Abstract
Identification of Streptococcus pneumoniae and its more than 90 serotypes is routinely conducted by culture and Quellung reactions. Quantitative polymerase chain reactions (qPCRs) have been developed for molecular detection, including a pan-pneumococcus lytA assay, and assays targeting 79 serotypes. Reactions require genomic DNA from every target to prepare standards, which can be time consuming. In this study, we have developed a synthetic DNA molecule as a surrogate for genomic DNA and present new single-plex qPCR reactions to increase molecular detection to 94 pneumococcal serotypes. Specificity of these new reactions was confirmed with a limit of detection between 2 and 20 genome equivalents/reaction. A synthetic DNA (NUversa, ∼8.2 kb) was then engineered to contain all available qPCR targets for serotyping and lytA. NUversa was cloned into pUC57-Amp-modified to generate pNUversa (∼10.2 kb). Standards prepared from pNUversa and NUversa were compared against standards made out of genomic DNA. Linearity [NUversa (R2 > 0.982); pNUversa (R2 > 0.991)] and efficiency of qPCR reactions were similar to those utilizing chromosomal DNA (R2 > 0.981). Quantification with plasmid pNUversa was affected, however, whereas quantification with synthetic NUversa was comparable to that of genomic DNA. Therefore, NUversa may be utilized as DNA standard in single-plex assays of the currently known 94 pneumococcal serotypes.
Keywords: Streptococcus pneumoniae, serotype, qPCR, NUversa
NUversa is a synthetic DNA molecule that can be utilized as a ‘universal’ standard in virtually all serotype-specific qPCR assays for pneumococcus published in the literature.
INTRODUCTION
Streptococcus pneumoniae (the pneumococcus) often causes life threatening infections, such as pneumonia, septicemia and meningitis (Klugman, Madhi and Albrich 2008; O’Brien et al.2009; van der Poll and Opal 2009). Pneumococcal disease (PD) kills ∼800 000 people, mostly children, every year worldwide (O’Brien et al.2009). To reduce the burden of PD, a 7-valent pneumococcal conjugate vaccine (PCV) was introduced in the USA in 2000 and was replaced by a 13-valent pneumococcal conjugate vaccine (PCV13) in 2010. Moreover, PCV has been introduced in many parts of the world including European countries. The introduction of these vaccines has reduced the burden of PD caused by vaccine serotypes on a global scale and has also decreased nasopharyngeal carriage of pneumococcal vaccine types in vaccinated populations (Simonsen et al.2014). There has been a modest increase of PD caused by non-vaccine types (NVT) since the introduction of PCV. In a phenomenon called serotype replacement, these strains have replaced vaccine-type (VT) strains in the nasopharynx, resulting in pneumococcal carriage rates similar to those observed prior to the introduction of vaccines (Singleton et al.2007; Weinberger, Malley and Lipsitch 2011; Feikin et al.2013).
The phenomenon of serotype replacement, and therefore the increase in prevalence of strains not included in current pneumococcal vaccines, might be of concern. NVT strains include more than 80 different serotypes. Monitoring the distribution of pneumococcal vaccine and non-vaccine serotypes is important for predicting the effectiveness of current vaccines, and might also be necessary for determining future vaccine formulations.
The Quellung reaction is the gold standard method for pneumococcal serotyping. Reactions utilize specific antibodies produced against the capsular polysaccharide (cps). In a positive reaction, antibodies produce ‘swelling’ of the pneumococcal capsule that can be observed under the microscope. While the Quellung reaction is proven, a number of molecular methods have been developed during last few years for molecular serotyping (Azzari et al.2010; Turner et al.2011; Azzari et al.2012; Pimenta et al.2013; Sakai et al.2015; Messaoudi et al.2016). Molecular methods are faster than the Quellung reactions, highly sensitive and specific, and adaptable to high-throughput platforms such as microarrays or TaqMan array cards (TACs). Satzke et al. recently evaluated molecular methods for molecular serotyping in a comprehensive, multi-center, comparative study (Satzke et al.2015). Real-time quantitative polymerase chain reaction (qPCR) proved to be highly specific and sensitive, with limits of detection (LOD) of reactions between ∼2 and ∼20 genome equivalents per reaction. qPCR reactions allow for the quantification of the bacterial load and can be utilized directly with DNA purified from nasopharyngeal specimens. The downside of qPCR reactions is that each reaction requires a specific DNA standard to construct the regression curve for the translation of real-time PCR results to genome equivalents; however, the exact whole genome size of many serotypes is frequently unavailable.
We, as well as others including the Centers for Disease Control and Prevention (CDC), have previously reported a series of serotype-specific single-plex and multiplex qPCR assays (Azzari et al.2010; Azzari et al.2012; Pimenta et al.2013; Sakai et al.2015; Messaoudi et al.2016; Pholwat et al.2016). Altogether, the reactions detect all 13 PCV types and 66 NVT strains, demonstrating high sensitivity and providing the absolute densities of individual serotypes in each sample. Although there is great progress with serotype coverage of qPCR reactions, there are still a number of serotypes for which molecular quantitative reactions are not available. In this study, we developed eleven novel single-plex qPCR assays for the detection and quantification of NVT strains. Furthermore, we engineered a synthetic DNA fragment containing sequences for lytA and for 94 pneumococcal serotypes, including targets for all qPCR assays developed thus far for pneumococcal serotyping. The synthetic DNA was cloned into a plasmid, hereafter called pNUversa, and was further characterized to be used as a standard for all qPCR reactions. Using pNUversa DNA as a standard allowed detection, but the plasmid's conformational structure affected quantification. A linearized pNUversa—a PCR product generated using pNUversa DNA—restored efficiency of quantitative reactions to levels similar to those achieved when using chromosomal DNA as a standard.
