Abstract
Integrative and conjugative elements (ICEs) are widespread mobile DNA that transmit both vertically, in a host-integrated state, and horizontally, through excision and transfer to new recipients. Different families of ICEs have been discovered with more or less restricted host ranges, which operate by similar mechanisms but differ in regulatory networks, evolutionary origin and the types of variable genes they contribute to the host. Based on reviewing recent experimental data, we propose a general model of ICE life style that explains the transition between vertical and horizontal transmission as a result of a bistable decision in the ICE–host partnership. In the large majority of cells, the ICE remains silent and integrated, but hidden at low to very low frequencies in the population specialized host cells appear in which the ICE starts its process of horizontal transmission. This bistable process leads to host cell differentiation, ICE excision and transfer, when suitable recipients are present. The ratio of ICE bistability (i.e. ratio of horizontal to vertical transmission) is the outcome of a balance between fitness costs imposed by the ICE horizontal transmission process on the host cell, and selection for ICE distribution (i.e. ICE ‘fitness’). From this emerges a picture of ICEs as elements that have adapted to a mostly confined life style within their host, but with a very effective and dynamic transfer from a subpopulation of dedicated cells.
Keywords: horizontal gene transfer, bistability, cellular differentiation, fitness cost
Integrative and conjugative elements impose a bistable life style on their host, enabling a small differentiated subpopulation of cells to transmit the element.
INTRODUCTION
History of discovery of genomic islands
Understanding prokaryotic evolution and adaptation is one of the most fascinating and challenging research topics in microbiology. Genomes of prokaryotic species kept in isolation and grown as pure cultures accumulate genetic changes over timescales of hundreds to thousands of generations (Barrick et al.2009; Barrick and Lenski 2013; Raeside et al.2014). These follow accidental paths that are the result of stochastic errors in DNA replication or repair mechanisms and the selective conditions imposed on mutant fitness. Evolutionary changes are also the consequence of the starting constellation of ‘mobile genetic elements’ in the genome. Activity of mobile elements can result in changes in the DNA sequence, for example, by excising, inserting or duplicating.
Prokaryotic species in their natural environment rarely live in isolation but rather in communities, and their genomes not only evolve as a result of their own starting configuration, but also as a consequence of in- and efflux of DNA (horizontal gene transfer, HGT) from and to other species in the community. Likely, at this point, no one really understands the dynamic magnitude of such in- and efflux in diverse prokaryotic communities, but we can appreciate some of the outcomes and mechanisms facilitating the process (Soucy, Huang and Gogarten 2015). Bioinformatic analyses on large-scale genome and metagenome sequencing projects have inferred that many if not most prokaryotic genes have at some point been horizontally exchanged between species (Lawrence and Ochman 1998; Koonin and Wolf 2008; Kloesges et al.2011; Caro-Quintero and Konstantinidis 2015), although barriers exist that can inhibit exchange between different species groups (Koonin and Wolf 2008; Popa and Dagan 2011). Early estimates suggested that 18% of Escherichia coli genes were acquired by HGT in the past 14 million years (Lawrence and Ochman 1998). Kloesges et al. (2011) concluded that at least 75% of all protein families in Proteobacteria have been subject to HGT during evolution. The recent outcomes of gene flow in communities have been most obvious for strongly selected phenotypes under specific environmental conditions. Examples include the evolution of E. coli O157:H7 virulent lineages (Zhang et al.2007), of multi-resistant Staphylococcus aureus in hospital environments (Senn et al.2016), the widespread distribution of carbapenemase genes in Gram-negative bacteria (Wendel et al.2016) or the distribution of the vanB vancomycin resistance determinant in Enterococcus faecalis (Bender et al.2016). Also large-scale pollution with toxic aromatic and halogenated compounds has led to selection and outgrowth of mutants capable of metabolizing them. Such metabolic inventions could be traced to existing genes in a community having been horizontally transferred and newly assembled in a single host (van der Meer et al.1992; Müller et al.2003, 2004; Sangwan et al.2014; Verma et al.2014).
Experimental studies with pure cultures have led to a detailed understanding of a number of mechanisms by which DNA is laterally transferred, although it is not unlikely that still further and other types of mechanisms of DNA mobility will be discovered in the future. HGT mechanisms have been classically divided into three categories: transformation (the uptake of DNA by a cell), transduction (transport of DNA through the action of phages or phage particles) and conjugation (efflux of DNA from a donor cell to a recipient with help of a dedicated protein machinery) (Canchaya et al.2003a, Chen, Christie and Dubnau 2005; Gogarten and Townsend 2005; Soucy, Huang and Gogarten 2015). The molecular machines and mechanisms facilitating HGT are encoded within the genomes of their hosts, frequently by the mobile DNA elements themselves (Christie 2016). Crucially, HGT is not ‘spontaneous’, but mobile DNA elements have evolved extensive and delicate regulatory systems controlling their activity, which are subject to evolutionary change and selection themselves. As an example, most natural prokaryotic species contain a variety of different integrated prophages. Some of those are intact and active, leading to lytic phage escape under stress conditions, while accidentally packaging host DNA, which can next be injected and possibly recombine with the DNA of a newly infected host. Other phages are no longer intact and their functionalities have eroded over time (Bobay, Touchon and Rocha 2014). The DNA of such ‘satellite’ phages can be accidentally packaged and mobilized by a coresiding phage during a lytic cycle, leading to inserted DNA in the genome of a new host without clear signatures on its origins (Canchaya et al.2003b, Touchon, Bobay and Rocha 2014).
Arguably, one of the important discoveries in the past two decades with respect to prokaryotic genome evolution and HGT was the appreciation that most bacterial genomes contain other integrated and potentially mobile DNA elements, which are not prophages (Dobrindt et al.2004). In the early 1990s, this was sort of a conundrum, because it was generally thought that chromosomal DNA is ‘stable’ and its DNA only moves via phages, via integrated conjugative plasmids (i.e. Hfr) or via recombination onto plasmids. However, various lines of evidence both from experimental model systems and from comparative genome projects support the conclusion that there are widespread specific mobile DNA elements integrated in bacterial chromosomes, which can transfer independently and are neither phage nor plasmid (Roos and van Passel 2011). Multigenome comparisons of closely related species led to the notion of ‘genomic islands’ or ‘regions of genome plasticity’, i.e. discrete, large (10 to 100 kb or more) regions of DNA in bacterial chromosomes, frequently unique to a single strain or subset of strains (Dobrindt et al.2004; Tsuru et al.2006; Boyd et al.2008; Mathee et al.2008; Konstantinidis et al.2009). The presence of such discrete DNA regions suggests incidental in- and efflux, but for most of them neither their origin nor the mechanisms of their mobility have been firmly established. In parallel to this, experimental work led to the discovery of conjugative DNA, which did not ‘fit’ the classical assignment of conjugative or mobilizable plasmids (Knapp et al.1986; Waldor, Tschäpe and Mekalanos 1996; Ravatn, Zehnder and van der Meer 1998, Sullivan and Ronson 1998). Characterization of a number of such elements has helped to understand their diversity and to clarify their transfer mechanisms.
Nomenclature and classifications
The term genomic islands (GIs) as originally proposed covers well the concept of a DNA region of likely foreign origin (i.e. exhibiting hallmarks of horizontally acquired DNA in a recent evolutionary past, different G+C content from the rest of the genome) that is present only in few strains of the same or closely related prokaryotic species (Dobrindt et al.2004). However, the term genomic island does not imply any specific functional mobility mechanism, and GIs encompass a variety of potentially very different types of elements. Based on observed functional gene content, they have been frequently subclassified in pathogenicity, resistance, catabolic or symbiosis islands (Juhas et al.2009). As far as currently understood, GIs only in some cases carry clearly recognizable DNA mobility functions. In other cases, the GI may constitute a past functional element that has eroded, may yet represent an unknown type of functional mobile DNA element, or may be the result of again some other HGT mechanism or illegitimate recombination event. Other names than GI have been used to describe the same concept of a chromosomal DNA region of foreign origin but without implicit functional implications, such as genomic islets or region of genomic plasticity (Mathee et al.2008). We think the term genomic island covers the concept adequately and can be used as such.
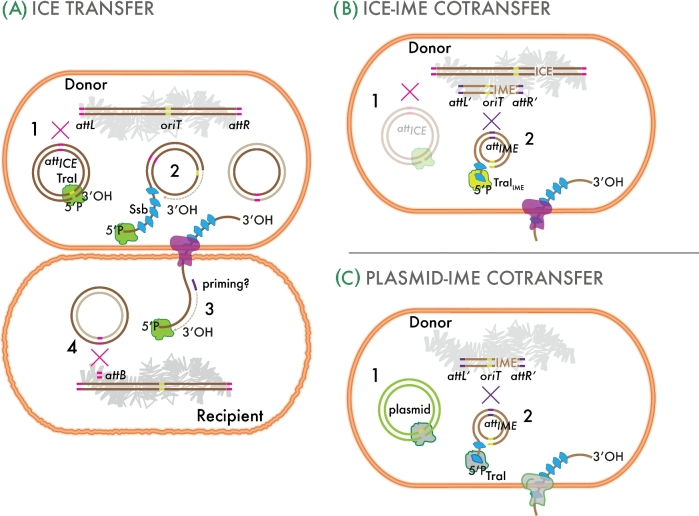
As the functional behavior of some GIs became clearer, other nomenclature started to appear and it is to be expected that further and other names will be proposed once detailed functional studies have been carried out. The major focus of this review are integrative and conjugative elements (ICEs) (Burrus et al.2002a), which, as their name implies, encompass DNA regions which are integrated in the prokaryotic chromosome but can also conjugate to other hosts (as explained in more detail in the paragraphs below, Fig. 1A). Conjugative transposons (or CTns), DNA elements first discovered in Gram-positive bacteria, can be viewed as ICEs; their maintenance relies on their integration into the chromosome of a host cell but they can also conjugate to new hosts and reintegrate (Burrus et al.2002a). ICEs and CTns can also erode over time within a host chromosome and accumulate changes leading to inactivation of their independent mobility. To complicate things more, such ‘degraded’ elements and likely others that perhaps were never completely independently mobile, can become mobilized by the machinery of intact ICEs, CTns or conjugative plasmids present in the same cell (Toussaint et al.2003; Daccord, Ceccarelli and Burrus 2010; Lee, Thomas and Grossman 2012; Carraro et al.2016b) (Fig. 1B and C). To acknowledge this possibility, some of such elements have been named integrative and mobilizable elements or IMEs (Burrus et al.2002b, Doublet et al.2005; Brochet et al.2008; Wozniak and Waldor 2010). Finally, quite recently it was shown that multiple chromosomal elements may regroup through recombination as ‘tripartite’ ICEs, and transfer as such (Haskett et al.2016).
Figure 1.
Generalized conceptual model of transfer of ICEs. (A) The ICE is integrated in the host chromosome (brown bars) but can excise by site-specific recombination (red cross) between the attachment ends (attL and attR), a process mediated by an ICE-specific integrase. The excised ICE molecule (1) undergoes single-strand cleavage at the origin of transfer (oriT), unwinds and reconstitutes by rolling-circle replication as a result of TraI relaxase activity (2). One single-stranded copy covered by single-strand DNA binding protein (Ssb) passes through a type IV conjugative channel (pink and grey membrane structure) or other into a recipient cell. (3) The double-stranded DNA is reconstituted and site specifically recombines with the recipient's attB attchment site to become re-integrated (4). (B) In certain cases, ICEs can mobilize other integrated elements (IME), which can excise by themselves (magenta cross), may have their own TraI relaxase, but rely on the transfer system of the ICE or even of a conjugative plasmid (C). After transfer, the IME can reintegrate into the recipient chromosome by site-specific recombination at its attIME site.
Scope of the review
Clearly, genome sequencing has drastically revolutionized our conception of the variety and extent of potentially mobile DNA elements that are integrated within prokaryotic host genomes. The easy access to rapid sequencing technology will further uncover the dynamic aspect of genome evolution in real-life settings, as recent epidemiological work demonstrates (Sentchilo et al.2013; Bianconi et al.2015; Senn et al.2016). A variety of reviews have deepened our understanding of the general importance of HGT (Koonin and Wolf 2008; Soucy, Huang and Gogarten 2015), of plasmid-type transfers both in uncultured and uncharacterized plasmids (Smillie et al.2010) and of evolution of viruses and plasmids from ‘capsidless’ genetic parasites (Koonin and Dolja 2014). Other reviews have highlighted GI evolution with ecological and pathogenic properties (Juhas et al.2009). Precise and sensitive bioinformatic methods have revealed the wide abundance of conjugation systems encoded on prokaryotic chromosomes, including conjugative systems of ICEs (Guglielmini et al.2011, 2014; Roos and van Passel 2011; Guglielmini, de la Cruz and Rocha 2013). Further recent reviews have extensively classified and described ICEs in terms of their basic genetic content, mechanistic properties (excision, transfer, comobilization, integration), or regulatory aspects and evolution (Wozniak and Waldor 2010; Bellanger et al.2014; Carraro and Burrus 2014; Johnson and Grossman 2015). Recent single cell approaches in combination with genetic tools, however, have uncovered most exciting and fascinating aspects of the life style of ICEs and their interactions with the host cell, which have not been subject to any authoritative reviewing.
The aim of this review is thus to highlight the ‘hidden life’ of ICEs, the way that they manipulate individual host cells at low to very low frequencies in order to promote their horizontal transmission. We build a framework to describe and understand their biological properties as well as ecological significance, rather than focusing on genomic comparisons or gene content. For clarity of understanding their life style, we will rehearse the basic general features of ICEs and their mobility. We will introduce the most well-studied model ICEs and their regulatory control systems, before turning to single cell studies and developing a framework to understand ICEs from an ecological perspective. From this emerges a picture of ICEs as elements that have adapted to a mostly confined life style within their host, but with a very effective and dynamic transfer from a subpopulation of dedicated cells that appear as a result of bistable regulatory decisions.
WHAT ARE ICEs, AND HOW DO WE INVESTIGATE THEM?
What are ICEs?
As their name implies, ICEs exhibit two different states: an integrative state, in which their DNA resides in the chromosome of the host, and a conjugative state, in which their DNA has excised from the chromosome of the host and can potentially conjugate to a new cell (Fig. 1A). Integration is the outcome of site-specific recombination between two direct repeats that are part of the attachment sites (att): attB in the host chromosome and attI (or attP) on the circular ICE. The integration reaction is catalyzed by the integrase and, as a result of the site-specific recombination, will lead to direct repeats (typically between 10 and 60 bp) forming on either end of the integrated element, that are now named attL (left end) and attR (right end). Frequent target sites are 3΄ end of tRNAs genes, but not exclusively (Burrus and Waldor 2003; Brochet et al.2009), and as more recent systematic work on ICEs in Streptococcus has shown (Ambroset et al.2015). The reverse reaction (excision) is again a site-specific recombination, now between the conserved repeats within attR and attL, leading to its liberation as a closed-circular DNA and assumed repair of the chromosomal attachment site (Fig. 1A). The integrase is essential for both the integration and excision reactions, but auxiliary proteins are frequently needed for optimal catalysis of one or the other direction (McLeod, Burrus and Waldor 2006). Such auxiliary proteins (like the excisionase or recombination directionality factor, or integration host factor) have specific binding sites near the actual recombined sequence (i.e. within the att region). Certain elements rely on a DDE transposase rather than an integrase to recombine with the chromosome, and their transfer requires replicative excision (e.g. TnGBS in streptococci (Guerillot et al.2013), or ICEA in Mycoplasma agalacticae (Dordet Frisoni et al.2013)).
The excised ICE DNA molecule is thought to be the major intermediate step for subsequent conjugative transfer. Based on analogies to plasmid conjugation, a single-strand nick is introduced in the ICE-circular DNA at the oriT sequence by a DNA relaxase (Fig. 1A). This leads to unwinding of a single-stranded DNA for transfer and simultaneous rolling circle-type replication of the remaining single-stranded DNA (Llosa et al.2002). The single-strand ICE-DNA for transfer is then guided by the coupling protein to a type IV conjugative (or a functionally similar) protein complex, during and after which it may be coated by single-stranded DNA-binding (Ssb) protein (Lee, Babic and Grossman 2010) (Fig. 1A). Many ICEs encode their own Ssb making it likely that this is used for such purpose during transfer (Beaber, Hochhut and Waldor 2002). Transferred single-strand ICE-DNA is supposed to enter a new recipient cell, whereupon it is replicated to form a double-stranded DNA, perhaps from a single-stranded DNA origin (sso) of replication (Wright, Johnson and Grossman 2015; Wright and Grossman 2016). The double-stranded ICE-DNA is finally again integrated into the new host chromosome through site-specific recombination by the integrase (Fig. 1A). At low frequencies, Hfr-like transfer of additional chromosomal regions can take place, probably as a result of incompletely excised ICE-DNA (Hochhut, Marrero and Waldor 2000; Daccord, Ceccarelli and Burrus 2010). Despite differing in details, all ICEs follow the same principle of alternating states of excision, transfer and integration. The regulatory systems that control ICE maintenance within and transfer from the host cell can also vary widely among different ICE types, as discussed further below.
How to find ICEs?
Although the term ICE refers to elements acting mechanistically similar, several types or families have been recognized, which do not share recent evolutionary ancestry (Guglielmini, de la Cruz and Rocha 2013). Many ICEs (like plasmids or phages) resemble mosaic elements and their classification remains dependent on the marker(s) that is (are) emphasized. One particular and thorough classification came up with eight (mosaic) groups covering conjugative systems of both plasmids and ICEs. This study used three classifiers on 1124 prokaryotic genomes and some 800 conjugative plasmids: (i) the relaxase (e.g. TraI), (ii) type IV secretion system proteins (e.g. VirB4) and (iii) the type IV coupling protein (e.g. TraG) (Guglielmini, de la Cruz and Rocha 2013; Guglielmini et al.2014). Other categorizations have based on the integrase gene and insertion site (van der Meer and Sentchilo 2003; Farrugia et al.2015). ICEs can be inferred as special cases of GIs, which are predicted from calculations of genome nucleotide statistics and database comparisons to frequently occurring elements or mobile genes, as well as from multi-genome comparisons of closely related strains (Langille, Hsiao and Brinkman 2010; Dhillon et al.2015). Suspected GI/ICE regions can be further examined manually to infer possible repeat boundaries, integration sites, integrase genes or presence of known conjugative genes as hallmarks for being a putative ICE (Ambroset et al.2015). Suspect ICE candidates in cultured strains may then be examined for genes conferring potential phenotypic markers that could be used to select their transfer to suitable recipient bacteria, which ideally do not contain any related ICE. Amplification of the junction boundary formed by ICE excision in the polymerase chain reaction (PCR) is frequently used as evidence to propose ICE functionality (Farrugia et al.2015; Rydzewski et al.2015; Deutsch, Utter and Fischetti 2016).
