Abstract
Two nutrient-controlled signalling pathways, the PKA and TOR pathway, play a major role in nutrient regulation of growth as well as growth-correlated properties in yeast. The relationship between the two pathways is not well understood. We have used Gap1 and Pho84 transceptor-mediated activation of trehalase and phosphorylation of fragmented Sch9 as a read-out for rapid nutrient activation of PKA or TORC1, respectively. We have identified conditions in which L-citrulline-induced activation of Sch9 phosphorylation is compromised, but not activation of trehalase: addition of the TORC1 inhibitor, rapamycin and low levels of L-citrulline. The same disconnection was observed for phosphate activation in phosphate-starved cells. The leu2 auxotrophic mutation reduces amino acid activation of trehalase, which is counteracted by deletion of GCN2. Both effects were also independent of TORC1. Our results show that rapid activation of the TOR pathway by amino acids is not involved in rapid activation of the PKA pathway and that effects of Gcn2 inactivation as well as leu2 auxotrophy all act independently of the TOR pathway. Hence, rapid nutrient signalling to PKA and TOR in cells arrested by nutrient starvation acts through parallel pathways.
Keywords: nutrient signalling, PKA pathway, TOR pathway, transceptor, Gcn2
Rapid nutrient signalling in yeast occurs through independent pathways.
ABBREVIATIONS
- PKA
Protein Kinase A
- PKB
Protein Kinase B
- TOR
Target of Rapamycin
- TORC1
Complex 1
INTRODUCTION
Nutrients are major regulators of metabolism and growth in all living cells. In the yeast Saccharomyces cerevisiae, many nutrient-induced signalling pathways have been identified (Broach 2012; Conrad et al.2014). The target of rapamycin (TOR) pathway is considered a major pathway for nitrogen regulation in yeast but there are more nitrogen-regulated pathways whose relationship with the TOR pathway is generally unclear (Cooper 2002; Magasanik and Kaiser 2002; Hinnebusch 2005; De Virgilio and Loewith 2006; Broach 2012; Ljungdahl and Daignan-ornier 2012). An important example is the protein kinase A (PKA) pathway, which is downregulated in nitrogen-starved cells concomitant with growth arrest and entry into stationary phase and rapidly reactivated upon re-addition of the missing nutrient. Inactivation of TOR complex 1 (TORC1) as well as inactivation of PKA causes arrest at the same nutrient starvation site in the G1 phase of the cell cycle, followed by storage carbohydrate accumulation, downregulation of ribosomal protein gene expression, induction of stress response element-controlled genes and increase in general stress resistance (Broach 2012; Conrad et al.2014). These observations suggest that the TOR and PKA pathways may interconnect through one or more regulatory mechanisms.
The TOR pathway was originally suggested to be a general nutrient-sensing pathway involved in sensing extracellular nutrients for regulation of cell growth (Loewith and Hall 2011). More recent research, however, suggests that the TOR pathway may function mainly in the sensing of intracellular nitrogen stores (Kim and Guan 2011; Loewith and Hall 2011; Zoncu et al.2011). This has been revealed by the discovery of TORC1 and its close relationship with the vacuolar/lysosomal compartment. TORC1 is located at the vacuolar membrane (Kunz et al.2000; Sturgill et al.2008; Binda et al.2009) and consists of Tor1 or Tor2, Kog1, Lst8 and Tco89 (Loewith et al.2002; Wedaman et al.2003; Reinke et al.2004). To exert its function, TORC1 interacts with the Ego complex consisting of the Ego1–3 proteins as well as Gtr1 and 2 (Dubouloz et al.2005; Sancak et al.2008; Efeyan, Zoncu and Sabatini 2012; Powis et al.2015), of which GTP-bound Gtr1 stimulates TORC1 in response to amino acids (Kim et al.2008; Sancak et al.2008). The regulation of TORC1 by the Ego complex is particularly sensitive to leucine levels (Binda et al.2009; Bonfils et al.2012; Kingsbury, Sen and Cardenas 2015).
The macrolide drug rapamycin inhibits TORC1 by binding to the highly conserved prolyl isomerase FKBP12, encoded by FPR1 in yeast (Schreiber 1991). The FKBP12–rapamycin complex prevents Tor1/2 from binding Kog1, leading to rapid degradation and inability to form a functional TORC1 complex (Hu et al.2016). The drug, and therefore the inhibition of the TORC1 complex, was long thought to mimic nitrogen starvation in S. cerevisiae. However, more recent research has shown that nitrogen starvation and inhibition of TORC1 by rapamycin differ in several aspects (Tate, Rai and Cooper 2015).
A well-known downstream target of the TORC1 pathway is the S6/protein kinase B (PKB) kinase, Sch9, which is responsible for inactivation of the major stress response regulators, Msn2/Msn4, induction of ribosomal protein genes, inhibition of autophagy and arrest in the G0 phase of the cell cycle (Pedruzzi et al.2003; Swinnen et al.2006). Upon activation of Tor, Sch9 is rapidly phosphorylated at five phosphorylation sites and has therefore been used as a convenient read-out for TORC1 activity (Urban et al.2007).
Another target of TORC1 considered to be involved in control of protein synthesis and cell growth is the protein kinase Gcn2 (Cherkasova and Hinnebusch 2003; Staschke et al.2010). It is activated by unloaded tRNA upon deprivation of amino acids, leading to phosphorylation of the α-subunit of eIF2, the main translation initiation factor. This leads to strong binding and inhibition by eIF2B, the guanine nucleotide exchange factor of eIF2. The resulting drop in GTP-bound eIF2 limits initiation of protein synthesis, causing a strong reduction in bulk protein synthesis under amino acid deprivation. The reduction in GTP-bound eIF2 also leads to activation of the general amino acid control system, resulting in enhanced expression of its main regulator Gcn4 (Mueller and Hinnebusch 1986; Hinnebusch 2005).
Activation of the PKA pathway by other essential nutrients in glucose-repressed cells is mediated by specific nutrient transceptors. These are high-affinity transporters that are strongly induced upon starvation for their substrate and function as transporter receptors for activation of PKA upon re-addition of the missing essential nutrient (Conrad et al.2014). The best established transceptors are Gap1 for amino acids, Mep2 for ammonium, Pho84 for phosphate and Sul1 and Sul2 for sulfate (Donaton et al.2003; Van Nuland et al.2006; Popova et al.2010; Kankipati et al.2015). The mechanism connecting the transceptors to PKA has not been identified yet, but does not involve cAMP signalling (Hirimburegama et al.1992; Conrad et al.2014). Trehalase, a well-known phosphorylation target of PKA (Schepers et al.2012), has been used as a very convenient and sensitive read-out for rapid activation of the PKA pathway (Donaton et al.2003; Popova et al.2010; Kankipati et al.2015).
Previous research on the relationship between the TORC1 and PKA pathways suggests a complex picture. Constitutive activation of the PKA pathway confers rapamycin resistance and overrides multiple responses caused by rapamycin inhibition of the TOR pathway, except for those dependent on the TORC1 effectors Tap42 and Sit4 (Schmelzle et al.2004). Transcriptional responses caused by complete inactivation of PKA can still be reinforced by rapamycin addition suggesting parallel functioning of both pathways (Zurita-Martinez and Cardenas 2005). Analysis of the rapamycin-sensitive phosphoproteome has revealed that TORC1 causes activation of PKA by inhibiting its regulatory subunit Bcy1 through an Sch9-dependent protein kinase cascade for part of the PKA targets studied (Soulard et al.2010).
