Abstract
The determination of cell size is a fundamental challenge for all living organisms. In a given growth condition, cell size for a particular bacterial species typically falls within a narrow distribution. Nonetheless, size can vary enormously across species, and the size of a single bacterium can even vary substantially across growth conditions. Recent phenomenological studies have revived classic interest in how cells maintain their size and how they adjust their size with changes in growth rate. However, the mechanisms by which cells establish a particular size are relatively enigmatic. Here, we review existing knowledge on how size in rod-shaped bacteria is shaped by nutrient, mechanical, and genetic factors. We also examine obstacles to accurate size measurement and recent technologies that help to overcome these hurdles. Finally, we discuss the relevance of cell size to bacterial physiology.
Keywords: cell size, bacteria, cell wall, systems biology, morphology, MreB
The authors review existing knowledge on how cell size is determined in rod-shaped bacteria, discuss technological advances that have enabled precise quantification of cell size, and explore how cell size is connected to bacterial physiology.
INTRODUCTION
Although the very name of the field of microbiology conjures up images of cells on the micron scale, the bacterial domain encompasses a menagerie of shapes with cell volumes ranging over more than 10 orders of magnitude (Young 2006). Nonetheless, individual species can maintain a characteristic morphology with remarkable robustness. For the model rod-shaped bacterium Escherichia coli, cells maintain their cross-sectional width throughout elongation and division to within 10% of 1 μm when grown in rich medium (Furchtgott et al.2011), and yet they change their size substantially when transitioning between nutrient-poor and nutrient-rich environments (Woldringh et al.1980). Do certain sizes afford selective advantages, or is size a consequence of other growth behaviors? And what are the mechanisms by which cells determine their size during steady-state growth and environmental transitions?
The peptidoglycan cell wall, a network of long sugar strands crosslinked by short peptides, has emerged as a common determinant of cell shape and size across the bacterial domain (Holtje 1998). The membrane and cell wall are inflated elastically by turgor pressure, the osmotic differential between the inside and outside of the cell. In many rod-shaped bacteria, wall growth is coordinated by the actin homolog MreB (Ursell et al.2014), which rotates approximately along the circumferential direction in a manner dependent on cell-wall synthesis (van Teeffelen et al.2011). The dynamics of MreB and associated wall synthesis enzymes have helped illuminate some molecular underpinnings of size determination.
The increasing sophistication of environmental control using microfluidics (Wang et al.2010; Rojas, Theriot and Huang 2014), as well as high-throughput image acquisition (Peters et al.2016; Shi et al.2017) and analysis (Ducret, Quardokus and Brun 2016; Paintdakhi et al. 2016; Stylianidou et al.2016; Ursell et al.2017), has enabled precise measurements of cell size with a resolution of tens of nanometers, which has in turn facilitated the development of molecular (Monds et al.2014) and phenomenological (Harris and Theriot 2016) factors involved in size control. Nonetheless, we are far from a complete understanding of the genetic factors that control cell size.
In this review, we discuss nutrient, mechanical, and genetic factors that affect cell size. We consider how the average cell size within a population can be modulated, rather than how cells maintain their size (homeostasis). We focus on rod-shaped organisms, for which size can be expressed in terms of width and length variables. Width is relatively constant across the cell cycle and across the population in many such organisms (at least for a given condition and growth phase) (Furchtgott et al.2011), and length is the only variable that substantially changes during the cell cycle. We also review recently developed tools to probe size regulation, and the importance of cell size for bacterial physiology.
NUTRIENT PERTURBATIONS TO CELL SIZE
It is clear that cell size is at least partly an emergent property of cellular physiology. Seminal work by Schaechter, Maaløe, and Kjeldgaard (1958) demonstrated that when Salmonella enterica serovar Typhimurium's growth rate was tuned by adjusting the nutrient content of the medium, mean cell volume during exponential growth increased with increasing growth rate, with a range of > 2-fold (Fig. 1A). This empirical relationship is now known as the Growth Law (Vadia and Levin 2015). The dependence of cell size on nutrient-dependent changes in growth rate has also been observed in Escherichia coli (Taheri-Araghi et al.2015), Bacillus subtilis (Weart et al.2007), and Caulobacter crescentus (Iyer-Biswas et al.2014). For both the Gram-negative (thin cell wall) E. coli (Taheri-Araghi et al.2015) and the Gram-positive (thick cell wall) B. subtilis (Peters et al.2016), the increase in size occurs through increases in both mean length and width. Thus, it appears that cell size can generally be modulated by nutrients.
Figure 1.
