Abstract
Protozoan predation is one of the main environmental factors constraining bacterial growth in aquatic environments, and thus has led to the evolution of a number of defence mechanisms that protect bacteria from predation. These mechanisms may also function as virulence factors in infection of animal and human hosts. Whole transcriptome shotgun sequencing of Vibrio cholerae biofilms during predation by the amoebae, Acanthamoeba castellanii, revealed that 131 transcripts were significantly differentially regulated when compared to the non-grazed control. Differentially regulated transcripts included those involved in biosynthetic and metabolic pathways. The transcripts of genes involved in tyrosine metabolism were down-regulated in the grazed population, which indicates that the tyrosine metabolic regulon may have a role in the response of V. cholerae biofilms to A. castellanii predation. Homogentisate 1, 2-dioxygenase (HGA) is the main intermediate of the normal L-tyrosine catabolic pathway which is known to auto-oxidize, leading to the formation of the pigment, pyomelanin. Indeed, a pigmented mutant, disrupted in hmgA, was more resistant to amoebae predation than the wild type. Increased grazing resistance was correlated with increased production of pyomelanin and thus reactive oxygen species (ROS), suggesting that ROS production is a defensive mechanism used by bacterial biofilms against predation by amoebae A. castellanii.
Keywords: protozoan predation, biofilms, environmental fitness, V. cholerae, grazing resistance, pigment production
Pigment production by the pathogenic bacterium that causes cholera protects the bacterium from being consumed by the natural predatory amoeba A. castellanii.
INTRODUCTION
Vibrio cholerae, the causative agent of cholera, persists in brackish and estuarine water systems (Colwell, Kaper and Joseph 1977; Huq et al.1990) where it is exposed to starvation conditions, fluctuations in temperature and salinity, and predators (Lutz et al.2013). The persistence of V. cholerae in the environment indicates its ability to respond to such stresses (Colwell and Huq 1994; Lutz et al2013; Sun et al.2015). Heterotrophic protists are the biggest consumers of bacteria in the environment, and are thus a major mortality factor for bacteria (Jürgens and Matz 2002).
In benthic marine, brackish and freshwater sediments, where V. cholerae naturally occurs, ciliates are the most abundant protists, while amoebae contribute most of the biomass (Lei et al.2014). V. cholerae shares an ecological niche with the model protozoa, Acanthamoeba castellanii and Tetrahymena pyriformis. The free-living amoeba, Acanthamoeba spp. have been isolated from various fresh and salt water sources (Khan 2006) where they feed on bacterial biofilms. V. cholerae and Acanthamoeba spp. were detected in water samples collected from different cholera endemic areas in Sudan (Shanan et al.2011). V. cholerae is often isolated from freshwater systems (Nair et al.1988) where T. pyriformis typically occurs, feeding on bacterioplankton (Elliott 1970). These predators are among the few axenic protozoan cultures available, making them ideal ecologically relevant model organisms.
Both clinical and environmental strains of V. cholerae have been shown to survive intracellularly within a range of amoeba (Thom, Warhurst and Drasar 1992; Abd, Weintraub and Sandström 2005; Abd et al.2007), and Van der Henst et al. (2016) showed that V. cholerae can grow inside A. castellanii. A study using laboratory microcosms of natural bacterioplankton communities from the Gulf of Mexico showed elimination of V. cholerae by ciliates and heterotrophic nanoflagellates (Martínez Pérez, Macek and Castro Galván 2004). In contrast, when V. cholerae biofilms were exposed to predation by flagellates, there was little effect on biofilm biomass, indicating that biofilms are protected from predation (Matz et al.2005).
Biofilms provide physical protection as well as a high cell density population that enables cell-to-cell communication, or quorum sensing (QS). QS has been shown to regulate antiprotozoal activities in V. cholerae biofilms including, the production of Vibrio polysaccharide that protects both early- and late-stage biofilms from predation by the surface-feeding nanoflagellate, Rhynchomonas nasuta and the amoeba A. castellanii (Lutz et al.2013; Sun, Kjelleberg and McDougald 2013). The extracellular protease, PrtV, provides grazing resistance against the flagellate Cafeteria roenbergensis and the ciliate, T. pyriformis (Vaitkevicius et al.2006). The type VI secretion system (T6SS) uses virulence-associated secretion proteins to deliver effector proteins that are cytotoxic to the amoebae, Dictyostelium discoideum and mammalian macrophages (Pukatzki et al.2006; Miyata et al.2011). Despite the fact that the early and late biofilms of a V. cholerae QS mutant were more susceptible to grazing by A. castellanii, C. roenbergensis and R. nasuta than the wild type, the biofilms are not completely eliminated by predation (Erken et al.2011; Lutz et al.2013), suggesting that other anti-predation strategies could be present.
Studies on bacterial prey and protozoan predators have shown several potential defences against grazing, including production of toxins, microcolony formation and changes in cell surface properties (Matz and Kjelleberg 2005). Examples of secondary metabolites that are active against protists include an alkaloid purple-pigmented metabolite, violacein, which acts as a chemical defence for several bacterial genera (Chromobacterium, Janthinobacterium, Pseudoalteromonas) (Matz et al.2004). Similarly, Pseudomonas fluorescens is known to employ the cyclic lipopeptide surfactants, massetolide and viscosin to protect itself against Naegleria americana (Mazzola et al.2009), in addition to 2,4-diacetylphloroglucinol (DAPG), pyrrolnitrin, hydrogen cyanide and pyoluteorin (Jousset et al.2010). Cell surface properties have also been shown to affect grazing resistance. For example, cell surface hydrophobicity affects grazing of picoplankton cells by nanoflagellates (Monger, Landry and Brown 1999). Moreover, Wildschutte et al. (2004) showed that differences in O-antigen are sufficient to allow for prey discrimination by protozoa grazing on different serotypes of Salmonella.
In order to study the factors contributing to grazing resistance of V. cholerae, the transcriptome of biofilms exposed to A. castellanii was analysed to identify genetic features that likely contribute to survival during predation. Here, we examine the effect of downregulation of genes involved in tyrosine degradation on grazing resistance of V. cholerae. A decrease in the activity of homogentisate 1, 2-dioxygenase (HmgA) leads to accumulation of homogentisic acid (HGA) that auto-oxidises to form pyomelanin (Turick et al.2010). Results show that the production of pyomelanin has a protective effect against predation by A. castellanii.
