Abstract
Zygosaccharomyces bailii is one of the most problematic spoilage yeast species found in the food and beverage industry particularly in acidic products, due to its exceptional resistance to weak acid stress. This article describes the annotation of the genome sequence of Z. bailii IST302, a strain recently proven to be amenable to genetic manipulations and physiological studies. The work was based on the annotated genomes of strain ISA1307, an interspecies hybrid between Z. bailii and a closely related species, and the Z. bailii reference strain CLIB 213T. The resulting genome sequence of Z. bailii IST302 is distributed through 105 scaffolds, comprising a total of 5142 genes and a size of 10.8 Mb. Contrasting with CLIB 213T, strain IST302 does not form cell aggregates, allowing its manipulation in the laboratory for genetic and physiological studies. Comparative cell cycle analysis with the haploid and diploid Saccharomyces cerevisiae strains BY4741 and BY4743, respectively, suggests that Z. bailii IST302 is haploid. This is an additional trait that makes this strain attractive for the functional analysis of non-essential genes envisaging the elucidation of mechanisms underlying its high tolerance to weak acid food preservatives, or the investigation and exploitation of the potential of this resilient yeast species as cell factory.
Keywords: Zygosaccharomyces bailii, genome sequence, food spoilage yeasts, weak acid tolerance, cellular aggregation
Annotated genome sequence of Zygosaccharomyces bailii IST302, a haploid strain amenable to genetic manipulations and physiological studies, and comparative genomic analysis with Z. bailii and Z. bailii-derived hybrid strains, Saccharomyces cerevisiae and Z. rouxii.
INTRODUCTION
Zygosaccharomyces bailii is found among the spoilage yeasts that frequently affect acidic foods and beverages, particularly mayonnaise, salad dressings, soft drinks, fruit concentrates and dairy products (reviewed in Stratford 2006; Sá-Correia et al. 2014). This yeast species ability to cause spoilage results from its remarkable capacity to tolerate stress induced by weak acids widely used as fungistatic preservatives, such as acetic, benzoic and sorbic acids (Ferreira, Loureiro-Dias and Loureiro 1997; Stratford et al.2013). In fact, Z. bailii may proliferate in the presence of weak acid concentrations above those legally permitted in food products (Stratford 2006). Moreover, Z. bailii is able to tolerate relatively high ethanol concentrations (Kalathenos, Sutherland and Roberts 1995) and high temperatures (Martorell et al.2007), and to vigorously ferment sugars (Thomas and Davenport 1985), being considered one of the most significant threats in wine industry (Loureiro and Malfeito-Ferreira 2003). Like other members of the Zygosaccharomyces genus, Z. bailii is fructophilic, i.e. prefers fructose over glucose when both sugars are present in the growth medium (Sousa-Dias et al.1996; Pina et al.2004). Therefore, acidic food and beverages that contain fructose are at particular risk of spoilage, given that Z. bailii specific growth rates and ethanol production are higher during growth in the presence of fructose compared with glucose. Although Z. bailii is unable to directly metabolize sucrose, acidic products containing this sugar are also susceptible to spoilage by this yeast species given that, at low pH, sucrose can be hydrolyzed in fructose and glucose (Pitt and Hocking 2009). These traits of Z. bailii physiology result in annual losses of millions of dollars (Pitt and Hocking 2009) making this species a serious concern in food industry. Regardless of its activity as spoilage yeast, Z. bailii has also gained attention for its potential in biotechnological processes, being proposed as a new cell factory for the production of organic acids and heterologous proteins (Branduardi et al.2004; Dato et al.2010).
Although there are plasmid vectors available for Z. bailii genetic manipulation (Branduardi, Dato and Porro 2014), the genetic engineering of this species has been hindered by the lack of stable haploid strains (Mollapour and Piper 2001; Rodrigues et al.2003) and, to date, only one auxotrophic mutant was constructed (Dato et al.2010). However, the parental strain of the derived mutant, previously considered as Z. bailii, was taxonomically reallocated to Z. parabailii (Suh et al.2013). Moreover, only recently it was possible to have access to the genome sequence of a Z. bailii strain, the reference strain CLIB 213T (Galeote et al.2013) and to Z. bailii-derived hybrid strain ISA1307 (Mira et al.2014). This hybrid strain has been used in several studies aiming at understanding Z. bailii physiology, in particular the mechanisms underlying acetic acid tolerance and toxicity (Sousa et al.1998; Rodrigues et al.2001; Guerreiro, Mira and Sá-Correia 2012; Palma et al.2015), but following its genome sequencing and annotation ISA1307 emerged as an interspecies hybrid between Z. bailii and a closely related species (Mira et al.2014). On the other hand, Z. bailii CLIB 213T is extremely difficult to manipulate in the laboratory mainly due to its strong aggregation phenotype and, for this reason, strains CBS 685 (NCYC 563) and NCYC 1766 were suggested as better representatives of the species (James and Stratford 2011). More recently, our laboratory has been working with Z. bailii IST302, which was found to be more amenable to genetic and physiological manipulations than strain CLIB 213T (Palma et al.2015, 2017). Therefore, the sequencing of its genome sequence was considered the next step.
In this work, we provide molecular evidences supporting the taxonomic identification of strain IST302 as Z. bailii, as well as its genome sequence and annotation and comparative analysis with CLIB 213T and other relevant yeast species. This study also envisaged the confirmation of the haploid nature of the genome, the characterization of relevant tolerance phenotypes and the search for potential molecular targets involved in Z. bailii remarkable weak acid tolerance and the aggregation phenotype. Collectively, the information gathered in this study points Z. bailii IST302 strain as highly attractive for the functional analysis of non-essential Z. bailii genes and the elucidation of mechanisms underlying its high tolerance to acetic acid and other weak acid food preservatives or to the investigation of Z. bailii potential as cell factory.
MATERIALS AND METHODS
Strains and growth medium
The prototrophic strains Zygosaccharomyces bailii IST302 (Palma et al.2015), the neotype strain Z. bailii ATCC58445T (= CLIB 213T) (Galeote et al.2013) and strain ISA1307, a Z. bailii-derived interspecies hybrid strain, were used in this work. Saccharomyces cerevisiae BY4741 (genotype MATa; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0) and BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0) were acquired from the EUROSCARF collection. Yeast strains were maintained in solid rich YPD growth medium that contains, per liter, 2 % glucose (Merck, Kenilworth, New Jersey), 1 % yeast extract (Difco Laboratories, Detroit, Michigan) and 2% peptone (Difco Laboratories, Detroit, Michigan) and 2% agar. For susceptibility assays, yeast strains ISA1307, IST302 and BY4741 were cultured in MM4 liquid medium that contains, per litre: 1.7 g yeast nitrogen base without amino acids or (NH4)2SO4 (Difco Laboratories, Detroit, Michigan), 20 g glucose (Merck, Kenilworth, New Jersey), 2.65 g (NH4)2SO4 (Merck), 20 mg methionine, 20 mg histidine, 60 mg leucine and 20 mg uracil (all from Sigma-Aldrich, St. Louis, Missouri).
Taxonomic identification of strain IST302
The nucleotide sequences of the genes from strain IST302 encoding the RNA polymerase II largest subunit Rpb1 and the RNA polymerase II second-largest subunit Rpb2, previously considered relevant for the identification of Z. bailii species among closely related species such as Z. parabailii and Z. pseudobailii (Suh et al.2013), were compared with the nucleotide sequences of RPB1 and RPB2 from several Z. bailii, Z. parabailii and Z. pseudobailii strains as described previously (Suh et al.2013). The nucleotide sequences of RPB1 and RPB2 genes from strain IST302 were obtained by whole-genome sequencing, as described hereafter.
