Abstract
Extensive research efforts have been dedicated to describing degradation of wood, which is a complex process; hence, microorganisms have evolved different enzymatic and non-enzymatic strategies to utilize this plentiful plant material. This review describes a number of fungal and bacterial organisms which have developed both competitive and mutualistic strategies for the decomposition of wood and to thrive in different ecological niches. Through the analysis of the enzymatic machinery engaged in wood degradation, it was possible to elucidate different strategies of wood decomposition which often depend on ecological niches inhabited by given organism. Moreover, a detailed description of low molecular weight compounds is presented, which gives these organisms not only an advantage in wood degradation processes, but seems rather to be a new evolutionatory alternative to enzymatic combustion. Through analysis of genomics and secretomic data, it was possible to underline the probable importance of certain wood-degrading enzymes produced by different fungal organisms, potentially giving them advantage in their ecological niches. The paper highlights different fungal strategies of wood degradation, which possibly correlates to the number of genes coding for secretory enzymes. Furthermore, investigation of the evolution of wood-degrading organisms has been described.
Keywords: lignin degradation, fungi, bacteria, ecology, evolution
A number of fungal and bacterial organisms have developed both competitive and mutualistic strategies based on the enzymatic machinery and low molecular compounds, which gives these organisms not only an advantage in wood degradation processes, but potentially giving them an advantage in their ecological niches.
ABBREVIATIONS
- CAZy:
Carbohydrate active enzymes
- DyP:
dye-decolorizing peroxidase
- ECM:
ectomycorrhizal fungi
- GLOX:
glyoxal oxidase
- GMC:
glucose-methanol-choline oxidase/dehydrogenase
- HTP:
heme thiolate peroxidase
- LDA:
lignin-degrading auxiliary enzymes
- LiP:
lignin peroxidase
- LMCO:
laccase-like multicopper oxidase
- LME:
lignin-modifying enzymes
- LPMO:
lytic polysaccharide monooxygenase
- MCO:
multicopper oxidase
- MnP:
manganese peroxidase
- POD:
ligninolytic class II peroxidases
INTRODUCTION
Evolution created lignocellulose, a main component of woody plants’ cell wall, as a compromise between mechanical functionality of land inhabiting plants and physical protection against herbivores and saprobionts. The chemical complexity of lignin has increased during the course of evolution from ancient pteridophytes and gymnosperms to the most evolved grasses (Barceló et al.2004). Rigid tubular structures are required in higher plants to transport water and nutrients yet, at the same time cellulose microfibrils together with lignin and hemicellulose form a natural biocomposite that allows trees to support their enormous weight. This natural biocomposite allows trees to withstand hostile weather—very strong winds and consequently live for many thousands of years. A prime example of this is the North American sequoias, which are considered the largest single living trees and one of the oldest organisms inhabiting our biosphere.
With cellulose fibrils being embedded in a lignin matrix, wooden plants cell walls have developed a very robust structural framework (Boer et al.2005). The expansion of land plants had significant and far-reaching consequences on the current appearance of terrestrial ecosystems, and was a major evolutionary event in the history of photosynthetic organisms. Most scientists agree that the successful colonization of the terrestrial realm involved a symbiotic association with certain fungi (Taylor, Krings and Taylor 2015b).
It is believed that the lignin biosynthetic pathway is highly conserved throughout the evolution of vascular plants. Molecular-clock analyses suggest that land plants form a monophyletic group and diverged from green algae about 700 million years ago (Ma) (Qiu and Palmer 1999). It is not surprising that the multifunctional basic peroxidases responsible for lignin synthesis appeared early in the evolution of land plants. It is likely that this class of enzymes conferred important adaptive traits to plants for their new life on land and the development of vascular tissues (Barceló et al.2004). This aromatic polymer (lignin) evolved c.a. 400–450 Ma in the Late Ordovician period. The earliest unequivocal fossil evidence of lignins has been found in pteridophytes, which are widely considered to be the first vascular plants and are likely to have played a key role in the colonization of the terrestrial landmasses (Barceló et al.2004; Weng and Chapple 2010). Phenylpropanoid pathways in plants were probably one of the most critical for the genesis of this aromatic polymer. However, monolignol biosynthesis evolved primarily for the generation of UV-protectant molecules which are more ancient than vascular plants. The ability to deposit polymers of phenylpropanoid units in the cell walls by early tracheophytes was evolutionarily very advantageous. Lignin strengthens the plant cell walls by adhesion of layers of cellulose microfibrils which allows plants to expand significantly in body size, enhance water transport, resistance to pathogens and slows the degradation of wood by microorganisms (Weng and Chapple 2010; Leisola, Pastinen and Axe 2012; Labeeuw et al.2015).
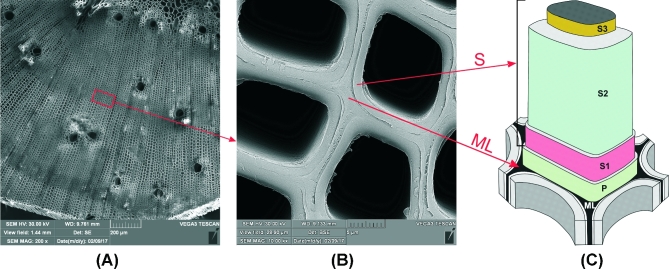
Figure 1A and B shows a picture of the cross section of a plants’ woody cell wall, whilst Fig. 1C shows a cartoon schematic of the cell wall structural components. The lignin and hemicellulose polymers function as the recalcitrant adhesive for cellulose microfibrils. The stratified structure and the cellulose fibers orientation in the S layers are non-parallel, which gives this biocomposite additional mechanical strength. Only recently were humans able to engineer a composite with such mechanical strength using glass or carbon fibers and epoxy resins. Although macroscopically many types of wood tend to have a similar appearance, there are significant differences between the structure and chemical composition of major polymeric components of softwood and hardwood (Table 1). Softwood (coniferous) and hardwood (deciduous) fibers mainly differ in the structure and composition of hemicellulose and lignin components. For instance, glucuronoxylan comprises 15% to 35% of the secondary cell wall polysaccharides in hardwood, while galactoglucomannan is the main hemicellulose component of secondary cell walls in softwood. Moreover, softwoods contain mainly guaiacyl lignin, whereas hardwoods contain varying ratios of syringyl and guaiacyl lignins (Ek, Gellerstedt and Henriksson 2009; Suzuki et al.2012). The recalcitrance of softwood lignocellulose to bioprocess technologies has been attributed to its higher lignin content, smaller pore size and fewer hemicellulose-derived acetyl groups in comparison with hardwood (Palonen et al.2004; MacDonald et al.2011).
Figure 1.
Scanning electron micrograph (SEM) of transverse section of xylem tissue of Pinus sylvestris. (A) Visible are both the early and the latewood tracheides, SEM – ×200; (B) SEM – ×10 000 of the indicated area of panels A and C—diagram of the woody cell walls layers (adopted from Côté 1967). ML, middle lamella; P, primary wall; S1, outer layer of the secondary wall; S2, middle layer of the secondary wall; S3, innermost layer of the secondary wall.
Table 1.
Chemical composition of plant material (Adapa, Tabil and Schoenau 2009; Brosse et al.2012; Rowell 1984; Hon and Shiraishi 2001; El-Tayeb et al.2012; Rytioja et al.2014; Kumar and Shukla 2016).
| Plant material | Chemical composition (% dry wt) | ||||||
|---|---|---|---|---|---|---|---|
| Cellulose | Lignin | Hemicelluloses | |||||
| Galactoglucomannan | Arabinoglucoronoxylan | Glucunoroxylan | Glucomamnan | Pectin, starch, ash, etc | |||
| Softwood | 33–42 | 27–32 | 5–18 | 7–14 | 1–3 | ||
| Hardwood | 38–51 | 21–31 | 15–35 | 2–5 | 1–4 | ||
| Straw | |||||||
| Barley | 33.25 | 17.13 | 20.36 | ||||
| Oat | 37.60 | 12.85 | 23.34 | ||||
| Wheat | 34.20 | 13.88 | 23.68 | ||||
| Switchgrass | 31–32 | 20.4–25.2 | 14.5–18.1 | ||||
| Miscanthus | 43–50.3 | 9.2–12.5 | 24.8–33.9 | ||||
| Corn stalks | 61.2 | 6.9 | 19.3 | ||||
| Cereal | 45–55 | 16–21 | 26–32 | ||||
SPECIES DECOMPOSING LIGNIN
Lignin degradation is caused by certain fungi as well as several bacterial species (Fig. 2) (Breen and Singleton 1999; Singh Arora and Kumar Sharma 2010; Bugg et al.2011a; Huang et al.2013). Fungi are more efficient in the breakdown of lignin than bacteria, in which delignification is slower and more limited (Table S2, Supporting Information) (Sigoillot et al.2012).
Figure 2.
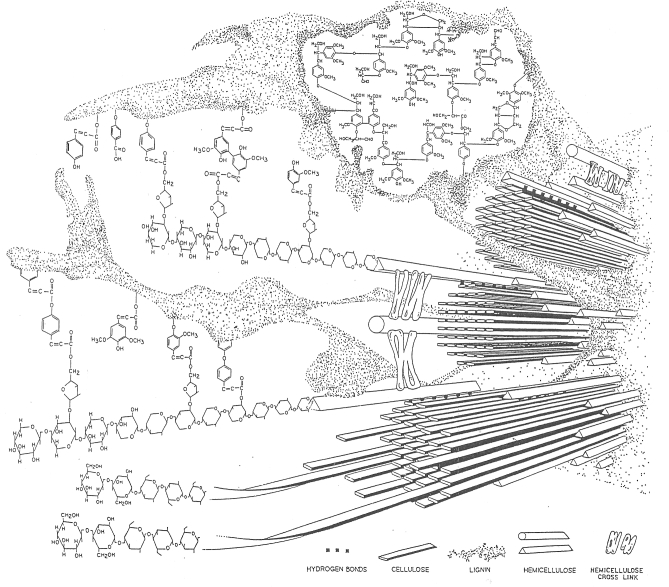
Relative quantities of secondary SW (grasses) components in this representative grass are 45–60% cellulose, 20–40% hemicellulose and 5–10% lignin (Bidlack 1990). Cellulose microfibrils consist of ∼40 of these bonded chains. Three or four glucan chains occur for each β1→4-D-xylopyranose chain of hemicellulose with an occasional L-arabinofuranose molecule linked α1→3 to the xylopyranose chain. The hemicellulosic xylans and arabinoxylans, represented by the actual molecules or as long, triangular rods, are hydrogen bonded to cellulose and ester or ether-bonded to non-core lignin. Ester bonds between hemicellulose and non-core lignin shown in the figure include linkage between the O-5 position of arabinose in arabinoxylan and ρ-coumaric, ferulic and diferulic acids (Scalbert et al.1985; Mueller-Harvey and Hartley 1986), as well as a hypothetical linkage between the O-3 position of xylose and cinnamic acid. Some of these lignin monomers, such as ferulic acid (Mueller-Harvey and Hartley 1986), may be so intimately associated with the hemicellulosic fraction that they fail to cross-link to lignin. Reproduced from Bidlack J, Malone M, Benson R: Molecular structure and component integration of secondary cell walls in plants. Proc Okla Acad Sci (1992, 72:51–56) with permission granted by author and the Oklahoma Academy of Science.
Microbial degradation of lignin has not been intensively studied in organisms other than fungi, but there are reports of bacteria that can break down lignin (Fig. 3). These lignin-degrading bacteria represent mainly three classes: Actinomycetes, α-Proteobacteria and γ-Proteobacteria (Bugg et al.2011a; Huang et al.2013). This ubiquitous group of microbes is widely distributed in natural ecosystems worldwide and occurs both in terrestrial and aquatic habitats (Srinivasan, Laxman and Deshpande 1991). Their apical and branching growth, production of aerial or substrate mycelia, and morphogenetic development resemble filamentous fungi. Due to the production of secondary metabolites and the use of extracellular enzymes, these bacterial species are considered major decomposers of lignocelluloses in soils (Tiquia, Wan and Tam 2002; Bibb 2005; Sonia, Naceur and Abdennaceur 2011; Saini et al.2015). In the prokaryotes, one of the most frequently identified and studied ligninolytic enzymes is laccase (Kuhad, Singh and Eriksson 1997; Tian et al.2014). Phylogenic analysis has shown that representatives of Archaea from four phyla have laccase genes. Yet the greatest numbers of strains with laccase genes were identified in Proteobacteria, Actinobacteria and Firmicutes. Within Cyanobacteria and Bacteroidetes, the number of strains with identifiable laccase genes was found to be substantially lower (Tian et al.2014). Recently, research of Rashid et al. (2015) showed that Sphingobacterium (phylum Bacteroides) produce manganese superoxide dismutase, which is able to oxidize lignin via hydroxyl radical mechanism.
Figure 3.
Ecology (occurrence) of lignin-degrading fungi. Polyporales—Cerrena unicolor (2), Phanerochaete chrysosporium (4), Postia placenta (8); Agaricales—Armillaria melea (1), Pleurotus spp. (5), Trametes versicolor (6), Agaricus bisporus (11); Russulales—Heterobasidion annosum (3); Boletales—Serpula lacrymans (9); Ustilaginales—Ustilago maydis (12); Xylariales—Xylaria spp. (10). White box—white rot fungi (WRF); brown box—brown rot fungi (BRF); green box—soft rot fungi (SRF); orange box—litter decomposers (LD); red box—parasite.
Many strains from the Brucella, Ochrobactrum, Sphingobium and Sphingomonas genera, belonging to the class α-Proteobacteria, decompose lignin. Within another group of ligninolytic bacteria (γ-Proteobacteria) the best known for their degrading abilities are Pseudomonas fluorescens (producing lignin peroxidase—LiP), Ps. putida (dye-decolorizing peroxidase—DyP, manganese peroxidase—MnP), Enterobacter lignolyticus (catalase/ peroxidase HPI and DyP) and Escherichia coli (laccase) (Tian et al.2014).
The representatives of Actinomycetes with the known ability to produce catabolic enzymes are Streptomyces viridosporus, S. paucinobilis and Rhodococcus jostii (Tian et al.2014). Several filamentous bacterial species from the genus Streptomyces have been identified as lignin degraders and less than 10 enzymes pertaining to lignin degradation have been characterized for them (Fernandes et al.2014). Within Actinomycetes, several species producing three enzymes (laccase, LiP, MnP) which are believed to play the most important role in biodegradation of lignin are known, e.g. S. coelicolor, S. griseus and S. psammoticus (Niladevi, Sheejadevi and Prema 2008; Le Roes-Hill, Rohland and Burton 2011). In S. cinnamomeus, laccase and LiP activity was observed (Jing and Wang 2012). The best-studied strain (S. viridosporus) produces several extracellular peroxidases and laccase (Bugg et al.2011a; Tian et al.2014). Laccase activity was also observed in S. ipomoea (Niladevi, Jacob and Prema 2008; Molina-Guijarro et al.2009; Fernandes et al.2014). All of these findings indicate that bacterial degradation of lignin may be more important than previously thought, particularly in soil.
The number of fungal species capable of decomposing lignin is not known exactly; however, Gilbertson reported that in North America, there are ∼1600 to 1700 species of wood-degrading fungi (Gilbertson 1980). Wood-degrading fungi mostly live as saprotrophs or weak parasites in natural and human-affected forest ecosystems. Lately, information about genes coding enzymes that degrade lignin has been found in Ustilago maydis, which is a biotrophic phytopathogen (Couturier et al.2012). Also, it has been shown that coprophilic fungi (e.g. Panaeolus papilionaceus, Coprinopsis friesii) produce ligninolytic enzymes such as laccases and/or peroxidases (Heinzkill et al.1998). Saprotrophic fungi play important functions in the biosphere, preventing accumulation of dead plant organic matter. Generally, wood-rotting fungi are primary lignin degraders and are the only group of microorganisms capable of mineralization of this abundant plant product accounting for about 25% of removable organic matter in the biosphere (Kuhad and Singh 2007; Huang et al.2013). Depending on the particular morphology of wood decomposition, the saprophytic fungi have been divided into three main groups: (i) white rot fungi, (ii) brown rot fungi and (iii) soft rot fungi. These groups can be further broken down into (iv) litter-decomposing and (v) dung-dwelling (coprophilic) fungi that also degrade lignin (Blanchette 1995; Liers et al.2011). All these types of fungi are able to decompose lignin, but only white rot degrade it completely to CO2 and H2O (Blanchette 1995).
Wood-decaying fungal Basidiomycota ecologically represent white and brown rot, litter-decomposing, plant-pathogenic and ectomycorrhizal (ECM) fungi, although over 90% of all known wood-rotting species belong to the white rot type. Species of Basidiomycetes occupy different ecological niches and grow on various substrates, for instance, deciduous trees, conifers, crops, forest litter, grassland soils and roots of plants (Rytioja et al.2014). The members of Polyporales and Agaricales, i.e. Ganoderma spp., Phlebia radiata, Lentinula edodes or Pleurotus spp., mostly represent white rot fungi. Among brown rot fungi, Gloeophyllum trabeum has been studied the most and along with Serpula lacrymans and Coniophora puteana (Boletales) plays a harmful role in relation to the wood used in construction (Blanchette 1995). In nature, white rot Basidiomycetes occur more often on hardwood of angiosperms than on softwood of gymnosperms, whereas brown rot fungi more often attack softwoods and are usually found in coniferous ecosystems (Gilbertson 1980; Hatakka 2005; Sigoillot et al.2012).
The details of the lignin degradation mechanism have been elucidated in the white rot fungus species Phanerochaete chrysosporium (currently Phanerodontia chrysosporium) which produces multiple isoenzymes of lignin and manganese peroxidases but does not produce laccase (Hatakka 2005; Huang et al.2013). The fungus occurs in the later stages of wood degradation, when available nitrogen is depleted by other microorganisms (Deacon 2006).