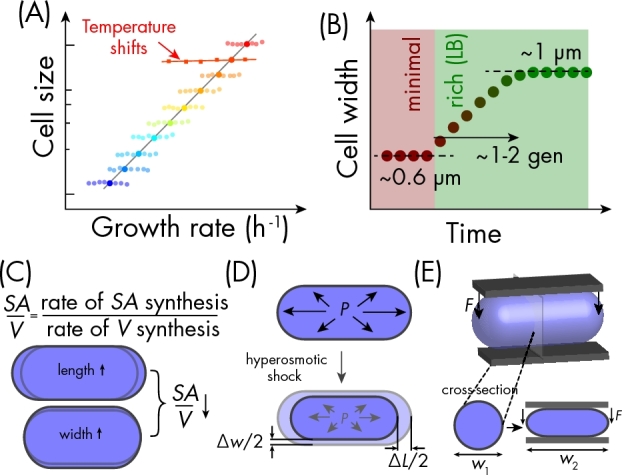
Environmentally induced changes in cell width. (A) For many rod-shaped bacteria, steady-state mean cell size (large circles) scales exponentially with nutrient-determined growth rate (schematic modeled after data from Taheri-Araghi et al.2015). By contrast, variations in growth rate within a population are not correlated with cell size (small circles), and cells maintain their mean size as temperature is varied (squares). (B) E. coli cell width increases rapidly upon nutrient upshift (Woldringh et al.1980). (C) Relative rates of surface area and volume synthesis determine cell size, specifically through the surface area-to-volume ratio (Harris and Theriot 2016). Increases in either cell length (top) or width (bottom) decrease surface area-to-volume ratio. (D) Hyperosmotic shock (e.g. by adding highly concentrated osmolytes to the medium) transiently reduces turgor pressure and causes reversible decreases in cell length and width in E. coli. (E) Mechanical compression (represented by downward force F) leads to an increase in cell width (Si et al.2015).
Upon nutrient upshift from minimal to rich medium, cell width starts increasing almost immediately, and plateaus after a few cell cycles (Fig. 1B) (Woldringh et al.1980). However, in E. coli, fluctuations in growth rate across a population at steady state were not coupled to changes in cell size (Fig. 1A) (Taheri-Araghi et al.2015). Moreover, in Salmonella Typhimurium (Schaechter, Maaløe and Kjeldgaard 1958) and C. crescentus (Iyer-Biswas et al.2014), size did not change when growth rate was tuned by changing temperature (Fig. 1A). These data indicate that size is not causally related to growth rate. Reinforcing this point, at the molecular level, MreB rotation speed increases with increasing temperature but is unaffected by changes in nutrient conditions (van Teeffelen et al.2011), indicating that the kinetics of cell-wall synthesis are differentially affected by changes in temperature vs nutrients.
A recent study introduced a model in which cell size is set by the ratio of the rate of surface area synthesis to the rate of volume synthesis (Fig. 1C) (Harris and Theriot 2016): nutrient upshift causes a relative increase in cytoplasmic synthesis that reduces the surface area-to-volume ratio (and hence increases cell width), whereas temperature affects both synthesis rates equally and hence does not affect size. While this model does not address the genetic factors that determine absolute cell size or the mechanism by which cells modulate surface area and volume synthesis to varying degrees, it nevertheless provides an exciting framework for predicting how steady-state cell size changes upon chemical or nutrient perturbations.
MECHANICAL PERTURBATIONS TO CELL SIZE
Although cell shape is generally thought to be defined by the molecular architecture of the cell wall, the wall is nevertheless elastic and can be deformed through applied mechanical stress. One way the environment interacts with the mechanical properties of the wall is through turgor pressure, which is defined by the difference in osmolarity between the surroundings and the cytoplasm. In Escherichia coli, a sudden increase in the osmolarity of the environment leads to a transient decrease in turgor pressure, which causes reversible shrinking of the wall as water leaves the cell and a decrease in both cell width and length of ∼10% (Fig. 1D) (Rojas, Theriot and Huang 2014). The magnitude of these changes is smaller than what can be achieved by changes in nutrients, and is reversible, since a subsequent hypoosmotic shock reverses the width/length changes (Rojas, Theriot and Huang 2014). Nonetheless, osmotic forces can affect cell size in a manner orthogonal to nutrients and/or growth rate.
