-99KE_KNH1135 and AG-01CM_0008BBY; although a higher sensitivity was observed with the ELISA for these isolates, they were still detected by the 183-bead Luminex assay (Figure 4). In addition, twelve isolates from the panel were tested with the 4F6-bead Luminex assay as well (Figure 7 and Table 3). Since mAb 4F6 was isolated from a subtype B patient (company data sheet), the detection of p24 for subtypes B and D (B-90TH_BK132, B-90US_873, D-00KE_NKU3006 and D-99UG_A03349M1) was found more sensitive and the concentration significantly higher with the 4F6-beads Luminex compared to the ELISA kit (p<0.0001). However, the concentrations measured for the other non-B isolates using the 4F6-beads Luminex assay were significantly lower than the concentration measured by the commercial ELISA kit (p<0.0025) (Table 3). A direct comparison of both Luminex assays (Figure 7 and Table 3) showed that the detection of p24 was more sensitive and the concentration significantly higher (p<0.0001) when using the 183-beads for all isolates tested except for one (D-00KE_NKU3006) for which the concentration was similar (p=0.034). Finally, 30 isolates were quantified by quantitative PCR as a standard method to monitor in vitro HIV-1 replication (Schupbach, 2003; Stefan et al., 2003) and compared to the 183-beads Luminex assay a significant correlation of HIV-1 RNA and p24 antigen was found (R = 0.92, p<0.0001).
Figure 6.
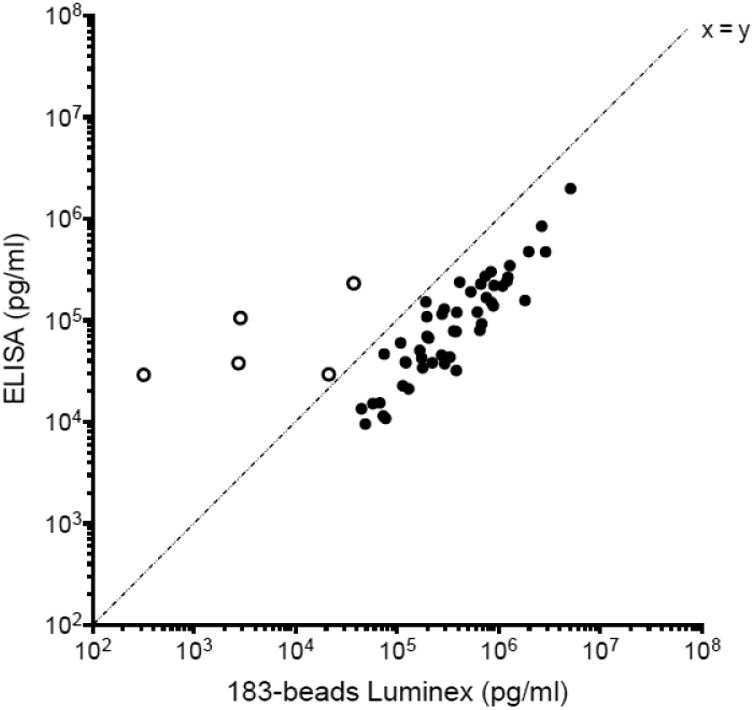
Comparison of p24 concentration of the 56 HIV-1 isolates between the capture Luminex assay and the commercial ELISA kit. The concentrations are expressed in pg/ml. The empty circles represent the isolates for which the commercial ELISA kit measured higher p24 concentration than the Luminex capture assay.
Figure 7.
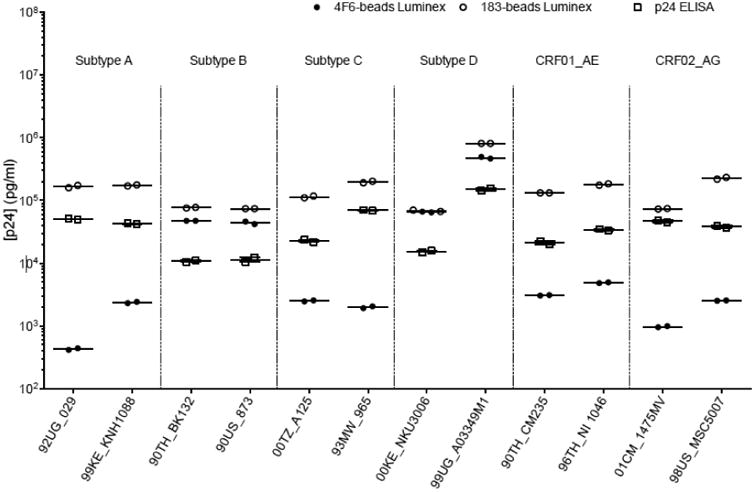
p24 capture Luminex assay using anti-p24gag HIV-1 mAb clone 4F6 (full circles), clone 183-H12-5C (empty circles)and ELISA kit (empty squares). The graph represents the p24 titration (in pg/ml) of 12 HIV-1 virus isolates representing six different subtypes (A, B, C, D, CRF01_AE, CRF01_AG).