We have investigated possible involvement of TORC1 in rapid nutrient transceptor-induced activation of the PKA pathway upon replenishment of the missing nutrient to starved cells, triggering resumption of cell growth. We show that rapamycin does not inhibit nutrient-induced activation of the PKA pathway under these conditions whereas it completely blocks phosphorylation of the TORC1 pathway target Sch9. Activation of the PKA target trehalase is more sensitive to low amino acid concentrations than phosphorylation of Sch9. We also show that deletion of GCN2 as well as leu2 auxotrophy affects nutrient-induced activation of PKA in opposite ways, independent of TORC1. We conclude that rapid nutrient-induced signalling to PKA and TORC1 in nutrient starvation arrested cells acts through parallel pathways.
EXPERIMENTAL PROCEDURES
Yeast strains, plasmids and culture conditions
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. To obtain the Sch9-HA tagged strain, a strain carrying Sch9 behind a galactose-inducible promoter was transformed with the pJU676 plasmid carrying the Sch9-HA construct (gift from Robbie Loewith). The transformants were grown on galactose and then switched to glucose to repress the endogenous SCH9 allele. Strain MC 63 was obtained by crossing BY4700 and BY4713 followed by tetrad dissection. The colonies obtained were tested for prototrophy. The GCN2 deletion strains were obtained by transformation and homologous recombination of a linear DNA fragment obtained by amplifying the KanMX4 cassette and flanking DNA sequences by PCR from the corresponding strain in the deletion strain collection (BY background).
Table 1.
S. cerevisiae strains used in this study.
| Strain | Genotype | Origin |
|---|---|---|
| LW201 | MATa his3 leu2 met15 ura3 trp::LEU2 Sch9::promGal-GST-SCH9 | Louwet (2010) |
| BY4741 | MATa ura3 his3 leu2 met15 | This work |
| MC63 | MATa (prototrophic BY4741) | This work |
| BY4712 | MATa leu2 | Brachmann et al. (1998) |
| BY4700 | MATa ura3 | Brachmann et al. (1998) |
| BY4706 | MATa met15 | Brachmann et al. (1998) |
| MC194 | MATa leu2 gcn2::KanMX4 | This work |
| MK37 | MATa ura3 his3 leu2 met15 gcn2::KanMX4 | Kimpe (2012) |
| MK257 | MATa ura3 his3 leu2 met15 gcn3::KanMX4 | Kimpe (2012) |
| JT b. 1398 | MATa ura3 his3 leu2 met15 gcn4::KanMX4 | Giaever et al. (2002) |
| MK167 | MATa ura3 his3 leu2 met15 sui2::SUI2S51A | Kimpe (2012) |
| MK227 | S288c ura3 gcn2::KanMX4 | Kimpe (2012) |
| RH2520 | S288c ura3 | Grundmann, Mosch and Braus (2001) |
| Plasmid | description | origin |
| pJU676 | pRS416, Sch9–5HA, URA3 | Urban et al. (2007) |
Strains were grown at 30°C with shaking in rich YP medium containing 2% glucose. For nutrient starvation, the cells were pregrown till an OD600 of 1.5, spun down and resuspended in appropriate nitrogen or phosphate starvation medium. They were then incubated further for 24 h in case of nitrogen starvation (0.17% yeast nitrogen base w/o amino acids, 4% glucose) or 72 h in case of phosphate starvation (0.57% yeast nitrogen base w/o phosphate, 4% glucose).
Sch9 chemical fragmentation and band shift assay
Sch9 chemical fragmentation was carried out as described by Urban et al. (2007). Cells were pregrown in 3 ml cultures overnight. These were used to inoculate 50 ml cultures of YPD and grown till an OD 1.0. The cultures were then transferred to a shaking water bath and left to incubate for 30 min. Rapamycin was added to a final concentration of 20 μg per ml. After 30-min incubation, one 10-ml sample was taken, and a source of nitrogen dissolved in fresh nitrogen starvation medium was added. Additional samples were taken after 5, 10 and 25 min of further incubation. All samples taken were immediately transferred into falcon tubes containing 1 ml 100% ice-cold TCA, shaken gently and kept on ice for at least 20 min. Cells were pelleted and washed twice with acetone before being dried in a speedvac for 5 min. After desiccation, 200 μl of glass beads and 100 μl of urea buffer were added. Samples were shaken vigorously in a fast prep three times for 20 s without intermittent cooling step. The samples were heated for 10 min to 65°C after which they were spun down at high speed to remove cell wall fragments. Finally, 30 μl of CHES buffer (pH 10.5) and 20 μl of NTCB were added and the samples left to incubate at room temperature overnight. The next day 100 μl of 2x sample buffer [62.5 mM Tris-HCl pH 6.8, 2.5% SDS, 0.002% bromophenol blue, 0.7135 M (5%) beta-mercaptoethanol, 10% glycerol] was added to all samples and 20 μl was loaded for western blotting of the Sch9 fragments. Blots were incubated with anti-HA peroxidase overnight and visualised.
Trehalase assay
Trehalase activity was determined as previously described (Pernambuco et al.1996) with minor adaptations. Nitrogen- or phosphate-deprived cells were cooled on ice for 30 min, harvested, washed with 25 mM MES buffer (pH 6.0) and resuspended in fresh starvation medium with 4% (w/v) glucose at a cell density of 25 mg cells wet weight per ml. Cells were pre-incubated at 30°C for 30 min before sample taking. At time point zero, the missing nutrient was added in the indicated final concentration.
Samples of 50 mg cells were taken at the indicated time points and added to 40 ml of ice-cold water. Cell pellets were resuspended in 500 μl ice-cold extraction buffer (50 mM MES/KOH, pH 7.0, 50 μM CaCl2). Crude cell extracts were prepared using a fastprep device, followed by centrifugation (5 min, 13 000 rpm, 4°C) and overnight dialysis at 4°C using 10 mM MES buffer pH 7.0 containing 50 μM CaCl2.
The amount of glucose liberated after 30-min incubation of the cell extract sample with trehalose buffer (250 mM trehalose, 25 mM MES/KOH, pH 7.0, 50 μM CaCl2) was determined using the glucose oxidase-peroxidase method by addition of GOD-PAP (Dialab). The absorbance at 505 nm was determined after 15 min incubation at 30°C. The residual glucose content in each sample after dialysis was also determined after boiling the samples prior to addition of trehalose buffer (blanks). Total amount of protein in each sample was determined via the standard Lowry method (Lowry et al.1951). The specific trehalase activity was expressed as nmol glucose min−1 (mg protein)−1.
Transport rate assay
Cells were resuspended in fresh starvation medium at a cell density of 80 mg/ml. Forty microliters of the cell suspension was pre-incubated at 30°C for 10 min. Ten microliters of a radioactive amino acid solution was added as indicated. After 1-min incubation, 5-ml ice-cold water was added and the cells were filtered through a glass microfiber filter (Whatman GF/C) pre-wet with a solution containing the same concentration of cold amino acid. Filters were immediately washed twice with 5-ml ice-cold water. For determination of the blanks, water was added prior to addition of the amino acid, immediately followed by filtration and washing. The radioactivity on the filter was determined in a liquid scintillation counter (Beckman Coulter LS6500). For each measurement, three samples and two blanks were taken in each experiment. Ten microliters of the labelled amino acid solution was used for determination of the protein content of crude cell extract using the Bradford method (Bradford 1976). Transport activity is expressed as nmol amino acid transported min−1 (mg protein)−1. The results of single time point determinations are based on three independent measurements and standard deviations are indicated by means of error bars.