Cell shape and size can also be perturbed by direct physical contacts with the environment that apply forces to the exterior of the cell. When confined to chambers with cross-sectional dimensions similar to the cell width (∼1 μm), filamentous E. coli cells adopted the bent shape of the chamber that persisted after cells were removed from the chamber (Takeuchi et al.2005). Compression by a polydimethylsiloxane layer prompted E. coli cells to grow in a ‘pancake’-like morphology, where the dimension perpendicular to the membrane could be decreased by 25%–50% with a concomitant increase in the perpendicular ‘width’ axis as the cells grew (Fig. 1E) (Si et al.2015). Although cells under large compression ended up with aberrant morphologies that no longer had a rod-like shape, they were still rod-like in the initial stages of growth. Intriguingly, when compressed by ∼50%, cells stopped growing altogether (Si et al.2015), suggesting that changes in cell size can in some cases exert feedback on growth rate. Thus, cells adapt their size and shape to mechanical forces from the environment. An attractive possibility is that intracellular structures, such as cytoskeletal filaments or DNA, could exert similar forces on the cell wall and thereby influence cell morphology.
GENETIC PERTURBATIONS THAT ALTER CELL WIDTH
Cell-wall synthesis enzymes
Recent studies have elaborated on genes connected to cell-wall synthesis that appear to be integral for width establishment and maintenance. Deletion of the Bacillus subtilis ponA gene encoding PBP1, a bifunctional peptidoglycan synthase, led to thinner cells (Fig. 2A) (Tocheva et al.2013); whether this mutant grew more slowly than wild-type cells was not reported. To interrogate the contribution of essential genes to cell shape in B. subtilis, a CRISPRi library was used to partially knockdown expression of these genes (Peters et al.2016). The strains had a wide range of lag times; when cell size was measured at a fixed time point after stationary-phase exit (and hence cells had a wide range of growth rates), average length and width across the population of cells from each strain varied substantially but were highly correlated, consistent with the Growth Law (Peters et al.2016). The only functional category of genes for which a significant fraction diverged from this expected relationship was cell-wall synthesis, suggesting that this class of genes plays a (perhaps to be expected) pivotal role in size determination.
Figure 2.
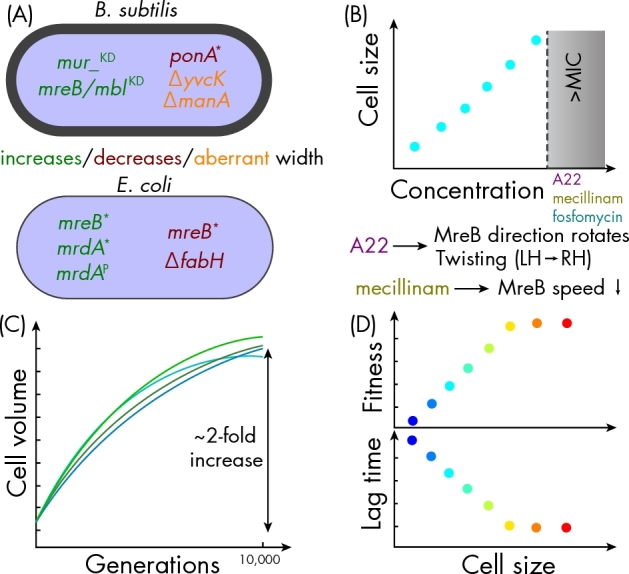
Molecular and evolutionary changes to cell width. (A) Genes with established connections to cell-width determination in B. subtilis (top) and E. coli (bottom). *: mutation in coding region; KD: CRISPRi knockdown; P: promoter mutation. Color indicates whether perturbation leads to increased, decreased, or aberrant cell width. (B) Sublethal treatments with A22 (targets MreB), mecillinam (PBP2), and fosfomycin (MurA) increase cell width. For A22, increases in cell width are correlated with rotation of the direction of MreB movement and a continuous transition from left-handed to right-handed twisting, while mecillinam causes MreB speed to decrease (Tropini et al.2014). (C) Cell volume was observed to increase concurrently with cell fitness in a long-term evolution experiment (Lenski and Travisano 1994). Each line in the schematic represents an individual evolved population. (D) The increased cell width of mreBA53X mutants is correlated with increased competitive fitness and decreased lag time (Monds et al.2014).
A recent study used three methods (possibly mechanistically linked) to vary cell width in Escherichia coli (Fig. 2B) (Tropini et al.2014): treatment with sublethal concentrations of A22 (inhibitor of MreB) or mecillinam (inhibitor of the essential cell-wall transpeptidase PBP2, encoded by mrdA), or heterologous expression of mrdA from Vibrio cholerae, another Gram-negative Gammaproteobacterium. A22 or mecillinam treatment caused approximately linear increases in cell width with drug concentration, suggesting that MreB and/or PBP2 may be directly involved in cell-width determination. Induction of E. coli mrdA in a strain constitutively expressing V. cholerae mrdA led to cell widths that inversely depended on the level of inducer, reflecting competition between the two enzyme variants that implicates PBP2 concentration and/or activity in width determination. MreB speed, directionality, and the ultrastructure of the cell wall revealed by cell twisting during growth (Wang et al.2012) changed systematically as width increased for all three perturbations, although E. coli cells appeared to modulate cell width in qualitatively different manners depending on how MreB or PBP2 function was disrupted (Tropini et al.2014). Thus, there may be multiple mechanisms by which perturbations to the wall synthesis machinery can lead to altered cell width.