METHODS
Bacterial strains utilized in this study
Strains and reference strains utilized in this study were either originally isolated from clinical specimens and serotyped by Quellung reactions at Emory University, purchased from the Statens Serum Institut (Copenhagen, Denmark), or kindly provided by Dr Lesley McGee and Dr Bernard Beall from the CDC. Strains included serotypes: 1, 2, 3, 4, 5, 6A, 6B, 6C, 6D, 7A, 7B, 7C, 7F, 8, 9A, 9L, 9N, 9V, 10A, 10B, 10C, 10F, 11A, 11B, 11C, 11D, 11E, 11F, 12A, 12B, 12F, 13, 14, 15A, 15B, 15C, 15F, 16A, 16F, 17A, 17F, 18A, 18B, 18C, 18F, 19A, 19B, 19C, 19F, 19‘F’ (atypical), 20, 21, 22A, 22F, 23A, 23B, 23F, 24A, 24B, 24F, 25A, 25F, 27, 28A, 28F, 29, 31, 32A, 32F, 33A, 33B, 33C, 33D, 33F, 34, 35A, 35B, 35C, 35F, 36, 37, 38, 39, 40, 41A, 41F, 42, 43, 44, 45, 46, 47A, 47F and 48. To assess specificity, the following non-pneumococcal species found in the upper airways of healthy individuals were also included: Streptococcus infantis, Streptococcus oralis, Streptococcus anguinosus, Streptococcus intermedius, Streptococcus sobrinus, Streptococcus pseudopneumoniae, Streptococcus mitis, Streptococcus parasanguinis, Streptococcus australis, Streptococcus mutans, Streptococcus peroris, Streptococcus oligofermentans, Streptococcus intestinalis, Streptococcus vestibularis, Streptococcus cristatus, Streptococcus salivarius, Streptococcus gordonii, Streptococcus sanguinis, Streptococcus sinensis and Dolosigranulum pigrum (Carvalho Mda et al.2012). DNA from Streptococcus pneumoniae reference strain TIGR4 (GenBank accession no. Z_AAGY00000000) (Tettelin et al.2001) was utilized as the standard for the pan-pneumococcus lytA qPCR assay (Carvalho Mda et al.2007).
Purification of DNA and preparation of DNA standards for validating qPCR assays
Strains were grown overnight on blood agar plates, and DNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) as follows. Pneumococci were resuspended in tris-ethylenediaminetetraacetic acid (TE) buffer (10 mM tris-HCl, 1 mM ethylenediaminetetraacetic acid, pH 8.0) containing 40 mg/ml lysozyme and 75 U/ml mutanolysin (Sigma-Aldrich Co., Saint Louis, Missouri, USA). This suspension was incubated at 37°C for 1 h after which 20 μl of proteinase K were added and incubated for 30 min at 56°C. Then, 4 μl of RNase A (100 mg/ml) (Qiagen, Hilden, Germany) were added to the suspension and incubated 5 min at room temperature. The following steps were conducted as outlined by the manufacturer. The eluted DNA was quantified using the Nanodrop system (Nanodrop Technologies, Wilmington, Delaware, USA). DNA was diluted with TE buffer to obtain DNA standards of the following amounts per reaction: 1 ng, 100 pg, 10 pg, 1 pg, 100 fg, 50 fg and 5 fg. Considering the genome size of the reference strain TIGR4, 2.16 Mb (Tettelin et al.2001), the approximate genome equivalent for each DNA standard was: 4.29 × 105, 4.29 × 104, 4.29 × 103, 4.29 × 102, 4.29 × 101, 2.14 × 101 and 2.14 genome equivalents, respectively. The efficiency (i.e. regression curves) of reactions using the above standards was evaluated in each run. Primers and probes were optimized for each quantitative reaction. The LOD was determined from the lowest concentration standards that achieved a positive reaction in each assay. The Ct-value cut-off to distinguish a positive reaction from a negative reaction was 40.
Development of novel qPCR assays for NVT Streptococcus pneumoniae
Novel qPCR assays for serotypes 10CF, 11BC, 16A, 17A, 17F, 19C, 24BF, 28AF, 32AF, 33C and 48 were developed. Systematic design of the new qPCR assays was essentially done as detailed in our previous study (Sakai et al.2015). Briefly, the pneumococcal cps locus was obtained from the GenBank Database. Accession numbers are listed in Table 1. Specificity in silico was analyzed using the National Center for Biotechnology Information-Basic Local Alignment Search Tool. Once specific sequences were selected within each locus, assays were designed utilizing software from Integrated DNA technologies (http://www.idtdna.com/site). All qPCR primers and probes were synthesized at Sigma-Aldrich Co. The assays were performed utilizing Platinum Quantitative PCR Super Mix-UDG (Life Technology, Carlsbad, California, USA) and 2.5 μl of pure DNA as template in a CFX96 real-time PCR-detection system (Bio-Rad, Hercules, California, USA).
Table 1.
Novel quantitative PCR assays for non-vaccine serotypes.