Relatively few good ICE models exist, which is mostly due to the difficulty to distinguish ICEs phenotypically, unless they provide very clear selectable markers (e.g. antibiotic resistance or metabolism of a specific carbon substrate). Second, ICE transfer from wild-type strains typically occurs at a low rate, ranging from 10–2 to 10–7 transconjugants per donor cell under laboratory conditions. This makes it often difficult to study the effects of gene mutations, complementations or other genetic manipulations on the ICE or in the host. Third, although genetic tools are available for most well-studied bacterial species, the lack of such tools for many environmental isolates makes it complicated to tackle the life style of their ICEs. In some cases, changing the host or creating regulatory mutations has helped to increase their transfer frequencies, enabling regular genetic, molecular and biochemical studies to decipher ICE biology.
EXPERIMENTAL MODEL ICEs: MECHANISMS AND REGULATION
ICEs and ICE models
Despite the large number of potential ICEs found by genome sequencing, actually only a little more than a handful of experimental model systems have been well characterized (Fig. 2). Many more ICEs have been discovered in very different species, which we will not specifically review in detail here because less is known on details of their life style. These include elements such as high-pathogenicity island in Yersinia pestis (Buchrieser, Prentice and Carniel 1998; Schubert, Rakin and Heesemann 2004), PAPI1 and pKLC in Pseudomonas aeruginosa (Mathee et al.2008; Kung, Ozer and Hauser 2010; Klockgether et al.2011; Mikkelsen et al.2013), dusA-integrated ICEs (Farrugia et al.2015) or large transferable pathogenicity islands in Streptomyces (Kers et al.2005). For an online search of existing GIs and ICEs, one can consult, for example, the ICEBerg database—even though this database has not been regularly updated (Bi et al.2012). In the following paragraphs, we will review the current state of knowledge on the regulation of ICE activity in the various model systems (Fig. 3). This is mostly done from a typical (population-level) genetic and biochemical perspective. But, as we will argue later, the architecture of the different ICE regulatory networks is such that it likely leads to bistable differentiation among individual cells, with only few cells in a population starting to display and follow the horizontal transmission pathway (hence, the ‘hidden life’ of ICE).
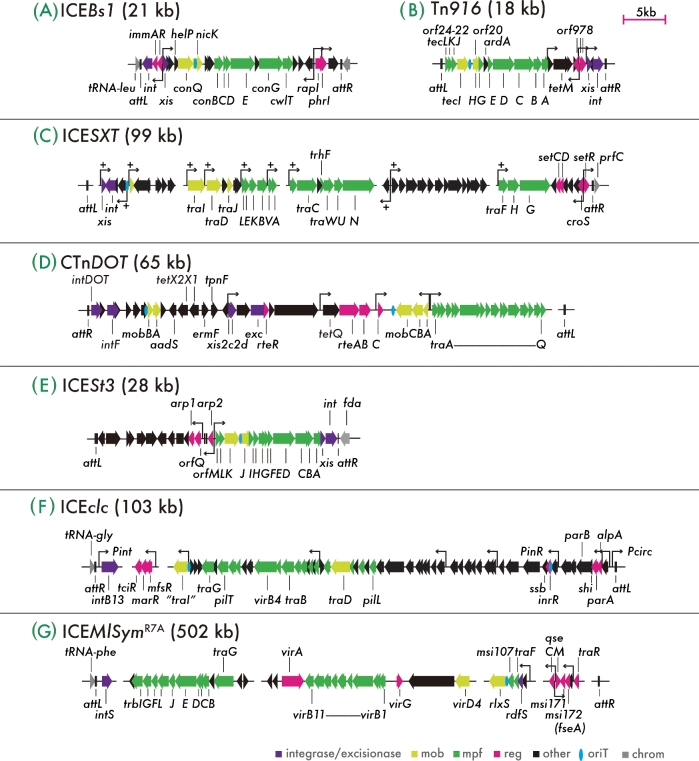
Figure 2.
Genetic organization of ICE models with relevant gene names. (A–G) ICEBs1 of B. subtilis, Tn916 of E. faecalis DS16, ICESXT of V. cholerae, CTnDOT of B. thetaiotaomicron, ICESt3 of S. thermophilus, ICEclc of P. knackmussii B13 and ICEMlSymR7A of M. loti R7A. Coding sequences of the ‘core’ ICE genes (i.e. important for its life style) are represented as thick arrows filled with different colors depending on (deduced or experimentally demonstrated) functions according to the color-scale below (mob, DNA mobilization; mpf, mating pair formation complex; reg, regulation; oriT, origin of transfer [blue ellipses]; chrom, chromosomal genes). Variable gene regions are omitted for clarity; their positions are indicated by non-connecting horizontal lines. Crucial promoters experimentally characterized are shown with bent arrows or with names (e.g. Pint in ICEclc). Plus signs on ICESXT indicate SetCD-regulated promoters. ICE ends, attR and attL, are indicated as vertical black lines. All ICEs are depicted to the same scale, and their lengths are shown within brackets. Note the double gene assignments in the Tn916 system (e.g. orf24-22 is tecLKJ, orf20 is tecH).
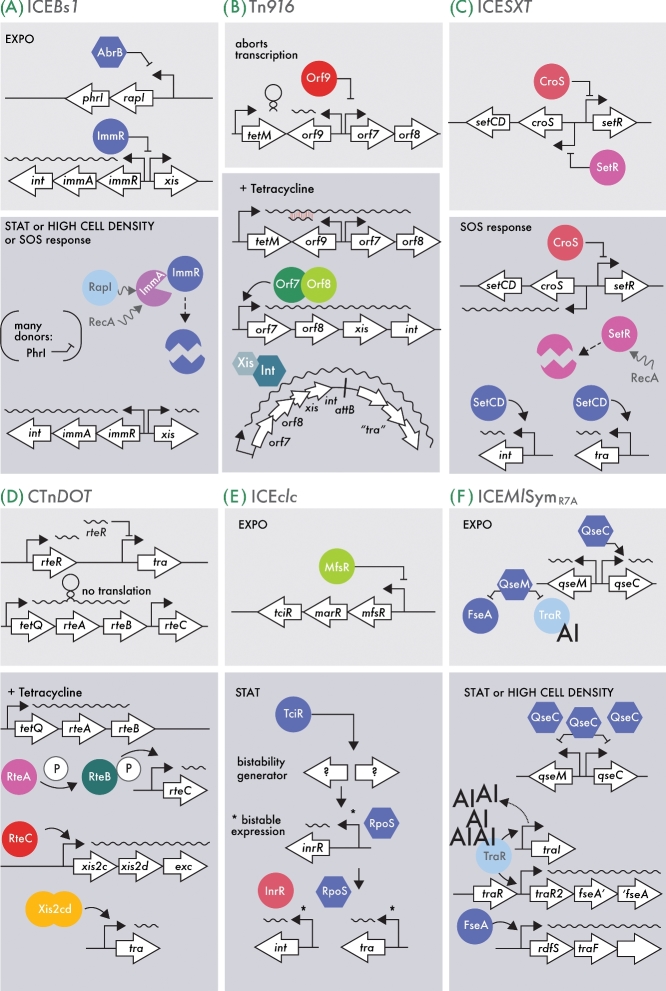
Figure 3.
Schematic outlines of the main regulatory networks controlling ICE transmission in the ICEBs1 (A), Tn916 (B), ICESXT (C), CTnDOT (D), ICEclc (E) and ICEMlSymR7A (F) models. Depicted are the integrated, silent (upper panels) and the activated state, leading to horizontal transmission (lower panels, dark background). Hooked arrows indicate promoters, other black arrows point to activation and blocked lines to repression; waved arrows (A) point to protein interactions. Open arrows indicate relevant genes; waved lines represent mRNAs, colored circles or hexagones point to key proteins in the network. Protein degradation is symbolized by broken circles, whereas protein phosphorylation is indicated by an encircled P. EXPO, exponential phase; STAT, stationary phase; AI, N-homoserinelactone autoinducer. (A) In short, the ICEBs1 balance is controlled by ImmR, which prevents transcription of the xis excisionase gene. When ImmR is degraded or overruled, xis is transcribed, leading to ICEBs1 excision. (B) Orf9 is the major repressor blocking Tn916 transmission. In the presence of tetracycline, an antisense mRNA prevents orf9 mRNA translation (red hybrid). In the absence of Orf9, the cascade of orf7/8 transcription can start, leading to xis, int and tra gene expression. The ICESXT element is controlled by a double-negative feedback loop implicating CroS and SetR (C). Under SOS response, SetR is preferentially degraded, liberating transcription of setCD for the master regulators, which then further activate the ICE excision and transfer system. (D) A small RNA named rteR and an RNA stem-loop structure are the major inhibitors for CTnDOT transmission. The presence of tetracycline liberates the inhibitory stem-loop, leading to transcription of rteA and rteB that elicit the activation cascade. (E) ICEclc is only activated in stationary-phase cells. Activation is dependent on the TciR transcription regulator, which stimulates an as yet unknown bistability generator, whose activation is transmitted to the downstream tra and int genes. (F) Two proteins QseC and QseM control ICEMlSymR7Atransmission. QseM inhibits downstream transfer factors, but is itself transcriptionally repressed by high intracellular amounts of QseC, upon which TraR, AI and FseA can elicit the transfer. For more details and references, see the main text.
ICEBs1
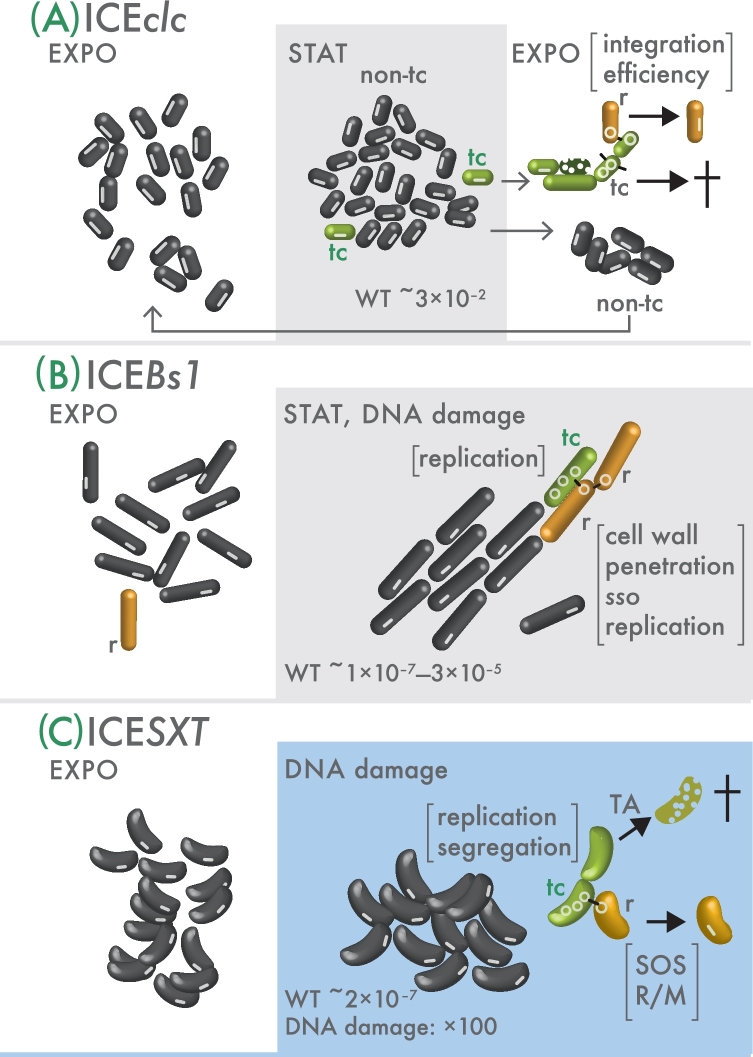
ICEBs1 is a 21-kb long ICE, thought to contain 25 protein-coding genes contained in three functional modules, but without clear phenotype on its host (Fig. 2A). ICEBs1 was originally found in Bacillus subtilis, although it can transfer to B. anthracis, B. licheniformis and Listeria monocytogenes in laboratory mating assays (Auchtung et al.2005). Excision of ICEBs1 occurs by site-specific recombination within two 60-bp direct repeats contained within attL and attR on either end of the integrated form of ICEBs1 (Auchtung et al.2005). The 60-bp region contains a 17-bp stem-loop sequence, which has two 5-bp inverted repeat sequences that may be important for the recombination reaction (Menard and Grossman 2013). Excision requires two genes on ICEBs1, int (ydcL), encoding a tyrosine recombinase similar to lambda phage integrase, and xis (sacV), encoding an excisionase (Lee et al.2007) (Fig. 2A). On the other hand, site-specific integration of the circular ICEBs1 form into the attB chromosomal attachment site requires only Int. The primary attB site is located within the gene trnS-leu2 for tRNALeu on the B. subtilis chromosome, but ICEBs1 can recombine at lower frequencies with secondary attachment sites composed of 17-bp sequences similar to attB (Menard and Grossman 2013).
The excised and circularized ICEBs1 molecule is processed for transfer by NicK, a relaxase encoded in the xis operon (Fig. 2A). NicK alone is able to catalyze a single-strand nick at the origin of transfer (oriT), which partially overlaps with the 5΄ end of the nicK gene (Lee and Grossman 2007). The host-encoded helicase PcrA and the ICEBs1-encoded helicase processivity factor HelP each assemble at the nicked oriT and cooperatively unwind the double-stranded circular DNA in a single direction (Lee, Babic and Grossman 2010; Thomas, Lee and Grossman 2013). A chromosomally encoded single-stranded DNA-binding protein (Ssb) stabilizes the unwound DNA, forming a nucleoprotein complex together with NicK on the nicked strand (Lee, Babic and Grossman 2010). At this stage, two concurring events take place: (i) a rolling circle-like replication restoring the double-stranded DNA molecule and increasing its copy number, and presumably (ii), the processing of the nicked strand toward the conjugation apparatus (Berkmen et al.2010; Lee, Babic and Grossman 2010). Interestingly, nicked oriT thus not only serves as an origin of transfer but also as an origin of replication for a rolling circle-like replication. Autonomous replication of ICEBs1 is dependent on the host cell machinery proteins PolC, DnaN, Ssb and PcrA, but also on the ICEBs1-encoded factors NicK and HelP (Lee, Babic and Grossman 2010; Thomas, Lee and Grossman 2013). ICEBs1 represented the first model for which plasmid-like autonomous replication could be demonstrated, which was unexpected since replication of the integrated form along with the chromosome was assumed to be the only mechanism for ICE maintenance (Burrus and Waldor 2004b). Autonomous replication is not essential for ICEBs1 transfer, but guarantees its stability in the donor, especially if cell division resumes before reintegration can occur (Lee, Babic and Grossman 2010; Auchtung et al.2016).
By analogy to plasmid transfer, it is believed that the nicked single-stranded nucleoprotein complex is directed toward the conjugative machinery, which translocates it into the recipient cell. Bioinformatic analysis predicts that the ICEBs1-encoded ConQ may act as the coupling protein (Lee and Grossman 2007), whereas ConB, ConC, ConD, ConE and ConG may be part of the translocation channel (DeWitt and Grossman 2014; Leonetti et al.2015) (Fig. 2A). CwlT may serve as cell wall hydrolase necessary for building an appropriate transfer pore (Lee and Grossman 2007; DeWitt and Grossman 2014). ConE and ConG are two ICEBs1 proteins essential for transfer and resemble the known mating pair formation proteins (Mpf) VirB4 and VirB6, respectively. ConE localizes mostly at the cell poles in dependence on the presence of ConB, where ICEBs1 replication foci are also observed (Berkmen et al.2010). Thus, it is believed that both ICEBs1 conjugation and replication occur predominantly at the cell poles, although transfer can occur from non-polar zones of the cell, too (Babic et al.2011). Polar localization of the conjugation machinery is thought to increase the efficiency at which the ICEBs1 transfers within chains of interconnected bacteria, a phenomenon predominant in biofilms (Babic et al.2011). Once in the recipient cell, the ICEBs1 single-stranded circular form is supposedly replicated. Since int expression is constitutive, Int directs (alone) the integration of the circular form into the extremity of a tRNALeu site (Lee and Grossman 2007). The ICEBs1 functional mating apparatus and the ConQ coupling protein may assist in mobilization of other residing plasmids with their own rolling-circle origin of replication serving as an origin of transfer, and their replicative relaxase interacting with ConQ (Lee, Thomas and Grossman 2012).
Regulation of ICEBs1 transmission relies mainly on the transcriptional repressor ImmR and its cognate protease ImmA. ImmR represses the activity of the Pxis-promoter, which drives transcription of xis together with genes coding for DNA processing and mating pair formation (Auchtung et al.2007). ImmR also autoregulates its own expression and that of immA and int, which are in the same operon on ICEBs1 (Fig. 2A), resulting in an apparent constitutive expression (Auchtung et al.2005, 2007) (Fig. 3A). Repression imposed by ImmR can be alleviated when ImmA is activated and proteolyses ImmR, starting the ICEBs1 excision and transfer process (Bose et al.2008). The interplay between the ImmR/ImmA regulatory system and host- and ICE-encoded factors determines the onset of transfer. The balance is influenced by SOS response, nutrient availability, cell density and the presence of non-ICEBs1-bearing cells, relayed to the ImmR/ImmA switch by the factors PhrI, RapI and AbrB (Auchtung et al.2005; Carraro and Burrus 2014) (Fig. 3A). PhrI and RapI act together as a quorum-sensing system monitoring the bacterial population bearing ICEBs1. PhrI is produced as a non-mature signal peptide, which is processed and secreted as an active pentapeptide outside the cell. The active form of PhrI acts as a density-dependent signal. When the number of ICEBs1-bearing cells is high (for example, in stationary phase), PhrI accumulates and is re-imported by the chromosomally encoded oligopeptide permease Opp (Auchtung et al.2005). RapI is an enhancer of ImmA proteolytic activity, but high PhrI levels inhibit RapI activity and ImmR maintains its repression (Bose et al.2008). For this reason, transfer rates are low at a high density of cells already containing ICEBs1. During exponential phase, the chromosomally encoded factor AbrB acts as a repressor for rapI transcription, while PhrI levels are too low to inhibit RapI directly, thereby preventing ICEBs1 transfer during active cell division (Fig. 3A).
In contrast, when nutrients are scarce and cell density is high, but the proportion of ICEBs1 carrying cells is low, both PhrI and AbrB fail to repress RapI activity (Fig. 3A) (Auchtung et al.2005). RapI then enhances ImmA-dependent proteolytic cleavage of ImmR, which causes derepression of the xis excisionase and transfer genes (Auchtung et al.2007; Bose et al.2008; Bose and Grossman 2011). Overexpression of RapI leads to a strong increase in the number of cells in a population excising ICEBs1 (Auchtung et al.2005). The onset of Xis production is the start for the Int-catalyzed excision reaction to liberate ICEBs1 from its chromosomal site, upon which transfer can take place (Lee et al.2007). In the transconjugant, ImmR confers immunity against secondary acquisition of ICEBs1 to the host cell by repressing expression of int on the newly acquired element (Auchtung et al.2007).