Within litter-decomposing Basidiomycota, ligninolytic enzymes have been detected, prime examples being Agaricus bisporus producing laccase and MnP and Marasmius quercophilus producing only laccase (Hatakka 2005). ECM fungi have genes encoding enzymes that degrade lignocellulose, but their ability to decompose lignocellulose is lower than in most other wood-decaying fungi. They are able to survive in the absence of host plants by assimilating reduced carbon compounds, similarly to free-living saprotrophs. Their degradation mechanism of lignocellulose is similar that of brown rot type of decay (e.g. Paxillus), while others use a white rot type mechanism (e.g. Cortinarius) (Kohler et al.2015; Lindahl and Tunlid 2015).
Representatives from Ascomycota mostly cause soft rot decay. The ability to degrade lignin by these fungi is limited (Cragg et al.2015). However, members of the Xylariales order (from the Daldinia, Hypoxylon, Xylaria genera) currently grouped with soft rot fungi have been previously included in the white rot fungi cluster (Hatakka 2005). Anamorphic fungi, such as the commonly occurring Alternaria alternata, also cause soft rot decay (Sigoillot et al.2012). Worrall, Anagnost and Zabel (1997) and later Pildain, Novas and Carmarán (2005) reported that a few Eutypella species produce white rot decay in the late stages of wood decay, but soft rot in the early stages. Other studies of microfungi (e.g. Penicillium chrysogenum, Fusarium solani and F. oxysporum) have shown lower degrees of lignin decomposition when compared to white rot fungi (Kirk and Farrell 1987). Production of extracellular laccases has been observed in the plant pathogenic, anamorphic fungus Botrytis cinerea, which causes soft rot like decay in many horticultural crop plants (e.g. Daucus and Cucumis) as well as the ‘noble rot’ and ‘grey rot’ of Vitis vinifera (Thurston 1994).
There is no information to be found in literature about laccase production in lower fungi (Zygomycota and Chytridiomycota) (Singh Arora and Kumar Sharma 2010), although recent research suggests that these fungi are involved in wood degradation in nature (Fukasawa, Osono and Takeda 2011). Zygomycota generally do not utilize cellulose and lignin. However, some representatives of this phylum can decompose lignin and are confined to outer layers of decaying plant tissue (Ingold 1978).
Investigation of fungal succession of woody colonization showed that Basidiomycota dominate in the earlier stages of wood decomposition, while Ascomycetes dominate in the later stages (Crawford, Carpenter and Harmon 1990; Fukasawa, Osono and Takeda 2011). Species in Xylariaceae (Ascomycota) are known as latent inhabitants of living wooden tissues and primary decomposers of partially degraded woody debris (Fukasawa, Osono and Takeda 2011). Additionally, the studies of some fungal species and their relationship with plants have been shown preference to a certain type of substrate (i.e. hardwood, softwood, sapwood, heartwood) (Hatakka 2005; Deacon 2006).
ENZYMES INVOLVED IN LIGNIN DEGRADATION
Enzymes involved in lignin degradation can generally be divided into two main groups: lignin-modifying enzymes (LME) and lignin-degrading auxiliary (LDA) enzymes. LDA enzymes are unable to degrade lignin on their own yet are necessary to complete the degradation process (da Silva Coelho-Moreira et al.2013).
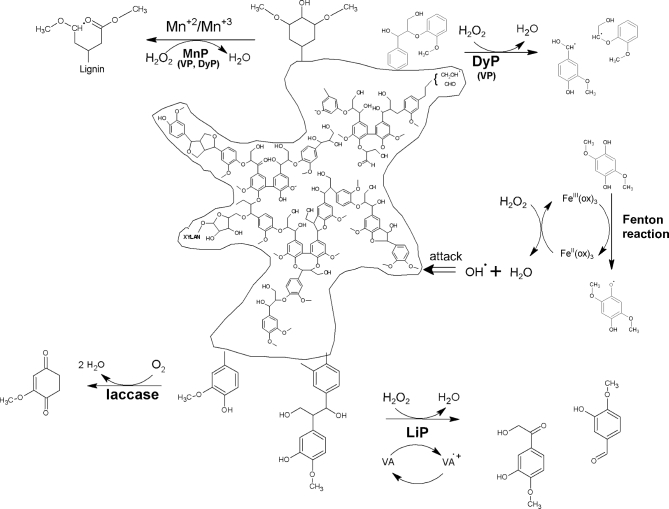
LME produced by different microorganisms are classified as phenol oxidase (laccases) and heme containing peroxidases (POD), namely lignin, manganese and multifunctional (versatile) peroxidase (Fig. 4).
Figure 4.
Comparison of lignin degradation by peroxidases and laccase (Hofrichter 2002; Brunow 2005; Desai and Nityanand 2011; Sánchez, Roció Sierra and Alméciga-Díaz 2011; Chen et al.2015). MnP—manganese peroxidase, VP—versatile peroxidase, LiP—lignin peroxidase, DyP—dye-decolorizing peroxidase, ox—oxalate, VA— veratryl alcohol. In parentheses, other enzymes capable to catalyze similar reaction are presented.
Recently, a new group of enzymes, heme-thiolate haloperoxidases, has been suggested to have a role in lignin degradation. This class of enzymes has yet to be classified to either the LME or LDA group. These enzymes are supposed to be the most versatile biocatalysts of the hemeprotein family. Heme-thiolate peroxidases (HTPs) share catalytic properties with at least three other classes of heme-containing oxidoreductases: classic plant and fungal peroxidases, cytochrome P450 monooxygenases and catalases (Hofrichter and Ullrich 2006). These enzymes were suggested to be involved in lignin degradation in culture of Ceriporiopsis subvermispora (Fernandez-Fueyo et al.2012b) but only were proved to oxidase aryl alcohols (Ullrich et al.2004). Up to date, there is no experimental evidence that these enzymes alone can degrade lignin, so we concluded that they should be considered as LDA enzymes.
Auxiliary enzymes allow the lignin degradation process through the sequential action of several proteins which may include oxidative generation of H2O2. This group includes glyoxal oxidase (GLOX; EC 1.2.3.5), aryl alcohol oxidases (AAO; EC 1.1.3.7), pyranose 2-oxidase (POX; EC 1.1.3.10), cellobiose dehydrogenase (CDH; EC 1.1.99.18) and glucose oxidase (EC 1.1.3.4). These enzymes are frequently found in white rot fungi secretomes (Levasseur et al.2008).
In nature, only Basidiomycota belonging to the aerobic white rot group are able to degrade lignin completely (Dashtban et al.2010). Anaerobic fungi lack the enzymatic machinery to catabolize/mineralize lignin. The enzymatic reaction that cleaves the aromatic ring requires oxygen or its partially reduced species (reactive oxygen species—ROS), and therefore cannot take place in an anaerobic environment (Berg and McClaugherty 2014).
MAJOR GROUPS OF LMEs
Lignin-modifying peroxidases (LMPs: LiP, MnP and VP) also called POD (Hammel and Cullen 2008) belong to class II peroxidases within the superfamily of catalase-peroxidases (formally called plant and fungal peroxidases) (Welinder 1992; Zamocky et al.2014). All members of this group contain protoporphyrin IX as a prosthetic group (Pollegioni, Tonin and Rosini 2015). This superfamily can be divided into three subclasses based on their amino acid sequences similarities and catalytic properties. Class I includes catalase-peroxidases, prokaryotic and organelle-localized eukaryotic heme peroxidases. Class II includes all extracellular fungal heme peroxidases and is classified as auxiliary activities (AA2) family in carbohydrate active enzyme (CAZy) (Lombard et al.2014). Class III comprises all secreted plant heme peroxidases (Zamocky, Furtmuller, Obinger 2009; Lundell, Makela and Hilden 2010). Very recently, first in fungi then later in bacteria, the new superfamily of heme peroxidases was identified. This newly identified group of enzymes is called dye-peroxidases (DyP), originally named ‘dye-decolorizing peroxidases’ (Sugano 2009). DyP may also participate in the degradation of lignocellulose and lignin, due to their apparent LMP-like enzymatic activities. The potential for cleavage of lignin substructure linkages was recently demonstrated for a DyP enzyme from the Basidiomycete Auricularia auricula-judae (Liers et al.2010; Lundell, Makela and Hilden 2010).
Lignin peroxidase (EC 1.11.1.14)
Lignin peroxidases were first discovered in Phanerochaete chrysosporium (Tien and Kirk 1983; Paszczynski, Huynh and Crawford 1986; Kirk and Farrell 1987) and later in Trametes versicolor (Johansson and Nyman 1993), Bjerkandera sp., and Phlebia tremellosa, which are well known white rot fungi (Aarti, Arasu and Agastian 2015). Activity of LiP has previously been detected in some bacteria, such as Acinetobacter calcoaceticus and Streptomyces viridosporus (Dashtban et al.2010). LiPs, capable of attacking lignin polymers, are relatively non-specific to their substrates. LiPs are known to oxidize different phenolic aromatic compounds and a variety of non-phenolic lignin model compounds along with many other organic molecules (Wong 2009). These enzymes are usually secreted by given microorganisms as a family of isozymes whose relative composition and isoelectric points (pI) vary depending on the growth medium and nutrient conditions (Santhanam et al.2012). The globular structure of LiP isolated from P. chrysosporium is composed of eight major and eight minor α-helices with limited β components and is organized into two domains that form an active center cavity composed of a heme-chelating single ferric ion (Choinowski et al.1999). The LiP contains two glycosylation sites, two Ca2+ binding sites and four disulfide bridges, all stabilizing the three-dimensional structure of this enzyme. Molecular mass of LiPs ranges from 35 to 48 kDa and it has a pI between 3.1 and 4.7. The high redox potential of LiPs (around 1.2 V at pH 3) makes these enzymes capable of oxidizing substrates that are not oxidized by other peroxidases (Sigoillot et al.2012). The catalytic cycle of LiP resembles the catalytic mechanism common to all peroxidases. In each cycle, the native enzyme is oxidized by H2O2 and generates a compound I intermediate that exists as a ferry oxo porphyrin radical cation [Fe(IV) = O.+]. Next, the enzyme is subjected to two single-electron reduction steps by the electron donor substrate, such as veratryl alcohol (VA), leading to a transient formation of compound II [Fe(IV) = O] and a VA radical cation (VA+). The compound II further oxidizes the second VA molecule, simultaneously returning to its native stage to initiate a new catalytic cycle of LiP. VA, similar to Mn(III), plays the role of being a small molecular weight redox transfer mediator between the enzyme and its polymeric substrate (Wong 2009).
Manganese-dependent peroxidase (EC 1.11.1.13)
Manganese peroxidase is another important LME secreted in multiple isoforms and was first detected in P. chrysosporium more than 30 years ago (Glenn and Gold 1985; Paszczynski, Huynh and Crawford 1986). These enzymes have also been found in other Basidiomycota species, including Panus tigrinus (Lisov, Leontievsky and Golovleva 2003), Lenzites betulinus (Hoshino et al.2002), Agaricus bisporus (Lankinen et al.2001), Bjerkandera sp. (Palma et al.2000) and Nematoloma frowardii (Hilden et al.2008). The details about the presence of MnP in bacteria, yeast and mold are emerging in the scientific literature. The MnP activity in Bacillus pumilus, Paenibacillus sp. (de Oliveira et al.2009) and Azospirillum brasilense (Kupriashina, Selivanov and Nikitina 2012) was studied under absence and presence of the inducers. MnP activity was also detected in the Actinobacterium S. psammoticus (Niladevi and Prema 2005). Similarly to LiP, MnP has a molecular structure composed of two α-helices domains with the heme sandwiched in between. MnP has five disulfide bridges and two Ca2+ ions, which maintain the structure of the active enzyme (Sutherland et al.1997). The Mn(II)-binding site consists of two glutamate and one aspartate γ-carboxylic groups and is located close to the porphyrin macrocycle (Wong 2009). The molecular mass of MnPs ranges from 38 to 62.5 kDa, which include 4%–18% glycans, and their pI ranges from 2.9 to 7.1 (Sigoillot et al.2012). The catalytic cycles of MnPs and LiPs are very similar, but Mn-dependent peroxidases are unique in utilizing Mn(II) as their reducing substrate, generating Mn(III), which diffuses from enzymes into the lignocellulose structure. Outside of the enzymes, Mn(III) is stabilized by chelation with organic acids such as oxalate, fumarate, glyoxylate and malate (Martinez et al.2005; Sigoillot et al.2012) and acts as a diffusible low molecular weight redox mediator that oxidizes organic substrates (Goszczynski et al.1994) via hydrogen or one electron abstraction. The organic acids also facilitate the release of Mn (III) from the active site of the enzyme (Niladevi 2009).
Addition of Mn(II) rapidly reduces compound I to compound II, thus forming Mn(III). A second Mn(II) is then used to reduce compound II back to the native enzyme. The conversion of compounds (I and II) can also be achieved by the addition of other electron donors, such as a variety of phenolic substances, but at a slower rate. Phenolic compounds are, however, not able to reduce efficiently MnP compound II back to the native enzyme. Mn(II) is an obligatory redox coupler for the enzyme to complete its catalytic cycle. Similar to the LiP, the excess of H2O2 can generate compound III which is an inactive form of the enzyme (Wariishi, Akileswaran and Gold 1988; Manavalan, Manavalan and Heese 2015).
Versatile peroxidase (EC 1.11.1.16)
Versatile peroxidases (VPs) combine the molecular architecture of LiP and MnP. VP oxidizes typical LiP substrates e.g. VA, methoxybenzenes and non-phenolic model lignin compounds, as well as Mn2+ (Hofrichter et al.2010; Garcia-Ruiz et al.2014). The structure of VP includes 11–12 helices, four disulfide bridges, two structural Ca2+ sites, a heme pocket and a Mn2+-binding site similar to that of MnP (Perez-Boada et al.2005). The substrate specificity of VPs is similar to that of LiPs, including oxidation of high and medium redox potential compounds. Yet VP also oxidizes azo-dyes and other non-phenolic compounds with high-redox potential in the absence of mediators (Garcia-Ruiz et al.2014). VP was detected in members of the genera Pleurotus [Pleurotus eryngii (Ruiz-Duenas et al.1999), Pl. ostreatus (Ruiz-Duenas et al.2001)] and Bjerkandera [Bjerkandera adusta (Heinfling et al.1998), B. fumosa (Rodakiewicz-Nowak, Jarosz-Wilkolazka and Luterek 2006)]. VPs are glycoproteins secreted as several isoenzymes with a molecular mass ranging between 40 and 45 kDa with a pI ranging between 3.4 and 3.9 (Mester and Field 1998). The basic catalytic cycle of VPs is similar to those of other peroxidases with the two intermediary compounds I and II, but is more complex due to a more diversified pool of potential substrates (Sigoillot et al.2012).
Dye-decolorizing peroxidase (EC 1.11.1.19)
Dye-decolorizing peroxidases constitute a new family of heme peroxidases, which are phylogenetically unrelated to the classical LME peroxidases, including LiP, MnP and VP (Zamocky et al.2015). The structure of DyP-type peroxidases reveals two domains that contain α-helices and anti-parallel β-sheets, and the heme cofactor is located at the cavity between the two domains (Colpa, Fraaije and van Bloois 2014). In view of the scant evolutionary relationship between DyPs and the classical LMP family, the observed similarities of the catalytic site (heme pocket) must be considered as a convergent type of evolution to provide similar reactivity of these enzymes (Linde et al.2015). The DyP-type peroxidases were first discovered in a culture of the fungus B. adusta (previously Geotrichum candidum) and were named after their ability to decolorize a wide range of dyes (Kim and Shoda 1999; Fernandez-Fueyo et al.2015). Ligninolytic activity of DyPs has been reported in other fungi, such as Termitomyces albuminosus (Johjima, Ohkuma and Kudo 2003), Auricularia auricula-judae (Liers et al.2010), Irpex lacteus (Salvachua et al.2013) and the bacteria Rhodococcus josti (Roberts et al.2011), Thermobifida fusca (van Bloois et al.2010) and Pseudomonas fluorescens (Rahmanpour and Bugg 2015). Recent genome sequence analysis revealed that DyPs are prominent in bacteria, whereas only a small number are reported in fungi and higher eukaryotes. Occurrence of DyP in archaea is even more limited, suggesting that these enzymes can be regarded as the bacterial equivalent of the fungal lignin-degrading peroxidases (Colpa, Fraaije and van Bloois 2014). Based on the phylogenetic analysis of genomic sequences from fungi, bacteria and archaea, DyPs have been classified into four subfamilies: A, B, C and D. Type A include bacterial DyPs that contain a Tat-dependent signal sequence, which suggests that they function outside of the cytoplasm. The type B subfamily, all putative cytoplasmic enzymes, is also found in Ascomycota and the only DyP from a non-fungal Eukaryota (the mycetozoon Dictyostelium discoideum). Type C includes sequences from bacteria, and two sequences from B. adusta which are involved in intracellular metabolism similar to type B. Type D contains primarily fungal variants of the enzyme from Ascomycota and Basidiomycota which are linked to dye decolorization (Colpa, Fraaije and van Bloois 2014). DyPs possess low substrate specificity and oxidize all of the typical peroxidase substrates (ABTS, DMP, Congo Red, Acid Red 151, Adlerol, Mordant Black 9, Reactive Black 5, etc.). In addition to typical peroxidase activity, it has been suggested that DyP has an additional hydrolase or oxygenase activity (Sugano et al.2009). These enzymes are active at low pH values (pH range 3–4) and are able to degrade different dyes, particularly anthraquinone dyes (Colpa, Fraaije and van Bloois 2014). Nevertheless, the physiological role of DyPs is still unclear; although evidence is accumulating that some bacterial variants of this enzyme are involved in lignin degradation and/or as a defense mechanism against oxidative stress (Liers et al.2013).