| Serotype(s) | Sequencea | Target region | Accession No. | Position | Size (bp) | Limit of detection genome equivalents (fg) | Concentration (nM) | Reaction efficiency (avg) | |
|---|---|---|---|---|---|---|---|---|---|
| 10CF | Forward | CGAGTTATGGATGTTCTTATTGGC | wcjG | CR931651 | 4808–4831 | 137 | 21.4 (50) | 400 | 103.4 |
| Reverse | CCCAACCCCACTCTGTATTG | 4925–4944 | 400 | ||||||
| Probe | ACAGGGCAAGACTGTGAATATTGTTCCA | 4837–4864 | 300 | ||||||
| 11BC | Forward | TCAAATTTGGCGTATTGCTTATCA | wzy | CR931654 | 10 998–11 021 | 106 | 2.1 (5) | 400 | 102.8 |
| Reverse | TGATTATGAGCATAGTTGATCCCC | 11 103–11 126 | 400 | ||||||
| Probe | TCCGTGGCAAGATTCTGGTGCTAAG | 11 079–11 103 | 200 | ||||||
| 16A | Forward | GCTAGCAGGAACTTTTCTAGGG | wcxR | CR931667 | 6675–6696 | 132 | 21.4 (50) | 200 | 92.9 |
| Reverse | TCCCTGTCCAAATCCGAAAC | 6787–6806 | 200 | ||||||
| Probe | CCCACGGGATGAATCCATTATGGCG | 6703–6727 | 200 | ||||||
| 17A | Forward | TGATTATGTCATTCGATTGCTTGG | wzy | CR931669 | 13 895–13 918 | 112 | 2.1 (5) | 400 | 93.5 |
| Reverse | AAATCCTAAAATTCCTGTTTGAAAAGC | 13 980–14 006 | 400 | ||||||
| Probe | ATTATGGGCGTGGGTTACCGTAGG | 13 941–13 964 | 200 | ||||||
| 17F | Forward | TGCTTTTGTGGGTAGGACAAG | wzx | CR931670 | 17 361–17 381 | 130 | 21.4 (50) | 400 | 97.8 |
| Reverse | TTATCCCATAAACCTGAGGCG | 17 470–17 490 | 400 | ||||||
| Probe | TGCAGGTGATATGCGGAGCCAAT | 17 440–17 462 | 200 | ||||||
| 19C | Forward | AATGGTTTTCAGATTACTTGATAGCTC | wchU | CR931677 | 19 294–19 320 | 115 | 2.1 (5) | 400 | 95.7 |
| Reverse | CGTTCCTTATGAGAGTGGTCAAG | 19 386–19 408 | 400 | ||||||
| Probe | TGTTCCTGCCCCCACATAATGAACT | 19 343–19 367 | 200 | ||||||
| 24BF | Forward | TCTGAAAGTAATTAGTAAGATTAACGGAAG | wzy | CR931688 | 15 038–15 067 | 133 | 2.1 (5) | 400 | 101.5 |
| Reverse | TCCATCTACTTTTAAAATAGCTCCAAC | 15 144–15 170 | 400 | ||||||
| Probe | CCACAGTCCCAAAATTGTCAGCAACC | 15 085–15 110 | 200 | ||||||
| 28AF | Forward | CAACTACAGGTATTTTTGATATCGGAG | wcxP | CR931692 | 12 047–12 073 | 141 | 2.1 (5) | 400 | 96.7 |
| Reverse | GTTTACTACGTTTGTGAAGCGC | 12 166–12 187 | 400 | ||||||
| Probe | AGAAAATAGTAGGTTGATTGGCGGTGCT | 12 075–12 102 | 200 | ||||||
| 32AF | Forward | GTACTTCCTGTTCTAGGCTTGG | wzy | CR931696 | 12 471–12 492 | 134 | 2.1 (5) | 400 | 101.7 |
| Reverse | CCCAGAGGAAAATAGCGTCTC | 12 584–12 604 | 400 | ||||||
| Probe | TTGTTCAAACCCAACCACTGCTCC | 12 492–12 515 | 200 | ||||||
| 33C | Forward | CAGAGACAGTTTCAGCAAATCTTAG | wzy | CR931700 | 11 270–11 294 | 147 | 2.1 (5) | 400 | 103 |
| Reverse | AGCCTACACCTCTTATAAACGTTG | 11 393–11 416 | 400 | ||||||
| Probe | CCGTGTCCTATCCACAAACTTGTCTTCC | 11 327–11 354 | 200 | ||||||
| 48 | Forward | CAGGTTTTGCTTCATATGGGAG | wzy | CR931722 | 13 043–13 064 | 133 | 2.1 (5) | 400 | 94.9 |
| Reverse | ATCGGCCAAAAGTTATCATTAGC | 13 153–13 175 | 400 | ||||||
| Probe | CGCTGCTTATGTGTATTACTCTCCCCTG | 13 068–13 095 | 200 |
Probes were labeled at 5΄ with FAM (6-carboxyfluorescein) and at 3΄ with BHQ1 (Black Hole Quencher-1).
Specificity of newly developed qPCR assays
Specificity was investigated as previously described (Sakai et al.2015). In short, two DNA libraries were prepared: one containing DNA from all known pneumococcal serotypes (N = 94) and a second library containing DNA from related bacterial species that inhabit the upper respiratory airways (N = 20). Purified DNA preparations were adjusted to 40 pg/μl, and 150 μl of each DNA was transferred to a 96-well microtiter plate and stored at –80°C until use. Specificity testing utilized 2.5 μl of DNA, equivalent to 100 pg (i.e. 4.29 × 104 genome equivalents), which was transferred from the libraries to each reaction plate. Reactions were run in a CFX96 real-time PCR-detection system.
Cloning and extraction of synthetic vector containing all qPCR target sequences for serotyping
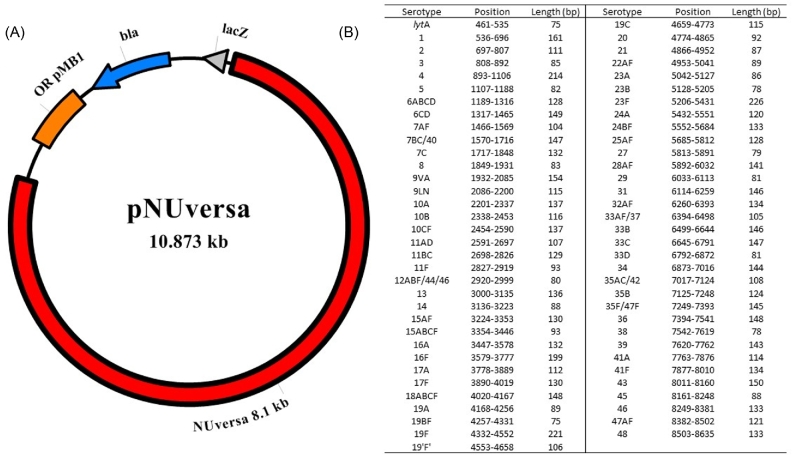
The qPCR target sequences of all serotype-specific single-plex assays (N = 66), as well as lytA, were sequentially assembled into a single sequence using the DNASTAR Lasergene software version 11.2.1 (DNASTAR Inc., Madison, Wisconsin, USA). The final nucleotide sequence (∼8.2 kb) was synthesized and then subcloned within pUC57-Amp-modified (GenScript, Piscataway, New Jersey, USA). The synthetic vector (∼10.9 kb), named pNUversa (Fig. 1), was transformed into TOP10 competent Escherichia coli (Invitrogen, Carlsbad, California, USA) according to the manufacturer's instructions. pNUversa was purified from E. coli stocks using the QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany), eluted in EB buffer and quantified using the Nanodrop system. pNUversa was utilized as template in PCR reactions to produce the synthetic product, hereafter called NUversa.
