Abstract
The release of large quantities of microorganisms to soil for purposes such as pest control or plant growth promotion may affect the indigenous soil microbial communities. In our study, we investigated potential effects of Metarhizium brunneum ART2825 on soil fungi and prokaryota in bulk soil using high-throughput sequencing of ribosomal markers. Different formulations of this strain, and combinations of the fungus with garlic as efficacy-enhancing agent, were tested over 4 months in a pot and a field experiment carried out for biological control of Agriotes spp. in potatoes. A biocontrol effect was observed only in the pot experiment, i.e. the application of FCBK resulted in 77% efficacy. Colony counts combined with genotyping and marker sequence abundance confirmed the successful establishment of the applied strain. Only the formulated applied strain caused small shifts in fungal communities in the pot experiment. Treatment effects were in the same range as the effects caused by barley kernels, the carrier of the FCBK formulation and temporal effects. Garlic treatments and time affected prokaryotic communities. In the field experiment, only spatial differences affected fungal and prokaryotic communities. Our findings suggest that M. brunneum may not adversely affect soil microbial communities.
Keywords: inundative release, biological control agent, amplicon sequencing, non-target effect, fungal inoculant, next-generation sequencing
Formulated Metarhizium brunneum has small and temporal effects on fungal communities in a pot experiment and no effects were detected in the field.
INTRODUCTION
Soil is a complex and dynamic environment providing habitats for a tremendous number and diversity of soil microorganisms (Nannipieri et al.2003). It has been estimated that 1 g of soil may harbor up to 1010 bacterial and 106 fungal cells and thousands of bacterial and fungal species (Torsvik, Goksoyr and Daae 1990; Bridge and Spooner 2001; Roesch et al.2007; Trevors 2009). Soil microorganisms provide a wealth of functions. They play a central role in nutrient cycling and the formation and maintenance of soil structure, they contribute to plant health and they are involved in the natural regulation of insects, pathogens and weeds (Kennedy 1999). All together these functions are vital for maintaining productivity in agriculture and it is important to understand which abiotic and biotic factors, including agricultural practices, may adversely affect microbial communities. The potential impacts of a number of factors including time, space and climate on microbial communities have been investigated in various systems (Lauber et al.2013; Tedersoo et al.2014; O’Brien et al.2016). Likewise, the effects of edaphic factors or anthropogenic activities, such as land use, soil compaction and pesticide applications have been studied (Lauber et al.2013; Hartmann et al.2014; Jacobsen and Hjelmsø 2014).
The ability of microorganisms to regulate insects, pathogens and weeds has been recognized as an important function with potential use in agriculture more than a century ago (Krassilstschik 1888; Prior 1996; Zimmermann 2007). Since then a variety of microorganisms has been identified and commercialized as microbial pesticides also known as biological control agents (BCA; Faria and Wraight 2007; Lugtenberg 2015). Microbial control usually implies application of large amounts of infective propagules of a BCA to soils under treatment. For instance, about 1012–1014 propagules of entomopathogenic fungi are applied per hectare translating into 105 conidia per cm2 of soil (Jaronski 2010). Such high loads of propagules may have unintended side effects leading to changes in soil microbial community structures. The European Union therefore has included an assessment of potential effects on indigenous soil microorganisms in the registration process of biological pesticides (Commission regulation No. 544/2011). Most studies assessing the effects of applied microorganisms on soil microbial communities have revealed only small or transient effects (Trabelsi and Mhamdi 2013; Kröber et al.2014; Zimmermann et al.2016), but little is known about potential effects of the application of entomopathogenic fungi (Hu and St Leger 2002; Rai and Singh 2002; Kirchmair et al.2008; Schwarzenbach, Enkerli and Widmer 2009; Hirsch et al.2013).
Agriotes spp. Eschscholtz (Elateridae) are major soil dwelling pests in the Holarctic (Kudryavtsev et al.1993; Vernon, Lagasa and Philip 2001) in various crops, such as cereals, different vegetables and potatoes (e.g. Miles 1942; Parker 1994; Blot and Brunel 1999). Control methods have included repeated tillage, crop rotation, pesticide application and biological control with varying degrees of success (reviewed in Ritter and Richter 2013; Traugott et al.2015). The progressive banning of chemical insecticides has resulted in an increased focus on biological alternatives for pest control such as the application of entomopathogenic fungi or nematodes (Ritter and Richter 2013). Studies with the entomopathogenic fungus Metarhizium brunneum ART2825 Petch (Hypocreales: Clavicipitaceae) have shown promising results in laboratory experiments in controlling Agriotesobscurus L., A. lineatus L. and A. sputator L. (Kölliker, Biasio and Jossi 2011; Eckard et al.2014). Improvements in formulation technologies and application strategies or co-applications with botanicals, chemicals or other BCAs have been shown to increase the efficacy of entomopathogenic fungi to control pest insects (Ansari, Shah and Butt 2010; Paula et al.2011; Behle, Jackson and Flor-Weiler 2013; Kabaluk, Lafontaine and Borden 2015). Formulations have been developed for entomopathogenic fungi in order to protect their spores during storage and distribution of the products, to enhance persistence in the field and/or to facilitate the application process (Glare and Moran-Diez 2016). Metarhizium (Metschn.) Sorokin has been formulated based on grains, e.g. sterile barley kernels (Aregger 1992), or was produced in form of microsclerotia (Jaronski and Jackson 2008). Application strategies including pheromone traps or CO2 lures have been used to enhance the efficacy of Metarhizium spp. against Agriotes spp. (Kabaluk, Lafontaine and Borden 2015; Brandl et al.2017). Also, several natural substances have been tested for controlling Agriotes spp. (Ritter and Richter 2013). Among those, garlic was shown to repel and reduce movement of A. obscurus larvae, which potentially may enhance the efficacy of M. brunneum by weakening the larvae and making them more susceptible to a fungal infection (Eckard et al.2017).
In this study, we investigated whether applications of the fungus M. brunneum ART2825 for controlling A. obscurus in potato production affect soil fungal and prokaryotic communities. The study relies on both an experiment in the greenhouse (pots) and a field experiment using different formulations of the fungus and garlic extract as potential efficacy-enhancing agent. Isolation and cultivation on selective medium, simple sequence repeat (SSR) genotyping, and high-throughput amplicon sequencing of ribosomal markers were used to monitor the applied fungus and observe changes in fungal and prokaryotic community structures over a period of 4 months.
MATERIALS AND METHODS
Rearing of Agriotes obscurus larvae
Lab-reared A. obscurus larvae were used for artificial infestation of substrates in the pot experiment. They were reared in a laboratory livestock established by the method of Kölliker, Jossi and Kuske (2009). Briefly, A. obscurus adults were collected from the field and placed into pots (ø 30 cm) containing 10–15 L soil rich in humus and were covered with a mesh bag until oviposition. Grass was repeatedly sown into the soil of the pots to guarantee food for the hatched larvae and the pots were kept moist. Five months after establishment, larvae were transferred into a pot containing fresh peat soil with sliced carrots as food source and stored at 10°C in the dark. Four weeks prior to experiments, each larva was placed individually into a cup with moist peat substrate and carrot slices and maintained at 22°C. Only healthy larvae were selected for subsequent infestation of pots.
Treatments
Nine and five different treatments were applied in six replicates in the pot and the field experiment, respectively. Different treatments and applied doses are listed in Table 1. The entomopathogenic fungus Metarhizium brunneum strain ART2825 was either applied as unformulated fungal spore powder (Fpowd) or was formulated as fungus colonized barley kernels (FCBK), as fungal capsules (Fcap) and as fungal granules (Fgran). The FCBK were produced in the laboratory as described by Aregger (1992). Batches (1.3 kg) of peeled barley kernels were autoclaved twice in plastic culture bags. Subsequently, the barley kernels were inoculated with culture broth of M. brunneum ART2825 in cornsteep medium (diluted to 107 spores/ml with water), which had been incubated at 22°C–24°C for 5 days. Following inoculation, the barley kernels were incubated for 4 to 5 weeks at 22°C–24°C. The Fpowd was produced by FYTOVITA spol s r. o. (Ostrožská Lhota, Czech Republic) using solid-state fermentation, and it was also used for the Fcap which were formulated by M. Przyklenk (University of Applied Sciences, Bielefeld, Germany) according to a modified protocol by Humbert et al. (2017). The Fcap included 8 × 107 spores/g capsules, autoclaved baker's yeast and calcium alginate. They were formed by dripping M. brunneum spore-alginate solution into a crosslinking solution which induced polymerization and formation of beads. The Fgran, a prototype produced by e-nema GmbH (Schwentinental, Germany), included the same components as the Fcap; however, an extruder and a fluid-bed dryer were used to form granules. Garlic capsules were produced by S. Gerike (University of Applied Sciences, Bielefeld, Germany) and consisted of 6% garlic oil (Neem Biotech Ltd., Abertillery, UK), calcium alginate, acetic acid and a chitosan coating. Garlic capsules were applied alone but also in combination with FCBK and Fcap in order to study potential synergistic effects of Metarhizium and garlic. The insecticide clothianidin (Insec; Cheyenne®, Philagro, Saint-Didier-au-Mont-d’Or, France) and sterile barley kernels (BK), which represent the carrier material in the FCBK formulation, were used as positive and negative controls, respectively. The pot experiment included the following nine treatments: FCBK, Fcap, Fpowd, Gcap, the combinations FCBK + Gcap and Fcap + Gcap, Insec, BK and untreated pots. In the field experiment, five treatments were applied: FCBK, Fcap, Fgran, Insec and untreated plots (Table 1).
Table 1.