Long-term evolution experiments (LTEEs) involving repeated passaging have served as a resource for identifying adaptive mutations. Quantification of single cells from the seminal LTEE performed by Richard Lenski and colleagues revealed that cell volume increased concurrently with changes in fitness, with a ∼2-fold increase in volume in all of the evolved lines after 10 000 generations (Fig. 2C) (Lenski and Travisano 1994). Thus, cell size appears to be linked with competitive fitness, which is determined by factors including growth rate, lag time, stationary-phase survival, and carrying capacity. Several mutations in the E. coli operon encoding PBP2 were identified in evolved lines (Fig. 2A). Two mutations upstream of mrdA resulted in a decrease in the cellular concentration of PBP2 when introduced into the ancestor, which led to an increase in cell volume and cell rounding (Philippe et al.2009). These mutations conferred increased fitness in competition over a passage cycle, but fitness was reduced during prolonged stationary phase, suggesting trade-offs between cell size and stationary-phase recovery and/or survival (Philippe et al.2009). Several other mutations in genes encoding cell-wall-related enzymes have been identified in LTEEs (Tenaillon et al.2012); it will be interesting to establish whether these mutations are causative for changes in cellular dimensions and/or are adaptive.
Cell-wall precursor synthesis
In B. subtilis, knockdown of the mur genes, which are involved in peptidoglycan precursor synthesis, resulted in wider cells, and the level of width increase in murB knockdown cells scaled with the level of murB repression (Fig. 2A) (Peters et al.2016). In both E. coli and Caulobacter crescentus, the drug fosfomycin, which inhibits one of the Mur enzymes, also caused a dose-dependent increase in width (Fig. 2B) (Harris and Theriot 2016). This dependence of width on the concentration of cell-wall precursor is consistent with model predictions of an altered ratio of peptidoglycan synthesis compared with the synthesis of other cytoplasmic components (Harris and Theriot 2016), and could result mechanistically from changes in the biochemical composition of the wall such as shorter glycan strands (Furchtgott et al.2011). As a probe of potential alterations to wall ultrastructure, it will be intriguing to probe whether perturbed cells exhibit more mechanical strain under hyperosmotic shock.
The actin-like cytoskeleton
In E. coli, a set of allelic variants involving single nucleotide polymorphisms of the A53 residue of MreB displayed a range of cell widths larger than the parental REL606 strain (Fig. 2D) (Monds et al.2014). These mutations also increased cell width in other E. coli genotypes such as BW25113 and MG1655 (unpublished). When grown on glucose, the mreBA53X mutants had a competitive fitness advantage relative to the parent that scaled with cell width up to a maximum fitness gain of ∼10%. Interestingly, fitness gains resulted from a width-dependent decrease in lag time (Fig. 2D) rather than any increase in maximal growth rate (Monds et al.2014). Thus, mutations that change cell width can also be adaptive, through a mechanism that somehow accelerates growth during the transition from starvation into fresh medium. mreBA53T cells were consistently shorter and larger in volume (due to the quadratic scaling of volume with width) than the ancestor when grown on a variety of carbon sources (Monds et al.2014), suggesting that the mechanism underlying cell size changes is largely decoupled from metabolic state. However, the fitness impact ranged from large increases in competitive index (∼10% on many carbon sources) to neutral (on lactose) to small decreases (∼3% on galactose) (Monds et al.2014). It remains to be seen whether fitness effects of cell size are exclusively associated with lag phase.
Selection for increased A22 resistance in E. coli uncovered mreB mutations that also affect cell size (Fig. 2A), with cell widths correlated with biophysical parameters such as MreB filament orientation (Ouzounov et al.2015). In B. subtilis, when growth phase was accounted for by measuring cell size at the same optical density, partial knockdowns of the homologs mreB and mbl resulted in increased width (Fig. 2A); interestingly, the distribution of cell widths in mreB knockdown cells also had a much longer tail than wild type, suggesting differential effects of mreB and mbl on B. subtilis cell width despite their common homology to actin (Peters et al.2016). Changes in the expression levels of RodZ, a bitopic protein that binds both MreB and the cell wall, led to increases in cell width (Shiomi, Sakai and Niki 2008). Deletion of rodZ led to rounding, and suppressors of the slow-growth phenotype of ΔrodZ cells were isolated in mreB, mrdA and mrdB (which encodes RodA, a recently discovered peptidoglycan polymerase (Cho et al.2016; Meeske et al.2016)). Many of these suppressors had rod-like shape but different cell sizes (Fig. 2A). Taken together, these data indicate a central role for the elongation machinery, and MreB in particular, in width determination.