Laccase (Lac, benzenediol: oxygen oxidoreductases; EC 1.10.3.2)
Laccases are considered to be among the most important components of the ligninolytic complex of wood-destroying microorganisms (Morozova et al.2007). All laccases oxidize a range of aromatic compounds, including phenolic moieties typically found in lignin, aromatic amines, benzenothiols and hydroxyindols using molecular oxygen as an electron acceptor. Inorganic/organic metal compounds are also substrates for laccases, and it has been reported that Mn(II) is oxidized by laccase to form Mn(III) and organometallic compounds (Fe(CN)6)2−) to (Fe(CN)6)3−) (Zimmerman et al.2008; Garcia-Ruiz et al.2014). Laccases are widely distributed in plants, fungi (Leontievsky et al.1997) and in some bacteria [A. lipoferum (Diamantidis et al.2000), Ba. subtilis (Martins et al.2002), S. lavendulae (Suzuki et al.2003) and Sinorhizobium meliloti (Pawlik et al.2016)] and even insects (Suderman et al.2006). The localization of laccase is associated with its different physiological functions and is dependent on the organism producing the enzyme. Laccases are extracellular, periplasmic and intracellular proteins, in contrast to the other ligninolytic enzymes mostly reported as extracellular proteins. In plants, laccases are intracellular enzymes that participate in the synthesis of lignin (Morozova et al.2007). The majority of fungi produce both intracellular and extracellular laccases. The intracellular laccases of fungi and periplasmic bacterial laccases most likely participate in the transformation of low molecular weight phenolic compounds in the cell (Baldrian 2006). For example, the CotA protein (laccase) is involved in dark pigment (melanin) formation in Ba. subtilis endospores’ coat, protecting the spores against harmful UV radiation (Hullo et al.2001).
Fungal laccases are involved in multiple processes, such as pathogenesis, detoxification, degradation of lignin (Youn, Hah and Kang 1995) and involvement in the development and morphogenesis of higher fungi (Leonowicz et al.2001; Morozova et al.2007). Laccases are metalloproteins belonging to the group of polyphenol oxidases containing copper atoms in the catalytic site and therefore called blue multicopper oxidases (Baldrian 2006). The active center of laccases is composed of three types of four copper atoms with differing electron paramagnetic resonance parameters (Morpurgo et al.1980). The type 1 copper (T1Cu) is characterized by a strong absorption around 600 nm and it is responsible for the intense blue color of purified laccase. The type 2 copper (T2Cu) is colorless and exhibits only weak absorption in the visible portion of the electromagnetic spectrum and type 3 copper (T3Cu) has an absorption peak at 330 nm. T2Cu and T3Cu create a 3-nuclear copper cluster T2/T3 consisting of one T2 copper ion and two T3 copper ions (Madhavi and Lele 2009). Such an arrangement of copper atoms distributed in three domains is present in most of the bacterial and fungal laccases. However, structural research in several Actinobacteria such as S. griseus, S. cyaneus, S. coelicolor, S. ipomoea, S. sviceus and T. fusca have revealed the presence of two Cu-binding domains rather than three. The enzymes with two-domain structures have been named small laccase or small laccase-like multicopper oxidases (LMCO) (Saini et al.2015). Laccases mainly react with free phenolic fragments of lignin due to the random polymer nature of lignin and laccases lower redox potential. However, in the presence of some low molecular mass mediators, laccase can also attack non-phenolic aromatic compounds with high redox potentials (Kunamneni et al.2007). Substrate oxidation by laccase is a single-electron reaction generating a free radical. The initial product of the reaction is typically an unstable phenoxy radical that can be converted to quinone in a second reaction catalyzed by the enzyme (Shraddha et al.2011; Fitigau, Peter and Boeriu 2013). Most fungi secrete several isoforms of laccases. These isozymes originate from the same or different genes encoding for the laccase enzyme. The number of isozymes found depends on species, growth conditions and the presence or lack of an inducer in the growth medium (de Souza et al.2004; Morozova et al.2007).
LDA ENZYMES
Glyoxal oxidase (EC 1.2.3.5)
Glyoxal oxidase generates extracellular hydrogen peroxide by the oxidation of a variety of simple dicarbonyl and hydroxycarbonyl compounds, especially glyoxal and methylglyoxal (Kersten and Cullen 1993; Yamada et al.2014). Based on an unusual free radical-coupled copper active site, GLOX was classified in the CAZy family among other copper radical oxidases in the family AA5 (Whittaker et al.1999; Daou et al.2016). Moreover, GLOX was suggested to regulate peroxidase activity and be activated in vitro by the presence of LiP (Kersten 1990). The mechanism of aldehyde oxidation by GLOX has not been studied in as great detail; however, it was suggested that GLOX reacts with the hydrated, gem-diol form of the substrate via an analogous mechanism to galactose oxidase (Yin et al.2015). The best-characterized GLOX (MW = 68 kDa) is an enzyme isolated from Phanerochaete chrysosporium; however, the genes coding for GLOX were found in many other white rot fungi (Kersten 1990). Kersten and Cullen (2014) suggested that along with both substrates mentioned above, glycol aldehyde (derived from lignin) may be a substrate for GLOX via these reactions: glycolaldehyde→glyoxal→glyoxylic acid→oxalate. There are two other possible sources for GLOX substrates: (i) MnP-dependent lipid peroxidation of linoleic acid (Watanabe et al.2001); (ii) abiotic oxidation of sugars and trioses in a Fenton system which produces glyoxal (Manini et al.2006; Kersten and Cullen 2014).
Aryl alcohol oxidase (EC 1.1.3.7)
Aryl alcohol oxidase, belonging to the glucose-methanol-choline oxidase/dehydrogenase family (GMC), was originally found in many Agaricales species (Hernandez-Ortega, Ferreira and Martinez 2012) and later in Aspergillus, Fusarium and even in some bacteria (Kumar and Goswami 2006; Tamboli et al.2011). This monomeric enzyme is composed of two domains with a non-covalently bound FAD cofactor and a molecular mass ranging from 66.7 up to 334 kDa (Guillen, Martinez and Martinez 1992; Bruckmann et al.2002; Fernandez et al.2009). AAO produces hydrogen peroxide by oxidative dehydrogenation of phenolic and non-phenolic aryl-alcohols, polyunsaturated (aliphatic) primary alcohols or aromatic secondary alcohols to their corresponding aldehydes (Ferreira et al.2010). Hernandez-Ortega, Ferreira and Martinez (2012) suggested that the H2O2 needed in lignolysis forms during the cycling reaction of AAO (extracellular enzyme) and aryl-alcohol dehydrogenase, which converts alcohol to aldehyde operating inside hyphae by NADP-dependent reaction. Recently described aryl-alcohol:quinone oxidoreductase from Pycnoporus cinnabarinus was shown to have homology with AAO. The enzyme was able to reduce radical intermediates (guaiacol, sinapic acid, etc.) generated by laccase (Mathieu et al.2016).
Heme-thiolate haloperoxidases
Two types of haloperoxidases were included in this recently discovered group of enzymes: chloroperoxidases (CPO, EC 1.11.1.10) and aromatic peroxygenases (APO, unspecific peroxygenases, EC 1.11.2.1). CPO was first discovered in the Ascomycete Caldariomyces fumago. Their reaction resembles that of cytochrome P450s, consisting of epoxidation and hydroxylation of substrates such as organic sulfides, olefins and aromatic rings; however, they are not susceptible to CPO-catalyzed oxygen transfer (Hofrichter and Ullrich 2006). Ortiz-Bermudez, Srebotnik and Hammel (2003) proved that CPO may not only chlorinate major structures in lignin but also cleave them.
APO has been discovered in cultures of Agrocybe aegerita (MW = 45 kDa) and has been shown to transfer oxygen to aromatic and aliphatic substrates similar to cytochrome P450, but instead of directly transferring molecular oxygen, these enzymes utilize hydrogen peroxide (Piontek et al.2010). Moreover, this enzyme has a surprisingly high reactivity toward benzylic C − H and phenolic substrates compared to typical peroxidases or model compounds due to the basic ferryl environment of their active center (Wang et al.2015). Piontek et al. (2013) proved that these enzymes, unlike the other classical peroxidases containing histidine imidazole, have a cysteine residue as the fifth ligand coordinating with the heme iron atom.
Glucose dehydrogenase (EC 1.1.99.10)
Flavin adenine dinucleotide (FAD)-dependent glucose dehydrogenase (GDH), an extracellularly secreted enzyme by fungi, is supposed to play important role in detoxification by reducing quinones produced by plants. Moreover, the enzyme was suggested to protect fungal cell from phenoxy radicals of the same origin (Sygmund et al.2011; Piumi et al.2014). GDH catalyzes the anomeric hydroxy group of glucose while the cofactor FAD serves as the primary electron acceptor. In comparison with GOX, GDH does not use oxygen as an external electron acceptor but instead uses phenoxy radicals, quinones, redox dyes and iron complexes such as ferricyanide and ferrocenium hexafluorophosphate (Piumi et al.2014). Although this enzyme is mostly studied in genus Aspergillus, recent studies showed that it may play important role in metabolism of Py. cinnabarinus (Piumi et al.2014; Rola et al.2015).
Pyranose 2-oxidase (EC 1.1.3.10)
Pyranose oxidase (pyranose: oxygen 2-oxidoreductase) catalyzes the C-2 oxidation of several aldopyranoses concomitant with the reduction of O2 to H2O2 (Daniel, Volc and Kubatova 1994; de Koker et al.2004). With a molecular weight of up to 300 kDa (homotetramer), POX is produced by a number of Basidiomycetes, especially in the order of Polyporales (Daniel, Volc and Kubatova 1994; de Koker et al.2004). This enzyme contains three conserved domains: the FAD-binding site, prostetic group—flavin attachment loop and the substrate-binding region. The active site loop was proved to be moving in the presence of oxygen facilitating release of sugar product (Prongjit et al.2009; Pitsawong et al.2010). POX is located within the periplasmic space, where it is associated with membrane-bound vesicles and other membrane structures including extracellular mucilage slime (de Koker et al.2004).
Cellobiose dehydrogenase (EC 1.1.99.18)
Cellobiose dehydrogenase (cellobiose: [acceptor] 1-oxi-doreductase) is secreted mainly by white rot fungi such as P. chrysosporium, Trametes versicolor, Py. sanguineus and I. lacteus. The brown rot fungi do not produce CDH with the only known exception being Coniophora puteana (Schmidhalter and Canevascini 1993; Kajisa et al.2004). CDH was also isolated from the soil plant pathogen Sclerotium rolfsii and detected in the ECM fungi Pisolithus tinctorius, Suillus variegatus and Cortinarius sp. (Baldrian and Valaskova 2008). The presence of CDH in bacteria has not been confirmed yet. Recent research shows that the biological function of CDHs is associated with the cellulose degradation system (Beeson et al.2015). However, CDHs can also reduce Fe3+ to Fe2+ and O2 to H2O2. Though, ferrous ion and hydrogen peroxide are Fenton reagents found to be major players in lignocellulose degradation by brown rot fungi (Henriksson, Johansson and Pettersson 2000).
CDH is an extracellular glycoprotein detected in the secretomes of white rot and some brown rot plant pathogenic and saprotrophic fungi from the dicaryotic phyla of Basidiomycota and Ascomycota (Zamocky et al.2006). CDH has a bipartite domain organization containing a C-terminal flavodehydrogenase domain with non-covalently bound FAD connected via a flexible linker region to an electron mediating, heme b (protoporphyrin IX) containing N-terminal cytochrome like domain (Hallberg et al.2002). As a flavocytochrome, CDH exhibits a unique architecture for extracellularly secreted enzymes (Kracher et al.2015). CDH was identified as the first non-hydrolytic enzyme involved in the breakdown of cellulose. It efficiently oxidizes cellobiose and other β-1,4-linked disaccharides and oligosaccharides at the C-1 position to their corresponding lactones using a wide spectrum of electron acceptors including quinones, phenoxyradicals, Fe3+, Cu2+, cytochrome c or triiodide ion (Henriksson, Johansson and Pettersson 2000). Lactones created in reactions catalyzed by CDH are converted into aldonic acids by spontaneous or enzymatic hydrolysis with lactonase (Beeson et al.2011). The oxidation of the substrate by CDH leads to the reduction of FAD to FADH2 (Zamocky et al.2006). CDH belongs to the AA3 enzymes group in the CAZy database and belongs to the GMC oxidoreductases family. Based on the available sequences CDHs were divided into three classes. Class I represents CDHs secreted by Basidiomycota with shorter amino acid sequences and the optimum acidic pH. Besides the catalytic domain, the enzyme contains conserved cellulose binding module (CBM) characteristic to cellulolytic enzymes. Enzymes belonging to class I show a strong preference for cellulose and cello-oligosaccharides substrates, while glucose and other monosaccharides are very weak substrates for class I CDH (Zamocky et al.2004). Class II that has the ability to accept glucose as a substrate and exhibit neutral pH optima is encompassing the larger and more complex Ascomycota CDHs. Class II enzymes are subdivided into class IIA enzymes containing a type 1 CBM and class IIB enzymes that lack such a domain (Zamocky et al.2006). The secretion of class III CDHs has not yet been confirmed (Harreither et al.2011).
LOW MOLECULAR WEIGHT SYSTEMS
Low molecular weight compounds play important roles in all stages of wood decay, acting as diffusible oxidative agents and electron shuttles for enzymatic systems. These low molecular weight fungal metabolites are important during the early stages of decay because fungal enzymes are too large to penetrate the intact wood cell wall. Therefore, the initial stages of decay involve direct and indirect non-enzymatic oxidative and reductive reactions of ROS, the phenoxyl radicals, organic acids and various transition metals coordination complexes as well as species produced in secondary radical cascades (Cowling 1961; Goodell et al.1997; Arantes and Milagres 2009). These diffusible mediators, rather than the enzymes themselves, are thought to react directly with lignin to generate radical sites within the polymer and initiate a cascade of bond scissions, ultimately leading to the depolymerization of lignin.
The main groups of these low molecular weight oxidants are extracellular aromatic compounds, which are the key electron carriers between enzymes and substrates. These extracellular aromatic compounds are present in plants such as flavonoids, tannins and lignans (Gross 2008). They can also originate from lignin degradation products such as benzoic acids (Chen, Chang and Kirk 1982, 1983), veratryl (3,4-dimethoxybenzyl) alcohol (Harvey, Schoemaker and Palmer 1986) and are found in humus (Kuiters and Denneman 1987; Suominen, Kitunen and Smolander 2003). Very recently, long range electron transfer mechanism was proved to play a crucial role in reactions catalyzed by LiP and VP while degrading lignin polymer. A buried tryptophan (Trp244), a neighbor phenylalanine (Phe198) and surphase tryptophan appeared to be the most important in this process (Acebes et al.2017). The same mechanism was already described for lytic polysaccharide monooxygenase (LPMO) in degradation of cellulose while insoluble high molecular weight lignin served as an electron reservoir. The electrons can be transferred by soluble low molecular weight compounds present in plant cell walls, or secreted by hyphae, for example, 3-hydroxyanthranilic acid produced by Pycnoporus cinnabarinus (Westereng et al.2015). Preferential oxidation of phenolic lignin units and side-chain oxidation by white rot fungi leads to the release of phenolic residues with oxidized side chains (Canas and Camarero 2010). In addition to the phenolic hydroxyl group(s), these compounds may contain additional functional groups, such as methoxyls, amines, ketones, aldehydes and carboxyls. These phenolic substances are oxidized by fungal oxidoreductases (peroxidases, dehydrogenases, laccase) to phenoxyl radicals, which could oxidize non-phenolic residues of lignin through the H-abstraction mechanism (d’Acunzo et al.2006). Studies of spruce wood lignin breakdown by Phanerochaete chrysosporium have identified low molecular weight products such as aromatic carboxylic acids and derivatives of benzoic acid, indicating that they had arisen from Cα–Cβ oxidative cleavage of lignin components and the compounds included a biphenyl dicarboxylic acid, and a diphenyl ether dicarboxylic acid, derived from the biphenyl and diphenyl ether components of lignin, respectively (Chen, Chang and Kirk 1982, 1983). Similar phenolic products have been observed after treatments of lignocellulose by bacterial lignin degraders. Prime examples include benzoic acids with hydroxyl(s), methoxyl(s), carboxyl(s) substitutions (Aneurinibacillus aneurinilyticus, Pseudonomas putida); benzaldehyde with hydroxyl, methoxy, trimethoxy substitutions (Bacillus sp., Streptomyces paucimobilis); and cinnamic acid with hydroxy, methoxy substitutions (Bacillus sp., Ps. putida) (Masai, Katayama and Fukuda 2007; Ahmad et al.2010; Bugg et al.2011a).
