Abstract
Pseudomonas aeruginosa is a major pathogen in the lungs of cystic fibrosis (CF) patients. However, it is now recognised that a diverse microbial community exists in the airways comprising aerobic and anaerobic bacteria as well as fungi and viruses. This rich soup of microorganisms provides ample opportunity for interspecies interactions, particularly when considering secreted compounds. Here, we discuss how P. aeruginosa-secreted products can have community-wide effects, with the potential to ultimately shape microbial community dynamics within the lung. We focus on three well-studied traits associated with worsening clinical outcome in CF: phenazines, siderophores and biofilm formation, and discuss how secretions can shape interactions between P. aeruginosa and other commonly encountered members of the lung microbiome: Staphylococcus aureus, the Burkholderia cepacia complex, Candida albicans and Aspergillus fumigatus. These interactions may shape the evolutionary trajectory of P. aeruginosa while providing new opportunities for therapeutic exploitation of the CF lung microbiome.
Keywords: interspecific interactions: multispecies interactions, microbiome, cystic fibrosis, Pseudomonas aeruginosa, microbial communities
How social interactions among microbes living in the cystic fibrosis lung may be influencing patient health.
INTRODUCTION
Individuals with cystic fibrosis (CF) suffer from a buildup of thick, viscous mucous in the airways, predisposing them to lifelong bacterial lung infections which are often fatal. Pseudomonas aeruginosa is the most common pathogen in CF, displaying high levels of antibiotic resistance and virulence—so that elimination is apparently impossible (Pressler et al.2011). Chronic infection with P. aeruginosa is associated with deterioration of pulmonary function, reduction in quality of life and premature death (Koch and Høiby 1993; Emerson et al.2002; Hart and Winstanley 2002).
The CF lung airways consist of polymicrobial infections that vary in their composition and diversity throughout a patient's lifetime. Diversity typically increases during the first decade of life, and decreases thereafter (Cox et al.2010; Klepac-Ceraj et al.2010). While Haemophilus influenzae and Staphylococcus aureus are present mainly in young children, by the age of 20, 60–70% of CF patients present intermittent colonisation by P. aeruginosa (Folkesson et al.2012). Earlier acquisition of P. aeruginosa has been associated with a more rapid decline in lung function and poorer clinical outcomes (Emerson et al.2002). In at least 50% of adult CF patients, P. aeruginosa has been reported as the dominant organism, displacing the resident microbial community (Valenza et al.2008). Furthermore, CF patients infected with P. aeruginosa are vulnerable to developing secondary infections, for example with the Burkholderia cepacia complex, predisposing patients to necrotising pneumonia, which is usually fatal (Sajjan et al.2001; Bragonzi et al.2012). Fungi and yeasts also inhabit the airways, where Aspergillus fumigatus and Candida albicans are the most prevalent fungi and yeast, respectively (Chotirmall and McElvaney 2014). Although their prevalence is likely underestimated and detection methods vary between diagnostic laboratories, both Aspergillus spp. and Candida spp. have been identified in up to 50% of CF patients (Pihet et al.2009; Chotirmall et al.2010).
The recent surge in the number of studies employing in-depth, parallel, next-generation sequencing of CF lung microbial communities has given a greater insight into what exactly lives in this complex ecosystem. Inhabiting microorganisms range from recognised pathogens such as Pseudomonas spp. and Burkholderia spp. to bacteria less understood in the context of CF such as Prevotella spp. and Veillonella spp. (Fodor et al.2012; Boutin et al.2015), and classically commensal microorganisms such as oral streptococci. A novel isolation method led to the detection of Candida dubliniensis in patients >30 years old with advanced stages of the disease, although the importance of this fungal pathogen in CF is not yet understood (Sahand et al. 2006; Chortimall et al.2010). Lower respiratory tract microbiome studies have also supported the identification of new proposed pathogens in the CF lung such as Ralstonia mannitolilytica, identified in seven patients in Canada and associated with accelerated disease progression and raised mortality (Coman et al.2017). In addition to identifying novel bacterial species, metagenomic studies have revealed a diverse viral community in the CF lung with over 450 viral genotypes identified (Lim et al.2014). Furthermore, some of these viruses have been linked to the onset of pulmonary exacerbations (periods of acute worsening of pulmonary symptoms) (Billard et al.2017).
Lung microbial diversity tends to decrease with increasing disease severity (as P. aeruginosa dominates the population) (Cox et al.2010; Fodor et al.2012; Frayman et al.2017). However, whether this association is linked to increased P. aeruginosa pathogenicity remains elusive. Lung community diversity can be highly patient specific and no universal indicator of the onset of exacerbation has been identified so far (Whelan et al.2017). Furthermore, during antibiotic treatment, limited changes in microbial community structure have been identified (Fodor et al.2012; Li et al.2016).
Through our progressive understanding of the complexities of polymicrobial communities, it is becoming increasingly clear that interactions between bacterial pathogens and the microbial community within which they reside can influence pathogenesis, antimicrobial resistance and disease progression (Hoffman et al.2006; Peters et al.2012; Antonic et al.2013; Baldan et al.2014; Fugère et al.2014; Beaume et al.2015). However, it is often difficult to elucidate whether these clinical changes are a cause or consequence of these interactions. In this review, we highlight the role of multispecies interactions in shaping P. aeruginosa virulence, and discuss examples where these interactions may be of paramount importance in predicting patient health. Secreted products by P. aeruginosa are likely to influence neighbouring microorganisms, and it is reasonable to suggest that community context may in turn shape the relative costs and benefits associated with these secretions. Crucially, this implies that the role of some CF microorganisms in disease may be subtle, acting through cross-species interactions rather than being recognised pathogens per se.
HOW MIGHT MULTISPECIES INTERACTIONS SHAPE P. aeruginosa VIRULENCE?
Over the course of chronic infections, P. aeruginosa CF isolates commonly display adaptive phenotypes such as conversion to mucoidy and loss of motility, as well as reduced expression of acute virulence factors and extracellular toxins (Smith et al.2006; Bragonzi et al.2009; Folkesson et al.2012; Lorè et al.2012; Davies et al.2016; Winstanley, O’Brien and Brockhurst 2016). Despite the general trend toward loss of virulence as P. aeruginosa becomes chronic, it is becoming increasingly clear that loss of virulence is not universal within a patient. Furthermore, P. aeruginosa isolates within patients are typically highly diverse with respect to the aforementioned phenotypic characteristics (Fothergill et al.2010; Mowat et al.2011; O’Brien et al.2017). Despite the potential for P. aeruginosa adaptive evolution to influence patient health, both the causes and consequences of these adaptive changes are not well understood. The ability of many microbial secretions to influence the fitness of other organisms either directly (e.g. bacteriocin-mediated killing) or indirectly (e.g. antibiotic degradation), with potential for positive (cooperation) or negative (competition) fitness consequences, suggests that microbial interactions may play an integral role in shaping P. aeruginosa evolution within the CF lung.
Here, we focus on four clinically relevant P. aeruginosa traits that may, in part, shape and be shaped by interactions with the natural microbial community. Crucially, these traits have potential to be ‘social’—that is, they may directly or indirectly influence the fitness of nearby cells (West et al.2007). This list is not exhaustive, but should be regarded as examples of microbial traits whose role cannot be fully understood without consideration of community context.
