Abstract
Benthic microbial ecosystems are widespread yet knowledge gaps still remain on the relationships between the diversity of species across kingdoms and productivity. Here, we ask two fundamental questions: (i) How does species diversity relate to the rates of primary and heterotrophic productivity? (ii) How do diel variations in light-energy inputs influence productivity and microbiome diversity? To answer these questions, microbial mats from a magnesium sulfate hypersaline lake were used to establish microcosms. Both the number and relatedness between bacterial and eukaryotic taxa in the microbiome were assayed via amplicon-based sequencing of 16S and 18S rRNA genes over two diel cycles. These results correlated with biomass productivity obtained from substrate-specific 13C stable isotope tracers that enabled comparisons between primary and heterotrophic productivity. Both bacterial and eukaryotic species richness and evenness were related only to the rates of 13C-labeled glucose and acetate biomass incorporation. Interestingly, measures of these heterotrophic relationships changed from positive and negative correlations depending on carbon derived from glucose or acetate, respectively. The bacterial and eukaryotic diversity of this ecosystem is also controlled, in part, from energy constraints imposed by changing irradiance over a diel cycle.
Keywords: diversity, productivity, microbial mat, stable isotope, hypersaline
Multikingdom productivity–diversity relationships in a magnesium sulfate, hypersaline mat are controlled by diel light cycling and heterotrophic growth.
INTRODUCTION
Benthic aquatic habitats support one of the most widespread ecosystems on earth (Stal 1994; Snelgrove 1999). They play critical roles in global biogeochemical cycling, and those operating in the euphotic zone are major sources of primary productivity (MacIntyre, Geider and Miller 1996; Fenchel and Glud 2000). Photosynthetic microbial mats have had a profound influence in shaping our modern aquatic and terrestrial ecosystems through atmospheric oxygenation (Canfield and Teske 1996; Sessions et al.2009). They inhabit a wide range of environments that include freshwater, marine and hypersaline ecosystems. Mats at the far end of the salinity spectrum harbor especially complex microbiomes with high species and functional diversity (Ley et al.2006; Allen et al.2009; Bernstein et al.2016; Mobberley et al.2017). While investigations of these unique microbial ecosystems have revealed new aspects of microbial life to the scientific community for many years, knowledge gaps still remain in our understanding of the relationships between microbiome diversity and productivity.
Here, we ask two scientific questions. How does species diversity relate to the rates of primary and heterotrophic productivity? Also, how do diel variations in light-energy inputs influence productivity and microbiome diversity? The relationships between species richness and productivity have been central and often controversial to the field of ecology (Mittelbach et al.2001). It is clear, however, that the rate of carbon and energy conversion into biomass can strongly influence the number of species in a given habitat (Waide et al.1999). In addition, positive relationships between energy inputs and species richness have been widely observed across ecosystems, giving rise to the ‘Species Energy’ theory (May 1975; Hurlbert and Stegen 2014). We present the results from a study designed to ask if species richness and evenness increased or decreased with either increasing productivity or increasing solar energy available for photosynthesis.
To perform this investigation, we developed microcosms derived from the Hot Lake microbial mat. Hot Lake is a magnesium sulfate (0.2–2 M) hypersaline lake in north-central Washington State (USA) that maintains a vibrant microbial mat in the euphotic zone. The inhabiting microbial mats grow in water with high concentrations (up to 240 mM-C and 220 mM-C) of dissolved inorganic and organic carbon, respectively (Anderson 1958; Zachara et al.2016). It harbors a complex microbiome with representatives from over 50 different bacterial phyla (Bernstein et al.2016). The Hot Lake mat has only been characterized with respect to prokarya and, until now, investigations have neglected to assay for eukaryotic diversity.
Here, we expanded our view of the microbiome structure to include multiple kingdoms via amplicon-based sequencing of 16S and 18S rRNA genes. This provided estimates for the number and relatedness of both bacterial and eukaryotic taxa and enabled us to compare species diversity over time points corresponding to maximum and minimum solar energy inputs over two consecutive diel cycles. These measurements were correlated and contextualized with direct measurements of biomass productivity. We tracked productivity by measuring the net rates of 13C incorporation into biomass. Substrate-specific stable isotope tracers—bicarbonate, acetate and glucose—were employed to assay autotrophic and heterotrophic productivity, respectively. Autotrophic biomass productivity is defined here as net primary productivity and is equivalent to the rate of autotrophic carbon assimilation minus the rates of respiration and autotrophically derived organic carbon lost from the system. This study presents unique results obtained from the Hot Lake, multikingdom microbiome and shows that there are contrasting types of relationships depending on the source of carbon being traced into biomass and multikingdom microbial diversity. These relationships between bacterial and eukaryotic diversity and biomass productivity are also at least partially controlled by dynamic solar energy inputs associated with diel cycles.