MATERIAL AND METHODS
Strains and growth conditions
Organisms used in this study are listed in Table 1. Bacterial strains were routinely grown in Luria-Bertani (LB) broth and on agar plates (Sambrook, Fritsch and Maniatis 1989) as appropriate, with carbenicillin (100 μg ml−1). A. castellanii was routinely passaged in 15 ml growth medium containing peptone-yeast-glucose (PYG) (20 g l−1 proteose peptone, 1 g l−1 yeast extract) supplemented with 1 litre 0.1 × M9 minimal medium (6 g l−1 Na2HPO4, 3 g l−1 KH2PO4, 0.5 g l−1 NaCl, 1 g l−1 NH4Cl) and 0.1 M sterile-filtered glucose in 25 cm2 tissue culture flasks with ventilated caps (Sarstedt Inc., Nümbrecht, Germany) and incubated statically at 30°C. A. castellanii was passaged 3 days prior to harvesting for experiments and enumerated microscopically using a haemocytometer.
Table 1.
Strains and plasmids used in this study.
| Strain | Properties | Reference/source |
|---|---|---|
| Bacterial strains | ||
| V. cholerae A1552 | Wild type, O1 El Tor, Inaba, smooth, Rifr | Valeru et al. (2009) |
| V. cholerae A1552 hmgA | O1 El Tor, Inaba, smooth, ΔhmgA, Rifr, Kmr | Valeru et al. (2009) |
| V. cholerae A1552 hmgA complement | O1 El Tor, Inaba, smooth, ΔhmgA, hmgA::pUC18, Rifr, Apr, Kmr | Valeru et al. (2009) |
| V. cholerae A1552 pUC18 | O1 El Tor, Inaba, smooth, pUC18, Rifr, Apr | This study |
| V. cholerae A1552 hmgA pUC18 | O1 El Tor, Inaba, smooth, ΔhmgA, pUC18, Rifr, Apr, Kmr | This study |
| Plasmids | ||
| pUC18 | Cloning vector, pMB1 ori, LAC pr, lacZ, Apr | Yanisch-Perron et al. (1985) |
| Protozoan strains | ||
| A. castellanii | Wild type | ATCC 30234 |
| T. pyriformis | Wild type | ATCC 205063 |
The browsing ciliate, T. pyriformis, was maintained as above but incubated statically at room temperature (RT). Prior to experiments, 500 μl of T. pyriformis was passaged in 20 ml of 0.5× NSS medium (8.8 g l−1 NaCl, 0.735 g l−1 Na2SO4, 0.04 g l−1 NaHCO3, 0.125 g l−1 KCl, 0.02 g l−1 KBr, 0.935 g l−1 MgCl2.6H2O, 0.205 g l−1 CaCl2.2H2O, 0.004 g l−1 SrCl2.6H2O and 0.004 g l−1 H3BO3) (Mårdén et al.1985) supplemented with 1% (v/v) of heat-killed Pseudomonas aeruginosa PAO1 (heat-killed bacteria [HKB]) in a 25 cm2 tissue culture flask, and further incubated at RT statically for 2 days before enumeration and use. This process is necessary to remove the nutrient media and to acclimatise the ciliate to phagotrophic feeding.
To prepare HKB, P. aeruginosa was grown overnight in LB at 37°C with shaking at 200 rpm and adjusted to OD600 = 1.0 (109 cells ml−1) in 0.5× NSS. The tubes were then transferred to a water bath at 65°C for 2 h, and then tested for viability by plating on LB agar plates at 37°C for 2 days. HKB stocks were stored at –20°C.
Transcriptomic profiling of continuous-culture biofilms
For the transcriptomic analysis, 3-day-old V. cholerae biofilms were exposed to grazing by A. castellanii in a continuous flow system. Briefly, three biological replicates of V. cholerae biofilms were cultivated on the interior surfaces of Silastic® laboratory tubing (Dow Corning, MI, USA) (3.2 mm diameter; length, 14 cm) in 0.5× Väätänen nine salts solution (VNSS) (1 g bacteriological peptone, 0.5 g yeast extract, 0.5 g D-glucose, 0.01 g FeSO4·7H2O and 0.01 g Na2HPO4) in 1 l of 0.5× NSS and fed at a flow rate of 9 ml h−1 using a continuous flow system at RT. After 3 days, washed cells of A. castellanii were re-suspended in 0.5× VNSS, injected into the tubing and incubated without flow for 2 h. A protist-free control biofilm was treated the same to exclude oxygen or starvation effects.
The V. cholerae biofilms on the walls of the tubing were washed by a flow of 2 volumes of 0.5× VNSS to remove planktonic bacteria, and immediately re-suspended in 2 volumes of RNAlater (Qiagen, Hilden, Germany) and harvested from the interior surface of the tubing by mechanical manipulation (manually squeezing out of the tubing). Total RNA was extracted by lysozyme digestion and use of the RNeasy plus mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. For the mRNA-Seq sample preparation, the Illumina standard kit was used, according to the manufacturer's protocol (Illumina, San Diego, CA, USA).
Transcriptome data analysis
Prior to RNA-Seq analysis, filters were applied to remove low-quality reads from all pair-end samples. Pair-end raw reads were trimmed with the BWA trimming mode at a threshold of Q13 (P = 0.05) as implemented by SolexaQA version 3.1.3 (Cox, Peterson and Biggs 2010). Low-quality 3’ ends of each read were filtered and reads that were less than 25 bp in length were discarded.
The trimmed reads were subsequently depleted of ribosomal RNA with SortMeRNA version 1.8 (Kopylova, Noé and Touzet 2012). Trimmed reads (102 bp) were first mapped to the A. castellanii contigs (GenBank accession ID GCA_000826485.1) using Bowtie (version 2.2.3) (Langmead and Salzberg 2012) with default parameters. Reads that were not mapped to amoeba contigs were then mapped to the reference genome, V. cholerae O1 biovar El Tor str. N16961 and the V. cholerae A1552 indel correction table (http://microbes.ucsc.edu/lists/vibrChol1/StrainA1552-list.html) using Bowtie2 with parameters set to -N 1. Cuffdiff (Cufflinks version 2.2.1) with default parameters finally used to identify differentially expressed transcripts of V. cholerae biofilms grazed by A. castellanii compared to ungrazed controls. Cuffdiff calculated the log fold change in fragments per kilobase of exon per million fragments mapped (FPKM), and then the significance of the fold change. A false discovery rate (FDR) adjusted P-value was calculated to give the statistical validity level of significance. Transcripts with an FDR-adjusted P-value of <0.05 were considered to be significantly differentially expressed (SDE) genes.
The lists of up- and downregulated SDE genes were placed into cluster of orthologous group (COG) categories by NCBI conserved domain search (Tatusov, Koonin and Lipman 1997). With the assistance of the Database for Annotation, Visualization and Integrated Discovery Bioinformatics Resources 6.7 (National Institute of Allergy and Infectious Diseases), the differentially expressed transcripts were further analysed using databases such as Gene Ontology Annotation Database (to analyse the biological processes, molecular functions and cellular components), KEGG Pathway (to analyse the metabolic pathways) and InterPro/UniProt (to analyse the protein domains).