Applied doses of the nine and five treatments applied in the pot and field experiment, respectively (n = 6).
| Treatment | Amount applied (g/pot or field plot) | |
|---|---|---|
| Pot | Field | |
| Fungus colonized barley kernels (FCBK) | 5.6 | 270 |
| Fungal capsules (Fcap) | 7 | 240 |
| Fungal granules (Fgran) | NI | 240 |
| Fungal spore powder (Fpowd) | 0.11 | NI |
| Garlic capsules (Gcap) | 14.4 | NI |
| Gcap and FCBK | 5.6 + 14.4 | NI |
| Gcap and Fcap | 7 + 14.4 | NI |
| Barley kernels (BK) | 5.6 | NI |
| Clothianidin (Insec) | 0.06 | 14 |
| Untreated | x | x |
The amount of fungal spores in the pot and in the field experiment were 1 × 1014 and 5 × 1013 spores/ha and clothianidin was applied at a rate of 11 kg/ha. All pots and plots included potato plants and the pest insect.
NI , treatment not included.
Set-up of the pot experiment
The pot experiment was conducted in a greenhouse at 20°C–25°C from April until September 2014. Each of the nine treatments (Table 1) was replicated six times resulting in 54 pots which were randomly arranged and kept at the same position during the experiment. Pots had a dimension of 22.5 × 25 × 26 cm and two mesh sealed holes (ø 2.5 cm) at the bottom for water drainage and for preventing the escape of A. obscurus larvae. Soil (3% humus, 22% clay, 38% silt) with a pH of 7.9 was collected from a field at Agroscope research station Reckenholz, Zürich (Switzerland). The field soil was homogenized with a cement mixer and filled into pots 4 weeks prior to application of the treatments. The pots were kept moist (16 ± 3% water content, no significant difference among treatments), and weeds were removed by hand prior to application of the treatments. Treatments were applied manually onto the soil surface, and then mixed into the upper 15 cm of the soil using a small gardening rake. Subsequently, two pre-sprouted seed potato tubers (Solanum tuberosum L.) of the cultivar ‘Celtiane’ were placed in each pot at a depth of 10 cm followed by the release of 10 late instar A. obscurus larvae into each pot.
Bulk soil samples were collected from pots before application of treatments and potato tubers on 14 May 2014 (week 0) and post application on 1 July 2014 (week 7) and 26 August 2014 (week 15). Each soil sample consisted of four soil cores (15 cm depth and 1.5 cm width) that were collected crosswise per pot and then mixed. The aboveground potato tissue was cut 5 cm above the soil surface after the third sampling at week 15, at the time when plants became senescent. Two weeks later, the pots were disassembled. Potatoes were harvested and washed, and the damage caused by A. obscurus, i.e. the number of holes per tuber, assessed and categorized according to standards provided by the European and Mediterranean Plant Protection Organization (Anonymous 2005). Released A. obscurus larvae were re-captured, counted and incubated individually in cups filled with peat substrate and carrot slices as food source at 21°C for 8 weeks to check for infection with Metarhizium spp.
Set-up of the field experiment
The field experiment was performed in an agricultural field located in Mellingen, Switzerland (47°24’24΄ N 8°16’12΄ E). The soil contained 2% humus, 21% clay and 32% silt at soil pH 7.3. The site is naturally infected with different wireworm species, predominantly of the genus Agriotes, and was planted with grass during three seasons preceding the experiment. All cultivation and farming steps were performed by the farmer owning the field except soil sampling, potato planting and potato harvesting. The experimental area was rectangular including 10 blocks with three plots per block (Fig. S1, Supporting Information). Each plot was approximately 3 m wide (four rows of potato plants) and 8.3 m long. The three plots forming a block were connected at the long side and blocks were separated by a 70 cm path. The entire experimental field, including a 3-m wide untreated belt surrounding the plots, measured ∼1600 m2.
Bulk soil samples were collected before application of treatments and potato tubers on 21 April 2015 (week 0) and post application on 24 June 2015 (week 9) and 11 August 2015 (week 16). Soil samples were obtained by collecting and combining 10 soil cores (15 cm depth and 2.5 cm diameter) from the inner two rows (five cores from each row) of each plot. One-meter buffer zones at both ends of each plot were not sampled to prevent potential carryover from neighboring plots. The field was ploughed in March 2015 and harrowed once after the first soil sampling in April 2015. Then, treatments were applied manually and integrated into the soil by harrowing for a second time. Subsequently, potato tubers of the cultivar ‘Celtiane’ were planted in rows which were piled immediately after application in order to prevent UV exposure of the products. Fifty-five kilogram per hectare of the fertilizer MgS Ammonsalpeter 25 (Agroline, Roggwil, Switzerland, 25% nitrogen, 5% magnesium, 8.5% sulfur) was applied, and the herbicide Titus (DuPont de Nemours International Sàrl, Le Grand-Saconnex, Switzerland, 25% rimsulfuron) + Exell (Stähler Suisse, Zofingen, Switzerland, 77% detergents, 22% ethylenglycolmonobuthylether), the pesticide Audienz (Omya AG, Oftringen, Switzerland, 44.2% spinosad) and the fungicide Mapro (ISK Biosciences GmbH, Bern, Switzerland, 38,8% fluazinam) were sprayed in May and June. The leaves of potato plants were herbicide treated, after an infection with the fungus Colletotrichum Corda had been detected in July, by applying Reglone (Syngenta AG, Basel, Switzerland, 17% diquat) for haulm destruction. After the third soil sampling in August, potato tubers of the inner two rows of each plot were harvested. Fifty potato tubers per plot were randomly selected, washed and Agriotes-caused damage scored.
Weather data were obtained from the closest meteorological station in Kuenten CH (6 km from the field site). During the sampling period, the daily mean temperature was 18.3°C and ranged between 7.6°C and 28°C. During this time, a total of 431.8 mm precipitation was recorded. Average humidity was 73.9% and ranged between 53.4% and 97.7%.
Processing of soil samples, isolation of Metarhizium CFU and identification of applied strain
Soil samples were homogenized and sieved with a 5-mm mesh, and aliquots were used for assessment of soil moisture content, for determination and isolation of Metarhizium spp. colony forming units (CFU) and for extraction of soil DNA (described below). The CFU determination of Metarhizium spp. was performed with slight modifications according to the protocol described by Schneider et al. (2012). Three times 20 g of soil per sample were dissolved in 100 ml pyrophosphate solution and plated onto selective medium agar plates resulting in three plates per sample. Metarhizium colonies were counted after 10 to 14 days.
After CFU assessment, isolates were selected from the plates for genetic identification using SSR marker-based genotyping. From the pot experiment, five to six isolates were selected from each treatment at week 0, one to two isolates were obtained from all fungal treatments at week 7 and six isolates were chosen from all fungal treatments at week 15. In addition, one to eight isolates per treatment were recovered from Metarhizium spp. infected A. obscurus larvae which were re-captured after the end of the pot experiment and incubated in the lab for detection of late fungal infections. One Metarhizium spp. colony per soil sample ( = plot) per sampling time point was selected from the field experiment.
Fungal tissues isolated from FCBK, Fcap and Fgran were used as positive controls. Isolates were transferred to potato agar plates and stored at 4°C until all isolates of the pot or the field experiment were collected. Subsequently, all isolates were plated onto filter paper, which was placed on potato agar plates. Mycelium of each isolate was scraped off the filter paper, and DNA was extracted according to the protocol described by Kepler et al. (2014). SSR analysis for genotyping of Metarhizium isolates was performed using SSR markers Ma2049, Ma2054 and Ma2063 (Set I) and Ma195, Ma307 and Ma2287 (Set V) (Mayerhofer et al.2015). DNA extracts were diluted 10 to 100 times, and PCR was performed as described in Mayerhofer et al. (2015). PCR products were visualized with an ABI 3130xl (Applied Biosystems, Foster City, CA, USA) using 36 cm capillaries and POP-7 polymer. GENESCAN 400 HD ROX was used as an internal size standard. Allele sizes were determined using the software GeneMarker® (SoftGenetics®, State College, PA, USA) and corrected relative to allele sizes of the reference strains M. anisopliae ART2062 (Metschn.) Sorokin, M. brunneum ARSEF7524 and M. robertsii ARSEF7532 J.F. Bisch.
DNA extraction from soil, PCR and Illumina sequencing
Soil genomic DNA was extracted from each replicate (pot or plot) per treatment and per sampling time point for both experiments. One half gram of each sample was placed into a 2-ml Eppendorf tube containing 0.5 g of glass beads (ø 0.1–0.11 mm; Sartorius, Tagelswangen, Switzerland), vortexed with 1.3 ml extraction buffer and stored at –20°C until further use. Soil DNA was extracted as described by Bürgmann et al. (2001) and modified by Hartmann et al. (2005). Soil DNA extracts were purified with the NucleoSpin® gDNA clean-up kit (Machery-Nagel, Düren, Germany) and stored at –20°C. DNA concentrations were measured using PicoGreen (Invitrogen, Carlsbad, CA, USA) with a Cary Eclipse fluorescence spectrophotometer (Varian, Inc., Palo Alto, CA, USA) and DNA extracts were diluted to 2 ng/μl with autoclaved dd H20. PCR was adopted from Frey et al. (2016) with small modifications. Fungal internal transcribed spacer region 2 (ITS2) was amplified using the primer pair ITS3 (5΄ CAHCGATGAAGAACGYRG 3΄)/ITS4 (5΄ TCCTSCGCTTATTGATATGC 3΄) (Tedersoo et al.2014). The prokaryotic variable region (V3-V4) of the small subunit of the ribosomal RNA (16S rRNA), targeting bacterial and archaeal sequences, was amplified with the modified version of primer pair 341F (5΄ CCTAYGGGDBGCWSCAG 3΄)/806R (5΄ GGACTACNVGGGTHTCTAAT 3΄) (Frey et al.2016). Forward and reverse primers for amplification of ITS2 and V3-V4 included adapter sequences CS1 (forward) and CS2 (reverse) at the 5΄ end of each primer to allow multiplexing with the Fluidigm Access Array System (Fluidigm, South San Francisco, CA, USA). Prior to PCR, 20 ng of soil genomic DNA was incubated with 45 μg BSA in 15 μl for 5 min at 90°C. PCR was performed in a volume of 50 μl containing the pre-incubated DNA, 1x PCR buffer containing 15 mM MgCl2 (Qiagen, Venlo, Netherlands), 0.4 μM of the forward and the reverse primer, 0.2 mM of each dNTP (Promega, Madison, WI, USA), 1 mM MgCl2 (Qiagen), additional 1.8 mg/ml BSA and 2 U of HotStartTaq® Plus DNA polymerase (Qiagen). PCR cycling conditions included one initial denaturation step at 95°C for 5 min, followed by 30 or 35 cycles (for prokaryota or fungi) of denaturation at 94°C for 40 s, annealing at 58°C for 40 s (both primer pairs) and elongation at 72°C for 1 min. PCR was finalized with elongation at 72°C for 10 min. The integrity and quality of the PCR products were checked on an agarose gel. PCR was repeated four times per sample, replicates were pooled and sent for sequencing on a Illumina MiSeq platform at the Génome Québec Innovation Center at the McGill University (Montréal, Canada). There, barcodes were added to the PCR products using Fluidigm Access Array technology to allow multiplex sequencing. Subsequently, PCR products were purified with AMPure XP beads (Beckman Coulter, Brea, CA, USA), and pair-end sequencing was performed using Illumina MiSeq v3 (Illumina Inc., San Diego, CA, USA). Raw sequences were deposited in the NCBI SRA database with the accession number PRJNA386024.