Growth curves
Growth curves were determined using cells from a 3-ml overnight culture to inoculate wells with 200 μl of YPD medium in a 96-well plate to a final OD600 of 0.1. OD was measured every 30 min using a Synergy H1 hybrid reader (Biotek). The cultures were shaken continuously. The growth curves shown are the average of three independent cultures per strain.
Reproducibility of the results
The number of independent repetitions for each experiment is indicated in the figure legends. A representative result is shown.
RESULTS
TORC1 is activated upon re-addition of single amino acids to nitrogen-starved cells but not upon addition of phosphate to phosphate-starved cells
Chemical fragmentation of Sch9 has been widely used to measure TORC1 activation in Saccharomyces cerevisiae (Urban et al.2007; Hosiner et al.2009; Huang et al.2013; Kingsbury et al.2014). In this assay, the Sch9 protein is fragmented by use of 2-nitro-5-thiocyanobenzoic acid (NTCB). NTCB selectively cyanylates cysteine residues and under alkaline conditions this is followed by chain cleavage at these residues liberating a fragment which shows a ladder on an SDS gel at about 50 kD depending on its phosphorylation state (Wu, Gage and Watson 1996). This fragment contains the major TORC1 phosphorylation sites. Due to its smaller size compared to the complete protein (91.7 kD), it provides a higher resolution for separation of the phosphorylated versus non-phosphorylated forms on an SDS gel and therefore a better recognition of the extent of Sch9 phosphorylation (Urban et al.2007).
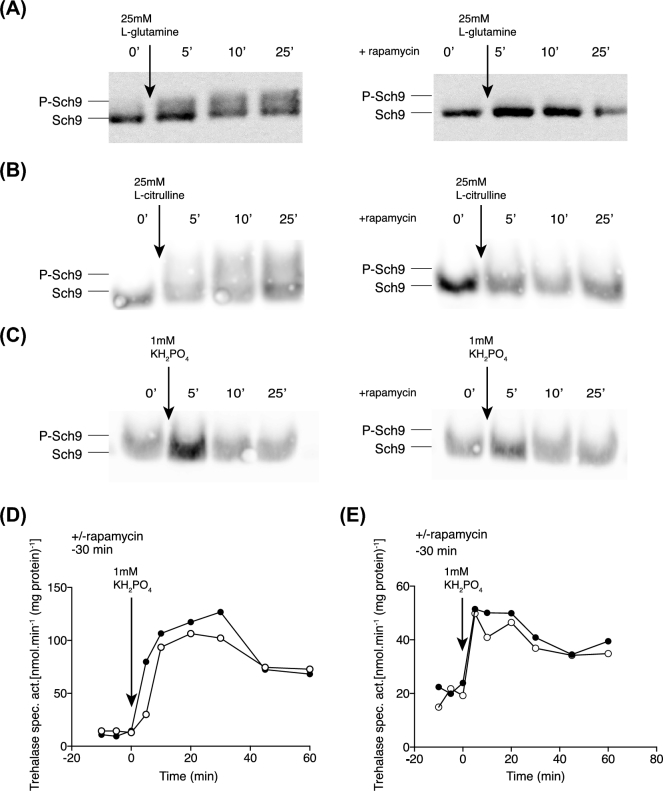
Whereas most research on activation of the TORC1 pathway has been conducted with cells grown on an unfavourable nitrogen source, like e.g. proline, research on nutrient transceptor activation of the PKA pathway has been mostly conducted with nitrogen-starved cells. The Gap1 amino acid transceptor is expressed under both conditions. Hence, we have first determined whether TORC1 shows regular activation upon addition of a nitrogen source to cells starved for nitrogen for 24 h using the Sch9 phosphorylation assay. As shown in Fig. 1A, addition of 25 mM L-glutamine to nitrogen starvation arrested cells causes rapid appearance of Sch9 phosphorylated forms within 5 min, indicating L-glutamine-induced activation of TORC1. Addition of rapamycin 30 min prior to the addition of L-glutamine completely abolished the appearance of the Sch9 phosphorylated forms, confirming that the observed band shifts are due to activation of TORC1.
Figure 1.
TOR is not involved in nutrient transceptor signalling to PKA. TORC1 and PKA activation using cells of strain LW201 expressing Sch9–5HA (pJU676). (A) Appearance of Sch9 phosphorylated forms after addition of 25 mM glutamine to nitrogen-starved cells, with or without 30 min prior incubation with rapamycin. Representative result of five repeats. (B) Appearance of Sch9 phosphorylated forms after addition of 25 mM L-citrulline to nitrogen-starved cells, with or without 30 min prior incubation with rapamycin. Representative result of four repeats. (C) Absence of the appearance of Sch9 phosphorylated forms after addition of 1 mM KH2PO4 to phosphate-starved cells, with or without 30 min prior incubation with rapamycin. Representative result of two repeats. (D) Trehalase activation after addition of 25 mM L-citrulline to nitrogen-starved cells without (closed circles) or with (open circles) 30 min pre-incubation with rapamycin. Representative result of three repeats. (E) Trehalase activation after addition of 1 mM KH2PO4 to phosphate-starved cells without (closed circles) or with (open circles) 30 min pre-incubation with rapamycin. Representative result of two repeats.
Whereas L-glutamine is known to be a potent activator of the TORC1 pathway, the effect of L-citrulline has not been documented. On the other hand, much research on Gap1 transceptor-mediated activation of the PKA pathway has been conducted with L-citrulline, because it is the only amino acid that is in low concentrations exclusively transported by Gap1 (Grenson, Hou and Crabeel 1970; Donaton et al.2003), allowing effects due to other transporters to be excluded. As shown in Fig. 1B, although somewhat less pronounced compared to L-glutamine, addition of 25 mM L-citrulline to nitrogen-starved cells also triggered within 5 min the appearance of Sch9 phosphorylated forms. Pre-addition of rapamycin for 30 min completely eliminated the observed effect confirming that L-citrulline also causes activation of TORC1 when added to nitrogen-starved cells.
Whereas activation of the PKA pathway by re-addition of an essential nutrient to cells starved for that nutrient has been observed with nitrogen sources, phosphate sources, sulfate sources, iron and zinc, most work on activation of the TORC1 pathway has been conducted only with nitrogen sources (Kim and Guan 2011; Loewith and Hall 2011). We next investigated whether addition of phosphate to phosphate-starved cells in the absence or presence of rapamycin would also trigger activation of the TORC1 pathway. As shown in Fig. 1C, there is no significant appearance of Sch9 phosphorylated forms after the addition of phosphate to phosphate-starved cells. Hence, there is also no difference in the absence or presence of rapamycin for 30 min prior to the addition of phosphate.
Rapamycin does not inhibit nutrient transceptor-mediated activation of the PKA pathway at concentrations that inhibit TORC1 signalling
Addition of rapamycin 30 min prior to the addition of 5 mM L-citrulline to nitrogen-starved cells (Fig. 1D) or the addition of 1 mM phosphate to phosphate-starved cells (Fig. 1E) did not prevent the typical activation of trehalase observed after re-addition of the essential nutrient to the starved cells and which we have used as a classical read-out for activation of PKA. This indicates that activation of TORC1 is not involved in Gap1 and Pho84 transceptor-mediated activation of the PKA pathway by amino acids and phosphate, respectively.
