Abstract
The natural mummification process of the human gut represents a unique opportunity to study the resulting microbial community structure and composition. While results are providing insights into the preservation of bacteria, fungi, pathogenic eukaryotes and eukaryotic viruses, no studies have demonstrated that the process of natural mummification also results in the preservation of bacteriophage DNA. We characterized the gut microbiome of three pre-Columbian Andean mummies, namely FI3, FI9 and FI12, and found sequences homologous to viruses. From the sequences attributable to viruses, 50.4% (mummy FI3), 1.0% (mummy FI9) and 84.4% (mummy FI12) were homologous to bacteriophages. Sequences corresponding to the Siphoviridae, Myoviridae, Podoviridae and Microviridae families were identified. Predicted putative bacterial hosts corresponded mainly to the Firmicutes and Proteobacteria, and included Bacillus, Staphylococcus, Clostridium, Escherichia, Vibrio, Klebsiella, Pseudomonas and Yersinia. Predicted functional categories associated with bacteriophages showed a representation of structural, replication, integration and entry and lysis genes. The present study suggests that the natural mummification of the human gut results in the preservation of bacteriophage DNA, representing an opportunity to elucidate the ancient phageome and to hypothesize possible mechanisms of preservation.
Keywords: ancient microbiomes, bacteriophages, microbiome, mummy, phageome, virome
Gut phageome of mummified human remains.
INTRODUCTION
The human gut microbiome is home to diverse communities comprised of bacteria, archaea and eukaryotes (Yatsunenko et al. 2012; Hoffmann et al. 2013); yet, an increasing number of studies have demonstrated that the human gut is also inhabited by diverse viral communities, many of which are bacteriophages (Minot et al. 2011, 2013). Bacteriophages play important roles in biogeochemical cycles (Fuhrman 1999) and in the evolution of their bacterial hosts (Ai, Meng and Zeng 2000; Bollback and Huelsenbeck 2001; Coberly et al. 2009; Minot et al. 2013; Cvirkaite-Krupovic, Carballido-Lopez and Tavares 2015); however, we are just beginning to understand the role of bacteriophages as part of the human microbiome (Sun and Relman 2013; Abeles and Pride 2014). Previous studies have demonstrated that bacteriophages are part of the human oral (Pride et al. 2012; Edlund et al. 2015), skin (Robles-Sikisaka et al. 2013; Denesvre et al. 2015), genitourinary tract (Santiago-Rodriguez et al. 2015c) and gut microbiomes (Breitbart et al. 2003; Minot et al. 2011; Hofer 2013; Cadwell 2015; Ray 2015). Bacteriophages also have major impacts on human health and disease (Willner et al. 2009, 2011; Ly et al. 2014; Landini et al. 2015; Norman et al. 2015; Santiago-Rodriguez et al. 2015e; Wang, Gao and Zhao 2015). In diseases such as periodontitis, the relative abundance of bacteriophages belonging to the Myoviridae family is higher in subjects with the disease compared to subjects with good periodontal health. While results may be influenced by the representation of myoviruses in databases, these bacteriophages are believed to shape oral bacterial communities by lysing their hosts, thus, are believed to promote periodontal disease (Ly et al. 2014; Santiago-Rodriguez et al. 2015e). Other more serious diseases, including inflammatory bowel disease (IBD), have also associated bacteriophages with a dysbiosis of the gut bacterial communities, probably resulting in the disease (Norman et al. 2015).
Human microbiomes dating to hundreds and thousands of years are just beginning to be characterized, and have also been associated with dietary shifts, dietary habits and periodontal health and disease (Adler et al. 2013; Cano et al. 2014; Warinner, Speller and Collins 2015; Weyrich, Dobney and Cooper 2015). Yet, very few studies have focused on the viral composition of ancient human samples. Previous studies have focused on viruses in ancient human specimens including retroviruses and those from the Flaviridae, Rhabdoviridae, Parvoviridae families (Emerman and Malik 2010; Patel, Emerman and Malik 2011; Aswad and Katzourakis 2012; Katzourakis 2013; Lavialle et al. 2013; Rivera-Perez et al. 2015). It is feasible to hypothesize that ancient microbiomes are also home to a community of bacteriophages homologous to those present in modern human microbiomes. A previous study characterizing the virome of fossilized fecal material from the14th century found that bacteriophages comprised a good proportion of the viral communities (Appelt et al. 2014).