Early and late biofilm grazing assay with A. castellanii
Overnight cultures of V. cholerae were adjusted so that 105 cells ml−1 in 0.5× VNSS were added to 24-well microtitre plates (Falcon, Becton Dickinson, NJ, USA) and incubated for 24 and 72 h with shaking at 60 rpm at RT. After incubation, fresh VNSS with or without A. castellanii (2 × 104 cells ml−1) was added, and the plates were incubated at RT with shaking at 60 rpm for 3 days. The cell density in each well was measured by spectrophotometry at OD600 nm (Wallac Victor2 1420 Multilabel Counter, Perkin Elmer Life Sciences, Billerica, MA, USA). In order to quantify the biofilm biomass, crystal violet (CV) assays were performed (O’Toole and Kolter 1998). Briefly, all the planktonic cells were removed by washing three times with 0.5× NSS before adding CV (0.3%) for 15 min. The wells were washed a further three times using 0.5× NSS to remove the unattached CV, and then the stain was solubilised using 96% ethanol and the OD490 nm was determined by spectrophotometry.
To determine if the cell-free supernatants from the hmgA mutant would provide protection against grazing by A. castellanii to the WT, the cell-free supernatant of 3-days-old established biofilms was acquired by centrifugation at 6000× g for 10 min and filtration (0.22-mm filters, Millex-GP, Millipore, Billerica, MA, USA). The cell-free supernatants were then added to 3-days-old established biofilms of the WT strain at a ratio of 50% with fresh VNSS with or without A. castellanii (2 × 104 cells ml−1) and incubated at RT with shaking at 60 rpm for 3 days. The biofilm biomass was then quantified by CV assays.
Reactive oxygen species and pyomelanin quantification
The pyomelanin in the aqueous phase was determined by spectrophotometry (OD405 nm) of the cell-free supernatant acquired by centrifugation at 6000× g for 10 min and filtration (0.22-mm filters, Millex-GP, Millipore, Billerica, MA, USA). In order to study the effect of nutrients released from A. castellanii on pyomelanin production by V. cholerae, A. castellanii was incubated in 0.5× VNSS with or without 1% (v/v) of HKB for 3 days at RT. Furthermore, to assess if phagocytosis by the amoeba predator is required for induction of pyomelanin production, A. castellanii was heat inactivated in 65°C for 15 min. The trophozoites were confirmed to be intact by microscopy, and the viability checked by addition to PYG and incubation at RT for 3 days. The cell-free supernatant or heat-killed A. castellanii was added to the 3-day-old established biofilm. The amount of pyomelanin in the cell-free supernatant after incubation for 3 days at RT was determined. Amount of pyomelanin was then normalised by using the corresponding biofilm biomass measured by CV assays (OD490 nm).
To assess the level of reactive oxygen species (ROS), 25 μM dihydroethidium (DHE) (Sigma-Aldrich, MO, USA), a fluorescent dye for detection of intracellular O2− was used (Owusu-Ansah, Yavari and Banerjee 2008). The biofilms were washed with 0.5× NSS after which 25 μM DHE in fresh 0.5× VNSS medium was added and incubated in the dark for 2 h. After incubation, the cells were washed with 0.5× NSS, and the ROS production was determined by spectrophotometry (518 and 605 nm for excitation and emission, respectively). The plates were incubated for 3 days before measurement of pyomelanin as described.
H2O2 treatment of V. cholerae biofilms
Overnight cultures were inoculated at a final concentration of 106 cells ml−1 in 0.5 × VNSS in 24-well plates incubated at RT with shaking at 60 rpm. After 3 days, the biofilms were treated with 30 mM H2O2 for 30 min, after which the H2O2 was removed and fresh 0.5× VNSS medium with or without A. castellanii (2 × 104 cells ml−1) was added. After co-incubation for 3 days, the V. cholerae biofilm biomass was quantified using the CV assay.
Catalase treatment of V. cholerae biofilms
Overnight cultures of V. cholerae were inoculated at a final concentration of 105 cells ml−1 in 0.5 × VNSS in 24-well plates incubated at RT with shaking at 60 rpm. After 3 days, the media with or without A. castellanii (2 × 104 cells ml−1) was refreshed and 0.1 mg ml−1 catalase (Sigma-Aldrich, MO, USA) was added. After co-incubation for 3 days, the V. cholerae biofilm biomass was quantified using the CV assay.
Quantitative reverse-transcriptase PCR validation of transcriptomic data
RNA was prepared from a late biofilm grazing assay with A. castellanii in 24-well plates. After removal of supernatant, RNAlater (Qiagen, Hilden, Germany) was added and cells were harvested from the wells by mechanical manipulation. RNA extraction was then performed as described previously. The concentration was measured using spectrophotometry (NanoDrop ND-1000; NanoDrop Technologies) after treatment with TURBO DNase (Ambion, ThermoFisher, Waltham, MA, USA). DNA was prepared from 500 ng RNA from each sample by iScript Reverse Transcription (Bio-Rad, Hercules, CA, USA). Quantitative reverse-transcriptase PCR (qRT-PCR) experiments were done using PowerUp SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) by QuantStudio 6 Flex Real-Time PCR System using the primers specific for VC1344, VC1345, VC1346 and VC1347 listed in Supplementary Table 1, Supporting Information. The expression was determined relative to the expression of the endogenous control gene gyrA using the comparative Ct (DDCt) method of RT-PCR.
T. pyriformis grazing assays
Microtitre plates containing 1-day-old biofilms were prepared as described above. After 24 h, the supernatants were removed and fresh 0.5× VNSS media with or without T. pyriformis was added (103 cells ml−1; determined by inverted microscopy) and the plates were incubated at RT with shaking at 60 rpm for 3 days. The cell density was measured by spectrophotometry at OD600 nm. Planktonic fractions were collected for enumeration of CFU ml−1 and biofilm biomass determined by CV staining and spectrophotometry (OD490 nm). Numbers of T. pyriformis were determined by microscopy at each sampling time, and pyomelanin was measured in the cell-free supernatants as described previously.
Vibrio cholerae–A. castellanii intracellular survival assay
To determine the role of hmgA in intracellular survival of V. cholerae internalised by A. castellanii, the number of internal V. cholerae was measured after 24 h. Briefly, A. castellanii (2 × 105 cells ml−1) in 0.5× NSS and 1% HKB were seeded in 24-well microtitre plates 1 day prior to the start of the experiment. After 24 h, the wells were washed gently with 0.5× NSS and V. cholerae (107 cells ml−1) in 0.5× NSS were added. Plates were incubated for 1 h statically at RT to allow ingestion of V. cholerae by A. castellanii. Extracellular bacteria were removed by washing wells three times with 0.5× NSS and treatment with gentamicin (300 μg ml−1) for 1 h at RT. The gentamicin was then removed by washing three times with 0.5× NSS. The cells were then incubated in 0.5× NSS at RT statically for 24 h, after which the amoeba cells were lysed by addition of 1% Triton X-100 in 0.5× NSS for 20 min. Bacteria were enumerated by drop plate colony counting.