Phenazine production
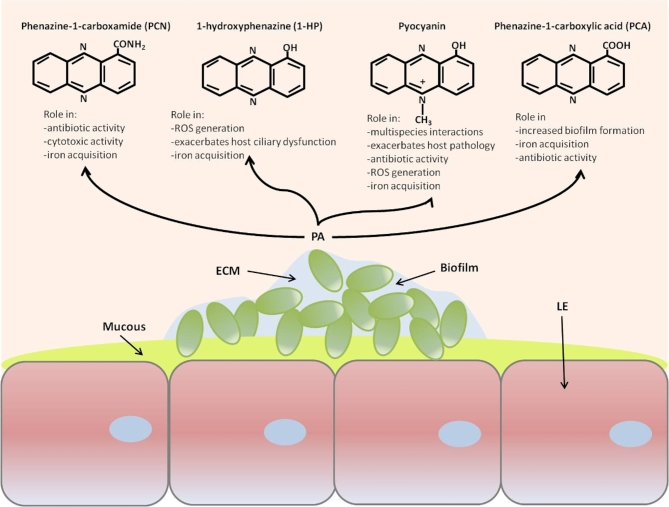
Phenazines are secondary metabolites produced by a variety of bacteria, notable for their broad-spectrum antibiotic properties and roles in virulence (Sorensen and Klinger 1987). Phenazine production is mediated by quorum sensing (QS), a method of bacterial cell–cell communication that allows the coordinated expression of genes in bacterial populations (Dietrich et al.2006). P. aeruginosa secretes four main classes of phenazines: pyocyanin, phenazine-1-carboxamide (PCN), 1-hydroxyphenazine (1-HP) and phenazine-1-carboxylic acid (PCA) (Fig. 1). One class of phenazine, pyocyanin, is a blue redox-active pigment that exerts a host inflammatory response, impairs ciliary function and induces oxidative stress within the lung (O’Malley et al.2003; Winstanley and Fothergill 2009). While the effects of pyocyanin on the host may influence other microorganisms indirectly, there is some evidence that pyocyanin can also have a direct role in shaping microbial communities. Pyocyanin can function as an iron-reducing agent, allowing iron-limited microorganisms to thrive (see below) (Cox 1986). Furthermore, the bactericidal effect of pyocyanin may reduce community diversity (Norman et al.2004) and select for a community of resistant species. Two recent studies (Korgaonkar and Whiteley 2011; Korgaonka et al.2013) reported that P. aeruginosa responds directly to cell wall fragments from Gram-positive bacteria by increasing production of multiple extracellular factors, including pyocyanin. Co-infection of P. aeruginosa with avirulent Gram-positive bacteria in both rat lung and Drosophila models resulted in increased lung damage and overall enhanced virulence, respectively (Duan et al.2003; Korgaonka et al.2013), although the exact mechanisms are unknown. Clinical isolates respond similarly: Whiley et al. (2014) reported enhanced P. aeruginosa pyocyanin production when co-cultured with oral viridans streptococci (Streptococcus oralis, Streptococcus mitis, Streptococcus gordonii and Streptococcus sanguinis), and these co-cultures exhibited increased pathogenicity in an insect host model compared with P. aeruginosa alone. However, in this case increased pathogenicity might also have arisen from other virulence-associated secretions, rather than pyocyanin per se.
Figure 1.
Schematic representation of phenazine production by Pseudomonas aeruginosa growing in a biofilm in the cystic fibrosis lung. PA = P. aeruginosa, ECM = extracellular matrix, LE = lung epithelium.
Studies in which animal models are infected with P. aeruginosa strains producing varying levels of pyocyanin reveal that pyocyanin production tends to lead to more virulent infections (Mahajan-Miklos et al.1999; Cao, Baldini and Rahme 2001; Lau et al.2004a,b; Courtney et al.2007; O’Brien et al.2017). In CF, periods of patient exacerbations have been linked with increased pyocyanin production in the lung (Fothergill et al.2007, 2010; Mowat et al.2011). However, not all patients with worsening symptoms harbour increased numbers of overproducing phenotypes (Nguyen and Singh 2006; Smith et al.2006), and the causality of this relationship remains unconvincing. Furthermore, why pyocyanin overproducers evolve and thrive in some scenarios and not others remains to be elucidated. Interestingly, while virulence is predictably lost over the course of CF infections, longitudinal studies of pyocyanin production have so far failed to detect any predictable evolutionary changes over the course of chronic infections (Jiricny et al.2014; Winstanley, O’Brien and Brockhurst 2016). We speculate that multispecies interactions can at least partly explain the observed fluctuations in pyocyanin production. If this is the case, assays for pyocyanin production by clinical isolates in media or even artificial sputum models that mimic abiotic conditions in the CF lung (e.g. Fothergill et al.2010; Mowat et al.2011; Jiricny et al.2014; O’Brien et al.2017) may not be sufficient indicators of what these strains are producing in vivo. Ultimately, by understanding whether community context matters for P. aeruginosa pyocyanin production, it may be possible to manipulate the lung microbiome to reduce the severity of clinical symptoms during CF-associated exacerbations.
Biofilm formation
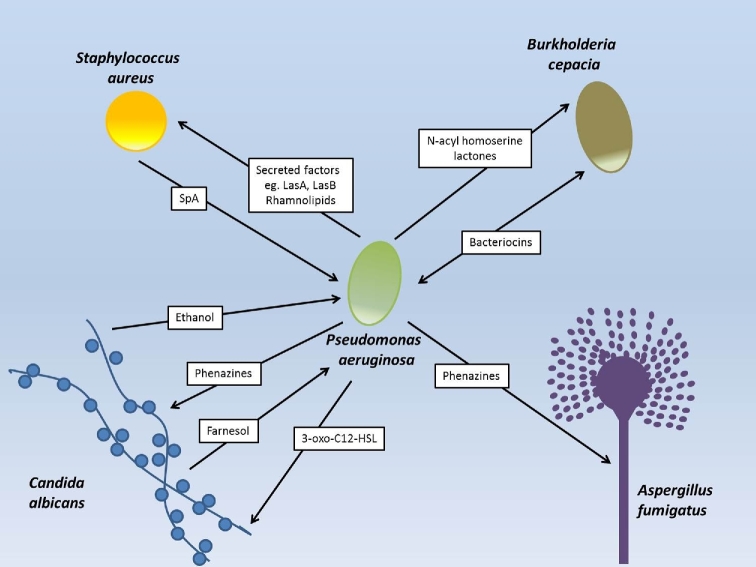
The intractability of P. aeruginosa in CF has been largely attributed to the presence of mucoid alginate-producing strains in the later stages of infection (Ramsey and Wozniak 2005; Sousa and Periera 2014; Winstanley, O’Brien and Brockhurst 2016). These strains form resilient biofilms, conferring enhanced resistance to antibiotics, phage and the host immune system, ultimately causing a decline in lung function (Høiby et al.2010; Høiby, Ciofu and Bjarnsholt 2010). While this transition to mucoidy is commonly viewed as a global response to environmental stress (e.g. Davies et al.2016), there is some evidence that multispecies social interactions may play a role. For instance, ethanol produced by C. albicans stimulates biofilm formation in P. aeruginosa (DeVault, Kimbara and Chakrabarty 1990), while a protein secreted by S. aureus, SpA, inhibits it (Armbruster et al.2016) (Fig. 2). Exopolysachharides can also impact on spatial organisation in polymicrobial biofilms (Chew et al.2014). One P. aeruginosa exopolysaccharide, Pel, is required for a close association in biofilms with S. aureus. However, another exopolysaccharide, Psl, allows P. aeruginosa to form a single species biofilm on top of S. aureus. Therefore, the type of exopolysaccharide produced by P. aeruginosa can impact the architecture of the biofilm and the ability of these two species to interact closely (Chew et al.2014).