Independently from the PhrI-RapI cascade, the SOS response can also induce excision of ICEBs1 (Auchtung et al.2005) (Fig. 3A). DNA damage such as induced by mitomycin C triggers RecA to enforce (in an as yet unknown mechanism) ImmA cleavage of ImmR, liberating xis and transfer gene expression. It was postulated that SOS-mediated control of ICEBs1 evolved in order to allow ICEBs1 to abandon damaged host cells (Auchtung et al.2005). Further global factors can impede ICEBs1 excision and transfer. For example, the general negative regulator of competence development Rok represses excision of ICEBs1 (Smits and Grossman 2010), whereas ClpP protease activity controls the amounts of ImmA (Bose and Grossman 2011).
Tn916
Tn916 is an 18-kb long ICE found in Enterococcus faecalis DS16 at multiple AT-rich sites on the chromosome (Franke and Clewell 1981) (Fig. 2B). Tn916 belongs to a wide family of ICEs, confers tetracycline resistance to its host (by the tetM gene) and can transfer to both Gram-positive and Gram-negative bacteria (Poyart, Celli and Trieu-Cuot 1995; Roberts and Mullany 2009). Excision of Tn916 is mediated by the IntTn916 tyrosine recombinase (Scott, Kirchman and Caparon 1988; Storrs et al.1991; Bringel, Van Alstine and Scott 1992), which further requires the Xis excisionase (Rudy et al.1997, Marra and Scott 1999; Hinerfeld and Churchward 2001b). Both proteins bind DNA at or close to the attR and attL sites (Rudy, Scott and Churchward 1997, Jia and Churchward 1999; Connolly, Iwahara and Clubb 2002). Interestingly, Xis-attL interaction promotes excision, contrary to Xis-attR binding that inhibits it (Hinerfeld and Churchward 2001b). IntTn916 does not require homology between the coupling sequences within attR and attL, and thus creates heteroduplexes (Caparon and Scott 1989; Jaworski and Clewell 1994; Lu and Churchward 1994; Lu and Churchward 1995; Taylor and Churchward 1997). Since the recombination event is not site specific but solely requires AT-rich regions, Tn916 has been used for random insertion mutagenesis in other Gram-positive hosts (Smidt et al.1999). For the integration reaction, IntTn916 catalyzes recombination in a homology-independent manner between attB and attP (Caparon and Scott 1989; Storrs et al.1991; Rudy and Scott 1994; Scott et al.1994). Renewed excision is prevented by Xis binding to attR and avoiding IntTn916 to associate efficiently (Hinerfeld and Churchward 2001b).
Tn916 excision is triggered by increased expression of xis and int, whose transcription is mostly silent in the integrated state (Su, He and Clewell 1992; Celli, Poyart and Trieu-Cuot 1997; Celli and Trieu-Cuot 1998). Enhanced xis and int transcription results from readthrough from the Porf7 promoter, which is located upstream of the orf7 and orf8 genes in front of xis and int (Fig. 3B). Transcription of Porf7-orf7-orf8 is repressed by the orf9 gene product, which is alleviated in the presence of tetracycline (Celli and Trieu-Cuot 1998). The regulatory cascade implicating tetracycline is complex and involves the formation of an antisense orf9 mRNA, presumably inhibiting efficient translation of the sense orf9 mRNA. The cascade starts at the tetracycline resistance gene tetM, which is located upstream and in the opposite orientation as orf9 (Fig. 3B). In absence of tetracycline, tetM transcription is prematurely terminated because of attenuation at the leader sequence (Su, He and Clewell 1992). In the presence of tetracycline, the ribosomes proceed past the mRNA leader sequence, preventing the exposure to the terminator and allowing RNA polymerase to continue transcription through tetM and the antisense orf9 strand. The antisense orf9 RNA inhibits translation of the regular orf9 mRNA, thus alleviating the repression of Orf9 on Porf7 (Fig. 3B). The produced Orf7 and Orf8 proteins activate their own transcription from Porf7, leading to continued transcription through the downstream-located xis and int genes, triggering Tn916 excision (Fig. 3B) (Su, He and Clewell 1992; Celli, Poyart and Trieu-Cuot 1997; Celli and Trieu-Cuot 1998).
Excision of Tn916-DNA leads to the physical association of the genes for conjugation (tecLKJIHGFEDCBA) downstream of the xis-int operon (Figs 2B and 3B) (Senghas et al.1988; Scott et al.1994). In the integrated form, the tec genes are promoterless and thus very poorly transcribed. However, the downstream association of the tec genes in the excised form allows extension of transcription from Pxis and Porf7 all the way through orf7-orf8, xis-int plus the tec operon (Celli and Trieu-Cuot 1998). The relaxase TecH (Orf20) catalyzes single-stranded cleavage at the oriT, which is located in the intergenic region upstream of tecH (Jaworski and Clewell 1995; Hinerfeld and Churchward 2001a, Rocco and Churchward 2006; Wright and Grossman 2016). The nicked single-stranded Tn916 is translocated into the recipient cell, where it is (again) assumed to reconstitute as a double-stranded circular molecule prior to its integration in the host chromosome (Scott et al.1994). The conjugation systems of Tn916 and ICEBs1 are distantly related (Burrus et al.2002b, Rocco and Churchward 2006; Wright and Grossman 2016). Similar to ICEBs1, replication of excised Tn916 relies on a rolling-circle mechanism involving oriT as replication origin, TecH (Orf20) and the helicase processivity factors Orf22 (TecK) and Orf23 (TecJ) as replisome, and a single-stranded origin of replication (sso) necessary for synthesis of the complementary strand (Wright and Grossman 2016). Successful transfer is further dependent on the Tn916-encoded protein ArdA, which sequesters the recipient's type I restriction/modification enzymes and helps avoiding digestion of the incoming restored double-stranded Tn916 DNA (Serfiotis-Mitsa et al.2008).
ICESXT-R391
ICESXT (commonly named SXT) is a 99.5-kb long genetic element first discovered in Vibrio cholerae O139 (Waldor, Tschäpe and Mekalanos 1996) (Fig. 2C). ICESXT confers resistance to chloramphenicol, streptomycin and sulfamethoxazole/trimethoprim (SXTR, hence the name). R391 is a closely related ICE with a size of 89 kb, originally found in Providencia rettgeri and conferring resistance against mercury and kanamycin (Burrus, Marrero and Waldor 2006). In its integrated form, ICESXT (and R391) resides in the 5΄ extremity of the gene coding for the peptide chain release factor 3 (prfC). However, ICESXT can integrate at lower frequencies at secondary positions when prfC is deleted (Hochhut and Waldor 1999; Burrus and Waldor 2003). ICESXT and R391 are the prototypical members of a large family of ICEs that share a common 47-kb long backbone (with more than 95% nucleotide identity) including 52 genes (Wozniak et al.2009) (Fig. 2C). The conserved backbone is disrupted by DNA regions that vary between members of the ICESXT/R391 family and code for auxiliary and unknown functions. The ICESXT/R391 family currently encompasses some 50 plus members among a wide spectrum of Gammaproteobacteria (Burrus, Marrero and Waldor 2006).
Similar as for lambdoid prophages and for ICEBs1, initiation of ICESXT excision and transfer is induced by the SOS response, triggered in V. cholerae by DNA damage through exposure to mitomycin C or ciprofloxacin (Beaber, Hochhut and Waldor 2004) (Fig. 3C). Induction of the SOS response through antibiotic exposure is particularly worrisome, and can lead to higher transfer rates and wider distribution of ICESXT/R391 elements in clinical strains of V. cholerae (Beaber, Hochhut and Waldor 2004). In the current hypothesis, two counteracting repressors (SetR and CroS) control the balance of activation of two downstream key regulatory genes of ICESXT transmission, setCD (Fig. 3C) (Beaber and Waldor 2004; Beaber, Hochhut and Waldor 2004; Poulin-Laprade and Burrus 2015). The action of the SOS response is a RecA-and LexA-dependent proteolysis of the ICESXT-encoded CI-like repressor SetR (Fig. 3C). This liberates the croS-setCD operon from SetR transcriptional repression (Beaber and Waldor 2004; Beaber, Hochhut and Waldor 2004; Poulin-Laprade and Burrus 2015). The resulting SetC and SetD combine to an FlhCD-like activator complex, which activates transcription of numerous genes including the integrase gene int, the tra genes for conjugative transfer and the excisionase gene xis (Figs 2C and 3C) (Beaber et al.2002, Burrus and Waldor 2003; O’Halloran, McGrath and Pembroke 2007; Poulin-Laprade, Carraro and Burrus 2015, Poulin-Laprade et al.2015). Int and Xis subsequently catalyze ICESXT excision, yielding a closed circular double-stranded form (Burrus and Waldor 2003).
Transfer of ICESXT is thought to proceed when the MOBH1-type relaxase TraI nicks a single strand at the oriT sequence, which is located some 40 kb upstream of traI (Beaber, Hochhut and Waldor 2002, Ceccarelli et al.2008) (Fig. 2C). Recognition of oriT is dependent on the ICESXT-encoded protein MobI, which lays encoded in the vicinity of oriT (Ceccarelli et al.2008; Daccord, Ceccarelli and Burrus 2010). The nicked oriT then serves as both the starting point for DNA processing and conjugation as well as for TraI-dependent reconstructive replication (i.e. the formation of double- from single-stranded DNA). Reconstructive replication may lead to an increase of ICESXT copy numbers in the cell after excision, which is crucial for maintaining its stability in the host (Carraro, Poulin and Burrus 2015). Excised (and multiplied) ICESXT/R391 molecules segregate among dividing daughter cells with help of a type II partitioning system, which is encoded on the ICE itself and is expressed simultaneously with its excision. Mutant data also suggested that the equilibrium of ICESXT excision and integration in a cell is dependent on the number of accumulating ICESXT copies. For example, in the absence of the mating pair protein TraG, excised ICESXT accumulates in the cell because of its inability to be conjugated (Carraro, Poulin and Burrus 2015).
Transfer of the single-stranded ICESXT is mediated by a type IV secretion system, encoded by four operons named traLEKB, traVA, s054/traC/trhF/traWUN and traFHG (Wozniak et al.2009; Poulin-Laprade, Carraro and Burrus 2015, Poulin-Laprade et al.2015) (Fig. 2C). These genes display relatively high homology and identical synteny to those of IncA/C conjugative plasmids (Wozniak et al.2009; Poulin-Laprade, Carraro and Burrus 2015). Further genes required for effective ICESXT transfer include s063 (Wozniak et al.2009), traJ and traD, likely acting as coupling proteins (Beaber, Hochhut and Waldor 2002, Poulin-Laprade et al.2015). Entry of ICESXT and R391 into a new host is subject to an exclusion system driven by the inner membrane proteins TraG and Eex, acting as donor and recipient exclusion factors, respectively (Marrero and Waldor 2005, 2007a). ICEs of the ICESXT/R391 family segregate into two exclusion groups: S (for ICESXT-like) and R (for R391-like). Exclusion largely but not completely prevents a transferring ICE of a given group (S or R) to settle into a recipient cell that already contains an ICE of the same group (Marrero and Waldor 2005, 2007a). After the entrance of ICESXT in the recipient cell, its single strand is replicated to a double-stranded DNA, which recombines into the prfC gene through Int activity (Burrus and Waldor 2003). Tandem arrays of ICESXT-R391 family members may originate at the same integration site (Hochhut et al.2001; Burrus and Waldor 2004a), which are unstable and lead to deletions through recombination (Burrus and Waldor 2004a). Novel hybrid ICEs may also arise from tandem ICESXT/R391 arrays through RecA-mediated recombination (Burrus and Waldor 2004a, Garriss, Waldor and Burrus 2009), or by the Bet and Exo lambda-Red-like homologous recombination system, encoded on ICESXT and R391 itself (Burrus and Waldor 2004a, Garriss, Waldor and Burrus 2009; Garriss, Poulin-Laprade and Burrus 2013). Similar as for ICEBs1, also ICESXT/R391 can mobilize other resident compatible genetic elements such as plasmids, GIs or even chromosomal regions, casting a new light on the impact that ICEs can have on HGT in general (Hochhut, Marrero and Waldor 2000; Osorio et al.2008; Daccord, Ceccarelli and Burrus 2010; Daccord et al.2012a, b; Poulin-Laprade et al.2015).
CTnDOT
CTnDOT is a 65-kb long ICE integrated at sequence-specific sites in the genome of Bacteroides thetaiotaomicron (Shoemaker, Barber and Salyers 1989) (Fig. 2D). CTnDOT belongs to a wider family of CTn-like ICEs, which are widely present in Bacteroides species (Shoemaker et al.2001; Bartha et al.2011). CTnDOT enables its host to resist to tetracycline and erythromycin (Shoemaker, Barber and Salyers 1989). Like many other mobile genetic elements, CTnDOT can mobilize other transposons in natural conditions via its mob and tra genes (Shoemaker et al.1993; Shoemaker, Wang and Salyers 1996a,b). As for ICESXT/R391-elements, CTnDOT-like hybrids can occur, such as CTn12256 (Wang et al.2011).
Two different protein complexes are responsible for the integration and excision reactions of CTnDOT, with IntDOT being the core site-specific recombinase. IntDOT interacts and assembles at the attL and attR sites with a variety of other factors into the excisive intasome complex (DiChiara, Salyers and Gardner 2005; Dichiara, Mattis and Gardner 2007; Keeton et al.2013a). The excisive intasome complex is composed of IntDOT, two additional proteins Xis2c and Xis2d, a protein named Exc and the chromosome-encoded factor BHFa (Cheng et al.2000, 2001; Keeton and Gardner 2012; Keeton et al.2013a, Ringwald and Gardner 2015). Xis2c and Xis2d are essential for the excision of CTnDOT and bind to attL and attR (Hopp, Gardner and Salyers 2015). Exc is a topoisomerase III essential to excision (Sutanto et al.2002, 2004), possibly by promoting the stability of the intasome at attR/L sites (Keeton and Gardner 2012). IntDOT itself catalyzes the asymmetric recombination between attR and attL, leading to the excised CTnDOT molecule, with the unusual formation of a heteroduplex region (Sutanto et al.2004; DiChiara, Salyers and Gardner 2005; Keeton and Gardner 2012; Keeton et al.2013a).
To promote site-specific insertion, it is thought that IntDOT assembles at attDOT along with the host factor BHFa, to form the integrative intasome nucleoprotein complex (Ringwald and Gardner 2015). This integrative intasome differs from it excisive counterpart by the absence of Xis2c, Xis2d and Exc. Recombination occurs with the attB site, which is composed of two essential core sites, termed B and B', whereas attDOT on CTnDOT contains the D and D' core sites (Laprise, Yoneji and Gardner 2013). Both B and D cores contain the GTANNTTT sequence, which is recognized by IntDOT. The complex interacts with attB and recombines both attDOT-attB sites, regardless of the heterology of the coupling pairs (Cheng et al.2002; Malanowska, Salyers and Gardner 2006; Malanowska et al.2007, 2009; Wood et al.2010).
Excised and circularized CTnDOT is mobilized and translocated to a recipient cell by the products of CTnDOT-encoded mob and tra genes, respectively (Bonheyo et al.2001a, b, Peed, Parker and Smith 2010) (Fig. 2D). In analogy to other conjugative systems, it is assumed that Mob and Tra proteins nick the CTnDOT circular form at its oriT, process and export a single-stranded DNA into a recipient cell. Besides their essential role in excision, Xis2c and Xis2d additionally promote the transcription of the tra genes via an unknown mechanism, which involves their binding to the Ptra promoter (Whittle, Shoemaker and Salyers 2002; Keeton et al.2013b). Reconstructive replication is thought to occur in both the donor and the recipient, and the double-stranded DNA molecule can subsequently reintegrate into the chromosomal attB site (Cheng et al.2000).
Excision and transfer of CTnDOT is under control of the regulators RteA, RteB and RteC (Stevens et al.1993) and rteR, a small RNA repressing tra gene expression (Jeters et al.2009; Waters and Salyers 2012) (Fig. 3D). RteA and RteB are part of an operon encompassing the tetQ gene, encoding a tetracycline resistance factor (TetQ) (Stevens et al.1993). Transcription of tetQ-rteA-rteB yields an mRNA with a leader sequence of three amino acids in between the PQ promoter and the tetQ start codon (Wang, Shoemaker and Salyers 2004; Wang et al.2005). Translational control is exerted through two possible hairpins in the leader mRNA, a relatively stable one composed of two sequences named Hp1 and Hp8, and a shorter less stable one, constituted of Hp1 and another sequence named Hp2. In the absence of tetracycline, the Hp1-Hp8 hairpin attenuates the translation of the tetQ-rteA-rteB mRNA by occluding the ribosome binding site for tetQ, which is contained within the Hp8 loop (Wang, Shoemaker and Salyers 2004; Wang et al.2005). In the presence of tetracycline or other ribosome-targeting antibiotics, repression on CTnDOT activation is overcome (Waters and Salyers 2013), presumably because the tetracycline-affected ribosomes tend to stall during leader peptide synthesis, favoring the formation of the Hp1-Hp2 hairpin instead of the Hp1-Hp8 hairpin (Wang et al.2005). This liberates the ribosome binding site for tetQ and enables translation of tetQ, rteA and rteB. RteA phosphorylates RteB, which subsequently binds to the PC promoter, stimulating the expression of rteC (Fig. 3D) (Moon et al.2005). RteC activates the transcription of the xis2c-xis2d-exc operon, by binding its promoter PE, starting the excision process (Whittle, Shoemaker and Salyers 2002; Moon et al.2005; Park and Salyers 2011).
ICESt1/3
ICESt1 and ICESt3 are 34.7- and 28-kb ICEs integrated in the 3΄ end of the fda gene in the genome of Streptococcus thermophilus CNRZ368 and CNRZ385, respectively (Burrus et al.2000, b, Pavlovic et al.2004) (Fig. 2E). ICESt3 transfer has been observed under laboratory conditions, whereas that of ICESt1 is at the limit of detection (Bellanger et al.2009). ICESt1 encompasses a secondary attL site named attL' that can be used for site-specific recombination and yields a truncated alternative ICE, termed ICESt2 (Pavlovic et al.2004). Similarly to other ICEs, ICESt3 can mobilize other inserted elements in cis (Burrus et al.2002b, Pavlovic et al.2004; Bellanger et al.2011). Excision of ICESt1/3 requires both the integrase Int and the excisionase Xis, but Int is sufficient for integration (Bellanger et al.2009). Excision and transfer of both ICESt1 and ICESt3 is under control of the repressor Arp1, a homolog to the phage λ CI repressor, and, possibly, Arp2, homologous to ImmR of ICEBs1 (Bellanger et al.2007, 2008, 2009; Carraro et al.2011). The addition of mitomycin C leads to increased ICESt1/3 excision, which may similarly as for ICEBs1 and ICESXT implicate a RecA-dependent autocleavage of, in this case, Arp1. The details of the activation cascade are not known yet, but, possibly, degradation of both Arp1 and Arp2 is necessary, leading to alleviation of the repression of mobility functions, and subsequent ICESt1/3 excision. Excision of both elements also increases in stationary-phase cells (Bellanger et al.2007; Carraro et al.2011).