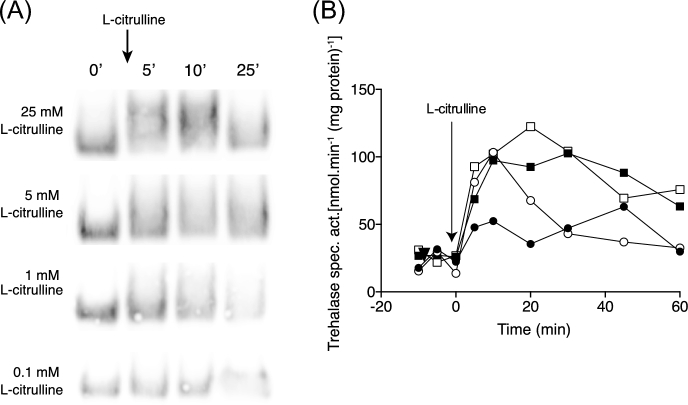
Nutrient transceptor activation of the PKA pathway is more sensitive to low levels of L-citrulline compared to activation of the TORC1 pathway
To gain further evidence for independence of nitrogen activation of the PKA and TORC1 pathways, we determined the activation of both upon addition of different concentrations of L-citrulline to nitrogen-starved cells. As shown in Fig. 2A, addition of 0.1 or 1 mM L-citrulline triggered no or just a very weak appearance of Sch9 phosphorylated forms, while with concentrations of 5 and 25 mM L-citrulline a strong response could be observed. On the other hand, as shown in Fig. 2B, 1, 5 and 25 mM L-citrulline caused a similar maximal activation of trehalase, with the 1 mM response declining more rapidly, presumably due to uptake and rapid depletion of this very low L-citrulline level. Even 0.1 mM L-citrulline triggered a small but reproducible increase in trehalase activity within the first 5 min. These results indicate that in nitrogen-starved cells the PKA pathway is more sensitive to re-addition of amino acids than the TORC1 pathway.
Figure 2.
Sch9 phosphorylation and trehalase activation with different concentrations of L-citrulline. TORC1 and PKA activation using cells of strain LW201 expressing Sch9–5HA (pJU676). (A) Appearance of Sch9 phosphorylated forms after addition of different concentrations of L-citrulline to nitrogen-starved cells. Samples were taken at the indicated time points (min). Representative result of three repeats. (B) Trehalase activation after addition of different concentrations of L-citrulline to nitrogen-starved cells: 0.1 mM (closed circles), 1 mM (open circles), 5 mM (closed squares) and 25 mM (open squares). Representative result of three repeats.
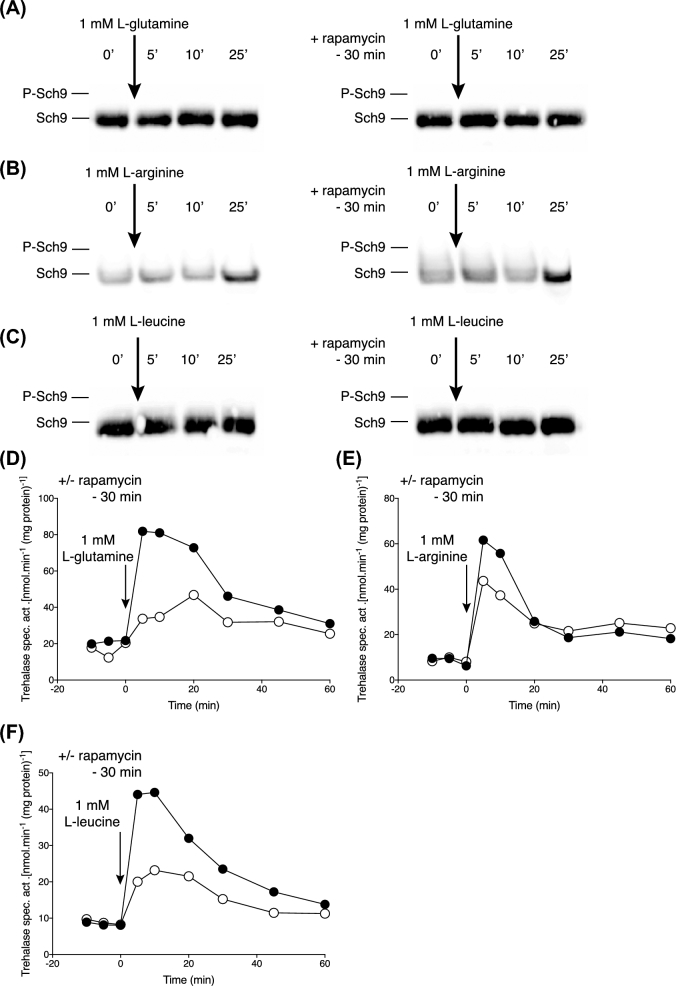
Rapamycin reduces PKA signalling with low concentrations of other amino acids but optimisation of rapamycin treatment abolishes this effect
We also tested whether 1 mM of other amino acids, i.e. L-glutamine, L-arginine and L-leucine, was able to cause activation of TORC1, as measured by Sch9 phosphorylation. We found that 1 mM L-glutamine, L-arginine or L-leucine (Fig. 3A–C) were unable to cause a clear increase in Sch9 phosphorylated forms. Hence, we could also not detect any significant difference between the condition with and without rapamycin (Fig. 3A–C).
Figure 3.
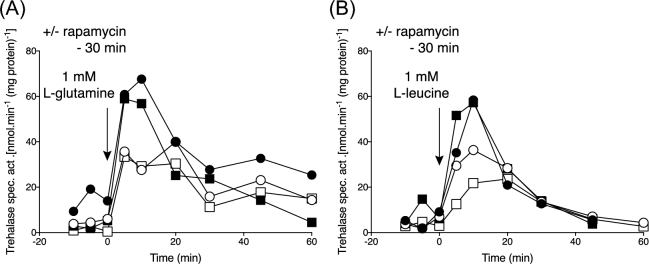
Sch9 phosphorylation and trehalase activation with low levels of amino acids not solely transported by Gap1. TORC1 and PKA activation using cells of strain LW201 expressing Sch9–5HA (pJU676). (A-C) Absence of the appearance of Sch9 phosphorylated forms after addition of 1 mM L-glutamine, L-arginine or L-leucine, with or without 8 min pre-incubation with rapamycin. Representative result of three repeats. (D-F) Trehalase activation after addition of 1 mM L-glutamine, L-arginine or L-leucine without (closed circles) or with (open cirlces) prior addition of rapamycin for 30 min. Representative result of three repeats.