Control of cell width beyond the wall synthesis machinery
Although cell-wall synthesis clearly plays a central role in size and shape determination, other processes linked to the physical architecture of the cell have been implicated in width control. In E. coli, deletion of fabH, which synthesizes a precursor of fatty acid biosynthesis and hence the membrane component of the cell envelope, results in a 28% decrease in width during growth in LB (Fig. 2A) (Yao et al.2012).
In B. subtilis, there are also hints that metabolic genes can directly affect width. Deletion of a transferase protein of largely unknown function (YvcK) led to width dysregulation and aberrant shapes on gluconeogenic carbon sources (Fig. 2A) (Gorke, Foulquier and Galinier 2005). Intriguingly, the shape phenotypes of ΔyvcK cells were suppressed by overexpression of MreB or deletion of ponA, which encodes PBP1 (Foulquier et al.2011). Deletion of manA, which encodes a mannose metabolic enzyme, changed the teichoic acid content of the cell wall and resulted in increased variance in both width and length at the population level and non-uniform width at the single-cell level in conditions that allowed faster growth rates (Elbaz and Ben-Yehuda 2010), suggesting that other components of the cell envelope may also modulate cell dimensions. The CRISPRi screen of the effect of essential gene knockdown on cell size also identified some unexpected genes as outliers to the length/width correlation, including many genes involved in replication such as nrdF, nrdE, dnaA, and dnaX, as well as a translational termination factor tufA (Peters et al.2016). In the latter case, EF-Tu has been suggested to interact with MreB (Soufo et al.2010), yet again suggesting a link between MreB and width determination. Several gene knockouts in E. coli yielded wide cells in a genome-scale morphological screen (http://shigen.nig.ac.jp/ecoli/strain/) (Ursell et al.2017). In each of these cases, it is uncertain whether genes besides the cell-wall synthesis machinery are directly involved in determining width as opposed to influencing the metabolic coupling between cell size and growth rate.
MODULATION OF CELL LENGTH BEYOND STEADY-STATE ELONGATION AND DIVISION
Unlike cell width, length by definition increases during elongation, and hence the mean length does not represent the length of individual growing cells or the range that they explore. Although atypical, some E. coli isolates can reach lengths >750 μm without increasing width (El-Hajj and Newman 2015). Many bacterial species have been shown to increase in cell volume exponentially in at least some growth conditions, including cells with different shapes (Zhou et al.2015) and growth patterns (Brown et al.2012). In these cases, mean length is determined independently of doubling time T or elongation rate g, which are linked at steady state by T = ln 2/g. Nonetheless, length can be altered via nutrients through regulation of the division machinery, which is achieved by a metabolic sensor that inhibits FtsZ in E. coli (Hill et al.2013) and B. subtilis (Weart et al.2007). Cells sometimes also shorten when entering stationary phase (Kolter, Siegele and Tormo 1993), and transiently become filamentous via inhibition of FtsZ by SulA in the SOS response (Bi and Lutkenhaus 1993). It was recently found that size homeostasis is achieved in E. coli (and perhaps many other species) by adding a constant volume Δ per cell cycle independent of initial length (Campos et al.2014; Taheri-Araghi et al.2015). However, the genetic determinants of Δ are unknown. Because elongation changes the proportion of the cell that is polar and hence alters the surface area-to-volume ratio, growth rate (Harris and Theriot 2016) must be considered along with regulatory mechanisms such as stress responses in length determination.
