Abstract
The nitrogen cycle in the marine environment is strongly affected by ammonia-oxidizing Thaumarchaeota. In some marine settings, Thaumarchaeotes can comprise a large percentage of the prokaryotic population. To better understand the biogeographic patterns of Thaumarchaeotes, we sought to investigate differences in their abundance and phylogenetic diversity between geographically distinct basins. Samples were collected from four marine basins (The Caspian Sea, the Great Australian Bight, and the Central and Eastern Mediterranean). The concentration of bacterial and archaeal 16S rRNA genes and archaeal amoA genes were assessed using qPCR. Minimum entropy decomposition was used to elucidate the fine-scale diversity of Thaumarchaeotes. We demonstrated that there were significant differences in the abundance and diversity of Thaumarchaeotes between these four basins. The diversity of Thaumarchaeotal oligotypes differed between basins with many oligotypes only present in one of the four basins, which suggests that their distribution showed biogeographic patterning. There were also significant differences in Thaumarchaeotal community structure between these basins. This would suggest that geographically distant, yet geochemically similar basins may house distinct Thaumarchaeaotal populations. These findings suggest that Thaumarchaeota are very diverse and that biogeography in part contributes in determining the diversity and distribution of Thaumarchaeotes.
Keywords: Thaumarchaeota, qPCR, minimum entropy decomposition, biogeography
Archaea are dominant in the water column in most deep marine basins; however, geographically distant, yet geochemically similar basins may house distinct ammonium-oxidizing Thaumarchaeaotal populations.
INTRODUCTION
Thaumarchaeota comprise a large proportion of the active microbial community in the oceans and play a significant role in marine nitrogen and carbon cycling (Karner, DeLong and Karl 2001; Francis et al.2005; Konneke et al.2005; Ingalls et al.2006; Brochier-Armanet et al.2008; Hollibaugh et al.2011; Pester, Schleper and Wagner 2011; Yakimov et al.2011; Baker, Lesniewski and Dick 2012). Thaumarchaeotes are primarily involved in nitrification, particularly in the process of ammonia oxidation (Wuchter et al.2006; Prosser and Nicol 2008). The phylogenetic diversity of marine Thaumarchaeotes has been investigated in a number of settings (Bergauer et al.2013; Hu et al.2013; Tolar, King and Hollibaugh 2013; Swan et al.2014). The ubiquitous nature of the Thaumarchaeotal phylum suggests that they are able to thrive in diverse environments and compete for available resources under vastly different conditions. Despite their cosmopolitan nature, a few studies have reported biogeographic patterns in the distribution of Thaumarchaeotes, which are partitioned by location and habitat (Biller et al.2012; Peng, Jayakumar and Ward 2013; Sintes et al.2013). These differences appear to occur most strongly between habitats (i.e. marine water column versus soils). These habitats show drastically different environmental characteristics and correspondingly great differences in the phylogenetic diversity of Thaumarchaeotes present. More recent studies have shown that biogeographic patterns in Thaumarchaetoal communities correspond in part to the availability of ammonia in these different locations (Sintes et al.2013)
Thaumarchaeotes have been extremely difficult to isolate, limiting our understanding of this phylum. Two recent studies have sought to investigate the genomic diversity of deep sea Thaumarchaeotes using single-cell genomics (Luo et al.2014; Swan et al.2014). These studies demonstrated that there are genomic similarities between Thaumarchaeotes collected at similar depths, which was in line with previously reported results indicating that there are depth-associated ecotypes of Thaumarchaeotes (Francis et al.2005). The Single Amplified Genomes obtained in these studies possessed many genes not encoded in the model cultured Thaumarchaeote—Nitrosopumilus maritimus (Luo et al.2014; Swan et al.2014). This finding suggests that there is greater genetic diversity within the Thaumarchaeota than previously identified through studies on the few cultured representatives such as N. maritimus.
Thaumarcheaotal abundance has been shown to vary relative to environmental conditions including temperature, dissolved oxygen and salinity among others (Hatzenpichler 2012). Therefore, different environmental conditions experienced in different oceanic basins may impact the abundance of Thaumarchaeotes. A number of studies have sought to determine the abundance of Thaumarchaeotes in order to better understand numerical importance of Thaumarchaeotes to the total archaeal and prokaryotic community. Diverse methods for determining Thaumarchaeotal abundance have been employed ranging from Fluorescent In Situ Hybridization (FISH) to quantitative Polymerase Chain Reaction (qPCR) methods (DeLong et al.1999; Wuchter et al.2006). qPCR methods have used primers targeting the 16S rRNA of Thaumarchaeotes, the archaeal ammonia monooxygenase (amoA) gene and the acetyl-CoA carboxylase alpha subunit (accA) gene (Yakimov, La Cono and Denaro 2009; Tolar, King and Hollibaugh 2013). Thaumarchaeotal abundance in different settings often has ranged from 101 to 104 cells/ml of seawater (Yakimov, La Cono and Denaro 2009; Amano-Sato et al.2013; Bergauer et al.2013; Hu et al.2013; Tolar, King and Hollibaugh 2013). Differences in abundance have previously been observed between different depths and water masses (Amano-Sato et al.2013) as well as between seasons (Bale et al.2013). Direct comparison of the Thaumarchaeotal abundances between different basins is complicated through different experimental factors, such as different extraction methodologies, or different qPCR primer pairs used between studies.
In this study, we characterized the abundance and high-resolution phylogenetic diversity of the Thaumarchaeotal community in four marine basins (The Caspian Sea, the Great Australian Bight (GAB), the Central and Eastern Mediterranean). These four basins represent distinct oceanographic settings and a range of nutrient loadings. These basins cover enclosed (Caspian), semi-enclosed (Central and Eastern Mediterranean) and open-ocean (GAB) settings. In terms of nutrient loadings, the Caspian Sea is highly eutrophic and anthropogenically impacted (Mahmoudi et al.2015), whereas the Central and Eastern Mediterranean are considered ultra-oligotrophic basins (Thingstad et al.2005). The GAB would be intermediate between these basins as it is not ultra-oligotroph nor eutrophic (Kämpf and Kavi 2017). Although previous studies have investigated the Thaumarchaeotal abundance in the Mediterranean (De Corte et al.2008; Yakimov, La Cono and Denaro 2009), little is known about the Thaumarchaeotal population in the Caspian Sea and the GAB. In addition to providing insights into the microbial community in previously unexplored locations, this set of samples will also provide insights into the abundance and diversity of Thaumarchaeotes in adjacent basins with similar physical and chemical parameters (Central and Eastern Mediterranean).