Figure 1.
pNUversa plasmid map and position of sequences. (A) The synthetic NUversa gene (8175 bp) was cloned into a pUC57-Amp modified cloning vector to generate pNUversa (10 873 bp). The origin of replication (OR pMB1), ampicillin resistance gene (bla) and lacZ gene where the multiple cloning site is located are indicated. (B) Left panel shows the list of targets (N = 67) for single-plex quantitative PCR reactions included within NUversa. Targets for assays detecting more than one serotype, or serogroup, are denoted by a serogroup number and serotypes, i.e. 6ABCD, or serotypes separated by a slash, i.e. 7BC/40. Sequences are listed as they appear in pNUversa, including nucleotide position within NUversa and length of the fragment in bp.
Long PCR and purification of the synthetic DNA (NUversa)
To produce the synthetic DNA fragment NUversa, long PCR was conducted with plasmid DNA from pNUversa as the template, platinum Taq DNA polymerase High Fidelity (Invitrogen, Carlsbad, California, USA) and the following primers whose complementary sequences are located upstream and downstream of NUversa: lytA re (5΄-TCGTGCGTTTTAATTCCAGCT-3΄: 200 nM) and Sero48 re (5΄-ATCGGCCAAAAGTTATCATTAGC-3΄: 200 nM). Reaction conditions were 30 s at 94°C for initial denaturation, followed by 35 cycles of 15 s at 94°C, 30 s at 60°C and 9 min at 68°C for amplification. PCR product was confirmed with electrophoresis in 1% agarose gel (Standard Low -mr Agarose, Bio-Rad Laboratories Inc., Hercules, California, USA) with SYBR safe staining (Invitrogen, Carlsbad, California, USA). The obtained NUversa product was purified with the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions, eluted with EB buffer, evaluated for quality and quantified using the Nanodrop system. The purified NUversa product was stored at –80°C until use.
Preparation of qPCR standards
To evaluate qPCR standards prepared with plasmid pNUversa DNA, and the synthetic NUversa PCR product, single-plex qPCR assays for the pan-pneumococcus lytA assay and serotype-specific qPCR assays targeting PCV13 serotypes were performed. Regression curves were obtained in each assay. Moreover, chromosomal DNA that was extracted from reference strains (TIGR4 and reference strains of serotype 1, 3, 4, 5, 6A, 7F, 9V, 14, 18C, 19A, 19F and 23F) was also included in these experiments. For quantification purposes, genome equivalents were estimated using the formula: molecules/mol × mol/g = molecules/g.
Utilizing Avogadro number and the average weight of a base pair (bp) [650 Da (g/mol)], the number of molecules were estimated as
#6.022 × 1023 (molecules/mole) × (gene amount (g)/(length of target gene (bp) × 650 (Da)).
#Avogadro number
Taking into account that the size of pNUversa and the NUversa PCR product were 10 873 bp and 8175 bp, respectively, genome equivalents contained in 100 pg and 10 pg of the vector and the PCR product were ∼8.52 × 106 and ∼1.13 × 106, respectively. DNA standards prepared by serial dilutions corresponded to [pNUversa] 8.52 × 106, 8.52 × 105, 8.52 × 104, 8.52 × 103, 8.52 × 102, 8.52 × 10, 8.52 and 4.26, genome equivalents or [NUversa] 1.13 × 106, 1.13 × 105, 1.13 × 104, 1.13 × 103, 1.13 × 102, 1.13 × 10 and 5.65, genome equivalents. Primer and probe sequences, and concentration, of VT single-plex qPCR assays were previously described (Pimenta et al.2013). qPCR assays were performed as mentioned above using Platinum Quantitative PCR SuperMix-UDG and the CFX96 real-time PCR-detection system.
RESULTS
Validation and optimization of quantitative PCR assays
Eleven newly designed qPCR assays (Table 1) were validated for their specificity using DNA from 94 pneumococcal serotypes and DNA from 20 non-pneumococcal species, most of which are inhabitants of the human upper airways, including several streptococcal species. Reactions were 100% specific as they only detected DNA in wells where the target pneumococcal serotype was added (not shown). Ct values of a typical positive reaction were between 20 and 30 cycles. Table 1 shows the optimal concentration that yielded a recommended reaction efficiency of between 90% and 110% and a correlation coefficient of nearly 1.000.
The LOD was also evaluated for each assay using an approach including serial dilution. Quantitative assays demonstrated an LOD of 5–50 fg per reaction, or ∼2–∼20 genome equivalents per reaction. Altogether, these new qPCR assays demonstrated high specificity, sufficient reaction efficiency to be utilized for quantification and detection of very low genome equivalents.
Efficiency of quantitative reactions using NUversa is similar to those obtained using genomic DNA
We engineered a synthetic DNA molecule containing targets for most, if not all, qPCR reactions available in the literature (N = 66) including the lytA target and those sequences presented in this study that together target 94 pneumococcal serotypes. The synthetic DNA molecule was cloned into plasmid pUC57-Amp-modified and this new plasmid was named pNUversa (Fig. 1). The sequence of pNUversa was deposited in the GenBank with accession number MF540153. Purified pNUversa was then utilized as a template in qPCR reactions to generate linear DNA, called NUversa, which was used as a standard in lytA-based qPCR reactions.