Data analysis
Statistical analysis was performed using GraphPad Prism version 7.01 for Windows, GraphPad Software, La Jolla California, USA (www.graphpad.com). Data that did not follow Gaussian distribution as determined by analysing the frequency distribution graphs was natural log transformed. Two-tailed Student's t-tests were used to compare means between experimental samples and controls. For experiments including multiple samples, one-way or two-way ANOVAs were used for the analysis, and Sidak's or Dunnett's Multiple Comparison Test provided the post-hoc comparisons of means when appropriate.
RESULTS AND DISCUSSION
The current study was designed to further elucidate antiprotozoal activities generated by V. cholerae biofilms. Heterotrophic protists are major predators of bacteria, and consequently, bacteria have evolved both pre- and post-ingestional defence strategies to resist predation (Matz and Kjelleberg 2005). Such defence strategies employed by V. cholerae include biofilm formation (Matz et al.2005), expression of the PrtV protease (Vaitkevicius et al.2006) and the T6SS (Pukatzki et al.2006). Although a QS mutant of V. cholerae was more sensitive to predation than the corresponding isogenic wild type, it was still partially resistant to grazing (Erken et al.2011), implying the existence of other QS-independent anti-protozoal mechanisms.
RNA-seq revealed differences between grazed and ungrazed biofilms
In order to identify other anti-predation strategies employed by V. cholerae biofilms, RNA-Seq was performed. Total RNA isolated from three biological replicates of biofilms exposed to grazing by A. castellanii was subjected to Illumina HiSeq 2000 sequencing. Between 108 and 127 million pairs of reads were generated with approximately 5 million reads per sample removed after quality filtering and trimming. Between 98.31% and 99.42% of reads were mapped to the V. cholerae N16961 genome and approximately 0.13% were mapped to A. castellanii contigs. The log2 fold change in FPKM varies from –1.964 to –0.724 for the downregulated transcripts, and from 0.797 to 3.535 for the upregulated transcripts.
SDE genes were considered at fold change of 2.0 and adjusted P-value of P < 0.05. Cuffdiff analysis of the transcriptome revealed that 71 transcripts were significantly upregulated and 60 were significantly downregulated in the grazed biofilm compared with the ungrazed control (see Supplementary Table S2, Supporting Information, for the complete list of differentially expressed genes).
A relatively large fraction of the upregulated transcripts correspond to genes involved in metabolism, in particular nucleic acid, amino acid, lipid and carbohydrate transfer and metabolism. These transcripts encode proteins associated with amino acid biosynthesis and metabolism, such as VC0027 (threonine metabolism), VC1061 (cysteine biosynthesis), hisD, hisG, hisH, VC1134, VC1135, VC1137, VC1138 and VC1139 (histidine metabolism), trpA (tryptophan biosynthesis), gltD and VC2373 (glutamate biosynthesis), glnA (glutamine biosynthesis) argC, VC2617, VC2641, VC2642, VC2643, and VC2508 (arginine metabolism and biosynthesis), VC1704 (cysteine and methionine metabolism), VC0162, VC0031 and VC0028 (isoleucine biosynthesis) and VC0392, VCA0604 and VCA0605 (aminotransferases). The increase in metabolism and energy production might be related to an increase in available nutrient resources since feeding will result in the release of nutrients by protozoa, either due to ‘sloppy feeding’ or excretion of waste products (Wang, Jiang and Weitz 2009).
The genes in the tyrosine catabolic pathway (VC1344 to VC1347) were downregulated in the grazed samples compared to ungrazed samples. These genes lead to the catabolism of tyrosine to fumarate and acetoacetate (Valeru et al.2009; Wang et al.2011), and are confirmed to be associated with a pigmented phenotype due to pyomelanin production (Ivins and Holmes 1980, 1981; Ruzafa, Sanchez-Amat and Solano 1995). The enzyme HmgA (VC1345) is involved in L-tyrosine catabolism in both prokaryotic and eukaryotic organisms (Fernández-Cañón and Peñalva 1995; Kotob et al.1995) and a null mutation in hmgA in V. cholerae leads to accumulation and auto-oxidation of HGA, which in turn will lead to production of pyomelanin (Fig. 1). The downregulation of VC1344–VC1347 during predation was confirmed by qPCR (log2 fold change of –1.51, –1.21, –1.67 and –1.45, respectively).
Figure 1.
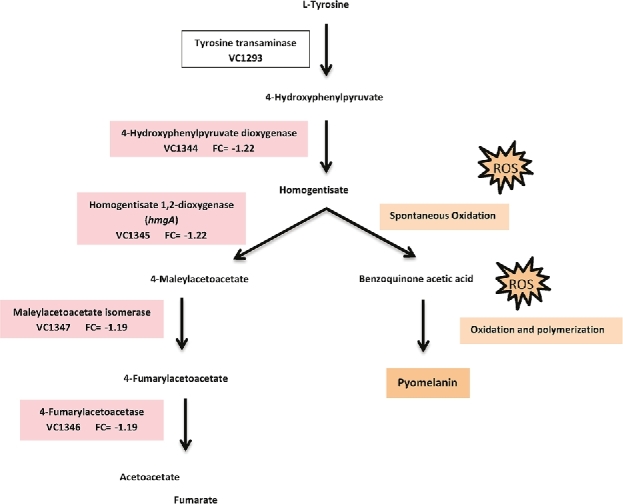
Differentially expressed transcripts in grazed compared to ungrazed biofilms involved in tyrosine degradation in V. cholerae. FC represents log2 fold changes. The pathway in V. cholerae is proposed by Valeru et al. (2009).
Melanin production by auto-oxidation of HGA has been shown to occur in many organisms, ranging from bacteria to humans. The gene encoding HmgA as well as the rest of the pathway is well conserved, and a BLAST search of the NCBI database revealed that many Vibrio spp., including non-pathogenic environmental Vibrio spp. have this pathway. The protective mechanisms of melanin are unclear, but melanin (charged polymers) present in the cell wall may serve as a physical or chemical barrier (Nosanchuk and Casadevall 1997; Jacobson 2000; Eisenman et al.2005). An hmgA mutant of V. cholerae exhibited greater UV and oxidative stress resistance, increased expression of a subunit of the toxin coregulated pilus and cholera toxin, and was enhanced in its ability to colonise the infant mouse (Valeru et al.2009). In contrast, a V. campbellii hmgA mutant did not show increased UV resistance and was less virulent than the wild-type strain, although the wild-type strain exhibited higher resistance to oxidative stress when incubated with supernatants from the hmgA mutant (Wang et al.2013).
