Abstract
Our ability to rewire cellular metabolism for the sustainable production of chemicals, fuels and therapeutics based on microbial cell factories has advanced rapidly during the last two decades. Especially the speed and precision by which microbial genomes can be engineered now allow for more advanced designs to be implemented and tested. However, compared to the methods developed for engineering cell factories, the methods developed for testing the performance of newly engineered cell factories in high throughput are lagging far behind, which consequently impacts the overall biomanufacturing process. For this purpose, there is a need to develop new techniques for screening and selection of best-performing cell factory designs in multiplex. Here we review the current status of the sourcing, design and engineering of biosensors derived from allosterically regulated transcription factors applied to the biotechnology work-horse budding yeast Saccharomyces cerevisiae. We conclude by providing a perspective on the most important challenges and opportunities lying ahead in order to harness the full potential of biosensor development for increasing both the throughput of cell factory development and robustness of overall bioprocesses.
Keywords: synthetic biology, transcription factor, allosteric regulation, transfer function
Design, engineering and application of biosensors based on transcription factors in yeast.
INTRODUCTION
Biotechnology has flourished in the last two decades as a manufacturing discipline for the future by engineering living cells to convert renewable carbon sources into products related to health, food and transportation (Peralta-Yahya et al.2012; Galanie et al.2015; Zhou et al.2016). Notwithstanding, there is enormous interest to further expand both the production portfolio and the productivity of the organismal hosts used for production, collectively referred to as cell factories (Van Dien 2013). The most commonly used organisms that have been established as cell factories include the gram-negative bacteria Escherichia coli and the budding yeast Saccharomyces cerevisiae. For both of these chassis, the accumulation of the genetic tools and model-guided efforts derived from genome-scale models have enabled the engineering of these chassis for the industrial production of fuels, platform chemicals and pharmaceuticals (Zhang, Jensen and Keasling 2015). Additionally, therapeutic products like opioids and penicillin have been refactored in microbial chassis (Galanie et al.2015; Awan et al.2017). These achievements highlight the versatility and importance of microbial biobased production.
Irrespective of host organisms, in order to replace current petrochemical pipelines for production of commodity and fine chemicals with biobased production using renewable carbon sources, it is critical that cell factories can be constructed in a cost-effective manner and that the cell factories perform at scale in relation to titres, rates and yields (Van Dien 2013). To support this, the ongoing sampling and parsing of biochemical and omics data continuously improve the ability of metabolic engineers to rationally perturb endogenous metabolism for the overproduction of metabolites, as well as the introduction of heterologous biosynthetic pathways for production of non-native compounds (Borodina and Nielsen 2014). Likewise, the decrease in cost of DNA sequencing and synthesis as well as the emergence of synthetic biology tools aims to support the rational engineering of cell factories by focusing on parts characterisation and standardized methods which are host-agnostic and can be used broadly to speed the engineering and selection of cell factory designs (Canton, Labno and Endy 2008). This includes advances in diversity generation of cell factory designs accommodated by genome engineering tools, such as the recent advances in CRISPR-derived genome engineering technologies, which allows unprecedented speed and targeted multiplexing of strain building procedures for an expanding number of chassis (Wang et al.2009; Garst et al.2016).
Despite these scientific advancements, there are still significant challenges to overcome in order to develop viable cell factories that enable economically feasible bioprocesses. Given the size ranges of genomes, numbers of metabolites and enzymatic reactions annotated for most commonly used production hosts, even targeting relatively narrow solution spaces in order to improve cell factory productivity, remains a daunting challenge by the sheer number of individual cell lines and microbial strains that must be screened in order to identify the best performing cell factory (Rogers and Church 2016). While in some cases visible phenotypes or growth-coupled production can support high-throughput screening of cell factory designs, testing cell factory performance often relies on analytical methods like mass spectrometry and chromatographic techniques for the identification and quantification of products of interest. Even for well-equipped research laboratories these methods have a maximum throughput in the range of thousands of samples per day, thus creating a costly, laborious and ineffective route to identify best-producing cells. As such there is a need to develop methods, which can enable testing of single cell factory designs in multiplex.
In recent years, the development, characterisation and application of metabolite biosensors from prokaryotes have spurred significant interest. Often used classes of biosensors rely on fluorescence resonance energy transfer, RNA-based aptamers or simple reporter assays (Dahl et al.2013; Ravasio et al.2014; Strachan et al.2014). Another class of biosensors applied for metabolic engineering includes allosterically regulated transcription factors (aTFs) which are abundantly present in prokaryotes (Fernandez-López et al.2015). In prokaryotes, these one-component protein-based biosensors are able to bind a target metabolite, and upon binding provide a conformational change allowing a change in the expression from a target promoter. In native hosts, this results in the actuation of a cellular response in relation to changing environmental cues (i.e. carbon source availability). Such a sensing-actuation mechanism is naturally abundant in prokaryotic signalling pathways as bacteria continuously exchange, receive and respond to both intra- and extracellular cues in order to coordinate cellular decision making. One particularly well-studied aTF is the tetracyclin-responsive transcriptional repressor TetR (Gossen and Bujard 1992). TetR has been widely adopted for controlling inducible expression perturbations for almost three decades (Gossen et al.1995; Guet et al.2002; Stanton et al.2013). Other more recent examples of the successful development and application of TF-based biosensors from prokaryotes include the in situ diagnosis of human gut microbiota, evolution-guided optimisation of biosynthetic pathways in E. coli and synthetic cell–cell communication devices (Kotula et al.2014; Raman et al.2014; Chen et al.2015).
Although most of the recently reported characterisations and applications using these naturally occurring biosensors based on aTFs have focused on their transplantation into other prokaryotic hosts, examples of on-boarding aTFs as biosensors for monitoring, selection and therapy in eukaryotic cells are expanding. In this review, we provide an overview of the aTFs, which have been successfully used in yeast. We put special attention to sourcing of aTFs, their molecular design and their applications in yeast. This review also highlights the various engineering efforts, which have been performed to optimize the performance of biosensors in order to make them applicable as small-molecule biosensors. Finally, we provide a perspective on future directions for further proof of principles and industrial applications using biosensors.