We first evaluated the quantitative nature of qPCR reactions targeting the lytA gene using NUversa DNA standards, and compared these results with those obtained utilizing the traditional standard genomic DNA from TIGR4. Efficiency of reactions from four independent experiments (i.e. each experiment performed in duplicate) with NUversa (94.1 ± 1.3) or genomic DNA (94.1 ± 3.2) fell within the recommended efficiency of >90% and <110% (Table 2). Regression curves showed a similar R2, 0.9993–1.000 when using TIGR4 genomic DNA and 0.991–0.997 when NUversa PCR product was utilized (Fig. 2). Standard deviations of Ct values obtained with the different standards ranged from 0.2555 to 0.3621 when utilizing TIGR4 genomic DNA, whereas those utilizing NUversa ranged from 0.4904 to 1.4602. Standard error of the means was 0.1278–0.1811 with TIGR4 genomic DNA and 0.2452–1.0325 with NUversa PCR product (Table 2). Further statistical analysis showed no significance when R2, the slope or y-intercept were compared by two-tailed t-Test (P > 0.05, all cases).
Table 2.
lytA quantitative PCR with TIGR4 genomic DNA and NUversa.
| Efficiency (%) | 95.5 | 98.0 | 90.9 | 92.2 | ||||
|---|---|---|---|---|---|---|---|---|
| Genomic DNA | Gen Eqa | Ct | Ct | Ct | Ct | Mean | SD | SEM |
| 1 ng | 4.29E+05 | 20.21 | 20.78 | 20.16 | 20.36 | 20.38 | 0.2819 | 0.1409 |
| 100 pg | 4.29E+04 | 23.62 | 24.32 | 23.65 | 23.82 | 23.85 | 0.3215 | 0.1607 |
| 10 pg | 4.29E+03 | 27.03 | 27.61 | 27.14 | 27.21 | 27.25 | 0.2555 | 0.1278 |
| 1000 fg | 4.29E+02 | 30.52 | 31.25 | 30.73 | 30.94 | 30.86 | 0.3125 | 0.1563 |
| 100 fg | 4.29E+01 | 34.47 | 34.36 | 34.22 | 34.90 | 34.49 | 0.2910 | 0.1455 |
| 50 fg | 2.14E+01 | 34.90 | 35.35 | 35.59 | 35.73 | 35.39 | 0.3621 | 0.1811 |
| 5 fg | 2.14E+00 | 38.20 | 38.68 | 38.98 | 38.60 | 38.61 | 0.3226 | 0.1613 |
| NUversa | Efficiency (%) | 92.8 | 93.3 | 94.7 | 95.8 | |||
| 10 pg | 1.13E+06 | 20.23 | 19.17 | 19.13 | 19.80 | 19.58 | 0.5336 | 0.2668 |
| 1000 fg | 1.13E+05 | 23.89 | 22.60 | 22.73 | 23.27 | 23.12 | 0.5880 | 0.2940 |
| 100 fg | 1.13E+04 | 27.37 | 26.02 | 26.23 | 26.57 | 26.55 | 0.5967 | 0.2983 |
| 10 fg | 1.13E+03 | 31.08 | 29.50 | 30.02 | 30.23 | 30.21 | 0.6567 | 0.3283 |
| 1000 ag | 1.13E+02 | 34.17 | 32.99 | 33.55 | 33.74 | 33.61 | 0.4904 | 0.2452 |
| 100 ag | 1.13E+01 | 37.87 | 36.69 | 36.60 | 37.62 | 37.19 | 0.6455 | 0.3228 |
| 50 ag | 5.65E+00 | Negative | Negative | 39.31 | 37.25 | 38.28 | 1.4602 | 1.0325 |
Genome equivalents.
Figure 2.
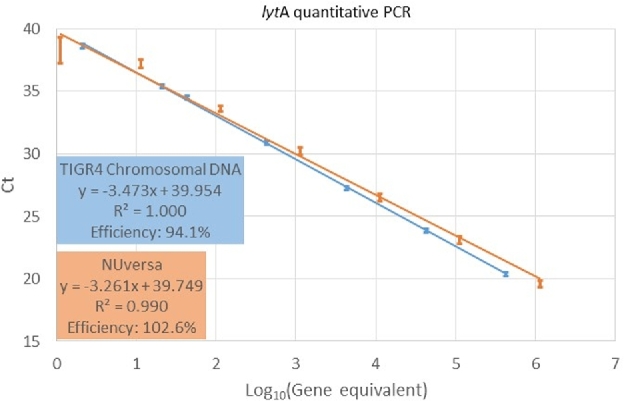
Linearity of qPCR reactions utilizing NUversa or genomic DNA. NUversa (orange) and genomic DNA (blue) were serially diluted to obtain seven standards (detailed in Methods) spanning 5 through 1.13 × 106 and 2 through 4.29 × 105 genome equivalents, respectively. Genome equivalent standards were utilized as template in qPCR reactions targeting the lytA gene. Plots represent the mean of cycles (Ct) of threshold values. Standard errors were calculated from four different replicates and each replicate included duplicate reactions. Regression equations (y), coefficient of determination (R2) and reaction efficiency are shown in the insets.