The copy number of ICESt3 dramatically increases after mitomycin C treatment, thus suggesting not only simple excision but some form of replication in cells with excised form (Carraro et al.2011). Further investigation using a mini-ICESt3 element with constitutive replication revealed that multiple copies of ICESt3 are the result of a plasmid-like rolling-circle replication that likely contributes to the stability of the element (Carraro et al.2016a). Additionally, ICESt1 encodes a novel type II GATC restriction/modification system, composed of Sth368IR and Sth368IM, which are responsible for the immunity to phage ΦST84 (Serfiotis-Mitsa et al.2008). The exact composition of the transfer system of ICESt1/3 has not been studied in detail, but bears similarity to those of Tn916 and ICEBs1 (Bellanger et al.2011).
ICEclc-ICEHin1056 elements
The 103-kb self-transmissible ICEclc element of Pseudomonas knackmussii B13 (Dorn et al.1974; Ravatn, Zehnder and van der Meer 1998) (Fig. 2F) and the 59.3-kb ICEHin1056 element of Haemophilus influenzae are members of a wider ICE family found in Beta- and Gammaproteobacteria, with little homology to the aforementioned ICE systems. ICEclc is a good experimental model due to its high rate of native self-transfer (10−2 per donor) (Miyazaki and van der Meer 2011b). It can complement some hosts to use 3-chlorobenzoate (3-CBA) and 2-aminophenol as carbon and energy sources (Dorn et al.1974). ICEclc is present in two identical copies integrated at two different sites in the chromosome of strain B13 (Miyazaki et al.2015), and is capable of self-transfer to a variety of hosts belonging to Beta- and Gammaproteobacteria (Ravatn, Zehnder and van der Meer 1998, Springael et al.2002; Gaillard et al.2008; Sentchilo et al.2009). ICEclc is integrated in the 3΄ end of genes for tRNAGly (Sentchilo et al.2009), whereas ICEHin1056 is integrated in genes for tRNALeu. ICEHin1056 confers resistance to ampicillin, tetracycline, chloramphenicol or other antibiotics to its host (Mohd-Zain et al.2004). ICEHin1056; ICEclc and other elements such as SPI-7 in Salmonella enterica subsp. enterica, or PAPI-1, pKLC102 and PAGI-3 of P. aeruginosa share a core region with low but consistent ortholog functions (as low as 25–30% amino acid identity between ortholog proteins), and have therefore been grouped into the same ICE family (Mohd-Zain et al.2004).
Excision of ICEclc occurs through site-specific recombination between two 18-bp direct repeats on either end of the integrated form (attR and attL), leading to a reconstituted attB site on the chromosome and the attP site on the closed circular ICEclc molecule (Sentchilo et al.2009). Excision (Miyazaki and van der Meer 2011b) and integration (Ravatn et al.1998b) are dependent on the IntB13 integrase, a member of the P4-family tyrosine recombinase but with unusual long length (Ravatn 1998). Excision is strongly reduced but not completely absent without attL sequence, suggesting low-frequency recombination at secondary sites (Delavat et al.2016). Recent data also suggest that excised ICEclc can temporarily replicate to form multiple copies in a donor cell, but the mechanism of this replication has not been elucidated (Delavat et al.2016). An excisionase has so far not been identified, although sequences crucial for efficient integration have been found outside the direct 18-bp recombination sites (Miyazaki and van der Meer 2013). ICEclc integrates in genes for tRNAGly, with higher frequency into those with perfect match to its own attP sequence (e.g. 4 of 6 in P. putida) (Sentchilo et al.2009). Since the attP carries an identical 18-bp sequence to the 3΄ end of the target gene, the target is restored upon integration (Ravatn et al.1998a). In the integrated state, the intB13 gene is transcribed from a weak promoter named Pint (Sentchilo, Zehnder and van der Meer 2003). However, upon recombination of both att ends, a stronger constitutive promoter named Pcirc, which in the integrated state faces outward from the ICE, is placed upstream of Pint (Fig. 5) (Sentchilo, Zehnder and van der Meer 2003). This leads to higher expression of intB13, which is thought to facilitate the reintegration of the element (Sentchilo et al.2009; Delavat et al.2016).
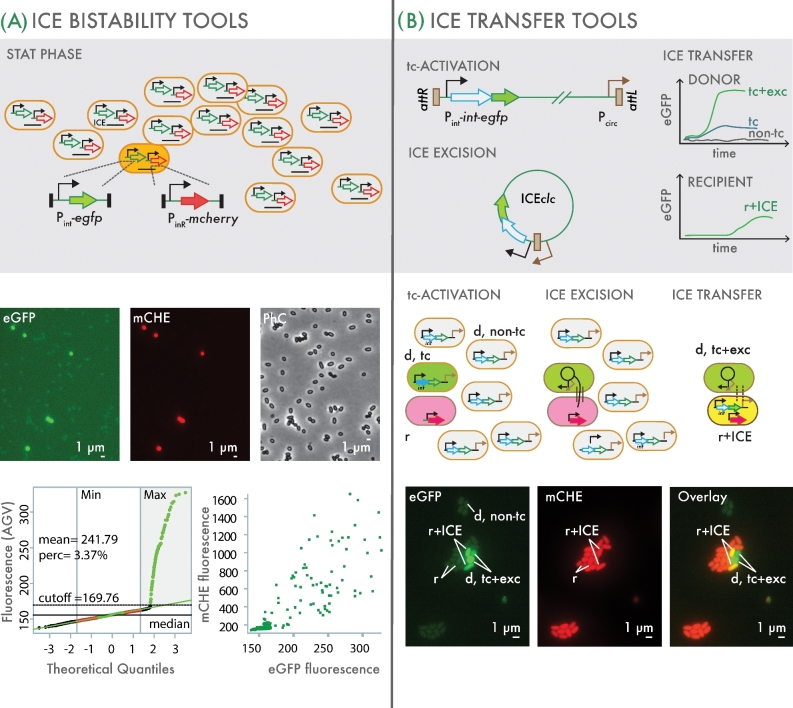
Figure 5.
Methods to detect ICE bistability and transfer. (A) Fusing ICE-core promoters (in this example Pint and PinR from ICEclc) to fluorescent reporter genes (like egfp or mcherry) allows observing ICE bistable gene expression (Minoia et al.2008; Reinhard and van der Meer 2013). The example schematically shows cells carrying ICEclc and additional single copy Pint- and PinR-reporter fusions, leading to the appearance of a subpopulation of fluorescent cells in stationary (STAT) phase. The subpopulation size can be deduced from quantile–quantile plots of observed versus expected fluorescence distribution (Reinhard and van der Meer 2013), or from scatter plots of dual fluorescent marker expression among individual cells (lower panel). (B) ICEclc transfer can be followed at single cell level by fusing the egfp gene downstream of the intB13 integrase gene (Delavat et al.2016). Donor cells (d) activating the ICEclc transfer competence (tc) program express eGFP through the Pint promoter, but become brighter fluorescent upon ICEclc excision (tc+exc) as a result of the Pcirc promoter being transcriptionally fused upstream of Pint. Silent donor cells (d, non-tc) are barely visible in background. ICEclc transfer (lower panel) can be detected from donor cells with an excised ICEclc (d, tc+exc) to recipient cells (r) expressing a different fluorescent protein (e.g. mCherry), as a result of combined colors (r+ICE). For further details, see the main text.
Still relatively little is known about the details of transfer of ICEHin1056 or ICEclc elements. Their predicted type MPFG conjugative system is evolutionary distinct from typical plasmid or ICE (e.g. ICESXT) type IV secretion systems (Juhas et al.2007; Guglielmini et al.2011; Guglielmini, de la Cruz and Rocha 2013) (Fig. 2F). Deletion mutation studies on ICEHin1056 identified a number of genes essential for efficient conjugation (Juhas et al.2007). Mutation analysis of ICEclc identified the gene for the relaxase, whose function as a nickase could be demonstrated in vivo and in vitro (Miyazaki and van der Meer 2011b, Miyazaki et al.2012). Intriguingly, two regions on ICEclc were experimentally identified that can act as an origin of transfer, raising the question as to how an excised ICEclc DNA can mechanistically wield two oriTs, which could interfere with each other's nicking and relaxation process (Fig. 2F) (Miyazaki et al.2012). The conjugative transfer machinery of an immobilized integrated ICEclc can also act in trans to promote transfer of an excisable but self-transfer-deficient ICEclc (Miyazaki and van der Meer 2011a).
ICEclc excision is stimulated when cells enter in stationary phase and have been cultured on 3-CBA as carbon substrate (Sentchilo et al.2003) (Fig. 3E). In contrast, typical inducers of SOS response such as chemical toxicity or UV do not result in measurably higher ICEclc excision rates (Sentchilo et al.2003). Excision correlates with increased expression from the Pint promoter in stationary phase, which controls intB13 transcription (Sentchilo, Zehnder and van der Meer 2003). Expression of Pint is dependent on the inrR gene, but the action exerted by InrR is unclear (Minoia et al.2008). Expression of inrR and intB13 is further dependent on the stationary-phase sigma factor RpoS (Miyazaki et al.2012). Expression of most of the core genes of ICEclc is also only apparent in stationary-phase cells (Gaillard et al.2010). More recently, a cluster of three regulator genes was found (i.e. mfsR, marR and tciR), which acts as global regulator for ICEclc activation (Fig. 2F). MfsR is a transcription repressor, which downregulates its own expression as well as the marR and tciR genes downstream in the same operon (Fig. 3E) (Pradervand et al.2014b). Although the function of MarR remains unclear, alleviation of MfsR repression causes overexpression of tciR and leads to an increase of ICEclc excision and transfer (Pradervand et al.2014b). Biochemical evidence and sequence comparisons showed that MfsR is the cognate repressor of a set of genes for a major facilitator superfamily efflux pump, also located on ICEclc but separated from mfsR by the insertion of the gene cluster coding for 2-aminophenol degradation (Pradervand et al.2014a). Further evidence was provided to subsume that the mfsR-efflux cluster was a recent innovation on ICEclc compared to related ICEs, and may have led to the increase of self-transfer frequencies from 10−7 (observed for most of the ICEs from this family) to its current 10−2 transconjugants per viable donor (Pradervand et al.2014b).
ICEMISymR7A
The 502-kb ICEMlSymR7A element is one of the largest ICEs known so far and encodes the major symbiosis factors in Mesorhizobium loti R7A (Sullivan et al.1995, 2002; Sullivan and Ronson 1998; Sullivan, Brown and Ronson 2013) (Fig. 2G). The balance between integrated and excised ICEMISymR7A is controlled by the regulators QseC and QseM (Fig. 3F). QseC is a transcriptional regulator which controls both its own and qseM expression. The qseC and qseM are adjacent but divergently oriented and control is exerted through binding of QseC to two operators (OR and OL) (Ramsay et al.2013). QseM is an allosteric inhibitor of the quorum-sensor regulator TraR, which induces the ICEMlSymR7A transfer genes. At low population density, excision is prevented because QseC preferentially binds the OL operator, which at low QseC concentrations in the cell stimulates qseC expression but does not impede qseM expression. At high population density and stationary phase, QseC accumulates in cells, causing QseC to bind simultaneously to both operators OL and OR. QseC binding at both operators represses further transcription of the qseC and qseM genes. Lower concentrations of inhibiting QseM lead to on average more active TraR, which can be co-activated by the N-acylhomoserine lactone produced by TraI (Fig. 3F). Coactivation of TraR stimulates both traI expression and that of the msi172 and msi171 genes in the same operon (Ramsay et al.2006, 2009, 2013). A small proportion of cotranscribed msi172-171 mRNAs is frameshifted upon translation, giving rise to a fusion protein named FseA (for Frameshifted excision activator), which subsequently activates transcription of the rdfS excisionase gene (Ramsay et al.2015). FseA on its turn can also be inhibited by QseM, but only when its cellular concentrations are sufficiently high (Fig. 3F) (Ramsay et al.2015). ICEMlSymR7A excision is then mediated by the integrase IntS with the support of the excisionase RdfS (Ramsay et al.2006). ICEMlSymR7A transfer requires a type IV secretion system (trb operon) and the putative relaxase Rlx (Fig. 2G). Rlx itself is essential for the maintenance of ICEMlSymR7A, perhaps through a similar partial replication of excised ICE as observed for ICEBs1, Tn916 and ICESXT, but this has not been proven (Ramsay et al.2006, 2009).
THE HIDDEN LIFE OF ICE
Bistability
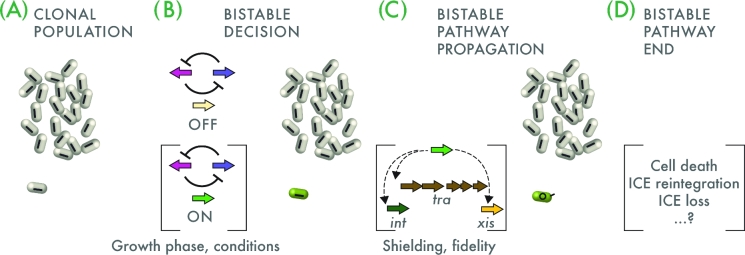
ICEs have an apparent dual mode of life; they can be in an integrated state but need to excise to transmit themselves to other cells (Fig. 1). The two states have very different requirements and thus it is likely that they cannot stably coexist simultaneously in an individual cell. Therefore, whereas both states can occur simultaneously at the level of a population, the individual cell can either have a quiescent, integrated ICE or an ICE which becomes activated, excises and transfers. In the case of an element relying on replication to excise (like TnGBS; Brochet et al.2009), both integrated and excised forms would be able to coexist in an individual cell. The occurrence of two stable (phenotypic) states in a clonal population of cells is more widely known as bistability (Fig. 4). Our hypothesis is thus that ICEs invoke a bistable decision at the level of vertical to horizontal transmission: at some point in time, an individual cell either follows the path of ICE excision or retains the ICE in its integrated state (Fig. 4B). Incidentally, once the cell has excised the ICE it is arguably no longer genetically identical to a cell with an integrated ICE, but this occurs only after the bistable decision between horizontal and vertical transmission is made. The regulatory networks that embed the ICE in the cell thus control under which conditions how many (and maybe which) individual cells will start the horizontal transmission mode or will remain silent (Fig. 4B). The proportion of cells with ICE going through horizontal compared to vertical transmission mode is prone to selection and may be different for the various ICE systems (and their hosts). Similarly, one would expect that those regulatory networks and events that are most successful in propagating ICE horizontal transmission are evolutionary selected at population level.
Figure 4.
Concept of bistability in ICE horizontal transmission. ICE regulatory architectures contain a bistability generator (e.g. double inhibitory cyan/magenta loop as shown), which permits at low frequency in a clonal population (A) to activate the ICE horizontal transmission process (B, green gene arrow, individual green cell). In most other cells, the ICE will remain in its OFF state. At this point, the ICE is still in its integrated state (represented by the black bar inside the cells). Growth phase or environmental conditions can change the frequency of the bistable decision outcome, as illustrated in the details of Fig. 3. (C) Activation of the ICE will continue as (bistable) propagation of the horizontal transmission pathway in the same individual cells (e.g. activation of tra transfer, int integrase and xis excisionase genes), resulting in ICE excision and transfer (represented by the black circle and small black stick in the green cell), when suitable donors are present. This may require specific mechanisms to ensure preventing downstream pathway expression in non-active cells (shielding), and a faithful progression of the different steps of the transmission cascade in activated cells (fidelity). Finally, the ICE bistable horizontal transmission pathway has to end (D), either by death of the donor cell, by ICE reintegration, loss or some other mechanism.
What is the current evidence that the ICE vertical to horizontal transmission modes are true bistable states? The most compelling (but indirect) argument is that (wild-type) ICE transfer rates, even under optimal donor–recipient conditions, are in the range of 10—2 and 10—7 per colony-forming donor cell. This suggests that somehow only very few donors reach a state of being able to transfer the ICE. As explained above, some regulatory mutations, such as overexpressing rapI in ICEBs1 (Auchtung et al.2005) or ΔmfsR in ICEclc (Pradervand et al.2014b), cause an increase of transfer rates to almost 100% per live donor cell. Low wild-type ICE transfer rates therefore must have their origin in the regulatory program of the ICE and its embedment within the host. Finally, direct evidence from single cell observations on at least one ICE system (ICEclc) showed that its behavior is indeed characterized by bistable and mutually exclusive states; very few cells developing the capacity to excise and transfer the ICE, the others remaining silent (Fig. 5) (Minoia et al.2008; Reinhard et al.2013; Delavat et al.2016).
Most ICE regulatory networks have not been characterized in terms of generating and maintaining bistability. However, their extensive control mechanisms and regulatory configurations are reminiscent of gene expression networks that can invoke bistability (Fig. 3). Bistable states arise as a (rare) result of variation in gene expression networks. Gene expression in prokaryotic networks is dynamic and, essentially because of noisy components and interactions, will lead to variation both in time and among individual cells (Thattai and van Oudenaarden 2004; Pedraza and van Oudenaarden 2005; Eldar and Elowitz 2010; Balazsi, van Oudenaarden and Collins 2011). Gene expression variation will thus lead to phenotypic variation, and the level of variation can differ for individual networks or promoters (Kussell and Leibler 2005), with variability being a selectable trait (Ackermann et al.2008; Freed et al.2008). In some cases, the variation in gene expression is not centered around a single mean phenotype, but can lead to two stable phenotypes—mostly resulting in two subpopulations of individual cells displaying either the one or the other phenotype (Ferrell 2002; Dubnau and Losick 2006). Importantly, such bistable states are an epigenetic result of the network functioning and do not involve modifications or mutations on the DNA (Kussell and Leibler 2005; Balazsi, van Oudenaarden and Collins 2011). Bistable phenotypes may endure for a particular time in individual cells and their offspring, or erode over time as a result of cell division or other mechanism, after which the ground state of the network reappears. One can thus distinguish different steps in a bistable network: (i) the bistability switch, which is at the origin of producing the different states; (ii) a propagation or maintenance mechanism; and (iii) a degradation mechanism (Fig. 4C and D).
As an example, competence formation in Bacillus subtilis starts when cells enter stationary phase, and leads to a subpopulation of some 10% of cells temporally being able to take up DNA by transformation, whereas the other cells remain non-competent. In the current hypothesis, bistability in competence formation is generated at the level of transcription of the major competence regulator ComK. ComK activates its own promoter but acts as a tetramer, so needs a certain threshold concentration in the cell to be able to cause activation. The threshold is not easily reached because ComK is rapidly degraded. However, ComK degradation is inhibited by another protein ComS, which accumulates in stationary-phase cells. Stochastic variations in individual cells in ComK and ComS levels and noise at the comK promoter thus determine the onset of comK transcription, which then reinforces itself because of the feedback mechanism (Maamar, Raj and Dubnau 2007). The outcome of bistable networks is non-intuitive and mathematical models can be helpful to understand and predict the system states (e.g. the ComK-circuit by Suel et al.2006).