BIOSENSOR CHARACTERISATION AND IDENTIFICATION
Characteristics of aTF-based biosensors
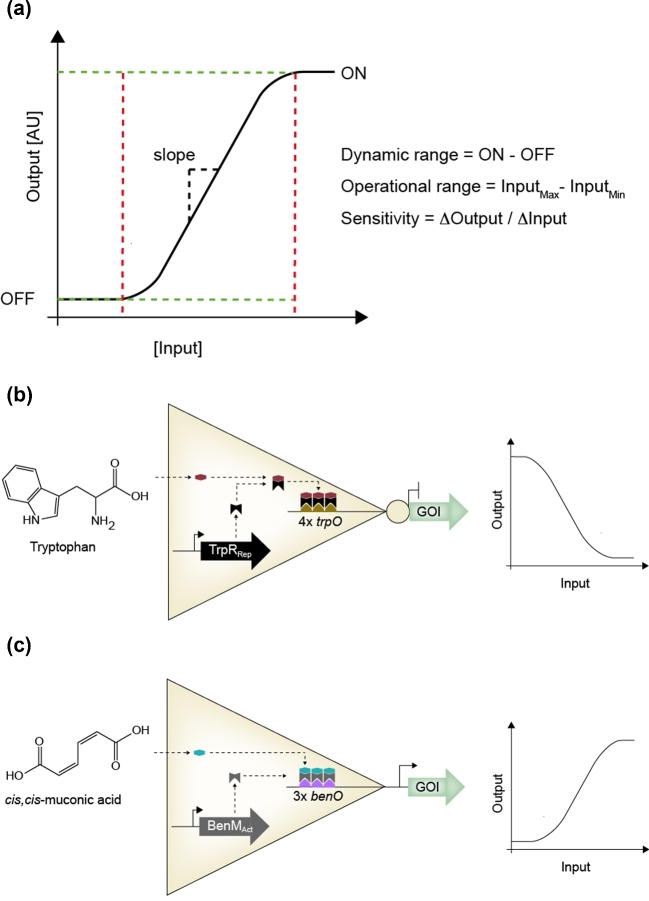
Before introducing the design and engineering of aTF-based biosensors, it is important first to introduce the main structural components and the functional characteristics associated with aTF biosensor performance. The most frequent structural characteristics of aTFs remain the DNA-binding domain (DBD) and the effector-binding domain (EBD), linked by a flexible loop-region, allowing for complex interdomain allostery in relation to binding of either ligand and/or DNA (Werten et al.2016). Also, it is important to highlight that monomeric aTFs exert a high degree of di- or oligomerisation (Fernandez-López et al.2015). The structure and propensity for oligomerisation together determine the two main characteristics of aTF functionality, namely (i) the ligand specificity and (ii) the transfer function describing the correlation between ligand concentration (input) and the biosensor output signal (Fig. 1a)(De Paepe et al.2017). The specificity of a biosensor describes the strength of the binding between ligand and aTF or the difference in biosensor output in relation to a target ligand compared to the output observed when introducing other potential ligands of interest. Inevitably, defining the specificity of a biosensor is of primary importance when the biosensor is expected to provide high signal-to-noise ratios within a complex cellular environment in which the aTF is exposed to several potential ligands (De Paepe et al.2017). The transfer function of a biosensor describes its quantitative performance (Fig. 1a). This includes the operational range, in which a significant change in biosensor output can be measured in relation to ligand concentration. Likewise, the dynamic output range refers to the maximum output, which can be obtained compared to the output observed in the absence of ligand binding. This is also referred to as the ON and OFF states, respectively, of the biosensor (Moser et al.2013). From these parameters, the sensitivity of the biosensor can be inferred as the difference in output of ON and OFF states divided by the difference in inducer concentration at the detection threshold and the maximum output, also referred to as the Hill's coefficient (Dietrich, McKee and Keasling 2010; Moser et al.2013). Sensitivity is largely dependent on the cooperativity between aTF monomer oligomerisation and ligand binding (Stefan and Le Novère 2013). It should also be mentioned that transfer functions can have qualitative differences depending on the mode of action of the biosensor, i.e. negative vs positive correlations between biosensor output and ligand concentration (Fig. 1b and c) (see also section ‘Classes of aTF-based biosensors: repressors vs activators’ and ‘Engineering new specificities and improving performances in existing aTFs’).
Figure 1.
The biosensor transfer function and design. (a) The transfer function of a biosensor enables the quantitative characterisation of biosensors showing the correlation between input effector concentration and the biosensor output. Transfer functions thereby provide information on product sensitivity, the operational range of detection and the dynamic output range as defined by the difference between minimum (OFF) and maximum (ON) biosensor outputs. (b) Expression of the allosterically regulated transcriptional repressor TrpRRep controls the expression of a gene of interest (GOI) by tryptophan-dependent binding to the promoter harbouring four TrpR binding sites (4xtrpO). To the right a schematic transfer function of TrpR output in relation to tryptophan concentration is illustrated. (c) Expression of the allosterically regulated transcriptional activator BenMAct controls the expression of a GOI by cis,cis-muconic acid-dependent binding to the promoter harbouring three BenM binding sites (3xbenO). To the right a schematic transfer function of BenM output in relation to cis,cis-muconic concentration is illustrated. AU; arbitrary units.
Knowledge about specificity and the transfer function is crucial in order to rationally engineer biosensors. In the remainder of the section ‘Biosensor characterisation and identification’, we review the basic components of aTF-based biosensors sourced from prokaryotes and the engineering strategies adopted for onboarding these in yeast.
Classes of aTF-based biosensors: repressors vs activators
Allosterically regulated TF-based biosensors can be divided in two groups: transcriptional activators and transcriptional repressors. The aTF regulates the transcription of the reporter gene in a ligand-dependent manner by localising on the operator site and sterically preventing the transcription, or by supporting the accessibility of the transcriptional machinery to the transcription start site (Fig. 1b and c).
The most prominent example of the repressing mechanism is the TetR repressor from E. coli which blocks the transcription of the tetA gene, encoding the tetracycline efflux pump (Hillen and Berens 1994; Møller et al.2016). By binding directly to TetR, tetracycline is able to induce a conformational switch, which prevents DNA binding and therefore allows the transcription of tetA. Subsequently, tetracycline is secreted from the cell and TetR is once again able to bind to the operator site and block the transcription of tetA (Hillen and Berens 1994). This system has been successfully transferred to mammalian and yeast cells in order to control gene expression by tetracycline and analogs thereof (Gossen and Bujard 1992; Garí et al.1997). Another mode of action for repressors is illustrated by tryptophan-binding TrpR from E. coli (Arvidson, Bruce and Gunsalus 1986; Jeeves et al.1999). Here, TrpR binds the co-repressing ligand Trp before binding its cognate DNA operator site, and thereby regulates aromatic amino acid metabolism (Fig. 1b)(Zhao et al.2015). However, co-repressors have so far not been adopted as biosensors in yeast.
In addition to repressors, recent studies have also highlighted successful transfer of aTF-based transcriptional activators into yeast. This was recently illustrated by Skjoedt et al. (2016), who employed BenM from Acinetobacter sp. ADP1, which is able to induce the transcription synergistically upon the detection of benzoate and cis,cis-muconic acid (CCM) (Craven et al.2009). In addition to BenM, Skjoedt et al. were able to show that several other activators from diverse bacterial species were able to function as aTF-based biosensors in yeast, allowing in vivo sensing of malonate, L-arginine and naringenin, in addition to CCM (Fig. 1c, Table 1). Interestingly, none of the biosensor designs required the expression of auxiliary transcription machineries (e.g. sigma factors). As such, it is now possible to engineer both prokaryotic transcriptional activators and repressors in eukaryotes, and the choice of aTF type ideally only relies on the biosensor design and the availability of biosensors for the effector of interest.
Table 1.
Examples of effector molecules and their aTF-based biosensors engineered in yeast.