The natural mummification process is also known to preserve ancient microbial DNA due to cold temperatures and low oxygen levels (Cano et al. 2000; Zink et al. 2000; Tito et al. 2012). Our previous study characterizing the gut microbiome of a pre-Columbian Andean mummy identified sequences associated with bacteria, archaea, fungi, pathogenic eukaryotes and eukaryotic viruses (Santiago-Rodriguez et al. 2015a); yet, no studies have demonstrated that the process of natural mummification also results in the preservation of bacteriophage DNA. Bacteriophage communities are usually characterized using viral metagenomics, which consists in the enrichment of viruses by CsCl gradient ultracentrifugation (Rosario et al. 2009; Walker 2010; Ly et al. 2014; Santiago-Rodriguez et al. 2015c). A previous study characterized the viral communities of a 14th century coprolite using viral metagenomics, but the method needs to be tested in mummified human specimens (Appelt et al. 2014). Shotgun metagenomics has also shown to provide information on microbial communities in ancient human samples (Adler et al. 2013). While shotgun metagenomics is not selective for bacteriophage DNA, it is useful in characterizing phage communities in modern samples (Belda-Ferre et al. 2012; Santiago-Rodriguez et al. 2015e). Therefore, by using metagenomics, we aim to: (i) determine the percentage of sequences associated with bacteriophages, (ii) identify bacteriophages sharing sequence homology to modern bacteriophages and (iii) determine predicted functional categories associated with bacteriophages in the gut of naturally preserved human mummies.
MATERIALS AND METHODS
Description of mummified human remains
The specimens studied are presently stored at the Museum of Anthropology and Ethnology of the University of Florence, Italy. Autopsies were performed by paleopathologists G. Fornaciari and colleagues, and specimens were collected from internal organs by cutting the skin and the ribs. The first mummy, FI3, was an adult male dating to the 14th—15th century that showed a good preservation of the skin with the adnexa and a massive presence of fungi and ectoparasites. The presence of microscopic, non-pathological fungi, including the genus Aspergillus (easily identifiable with Periodic Acid Schiff staining), is a very common finding in mummies as a post-mortem invasion phenomenon. DNA was extracted from abdominal viscera. The second mummy, FI9, was a female of estimated 18–23 years of age, dating to the 11th century A.D. DNA was extracted from the descending colon, but the ascending and transverse colon, as well as paleofeces were previously characterized by our group (Santiago-Rodriguez et al. 2015a). The third mummy, FI12, was an adult female, estimated age 20–25 years, and autopsy showed that she was afflicted by bronchopneumonia. An exact date for the mummy is unknown, but it is evident that she belonged to the Inca culture from the fetal position found in the burial basket. DNA was extracted from the transverse colon.
Avoidance of contamination
We employed the standard precautions for ancient DNA work including the use of sterile gloves, pretreatment of mortars, pestles and homogenizers with HCl, use of UV-irradiated safety cabinets, dedicated gel trays, tanks and reagents. The autopsy was performed by paleopathologists wearing sterile surgical coats, sterile latex gloves, sterile masks, headdresses and overshoes. The outermost portions of the specimens were not used to eliminate the risk of surface contamination, and one replicate per sample was obtained for further analyses. The mummified specimens were immediately kept and sealed in sterile containers, reducing the possibility of subsequent contamination. The samples were stored aseptically in hermetic plastic containers in a dry environment with silica gel at 18°C –20°C. DNA extraction and further precautions were performed as described previously (Santiago-Rodriguez et al. 2015a) and are detailed in Methods (Supporting Information).
Metagenome analyses for viruses
DNA library preparation for metagenome sequencing was performed at the Next-Generation sequencing provider Molecular Research Laboratory (MRDNA) (www.mrdnalab.com; Shallowater, TX, USA) under strict procedures to eliminate cross-contamination with modern DNA as described previously (Santiago-Rodriguez et al. 2015a). Libraries were sequenced using Illumina MiSeq following Truseq DNA library preparation protocol, and sequence files were processed as described previously (Santiago-Rodriguez et al. 2015a). Data were then uploaded and annotated using the MG-RAST pipeline and taxonomic assignments were determined using the SEED database with a minimum e-value of 80% (Meyer et al. 2008). To determine the percentage of sequences associated with viruses and the predicted putative hosts at the phylum level, data were acquired from the Virus category. Sequences were also mapped to a virus database that included both prokaryotic and eukaryotic viruses (www.phantome.org; ftp://ftp.ncbi.nih.gov/genomes/Viruses/). Mapping was performed using CLC Genomics Workbench with the following parameters: no masking, mismatch cost = 2, insertion cost = 3, deletion cost = 3, with an 80% identity over a minimum of 50% of the read length. Mapped reads were also retrieved from CLC Genomics Workbench as a SAM file and processed using mapDamage for further ancient DNA authentication as described previously (Ginolhac et al. 2011). Predicted functional categories associated with bacteriophages were analyzed using MG-RAST with a minimum e-value of 80%.
16S rRNA gene analyses
16S rRNA gene data from these mummies were used to associate the phages predicted putative hosts with the bacterial taxonomy at the phylum level. SourceTracker analyses were also performed to identify possible sources of contamination (Knights et al. 2011; Santiago-Rodriguez et al. 2015a,b). 16S rRNA gene methods and analyses are described in Methods (Supporting Information).