TECHNOLOGIES AND CHALLENGES FOR MEASURING CELL SIZE
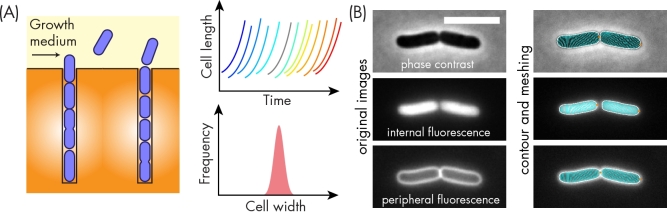
Since cell width responds to changes in nutrient conditions and growth phase, it is unclear whether cell width is encoded by a specific set of pathways or whether it can generally be modulated by perturbations to many cellular processes. Any comparison between strains must control for growth environment. Preferably, cellular dimensions would be measured in chemostat-like conditions, such as in a microfluidic flow cell (Fig. 3A) (Wang et al.2010; Iyer-Biswas et al.2014; Rojas, Theriot and Huang 2014), to ensure steady-state growth, and across a wide dynamic range of growth rates. Even then, methodological differences must be considered: some studies utilize phase-contrast (Campos et al.2014) imaging while others use surface (Rojas, Theriot and Huang 2014) or interior (Wang et al.2010; Taheri-Araghi et al.2015) fluorescence microscopy measurements (Fig. 3B). Furthermore, the details of contour extraction by image-analysis packages and subsequent width/length quantifications can have qualitative effects on basic conclusions. For example, two separate analyses (Iyer-Biswas et al.2014; Sauls, Li and Jun 2016) of the same dataset of birth and division volumes of Caulobacter crescentus cells supported different cell-size homeostasis models. As a result, few studies have quantified the average cellular dimensions of particular genotypes in a manner that can be directly compared with other strains or mutants, and fewer still have elaborated on whether genetic perturbations result in tunable changes in cell width (as might be expected if the gene of interest modulates width directly rather than indirectly).
Figure 3.
Technologies for quantifying cellular dimensions. (A) Microfluidic flow cells permit monitoring of steady-state growth and dimensions. In the schematic, rod-shaped cells grow in narrow channels until they reach the flow path of the growth medium, whereupon they are carried away (Wang et al.2010). Such devices enable tracking of cell length from birth to division for thousands of individual cells (top right) and the measurement of the distribution of cell widths (bottom right); the latter is typically narrow for organisms such as E. coli. (B) Examples of phase-contrast and peripheral and internal fluorescence modalities for cell contour measurement via image analysis. On the left are raw images for a pair of E. coli daughter cells from a recent division. The cells expressed cytoplasmic GFP (middle), and the membrane was labeled with the dye FM4–64 (bottom). On the right are the contours (white) extracted from each image using the software package Morphometrics (Ursell et al.2017), with mesh lines (cyan) that can be used to compute the length of the midline starting from one of the poles (orange dots) and the distribution of widths along that midline.
DISCUSSION
Cell size is clearly a readout of physiological parameters such as growth rate, and is also potentially a regulator of behaviors such as lag phase. The studies we have reviewed here exemplify how cell size is carefully controlled in part by synthesis of the cell envelope. Is there a single protein such as MreB that is the primarily molecular ruler for determining parameters like cell width, or are there many connected factors? Although it is difficult to absolutely define such a factor, one quality that such an agent might possess would be tunability, with mutations to the same protein yielding a range of size phenotypes. These phenotypes might be expected to be roughly independent of the genetic background, although different species may rely on radically different molecular mechanisms to establish, sense and maintain size.
Screening for other mutants with subtle size changes may be facilitated by flow cytometry-based screens, which to date have been applied to identify shape mutants (Laubacher et al.2013; Sycuro et al.2013). Cell shape affects many behaviors, such as adhesion, predation, and motility (Young 2006); is cell size similarly important? Cell size has been linked to immune evasion (Champion and Mitragotri 2006), and it will be exciting to systematically probe other behaviors with size mutants. To identify other genetic factors, mutants must be compared to the corresponding wild-type strain in the same environmental conditions and the same growth phase (or in steady-state growth). Size changes may be environment-specific; it is tempting to speculate that envelope-related mutations will induce similar changes relative to wildtype in all environments, whereas metabolic mutations may cause size changes indirectly and only in particular growth conditions.
Fitness advantages associated with faster growth can be exploited in LTEEs to isolate mutations that tune cell size; one such experiment carried out at 42 °C identified mutations in MreB and PBP2 (Tenaillon et al.2012), although it was not investigated as to whether these mutations are adaptive and/or cause size changes. Since fitness can improve via increases in growth rate, size increases may not be surprising. Nevertheless, some mutations alter cell size independent of volume growth rate (Monds et al.2014), suggesting that the mutated genes specifically affect surface synthesis. Conversely, coupled measurements of growth rate and size could provide insight into the relative adaptation of surface and cytoplasmic synthesis rates to environmental conditions such as osmolarity, pH, oxygen, metabolites, antibiotics, and growth phase.