Cell cycle analysis and quantification of Zygosaccharomyces bailii IST302 total genomic DNA by flow cytometry
Zygosaccharomyces bailii IST302 cells were batch cultured in YPD growth medium, at 30°C, until mid-exponential phase (OD600nm = 0.6±0.1). A total of 107 cells were harvested by centrifugation, washed twice with distilled water and fixed at least one night in 1 mL of 70 % ethanol (vol/vol). The remaining cell culture was induced to arrest at the G1 phase with 8-hydroxyquinoline (Sigma-Aldrich, St. Louis, Missouri) at a final concentration of 100 μg/mL for 24 h or at the G2 phase with nocodazole (Sigma-Aldrich, St. Louis, Missouri) at a final concentration of 5 μg/mL for 3 h. Arrested cells were harvested, washed and fixed with ethanol, as previously described. Quantification of total genomic DNA from Z. bailii IST302 by flow cytometry was performed using a SYBR Green I-based staining protocol, as described before (Fortuna et al.2000). Cells fixed with ethanol were collected by centrifugation, washed with 50 mM of sodium citrate buffer (pH 7.5) and resuspended in 750 μL of this same buffer supplemented with 250 μL of RNAse A (1 mg/mL) (Sigma-Aldrich, St. Louis, Missouri). After 1 h of incubation at 50°C, 50 μL of proteinase K (20 mg/mL) (NZYtech, Lisbon, Portugal) were added to the cell suspension and the mixture was incubated at 50°C for another hour. Cells were subsequently stained overnight at 4°C using SYBR® Green I at a final concentration of 500-fold dilution of the commercial SYBR® Green I (Life Technologies, Carlsbad, California). Triton X-100 was added to the stained samples at a final concentration of 0.25% (v/v). Samples were vortexed, sonicated in a Branson Sonifier 250 (3-4 pulses with at output power of 3 and 30% duty cycle) and analyzed in a FACScalibur (Becton Dickson, Franklin Lakes, New Jersey) flow cytometer. A minimum of 50000 cells per sample were acquired at low flow rate using CellQuest software (Becton Dickson, Franklin Lakes, New Jersey). Analysis of the acquired data was performed using the FlowJo® v10.0.8 software. The fluorescence intensities of the cell cycle peak G0/G1 for S. cerevisiae BY4741 and BY4743 were used to build a calibration curve in order to estimate the genome size of Z. bailii IST302 strain. Fluorescence intensities for the hybrid strain ISA1307 were used for comparison purposes.
Karyotyping of Zygosaccharomyces bailii IST302 strain
DNA for pulsed field gel electrophoresis (PFGE) was prepared in plugs as previously described (Maringele and Lydall 2006). Strains IST302 and CLIB 213T were cultivated overnight at 30°C at 250 rpm in YPD growth medium. Yeast cells were harvested by centrifugation in order to obtain a cell pellet of ∼50 μL volume. Cell pellets were washed twice with 0.05 M EDTA, pH 8.0 and resuspended in 100 μL of 0.05 M EDTA, pH 8.0. Plugs were formed by mixing the suspension of cells with a total of 50 μL of a Zymolyase solution [46 μL SCE buffer (1 M sorbitol, 0.1 M sodium citrate, and 60 mM EDTA), 1.5 μL zymolyase (NZYtech, Lisbon, Portugal) and 2.5 μL β-mercaptoethanol] was added to each sample followed by the addition of 300 μL of low-melting point agarose solution (1% low melting point agarose in 10 mL EDTA 0,125 mM, pH 7.0). The mixture was pipetted into wells from the plug molds and placed at 4°C during 30 min. The plugs were then carefully removed and then incubated overnight at 37°C, 50 rpm, in ETB buffer (EDTA 0.05 M, pH 8.0, Tris-HCl 0.1 M, pH 8.0 and 5 % (vol/vol) β-Mercaptoethanol). After this incubation step, plugs were washed three times in TE buffer (10 mM Tris, pH 8.0 and 1 mM EDTA, pH 8.0) and incubated overnight at 37°C in a solution containing 0.05 M EDTA, 1 mg/mL of proteinase K (NZYtech, Lisbon, Portugal), 1 mg/mL RNAse (Sigma-Aldrich, St. Louis, Missouri) and 1 % sodium-N-lauryl sarcosinate. The plugs were washed with EDTA 50 mM at 37°C, 100 rpm, during 15 mins and incubated at room temperature for 1 h in TE buffer, pH 8.0. PFGE was performed in a CHEF- Pharmacia LKB Gene Navigator system. PFGE gels were run in 0.5 % Tris borate–EDTA buffer at 13°C with a voltage of 3 V/cm and switch times of 600 s for 48 h and 300 s for 96 h in a 1 % pulsed-field gel agarose (NZYtech, Lisbon, Portugal).
Genome sequencing, assembly and annotation
Zygosaccharomyces bailii IST302 genome sequencing and assembly was performed as described previously (Mira et al.2014) using a whole-genome shotgun approach that explored paired-end Illumina sequencing (Illumina Hiseq 2000 system, CD Genomics, New York, USA). Short reads were assembled using SOAPdenovo (http://soap.genomics.org.cn) (Li et al.2010). The obtained scaffolds were sequentially ordered based on their level of synteny to the genomes of Z. rouxii CBS 732T, S. cerevisiae S288c, Z. bailii-derived hybrid strain ISA1307 and Z. bailii CLIB 213T. To predict genes on the scaffolds, two ab initio gene predictor algorithms were used: GeneMark-S and GenMark-ES version 2.3 (Ter-Hovhannisyan et al.2008). Gene models differently predicted by the algorithms were manually curated based on the structure obtained for Z. rouxii CBS 732T, S. cerevisiae S288c, Z. bailii-derived hybrid strain ISA1307 and Z. bailii CLIB 213T homologs. The genomes were analyzed using the PEDANT system (Walter et al.2009) to allow comparative feature analysis. To avoid misleading ortholog information based on similarity and bidirectional best hits, QuartetS (Yu et al. 2011) was applied to retrieve a reliable ortholog matrix, which was used for comparative analysis of the genes encoding multidrug/multixenobiotic resistance (MDR/MXR) transporters, of the genes involved in life cycle and meiosis and of the genes involved in cellular aggregation in Z. bailii IST302, Z. bailii CLIB 213T, Z. rouxii CBS732T and S. cerevisiae S288c. The identification of the centromeres was performed manually based on previously identified point centromere sequences and on the collinear conservation of flanking genes of hemiascomycetous yeasts, in particular of Z. rouxii (Pribylova et al.2007; Souciet et al.2009).
The genome sequence and annotation of Z. bailii IST302 has been deposited in ENA (http://www.ebi.ac.uk/ena/data/view/FUGC01000001-FUGC01000105).