ITS2 sequence of the applied strain
The sequence of the ITS2 region of M.brunneum ART2825 was determined with Sanger sequencing using the primer pair ITS3/ITS4 lacking adapter sequences CS1 and CS2 and the BigDye® Terminator v3.1 cycle sequencing kit (Applied Biosystems). Sequences were visualized using a capillary electrophoresis device (ABI 3130xl Genetic Analyzer, Applied Biosystems) and assembled using DNA Baser 3.4.5 (Heracle BioSoft, Pitesti, Romania).
Sequence processing and taxonomic classification
Sequences were processed and classified using a customized pipeline (Frey et al.2016) mostly based on UPARSE within USEARCH v8 (Edgar 2010, 2013). Overlapping paired-end reads were merged using fastq_mergepairs (Edgar and Flyvbjerg 2015) with a minimal overlap of 50 bp and a minimal merge length of 150 bp for fungal and 300 bp for prokaryotic sequences. Substitution errors were removed using the BayesHammer algorithm implemented in SPAdes 3.5 (Nikolenko, Korobeynikov and Alekseyev 2013; Nurk et al.2013) and primers were removed with Cutadapt 1.8.1 allowing one mismatch (Martin 2011). Quality control was performed using fastq_filter in USEARCH discarding reads with expected total error greater than one (Edgar and Flyvbjerg 2015). Dereplication and clustering into OTUs with 97% identity was performed using derep_fulllength and cluster_otus within USEARCH with concurrent removal of singletons and chimera (Edgar 2013). The eukaryotic or prokaryotic centroids were searched for ribosomal signatures with ITSx (Bengtsson-Palme et al.2013) or Metaxa2 (Bengtsson-Palme et al.2015), respectively, and only sequences which included these signatures were kept in the dataset. The algorithm usearch_global was used to map sequences to the centroids (maxdiffs 0, maxaccepts 0, top_hit_only). Eukaryotic sequences were compared to a custom-made NCBI Genbank database (Benson et al.2015) and the UNITE database (Abarenkov et al.2010) for taxonomic classification using the naïve Bayesian classifier implemented in MOTHUR v.1.35.1 (Schloss et al.2009). Sequences that were assigned as Metazoa, Viridiplantae, Protista and unclassified were removed from the dataset. The GREENGENES database (DeSantis et al.2006; McDonald et al.2012) was used for taxonomic classification of prokaryota. Subsequently, only archaeal and bacterial sequences were kept in the dataset.
Statistical analyses
The abundance of Metarhizium spp. was assessed by counting CFU and calculating CFU g−1 soil dry weight. Metarhizium CFU g−1 soil dry weight in three replicates per soil sample were averaged using the median per sample. Significance of differences was assessed with a Kruskal–Wallis rank sum test (Hollander and Wolfe 1973) followed by Dunn's Kruskal-Wallis multiple comparison test in the FSA package (Dunn 1964; Ogle 2016) with Benjamini-Hochberg (BH) P-value adjustment (Benjamini and Hochberg 1995) implemented in R version 3.3.0 used with Rstudio version 0.98.994 (R-Development-Core-Team 2008; RStudio-Team 2015). Correlations were calculated using the Pearson correlation coefficient in R. Efficacy of the treatments in the pot experiment was determined by calculating the % control based on percentage of undamaged potato tubers compared to the control (Abbott 1987). Saturation of sequencing was checked using intrasample rarefaction curve analysis (rarefaction.single in MOTHUR) with a re-sampling without replacement approach and plotted in R. Observed OTU richness and the inverse Simpson index representing effective number of species of soil fungal and prokaryotic communities were calculated with ‘summary.single’ in MOTHUR (Simpson 1949; Jost 2006). This includes an iterative subsampling procedure (9999 times) to the sampling depth of the sample with the fewest sequences (pot experiment: 7425 fungal and 9088 prokaryotic sequences, field experiment: 2101 fungal and 10 896 prokaryotic sequences). Dissimilarities in the fungal or prokaryotic communities between pairs of samples were assessed using Bray-Curtis (BC) dissimilarity matrices with iterative subsampling (9999) which were calculated with dist.shared in MOTHUR. Significance of differences of the fungal and prokaryotic communities among treatments and sampling time points was assessed with overall and pairwise ANOSIM (Spearman rank correlation and 9999 iterations) based on BC dissimilarities implemented in PRIMER v7 (Clarke 1993; Clarke and Gorley 2015) and with overall PERMANOVA based on BC dissimilarities using the function adonis within the R package vegan (Oksanen et al.2016) followed by assessment of pairwise differences using the function pairwise.perm.manova within the R package RVAideMemoire (Hervé 2017). Unconstrained ordinations were determined in R using non-metric multidimensional scaling (NMDS) based on BC dissimilarities with the function metaMDS within the R package vegan (Faith, Minchin and Belbin 1987; Minchin 1987; Oksanen et al.2016). Significant differences of relative sequence abundance of each OTU across sampling time points, treatments and interactions among sampling time points and treatments were assessed with PERMANOVA based on Euclidian distance using the function adonis followed by BH P-value correction. Pairwise differences were calculated for each OTU with a significant overall PERMANOVA pseudo F-statistic per sampling time point using the R function pairwise.perm.manova. In addition, the contribution of single OTUs to BC dissimilarities was calculated using the SIMPER (similarity percentage) routine in Primer 7v (Clarke 1993) with the 100 most abundant OTUs (relative abundance and square root transformation) per sampling time point. Only significant pairwise comparisons of a treatment and the control assessed with pairwise ANOSIM and pairwise PERMANOVA were selected for the SIMPER analyses.
RESULTS
Abundance of the applied Metarhizium strain and efficacy of biocontrol treatments in pots
The abundance of Metarhizium spp. increased significantly in all fungus-treated pots from a median of 56–144 CFU g−1 soil dry weight before application to 5569–17 596 CFU g−1 soil dry weight in BCA-treated pots at week 7 and remained high until the end of the experiment (Fig. 1A). In contrast, the abundance of Metarhizium spp. in pots not treated with the fungus remained low with a median of 0–153 CFU during the entire experiment.
Figure 1.
Abundance of Metarhizium spp. CFU g−1 soil dry weight per treatment and sampling time in the pot (A) and the field (B) experiment and abundance of the OTU including the sequence of the applied strain per treatment and sampling time in the pot (C) and field experiment (D). Asterisk indicates significant differences to untreated control at the corresponding sampling time point (n = 6; P ≤ 0.05).
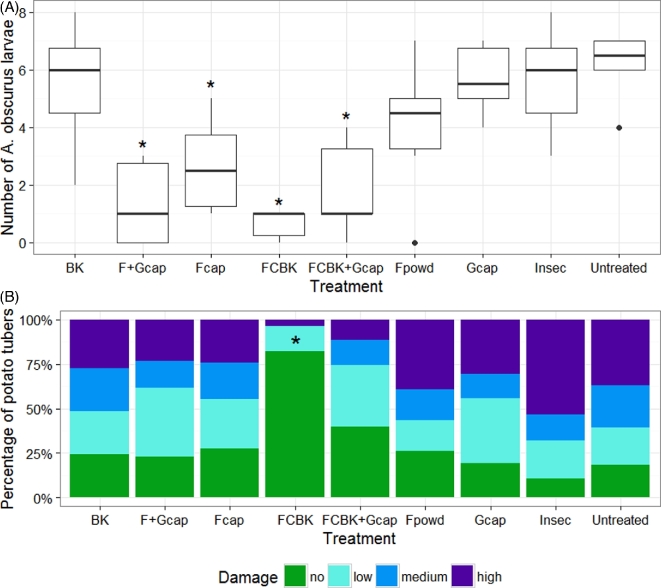
SSR marker-based genotyping revealed that 92% of the isolates (n = 39) selected from soil of fungus-treated pots after applications had the genotype of the applied strain (Table S1, Supporting Information). The applied strain was already detected in pot substrates before treatment (week 0), but only one out of five isolates from control pots and two out of six isolates from pots treated with Fcap + Gcap revealed the genotype of the applied strain. The abundance of Metarhizium spp. in these pots was 57 and 141 CFU g−1 soil dry weight. Except for Fpowd-treated pots, significantly fewer Agriotesobscurus larvae were retrieved from fungus-treated pots, as compared to the untreated controls at the end of the experiment (Fig. 2A). A median re-capture rate of six A. obscurus larvae out of ten released ones in the untreated pots was within the range of what was observed in previous experiments (unpublished data). The lowest number of A. obscurus larvae was found in the FCBK-treated pots with a median of one larva per pot. From 18 mycosed A. obscurus cadavers obtained from pots treated with FCBK, Fcap, Fpowd, FCBK + Gcap and Fcap + Gcap 82.4% were infected by the applied strain (Table S1; for information on treatments, see Table 1). One A. obscurus larva originating from an Insec-treated pot was also infected with the applied strain. Treatments with Gcap, BK or Insec did not result in decreased numbers of A. obscurus larvae and the combinations of FCBK or Fcap with Gcap did not enhance efficacy of treatments (no further decrease in the number of A. obscurus larvae). The number of A. obscurus larvae was moderately and significantly correlated with the percentage of damaged potatoes (r = 0.46, P < 0.001). The mean percentage of undamaged potato tubers ranged from 10% to 81% (Fig. 2B). FCBK was the only treatment resulting in a significantly higher number of undamaged potato tubers as compared to the control (Fig. 2B) yielding an efficacy (undamaged potatoes compared to the control) of 77%. There were no significant differences in percentages of low, medium or highly damaged potato tubers among treatments. The combined treatments of fungus and garlic, Gcap, BK and Insec did not exhibit an effect on potato tuber damage.