| Transcription factor name | NLS | Additional modules | Effector | Dynamic range (x-fold) | Operational range (M) | Reference | |
|---|---|---|---|---|---|---|---|
| FadR | NO | NO | Myristic acid | 1.4 | 0.25 × 10−3 | Teo, Hee and Chang (2013) | |
| FapR | SV40 | NO | Malonyl-CoA | 2 | 0–13.5 ×10−6 (cerulenin) | David, Nielsen and Siewers (2016) | |
| FapR | SV40 | NO | Malonyl-CoA | 4.17 | 0–35.8 × 10−6 (cerulenin) | Li et al. (2015) | |
| MetJ | NO | B42 | S-Adenosylmethionine | ∼70 | Not specified | Umeyama, Okada and Ito (2013) | |
| XylR | SV40 | NO | Xylose | ∼4 | 26.6–666.1 × 10−3 | Teo and Chang (2015) | |
| Repressors | XylR | SV40 | NO | Xylose | 14 | 0–33.3 × 10−3 | Wang, Li and Zhao (2016) |
| XylR | SV40 | NO | Xylose | 6.3 | 0–133.3 × 10−3 | ||
| XylR | SV40 | SSN6 | Xylose | >25 | 1.33–13.3 × 10−6 | Hector and Mertens (2017) | |
| Ada (N-terminal domain) | NO | Gal4-AD | MeI—methyl iodide | 5.2x ± 0.4 | 2.8 × 10−5– 4 × 10−3 | Moser et al. (2013) | |
| MMS | 6.4x ± 1.9 | 28–340 × 10−6 | |||||
| DMS | 6.9x ± 2.0 | 2–150 × 10−6 | |||||
| MNNG | 3.8x ± 0.2 | 2–64 × 10−6 | |||||
| BenM | NO | NO | cis,cis-muconic acid | 10 | 0.2–1.4 × 10−3 | Skjoedt et al. (2016) | |
| CamR | SV40 | 3xVP16 | camphor | ∼50 | Not specified | Ikushima, Zhao and Boeke (2015) | |
| CymR | SV40 | VP16 | p-Cumate | Not specified | Not specified | Ikushima and Boeke (2017) | |
| Erg20 | NO | Gal4-DBD/Gal4-AD | IPP | ∼2 | ND | Chou and Keasling (2013) | |
| Activators | FdeR | NO | NO | Naringenin | 1.7 | 5 × 10−8– 2 × 10−7 | Skjoedt et al. (2016) |
| Gal4(DBD)-DIG-VP16 | NO | ND | Digoxin | 60 | 10−6 –∼500 × 10−6 | Feng et al. (2015) | |
| Gal4(DBD)-PRO-VP16 | progesterone | 60 | ∼10−6–10−5 | ||||
| hAR | NO | NO | 5α-Dihydrotestosterone | ∼100–500 | ∼10−9–10−7 | Bovee et al. (2007) | |
| hAR | NO | NO | 17β-Testosterone | ∼100–500 | ∼10−9–10−7 | ||
| hAR | NO | NO | 17β-Boldenone | ∼100–500 | ∼10−8–10−6 | ||
| hAR | NO | NO | 17β-Estradiol | ∼300 | ∼10−7–10−5 | ||
| hER-α | NO | NO | 17β-Estradiol | 4 | 45 × 10−12–2.8 × 10−9 | Sanseverino et al. (2005) | |
| LexADBD-ER- | NO | Different ADs | β-Estradiol | From 15 to 60 (depending on the design) | 1–250 × 10−9 (depending on design) | Ottoz, Rudolf and Stelling (2014) | |
| LexADBD-VL/NLS-VH-VP16 | SV40 | ND | Bisphenol A | 38.4 ± 6.41 | ∼4.4 × 10−8–43.8 × 10−8 | Gion et al. (2009) | |
| PhlF | NO | 3xVP16 | DAPG | ∼50 | Not specified | Ikushima and Boeke (2017) | |
| TetR | NO | VP16 | Doxycycline | 2000 | ∼10−12–10−9 | Garí et al. (1997) | |
| Zif268-hER(mutated)-VP16 | NO | Zif268(DBD)/VP16(AD) | 4΄-4΄-Dihydroxybenzyl | ∼50 | ∼10−8–10−6 | McIsaac et al. (2014) |
Biosensor prospecting
In the last three decades, a great number of interactions between TFs and genes have been discovered, especially from studies of adaptation of microorganisms to different environmental conditions by sensing the presence/absence of metabolites, toxic compounds or other chemicals (Junker, Kiewitz and Cook 1997; Brzostowicz et al.2003). Such studies have allowed for the development of different predictive tools and databases, which can be used for identifying an aTF suitable for user-defined purposes.
In order to mine publicly available literature for new aTFs to be tested and characterized for use as biosensors in yeast, a minimum amount of information should be considered as a starting point, including if the gene sequence and the operator of the aTF are known. Also, the chemistry of the effector needs to be taken into account: (i) is the effector toxic; (ii) can it be internalised at a pH that is compatible with yeast growth during biosensor characterisation; (iii) does it need any complementary genes, such as transporters (Skjoedt et al.2016).
In addition to literature mining for known aTFs, public databases are available for browsing the genomes of a vast number of prokaryotes (e.g. www.pseudomonas.com). Here the approach to identify new aTFs can harness the fact that in prokaryotes, aTFs are often encoded divergently and in close genomic proximity of the genes, which they are regulating (Collier et al.1998). Additionally, as DBDs from major aTF families are often conserved, this enables searching genome sequence databases for conserved regions of annotated genes (Maddocks and Oyston 2008; Jain 2015). Ideally, in accordance with the divergent expression of genes encoding the aTF and their target operon, the operator site can be inferred from mining the genomic sequence upstream of the aTF expression cassette. Such a method can enable the search for any sequence that is homologous to known aTF domains (DBD or EBD), which are encoded in proximity of the genes connected to the metabolic reactions of the candidate ligand. Hence, by way of such in silico approach, new biosensor candidates can be prospected.
Complementary to the in silico approach, expression databases (e.g. https://www.ebi.ac.uk/arrayexpress/help/GEO_data.html) can be used to search for differentially expressed genes in relation to environmental cues or chemical stimuli of interest, as both genes encoding aTFs and the genes of divergently expressed operons can enable the shortlisting of both candidate aTFs and potentially their operators (Strachan et al.2014).
Alternatively, it is also possible to use different predictive tools and databases for transcriptional regulation in order to identify aTFs of interest. One example is RegulonDB (http://regulondb.ccg.unam.mx), which contains a list of TFs in E. coli with information on operator sequences, and the genes regulated by the TF (Gama-Castro et al.2016). RegulonDB also allows querying for a known metabolite, gene or TF, and visualise the connected regulatory systems. Another data mining tool, named SensiPath, allows searching sensing-enabling metabolic pathways (http://sensipath.micalis.fr/)(Delépine et al.2016). This tool enables the linking of a specific metabolite, which otherwise has no known target aTF, to a chemical that is a maximum of two enzymatic steps away from the candidate compound which can be detected by an annotated aTF. Beyond the identification of a proxy for high-throughput screening of your metabolite of interest, this method is also useful as a mean to relieve potential toxicity of your metabolite of interest, by identification of enzymatic steps enabling the conversion of it to a non-toxic chemical.
In general, querying any of these databases can be very useful, but in order to identify robust starting points for biosensor design based on aTF and operator sequences it is advisable to perform a combination of approaches, and search through several of the mentioned databases.
BIOSENSOR DESIGN ENGINEERING
Once an aTF has been selected, the next step is the design of a functional transcriptional unit with an optimal transfer function. To accomplish this, several factors must be considered. In this section, we review the basic principles of engineering aTFs with focus on designs in eukaryotes, especially yeasts.
aTF selection and aTF expression tuning
Selection of the aTF is a natural first step in the design of a biosensor. This can include the need for discriminating between seemingly close homologues and tuning the strength of the promoter controlling the expression of the aTF (Teo, Hee and Chang 2013; Teo and Chang 2015; Wang, Li and Zhao 2016).
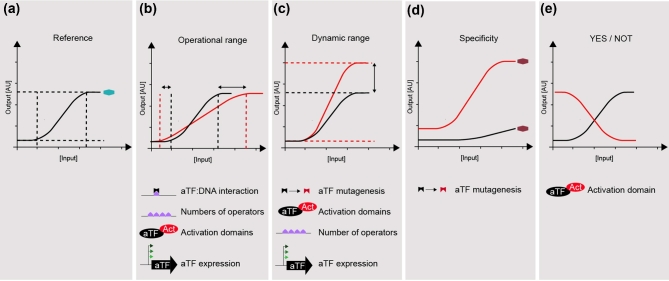
One example highlighting the importance of aTF selection has been reported by Teo et al. (2013). In this study, two variants of FadR from E. coli and Vibrio cholera were tested in yeast in order to develop a fatty acid biosensor. While both repressors were unable to sufficiently repress the transcription of the reporter when cloned under the control of the weak CYC1 promoter, expressing them under the control of the strong constitutive TEF1 promoter enabled repression of the reporter gene controlled by each of the two FadR variant, albeit with significant difference in the level of repression. While strong expression of the E. coli FadR enabled repression of the reporter fluorescence by almost 90%, FadR from V. cholerae was only able to reduce expression by 50% compared to a reference strain without FapR. As such, the promoter driving the expression of the aTF can impact the OFF state of the biosensor. Additionally, the operational range can be influenced by the choice of promoter driving the expression of the gene encoding the aTF. In fact, a weaker promoter controlling the aTF was shown to enable a change in the operational range, with lower expression driving the operational range towards lower concentrations of the effector and stronger promoter driving the operational range towards higher effector concentrations (Teo, Hee and Chang 2013) (Fig. 2a and b). Still, strong expression is generally the first choice for controlling the expression of genes encoding repressor-based aTFs in order to support lower OFF states (Teo, Hee and Chang 2013; Li et al.2015; David, Nielsen and Siewers 2016; Hector and Mertens 2017) Complementary to this, when transcriptional activators are employed, a weaker promoter driving the expression of the aTF may be preferred since a strong promoter can lead to a high OFF state expression of the reporter gene or prolonged lag phase during growth (Teo and Chang 2015; Skjoedt et al.2016). As such, the promoter controlling the aTF should be chosen carefully in order to ensure control of both OFF state, operational range and potential growth effects.