Susceptibility assays
Susceptibility of Z. bailii IST302, ISA1307 hybrid strain and S. cerevisiae BY4741 to several growth inhibitory compounds was assessed by spot assays. Yeast cells were grown in MM4 medium at the appropriate pH until exponential phase (OD600nm of 0.5 ± 0.05) and then reinoculated at an OD600nm of 0.05, in 50 mL of fresh medium. When the cultures reached an OD600nm of 0.5 ± 0.05, cells were resuspended in sterile water to obtain suspensions with ∼5 × 105 cell/mL. These cell suspensions and three subsequent dilutions (1:5; 1:10; 1:20) were applied as 4 μL spots on MM4 plates supplemented with adequate concentrations of acetic, benzoic, formic, lactic, propionic or sorbic acids; of the herbicides 2,4-dichlorophenoxyacetic acid (2,4-D), 2-methyl-4-chlorophenoxyacetic acid (MCPA) or alachlor; and of the fungicides itraconazole, fluconazole, miconazole, tioconazole, clotrimazole or mancozeb. Agar plates were incubated at 30°C for 3 days. Due to the different pKa of the acids, yeast susceptibility to acetic, benzoic, sorbic and propionic acids was tested in MM4 medium at pH 4.0, whereas susceptibility to formic and lactic acids was tested in MM4 at pH 3.5.
Sporulation of Zygosaccharomyces bailii IST302
Zygosaccharomyces bailii IST302 cells were grown in a presporulation medium (0.8% yeast extract, 0.3% peptone, 10% glucose and 2% agar) during 2 days before being transferred to sporulation media (0.3% malt extract, 0.5% peptone and 2% agar). The formation of spores was observed on a Zeiss ® Axioplan microscope (×1000 magnification) after 2 and 4 days of incubation at 30°C.
Microscopic observation of the morphology of yeast colonies and cells
Yeast cells were plated in YPD with a targeted density of 20–50 CFU/plate. Colonies were observed and photographed in a Carl Zeiss Stemi 2000-C stereomicroscope after 4–5 days of growth at 30°C.
Cells of Z. bailii IST302, Z. bailii CLIB 213T and of ISA1307 hybrid strain were cultured in YPD medium for 24 h and used to inoculate a fresh YPD medium with an initial OD600nm of 0.05±0.01. After 24 h of growth, cells were harvested in 1.5 mL tubes by centrifugation at 8000 rpm and suspended in 0.05 M EDTA (pH 8.0), followed by vigorous agitation. These cells were observed on a Zeiss ® Axioplan microscope (×400 magnification). Cells suspended in 0.05 M EDTA (pH 8.0) were subsequently washed twice with sterile water, suspended in 0.01 M CaCl2, vigorously mixed and finally observed under the microscope.
RESULTS AND DISCUSSION
Taxonomic identification of strain IST302
The molecular differences registered among Zygosaccharomyces bailii and closely related species (Suh et al.2013), together with the genomic complexity of the hybrid strain ISA1307 (Mira et al.2014) that was first considered as Z. bailii, support the notion of the remarkable genomic diversity among Z. bailii and Z. bailii closely related species. This fact recommended the careful examination of the taxonomic position and genomic characterization of IST302 strain. The preliminary taxonomic identification of this strain as Z. bailii was based on the comparison of the large subunit 26S ribosomal DNA partial sequence with other DNA sequences from Z. bailii strains using the Basic Local Alignment Search Tool (BLAST) of the National Center for Biotechnology Information (NCBI) (Palma et al.2015). However, considering the reallocation of yeast strains formerly identified as Z. bailii to the new species Z. parabailii and Z. pseudobailii (Suh et al.2013), we have also compared the partial sequences of RBP1 and RBP2 genes from strain IST302 with their homologs from several strains of the three species, as recommended by Suh et al. (2013). Results show that nucleotide sequence variation percentage is low when RBP1 and RBP2 nucleotide sequences from strain IST302 are compared with the corresponding sequences from Z. bailii strains (0.1%–0.7%), whereas significant sequence variation is observed compared with Z. parabailii (3.8%–4.1%) and Z. pseudobailii (5.8%–6.8%) strains (Fig. 1). Collectively, these results confirm IST302 strain as Z. bailii.
Figure 1.
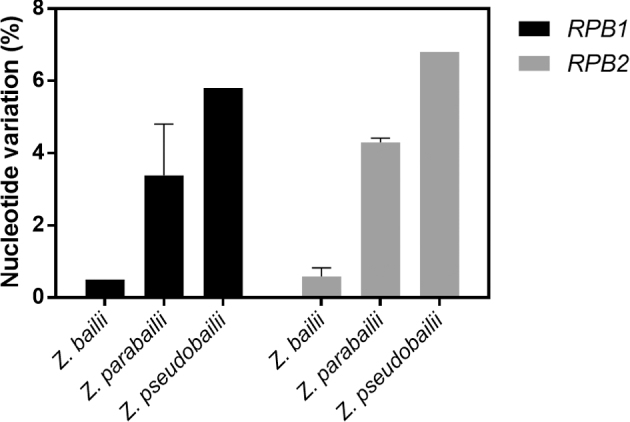
Nucleotide variation in RBP1 and RBP2 genes from strain IST302 when compared with the partial sequence of RBP1 and RBP2 from Z. bailii, Z. parabailii and Z. pseudobailii strains. Nucleotide variations were calculated using EMBOSS Water local alignment tool from EMBL-EBI packages. Values are presented as percentage of nucleotide variations and are the mean of results obtained by comparing RBP1 and RBP2 sequences from strain IST302 (ZBIST_2664 and ZBIST_4878, respectively) with their homologs from six strains of Z. bailii, seven strains of Z. parabailii and two strains of Z. pseudobailii according to Suh et al. (2013).
Tolerance of Zygosaccharomyces bailii IST302 to weak acids, drugs and pesticides
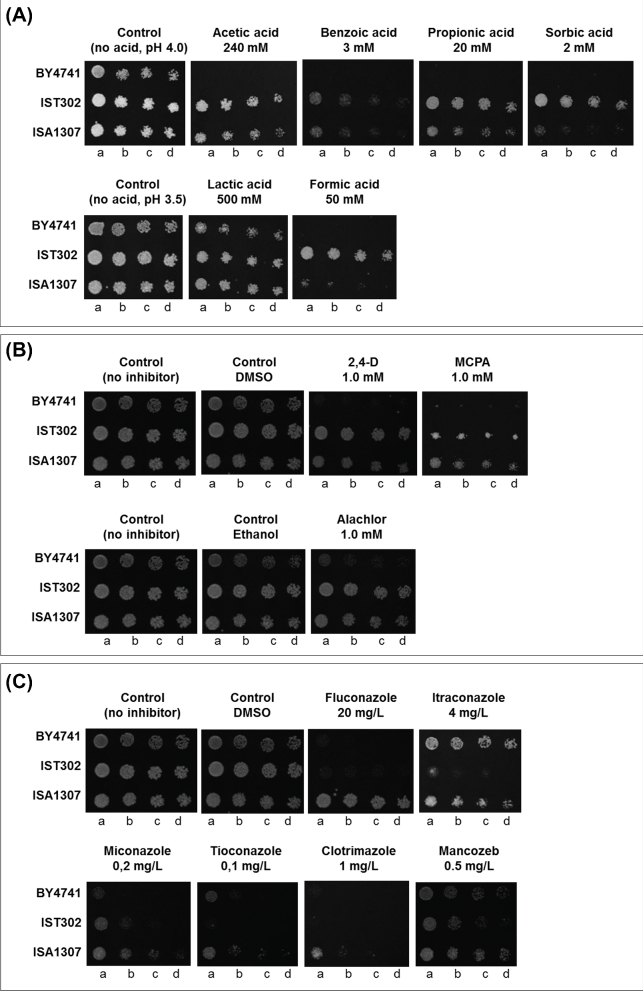
The tolerance of Z. bailii IST302 to several compounds was assessed by spot assays (Fig. 2). Compared with Saccharomyces cerevisiae BY4741, Z. bailii IST302 is much more tolerant to weak acids (acetic, benzoic, sorbic, propionic and formic acids), to the lipophilic weak acid herbicides 2,4-D (2,4-D-dichlorophenoxyacetic acid) and MCPA (methyl-chlorophenoxyacetic acid) and also to the herbicide alachlor, but not to the antifungals tested (Fig. 2A and B). In general, Z. bailii IST302 is slightly more tolerant to weak acids than the hybrid strain ISA1307. Interestingly, the hybrid strain ISA1307 is significantly more tolerant to all the antifungal drugs tested than Z. bailii IST302 and S. cerevisiae BY4741 (Fig. 2C). The standardization of the number of viable/total cells was not possible for Z. bailii CLIB 213T due to the difficulty to calculate viable cell concentration or culture optical density. For this reason, only the susceptibility of Z. bailii IST302, the hybrid strain ISA1307 and S. cerevisiae BY4741 to several compounds could be compared under equivalent experimental conditions.