Pigment production has also been demonstrated to provide a range of functions in many different microorganisms. For example, melanin can protect the pathogenic fungus, Cryptococcus neoformans, from antibody-mediated phagocytosis by macrophages (Wang, Aisen and Casadevall 1995), as well as from digestion of phagocytosed cells by the amoeba A. castellanii (Steenbergen, Shuman and Casadevall 2001). Melanised C. neoformans are significantly less susceptible to hydrolytic enzymes commonly used by environmental predators than non-melanised cells (Rosas and Casadevall 2001). Melanin production in the fungus, Paracoccidioides brasiliensis, increases protection from phagocytosis by macrophages and intracellular resistance, and decreased drug susceptibility (da Silva et al.2006), while in the yeast Exophiala (Wangiella) dermatitidis, melanin production prevented killing by the phagolysosomal oxidative burst of human neutrophils (Schnitzler et al.1999).
Pyomelanin production increases the grazing resistance of V. cholerae biofilms
In order to determine the role of pyomelanin in grazing resistance of biofilms, V. cholerae wild type and hmgA mutant strains were allowed to form biofilms and after 1 or 3 days, and either T. pyriformis or A. castellanii were added. After 3 days of grazing, CFU and CV measurements determined planktonic cell and biofilm biomass, respectively.
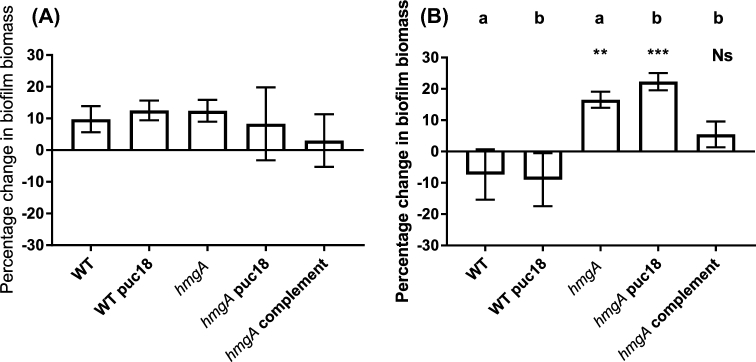
The grazing resistance of early biofilms (1-day-old) of the V. cholerae hmgA mutant and wild-type strains in the presence of A. castellanii were not significantly different (Fig. 2A). In contrast, when late biofilms (3-days-old) were exposed to grazing by A. castellanii, the hmgA mutant was significantly more grazing resistant than the wild type. The biofilm biomass of the grazed wild type was reduced by 7.3% compared to the non-grazed control, whereas the hmgA mutant biofilm biomass increased by 16.5% after grazing (Fig. 2B). Control ungrazed biofilms of the WT and hmgA mutant strains were not significantly different, indicating that the biofilm growth for both strains was similar (Supplementary Fig. 1, Supporting Information). Furthermore, the cell-free supernatant of the hmgA mutant does not show toxicity towards A. castellanii trophozoites compared to the WT (Supplementary Fig. 2, Supporting Information).
Figure 2.
Biofilm biomass of early (A) and late (B) biofilms of V. cholerae A1552 exposed to grazing by A. castellanii for 72 h. Biofilm biomass was determined by CV staining. Data were natural log transformed and the percentage change of biofilm biomass was calculated by removing the biomass of ungrazed samples from the grazed samples divided by the ungrazed. The experiment was run in three replicates and repeated three times separately. Error bars represent standard deviation. Small letters indicate different statistical groups derived from one-way ANOVA and Dunnett's multiple comparisons tests. Statistical significance is indicated by **P < 0.001; ***, P < 0.0001 and Ns, not significant.
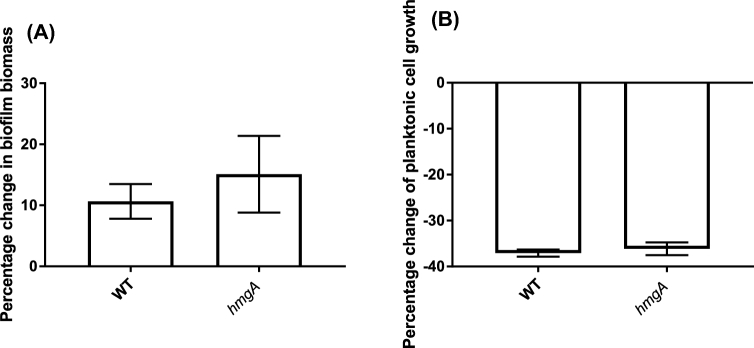
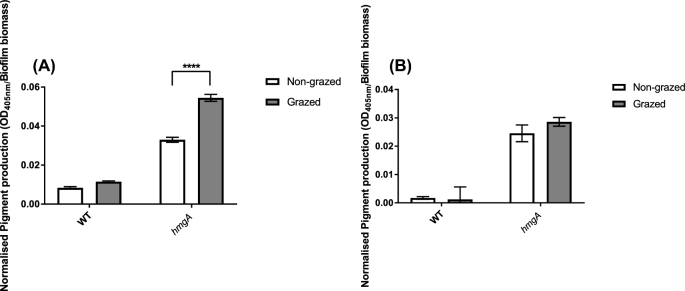
The resistance of planktonic cells of the hmgA mutant to predation by T. pyriformis was also investigated, as A. castellanii cannot feed efficiently on planktonic cells (Huws, McBain and Gilbert 2005). T. pyriformis is a filter-feeding ciliate that can feed effectively on early biofilms as well as planktonic cells (Parry 2004). The use of a second type of grazer with different feeding mechanisms and niche is important for establishment of the generality of a grazing resistance mechanism. The early biofilm (1-day-old) biomass (Fig. 3A) and numbers of planktonic cells (Fig. 3B) of the hmgA mutant and wild-type strains in the presence of T. pyriformis were not significantly different. Interestingly, a further increase in pyomelanin production by the hmgA mutant was observed after 3 days of grazing by A. castellanii but not when exposed to grazing by T. pyriformis (Fig. 4), which is consistent with the increased grazing resistance of the mature biofilm against A. castellanii.
Figure 3.
Early biofilms of V. cholerae A1552 exposed to T. pyriformis for 72 h. Biofilm biomass was determined by CV staining (A) and the planktonic cells in the supernatant enumerated by the drop plate method (B). Data were natural log transformed and the percentage change in biofilm biomass was calculated by removing the ungrazed biomass from the grazed biofilm biomass divided by the ungrazed. The experiment was run in three replicates and repeated three times separately. Error bars represent standard deviation. Statistical analysis was performed using Student's t-test that revealed no significant difference, P = 0.3 (A) and P = 0.1 (B).
Figure 4.