MATERIALS AND METHODS
Sampling and field-based microcosms
Microbial mat sections were excised from the native benthic mat Hot Lake, in north-central Washington State (USA); 48.973062°N, 119.476876°W (Anderson 1958; Moran et al.2014; Zachara et al.2016). Each mat sample was ∼1 cm thick and taken from a water depth of 20–30 cm on 17 September 2014. The mat samples were cut and carefully removed from the underlying sediment. The mat overlays gypsum-dominated sediment which was also independently sampled. Immediately after sampling, microcosms were constructed to mimic the native benthic orientation with no or very little mixing of the overlaying bulk water. Each microcosm consisted of the excised mat incubated ex vivo on a 2-cm layer of homogenized sediment submerged under 8 cm native Hot Lake water column. The treatments were performed by amending the Hot Lake water with 13C-enriched substrates; bicarbonate (2.25 mmoles; 50% 13C from Cambridge Isotope Laboratories, Tewksbury, MA, USA), glucose (5 mmoles; 100% 1-6 13C from Cambridge Isotope Laboratory) or acetate (5 mmoles,1,2 99% 13C from ICON Isotopes, Dexter, MI, USA). One microcosm incubation was constructed for each treatment. The incubations were performed in 740 mL glass containers (17 cm diameter) exposed at the surface to ambient air and light for 48 h and placed in a temperature controlled water bath held at 22°C corresponding to the lake's water temperature at the sampling position. Sections of the mat (∼3 × 3 cm) were excised from each incubation every 6 h. Subsamples were rinsed in incubation medium devoid of added substrate for 30 min to remove unincorporated substrate, and then placed on dry ice. Later, the frozen subsamples were lyophilized and then homogenized before being aliquoted into three biological replicates per sample type (amplicon and isotope analyses described below). Given the proximity of the experiment to the vernal equinox, we were able to time these samples such that the 6-h intervals correlated to solar noon, dusk, solar midnight and dawn for two sequential days. Weather conditions were recorded from a remote, automated station ∼1.5 km from Hot Lake. These data were provided by the US Bureau of Land Management & Boise Interagency Fire Center and hosted by MesoWest, a project of the Department of Atmospheric Sciences at the University of Utah (http://mesowest.utah.edu).
Stable isotopes and productivity measurements
Mat samples were lyophilized then homogenized using a mortar and pestle. The δ13C content was measured using a Costech Analytical (Valencia, CA) Elemental Analyzer (EA, ECS 4010 CHNSO Analyzer) coupled to a Thermo Scientific (Bremen, Germany) Delta V Plus isotope ratio mass spectrometer. A combustion oven (loaded with cobaltic oxide and chromium oxide catalyst) was maintained at a temperature of 1020°C along with a reduction oven (loaded with copper catalyst) held at 650°C. In-house glutamic acid isotope standards were calibrated against USGS 40 and USGS 41 (δ13C of –26.39 ‰ and +37.63 ‰ respectively) as a basis for a two-point data correction (Coplen et al.2006). The high δ13C content of some samples caused isotopic memory, which we erased using a series of up to five runs of our in-house standard following a set of labeled samples. Net primary and heterotrophic productivity was defined as the rate of 13C incorporation into biomass over time (Δδ13C/Δt) with respect to labeled carbon derived from inorganic or organic substrates, respectively. Productivity was calculated from the isotope enrichment profiles using finite differences to estimate the first derivative (Eberly 2008). A centered difference approximation (Equation 1) was used on all points except the initial and end time points that were approximated using Euler's method (Equation 2) noting that the initial time point utilized the non-treated labeled, native mat for its corresponding δ13C(t-h) value (h equals one 6 h time step).
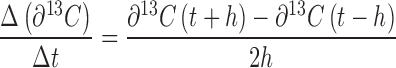 |
(1) |
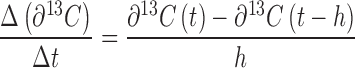 |
(2) |
Amplicon sequencing
Genomic DNA was extracted using the MoBio PowerSoil DNA isolation kit (Qiagen, Carlsbad, CA) in accordance with the Earth Microbiome Project protocols (Gilbert et al.2010). Sequencing was performed on an Illumina MiSeq instrument (Illumina, San Diego, CA). Three separate 16S and 18S rRNA amplification reactions were performed for each extraction. The 16S primers targeted the V4 hypervariable region of the 16S SSU rRNA using the V4 forward primer (515F) and V4 reverse primer (806R) with 0–3 random bases and the Illumina sequencing primer binding site (Caporaso et al.2010). The 18S primers targeted the V9 hypervariable region of the 18S SSU rRNA (Amaral-Zettler et al.2009).
Amplicon analysis
Illumina reads were processed with QIIME 1.9.1 and VSEARCH 1.9.10, an open-source implementation of USEARCH (Caporaso et al.2010; Edgar 2010; Rognes et al.2016). Using VSEARCH, overlapping 16S reads were paired, filtered to a maximum expected error of 1 bp per read and labeled. Reads were pooled, de-replicated and chimera-checked with the VSEARCH implementation of UCHIME de novo (Edgar et al.2011) followed by UCHIME-ref using the RDP Gold database (Wang et al.2007). After discarding chimeras and singletons, reads were clustered into OTUs at 97% similarity and an OTU table was constructed by mapping all labeled reads to these clusters. Taxonomy was assigned to each OTU centroid using the May 2013 version of the Greengenes database and a last common ancestor approach as implemented in QIIME 1.9.1 (McDonald et al.2012). The same pipeline was used to process 18S genes, with a few changes; reads were filtered to a maximum expected error of 0.1 bp per read and no reference-based chimera checking was used and the 18S component of the SILVA database v123 (22-08-2016) was used for taxonomy assignment, respectively (Quast et al.2013).
Diversity analysis
Downstream analysis was completed in R (Team 2000), using the phyloseq (McMurdie and Holmes 2013) and vegan packages (Oksanen et al.2013). Samples were rarified to an even depth of 22 000 reads per sample, and 16S amplicons not classified as Bacteria were removed. Bray-Curtis dissimilarities were used to characterize beta diversity via canonical analysis of principle components, while counts of unique OTUs and Simpson's E (evenness) were used to characterize alpha diversity (Hamady, Lozupone and Knight 2010).