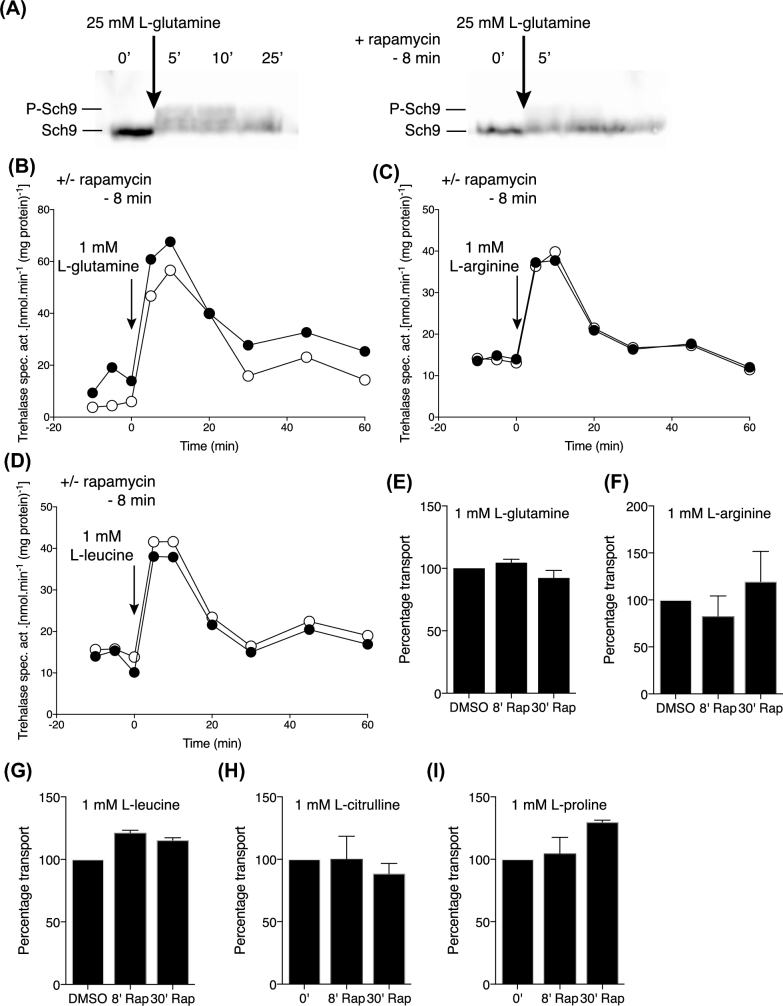
Unexpectedly, activation of trehalase upon addition of 1 mM L-glutamine, L-arginine or L-leucine to nitrogen-starved cells was partially reduced upon pre-addition of rapamycin for 30 min (Fig. 3D–F). This was puzzling since TORC1 does not appear to be activated under this condition (Fig. 3A–C). Although in general, a 30-min treatment with rapamycin is used for inhibition of TORC1 (Urban et al.2007; Stauffer and Powers 2015), a work by Stracka et al. (2014) suggests that a much shorter period of only 8 min is enough to prevent appearance of Sch9 phosphorylated forms and therefore activation of TORC1. The authors showed that the dephosphorylation of Sch9 already occurred 5 min after the addition of rapamycin and reached its full extend after only 8 min. This suggested that the partial inhibition of amino-acid induced trehalase activation may have been due to a side effect of TORC1 inhibition rather than indicating a direct requirement of TORC1 activation in nutrient transceptor-induced activation of PKA.
To evaluate this possibility, we first tested whether 8-min rapamycin treatment was indeed enough to prevent the appearance of Sch9 phosphorylated forms upon addition of a high concentration, 25 mM, of L-glutamine. The result indicated strong inhibition of TORC1 activation after the addition of rapamycin for 8 min (Fig. 4A). We next tested the effect of pre-incubation with rapamycin for 8 min on activation of trehalase with 1 mM L-glutamine, L-arginine and L-leucine, and we found that in this case trehalase activation was not affected at all (Fig. 4B–D). These results confirmed that amino acid activation of TORC1 does not play a role in the rapid amino acid transceptor-induced activation of PKA. Since Gap1 and other amino acid transporters are to some extend controlled by TORC1 (Schmidt et al.1998; Crapeau, Merhi and Andre 2014), we hypothesised that the inhibition of trehalase activation by the 30-min rapamycin treatment might be related to an effect on amino acid transport activity. However, neither 8 nor 30 min of rapamycin treatment caused any significant reduction in the uptake activity of 1 mM L-glutamine, 1 mM L-arginine or 1 mM L-leucine (Fig. 4E–G). Hence, the observed inhibition of amino acid activation of trehalase after pre-addition of rapamycin for 30 min must be due to another indirect effect. Rapamycin treatment also did not affect the uptake of amino acids that only serve as poor nitrogen sources, such as L-citrulline and L-proline (Fig. 4H–I).
Figure 4.
Differential effect of short rapamycin treatment on Sch9 phosphorylation, trehalase activation and amino acids not solely transported by Gap1. TORC1 and PKA activation using cells of strain LW201 cells expressing Sch9–5HA (pJU676). (A) Inhibition of the appearance of Sch9 phosphorylated forms after addition of 25 mM L-glutamine by treatment with rapamycin for 8 min. Representative result of three repeats. (B-D) Trehalase activation after addition of 1 mM L-glutamine, L-arginine or L-leucine without (closed circles) or with (open cirlces) 8 min pre-incubation with rapamycin. Representative result of two repeats. (E-I) Uptake of 1 mM L-glutamine, L-arginine, L-leucine, L-citrulline or L-proline after pre-incubation with rapamycin for 8 or 30 min. Representative result of two repeats.
Gcn2 is not involved in crosstalk between TORC1 and nutrient transceptor signalling to PKA
Gcn2, an eIF2 kinase, is considered a downstream target of TORC1, which is itself a known stimulator of protein synthesis and cell growth (Cherkasova and Hinnebusch 2003; Staschke et al.2010; Rodland et al.2014). Hence, inactivation of TORC1 likely downregulates the translation initiation factor eIF2. Since nutrient transceptor signalling to the PKA pathway is correlated with the start-up of cell growth, there may be crosstalk between eIF2 and the transceptor signalling mechanism, and therefore indirectly also with TORC1. Hence, we tested whether deletion of GCN2 affects amino acid signalling to PKA. As is shown in Fig. 5A for L-glutamine and Fig. 5B for L-leucine, deletion of GCN2 did not affect amino acid activation of trehalase, nor did it counteract the partial inhibition by a 30-min rapamycin pretreatment. This indicates that absence of Gcn2 does not compromise amino acid transceptor signalling to PKA and that inactivation of TORC1 does not reduce PKA signalling through Gcn2 and must therefore act via another indirect mechanism.
Figure 5.
GCN2 deletion does not abolish the reduction in trehalase activation caused by pre-incubation with rapamycin. Trehalase activation after addition of 1 mM L-glutamine (A) or 1 mM L-leucine (B) without (closed symbols) or with (open symbols) 30 min pre-incubation with rapamycin in a wild-type (S288C) (open and closed circles) or gcn2Δ (MK227) (open and closed squares) strain. Representative result of two repeats.