Amount of pyomelanin produced by non-grazed or grazed established biofilms after 3 days exposure to A. castellanii (A) or T. pyriformis (B). Pyomelanin secreted by the biofilm into the supernatant was measured by optical density (OD 405 nm) of the cell-free supernatant obtained from the biofilms. Amount of pyomelanin was then normalised by using the corresponding biofilm biomass measured by CV assays (OD 490 nm). Experiments were run in triplicates and repeated three times on different days. Error bars represent the standard deviation of three replicates. Bars indicate different statistical groups derived from two-way ANOVA and Sidak's multiple comparisons test. Statistical significance is indicated by ****, P < 0.0001.
Addition of more nutrients to WT and hmgA mutant strains did not result in the same increase in pyomelanin levels as active grazing by A. castellanii. This indicates that the extra nutrients in the A. castellanii culture are not responsible for induction of pyomelanin production in the hmgA mutant when exposed to grazing by A. castellanii. Furthermore, the addition of cell-free supernatants from A. castellanii with or without HKB and heat-killed A. castellanii cells did not induce overproduction of pyomelanin, supporting our hypothesis that active phagocytosis by A. castellanii is required (Supplementary Fig. 3, Supporting Information).
Pyomelanin and production of ROS
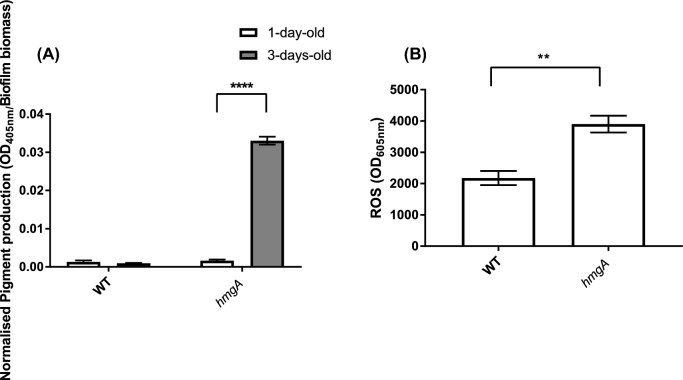
The increases in grazing resistance of the hmgA mutant biofilms (Fig. 2B) correlated with an increase in pigment production. On day 1, the pyomelanin concentration in both the supernatant of wild type and hmgA mutant strains was low (normalised pigment production [OD405nm/Biofilm biomass] = 0.0013 and 0.0009, respectively) (Fig. 5A). However, after 3 days the pyomelanin concentration in supernatants of the hmgA mutant was 20-fold higher than those of the wild type (normalised pigment production [OD405nm/Biofilm biomass] = 0.0016 and 0.033, respectively).
Figure 5.
Amount of pyomelanin produced by biofilms of V. cholerae A1552 wild type and hmgA mutant strains after 1 and 3 days (A). Amount of ROS in the cell-free supernatant of 3 days-old-biofilms of V. cholerae A1552 wild type and hmgA mutant (B). Pyomelanin secreted by the biofilm into the supernatant was measured by optical density (OD 405 nm) of the cell-free supernatant obtained from the biofilms. Amount of pyomelanin was then normalised by using the corresponding biofilm biomass measured by CV assays (OD 490 nm). Error bars represent the standard deviation of three replicates. Statistical analysis was performed using two-way ANOVA and Sidak's multiple comparisons test (A) and Student's t-test (B). Statistical significance is indicated by **, P < 0.001, ****, P < 0.0001.
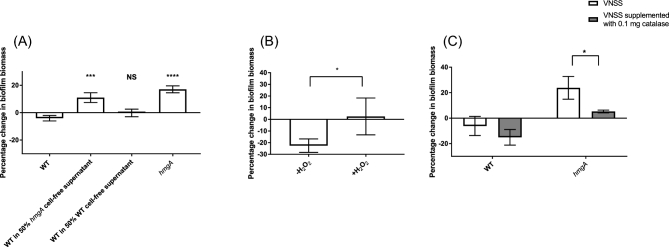
To determine if the hmgA cell-free supernatant would provide protection against predation by A. castellanii to the WT strain, the cell-free supernatant of 3-days-old established hmgA mutant biofilms were added to the A. castellanii grazing assays. The addition of undiluted cell-free supernatants of V. cholerae to pre-established biofilms led to their dispersal due to lack of nutrients and accumulation of waste. Therefore, cell-free supernatants diluted with fresh VNSS was used, and results showed that at a concentration of 50%, the cell-free supernatant of the hmgA strain significantly increased the resistance of the wild-type biofilm to grazing by A. castellanii (Fig. 6A).
Figure 6.
Effect of cell-free supernatants (A), H2O2 (B) and catalase (C) on grazing resistance to A. castellanii. The cell-free supernatant of 3-days-old V. cholerae biofilms were added to 3-days-old biofilms at a concentration of 50% in fresh VNSS and incubated with A. castellanii for 3 days. Biofilm biomass was determined by CV staining. Experiments were run in triplicate and repeated three times on different days. Error bars represent the standard deviation of three replicates. Statistical analysis was performed using one-way ANOVA and Dunnett's multiple comparisons test comparing all to the WT (A), Student's t-test (B) and two-way ANOVA and Sidak's multiple comparisons test (C). Statistical significance is indicated by *, P < 0.05; ***, P < 0.001 and ****, P < 0.0001 and Ns, not significant.
In order to further investigate the relationship between grazing and pigment production, the amount of O2− generated in biofilms of the V. cholerae A1552 wild type, hmgA mutant and complemented strain was monitored, as it has been suggested that ROS are generated during pigment production (Valeru et al.2009). Notably, when pigment production increased on day 3 in the hmgA mutant, the ROS level also increased (Fig. 5B). Biofilms of the hmgA mutant strain generated 79% more O2− than the A1552 biofilms (P = 0.001).
Hydrogen peroxide increased the grazing resistance of V. cholerae
Previous studies have shown that exposure of V. cholerae to H2O2 induced oxidative stress responses and virulence factor expression (Valeru et al.2009). The auto-oxidation of HGA can generate superoxide radicals and H2O2 in eukaryotic cells at physiological pH (Martin and Batkoff 1987). Here, the effect of addition of H2O2 as a substitute for pyomelanin-associated ROS on resistance of V. cholerae biofilms to amoebae grazing was tested in order to further confirm the pyomelanin/ROS-mediated grazing resistance.
V. cholerae biofilms were pre-grown for 3 days, followed by exposure to H2O2 for 30 min (Fig. 6B). A. castellanii was added after H2O2 exposure and the culture incubated for 3 days. The health and number of A. castellanii was monitored and there was no difference between the total numbers of amoebae when co-incubated with the V. cholerae biofilms that had been exposed to H2O2 compared to the controls with HKB. After 3 days of grazing by A. castellanii, the biomass of the untreated V. cholerae biofilms were significantly reduced, while biofilms that had been treated with H2O2 for 30 min were not reduced (P = 0.0246) (Fig. 6B). In addition, pre-grown 3-days-old biofilms were treated with 0.1 mg ml−1 catalase to reduce ROS of the hmgA mutant biofilm. After 3 days of grazing by A. castellanii, the biomass of the treated V. cholerae hmgA biofilm was significantly reduced compared to the untreated biofilm (P = 0.0176) (Fig. 6C). Taken together, our results suggest that the production of pyomelanin results in production of ROS, which in turn, results in an increase in grazing resistance.