Statistics
This study employed a nested design, with two kingdoms (measured by 16S and 18S amplicons) and three substrate treatments (13C-labeled bicarbonate, glucose, acetate). Time course measurements (eight time points) pertaining to each of these blocks were collected via non-matched, destructive sampling methods over two full diurnal cycles. A one-way ANOVA was used as an initial test if alpha diversity was the same in all time points in each block (Table S1, Supporting Information). Linear relationships between alpha diversity with irradiance and productivity were quantified by fitting a linear model within each block to obtain a positive or negative slope and P-value. This P-value is the probability of observing any value equal or larger than the calculated t-statistic, which is a measure for the number of standard deviations that the estimated slope coefficient is from zero. We assigned P-values ≤ 0.05 as significant evidence that enables the null hypothesis to be rejected and conclude that there is a relationship between irradiance/productivity and richness/evenness.
Data repository and reproducible analyses
Genetic sequencing data is available on Zenodo for both 16S amplicons and 18S amplicons as records 803376 and 803476. Feature abundance tables of amplicon along with environmental measurements and scripts used for analysis and graphing are available on GitHub: https://github.com/pnnl/bernstein-2017-productivity-and-diversity-2/.
RESULTS
The system and its components
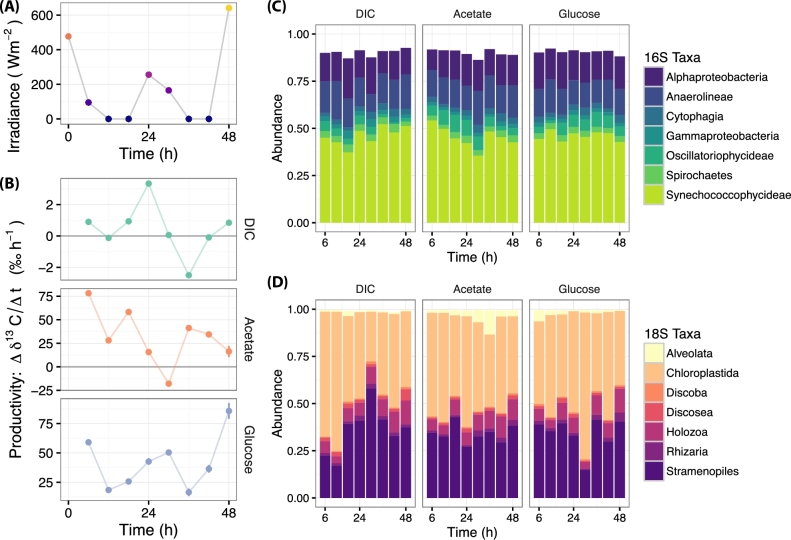
Both net primary productivity (NPP) and net heterotrophic productivity (NHP) changed with time over the course of two diel cycles (Fig. 1A and B). As expected, NPP showed local maxima and minima corresponding with solar noon (13:00) and night, respectively. The temporal pattern and magnitude of NHP varied with the substrate type. NHP as measured with 13C-labeled glucose showed strong positive correlation (r = 0.869; P < 0.001) with solar irradiance as compared to a weaker, negative correlation observed from the labeled acetate treatment (r = –0.394; P = 0.057) (Fig. S1, Supporting Information). These results show that solar irradiance not only affects photoautotrophic productivity but also the rate of organic carbon assimilation into biomass. Interestingly, the degree to which NHP shares a relationship with the light energy input to the system depends on the substrate from which organic carbon is derived.
Figure 1.
Light energy input, productivity output and the microbiome structure. (A) The solar irradiance (W m−2) incident to the system over two diel cycles. The different solar maxima correspond to variable weather patterns and cloud cover. (B) Primary and heterotrophic productivity measured as the change in 13C enrichment over time with respect to labeled bicarbonate (DIC), acetate or glucose treatments; each data point represents the mean of three biological replicates with error bars spanning ± 1 standard error. (C) Relative abundance of common bacterial taxa—at the class level—separated by time and treatment. (D) Relative abundance of common eukaryotic taxa separated by time and treatment.
The Hot Lake microbial mat harbors very high bacterial diversity (Bernstein et al.2016). In this current study, we identified over 3000 16S OTUs representing more than 30 phyla. The bacterial component of the microbiome is dominated by cyanobacteria of the class Synechococcophycideae, which contains the most abundant 16S OTUs identified as Pseudanabaenaceae at the family level (Fig. 1C). Oscillatoriophycideae represented another prevalent class of cyanobacteria with the most common class of OTUs identified as Phormidiaceae. These highly abundant cyanobacteria have been characterized and enriched from the Hot Lake ecosystem in previous studies (Cole et al.2014; Bernstein et al.2016) and are known to be filamentous and commonly associated with Alphaproteobacteria and other heterotrophic phyla identified from this current set of experiments. The system is also abundant in anoxygenic phototrophs described by the class Anaerolineae, which contains abundant OTUs with family assignments as Anerolineaceae that have been associated with anaerobic photoheterotrophic species (Yamada et al.2006; Narihiro and Kamagata 2013).