Gcn2 affects amino acid transceptor signalling to PKA in an auxotrophic strain
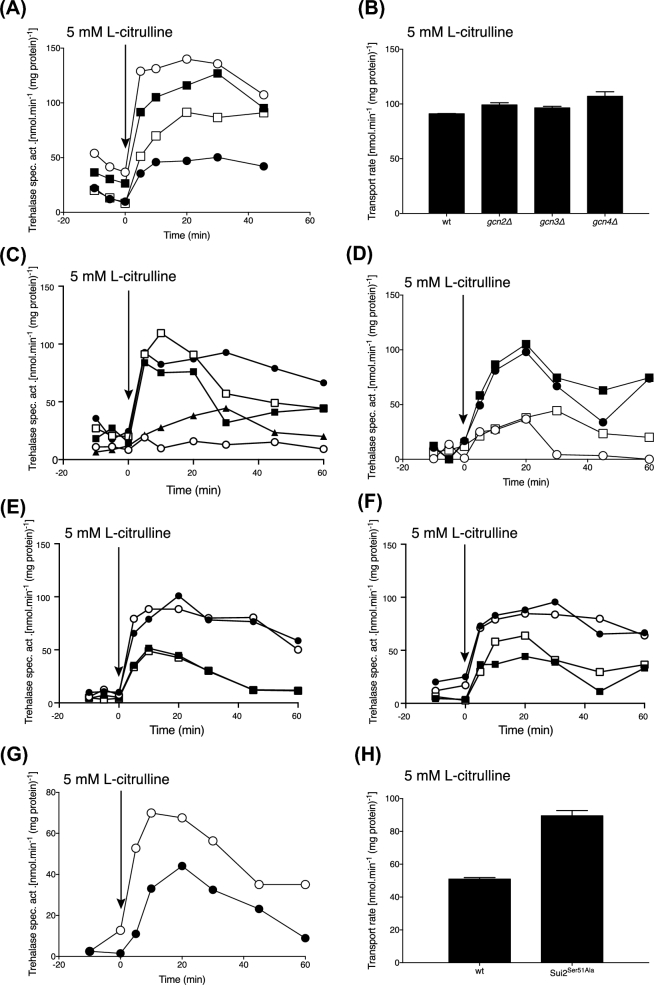
While deletion of GCN2 did not affect amino acid transceptor signalling to PKA in a prototrophic strain (Fig. 5A and B), we surprisingly noticed that in an auxotrophic strain deletion of GCN2 caused a strong increase in L-citrulline-induced activation of trehalase (Fig. 6A). However, in the auxotrophic parent strain, the extent of trehalase activation was significantly lower compared to that in the corresponding prototrophic strain. Hence, deletion of GCN2 counteracted the reduction caused by the auxotrophic mutations. A similar effect, although somewhat less pronounced, was observed upon deletion of GCN3, encoding the α-subunit of eIF2B which is phosphorylated by Gcn2, leading to inhibition of eIF2. Also deletion of GCN4, the main regulator of general amino acid control, which is induced upon inhibition of eIF2, counteracted the reduction to a lower extent. This counteracting effect was not due to enhanced expression of Gap1 in the plasma membrane since neither of the three GCN gene deletions caused any increase in L-citrulline uptake (Fig. 6B). Also the expression level of the genes encoding Gap1, the Tpk1 catalytic subunit of PKA and neutral trehalase (Nth1) was not enhanced in the GCN deletion strains, as measured by real-time PCR (not shown).
Figure 6.
Trehalase activation in gcnΔ and auxotrophic mutants. (A) Trehalase activation after addition of 5 mM L-citrulline to nitrogen-starved cells of BY4741 (closed circles), gcn2Δ (open circles), gcn3Δ (closed squares) and gcn4Δ (open squares). Representative result of six repeats. (B) Uptake of 5 mM L-citrulline by nitrogen-starved cells of the BY4742 wild-type, gcn2Δ, gcn3Δ and gcn4Δ strains. Representative result of four repeats. (C) Trehalase activation after addition of 5 mM L-citrulline to nitrogen-starved cells of the prototrophic strain BY4741 (closed circles) compared to the leu2Δ (BY4712) (open circles), ura3Δ (BY4700) (closed squares), met15Δ (BY4706) (open squares) and multiple auxotrophic BY4741 strain (closed triangles). Representative result of four repeats. (D) Trehalase activation after addition of 5 mM L-citrulline to nitrogen-starved cells of the prototrophic (open circles), leu2Δ (BY4712) (open circles), leu2Δ gcn2Δ (MC194) (closed squares) and the multiple auxotrophic BY4741 strain (open squares). Representative result of three repeats. (E) Trehalase activation after addition of 5 mM L-citrulline to nitrogen-starved cells of the BY4741 wild-type (squares) and the gcn2Δ strain (circles) with (open symbols) or without (closed symbols) prior incubation with rapamycin. Representative result of three repeats. (F) Trehalase activation after addition of 5 mM L-citrulline to nitrogen-starved cells of the BY4741 wild-type (squares) and gcn2Δ strain (circles) with (open symbols)) or without (closed symbols) prior incubation with 30 μg/mL of cycloheximide. Representative result of three repeats. (G) Trehalase activation after addition of 5 mM L-citrulline to nitrogen-starved cells of a strain containing the wild-type SUI2 allele (BY4741) (closed circles) or the mutated sui2-S51A allele (MK167) (open circles). Representative result of three repeats. (H) Uptake of 5 mM L-citrulline by nitrogen-starved cells of a strain containing the wild-type SUI2 allele (BY4741) or the mutated sui2-S51A allele (MK167). Representative result of three repeats.
Next, we first investigated what auxotrophic mutations of the BY4741 strain were responsible for the reduced amino acid transceptor activation of PKA. For that purpose, strains carrying single auxotrophic mutations, leu2Δ, ura3Δ and met15Δ were examined. As shown in Fig. 6C, the reduced trehalase activation was apparently caused mainly by the leucine auxotrophy. None of the auxotrophic mutations, including the leu2Δ mutation, caused a reduction in the start-up growth rate on complete medium, indicating that the reduced trehalase activation is not due to defective uptake of the auxotrophic supplements, especially leucine (not shown).
Since leucine is known to be a major activator of TORC1 and leucine deprivation is considered to result in TORC1 inhibition (Binda et al.2009; Bonfils et al.2012), we tested the effect of GCN2 deletion in a leu2 auxotrophic strain. As shown in Fig. 6D, deletion of GCN2 in a leu2Δ auxotrophic strain was clearly able to restore L-citrulline-induced activation of trehalase up to the level in a prototrophic yeast strain. This result suggests that the activation state of eIF2 in some way positively affects nutrient transceptor signalling to PKA. We reasoned that the inhibition caused by leu2Δ and its suppression by gcn2Δ might act through TORC1. However, both effects were insensitive to rapamycin and to the presence of the protein synthesis (elongation) inhibitor cycloheximide (Fig. 6E and F). Hence, both effects did not require TORC1 nor protein synthesis. Moreover, mutagenesis of the Gcn2 target phosphorylation site in the α-subunit of eIF2 (Sui2) was able to counteract the leu2Δ inhibitory effect although to a somewhat lower extent compared to gcn2Δ (Fig. 6G). However, in such a strain, the uptake of 5 mM L-citrulline was clearly increased (Fig. 6H) while in a gcn2Δ strain the transport was unaffected (Fig. 6B). This suggests that both effects may act in a different way, although up to now Sui2 is the only known target of Gcn2. Hence, these results suggest that Gcn2 may have other targets besides Sui2 and that the effect of gcn2Δ on restoration of transceptor signalling to PKA in a leu2Δ mutant may not act through its eIF2 target.