In this study, abundance of Thaumarchaeotes was determined using qPCR of the archaeal amoA gene. The abundance of bacteria and archaea were determined using qPCR with domain-specific 16S rRNA primers to better understand the proportion of Thaumarchaeota in relation to the total bacterial and archaeal abundance. The majority of previous studies investigating the phylogenetic diversity of Thaumarchaeota have determined diversity using binning of marker genes based on % similarity. These binning approaches are limited and lack higher resolution insights into the diversity of particular groups of microbes. We sought to investigate the phylogenetic diversity of Thaumarchaeotes, using minimum entropy decomposition (MED) (Eren et al.2015) of the 16S rRNA sequence. MED uses Shannon entropy of highly divergent regions of a marker gene sequence alignment to distinguish between ecologically relevant groups of sequences. The combination of high-resolution community analysis and abundance comparisons will provide further insights into the diversity and abundance of this ubiquitous phylum in the oceans.
MATERIALS AND METHODS
Sample collection and site characterization
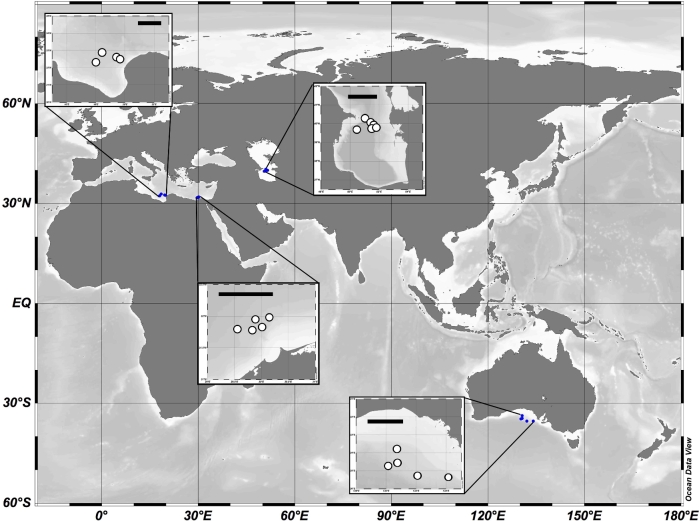
Water samples were collected from the following four deep-sea basins: Eastern Mediterranean (Nile Deep Sea Fan), Central Mediterranean (Sirte Basin), Caspian Sea (Southern Caspian) and GAB (Bight Basin) (Fig. 1). Water was collected from five stations in the Eastern Mediterranean between 11 and 15 October 2012. Four depths were sampled at each station, representing near surface, one-third of water depth, two-thirds of water depth and near bottom. Five stations were sampled in the GAB between 7 and 14 April 2013. Four depths were sampled at each location, representing near surface, one-third of water depth, two-thirds of water depth and near bottom. Six stations were sampled in the Caspian Sea between 27 July and 2 August 2013. Two to four depths were sampled at each location depending on water depth. Samples were collected from four stations in the Central Mediterranean between 29 January and 5 February 2014. Four depths were sampled at each station, representing near surface, one-third of water depth, two-thirds of water depth and near bottom. The majority of samples collected represent mesopelagic and bathypelagic depths. In all, 75 samples were collected as part of this study (full geochemical data are shown in Table S1, Supporting Information). In situ physical and chemical parameters were measured using a Valeport CTD equipped with sensors for temperature, salinity, dissolved oxygen and depth. Water was collected in Niskin bottles at each sampling depth and stored at –20°C for analysis of inorganic nutrients. Samples for microbial community analysis were collected by filtering seawater through a 142-mm, 0.2-μm nylon filter. Samples were collected in Eastern Mediterranean as described in Techtmann et al. (2015). Between 62 and 158 L per sample were filtered in the Eastern Mediterranean. Microbial samples were collected from the Caspian Sea using a McLane Pump Large Volume water sampler (McLane Labs, Falmouth MA) to filter 10.3 and 27 L of water per sample. Samples from the Central Mediterranean and the GAB were collected by recovering 20 L of water from a Niskin bottle and filtering 20 L per sample on deck. In all cases, the filters were immediately stored at –20°C until shipment to the lab, at which time the samples were stored at –80°C until processing. Differences in the methods of sample collection and volumes are due to the resources available on the cruise. For cruises where the in situ pump was available, the Large Volume water sampler was employed. When using the Large Volume water sampler, the sample collection was determined based on time. This led to varying volumes being filtered through the pump depending on the particulate load in the water leading to clogging of the filter, which resulted in decreased flow rate and overall less volume. Approximately 20 L of water was filtered for all basins other than the Eastern Mediterranean. The cell abundance in the Mediterranean is quite low due to its ultra-oligotrophic nature (Techtmann et al.2015). This low biomass required larger volumes of water to collect similar amounts of biomass. On some cruises, the Large Volume water sampler was not available and thus samples were collected using Niskin bottles and on-board filtration was required. For cruises where Niskin bottle sampling was performed, 20 L of water was filtered as a standard volume.
Figure 1.
Map of sampling locations. Four basins were sampled. Sampling stations are shown as white circles in the zoomed-in regions. Scale bars in the zoomed-in basin maps correspond to 100 km.