Table 3. Mean concentrations of p24 measured by p24 capture Luminex using the clone 4F6 or the clone 183-H12-5C and the p24 ELISA kit.Values are expressed in pg/ml.
| 183-beads Luminex | 4F6-beads Luminex | ELISA | (4F6 vs ELISA) p values | (4F6 vs 183) p values | |
|---|---|---|---|---|---|
| A - 92UG_029 | 167,480 | 428 | 50,603 | 2.70E-04 | 3.00E-06 |
| A - 99KE_KNH1088 | 175,322 | 2,360 | 42,473 | 3.20E-04 | 4.20E-06 |
| B - 90TH_BK132 | 77,133 | 48,228 | 10,813 | 7.60E-07 | 7.80E-09 |
| B - 90US_873 | 73,078 | 45,370 | 11,448 | 5.50E-05 | 1.60E-06 |
| C - 00TZ_A125 | 113,857 | 2,506 | 22,761 | 2.50E-03 | 3.90E-06 |
| C - 93MW_965 | 197,434 | 1,986 | 69,566 | 1.90E-04 | 6.10E-07 |
| D - 00KE_NKU3006 | 67,678 | 63,166 | 15,395 | 2.40E-05 | 3.60E-02 |
| D - 99UG_A03349M1 | 848,124 | 487,723 | 149,826 | 9.80E-06 | 1.00E-05 |
| CRF01_AE - 90TH_CM235 | 130,108 | 3,068 | 21,181 | 5.10E-03 | 1.20E-07 |
| CRF01_AE - 96TH_NI 1046 | 178,372 | 4,889 | 34,077 | 1.00E-03 | 4.70E-07 |
| CRF02_AG - 01CM_1475MV | 74,598 | 973 | 46,679 | 1.60E-03 | 2.50E-07 |
| CRF02_AG - 98US_MSC5007 | 222,968 | 2,536 | 38,273 | 1.70E-03 | 9.20E-06 |
4. Discussion
The assay developed by Biancotto et al. (Biancotto et al., 2009) using mAb 4F6 as a C-Ab has demonstrated a great ability to measure concentrations of HIV-1 belonging to B subtypes (Balzarini et al., 2013; Introini et al., 2013; Merbah et al., 2012; Parrish et al., 2013; Saba et al., 2010; Vanpouille et al., 2012); however, its capacity to measure p24 from multiple non-B samples was more limited and opened the door for an assay using a more universal C-Ab. Biancotto et al. have established the multiple benefits of using a p24 capture Luminex assay: lower cost, a broad dynamic range, and high sensitivity. Additionally, the multitasking flow cytometry platform provides also an opportunity to multiplex with up to 499 additional analytes using 50μl of sample. A major obstacle in developing a universal HIV-1 p24 Luminex assay lies in the fact that a majority of available anti-p24 mAbs were isolated from patients infected with HIV-1 subtype B. In this study, the limitation of using subtype B-derived antibodies was mitigated by screening the literature for cross-reactive antibodies available in the public domain. The three Abs selected here were evaluated for multi-subtype detection. All three Abs (two mAbs (clones 39/5.4A and 183-H12-5C) and HIV-IG) were able to detect p24 from subtypes A, B, C, D and CRF01_AE and CRF02_AG by ELISA, but only the clone 183-H12-5C was able to bind p24 protein using the Luminex assay. One possible explanation for the differences observed in binding affinity is the change of antibody structure after coupling to the beads. This coupling procedure is achieved via carbodiimide reactions involving the primary amino groups on the antibodies, located on lysine residues and the N-terminus of each polypeptide chain, and the carboxyl functional groups on the bead surface. Therefore, the orientation of the antibody as well as the availability of the antigen-binding site cannot be controlled during the coupling procedure. This could explain why these antibodies did not detect p24 once coupled to the beads. The p24 Luminex capture assay using the clone 183-H12-5C was able to detect and measure the p24 concentration of the 56 isolates from the international HIV-1 panel (Brown et al., 2005) representing subtypes A, B, C, D, CRF01_AE and CRF02_AG. In a direct comparison, 91% of the isolates tested with the Luminex capture assay outperformed the commercial p24 ELISA kit; reduced sensitivity was observed in four out of nine subtype A, and one out of ten CRF02_AG isolates. The gag gene coding for p24 of CRF02_AG belongs to HIV-1 subtype A that is the most diverse among HIV-1 subtypes. Hence, it is possible that 183-H12-5C which was subtype B derived antibody does not fully bind to all of the subtype A lineages. The variations within the gag gene and consequently within p24 from different clades explain the difference in sensitivity