The Hot Lake microbial mat also harbors high eukaryotic diversity. Representatives were identified across 12 phyla from over 2000 18S OTUs. The most abundant eukaryotic taxon was Chloroplastidia, a group which contains green algae and plants (Fig. 1D). Within this group, we identified OTUs specifically belonging to Alismatales, an order that contains hypersaline-adapted seagrasses (Asem et al.2014). Stramenopiles were also highly abundant. Within this group, the most prevalent OTUs belonged to golden algae (Chrysophyceae), protists (Blastocyctis and Thraustochytriaceae) and diatoms (Bacillariophyceae). Another abundant taxon was Holozoa, a group of single celled animals that includes the well-known Choanoflagelletes (Codonosigidae sp.). Although less abundant, OTUs belonging to the taxonomic group Alveolata were identified, which includes photosynthetic Dinoflagellates and parasitic organisms such as Apicomplexa.
Diel influence on multikingdom diversity
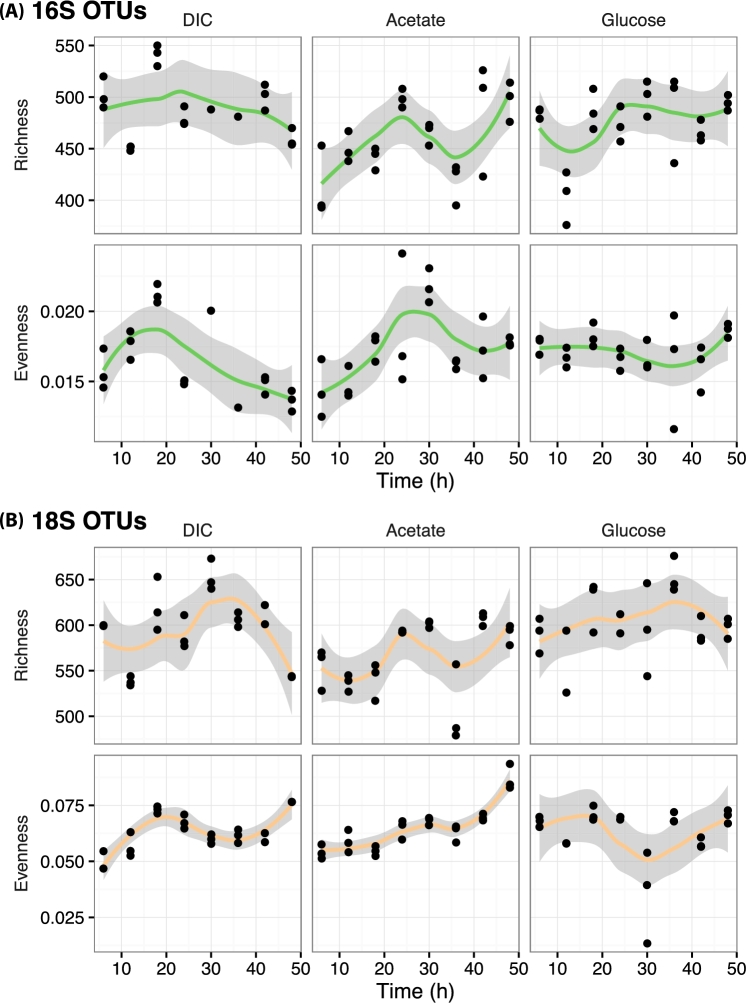
The alpha diversity of both prokaryotes and eukaryotes changed within the two diel cycle frame of observation. Species richness and evenness showed significant change (P < 0.05; Table S1, Supporting Information) for all substrate treatments with the exception of the bacterial evenness (P = 0.516) and eukaryotic richness (P = 0.079) corresponding to the glucose treatments (Fig. 2). Non-linear trend lines, established via locally weighted scatterplot smoothing functions (Cleveland and Devlin 1988), showed local maxima for bacterial richness were observed under all treatments at solar noon (13:00) corresponding to 24 h of incubation (Fig. 2A). This result was most pronounced under the labeled acetate and glucose treatments as compared to dissolved inorganic carbon (DIC). With the exception of the acetate treatment, the eukaryote species richness and evenness did not share the consistent localized maxima at the 24-h (solar noon) sampling time. In general, a periodic time profile could be observed indicating that organismal diversity in this ecosystem changes on the time scale of a single diel cycle.
Figure 2.
Alpha diversity trends over the 48 h, outdoor incubation. The data sets are parsed by each substrate treatment: bicarbonate (DIC), acetate and glucose. (A) Changes in bacterial species richness and evenness. (B) Changes in microeukaryotic species richness and evenness. Each data set is fit to a locally weighted scatterplot smoothing (loess) function and shaded regions represent the 95% confidence bounds.