Geochemical measurements
Dissolved organic carbon (DOC) and inorganic nutrients were measured at the SOEST Laboratory for Analytical Biogeochemistry (University of Hawaii). DOC was measured using a Shimadzu High-Temperature TOC-L Combustion Analyzer (Shimadzu, Japan). DOC is reported as non-purgeable organic carbon (NPOC). Quality control testing for NPOC was conducted using purchased Deep Seawater Reference Material from the RSMAS Consensus Reference Materials Project (http://yyy.rsmas.miami.edu/groups/biogeochem/CRM.html). Ammonia was measured fluorometrically following the method of Kerouel and Aminot (1997). Nitrate and nitrite were analyzed via the diazo reaction based on the methods of Armstron, Stearns and Strickla (1967) and Grasshoff, Ehrhardt and Kremling (1983). Silicate measurement is based on the reduction of silicomolybdate in acidic solution to molybdenum blue by ascorbic acid (Grasshoff, Ehrhardt and Kremling 1983). Orthophosphate concentrations were determined based on the colorimetric method of Murphy and Riley (1962). For all compounds measured, concentrations were determined by the creation of a six-point standard curve made with known concentrations of the analyte of interest. The lower limit of the detection for the analytes is as follows: nitrite 0.01 μM, nitrate 0.01 μM, phosphate 0.008 μM, silicate 0.065 μM, ammonia 0.02 μM.
DNA extraction
DNA was extracted from the filters using the modified Miller method as described in Techtmann et al. (2015). Quality of DNA was determined by measuring the 260/280 and 260/230 ratios using the Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA, USA). DNA concentrations were determined using the Qubit Fluorometric assay (Life Technologies, Carlsbad, CA, USA).
Preparation of standards for qPCR assays
Bacterial and archaeal 16S rRNA gene concentration was determined to identify the relative contribution of archaea to the total prokaryotic community in these samples. Bacterial and archaeal 16S rRNA gene abundance was determined using qPCR according to the methods previously described in Jorgensen et al. (2012) using the following primer pairs: Bact341 and Uni519R for bacteria as well as Uni519F and Arch908R for archaea. Archaeal amoA copy number was determined using qPCR as previously described in Wuchter et al. (2006).
Standards were prepared by amplifying the gene of interest from environmental DNA using the appropriate primer pair. Each amplification was performed under the following conditions: initial denaturation at 98°C for 2 min; 40 cycles of 98°C for 15 s, 30 s at the appropriate annealing temperature of the primer pair and 72°C for 2 min, followed by a final extension at 72°C for 5 min. PCR reactions included Phusion master mix (Thermo Scientific), 0.4 μM forward and reverse primers, and 1 μl of DNA template. Amplified products were run on a 1.2% agarose gel stained with SYBR Safe (Thermo Scientific. Amplicons were gel purified using the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI).
Purified PCR products were cloned into the pCR4-TOPO® TA Vector (Life Technologies) using the Topo TA cloning kit (Life Technologies). Plasmids were purified from transformants and sequenced using vector primers (M13 forward and reverse) to confirm the correct insert. Plasmids containing the proper insert were linearized by digesting the plasmid with Not1. Linearized plasmids were gel purified using the Wizard SV Gel and PCR Clean-Up System (Promega). The purified products were quantified using Qubit (Life Technologies) and used as standards for qPCR.
qPCR quantification of gene concentrations
qPCR was performed on an iCycler thermocycler (Bio-Rad, Hercules, CA). Six-point standard curves were performed in triplicate with concentrations ranging from 2 × 10−4 to 20 pM. Environmental DNA was diluted 1:10 to account for potential inhibitors, and 1 μl of diluted environmental DNA was used in 20 μl qPCR reactions. The copy numbers of bacterial 16S rRNA, archaeal 16S rRNA and archaeal amoA in environmental DNA were determined in duplicate. Total prokaryotic abundance was determined by adding the copy number of bacterial and archaeal 16S rRNA genes together. Gene copy number was corrected for the dilution factor and then normalized to the total volume of seawater that was passed through that filter to the determine the number of gene copies per ml of seawater according to the following equation: copies number per μl of DNA was multiplied by the total volume of extract (μl of DNA) and then divided by the volume of seawater filtered (ml). An analysis of vairance (ANOVA) was performed on log-transformed copy number data in order to determine if there was a significant difference in the abundance of these genes between basins. The ANOVA analysis was performed comparing samples collected from the same depth strata (epipelagic (0–200 m), mesopelagic (200–1000 m) or bathypelagic (1000–4000 m)). The Tukey honest significant difference (HSD) test was used as a post hoc test to distinguish which basins were significantly different from each other (Table S3, Supporting Information).
16S rRNA sequencing and analysis
The V4 region of the 16S rRNA gene was amplified using Phusion DNA polymerase (Thermo Scientific) with universal primers 515f and barcoded 806r from all 75 samples collected as part of this project. These primers were able to amplify both bacterial and archaeal sequences. Sequencing was performed on the Illumina MiSeq according to the protocol in Caporaso et al (2012). The resulting DNA sequences were analyzed using the QIIME version 1.8.0-dev pipeline (Caporaso et al.2010). Paired-end raw reads were assembled using fastq-join (Aronesty 2011). The assembled sequences were demultiplexed and quality filtered in QIIME to remove reads with phred scores below 20 (–q 19). Chimera detection was then performed on assembled reads using UCHIME (Edgar 2010; Edgar et al.2011). The taxonomy for each read was assigned using Ribosmal Database Project (RDP) (Wang et al.2007) retrained with the May 2013 Greengenes release. The total number of sequences assigned to the Thaumarchaeota was 2630 432. While the sequencing data included both bacterial and archaeal sequences, only reads classified as Thaumarchaeota were used for further analysis. MED was performed on reads classified as Thaumarchaeota (Eren et al.2015). MED, like oligotyping, applies Shannon entropy to different nucleotide positions in a sequence alignment. MED works on the premise that not all nucleotide positions contribute equally to partitioning into ecologically meaningful bins. Positions with high entropy are indicative of ecologically meaningful differences and not due to sequencing artifacts. The total pool of nucleotides is decomposed into nodes based on the nucleotide present in the position with the highest Shannon entropy. This process is repeated iteratively until a minimum entropy threshold is achieved. This approach will distinguish between ecologically meaningful bins even for highly similar sequences, which in some cases only vary by one nucleotide. The original quality-trimming step resulted in reads with different ending positions. The data were re-trimmed to 250 bases so that all of the Thaumarchaeotal reads had the same starting and ending positions. The dataset used for MED analysis was deposited in MG-RAST Accession numbers (mgm4642815.3 - mgm4642887.3). MED was performed using the decompose command from the Oligotyping version 1.8. The minimum substantive abundance of an oligotype was set at 526 and the maximum variation allowed in each node was set at 3 nucleotides. This removed 351 057 sequences. A total of 2279 432 sequences passed quality filtering as part of the MED pipeline. MED resulted in 332 final nodes after refinement. The MED MATRIX-COUNTS file of Thaumarchaeotal oligotypes was converted into a biom file and rarified to 1163 sequences to account for differences in number of recovered Thaumarchaeotal reads. Alpha diversity was determined using the alpha_diversity.py command in QIIME. Faith's phylogenetic diversity was determined by aligning the representative sequences for each of the 332 nodes using pynast and building a phylogenetic tree using the fast tree implementation in QIIME. This tree file was used to calculate phylogenetic diversity using the core_diversity.py script in QIIME.