Figure 2.
Number of A. obscurus larvae per treatment retrieved from pots initially receiving 10 larvae (A) and levels of potato tubers damage in percentage of total number of tubers harvested per treatment (B): no (0 holes per tuber; SE = 7%–13%), low (1–2 holes per tuber; SE = 4%–10%), medium (3–5 holes per tuber; SE = 0%–10%), high (>4 holes per tuber; SE = 4%–14%), n = 2 to 10 potato tubers per pot. Asterisk indicates significant differences to untreated control (P ≤ 0.05).
Abundance of the applied Metarhizium strain and efficacy of biocontrol treatments in the field
Abundance of Metarhizium spp. increased after the application of FCBK from a median of 1304 to 6969 CFU g−1 soil dry weight 9 weeks after application and slightly decreased to 4261 CFU g−1 soil dry weight at week 16 (Fig. 1B). The application of Fgran or Fcap did not yield significantly increased Metarhizium abundances. The applied strain was not detected in any of the treatments at week 0 as shown by SSR-based genotyping (Table S1). In all fungus-treated field plots, 36% of the isolates (n = 36) had the genotype of the applied strain; however, after the application of FCBK 91.7% of the isolates (n = 12) were identified as the applied strain, whereas after the application of Fcap and Fgran only 0 and 16.7% of the isolates (n = 12) had the genotype of the applied strain. Of the 50 potato tubers analyzed per plot, a median of 42% to 51% was damaged and the number of damaged potato tubers did not differ among treatments (Fig. S2, Supporting Information).
Soil microbial communities of the pots
After quality filtering, a mean of 22 406 ± 8505 fungal and 19 706 ± 3418 prokaryotic sequences per sample was obtained in the pot experiment and clustered into a mean of 433 ± 51 fungal and 2795 ± 245 prokaryotic OTUs per sample, respectively. Rarefaction curve analysis revealed that fungi were sampled more exhaustively but with a higher variation among samples than prokaryota (Fig. S3A and B, Supporting Information). The fungal community across all pots was dominated by Ascomycota (84%) followed by Zygomycota (7%), Basidiomycota (6%), Chytridiomycota (1%), Glomeromycota (0.4%) and Blastocladiomycota (0.004%), besides unclassified fungi (0.01%; Fig. S3C). A total of 45 bacterial and three archaeal phyla were detected across all pots. The most abundant phyla (>10%) were Proteobacteria (22%), Actinobacteria (20%), Chloroflexi (14%) and Acidobacteria (13%; Fig. S3D).
Abundance of the applied strain and effects of treatments on soil microorganisms in pots
Within the fungal sequence dataset of the pot experiment, two OTUs were assigned to the genus Metarhizium. OTU 3 and OTU 1703 were classified as M.brunneum and M.flavoviride var. flavoviride W. Gams & Rozsypal with a sequence abundance of 87 207 and 14, respectively. OTU 3 occurred in 158 of the 162 samples and included 2641 unique sequences. The most abundant unique sequence (33 693 sequences) exactly matched the ITS2 sequence of the applied strain (GenBank Acc. N. KY786031). The abundance of OTU 3 was significantly increased in all M.brunneum-treated pots at week 7 and 15, and this increase correlated with the increase of Metarhizium spp. CFU g−1 soil dry weight (r = 0.65, n = 162, P < 0.001; Fig. 1A and C). OTU 3 was removed from the fungal dataset in order to avoid analytical bias of the abundance of OTU 3 on statistical tests used to assess treatment effects on the community structure of soil fungi.
OTU richness of the fungal communities did not differ among treatments at week 0 and at week 15. However, at week 7, OTU richness was significantly lower in BK and the FCBK-treated pots as compared to the untreated pots at week 7 (Fig. S4A, Supporting Information). No significant differences in OTU richness were observed among prokaryotic communities of the different treatments compared to untreated pots at the respective sampling time points (Fig. S4B). Similar results were obtained using the inverse Simpson index (data not shown). Overall ANOSIM analyses based on BC dissimilarities of the fungal communities across all pots revealed no differences among treatments at week 0; however, small but significant differences were detected at weeks 7 and 15 (Table 2). Pairwise ANOSIM tests of treatments compared to untreated pots and the NMDS analyses revealed that the fungal communities were moderately affected (R > 0.4) by the addition of BK, FCBK and Fcap at week 7 (Table S2, Supporting Information; Fig. 3A). Also, the fungal communities in these three treatment groups differed among each other (mean pairwise ANOSIM R-value of 0.62 ± 0.09) at week 7 (Table S2). At week 15, the fungal communities in pots treated with BK, FCBK, FCBK + Gcap, Fcap and Fcap + Gcap differed significantly from the untreated pots and among each other (mean pairwise ANOSIM R-value of 0.41 ± 0.09; Table S2). While prokaryotic communities in the pots did not differ among the treatments at week 0, small changes were detected at week 7 and week 15 (Table 2). Pairwise comparisons of the prokaryotic communities of different treatments compared to untreated pots at the respective sampling time point and the NMDS plot revealed that all treatments including garlic (Gcap, FCBK + Gcap, Fcap + Gcap) affected the prokaryotic communities at week 7 and 15 (Table S2, Fig. 3B). However, there were no differences in pairwise ANOSIM
Table 2.
Differences in the fungal and the prokaryotic community structures among treatments at different sampling time points (n = 6), treatments with (n = 18) and without garlic (n = 36), of untreated pots or plots (n = 6) over time and among blocks across the long side of the field (n = 9) in the pot and/or field experiment assessed with overall ANOSIM (analysis of similarity) based on Bray Curtis dissimilarity.
| Fungi | Prokaryota | ||
|---|---|---|---|
| Experiment | Overall test | ANOSIM R | ANOSIM R |
| Pot | Among treatments at week 0 | 0.03 | 0.08** |
| Pot | Among treatments at week 7 | 0.31*** | 0.38*** |
| Pot | Among treatments at week 15 | 0.26*** | 0.29*** |
| Pot | Untreated over time | 0.22** | 0.63*** |
| Field | Among treatments at week 0 | –0.1 | –0.09 |
| Field | Among treatments at week 9 | 0.04 | –0.05 |
| Field | Among treatments at week 16 | 0.02 | –0.08 |
| Field | Untreated over time | 0.12* | –0.04 |
| Field | Among 10 blocks along the field | 0.49*** | 0.61*** |
* P ≤ 0.05
* * P ≤ 0.01
* * * P ≤ 0.001
NA , not assessed.
Figure 3.
Unconstrained ordination of soil samples based on Bray-Curtis dissimilarities of fungal communities per treatment at week 7 in the pot experiment (A, stress = 0.14), of prokaryotic communities for treatments with and without garlic at week 7 and 15 in the pot experiment (B, stress = 0.2), of fungal (C, stress = 0.11) and prokaryotic (D, stress = 0.13) communities in the untreated pots at different sampling time points and of fungal (E, stress = 0.2) and prokaryotic (F, stress = 0.09) communities along a spatial gradient across the long side of the field.
comparisons among the three garlic treatments (Table S2). Corresponding results were obtained for overall and pairwise analyses of fungal and prokaryotic communities using PERMANOVA (Table S3, Supporting Information).
Assessing differences in relative sequence abundance of each fungal OTU among treatments revealed only 0.2% (7) of the fungal OTUs with a significant overall PERMANOVA pseudo F-value for the factor treatment. The relative abundance of five of these seven fungal OTUs changed significantly between untreated and either BK, FCBK, Fcap, FCBK + Gcap or Fcap + Gcap assessed with pairwise PERMANOVA (Fig. 4). Similarity percentage analyses (SIMPER) based on BC dissimilarities were performed to identify fungal and prokaryotic OTUs contributing to differences of microbial community structures of single treatments and untreated control pots with a significant pairwise comparison assessed with ANOSIM and PERMANOVA (Tables S4 and S5, Supporting Information). Data from PERMANOVA and SIMPER analyses revealed that fungal OTU 1, which was classified as member of the family Bionectriaceae, increased significantly in the FCBK-treated pots at week 7 and in the FCBK and FCBK + Gcap-treated pots at week 15 (Fig. 4) and contributed 12.3% and 10.6% to the differences between FCBK-treated pots and untreated pots at week 7 and 15, and 2% to the differences between FCBK + Gcap -treated and untreated pots at week 15 (Table S4). Fungal OTU 11, classified as Rhizopus oryzae, increased significantly in the BK-treated pots at week 7 and in the BK and Fcap + Gcap-treated pots at week 15 (Fig. 4) and accounted for 12% and 9.6% of the differences between BK-treated and untreated pots at week 7 and week 15 and 0.8% of the differences between Fcap + Gcap-treated and untreated pots at week 15. Fungal OTU 13, which was identified as member of the family Nectriaceae, increased significantly in FCBK + Gcap-treated pots at week 15 (Fig. 4) and contributed 2.8% to the differences between FCBK + Gcap -treated and untreated pots at week 15. Fungal OTU 45, identified as Mortierella spp., increased significantly in Fcap-treated pots at week 7 (Fig. 4) and contributed 1.5% to the differences between Fcap-treated and untreated pots at week 7 (Table S4). The unclassified fungal OTU 291 increased significantly in FCBK, FCBK + Gcap and Gcap-treated pots at week 7 (Fig. 4); however, it was not among the 100 most abundant OTUs which were used for the SIMPER analyses.