The synthetic DNA (NUversa), but not the plasmid, can be utilized as standards for serotype-specific qPCR reactions
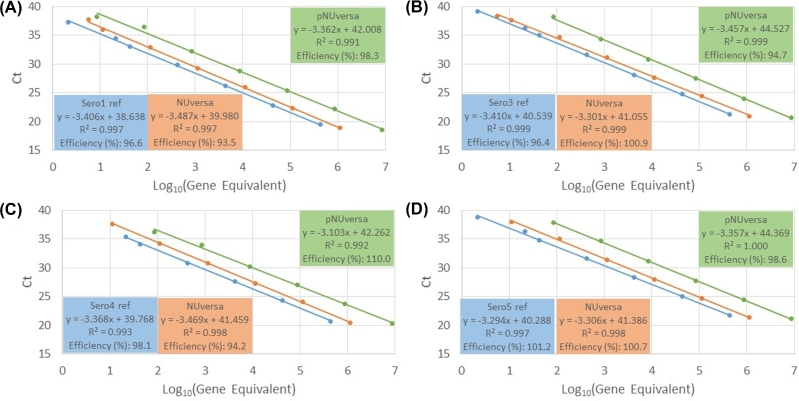
Given that current real-time platforms are utilizing plasmid DNA as standards, we next investigated whether NUversa and pNUversa (i.e. the plasmid encoding NUversa) could be utilized as DNA standards for serotype-specific single-plex qPCR reactions. Serial dilutions of genomic DNA, pNUversa or NUversa were made and utilized as template in serotype-specific qPCR reactions targeting PCV13 serotypes, (i.e. 1, 3, 4, 5, 6A, 6B 7F, 9V, 14, 18C, 19A, 19F and 23F). Because most strains from where the genomic DNA was extracted have not been genome sequenced, the approximate genome equivalent was estimated using the whole genome size of serotype 4 strain TIGR4 (Tettelin et al.2001). As shown in Table 3, Fig. 3 and Supplementary Fig. S1, reaction efficiency of most qPCR assays using any of the standards fell within the recommended efficiency of >90% and <110%. The y-intercept, however, was only similar in reactions utilizing genomic DNA and NUversa, whereas reactions that utilized the plasmid pNUversa were statistically different to those with genomic DNA and those with NUversa (Table 3).
Table 3.
Comparison of regression curve parameters obtained with serotype/serogroup-specific qPCR using genomic DNA, NUversa and pNUversa as standards.
| Serotype/serogroup-specific qPCR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 4 | 5 | 6ABCD | 7AF | 9VA | 14 | 18ABCF | 19F | 19A | 23F | Average | SD | bt-Test | ||
| Efficiency (%) | aChromosomal DNA | 96.6 | 96.4 | 98.1 | 101.2 | 98.5 | 100.1 | 98.2 | 91.2 | 99.0 | 101.1 | 91.6 | 96.0 | 97.3 | 3.247 | |
| NUversa linear | 93.5 | 100.9 | 94.2 | 100.7 | 91.7 | 94.1 | 107.5 | 100.0 | 95.7 | 95.6 | 94.8 | 95.8 | 97.0 | 4.417 | 0.859 | |
| pNUversa | 98.3 | 94.7 | 110.0 | 98.6 | 97.1 | 96.0 | 101.5 | 94.6 | 106.4 | 96.8 | 90.9 | 100.5 | 98.8 | 5.268 | 0.327 | |
| The coefficient of | aChromosomal DNA | 0.997 | 0.999 | 0.993 | 0.997 | 0.999 | 0.997 | 0.992 | 0.994 | 0.994 | 0.982 | 0.981 | 0.999 | 0.994 | 0.006 | |
| determination (R2) | NUversa linear | 0.997 | 0.999 | 0.998 | 0.998 | 0.990 | 0.993 | 0.987 | 0.982 | 0.966 | 0.997 | 0.988 | 0.999 | 0.991 | 0.010 | 0.438 |
| pNUversa | 0.991 | 0.999 | 0.992 | 1.000 | 0.994 | 0.992 | 0.999 | 0.997 | 0.995 | 0.997 | 0.995 | 0.999 | 0.996 | 0.003 | 0.298 | |
| Slope | aChromosomal DNA | –3.406 | –3.410 | –3.368 | –3.294 | –3.357 | –3.319 | –3.366 | –3.554 | –3.347 | –3.295 | –3.540 | –3.422 | –3.390 | 0.084 | |
| NUversa linear | –3.487 | –3.301 | –3.469 | –3.306 | –3.539 | –3.471 | –3.154 | –3.321 | –3.429 | –3.432 | –3.452 | –3.427 | –3.399 | 0.108 | 0.825 | |
| pNUversa | –3.362 | –3.457 | –3.103 | –3.357 | –3.393 | –3.422 | –3.287 | –3.458 | –3.178 | –3.402 | –3.562 | –3.310 | –3.358 | 0.126 | 0.353 | |
| y-intercept | aChromosomal DNA | 38.638 | 40.539 | 39.768 | 40.288 | 40.793 | 39.601 | 40.084 | 38.990 | 38.959 | 39.667 | 40.468 | 40.245 | 39.837 | 0.691 | |
| NUversa linear | 39.980 | 41.055 | 41.459 | 41.386 | 41.912 | 40.813 | 39.744 | 38.545 | 39.631 | 38.976 | 40.323 | 39.795 | 40.302 | 1.040 | 0.081 | |
| pNUversa | 42.008 | 44.527 | 42.262 | 44.369 | 43.615 | 43.144 | 42.894 | 41.956 | 41.524 | 41.889 | 44.880 | 42.240 | 42.942 | 1.157 | 2.49E-08 | |
Serotypes of strains for chromosomal DNA preps were 1, 3, 4, 5, 6A, 7F, 9V, 14, 18C, 19F, 19A and 23F
Set of parameter values from NUversa and pNUversa reactions were statistically compared to values from chromosomal DNA reaction with unpaired, two-tailed t-test.
Figure 3.
Linearity of qPCR reactions utilizing genomic DNA, NUversa or plasmid pNUversa. Genomic DNA (blue), NUversa (orange) or pNUversa (green), were serially diluted to obtain seven (genomic DNA and NUversa) or eight (pNUversa) standards (detailed in Methods) spanning 2 through 4.29 × 105, 5 through 1.13 × 106 and 4 through 8.52 × 106 genome equivalents, respectively. Genome equivalent standards were utilized as template in serotype-specific qPCR reactions targeting (A) serotype 1, (B) serotype 3, (C) serotype 4 or (D) serotype 5. Plots represent the mean of cycles of threshold values obtained from duplicate reactions. Regression equations, coefficient of determination (R2) and reaction efficiency are shown in the insets.