DISCUSSION
Whereas most research on the regulation of the PKA and TOR pathways has concentrated on targets in steady-state growing cultures, our work has focussed on the rapid responses of PKA pathway targets triggered during start-up of growth by re-addition of an essential nutrient to cells starved for that nutrient. Since the TORC1 pathway has been proposed to control nutrient regulation of cell growth and other cellular properties (Loewith and Hall 2011), we have investigated in this work a possible role of the TORC1 pathway in nutrient transceptor-induced activation of the PKA pathway. Previous work has revealed a similar core of genes transcriptionally regulated upon glucose, nitrogen and phosphate starvation, and a major influence of the PKA pathway as opposed to a minor influence of the TORC1 pathway on this core transcriptional response (Conway, Grunwald and Heideman 2012).
Phosphate addition to phosphate-starved cells failed to show any increase in Sch9 phosphorylated fragments (Fig. 1C), apparently indicating that under these conditions phosphate does not trigger activation of the TORC1 pathway. Therefore, we focussed on amino acid signalling, triggered by re-addition of an amino acid to nitrogen-starved cells. Under these conditions, major amino acids such as glutamine trigger a rapid increase in Sch9 phosphorylated fragments (Fig. 1A), consistent with many reports on rapid nitrogen activation of the TORC1 pathway (Loewith and Hall 2011). We have now shown that low concentrations of L-citrulline, which are nearly exclusively taken up by the Gap1 amino acid transceptor, also trigger a rapid increase in Sch9 phosphorylated fragments. These results indicate activation of TORC1 upon amino acid transport and/or sensing by Gap1, and raised the question whether Gap1-mediated activation of the TORC1 pathway plays a role in Gap1-mediated activation of the PKA pathway.
Whereas pre-addition of rapamycin for 30 min effectively blocked the L-glutamine and L-citrulline-induced increase in Sch9 phosphorylated fragments, it had no significant effect on L-citrulline-induced activation of trehalase. On the other hand, activation of trehalase with 1 mM L-glutamine, L-arginine or L-leucine was surprisingly partially reduced by the same rapamycin treatment. When we reduced the pretreatment with rapamycin to 8 min, this partial reduction was abolished, despite the fact that pre-addition of rapamycin for 8 min was enough to largely block the strong increase in Sch9 phosphorylated fragments observed after addition of 25 mM L-glutamine. From these results, we conclude that TORC1 signalling is not required for Gap1 transceptor induced activation of the PKA pathway and that the partial inhibition after 30 min rapamycin treatment of trehalase activation with some amino acids is likely due to an indirect effect on a common target in the signalling pathway. This is consistent with the results of Zurita-Martinez and Cardenas (2005), who concluded that the TOR and PKA signalling pathways mainly act in parallel but are also subjected to crosstalk. One possible mechanism is the inhibition of PKA by phosphorylation of its Bcy1 regulatory subunit on several sites through an Sch9-dependent protein kinase cascade upon rapamycin inhibition of TORC1 (Soulard et al.2010). The extent of this inhibition, in contrast to Sch9 phosphorylation, could be dependent on the duration of rapamycin treatment, since PKA is located more downstream in the TORC1 signalling cascade and more extensive phosphorylation may lead to stronger inhibition. This would explain why inhibition of amino acid activation of trehalase is only observed after a pretreatment of 30 min with rapamycin and not after a pretreatment of 8 min.
Although it has been reported that Gap1 is downregulated by endocytosis and degradation upon rapamycin treatment (Crapeau, Merhi and Andre 2014), under our conditions rapamycin addition to nitrogen starvation arrested cells did not affect short-term amino acid uptake significantly. Hence, the partial inhibition of amino acid-induced activation of trehalase by rapamycin does not seem to be caused by downregulation of Gap1.
We discovered that in auxotrophic strains inactivation of Gcn2 or the downstream regulators Gcn3 and Gcn4, as well as absence of Sui2 phosphorylation, counteracted the strong reduction in amino acid induced activation of trehalase observed in an auxotrophic strain compared to the corresponding prototrophic strain. Higher activation of trehalase in these strains would be consistent with a higher level of GTP-bound eIF2 acting as a stimulator of amino acid-induced activation of trehalase. Auxotrophic amino acid biosynthesis mutations can lead to lower intracellular levels of the respective amino acids and thus also to reduced tRNA amino acylation. As a result, Gcn2 may be more active and eIF2 more downregulated, which is consistent with the counteracting effect of GCN2 deletion on amino acid-induced activation of trehalase.
Further investigation revealed that the leu2Δ mutation was the main cause of the reduced activation of trehalase in the auxotrophic strains and thus apparently the main target counteracted by inactivation of Gcn2 or its downstream regulators. Although recent research has shown that besides the inability to synthesise leucine, leucine auxotrophic strains also show a pronounced decrease in leucine uptake (Cohen and Engelberg 2007), we did not observe a decrease in the start-up rate of growth, suggesting that the effect on trehalase activity is due to a signalling function of leucine rather than to its requirement as nutrient. Since leucine is known to be a potent activator of the TORC1 pathway (Binda et al.2009; Bonfils et al.2012), we considered that this might indicate crosstalk with the TORC1 pathway. However, addition of rapamycin, or blocking protein synthesis, had no effect on the restoration of amino acid-induced trehalase activation by inactivation of Gcn2. Moreover, deletion of GCN2 did not prevent proper downregulation and reactivation of trehalase, consistent with TORC1 not being involved in rapid nutrient transceptor-induced signalling to PKA.
We can conclude that for the rapid nutrient transceptor-induced activation of the PKA pathway in nutrient-starved cells, the TORC1 pathway does not appear to be directly involved as a signal transducer but that crosstalk of the TORC1 pathway with the PKA pathway may affect nutrient transceptor signalling at later time points.
Acknowledgments
We thank Willy Verheyden and Renata Wicik for technical help with the trehalase and Sch9 phosphorylation experiments and Nico Vangoethem for help with the preparation of the figures. We are grateful also to Dr Robbie Loewith (Geneva) for providing the pJU676 plasmid.
AUTHOR CONTRIBUTIONS
Conceived and designed the experiments: MC, MK, GVZ, JMT. Performed the experiments: MC, HNK, MK, ZZ. Analysed the data: MC, HNK, MK, GVZ, JMT. Wrote the paper: MC, GVZ, JMT. Led the research: JMT.
FUNDING
This work has been supported by PhD fellowships from the Research Fund of the KU Leuven Doctorandusbeurzen voor internationale samenwerking met landen buiten de EER (DBOF) to HNK, Fund for Scientific Research—Flanders to MK and CSC (China) to ZZ, and by grants from the Fund for Scientific Research—Flanders, Interuniversity Attraction Poles Network P7/40 and the Research Fund of the KU Leuven (Concerted Research Actions) to JMT.
Conflict of interest: None declared.