The second group of low molecular weight oxidants includes ROS such as hydroxyl radical (OH•), superoxide anion radical (O2‐•) and hydrogen peroxide (H2O2). The main source of these ROS are enzymatic reactions (e.g. glucose oxidase, CDH, laccase, catalase and quinone redox cycling) and Fenton reactions detected mainly in brown rot fungi (Guillen et al.2000; Martinez et al.2009). The hydroxyl radical is the most powerful non-specific oxidant present in biological systems, with a very short half-life, playing very important roles in the initial non-enzymatic attack of the wood cell wall in brown rot fungi (Halliwell and Gutteridge 1999; Goodell 2003). These hydroxyl radicals are powerful oxidants that can both depolymerize polysaccharides by hydrogen abstraction and attack lignin by demethylation/demethoxylation (Niemenmaa, Uusi-Rauva and Hatakka 2008; Arantes and Milagres 2009). For the Fenton reaction, Fe(II) ions and hydrogen peroxide are needed. The main source of Fe(II) ions are insoluble Fe(III) oxyhydroxide complexes, which must be solubilized and reduced to ferrous iron. The reductive solubilization is most likely facilitated by oxalic acid, accumulated extracellularly by both brown and white rot fungi, leading to a lowering of the pH in close proximity to hyphae in the lumens of decaying wood cells (Goodell 2003). It is believed that the main source of hydrogen peroxide in brown rot fungi is enzymatic oxidation of methanol (by alcohol oxidases). Methanol is generated through demethylation of the lignin polymer (Postia placenta, Gloeophyllum trabeum) (Daniel et al.2007). In white rot fungi, H2O2 is generated by different oxidases, such as GLOX or AAO (Guillen et al.2000). During this stage, specific mechanisms of ferric iron chelation and reduction to its ferrous state are required and most likely involve extracellular low molecular weight phenolic compounds. The first group is made up of plant-derived lignin degradation products such as syringyl-type phenolic compounds (Eggert et al.1996; Nousiainen et al.2009). The next group are phenolic compounds produced by fungi, such as 2,5-dimethoxy-1,4-benzoquinone and 4,5-dimethoxy-1,2-benzoquinone (Po. placenta, G. trabeum, Serpula lacrymans) (Enoki, Itakura and Tanaka 1997; Paszczynski et al.1999; Newcombe et al.2002; Shimokawa et al.2004; Suzuki et al.2006). This phenolate and hydroquinone aromatic compounds substituted with electron-donating functional groups, such as –OCH3 oxidize rapidly and in the presence of ferric ions generate ferrous ions (Suzuki et al.2006). A redox cycling process has been proposed in G. trabeum, using two quinones that are produced extracellularly, 2,5-dimethoxy-1,4-benzoquinone and 4,5-dimethoxy-1,2-benzoquinone (Jensen et al.2001). Each hydroquinone is able to reduce chelated Fe(III) to Fe(II). The ferrous iron-oxalate complex then reacts with hydrogen peroxide to generate hydroxyl radicals, which attack lignin. The oxidized quinone is then recycled by reduction of O2 or Fe3+ to the hydroquinone. Besides the role as Fe (II) chelator, oxalic acid has other roles both in cellulose and lignin degradation, e.g. chelation and stabilization of Mn(II/III), acting as phenolic oxidants and as a buffer in the natural soil environment (Perez and Jeffries 1993; Hyde and Wood 1997; Shimada et al.1997). Oxalic acid was detected in cultures of brown and white rot fungi but the main producers of this simple dicarboxylic acid are brown rot fungi (Shimada et al.1997; Graz and Jarosz-Wilkolazka 2011). The low molecular Fe(III) reductants can diffuse through the woods cell wall and mediate the Fenton reaction (Goodell et al.1997; Varela and Tien 2003; Wang and Gao 2003). There is also an alternative mechanism for ferric iron reduction via the activity of cellulose dehydrogenase but this enzyme has not been found in brown rot fungi (Mason, Nicholls and Wilson 2003). The other postulated mechanism of Fe(III) reduction involves extracellular low molecular weight NADH-dependent glycopeptides identified in cultures of Po. placenta (brown rot) (Hammel et al.2002; Goodell 2003). During ligninolysis, other substrate-derived oxidants are produced such as alkoxyl (•OR) and peroxyl (•OOR) radicals but they are less reactive than Fenton reagents (Hammel et al.2002). Superoxide anion radicals produced by white rot fungi can be converted to H2O2 via both the dismutation and the oxidation of Mn(II) to Mn(III) and can also be involved in the Haber-Weiss reaction (Barr et al.1992). Furthermore, by reacting with phenoxyl radicals formed during lignin degradation, superoxide ions contribute to oxidative depolymerization of lignin and its degradation products (Guillen et al.2000).
GENOMICS ANALYSIS
Genomic analysis suggests that brown rot evolved from white rot fungi. Over time, brown rot lost genes coding for ligninolytic class II peroxidases and the cellobiohydrolase-encoding genes are generally absent or lacking a CBM1 domain (Table S3, Supporting Information (Riley et al.2014). Moreover, the genes GH74 xyloglucanase, GH10 endo-xylanase, GH79 β-glucuronidase, CE1 acetyl xylan esterase and CE15 glucuronoyl methylesterase tend to appear more frequently in white rot fungi compared to brown rot Basidiomycetes. A key component of redox cycles supporting Fenton chemistry, quinone reductase activity, was found only in brown rot fungi (Alfaro et al.2014) even if its gene homologs were detected in the white rot fungi (Floudas et al.2012). These findings may indicate that hydrolytic and oxidative reactions of cellulose degradation are characteristic only of white rot fungi (Hori et al.2013; Alfaro et al.2014). However, there are exceptions, Fistulina hepatica (brown rot) still possesses genes related to cellulose degradation described as characteristically plesiomorphic and are shared with its white rot ancestors.
Some species of Coniophora and Serpula are able to degrade cellulose similarly to white rot species (Nilsson 1974; Nilsson and Ginns 1979; Redhead and Ginns 1985; Floudas et al.2015). The genes encoding DyP and HTP are less numerous as compared to white rot fungi, but generic peroxidase genes are numerous in brown rot genomes, which are characterized by low a redox potential and are incapable of degrading lignin (Ruiz-Duenas et al.2013). Certain fungi, for example, Schizophyllum commune, lack genes encoding POD and DyP and have only two laccase genes, which is more similar to brown rot fungi than white rot fungi (Ohm et al.2010; Floudas et al.2015). The historical line between white rot and brown rot is blurred now as some researchers suggested that Sc. commune is an evolutionary intermediate between white and brown rot species (Riley et al.2014). A number of so-called litter decomposers (LDs) are capable of degrading lignocellulose. One of the LD fungi, Agaricus bisporus, has only two MnPs, which are both upregulated when grown on straw medium. It is possible that lignin degradation by Ag. bisporus MnPs is supported by 24 HTP genes, which is probably the highest number described to date (Morin et al.2012). Kohler et al. (2015) showed that a higher number of POD or LPMO genes are present within white rot fungi, which evolved the ability to degrade wood first during the evolution of life on Earth. The taxonomic group Boletales have lower numbers of these genes and consequently their mode of wood degradation became more like that of brown rot. It seems that we have the possibility to observe the real time evolution of wood-decaying mechanisms in the genomes of these transitory species.
Until recently, it was assumed that efficient lignin degraders have different combinations of PODs (Floudas et al.2012; Fernandez-Fueyo et al.2012a; Hori et al.2014). While POD genes were found in brown rot fungi, GLOX genes were not (Floudas et al.2012), implying that this enzyme might have an important role in lignin degradation, similar to POD or DyP. Hernandez-Ortega, Ferreira and Martinez (2012) suggested that AAO may be the main enzyme generating hydrogen peroxide in white rot fungi. Floudas et al. (2012) proved the existence of AAO genes in 24 analyzed white rot fungal genomes (except Auricularia delicata), whereas other H2O2-producing GMC are less abundant. However, it seems that gene annotation may be misleading in some cases. Mathieu et al. (2016) described the gene in Pycnoporus cinnabarinus with sequence homology to fungal AAO which was later expressed heterologously showing biochemical properties of aryl alcohol quinone oxidoreductase. The number of laccase genes is fewer in genomes of brown rot fungi. However, even among white rot fungi the number of laccase genes differs significantly among species, e.g Phanerochaete chrysosporium (0), Phlebia radiata (2), Cerena sp. (9) and Pleurotus sp. (12) (Janusz et al.2013; Kuuskeri et al.2016; Yang et al.2016) suggesting that laccase role in lignin degradation is not clear. It is well known that P. chrysosporium has 15 POD genes, composed only of MnP and LiP. On the other hand, VPs are common in the Pleurotus genus (Ruiz-Duenas et al.2001; Gao et al.2016) suggesting that different fungi use different strategies for wood degradation. However, the species mentioned above are ecologically different. Interestingly, being a saprotroph, P. chrysosporium only degrades dead wood, whereas Pleurotus sp. is rather parasitic compared to other saprotrophic species. Another conifer degrading fungus, Heterobasidion annosum, has no genes coding for LiP, VP or GLOX; therefore, implying its ability to degrade lignin is based on eight MnPs and 13 laccase genes (Olson et al.2012). Fernandez-Fueyo et al. (2014) proved that the ability of Pleurotus ostreatus to degrade lignocellulose at different pHs and temperatures may rely on a number of MnP and VP isoenzymes.
The GLOX enzyme seems to be highly important in LDs as it is upregulated in the fungus Ag. bisporus when grown in compost medium. Other potential H2O2-generating extracellular enzymes include various glucose-methanolcholine oxidoreductases and likely include an AAO and a highly expressed methanol oxidase (Morin et al.2012). Comparing genomes of P. chrysosporium and P. carnosa CAZymes belong to families GH5, GH79, CE16, PL14 and genes encoding MnP and LiP enzymes identified as responsible for efficient softwood degradation by P. chrysosporium (Suzuki et al.2012). Phlebia radiata employs oxidoreductases (lignin attacking and auxiliary) during the first weeks of lignin degradation, followed by the production of cellulose and hemicellulose-degrading enzymes (Kuuskeri et al.2016). It is possible that fungi are capable of applying flexible biochemical mechanisms to degrade different kinds of wood. Liers et al. (2011) showed that 7/11fungal species produced mainly MnP and VP, 8/11 produced laccase and only 3/11 synthesized AAO when growing on Fagus sylvatica wood. Only one fungus produced DyP and LiP was not detected in any (Liers et al.2011). The surprisingly efficient lignin-degrading fungus, P. chrysosporium, has no DyP genes in its genome, whereas several DyP genes are highly expressed in Trametes versicolor and Phlebiopsis gigantea (Floudas et al.2012; Hori et al.2014).
In the fungal kingdom, plant pathogens were determined to have the largest secretomes, animal pathogens the smallest and saprotrophs have intermediate secretomes (Krijger et al.2014). Possessing a great number of genes coding for wood-degrading enzymes, pathogenic and saprotrophic fungi are able to easily adapt to attack vast numbers of different tree species (https://nt.ars-grin.gov/fungaldatabases/). Raffaello et al. (2014) proved that a number of all uniquely upregulated genes in H. annosum are greater than a number of genes coding for wood-degrading enzymes. Moreover, the number of upregulated genes during sapwood degradation is almost three times higher than when H. annosum was cultivated on hardwood (Raffaello et al.2014) which suggest that we have long way to go to fully understand this process. Besides the wood-degrading enzymes found in Ophiostoma piceae, during growth on natural substrates, there is upregulated expression of genes coding for the following transporters, allantois, urea, hexose, iron and sugars in addition to major facilitator superfamily transporters (Haridas et al.2013). These findings suggest that fungal metabolism should be analyzed more globally when grown on natural substrates.
Based on genomic research, bacterial strategies to degrade lignin seem to be equally sophisticated as those of fungi. Klebsiella sp. strain BRL6–2 was proved to have genes coding for glutathione peroxidases, DyP-type peroxidases and catalases/peroxidases, catalase genes and no laccase genes. In contrast, microbiome analysis showed that laccase seems to be responsible for lignin degradation by Pseudomonas putida in guts of giant panda (Fang et al.2012). Moreover, multiple cytochrome oxidase genes were found in its genome as was previously observed for a related isolate Enterobacter lignolyticus SCF1 (Deangelis et al.2013; Woo et al.2014). In the Rhodococcus jostii RHA1 genome sequence, 26 peripheral pathways and 8 central pathways are involved in the catabolism of aromatic compounds including modifications by monooxygenases and dioxygenases (McLeod et al.2006; Martinkova et al.2009). The genome of Amycolatopsis sp. 75iv2 (formerly Streptomyces setonii and S. griseus 75vi2) was proved to comprise genes coding for heme peroxidases, laccases and cytochrome P450 (Brown et al.2011). Beside a suite of oxidative enzymes that could modify lignin for breakdown by hydroxylation or demethylation, one interesting example, a gene encoding a non-heme iron dioxygenase fused to a carbohydrate-binding domain was found in Streptomyces sp. SirexAA-E (Bianchetti et al.2013). Rhodococcus jostii RHA1, known producer of DyP, was proved to be able to degrade lignin without hydrogen peroxide via the β-ketoadipate pathway (Ahmad et al.2010).
It seems like bacteria may employ similar sets of enzymes to cope with so complex polymer as lignin is. Moreover, it is possible that bacteria may evolve other lignolytic enzymes beyond the canonical enzymes used by fungi, and therefore their strategies to decompose lignin may differ significantly comparing to fungi.
EVOLUTION
It is believed that the increase in fungal presence at particular biostratigraphic levels coincides with periods of rapid fungal evolution and diversity (Taylor, Krings and Taylor 2015a). The application of evolutionary genomics together with evolutionary and ecological genetics and the evaluation of fossils have allowed scientists to determine the oldest fungal ancestor—chytrid-like form. Molecular clock data obtained using fossilized fungi suggest that terrestrial fungi diverged from Chytridiomycota ∼550 Ma (Taylor, Krings and Taylor 2015b). Employing an approach that did not utilize fossils, the fungi appear to be much older, with the origin of the soil-born Glomeromycota appearing to have arisen between 1400 and 1200 Ma and the separation of Ascomycota and Basidiomycota around 1200 Ma (Heckman et al.2001; Taylor, Krings and Taylor 2015b) (Fig. 5). According to Lücking et al. (2009), the saprophytic Ascomycota and Basidiomycota originated much later at around 500 Ma, more or less parallel to the first appearance of primitive land plants. The diversification of the higher Ascomycetes, the Pezizomycotina, is estimated to be around 320 Ma, which correlates with the diversification of vascular plants as documented by the fossil record (Lücking et al.2009). Further molecular clock analyses revealed that the Pezizomycotina and Agaricomycetes are ∼400 and 300 million years old, respectively (Kohler et al.2015). To place the origin of lignin degradation in the context of geologic time, Floudas et al. (2012) exploited Bayesian relaxed molecular clock analyses with fossil-based calibrations, which revealed the mean age of the Agaricomycetes at c.a. 290 Ma. Using this approach, the mean age of Agaricomycotina was determined at c.a. 430 to 470 Ma, which coincide with earlier findings (Floudas et al.2012).
Figure 5.
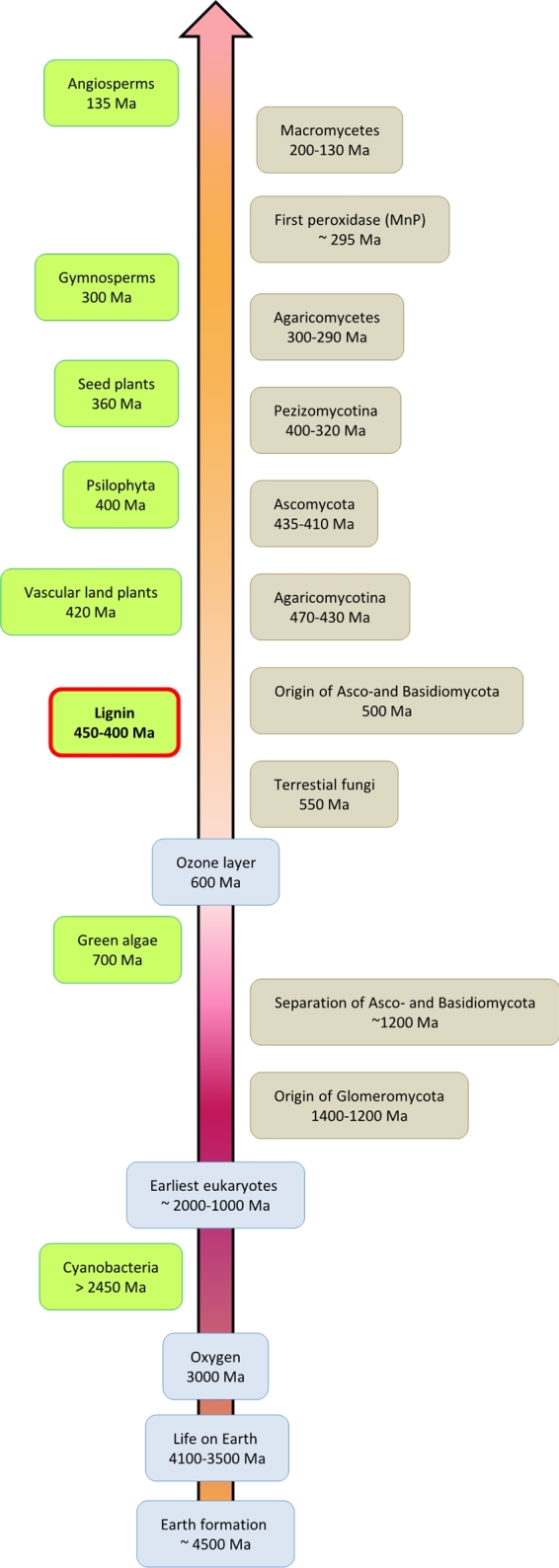
Timetree of plants and fungi combined and simplified from various sources (Heckman et al.2001; Hullo et al.2001; Lücking et al.2009; Moore, Robson and Trinci 2011; Floudas et al.2012; Schirrmeister et al.2013; Taylor, Krings and Taylor 2015); left part of the figure demonstrates the history of the evolution of plants (marked with green); right part of the figure demonstrates the history of the evolution of fungi (marked with beige); numbers below descriptions represent time of the specific evolutionary event expressed as millions years ago (Ma).