Statistical analysis on 16S rRNA sequencing data
To test the hypothesis that there were significant differences in the alpha diversity of Thaumarchaeotes between basins, a one-way ANOVA was performed comparing the alpha diversity metrics from the Thaumarchaeota oligotype table for samples grouped according to basin. This test was done on diversity as determined by Faith's phylogenetic diversity, Shannon, Simpson and Chao1 diversity metrics. Tukey HSD was used as a post hoc test to determine which basins were significantly different from each other. P values of less than 0.05 were considered significant. To test the hypothesis that there were significant differences in the community structure of Thaumarchaeotes, Bray-Curtis dissimilarity was calculated using the vegdist command in the vegan package in R (Oksanen et al.2013; R Core Team 2013). Non-metric multidimensional scaling (NMDS) was performed on the Bray-Curtis dissimilarity matrix using the metaMDS command implemented in the vegan package. Permutational analysis of variance (PERMANOVA) analysis was used to identify if there were significant differences in Thaumarchaeotal community structure between basins using the adonis function in the vegan package (Oksanen et al.2013). Pairwise PERMANOVAs were performed on subsets of the dissimilarity matrix in order to identify which basins showed significantly different Thaumarchaeotal populations. To identify how environmental conditions shape Thaumarchaeotal community structure, environmental variables were fit to the Bray-Curtis dissimilarity matrix using the envfit function in the vegan package. Variables that fit the data with P values of less than 0.05 were plotted.
A fine-scale analysis was undertaken to examine how particular oligotypes are affected by geographic location and geochemical variables. The MetagenomeSeq package (Paulson et al.2013), as implemented in QIIME, was used to identify which oligotypes were differentially abundant between basins. All basins were compared to each other and oligotypes that were significantly different (corrected P value < 0.05) in one basin compared to the other three were considered to be differentially abundant.
RESULTS
These basins represent distinct physical and chemical contexts
Temperature, salinity, nutrient concentrations and organic carbon loads were different between these sites (Table 1 and Table S1). The temperature of the Mediterranean deep-water samples (both the Central and Eastern basins) was between 13.7°C and 14.5°C with the average being 13.9°C, while the temperature of deep water of most of the other sampled locations was between 2.5°C and 6°C. The salinity of the Caspian was 11 psu compared to the Mediterranean samples, which had a salinity of 39 psu. Inorganic phosphate concentrations were also much lower in the Mediterranean relative to the GAB and Caspian. There was an increase in phosphate concentrations with depth except in the Central Mediterranean (Table S1). Nitrate and silicate were elevated in the GAB relative to the other locations. Nitrite was below detection in all basins but the Caspian, where it was low, but above the detection limit. Total organic carbon was elevated in the Caspian where TOC concentrations were almost 10 times that of the other basins.
Table 1.
Physical and chemical parameters for each basin.
| E. Med | C. Med | GAB | Caspian | |
|---|---|---|---|---|
| Temp surface | 20.3 (17.5–26.3) | 17.3 (16.7–17.7) | 16.6 (12.7–19.9) | 20.2 (11.2–25.1) |
| Temp deep (°C) | 13.9 (13.7–14.5) | 13.7 (13.7–13.8) | 2.9 (2.3–4.3) | 6.7 (6.1–7.2) |
| Dissolved oxygen—surface (mg/L) | 9.5 (9.3–11.5) | 6.8 (5.1–7.5) | 7.8 (7.6–8.0) | 8.4 (8.1–8.5) |
| Dissolved oxygen—deep (mg/L) | 6.4 (5.9–6.9) | 5.5 (4.2–6.1) | 5.9 (5.6–6.4) | 2.6 (0.5–5.2) |
| Salinity (psu) | 38.9 (38.5–39.5) | 38.6 (38.2–38.9) | 34.8 (34.4–35.7) | 11.3 (11.2–11.4) |
| Nitrate (μM) | 4.09 (0.01–6.95) | 3.11 (0.027–5.74) | 18.6 (0.01–33.1) | 4.5 (0.08–12.4) |
| Ammonia (μM) | 0.03 (<0.02–0.131) | 0.05 (0.027–5.74) | <0.02 | 0.02 (<0.02–0.04) |
| Nitrite (μM) | <0.01 | <0.01 | <0.01 | 0.03 (<0.01–0.1) |
| Inorganic P (μM) | 0.231 (0.008–0.437) | 0.151 (0.008–0.224) | 1.44 (0.013–2.45) | 0.42 (0.01–1.65) |
| Silicate (μM) | 5.17 (0.377–9.98) | 3.87 (0.449–7.56) | 30.5 (0.065–90.8) | 25.6 (0.24–84.7) |
| TOC (mg/L) | 0.952 (0.68–1.482) | 0.77 (0.58–1.07) | 0.952 (0.68–1.196) | 6.9 (5.7–7.9) |
| Total nitrogen (mg/L) | 0.13 (0.09–0.19) | 0.09 (0.07–0.13) | 0.33 (0.07–0.54) | 0.43 (0.34–0.50) |
The first number is the average for each parameter. The numbers in the parentheses are the range of values for each environmental parameter. Surface samples (<60 m) and deep-water samples (>60 m) were grouped together for comparison temperature and dissolved oxygen.