Figure 2.
Engineering the biosensor transfer function of allosterically regulated transcription factors. (a) The transfer function of a reference biosensor. (b) The operational range can be engineered by (i) changing the affinity of the interaction between the aTF and DNA, (ii) changing the number of operators bound by the cognate aTF, (iii) addition of an transcription activation domain and (iv) tuning the expression of the gene encoding the aTF. (c) The dynamic range of a biosensor can be engineered by (i) mutating the aTF followed by selection, (ii) addition of transcription activation domains, (iii) changing the number of operators bound by the aTF and (iv) tuning the expression of the gene encoding the aTF. (d) The effector specificity of an aTF towards new ligands can be changed by mutating the aTF followed by selection, (e) A logic gate with a positive correlation between input and output (YES) can be turned into a NOT gate by addition of an activation domain. In (a–e), all black lines correspond to the transfer function of the reference biosensor, whereas red lines indicate transfer function of engineered biosensors.
Engineering the reporter promoter
The architecture of the promoter controlling the output of the biosensor is another essential engineering target. While native promoters are often well characterised and therefore can be advantageous to use for control over biosensor output over non-native ones, it is important to take into consideration the native regulation of the promoter. Several groups have engineered native yeast promoters for the design of biosensors. A common strategy has been to insert aTF operator sites (see section ‘Operator sequence’) into a native yeast promoter chassis in which upstream activation sequences or upstream repression sequences have been deleted whenever present, in order to minimise the complexity of native regulation (Ikushima, Zhao and Boeke 2015; Li et al.2015; Skjoedt et al.2016; Wang, Li and Zhao 2016; Ikushima and Boeke 2017). In the example by Skjoedt et al. (2016) engineering of the promoter driving the reporter gene included testing of four different ‘crippled’ variants of the CYC1 promoter and identified the shortest variant (209 bp) to be the one allowing for the lowest OFF state, while still maintaining approximately 4-fold dynamic output range in the presence of CCM. In general, the reporter promoter should be strong enough to allow for a detectable output but, overexpressing the reporter gene can cause ‘leaky’ expression, which is of particular importance when the biosensor is to be coupled to selection. In addition to controlling ON and OFF states, when carefully engineered, the reporter promoter design can also tune both the dynamic output range and the operational range (Fig. 2b and c)(McIsaac et al.2014; Skjoedt et al.2016).
Operator sequence
The operator is a DNA sequence that is recognised by the DBD of the aTF. Its presence is essential for aTF binding to DNA, and it impacts both the dynamic and operational range. Adding operator sequences to a native or truncated promoter intended to control the biosensor output has generally proved to have a negative impact on the basal expression (Li et al.2015; David, Nielsen and Siewers 2016; Skjoedt et al.2016; Hector and Mertens 2017). Likewise, the position of the operator is important, and due to the difficulty predicting the effect operator positioning will have on the reporter gene, several groups have mined different designs in order to find the best positions. In one example, Hector and Mertens (2017) tested three different operator designs: (i) one operator sequence immediately downstream of the TATA element, (ii) two operators after the TATA element and (iii) two operators flanking the TATA sequence. The addition of the first operator immediately downstream of the TATA sequence decreased the promoter basal activity by almost 50%, and allowed for a 4-fold de-repression when the effector was added. The addition of a second operator downstream of the TATA sequence caused an even further decrease in the promoter activity by stronger repression. Finally, the best performing design, two operators flanking the TATA element, had a basal expression comparable to the promoter with only one operator, but allowed for approximately 8-fold de-repression. This is a clear example of how the same number of operators can have significantly different outputs when positioned differently. However, the position of operator(s) is not the only design parameter impacting reporter promoter output. Wang et al. (2016) showed that different operator sequences can also perturb the operational range of biosensors. This was particularly exemplified by exchanging the operator site of XylR from Staphylococcus xylosus with the operator sequence of the homologous from Bacillus subtilis. Swapping between these two operators changed the operational range from 0 to 20 g L−1 of xylose, to be saturated at <5 g L−1 (Table 1)(Wang, Li and Zhao 2016). By designing a degenerate operator sequence, the authors identified several operational ranges, and were also able to achieve an increase in dynamic range. This behaviour brings new importance to the interaction between the aTF DBD and the operator of choice, and provides additional means to developing biosensors with sought-for transfer function characteristics.
Nuclear localisation signal
The transport of molecules between the nucleus and cytoplasm is made possible by the nuclear pore complex (NPC). NPCs function as canals and allow free diffusion of molecules and small proteins in and out of the nucleus. Even though the NPCs are large enough to allow free diffusion of macromolecules smaller than 50 kDa, they function as a barrier by preventing the diffusion of larger proteins to the nucleus, unless a nuclear localisation signal (NLS) is encoded in their primary structure (Hahn et al.2008). A common choice of NLS to support nuclear localisation of aTFs is the simian virus 40 (SV40) NLS (Feng et al.2015; Ikushima, Zhao and Boeke 2015; Li et al.2015; Garst et al.2016; Hector and Mertens 2017; Ikushima and Boeke 2017), and it has been shown that the absence of the NLS can lead to a non-functional biosensor (Li et al.2015). Specifically, Li and co-workers compared the ability of FapR from B. subtilis to repress the transcription of the reporter gene when expressed in presence or absence of the NLS and showed that the strain harbouring FapR with NLS showed 30% of the fluorescence intensity of the control strain not expressing FapR, while the strain harbouring the repressor without the NLS showed fluorescence comparable to the strain without the repressor. Yet, the presence of the NLS does not seem strictly necessary when employing aTFs, which have a molecular mass lower or comparable to the nuclear pore diffusion limit (Teo, Hee and Chang 2013; Ikushima, Zhao and Boeke 2015; Skjoedt et al.2016), but NLS should indeed be considered a design parameter when engineering aTF-based biosensors in yeast.
Module transferability
The simple road-block of RNA polymerase progression provided by DNA-bound aTF repressors in the absence of effectors allows engineering of transcriptional activation by addition of one or several activator domains. Here, the strategy is to add single or multiple VP16 activation domain(s) from herpes simplex virus to the repressor (Gion et al.2009; Ottoz, Rudolf and Stelling 2014; Feng et al.2015; Ikushima, Zhao and Boeke 2015; Ikushima and Boeke 2017) (Table 1). In the seminal study on TetR, a TetR-dependent expression system was developed in mammalian cells in which the aTF was converted from a repressor to an activator by fusing a VP16 activator domain on the C-terminal of TetR (Gossen and Bujard 1992) (Fig. 2e). This system, referred to as Tet-Off, relies on the ability of the TF to bind a synthetic promoter, where seven operator sequences were linked to a minimal promoter fragment derived from the CMV promoter, and thereby induce the transcription of the reporter gene in the absence of effectors. The activity of the aTF can then be relieved by tetracyclin or doxycycline administration (Loew et al.2010). Another variant, referred to as Tet-On, where transcription of the reporter by TetR fused to VP16 is only achieved in the presence of tetracycline or doxycycline, was later developed through random mutagenesis (Gossen et al.1995) (Fig. 2e). Both these systems were employed in yeast in order to control gene expression without perturbing the cellular metabolism, as in the case of galactose or methionine addition (Bellí et al.1998; Urlinger et al.2000; Giuraniuc, MacPherson and Saka 2013; Roney et al.2016).