It should be noted, however, that the accumulation of the vast coal deposits around the globe formed during the Carboniferous–Permian age (359–252 Ma) has historically been hypothesized as resulting from the inability of the saprotrophic organisms to degrade plant material (including lignin and cellulose) in the anoxic conditions that existed in the swamp ecosystems (Taylor, Krings and Taylor 2015a). However, molecular clock assumptions together with a comparative analysis of 31 saprophytic fungal genomes placed the origin of lignin degradation near the end of the Carboniferous period (359–299 Ma). Nevertheless, there is also evidence that by the Late Devonian (385–359 Ma) there were fungi associated with lignin degradation (Arnold 1931; Stubblefield and Banks 1983; Taylor, Krings and Taylor 2015a). In addition, there are extensive coal deposits that formed during the Permian (298–252 Ma) and Cretaceous (145–66 Ma), as well as in other time periods, and their presence seems to refute the hypothesis that Carboniferous coals formed due to a lack of wood-decaying organisms at that time (Taylor, Krings and Taylor 2015a). Researchers concluded that lignin degradation evolved only once and within the Agaricomycotina some members lost the ability altogether and evolved other methods to degrade plant organic matter, e.g. brown rot and ECM (Floudas et al.2012). It has been suggested that diversification in fungal nutritional preferences occurred alongside diversification of angiosperms and gymnosperms. Additionally, the chronology of divergences in the fungal nutritional lifestyle is compatible with the predicted major diversification in conifers, suggesting that the boreal forest (taiga) biome may have its origin in parallel genetic coevolution of plants and fungi (Eastwood et al.2011). Genome information indicates that brown rot fungi evolved several times from ancestral white rot species, which makes this group rather genetically heterogeneous, and to present some of them resemble white rot fungi (Floudas et al.2012). Thus, the brown rot classifier denotes the properties of decay in plant tissues rather than phylogeny (Kaffenberger and Schilling 2015).
Recently, it has been suggested that Agaricomycetes capable of lignin degradation may have been present before the Carboniferous and lignin degradation was likely never restricted to this class of fungi (Kohler et al.2015; Nelsen et al.2016). The Carboniferous−Permian peak of the coal accumulation was probably the result of a unique combination of tropical conditions and tectonics during the assembly of Pangea and fungal community composition bared no direct relevance. This hypothesis is based on the fact that lignin modification is known to occur via other enzymatic mechanisms in other fungal lineages and these alternate mechanisms may have evolved deeper in the phylogeny (Nelsen et al.2016). Evolutionary and functional analysis of mycorrhizal Basidiomycota and Ascomycota, using comparative analysis of 18 newly sequenced genomes, revealed that mycorrhizal symbioses arose repeatedly during fungal evolution from functionally diverse saprotrophic ancestors. During the process, distinct suites of plant cell-wall-degrading enzymes were retained thus suggesting that ECM fungi possess diverse abilities to decompose lignocellulose (Kohler et al.2015). The hypothesis concerning multiple ECM origins has been questioned by Wolfe, Tulloss and Pringle (2012), who demonstrated a single origin of ECM mutualism in Amanita. However, Wolfe et al.’s theory is supported by the loss of various crucial cellulase genes, which suggests that Amanita can no longer decompose complex organic matter and thus must rely on carbon sources from the host plant via ECM symbiosis (Wolfe, Tulloss and Pringle 2012). It was demonstrated that the loss of aggressive ligninolysis in Boletales might have permitted the brown rot species transition to biotrophic ECM symbiont, which is promoted in soils impoverished of nitrogen by brown rot enrichment and by the nutritional advantage conferred by the connection to a mycorrhizal network (Eastwood et al.2011). Comparative analysis of Fistulina hepatica and Cylindrobasidium torrendii proved that brown rot fungi evolved from white rot as a result of multiple genes losses (Floudas et al.2015). According to Martinez et al. (2009), brown rot fungi diverged from white rot fungi by losing the capacity to degrade lignin, particularly by losing extracellular peroxidases. The transition from white rot toward brown rot could have taken place multiple times across the Agaricomycotina lineage (Floudas et al.2012). Transitions back to a white rot lifestyle, from either a brown rot or a mycorrhizal fungi, have also been observed, which imply that fungi with non-ligninolytic properties would have retained genes for lignin-degrading enzymes (Morgenstern, Klopman and Hibbett 2008). Analysis of the functional diversity of forest fungi revealed that brown rot fungi have the fewest oxidoreductases, not because of gene losses but because of gene duplications in white rot species, and alternative mechanism evolved (Eastwood et al.2011). Some fungal wood degraders act in an unclassified and diverse way (dual life style between white rot and brown rot), which indicate that wood decay mechanisms as well as fungal classification based on decay strategy are not that reliable (Floudas et al.2015).
Reconstruction of the evolution of degradation capabilities was proposed based on phylogenetic analyses applied for different genes encoding for plant cell-wall-degrading enzymes (Morgenstern, Klopman and Hibbett 2008; Castilho et al.2009; Floudas et al.2012; Cázares-García, Vázquez-Garcidueñas and Vázquez-Marrufo 2013; Ruiz-Duenas et al.2013; Kohler et al.2015). Establishing a topological hierarchy regarding the genetic and evolutionary relationships of laccase genes based on their amino acid sequences suggests that fungal laccases of the Ascomycetes and Basidiomycetes clearly belong to two distinct clusters (Castilho et al.2009). Contrary to other findings, lignin is the major precursor of coal so the finding that the origin of AA2 genes roughly coincided with the end of Carboniferous supported the view that evolution of white rot fungi impacted the global carbon cycle (Nagy et al.2015). Furthermore, the reconstruction of the evolution of 18 gene families showed an increase in the copy number and the diversification of genes responsible for lignocellulose decomposition after the divergence of Auricularies, which refines previous hypotheses on the origin of white rot (Floudas et al.2012). The authors speculate that the diversity of oxidative enzymes present in white rot fungi emerged after the origins of the first white rot species (Nagy et al.2015). A detailed analysis using amino acid sequences data generated from 11 species of wood-decaying Agaricomycetes revealed a detailed picture of the fungal peroxidases phylogeny (Morgenstern, Klopman and Hibbett 2008). This distinctive and ecologically important class of proteins has diversified extensively escpecially in Basidiomycetes, after the split of Basidiomycetous and Ascomycetous lineages of fungi. The sequences of extracellular fungal peroxidases including MnPs, LiPs and VPs constitute a monophyletic gene family within the superfamily of plant and microbial peroxidases. LiPs evidently occur and diversified only in members of the Polyporales, which harbors many white rot taxa. According to Morgenstern, Klopman and Hibbett (2008), MnPs and VPs are more widespread and may have multiple origins. MnPs, LiPs and VPs were identified in only three orders, although their degenerate forms were found in different Pezizomycota (Passardi et al.2007). The study provided by Zhou, Wei and Dai (2014) gave essential evidence for the explanation of the evolutionary dynamics of peroxidases and thus gaining insights into the cause for the alternation of white rot and brown rot fungi in their common lineage; these enzymes originated many times at the level of orders independently, and thus they did not exist in all taxonomic groups of Basidiomycetes. According to the study, the occurrence of enzymes is limited to Russulales, Agaricales and Polyporales. Also, the evolutionary order of peroxidase appearance was proposed; peroxidases without any obvious lignin-degrading activity and MnPs were the primary groups of enzymes to evolve, following VPs and LiPs, which secured complete lignin degradation. This hypothesis also indicates that the gene duplication events probably occurred before the divergence of Ascomycetes and Basidiomycetes and the duplicated enzymes were passed down to newborn species or genera (Zhou, Wei and Dai 2014). It has been proven that MnPs are phylogenetically older than LiPs and thus duplicated and diversified before the split of major lineages of Agaricomycetes. However, the VPs of Agaricales and Polyporales do not exhibit a close relationship but seem to have evolved independently in different clades, which suggests that VPs may have arisen relatively early in the evolutionary history of fungal peroxidases. According to the organismal phylogeny calibrated with the split between Ascomycota and Basidiomycota, the first ligninolytic MnP arose at about 295 Ma, which is slightly earlier than (and therefore consistent with) the oldest definitive white rot fossils from the Permian (c.a. 260 Ma) and Triassic (c.a. 230 Ma) (Floudas et al.2012). It has been confirmed that a common ancestor of Agaricales possesses a sophisticated enzymatic system for utilization of crystalline cellulose and cellobiose, and all oxidative gene families, including POD and DyP, similar to contemporary white rot species of Agaricales (Floudas et al.2015).
Over the course of the evolutionary process, laccases have maintained a high degree of similarity in terms of amino acid sequence and three-dimensional structure. In order to track the evolutionary history of fungal laccases, the reconstruction of the gene loci evolution inferred from the comparative analysis of 48 different sequences was applied. The phylogenetic relations indicated that most laccases from the Ascomycota and the Basidiomycota divisions formed independent clades, consistent with taxonomy. It has been shown that a single monophyletic branch exists for fungal laccases and that laccase isozyme genes may have evolved independently, possibly through duplication–divergence events. Bearing in mind that the enzyme activity is linked to three different copper sites, it must have been a very early biological event. Therefore, laccases are probably ancient enzymes from an evolutionary point of view (Valderrama et al.2003). Also, the reconstruction of LMCO phylogeny in Trichoderma reesei, a model organism of cellulose and hemicellulose degradation, has been undertaken by Levasseur et al. (2010). LMCO and classical laccases were clearly divided into two distinct groups containing the laccases of Basidiomycota and Ascomycota, having two different evolutionary histories leading to high-sequence divergence between homologs. A comparative in silico analysis of the identified genes of laccase in Trichoderma performed elsewhere (Cázares-García, Vázquez-Garcidueñas and Vázquez-Marrufo 2013) allows for clear differentiation between extracellular and intracellular enzymes. Additionally, phylogenetic analysis supports the hypothesis of two laccase genes in the common Trichoderma ancestor with later events of retention, loss or gain of laccase genes can be extended to the Ascomycetes group. A global phylogenetic analysis of more than 350 MCOs from fungi, insects, plants and bacteria created a clear classification of this enzyme family. Within the largest group of enzymes, formed by the Basidiomycetous and Ascomycetous laccases sensu stricto, the gene phylogeny does not strictly follow the species phylogeny but enzyme similarities seem to group at least partially according to the lifestyle of the corresponding species. One of the most obvious features of the constructed tree was that the sequences of laccase sensu stricto clustered according to the taxonomical association of the corresponding species (Hoegger et al.2006). The fungal laccases were clearly separated into two clusters, as has been shown earlier elsewhere (Castilho et al.2009). The LMCO classification proposed by Hoegger et al. (2006) lead to the creation of a database for a systematic sequence-based classification and analysis of the multicopper oxidase protein family—LccED (Laccase and Multicopper Oxidase Engineering Database). In the beginning, the classification was based on 10 protein superfamilies (named from A to J) and one more superfamily (K), established for two-domain bacterial LMCO (Fernandes et al.2014). Presently, LccED lists the number of these superfamilies growth to have reached 16 (https://lcced.biocatnet.de/). This classification seems to be an important step toward understanding the evolutional relations between this large group of fungal enzymes. Phylogenetic distribution of lignin-degrading prokaryotes, widely discussed by Tian et al. (2014), demonstrated clear clustering of the laccase-encoding species.
Until recently, the enzymes responsible for degradation of lignin in prokaryotes were poorly understood (Bugg et al.2011b; Tian et al.2014), but peroxidases from the DyP family have been shown to be active in lignin degradation. Very few MnPs were described for bacteria, whereas most of the identified LiP sequences are putative. Bacterial catabolic pathways are still poorly understood, and our current knowledge is not sufficiently advanced enough to establish a phylogenetic overview of all ligninolytic prokaryotic species (Tian et al.2014).
Phylogenetic studies proved to be an important tool for the characterization of proteins responsible for wood degradation and it became the basis for its division into families and subfamilies (Hoegger et al.2006; Passardi et al.2007; Morgenstern, Klopman and Hibbett 2008; Lundell, Makela and Hilden 2010; Zhou, Wei and Dai 2014; Zamocky et al.2015). However, there is a huge knowledge gap concerning lignin degradation. The role of laccases in decomposition of lignin is still questionable, since there are non-lignin-degrading fungi which may contain several laccase encoding genes (Hatakka and Hammel 2011). Recently, several novel clades with putative MnP and LiP members were described in various Ascomycetes unable to decay wood (Zamocky et al.2015). Also, lignin is degraded after nutrient depletion and lignin degradation is too slow to serve as a source of metabolic energy (Leisola, Pastinen and Axe 2012). Genome studies of wood-inhabiting Basidiomycetes show that there is a need for a more detailed classification of the rot types, since some fungi do not fulfill the traditional criteria for dichotomous grouping (Riley et al.2014; Rytioja et al.2014; Floudas et al.2015). Additionally, the presence of extensive coal deposits formed during the Permian and Cretaceous periods seems to refute the hypothesis that Carboniferous coals formed due to a lack of wood-decaying organisms at that time (Taylor, Krings and Taylor 2015a).
CONCLUDING REMARKS
Even if fungal organisms are considered to be one of the first eukaryotic ones, their evolution could not be perceived as finished. They are still evolving new sophisticated metabolic solutions to grow faster, cover more areas, spend less energy and adopt novel ecological niches. Many of them easily adapted to live in an environment near humans, which may be harsh conditions for other species. For many years, white rot fungi were suggested to be the most efficient wood degraders. However, recent data suggest that Nature may have an alternative solution—brown rot fungi, which are capable of depolymerizing holocellulose and extensively modifying lignin. They are thought to have evolved from ancestral forms of saproptrophic white rot ancestors, possessing the whole lignocellulose-degrading system, both enzymatic and non-enzymatic (Martinez et al.2009). The evolution of brown rot saprotrophy was accompanied by reductions and losses in key enzymes implicated in biomass breakdown in white rot, especially cellulases and LMEs. Low molecular weight compounds most likely originated from microbial metabolism or from plant biomass and act as diffusible oxidizing agents and electron shuttles. Therefore, non-enzymatic degradation processes are expected to be considerably less energetically expensive than biosynthesis and secretion of complex enzymatic proteins.
Lignin degradation is a complex process and multidimensional scientific survey which must be undertaken to understand its mechanisms and evolution. This must include the vast distribution among phylogenetic lineages and wood-decaying mechanisms. It seems that the diversity in the fungal kingdom allowed these microrganisms to be successful in inhabitation of different ecological niches from rotting plant material in water, to forests and even to houses made of wood material. It is hard to perceive how the evolution of wood roting fungi will look in the future. We believe that fungal organisms are well adapted and prepared for changes in the environment. However, rapidly changing climate may have had the greatest impact on the evolution of fungi.
Supplementary Material
Supplementary data are available at FEMSRE online.
Acknowledgements
The coauthor, A. Paszczynski, acknowledges support for his sabbatical leave from both the University of Idaho and Maria Curie-Skłodowska University.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSRE online.
FUNDING
This work was supported by the National Science Centre (Poland) projects (project DEC-2013/09/B/NZ9/01829; 2014/13/B/NZ9/02106 and project 2015/17/D/NZ9/02066) and by the research program BS/UMCS.
The SEM images were taken using scanning electron microscope purchased thanks to the financial support of the European Regional Development Fund under the project: ‘Increase in R & D potential of Faculties of Chemistry, and Biology and Earth Sciences UMCS in Lublin’ (POPW.01.03.00-06-017/09-00).
Conflict of interest. None declared.