Abundance of Thaumarchaeotes is different between basins
Total bacterial and total archaeal abundance was determined by qPCR of the bacterial and archaeal 16S rRNA genes. Abundance of Thaumarchaeota was determined by using qPCR to quantify the archaeal amoA gene. Analysis of bacterial, archaeal and Thaumarchaeotal abundance was compared across all basins at different depth classes (Fig. 2, Table 2, and Tables S2 and S3, Supporting Information). Bacterial 16S rRNA copy number ranged from 102 to 107 copies per ml of seawater. Bacterial abundance was significantly different between each basin at the three depth strata collected (ANOVA—Table 2). For the most part, bacterial abundance decreases with depth. In the epipelagic, bacterial abundance was highest in the Eastern Mediterranean, followed by the GAB, and then the Caspian with the lowest epipelagic bacterial abundance in the Central Mediterranean (Fig. 2). In the mesopelagic, the highest bacterial abundance was in the Central Mediterranean followed by the Caspian and the GAB with the Eastern Mediterranean showing the lowest abundance. Bacterial abundance in the bathypelagic was highest in the GAB followed by the Central Mediterranean, with the Eastern Mediterranean having the lowest bathypelagic bacterial abundance.
Figure 2.
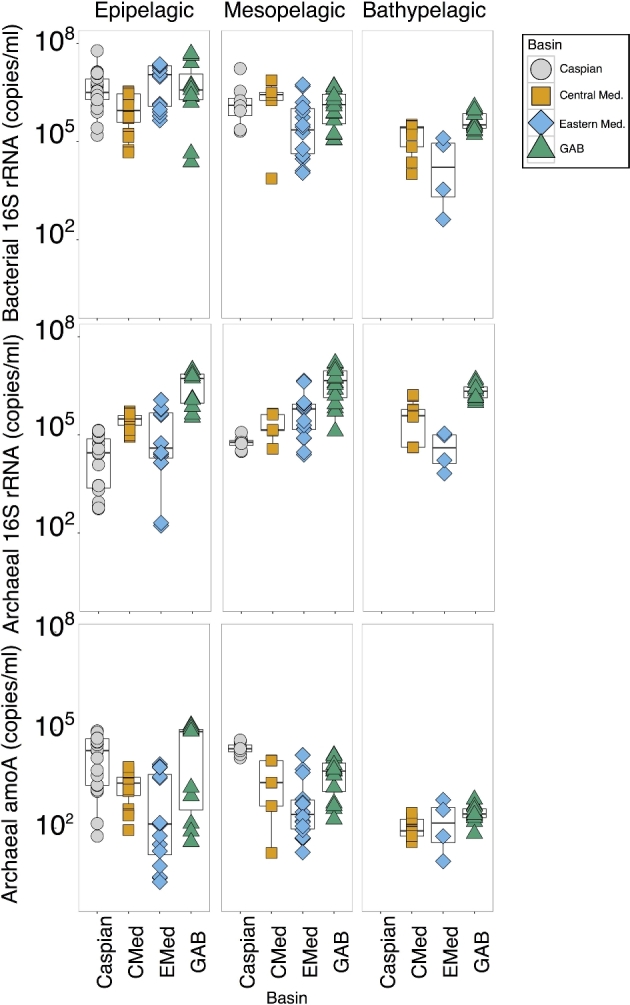
Abundance of genes by qPCR. Box plot representing copy number of bacterial 16S rRNA, archaeal 16S rRNA and archaeal amoA with samples separated according to basin and depth. Samples from each basin were separated into three depth categories—epipelagic (0–200 m), mesopelagic (200–1000 m) and bathypelagic (1000–4000 m). Shapes and colors of the sample points correspond to the basin from which they were collected.
Table 2.
Statistical analysis of abundance of microbes between basins at different depths.
| Bacterial 16S rRNA | Archaeal 16S rRNA | Archaeal amoA | ||||
|---|---|---|---|---|---|---|
| F stat | P value | F stat | P value | F stat | P value | |
| Epipelagic | 4.363 | 0.001 | 22.96 | 3 × 10−10 | 9.768 | 2.09 × 10−5 |
| Mesopelagic | 3.072 | 0.037 | 20.08 | 2.75 × 10−8 | 11.37 | 1.28 × 10−5 |
| Bathypelagic | 10.75 | 0.0005 | 23.47 | 3.5 × 10−6 | 3.115 | 0.064 |
ANOVA was used to compare the abundance of bacterial and archaeal 16S rRNA and archaeal amoA gene abundance between basins at different depths. P values in bold are considered significant.
Archaeal abundance was significantly different between basins at all three depth categories sampled (Table 2). Archaeal abundance peaked in the mesopelagic and decreased in the bathypelagic in most basins. Overall, the GAB showed the highest archaeal abundance across all three depth categories. Samples from the Mediterranean showed intermediate archaeal abundances in all three depth categories. The Caspian showed the lowest abundance of archaea.
Thaumarchaeotal abundance as defined by archaeal amoA abundance was significantly different between basins in the epi- and mesopelagic, but there was no significant difference in the archaeal amoA abundance in the bathypelagic (Table 2). Overall archaeal amoA abundance was high in the epi- and mesopelagic and decreased dramatically in the bathypelagic. Epipelagic samples from the GAB showed the highest archaeal amoA abundance followed closely by the Caspian. The Central and Eastern Mediterranean had the lowest archaeal amoA abundance with the Eastern Mediterranean showing the lowest abundance of all four basins. In the mesopelagic, the Caspian had the highest abundance of archaeal amoA followed by the GAB and the Central Mediterranean with the Eastern Mediterranean showing the lowest abundance in the mesopelagic. All of the samples from the bathypelagic showed similar abundance of archaeal amoA.