Figure 4.
Relative sequence abundance of fungal OTUs among different treatments and time points (n = 6). Asterisk indicates a significant difference between treatments and untreated pots at the respective sampling time point (P < 0.05). OTU 1, OTU 11, OTU 13 and OTU 45 were classified as Bionectriaceae, R.oryzae, Nectriaceae and Mortierella spp., respectively. OTU 291 was an unclassified fungal OTU.
Overall, PERMANOVA of relative sequence abundance per OTU revealed that 0.46% (44) of the prokaryotic OTUs were significantly affected by treatments (data not shown) and of these 36 were significantly different between any treatment and untreated pots. None of these 36 OTUs were significantly different between untreated and FCBK or Fpowd-treated pots, two changed significantly in pots treated with Fcap, one changed significantly after the addition of Insec and 33 were significantly different between untreated and any treatment including garlic (Fig. S5, Supporting Information). Only 10 of the OTUs detected with PERMANOVA were among the 100 most abundant OTUs investigated with SIMPER analyses, and they contributed between 0.4% and 5.41% to the respective differences (Table S5).
Changes of the microbial communities over time in pots
OTU richness of fungal communities in the untreated pots did not change over time. In contrast, OTU richness of soil prokaryotic communities in the untreated pots increased significantly and continuously from week 0 to week 15 (Fig. S4A and B, Supporting Information). Similarly, a significant increase of prokaryotic OTU richness was observed in BK, Fcap + Gcap, FCBK + Gcap, Fpowd and Insec-treated pots and it tended to increase also in all other treatments (Fcap, FCBK and Gcap). Overall ANOSIM values, overall PERMANOVA and NMDS revealed that fungal and prokaryotic community structures of the untreated pots differed among the sampling time points (Table 2, Fig. 3C and D, Table S3). Fungal community structures in the untreated pots changed slightly but significantly between week 0 and 15 (pairwise ANOSIM R = 0.45, P = 0.006), and similar changes over time were observed in FCBK, BK, Fcap and Fcap + Gcap-treated pots (mean pairwise ANOSIM R-value of 0.42 ± 0.13; Table S2). The prokaryotic communities of the untreated pots underwent a continuous significant shift across the three sampling time points which was shown by NMDS (Fig. 3D) and pairwise ANOSIM comparisons of week 0 to 7 and 0 to 15 resulting in R-values of 0.65 and 0.43, respectively (Table S2). Corresponding significant changes over time of the prokaryotic community structures were observed in all treated pots (mean pairwise ANOSIM R-values of 0.63 ± 0.16 week 0 to 7 and 0.4 ± 0.14 week 7 to 15; Table S2). Assessing differences in relative sequence abundance of fungal and prokaryotic OTUs showed that 99 fungal and 776 prokaryotic OTUs were significantly affected by time (data not shown).
Soil microbial communities in the field
A mean of 19 610 ± 12 252 fungal sequences per sample were obtained for 89 field samples (excluding one sample with a sequence abundance of only 360) and clustered into a mean of 435 ± 98 OTUs per sample. The 90 field samples included a mean of 17 322 ± 2437 prokaryotic sequences which were clustered into 1767 ± 132 OTUs per sample. Rarefaction analyses revealed that sampling the fungal diversity was closer to saturation than the prokaryotic sampling; however, variation was lower among prokaryotic samples (Fig. S6A and B, Supporting Information). The following six fungal phyla were detected in descending abundance in the soil of the field experiment: Ascomycota (79%), Basidiomycota (11%), Zygomycota (4%), Chytridiomycota (1%), Glomeromycota (0.7%) and Blastocladiomycota (0.03%) with 0.2% unclassified fungal sequences (Fig. S6C). Forty-five bacterial phyla were detected across the field samples. Bacterial phyla with an abundance of at least 10% comprised Proteobacteria (23%), Actinobacteria (17%), Chloroflexi (11%), Verrucomicrobia (11%) and Planctomycetes (11%) (Fig. S6D). The archaeal phylum Crenarchaeota (3%) was the only one of three archaeal phyla representing more than 1% prokaryotic sequences.
Abundance of the applied strain and effects of treatments on microbial communities in the field
Three OTUs were classified as Metarhizium within the fungal sequence dataset of the field samples. OTU 1 (including 73 521 sequences), OTU 2930 (including 3 sequences) and OTU 2732 (including 4 sequences) were assigned to M. brunneum, M. anisopliae and Metarhizium spp., respectively. OTU 1 included 4871 unique sequences, and the unique sequence which exactly matched the ITS2 region of the applied strain was detected 6735 times (data not shown). The relative abundance of OTU 1 was significantly higher in FCBK-treated field plots 9 and 16 weeks after the treatment (Fig. 1D). None of the other treatments resulted in increased OTU 1 abundance. There was a positive correlation between the relative abundance of OTU 1 and the number of Metarhizium spp. CFU g−1 soil dry weight (r = 0.66, n = 90, P < 0.001). OTU 1 was deleted from the fungal dataset in order to avoid analytical bias on statistical tests when assessing changes in fungal communities. There were no significant differences in OTU richness or the inverse Simpson index of the fungal and prokaryotic communities among treatments at different sampling time point (data not shown). Overall ANOSIM and pairwise PERMANOVA showed that neither fungal nor prokaryotic communities in the field were affected by the treatments compared to the untreated plots at the respective sampling time points (Table 2, Table S6, Supporting Information).
Changes of the microbial communities over time and space in the field
Fungal and prokaryotic communities in the untreated plots did not differ in their OTU richness and inverse Simpson index (data not shown), and in their community structures based on BC dissimilarities over time assessed with ANOSIM (Table 2). However, community structure analyses based on BC dissimilarities assessed with overall PERMANOVA revealed a significant time effect on fungal and prokaryotic communities (Table S6). ANOSIM and PERMANOVA analyses revealed significant spatial effects (Table 2, Table S6). Fungal and prokaryotic communities both changed gradually from one end to the middle of the field (about 45 m) and became similar again towards the other end of the field (Fig. 3E and F). Fungal OTU richness differed significantly between the middle (36 and 45 m) and the end of the field (72 and 81 m, Fig. S4C). The community structure (based on BC dissimilarities and visualized by NMDS) of fungal communities differed among blocks (including three plots each) along the long side of the field, i.e. among blocks from the middle section of the field compared to blocks from both ends (Table 2, Fig. 3E, Table S2). Corresponding spatial changes were also detected for the prokaryotic community structures (Table 2, Fig. 3F; Fig. S4D, Table S2).
DISCUSSION
Risk assessment of any environmental hazard, i.e. an agent or activity causing a hazard, includes the assessment of exposure to the hazard and effects on the population or individual exposed to the hazard (Brown 1985; U.S. Interagency Staff Group on Carcinogenesis 1986). In this study, exposure was defined as a significant increase of Metarhiziumbrunneum ART2825 abundance. Exposure analysis was performed with a cultivation-dependent approach (i.e. determination of Metarhizium spp. CFU followed by identification of the genotype of the applied strain) and with a cultivation-independent approach (i.e. assessment of the OTU of the applied strain within the amplicon sequences). With both approaches, significant exposure to the applied fungal strain was demonstrated both in the pot experiment and in FCBK-treated field plots. Isolates of the genotype of M. brunneum ART2825 were detected at low frequency (6%) in pots before application (untreated and Fcap + Gcap) and isolated from a larvae from an Insec-treated pot and very likely represent natural occurrence of the strain, since the soil used in the pot experiment originated from a field at Agroscope Reckenholz where M. brunneum ART2825 has originally been isolated from an Agriotesobscurus larva (Kölliker, Biasio and Jossi 2011; Eckard et al.2014). Although the applied strain established in all fungal-treated pots and in FCBK-treated plots, the biocontrol effect was limited. Only the application of FCBK lead to a 77% efficacy (increase of undamaged potato tubers compared to the control) and a significant reduction of A. obscurus larvae in the pot experiment, which corroborated previous laboratory experiments (Kölliker, Biasio and Jossi 2011; Eckard et al.2014). The number of A. obscurus larvae was significantly reduced in Fcap, F + Gcap and FCBK + Gcap-treated pots compared to untreated pots; however, this did not result in reduced potato tuber damage. The inconsistent results of potato tuber damage and number of Agriotes larvae might result from feeding interruptions prior and post molting, which may be uncoordinated within a population (Furlan 1998, 2004; Sufyan, Neuhoff and Furlan 2014) and differences in foraging behavior of A. obscurus larvae possibly due to different volatile organic compounds (reviewed in Barsics et al.2014) emitted from treatments. In contrast to the pot experiment, no biocontrol success was achieved in the field in any of the treatments within one season of fungal applications. This might be explained by unfavorable conditions for the fungus possibly created by non-optimal soil moisture, soil texture, soil temperature or antagonistic microbes (Jaronski 2007). In addition, the applied strain may not be able to provide sufficient protection against all Agriotes species present in the field that have been shown to be difficult to control (Blackshaw and Vernon 2008; Sufyan, Neuhoff and Furlan 2013; Sufyan, Neuhoff and Furlan 2014). In other field studies using Metarhizium spp. to control Agriotes larvae, varying degrees of success have been reported (Kabaluk et al.2005; Ritter, Katroschan and Richter 2011). For instance, M. brunneum ART2825 formulated as FCBK was applied to protect lettuce from A. sputator and A. ustulatus and showed 21% and 65% reduction of the two pest insects, respectively (Ritter, Katroschan and Richter 2011). However, insignificant reduction in potato tuber damage was detected after the application of M. anisopliae granules (Kabaluk et al.2005). In the pot trial of this study, the combined treatments of M. brunneum ART2825 and garlic capsules (FCBK + Gcap, Fcap + Gcap) did not enhance efficacy, an observation which was also made in a laboratory experiment using two-dimensional terraria (Eckard et al.2017). One reason for insufficient control of Agriotes larvae may be a repelling effect of Metarhizium spp. on Agriotes spp. (Kabaluk et al.2005). The use of attractants such as CO2-emitting capsules or pheromone pitfalls, as tested in other studies, may help to overcome possible repelling effects (Kabaluk, Lafontaine and Borden 2015; Brandl et al.2017). The application of the insecticide clothianidin was neither successful in the pot experiment nor in the field experiment. This is in accordance with results obtained from bioassay experiments, where A. obscurus larvae have been exposed to clothianidin-treated wheat seedlings (van Herk et al.2008). In this bioassay, over 70% of the larvae were moribund following a similar insecticide treatment but most recovered 14 days after application. However, even though efficacy of the treatments was limited in our study, criteria for exposure were nevertheless achieved and allowed an assessment of effects of M. brunneum ART2825 on soil microorganisms in the pot and in the field experiment.