Figure 2.
Summary of discussed interactions between Pseudomonas aeruginosa and other microbial inhabitants of the cystic fibrosis lung. Arrows depict the direction of the interaction. Note that we have omitted interactions driven by iron acquisition in this figure because the ability of siderophores to shape interactions is likely to be driven mainly by indirect effects of iron limitation.
Viruses of bacteria (phages) have also been described in the CF lung (Lim et al.2013), and are a promising novel way of eliminating drug-resistant pathogens (Waters et al.2017). Interactions between P. aeruginosa and lytic phages (which lyse the bacterial cell upon infection) may drive the transition to mucoidy by enhancing resistance to phage infection (Miller and Rubero 1984; Scanlan and Buckling 2012). Conversely, evolving P. aeruginosa with temperate phages (which can either complete the lytic cycle or integrate into the bacterial chromosome as a prophage), can reduce biofilm formation by accelerating the loss of biofilm-dependent type IV pili (Davies et al.2016). While understanding how the abiotic and biotic environment interact to promote mucoidy is no easy task, it is an endeavour worth pursuing. Mucoid variants of P. aeruginosa are highly problematic in the clinic, and novel therapeutics aimed at disrupting mucoidy are highly sought after (Romling and Balsalobre 2012; Gnanadhas et al.2015a,b).
Iron-acquisition
Iron is an essential nutrient for many microorganisms, yet in the early stages of CF lung infection the availability of iron for inhabiting microbiota is highly restricted (Tyrrell and Callaghan 2016). P. aeruginosa can overcome this by producing iron-chelating siderophores that can acquire otherwise sequestered ferric iron. Due to their capacity to enhance bacterial growth, siderophores are viewed as virulence factors (Buckling et al.2007). A wide body of research suggests that iron uptake strategy in Pseudomonads can be influenced by social context, because non-producers can exploit producers, and gain a fitness advantage (e.g. Griffin, West and Buckling 2004; Harrison and Buckling 2005, 2007; O’Brien, Rodrigues and Buckling 2013; Andersen et al.2015). However, most of these studies are limited to intraspecific interactions in spatially homogeneous environments (but see Luján et al.2015, and Harrison et al. 2017).
In the CF lung, many species compete for iron simultaneously, and this competition can indirectly shape iron-uptake strategies in P. aeruginosa. For instance, competition between P. aeruginosa and B. cepacia induces P. aeruginosa genes normally expressed under iron-limited conditions (including siderophores). This is because a B. cepacia siderophore, ornibactin (which P. aeruginosa cannot use), restricts iron availability to P. aeruginosa (Weaver and Kolter 2004). A similar phenomenon was observed using experimental evolution, whereby P. aeruginosa was evolved in the presence and absence of S. aureus (Harrison et al.2008). In this case, P. aeruginosa upregulated siderophore production in response to S. aureus, which acted as an iron competitor. Conversely, P. aeruginosa can obtain iron by lysing S. aureus cells (Mashburn et al.2005), although Harrison et al. (2008) suggest that this benefit depends on the degree of competition between the two strains. Interestingly, when multiple species compete for iron, subsequent iron limitation may also reduce the ability of P. aeruginosa to form biofilms (Singh et al.2002; O’May et al.2009). This is in line with what we observe in longitudinal studies of CF isolates, whereby iron becomes more available, and biofilms become more common over the course of infection (Hunter et al.2013; Tyrrell and Callaghan 2016; Winstanley, O’Brien and Brockhurst 2016). However, this correlation can be of course open to different interpretations.
There is some evidence to suggest that the requirement for siderophores is reduced in the later stages of infection, as freely available ferrous (Fe2+) tends to dominate over ferric iron (Hunter et al.2013). Furthermore, as host cells are damaged they release iron in the form of haem and haemoglobin, from which P. aeruginosa can sequester iron using the haem assimilation system (Has) and Phu (Pseudomonas haem uptake) systems. Indeed, over the course of chronic infections there is some evidence that siderophores are lost and replaced with haem utilisation (Marvig et al.2014). Finally, the role of pyocyanin in iron acquisition per se is poorly understood, although one study suggests that a different phenazine, PCA, assists in biofilm development by promoting ferrous iron (Wang et al.2011). Crucially, the role of various iron-uptake systems in shaping microbial communities may differ depending on the predominant form of acquisition. While siderophore sharing is generally species specific (Buckling et al.2007, but see Barber and Elde 2015), other acquisition mechanisms such as pyocyanin-mediated reduction is unlikely to be limited to conspecifics, and so understanding how they might be shaped by community interactions is not straightforward.
CASE STUDIES
While the scope for interactions within the CF lung is clearly vast, we highlight interactions between P. aeruginosa and four commonly encountered species: the Gram-positive bacteria S. aureus, the B. cepacia complex (Gram-negative), a filamentous fungi (A. fumigatus) and C. albicans (a yeast) to display the breadth and diversity of interactions with P. aeruginosa.
P. aeruginosa and S. aureus
P. aeruginosa and S. aureus display a striking negative correlation with one another as CF patients age (Cystic Fibrosis Foundation 2011), suggesting that P. aeruginosa can displace S. aureus in the later stages of infection. P. aeruginosa secretes a wealth of S. aureus-killing exoproducts, such as pyocyanin, elastase, protease, rhamnolipids, 4-hydroxy-2-alkylquinoline (HAQ) and 4-hydroxy-2-heptylquinoline-N-oxide (HQNO) (Mashburn et al.2005; Palmer et al.2005; Hoffman et al.2006; Mitchell et al.2010; Korgaonkar and Whiteley 2011; Cardozo et al.2013; Korgaonkar et al.2013; DeLeon et al.2014). P. aeruginosa can also harm S. aureus indirectly by manipulating the innate immunity of the host, such as inducing the production of S. aureus-killing phospholipase sPLA2-IIA by bronchial epithelial cells (Pernet et al.2014). This interaction between the host and P. aeruginosa enhances the clearance of S. aureus without significantly affecting the growth of P. aeruginosa. It is of course debatable whether the upregulation of sPLA2-IIA by P. aeruginosa has evolved as a competitor-killing mechanism, or if it is simply a response by the host to which P. aeruginosa is resistant. Nonetheless, sPLA2-IIA is the most potent known antibacterial enzyme in mammals, especially targeting Gram-positive bacteria, suggesting that interactions between P. aeruginosa and the host can shape bacterial communities more widely (Qu and Lehrer 1998; Nevalainen, Graham and Scott 2008). Finally, one recent study that experimentally evolved P. aeruginosa in the presence and absence of S. aureus, demonstrated that adaptation to S. aureus was mediated by inactivation of virulence-associated lipopolysaccharide (LPS) in P. aeruginosa. Crucially, this adaptation also conferred enhanced resistance to beta-lactam antibiotics, despite the fact that evolution took place in their absence (Tognon et al.2017).