Thaumarchaeotal phylotype diversity was significantly different between basins
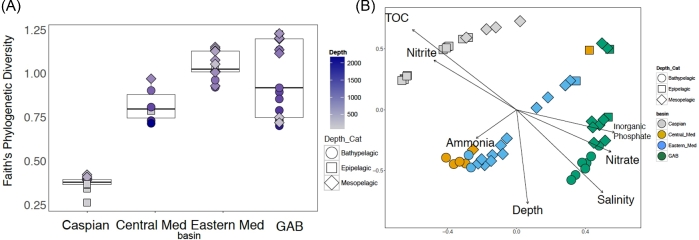
MED analysis resulted in the identification of 332 oligotypes across all of the four basins. Alpha diversity of Thaumarchaeotal oligotypes as determined by a number of alpha diversity metrics (Faith's phylogenetic diversity, Shannon, Simpson and Chao1) was significantly different between basins for all tested metrics (Fig. 3A, Table S5, Supporting Information) (ANOVA P value of Faith's PD < 0.0001). The diversity of Thaumarchaeota was the lowest in the Caspian, followed by the Central Mediterranean, GAB and samples from the Eastern Mediterranean showed the highest average Thaumarchaeotal diversity of the four basins.
Figure 3.
Diversity of Thaumarchaeotal Oligotypes. (A) Faith's phylogenetic diversity for the Thaumarchaeotal diversity of each sample. Each sample from a basin is shown as a point. Squares represent epipelagic samples (0–200 m). Diamonds represent mesopelagic samples (200–1000 m) and circles represent bathypelagic samples (1000–4000 m). Points are colored based on the depth of sample collection. (B) NMDS of a Bray Curtis dissimilarity matrix prepared from the Thaumarchaeotal oligotype table. Samples from the Caspian Sea are shown in gray, samples from the Central Mediterranean are shown in orange, samples from the Great Australian Bight are shown in Green and samples from the Eastern Mediterranean are shown in blue. Squares represent epipelagic samples. Diamonds represent mesopelagic samples and circles represent bathypelagic samples Environmental factors were fit to this data and environmental parameters that significantly fit the data are plotted as vectors.
To determine if there were differences in the observed structure of the Thaumarchaeotal communities in these basins, NMDS was performed on Bray-Curtis dissimilarity (Fig. 3B). The Thaumarchaeotal community structure was similar in samples collected from the same basin. Samples from the Caspian had the most distinct set of Thaumarchaeotal oligotypes, and thus these samples clustered distantly from other basins in the NMDS analysis. PERMANOVA analysis indicated that there were statistically significant differences in the Thaumarchaeotal community between basins (P value 0.001). Pairwise PERMANOVAs between each basin indicated that there were significant differences in the structure of the Thaumarchaeotal community between each most basins (Table S6, Supporting Information). The difference between the Central and Eastern Mediterranean communities is slightly significant but is above the 0.05 threshold (pairwise PERMANOVA P value 0.055). The samples from these basins showed a trend where samples from the similar depths clustered together and were distinct from other depths (Fig. 3).
Bray-Curtis dissimilarity is affected by both presence and abundance of an oligotype. It is therefore possible that the same Thaumarchaeotal oligotypes were present in each basin, but the observed differences in Thaumarchaeotal community structure were due to differences in abundance of these oligotypes. To determine if there were distinct Thaumarchaeotal oligotypes in each basin, NMDS was performed on a Bray Curtis dissimilarity matrix constructed from a Thaumarchaeotal oligotype table that was converted to presence/absence. The same trends were observed with the presence/absence data as with the Thaumarchaeotal oligotype abundance data, showing that there were significantly different Thaumarchaeotal populations in each basin (PERMANOVA P value 0.001) (Fig. S3, Supporting Information). Thaumarchaeotal community structure in Mediterranean was marginally distinct between the Central and Eastern Basins, despite no significant difference in the geochemistry between the Central and Eastern Mediterranean (Fig. S4 and Tables S1 and S7, Supporting Information). While there may differences in the Thaumarchaeotal community structure based on the season of sampling, the majority of samples in this study came from deep water, which may be less impacted by seasonality. Additionally, our results indicate that the Thaumarchaeotal community structure for samples from the two Mediterranean basins (Central and Eastern) were most similar to each other despite these samples being collected during different seasons (late fall and winter).
DISCUSSION
Thaumarchaeotes are present throughout the oceans and play an important role in the global nitrogen and carbon cycles. We therefore examined if differences in geochemistry or geographic isolation resulted in differences in Thaumarchaeotal phylogenetic diversity in four marine basins. In general, the bacterial and archaeal abundance followed trends reported previously with bacterial abundance decreasing with depth and archaeal abundance increasing with depth (Karner, DeLong and Karl 2001). Thaumarchaeotal abundance was significantly different between basins at both the epi- and mesopelagic depths. At both depths, the samples from the Mediterranean were lower in abundance than the GAB and the Caspian. While the copy numbers of archaeal amoA genes in the Mediterranean were lower than the other basins sampled in this study, others have reported similar archaeal amoA abundances for the Central and Eastern Mediterranean (De Corte et al.2008; Yakimov, La Cono and Denaro 2009). In the GAB and the Mediterranean basins, the Thaumarchaeotes appear to be a small portion of the archaeal community. This finding suggests that while Thaumarchaeotes are a substantial portion of the microbial community in the GAB and Mediterranean basins, there are other important archaeal groups in these basins.
qPCR-based methods for determining bacterial and archaeal abundance using 16S rRNA genes in some cases may overestimate abundances, as many bacteria and archaea have multiple 16S rRNA operons within their genomes (Acinas et al.2004). While all sequenced Thaumarchaeotes have a single rRNA operon, many Euryarchaeotes have been shown to possess multiple rRNA operons (Lee, Bussema and Schmidt 2009; Stoddard et al.2015). Previous microbial community analysis in the Mediterranean has indicated that Euryarchaeotes represent substantial proportion of the recovered reads from these samples (Techtmann et al.2015). Therefore, the abundance of Euryarchaeotes in these samples may result in overestimates of the total archaeal abundance and may indicate that Thaumarchaeotes represent and even more substantial portion of the archaeal community in these three basins.