The dynamic range of cell size for any given organism is unknown. For Escherichia coli, cell width decreases to ∼0.6 μm in minimal medium (Woldringh et al.1980), and treatment with sublethal concentrations of A22 increases cell width up to ∼2 μm in a dose-dependent manner (Tropini et al.2014). Does this range represent fundamental limits to E. coli width? This question could be partially answered via A22 treatment of mutants with increased cell width to determine whether they have the same limiting width. Comparative studies of other species may also reveal mechanisms of width regulation. For instance, Agrobacterium tumefaciens cells have a tapered morphology, with the growing tip increasing in width substantially throughout the cell cycle (Brown et al.2012).
The ability to tune cell size independent of growth rate could also be beneficial for biotechnology, in which the yield of heterologous proteins may be higher in bigger or smaller cells. Meta-analyses comparing size measurements across genome-scale mutant libraries (Baba et al.2006) to chemical genomics screens (Nichols et al.2011) will indicate how size generally sensitizes cells to certain environments or treatments (Ursell et al. 2017). The trade-offs in fitness observed in different conditions and methods of perturbation already observed also highlight the importance of cell size to bacterial evolution. The ability to tune cell size in a multitude of ways will be critical to uncover how cell size affects fundamental physiological properties, including the composition of the proteome and the envelope, DNA/RNA abundance, and subcellular patterning of lipids and proteins. Current research directions present exciting synergistic opportunities for biophysical, mechanistic, phenomenological, and physiological studies of bacterial cell size that will critically impact microbiology and the emerging nexus between cell and evolutionary biology.
Acknowledgments
The authors thank the Huang lab, Alexandre Colavin, Leigh Harris, and Petra Levin for helpful discussions.
FUNDING
This work was supported in part by a Stanford Graduate Fellowship and a National Science Foundation (NSF) Graduate Research Fellowship (to SC), the Allen Discovery Center at Stanford on Systems Modeling of Infection (to KCH), the Stanford Center for Systems Biology under NIH grant P50-GM107615 (to KCH), and NSF CAREER Award MCB-1149328 (to KCH).
Conflict of interest. None declared.
REFERENCES
- Baba T, Ara T, Hasegawa M et al. . Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2006;2:2006–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol 1993;175:1118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, de Pedro MA, Kysela DT et al. . Polar growth in the Alphaproteobacterial order Rhizobiales. P Natl Acad Sci USA 2012;109:1697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M, Surovtsev IV, Kato S et al. . A constant size extension drives bacterial cell size homeostasis. Cell 2014;159:1433–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion JA, Mitragotri S. Role of target geometry in phagocytosis. P Natl Acad Sci USA 2006;103:4930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Wivagg CN, Kapoor M et al. . Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol 2016;1:16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nature Microbiol 2016;1:16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hajj ZW, Newman EB. An Escherichia coli mutant that makes exceptionally long cells. J Bacteriol 2015;197:1507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz M, Ben-Yehuda S. The metabolic enzyme ManA reveals a link between cell wall integrity and chromosome morphology. PLos Genet 2010;6:e1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulquier E, Pompeo F, Bernadac A et al. . The YvcK protein is required for morphogenesis via localization of PBP1 under gluconeogenic growth conditions in Bacillus subtilis. Mol Microbiol 2011;80:309–18. [DOI] [PubMed] [Google Scholar]
- Furchtgott L, Wingreen NS, Huang KC. Mechanisms for maintaining cell shape in rod-shaped Gram-negative bacteria. Mol Microbiol 2011;81:340–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorke B, Foulquier E, Galinier A. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology 2005;151:3777–91. [DOI] [PubMed] [Google Scholar]
- Harris LK, Theriot JA. Relative rates of surface and volume synthesis set bacterial cell size. Cell 2016;165:1479–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill NS, Buske PJ, Shi Y et al. . A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLos Genet 2013;9:e1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol R 1998;62:181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Biswas S, Wright CS, Henry JT et al. . Scaling laws governing stochastic growth and division of single bacterial cells. P Natl Acad Sci USA 2014;111:15912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R, Siegele DA, Tormo A. The stationary phase of the bacterial life cycle. Ann Rev Microbiol 1993;47:855–74. [DOI] [PubMed] [Google Scholar]
- Laubacher ME, Melquist AL, Chandramohan L et al. . Cell sorting enriches Escherichia coli mutants that rely on peptidoglycan endopeptidases to suppress highly aberrant morphologies. J Bacteriol 2013;195:855–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE, Travisano M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. P Natl Acad SciUSA 1994;91:6808–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Riley EP, Robins WP et al. . SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 2016;537:634–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monds RD, Lee TK, Colavin A et al. . Systematic perturbation of cytoskeletal function reveals a linear scaling relationship between cell geometry and fitness. Cell Rep 2014;9:1528–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RJ, Sen S, Choo YJ et al. . Phenotypic landscape of a bacterial cell. Cell 2011;144:143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzounov N, Nguyen J, Bratton B et al. . MreB helical pitch angle determines cell diameter in Escherichia coli. arXiv:150307789, 2015. [Google Scholar]