Crucially, any counter adaptation by S. aureus to resist killing by P. aeruginosa can in turn shape the pathogenicity of S. aureus. Small colony variants of S. aureus (SCVs) arise by mutations in metabolic genes (Melter and Radojevic 2010) and experience reduced killing by P. aeruginosa HQNO's compared to their wild-type counterparts (Hoffman et al.2006; Biswas et al.2009; Filkins et al.2015). From a clinical perspective, SCVs display enhanced resistance to antibiotics (Wolter, Kotsiou and McCormack 1995), greater persistence (Hoffman et al.2006) and correlate with worsening symptoms in CF (Wolter et al.2013). Moreover, HQNO has been identified in CF patients harbouring P. aeruginosa, but not in uninfected individuals, suggesting that HQNO-mediated interactions between these two species have potential to directly influence disease progression (Hoffman et al.2006).
P. aeruginosa and A. fumigatus
Aspergillus fumigatus is the most common fungus found in the CF airways (Nagano et al.2007; Pihet et al.2009), and its presence is associated with a diversity of clinical phenotypes ranging from no obvious respiratory decline, to Aspergillus bronchitis and bronchiectasis (Shoseyov et al.2006; Agarwal et al.2013; Chotirmall and McElavaney 2014). Infection tends to occur subsequent to P. aeruginosa colonisation, resulting in co-infections that trigger more severe clinical outcomes compared with P. aeruginosa or A. fumigatus alone (Amin et al.2010; Ferreira et al.2015; Reece et al.2017).
Several lines of evidence suggest that these two species interact extensively in the CF lung. P. aeruginosa has been classically viewed as inhibiting A. fumigatus growth by producing an array of phenazines which kill fungi at high concentrations (Kerr 1994; Moree et al.2012; Briard et al.2015). However, in the CF lung, the phenazines pyocyanin and phenazine-1-carboxylate have been found in vivo at concentrations in the range of 1–100 μM, which Briard et al. (2015) demonstrated to be subinhibitory against A. fumigatus. Furthermore, at these concentrations, these phenazines actually functioned as iron-reducing agents, liberating bioavailable iron and subsequently, fungal growth in iron-starved environments. Another phenazine, 1-HP, stimulated siderophore production in A. fumigatus and growth as a consequence Briard et al. (2015). Accordingly, there is a generally high percentage of Fe2+ once phenazine levels rise above 50 μM in sputum (Hunter et al. 2012, 2013). However, concentrations of phenazines may in reality vary within the lung, particularly in the lower respiratory tract where mucous is more concentrated. These findings may explain why A. fumigatus infections tend to occur after P. aeruginosa colonisation—P. aeruginosa creates an iron-rich environment in which A. fumigatus can thrive. However, an alternative explanation is that co-infection reduces the pro-inflammatory response exerted by the host, potentially enabling both strains to benefit (Reece et al.2017). Furthermore, damaged lungs per se may permit better colonisation by pathogens and increased virulence as a consequence.
P. aeruginosa and C. albicans
Despite being the second most common fungus in CF, the role of C. albicans in CF is not fully understood. In practice, this means that a positive result for C. albicans in the clinic tells us little about patient prognosis. While invasive airway infections by C. albicans alone remain rare, its pathogenic effects may be experienced through interactions with other species. For instance, in mixed biofilms with C. albicans, P. aeruginosa upregulates its production of virulence-associated secretions such as pyoverdine, phenazines and rhamnolipids, relative to single-species biofilms (Hogan and Kolter 2002; Hogan, Vik and Kolter 2004; Cugini et al.2007; Gibson, Sood and Hogan 2009). Enhanced phenazine production by P. aeruginosa in turn upregulates C. albicans ethanol production, as the phenazines exert a switch from respiration to fermentation (Morales et al.2013). As mentioned previously, ethanol increases P. aeruginosa biofilm formation, resulting in mucoid phenotypes with enhanced growth rate (DeVault, Kimbara and Chakrabarty 1990; Morales et al.2013). This phenazine-mediated switch to fermentation in C. albicans may have consequences for microbiome diversity and composition. Ethanol has also been shown to enhance growth, virulence and biofilm formation in other lung pathogens such as S. aureus (Korem, Gov and Rosenberg 2010) and Acinetobacter baumanii (Nwugo et al.2012), although the exact mechanisms have not yet been elucidated. Ethanol is also an immunosuppressant, negatively influencing the host immune response (Greenberg et al.1999; Goral, Karavitis and Kovacs 2008). In a rat model system, ethanol inhibits lung clearance of P. aeruginosa by inhibiting macrophage recruitment (Greenberg et al.1999). Hence, ethanol may indirectly shape microbial communities by interfering with pathogen clearance. Another fermentation product, acetic acid, is also likely to reduce extracellular pH, which may favour the presence of low-pH-adapted microorganisms (Morales et al.2013).
Signalling can occur between these two species, influencing one another's gene expression and virulence (Fig. 2). The P. aeruginosa signal molecule 3-oxo-C12HSL promotes the yeast form of C. albicans, so that when levels of this signal drop (such as during chronic infection; Bjarnsholt et al.2010), the invasive, filamentous form of the fungus may be triggered (McAlester, O’Gara and Morrissey 2008). Conversely, C. albicans secrete the alcohol farnesol that suppresses the P. aeruginosa signal molecule PQS and consequently, pyocyanin production, while inducing quinolone and phenazine expression (Cugini et al.2007; Cugini, Morales and Hogan 2010). Finally, C. albicans can reduce the expression of the siderophores pyoverdine and pyochelin in P. aeruginosa leading to decreased virulence (Lopez-Medina et al.2015), although exact mechanisms have yet to be elucidated. Clearly, these interactions, and their effects on gene expression, are complex, and we are far from understanding how they will combine to influence host health.
P. aeruginosa and B. cepacia complex
Secondary bacterial infections with the B. cepacia complex are associated with cepacia syndrome—a rapidly progressing and fatal pneumonia (Huang et al.2001; Lambiase et al.2006). Members of the B. cepacia complex form mucoid biofilms with P. aeruginosa, engaging in an intimate network of interactions, and possibly even exchanging genetic material (Eberl and Tümmler 2004).
Competition between these two species is rife. In one study that screened 66 P. aeruginosa and B. cenocepacia CF clinical isolates, 81% of P. aeruginosa and 57% of B. cenocepacia strains produced bacteriocin-like toxins, conferring inhibitory activity toward the other species (Bakkal et al.2010). Populations of Burkholderia have been found to invade populations of P. aeruginosa (Schwab et al.2014) and vice versa (Bragonzi et al.2012; Costello et al.2014), suggesting that the outcome of competition is highly context dependent. Interactions between these two species may also occur in more subtle ways: one class of signal molecules produced by P. aeruginosa, N-acyl homoserine lactones, can stimulate the production of siderophores, lipase and protease production in Burkholderia (McKenney et al.1995; Riedel et al.2001; Lewenza, Visser and Sokol 2002; Costello et al.2014). Moreover, alginate production by P. aeruginosa can aid B. cenocepacia survival by inhibiting the host immune response (Chattoraj et al.2010). Despite the role of both species as harmful pathogens, how their interactions may influence virulence is not well understood.