RESULTS
Metagenome and 16S rRNA gene high-throughput sequencing data
For the metagenome analyses, a total of 16 805 260 (mummy FI3), 146 081 692 (mummy FI9) and 16 537 474 (mummy FI12) sequences, with an average length of 100 bp were analyzed. For the 16S rRNA gene analyses, a total of 79 752 (mummy FI3), 8731 (mummy FI9) and 57 979 (mummy FI12) sequences with an average length of 270 bp were acquired, but data were rarefied to 8000 sequences to minimize the effect of disparate sequence number in the results (Santiago-Rodriguez et al. 2015a). We utilized SourceTracker using 135 human (45 oral, 45 skin and 45 gut), and 45 soil microbiomes to identify possible sources of contamination in the mummified gut tissues (Fig. S1, Supporting Information) (Santiago-Rodriguez et al. 2015a). Given that the samples were obtained from the mummies’ colon, it is expected some of the ancient sequences to match modern gut microbiomes, as in the case of mummy FI3. However, given that the sequences did not match the most likely sources of contamination (shown as unknown) in any of the mummies, namely skin and soil microbiomes, suggests that no external sources of contamination contributed to the findings reported in the present study (Santiago-Rodriguez et al. 2015a,b). We also performed mapDamage analyses for mummy FI3 mapped viral reads, but did not note the typical DNA damage pattern at the 5′ or 3′ ends as described for eukaryotic genomes (Knapp et al. 2012; Der Sarkissian et al. 2014) (Fig. S2, Supporting Information).
Natural mummification preserves bacteriophage DNA
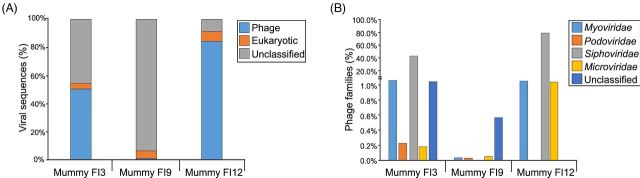
Metagenome analyses showed that the mummified guts included in the present study had sequence homology to viral genomes, with a proportion corresponding to bacteriophages. A total of 2198 (mummy FI3), 74 052 (mummy FI9) and 275 (mummy FI12) sequences were homologous to viruses. Approximately 50.4%, 4.0% and 45.6% of the viral sequences in mummy FI3 were homologous to bacteriophages, eukaryotic viruses and unclassified viruses, respectively. Mummy FI9 had the majority of the viral sequences (93.7%) not matching bacteriophages or eukaryotic viruses (unclassified). Approximately 84.4%, 6.9% and 8.7% of the viral sequences in mummy FI12 were homologous to bacteriophages, eukaryotic viruses and unclassified viruses, respectively (Fig. 1A). Analysis of the phage families showed that 42.9% (mummy FI3), 0.29% (mummy FI9) and 79.9% (mummy FI12) of the sequences were homologous to siphoviruses. Approximately 4.6% (mummy FI3), 0.04% (mummy FI9) and 3.6% (mummy FI12) of the sequences were homologous to myoviruses. Podoviruses represented 0.2% and 0.03% of the viral sequences in mummies FI3 and FI9, respectively. No podovirus-homologous sequences were present in mummy FI12. Microviruses contributed 0.2%, 0.05% and 1.8% of the viral sequences in mummies FI3, FI9 and FI12, respectively. The remaining sequences associated with bacteriophages could not be classified (Fig. 1B).
Figure 1.
Panel (A) shows the percentage of sequences homologous to phages, eukaryotic viruses and unclassified viruses. Percentage was calculated based on the total number of sequences corresponding to viruses. Panel (B) shows the percentage of sequences corresponding to phage families. Families included the Siphoviridae, Myoviridae, Podoviridae, Microviridae and unclassified, and were determined based on sequence homology to known phages.
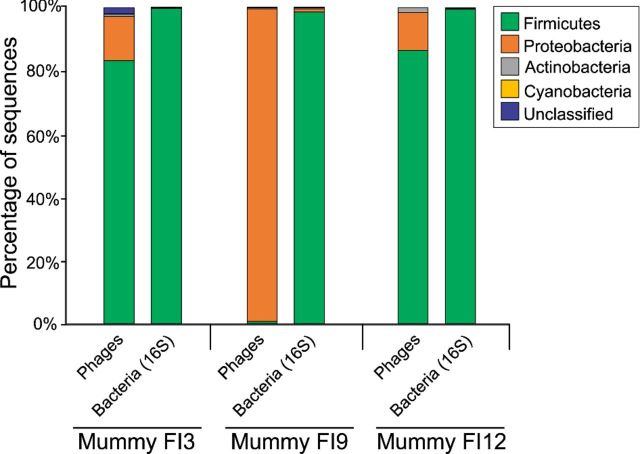
Analyses of predicted putative hosts at the phylum level showed that the majority (80.4%) of the sequences associated with bacteriophages in mummies FI3 and FI12 were homologous to those having Firmicutes as the bacterial hosts. Other putative hosts at the phylum level included the Proteobacteria, Actinobacteria or Cyanobacteria. Notably, mummy FI9 had the majority of the bacteriophage sequences sharing homology to those infecting Proteobacteria (93.6%) (Fig. 2). 16S rRNA gene analyses showed that the Firmicutes were the most represented bacterial group in mummies FI3 (99.9%), FI9 (98.5%) and FI12 (99.4%) (Fig. 2).
Figure 2.