- Paintdakhi A, Parry B, Campos M et al. . Oufti: An integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol Microbiol 2016;99:767–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Colavin A, Shi H et al. . A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 2016;165:1493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe N, Pelosi L, Lenski RE et al. . Evolution of penicillin-binding protein 2 concentration and cell shape during a long-term experiment with Escherichia coli. J Bacteriol 2009;191:909–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas E, Theriot JA, Huang KC. Response of Escherichia coli growth rate to osmotic shock. P Natl Acad Sci USA 2014;111:7807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauls JT, Li D, Jun S. Adder and a coarse-grained approach to cell size homeostasis in bacteria. Curr Opin Cell Biol 2016;38:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaechter M, Maaløe O, Kjeldgaard N. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol 1958;19:592–606. [DOI] [PubMed] [Google Scholar]
- Shi H, Colavin A, Lee TK et al. . Strain Library Imaging Protocol for high-throughput, automated single-cell microscopy of large bacterial collections arrayed on multiwell plates. Nat Protoc 2017;12:429–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Sakai M, Niki H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J 2008;27:3081–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si F, Li B, Margolin W et al. . Bacterial growth and form under mechanical compression. Sci Rep 2015;5:11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufo HJD, Reimold C, Linne U et al. . Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein. P Natl Acad SciUSA 2010;107:3163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianidou S, Brennan C, Nissen SB et al. . SuperSegger: robust image segmentation, analysis and lineage tracking of bacterial cells. Mol Microbiol 2016;102:690–700. [DOI] [PubMed] [Google Scholar]
- Sycuro LK, Rule CS, Petersen TW et al. . Flow cytometry-based enrichment for cell shape mutants identifies multiple genes that influence Helicobacter pylori morphology. Mol Microbiol 2013;90:869–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri-Araghi S, Bradde S, Sauls JT et al. . Cell-size control and homeostasis in bacteria. Curr Biol 2015;25:385–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi S, DiLuzio WR, Weibel DB et al. . Controlling the shape of filamentous cells of Escherichia coli. Nano Lett 2005;5:1819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Rodriguez-Verdugo A, Gaut RL et al. . The molecular diversity of adaptive convergence. Science 2012;335:457–61. [DOI] [PubMed] [Google Scholar]
- Tocheva EI, Lopez-Garrido J, Hughes HV et al. . Peptidoglycan transformations during Bacillus subtilis sporulation. Mol Microbiol 2013;88:673–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropini C, Lee TK, Hsin J et al. . Principles of bacterial cell-size determination revealed by cell-wall synthesis perturbations. Cell Rep 2014;9:1520–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell T, Lee TK, Shiomi D et al. . Rapid, precise quantification of bacterial cellular dimensions across a genomic-scale knockout library. BMC Biol, 2017;15:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell TS, Nguyen J, Monds RD et al. . Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. P Natl Acad Sci USA 2014;111:E1025–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadia S, Levin PA. Growth rate and cell size: a re-examination of the growth law. Curr Opin Microbiol 2015;24:96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeffelen S, Wang S, Furchtgott L et al. . The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. P Natl Acad Sci USA 2011;108:15822–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Robert L, Pelletier J et al. . Robust growth of Escherichia coli. Curr Biol 2010;20:1099–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Furchtgott L, Huang KC et al. . Helical insertion of peptidoglycan produces chiral ordering of the bacterial cell wall. P Natl Acad Sci USA 2012;109:E595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weart RB, Lee AH, Chien A-C et al. . A metabolic sensor governing cell size in bacteria. Cell 2007;130:335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh CL, Grover NB, Rosenberger RF et al. . Dimensional rearrangement of rod-shaped bacteria following nutritional shift-up. II. Experiments with Escherichia coli B/r. J Theor Biol 1980;86:441–54. [DOI] [PubMed] [Google Scholar]
- Yao Z, Davis RM, Kishony R et al. . Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. P Natl Acad Sci USA 2012;109:E2561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD. The selective value of bacterial shape. Microbiol Mol Biol R 2006;70:660–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Halladin DK, Rojas ER et al. . Mechanical crack propagation drives millisecond daughter cell separation in Staphylococcus aureus. Science 2015;348:574–8. [DOI] [PMC free article] [PubMed] [Google Scholar]