Another well-known example of bistability decision is the phage lambda switch. Individual phage attaching to an Escherichia coli cell delivers their DNA inside, which can then either lead to immediate propagation of new phage and cell lysis or to integration of the phage DNA into the E. coli chromosome (Zeng et al.2010). Although the network is detailed and controlled at a variety of different levels (Arkin, Ross and McAdams 1998), the switch comes down to a final balance of two transcription factors CI and Cro. In case CI levels are high, the phage PL and PR promoters will be repressed and the phage will integrate; in case CI levels are low, the Cro feedback loop will emerge, leading to repression of the PRM promoter and activation of the lytic state (Bednarz et al.2014). Whereas the lytic decision leads to a dramatic series of events culminating in cell death, phage lysogeny can remain for hundreds of generations, illustrating additional mechanisms to ensure maintenance of the chosen bistable state (Arkin, Ross and McAdams 1998). Finally, lysogeny can revert to the lytic state, when, under influence of the host factor RecA, CI is degraded. In that situation, the prophage will excise and start production of phage particles, DNA packaging and finally, lyse the host cell. ICEs have a mechanistic behavior very akin to prophage excision and integration (Fig. 1A).
How bistability is generated in the various ICEs is not well understood. Some systems, such as ICESXT, carry analogous double-negative feedback loops as known from phage lambda (i.e. CroS and SetR, similar to Cro and CI, respectively), which control ICE activation (Beaber, Hochhut and Waldor 2004; Poulin-Laprade and Burrus 2015) (Fig. 3C). By analogy, one could thus assume that this loop must generate bistability, which is then transmitted to the transfer functions by means of the master activator complex SetCD (Fig. 3C). Also the ICESt1 and ICESt3 elements carry double-negative feedback systems formed by the Arp1 and Arp2 repressors (Bellanger et al.2007, 2008, 2009; Carraro et al.2011) (Fig. 2E), which could potentially produce a bistable switch analogous to phage λ. Other ICE regulatory systems invoke phosphorylation pathways, positive feedback loops or involve protein degradation (Fig. 3A, B, D, F), configurations which are predicted or known from other systems to generate bistability (Ferrell 2002; Dubnau and Losick 2006; Tiwari et al.2011). One could thus imagine ICE regulatory networks having a basic propensity for yielding bistability, on top of which further regulatory signals are integrated (Fig. 4B). Once initiated, the horizontal transmission pathway needs to be faithfully followed in the activated cells, but should remain silent in the other cells (Fig. 4C). Finally, mechanisms should exist which can end the bistable horizontal transmission pathway, either through cell death, ICE reintegration or other (Fig. 4D). It will be crucial to unravel these bistability generators at molecular level and understand how the ICE bistable horizontal transmission is faithfully followed in individual cells.
Transfer competence
In our own research on ICEclc in Pseudomonas, we have tried to demonstrate the existence of ICE bistability and how it controls ICE transfer. Instrumental for this work were single cell observations, coupled with genetic manipulations, cell staining and fluorescence reporter proteins (Fig. 5). Fluorescence reporters transcriptionally fused to key ICEclc promoters such as Pint (of the intB13 integrase promoter) or PinR (for the integrase expression regulator InrR, Fig. 3E), introduced in single copy in the host chromosome but outside the ICE, showed the remarkable appearance of a subset of cells with clear expression against a large background of cells without any noticeable fluorescence (Fig. 5A). That this is the typical characteristic of a bimodal expression state can be seen by a quantile–quantile analysis, which plots the observed fluorescence intensities of individual cells against their expected (normal) distribution (Fig. 5A). A single (normal) distribution would result in a straight line, but reporter protein fluorescence values from both Pint- and PinR-fusions produce the typical ‘hockey stick’ of two separate normal distributed populations (Fig. 5A) (Reinhard and van der Meer 2013). This analysis enabled to correctly quantify the subpopulation of cells with active ICE promoters, which at this point have not yet excised the ICE (Delavat et al.2016). It showed that the onset of ICEclc activation occurs in clonal stationary-phase cells of Pseudomonas knackmussii or P. putida and that the subpopulation of ICE-active cells is highest (3%–5%) when they have been cultured on 3-CBA as sole carbon and energy source (Miyazaki et al.2012; Reinhard and van der Meer 2013). Double labeling with both single copy Pint-egfp and PinR-mcherry fusions then showed that essentially the same individual cells express both promoters (Fig. 5A). This suggested that the expression states of the ICEclc promoters in such cells are not only accidentally bimodal, but are representative for a coordinated bistable program that, once initiated, proceeds in the same individual cells. Evidence was also obtained that bistable activation of the Pint and PinR-promoters at least partially originates from cellular variation in RpoS (Miyazaki et al.2012). The influence of varying intrinsic and extrinsic factors is analogous to noisy gene expression in other networks and is assumed to be at the origin of the bistability development (Elowitz et al.2002; Pedraza and van Oudenaarden 2005; Rosenfeld et al.2005).
To further confirm that ICEclc actually induces and coordinates a bistability program, which ‘locks‘ cells in a particular state, we used time-lapse imaging combined with specific incubation chambers that allowed tracking of single cell growth on agarose surfaces during multiple days (Reinhard and van der Meer 2010; Reinhard et al.2013; Delavat et al.2016). The use of such sterilizable, closed chambers has been essential to achieve long-term growth and maintain stationary-phase conditions long enough (1–3 days) to allow ICEclc activation. In addition, we developed a fluorescence reporter tool to distinguish ICE activation, excision and transfer at single cell level (Fig. 5B) (Delavat et al.2016). This reporter is based on the particularity of ICEclc to transcriptionally fuse the attL-proximal Pcirc promoter, which faces outwards in the integrated state (Fig. 2F), upstream of the integrase gene (intB13) upon excision and formation of a circular intermediate (Fig. 5B) (Sentchilo, Zehnder and van der Meer 2003). This promoter is stronger than the bistable Pint promoter directly upstream of the intB13 gene (Sentchilo, Zehnder and van der Meer 2003). For the reporter tool, we thus placed an egfp gene downstream in the same transcriptional unit as the intB13 integrase gene on ICEclc (Delavat et al.2016). We hypothesized that fluorescence would be visible in the subpopulation of cells activating ICEclc in stationary phase, but would increase even more when such cells would actually excise the ICE. This would be the result of the stronger Pcirc promoter being fused upstream of intB13 and egfp (Fig. 5B). The tool would also permit to identify ICEclc transfer, because it would cause fluorescent protein expression in transconjugant cells. Two recipient strains were produced that could be distinguished microscopically from donor cells, either on the basis of constitutive fluorescent markers (Fig. 5B) or by activation of fluorescent protein expression upon insertion of ICEclc into an artificial trap (Sentchilo et al.2009).
Experimental observations with the reporter tools indeed confirmed that only cells that previously activate ICEclc can subsequently excise, and only cells in which excision has taken place are capable of ICE transfer to a recipient (Delavat et al.2016) (Fig. 5B). We had observed previously that all donor cells in which ICEclc starts to become active (i.e. that show visible fluorescent protein expression from Pint and/or PinR promoters) display different cell morphology than non-active donor cells, divide slower, less and eventually die (Reinhard et al.2013). Coordinately, these results were strong evidence to propose that donor cells, once the ICE is activated, become locked in a bistable program that we named transfer competence development. The sole purpose of this differentiation program seems to be to prepare the donor cell for possible ICE transfer. As far as we could observe in time-lapse microscopy, there is no escape from this program and individual transfer competent donor cells do not return to normal exponential growth (Fig. 6A). On the other hand, PCR data on larger population sizes provided evidence that ICEclc is at low frequencies occupying different tRNAgly-gene integration sites. This could be the result of donor-to-donor ICEclc transfer and integration, or of intracellular excision and reintegration at a different insertion site (Sentchilo et al.2009). In the latter case, one would have to conclude that low-frequency escape of the bistable transfer competence program is possible.
Figure 6.
Inferred ICE–host cell adaptations selected for optimal ICE transmission. (A) ICEclc in P. knackmussii or P. putida remains integrated in exponentially growing cells (EXPO, white bar inside black cells), but is activated in ∼3% of individual cells during stationary phase (STAT, green cells). Such cells become transfer competent (tc), but only excise ICEclc once they are provided with new nutrients (EXPO, white circles inside green cells). tc cells can divide a few times to produce a microcolony that improves the transfer probability, but individual cells in such microcolony show highly variable ICE and cell fates (illustrated here as lysing cells [with holes], ICE replication, single or multiple ICE transfer [small sticks pointing out from cells], or ICE loss). Eventually, tc cells perish and are overgrown by the non-tc cells, and the cycle repeats itself in a next stationary phase (Reinhard et al.2013; Delavat et al.2016). ICEclc transferred to a new recipient (r, light brown cell) may integrate depending on the availability and sequence match with the chromosomal attachment site (i.e. integration efficiency). (B) ICEBs1 in B. subtilis remains integrated in exponentially growing cells (EXPO) and when the density of ICE-free cells (r, light brown) is low. Stationary phase (STAT) conditions and high density of ICE-free cells, or occurrence of DNA damage, stimulate the process of ICEBs1 horizontal transmission from donors (tc, green cell) to ICE-free cells. ICEBs1 replicates in the donor to avoid loss upon donor cell division. Optimal conjugation further requires specific hydrolysis of the recipient cell wall and efficient ICEBs1 replication from its single-stranded origin (sso) aids in maintenance in the recipient. Transfer can directly continue in cell chains (r to r). Wild-type ICEBs1 transfer rates estimated to between 10–7 and 3×10–5 per colony-forming cell (A. Grossman, personal communication). (C) ICESXT transfer in V. cholerae cells is stimulated ca 100-fold by DNA damage induced SOS response from a background level of around 2×10–7 per cell (Waldor, Tschäpe and Mekalanos 1996; Beaber, Hochhut and Waldor 2004). ICESXT replication and partitioning ensure proper segregation among dividing donor cells with excised ICE (tc, green cell; excised ICE as circles), but TA systems may specifically inhibit any ICE-free daughter cell (punctured green cell in illustration). Successful conjugation and integration in a recipient (r, light brown cell) further depends on the recipient's SOS response, exclusion mechanisms or defense systems such as restriction-modification (R/M). Note that cases (B) and (C) are inferred from bistability assumptions but have no current support from single cell observations. For further details and references, see the main text.
Although there is currently little direct evidence from single cell observations, apart from anecdotal evidence of subpopulations having high variation in expression components of the ICE (Ramsay et al.2015), we hypothesize that most if not all (wild-type) ICE systems go through a bistable activation state, followed by ICE excision and possibly, by a dedicated transfer competence development program (Fig. 4). The bistable switch likely has a background (stochastic) state inherent to its molecular architecture, on top of which various regulatory signals can be integrated (Fig. 3) that increase or decrease the frequency of the occurrence of the decision to initiate horizontal transmission among individual cells in the population. The reasons, as outlined above, are that all ICE regulatory networks are similar in architecture to systems generating bistability, that excision rates as measured by PCR and transfer rates are low, which is consistent with small subpopulations of dedicated donor cells being formed. The concept of ICE bistability makes it easier to understand various aspects of their life style and their connections to the host, and the selective forces that must act on both the vertical and horizontal transmission modes.
The vertical/horizontal transmission mode balance
From an evolutionary perspective, all ICEs are in a state of semi-equilibrium with their host. Once inserted in the host's chromosome, the ICE-DNA will be faithfully copied in every dividing cell (vertical transmission). As long as the integrated ICE does not pose a major fitness cost on the host, or even provides selective benefit, it may endure. In contrast, in order to distribute itself among ICE-free cells of its own or other species, the ICE must excise from the host chromosome and induce the donor cell to produce the DNA transfer machinery (Fig. 6). What does the horizontal transmission mode entail for the host? Possibly, the process of horizontal transmission and its associated factors are very costly or even dangerous to produce for the host. ICE transfer does not seem to be as ‘deadly’ as a lytic phage propagation, but apparently it is sufficiently disadvantageous to have been selectively regulated to very low levels (10–2 to 10–7 per viable donor) such as not to endanger survival of the host. Consequently, one would expect an important interplay between the host (avoiding too much ICE-caused damage) and the ICE (with its genetic makeup for vertical and horizontal transmission modes). Possibly, the ICE is further influenced by other co-residing mobile elements or prophages that the host cell needs to deal with (Croucher et al.2016).
What do we know about the fitness costs of ICEs in their integrated state? This is difficult to measure, because one would have to deactivate the horizontal transmission system of the ICE, which is mostly an integral part of the ICE. However, there is some information both from phylogenomic and from experimental studies. Initial studies on diversity of pathogenicity islands in uropathogenic E. coli (Hacker et al.1997; Hacker and Kaper 1999; Hacker and Carniel 2001) proposed that integrated ICE eventually erode and lose (part of) their mobility functions. One could argue that the mere presence of 100 kb DNA in a host chromosome is a burden on fitness, because of the larger investment in DNA replication (e.g. on a 6 Mbp chromosome, 100 kb ‘extra’ DNA to replicate would require a 1.6% surplus of replication and biosynthesis effort). Experimental observations on a variety of P. aeruginosa hosts with or without ICEclc indicated that its cost on competitive reproductive fitness under non-selective conditions remained below 1% and further, that ICEclc remained transcriptionally ‘isolated’ from the host (Gaillard et al.2008). This 1% cost is in rough agreement with the maximum observed proportion of ICEclc transfer competent cells. It suggests that ICEclc limits fitness costs on the whole population by restricting its activity to a small subpopulation only. In contrast, ICEclc regulatory mutants causing massively increased subpopulation activation have an immediate and high fitness cost (Pradervand et al.2014b). The tendency that stealthy elements are more favorable for the host is supported by other observations showing that genes with high expression levels have a tendency to be less frequently transferred, which is thought to be linked to disturbance of general cellular transcription and translation systems (Park and Zhang 2012) or increasing consumption of cellular resources (Baltrus 2013). There is also evidence that the host can actively silence horizontally acquired gene regions to avoid their interference (Lucchini et al.2006).
What is the fitness cost of ICE horizontal transmission? It is in the genetic makeup of the ICE to transfer horizontally; therefore, we have to assume that the ICE itself has a fitness benefit to be distributed to new recipient cells. What about the host cell? What does ICE excision and transfer require from a host cell? Unfortunately, there are not many experimental studies which have addressed this question and it is difficult to generalize for all ICEs. Experimental observations on the ICEclc system suggest that excision and transfer of the ICE not only requires development of specialized differentiated transfer competent cells, but also creates a strong imbalance in the ICE-host partnership. We call it ‘imbalance’, because of the highly variable individual observed cell and ICE fates (Fig. 6A). First of all, we observed that, although ICEclc is activated in stationary-phase cells, it mostly does not excise nor transfer in that state (Delavat et al.2016). Only upon restimulating of such ‘pre-activated’ cells by nutrient addition the ICE excision and transfer process starts (Fig. 6A) (Delavat et al.2016). This suggests that transfer is an energetically costly process that demands many resources of an individual donor cell. In addition, transfer-competent and nutrient-stimulated donor cells re-initiate (limited) cell division, which are characterized by highly variable ICE fates. Some cells show ICE replication upon cell division, others lose the ICE, some cells transfer once, and again others transfer twice or even three times (Delavat et al.2016). Most ICEclc transfer competent cells do not divide more than two times, can persist for a while but eventually perish (Reinhard et al.2013). Since cells with the ICE in its integrated state divide normally, the transfer competent cells proportionally disappear from the population (Reinhard et al.2013; Delavat et al.2016). Moreover, even though mutants can be obtained with strongly increased ICEclc transfer rates (i.e. the aforementioned deletion of the mfsR master regulator), they are extremely unstable under non-selective conditions and undergo massive cell death (Pradervand et al.2014b). There is experimental evidence that impairment of cell division is determined by ICEclc itself through the activity of the shi gene (Reinhard et al.2013). In contrast to the ICEclc system, single cell observations of B. subtilis with the hypertransferable ICEBs1-rapI overexpression mutant did not indicate any visible cell damage or fitness cost (Babic et al.2011). However, deletions of the key regulator immR on ICEBs1 result in constitutive expression of most ICEBs1 genes, very frequent ICE loss and more frequent cellular lysis (Auchtung et al.2007), suggesting that deregulated expression of ICEBs1 genes is also leading to fitness impairment of the host. Modeling the ICEclc behavior suggested that horizontal transmission of ICEclc with massive fitness cost can only be sustained when the proportion of cells that activates the ICE in stationary phase remains around 1% or lower (Delavat et al.2016). ICEclc excision and transfer rates thus seem to be governed by a balance to maintain host and ICE fitness, i.e. sacrificing a small proportion of host cells, while maintaining a low but sufficient rate of horizontal transmission of the ICE. Given the low transfer frequencies of wild-type ICEs, this balance between limiting fitness cost and ensuring horizontal transmission may be more general (Fig. 6).
Stability and addiction
The ICE faces the risk of elimination in those cells in which it starts the excision and transfer, in particular when they divide. As our experimental observations confirm, ICEclc-free donor cells indeed appear at this stage but are impaired for further division (Delavat et al.2016). ICEs have evolved various systems to counteract this potential loss. The first of these is a limited replication and ICE partitioning system during the excised state, guaranteeing segregation of ICE molecules in case of donor cell division (Fig. 6B and C). The second system consists of an active killing of ICE-free donor cells appearing (Fig. 6C). Importantly, the existence of both systems was inferred from genetic studies at population level and, therefore, we do not precisely understand how they act at individual cell level.
Recent evidence on various models has now clearly demonstrated that ICEs transiently replicate in cells upon excision (Lee, Babic and Grossman 2010; Thomas, Lee and Grossman 2013; Wright and Grossman 2016; Carraro et al.2016a). In case the donor cell with excised ICE is dividing, the increased copy number may reduce the chance of daughter cells being devoid of any ICE (Fig. 6B). Additionally, increased ICE copy numbers in donor cells may also permit multiple transfer events, as single cell observations on the ICEclc system indicated (Delavat et al.2016) (Fig. 6A). Replication of excised ICE is the result of a double use of the oriT by the relaxase as a double-stranded origin of replication as well as the processing start of the ICE-DNA for transfer. In this light, it is puzzling why some ICE (e.g. ICEclc) even have double oriT sequences, but maybe this permits individual ICE molecules in a single donor cell to transfer independently (Miyazaki and van der Meer 2011a, b). In addition to the double-stranded origin of replication, recent work on ICEBs1 and Tn916 showed that ICE can have a single-stranded origin of replication (sso) (Wright, Johnson and Grossman 2015; Wright and Grossman 2016). The role of this sso may be to increase the chance of the (single-stranded) ICE to be correctly reconstituted in the recipient cell and facilitating its subsequent integration. Some ICEs, such as ICESXT, also further reduce the chance of appearance of ICE-free daughters in the excision stage by deploying active partition systems, similar to plasmids (Carraro and Burrus 2015; Carraro, Poulin and Burrus 2015).