Figure 2.
Zygosaccharomyces bailii IST302 is more tolerant to weak acid food preservatives and herbicides, but not to antifungals, when compared with S. cerevisiae BY4741. Growth of Z. bailii IST302, S. cerevisiae BY4741 and the hybrid strain ISA1307 was compared by spot assays in MM4 medium supplemented, or not, with the appropriate concentrations of (A) weak organic acids, (B) herbicides and (C) antifungal drugs. Three control plates are shown, with and without the solvents used to dissolve the different drugs when necessary. Cell suspensions with ∼5 × 105 cells/mL (lane a) and subsequent dilutions of 1:5, 1:10 and 1:20 (lanes b, c and d, respectively) were spotted onto the surface of MM4 solid medium. The images shown were taken after 3 days of incubation at 30°C and are representative of at least three independent experiments.
Estimation of total DNA content and karyotyping of Zygosaccharomyces bailii IST302
The chromosomal profiles of Z. bailii IST302 and of the reference strain Z. bailii CLIB 213T were compared by PFGE (Fig. 3). Karyotyping showed differences in the number and size of the chromosomes between Z. bailii IST302 and CLIB 213T strains. Following the separation of chromosomes by PFGE, strain IST302 originated a total of seven DNA bands, whereas only six DNA bands are observed for CLIB 213T. Based on the molecular size of chromosomes from the marker species Hansenula wingei, it was possible to estimate the total DNA content of Z. bailii IST302 and CLIB 213T as 11.65 and 9.5 Mb, respectively. Given that the DNA content calculated from CLIB 213T is lower than the DNA sequenced (Galeote et al.2013), it is possible that two comigrating chromosomes could have not been separated using the PFGE conditions tested. In fact, the chromosomal DNA band of 1.8 Mb is more intense than expected for a single band and possibly represents two chromosomes of the same size.
Figure 3.
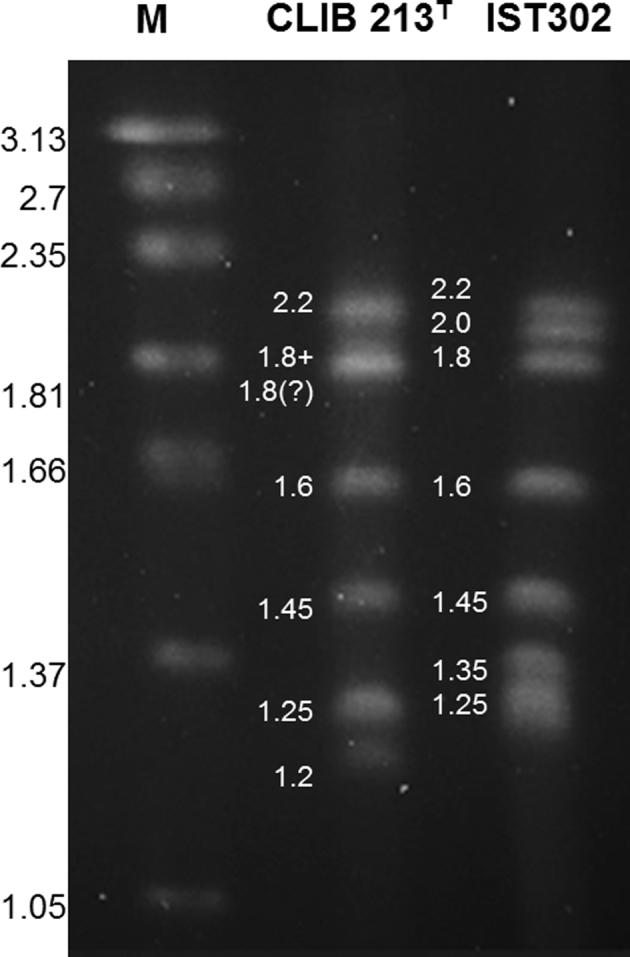
Karyotype profiles of Z. bailii IST302 and CLIB 213T strains. Separation of the chromosomes was performed by PFGE as described in Materials and Methods section. The size of Z. bailii IST302 and CLIB 213T chromosomes was estimated from a comparison with the size of the chromosomes from Hansenula wingei that were used as the molecular size standards (M).
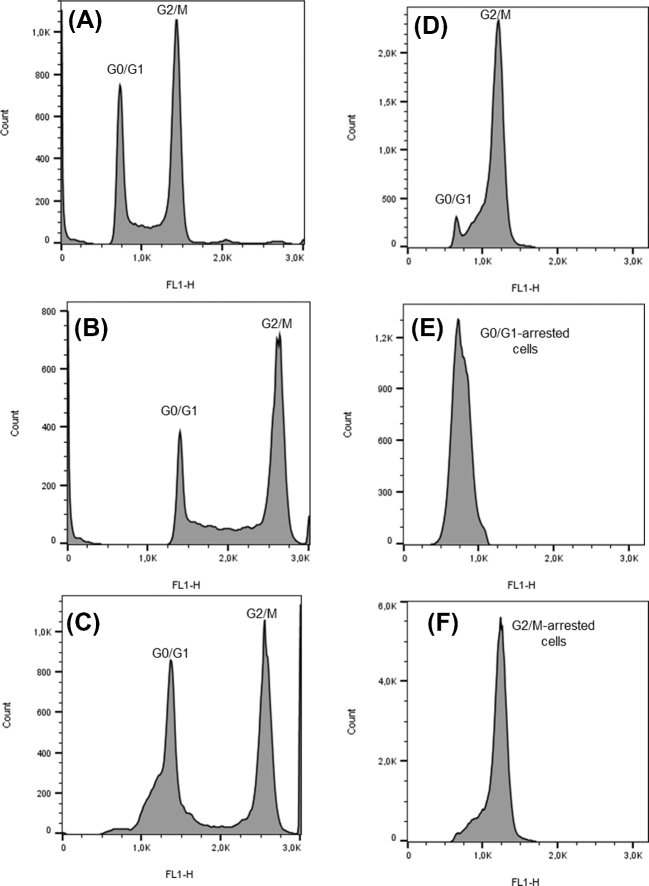
Flow cytometry was also used to estimate the genome size of Z. bailii IST302, as well as its ploidy. Saccharomyces cerevisiae BY4741 was used as the haploid reference strain and S. cerevisiae BY4743 as the diploid reference. The hybrid strain ISA1307, whose genome size and ploidy were recently estimated (Mira et al.2014), was also included in this analysis for comparison purposes. Zygosaccharomyces bailii CLIB 213T was excluded from this analysis due to its strong cellular aggregation phenotype. As anticipated, the fluorescent light intensity of G0/G1 or G2/M phase cells from the reference haploid strain S. cerevisiae BY4741 is half the fluorescent light intensity of the diploid BY4743 strain, since strain BY4741 possesses half the DNA content of strain BY4743 (Fig. 4A and B). Flow cytometry data from strain ISA1307 (Fig. 4C) shows two fluorescence peaks (G0/G1 and G2/M) with a DNA content similar to one of the diploid S. cerevisiae BY4743 (Fig. 4B). Interestingly, strain IST302 (Fig. 4D) shows two fluorescence peaks (G0/G1 and G2/M) with a DNA content similar to the one observed for S. cerevisiae BY4741 haploid strain (Fig. 4A). Given the unexpected high proportion of Z. bailii IST302 cells in G2/M phase, it was decided to treat cells with 8-hydroxyquinoline and nocodazole in order to arrest cells in G0/G1 (Fig. 4E) and in G2/M (Fig. 4F) cell cycle phases, respectively, thus confirming the fluorescent peak corresponding to each phase. Results suggest that Z. bailii IST302 has a haploid DNA content (Fig. 4D). Based on the fluorescent intensity of G0/G1 subpopulation from S. cerevisiae haploid and diploid strains, the size the genome of Z. bailii IST302 was estimated as having ∼11 Mb. The life cycle of the strain will be discussed further in this work.