REFERENCES
- Aarti C, Arasu MV, Agastian P. Lignin degradation: A microbial approach. Indian J Biol Sci 2015;1:119–27. [Google Scholar]
- Acebes S, Ruiz-Duenas FJ, Toubes M et al. Mapping the long-range electron transfer route in ligninolytic peroxidases. J Phys Chem B 2017;121:3946–54. [DOI] [PubMed] [Google Scholar]
- Adapa P, Tabil L, Schoenau G. Compaction characteristics of barley, canola, oat and wheat straw. Biosyst Eng 2009;104:335–44. [Google Scholar]
- Ahmad M, Taylor CR, Pink D et al. Development of novel assays for lignin degradation: comparative analysis of bacterial and fungal lignin degraders. Mol BioSyst 2010;6:815–21. [DOI] [PubMed] [Google Scholar]
- Alfaro M, Oguiza JA, Ramirez L et al. Comparative analysis of secretomes in basidiomycete fungi. J Proteomics 2014;102:28–43. [DOI] [PubMed] [Google Scholar]
- Arantes V, Milagres AME. Relevance of low molecular weight compounds produced by fungi and involved in wood biodegradation. Quimica Nova 2009;32:1586–95. [Google Scholar]
- Arnold CA. On Callixylon Newberryi (Dawson) Elkins Et Wieland. Detroit, USA: University of Michigan Press, 1931. [Google Scholar]
- Baldrian P. Fungal laccases - occurrence and properties. FEMS Microbiol Rev 2006;30:215–42. [DOI] [PubMed] [Google Scholar]
- Baldrian P, Valaskova V. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev 2008;32:501–21. [DOI] [PubMed] [Google Scholar]
- Barceló AR, Gómez Ros LV, Gabaldón C et al. Basic peroxidases: The gateway for lignin evolution? Phytochem Rev 2004;3:61–78. [Google Scholar]
- Barr DP, Shah MM, Grover TA et al. Production of hydroxyl radical by lignin peroxidase from Phanerochaete chrysosporium. Arch Biochem Biophys 1992;298:480–5. [DOI] [PubMed] [Google Scholar]
- Beeson WT, Iavarone AT, Hausmann CD et al. Extracellular aldonolactonase from Myceliophthora thermophila. Appl Environ Microb 2011;77:650–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeson WT, Vu VV, Span EA et al. Cellulose degradation by polysaccharide monooxygenases. Annu Rev Biochem 2015;84:923–46. [DOI] [PubMed] [Google Scholar]
- Berg B, McClaugherty C. Decomposer organisms. In: Plant Litter: Decomposition, Humus Formation, Carbon Sequestration. Berlin, Heidelberg: Springer Berlin Heidelberg, 2014;35–52. [Google Scholar]
- Bianchetti CM, Harmann CH, Takasuka TE et al. Fusion of dioxygenase and lignin-binding domains in a novel secreted enzyme from cellulolytic Streptomyces sp. SirexAA-E. J Biol Chem 2013;288:18574–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb MJ. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 2005;8:208–15. [DOI] [PubMed] [Google Scholar]
- Bidlack JE. Cell-wall components and lignin biosynthesis in forages volume Ph.D. Dissertation. Ames, IA: Iowa State University, 1990. [Google Scholar]
- Blanchette RA. Degradation of the lignocellulose complex in wood. Can J Bot 1995;73:999–1010. [Google Scholar]
- Boer WD, Folman LB, Summerbell RC et al. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev 2005;29:795–811. [DOI] [PubMed] [Google Scholar]
- Breen A, Singleton FL. Fungi in lignocellulose breakdown and biopulping. Curr Opin Biotechnol 1999;10:252–8. [DOI] [PubMed] [Google Scholar]
- Brosse N, Dufour A, Meng X et al. Miscanthus: a fast-growing crop for biofuels and chemicals production. Biofuels Bioprod Bior 2012;6:580–98. [Google Scholar]
- Brown ME, Walker MC, Nakashige TG et al. Discovery and characterization of heme enzymes from unsequenced bacteria: application to microbial lignin degradation. J Am Chem Soc 2011;133:18006–9. [DOI] [PubMed] [Google Scholar]
- Bruckmann M, Termonia A, Pasteels JM et al. Characterization of an extracellular salicyl alcohol oxidase from larval defensive secretions of Chrysomela populi and Phratora vitellinae (Chrysomelina). Insect Biochem Mol Biol 2002;32:1517–23. [DOI] [PubMed] [Google Scholar]
- Brunow G. Methods to reveal the structure of lignin. Alexander S, (ed.)Biopolymers Online, Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA, 2005. [Google Scholar]
- Bugg TD, Ahmad M, Hardiman EM et al. Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep 2011a;28:1883–96. [DOI] [PubMed] [Google Scholar]
- Bugg TDH, Ahmad M, Hardiman EM et al. The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol 2011b;22:394–400. [DOI] [PubMed] [Google Scholar]
- Canas AI, Camarero S. Laccases and their natural mediators: biotechnological tools for sustainable eco-friendly processes. Biotechnol Adv 2010;28:694–705. [DOI] [PubMed] [Google Scholar]
- Castilho FJD, Torres RA, Barbosa AM et al. On the diversity of the laccase gene: a phylogenetic perspective from Botryosphaeria rhodina (Ascomycota: Fungi) and other related taxa. Biochem Genet 2009;47:80–91. [DOI] [PubMed] [Google Scholar]
- Cázares-García SV, Vázquez-Garcidueñas MS, Vázquez-Marrufo G. Structural and phylogenetic analysis of laccases from Trichoderma: a bioinformatic approach. PLoS One 2013;8:e55295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chang H, Kirk TK. Aromatic acids produced during degradation of lignin in spruce wood by Phanerochaete chrysosporium. Holzforschung 1982;36:3–9. [Google Scholar]
- Chen C, Shrestha R, Jia K et al. Characterization of Dye-decolorizing peroxidase (DyP) from Thermomonospora curvata reveals unique catalytic properties of A-type DyPs. J Biol Chem 2015;290:23447–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Chang H, Kirk TK. Carboxylic acids produced through oxidative cleavage of aromatic rings during degradation of lignin in spruce wood by Phanerochaete chrysosporium. J Wood Chem Tech 1983;3:35–57. [Google Scholar]
- Choinowski T, Blodig W, Winterhalter KH et al. The crystal structure of lignin peroxidase at 1.70 A resolution reveals a hydroxy group on the cbeta of tryptophan 171: a novel radical site formed during the redox cycle. J Mol Biol 1999;286:809–27. [DOI] [PubMed] [Google Scholar]
- Colpa DI, Fraaije MW, van Bloois E. DyP-type peroxidases: a promising and versatile class of enzymes. J Ind Microbiol Biot 2014;41:1–7. [DOI] [PubMed] [Google Scholar]
- Côté WA. Wood Ultrastructure: An Atlas of Electron Micrographs. Seattle: University of Washington Press, 1967. [Google Scholar]
- Couturier M, Navarro D, Olive C et al. Post-genomic analyses of fungal lignocellulosic biomass degradation reveal the unexpected potential of the plant pathogen Ustilago maydis. BMC Genomics 2012;13:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling EB. Comparative Biochemistry of the Decay of Sweetgum Sapwood by White-Rot and Brown-Rot Fungi. Washington: U. S. Department of Agriculture; 1961. [Google Scholar]
- Cragg SM, Beckham GT, Bruce NC et al. Lignocellulose degradation mechanisms across the Tree of Life. Curr Opin Chem Biol 2015;29:108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford RH, Carpenter SE, Harmon ME. Communities of filamentous fungi and yeast in decomposing logs of Pseudotsuga menziesii. Mycologia 1990;82:759–65. [Google Scholar]
- da Silva Coelho-Moreira J, Maciel GM, Castoldi R et al. Involvement of lignin-modifying enzymes in the degradation of herbicides. In: Price A. (ed.) Herbicides - Advances in Research. Croatia: In Tech, 2013. [Google Scholar]
- d’Acunzo F, Galli C, Gentili P et al. Mechanistic and steric issues in the oxidation of phenolic and non-phenolic compounds by laccase or laccase-mediator systems. The case of bifunctional substrates. New J Chem 2006;30:583–91. [Google Scholar]
- Daniel G, Volc J, Filonova L et al. Characteristics of Gloeophyllum trabeum alcohol oxidase, an extracellular source of H2O2 in brown rot decay of wood. Appl Environ Microb 2007;73:6241–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel G, Volc J, Kubatova E. Pyranose oxidase, a major source of H2O2 during wood degradation by Phanerochaete chrysosporium, Trametes versicolor, and Oudemansiella mucida. Appl Environ Microb 1994;60:2524–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou M, Piumi F, Cullen D et al. Heterologous production and characterization of two glyoxal oxidases from Pycnoporus cinnabarinus. Appl Environ Microb 2016;82:4867–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashtban M, Schraft H, Syed TA et al. Fungal biodegradation and enzymatic modification of lignin. Int J Biochem Mol Biol 2010;1:36–50. [PMC free article] [PubMed] [Google Scholar]
- de Koker TH, Mozuch MD, Cullen D et al. Isolation and purification of pyranose 2-oxidase from Phanerochaete chrysosporium and characterization of gene structure and regulation. Appl Environ Microb 2004;70:5794–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira PL, Duarte MC, Ponezi AN et al. Purification and partial characterization of manganese peroxidase from Bacillus pumilus and Paenibacillus sp. Braz J Microbiol 2009;40:818–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza CG, Tychanowicz GK, de Souza DF et al. Production of laccase isoforms by Pleurotus pulmonarius in response to presence of phenolic and aromatic compounds. J Basic Microbiol 2004;44:129–36. [DOI] [PubMed] [Google Scholar]
- Deacon JW. Fungal Biology: 4th edn Malden, MA: Blackwell Publisher, 2006. [Google Scholar]
- Deangelis KM, Sharma D, Varney R et al. Evidence supporting dissimilatory and assimilatory lignin degradation in Enterobacter lignolyticus SCF1. Front Microbiol 2013;4:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai SS, Nityanand C. Microbial laccases and their applications: a review. Asian J Biotechnol 2011;3:98–124. [Google Scholar]
- Diamantidis G, Effosse A, Potier P et al. Purification and characterization of the first bacterial laccase in the rhizospheric bacterium Azospirillum lipoferum. Soil Biol Biochem 2000;32:919–27. [Google Scholar]
- Eastwood DC, Floudas D, Binder M et al. The plant cell wall–decomposing machinery underlies the functional diversity of forest fungi. Science 2011;333:762–5. [DOI] [PubMed] [Google Scholar]
- Eggert C, Temp U, Dean JF et al. A fungal metabolite mediates degradation of non-phenolic lignin structures and synthetic lignin by laccase. FEBS Lett 1996;391:144–8. [DOI] [PubMed] [Google Scholar]
- Ek M, Gellerstedt G, Henriksson G. Wood Chemistry and Wood Biotechnology. Berlin: Walter De Gruyter Inc, 2009. [Google Scholar]
- El-Tayeb TS, Abdelhafez AA, Ali SH et al. Effect of acid hydrolysis and fungal biotreatment on agro-industrial wastes for obtainment of free sugars for bioethanol production. Braz J Microbiol 2012;43:1523–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoki A, Itakura S, Tanaka H. The involvement of extracelluar substances for reducing molecular oxygen to hydroxyl radical and ferric iron to ferrous iron in wood degradation by wood decay fungi. J Biotechnol 1997;53:265–72. [Google Scholar]
- Fang W, Fang Z, Zhou P et al. Evidence for lignin oxidation by the giant panda fecal microbiome. PLoS One 2012;7:e50312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes TAR, da Silveira WB, Passos FML et al. Laccases from Actinobacteria. What we have and what to expect. Adv Microbiol 2014;4:285–96. [Google Scholar]
- Fernandez IS, Ruiz-Duenas FJ, Santillana E et al. Novel structural features in the GMC family of oxidoreductases revealed by the crystal structure of fungal aryl-alcohol oxidase. Acta Crystallogr D 2009;65:1196–205. [DOI] [PubMed] [Google Scholar]
- Fernandez-Fueyo E, Castanera R, Ruiz-Duenas FJ et al. Ligninolytic peroxidase gene expression by Pleurotus ostreatus: differential regulation in lignocellulose medium and effect of temperature and pH. Fungal Genet Biol 2014;72:150–61. [DOI] [PubMed] [Google Scholar]
- Fernandez-Fueyo E, Linde D, Almendral D et al. Description of the first fungal dye-decolorizing peroxidase oxidizing manganese(II). Appl Microbiol Biot 2015;99:8927–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Fueyo E, Ruiz-Duenas FJ, Ferreira P et al. Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. P Natl Acad Sci USA 2012a;109:5458–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Fueyo E, Ruiz-Duenas FJ, Miki Y et al. Lignin-degrading peroxidases from genome of selective ligninolytic fungus Ceriporiopsis subvermispora. J Biol Chem 2012b;287:16903–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira P, Hernandez-Ortega A, Herguedas B et al. Kinetic and chemical characterization of aldehyde oxidation by fungal aryl-alcohol oxidase. Biochem J 2010;425:585–93. [DOI] [PubMed] [Google Scholar]
- Fitigau IF, Peter F, Boeriu CG. Oxidative polymerization of lignins by laccase in water-acetone mixture. Acta Biochim Pol 2013;60:817–22. [PubMed] [Google Scholar]
- Floudas D, Binder M, Riley R et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 2012;336:1715–9. [DOI] [PubMed] [Google Scholar]
- Floudas D, Held BW, Riley R et al. Evolution of novel wood decay mechanisms in Agaricales revealed by the genome sequences of Fistulina hepatica and Cylindrobasidium torrendii. Fungal Genet Biol 2015;76:78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa Y, Osono T, Takeda H. Wood decomposing abilities of diverse lignicolous fungi on nondecayed and decayed beech wood. Mycologia 2011;103:474–82. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zheng L, Li JJ et al. Insight into the impact of two structural calcium ions on the properties of Pleurotus eryngii versatile ligninolytic peroxidase. Arch Biochem Biophys 2016;612:9–16. [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz E, Mate DM, Gonzalez-Perez D et al. Directed evolution of ligninolytic oxidoreductases: from functional expression to stabilization and beyond. In: Fessner W. (ed.) Cascade Biocatalysis. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA, 2014, 1–22. [Google Scholar]
- Gilbertson RL. Wood-rotting fungi of North-America. Mycologia 1980;72:1–49. [Google Scholar]
- Glenn JK, Gold MH. Purification and characterization of an extracellular Mn(II)-dependent peroxidase from the lignin-degrading basidiomycete, Phanerochaete chrysosporium. Arch Biochem Biophys 1985;242:329–41. [DOI] [PubMed] [Google Scholar]
- Goodell B. Brown-rot fungal degradation of wood: Our evolving view. Goodell Barry, Darrel D. Nicholas, Tor P. Schultz, (eds).Wood Deterioration and Preservation. Washington, DC: American Chemical Society; 2003;845:97–118. [Google Scholar]
- Goodell B, Jellison J, Liu J et al. Low molecular weight chelators and phenolic compounds isolated from wood decay fungi and their role in the fungal biodegradation of wood. J Biotechnol 1997;53:133–62. [Google Scholar]
- Goszczynski S, Paszczynski A, Pasti-Grigsby MB et al. New pathway for degradation of sulfonated azo dyes by microbial peroxidases of Phanerochaete chrysosporium and Streptomyces chromofuscus. J Bacteriol 1994;176:1339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graz M, Jarosz-Wilkolazka A. Oxalic acid, versatile peroxidase secretion and chelating ability of Bjerkandera fumosa in rich and limited culture conditions. World J Microbiol Biot 2011;27:1885–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross GG. From lignins to tannins: forty years of enzyme studies on the biosynthesis of phenolic compounds. Phytochemistry 2008;69:3018–31. [DOI] [PubMed] [Google Scholar]
- Guillen F, Gomez-Toribio V, Martinez MJ et al. Production of hydroxyl radical by the synergistic action of fungal laccase and aryl alcohol oxidase. Arch Biochem Biophys 2000;383:142–7. [DOI] [PubMed] [Google Scholar]
- Guillen F, Martinez AT, Martinez MJ. Substrate specificity and properties of the aryl-alcohol oxidase from the ligninolytic fungus Pleurotus eryngii. Eur J Biochem 1992;209:603–11. [DOI] [PubMed] [Google Scholar]
- Hallberg BM, Henriksson G, Pettersson G et al. Crystal structure of the flavoprotein domain of the extracellular flavocytochrome cellobiose dehydrogenase. J Mol Biol 2002;315:421–34. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine: 3rd edn Oxford, NY: Clarendon Press; 1999. [Google Scholar]
- Hammel KE, Cullen D. Role of fungal peroxidases in biological ligninolysis. Curr Opin Plant Biol 2008;11:349–55. [DOI] [PubMed] [Google Scholar]
- Hammel KE, Kapich AN, Jensen KA et al. Reactive oxygen species as agents of wood decay by fungi. Enzyme Microb Technol 2002;30:445–53. [Google Scholar]
- Haridas S, Wang Y, Lim L et al. The genome and transcriptome of the pine saprophyte Ophiostoma piceae, and a comparison with the bark beetle-associated pine pathogen Grosmannia clavigera. BMC Genomics 2013;14:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harreither W, Sygmund C, Augustin M et al. Catalytic properties and classification of cellobiose dehydrogenases from ascomycetes. Appl Environ Microb 2011;77:1804–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey PJ, Schoemaker HE, Palmer JM. Veratryl alcohol as a mediator and the role of radical cations in lignin biodegradation by Phanerochaete chrysosporium. FEBS Lett 1986;195:242–6. [Google Scholar]
- Hatakka A. Biodegradation of lignin . Alexander S, (ed.)Biopolymers Online, Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA, 2005. [Google Scholar]
- Hatakka A, Hammel KE. Fungal biodegradation of lignocelluloses. In: Hofrichter M. (ed.) Industrial Applications. Berlin, Heidelberg: Springer Berlin Heidelberg, 2011, 319–40. [Google Scholar]
- Heckman DS, Geiser DM, Eidell BR et al. Molecular evidence for the early colonization of land by fungi and plants. Science 2001;293:1129–33. [DOI] [PubMed] [Google Scholar]
- Heinfling A, Martinez MJ, Martinez AT et al. Purification and characterization of peroxidases from the dye-decolorizing fungus Bjerkandera adusta. FEMS Microbiol Lett 1998;165:43–50. [DOI] [PubMed] [Google Scholar]