In contrast, the Caspian Sea had the lowest concentration of archaeal 16S rRNA gene copies and the highest concentration of archaea amoA genes (Fig. 2 and Table S2). This finding would suggest that the majority of archaea in the waters of the Caspian are Thaumarchaeota and that there are very few other archaeal groups in the Caspian water column. In contrast to this, a recent study of the microbial community in sediments of the Caspian Sea demonstrated that while Thaumarchaeota accounted for a large number of archaeal 16S rRNA sequences recovered from surface sediments, similar numbers of sequences recovered from other archaeal groups were also recovered (Mahmoudi et al.2015). This finding suggests that Thaumarchaeota are not as dominant in the sediments of the Caspian as they are in the water column. Other studies in the water column of the Black and Baltic Seas have indicated a similar dominance of Thaumarchaeotes in relation to the total archaeal community (Lam et al.2007; Labrenz et al.2010). It is believed that the high numbers of Thaumarchaeotes in the ocean is a result of their ability to thrive in oligotrophic conditions and make use of the very low levels of ammonia found in oceans (Pester, Schleper and Wagner 2011; Hatzenpichler 2012). It is therefore notable that the most eutrophic of the four basins sampled in this study (the Caspian Sea) had the highest abundance of archaeal amoA genes. The ability of Thaumarchaeotes to thrive under eutrophic conditions was previously demonstrated by their presence at high abundance in wastewater treatment systems (Mussmann et al.2011). While the Thaumarchaetoes in wastewater treatment systems expressed the amoA gene, they were not obligate autotrophic ammonia oxidizers. Therefore, it is possible that the Thaumarchaeotes in the eutrophic Caspian may also be more generalized with physiologies extending beyond obligate autotrophic ammonia oxidation.
Despite the high abundance of Thaumarchaeotes in the Caspian, our findings demonstrated that the Caspian Sea had the lowest phylogenetic diversity of Thaumarchaeotes of the four basins sampled, with 30 times fewer oligotypes than found in samples from the GAB. A similar phenomenon was seen in the Baltic Sea, whose Thaumarchaeotal population was dominated by one phylotype (Labrenz et al.2010). Both the Baltic and Caspian are characterized by low salinity and anoxic bottom water (Labrenz et al.2010; Mahmoudi et al.2015). Therefore, it is possible that these conditions select for a very low diversity group of Thaumarchaeotes capable of thriving under low salinity and anoxic conditions.
Previous studies have indicated biogeographic distinctions in marine Thaumarchaeotal populations (Biller et al.2012; Peng, Jayakumar and Ward 2013; Sintes et al.2013; Doxey et al.2015). These studies have examined settings with distinct geochemical conditions (i.e. soil versus marine water column). Here we demonstrated that even in adjacent basins with similar physical and chemical parameters (Central and Eastern Mediterranean), there are key distinctions in the oligotypes of Thaumarchaeotes. This finding indicates that while geochemistry can affect Thaumarchaeotal populations as previously reported (Hatzenpichler 2012; Sintes et al.2013), there are other factors such as geographical separation that allow for establishment of distinct Thaumarchaeotal populations under similar physical and chemical conditions.
We further sought to understand Thaumarchaeotal diversity using MED to parse out fine-scale differences in the Thaumarchaeotal diversity between basins. We have identified key Thaumarchaeotal oligotypes that were enriched in one basin relative to the others (Table S4, Supporting Information). In many cases, the indicator oligotypes are highly similar to each other, yet representative of only one basin. In many cases, these indicator oligotypes would be grouped into a single OTU if classified according to standard % identity cutoffs (97%). This highlights the utility of high-resolution methods such as MED for studies of biogeographic pattering of microbial populations. The finding that there are significant differences between geographically adjacent and geochemically similar basins as well as the presence of basin-specific oligotypes expands our understanding of the biogeographic patterns in Thaumarchaeotal diversity. Further work is required to elucidate the genetic differences between these closely related oligotypes, which enable them to colonize these different settings.
CONCLUSIONS
Thaumarchaeotes are ubiquitous microbes that play an essential role in the nitrogen cycle of many environments. Here we demonstrated that Thaumarchaeota are present at significantly different abundances in these distinct basins. The use of high-resolution approaches, namely MED, to analyze 16S rRNA data has provided insights into the biogeographic patterns of Thaumarchaeotal diversity. The ubiquity of Thaumarchaeotes makes it tempting to infer a common function and low diversity for this phylum. Here we show that there is great diversity in the Thaumarchaeotal phylum in three of the four basins sampled. The high abundance and low diversity of the Caspian suggest that there are distinct taxa capable of dominating under very selective conditions. This work expands our understanding of the extent and diversity of Thaumarchaeotes across a range of environmental conditions and marine basins.
Supplementary Material
Supplementary data are available at FEMSEC online.
Acknowledgements
The authors are grateful to BP and its partners for support in the sampling effort.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSEC online.
FUNDING
This research was supported by contract A13-0119-001 Deep Sea Basin Microbiology between the University of Tennessee and BP.
Conflict of interest. None declared.
References
- Acinas SG, Marcelino LA, Klepac-Ceraj V et al. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol 2004;186:2629–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano-Sato C, Akiyama S, Uchida M et al. Archaeal distribution and abundance in water masses of the Arctic Ocean, Pacific sector. Aquat Microb Ecol 2013;69:101–12. [Google Scholar]
- Armstron FAJ, Stearns CR, Strickla JD. Measurement of upwelling and subsequent biological processes by means of technicon autoanalyzer and associated equipment. Deep-Sea Res 1967;14:381. [Google Scholar]
- Aronesty E. ea-utils: Command-Line Tools for Processing Biological Sequencing Data. 2011. https://code.google.com/p/ea-utils/.