Figure 4.
Cell cycle analysis histograms of S. cerevisiae BY4741 (haploid) (A) and BY4743 (diploid) (B), of the hybrid strain ISA1307 (C) and of Z. bailii IST302 (D–F). Fluorescent intensities of G0/G1 peaks of the cell cycle histograms were estimated by flow cytometry. The mean fluorescence values (FL1-H) obtained for S. cerevisiae haploid (A) and diploid (B) strains were used to build a calibration curve that was used to estimate the size of the genome of Z. bailii IST302 (D). Zygosaccharomyces bailii hybrid strain ISA1307 was used for comparison purposes (C). Zygosaccharomyces bailii IST302 cells were arrested in G0/G1 with 8-hydroxyquinoline (E) or in G2/M with nocodazole (F) in order to confirm the G0/G1 and G2/M peaks and determine the ploidy of the strain.
Assembly and annotation of Zygosaccharomyces bailii IST302 genome sequence
The genome sequence of Z. bailii IST302 was obtained by paired-end Illumina sequencing. A total of 13 million reads were acquired and assembled into 105 scaffolds, resulting in an overall sequence coverage of ×120. A summary of genome assembly statistics and its comparison with Z. bailii CLIB 213T and the Z. bailii-derived interspecies hybrid ISA1307 is presented in Table 1. The sum of all scaffolds size is 10 772 966 bp, which corresponds to 98% of the genome size estimated by flow cytometry and 93% of the genome size estimated by PFGE. The annotation of Z. bailii IST302 genome sequence was based on the combination of two ab initio gene predictor algorithms and the gene structure of Z. rouxii CBS 732T, S. cerevisiae S288c, Z. bailii-derived hybrid strain ISA1307 and Z. bailii CLIB 213T. A total of 5142 genes were predicted to be encoded in the genome of Z. bailii IST302 (Table 2), whereas 5084 putative protein-coding genes were predicted to be encoded in the genome of Z. bailii CLIB 213T (Galeote et al.2013). About 80% of the annotated genes are located in the first 25 scaffolds and 47% of the scaffolds contain zero or one gene annotated.
Table 1.
Genome assembly statistics of Z. bailii IST302.
| IST302 | ISA1307 (Mira et al. 2014) | CLIB 213T (Galeote et al. 2013) | |
|---|---|---|---|
| Total reads | 13 024 866 | 120 000 000 | 50 868 918 |
| No. of scaffolds | 105 | 154 | 56 |
| Coverage | ×120 | ×600 | ×250 |
| N50 (bp) | 432084 | 232974 | 932251 |
| Maximum contig length (bp) | 1149457 | 806952 | 1686157 |
| Minimum contig length (bp) | 1051 | 2160 | |
| Average contig length (bp) | 102599 | 137280 | |
| Assembly size (bp) | 10772966 | 21141152 | 10361356 |
Table 2.
Comparison of Z. bailii and Z. bailii-related strains genome features.
| No. of chromosomes | Ploidy | Genome | Average GC | Total no. | |
|---|---|---|---|---|---|
| Strain | separated by PFGE | ratio | size (Mb) | content (%) | of CDS |
| Z. bailii IST302 | 7 | ∼n | ∼11.0 | 42.2 | 5142 |
| Z. bailii CLIB 213T | 6 | nd | ∼10.4 | 42.5 | 5084 |
| Z. bailii hybrid strain ISA1037 | 13 | ∼2n | 22.0 | 42.4 | 9931 |
nd. Not determined
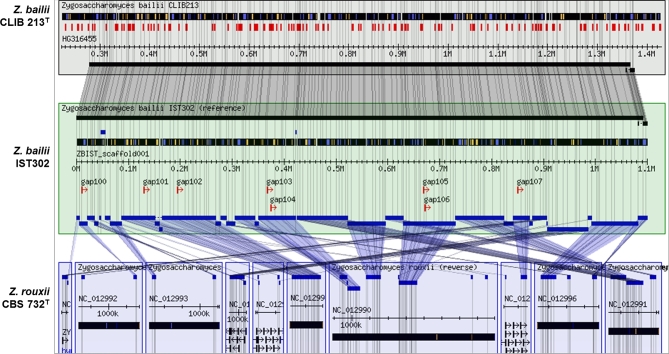
As expected, a high degree of conservation is observed between Z. bailii IST302 and CLIB 213T strains. However, the comparison of these Z. bailii strains with the taxonomically related yeast species Z. rouxii reveals numerous chromosomal rearrangements (Fig. 5).
Figure 5.
Multigenome alignment of genomic regions of Z. bailii IST302, Z. bailii CLIB 213T and Z. rouxii CBS 732T. Zygosaccharomyces bailii IST302 is presented as the central reference strain to which the others are compared. Conserved synteny blocks are presented in shaded boxes. The image was obtained using the multigenome alignment Gbrowse_syn (McKay, Vergara and Stajich 2010).
A total of seven centromere-like sequences were identified in the genome sequence of IST302 (Supplementary file 1, Supporting Information), based on previously identified point centromere sequences and on the collinear conservation of flanking genes of hemiascomycetous yeasts, in particular of Z. rouxii (Pribylova et al.2007; Souciet et al.2009). This result is in agreement with the seven chromosomal bands identified during the karyotyping analysis (Fig. 3). Interestingly, seven centromere-like sequences were also identified in the genome sequence of CLIB 213T strain (Supplementary file 2, Supporting Information), thus reinforcing that the separation of the chromosomes by PFGE (Fig. 3) was not complete for two of the chromosomes that possess highly similar sizes. It is also likely that, given the different sizes of both Z. bailii strains chromosomes and the conservation of collinearity, several genetic events might have occurred leading to different chromosome sizes. Strain-specific diversity of chromosome copy number and/or size has also been reported in other yeast species, such as Z. rouxii (Solieri et al.2008), S. cerevisiae (Zhu, Sherlock and Petrov 2016), Debaryomyces hansenii (Jacques et al.2010), Candida albicans (Rustchenko-Bulgac 1991), C. glabrata (Muller et al.2009) and Dekkera bruxellensis (Hellborg and Piskur 2009).