We investigated the effect of pyomelanin on survival of V. cholerae intracellularly in A. castellanii. The total number of V. cholerae cells associated with A. castellanii (extracellular and intracellular), as well as the number of intracellular V. cholerae was determined, and there was no difference between wild type and hmgA mutant strains (Supplementary Fig. 4, Supporting Information).
Pigmented hypertoxinogenic strains of V. cholerae have been previously reported both in random mutagenesis experiments (Mekalanos, Sublett and Romig 1979; Parker et al.1979; Ivins and Holmes 1980) as well as in environmental isolates. For example, V. cholerae, ATCC 14035, Serotype Ogawa serovar O1 strain isolated originally from a stool sample produced a reddish brown pigment when grown in low-nutrient condition media supplemented with L-glutamic acid and L-tyrosine (Ruzafa, Sanchez-Amat and Solano 1995). In addition, six nontoxigenic serogroup O139 (water isolates) and one toxigenic O1 (clinical isolate) strains isolated from different years and from different provinces of China were pigmented. All the O139 strains had the same 15-bp deletion in hmgA, and a 10-bp deletion was found in the VC1345 gene of the O1 strain, indicating that the mutation of this gene may provide a fitness advantage in the environment (Wang et al.2011).
Overall, this study demonstrates that V. cholerae O1 El Tor alters its transcriptome in the presence of the predator, A. castellanii. One metabolic pathway that was downregulated under grazing pressure was the tyrosine catabolic pathway, resulting in accumulation of pyomelanin. Experiments with a pyomelanin-overproducing mutant demonstrate that it is more resistant to predation by A. castellanii than the isogenic wild type. Furthermore, the hmgA mutant produces more ROS, which may account for the increased grazing resistance of the hmgA mutant, as V. cholerae biofilms pre-treated with H2O2 were also more grazing resistant.
This project provides insight into the genes involved in defence against protozoan grazing of V. cholerae. Data presented here show that the expression of pyomelanin aids in protection of V. cholerae from grazing in the environment and previous reports have shown that it also plays a role in virulence factor expression and colonisation ability (Valeru et al.2009). This further supports our hypothesis that predation is a major selective factor for maintenance of virulence genes in the environment and thus melanin production may be one such dual use virulence factor.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSEC online.
Supplementary Material
Supplementary data are available at FEMSEC online.
Acknowledgements
The authors would like to acknowledge two anonymous reviewers for helpful comments.
FUNDING
This study was supported by the Australian Research Council [DP140102247 and DP170100453], The ithree Institute at the University of Technology Sydney and the Singapore Centre for Environmental Life Sciences Engineering, Nanyang Technological University, Singapore.
Conflict of interest. None declared.
REFERENCES
- Abd H, Saeed A, Weintraub A et al. Vibrio cholerae O1 strains are facultative intracellular bacteria, able to survive and multiply symbiotically inside the aquatic free-living amoeba Acanthamoeba castellanii. FEMS Microbiol Ecol 2007;60:33–9. [DOI] [PubMed] [Google Scholar]
- Abd H, Weintraub A, Sandström G. Intracellular survival and replication of Vibrio cholerae O139 in aquatic free-living amoebae. Environ Microbiol 2005;7:1003–8. [DOI] [PubMed] [Google Scholar]
- Colwell RR, Huq A. Environmental reservoir of Vibrio cholerae the causative agent of cholera. Ann NY Acad Sci 1994;740:44–54. [DOI] [PubMed] [Google Scholar]
- Colwell RR, Kaper J, Joseph SW. Vibrio cholerae, Vibrio parahaemolyticus, and other vibrios: occurrence and distribution in Chesapeake Bay. Science 1977;198:394–6. [PubMed] [Google Scholar]
- Cox MP, Peterson DA, Biggs PJ. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 2010;11:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva MB, Marques AF, Nosanchuk JD et al. Melanin in the dimorphic fungal pathogen Paracoccidioides brasiliensis: effects on phagocytosis, intracellular resistance and drug susceptibility. Microbes Infect 2006;8:197–205. [DOI] [PubMed] [Google Scholar]
- Eisenman HC, Nosanchuk JD, Webber JBW et al. Microstructure of cell wall-associated melanin in the human pathogenic fungus Cryptococcus neoformans. Biochemistry 2005;44:3683–93. [DOI] [PubMed] [Google Scholar]
- Elliott AM. The distribution of Tetrahymena pyriformis. J Eukaryot Microbiol 1970;17:162–8. [Google Scholar]
- Erken M, Weitere M, Kjelleberg S et al. In situ grazing resistance of Vibrio cholerae in the marine environment. FEMS Microbiol Ecol 2011;76:504–12. [DOI] [PubMed] [Google Scholar]
- Fernández-Cañón JM, Peñalva MA. Molecular characterization of a gene encoding a Homogentisate Dioxygenase from Aspergillus nidulans and identification of its human and plant homologues. J Biol Chem 1995;270:21199–205. [DOI] [PubMed] [Google Scholar]
- Huq A, Colwell RR, Rahman R et al. Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent-monoclonal antibody and culture methods. Appl Environ Microb 1990;56:2370–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huws SA, McBain AJ, Gilbert P. Protozoan grazing and its impact upon population dynamics in biofilm communities. J Appl Microbiol 2005;98:238–44. [DOI] [PubMed] [Google Scholar]
- Ivins BE, Holmes RK. Isolation and characterization of melanin-producing (mel) mutants of Vibrio cholerae. Infect Immun 1980;27:721–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivins BE, Holmes RK. Factors affecting phaeomelanin production by a melanin-producing (mel) mutant of Vibrio cholerae. Infect Immun 1981;34:895–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson ES. Pathogenic roles for fungal melanins. Clin Microbiol Rev 2000;13:708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousset A, Rochat L, Scheu S et al. Predator-prey chemical warfare determines the expression of biocontrol genes by rhizosphere-associated Pseudomonas fluorescens. Appl Environ Microb 2010;76:5263–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens K, Matz C. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Anton Leeuw 2002;81:413–34. [DOI] [PubMed] [Google Scholar]
- Khan NA. Acanthamoeba: biology and increasing importance in human health. FEMS Microbiol Rev 2006;30:564–95. [DOI] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012;28:3211–7. [DOI] [PubMed] [Google Scholar]
- Kotob SI, Coon SL, Quintero EJ et al. Homogentisic acid is the primary precursor of melanin synthesis in Vibrio cholerae, a Hyphomonas strain, and Shewanella colwelliana. Appl Environ Microb 1995;61:1620–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y-L, Stumm K, Wickham SA et al. Distributions and biomass of benthic ciliates, foraminifera and amoeboid protists in marine, brackish, and freshwater sediments. J Eukaryot Microbiol 2014;61:493–508. [DOI] [PubMed] [Google Scholar]