There has been a recent drive toward developing novel therapeutics using products secreted by naturally occurring competitors to target specific pathogens in not just CF (e.g. Brown et al.2009) but halitosis (Burton et al.2006) and Clostridium difficile (Rea et al.2013) infections. In particular, one Burkholderia bacteriocin named Tailocin has been proposed as a potential therapeutic with activity against P. aeruginosa (Yao et al.2017). In order to fully appreciate how P. aeruginosa populations will respond to these classes of drugs it is vital that we understand how the species involved interact naturally on both ecological and evolutionary timescales.
OUTLOOK
There is tantalising evidence that interactions within and among species can alter virulence properties of P. aeruginosa in the short term and potentially shape the evolutionary trajectory of this pathogen in the long term. While our knowledge of how P. aeruginosa responds to other species individually is growing, the consequences of these interactions for virulence in a complex multispecies community remains unclear. Moreover, experimental evolution studies in complex environments containing the natural microbiota are required to decipher whether ecological responses drive selection for evolutionary change. On the one hand, multispecies infections may constrain the rate of evolutionary change if trade-offs are required to adapt to multiple species. On the other hand, increasing the number of interacting species may result in even more rapid evolution, as selection acts on increasing numbers of traits.
Understanding how virulence-associated secretions are shaped by the lung microbiome opens doors for novel therapeutic approaches already being exploited in gut microbiome research (Bakken et al.2011; Hamilton et al.2012; Lee et al.2016). For instance, an increased understanding of community dynamics could allow us to establish ‘infection-resistant’ communities to prevent initial colonisation of recognised pathogens or by replacing pathogens with commensal communities. This approach has already proved successful in treating Clostridium difficile gut infections with ‘healthy’ gut communities (Bakken et al.2011; Hamilton et al.2012; Lee et al.2016), and Streptococcus mutans dental caries with lactobacillus communities (Gungor, Kirzioglu and Kivanc 2015). Furthermore, through an increased understanding of the ecology of these lung communities, it may be possible to suppress P. aeruginosa indirectly by manipulating clinically relevant interactions.
However, there are many challenges. As a field, we are not clear on what a ‘normal’ or ‘healthy’ community might look like against a genetic background of CF. Furthermore, spatial structure in the lung, at both molecular and geographical scales, will impact the ability of colonising species to interact. However, the relevance of this structure for cell–cell interactions, as well as the extent to which sputum samples capture this structure, is unknown. Finally, the vast genotypic and phenotypic variation observed in P. aeruginosa populations from the same sputum sample (Mowat et al.2011; O’Brien et al.2017) suggests that in order to fully understand and characterise these fascinating populations, interactions should be considered on both a species and strain level. Clearly this is a hugely overwhelming task, but employing novel model systems that incorporate natural or semi-natural microbial communities allow us to make small steps toward achieving this goal (e.g. Harrison et al.2014).
FUNDING
S.O'B. was supported by the Wellcome Trust (ref: 105 624) through the Centre for Chronic Diseases and Disorders (C2D2) at the University of York, and is currently supported by an Adaptation to a Changing Environment (ACE) research fellowship from ETH Zürich. J.L.F would like to acknowledge support from the Leverhulme Trust and the Medical Research Foundation.
Conflict of interest. None declared.
REFERENCES
- Agarwal R, Chakrabarti A, Shah A et al. Allergic bronchopulmonary aspergillosis: review of literature and proposal of new diagnostic and classification criteria. Clin Exp Allergy 2013;43:850–73. [DOI] [PubMed] [Google Scholar]
- Amin R, Dupuis A, Aaron SD et al. The effect of chronic infection with Aspergillus fumigatus on lung function and hospitalization in cystic fibrosis patients. Chest 2010;137:171–6. [DOI] [PubMed] [Google Scholar]
- Andersen SB, Marvig RL, Molin S et al. Long-term social dynamics drive loss of function in pathogenic bacteria. Proc Natl Acad Sci U S A 2015;112:10756–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonic V, Stojadinovic A, Zhang B et al. Pseudomonas aeruginosa induces pigment production and enhances virulence in a white phenotypic variant of Staphylococcus aureus. Infect Drug Resist 2013;6:175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster CR, Wolter DJ, Mishra M et al. Staphylococcus aureus protein A mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa. MBio 2016;7:e00538–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkal S, Robinson SM, Ordonez CL et al. Role of bacteriocins in mediating interactions of bacterial isolates taken from cystic fibrosis patients. Microbiology 2010;156:2058–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken JS, Borody T, Brandt LJ et al. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol 2011;9:1044–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldan R, Cigana C, Testa F et al. Adaptation of Pseudomonas aeruginosa in cystic fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PLoS One 2014;9:e89614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber MF, Elde NC. Buried treasure: evolutionary perspectives on microbial iron piracy. Trends Genet 2015;31:627–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaume M, Köhler T, Fontana T et al. Metabolic pathways of Pseudomonas aeruginosa involved in competition with respiratory bacterial pathogens. Front Microbiol 2015;6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billard L, Le Berre R, Pilorgé L et al. Viruses in cystic fibrosis patients’ airways. Crit Rev Microbiol 2017, DOI: 10.1080/1040841X.2017.1297763. [DOI] [PubMed] [Google Scholar]
- Biswas L, Biswas R, Schlag M et al. Small-colony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosa. Appl Environ Microbiol 2009;75:6910–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T, Jensen PØ, Jakobsen TH et al. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One 2010;5, DOI: 10.1371/journal.pone.0010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin S, Graeber SY, Weitnauer M et al. Comparison of microbiomes from different niches of upper and lower airways in children and adolescents with cystic fibrosis. PLoS One 2015;10:e0116029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragonzi A, Farulla I, Paroni M et al. Modelling co-infection of the cystic fibrosis lung by Pseudomonas aeruginosa and Burkholderia cenocepacia reveals influences on biofilm formation and host response. PLoS One 2012;7:e52330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragonzi A, Paroni M, Nonis A et al. Pseudomonas aeruginosa microevolution during cystic fibrosis lung infection establishes clones with adapted virulence. Am J Respir Crit Care Med 2009;180:138–45. [DOI] [PubMed] [Google Scholar]
- Briard B, Bomme P, Lechner BE et al. Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci Rep 2015;5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SP, West SA, Diggle SP et al. Social evolution in micro-organisms and a Trojan horse approach to medical intervention strategies. Philos Trans R Soc Lond B Biol Sci 2009;364:3157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A, Harrison F, Vos M et al. Siderophore-mediated cooperation and virulence in Pseudomonas aeruginosa. FEMS Microbiol Ecol 2007;62:135–41. [DOI] [PubMed] [Google Scholar]