Several ICEs of the ICESXT-R391 family carry genes for toxin/anti-toxin (TA) systems, notably MosT/MosA and S044/S045 in ICESXT and HipA/B in R391 (Dziewit et al.2006; Wozniak and Waldor 2009; Carraro, Poulin and Burrus 2015). Expression of the mosAT operon increases upon ICESXT excision, which was interpreted as a mechanism to avoid cellular loss of ICESXT in the excised state (Wozniak and Waldor 2009). MosT is a bacteriostatic toxin, which is countered by the antitoxin MosA. Presumably, MosT is more stable than MosA and therefore, accidental loss of ICESXT in dividing cells would result in a rapid decline in MosA concentration, causing the enduring MosT to damage the cell (Fig. 6C). Upon reintegration of ICESXT, expression of mosAT returns to a relative silent state driven by MosA autorepression (Wozniak and Waldor 2009). The second TA system (S044/S045) is present in some ICESXT variants, and consists of a growth-inhibiting toxin (S044) and its neutralizing antitoxin (S045) (Dziewit et al.2006). Single cell observations on the ICEclc system suggest that ICE-free daughter cells from active donors do arise (Delavat et al.2016). However, since cell division becomes restricted in all ICEclc donor cells through the shi system, it is not known whether ICE-free daughter cells arise from a failure in some partitioning or maintenance event (Reinhard et al.2013). There are currently no single cell observations on the fate of donor cells in other ICE systems to confirm the action of TA and partitioning systems at the stage of appearance of excised ICE copies. In some other ICEs, it was speculated that their TA systems may act to limit damage caused to cells simultaneously infected by phages. This so-called TA abortive infection system, which is widespread among both plasmids and ICEs, would induce phage-infected cells to undergo suicide, in order to limit phage propagation (Dy et al.2014).
Can the host cell ‘ever’ get rid of an ICE? This is largely unknown. Population-level and longitudinal studies on chronic (eukaryotic) host–pathogen infections suggest that ICEs (and GIs) slowly erode and become immobile (Hacker et al.1997; Hacker and Kaper 1999; Hacker and Carniel 2001), and at low frequencies are lost completely from their host (Marvig et al.2015). Recent experimental and modeling work further suggests that invasion and maintenance of mobile genetic elements (including ICEs) in host genomes is to be seen as an intragenomic ‘arms race’ (Croucher et al.2016). That study further showed how Gram-positive (naturally competent) hosts can prevent maintenance and spread of mobile DNA elements through efficient uptake and recombination with DNA of their own kin containing the ‘mobile-DNA-free’ locus. The mobile elements on their turn counteract this by inserting into chromosomal loci responsible for the production of the transformation machinery (Croucher et al.2016), or by production of extracellular nucleases, which inhibit transformation (Dalia et al.2015).
ECOLOGICAL SIGNIFICANCE
ICE transfer in the environment
The observed wild-type ICE transfer rates should reflect more or less the outcome of natural selective conditions on maximizing horizontal transmission rates while minimizing host fitness cost (Fig. 6A–C). ICE transfer, however, has never really been studied under environmental conditions directly or in mixed microbial communities as they occur in e.g. eukaryotic hosts. Consequently, we can only infer indirectly how the various ICE–host systems have been shaped by evolutionary selective forces. Recent, primarily medical, phylogenomic studies on pathogen–host interactions suggest very little in vivo ICE transfer, occasional acquisition, but more frequent partial or complete deletion (Dettman et al.2013; Marvig et al.2015; Croucher et al.2016). Older literature showed effective transfer of ICEclc in seeded mixed bioreactors (Springael et al.2002) and sporadic natural isolation of highly similar ICEs in different environments suggest robust transfer (Gaillard et al.2006), but true rates have not been reported.
In addition to the intrinsic low ICE excision frequencies resulting from its bistability mechanism, both host and environmental factors impact transfer rates. As illustrated in Fig. 3, ICE excision and transfer frequencies can be influenced by, e.g., carbon sources, growth phase conditions, cell density and presence of autoinducer, presence of antibiotics or factors and conditions eliciting the SOS response in cells. These might constitute selected evolved reactions from the ICE to ‘save’ itself from host cells that are in bad shape (e.g. SOS response), have been subject to some form of other biochemical damage (Reinhard and van der Meer 2014) or have lost their ICE (Fig. 6C) (Babic et al.2011). These concepts are still very difficult to study at single cell level and in many cases we simply do not know the actual reason for the onset of ICE activation, which might even be completely stochastic (Reinhard and van der Meer 2014). Transfer rates in the environment are further controlled by a variety of processes, such as contact possibilities of donor to recipient cells, donor cells not reaching the appropriate physiological state for transfer because of poor environmental conditions (e.g. temperature, limited nutrients) or recipient cells not being permissive to accept and integrate the ICE. Incidentally, we showed that the ICE might even have evolved to cope with regularly encountered recipient cell shortage in the environment. For example, although division of ICEclc transfer competent cells becomes strongly impaired, their limited division (two to three times) allows them to form small microcolonies that have a higher probability to encounter potential sparse recipient cells (Reinhard and van der Meer 2014) (Fig. 6A). Transfer from such microcolonies is, on average, more efficient (Delavat et al.2016), and when the process of limited cell division of transfer competent donor cells is specifically eliminated through mutation of ICEclc factors, transfer rates decrease (Reinhard et al.2013). Since ICE fitness is selected by its propensity to become transferred (as the example of the acquisition of the mfsR regulatory system in ICEclc surmises) (Pradervand et al.2014a, b), it seems reasonable to assume that other ICEs have evolved systems enabling optimal transfer under the regular living conditions of their respective hosts, such as efficient ICEBs1 transfer in B. subtilis cell chains (Babic et al.2011) (Fig. 6B).
ICE beneficial functions
In addition to forcing the host to maintain the ICE (through, e.g. its TA addiction mechanism and partitioning), the ICE can also entice the host by providing selective advantages. These can be provided through the many auxiliary genes the ICE is carrying apart from its strictly necessary ‘core’ regions, which encode the ICE’s crucial life-style functions. Fitness advantages of ICE-provided genes can be obvious, such as encoded resistance to antibiotics or heavy metals, plant symbiosis or the ability to use atypical pollutants as carbon source. Less obvious is the role of the many ICE-located genes with unknown functions. Even these may have a benefit for the host, which was recently demonstrated in a study showing growth rate reduction of Streptococcus thermophilus deleted for an (obscure) 102-kb GI in comparison to the wild type containing the GI (Selle, Klaenhammer and Barrangou 2015). The sizes of the flexible gene regions vary widely within the same ICE family (Osorio et al.2008; Wozniak et al.2009; Bordeleau et al.2010; Miyazaki et al.2015), illustrating how effectively the ICE can acquire and distribute a large and variable ‘cargo’. Importantly, most ICEs have a broad host range and can therefore horizontally transmit their cargo among very different species groups.
Barriers of ICE transfer
The fact that ICE families colonize very different hosts and that ICE variants within families can be found in bacteria isolated from contaminated soil as well as in strains isolated from cystic fibrosis patients highlights their active and efficient transfer as well as broad range of adaptation properties. Nevertheless, not all ICE families are found in all possible bacterial hosts, indicating that barriers to ICE transfer must exist, but it is likely that ICEs have evolved strategies to overcome such barriers. Because ICEs normally reside in the chromosome of their host and only transiently occur in extrachromosomal form, their dissemination is dependent on their successful integration within the genome of its host (Fig. 6A). ICEs integrate site specifically and mostly at conserved chromosomal target sites (such as the 3΄ end of tRNA or other conserved genes) (Ghinet et al.2011; Ambroset et al.2015), which they restore perfectly to avoid mutation. This strategy helps to broaden the range of potential hosts in which the ICEs can potentially integrate (Sentchilo et al.2009). Dead-end hosts may occur, where the ICE accidentally integrated because of limited integration site similarity, but from where it can no longer efficiently express itself, excise or transfer (Burrus and Waldor 2003; Gaillard et al.2008; Menard and Grossman 2013). The chances of successful ICE transfer and integration diminish the more distant becomes the recipient genome from the host. Multigenome comparisons indicated that the frequency of HGT occurrence is drastically smaller among hosts with more than 5% dissimilarity in their GC content (Popa and Dagan 2011). Moreover, multiple GIs within a single host tend to have a similar relative dinucleotide frequencies (Roos and van Passel 2011), suggesting some preference for donor genome constitution. Other ICEs like Tn916 have a very low site specificity and integrate mainly in AT-rich sites (Franke and Clewell 1981). This strategy has the advantage of allowing efficient integration into a variety of hosts (Poyart, Celli and Trieu-Cuot 1995; Roberts and Mullany 2009), but with the risk of integrating within coding sequences, disrupting important host functions and reducing its efficiency to propagate. The additional advantage of the strategy evolved by Tn916 may be to insert within other mobile genetic elements (Ding et al.2009; Chancey et al.2015), which, on average, tend to have a higher AT content than their host (Rocha and Danchin 2002), and ensure cis mobilization.
ICE conjugative transfer itself requires adhesion of the type IV secretion system of the donor cell onto the cell envelope of the recipient cell. Conjugation efficiencies may thus vary from species to species depending on how well the adhesion and conjugative pore are forming. The role of cell envelopes of donor and recipients in ICE conjugation efficiency is best known from ICEBs1 in Bacillus subtilis. ICEBs1 transfer is dependent on the secreted cell-wall hydrolase CwlT, which affects peptidoglycan synthesis (DeWitt and Grossman 2014) (Fig. 6B). Mutations in the recipient causing altered cell envelope, such as for phospholipid biosynthesis, also reduced ICE transfer rates, but no single mutation in the recipient completely abolished transfer (Johnson and Grossman 2014, 2016). Transfer efficiencies may also be affected by exclusion through coresiding ICE in the recipient, as shown for the S and R groups from ICESXT and R391 (Marrero and Waldor 2007a, b).
Finally, the ICE is vulnerable even after transfer into the recipient cell. Since it consists of single-stranded DNA, this may elicit the SOS response in the recipient (Baharoglu, Bikard and Mazel 2010). ICE-DNA may also be considered as foreign DNA by the host, in analogy to phage DNA, and become subject to restriction digestion (Wilson and Murray 1991) or CRISPR-mediated DNA cleavage (Lopez-Sanchez et al.2012; Zhang et al.2013) (Fig. 6C). Incidentally, however, ICE may also contain CRISPR-Cas types themselves (van Belkum et al.2015).
CONCLUSIONS AND FUTURE WORK
The genomic age has revolutionized our understanding of (prokaryotic) genome plasticity and it has become evident that ICEs have colonized most bacterial phyla and are key players in bacterial evolution. Despite their genetic, regulatory and mechanistic diversity (Figs 2 and 3), ICEs have developed similar effective dissemination strategies that are characterized by a bistable dual life style (Fig. 5). In their integrated state, ICEs tend to reduce expression from their core functions, to avoid fitness cost on the host and ensure maximum vertical transmission. The variable genes carried by most ICEs, which are not directly involved in its core functions, can provide benefit to the host and thus pose a selective advantage to maintain the ICE. At low to very low frequencies, and typically tightly embedded in specific growth conditions or upon external signals, the ICE activates its excision and transfer machinery to become horizontally transmitted, in the process of which the individual donor cell may not survive (Figs 3, 4 and 6). The low-frequency activation is thus likely a consequence of selection to avoid the damage caused by a transferring ICE on the host cell, and is the best ‘compromise’ for sufficient ICE horizontal transmission and host population survival.
Finally, what are ICEs really? Historically, phages and plasmids were strictly separated. But the appearance of GIs and ICEs blurred the characteristics of both groups, with them sharing both phage and plasmid (conjugation) properties (Fig. 1) (Juhas et al.2009; Carraro, Poulin and Burrus 2015). Perhaps there is a continuous scale of mobile DNA elements, with only rough boundaries between them and with many overlapping functionalities. Lytic phages on one end of the scale, as capsid-based entities that encapsulate their genetic information (either DNA or RNA) and that can infect bacteria through direct injection of their genetic material. Inside bacteria, the phages hijack the host cell machinery, multiply in the cytoplasm and finally lyse the host to liberate new phage. Other phages inducing a lysogenic life cycle, where the phage-DNA integrates into the genome of its host and only sporadically starts the lytic cycle. ICEs lay in the middle of scale, still deploying an integrative state, but having lost a lytic cycle and adopted conjugation as a transfer mode. ICE horizontal transmission still seems to harm the host cell, not as dramatically as a lytic phage, but sufficient to be kept at a low level. On the other end of the scale, the conjugative plasmids, which remain extrachromosomal and replicate independently of the host genome, and which can transfer under more relaxed conditions without damaging the donor cell. Multiple observations show that the frontier between these groups of elements is thinner than previously thought (Carraro, Poulin and Burrus 2015), with mechanistical, structural and genetic similarities between them, suggesting old evolutionary relationships. Still many different other types of mobile elements exist, having lost one or the other feature but profiting to some extent from coresiding phage, ICE or plasmid (Fig. 1B and C). Examples are plentiful, such as capsid-less viruses being packaged by other viruses, satellite prophages mobilized by lysogenic phages, plasmids being comobilized by other plasmids or ICEs, or GIs and IMEs mobilized by ICEs, plasmids or by phages. This has led some authors to propose the general concept of a genetic replicator, which represents all elements carrying genetic information (chromosome, plasmids, transposons, ICEs, temperate viruses and lytic viruses). These replicators may be classified based on their horizontal transfer potential and the relationship (from mutualistic to parasitic) with the host cell vehicle (Jalasvuori and Koonin 2015; Jalasvuori, Mattila and Hoikkala 2015).
What are important open questions to further understand the life style of ICEs? First, we postulate that all ICEs should be governed by a key bistable decision that determines the change between vertical and horizontal transmission. So far, this has been experimentally observed only for a single system (ICEclc). Therefore, it would be crucial to extend single cell observations to other ICE systems. Second, given the extensive diversity of regulation mechanisms (Fig. 3), it will be interesting to model and understand the mechanisms producing and maintaining bistability (Fig. 4). Third, single cell observations and modeling will be key to understand further ICE–host fate decisions, and the roles played by ecological selective forces on ICE–host evolution (Fig. 6). Possibly, the roles of other mobile DNAs in shaping the host–ICE partnership have been underestimated, and a more complete theory on host-mobile-DNA is necessary (e.g. Croucher et al.2016).
Acknowledgments
The authors like to thank Sandra Sulser and Friedrich Reinhard for their help in drafting parts of the initial text.
FUNDING
This work was supported by Swiss National Science Foundation Grants 31003A_144141/1 and 310030B_156926/1 (to JRvdM).
Conflict of interest.None declared.