- Heinzkill M, Bech L, Halkier T et al. Characterization of laccases and peroxidases from wood-rotting fungi (family Coprinaceae). Appl Environ Microb 1998;64:1601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson G, Johansson G, Pettersson G. A critical review of cellobiose dehydrogenases. J Biotechnol 2000;78:93–113. [DOI] [PubMed] [Google Scholar]
- Hernandez-Ortega A, Ferreira P, Martinez AT. Fungal aryl-alcohol oxidase: a peroxide-producing flavoenzyme involved in lignin degradation. Appl Microbiol Biot 2012;93:1395–410. [DOI] [PubMed] [Google Scholar]
- Hilden KS, Bortfeldt R, Hofrichter M et al. Molecular characterization of the basidiomycete isolate Nematoloma frowardii b19 and its manganese peroxidase places the fungus in the corticioid genus Phlebia. Microbiology 2008;154:2371–9. [DOI] [PubMed] [Google Scholar]
- Hoegger PJ, Kilaru S, James TY et al. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J 2006;273:2308–26. [DOI] [PubMed] [Google Scholar]
- Hofrichter M. Review: lignin conversion by manganese peroxidase (MnP). Enzyme Microb Technol 2002;30:454–66. [Google Scholar]
- Hofrichter M, Ullrich R. Heme-thiolate haloperoxidases: versatile biocatalysts with biotechnological and environmental significance. Appl Microbiol Biot 2006;71:276–88. [DOI] [PubMed] [Google Scholar]
- Hofrichter M, Ullrich R, Pecyna MJ et al. New and classic families of secreted fungal heme peroxidases. Appl Microbiol Biot 2010;87:871–97. [DOI] [PubMed] [Google Scholar]
- Hon DNS, Shiraishi N. Wood and Cellulosic Chemistry: 2nd edn New York: Marcel Dekker, 2001. [Google Scholar]
- Hori C, Gaskell J, Igarashi K et al. Genomewide analysis of polysaccharides degrading enzymes in 11 white- and brown-rot Polyporales provides insight into mechanisms of wood decay. Mycologia 2013;105:1412–27. [DOI] [PubMed] [Google Scholar]
- Hori C, Ishida T, Igarashi K et al. Analysis of the Phlebiopsis gigantea genome, transcriptome and secretome provides insight into its pioneer colonization strategies of wood. PLoS Genet 2014;10:e1004759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino F, Kajino T, Sugiyama H et al. Thermally stable and hydrogen peroxide tolerant manganese peroxidase (MnP) from Lenzites betulinus. FEBS Lett 2002;530:249–52. [DOI] [PubMed] [Google Scholar]
- Huang XF, Santhanam N, Badri DV et al. Isolation and characterization of lignin-degrading bacteria from rainforest soils. Biotechnol Bioeng 2013;110:1616–26. [DOI] [PubMed] [Google Scholar]
- Hullo MF, Moszer I, Danchin A et al. CotA of Bacillus subtilis is a copper-dependent laccase. J Bacteriol 2001;183:5426–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde SM, Wood PM. A mechanism for production of hydroxyl radicals by the brown-rot fungus Coniophora puteana: Fe(III) reduction by cellobiose dehydrogenase and Fe(II) oxidation at a distance from the hyphae. Microbiology 1997;143:259–66. [DOI] [PubMed] [Google Scholar]
- Ingold CT. Biology of Mucor and Its Allies. London, UK: Hodder & Stoughton Educational, 1978. [Google Scholar]
- Janusz G, Kucharzyk KH, Pawlik A et al. Fungal laccase, manganese peroxidase and lignin peroxidase: gene expression and regulation. Enzyme Microb Technol 2013;52:1–12. [DOI] [PubMed] [Google Scholar]
- Jensen KA, Houtman CJ, Ryan ZC et al. Pathways for extracellular fenton chemistry in the brown rot basidiomycete Gloeophyllum trabeum. Appl Environ Microb 2001;67:2705–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing D, Wang J. Controlling the simultaneous production of laccase and lignin peroxidase from Streptomyces cinnamomensis by medium formulation. Biotechnol Biofuels 2012;5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson T, Nyman PO. Isozymes of lignin peroxidase and manganese(II) peroxidase from the white-rot basidiomycete Trametes versicolor .1. Isolation of enzyme forms and characterization of physical and catalytic properties. Arch Biochem Biophys 1993;300:49–56. [DOI] [PubMed] [Google Scholar]
- Johjima T, Ohkuma M, Kudo T. Isolation and cDNA cloning of novel hydrogen peroxide-dependent phenol oxidase from the basidiomycete Termitomyces albuminosus. Appl Microbiol Biot 2003;61:220–5. [DOI] [PubMed] [Google Scholar]
- Kaffenberger JT, Schilling JS. Comparing lignocellulose physiochemistry after decomposition by brown rot fungi with distinct evolutionary origins. Environ Microbiol 2015;17:4885–97. [DOI] [PubMed] [Google Scholar]
- Kajisa T, Yoshida M, Igarashi K et al. Characterization and molecular cloning of cellobiose dehydrogenase from the brown-rot fungus Coniophora puteana. J Biosci Bioeng 2004;98:57–63. [DOI] [PubMed] [Google Scholar]
- Kersten P, Cullen D. Copper radical oxidases and related extracellular oxidoreductases of wood-decay Agaricomycetes. Fungal Genet Biol 2014;72:124–30. [DOI] [PubMed] [Google Scholar]
- Kersten PJ. Glyoxal oxidase of Phanerochaete chrysosporium: its characterization and activation by lignin peroxidase. P Natl Acad Sci USA 1990;87:2936–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten PJ, Cullen D. Cloning and characterization of cDNA encoding glyoxal oxidase, a H2O2-producing enzyme from the lignin-degrading basidiomycete Phanerochaete chrysosporium. P Natl Acad Sci USA 1993;90:7411–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Shoda M. Purification and characterization of a novel peroxidase from Geotrichum candidum dec 1 involved in decolorization of dyes. Appl Environ Microb 1999;65:1029–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk TK, Farrell RL. Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol 1987;41:465–505. [DOI] [PubMed] [Google Scholar]
- Kohler A, Kuo A, Nagy LG et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet 2015;47:410–5. [DOI] [PubMed] [Google Scholar]
- Kracher D, Zahma K, Schulz C et al. Inter-domain electron transfer in cellobiose dehydrogenase: modulation by pH and divalent cations. FEBS J 2015;282:3136–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger J-J, Thon MR, Deising HB et al. Compositions of fungal secretomes indicate a greater impact of phylogenetic history than lifestyle adaptation. BMC Genomics 2014;15:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhad RC, Singh A. Lignocellulose Biotechnology: Future Prospects. New Delhi: I.K.International (P) Ltd., 2007. [Google Scholar]
- Kuhad RC, Singh A, Eriksson KE. Microorganisms and enzymes involved in the degradation of plant fiber cell walls. Adv Biochem Eng Biot 1997;57:45–125. [DOI] [PubMed] [Google Scholar]
- Kuiters AT, Denneman CAJ. Water-soluble phenolic substances in soils under several coniferous and deciduous tree species. Soil Biol Biochem 1987;19:765–9. [Google Scholar]
- Kumar AK, Goswami P. Functional characterization of alcohol oxidases from Aspergillus terreus MTCC 6324. Appl Microbiol Biot 2006;72:906–11. [DOI] [PubMed] [Google Scholar]
- Kumar S, Shukla SK. A review on recent gasification methods for biomethane gas production. Int J Energy Eng 2016;6:32–43. [Google Scholar]
- Kunamneni A, Ballesteros A, Plou FJ et al. Fungal laccase—a versatile enzyme for biotechnological applications, Méndez-Vilas A, (ed.)Communicating Current Research and Educational Topics and Trends in Applied Microbiology. Badajoz, Spain: Formatex Research Center; 2007, 233–45. [Google Scholar]
- Kupriashina MA, Selivanov N, Nikitina VE. Isolation and purification of Mn-peroxidase from Azospirillum brasilense Sp245. Prikl Biokhim Mikrobiol 2012;48:23–6. [PubMed] [Google Scholar]
- Kuuskeri J, Hakkinen M, Laine P et al. Time-scale dynamics of proteome and transcriptome of the white-rot fungus Phlebia radiata: growth on spruce wood and decay effect on lignocellulose. BiotechnolBiofuels 2016;9:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeeuw L, Martone PT, Boucher Y et al. Ancient origin of the biosynthesis of lignin precursors. Biol Direct 2015;10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankinen VP, Bonnen AM, Anton LH et al. Characteristics and N-terminal amino acid sequence of manganese peroxidase from solid substrate cultures of Agaricus bisporus. Appl Microbiol Biot 2001;55:170–6. [DOI] [PubMed] [Google Scholar]
- Le Roes-Hill M, Rohland J, Burton S. Actinobacteria isolated from termite guts as a source of novel oxidative enzymes. Anton Leeuw 2011;100:589–605. [DOI] [PubMed] [Google Scholar]
- Leisola M, Pastinen O, Axe DD. Lignin-designed randomness. BIO-Complexity 2012;2012:1–11.22997604 [Google Scholar]
- Leonowicz A, Cho NS, Luterek J et al. Fungal laccase: properties and activity on lignin. J Basic Microbiol 2001;41:185–227. [DOI] [PubMed] [Google Scholar]
- Leontievsky A, Myasoedova N, Pozdnyakova N. et al. `Yellow' laccase of Panus tigrinus oxidizes non-phenolic substrates without electron-transfer mediators. FEBS Lett 1997;413:446–8. [DOI] [PubMed] [Google Scholar]
- Levasseur A, Piumi F, Coutinho PM et al. FOLy: an integrated database for the classification and functional annotation of fungal oxidoreductases potentially involved in the degradation of lignin and related aromatic compounds. Fungal Genet Biol 2008;45:638–45. [DOI] [PubMed] [Google Scholar]
- Levasseur A, Saloheimo M, Navarro D et al. Exploring laccase-like multicopper oxidase genes from the ascomycete Trichoderma reesei: a functional, phylogenetic and evolutionary study. BMC Biochem 2010;11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liers C, Arnstadt T, Ullrich R et al. Patterns of lignin degradation and oxidative enzyme secretion by different wood- and litter-colonizing basidiomycetes and ascomycetes grown on beech-wood. FEMS Microbiol Ecol 2011;78:91–102. [DOI] [PubMed] [Google Scholar]
- Liers C, Bobeth C, Pecyna M et al. DyP-like peroxidases of the jelly fungus Auricularia auricula-judae oxidize nonphenolic lignin model compounds and high-redox potential dyes. Appl Microbiol Biot 2010;85:1869–79. [DOI] [PubMed] [Google Scholar]
- Liers C, Pecyna MJ, Kellner H et al. Substrate oxidation by dye-decolorizing peroxidases (DyPs) from wood- and litter-degrading agaricomycetes compared to other fungal and plant heme-peroxidases. Appl Microbiol Biot 2013;97:5839–49. [DOI] [PubMed] [Google Scholar]
- Lindahl BD, Tunlid A. Ectomycorrhizal fungi - potential organic matter decomposers, yet not saprotrophs. New Phytol 2015;205:1443–7. [DOI] [PubMed] [Google Scholar]
- Linde D, Ruiz-Duenas FJ, Fernandez-Fueyo E et al. Basidiomycete DyPs: Genomic diversity, structural-functional aspects, reaction mechanism and environmental significance. Arch Biochem Biophys 2015;574:66–74. [DOI] [PubMed] [Google Scholar]
- Lisov AV, Leontievsky AA, Golovleva LA. Hybrid Mn-peroxidase from the ligninolytic fungus Panus tigrinus 8/18. Isolation, substrate specificity, and catalytic cycle. Biochemistry (Mosc) 2003;68:1027–35. [DOI] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E et al. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 2014;42:D490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücking R, Huhndorf S, Pfister DH et al. Fungi evolved right on track. Mycologia 2009;101:810–22. [DOI] [PubMed] [Google Scholar]
- Lundell TK, Makela MR, Hilden K. Lignin-modifying enzymes in filamentous basidiomycetes–ecological, functional and phylogenetic review. J Basic Microbiol 2010;50:5–20. [DOI] [PubMed] [Google Scholar]
- MacDonald J, Doering M, Canam T et al. Transcriptomic responses of the softwood-degrading white-rot fungus Phanerochaete carnosa during growth on coniferous and deciduous wood. Appl Environ Microb 2011;77:3211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod MP, Warren RL, Hsiao WW et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. P Natl Acad Sci USA 2006;103:15582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavi V, Lele SS. Laccase: properties and applications. BioResources 2009;4:1694–17. [Google Scholar]
- Manavalan T, Manavalan A, Heese K. Characterization of lignocellulolytic enzymes from white-rot fungi. Curr Microbiol 2015;70:485–98. [DOI] [PubMed] [Google Scholar]
- Manini P, La Pietra P, Panzella L et al. Glyoxal formation by Fenton-induced degradation of carbohydrates and related compounds. Carbohydr Res 2006;341:1828–33. [DOI] [PubMed] [Google Scholar]
- Martinez AT, Speranza M, Ruiz-Duenas FJ et al. Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int Microbiol 2005;8:195–204. [PubMed] [Google Scholar]
- Martinez D, Challacombe J, Morgenstern I et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. P Natl Acad Sci USA 2009;106:1954–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinkova L, Uhnakova B, Patek M et al. Biodegradation potential of the genus Rhodococcus. Environ Int 2009;35:162–77. [DOI] [PubMed] [Google Scholar]
- Martins LO, Soares CM, Pereira MM et al. Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J Biol Chem 2002;277:18849–59. [DOI] [PubMed] [Google Scholar]
- Masai E, Katayama Y, Fukuda M. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotech Bioch 2007;71:1–15. [DOI] [PubMed] [Google Scholar]
- Mason MG, Nicholls P, Wilson MT. Rotting by radicals–the role of cellobiose oxidoreductase? Biochem Soc Trans 2003;31:1335–6. [DOI] [PubMed] [Google Scholar]
- Mathieu Y, Piumi F, Valli R et al. Activities of secreted aryl alcohol quinone oxidoreductases from Pycnoporus cinnabarinus provide insights into fungal degradation of plant biomass. Appl Environ Microb 2016;82:2411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mester T, Field JA. Characterization of a novel manganese peroxidase-lignin peroxidase hybrid isozyme produced by Bjerkandera species strain BOS55 in the absence of manganese. J Biol Chem 1998;273:15412–7. [DOI] [PubMed] [Google Scholar]
- Molina-Guijarro JM, Perez J, Munoz-Dorado J et al. Detoxification of azo dyes by a novel pH-versatile, salt-resistant laccase from Streptomyces ipomoea. Int Microbiol 2009;12:13–21. [PubMed] [Google Scholar]
- Moore D, Robson GD, Trinci APJ. 21st Century Guidebook to Fungi. Cambridge: Cambridge University Press, 2011. [Google Scholar]
- Morgenstern I, Klopman S, Hibbett DS. Molecular evolution and diversity of lignin degrading heme peroxidases in the Agaricomycetes. J Mol Evol 2008;66:243–57. [DOI] [PubMed] [Google Scholar]
- Morin E, Kohler A, Baker AR et al. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. P Natl Acad Sci USA 2012;109:17501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova OV, Shumakovich GP, Gorbacheva MA et al. “Blue” laccases. Biochemistry (Mosc) 2007;72:1136–50. [DOI] [PubMed] [Google Scholar]
- Morpurgo L, Graziani MT, Finazzi-Agro A et al. Optical properties of japanese-lacquer-tree (Rhus vernicifera) laccase depleted of type 2 copper(II). Involvement of type-2 copper(II) in the 330nm chromophore. Biochem J 1980;187:361–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Harvey I, Hartley RD. Linkage of ρ-coumaroyl and feruloyl groups to cell-wall polysaccharides of barley straw. Carbohydr Res 1986;148:71–85. [Google Scholar]
- Nagy LG, Riley R, Tritt A et al. Comparative genomics of early-diverging mushroom-forming fungi provides insights into the origins of lignocellulose decay capabilities. Mol Biol Evol 2015;33:959–70. [DOI] [PubMed] [Google Scholar]
- Nelsen MP, DiMichele WA, Peters SE et al. Delayed fungal evolution did not cause the Paleozoic peak in coal production. P Natl Acad Sci USA 2016;113:2442–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcombe D, Paszczynski A, Gajewska W et al. Production of small molecular weight catalysts and the mechanism of trinitrotoluene degradation by several Gloeophyllum species. Enzyme Microb Tech 2002;30:506–17. [Google Scholar]
- Niemenmaa O, Uusi-Rauva A, Hatakka A. Demethoxylation of [O14CH3]-labelled lignin model compounds by the brown-rot fungi Gloeophyllum trabeum and Poria (Postia) placenta. Biodegradation 2008;19:555–65. [DOI] [PubMed] [Google Scholar]
- Niladevi KN. Ligninolytic enzymes. In: Singh-Nee Nigam P, Pandey A (eds.) Biotechnology for Agro-Industrial Residues Utilisation. Berlin, Germany: Springer; 2009. [Google Scholar]
- Niladevi KN, Jacob N, Prema P. Evidence for a halotolerant-alkaline laccase in Streptomyces psammoticus: purification and characterization. Proc Biochem 2008;43:654–60. [Google Scholar]
- Niladevi KN, Prema P. Mangrove Actinomycetes as the source of ligninolytic enzymes. Actinomycetologica 2005;19:40–7. [Google Scholar]
- Niladevi KN, Sheejadevi PS, Prema P. Strategies for enhancing laccase yield from Streptomyces psammoticus and its role in mediator-based decolorization of azo dyes. Appl Biochem Biotech 2008;151:9–19. [DOI] [PubMed] [Google Scholar]
- Nilsson T. Comparative study on the cellulolytic activity of white-rot and brown-rot fungi. Mater Organismen 1974;9:173–98. [Google Scholar]
- Nilsson T, Ginns J. Cellulolytic activity and the taxonomic position of selected brown-rot fungi. Mycologia 1979;71:170–7. [Google Scholar]
- Nousiainen P, Maijala P, Hatakka A et al. Syringyl-type simple plant phenolics as mediating oxidants in laccase catalyzed degradation of lignocellulosic materials: model compound studies. Holzforschung 2009;63:699–704. [Google Scholar]
- Ohm RA, de Jong JF, Lugones LG et al. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol 2010;28:957–63. [DOI] [PubMed] [Google Scholar]
- Olson A, Aerts A, Asiegbu F et al. Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol 2012;194:1001–13. [DOI] [PubMed] [Google Scholar]
- Ortiz-Bermudez P, Srebotnik E, Hammel KE. Chlorination and cleavage of lignin structures by fungal chloroperoxidases. Appl Environ Microb 2003;69:5015–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma C, Martinez AT, Lema JM et al. Different fungal manganese-oxidizing peroxidases: a comparison between Bjerkandera sp and Phanerochaete chrysosporium. J Biotechnol 2000;77:235–45. [DOI] [PubMed] [Google Scholar]