- Burton JP, Chilcott CN, Moore CJ et al. A preliminary study of the effect of probiotic Streptococcus salivarius K12 on oral malodour parameters. J Appl Microbiol 2006;100:754–64. [DOI] [PubMed] [Google Scholar]
- Cao H, Baldini RL, Rahme LG. Common mechanisms for pathogens of plants and animals. Annu Rev Phytopathol 2001;39:259–84. [DOI] [PubMed] [Google Scholar]
- Cardozo VF, Oliveira AG, Nishio EK et al. Antibacterial activity of extracellular compounds produced by a Pseudomonas strain against methicillin-resistant Staphylococcus aureus (MRSA) strains. Ann Clin Microbiol Antimicrob 2013;12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattoraj SS, Murthy R, Ganesan S et al. Pseudomonas aeruginosa alginate promotes Burkholderia cenocepacia persistence in cystic fibrosis transmembrane conductance regulator knockout mice. Infect Immun 2010;78:984–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew SC, Kundukad B, Seviour T et al. Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. MBio 2014;5:e01536–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotirmall SH, McElavaney NG. Fungi in the cystic fibrosis lung: bystanders or pathogens? Int J Biochem Cell Biol 2014;52:161–73. [DOI] [PubMed] [Google Scholar]
- Chotirmall SH, O’Donoghue E, Bennett K et al. Sputum Candida albicans presages FEV1 decline and hospital-treated exacerbations in cystic fibrosis. Chest 2010;138:1186–95. [DOI] [PubMed] [Google Scholar]
- Coman I, Bilodeau L, Lavoie A et al. Ralstonia mannitolilytica in cystic fibrosis: a new predictor of worse outcomes. Respir Med Case Rep 2017;20:48–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello A, Reen FJ, O’Gara F et al. Inhibition of co-colonizing cystic fibrosis-associated pathogens by Pseudomonas aeruginosa and Burkholderia multivorans. Microbiology 2014;160:1474–87. [DOI] [PubMed] [Google Scholar]
- Courtney JM, Bradley J, Mccaughan J et al. Predictors of mortality in adults with cystic fibrosis. Pediatr Pulmonol 2007;42:525–32. [DOI] [PubMed] [Google Scholar]
- Cox CD. Role of pyocyanin in the acquisition of iron from transferrin. Infect Immun 1986;52:263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MJ, Allgaier M, Taylor B et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One 2010;5:e11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugini C, Calfee MW, Farrow JM et al. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol Microbiol 2007;65:896–906. [DOI] [PubMed] [Google Scholar]
- Cugini C, Morales DK, Hogan DA. Candida albicans-produced farnesol stimulates Pseudomonas quinolone signal production in LasR-defective Pseudomonas aeruginosa strains. Microbiology 2010;156:3096–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cystic Fibrosis Foundation. Cystic Fibrosis Foundation Patient Registry 2010 Annual Report, 2011.
- Davies EV, James CE, Williams D et al. Temperate phages both mediate and drive adaptive evolution in pathogen biofilms. Proc Natl Acad Sci U S A 2016;113:8266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeon S, Clinton A, Fowler H et al. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun 2014;82:4718–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVault JD, Kimbara K, Chakrabarty AM. Pulmonary dehydration and infection in cystic fibrosis: evidence that ethanol activates alginate gene expression and induction of mucoidy in Pseudomonas aeruginosa. Mol Microbiol 1990;4:737–45. [DOI] [PubMed] [Google Scholar]
- Dietrich LE, Price-Whelan A, Petersen A et al. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol 2006;61:1308–21. [DOI] [PubMed] [Google Scholar]
- Duan K, Dammel C, Stein J et al. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol 2003;50:1477–91. [DOI] [PubMed] [Google Scholar]
- Eberl L, Tümmler B. Pseudomonas aeruginosa and Burkholderia cepacia in cystic fibrosis: Genome evolution, interactions and adaptation. Int J Med Microbiol 2004;294:123–31. [DOI] [PubMed] [Google Scholar]
- Emerson J, Rosenfeld M, McNamara S et al. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr Pulmonol 2002;34:91–100. [DOI] [PubMed] [Google Scholar]
- Ferreira JAG, Penner JC, Moss RB et al. Inhibition of Aspergillus fumigatus and its biofilm by Pseudomonas aeruginosa is dependent on the source, phenotype and growth conditions of the bacterium. PLoS One 2015;10:e0134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filkins LM, Graber JA, Olson DG et al. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol 2015;197:2252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor AA, Klem ER, Gilpin DF et al. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One 2012;7:e45001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkesson A, Jelsbak L, Yang L et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 2012;10:841–51. [DOI] [PubMed] [Google Scholar]
- Fothergill JL, Mowat E, Ledson MJ et al. Fluctuations in phenotypes and genotypes within populations of Pseudomonas aeruginosa in the cystic fibrosis lung during pulmonary exacerbations. J Med Microbiol 2010;59:472–81. [DOI] [PubMed] [Google Scholar]
- Fothergill JL, Panagea S, Hart CA et al. Widespread pyocyanin over-production among isolates of a cystic fibrosis epidemic strain. BMC Microbiol 2007;7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayman KB, Armstrong DS, Carzino R et al. The lower airway microbiota in early cystic fibrosis lung disease: a longitudinal analysis. Thorax 2017, DOI: 10.1136/thoraxjnl-2016-209279. [DOI] [PubMed] [Google Scholar]
- Fugère A, Séguin DL, Mitchell G et al. Interspecific small molecule interactions between clinical isolates of Pseudomonas aeruginosa and Staphylococcus aureus from adult cystic fibrosis patients. PLoS One 2014;9:e86705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J, Sood A, Hogan DA. Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. Appl Environ Microbiol 2009;75:504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanadhas DP, Elango M, Datey A et al. Chronic lung infection by Pseudomonas aeruginosa biofilm is cured by L-methionine in combination with antibiotic therapy. Sci Rep 2015a;5:16043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanadhas DP, Elango M, Janardhanraj S et al. Successful treatment of biofilm infections using shock waves combined with antibiotic therapy. Sci Rep 2015b;5:17440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goral J, Karavitis J, Kovacs EJ. Exposure-dependent effects of ethanol on the innate immune system. Alcohol 2008;42:237–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg SS, Zhao X, Hua L et al. Ethanol inhibits lung clearance of Pseudomonas aeruginosa by a neutrophil and nitric oxide-dependent mechanism, in vivo. Alcohol Clin Exp Res 1999;23:735–44. [PubMed] [Google Scholar]
- Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature 2004;430:1024–7. [DOI] [PubMed] [Google Scholar]
- Gungor OE, Kirzioglu Z, Kivanc M. Probiotics:can they be used to improve oral health? Benef Microbes 2015;6:647–56. [DOI] [PubMed] [Google Scholar]
- Hamilton MJ, Weingarden AR, Sadowsky MJ et al. Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. Am J Gastroenterol 2012;107:761–7. [DOI] [PubMed] [Google Scholar]
- Harrison F, Buckling A. Hypermutability impedes cooperation in pathogenic bacteria. Curr Biol 2005;15:1968–71. [DOI] [PubMed] [Google Scholar]