The life cycle of Zygosaccharomyces bailii IST302
The analysis of Z. bailii IST302 genes involved in life cycle and meiosis (Table 3 and Supplementary file 3, Supporting Information) revealed that this strain holds homologs of S. cerevisiae mating pheromone alpha- and a- factors, MF(ALPHA)1 and MFA1, respectively, and a homolog of S. cerevisiae MATALPHA1 that codes for a transcriptional co-activator of mating type specific genes. Neither MATALPHA2 nor MATA1/2 genes were detected in the genome sequence of this strain, suggesting that this haploid strain is of alpha mating type. Remarkably, although a homolog of the endonuclease encoding gene HO, responsible for mating type switching in S. cerevisiae, is present in Z. bailii IST302 genome, the silent cassettes HML/HMR were not identified in the genome sequence of this strain, neither in CLIB 213T (Table 3). In the context of evolution of hemiascomycete yeasts, the absence of silent cassettes is unexpected given that Z. rouxii, a close relative of Z. bailii, possesses those cassettes (Butler et al.2004; Fabre et al.2005; Watanabe, Uehara and Mogi 2013). Although the hypothesis that HML/HMR cassettes were not sequenced cannot be discarded, the fact that they were not identified in this study nor in the genome sequences of CLIB 213T (Galeote et al.2013) and ISA1307 (Mira et al.2014) strongly suggests that Z. bailii is unable to undergo mating type switching. Other genes involved in meiosis are also absent from the genome sequence of IST302, as well as in CLIB 213T (Supplementary file 3), as, for example, the homologs of S. cerevisiae gene IME1 that codes for a master regulator of meiosis, and of other genes involved in chromosome cohesion/recombination (SPO13, SPO22, MER3, REC104, MLH2, MSH4 and MSH5), spore assembly (MCP54, SMA1, SPO20, SPO74 and SPO77) or in the formation of the synaptonemal complex (ZIP2, ZIP3, SPO16). These genes are also absent from the genome sequence of the hybrid strain ISA1307 (Mira et al. 2014). The life cycle of Z. bailii and Z. bailii-related strains has been investigated based on their ability to sporulate when grown in specific media (Mollapour and Piper 2001; Rodrigues et al. 2003). The peculiar formation of mitotic, but not meiotic, spores was described both in diploid Z. bailii strains (Mollapour and Piper 2001) and in the hybrid strain ISA1307 (Rodrigues et al. 2003). Dato et al. (2008) also described the presence of binucleate cells in early-stationary phase. We investigated the ability of Z. bailii IST302 to form spores and, surprisingly, when grown in a sporulation medium we observed the formation of protrusions between two cells (Fig. 6B–F) before the development of two mitotic spores per cell (Fig. 6D and E), apparently without the occurrence of karyogamy as previously described (Mollapour and Piper 2001; Rodrigues et al. 2003). Interestingly, after conjugation and formation of two spores per cell, one of the cells seems to donate each of the two spores to the other cell (Fig. 6G–I) before spore release (Fig. 6J). Altogether, our results suggest that Z. bailii IST302 haploid cells cannot undergo mating type switching, but under nutrient deprivation are able to form vegetative spores. This study provided additional information for the understanding of Z. bailii and Z. bailii-related strains life cycle complexity, although more studies involving other strains are required to disclose the peculiar life cycle of this species.
Table 3.
ORFs identified in the genomes of Z. bailii IST302, CLIB 213T and Z. rouxii CBS 732T encoding the homologs of S. cerevisiae genes involved in mating type.
| S. cerevisiae S288C | Biological function | Z. bailii IST302 | Z. bailii CLIB 213T | Z. rouxii CBS 732T |
|---|---|---|---|---|
| MF(ALPHA)1/2 (YPL187W / YGL089C) | Mating pheromone alpha-factor | ZBIST_1222 | (Several gaps in the genome sequence at this location) | ZYRO0G08184g |
| MFA1/2 (YDR461W / YNL145W) | Mating pheromone a-factor | ZBIST_2952 | - | - |
| HMRA1/2 (YCR097W / YCR096C) | Silenced copy of a1/a2 at HMR | - | - | ZYRO0C18348g |
| HMLALPHA1/2 (YCL066W/ YCL067C) | Silenced copy of ALPHA1/2 at HML | - | - | ZYRO0F15818g |
| MATALPHA1 (YCR040W) | Transcriptional co-activator involved in regulation of mating type specific gene expression | ZBIST_5098 | BN860_00122g_l | ZYRO0F15840g |
| MATALPHA2 (YCR039C) | Transcriptional repressor of a-specific genes in haploids | - | - | - |
| MATA1 | Homeodomain protein involved in transcriptional regulation of mating type specific genes | - | - | - |
| MATA2 | Protein of unknown function; identical to the C-terminal of the MATalpha2 protein | - | - | - |
| HO (YDL227C) | Site-specific endonuclease | ZBIST_2381 | BN860_01090g_n | ZYRO0C10428g |
Figure 6.
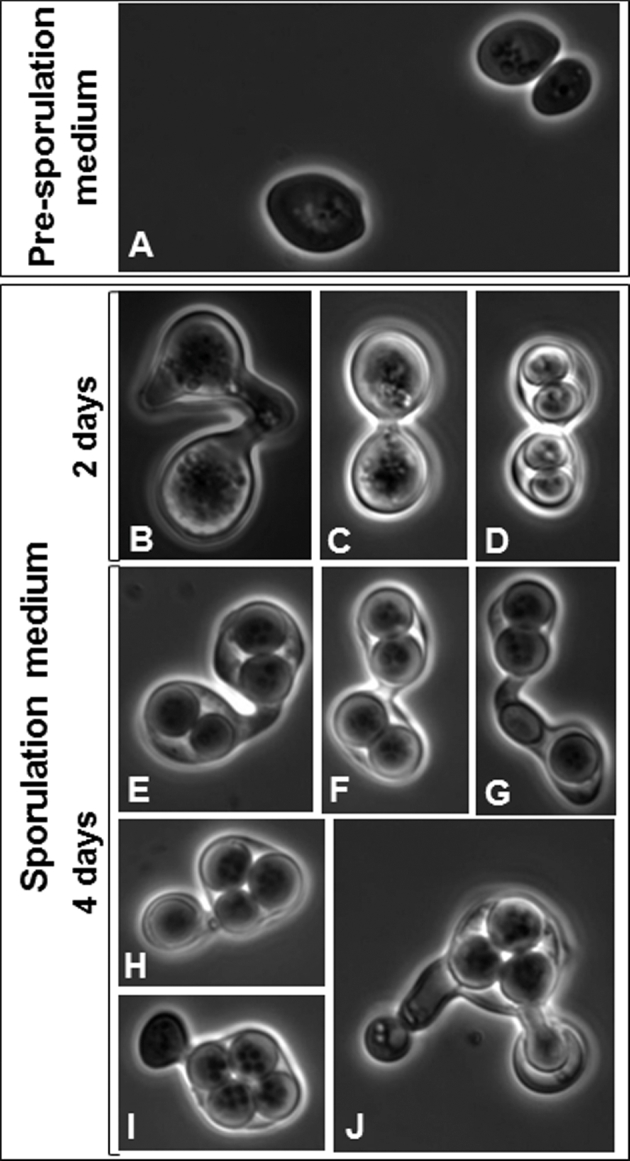
Microscopic observation of Z. bailii IST302 sporulation. Zygosaccharomyces bailii IST302 cells were grown in a pre-sporulation medium (A) for 2 days and transferred to a sporulation medium (B–J). The formation of spores was observed on a Zeiss ® Axioplan microscope (×1000 magnification) after 2 (B-D) and 4 (E-J) days of incubation at 30°C.