The activator domain is not the only module that can be added to aTFs. As shown by several groups, both DBDs and whole proteins can be added (Chou and Keasling 2013; Moser et al.2013). Moser et al. (2013), while developing a sensor for strong methylating compounds, exploited the characteristic of the Ada protein from E. coli to detect methyl groups on DNA and to transfer them to a cysteine residue on its amino acidic sequence, which then leads to its activation as an aTF in yeast. To accomplish this, they fused the N-terminal domain of the Ada protein, which is the region that allows for both DNA binding and methyltransferase activity, to the Gal4 transactivation domain (Gal4-AD) in order to induce transcriptional activation in yeast upon detection of methyl groups (Moser et al.2013).
In another study, Feng et al. (2015) fused the DBD of Gal4 to a synthetic EBD and to a VP16 module to create sensors for digoxin, which they later rationally engineered to induce a specificity switch towards progesterone (Fig. 2d). Both sensors could induce the transcription of the reporter gene by 60-fold upon the detection of the ligand (Table 1).
Finally, another hybrid biosensor design has been reported by Chou and Keasling (2013) illustrating the flexibility supported by the modular transferability of TF protein domains. Here, the authors exploited the ability of an isopentenyl diphosphate (IPP) isomerase (idi) to dimerise in order to create an IPP sensor in yeast (Chou and Keasling 2013). To do so, they first created two chimeric proteins: the first one consisted of the Gal4 DBD fused to the IPP isomerase, while the second chimeric protein was IPP isomerase fused to the Gal4 AD. The sensor then relied on the ability of the IPP isomerase dimer to bring the two Gal4 domains into close proximity to activate transcription of a reporter gene controlled by a GAL promoter. After validating the sensor, they replaced the IPP isomerase module with IPP-binding Erg20 to further improve the dynamic range of the IPP biosensor in yeast (Chou and Keasling 2013).
In summary, the addition of new modules can allow for the creation of new biosensors when naturally occurring ones for a specific target are unknown. By adding a DBD to a synthetically designed EBD, or by adding an AD, it is possible to induce a stronger response upon the detection of the ligand, since the de-repression level allowed by repressor-based aTFs is generally lower than the induction of aTFs fused with activation domains (Table 1). Also, by testing different modules, such as different ADs or EBDs, it is possible to improve the sensor's response, leading to an improved dynamic range (Chou and Keasling 2013; Ottoz, Rudolf and Stelling 2014) (Fig. 2c).
Engineering new specificities and improving performances in existing aTFs
Many aTFs have been identified based on traditional forward engineering (Shamanna and Sanderson 1979; Neidle, Hartnett and Ornston 1989). However, it still remains a challenge to infer ligand specificity of an aTF from such studies. Even after exhaustive database mining, it may still not be possible to correlate an aTF to a specific compound of interest (see section ‘Biosensor prospecting’). In such cases, it may be necessary to engineer a preexisting aTF with new ligand specificity of interest. However, one of the challenges researchers are facing when aiming to engineer new specificities into existing aTF-based biosensors is the need to simultaneously maintain critical properties like DNA binding, allosteric signal transduction, dimerisation and general protein structure, as all of these functionalities are crucial for ligand-induced transcriptional regulation (Raman et al.2014). However, learning from nature's rich diversity of aTFs, several studies have shown that both rational and directed evolution approaches can be used to successfully engineer new ligand specificities into existing aTF-based biosensors (Fig. 2d).
As for directed evolution, several examples have been reported in bacteria using random mutagenesis based on error-prone PCR (epPCR) coupled with ligand-specific selection in order to affinity-mature aTFs for new ligand specificities (Cebolla, Sousa and De Lorenzo 1997; Collins, Arnold and Leadbetter 2005; Taylor et al.2015; Xiong et al.2017). While most of these examples refer to studies in bacteria, a few studies highlight the power of directed evolution for improving the biosensor performance of aTFs when engineer in yeast. In relation to ligand specificity, Chockalingam et al. (2005) adopted directed evolution to alter the specificity of the human estrogen receptor alpha (hER-alpha) from 17-beta estradiol to 4,4΄-dihydrobenzyl (DHB) by site saturation mutagenesis on ligand contacting residues, and epPCR on the whole receptor. Library variants with improved response to DHB relative to parental hER-alpha were selected based on growth of the host yeast cells in medium containing an appropriate concentration of DHB. Taken together, these studies have demonstrated that mutating residues involved in the binding site can change ligand specificity without affecting allosteric function. However, it should be noted that the specific residues that give rise to allostery are generally unknown and do not tend to be localised in certain parts of the protein (Süel et al.2003). Furthermore, mutations in EBD tend to alter allosteric communication with the DBD and mutations can have far-reaching effects (Raman et al.2014; Taylor et al.2015).
In addition to studies related to changes in specificity, directed evolution can also be used to improve the transfer function of an existing aTF. By increasing aTFs specificity and sensitivity towards the ligand and altering the binding affinity to the operator sequence, it is possible to modify most of the characteristics of the transfer function exemplified in Fig. 2. For instance, Skjoedt et al. (2016) showed that directed evolution on BenM's EBD via epPCR, yeast plasmid gap repair and fluorescence-activated cell sorting (FACS) enabled the identification of aTF variants with >10-fold increase in reporter gene output upon the presence of CCM, while wild-type BenM was only able to induce 4-fold (Fig. 2c). In another example, yet performed in bacteria, Richards et al. (2017) recently showed that by random mutagenesis it is possible to induce a YES/NOT logic gate inversion in a LacI variant which was previously made insensitive to the ligand by removing the allosteric communication between the EBD and the DBD (Fig. 2e). These variants, generated via epPCR and transformed in E. coli, also show different responses to increasing concentrations of the effector and different operational ranges.
In addition to random mutagenesis, computational approaches are also being developed and have been employed to create sensors with new specificities. An example that clearly states the potential of this method comes from De Los Santos et al. (2016), where the authors were able to induce a vanillin response into QacR from S. aureus by computational design, an aTF that is induced by a broad range of structurally dissimilar compounds. Similarly, Taylor et al. (2015) adopted both random epPCR-based and Rosetta-guided mutagenesis to change ligand specificity of LacI.
To summarise, by using a combination of computational, random and site saturation mutagenesis methods, coupled with adequate selection, it is possible to create sensors with improved or novel characteristics, and therefore allow a broadening of the range of applications where aTFs can be employed.
APPLICATIONS OF TF-BASED BIOSENSORS IN YEAST
TF-based biosensors described in section ‘Biosensor design engineering’ have been implemented in yeast for different applications that range from the detection of pollutants to actuators on native yeast metabolism to improve overall cell factory performances. In this section, we review applications of aTF biosensors in yeast, while a list of aTF biosensors currently known to have been employed in yeast is found in Table 1.
aTF biosensor-reporter systems in yeast
Industrially relevant organisms like Sa. cerevisiae are able to metabolise glucose, which is the most abundant sugar in the lignocellulosic biomass. However, in order to bring down cost of biobased production from yeast, it is important to improve its feedstock utilisation. In addition to glucose, xylose represents an important fraction of available sugars in lignocellulosic biomass (Hector and Mertens 2017). In order to create a sensor that could allow for protein expression or gene circuits development induced by xylose, various groups have developed xylose biosensors based on XylR, a xylose-responsive transcriptional regulator found in bacteria (Table 1) (Teo and Chang 2015; Wang, Li and Zhao 2016; Hector and Mertens 2017).
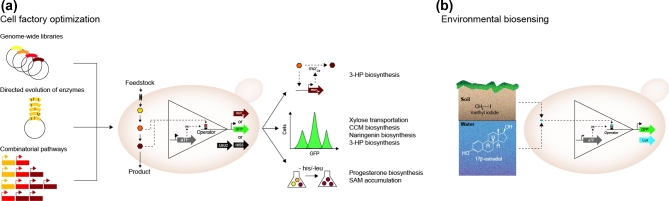
Wang et al. (2016) developed a xylose-dependent biosensor based on S. xylosus XylR, which is characterised by a broad operational range (Table 1). To demonstrate the utility of the sensor, the authors used directed evolution to improve the xylose transportation capacity of hexose transporter HXT14 from Sa. cerevisiae. Here, a mutant library of the gene encoding HXT14 was created via epPCR and the library was introduced in the strain already containing the xylose biosensor. After the addition of xylose, cells with higher fluorescence were selected by FACS, thereby allowing the identification of a xylose transporter with a 6.5-fold higher transportation capacity (Fig. 3a).