REFERENCES
- Ackermann M, Stecher B, Freed NE et al. Self-destructive cooperation mediated by phenotypic noise. Nature 2008;454:987–90. [DOI] [PubMed] [Google Scholar]
- Ambroset C, Coluzzi C, Guedon G et al. New insights into the classification and integration specificity of Streptococcus Integrative Conjugative Elements through extensive genome exploration. Front Microbiol 2015;6:1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin A, Ross J, McAdams HH. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics 1998;149:1633–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Aleksanyan N, Bulku A et al. Biology of ICEBs1, an integrative and conjugative element in Bacillus subtilis. Plasmid 2016;86:14–25. [DOI] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Garrison KL et al. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol Microbiol 2007;64:1515–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Monson RE et al. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. P Natl Acad Sci USA 2005;102:12554–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babic A, Berkmen MB, Lee CA et al. Efficient gene transfer in bacterial cell chains. MBio 2011;2:e00027–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharoglu Z, Bikard D, Mazel D. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet 2010;6:e1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: from microbes to mammals. Cell 2011;144:910–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus DA. Exploring the costs of horizontal gene transfer. Trends Ecol Evol 2013;28:489–95. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Lenski RE. Genome dynamics during experimental evolution. Nat Rev Genet 2013;14:827–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Yu DS, Yoon SH et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 2009;461:1243–7. [DOI] [PubMed] [Google Scholar]
- Bartha NA, Soki J, Urban E et al. Investigation of the prevalence of tetQ, tetX and tetX1 genes in Bacteroides strains with elevated tigecycline minimum inhibitory concentrations. Int J Antimicrob Ag 2011;38:522–5. [DOI] [PubMed] [Google Scholar]
- Beaber JW, Burrus V, Hochhut B et al. Comparison of SXT and R391, two conjugative integrating elements: definition of a genetic backbone for the mobilization of resistance determinants. Cell Mol Life Sci 2002;59:2065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol 2002;184:4259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 2004;427:72–4. [DOI] [PubMed] [Google Scholar]
- Beaber JW, Waldor MK. Identification of operators and promoters that control SXT conjugative transfer. J Bacteriol 2004;186:5945–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarz M, Halliday JA, Herman C et al. Revisiting bistability in the lysis/lysogeny circuit of bacteriophage lambda. PLoS One 2014;9:e100876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger X, Morel C, Decaris B et al. Derepression of excision of integrative and potentially conjugative elements from Streptococcus thermophilus by DNA damage response: implication of a cI-related repressor. J Bacteriol 2007;189:1478–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger X, Morel C, Decaris B et al. Regulation of excision of integrative and potentially conjugative elements from Streptococcus thermophilus: role of the arp1 repressor. J Mol Microb Biotech 2008;14:16–21. [DOI] [PubMed] [Google Scholar]
- Bellanger X, Payot S, Leblond-Bourget N et al. Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev 2014;38:720–60. [DOI] [PubMed] [Google Scholar]
- Bellanger X, Morel C, Gonot F et al. Site-specific accretion of an integrative conjugative element together with a related genomic island leads to cis mobilization and gene capture. Mol Microbiol 2011;81:912–25. [DOI] [PubMed] [Google Scholar]
- Bellanger X, Roberts AP, Morel C et al. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J Bacteriol 2009;191:2764–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender JK, Kalmbach A, Fleige C et al. Population structure and acquisition of the vanB resistance determinant in German clinical isolates of Enterococcus faecium ST192. Sci Rep 2016;6:21847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkmen MB, Lee CA, Loveday EK et al. Polar positioning of a conjugation protein from the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol 2010;192:38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi D, Xu Z, Harrison EM et al. ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res 2012;40:D621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianconi I, Jeukens J, Freschi L et al. Comparative genomics and biological characterization of sequential Pseudomonas aeruginosa isolates from persistent airways infection. BMC Genomics 2015;16:1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay LM, Touchon M, Rocha EP. Pervasive domestication of defective prophages by bacteria. P Natl Acad Sci USA 2014;111:12127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonheyo G, Graham D, Shoemaker NB et al. Transfer region of a Bacteroides conjugative transposon, CTnDOT. Plasmid 2001a;45:41–51. [DOI] [PubMed] [Google Scholar]
- Bonheyo GT, Hund BD, Shoemaker NB et al. Transfer region of a Bacteroides conjugative transposon contains regulatory as well as structural genes. Plasmid 2001b;46:202–9. [DOI] [PubMed] [Google Scholar]
- Bordeleau E, Brouillette E, Robichaud N et al. Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ Microbiol 2010;12:510–23. [DOI] [PubMed] [Google Scholar]
- Bose B, Auchtung JM, Lee CA et al. A conserved anti-repressor controls horizontal gene transfer by proteolysis. Mol Microbiol 2008;70:570–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose B, Grossman AD. Regulation of horizontal gene transfer in Bacillus subtilis by activation of a conserved site-specific protease. J Bacteriol 2011;193:22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd EF, Cohen AL, Naughton LM et al. Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol 2008;8:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringel F, Van Alstine GL, Scott JR. Conjugative transposition of Tn916: the transposon int gene is required only in the donor. J Bacteriol 1992;174:4036–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet M, Couve E, Glaser P et al. Integrative conjugative elements and related elements are major contributors to the genome diversity of Streptococcus agalactiae. J Bacteriol 2008;190:6913–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet M, Da Cunha V, Couve E et al. Atypical association of DDE transposition with conjugation specifies a new family of mobile elements. Mol Microbiol 2009;71:948–59. [DOI] [PubMed] [Google Scholar]
- Buchrieser C, Prentice M, Carniel E. The 102-kilobase unstable region of Yersinia pestis comprises a high- pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J Bacteriol 1998;180:2321–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V, Marrero J, Waldor MK. The current ICE age: Biology and evolution of SXT-related integrating conjugative elements. Plasmid 2006;55:173–83. [DOI] [PubMed] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B et al. Conjugative transposons: the tip of the iceberg. Mol Microbiol 2002a;46:601–10. [DOI] [PubMed] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B et al. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 2002b;48:77–97. [DOI] [PubMed] [Google Scholar]
- Burrus V, Roussel Y, Decaris B et al. Characterization of a novel integrative element, ICESt1, in the lactic acid bacterium Streptococcus thermophilus. Appl Environ Microb 2000;66:1749–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V, Waldor MK. Control of SXT integration and excision. J Bacteriol 2003;185:5045–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V, Waldor MK. Formation of SXT tandem arrays and SXT-R391 hybrids. J Bacteriol 2004a;186:2636–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol 2004b;155:376–86. [DOI] [PubMed] [Google Scholar]
- Canchaya C, Fournous G, Chibani-Chennoufi S et al. Phage as agents of lateral gene transfer. Curr Opin Microbiol 2003a;6:417–24. [DOI] [PubMed] [Google Scholar]
- Canchaya C, Proux C, Fournous G et al. Prophage genomics. Microbiol Mol Biol R 2003b;67:238–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparon MG, Scott JR. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell 1989;59:1027–34. [DOI] [PubMed] [Google Scholar]
- Caro-Quintero A, Konstantinidis KT. Inter-phylum HGT has shaped the metabolism of many mesophilic and anaerobic bacteria. ISME J 2015;9:958–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro N, Burrus V. Biology of three ICE families: SXT/R391, ICEBs1, and ICESt1/ICESt3. Microbiol Spectr 2014;2:MDNA3-0008-2014. [DOI] [PubMed] [Google Scholar]
- Carraro N, Burrus V. The dualistic nature of integrative and conjugative elements. Mob Genet Element 2015;5:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro N, Libante V, Morel C et al. Differential regulation of two closely related integrative and conjugative elements from Streptococcus thermophilus. BMC Microbiol 2011;11:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro N, Libante V, Morel C et al. Plasmid-like replication of a minimal Streptococcal Integrative and Conjugative Element. Microbiology 2016a; Jan 29. doi: 10.1099/mic.0.000219 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Carraro N, Poulin D, Burrus V. Replication and active partition of Integrative and Conjugative Elements (ICEs) of the SXT/R391 family: the line between ICEs and conjugative plasmids is getting thinner. PLoS Genet 2015;11:e1005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro N, Rivard N, Ceccarelli D et al. IncA/C conjugative plasmids mobilize a new family of multidrug resistance islands in clinical Vibrio cholerae non-O1/non-O139 isolates from Haiti. MBio 2016b;7:pii: e00509-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli D, Daccord A, Rene M et al. Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J Bacteriol 2008;190:5328–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, Poyart C, Trieu-Cuot P. Use of an excision reporter plasmid to study the intracellular mobility of the conjugative transposon Tn916 in gram-positive bacteria. Microbiology 1997;143(Pt 4):1253–61. [DOI] [PubMed] [Google Scholar]
- Celli J, Trieu-Cuot P. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol Microbiol 1998;28:103–17. [DOI] [PubMed] [Google Scholar]
- Chancey ST, Agrawal S, Schroeder MR et al. Composite mobile genetic elements disseminating macrolide resistance in Streptococcus pneumoniae. Front Microbiol 2015;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Christie PJ, Dubnau D. The ins and outs of DNA transfer in bacteria. Science 2005;310:1456–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Paszkiet BJ, Shoemaker NB et al. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J Bacteriol 2000;182:4035–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Sutanto Y, Shoemaker NB et al. Identification of genes required for excision of CTnDOT, a Bacteroides conjugative transposon. Mol Microbiol 2001;41:625–32. [DOI] [PubMed] [Google Scholar]
- Cheng Q, Wesslund N, Shoemaker NB et al. Development of an in vitro integration assay for the Bacteroides conjugative transposon CTnDOT. J Bacteriol 2002;184:4829–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ. The mosaic type IV secretion systems. EcoSal Plus 2016;7:doi: 10.1128/ecosalplus.ESP-0020-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly KM, Iwahara M, Clubb RT. Xis protein binding to the left arm stimulates excision of conjugative transposon Tn916. J Bacteriol 2002;184:2088–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Mostowy R, Wymant C et al. Horizontal DNA transfer mechanisms of bacteria as weapons of intragenomic conflict. PLoS Biol 2016;14:e1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daccord A, Ceccarelli D, Burrus V. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol Microbiol 2010;78:576–88. [DOI] [PubMed] [Google Scholar]
- Daccord A, Ceccarelli D, Rodrigue S et al. Comparative analysis of mobilizable genomic islands. J Bacteriol 2012a;195:606–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daccord A, Mursell M, Poulin-Laprade D et al. Dynamics of the SetCD-regulated integration and excision of genomic islands mobilized by integrating conjugative elements of the SXT/R391 family. J Bacteriol 2012b;194:5794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalia AB, Seed KD, Calderwood SB et al. A globally distributed mobile genetic element inhibits natural transformation of Vibrio cholerae. P Natl Acad Sci USA 2015;112:10485–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delavat F, Mitri S, Pelet S et al. Highly variable individual donor cell fates characterize robust horizontal gene transfer of an integrative and conjugative element. P Natl Acad Sci USA 2016;113:E3375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman JR, Rodrigue N, Aaron SD et al. Evolutionary genomics of epidemic and nonepidemic strains of Pseudomonas aeruginosa. P Natl Acad Sci USA 2013;110:21065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch DR, Utter B, Fischetti VA. Uncovering novel mobile genetic elements and their dynamics through an extra-chromosomal sequencing approach. Mob Genet Elements 2016;6:e1189987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt T, Grossman AD. The bifunctional cell wall hydrolase CwlT is needed for conjugation of the integrative and conjugative element ICEBs1 in Bacillus subtilis and B. anthracis. J Bacteriol 2014;196:1588–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon BK, Laird MR, Shay JA et al. IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res 2015;43:W104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichiara JM, Mattis AN, Gardner JF. IntDOT interactions with core- and arm-type sites of the conjugative transposon CTnDOT. J Bacteriol 2007;189:2692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiChiara JM, Salyers AA, Gardner JF. In vitro analysis of sequence requirements for the excision reaction of the Bacteroides conjugative transposon, CTnDOT. Mol Microbiol 2005;56:1035–48. [DOI] [PubMed] [Google Scholar]
- Ding F, Tang P, Hsu MH et al. Genome evolution driven by host adaptations results in a more virulent and antimicrobial-resistant Streptococcus pneumoniae serotype 14. BMC Genomics 2009;10:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U, Hochhut B, Hentschel U et al. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2004;2:414–24. [DOI] [PubMed] [Google Scholar]
- Dordet Frisoni E, Marenda MS, Sagne E et al. ICEA of Mycoplasma agalactiae: a new family of self-transmissible integrative elements that confers conjugative properties to the recipient strain. Mol Microbiol 2013;89:1226–39. [DOI] [PubMed] [Google Scholar]
- Dorn E, Hellwig M, Reineke W et al. Isolation and characterization of a 3-chlorobenzoate degrading Pseudomonad. Arch Microbiol 1974;99:61–70. [DOI] [PubMed] [Google Scholar]
- Doublet B, Boyd D, Mulvey MR et al. The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol 2005;55:1911–24. [DOI] [PubMed] [Google Scholar]
- Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol 2006;61:564–72. [DOI] [PubMed] [Google Scholar]
- Dy RL, Przybilski R, Semeijn K et al. A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res 2014;42:4590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziewit L, Jazurek M, Drewniak L et al. The SXT conjugative element and linear prophage N15 encode toxin-antitoxin-stabilizing systems homologous to the tad-ata module of the Paracoccus aminophilus plasmid pAMI2. J Bacteriol 2006;189:1983–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature 2010;467:167–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED et al. Stochastic gene expression in a single cell. Science 2002;297:1183–6. [DOI] [PubMed] [Google Scholar]
- Farrugia DN, Elbourne LD, Mabbutt BC et al. A novel family of integrases associated with prophages and genomic islands integrated within the tRNA-dihydrouridine synthase A (dusA) gene. Nucleic Acid Res 2015;43:4547–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE., Jr Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol 2002;14:140–8. [DOI] [PubMed] [Google Scholar]
- Franke AE, Clewell DB. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of "conjugal" transfer in the absence of a conjugative plasmid. J Bacteriol 1981;145:494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed NE, Silander OK, Stecher B et al. A simple screen to identify promoters conferring high levels of phenotypic noise. PLoS Genet 2008;4:e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard M, Pernet N, Vogne C et al. Host and invader impact of transfer of the clc genomic island into Pseudomonas aeruginosa PAO1. P Natl Acad Sci USA 2008;105:7058–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard M, Pradervand N, Minoia M et al. Transcriptome analysis of the mobile genome ICEclc in Pseudomonas knackmussii B13. BMC Microbiol 2010;10:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard M, Vallaeys T, Vorhölter FJ et al. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J Bacteriol 2006;188:1999–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriss G, Poulin-Laprade D, Burrus V. DNA-damaging agents induce the RecA-independent homologous recombination functions of integrating conjugative elements of the SXT/R391 family. J Bacteriol 2013;195:1991–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriss G, Waldor MK, Burrus V. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet 2009;5:e1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghinet MG, Bordeleau E, Beaudin J et al. Uncovering the prevalence and diversity of integrating conjugative elements in actinobacteria. PLoS One 2011;6:e27846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol 2005;3:679–82. [DOI] [PubMed] [Google Scholar]
- Guerillot R, Da Cunha V, Sauvage E et al. Modular evolution of TnGBSs, a new family of integrative and conjugative elements associating insertion sequence transposition, plasmid replication, and conjugation for their spreading. J Bacteriol 2013;195:1979–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini J, de la Cruz F, Rocha EP. Evolution of conjugation and type IV secretion systems. Mol Biol Evol 2013;30:315–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini J, Neron B, Abby SS et al. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res 2014;42:5715–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini J, Quintais L, Garcillan-Barcia MP et al. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 2011;7:e1002222, doi: 10.1371/journal.pgen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J, Blum-Oehler G, Muhldorfer I et al. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol 1997;23:1089–97. [DOI] [PubMed] [Google Scholar]
- Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity: a Darwinian view of the evolution of microbes. EMBO Rep 2001;2:376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J, Kaper JB. The concept of pathogenicity islands. In: Kaper JBHacker J. (eds) Pathogenicitiy Islands and Other Mobile Virulence Elements. Washington, DC: American Society for Microbiology, 1999, 1–12. [Google Scholar]
- Haskett TL, Terpolilli JJ, Bekuma A et al. Assembly and transfer of tripartite integrative and conjugative genetic elements. P Natl Acad Sci USA 2016;113:12268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinerfeld D, Churchward G. Specific binding of integrase to the origin of transfer (oriT) of the conjugative transposon Tn916. J Bacteriol 2001a;183:2947–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinerfeld D, Churchward G. Xis protein of the conjugative transposon Tn916 plays dual opposing roles in transposon excision. Mol Microbiol 2001b;41:1459–67. [DOI] [PubMed] [Google Scholar]
- Hochhut B, Beaber JW, Woodgate R et al. Formation of chromosomal tandem arrays of the SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J Bacteriol 2001;183:1124–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochhut B, Marrero J, Waldor MK. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J Bacteriol 2000;182:2043–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochhut B, Waldor MK. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol 1999;32:99–110. [DOI] [PubMed] [Google Scholar]
- Hopp CM, Gardner JF, Salyers AA. The Xis2d protein of CTnDOT binds to the intergenic region between the mob and tra operons. Plasmid 2015;81:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalasvuori M, Koonin EV. Classification of prokaryotic genetic replicators: between selfishness and altruism. Ann N Y Acad Sci 2015;1341:96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalasvuori M, Mattila S, Hoikkala V. Chasing the origin of viruses: Capsid-forming genes as a life-saving preadaptation within a community of early replicators. PLoS One 2015;10:e0126094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski DD, Clewell DB. Evidence that coupling sequences play a frequency-determining role in conjugative transposition of Tn916 in Enterococcus faecalis. J Bacteriol 1994;176:3328–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski DD, Clewell DB. A functional origin of transfer (oriT) on the conjugative transposon Tn916. J Bacteriol 1995;177:6644–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeters RT, Wang GR, Moon K et al. Tetracycline-associated transcriptional regulation of transfer genes of the Bacteroides conjugative transposon CTnDOT. J Bacteriol 2009;191:6374–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Churchward G. Interactions of the integrase protein of the conjugative transposon Tn916 with its specific DNA binding sites. J Bacteriol 1999;181:6114–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Grossman AD. Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis. Mol Microbiol 2014;93:1284–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Grossman AD. Integrative and Conjugative Elements (ICEs): What they do and how they work. Annu Rev Genet 2015;49:577–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Grossman AD. The composition of the cell envelope affects conjugation in Bacillus subtilis. J Bacteriol 2016;198:1241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Crook DW, Dimopoulou ID et al. Novel type IV secretion system involved in propagation of genomic islands. J Bacteriol 2007;189:761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, van der Meer JR, Gaillard M et al. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 2009;33:376–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton CM, Gardner JF. Roles of Exc protein and DNA homology in the CTnDOT excision reaction. J Bacteriol 2012;194:3368–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton CM, Hopp CM, Yoneji S et al. Interactions of the excision proteins of CTnDOT in the attR intasome. Plasmid 2013a;70:190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton CM, Park J, Wang GR et al. The excision proteins of CTnDOT positively regulate the transfer operon. Plasmid 2013b;69:172–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kers JA, Cameron KD, Joshi MV et al. A large, mobile pathogenicity island confers plant pathogenicity on Streptomyces species. Mol Microbiol 2005;55:1025–33. [DOI] [PubMed] [Google Scholar]
- Klockgether J, Cramer N, Wiehlmann L. et al. Pseudomonas aeruginosa genomic structure and diversity. Front Microbiol 2011;2:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloesges T, Popa O, Martin W et al. Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol Biol Evol 2011;28:1057–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S, Hacker J, Jarchau T et al. Large, unstable inserts in the chromosome affect virulence properties of uropathogenic Escherichia coli O6 strain 536. J Bacteriol 1986;168:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Serres MH, Romine MF et al. Comparative systems biology across an evolutionary gradient within the Shewanella genus. P Natl Acad Sci USA 2009;106:15909–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Dolja VV. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol Mol Biol R 2014;78:278–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 2008;36:6688–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung VL, Ozer EA, Hauser AR. The accessory genome of Pseudomonas aeruginosa. Microbiol Mol Biol R 2010;74:621–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science 2005;309:2075–8. [DOI] [PubMed] [Google Scholar]
- Langille MG, Hsiao WW, Brinkman FS. Detecting genomic islands using bioinformatics approaches. Nat Rev Microbiol 2010;8:373–82. [DOI] [PubMed] [Google Scholar]
- Laprise J, Yoneji S, Gardner JF. IntDOT interactions with core sites during integrative recombination. J Bacteriol 2013;195:1883–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. Molecular archaeology of the Escherichia coli genome. P Natl Acad Sci USA 1998;95:9413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, Auchtung JM, Monson RE et al. Identification and characterization of int (integrase), xis (excisionase) and chromosomal attachment sites of the integrative and conjugative element ICEBs1 of Bacillus subtilis. Mol Microbiol 2007;66:1356–69. [DOI] [PubMed] [Google Scholar]