- Harrison F, Buckling A. High relatedness selects against hypermutability in bacterial metapopulations. Proc Biol Sci 2007;274:1341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison F, McNally A, da Silva AC et al. Optimised chronic infection models demonstrate that siderophore ‘cheating’ in Pseudmonas aeruginosa is context specific. ISME J 2017, DOI: 10.1038/ismej.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison F, Muruli A, Higgins S et al. Development of an ex vivo porcine lung model for studying growth, virulence and signaling of Pseudomonas aeruginosa. Infect Immun 2014;82:3312–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison F, Paul J, Massey RC et al. Interspecific competition and siderophore-mediated cooperation in Pseudomonas aeruginosa. ISME J 2008;2:49–55. [DOI] [PubMed] [Google Scholar]
- Hart CA, Winstanley C. Persistent and aggressive bacteria in the lungs of cystic fibrosis children. Br Med Bull 2002;61:81–96. [DOI] [PubMed] [Google Scholar]
- Hoffman LR, Deziel E, D’Argenio DA et al. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 2006;103:19890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 2002;296:2229–32. [DOI] [PubMed] [Google Scholar]
- Hogan DA, Vik Å, Kolter R. A Pseudomonas aeruginosa quorumsensing molecule influences Candida albicans morphology. Mol Microbiol 2004;54:1212–23. [DOI] [PubMed] [Google Scholar]
- Høiby N, Bjarnsholt T, Givskov M et al. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 2010;35:322–32. [DOI] [PubMed] [Google Scholar]
- Høiby N, Ciofu O, Bjarnsholt T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol 2010;5:1663–74. [DOI] [PubMed] [Google Scholar]
- Huang CH, Jang TN, Liu CY et al. Characteristics of patients with Burkholderia cepacia bacteremia. J Microbiol Immunol Infect 2001;34:215–9. [PubMed] [Google Scholar]
- Hunter RC, Asfour F, Dingemans J et al. Ferrous iron is a significant component of bioavailable iron in cystic fibrosis airways. MBio 2013;4:e00557–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RC, Klepac-Ceraj V, Lorenzi MM et al. Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. Am J Respir Cell Mol Biol 2012;47:738–45. [DOI] [PubMed] [Google Scholar]
- Jiricny N, Molin S, Foster K et al. Loss of social behaviours in populations of Pseudomonas aeruginosa infecting lungs of patients with cystic fibrosis. PLoS One 2014;9:e83124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J. Inhibition of fungal growth by Pseudomonas aeruginosa and Pseudomonas cepacia isolated from patients with cystic fibrosis. J Infect 1994;28:305–10. [DOI] [PubMed] [Google Scholar]
- Klepac-Ceraj V, Lemon KP, Martin TR et al. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ Microbiol 2010;12:1293–303. [DOI] [PubMed] [Google Scholar]
- Koch C, Høiby N. Pathogenesis of cystic fibrosis. Lancet 1993;341:1065–9. [DOI] [PubMed] [Google Scholar]
- Korem M, Gov Y, Rosenberg M. Global gene expression in Staphylococcus aureus following exposure to alcohol. Microb Pathog 2010;48:74–84. [DOI] [PubMed] [Google Scholar]
- Korgaonkar A, Trivedi U, Rumbaugh KP et al. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A 2013;110:1059–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar AK, Whiteley M. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J Bacteriol 2011;193:909–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambiase A, Raia V, Del Pezzo M et al. Microbiology of airway disease in a cohort of patients with cystic fibrosis. BMC Infect Dis 2006;6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau GW, Hassett DJ, Ran H et al. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med 2004a;10:599–606. [DOI] [PubMed] [Google Scholar]
- Lau GW, Ran H, Kong F et al. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect Immun 2004b;72:4275–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Steiner T, Petrof EO et al. Frozen vs fresh fecal microbiota transplantation and clinical resolution of diarrhea in patients with recurrent Clostridium difficile infection: a randomized clinical trial. JAMA 2016;315:142–9. [DOI] [PubMed] [Google Scholar]
- Lewenza S, Visser MB, Sokol PA. Interspecies communication between Burkholderia cepacia and Pseudomonas aeruginosa. Can J Microbiol 2002;48:707–16. [DOI] [PubMed] [Google Scholar]
- Li J, Hao C, Ren L et al. Data mining of lung microbiota in cystic fibrosis patients. PLoS One 2016;11:e0164510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YW, Evangelista JS, Schmieder R et al. Clinical insights from metagenomic analysis of sputum samples from patients with cystic fibrosis. J Clin Microbiol 2014;52:563–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YW, Schmieder R, Haynes M et al. Metagenomics and metatranscriptomics: windows on CF-associated viral and microbial communities. J Cyst Fibros 2013;12:154–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Medina E, Fan D, Coughlin LA et al. Candida albicans inhibits Pseudomonas aeruginosa virulence through suppression of pyochelin and pyoverdine biosynthesis. PLoS Pathog 2015;11:e1005129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorè NI, Cigana C, De Fino I et al. Cystic fibrosis-niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts. PLoS One 2012;7:e35648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan AM, Gomez P, Buckling A. Siderophore cooperation of the bacterium Pseudomonas fluorescens in soil. Biol Lett 2015;11:20140934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlester G, O’Gara F, Morrissey JP. Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J Med Microbiol 2008;57:563–9. [DOI] [PubMed] [Google Scholar]
- McKenney D, Brown KE, Allison DG. Influence of Pseudomonas aeruginosa exoproducts on virulence factor production in Burkholderia cepacia: evidence of interspecies communication. J Bacteriol 1995;177:6989–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Tan MW, Rahme LG et al. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis eleganspathogenesis model. Cell 1999;96:47–56. [DOI] [PubMed] [Google Scholar]
- Marvig RL, Damkiær S, Hossein Khademi SM et al. Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin. MBio 2014;5, DOI: 10.1128/mBio.00966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn LM, Jett AM, Akins DR et al. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol 2005;187:554–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melter O, Radojevič B. Small colony variants of Staphylococcus aureus–review. Folia Microbiol (Praha) 2010;55:548–58. [DOI] [PubMed] [Google Scholar]
- Miller RV, Rubero VJ. Mucoid conversion by phages of Pseudomonas aeruginosa strains from patients with cystic fibrosis. J Clin Microbiol 1984;19:717–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell G, Séguin D, Asselin A-E et al. Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol 2010;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales DK, Grahl N, Okegbe C et al. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. MBio 2013;4:e00526–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moree WJ, Phelan VV, Wu CH et al. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc Natl Acad Sci U S A 2012;109:13811–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat E, Paterson S, Fothergill JL et al. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am J Respir Crit Care Med 2011;183:1674–9. [DOI] [PubMed] [Google Scholar]
- Nagano Y, Millar BC, Johnson E et al. Fungal infections in patients with cystic fibrosis. Rev Med Microbiol 2007;18:11–6. [Google Scholar]
- Nevalainen TJ, Graham GG, Scott KF. Antibacterial actions of secreted phospholipases A2. Biochim Biophys Acta 2008;1781:1–9. [DOI] [PubMed] [Google Scholar]