Bacteriophage predicted putative bacterial host at the phylum level was determined based on sequence homology. Predicted putative hosts included the Firmicutes, Proteobacteria, Actinobacteria, Cyanobacteria and unclassified. Figure also shows the bacterial phylum based on analysis of the 16S rRNA gene variable region V4.
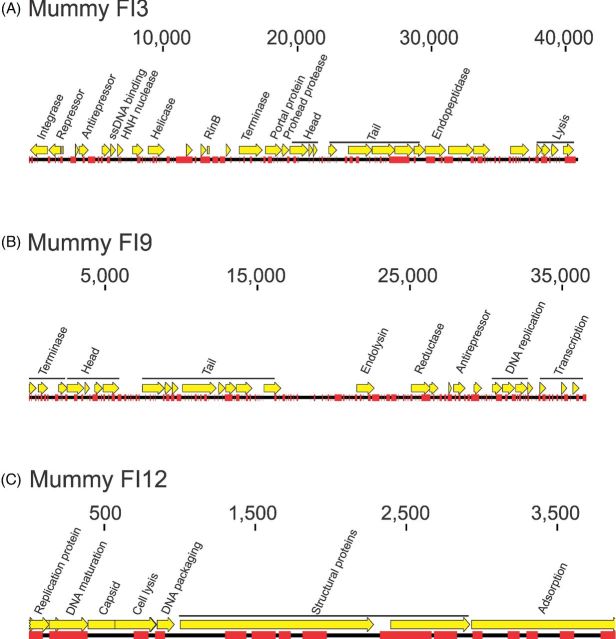
Mapping results also demonstrated that reads in the mummified guts corresponded to phage homologs. Table 1 shows examples of the mapping results to presumptive bacteriophages with the highest number of reads and coverage. Examples include Staphylococcus, Cronobacter and Brochothrix phages in mummy FI3, Lactobacillus and Staphylococcus phages in mummy FI9, and Bacillus and Cronobacter phages in mummy FI12. Reads also mapped across bacteriophage genomes, although not broadly, in mummies FI3 (Fig. 3A), FI9 (Fig. 3B) and FI12 (Fig. 3C). Examples shown include Staphylococcus bacteriophage StB20 (Fig. 3A), Lactobacillus phage AQ113 (Fig. 3B) and Enterobacteria phage phiX174 sensu lato (Fig. 3C). Regions that mapped to modern Enterobacteria phage phiX174 in mummies FI3 (Panel A), FI9 (Panel B) and FI12 (Panel C) are shown in Fig. S3 (Supporting Information). Nucleotide differences between modern and ancient sequences are shown in red, and indicate that ancient sequences do not correspond to the standard spike-in control used in Illumina sequencing.
Table 1.
Examples of bacteriophage sequences mapping known bacteriophage genomes in mummies FI3, FI9 and FI12. Bacterial taxonomic classification at the family level was determined using 16S rRNA gene analyses. Family and relative abundance percentages are shown.
| Presumptive bacteriophage | Phage family | Number of reads | Average coverage | Accession number | Reference length (bp) |
|---|---|---|---|---|---|
| Mummy FI3 | |||||
| Acinetobacter phage 133 | Myoviridae | 48 | 7.51E-03 | NC_015250.1 | 159 801 |
| Aeromonas phage 65 | Myoviridae | 93 | 9.62E-03 | NC_015251.1 | 235 229 |
| Bacillus phage 0305phi8-36 | Myoviridae | 110 | 0.01 | NC_009760.1 | 218 948 |
| Brochothrix phage A9 | Myoviridae | 295 | 0.1 | NC_015253.1 | 127 065 |
| Campylobacter phage CP21 | Myoviridae | 101 | 0.02 | NC_019507.1 | 182 833 |
| Cellulophaga phage phiST | Siphoviridae | 67 | 0.03 | NC_020842.1 | 79 114 |
| Clostridium phage CDMH1 | Myoviridae | 60 | 0.05 | NC_024144.1 | 54 279 |
| Cronobacter phage vB_CsaM_GAP32 | Myoviridae | 411 | 0.05 | NC_019401.1 | 358 663 |
| Cyanophage P-RSM6 | Myoviridae | 54 | 6.28E-03 | NC_020855.1 | 192 497 |
| Enterobacteria phage phiX174 sensu lato | Microviridae | 21 | 0.26 | NC_001422.1 | 5386 |
| Erwinia phage Ea35-70 | Myoviridae | 69 | 5.41E-03 | NC_023557.1 | 271 084 |
| Klebsiella phage K64-1 DNA | Myoviridae | 54 | 4.77E-03 | NC_027399.1 | 346 602 |
| Mycobacterium phage Myrna | Myoviridae | 135 | 0.02 | NC_011273.1 | 164 602 |
| Pelagibacter phage HTVC008M | Myoviridae | 77 | 0.01 | NC_020484.1 | 147 284 |
| Prochlorococcus phage P-HM2 | Myoviridae | 98 | 0.01 | NC_015284.1 | 183 806 |
| Pseudomonas phage 201phi2-1 | Myoviridae | 93 | 6.85E-03 | NC_010821.1 | 316 674 |
| Pseudomonas phage PaBG | Myoviridae | 176 | 0.03 | NC_022096.1 | 258 139 |
| Sphingomonas phage PAU | Myoviridae | 90 | 0.01 | NC_019521.1 | 219 372 |
| Staphylococcus phage StB20 | Siphoviridae | 1127 | 2.5 | NC_019915.1 | 40 917 |
| Staphylococcus phage Twort | Myoviridae | 172 | 0.06 | NC_007021.1 | 130 706 |
| Synechococcus phage ACG-2014f | Myoviridae | 219 | 0.04 | NC_026927.1 | 228 143 |
| Synechococcus phage S-SKS1 | Siphoviridae | 149 | 0.02 | NC_020851.1 | 208 007 |
| Vibrio phage KVP40 | Myoviridae | 65 | 5.93E-03 | NC_005083.2 | 244 834 |
| Yersinia phage phiR1-37 | Myoviridae | 106 | 0.01 | NC_016163.1 | 262 391 |
| Mummy FI9 | |||||