Furthermore, the 183 bead p24 Luminex capture assay successfully detected HIV-1 p24 antigen from viral stocks, the supernatants of cultured, HIV-infected cells (cell lines, PBMCs, macrophages) as well as supernatant from CRF01_AE infected human tissue explants which requires a highly sensitive assay due to the low level of viral replication. The sensitivity of this assay may be improved in the future by decreasing the number of beads per well as shown by Biancotto et al. (Biancotto et al., 2009).
The focus of much of the world's vaccine studies and HIV-1 research is located in areas where non-B subtypes are prevalent. It is crucial to generate assays that combine low cost and broad reactivity across subtypes. With the added benefit of multiplexing, and conservation of sample, the p24 Luminex capture assay presented here offers a valuable and timely new tool for the field.
Highlights.
Identified a cross-reactive antibody to p24 of HIV-1 main subtypes and 2 CRFs.
Developed a p24 Luminex capture assay and tested against 56 HIV-1 isolates.
Compared the performance of the assay with other quantitative methods.
The assay is highly sensitive with the limit of detection at 3.7pg/ml.
Successfully tested to detect virus production in tissue culture supernatant.
Acknowledgments
We are thankful to Victoria Polonis for providing us with the 56 HIV-1 isolates.
We also want to thank Mangala Rao for the participation of her team in performing the p24 ELISA.
HIV-1 p24 Monoclonal Antibody (183-H12-5C): The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 p24 Monoclonal Antibody (183-H12-5C) from Dr. Bruce Chesebro and Kathy Wehrly. HIV Immunoglobulin (HIV-IG): The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Catalog #3957, HIV-IG from NABI and NHLBI.
This work was supported by a cooperative agreement (W81XWH-11-2-0174) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense (DoD). The views expressed are those of the authors and should not be construed to represent the positions of the United States Departments of the Army, Defense, or Health and Human Services.
Abbreviations
- C-Ab
capture antibody
- FI
fluorescence intensity
- HIV-IG
mix of human HIV immunoglobulins derived from pooled plasma of asymptomatic HIV-positive donors
- IG-beads
beads coated with capture HIV-IG
- mAb
monoclonal antibody
- 183-beads
beads coated with the anti-p24 capture monoclonal antibody clone 183-H12-5C
- 39/5.4A-beads
beads coated with the anti-p24 capture monoclonal antibody clone 39/5.4A
- 4F6-beads
beads coated with the anti-p24 capture monoclonal antibody clone 4F6
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balzarini J, Andrei G, Balestra E, Huskens D, Vanpouille C, Introini A, Zicari S, Liekens S, Snoeck R, Holy A, Perno CF, Margolis L, Schols D. A multi-targeted drug candidate with dual anti-HIV and anti-HSV activity. PLoS Pathog. 2013;9:e1003456. doi: 10.1371/journal.ppat.1003456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancotto A, Brichacek B, Chen SS, Fitzgerald W, Lisco A, Vanpouille C, Margolis L, Grivel JCC. A highly sensitive and dynamic immunofluorescent cytometric bead assay for the detection of HIV-1 p24. Journal of virological methods. 2009;157:98–101. doi: 10.1016/j.jviromet.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BK, Darden JM, Tovanabutra S, Oblander T, Frost J, Sanders-Buell E, Souza MSd, Birx DL, McCutchan FE, Polonis VR. Biologic and Genetic Characterization of a Panel of 60 Human Immunodeficiency Virus Type 1 Isolates, Representing Clades A, B, C, D, CRF01_AE, and CRF02_AG, for the Development and Assessment of Candidate Vaccines. Journal of Virology. 