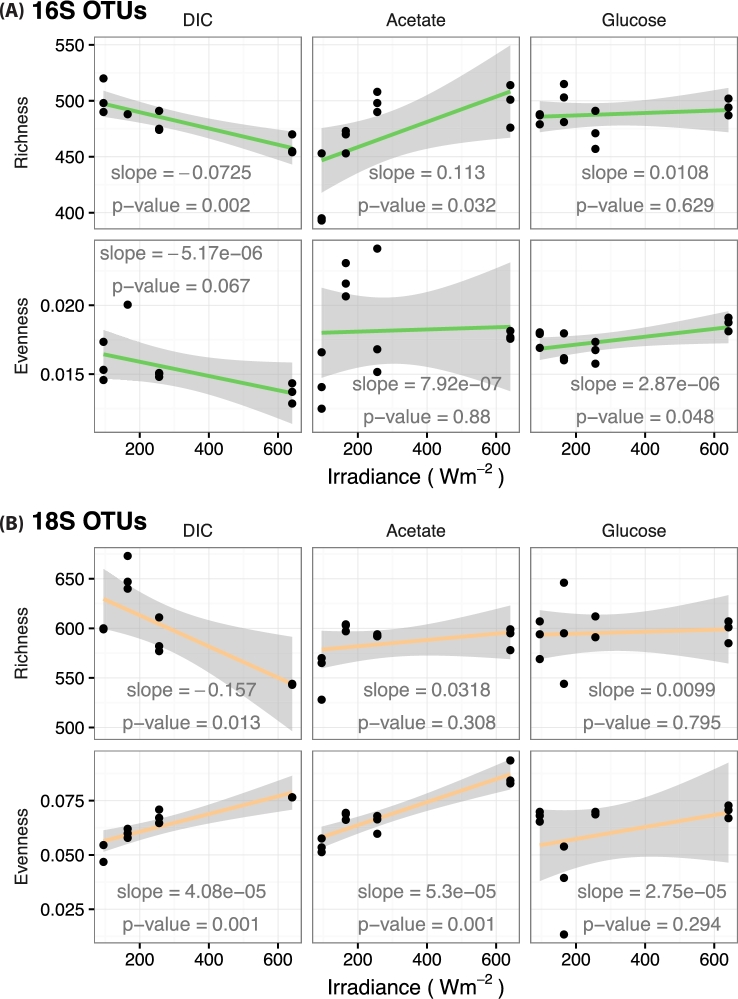
Further evidence for the control of diel light cycling over alpha diversity was evinced by evaluating changes in species richness and evenness with respect to solar incident irradiances (Fig. 3). We employed linear models to perform a simple test designed to ask if alpha diversity increased or decreased with increasing solar irradiance. The slopes describing these relationships showed that alpha diversity changed with solar energy input to the system (Fig. 3). Significant, negative relationships (P ≤ 0.013) were observed between bacterial and eukaryotic species richness and irradiance under DIC treatments. In contrast, a significant, positive relationship (P = 0.032) between bacterial species richness and irradiance was observed under the acetate treatment highlighting that substrate type and availability also influenced the species diversity of the system. Light energy input was found to increase the level of similarity between abundances of organisms in this ecosystem. Only positive correlations were found to be statistically significant (P ≤ 0.05) between evenness and solar irradiance. Specifically, bacterial evenness increased with increasing irradiance under the glucose treatment and eukaryotic evenness increased under the DIC and acetate treatments. The collection of these results in combination with the dynamic alpha diversity profiles (Fig. 2) is evidence that species diversity, of this ecosystem, is controlled in part by solar irradiance.
Figure 3.
Alpha diversity trends with light energy input to the system. The data sets are parsed by each substrate treatment: bicarbonate (DIC), acetate and glucose. (A) Changes in bacterial species richness and evenness corresponding to solar irradiance. (B) Changes in microeukaryotic species richness and evenness corresponding to solar irradiance. Broad assignments of increasing or decreasing richness and evenness correspond to positive or negative slopes, respectively. Shaded regions represent the 95% confidence bounds of the linear model.
Diversity–productivity relationships
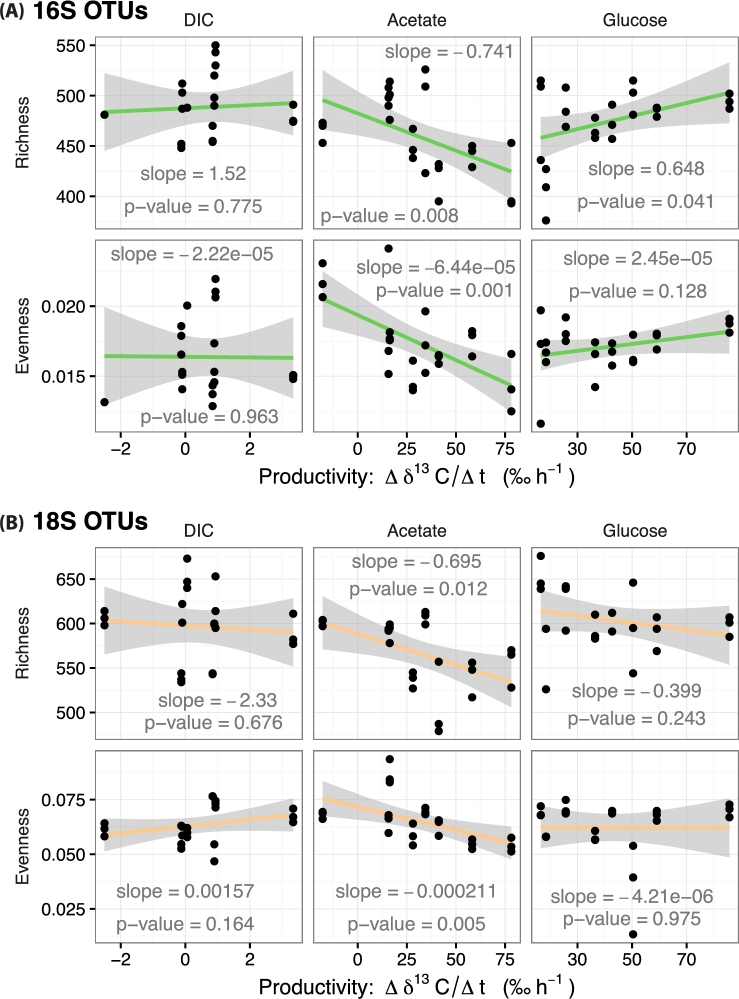
We also employed linear models to test if biomass productivity increased or decreased with species richness and evenness across both kingdoms. This test showed that NHP, as measured by conversation of 13C-labeled glucose and acetate in into biomass, changed with diversity but not NPP (Fig. 4). The slopes describing the relationships between alpha diversity and NPP were not significant (P > 0.05), indicating that we cannot reject the null hypothesis that changes in NPP are constant with changes in species richness and evenness. This observation held true for both bacterial and eukaryotic components of the microbiome. Measurements of NHP, as assayed by incorporation of acetate-derived 13C, decreased with increasing bacterial and eukaryotic species richness and evenness (P ≤ 0.012). In contrast, NHP—as assayed by uptake of 13C labeled glucose—showed a positive and statistically significant relationship (P = 0.041) with bacterial species richness (Fig. 4A). Linear regression between eukaryotic alpha diversity and heterotrophic productivity, as assayed by bioconversion of 13C-labeled glucose, did not result in statistically significant slopes. These analyses collectively show that biomass productivity is influenced by both the number and relatedness of species but that there are contrasting types of relationships depending on the source of carbon being traced into biomass.