- Lee CA, Babic A, Grossman AD. Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol 2010;75:268–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, Grossman AD. Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol 2007;189:7254–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, Thomas J, Grossman AD. The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J Bacteriol 2012;194:3165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonetti CT, Hamada MA, Laurer SJ et al. Critical components of the conjugation machinery of the Integrative and Conjugative Element ICEBs1 of Bacillus subtilis. J Bacteriol 2015;197:2558–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M, Gomis-Ruth FX, Coll M et al. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol 2002;45:1–8. [DOI] [PubMed] [Google Scholar]
- Lopez-Sanchez MJ, Sauvage E, Da Cunha V et al. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol Microbiol 2012;85:1057–71. [DOI] [PubMed] [Google Scholar]
- Lu F, Churchward G. Conjugative transposition: Tn916 integrase contains two independent DNA binding domains that recognize different DNA sequences. EMBO J 1994;13:1541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Churchward G. Tn916 target DNA sequences bind the C-terminal domain of integrase protein with different affinities that correlate with transposon insertion frequency. J Bacteriol 1995;177:1938–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini S, Rowley G, Goldberg MD et al. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog 2006;2:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science 2007;317:526–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod SM, Burrus V, Waldor MK. Requirement for Vibrio cholerae integration host factor in conjugative DNA transfer. J Bacteriol 2006;188:5704–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanowska K, Cioni J, Swalla BM et al. Mutational analysis and homology-based modeling of the IntDOT core-binding domain. J Bacteriol 2009;191:2330–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanowska K, Salyers AA, Gardner JF. Characterization of a conjugative transposon integrase, IntDOT. Mol Microbiol 2006;60:1228–40. [DOI] [PubMed] [Google Scholar]
- Malanowska K, Yoneji S, Salyers AA et al. CTnDOT integrase performs homology-dependent and homology-independent strand exchanges. Nucleic Acids Res 2007;38:5861–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra D, Scott JR. Regulation of excision of the conjugative transposon Tn916. Mol Microbiol 1999;31:609–21. [DOI] [PubMed] [Google Scholar]
- Marrero J, Waldor MK. Interactions between inner membrane proteins in donor and recipient cells limit conjugal DNA transfer. Dev Cell 2005;8:963–70. [DOI] [PubMed] [Google Scholar]
- Marrero J, Waldor MK. Determinants of entry exclusion within Eex and TraG are cytoplasmic. J Bacteriol 2007a;189:6469–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero J, Waldor MK. The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J Bacteriol 2007b;189:3302–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvig RL, Sommer LM, Jelsbak L et al. Evolutionary insight from whole-genome sequencing of Pseudomonas aeruginosa from cystic fibrosis patients. Future Microbiol 2015;10:599–611. [DOI] [PubMed] [Google Scholar]
- Mathee K, Narasimhan G, Valdes C et al. Dynamics of Pseudomonas aeruginosa genome evolution. P Natl Acad Sci USA 2008;105:3100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard KL, Grossman AD. Selective pressures to maintain attachment site specificity of integrative and conjugative elements. PLoS Genet 2013;9:e1003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen H, Hui K, Barraud N et al. The pathogenicity island encoded PvrSR/RcsCB regulatory network controls biofilm formation and dispersal in Pseudomonas aeruginosa PA14. Mol Microbiol 2013;89:450–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoia M, Gaillard M, Reinhard F et al. Stochasticity and bistability in horizontal transfer control of a genomic island in Pseudomonas. P Natl Acad Sci USA 2008;105:20792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki R, Bertelli C, Benaglio P et al. Comparative genome analysis of Pseudomonas knackmussii B13, the first bacterium known to degrade chloroaromatic compounds. Environ Microbiol 2015;17:91–104. [DOI] [PubMed] [Google Scholar]
- Miyazaki R, Minoia M, Pradervand N et al. Cellular variability of RpoS expression underlies subpopulation activation of an integrative and conjugative element. PLoS Genet 2012;8:e1002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki R, van der Meer JR. A dual functional origin of transfer in the ICEclc genomic island of Pseudomonas knackmussii B13. Mol Microbiol 2011a;79:743–58. [DOI] [PubMed] [Google Scholar]
- Miyazaki R, van der Meer JR. A new large-DNA-fragment delivery system based on integrase activity from an integrative and conjugative element. Appl Environ Microb 2013;79:4440–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki R, van der Meer JR. How can a dual oriT system contribute to efficient transfer of an integrative and conjugative element? Mobile Genetic Elements 2011b;1:82–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Zain Z, Turner SL, Cerdeño-Tárraga AM et al. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J Bacteriol 2004;186:8114–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Shoemaker NB, Gardner JF et al. Regulation of excision genes of the Bacteroides conjugative transposon CTnDOT. J Bacteriol 2005;187:5732–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller TA, Byrde SM, Werlen C et al. Genetic analysis of phenoxyalkanoic acid degradation in Sphingomonas herbicidovorans MH. Appl Environ Microb 2004;70:6066–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller TA, Werlen C, Spain J et al. Evolution of a chlorobenzene degradative pathway among bacteria in a contaminated groundwater mediated by a genomic island in Ralstonia. Environ Microbiol 2003;5:163–73. [DOI] [PubMed] [Google Scholar]
- O’Halloran JA, McGrath BM, Pembroke JT. The orf4 gene of the enterobacterial ICE, R391, encodes a novel UV-inducible recombination directionality factor, Jef, involved in excision and transfer of the ICE. FEMS Microbiol Lett 2007;272:99–105. [DOI] [PubMed] [Google Scholar]
- Osorio CR, Marrero J, Wozniak RAF et al. Genomic and functional analysis of ICEPdaSpa1, a fish-pathogen-derived SXT-related integrating conjugative element that can mobilize a virulence plasmid. J Bacteriol 2008;190:3353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Zhang J. High expression hampers horizontal gene transfer. Genome Biol Evol 2012;4:523–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Salyers AA. Characterization of the Bacteroides CTnDOT regulatory protein RteC. J Bacteriol 2011;193:91–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic G, Burrus V, Gintz B et al. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology 2004;150:759–74. [DOI] [PubMed] [Google Scholar]
- Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science 2005;307:1965–9. [DOI] [PubMed] [Google Scholar]
- Peed L, Parker AC, Smith CJ. Genetic and functional analyses of the mob operon on conjugative transposon CTn341 from Bacteroides spp. J Bacteriol 2010;192:4643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa O, Dagan T. Trends and barriers to lateral gene transfer in prokaryotes. Curr Opin Microbiol 2011;14:615–23. [DOI] [PubMed] [Google Scholar]
- Poulin-Laprade D, Burrus V. A lambda Cro-like repressor is essential for the induction of conjugative transfer of SXT/R391 elements in response to DNA damage. J Bacteriol 2015;197:3822–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin-Laprade D, Carraro N, Burrus V. The extended regulatory networks of SXT/R391 integrative and conjugative elements and IncA/C conjugative plasmids. Front Microbiol 2015;6:837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin-Laprade D, Matteau D, Jacques PE et al. Transfer activation of SXT/R391 integrative and conjugative elements: unraveling the SetCD regulon. Nucleic Acids Res 2015;43:2045–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart C, Celli J, Trieu-Cuot P. Conjugative transposition of Tn916-related elements from Enterococcus faecalis to Escherichia coli and Pseudomonas fluorescens. Antimicrobiol Agents Ch 1995;39:500–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradervand N, Delavat F, Sulser S et al. The TetR-type MfsR protein of the integrative and conjugative element ICEclc controls both a putative efflux system and initiation of ICE transfer. J Bacteriol 2014a;196:3971–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradervand N, Sulser S, Delavat F et al. An operon of three transcriptional regulators controls horizontal gene transfer of the Integrative and Conjugative Element ICEclc in Pseudomonas knackmussii B13. PLoS Genet 2014b;10:e1004441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeside C, Gaffe J, Deatherage DE et al. Large chromosomal rearrangements during a long-term evolution experiment with Escherichia coli. MBio 2014;5:e01377–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay JP, Major AS, Komarovsky VM et al. A widely conserved molecular switch controls quorum sensing and symbiosis island transfer in Mesorhizobium loti through expression of a novel antiactivator. Mol Microbiol 2013;87:1–13. [DOI] [PubMed] [Google Scholar]
- Ramsay JP, Sullivan JT, Jambari N et al. A LuxRI-family regulatory system controls excision and transfer of the Mesorhizobium loti strain R7A symbiosis island by activating expression of two conserved hypothetical genes. Mol Microbiol 2009;73:1141–55. [DOI] [PubMed] [Google Scholar]
- Ramsay JP, Sullivan JT, Stuart GS et al. Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol Microbiol 2006;62:723–34. [DOI] [PubMed] [Google Scholar]
- Ramsay JP, Tester LG, Major AS et al. Ribosomal frameshifting and dual-target antiactivation restrict quorum-sensing-activated transfer of a mobile genetic element. P Natl Acad Sci USA 2015;112:4104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravatn R. Horizontal transfer of a 105-kb integrative element harbouring the chlorocatechol degradation genes from Pseudomonas sp. strain B13. Thesis, Swiss Federal Institute of Technology Zürich, Zürich, 1998. [Google Scholar]
- Ravatn R, Studer S, Springael D et al. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J Bacteriol 1998a;180:4360–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravatn R, Studer S, Zehnder AJB et al. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J Bacteriol 1998b;180:5505–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravatn R, Zehnder AJB, van der Meer JR. Low-frequency horizontal transfer of an element containing the chlorocatechol degradation genes from Pseudomonas sp. strain B13 to Pseudomonas putida F1 and to indigenous bacteria in laboratory-scale activated–sludge microcosms. Appl Environ Microb 1998;64:2126–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard F, van der Meer JR. Microcolony growth assays. In: Timmis KN, de Lorenzo V, McGenity T et al. (eds) Handbook of Hydrocarbon and Lipid Microbiology, Vol. 5 Berlin Heidelberg: Springer, 2010, 3562–70. [Google Scholar]
- Reinhard F, Miyazaki R, Pradervand N et al. Cell differentiation to "mating bodies" induced by an integrating and conjugative element in free-living bacteria. Curr Biol 2013;23:255–9. [DOI] [PubMed] [Google Scholar]
- Reinhard F, van der Meer JR. Improved statistical analysis of low abundance phenomena in bimodal bacterial populations. PLoS One 2013;8:e78288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard F, van der Meer JR. Life history analysis of integrative and conjugative element activation in growing microcolonies of Pseudomonas. J Bacteriol 2014;196:1425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringwald K, Gardner J. The Bacteroides thetaiotaomicron protein Bacteroides host factor A participates in integration of the integrative conjugative element CTnDOT into the chromosome. J Bacteriol 2015;197:1339–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol 2009;17:251–8. [DOI] [PubMed] [Google Scholar]
- Rocco JM, Churchward G. The integrase of the conjugative transposon Tn916 directs strand- and sequence-specific cleavage of the origin of conjugal transfer, oriT, by the endonuclease Orf20. J Bacteriol 2006;188:2207–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet 2002;18:291–4. [DOI] [PubMed] [Google Scholar]
- Roos TE, van Passel MW. A quantitative account of genomic island acquisitions in prokaryotes. BMC Genomics 2011;12:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U et al. Gene regulation at the single-cell level. Science 2005;307:1962–5. [DOI] [PubMed] [Google Scholar]
- Rudy C, Taylor KL, Hinerfeld D et al. Excision of a conjugative transposon in vitro by the Int and Xis proteins of Tn916. Nucleic Acids Res 1997;25:4061–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy CK, Scott JR. Length of the coupling sequence of Tn916. J Bacteriol 1994;176:3386–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy CK, Scott JR, Churchward G. DNA binding by the Xis protein of the conjugative transposon Tn916. J Bacteriol 1997;179:2567–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydzewski K, Tlapak H, Niehaus IP et al. Identification and characterization of episomal forms of integrative genomic islands in the genus Francisella. Int J Med Microbiol 2015;305:874–80. [DOI] [PubMed] [Google Scholar]
- Sangwan N, Verma H, Kumar R et al. Reconstructing an ancestral genotype of two hexachlorocyclohexane-degrading Sphingobium species using metagenomic sequence data. ISME J 2014;8:398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S, Rakin A, Heesemann J. The Yersinia high-pathogenicity island (HPI): evolutionary and functional aspects. Int J Med Microbiol 2004;294:83–94. [DOI] [PubMed] [Google Scholar]
- Scott JR, Bringel F, Marra D et al. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol Microbiol 1994;11:1099–108. [DOI] [PubMed] [Google Scholar]
- Scott JR, Kirchman PA, Caparon MG. An intermediate in transposition of the conjugative transposon Tn916. P Natl Acad Sci USA 1988;85:4809–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selle K, Klaenhammer TR, Barrangou R. CRISPR-based screening of genomic island excision events in bacteria. P Natl Acad Sci USA 2015;112:8076–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senghas E, Jones JM, Yamamoto M et al. Genetic organization of the bacterial conjugative transposon Tn916. J Bacteriol 1988;170:245–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn L, Clerc O, Zanetti G et al. The stealthy superbug: the role of asymptomatic enteric carriage in maintaining a long-term hospital outbreak of ST228 methicillin-resistant Staphylococcus aureus. MBio 2016;7:doi: 10.1128/mBio.02039-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentchilo V, Czechowska K, Pradervand N et al. Intracellular excision and reintegration dynamics of the ICEclc genomic island of Pseudomonas knackmussii sp. strain B13. Mol Microbiol 2009;72:1293–306. [DOI] [PubMed] [Google Scholar]
- Sentchilo V, Mayer AP, Guy L et al. Community-wide plasmid gene mobilization and selection. ISME J 2013;7:1173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentchilo VS, Ravatn R, Werlen C et al. Unusual integrase gene expression on the clc genomic island of Pseudomonas sp. strain B13. J Bacteriol 2003;185:4530–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentchilo VS, Zehnder AJB, van der Meer JR. Characterization of two alternative promoters and a transcription regulator for integrase expression in the clc catabolic genomic island of Pseudomonas sp. strain B13. Mol Microbiol 2003;49:93–104. [DOI] [PubMed] [Google Scholar]
- Serfiotis-Mitsa D, Roberts GA, Cooper LP et al. The Orf18 gene product from conjugative transposon Tn916 is an ArdA antirestriction protein that inhibits type I DNA restriction-modification systems. J Mol Biol 2008;383:970–81. [DOI] [PubMed] [Google Scholar]
- Shoemaker NB, Barber RD, Salyers AA. Cloning and characterization of a Bacteroides conjugal tetracycline-erythromycin resistance element by using a shuttle cosmid vector. J Bacteriol 1989;171:1294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker NB, Vlamakis H, Hayes K et al. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl Environ Microb 2001;67:561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker NB, Wang GR, Salyers AA. NBU1, a mobilizable site-specific integrated element from Bacteroides spp., can integrate nonspecifically in Escherichia coli. J Bacteriol 1996a;178:3601–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker NB, Wang GR, Salyers AA. The Bacteroides mobilizable insertion element, NBU1, integrates into the 3΄ end of a Leu-tRNA gene and has an integrase that is a member of the lambda integrase family. J Bacteriol 1996b;178:3594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker NB, Wang GR, Stevens AM et al. Excision, transfer, and integration of NBU1, a mobilizable site-selective insertion element. J Bacteriol 1993;175:6578–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidt H, Song D, van Der Oost J et al. Random transposition by Tn916 in Desulfitobacterium dehalogenans allows for isolation and characterization of halorespiration-deficient mutants. J Bacteriol 1999;181:6882–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie C, Garcillan-Barcia MP, Francia MV et al. Mobility of plasmids. Microbiol Mol Biol R 2010;74:434–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits WK, Grossman AD. The transcriptional regulator Rok binds A+T-rich DNA and is involved in repression of a mobile genetic element in Bacillus subtilis. PLoS Genet 2010;6:e1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nat Rev Genet 2015;16:472–82. [DOI] [PubMed] [Google Scholar]
- Springael D, Peys K, Ryngaert A et al. Community shifts in a seeded 3-chlorobenzoate degrading membrane biofilm reactor: indications for involvement of in situ horizontal transfer of the clc-element from inoculum to contaminant bacteria. Environ Microbiol 2002;4:70–80. [DOI] [PubMed] [Google Scholar]
- Stevens AM, Shoemaker NB, Li LY et al. Tetracycline regulation of genes on Bacteroides conjugative transposons. J Bacteriol 1993;175:6134–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrs MJ, Poyart-Salmeron C, Trieu-Cuot P et al. Conjugative transposition of Tn916 requires the excisive and integrative activities of the transposon-encoded integrase. J Bacteriol 1991;173:4347–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YA, He P, Clewell DB. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob Agents Ch 1992;36:769–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suel GM, Garcia-Ojalvo J, Liberman LM et al. An excitable gene regulatory circuit induces transient cellular differentiation. Nature 2006;440:545–50. [DOI] [PubMed] [Google Scholar]
- Sullivan JT, Brown SD, Ronson CW. The NifA-RpoN regulon of Mesorhizobium loti strain R7A and its symbiotic activation by a novel LacI/GalR-family regulator. PLoS One 2013;8:e53762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JT, Patrick HN, Lowther WL et al. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. P Natl Acad Sci USA 1995;92:8985–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JT, Ronson CW. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. P Natl Acad Sci USA 1998;95:5145–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JT, Trzebiatowski JR, Cruickshank RW et al. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J Bacteriol 2002;184:3086–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutanto Y, DiChiara JM, Shoemaker NB et al. Factors required in vitro for excision of the Bacteroides conjugative transposon, CTnDOT. Plasmid 2004;52:119–30. [DOI] [PubMed] [Google Scholar]
- Sutanto Y, Shoemaker NB, Gardner JF et al. Characterization of Exc, a novel protein required for the excision of Bacteroides conjugative transposon. Mol Microbiol 2002;46:1239–46. [DOI] [PubMed] [Google Scholar]
- Taylor KL, Churchward G. Specific DNA cleavage mediated by the integrase of conjugative transposon Tn916. J Bacteriol 1997;179:1117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A. Stochastic gene expression in fluctuating environments. Genetics 2004;167:523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Lee CA, Grossman AD. A conserved helicase processivity factor is needed for conjugation and replication of an integrative and conjugative element. PLoS Genet 2013;9:e1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari A, Ray JC, Narula J et al. Bistable responses in bacterial genetic networks: designs and dynamical consequences. Math Biosci 2011;231:76–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Bobay LM, Rocha EP. The chromosomal accommodation and domestication of mobile genetic elements. Curr Opin Microbiol 2014;22:22–9. [DOI] [PubMed] [Google Scholar]
- Toussaint A, Merlin C, Monchy S et al. The biphenyl- and 4-chlorobiphenyl-catabolic transposon Tn4371, a member of a new family of genomic islands related to IncP and Ti plasmids. Appl Environ Microb 2003;69:4837–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuru T, Kawai M, Mizutani-Ui Y et al. Evolution of paralogous genes: reconstruction of genome rearrangements through comparison of multiple genomes within Staphylococcus aureus. Mol Biol Evol 2006;23:1269–85. [DOI] [PubMed] [Google Scholar]
- van Belkum A, Soriaga LB, LaFave MC et al. Phylogenetic distribution of CRISPR-Cas systems in antibiotic-resistant Pseudomonas aeruginosa. MBio 2015;6:e01796–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer JR, Sentchilo VS. Genomic islands and evolution of catabolic pathways in bacteria. Curr Opin Biotechnol 2003;14:248–54. [DOI] [PubMed] [Google Scholar]
- van der Meer JR, de Vos WM, Harayama S et al. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev 1992;56:677–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma H, Kumar R, Oldach P et al. Comparative genomic analysis of nine Sphingobium strains: insights into their evolution and hexachlorocyclohexane (HCH) degradation pathways. BMC Genomics 2014;15:1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor MK, Tschäpe H, Mekalanos JJ. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol 1996;178:4157–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GR, Shoemaker NB, Jeters RT et al. CTn12256, a chimeric Bacteroides conjugative transposon that consists of two independently active mobile elements. Plasmid 2011;66:93–105. [DOI] [PubMed] [Google Scholar]
- Wang Y, Rotman ER, Shoemaker NB et al. Translational control of tetracycline resistance and conjugation in the Bacteroides conjugative transposon CTnDOT. J Bacteriol 2005;187:2673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shoemaker NB, Salyers AA. Regulation of a Bacteroides operon that controls excision and transfer of the conjugative transposon CTnDOT. J Bacteriol 2004;186:2548–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JL, Salyers AA. The small RNA RteR inhibits transfer of the Bacteroides conjugative transposon CTnDOT. J Bacteriol 2012;194:5228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JL, Salyers AA. Regulation of CTnDOT conjugative transfer is a complex and highly coordinated series of events. MBio 2013;4:e00569–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel AF, Ressina S, Kolbe-Busch S et al. Species diversity of environmental GIM-1-producing bacteria collected during a long-term outbreak. Appl Environ Microb 2016;82:3605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle G, Shoemaker NB, Salyers AA. Characterization of genes involved in modulation of conjugal transfer of the Bacteroides conjugative transposon CTnDOT. J Bacteriol 2002;184:3839–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GG, Murray NE. Restriction and modification systems. Annu Rev Genet 1991;25:585–627. [DOI] [PubMed] [Google Scholar]
- Wood MM, Dichiara JM, Yoneji S et al. CTnDOT integrase interactions with attachment site DNA and control of directionality of the recombination reaction. J Bacteriol 2010;192:3934–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RA, Fouts DE, Spagnoletti M et al. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet 2009;5:e1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RA, Waldor MK. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet 2009;5:e1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 2010;8:552–63. [DOI] [PubMed] [Google Scholar]
- Wright LD, Grossman AD. Autonomous replication of the conjugative transposon Tn916. J Bacteriol 2016;198:3355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright LD, Johnson CM, Grossman AD. Identification of a single strand origin of replication in the Integrative and Conjugative Element ICEBs1 of Bacillus subtilis. PLoS Genet 2015;11:e1005556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Skinner SO, Zong C et al. Decision making at a subcellular level determines the outcome of bacteriophage infection. Cell 2010;141:682–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Rho M, Tang H et al. CRISPR-Cas systems target a diverse collection of invasive mobile genetic elements in human microbiomes. Genome Biol 2013;14:R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Laing C, Steele M et al. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics 2007;8:121. [DOI] [PMC free article] [PubMed] [Google Scholar]