- Lutz C, Erken M, Noorian P et al. Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front Microbiol 2013;4:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mårdén P, Tunlid A, Malmcrona-Friberg K et al. Physiological and morphological changes during short term starvation of marine bacterial islates. Arch Microbiol 1985;142:326–32. [Google Scholar]
- Martin JP, Batkoff B. Homogentisic acid autoxidation and oxygen radical generation: implications for the etiology of alkaptonuric arthritis. Free Radic Biol Med 1987;3:241–50. [DOI] [PubMed] [Google Scholar]
- Martínez Pérez ME, Macek M, Castro Galván MT. Do protozoa control the elimination of Vibrio cholerae in brackish water? Int Rev Hydrobiol 2004;89:215–27. [Google Scholar]
- Matz C, Kjelleberg S. Off the hook–how bacteria survive protozoan grazing. Trends Microbiol 2005;13:302–7. [DOI] [PubMed] [Google Scholar]
- Matz C, Deines P, Boenigk J et al. Impact of violacein-producing bacteria on survival and feeding of bacterivorous nanoflagellates. Appl Environ Microb 2004;70:1593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C, McDougald D, Moreno AM et al. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. P Natl Acad Sci USA 2005;102:16819–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzola M, de Bruijn I, Cohen MF et al. Protozoan-induced regulation of cyclic lipopeptide biosynthesis is an effective predation defense mechanism for Pseudomonas fluorescens. Appl Environ Microb 2009;75:6804–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekalanos J, Sublett R, Romig W. Genetic mapping of toxin regulatory mutations in Vibrio cholerae. J Bacteriol 1979;139:859–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata ST, Kitaoka M, Brooks TM et al. Vibrio cholerae requires the Type VI Secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun 2011;79:2941–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monger BC, Landry MR, Brown SL. Feeding selection of heterotrophic marine nanoflagellates based on the surface hydrophobicity of their picoplankton prey. Limnol Oceanogr 1999;44:1917–27. [Google Scholar]
- Nair GB, Sarkar B, De S et al. Ecology of Vibrio cholerae in the freshwater environs of Calcutta, India. Microb Ecol 1988;15:203–15. [DOI] [PubMed] [Google Scholar]
- Nosanchuk JD, Casadevall A. Cellular charge of Cryptococcus neoformans: contributions from the capsular polysaccharide, melanin, and monoclonal antibody binding. Infect Immun 1997;65:1836–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 1998;28:449–61. [DOI] [PubMed] [Google Scholar]
- Owusu-Ansah E, Yavari A, Banerjee U. A protocol for in vivo detection of reactive oxygen species. Protoc Exch 2008, DOI: 10.1038/nprot.2008.23. [Google Scholar]
- Parker C, Gauthier D, Tate A et al. Expanded linkage map of Vibrio cholerae. Genetics 1979;91:191–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry JD. Protozoan grazing of freshwater biofilms. Adv Appl Microbiol 2004;54:167–96. [DOI] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. P Natl Acad Sci USA 2006;103:1528–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas Á, Casadevall A. Melanization decreases the susceptibility of Cryptococcus neoformans to enzymatic degradation. Mycopathologia 2001;151:53–6. [DOI] [PubMed] [Google Scholar]
- Ruzafa C, Sanchez-Amat A, Solano F. Characterization of the melanogenic system in Vibrio cholerae, ATCC 14035. Pigment Cell Melanoma Res 1995;8:147–52. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. NY: Cold Spring Harbor Laboratory Cold Spring Harbor, 1989. [Google Scholar]
- Schnitzler N, Peltroche-Llacsahuanga H, Bestier N et al. Effect of melanin and carotenoids of Exophiala (Wangiella) dermatitidis on phagocytosis, oxidative burst, and killing by human neutrophils. Infect Immun 1999;67:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanan S, Abd H, Hedenström I et al. Detection of Vibrio cholerae and Acanthamoeba species from same natural water samples collected from different cholera endemic areas in Sudan. BMC Res Notes 2011;4:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen J, Shuman H, Casadevall A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci USA 2001;98:15245–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Kjelleberg S, McDougald D. Relative contributions of Vibrio polysaccharide and quorum sensing to the resistance of Vibrio cholerae to predation by heterotrophic protists. PloS One 2013;8:e56338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Tay QX, Kjelleberg S et al. Quorum sensing-regulated chitin metabolism provides grazing resistance to Vibrio cholerae biofilms. ISME J 2015;9:1812–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science 1997;278:631–7. [DOI] [PubMed] [Google Scholar]
- Thom S, Warhurst D, Drasar BS. Association of Vibrio cholerae with fresh water amoebae. J Med Microbiol 1992;36:303–6. [DOI] [PubMed] [Google Scholar]
- Turick CE, Knox AS, Becnel JM et al. Properties and Function of Pyomelanin In: Elnashar M. (ed). Biopolymers. In Tech, 2010. [Google Scholar]
- Vaitkevicius K, Lindmark B, Ou G et al. A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. P Natl Acad Sci USA 2006;103:9280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeru SP, Rompikuntal PK, Ishikawa T et al. Role of melanin pigment in expression of Vibrio cholerae virulence factors. Infect Immun 2009;77:935–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Henst C, Scrignari T, Maclachlan C et al. An intracellular replication niche for Vibrio cholerae in the amoeba Acanthamoeba castellanii. ISME J 2016;10:897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Aisen P, Casadevall A. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect Immun 1995;63:3131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jiang L, Weitz JS. Bacterivorous grazers facilitate organic matter decomposition: a stoichiometric modeling approach. FEMS Microbiol Ecol 2009;69:170–9. [DOI] [PubMed] [Google Scholar]
- Wang R, Wang H, Zhou H et al. Characters of homogentisate oxygenase gene mutation and high clonality of the natural pigment-producing Vibrio cholerae strains. BMC Microbiol 2011;11:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Lin B, Mostaghim A et al. Vibrio campbellii hmgA-mediated pyomelanization impairs quorum sensing, virulence, and cellular fitness. Front Microbiol 2013;4:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildschutte H, Wolfe DM, Tamewitz A et al. Protozoan predation, diversifying selection, and the evolution of antigenic diversity in Salmonella. P Natl Acad Sci USA 2004;101:10644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 1985;33:103–119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at FEMSEC online.