Figure 4.
Alpha diversity trends with microbial biomass productivity. The data sets are parsed by primary productivity (13C-labeled bicarbonate; DIC) and heterotrophic productivity (13C-labeled acetate and glucose). (A) Changes in bacterial species richness and evenness with system-wide biomass productivity. (B) Changes in eukaryote species richness and evenness with system-wide biomass productivity. Broad assignments of increasing or decreasing richness and evenness correspond to positive or negative slopes, respectively. Shaded regions represent the 95% confidence bounds of the linear model.
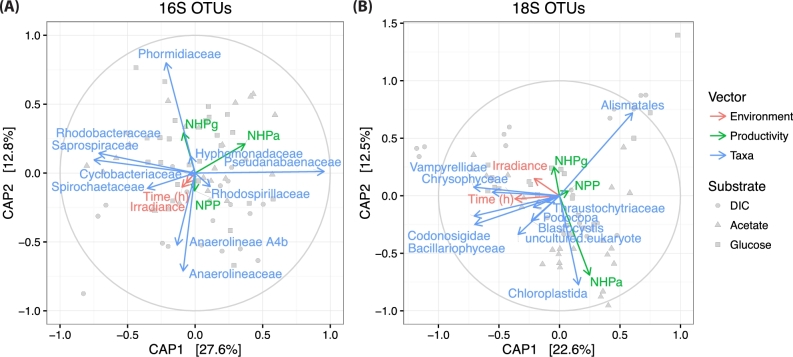
The key biological and environmental properties associated with beta diversity were assessed by a canonical analysis of principle coordinates considering Bray-Curtis dissimilarities between rarified samples (Fig. 5). Dissimilarity between the 10 most common taxa (from both kingdoms) were assessed by ordination with respect to NPP, NHP, time and incident solar irradiance over the course of the two consecutive diel cycles. Interestingly, the abundances of cyanobacteria (Pseudanabaenaceae and Phormidiaceae) ordinated in closer alignment with each measure of NHP as compared to NPP (Fig. 5A). Of the most common eukaryotic taxa, those belonging to Chloroplastida showed the closest alignment with each measure of net biomass productivity (Fig. 5B). Specifically, Alismatales, an order that contains hypersaline-adapted seagrasses (Asem et al.2014), showed the closest alignment with NPP. These results indicate that while bacterial and eukaryotic alpha diversity did not correlate significantly with NPP (Fig. 4), the abundances of OTUs associated with photoautotrophic taxa still covaried with measures of productivity.
Figure 5.
Beta diversity of bacteria and eukaryotes. Canonical analysis of principle coordinates of Bray-Curtis dissimilarities between rarified samples. Each vector has a magnitude (length) and a sign (direction) corresponding to its contributions to the principle components. Red vectors are assigned to the independent variables, time and irradiance. Green vectors are assigned to net biomass productivity: net primary productivity (NPP), net heterotrophic productivity measured with acetate (NHPa) and heterotrophic productivity measured with glucose (NHPg). Blue vectors are assigned to the 10 most common taxa. (A) An ordination of bacterial samples captures more than 40% of the variance observed. (B) An ordination of the eukaryote samples captures more than 33% of the variance observed. Each data point represents the microbiome structure observed for different treatments that are shown by symbol shapes.
DISCUSSION
The goal of this study was to interrogate two scientific questions: (i) How does species diversity relate to the rates of primary and heterotrophic productivity? (ii) How do light energy inputs over diel cycles influence productivity and microbiome diversity? The results showed relationships between both bacterial and eukaryotic species richness and evenness with heterotrophic biomass productivity. This result is supported by statistically significant correlations between OTU richness and evenness to the rates of 13C labeled carbon incorporated into biomass from either acetate or glucose. These substrates were chosen to represent substrates corresponding to reducing sugars that can be fermented and organic acids that must be oxidized using external terminal electron acceptors. Significant Pearson's correlations were not observed between either alpha diversity metric to the rates of bicarbonate incorporation. Interestingly, measures of these relationships changed from positive and negative correlations depending on carbon derived from glucose and acetate, respectively. Likely, these contrasting relationships observed between glucose and acetate treatments stem from the different metabolic strategies that microbes have available to harvest both carbon and energy for growth. This study also confirmed that relationships exist between the rate of light-energy input to the system and species diversity. Bacterial and eukaryotic diversity of this ecosystem is partially controlled by solar irradiance and therefore varies over the diel. Evidence of diel and/or light energy control over taxa associated with photoautotrophic and heterotrophic ecosystem processes was apparent by observing statistically significant correlations between solar incident irradiance and heterotrophic productivity.