Genes encoding MDR/MXR transporters
Having in mind that when S. cerevisiae cells are exposed to weak acids their ability to decrease the counterions’ intracellular accumulation is essential to cope with their deleterious effects (reviewed by Piper et al. 2001; Mira, Teixeira and Sá-Correia 2010), we searched for transporters associated with multidrug/multixenobiotic resistance (MDR/MXR) belonging to the ATP-binding cassette (ABC) superfamily and to the Major Facilitator Superfamily (MFS) (Higgins 2007; Sá-Correia et al.2009) proposed, or hypothesized, to be responsible for the active efflux of weak acid counterions or other toxicants. For this, homologs of several S. cerevisiae ABC and MFS proteins previously implicated in the MDR/MXR phenomenon were identified in the genome of Z. bailii IST302 (Tables 4–6). Two homologs of S. cerevisiae ABC protein Pdr12, implicated in the active efflux of propionate, sorbate or benzoate anions (Piper et al.1998; Holyoak et al.1999), were identified in the genome of Z. bailii IST302 and in Z. rouxii CBS 732T as well (Table 4). Moreover, a total of four homologs of the Pdr18/Snq2 paralog pair were identified in Z. bailii IST302 and ISA1307 (Mira et al.2014), whereas only two Pdr18/Snq2 homologs were detected in Z. bailii CLIB 213T and Z. rouxii (Table 4). Interestingly, Pdr18 was implicated in the incorporation of ergosterol in yeast plasma membrane and found to play a role in S. cerevisiae tolerance to the lipophilic weak acid herbicide 2,4-D (Cabrito et al.2011), ethanol (Teixeira et al.2012) and acetic acid (Godinho, unpublished results). The paralog pair Tpo2/Tpo3 (Fernandes et al.2005), Aqr1 (Tenreiro et al.2002) and Azr1 (Tenreiro et al.2000) are the sole MDR/MXR transporters of the MFS so far implicated in S. cerevisiae tolerance to acetic acid, presumably by catalyzing the active efflux of acetate. Remarkably, Z. bailli IST302 genome holds only a single homolog of Tpo2 and Tpo3, and one homolog of Aqr1 and one of Azr1 (Tables 5 and 6).
Table 4.
ORFs identified in the genomes of Z. bailii IST302, CLIB 213T and Z. rouxii CBS 732T encoding the homologs of S. cerevisiae MDR/MXR transporters of ABC superfamily. The homologs were organized according to the taxonomic clusters previously defined (Seret et al.2009).
| ABC family | Total | ||||||
|---|---|---|---|---|---|---|---|
| S. cerevisiae S288c | PDR5 | SNQ2 | PDR12 | PDR11 | YOL075c | ADP1 | 10 |
| PDR15 | PDR18 | AUS1 | |||||
| PDR10 | |||||||
| Z. bailii IST302 | ZBIST_3521 | ZBIST_0908 | ZBIST_5124 | - | - | ZBIST_3422 | 12 |
| ZBIST_4874 | ZBIST_0076 | ZBIST_5081 | |||||
| ZBIST_4875 | ZBIST_4998 | ||||||
| ZBIST_4873 | ZBIST_3502 | ||||||
| ZBIST_4996 | |||||||
| Z. bailii CLIB 213T | BN860_00584g_q | BN860_07778g_c | BN860_00100g_t | - | - | BN860_01948g_i | 9 |
| BN860_04478g_f | BN860_04852g_g | ||||||
| BN860_04500g_f | |||||||
| BN860_04456g_f | |||||||
| BN860_00276g_c | |||||||
| Z. rouxii CBS 732T | ZYRO0D17710g | ZYRO0B14762g | ZYRO0F08888g | - | ZYRO0B05588g | ZYRO0E04092g | 10 |
| ZYRO0D11858p | ZYRO0A04114g | ZYRO0F08866p | |||||
| ZYRO0D11836p | |||||||
| ZYRO0D11880g | |||||||
Table 6.
ORFs identified in the genomes of Z. bailii IST302, CLIB 213T and Z. rouxii CBS 732T encoding the homologs of S. cerevisiae MDR/MXR transporters of the DAG family from MFS. The homologs were organized according to the taxonomic clusters previously defined (Dias and Sá-Correia 2013).
| DAG family | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subfamily | DHA2 | ARN | GEX | Total | |||||||
| S. cerevisiae S288c | SGE1 | VBA1 | VBA4 | ATR1 | YOR378W | Others | ARN4 | ARN3 | ARN1 | GEX1 | 16 |
| AZR1 | VBA2 | YMR279C | ARN2 | GEX2 | |||||||
| VBA3 | |||||||||||
| VBA5 | |||||||||||
| Z. bailii IST302 | ZBIST_3880 | ZBIST_0896 | ZBIST_0616 | - | ZBIST_4630 | ZBIST_3876 | - | ZBIST_4766 | ZBIST_2552 | - | 12 |
| ZBIST_0332 | ZBIST_3511 | ZBIST_4135 | |||||||||
| ZBIST_0774 | |||||||||||
| ZBIST_0773 | |||||||||||
| Z. bailii CLIB 213T | BN860_00188g_k | BN860_07514g_c | BN860_01134g_l | - | BN860_01552g | BN860_00386g_g | - | - | BN860_04984g_f | - | 11 |
| BN860_10814g_c | BN860_00320g_q | BN860_00144g_p | |||||||||
| BN860_04742g_g | |||||||||||
| BN860_04720g_f | |||||||||||
| Z. rouxii CBS 732T | - | ZYRO0G03234g | ZYRO0C01430g | - | - | - | - | - | ZYRO0G07414g | - | 6 |
| ZYRO0A00330g | |||||||||||
| ZYRO0G21868g | |||||||||||
| ZYRO0G21890p | |||||||||||
Table 5.
ORFs identified in the genomes of Z. bailii IST302, CLIB 213T and Z. rouxii CBS 732T encoding the homologs of S. cerevisiae MDR/MXR transporters of the DHA1 family from MFS. The homologs were organized according to the taxonomic clusters previously defined (Dias and Sá-Correia 2013).