| Enterobacteria phage phiX174 sensu lato | Microviridae | 416 | 1.86 | NC_001422 | 5386 |
| Lactobacillus phage AQ113 | Myoviridae | 4215 | 0.05 | NC_019782 | 36 566 |
| Staphylococcus phage StB20 | Siphoviridae | 7695 | 0.04 | NC_019915 | 40 917 |
| Mummy FI12 | |||||
| Bacillus phage G | Myoviridae | 263 | 0.41 | NC_023719.1 | 497 513 |
| Cronobacter phage vB_CsaM_GAP32 | Myoviridae | 168 | 0.07 | NC_019401.1 | 358 663 |
| Enterobacteria phage phiX174 sensu lato | Microviridae | 47 | 1.86 | NC_001422.1 | 5386 |
| Pseudomonas phage phiKZ | Myoviridae | 51 | 0.06 | NC_004629.1 | 280 334 |
Figure 3.
Examples of mapping results in mummies FI3 (Panel A), FI9 (Panel B) and FI12 (Panel C). Examples included presumptive Staphylococcus phage StB20 (mummy FI3), Lactobacillus phage AQ113 (mummy FI9) and Enterobacteria phage phiX174 sensu lato (mummy FI12). Reads mapping to the phage genomes are shown in red.
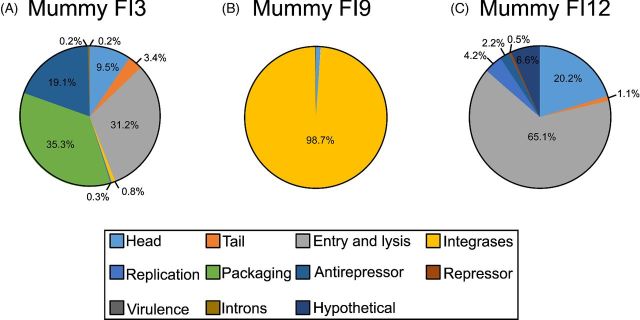
Predicted functional categories associated with bacteriophage genes were divided into structure (head and tail), entry and lysis, integrases, replication, packaging, antirepressors, repressors, virulence, introns and hypothetical proteins. The majority of the bacteriophage categories in mummy FI3 corresponded to packaging (35.3%), and entry and lysis (31.2%) (Fig. 4A). Mummy FI9 had the majority of the bacteriophage categories (98.7%) corresponding to integrases (Fig. 4B). Most of the bacteriophage categories in mummy FI12 corresponded to the entry and lysis (65.1%) (Fig. 4C).
Figure 4.
Functional categories attributed to bacteriophages. Categories included structure (head and tail), entry and lysis, integrases, replication, packaging, antirepressors, repressors, virulence genes, introns and hypothetical proteins.
DISCUSSION
Our data reconsistent with authentic ancient DNA, as shown with the high level of fragmentation (Ubaldi et al. 1998) and the SourceTracker analyses (Santiago-Rodriguez et al. 2015a). We also performed the mapDamage analyses to assess patterns of DNA damage that could be consistent with ancient DNA (Der Sarkissian et al. 2014), but did not note these patterns with the phageomes tested. While mapDamage has proven to be useful in determining DNA damage in eukaryotic genomes (Knapp et al. 2012), the method still needs to be further tested in microbiomes. Damage artifacts are often used to authenticate ancient DNA, but these may represent a challenge when characterizing ancient microbiomes. DNA damage analyses may not always provide reliable information of ancient microbiomes as nucleotide differences could also represent a novel microorganism (Warinner, Speller and Collins 2015). In addition, ancient DNA originating from microorganisms or eukaryotes would possibly need to be analyzed differently as different degrees of damage may be associated with specific taphonomic conditions.
Little is known about bacteriophage DNA preservation in ancient human specimens (Appelt et al. 2014). Our study adds to the knowledge of ancient viruses by showing that the natural mummification process of the human gut results in the preservation of bacteriophage DNA. While the recovering and sequencing methods differed, the relative abundances of siphoviruses, myoviruses, podoviruses and microviruses were virtually similar to previously reported viromes in extant human guts and coprolites (Breitbart et al. 2003; Appelt et al. 2014). Of interest was also the presence of sequences that were not homologous to known bacteriophages, consistent with their rapid evolution in modern human guts (Minot et al. 2013). These results are also consistent with previous studies showing that a proportion of viral sequences in the human gut usually cannot be assigned to existing reference genomes (Breitbart et al. 2003; Minot et al. 2011; Muniesa and Jofre 2014).