Application of M. brunneum ART2825 formulated as FCBK and Fcap (but not the application of fungal spore powder alone) resulted in slight changes of the fungal communities in the pot trial, suggesting that the observed effects on microbial communities were caused by compounds of the formulations rather than by the fungus itself. These small changes between untreated pots and FCBK-treated pots were reflected in a significant increase of only two OTUs which were classified as a member of Nectriaceae and an unclassified fungus. The taxonomic classification of these OTUs allowed very limited assumptions of their functions and possible interactions with the applied strain. The changes between untreated pots and pots treated with Fcap were reflected in a significant increase of only one OTU classified as Mortierella spp. which are known for their saprophytic life style. This fungus may profit from the alginate carrier; however, it increased with a strong variation as observed in most treatments and sampling time points. In a similar pot experiment, aimed at controlling Diabrotica v. virgifera LeConte, the application of FCBK and similar fungal capsules did not affect the fungal communities (J. Mayerhofer in preparation). This may suggest that the impact of FCBK and Fcap application on fungal communities is context dependent involving also soil specific or environmental factors. Application of FCBK had also no effect on fungal and prokaryotic communities in the field, and all fungal treatments had no effect on prokaryotic communities in the pot experiment. Our results are in agreement with other studies involving entomopathogenic fungi, e.g. M.anisopliae or Beauveria bassiana (Bals.-Criv.) Vuill., which detected no, small or only transient effects on soil microorganisms (Hu and St Leger 2002; Rai and Singh 2002; Kirchmair et al.2008; Schwarzenbach, Enkerli and Widmer 2009; Hirsch et al.2013). Likewise, the release of other microorganisms for control of phytopathogens, weeds or nematodes resulted in small or transient effects on soil microbial communities (Grosch et al.2006; Rousidou et al.2013; Zimmermann et al.2016). Furthermore, microorganisms released as biofertilizers, phytostimulators or plant growth promotors had no effects on bacterial communities in the rhizosphere (Lerner et al.2006; García de Salamone et al.2010; Kröber et al.2014) or only moderate effects on bacteria and fungi in the rhizosphere or the bulk soil (van Dillewijn, Villadas and Toro 2002; Trabelsi et al.2011; Schmidt et al.2012).
The untreated control pots and field plots allowed to assess changes of the microbial communities over time including seasonal and environmental changes, and effects caused by the plants on the resident microbial soil communities. In our study, time-related effects on the fungal and the prokaryotic soil communities in the pot experiment were similar or greater than the treatment effects. This is in agreement with other studies showing that seasonal changes of soil microbial composition in relation to developmental stage of the plant exceed treatment effects of applied fungi and bacteria in bulk soil (Savazzini, Longa and Pertot 2009) as well as in the rhizosphere in the field (van Dillewijn, Villadas and Toro 2002; Grosch et al.2006; Zimmermann et al.2016). In the field experiment of our study, the assessment of temporal changes using ANOSIM and overall PERMANOVA showed contradicting results which may have resulted from different sensitivities of the tests. However, the fungal and prokaryotic community structures varied spatially across the field. We suspect that this variation may be related to differences in edaphic factors across the field. Humus, clay and silt content as well as soil pH were assessed, but results did not yield sufficient resolution to support this hypothesis. In other studies, soil edaphic factors including pH, organic carbon, texture, soil moisture and land management have been shown to influence soil microorganism at the agricultural plot scale (Chen et al.2007; Philippot et al.2009; Rousk et al.2010; Naveed et al.2016).
Entomopathogenic fungi are formulated for applications in order to increase persistence, efficacy or shelf life of the fungi (Burges 1998). Metarhiziumbrunneum ART2825 was applied in form of FCBK, Fcap and Fpowd in the pot experiment. The addition of BK, the non-fungal component of the FCBK, also affected fungal communities. These effects were mainly due to an increased abundance of Rhizopus oryzae, a well-known degrader of organic matter. The increase of the relative sequence abundance of R.oryzae varied among replicates as shown by a large dispersion which may indicate that the response of soil microbial communities was pot specific over time and may indicate the introduction of responsive microorganism by the addition of potato tubers. Surprisingly, R.oryzae was not enhanced in the FCBK-treated pots. Possibly, because the niche ‘BK’ was already occupied by the applied strain preventing R. oryzae to colonize this nutrient source. The results of this study suggest that the formulations may have been responsible for these effects. Similarly, the effects caused by a biological nematicide containing the fungus Paecilomyces lilacinus (Thom) Samson formulated with glucose and skimmed milk were triggered by the formulation only (Rousidou et al.2013). The prokaryotic communities reacted to the application of Gcap and the combinations of FCBK + Gcap and Fcap + Gcap in a very similar way but not after application of fungal products only, suggesting that the observed effects were due to the application of Gcap. The effects of Gcap on soil prokaryotes resulted either from garlic oil, parts of the formulation (not studied separately) or the combination of both. Garlic has been used traditionally as an antimicrobial agent in medicine and for human consumption, but more recently also to protect plants against soil-borne fungal and bacterial diseases (Lawson 1998; Curtis et al.2004; Sealy, Evans and Rothrock 2007). A possible mechanism explaining the effect of garlic may be interference with quorum sensing, a common regulatory process between bacterial cells coupling gene expression to cell density as suggested by others (Gonzalez and Keshavan 2006; Bodini et al.2009; Dessaux, Chapelle and Faure 2011).
The systemic neonicotinoid clothianidin used in our study did not affect the fungal and the prokaryotic soil community structures, both in the pot and in the field experiment. However, the concentration of clothianidin was not monitored and therefore exposure to the compound was not confirmed. It is possible that the insecticide has been partially or completely degraded before effects became manifest although half-life of this chemical in soil is supposed to range between 20 and 1000 weeks (Simon-Delso et al.2015). Pesticide-degrading microorganisms can reduce the clothianidin concentration as it is degradable aerobically and anaerobically by microbes (Mulligan et al.2016). Moreover, studies with other systemic neonicotinoids have documented effects on soil fungal and bacterial communities, confirming that clothianidin can potentially have adverse effects on microbial communities (Singh and Singh 2005; Cai et al.2015, 2016; Zaller et al.2016).
The release of microorganisms to soil for pest control offers great potential and benefits for agriculture. Particularly, entomopathogenic fungi provide an alternative to chemical pesticides or may allow to reduce application of such chemicals and their release to the environment. Registration of entomopathogenic fungi for pest control requires knowledge on possible effects on soil microbial communities. This study showed that M.brunneum ART2825 formulated as FCBK and Fcap, in contrast to the application of fungal spores only, can cause small changes in fungal communities. However, changes were in the same range or even smaller than changes caused by BK (the non-fungal compound of the formulation FCBK), or natural fluctuations in community structures. Amplicon sequencing proved to be a powerful tool for simultaneously assessing exposure to the released strain and effects on the community structure of soil microorganisms. Future investigation should focus on specific functional groups (such as Rhizobia, or mycorrhizal fungi) or use meta-proteomics or transcriptomics approaches to assess possible effects at the functional level. This may provide complementary knowledge on the effects of BCAs on microbial communities.
Supplementary Material
Acknowledgements
We would like to thank Christian Schweizer and Christian Gees for support in sampling and sample preparation. The authors declare no conflict of interest.
FUNDING
This study was funded by means of the 7th Framework Programme of the European Union in the frame of the EU-project “Innovative Biological products for Soil pest control” [INBIOSOIL Grant Agreement No. 282767].
Conflict of Interest. None declared.