REFERENCES
- Binda M, Peli-Gulli MP, Bonfils G et al. . The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell 2009;35:563–73. [DOI] [PubMed] [Google Scholar]
- Bonfils G, Jaquenoud M, Bontron S et al. . Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell 2012;46:105–10. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ et al. . Designer deletion strains derived from Saccharomyces cerevisiae S288c: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998;14:115–32. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–54. [DOI] [PubMed] [Google Scholar]
- Broach JR. Nutritional control of growth and development in yeast. Genetics 2012;192:73–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Gene Dev 2003;17:859–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R, Engelberg D. Commonly used Saccharomyces cerevisiae strains (e.g. BY4741, W303) are growth sensitive on synthetic complete medium due to poor leucine uptake. FEMS Microbiol Lett 2007;273:239–43. [DOI] [PubMed] [Google Scholar]
- Conrad M, Schothorst J, Kankipati HN et al. . Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 2014;38:254–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway MK, Grunwald D, Heideman W. Glucose, nitrogen, and phosphate repletion in Saccharomyces cerevisiae: common transcriptional responses to different nutrient signals. G3 (Bethesda) 2012;2:1003–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol Rev 2002;26:223–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crapeau M, Merhi A, Andre B. Stress conditions promote yeast Gap1 permease ubiquitylation and down-regulation via the arrestin-like Bul and Aly proteins. J Biol Chem 2014;289:22103–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C, Loewith R. Cell growth control: little eukaryotes make big contributions. Oncogene 2006;25:6392–415. [DOI] [PubMed] [Google Scholar]
- Donaton MC, Holsbeeks I, Lagatie O et al. . The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Mol Microbiol 2003;50:911–29. [DOI] [PubMed] [Google Scholar]
- Dubouloz F, Deloche O, Wanke V et al. . The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell 2005;19:15–26. [DOI] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends Mol Med 2012;18:524–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L et al. . Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002;418:387–91. [DOI] [PubMed] [Google Scholar]
- Grenson M, Hou C, Crabeel M. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. IV. Evidence for a general amino acid permease. J Bacteriol 1970;103:770–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann O, Mosch HU, Braus GH. Repression of GCN4 mRNA translation by nitrogen starvation in Saccharomyces cerevisiae. J Biol Chem 2001;276:25661–71. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 2005;59:407–50. [DOI] [PubMed] [Google Scholar]
- Hirimburegama K, Durnez P, Keleman J et al. . Nutrient-induced activation of trehalase in nutrient-starved cells of the yeast Saccharomyces cerevisiae: cAMP is not involved as second messenger. J Gen Microbiol 1992;138:2035–43. [DOI] [PubMed] [Google Scholar]
- Hosiner D, Lempiainen H, Reiter W et al. . Arsenic toxicity to Saccharomyces cerevisiae is a consequence of inhibition of the TORC1 kinase combined with a chronic stress response. Mol Biol Cell 2009;20:1048–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Guo S, Yan G et al. . Ubiquitin regulates TORC1 in yeast Saccharomyces cerevisiae. Mol Microbiol 2016;100:303–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Liu J, Withers BR et al. . Reducing signs of aging and increasing lifespan by drug synergy. Aging Cell 2013;12:652–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankipati HN, Rubio-Texeira M, Castermans D et al. . Sul1 and Sul2 sulfate transceptors signal to protein kinase A upon exit of sulfur starvation. J Biol Chem 2015;290:10430–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L et al. . Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 2008;10:935–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guan KL. Amino acid signaling in TOR activation. Annu Rev Biochem 2011;80:1001–32. [DOI] [PubMed] [Google Scholar]
- Kimpe M. Gap1 amino acid signaling and translation initiation in Saccharomyces cerevisiae. PhD Thesis, KU Leuven, 2012. [Google Scholar]
- Kingsbury JM, Sen ND, Cardenas ME. Branched-chain aminotransferases control TORC1 signaling in Saccharomyces cerevisiae. PLoS Genet 2015;11:e1005714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury JM, Sen ND, Maeda T et al. . Endolysosomal membrane trafficking complexes drive nutrient-dependent TORC1 signaling to control cell growth in Saccharomyces cerevisiae. Genetics 2014;196:1077–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Schneider U, Howald I et al. . HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J Biol Chem 2000;275:37011–20. [DOI] [PubMed] [Google Scholar]
- Ljungdahl PO, Daignan-Fornier B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 2012;190:885–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011;189:1177–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S et al. . Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 2002;10:457–68. [DOI] [PubMed] [Google Scholar]
- Louwet W. Role of PKA and Sch9 in nutrient-induced signaling in Saccharomyces cerevisiae. PhD Thesis, KU Leuven; 2010. [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL et al. . Protein measurement with the Folin phenol reagent. J Biol Chem 1951;193:265–75. [PubMed] [Google Scholar]
- Magasanik B, Kaiser CA. Nitrogen regulation in Saccharomyces cerevisiae. Gene 2002;290:1–18. [DOI] [PubMed] [Google Scholar]
- Mueller PP, Hinnebusch AG. Multiple upstream AUG codons mediate translational control of GCN4. Cell 1986;45:201–7. [DOI] [PubMed] [Google Scholar]
- Pedruzzi I, Dubouloz F, Cameroni E et al. . TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell 2003;12:1607–13. [DOI] [PubMed] [Google Scholar]
- Pernambuco MB, Winderickx J, Crauwels M et al. . Glucose-triggered signalling in Saccharomyces cerevisiae: different requirements for sugar phosphorylation between cells grown on glucose and those grown on non-fermentable carbon sources. Microbiology 1996;142:1775–82. [DOI] [PubMed] [Google Scholar]
- Popova Y, Thayumanavan P, Lonati E et al. . Transport and signaling through the phosphate-binding site of the yeast Pho84 phosphate transceptor. P Natl Acad Sci USA 2010;107:2890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis K, Zhang T, Panchaud N et al. . Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling. Cell Res 2015;25:1043–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A, Anderson S, McCaffery JM et al. . TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem 2004;279:14752–62. [DOI] [PubMed] [Google Scholar]
- Rodland GE, Tvegard T, Boye E et al. . Crosstalk between the Tor and Gcn2 pathways in response to different stresses. Cell Cycle 2014;13:453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD et al. . The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008;320:1496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers W, Van Zeebroeck G, Pinkse M et al. . In vivo phosphorylation of Ser21 and Ser83 during nutrient-induced activation of the yeast protein kinase A (PKA) target trehalase. J Biol Chem 2012;287:44130–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T, Beck T, Martin DE et al. . Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol Cell Biol 2004;24:338–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Beck T, Koller A et al. . The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J 1998;17:6924–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SL. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 1991;251:283–7. [DOI] [PubMed] [Google Scholar]
- Soulard A, Cremonesi A, Moes S et al. . The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol Biol Cell 2010;21:3475–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staschke KA, Dey S, Zaborske JM et al. . Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J Biol Chem 2010;285:16893–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer B, Powers T. Target of rapamycin signaling mediates vacuolar fission caused by endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol Biol Cell 2015;26:4618–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracka D, Jozefczuk S, Rudroff F et al. . Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins. J Biol Chem 2014;289:25010–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill TW, Cohen A, Diefenbacher M et al. . TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell 2008;7:1819–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen E, Wanke V, Roosen J et al. . Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div 2006;1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Rai R, Cooper TG. Nitrogen starvation and TorC1 inhibition differentially affect nuclear localization of the Gln3 and Gat1 transcription factors through the rare glutamine tRNACUG in Saccharomyces cerevisiae. Genetics 2015;199:455–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A et al. . Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 2007;26:663–74. [DOI] [PubMed] [Google Scholar]
- Van Nuland A, Vandormael P, Donaton M et al. . Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol Microbiol 2006;59:1485–505. [DOI] [PubMed] [Google Scholar]
- Wedaman KP, Reinke A, Anderson S et al. . Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol Biol Cell 2003;14:1204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Gage DA, Watson JT. A strategy to locate cysteine residues in proteins by specific chemical cleavage followed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Biochem 1996;235:161–74. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A et al. . mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011;334:678–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Martinez SA, Cardenas ME. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot Cell 2005;4:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]