While we have demonstrated that phage DNA is preserved in mummified gut tissue, the preservation processes remain to be elucidated. A possible explanation for bacteriophage DNA preservation in mummified gut tissue may include the replication cycles. A proportion of the sequences associated with bacteriophages in the mummies guts corresponded to integrases, antirepressors and repressors. While these genes are known to be markers of lysogeny and may suggest the presence of prophages, it is difficult to demonstrate with our current data that temperate bacteriophages were in a prophage state in the mummified gut tissues. However, it is reasonable to hypothesize that prophages were preserved along with their bacterial host genomes. Of interest is also the possibility that the process of natural mummification resulted in the induction of prophages due to desiccation, which is known to trigger the lytic cycle (Brovko 2007).
The detection of strictly virulent bacteriophage DNA is intriguing as it may suggest that there may be other mechanisms supporting bacteriophage preservation during the process of natural mummification of the human gut. It is known that lytic bacteriophages are persistent members of the human microbiome and can be detected up to 60 days in the oral cavity, suggesting that they may attach to mucous layers (Barr et al. 2013; Abeles and Pride 2014; Edlund et al. 2015). Although similar studies have not been carried out in ancient samples, it is feasible that mucous layer(s) in mummified human specimens may also act as a reservoir of lytic bacteriophages (Fornaciari 1993; Corthals et al. 2012). Intact capsids may aid in the preservation of bacteriophage DNA, but this still needs to be demonstrated with mummified gut tissue using electron microscopy (Appelt et al. 2014). Another possible explanation for the detection of bacteriophages in the mummified guts is their seemingly high proportions in the human gut, where concentrations may range between 107 and 1010 per gram of feces (Muniesa and Jofre 2014). This relatively high initial concentration may aid in the detection of bacteriophage DNA even if some inactivation has occurred during natural mummification.
Given that lysogenic bacteriophages co-evolve with their bacterial hosts, culture-independent methods using sequence homology have shown to be relatively accurate in predicting putative bacterial hosts up to the genus level in modern human viromes (Minot et al. 2011; Ly et al. 2014). While the same techniques have shown to not possess this same specificity with strictly lytic bacteriophages, they are still useful in providing insights into predicted putative bacterial hosts (Ly et al. 2014). Previous studies have also associated predicted phage putative hosts with 16S rRNA gene data (Pride et al. 2012; Ly et al. 2014; Abeles et al. 2015; Santiago-Rodriguez et al. 2015c,d), but phageome relative abundances do not always mirror those of their bacterial hosts (Edlund et al. 2015). This may be due to different dynamic relationships present for different host/phage pairs (Abeles et al. 2014, 2015; Ly et al. 2014; Santiago-Rodriguez et al. 2015c). While associations between the phageome and 16S rRNA gene data exhibit limitations, results may still provide insights into phage–host interactions.
Other possible reasons for specific bacteriophages being preserved in ancient gut phageomes may include differences in gender (Abeles et al. 2014), dietary habits (Minot et al. 2011), and health status (Cadwell 2015), which are known to affect the phageome in extant human guts. We can only speculate that differences in the mummies gender, dietary habits, culture and health status may influence their phageomes (Santiago-Rodriguez et al. 2015a). This may have been the case for mummy FI9, where, although the metagenome analyses generated >140 million sequences (compared to >16 million sequences for mummies FI3 and FI12), we can only hypothesized that differences in her phageome may be associated to the mentioned factors. Ideally, virome metagenomics would better capture these trends when compared to shotgun metagenomics. While we are in the process of developing a method to study bacteriophages and other viruses in ancient specimens using viral metagenomics, from the data it is evident that shotgun metagenomics provide insights on bacteriophage sequences in ancient human gut microbiomes. Results also provide insights into bacteriophage community structure and composition in the gut of naturally preserved mummies.
Supplementary Material
Acknowledgments
This study was partially funded by a fellowship awarded to T SR from the Howard Hughes Medical Institute through the Life Sciences Research Foundation.
SUPPLEMENTARY DATA
Conflict of interest. None declared.