REFERENCES
- Abarenkov K, Henrik Nilsson R, Larsson K-H. et al. The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytol 2010;186:281–5. [DOI] [PubMed] [Google Scholar]
- Abbott WS. Abbotts formula - a method of computing the effectiveness of an insecticide. J Am Mosquito Contr 1987;3:302–3. [PubMed] [Google Scholar]
- Anonymous Wireworms. EPPO Bulletin 2005;35:179–82. [Google Scholar]
- Ansari MA, Shah FA, Butt TM. The entomopathogenic nematode Steinernema kraussei and Metarhizium anisopliae work synergistically in controlling overwintering larvae of the black vine weevil, Otiorhynchus sulcatus, in strawberry growbags. Biocontrol Sci Technol 2010;20:99–105. [Google Scholar]
- Aregger E. Conidia production of the fungus Beauveria brongniartii on barley and quality evaluation during storage at 2°C. J Invertebr Pathol 1992;59:2–10. [Google Scholar]
- Barsics F, Haubruge E, Francis F. et al. The role of olfaction in wireworms: a review on their foraging behavior and sensory apparatus. Biotechnol Agron Soc 2014;18:524–35. [Google Scholar]
- Behle RW, Jackson MA, Flor-Weiler LB. Efficacy of a granular formulation containing Metarhizium brunneum F52 (Hypocreales: Clavicipitaceae) microsclerotia against nymphs of Ixodes scapularis (Acari: Ixoididae). J Econ Entomol 2013;106:57–63. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Hartmann M, Eriksson KM. et al. METAXA2: Improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Resour 2015;15:1403–14. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Ryberg M, Hartmann M. et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 2013;4:914–9. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 1995;57:289–300. [Google Scholar]
- Benson DA, Clark K, Karsch-Mizrachi I. et al. GenBank. Nucleic Acids Res 2015;43:D30–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw RP, Vernon RS. Spatial relationships between two Agriotes click-beetle species and wireworms in agricultural fields. Agr For Entomol 2008;10:1–11. [Google Scholar]
- Blot Y, Brunel E. Survey on the infection of wheat and maize by larvae of wireworms of Agriotes and Athous genera (Coleoptera: Elateridae) in some areas of West France. Ann Soc Entomol Fr 1999;35:453–7. [Google Scholar]
- Bodini SF, Manfredini S, Epp M. et al. Quorum sensing inhibition activity of garlic extract and p-coumaric acid. Lett Appl Microbiol 2009;49:551–5. [DOI] [PubMed] [Google Scholar]
- Brandl MA, Schumann M, Przyklenk M. et al. Wireworm damage reduction in potatoes with an attract-and-kill strategy using Metarhizium brunneum. J Pest Sci 2017;90:479–93. [Google Scholar]
- Bridge P, Spooner B. Soil fungi: diversity and detection. Plant Soil 2001;232:147–54. [Google Scholar]
- Brown SL. Quantitative risk assessment of environmental hazards. Annu Rev Public Health 1985;6:247–67. [DOI] [PubMed] [Google Scholar]
- Burges HD. Formulations of mycoinsecticides. In: Burges HD. (ed.) Formulation of Microbial Pesticides: Beneficial Microorganisms, Nematodes and Seed Treatments. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998;131–85. [Google Scholar]
- Bürgmann H, Pesaro M, Widmer F. et al. A strategy for optimizing quality and quantity of DNA extracted from soil. J Microbiol Methods 2001;45:7–20. [DOI] [PubMed] [Google Scholar]
- Cai Z, Rong Y, Chen J. et al. Effects of the novel cis-nitromethylene neonicotinoid insecticide Paichongding on enzyme activities and microorganisms in yellow loam and Huangshi soils. Environ Sci Pollut R 2016;23:7786–93. [DOI] [PubMed] [Google Scholar]
- Cai Z, Wang J, Ma J. et al. Anaerobic degradation pathway of the novel chiral insecticide paichongding and its impact on bacterial communities in soils. J Agr Food Chem 2015;63:7151–60. [DOI] [PubMed] [Google Scholar]
- Chen MM, Zhu YG, Su YH. et al. Effects of soil moisture and plant interactions on the soil microbial community structure. Eur J Soil Biol 2007;43:31–8. [Google Scholar]
- Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 1993;18:117–43. [Google Scholar]
- Clarke KR, Gorley RN. PRIMER v7: User Manual/Tutorial. Plymouth: PRIMER-E, 2015, 296. [Google Scholar]
- Curtis H, Noll U, Störmann J. et al. Broad-spectrum activity of the volatile phytoanticipin allicin in extracts of garlic (Allium sativum L.) against plant pathogenic bacteria, fungi and Oomycetes. Physiol Mol Plant Pathol 2004;65:79–89. [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb 2006;72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaux Y, Chapelle E, Faure D. Quorum sensing and quorum quenching in soil ecosystems. In: Witzany G. (ed.) Biocommunication in Soil Microorganisms. Berlin, Heidelberg: Springer Berlin Heidelberg, 2011, 339–67. [Google Scholar]
- Dunn OJ. Multiple comparisons using rank sums. Technometrics 1964;6:241–52. [Google Scholar]
- Eckard S, Ansari MA, Bacher S. et al. Virulence of in vivo and in vitro produced conidia of Metarhizium brunneum strains for control of wireworms. Crop Protect 2014;64:137–42. [Google Scholar]
- Eckard S, Bacher S, Enkerli J. et al. A simple in vitro method to study interactions between soil insects, entomopathogenic fungi, and plant extracts. Entomol Exp Appl 2017;163:315–27. [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 2013;10:996–8. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Flyvbjerg H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015;31:3476–82. [DOI] [PubMed] [Google Scholar]
- Faith DP, Minchin PR, Belbin L. Compositional dissimilarity as a robust measure of ecological distance. Vegetatio 1987;69:57–68. [Google Scholar]
- Faria MRd, Wraight SP. Mycoinsecticides and Mycoacaricides: A comprehensive list with worldwide coverage and international classification of formulation types. Biol Control 2007;43:237–56. [Google Scholar]
- Frey B, Rime T, Phillips M. et al. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol Ecol 2016;92. [DOI] [PubMed] [Google Scholar]
- Furlan L. The biology of Agriotes ustulatus Schäller (Col., Elateridae). II. Larval development, pupation, whole cycle description and practical implications. J Appl Entomol 1998;122:71–8. [Google Scholar]
- Furlan L. The biology of Agriotes sordidus Illiger (Col. Elateridae). J Appl Entomol 2004;128:696–706. [Google Scholar]
- García de Salamone IE, Di Salvo LP, Escobar Ortega JS. et al. Field response of rice paddy crop to Azospirillum inoculation: physiology of rhizosphere bacterial communities and the genetic diversity of endophytic bacteria in different parts of the plants. Plant Soil 2010;336:351–62. [Google Scholar]
- Glare TR, Moran-Diez ME. Microbial-Based Biopesticides - Methods and Protocols. New York: Springer Protocols, 2016. [DOI] [PubMed] [Google Scholar]
- Gonzalez JE, Keshavan ND. Messing with bacterial quorum sensing. Microbiol Mol Biol R 2006;70:859–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosch R, Scherwinski K, Lottmann J. et al. Fungal antagonists of the plant pathogen Rhizoctonia solani: selection, control efficacy and influence on the indigenous microbial community. Mycol Res 2006;110:1464–74. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Frey B, Kölliker R. et al. Semi-automated genetic analyses of soil microbial communities: comparison of T-RFLP and RISA based on descriptive and discriminative statistical approaches. J Microbiol Methods 2005;61:249–360. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Niklaus PA, Zimmermann S. et al. Resistance and resilience of the forest soil microbiome to logging-associated compaction. ISME J 2014;8:226–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé M. RVAideMemoire: Testing and Plotting Procedures for Biostatistics. R package version 0.9–66. 2017. https://CRAN.R-project.org/package=RVAideMemoire. [Google Scholar]
- Hirsch J, Galidevara S, Strohmeier S. et al. Effects on diversity of soil fungal community and fate of an artificially applied Beauveria bassiana strain assessed through 454 pyrosequencing. Microb Ecol 2013;66:608–20. [DOI] [PubMed] [Google Scholar]
- Hollander M, Wolfe DA. Nonparametric Statistical Methods. New York: John Wiley & Sons, 1973. [Google Scholar]
- Hu G, St Leger J. Field studies using a recombinant mycoinsecticide (Metarhizium anisopliae) reveal that it is rhizosphere competent. Appl Environ Microb 2002;68:6383–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert P, Przyklenk M, Vemmer M. et al. Calcium gluconate as cross-linker improves survival and shelf life of encapsulated and dried Metarhizium brunneum and Saccharomyces cerevisiae for the application as biological control agents. J Microencapsul 2017;34:47–56. [DOI] [PubMed] [Google Scholar]
- Jacobsen CS, Hjelmsø MH. Agricultural soils, pesticides and microbial diversity. Curr Opin Biotechnol 2014;27:15–20. [DOI] [PubMed] [Google Scholar]
- Jaronski ST. Soil ecology of the entomopathogenic Ascomycetes: a critical examination of what we (think) we know. In: Ekesi S, Mainiania NK (eds.) Use of Entomopathogenic Fungi in Biological Pest Management. Trivandrum India: Research Signpost, 2007, 91–143. [Google Scholar]
- Jaronski ST. Ecological factors in the inundative use of fungal entomopathogens. BioControl 2010;55:159–85. [Google Scholar]
- Jaronski ST, Jackson MA. Efficacy of Metarhizium anisopliae microsclerotial granules. Biocontrol Sci Technol 2008;18:849–63. [Google Scholar]
- Jost L. Entropy and diversity. Oikos 2006;113:363–75. [Google Scholar]
- Kabaluk TJ, Goettel MS, Erlandson MA. et al. Metarhizium anisopliae as a biological control for wireworms and a report of some other naturally-occurring parasites. IOBC/WPRS Bull 2005;28:109–15. [Google Scholar]
- Kabaluk TJ, Lafontaine JP, Borden JH. An attract and kill tactic for click beetles based on Metarhizium brunneum and a new formulation of sex pheromone. J Pest Sci 2015;88:707–16. [Google Scholar]
- Kennedy AC. Bacterial diversity in agroecosystems. Agr Ecosyst Environ 1999;74:65–76. [Google Scholar]
- Kepler RM, Humber RA, Bischoff JF. et al. Clarification of generic and species boundaries for Metarhizium and related fungi through multigene phylogenetics. Mycologia 2014;106:811–29. [DOI] [PubMed] [Google Scholar]
- Kirchmair M, Neuhauser S, Huber L. et al. The impact of soil treatment on soil mycobiota. IOBC/WPRS Bull 2008;31:239–44. [Google Scholar]
- Kölliker U, Biasio L, Jossi W. Potential control of Swiss wireworms with entomopathogenic fungi. IOBC/WPRS Bull 2011;66:517–20. [Google Scholar]
- Kölliker U, Jossi W, Kuske S. Optimised protocol for wireworm rearing. IOBC/WPRS Bull 2009;45:457–60. [Google Scholar]
- Krassilstschik J. La production industrielle des parasites végétaux pour la destruction des insectes nuisibles. Bull Biol Fr Belg 1888;19:461–72. [Google Scholar]
- Kröber M, Wibberg D, Grosch R. et al. Effect of the strain Bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing. Front Microbiol 2014;5, DOI: 10.3389/fmicb.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryavtsev I, Siirde K, LÄÄts K. et al. Determination of distribution of harmful click beetle species (Coleoptera, Elateridae) by synthetic sex pheromones. J Chem Ecol 1993;19:1607–11. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Ramirez KS, Aanderud Z. et al. Temporal variability in soil microbial communities across land-use types. ISME J 2013;7:1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson LD. Garlic: A Review of Its Medicinal Effects and Indicated Active Compounds. American Chemical Society, 1998. [Google Scholar]
- Lerner A, Herschkovitz Y, Baudoin E. et al. Effect of Azospirillum brasilense inoculation on rhizobacterial communities analyzed by denaturing gradient gel electrophoresis and automated ribosomal intergenic spacer analysis. Soil Biol Biochem 2006;38:1212–8. [Google Scholar]
- Lugtenberg B. Principles of Plant-Microbe Interactions: Microbes for Sustainable Agriculture. Cham: Springer International Publishing, 2015. [Google Scholar]
- McDonald D, Price MN, Goodrich J. et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 2011;17:10–2. [Google Scholar]
- Mayerhofer J, Lutz A, Widmer F. et al. Multiplexed microsatellite markers for seven Metarhizium species. J Invertebr Pathol 2015;132:132–4. [DOI] [PubMed] [Google Scholar]
- Miles HW. Wireworms and agriculture, with special reference to Agriotes obscurus L. Ann Appl Biol 1942;29:176–80. [Google Scholar]
- Minchin PR. An evaluation of the relative robustness of techniques for ecological ordination. Vegetatio 1987;69:89–107. [Google Scholar]
- Mulligan RA, Tomco PL, Howard MW. et al. Aerobic versus anaerobic microbial degradation of clothianidin under simulated california rice field conditions. J Agr Food Chem 2016;64:7059–67. [DOI] [PubMed] [Google Scholar]
- Nannipieri P, Ascher J, Ceccherini MT. et al. Microbial diversity and soil functions. Eur J Soil Sci 2003;54:655–70. [Google Scholar]
- Naveed M, Herath L, Moldrup P. et al. Spatial variability of microbial richness and diversity and relationships with soil organic carbon, texture and structure across an agricultural field. Appl Soil Ecol 2016;103:44–55. [Google Scholar]
- Nikolenko SI, Korobeynikov AI, Alekseyev MA. BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics 2013;14:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Bankevich A, Antipov D. et al. Assembling genomes and mini-metagenomes from highly chimeric reads. In: Deng M, Jiang R, Sun F. et al. (eds.) Research in Computational Molecular Biology: 17th Annual International Conference, RECOMB 2013, Beijing, China, April 7–10, 2013 Proceedings. Berlin, Heidelberg: Springer Berlin Heidelberg, 2013;158–70. [Google Scholar]
- O’Brien SL, Gibbons SM, Owens SM. et al. Spatial scale drives patterns in soil bacterial diversity. Environ Microbiol 2016;18:2039–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle DH. FSA: Fisheries Stock Analysis. R package version 0.8.7, 2016. https://CRAN.R-project.org/package=FSA.