Figure 3.
Applications of aTF-based biosensors in yeast. (a) Biosensors based on aTFs have allowed the screening of cDNA libraries, protein variants and combinatorial pathway designs in high throughput, by coupling accumulation of carbon sources and metabolites with expression of reporters, selectors or actuators. Such efforts have allowed the screening of cell factories with improved carbon source uptake and productivity of various chemicals. (b) Environmental biosensing using aTF-based biosensors in yeast has been applied for monitoring of hazardous methylating and estrogenic compounds.
Another example of biosensor used as a screening system comes from Li et al. (2015) where authors used an aTF-based biosensor to screen a genome-wide overexpression cDNA library to identify enzymes that could improve 3-hydroxypropionic acid (3-HP)(Fig. 3a). The sensor is based on FapR (Table 1) from B. subtilis, a TF that is able to detect malonyl-CoA, a key intermediate for the biosynthesis of several industrially relevant compounds such as 3-HP, an attractive value-added chemical. This approach allowed them to identify two enzymes, PMP1 and TPI1, which lead to higher GFP de-repression. The first gene encodes for a plasma membrane that regulates a plasma membrane proton ATPase, while the latter encodes a triose phosphate isomerase. The overexpression of these two genes singularly lead to a 116% and 120% increase in 3-HP production respectively, compared to the control strain. Interestingly, the overexpression of both enzymes simultaneously led to lower titres, comparable to the control strain (Li et al.2015).
More recently, BenM from Acinetobacter sp. ADP1 and FdeR from Herbaspirillum seropedicae (Table 1) were used to screen cell factories in order to identify best-performing pathway designs for naringenin and CCM, the latter being an important precursor for bioplastic production (Curran et al.2013; Skjoedt et al.2016). Subsequently, the production strains were tested by HPLC and flow cytometry. Here, the authors observed a strong correlation (r = 0.98) between the fluorescence intensity and biobased production of CCM and narigenin, demonstrating the ability of these biosensor as orthogonal screening systems for high-producing strains (Fig. 3a)(Skjoedt et al.2016).
Biosensors can be employed outside the field of cell factory design, and can be used as tools to screen for environmental contaminants found in water and soil. One of the earliest examples of applying aTFs as environmental screening tools is founded on the human estrogen receptor alpha (Sanseverino et al.2005). This biosensor allowed the detection of chemicals with estrogenic activity, which are able to damage the endocrine system in vertebrates. To detect those chemicals Sanseverino et al. (2005) developed a biosensor based on hER-alpha and the bacterial lux cassette (luxCDABE)(Table 1), which allowed for fast and easy detection of estrogenic compounds within hours of sampling (Fig. 3b). Apart from the almost real-time monitoring potential, this system allows for, there are still different limitations, the most important of which is that yeast cells respond differently when subjected to different estrogenic molecules and may not be able to detect specific estrogenic compounds (Sanseverino et al.2005).
Another class of toxic molecules that are common both in industry and in agriculture is represented by methylating chemicals, and in order to detect the presence of these compounds in the environment, Moser et al. (2013) developed a system that relies on the E. coli Ada protein to be directly methylated and/or to detect DNA methylation, and to transduce a quantifiable signal via a fluorescent output. This sensor is particularly interesting because it is able to detect methyl iodide (MeI) which is used in agriculture as a controversial fumigant. To validate the sensor, the authors added an aqueous solution of MeI to a soil sample and monitored fluorescence as a proxy for MeI levels over time using the biosensor (Fig. 3b)(Moser et al.2013)
aTF biosensor selection in yeast
Biosensors as a tool to screen libraries based on fluorescence have proved very useful. However, screening large libraries based on their fluorescence levels requires expensive instrumentation and, in order to screen for even larger libraries, the coupling of the biosensor to a selection assay has proved very useful (Dietrich, McKee and Keasling 2010). Two such examples come from Umeyama, Okada and Ito (2013) and Feng et al. (2015). The first example relies on the MetJ repressor fused to a B42 activation domain to induce the transcription of HIS3 LEU2 and Venus genes in the presence of S-adenosylmethionine (SAM). Additionally, the system is repressed by the Dox-responsive repressor TetR to allow for further tuning of the sensor-selector system (Fig 3a). By this dual-input system, GAL11 was identified as an enhancer of SAM accumulation. In the second example, the authors developed a system where a hybrid synthetic TF based on the DBD of Gal4, an evolved progesterone-responsive EBD and a VP16 module was used to control progesterone-dependent expression of HIS3 (Fig. 3a). This is particularly interesting because yeast-based platforms for the biosynthesis progesterone have already been developed (Duport et al.1998). Progesterone is synthesised by the conversion of pregnenolone to progesterone by the enzyme 3β-hydroxysteroid dehydrogenase (3β-HSD). To develop a selection assay, they added a His3-inhibitor to plates supplemented with pregnenolone, so that the cells harbouring the wild-type 3β-HSD were not able to produce enough progesterone to complement the histidine auxotrophy. Next, they mutagenised the 3β-HSD by epPCR and selected clones that could grow on the selective medium. Two of the surviving clones, harbouring a single amino acid mutation, were analysed for progesterone production by gas chromatography and mass spectroscopy. Both the clones were able to produce twice as much progesterone compared to the cells with the wild-type enzyme (Feng et al.2015).
aTF biosensor actuation in yeast
Apart from providing a detectable output, such as fluorescence or auxotrophy complementation, biosensors can be directly plugged into the metabolism by cloning an enzyme under the control of the aTF. A clear example of this method comes from David et al. (2016). Here FapR from B. subtilis was used to dynamically control the flux at the malonyl-CoA node and direct the precursors towards 3-HP production instead of producing fatty acids. Since 3-HP can be produced from Malonyl-CoA by one enzymatic step via a Malonyl-CoA reductase (MCRCa) from Chloroflexus aurantiacus, by putting the expression of this enzyme under the control of FapR, the authors were able to create a self-regulatory system that increased the expression of MCRCa at a level related to Malonyl-CoA availability (Fig. 3a). The combination of this approach with the development of a two-stage dynamic system where 3-HP is produced under glucose limiting conditions led ultimately to a 10-fold increase in 3-HP productivity, reaching a final titre of 1 g L−1(David et al.2016). The ability to control the flux at different nodes as shown in this study can be extremely useful to prevent metabolic imbalances and to increase productivity, as also evidenced from synthetic sensor–actuator systems in bacteria (Kobayashi et al.2004; Xu et al.2014). To date, however, the use of FapR is the only example of dynamic regulation of metabolism reported in yeast.
CONCLUSIONS AND PERSPECTIVES
In order to improve the applicability of biosensors, it is critical to expand the chemical space that biosensors are able to detect, or can be engineered to detect (Delépine et al.2016). From the studies performed on biosensor development, it is clear that achieving an optimal tuning between the elements composing the biosensor can indeed be performed by both rational and randomised approaches, as demonstrated using targeted engineering of modular domains and directed evolution, respectively (Fig. 2). However, forward engineering of aTFs with novel specificities and enhanced transfer functions is generally not feasible to date. In terms of transfer function, the goal is to gain enough understanding to design a functional biosensor by correlating the level of expression of the aTF to the synthetic promoter, containing a specific operator design, and operator variants that have different strengths, controlling the reporter gene, allowing for both relevant dynamic and operational ranges. Similarly, this also applies to engineering of new specificities at the aTF level. Here a deeper understanding of the structure to function knowledge is needed as not only operational and dynamic ranges depend on the aTF protein structure but also the specificity. Gathering more data focusing on the structure to function relationships of allostery is a prominent path towards increasing our understanding of aTF functionality and the underlying design principles (Taylor et al.2015; Richards, Meyer and Wilson 2017). Emerging and future studies based on laboratory evolution coupled with next-generation sequencing are expected to enable the build-up of data-sets supporting the construction of machine-learning algorithms with predictive power on aTF biosensor designs in relation to both transfer function and ligand specificity.