REFERENCES
- Abeles SR, Ly M, Santiago-Rodriguez TM, et al. Effects of long term antibiotic therapy on human oral and fecal viromes. PLoS One. 2015;10:e0134941. doi: 10.1371/journal.pone.0134941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles SR, Pride DT. Molecular bases and role of viruses in the human microbiome. J Mol Biol. 2014;426:3892–906. doi: 10.1016/j.jmb.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles SR, Robles-Sikisaka R, Ly M, et al. Human oral viruses are personal, persistent and gender-consistent. ISME J. 2014;8:1753–67. doi: 10.1038/ismej.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler CJ, Dobney K, Weyrich LS, et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat Genet. 2013;45:450–5. doi: 10.1038/ng.2536. 455e451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai Y, Meng F, Zeng Y. The evolution of pathogen-host interactions mediated by bacteriophages. Wei Sheng Wu Xue Bao. 2000;40:657–60. [PubMed] [Google Scholar]
- Appelt S, Fancello L, Le Bailly M, et al. Viruses in a 14th-century coprolite. Appl Environ Microb. 2014;80:2648–55. doi: 10.1128/AEM.03242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswad A, Katzourakis A. Paleovirology and virally derived immunity. Trends Ecol Evol. 2012;27:627–36. doi: 10.1016/j.tree.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Barr JJ, Auro R, Furlan M, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. P Natl Acad Sci USA. 2013;110:10771–6. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, et al. The oral metagenome in health and disease. ISME J. 2012;6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollback JP, Huelsenbeck JP. Phylogeny, genome evolution, and host specificity of single-stranded RNA bacteriophage (family Leviviridae) J Mol Evol. 2001;52:117–28. doi: 10.1007/s002390010140. [DOI] [PubMed] [Google Scholar]
- Breitbart M, Hewson I, Felts B, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–3. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brovko L. Identification of microorganisms and specific detection of bacteria using bioluminescence. In: Brovko L, editor. Bioluminescence for Food and Environmental Microbiology Safety. Bellingham, Washington, USA: SPIE Press; 2007. pp. 43–50. [Google Scholar]
- Cadwell K. Expanding the role of the virome: commensalism in the gut. J Virol. 2015;89:1951–3. doi: 10.1128/JVI.02966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano RJ, Rivera-Perez J, Toranzos GA, et al. Paleomicrobiology: revealing fecal microbiomes of ancient indigenous cultures. PLoS One. 2014;9:e106833. doi: 10.1371/journal.pone.0106833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano RJ, Tiefenbrunner F, Ubaldi M, et al. Sequence analysis of bacterial DNA in the colon and stomach of the Tyrolean Iceman. Am J Phys Anthropol. 2000;112:297–309. doi: 10.1002/1096-8644(200007)112:3<297::AID-AJPA2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Coberly LC, Wei W, Sampson KY, et al. Space, time, and host evolution facilitate coexistence of competing bacteriophages: theory and experiment. Am Nat. 2009;173:E121–38. doi: 10.1086/597226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthals A, Koller A, Martin DW, et al. Detecting the immune system response of a 500 year-old Inca mummy. PLoS One. 2012;7:e41244. doi: 10.1371/journal.pone.0041244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvirkaite-Krupovic V, Carballido-Lopez R, Tavares P. Virus evolution toward limited dependence on nonessential functions of the host: the case of bacteriophage SPP1. J Virol. 2015;89:2875–83. doi: 10.1128/JVI.03540-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denesvre C, Dumarest M, Remy S, et al. Chicken skin virome analyzed by high-throughput sequencing shows a composition highly different from human skin. Virus Genes. 2015;51:209–16. doi: 10.1007/s11262-015-1231-8. [DOI] [PubMed] [Google Scholar]
- Der Sarkissian C, Brotherton P, Balanovsky O, et al. Mitochondrial genome sequencing in Mesolithic North East Europe Unearths a new sub-clade within the broadly distributed human haplogroup C1. PLoS One. 2014;9:e87612. doi: 10.1371/journal.pone.0087612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund A, Santiago-Rodriguez TM, Boehm TK, et al. Bacteriophage and their potential roles in the human oral cavity. J Oral Microbiol. 2015;7:27423. doi: 10.3402/jom.v7.27423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerman M, Malik HS. Paleovirology–modern consequences of ancient viruses. PLoS Biol. 2010;8:e1000301. doi: 10.1371/journal.pbio.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornaciari G. Adenocarcinoma in the mummy of Ferrante I of Aragon, King of Naples (1431–1494) Paleopathol Newsl. 1993:5–8. [PubMed] [Google Scholar]
- Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–8. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- Ginolhac A, Rasmussen M, Gilbert MT, et al. mapDamage: testing for damage patterns in ancient DNA sequences. Bioinformatics. 2011;27:2153–5. doi: 10.1093/bioinformatics/btr347. [DOI] [PubMed] [Google Scholar]
- Hofer U. Viral evolution: variation in the gut virome. Nat Rev Microbiol. 2013;11:596. doi: 10.1038/nrmicro3092. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Dollive S, Grunberg S, et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A. Paleovirology: inferring viral evolution from host genome sequence data. Philos T Roy Soc B. 2013;368:20120493. doi: 10.1098/rstb.2012.0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp M, Horsburgh KA, Prost S, et al. Complete mitochondrial DNA genome sequences from the first New Zealanders. P Natl Acad Sci USA. 2012;109:18350–4. doi: 10.1073/pnas.1209896109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D, Kuczynski J, Charlson ES, et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8:761–3. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landini MM, Borgogna C, Peretti A, et al. Identification of the skin virome in a boy with widespread human papillomavirus-2-positive warts that completely regressed after administration of tetravalent human papillomavirus vaccine. Br J Dermatol. 2015;173:597–600. doi: 10.1111/bjd.13707. [DOI] [PubMed] [Google Scholar]