Energy and carbon-substrate availability constrain the productivities and biological carrying capacities of all ecosystems. In shallow water benthic ecosystems, such as the Hot Lake mat, the relationships between photosynthetically driven primary producers and heterotrophic members underpin the microbial food web. Many studies have been performed to delineate the food web dynamics and carbon flow between bacteria, protists, meiofauna and macrofauna (Miller, Geider and MacIntyre 1996; Sundbäck et al.1996; Al-Zaidan et al.2006). However, major knowledge gaps still remain in our understanding of the ecological driving forces that control a microbial ecosystem's capacity for primary and heterotrophic productivity. For instance, this current study discovered that different solar energy–productivity relationships (positive verse negative) can be observed depending on the source of carbon (Fig. S1). While differences exist between autotrophic (inorganic) and heterotrophic substrates, shifts in the relationships also occurred when the source of organic carbon changed between acetate and glucose. Again, these observed differences likely arise from the metabolic strategies and energy requirements for using respective glucose or acetate substrates to make biomass. A striking result obtained from this study was that heterotrophic productivity (regardless of the substrate source) is at least partially controlled by light energy inputs to the system and the diel cycle.
Extensive investigations in the field of microbial ecology have concluded that there is not a canonical relationship or ubiquitous ‘curve shape’ for microbial diversity–productivity relationships (Smith 2007). This current study found that there were statistically significant, linear relationships between NHP and species richness. Our analysis also showed the lack of a statistically significant correlation between NPP and species richness and evenness, regardless of kingdom. However, other patterns could be inferred from this data such as the monomodal or ‘hump-shaped’ curve that is a frequently observed response between diversity and productivity (Rosenzweig 1995). While further interpretation of these relationships is certainly an option, the adoption of more complex models to describe productivity–diversity relationships is beyond the scope of this study. The goal of this study was to simply determine if the rates of carbon and energy conversion into biomass increased or decreased the number and relatedness between bacteria and eukaryotic taxa.
Of the few studies that have investigated microbial productivity–diversity relationships, fewer still have explicitly considered the distinct contributions from primary producers and heterotrophic consumers. One example of note, however, investigated relationships in chemolithoautotrophic karst ecosystems by comparing 16S sequence-based analyses with the incorporation of carbon derived from 13C-labeled substrates: bicarbonate, acetate and leucine (Porter et al.2009). They found positive relationships between prokaryotic richness and chemoautotrophic productivity, and no detectable relationships (random patterns) for heterotrophic productivity and a primary energy input to the system in the form of sulfide. These are essentially the exact opposite findings as those that we report in this current study highlighting again that canonical relationships between productivity and microbial diversity are unlikely to exist. The relationships that underpin each microbial ecosystem have a specific context, and seemingly similar ecosystems can have very different responses.
General conclusions
Microbial productivity is influenced by both the number and relatedness of species existing within the ecosystem. However, within a multikingdom microbiome, such as the Hot Lake mat studied here, there are contrasting types of relationships depending on the source of carbon being traced into biomass. These relationships between species diversity and biomass productivity are also at least partially controlled by dynamic environmental factors. Specifically, solar energy inputs across diel cycles.
Supplementary Material
Supplementary data are available at FEMSEC online.
Acknowledgements
The authors would like to thank Joe Brown, William Nelson and Charles Resch for valuable discussions and technical assistance. The authors would also like to acknowledge the U.S. Bureau of Land Management, Wenatchee Field Office, for their assistance in authorizing this research and providing access to the Hot Lake Research Natural Area.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSEC online.
FUNDING
This work was supported by the U.S. Department of Energy (DOE), Office of Biological and Environmental Research (BER), as part of BER's Genomic Science Program (GSP). This contribution originates from the GSP Foundational Scientific Focus Area (FSFA) at the Pacific Northwest National Laboratory (PNNL). A portion of this study was supported by PNNL's institutional computing resource. PNNL is operated for DOE by Battelle Memorial Institute under contract DE-AC05-76RL01830.
Conflict of interest. None declared.