In terms of application, it is not hard to imagine that in the near future, biosensors will allow for complex gene circuit regulations by aTFs acting as core unit actuators of yeast cell factories. Here, dynamic shunting of competing pathway fluxes, decoupling of growth and production phases, and improved overall robustness of cell factories performing under industrial bioprocesses are but a few examples for which carefully engineered aTFs can be applied. Indeed, even though dynamic regulation of metabolic pathways for improved biobased production in E. coli has been reported by several groups (Dahl et al.2013; Xiao et al.2016), the recent study by David, Nielsen and Siewers (2016) on 3-HP production in yeast is expected to lead further development of synthetic control over cell metabolism by the use of product-activated biosensors.
Overall, inspired by nature, it is expected that further development in the field of synthetic biology will allow for more complex aTF-controlled regulations in order to improve biobased production of both monoculture and mixed cultures, ultimately enabling higher throughput of the iterative design-build-test cycle and overall cost-reduced biomanufacturing processes.
FUNDING
This work was supported by the Novo Nordisk Foundation and by the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie action PAcMEN (grant agreement No. 722287).
Conflict of interest.. None to declared.
REFERENCES
- Arvidson DN, Bruce C, Gunsalus RP. Interaction of the Escherichia coli trp aporepressor with its ligand, L-tryptophan. J Biol Chem 1986;261:238–43. [PubMed] [Google Scholar]
- Awan AR, Blount BA, Bell DJ et al. . Biosynthesis of the antibiotic nonribosomal peptide penicillin in baker's yeast. Nat Commun 2017;8:15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellí G, Garí E, Piedrafita L et al. . An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res 1998;26:942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodina I, Nielsen J. Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals. Biotechnol J 2014;9:609–20. [DOI] [PubMed] [Google Scholar]
- Bovee TFH, Helsdingen RJR, Hamers ARM et al. . A new highly specific and robust yeast androgen bioassay for the detection of agonists and antagonists. Anal Bioanal Chem 2007;389:1549–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzostowicz PC, Reams AB, Clark TJ et al. . Transcriptional cross-regulation of the catechol and protocatechuate branches of the beta-ketoadipate pathway contributes to carbon source-dependent expression of the Acinetobacter sp. strain ADP1 pobA gene. Appl Environ Microb 2003;69:1598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol 2008;26:787–93. [DOI] [PubMed] [Google Scholar]
- Cebolla A, Sousa C, De Lorenzo V. Effector specificity mutants of the transcriptional activator NahR of naphthalene degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J Biol Chem 1997;272:3986–92. [DOI] [PubMed] [Google Scholar]
- Chen Y, Kim JK, Hirning AJ et al. . Emergent genetic oscillations in a synthetic microbial consortium. Science (80-) 2015;349:986–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chockalingam K, Chen Z, Katzenellenbogen JA et al. . Directed evolution of specific receptor–ligand pairs for use in the creation of gene switches. P Natl Acad Sci USA 2005;102:5691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Keasling JD. Programming adaptive control to evolve increased metabolite production. Nat Commun 2013;4:2595. [DOI] [PubMed] [Google Scholar]
- Collier LS, Gaines GL, Neidle EL et al. . Regulation of benzoate degradation in Acinetobacter sp. strain ADP1 by BenM, a LysR-type transcriptional activator. J Bacteriol 1998;180:2493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CH, Arnold FH, Leadbetter JR. Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones. Mol Microbiol 2005;55:712–23. [DOI] [PubMed] [Google Scholar]
- Craven SH, Ezezika OC, Haddad S et al. . Inducer responses of BenM, a LysR-type transcriptional regulator from Acinetobacter baylyi ADP1. Mol Microbiol 2009;72:881–94. [DOI] [PubMed] [Google Scholar]
- Curran KA, Leavitt JM, Karim AS et al. . Metabolic engineering of muconic acid production in Saccharomyces cerevisiae. Metab Eng 2013;15:55–66. [DOI] [PubMed] [Google Scholar]
- Dahl RH, Zhang F, Alonso-Gutierrez J et al. . Engineering dynamic pathway regulation using stress-response promoters. Nat Biotechnol 2013;31:1039–46. [DOI] [PubMed] [Google Scholar]
- David F, Nielsen J, Siewers V. Flux control at the malonyl-CoA node through hierarchical dynamic pathway regulation in Saccharomyces cerevisiae. ACS Synth Biol 2016;5:224–33. [DOI] [PubMed] [Google Scholar]
- Delépine B, Libis V, Carbonell P et al. . SensiPath: computer-aided design of sensing-enabling metabolic pathways. Nucleic Acids Res 2016;44:W226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich JA, McKee AE, Keasling JD. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu Rev Biochem 2010;79:563–90. [DOI] [PubMed] [Google Scholar]
- De Los Santos ELC, Meyerowitz JT, Mayo SL et al. . Engineering transcriptional regulator effector specificity using computational design and in vitro rapid prototyping: developing a vanillin sensor. ACS Synth Biol 2016;5:287–95. [DOI] [PubMed] [Google Scholar]
- De Paepe B, Peters G, Coussement P et al. . Tailor-made transcriptional biosensors for optimizing microbial cell factories. J Ind Microbiol Biot 2017;44:623–45. [DOI] [PubMed] [Google Scholar]
- Duport C, Spagnoli R, Degryse E et al. . Self-sufficient biosynthesis of pregnenolone and progesterone in engineered yeast. Nat Biotechnol 1998;16:186–9. [DOI] [PubMed] [Google Scholar]
- Feng J, Jester BW, Tinberg CE et al. . A general strategy to construct small molecule biosensors in eukaryotes. Elife 2015;4:e10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-López R, Ruiz R, de la Cruz F et al. . Transcription factor-based biosensors enlightened by the analyte. Front Microbiol 2015;6:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanie S, Thodey K, Trenchard IJ et al. . Complete biosynthesis of opioids in yeast. Science (80-) 2015;349:1095–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Castro S, Salgado H, Santos-Zavaleta A et al. . RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res 2016;44:D133–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garí E, Piedrafita L, Aldea M et al. . A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 1997;13:837–48. [DOI] [PubMed] [Google Scholar]
- Garst AD, Bassalo MC, Pines G et al. . Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat Biotechnol 2016;35:48–55. [DOI] [PubMed] [Google Scholar]
- Gion K, Sakurai Y, Watari A et al. . Designed recombinant transcription factor with antibody-variable regions. Anal Chem 2009;81:10162–6. [DOI] [PubMed] [Google Scholar]
- Giuraniuc CV, MacPherson M, Saka Y. Gateway vectors for efficient artificial gene assembly in vitro and expression in yeast Saccharomyces cerevisiae. PLoS One 2013;8, DOI: 10.1371/journal.pone.0064419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. P Natl Acad Sci USA 1992;89:5547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G et al. . Transcriptional activation by tetracyclines in mammalian cells. Science (80-) 1995;268:1766–9. [DOI] [PubMed] [Google Scholar]
- Guet CC, Elowitz MB, Hsing W et al. . Combinatorial synthesis of genetic networks. Science (80-) 2002;296:1466–70. [DOI] [PubMed] [Google Scholar]
- Hahn S, Maurer P, Caesar S et al. . Classical NLS proteins from Saccharomyces cerevisiae. J Mol Biol 2008;379:678–94. [DOI] [PubMed] [Google Scholar]
- Hector RE, Mertens JA. A synthetic hybrid promoter for xylose-regulated control of gene expression in Saccharomyces yeasts. Mol Biotechnol 2017;59:24–33. [DOI] [PubMed] [Google Scholar]
- Hillen W, Berens C. Mechanisms underlying expression of TN10 encoded tetracycline resistance. Annu Rev Microbiol 1994;48:345–69. [DOI] [PubMed] [Google Scholar]
- Ikushima S, Boeke JD. New orthogonal transcriptional switches derived from Tet repressor homologues for Saccharomyces cerevisiae regulated by 2,4-diacetylphloroglucinol and other ligands. ACS Synth Biol 2017;6:497–506. [DOI] [PubMed] [Google Scholar]