- Baker BJ, Lesniewski RA, Dick GJ. Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume. ISME J 2012;6:2269–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale NJ, Villanueva L, Hopmans EC et al. Different seasonality of pelagic and benthic Thaumarchaeota in the North Sea. Biogeosciences 2013;10:7195–206. [Google Scholar]
- Bergauer K, Sintes E, van Bleijswijk J et al. Abundance and distribution of archaeal acetyl-CoA/propionyl-CoA carboxylase genes indicative for putatively chemoautotrophic Archaea in the tropical Atlantic's interior. FEMS Microbiol Ecol 2013;84:461–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Mosier AC, Wells GF et al. Global biodiversity of aquatic ammonia-oxidizing archaea is partitioned by habitat. Front Microbiol 2012;3, DOI: 10.3389/fmicb.2012.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier-Armanet C, Boussau B, Gribaldo S et al. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 2008;6:245–52. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Corte D, Yokokawa T, Varela MM et al. Spatial distribution of Bacteria and Archaea and amoA gene copy numbers throughout the water column of the Eastern Mediterranean Sea. ISME J 2008;3:147–58. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Taylor LT, Marsh TL et al. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microb 1999;65:5554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxey AC, Kurtz DA, Lynch MDJ et al. Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J 2015;9:461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Morrison HG, Lescault PJ et al. Minimum entropy decomposition: Unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J 2015;9:968–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis CA, Roberts KJ, Beman JM et al. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. P Natl Acad Sci USA 2005;102:14683–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasshoff K, Ehrhardt M, Kremling K. Methods of Seawater Analysis, 2nd Edition Weinheim, Deerfield Beach, Florida; Basel, Verlag Chemie, Germany, 1983. 419 p. [Google Scholar]
- Hatzenpichler R. Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl Environ Microb 2012;78:7501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollibaugh JT, Gifford S, Sharma S et al. Metatranscriptomic analysis of ammonia-oxidizing organisms in an estuarine bacterioplankton assemblage. ISME J 2011;5:866–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu AY, Yang Z, Yu CP et al. Dynamics of autotrophic marine planktonic Thaumarchaeota in the east china sea. PLoS One 2013;8, DOI: 10.1371/journal.pone.0061087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalls AE, Shah SR, Hansman RL et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. P Natl Acad Sci USA 2006;103:6442–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen SL, Hannisdal B, Lanzen A et al. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. P Natl Acad Sci USA 2012;109:E2846–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämpf J, Kavi A. On the “hidden” phytoplankton blooms on Australia's southern shelves. Geophys Res Lett 2017;44:1466–73. [Google Scholar]
- Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 2001;409:507–10. [DOI] [PubMed] [Google Scholar]
- Kerouel R, Aminot A. Fluorometric determination of ammonia in sea and estuarine waters by direct segmented flow analysis. Mar Chem 1997;57:265–75. [Google Scholar]
- Konneke M, Bernhard AE, de la Torre JR et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 2005;437:543–6. [DOI] [PubMed] [Google Scholar]
- Labrenz M, Sintes E, Toetzke F et al. Relevance of a crenarchaeotal subcluster related to Candidatus Nitrosopumilus maritimus to ammonia oxidation in the suboxic zone of the central Baltic Sea. ISME J 2010;4:1496–508. [DOI] [PubMed] [Google Scholar]
- Lam P, Jensen MM, Lavik G et al. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. P Natl Acad Sci USA 2007;104:7104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ZMP, Bussema C, Schmidt TM. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res 2009;37:D489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo HW, Tolar BB, Swan BK et al. Single-cell genomics shedding light on marine Thaumarchaeota diversification. ISME J 2014;8:732–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi N, Robeson MS, Castro HF et al. Microbial community composition and diversity in Caspian Sea sediments. FEMS Microbiol Ecol 2015;91:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J, Riley JP. A modified single solution method for determination of phosphate in natural waters. Anal Chim Acta 1962;26:31–36. [Google Scholar]
- Mussmann M, Brito I, Pitcher A et al. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. P Natl Acad Sci USA 2011;108:16771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R et al. vegan: Community Ecology Package. 2013. https://cran.r-project.org/web/packages/vegan/vegan.pdf.
- Paulson JN, Stine OC, Bravo HC et al. Differential abundance analysis for microbial marker-gene surveys. Nat Methods 2013;10:1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng XF, Jayakumar A, Ward BB. Community composition of ammonia-oxidizing archaea from surface and anoxic depths of oceanic oxygen minimum zones. Front Microbiol 2013;4, DOI: 10.3389/fmicb.2013.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pester M, Schleper C, Wagner M. The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 2011;14:300–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser JI, Nicol GW. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 2008;10:2931–41. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing Vienna, Austria: R Foundation for Statistical Computing, 2013. [Google Scholar]
- Sintes E, Bergauer K, De Corte D et al. Archaeal amoA gene diversity points to distinct biogeography of ammonia-oxidizing Crenarchaeota in the ocean. Environ Microbiol 2013;15:1647–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard SF, Smith BJ, Hein R et al. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res 2015;43:D593–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan BK, Chaffin MD, Martinez-Garcia M et al. Genomic and metabolic diversity of marine group I Thaumarchaeota in the mesopelagic of two subtropical gyres. PLoS One 2014;9:e95380, DOI: 10.1371/journal.pone.0095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Techtmann SM, Fortney JL, Ayers KA et al. The unique chemistry of eastern mediterranean water masses selects for distinct microbial communities by depth. PLoS One 2015;10, DOI: 10.1371/journal.pone.0120605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thingstad TF, Krom MD, Mantoura RFC et al. Nature of phosphorus limitation in the ultraoligotrophic Eastern Mediterranean. Science 2005;309:1068–71. [DOI] [PubMed] [Google Scholar]
- Tolar BB, King GM, Hollibaugh JT. An analysis of Thaumarchaeota populations from the Northern Gulf of Mexico. Front Microbiol 2013;4, DOI: 10.3389/fmicb.2013.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchter C, Abbas B, Coolen MJL et al. Archaeal nitrification in the ocean. P Natl Acad Sci USA 2006;103:12317–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimov MM, La Cono V, Denaro R. A first insight into the occurrence and expression of functional amoA and accA genes of autotrophic and ammonia-oxidizing bathypelagic Crenarchaeota of Tyrrhenian Sea. Deep-Sea Res Pt II 2009;56:748–54. [Google Scholar]
- Yakimov MM, La Cono V, Smedile F et al. Contribution of crenarchaeal autotrophic ammonia oxidizers to the dark primary production in Tyrrhenian deep waters (Central Mediterranean Sea). ISME J 2011;5:945–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at FEMSEC online.