2005;79:6089–6101. doi: 10.1128/JVI.79.10.6089-6101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauveau L, Puigdomenech I, Ayinde D, Roesch F, Porrot F, Bruni D, Visseaux B, Descamps D, Schwartz O. HIV-2 infects resting CD4+ T cells but not monocyte-derived dendritic cells. Retrovirology. 2015;12:2. doi: 10.1186/s12977-014-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coplan PM, Gupta SB, Dubey SA, Pitisuttithum P, Nikas A, Mbewe B, Vardas E, Schechter M, Kallas EG, Freed DC, Fu TMM, Mast CT, Puthavathana P, Kublin J, Brown Collins K, Chisi J, Pendame R, Thaler SJ, Gray G, McIntyre J, Straus WL, Condra JH, Mehrotra DV, Guess HA, Emini EA, Shiver JW. Cross-reactivity of anti-HIV-1 T cell immune responses among the major HIV-1 clades in HIV-1-positive individuals from 4 continents. The Journal of infectious diseases. 2005;191:1427–1434. doi: 10.1086/428450. [DOI] [PubMed] [Google Scholar]
- Hemelaar J, Gouws E, Ghys PD, Osmanov S, Isolation WUNfH, Characterisation Global trends in molecular epidemiology of HIV-1 during 2000-2007. AIDS (London, England) 2011;25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SW, Wang SF, Lin YT, Yen CH, Lee CH, Wong WW, Tsai HC, Yang CJ, Hu BS, Lin YH, Wang CT, Wang JJ, Hu Z, Kuritzkes DR, Chen YH, Chen YM. Patients infected with CRF07_BC have significantly lower viral loads than patients with HIV-1 subtype B: mechanism and impact on disease progression. PLoS One. 2014;9:e114441. doi: 10.1371/journal.pone.0114441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeau LM, Binder GK, Lu X, Slepushkin V, Merling R, Echeagaray P, Pereira M, Slepushkina T, Barnett S, Dropulic LK, Carroll R, Levine BL, June CH, Dropulic B. Efficient lentiviral vector-mediated control of HIV-1 replication in CD4 lymphocytes from diverse HIV+ infected patients grouped according to CD4 count and viral load. Mol Ther. 2004;9:902–13. doi: 10.1016/j.ymthe.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Introini A, Vanpouille C, Lisco A, Grivel JC, Margolis L. Interleukin-7 facilitates HIV-1 transmission to cervico-vaginal tissue ex vivo. PLoS Pathog. 2013;9:e1003148. doi: 10.1371/journal.ppat.1003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RB, Garrison KE, Mujib S, Mihajlovic V, Aidarus N, Hunter DV, Martin E, John VM, Zhan W, Faruk NF, Gyenes G, Sheppard NC, Priumboom-Brees IM, Goodwin DA, Chen L, Rieger M, Muscat-King S, Loudon PT, Stanley C, Holditch SJ, Wong JC, Clayton K, Duan E, Song H, Xu Y, SenGupta D, Tandon R, Sacha JB, Brockman MA, Benko E, Kovacs C, Nixon DF, Ostrowski MA. HERV-K-specific T cells eliminate diverse HIV-1/2 and SIV primary isolates. The Journal of clinical investigation. 2012;122:4473–4489. doi: 10.1172/JCI64560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroishi A, Saito A, Shingai Y, Shioda T, Nomaguchi M, Adachi A, Akari H, Nakayama EE. Modification of a loop sequence between alpha-helices 6 and 7 of virus capsid (CA) protein in a human immunodeficiency virus type 1 (HIV-1) derivative that has simian immunodeficiency virus (SIVmac239) vif and CA alpha-helices 4 and 5 loop improves replication in cynomolgus monkey cells. Retrovirology. 2009;6:70. doi: 10.1186/1742-4690-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merbah M, Arakelyan A, Edmonds T, Ochsenbauer C, Kappes JC, Shattock RJ, Grivel JCC, Margolis LB. HIV-1 expressing the envelopes of transmitted/founder or control/reference viruses have similar infection patterns of CD4 T-cells in human cervical tissue ex vivo. PloS one. 2012;7 doi: 10.1371/journal.pone.0050839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP, Jr, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A. 2013;110:6626–33. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JC, Sherman GG, Coovadia AH, Stevens WS, Meyers TM. Ultrasensitive Human Immunodeficiency Virus Type 1 p24 Antigen Assay Modified for Use on Dried Whole-Blood Spots as a Reliable, Affordable Test for Infant Diagnosis. Clinical and Vaccine Immunology. 2006;13:152155. doi: 10.1128/CVI.13.1.152-155.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa Borges A, Wieczorek L, Johnson B, Benesi AJ, Brown BK, Kensinger RD, Krebs FC, Wigdahl B, Blumenthal R, Puri A, McCutchan FE, Birx DL, Polonis VR, Schengrund CLL. Multivalent dendrimeric compounds containing carbohydrates expressed on immune cells inhibit infection by primary isolates of HIV-1. Virology. 2010;408:80–88. doi: 10.1016/j.virol.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba E, Grivel JC, Vanpouille C, Brichacek B, Fitzgerald W, Margolis L, Lisco A. HIV-1 sexual transmission: early events of HIV-1 infection of human cervico-vaginal tissue in an optimized ex vivo model. Mucosal Immunol. 2010;3:280–90. doi: 10.1038/mi.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach J. Viral RNA and p24 Antigen as Markers of HIV Disease and Antiretroviral Treatment Success. International Archives of Allergy and Immunology. 2003;132:196–209. doi: 10.1159/000074552. [DOI] [PubMed] [Google Scholar]
- Stefan AK, Susanne K, Brigitte R, Christian K, Martine P, Oliver GO, Dieter H, Willi KR. Comparison of TaqMan™ real-time PCR and p24 Elisa for quantification of in vitro HIV-1 replication. Journal of Virological Methods. 2003;107:169–175. doi: 10.1016/s0166-0934(02)00230-6. [DOI] [PubMed] [Google Scholar]
- Stoddart CA, Galkina SA, Joshi P, Kosikova G, Moreno ME, Rivera JM, Sloan B, Reeve AB, Sarafianos SG, Murphey-Corb M, Parniak MA. Oral administration of the nucleoside EFdA (4′-ethynyl-2-fluoro-2′-deoxyadenosine) provides rapid suppression of HIV viremia in humanized mice and favorable pharmacokinetic properties in mice and the rhesus macaque. Antimicrobial agents and chemotherapy. 2015;59:4190–4198. doi: 10.1128/AAC.05036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers MF, Henderson LE, Chance MR, Bess JW, South TL, Blake PR, Sagi I, Perez-Alvarado G, Sowder RC, Hare DR. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein science : a publication of the Protein Society. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Zhao J, Wang A, Viswanath R, Harma H, Little RF, Yarchoan R, Stramer SL, Nyambi PN, Lee S, Wood O, Wong EY, Wang X, Hewlett IK. Characterization of immune responses to capsid protein p24 of human immunodeficiency virus type 1 and implications for detection. Clinical and vaccine immunology : CVI. 2010;17:1244–1251. doi: 10.1128/CVI.00066-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanpouille C, Lisco A, Introini A, Grivel JCC, Munawwar A, Merbah M, Schinazi RF, Derudas M, McGuigan C, Balzarini J, Margolis L. Exploiting the anti-HIV-1 activity of acyclovir: suppression of primary and drug-resistant HIV isolates and potentiation of the activity by ribavirin. Antimicrobial agents and chemotherapy. 2012;56:2604–2611. doi: 10.1128/AAC.05986-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt VM, Simon MN. Mass determination of rous sarcoma virus virions by scanning transmission electron microscopy. Journal of virology. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Zhong Z, Zariffard M, Spear GT, Qiao L. Bovine papillomavirus-like particles presenting conserved epitopes from membrane-proximal external region of HIV-1 gp41 induced mucosal and systemic antibodies. Vaccine. 2013;31:5422–5429. doi: 10.1016/j.vaccine.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]