- Ikushima S, Zhao Y, Boeke JD. Development of a tightly controlled off switch for Saccharomyces cerevisiae regulated by camphor, a low-cost natural product. G3 2015;5:1983–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D. Allosteric control of transcription in GntR family of transcription regulators: a structural overview. IUBMB Life 2015;67:556–63. [DOI] [PubMed] [Google Scholar]
- Jeeves M, Evans PD, Parslow RA et al. . Studies of the Escherichia coli Trp repressor binding to its five operators and to variant operator sequences. Eur J Biochem 1999;265:919–28. [DOI] [PubMed] [Google Scholar]
- Junker F, Kiewitz R, Cook AM. Characterization of the p-toluenesulfonate operon tsaMBCD and tsaR in Comamonas testosteroni T-2. J Bacteriol 1997;179:919–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Kaern M, Araki M et al. . Programmable cells: interfacing natural and engineered gene networks. P Natl Acad Sci USA 2004;101:8414–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotula JW, Kerns SJ, Shaket LA et al. . Programmable bacteria detect and record an environmental signal in the mammalian gut. P Natl Acad Sci USA 2014;111:4838–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Si T, Wang M et al. . Development of a synthetic malonyl-CoA sensor in Saccharomyces cerevisiae for intracellular metabolite monitoring and genetic screening. ACS Synth Biol 2015;4:1308–15. [DOI] [PubMed] [Google Scholar]
- Loew R, Heinz N, Hampf M et al. . Improved Tet-responsive promoters with minimized background expression. BMC Biotechnol 2010;10:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac RS, Gibney PA, Chandran SS et al. . Synthetic biology tools for programming gene expression without nutritional perturbations in Saccharomyces cerevisiae. Nucleic Acids Res 2014;42:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks SE, Oyston PCF. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008;154:3609–23. [DOI] [PubMed] [Google Scholar]
- Møller TSB, Overgaard M, Nielsen SS et al. . Relation between tetR and tetA expression in tetracycline resistant Escherichia coli. BMC Microbiol 2016;16, DOI: 10.1186/s12866-016-0649-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser F, Horwitz A, Chen J et al. . Genetic sensor for strong methylating compounds. ACS Synth Biol 2013;2:614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle EL, Hartnett C, Ornston LN. Characterization of Acinetobacter calcoaceticus catM, a repressor gene homologous in sequence to transcriptional activator genes. J Bacteriol 1989;171:5410–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottoz DSM, Rudolf F, Stelling J. Inducible, tightly regulated and growth condition-independent transcription factor in Saccharomyces cerevisiae. Nucleic Acids Res 2014;42:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta-Yahya PP, Zhang F, del Cardayre SB et al. . Microbial engineering for the production of advanced biofuels. Nature 2012;488:320–8. [DOI] [PubMed] [Google Scholar]
- Raman S, Rogers JK, Taylor ND et al. . Evolution-guided optimization of biosynthetic pathways. P Natl Acad Sci USA 2014;111:17803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasio D, Walther A, Trost K et al. . An indirect assay for volatile compound production in yeast strains. Sci Rep 2014;4:3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DH, Meyer S, Wilson CJ. Fourteen ways to reroute cooperative communication in the lactose repressor: engineering regulatory proteins with alternate repressive functions. ACS Synth Biol 2017;6:6–12. [DOI] [PubMed] [Google Scholar]
- Rogers JK, Church GM. Multiplexed engineering in biology. Trends Biotechnol 2016;34:198–206. [DOI] [PubMed] [Google Scholar]
- Roney IJ, Rudner AD, Couture J-F et al. . Improvement of the reverse tetracycline transactivator by single amino acid substitutions that reduce leaky target gene expression to undetectable levels. Sci Rep 2016;6:27697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanseverino J, Gupta RK, Layton AC et al. . Use of Saccharomyces cerevisiae BLYES expressing bacterial bioluminescence for rapid, sensitive detection of estrogenic compounds. Appl Environ Microb 2005;71:4455–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamanna DK, Sanderson KE. Genetics and regulation of D-xylose utilization in Salmonella typhimurium LT2. J Bacteriol 1979;139:71–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjoedt ML, Snoek T, Kildegaard KR et al. . Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast. Nat Chem Biol 2016;12:951–8. [DOI] [PubMed] [Google Scholar]
- Stanton BC, Nielsen AAK, Tamsir A et al. . Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat Chem Biol 2013;10:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan MI, Le Novère N. Cooperative Binding. PLoS Comput Biol 2013;9:e1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan CR, Singh R, VanInsberghe D et al. . Metagenomic scaffolds enable combinatorial lignin transformation. P Natl Acad Sci USA 2014;111:10143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süel GM, Lockless SW, Wall MA et al. . Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol 2003;10:59–69. [DOI] [PubMed] [Google Scholar]
- Taylor ND, Garruss AS, Moretti R et al. . Engineering an allosteric transcription factor to respond to new ligands. Nat Methods 2015;13:177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo WS, Chang MW. Bacterial XylRs and synthetic promoters function as genetically encoded xylose biosensors in Saccharomyces cerevisiae. Biotechnol J 2015;10:315–22. [DOI] [PubMed] [Google Scholar]
- Teo WS, Hee KS, Chang MW. Bacterial FadR and synthetic promoters function as modular fatty acid sensor- regulators in Saccharomyces cerevisiae. Eng Life Sci 2013;13:456–63. [Google Scholar]
- Umeyama T, Okada S, Ito T. Synthetic gene circuit-mediated mo-nitoring of endogenous metabolites: identification of GAL11 as a novel multicopy enhancer of S-adenosylmethionine level in yeast. ACS Synth Biol 2013;2:425–30. [DOI] [PubMed] [Google Scholar]
- Urlinger S, Baron U, Thellmann M et al. . Exploring the sequence space for tetracycline- dependent transcriptional activators: novel mutations yield expanded range and sensitivity. P Natl Acad Sci USA 2000;97:7963–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dien S. From the first drop to the first truckload: commercialization of microbial processes for renewable chemicals. Curr Opin Biotechnol 2013;24:1061–8. [DOI] [PubMed] [Google Scholar]
- Wang HH, Isaacs FJ, Carr PA et al. . Programming cells by multiplex genome engineering and accelerated evolution. Nature 2009;460:894–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Li S, Zhao H. Design and engineering of intracellular-metabolite-sensing/regulation gene circuits in Saccharomyces cerevisiae. Biotechnol Bioeng 2016;113:206–15. [DOI] [PubMed] [Google Scholar]
- Werten S, Schneider J, Palm GJ et al. . Modular organisation of inducer recognition and allostery in the tetracycline repressor. FEBS J 2016;283:2102–14. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Bowen CH, Liu D et al. . Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nat Chem Biol 2016;12:339–44. [DOI] [PubMed] [Google Scholar]
- Xiong D, Lu S, Wu J et al. . Improving key enzyme activity in phenylpropanoid pathway with a designed biosensor. Metab Eng 2017;40:115–23. [DOI] [PubMed] [Google Scholar]
- Xu P, Li L, Zhang F et al. . Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. P Natl Acad Sci USA 2014;111:11299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jensen MK, Keasling JD. Development of biosensors and their application in metabolic engineering. Curr Opin Chem Biol 2015;28:1–8. [DOI] [PubMed] [Google Scholar]
- Zhao G, Hu T, Li J et al. . A novel strategy to analyze l-tryptophan through allosteric Trp repressor based on rolling circle amplification. Biosens Bioelectron 2015;71:103–7. [DOI] [PubMed] [Google Scholar]
- Zhou YJ, Buijs NA, Zhu Z et al. . Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat Commun 2016;7, DOI: 10.1038/ncomms11709. [DOI] [PMC free article] [PubMed] [Google Scholar]