DISCUSSION
In this study, NUversa, a synthetic DNA molecule was thoroughly characterized for its use as a standard in quantitative single-plex reactions for PCV13 pneumococcal serotypes including the pan-pneumococcus lytA assay. NUversa has the potential to be utilized in reactions targeting most pneumococcal serotypes. We also developed, tested and optimized eleven new real-time qPCR assays for the detection and quantification of 16 pneumococcal serotypes/serogroups belonging to NVT. While these NVT strains are less likely to be isolated from cases of PD, the emergence of NVT due to serotype replacement after vaccine introduction warrants development of assays for the detection of potential emergent strains and their use in epidemiological surveillance programs (Wyres et al.2013; Mosser et al.2014; van der Linden et al.2015).
To the best of our knowledge, many of the molecular assays (i.e. conventional PCR, qPCR, etc.) for the detection of target pneumococcal serotypes that were presented in this study have not previously been available. These new reactions, along with other qPCR reactions published by other groups, including the CDC, complete the coverage of the 94 pneumococcal serotypes for which an antiserum is available. We acknowledge that single-plex reactions for pneumococcal serotyping have a limited use in epidemiological studies because of the associated cost, need for increased amounts of DNA sample and increased time required to perform single-plex assays. However, validation studies conducted in this manuscript will allow qPCR assays, primers and probes described here to be incorporated into high-throughput technology such as multiplex qPCR schemes (Pimenta et al.2013) or a recently developed TAC for pneumococcal serotyping (Pholwat et al.2016). The TAC, for example, detects 78 different pneumococcal serotypes. It utilizes either DNA from Streptococcus pneumoniae isolates or DNA purified from clinical specimens as a template. This method yields fast and comprehensive serotyping in comparison to the standard method of Quellung reactions, but currently misses detection of >10 pneumococcal serotypes. The new reactions developed and validated here can be added to the TAC method, which will increase the number of molecular targets to 94 pneumococcal serotypes.
Another contribution of the studies presented within this manuscript was the characterization of NUversa and pNUversa. NUversa includes sequences from 67 qPCR targets that together detect 94 serotypes and performs as well as genomic DNA when utilized as a DNA standard in qPCR lytA-based reactions. This synthetic DNA can be utilized as a positive control or as a DNA standard for quantitative reactions in any laboratory where single-plex qPCR is performed. Moreover, with further optimization, NUversa could also be used as a control in multiplex reactions. Supporting this, we tested NUversa in two different multiplex reactions published by the CDC (Pimenta et al.2013), and NUversa was detected by all reactions within these multiplex reactions (not shown).
Using NUversa will allow a faster turnaround of data and findings, and it will be particularly helpful in resource limited settings, where maintaining a strain library required for current methods is prohibitively expensive and time consuming. To calculate genome equivalents per mass units (i.e. in silico), the genome size of the strain utilized to prepare DNA standards is required. A reference, genome-sequenced, TIGR4 strain has been classically utilized to prepare standards for quantitative reactions targeting the pan-pneumococcus lytA qPCR assay (Tettelin et al.2001; Carvalho Mda et al.2007). This strain has been widely distributed among research laboratories, and it is accessible to clinical laboratories. However, widely accessible pneumococcal strains belonging to all known serotypes to be utilized as a standard for serotype-specific single-plex qPCR reactions represent a challenge. Given that NUversa also contains the target for the pan-pneumococcus lytA assay, quantification using NUversa will be standardized in all studies, avoiding the use of subjective quantitative numbers obtained from DNA of poorly characterized strains as a DNA standard for constructing the standard curve. pNUversa, the plasmid encoding the NUversa synthetic sequence, was designed in a manner that allows for more sequences to be cloned into it if more serotypes are discovered in the years to come, or if new improved reactions, i.e. targets, become available for the current serotypes.
Different multiplex molecular platforms for the detection of human pathogens have been developed during the last few years (Kodani and Winchell 2012; Pholwat et al.2015). These platforms commonly utilize a plasmid containing targets for reactions detected by the systems. According to Hou et al., however, a circular plasmid is not recommended as a quantification standard for real-time PCR assays because it frequently forms supercoiled structures; this unknown conformational structure may restrict polymerase reaction and cause delayed Ct of corresponding standards (Hou et al.2010).
In the current study, detection of target sequences in a plasmid (pNUversa) did not pose major issues but the quantitative nature of this system was proven to be inaccurate and required a synthetic linear DNA molecule. Absolute quantitative assays for PCV13 serotypes were conducted to compare the reactivity of chromosomal DNA and two types of synthetic genes: a circular plasmid and NUversa PCR product. Whereas the regression curves generated from chromosomal DNA were highly similar to those from NUversa PCR product, a significant discrepancy was found between the y-intercepts of the chromosomal DNA and the y-intercept of circular pNUversa reactions. These high y-intercept values with pNUversa caused apparent overestimation of quantification values when compared to both chromosomal DNA and NUversa. Using a plasmid as standard for quantification purposes, at least in this case, overestimates the quantity of corresponding Ct values. Together, these data indicate that synthetic NUversa is as efficient as genomic DNA to quantify genome equivalents in serotype-specific qPCR reactions. We, however, did not perform qPCR assays for the remaining NVT that can be a limitation of our study. Another limitation includes the diversity of normal flora strains utilized for the validation studies. Whereas specificity was investigated using DNA from all 94 serotypes, 20 different normal flora streptococci and other respiratory bacteria, we did not include all potential bacterial species that can colonize the human upper airways.
In this study, the whole genome size of serotype reference strains was estimated utilizing the genome size of TIGR4. However, this estimation may cause inaccurate quantification since the whole genome size might vary between serotypes. For further assessment of this synthetic DNA, lytA qPCR assays with TIGR4 chromosomal DNA and NUversa PCR product were compared. Since the genome sizes of both DNA preparations were available, specific genome equivalents for each standard could be estimated. The obtained regression curves from these two sources of standards were highly similar and no significant (i.e. statistically) discrepancy was observed. This result emphasized the significance of linearity and accurate gene size for absolute quantification, and demonstrated that NUversa PCR product can be utilized instead of chromosomal DNA in the assay (Hou et al.2010).