- Palonen H, Thomsen AB, Tenkanen M et al. Evaluation of wet oxidation pretreatment for enzymatic hydrolysis of softwood. Appl Biochem Biotech 2004;117:1–17. [DOI] [PubMed] [Google Scholar]
- Passardi F, Theiler G, Zamocky M et al. PeroxiBase: The peroxidase database. Phytochemistry 2007;68:1605–11. [DOI] [PubMed] [Google Scholar]
- Paszczynski A, Crawford R, Funk D et al. De novo synthesis of 4,5-dimethoxycatechol and 2,5-dimethoxyhydroquinone by the brown rot fungus Gloeophyllum trabeum. Appl Environ Microb 1999;65:674–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszczynski A, Huynh VB, Crawford R. Comparison of ligninase-I and peroxidase-M2 from the white-rot fungus Phanerochaete chrysosporium. Arch Biochem Biophys 1986;244:750–65. [DOI] [PubMed] [Google Scholar]
- Pawlik A, Wojcik M, Rulka K et al. Purification and characterization of laccase from Sinorhizobium meliloti and analysis of the lacc gene. Int J Biol Macromol 2016;92:138–47. [DOI] [PubMed] [Google Scholar]
- Perez J, Jeffries TW. Role of organic acid chelators in manganese regulation of lignin degradation by Phanerochaete chrysosporium. Appl Biochem Biotech 1993;39-40:227–38. [DOI] [PubMed] [Google Scholar]
- Perez-Boada M, Ruiz-Duenas FJ, Pogni R et al. Versatile peroxidase oxidation of high redox potential aromatic compounds: site-directed mutagenesis, spectroscopic and crystallographic investigation of three long-range electron transfer pathways. J Mol Biol 2005;354:85–402. [DOI] [PubMed] [Google Scholar]
- Pildain MB, Novas MV, Carmarán CC. Evaluation of anamorphic state, wood decay and production of lignin-modifying enzymes for diatrypaceous fungi from Argentina. J Agr Technol 2005;1:81–96. [Google Scholar]
- Piontek K, Strittmatter E, Ullrich R et al. Structural basis of substrate conversion in a new aromatic peroxygenase: cytochrome P450 functionality with benefits. J Biol Chem 2013;288:34767–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontek K, Ullrich R, Liers C et al. Crystallization of a 45 kDa peroxygenase/peroxidase from the mushroom Agrocybe aegerita and structure determination by SAD utilizing only the haem iron. Acta Crystallogr F 2010;66:693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitsawong W, Sucharitakul J, Prongjit M et al. A conserved active-site threonine is important for both sugar and flavin oxidations of pyranose 2-oxidase. J Biol Chem 2010;285:9697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piumi F, Levasseur A, Navarro D et al. A novel glucose dehydrogenase from the white-rot fungus Pycnoporus cinnabarinus: production in Aspergillus niger and physicochemical characterization of the recombinant enzyme. Appl Microbiol Biot 2014;98:10105–18. [DOI] [PubMed] [Google Scholar]
- Pollegioni L, Tonin F, Rosini E. Lignin-degrading enzymes. FEBS J 2015;282:1190–213. [DOI] [PubMed] [Google Scholar]
- Prongjit M, Sucharitakul J, Wongnate T et al. Kinetic mechanism of pyranose 2-oxidase from Trametes multicolor. Biochemistry 2009;48:4170–80. [DOI] [PubMed] [Google Scholar]
- Qiu Y-L, Palmer JD. Phylogeny of early land plants: insights from genes and genomes. Trends Plant Sci 1999;4:26–30. [DOI] [PubMed] [Google Scholar]
- Raffaello T, Chen H, Kohler A et al. Transcriptomic profiles of Heterobasidion annosum under abiotic stresses and during saprotrophic growth in bark, sapwood and heartwood. Environ Microbiol 2014;16:1654–67. [DOI] [PubMed] [Google Scholar]
- Rahmanpour R, Bugg TD. Characterisation of Dyp-type peroxidases from Pseudomonas fluorescens Pf-5: Oxidation of Mn(II) and polymeric lignin by Dyp1B. Arch Biochem Biophys 2015;574:93–8. [DOI] [PubMed] [Google Scholar]
- Rashid GM, Taylor CR, Liu Y et al. Identification of manganese superoxide dismutase from Sphingobacterium sp. T2 as a novel bacterial enzyme for lignin oxidation. ACS Chem Biol 2015;10:2286–94. [DOI] [PubMed] [Google Scholar]
- Redhead SA, Ginns JH. A reappraisal of Agaric genera associated with brown rots of wood. T Mycol Soc Jpn 1985;26:349–81. [Google Scholar]
- Riley R, Salamov AA, Brown DW et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. P Natl Acad Sci USA 2014;111:9923–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JN, Singh R, Grigg JC et al. Characterization of dye-decolorizing peroxidases from Rhodococcus jostii RHA1. Biochemistry 2011;50:5108–19. [DOI] [PubMed] [Google Scholar]
- Rodakiewicz-Nowak J, Jarosz-Wilkolazka A, Luterek J. Catalytic activity of versatile peroxidase from Bjerkandera fumosa in aqueous solutions of water-miscible organic solvents. Appl Catal A-Gen 2006;308:56–61. [Google Scholar]
- Rola B, Pawlik A, Frac M et al. The phenotypic and genomic diversity of Aspergillus strains producing glucose dehydrogenase. Acta Biochim Pol 2015;62:747–55. [DOI] [PubMed] [Google Scholar]
- Rowell RM. The Chemistry of Solid Wood. Advances in Chemistry Series. Washington, D.C.: American Chemical Society, 1984. [Google Scholar]
- Ruiz-Duenas FJ, Camarero S, Perez-Boada M et al. A new versatile peroxidase from Pleurotus. Biochem Soc Trans 2001;29:116–22. [DOI] [PubMed] [Google Scholar]
- Ruiz-Duenas FJ, Guillen F, Camarero S et al. Regulation of peroxidase transcript levels in liquid cultures of the ligninolytic fungus Pleurotus eryngii. Appl Environ Microb 1999;65:4458–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Duenas FJ, Lundell T, Floudas D et al. Lignin-degrading peroxidases in Polyporales: an evolutionary survey based on 10 sequenced genomes. Mycologia 2013;105:1428–44. [DOI] [PubMed] [Google Scholar]
- Rytioja J, Hilden K, Yuzon J et al. Plant-polysaccharide-degrading enzymes from Basidiomycetes. Microbiol Mol Biol R 2014;78:614–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini A, Aggarwal NK, Sharma A et al. Actinomycetes: a source of lignocellulolytic enzymes. Enzyme Res 2015;2015:279381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvachua D, Prieto A, Martinez AT et al. Characterization of a novel dye-decolorizing peroxidase (DyP)-type enzyme from Irpex lacteus and its application in enzymatic hydrolysis of wheat straw. Appl Environ Microb 2013;79:4316–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez O, Roció Sierra R, Alméciga-Díaz CJ. Delignification process of agro-industrial wastes an alternative to obtain fermentable carbohydrates for producing fuel. In: Manzanera M. (ed.) Alternative Fuel. London, UK: InTech, 2011. [Google Scholar]
- Santhanam N, Badri DV, Decker SR et al. Lignocellulose decomposition by microbial secretions. In: Vivanco MJ, Baluška F (eds.) Secretions and Exudates in Biological Systems. Berlin, Heidelberg: Springer Berlin Heidelberg, 2012, 125–53. [Google Scholar]
- Scalbert A, Monties B, Lallemand JY et al. Ether linkage between phenolic-acids and lignin fractions from wheat straw. Phytochemistry 1985;24:1359–62. [Google Scholar]
- Schirrmeister BE, de Vos JM, Antonelli A et al. Evolution of multicellularity coincided with increased diversification of cyanobacteria and the Great Oxidation Event. P Natl Acad Sci USA 2013;110:1791–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhalter DR, Canevascini G. Isolation and characterization of the cellobiose dehydrogenase from the brown-rot fungus Coniophora puteana (Schum ex Fr.) Karst. Arch Biochem Biophys 1993;300:559–63. [DOI] [PubMed] [Google Scholar]
- Shimada M, Akamtsu Y, Tokimatsu T et al. Possible biochemical roles of oxalic acid as a low molecular weight compound involved in brown-rot and white-rot wood decays. J Biotechnol 1997;53:103–13. [Google Scholar]
- Shimokawa T, Nakamura M, Hayashi N et al. Production of 2,5-dimethoxyhydroquinone by the brown-rot fungus Serpula lacrymans to drive extracellular Fenton reaction. Holzforschung 2004;58:305–10. [Google Scholar]
- Shraddha, Shekher R, Sehgal S et al. Laccase: microbial sources, production, purification, and potential biotechnological applications. Enzyme Res 2011;2011:217861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigoillot JC, Berrin JG, Bey M et al. Fungal strategies for lignin degradation. In: Jouanin L, Lapierre C (eds.) Lignins: Biosynthesis, Biodegradation and Bioengineering vol.61, London, UK: Academic Press, Elsevier; 2012, 263–308. [Google Scholar]
- Singh Arora D, Kumar Sharma R. Ligninolytic fungal laccases and their biotechnological applications. Appl Biochem Biotech 2010;160:1760–88. [DOI] [PubMed] [Google Scholar]
- Sonia MT, Naceur J, Abdennaceur H. Studies on the ecology of actinomycetes in an agricultural soil amended with organic residues: I. identification of the dominant groups of Actinomycetales. World J Microbiol Biot 2011;27:2239–49. [Google Scholar]
- Srinivasan MC, Laxman RS, Deshpande MV. Physiology and nutritional aspects of actinomycetes: an overview. World J Microbiol Biot 1991;7:171–84. [DOI] [PubMed] [Google Scholar]
- Stubblefield SP, Banks HP. Fungal remains in the Devonian Trimerophyte Psilophyton dawsonii. Am J Bot 1983;70:1258–61. [Google Scholar]
- Suderman RJ, Dittmer NT, Kanost MR et al. Model reactions for insect cuticle sclerotization: cross-linking of recombinant cuticular proteins upon their laccase-catalyzed oxidative conjugation with catechols. Insect Biochem Mol Biol 2006;36:353–65. [DOI] [PubMed] [Google Scholar]
- Sugano Y. DyP-type peroxidases comprise a novel heme peroxidase family. Cell Mol Life Sci 2009;66:1387–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano Y, Matsushima Y, Tsuchiya K et al. Degradation pathway of an anthraquinone dye catalyzed by a unique peroxidase DyP from Thanatephorus cucumeris Dec 1. Biodegradation 2009;20:433–40. [DOI] [PubMed] [Google Scholar]
- Suominen K, Kitunen V, Smolander A. Characteristics of dissolved organic matter and phenolic compounds in forest soils under silver birch (Betula pendula), Norway spruce (Picea abies) and Scots pine (Pinus sylvestris). Eur J Soil Sci 2003;54:287–93. [Google Scholar]
- Sutherland GR, Zapanta LS, Tien M et al. Role of calcium in maintaining the heme environment of manganese peroxidase. Biochemistry 1997;36:3654–62. [DOI] [PubMed] [Google Scholar]
- Suzuki H, MacDonald J, Syed K et al. Comparative genomics of the white-rot fungi, Phanerochaete carnosa and P. chrysosporium, to elucidate the genetic basis of the distinct wood types they colonize. BMC Genomics 2012;13:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MR, Hunt CG, Houtman CJ et al. Fungal hydroquinones contribute to brown rot of wood. Environ Microbiol 2006;8:2214–23. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Endo K, Ito M et al. A thermostable laccase from Streptomyces lavendulae REN-7: purification, characterization, nucleotide sequence, and expression. Biosci Biotech Bioch 2003;67:2167–75. [DOI] [PubMed] [Google Scholar]
- Sygmund C, Staudigl P, Klausberger M et al. Heterologous overexpression of Glomerella cingulata FAD-dependent glucose dehydrogenase in Escherichia coli and Pichia pastoris. Microb Cell Fact 2011;10:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamboli DP, Telke AA, Dawkar VV et al. Purification and characterization of bacterial aryl alcohol oxidase from Sphingobacterium sp. ATM and its uses in textile dye decolorization. Biotechnol Bioproc E 2011;16:661. [Google Scholar]
- Taylor TN, Krings M, Taylor EL. 2 - How fungal fossils are formed and studied. In: Taylor T, Krings M, Taylor E (eds.) Fossil Fungi. San Diego: Academic Press, 2015a, 13–26. [Google Scholar]
- Taylor TN, Krings M, Taylor EL. 3 - How old are the fungi? In: Taylor T, Krings M, Taylor E (eds.) Fossil Fungi. San Diego: Academic Press, 2015b, 27–39. [Google Scholar]
- Thurston CF. The structure and function of fungal laccases. Microbiology 1994;140:19–26. [Google Scholar]
- Tian JH, Pourcher AM, Bouchez T et al. Occurrence of lignin degradation genotypes and phenotypes among prokaryotes. Appl Microbiol Biotechnol 2014;98:9527–44. [DOI] [PubMed] [Google Scholar]
- Tien M, Kirk TK. Lignin degrading enzyme from the Hymenomycete Phanerochaete chrysosporium Burds. Science 1983;221:661–3. [DOI] [PubMed] [Google Scholar]
- Tiquia SM, Wan JHC, Tam NFY. Microbial population dynamics and enzyme activities during composting. Compost Sci Util 2002;10:150–61. [Google Scholar]
- Ullrich R, Nuske J, Scheibner K et al. Novel haloperoxidase from the agaric basidiomycete Agrocybe aegerita oxidizes aryl alcohols and aldehydes. Appl Environ Microb 2004;70:4575–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderrama B, Oliver P, Medrano-Soto A et al. Evolutionary and structural diversity of fungal laccases. Anton Leeuw 2003;84:289–99. [DOI] [PubMed] [Google Scholar]
- van Bloois E, Torres Pazmino DE, Winter RT et al. A robust and extracellular heme-containing peroxidase from Thermobifida fusca as prototype of a bacterial peroxidase superfamily. Appl Microbiol Biot 2010;86:1419–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela E, Tien M. Effect of pH and oxalate on hydroquinone-derived hydroxyl radical formation during brown rot wood degradation. Appl Environ Microb 2003;69:6025–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Gao PJ. Function and mechanism of a low-molecular-weight peptide produced by Gloeophyllum trabeum in biodegradation of cellulose. J Biotechnol 2003;101:119–30. [DOI] [PubMed] [Google Scholar]
- Wang X, Ullrich R, Hofrichter M et al. Heme-thiolate ferryl of aromatic peroxygenase is basic and reactive. P Natl Acad Sci USA 2015;112:3686–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wariishi H, Akileswaran L, Gold MH. Manganese peroxidase from the basidiomycete Phanerochaete chrysosporium: spectral characterization of the oxidized states and the catalytic cycle. Biochemistry 1988;27:5365–70. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Shirai N, Okada H et al. Production and chemiluminescent free radical reactions of glyoxal in lipid peroxidation of linoleic acid by the ligninolytic enzyme, manganese peroxidase. Eur J Biochem 2001;268:6114–22. [DOI] [PubMed] [Google Scholar]
- Welinder K. Plant peroxidases: structure-function relationships. In: Penel C, Gaspar T, Greppin H (eds.) Plant Peroxidases, Topics and Detailed Literature on Molecular, Biochemical and Physiological Aspects. Geneve: Université de Genève, 1992. [Google Scholar]
- Weng JK, Chapple C. The origin and evolution of lignin biosynthesis. New Phytol 2010;187:273–85. [DOI] [PubMed] [Google Scholar]
- Westereng B, Cannella D, Wittrup Agger J et al. Enzymatic cellulose oxidation is linked to lignin by long-range electron transfer. Sci Rep 2015;5:18561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker MM, Kersten PJ, Cullen D et al. Identification of catalytic residues in glyoxal oxidase by targeted mutagenesis. J Biol Chem 1999;274:36226–32. [DOI] [PubMed] [Google Scholar]
- Wolfe BE, Tulloss RE, Pringle A. The irreversible loss of a decomposition pathway marks the single origin of an ectomycorrhizal symbiosis. PLoS One 2012;7:e39597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DW. Structure and action mechanism of ligninolytic enzymes. Appl Biochem Biotech 2009;157:174–209. [DOI] [PubMed] [Google Scholar]
- Woo HL, Ballor NR, Hazen TC et al. Complete genome sequence of the lignin-degrading bacterium Klebsiella sp. strain BRL6-2. Stand Genomic Sci 2014;9:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall JJ, Anagnost SE, Zabel RA. Comparison of wood decay among diverse lignicolous fungi. Mycologia 1997;89:199–219. [Google Scholar]
- Yamada Y, Wang J, Kawagishi H et al. Improvement of ligninolytic properties by recombinant expression of glyoxal oxidase gene in hyper lignin-degrading fungus Phanerochaete sordida YK-624. Biosci Biotech Bioch 2014;78:2128–33. [DOI] [PubMed] [Google Scholar]
- Yang J, Xu X, Ng TB et al. Laccase gene family in Cerrena sp. HYB07: sequences, heterologous expression and transcriptional analysis. Molecules 2016;21:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin DT, Urresti S, Lafond M et al. Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family. Nat Commun 2015;6:10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn HD, Hah YC, Kang SO. Role of laccase in lignin degradation by white-rot fungi. FEMS Microbiol Lett 1995;132:183–8. [Google Scholar]
- Zamocky M, Furtmuller PG, Obinger C. Two distinct groups of fungal catalase/peroxidases. Biochem Soc Trans 2009;37:772–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamocky M, Gasselhuber B, Furtmuller PG et al. Turning points in the evolution of peroxidase-catalase superfamily: molecular phylogeny of hybrid heme peroxidases. Cell Mol Life Sci 2014;71:4681–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamocky M, Hallberg M, Ludwig R et al. Ancestral gene fusion in cellobiose dehydrogenases reflects a specific evolution of GMC oxidoreductases in fungi. Gene 2004;338:1–14. [DOI] [PubMed] [Google Scholar]
- Zamocky M, Hofbauer S, Schaffner I et al. Independent evolution of four heme peroxidase superfamilies. Arch Biochem Biophys 2015;574:108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamocky M, Ludwig R, Peterbauer C et al. Cellobiose dehydrogenase–a flavocytochrome from wood-degrading, phytopathogenic and saprotropic fungi. Curr Protein Pept Sci 2006;7:255–80. [DOI] [PubMed] [Google Scholar]
- Zhou L-W, Wei Y-L, Dai Y-C. Phylogenetic analysis of ligninolytic peroxidases: preliminary insights into the alternation of white-rot and brown-rot fungi in their lineage. Mycology 2014;5:29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman AR, Kang DH, Ahn MY et al. Influence of a soil enzyme on iron-cyanide complex speciation and mineral adsorption. Chemosphere 2008;70:1044–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at FEMSRE online.