- Nguyen D, Singh PK. Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc Natl Acad Sci U S A 2006;103:8305–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman RS, Moeller P, McDonald TJ et al. Effect of pyocyanin on a crude-oil-degrading microbial community. Appl Environ Microbiol 2004;70:4004–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwugo CC, Arivett BA, Zimbler DL et al. Effect of ethanol on differential protein production and expression of potential virulence functions in the opportunistic pathogen Acinetobacter baumannii. PLoS One 2012;7:e51936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien S, Rodrigues AMM, Buckling A. The evolution of bacterial mutation rates under simultaneous selection by interspecific and social parasitism. Proc Biol Sci 2013;280:20131913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien S, Williams D, Fothergill JL et al. High virulence sub-populations in Pseudomonas aeruginosa long-term cystic fibrosis airway infections. BMC Microbiol 2017;17:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley YQ, Abdalla MY, McCormick ML et al. Subcellular localization of Pseudomonas pyocyanin cytotoxicity in human lung epithelial cells. Am J Physiol Lung Cell Mol Physiol 2003;284:L420–30. [DOI] [PubMed] [Google Scholar]
- O’May CY, Sanderson K, Roddam LF et al. Iron-binding compounds impair Pseudomonas aeruginosa biofilm formation, especially under anaerobic conditions. J Med Microbiol 2009;58:765–73. [DOI] [PubMed] [Google Scholar]
- Palmer KL, Mashburn LM, Singh PK et al. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol 2005;187:5267–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet E, Guillemot L, Burgel P-R et al. Pseudomonas aeruginosa eradicates Staphylococcus aureus by manipulating the host immunity. Nat Commun 2014;5:5105. [DOI] [PubMed] [Google Scholar]
- Peters BM, Jabra-Rizk MA, O’May GA et al. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev 2012;25:193–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihet M, Carrere J, Cimon B et al. Occurrence and relevance of filamentous fungi in respiratory secretions of patients with cystic fibrosis–a review. Med Mycol 2009;47:387–97. [DOI] [PubMed] [Google Scholar]
- Pressler T, Bohmova C, Conway S et al. Chronic Pseudomonas aeruginosa infection definition: EuroCareCF Working Group report. J Cyst Fibros 2011;10:S75–8. [DOI] [PubMed] [Google Scholar]
- Qu XD, Lehrer RI. Secretory phospholipase A2 is the principal bactericide for staphylococci and other gram-positive bacteria in human tears. Infect Immun 1998;66:2791–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol 2005;56:309–22. [DOI] [PubMed] [Google Scholar]
- Rea MC, Alemayehu D, Ross RP et al. Gut solutions to a gut problem: Bacteriocins, probiotics and bacteriophage for control of Clostridium difficile infection. J Med Microbiol 2013;62:1369–78. [DOI] [PubMed] [Google Scholar]
- Reece E, Segurado R, Jackson A et al. Co-colonisation with Aspergillus fumigatus and Pseudomonas aeruginosa is associated with poorer health in cystic fibrosis patients: an Irish registry analysis. BMC Pulm Med 2017;17:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel K, Hentzer M, Geisenberger O et al. N-Acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 2001;147:3249–62. [DOI] [PubMed] [Google Scholar]
- Römling U, Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med 2012;272:541–61. [DOI] [PubMed] [Google Scholar]
- Sahand IH, Moragues MD, Robert R et al. Evaluation of Bichro-Dubli Fumouze to distinguish Candida dubliniensis from Candida albicans. Diagn Microbiol Infect Dis 2006;55:165–7. [DOI] [PubMed] [Google Scholar]
- Sajjan U, Thanassoulis G, Cherapanov V et al. Enhanced susceptibility to pulmonary infection with Burkholderia cepacia in Cftr−/− mice. Infect Immun 2001;69:5138–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan PD, Buckling A. Co-evolution with lytic phage selects for the mucoid phenotype of Pseudomonas fluorescens SBW25. ISME J 2012;6:1148–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab U, Abdullah LH, Perlmutt OS et al. Localization of Burkholderia cepacia complex bacteria in cystic fibrosis lungs and interactions with Pseudomonas aeruginosa in hypoxic mucus. Infect Immun 2014;82:4729–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoseyov D, Brownlee KG, Conway SP et al. Aspergillus bronchitis in cystic fibrosis. Chest 2006;130:222–6. [DOI] [PubMed] [Google Scholar]
- Singh PK, Parsek MR, Greenberg EP et al. A component of innate immunity prevents bacterial biofilm development. Nature 2002;417:552–5. [DOI] [PubMed] [Google Scholar]
- Smith EE, Buckley DG, Wu Z et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 2006;103:8487–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen RU, Klinger JD. Biological effects of Pseudomonas aeruginosa phenazine pigments. Antibiot Chemother 1987;39:113–24. [DOI] [PubMed] [Google Scholar]
- Sousa AM, Pereira MO. Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs—a review. Pathogens 2014;3:680–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognon M, Kohler T, Gdaniec BG et al. Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. ISME J 2017, DOI: 10.1038/ismej.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell J, Callaghan M. Iron acquisition in the cystic fibrosis lung and potential for novel therapeutic strategies. Microbiology 2016;162:191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenza G, Tappe D, Turnwald D et al. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cyst Fibros 2008;7:123–7. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wilks JC, Danhorn T et al. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol 2011;193:3606–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters EM, Neill DR, Kaman B et al. Phage therapy is highly effective against chronic lung infections with Pseudomonas aeruginosa. Thorax 2017;72:666–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VB, Kolter R. Burkholderia spp. Alter Pseudomonas aeruginosa physiology through iron sequestration. J Bacteriol 2004;186:2376–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Diggle SP, Buckling A et al. The social lives of microbes. Annu Rev Ecol Evol Syst 2007;38:53–77. [Google Scholar]
- Whelan FJ, Heirali AA, Rossi L et al. Longitudinal sampling of the lung microbiota in individuals with cystic fibrosis. PLoS One 2017;12:e0172811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley RA, Sheikh NP, Mushtaq N et al. Differential potentiation of the virulence of the Pseudomonas aeruginosa cystic fibrosis Liverpool epidemic strain by oral commensal streptococci. J Infect Dis 2014;209:769–80. [DOI] [PubMed] [Google Scholar]
- Winstanley C, Fothergill JL. The role of quorum sensing in chronic cystic fibrosis Pseudomonas aeruginosa infections. FEMS Microbiol Lett 2009;290:1–9. [DOI] [PubMed] [Google Scholar]
- Winstanley C, O’Brien S, Brockhurst MA. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol 2016;24:327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter DJ, Emerson JC, McNamara S et al. Staphylococcus aureus small-colony variants are independently associated with worse lung disease in children with cystic fibrosis. Clin Infect Dis 2013;57:384–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter JM, Kotsiou G, McCormack JG. Mixed morphotype testing of Pseudomonas aeruginosa cultures from cystic fibrosis patients. J Med Microbiol 1995;42:220–4. [DOI] [PubMed] [Google Scholar]
- Yao GW, Duarte I, Le TT et al. A broad-host-range tailocin from Burkholderia cenocepacia. Appl Environ Microbiol 2017;83:e03414–16. [DOI] [PMC free article] [PubMed] [Google Scholar]