REFERENCES
- Al-Zaidan ASY, Kennedy H, Jones DA et al. Role of microbial mats in Sulaibikhat Bay (Kuwait) mudflat food webs: evidence from δ13C analysis. Mar Ecol Prog Ser 2006;308:27–36. [Google Scholar]
- Allen MA, Goh F, Burns BP et al. Bacterial, archaeal and eukaryotic diversity of smooth and pustular microbial mat communities in the hypersaline lagoon of Shark Bay. Geobiology 2009;7:82–96. [DOI] [PubMed] [Google Scholar]
- Amaral-Zettler LA, McCliment EA, Ducklow HW et al. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One 2009;4:e6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GC. Some limnological features of a shallow saline meromictic lake. Limnol Oceanogr 1958;3:259–70. [Google Scholar]
- Asem A, Eimanifar A, Djamali M et al. Biodiversity of the hypersaline Urmia Lake National Park (NW Iran). Diversity 2014;6:102–32. [Google Scholar]
- Bernstein HC, Brislawn C, Renslow RS et al. Trade-offs between microbiome diversity and productivity in a stratified microbial mat. ISME J 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield DE, Teske A. Late proterozoic rise in atomospheric oxygen concentration inferred from phylogenetic and sulphur-isotope studies. Nature 1996;382:127. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland WS, Devlin SJ. Locally weighted regression: an approach to regression analysis by local fitting. J Am Stat Assoc 1988;83:596–610. [Google Scholar]
- Cole JK, Hutchison JR, Renslow RS et al. Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: model systems for the study of autotroph-heterotroph interactions. Front Microbiol 2014;5:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coplen TB, Brand WA, Gehre M et al. New guidelines for δ13C measurements. Anal Chem 2006;78:2439–41. [DOI] [PubMed] [Google Scholar]
- Eberly D. Derivative Approximation by Finite Differences. Magic Software, Inc, 2008. [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–61. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenchel T, Glud RN. Benthic primary production and O2-CO2 dynamics in a shallow-water sediment: Spatial and temporal heterogeneity. Ophelia 2000;53:159–71. [Google Scholar]
- Gilbert JA, Meyer F, Antonopoulos D et al. Meeting report: the terabase metagenomics workshop and the vision of an Earth microbiome project. Stand Genomic Sci 2010;3:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 2010;4:17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbert AH, Stegen JC. When should species richness be energy limited, and how would we know? Ecol Lett 2014;17:401–13. [DOI] [PubMed] [Google Scholar]
- Ley RE, Harris JK, Wilcox J et al. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl Environ Microb 2006;72:3685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6:610–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre HL, Geider RJ, Miller DC. Microphytobenthos: the ecological role of the “secret garden” of unvegetated, shallow-water marine habitats. I. Distribution, abundance and primary production. Estuaries 1996;19:186–201. [Google Scholar]
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May RM. Patterns of species abundance and diversity. In: Cody ML, Diamond JM (eds). Ecology and Evolution Communities. Cambridge, MA, USA: Harvard University Press, 1975, 81–120. [Google Scholar]
- Miller DC, Geider RJ, MacIntyre HL. Microphytobenthos: the ecological role of the “secret garden” of unvegetated, shallow-water marine habitats. II. Role in sediment stability and shallow-water food webs. Estuaries 1996;19:202–12. [Google Scholar]
- Mittelbach GG, Christopher F, Scheiner SM et al. What is the observed relationship between species richness and productivity?. Ecology 2001;82:2381–96. [Google Scholar]
- Mobberley JM, Lindemann SR, Bernstein HC et al. Organismal and spatial partitioning of energy and macronutrient transformations within a hypersaline mat. FEMS Microbiol Ecol 2017;93, DOI: 10.1093/femsec/fix028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JJ, Doll CG, Bernstein HC et al. Spatially tracking 13C‐labelled substrate (bicarbonate) accumulation in microbial communities using laser ablation isotope ratio mass spectrometry. Environ Microbiol Rep 2014;6:786–91. [DOI] [PubMed] [Google Scholar]
- Narihiro T, Kamagata Y. Cultivating yet-to-be cultivated microbes: the challenge continues. Microb Environ 2013;28:163–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R et al. Package ‘vegan’. Community Ecology Package, Version 2, 2013. [Google Scholar]
- Porter ML, Engle AS, Kane TC et al. Productivity-diversity relationships from chemolithoautotrophically based sulfidic karst systems. Int J Speleol 2009;38:4. [Google Scholar]
- Quast C, Pruesse EY, Yilmaz P et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R Language Definition. R Foundation for Statistical Computing: Vienna, Austria, 2000. [Google Scholar]
- Rognes T, Flouri T, Nichols B et al. VSEARCH: a versatile open source tool for metagenomics. Peer J 2016;4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig ML. Species Diversity in Space and Time. Cambridge, UK: Cambridge University Press, 1995. [Google Scholar]
- Sessions AL, Doughty DM, Welander PV et al. The continuing puzzle of the great oxidation event. Curr Biol 2009;19:R567–74. [DOI] [PubMed] [Google Scholar]
- Smith VH. Microbial diversity–productivity relationships in aquatic ecosystems. FEMS Microbiol Ecol 2007;62:181–6. [DOI] [PubMed] [Google Scholar]
- Snelgrove PVR. Getting to the bottom of marine biodiversity: sedimentary habitats: ocean bottoms are the most widespread habitat on earth and support high biodiversity and key ecosystem services. Bioscience 1999;49:129–38. [Google Scholar]
- Stal LJ. Microbial mats in coastal environments. In: Microbial Mats. Berlin, Heidelberg: Springer, 1994, 21–32. [Google Scholar]
- Sundbäck K, Nilsson P, Nilsson C et al. Balance between autotrophic and heterotrophic components and processes in microbenthic communities of sandy sediments: a field study. Estuar Coast Shelf S 1996;43:689–706. [Google Scholar]
- Waide RB, Willig MR, Steiner CF et al. The relationship between productivity and species richness. Annu Rev Ecol Syst 1999;30:257–300. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Sekiguchi Y, Hanada S et al. Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Inte J Syst Evol Micr 2006;56:1331–40. [DOI] [PubMed] [Google Scholar]
- Zachara JM, Moran JJ, Resch CT et al. Geo-and biogeochemical processes in a heliothermal hypersaline lake. Geochim Cosmochim Acta 2016;181:144–63. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at FEMSEC online.