In summary, the synthetic NUversa DNA and the new single-plex reactions included in this study will be useful to many in the field conducting surveillance of the pneumococcus and pneumococcal serotypes by qPCR. Integrating NUversa and these qPCR assays with other previously reported high-throughput systems, such as the TAC system, will improve investigations of disease trends in this age of serotype replacement post-vaccination. These advancements will greatly contribute to the ongoing monitoring and evaluation of pneumococcal immunization programs around the world.
Supplementary Material
Supplementary data are available at FEMSLE online.
Acknowledgements
Authors are thankful to Dr Lesley McGee and Dr Bernard Beall from the CDC for providing several pneumococcal serotypes and all other streptococci utilized in this study. We also want to thank Dr Catherine Satzke, Dr Eileen Dunne and Professor Kim Mulholland, from Murdoch Childrens Research Institute, for stimulating discussion and providing support and advice to develop new technology for pneumococcal serotyping.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSLE online.
FUNDING
This work was supported by a subcontract granted to JEV (No. 26180) by Murdoch Childrens Research Institute, which received funds from the Bill and Melinda Gates Foundation (Grant 52099).
Conflict of interest. None declared.
REFERENCES
- Azzari C, Moriondo M, Cortimiglia M et al. . Potential serotype coverage of three pneumococcal conjugate vaccines against invasive pneumococcal infection in Italian children. Vaccine 2012;30:2701–5. [DOI] [PubMed] [Google Scholar]
- Azzari C, Moriondo M, Indolfi G et al. . Realtime PCR is more sensitive than multiplex PCR for diagnosis and serotyping in children with culture negative pneumococcal invasive disease. PLoS One 2010;5:e9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho Mda G, Bigogo GM, Junghae M et al. . Potential nonpneumococcal confounding of PCR-based determination of serotype in carriage. J Clin Microbiol 2012;50:3146–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho Mda G, Tondella ML, McCaustland K et al. . Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 2007;45:2460–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feikin DR, Kagucia EW, Loo JD et al. . Serotype-specific changes in invasive pneumococcal disease after pneumococcal conjugate vaccine introduction: a pooled analysis of multiple surveillance sites. PLoS Med 2013;10:e1001517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Zhang H, Miranda L et al. . Serious overestimation in quantitative PCR by circular (supercoiled) plasmid standard: microalgal pcna as the model gene. PLoS One 2010;5:e9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klugman KP, Madhi SA, Albrich WC. Novel approaches to the identification of Streptococcus pneumoniae as the cause of community-acquired pneumonia. Clin Infect Dis 2008;47:S202–6. [DOI] [PubMed] [Google Scholar]
- Kodani M, Winchell JM. Engineered combined-positive-control template for real-time reverse transcription-PCR in multiple-pathogen-detection assays. J Clin Microbiol 2012;50:1057–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi M, Milenkov M, Albrich WC et al. . The relevance of a novel quantitative assay to detect up to 40 major Streptococcus pneumoniae serotypes directly in clinical nasopharyngeal and blood specimens. PLoS One 2016;11:e0151428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser JF, Grant LR, Millar EV et al. . Nasopharyngeal carriage and transmission of Streptococcus pneumoniae in American Indian households after a decade of pneumococcal conjugate vaccine use. PLoS One 2014;9:e79578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien KL, Wolfson LJ, Watt JP et al. . Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 2009;374:893–902. [DOI] [PubMed] [Google Scholar]
- Pholwat S, Liu J, Stroup S et al. . Integrated microfluidic card with TaqMan probes and high-resolution melt analysis to detect tuberculosis drug resistance mutations across 10 genes. MBio 2015;6:e02273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pholwat S, Sakai F, Turner P et al. . Development of a TaqMan array card for pneumococcal serotyping on isolates and nasopharyngeal samples. J Clin Microbiol 2016;54:1842–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta FC, Roundtree A, Soysal A et al. . Sequential triplex real-time PCR assay for detecting 21 pneumococcal capsular serotypes that account for a high global disease burden. J Clin Microbiol 2013;51:647–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai F, Chochua S, Satzke C et al. . Single-plex quantitative assays for the detection and quantification of most pneumococcal serotypes. PLoS One 2015;10:e0121064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satzke C, Dunne EM, Porter BD et al. . The Pneucarriage Project: a multi-centre comparative study to identify the best serotyping methods for examining pneumococcal carriage in vaccine evaluation studies. PLoS Med 2015;12:e1001903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen L, Taylor RJ, Schuck-Paim C et al. . Effect of 13-valent pneumococcal conjugate vaccine on admissions to hospital 2 years after its introduction in the USA: a time series analysis. Lancet Resp Med 2014;2:387–94. [DOI] [PubMed] [Google Scholar]
- Singleton RJ, Hennessy TW, Bulkow LR et al. . Invasive pneumococcal disease caused by nonvaccine serotypes among alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA 2007;297:1784–92. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Nelson KE, Paulsen IT et al. . Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 2001;293:498–506. [DOI] [PubMed] [Google Scholar]
- Turner P, Hinds J, Turner C et al. . Improved detection of nasopharyngeal cocolonization by multiple pneumococcal serotypes by use of latex agglutination or molecular serotyping by microarray. J Clin Microbiol 2011;49:1784–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden M, Falkenhorst G, Perniciaro S et al. . Effects of infant pneumococcal conjugate vaccination on serotype distribution in invasive pneumococcal disease among children and adults in Germany. PLoS One 2015;10:e0131494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Poll T, Opal SM. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet 2009;374:1543–56. [DOI] [PubMed] [Google Scholar]
- Weinberger DM, Malley R, Lipsitch M. Serotype replacement in disease after pneumococcal vaccination. Lancet 2011;378:1962–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyres KL, Lambertsen LM, Croucher NJ et al. . Pneumococcal capsular switching: a historical perspective. J Infect Dis 2013;207:439–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at FEMSLE online.