- Oksanen J, Blanchet GF, Friendly M. et al. vegan: Community Ecology Package. R package version 2.4-0, 2016. https://CRAN.R-project.org/package=vegan.
- Parker WE. Evaluation of the use of food baits for detecting wireworms (Agriotes spp., Coleoptera: Elateridae) in fields intended for arable crop production. Crop Protect 1994;13:271–6. [Google Scholar]
- Paula AR, Carolino AT, Paula CO. et al. The combination of the entomopathogenic fungus Metarhizium anisopliae with the insecticide Imidacloprid increases virulence against the dengue vector Aedes aegypti (Diptera: Culicidae). Parasite Vector 2011; DOI: 10.1186/1756-3305-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L, Čuhel J, Saby NPA. et al. Mapping field‐scale spatial patterns of size and activity of the denitrifier community. Environ Microbiol 2009;11:1518–26. [DOI] [PubMed] [Google Scholar]
- Prior C. History, use and future of microbial insecticides in urban pest control. In: Proceedings of the Second International Conference on Urban Pest . Ascot, UK: International Institute of Biological control, 1996. [Google Scholar]
- R-Development-Core-Team R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2008. [Google Scholar]
- Rai V, Singh DK. Impact of fungal biopesticide (Beauveria bassiana and B. brongniartii) on soil fertility in groundnut field. Pesticide Res J 2002;14:83–91. [Google Scholar]
- Ritter C, Katroschan KU, Richter E. Alternative methods to control wireworms (Agriotes spp. Coleoptera: Elateridae) in vegetable production - potential of calcium cyanamide and Metarhizium anisopliae. IOBC/WPRS Bull 2011;66:521–4. [Google Scholar]
- Ritter C, Richter E. Control methods and monitoring of Agriotes wireworms (Coleoptera: Elateridae). J Plant Dis Prot 2013;120:4–15. [Google Scholar]
- Roesch LF, Fulthorpe RR, Riva A. et al, Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 2007;1:283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousidou C, Papadopoulou ES, Kortsinidou M. et al, Bio-pesticides: Harmful or harmless to ammonia oxidizing microorganisms? The case of a Paecilomyces lilacinus-based nematicide. Soil Biol Biochem 2013;67:98–105. [Google Scholar]
- Rousk J, Baath E, Brookes PC. et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 2010;4:1340–51. [DOI] [PubMed] [Google Scholar]
- RStudio-Team RStudio: Integrated Development for R. Boston: RStudio, Inc., 2015. [Google Scholar]
- Savazzini F, Longa CMO, Pertot I. Impact of the biocontrol agent Trichoderma atroviride SC1 on soil microbial communities of a vineyard in northern Italy. Soil Biol Biochem 2009;41:1457–65. [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CS, Alavi M, Cardinale M. et al. Stenotrophomonas rhizophila DSM14405 T promotes plant growth probably by altering fungal communities in the rhizosphere. Biol Fertil Soils 2012;48:947–60. [Google Scholar]
- Schneider S, Widmer F, Jacot K. et al. Spatial distribution of Metarhizium clade 1 in agricultural landscapes with arable land and different semi-natural habitats. Appl Soil Ecol 2012;52:20–8. [Google Scholar]
- Schwarzenbach K, Enkerli J, Widmer F. Effects of biological and chemical insect control agents on fungal community structures in soil microcosms. Appl Soil Ecol 2009;42:54–62. [Google Scholar]
- Sealy R, Evans MR, Rothrock C. The effect of a garlic extract and root substrate on soilborne fungal pathogens. HortTechnology 2007;17:169–73. [Google Scholar]
- Simon-Delso N, Amaral-Rogers V, Belzunces LP. et al. Systemic insecticides (neonicotinoids and fipronil): trends, uses, mode of action and metabolites. Environ Sci Pollut R 2015;22:5–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson EH. Measurment of diversity. Nature 1949;163:688. [Google Scholar]
- Singh J, Singh DK. Bacterial, Azotobacter, actinomycetes, and fungal population in soil after diazinon, imidacloprid, and lindane treatments in groundnut (Arachis hypogaea L.) fields. J Environ Sci Heal B 2005;40:785–800. [DOI] [PubMed] [Google Scholar]
- Sufyan M, Neuhoff D, Furlan L. Effect of male mass trapping of Agriotes species on wireworm abundance and potato tuber damage. Bull Insectol 2013;66:135–42. [Google Scholar]
- Sufyan M, Neuhoff D, Furlan L. Larval development of Agriotes obscurus under laboratory and semi-natural conditions. Bull Insectol 2014;67:227–35. [Google Scholar]
- Tedersoo L, Bahram M, Põlme S. et al. Global diversity and geography of soil fungi. Science 2014;346, DOI: 10.1126/science.1256688. [DOI] [PubMed] [Google Scholar]
- Torsvik V, Goksoyr J, Daae FL. High diversity in DNA of soil bacteria. Appl Environ Microb 1990;56:782–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabelsi D, Mengoni A, Ben Ammar H. et al. Effect of on-field inoculation of Phaseolus vulgaris with rhizobia on soil bacterial communities. FEMS Microbiol Ecol 2011;77:211–22. [DOI] [PubMed] [Google Scholar]
- Trabelsi D, Mhamdi R. Microbial inoculants and their impact on soil microbial communities: A review. BioMed Res Int 2013; DOI: 10.1155/2013/863240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traugott M, Benefer CM, Blackshaw RP. et al. Biology, ecology, and control of elaterid beetles in agricultural land. Annu Rev Entomol 2015;60:313–34. [DOI] [PubMed] [Google Scholar]
- Trevors JT. One gram of soil: a microbial biochemical gene library. A Van Leeuw J Microb 2009;97:99. [DOI] [PubMed] [Google Scholar]
- U.S. Interagency Staff Group on Carcinogens Chemical carcinogens: a review of the science and its associated principles. Environ Health Perspect 1986;67:201–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dillewijn P, Villadas PJ, Toro N. Effect of a Sinorhizobium meliloti strain with a modified puta gene on the rhizosphere microbial community of alfalfa. Appl Environ Microb 2002;68:4201–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Herk WG, Vernon RS, Moffat C. et al. Response of the pacific coast wireworm, Limonius canus, and the dusky wireworm, Agriotes obscurus (Coleopotera: Elateridae), to insecticide-treated wheat seeds in a soil bioassay. Phytoprotection 2008;89:7–19. [Google Scholar]
- Vernon B, Lagasa E, Philip H. Geographic and temporal distribution of Agriotes obscurus and A. lineatus (Coleoptera: Elateridae) in British Columbia and Washington as determined by pheromone trap surveys. J Entomol Soc B C 2001;98:257–65. [Google Scholar]
- Zaller JG, König N, Tiefenbacher A. et al. Pesticide seed dressings can affect the activity of various soil organisms and reduce decomposition of plant material. BMC Ecol 2016;16:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann G. Review on safety of the entomopathogenic fungus Metarhizium anisopliae. Biocontrol Sci Technol 2007;17:879–920. [Google Scholar]
- Zimmermann J, Musyoki MK, Cadisch G. et al. Proliferation of the biocontrol agent Fusarium oxysporum f. sp. strigae and its impact on indigenous rhizosphere fungal communities in maize under different agro-ecologies. Rhizosphere 2016;1:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.