- Lavialle C, Cornelis G, Dupressoir A, et al. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos T Roy Soc B. 2013;368:20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly M, Abeles SR, Boehm TK, et al. Altered oral viral ecology in association with periodontal disease. MBio. 2014;5:e01133–14. doi: 10.1128/mBio.01133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S, Bryson A, Chehoud C, et al. Rapid evolution of the human gut virome. P Natl Acad Sci USA. 2013;110:12450–5. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S, Sinha R, Chen J, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–25. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniesa M, Jofre J. Identifying and analyzing bacteriophages in human fecal samples: what could we discover? Future Microbiol. 2014;9:879–86. doi: 10.2217/fmb.14.47. [DOI] [PubMed] [Google Scholar]
- Norman JM, Handley SA, Baldridge MT, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–60. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MR, Emerman M, Malik HS. Paleovirology - ghosts and gifts of viruses past. Curr Opin Virol. 2011;1:304–9. doi: 10.1016/j.coviro.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pride DT, Salzman J, Haynes M, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012;6:915–26. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K. Gut microbiota: the gut virome and bacterial microbiome-the early years. Nat Rev Gastroentero. 2015 doi: 10.1038/nrgastro.2015.169. [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JI, Cano RJ, Narganes-Storde YM, et al. Retroviral DNA sequences as a means for determining ancient diets. PLosOne. 2015 doi: 10.1371/journal.pone.0144951. Submittted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles-Sikisaka R, Ly M, Boehm T, et al. Association between living environment and human oral viral ecology. ISME J. 2013;7:1710–24. doi: 10.1038/ismej.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K, Nilsson C, Lim YW, et al. Metagenomic analysis of viruses in reclaimed water. Environ Microbiol. 2009;11:2806–20. doi: 10.1111/j.1462-2920.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- Santiago-Rodriguez TM, Fornaciari G, Luciani S, et al. Gut Microbiome of an 11th Century A.D. Pre-Columbian Andean Mummy. PLoS One. 2015a;10:e0138135. doi: 10.1371/journal.pone.0138135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Rodriguez TM, Fornaciari G, Luciani S, et al. Taxonomic and predicted functional profiles of the gut microbiome of naturally-preserved pre-Columbian Andean mummies. mBio. 2015b Under review. [Google Scholar]
- Santiago-Rodriguez TM, Ly M, Bonilla N, et al. The human urine virome in association with urinary tract infections. Front Microbiol. 2015c;6:14. doi: 10.3389/fmicb.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Rodriguez TM, Ly M, Daigneault MC, et al. Chemostat culture systems support diverse bacteriophage communities from human feces. Microbiome. 2015d;3:58. doi: 10.1186/s40168-015-0124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Rodriguez TM, Naidu M, Abeles SR, et al. Transcriptome analysis of bacteriophage communities in periodontal health and disease. BMC Genomics. 2015e;16:549. doi: 10.1186/s12864-015-1781-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CL, Relman DA. Microbiota's ‘little helpers’: bacteriophages and antibiotic-associated responses in the gut microbiome. Genome Biol. 2013;14:127. doi: 10.1186/gb-2013-14-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tito RY, Knights D, Metcalf J, et al. Insights from characterizing extinct human gut microbiomes. PLoS One. 2012;7:e51146. doi: 10.1371/journal.pone.0051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubaldi M, Luciani S, Marota I, et al. Sequence analysis of bacterial DNA in the colon of an Andean mummy. Am J Phys Anthropol. 1998;107:285–95. doi: 10.1002/(SICI)1096-8644(199811)107:3<285::AID-AJPA5>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Walker A. Gut metagenomics goes viral. Nat Rev Microbiol. 2010;8:841. doi: 10.1038/nrmicro2476. [DOI] [PubMed] [Google Scholar]
- Wang J, Gao Y, Zhao F. Phage-bacteria interaction network in human oral microbiome. Environ Microbiol. 2015 doi: 10.1111/1462-2920.12923. [DOI] [PubMed] [Google Scholar]
- Warinner C, Speller C, Collins MJ. A new era in palaeomicrobiology: prospects for ancient dental calculus as a long-term record of the human oral microbiome. Philos T Roy Soc B. 2015;370:20130376. doi: 10.1098/rstb.2013.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyrich LS, Dobney K, Cooper A. Ancient DNA analysis of dental calculus. J Hum Evol. 2015;79:119–24. doi: 10.1016/j.jhevol.2014.06.018. [DOI] [PubMed] [Google Scholar]
- Willner D, Furlan M, Haynes M, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PloS One. 2009;4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner D, Furlan M, Schmieder R, et al. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. P Natl Acad Sci USA. 2011;108:4547–53. doi: 10.1073/pnas.1000089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink A, Reischl U, Wolf H, et al. Molecular evidence of bacteremia by gastrointestinal pathogenic bacteria in an infant mummy from ancient Egypt. Arch Pathol Lab Med. 2000;124:1614–8. doi: 10.5858/2000-124-1614-MEOBBG. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.