| DHA1 family | Total | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. cerevisiae S288c | DTR1 | QDR1 | AQR1 | QDR3 | HOL1 | TPO2 | TPO1 | YHK8 | TPO4 | FLR1 | Others | 12 |
| QDR2 | TPO3 | |||||||||||
| Z. bailii IST302 | ZBIST_2327 | ZBIST_5047 | - | ZBIST_0934 | ZBIST_3441 | ZBIST_0758 | ZBIST_1684 | - | - | ZBIST_3176 | ZBIST_0204 | 21 |
| ZBIST_4184 | ZBIST_1685 | ZBIST_3177 | ZBIST_4991 | |||||||||
| ZBIST_2985 | ZBIST_0576 | |||||||||||
| ZBIST_3175 | ZBIST_3170 | |||||||||||
| ZBIST_3171 | ZBIST_4947 | |||||||||||
| ZBIST_3173 | ZBIST_5091 | |||||||||||
| ZBIST_3879 | ||||||||||||
| Z. bailii CLIB 213T | BN860_05512g | BN860_01244g_g | - | BN860_08350g_c | BN860_02432g_i | BN860_04368g_h | BN860_06810g_b | - | - | BN860_00232g_r | BN860_07888g_d | 19 |
| BN860_00254g_r | BN860_00232g_c | |||||||||||
| BN860_00166g_k | BN860_00188g_t | |||||||||||
| BN860_00210g_q | BN860_00122g_t | |||||||||||
| BN860_00144g_u | BN860_00276g_b | |||||||||||
| BN860_00188g_r | BN860_07470g | |||||||||||
| BN860_00232g_j | ||||||||||||
| Z. rouxii CBS 732T | ZYRO0F14652g | ZYRO0A01474g | - | ZYRO0A03564g | ZYRO0E02288g | ZYRO0G19646g | ZYRO0B00990g | ZYRO0G16302g | - | ZYRO0E09966g | ZYRO0E10230g | 21 |
| ZYRO0F08228g | ZYRO0E09922p | ZYRO0G15422g | ||||||||||
| ZYRO0F04642g | ZYRO0E10054g | |||||||||||
| ZYRO0E09988g | ZYRO0F02090g | |||||||||||
| ZYRO0E09900g | ZYRO0D00286g | |||||||||||
| ZYRO0B16808p | ZYRO0F02178p | |||||||||||
| ZYRO0A13618g | ||||||||||||
The Z. bailii strains tested are remarkably more tolerant to the lipophilic weak acid herbicides 2,4-D and MCPA, as well as to the herbicide alachlor of a different chemical family, when compared to S. cerevisiae. The susceptibility of S. cerevisiae to 2,4-D and MCPA was found to significantly decrease in a mutant with the PDR5 gene deleted (Teixeira and Sá-Correia 2002). This gene encodes a plasma membrane ATP transporter involved in pleiotropic drug resistance. Interestingly, five homologs of the Pdr5/Pdr15/Pdr10 taxonomic cluster were identified in Z. bailii IST302 (Table 4) three of them are likely to be the result of two gene duplications (ZBIST_4873, ZBIST_4874 and ZBIST_4875). Moreover, in Z. bailii IST302 two paralogs are predicted to encode Tpo1-like transporters (Table 5). The MFS transporter Tpo1 was also associated with S. cerevisiae tolerance to 2,4-D and MCPA (Teixeira and Sá-Correia 2002). As highlighted before (Mira et al.2014), the total number of MFS-MDR/MXR homologs identified in Z. bailii and Z. rouxii is remarkably higher when compared with S. cerevisiae S288c genome. However, one of the most remarkable differences in the number of genes encoding these transporters between these Zygosaccharomyces species and S. cerevisiae is the number of Flr1 homologs. Zygosaccharomyces bailii and Z. rouxii do possess seven Flr1-like transporters and the hybrid strain ISA1307 holds 12 homologs of S. cerevisiae Flr1 (Mira et al.2014). Saccharomyces cerevisiae Flr1 is responsible for resistance to several drugs, such as the fungicide fluconazole (Alarco et al.1997; Brôco et al.1999) and mancozeb (Teixeira et al.2008). However, no significant difference in susceptibility to agricultural fungicide mancozeb could be registered in Z. bailii and S. cerevisiae strains. Interestingly, the hybrid strain ISA1307 is significantly more tolerant to all the antifungal drugs tested than Z. bailii IST302 or S. cerevisiae BY4741. Among the antifungal drugs tested, Z. bailii IST302 is only mildly more tolerant to fluconazole and miconazole when compared to S. cerevisiae. The antifungal activity of azole drugs relies on the inhibition of ergosterol biosynthetic enzymes (Hitchcock et al.1990; White, Marr and Bowden 1998). Differences in the basal content of ergosterol of Z. bailii and S cerevisiae strains (Lindberg et al.2013), among other reasons (White, Marr and Bowden 1998; Prasad, Shah and Rawal 2016), could justify the moderate differences observed in the susceptibility phenotypes of these two yeast species.
Although interesting, this genomic study involving the comparison of the number of MDR/MXR transporters homologous to those functionally related with resistance to weak acids and other inhibitory compounds in S. cerevisiae has limitations. In fact, the specificity and efficiency of these transporters as well as their expression levels have to be first investigated in Z. bailii cells challenged with those compounds before taking conclusions. Moreover, it was reported that other mechanisms are likely involved in Z. bailii tolerance to drugs/xenobiotics. For example, studies suggest that Z. bailii tolerance to acetic acid is related with a number of alternative physiological strategies that include the capacity to tolerate short-term intracellular pH changes (Arneborg, Jespersen and Jakobsen 2000; Dang et al.2012), to co-consume acetic acid and glucose (Sousa et al.1998; Guerreiro, Mira and Sá-Correia 2012; Rodrigues et al.2012) and to exhibit a reduced permeability of plasma membrane to acetic acid due to the higher basal level of complex sphingolipids (Lindahl et al.2016).
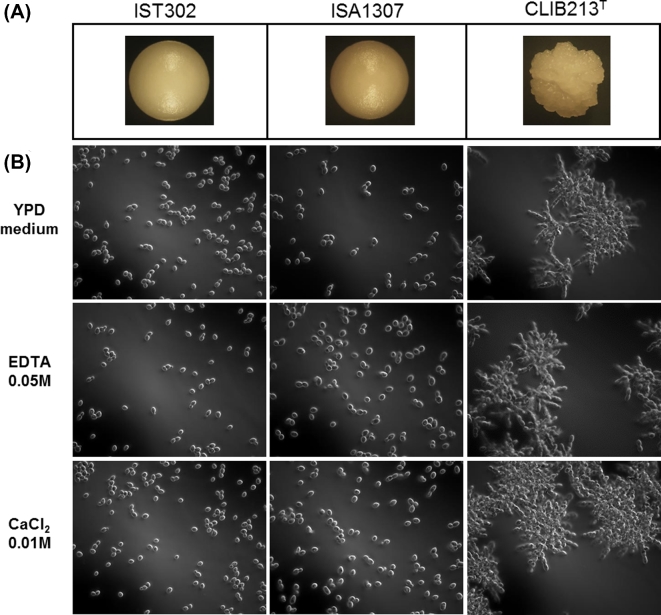
Zygosaccharomyces bailii IST302 does not form cell aggregates
Two of the most striking differences between Z. bailii IST302 and CLIB 213T are the morphology of their colonies (Fig. 7A) and the formation of large cellular aggregates (Fig. 7B) by Z. bailii CLIB 213T. When cultivated either in minimal or rich media, CLIB 213T shows a rough colony morphology (Fig. 7A) and forms cell aggregates that are visible with naked eye and under bright-field microscopy (Fig. 7B). Differently, Z. bailii IST302 shows regular and smooth colonies (Fig. 7A) and absence of cellular aggregation (Fig. 7B), which makes this strain very easy to manipulate for physiological studies when compared to the reference CLIB 213T strain. In order to investigate whether this type of cellular aggregation was due to flocculation, the dispersion of Z. bailii CLIB 213T cell aggregates was attempted by suspending cells in 0.05 M EDTA (pH 8.0) (Stratford 1989), but no significant dispersion of the aggregates was observed in the presence of the chelating agent, nor the addition of CaCl2 significantly increased cellular aggregation (Fig. 7B). This result suggests that the aggregation of Z. bailii CLIB 213T cells results from a Ca2+-independent mechanism. Zygosaccharomyces bailii IST302 and the Z. bailii hybrid strain ISA1307 were found to be non-flocculent yeast strains given that the suspension of each of these strains in 0.01 M CaCl2 did not induce a flocculation phenotype (Stratford 1989). Zygosaccharomyces bailii CLIB 213T was also cultivated in YP medium containing fructose or mannose instead of glucose (Soares et al. 2004), but cellular aggregation was still detected (results not shown). Interestingly, the cellular aggregation phenotype of CLIB 213T is similar to that described and identified by Ratcliff et al. (2012) as the ‘snowflake’ yeast phenotype which in S. cerevisiae was found to be related to specific mutations in Ace2 transcription factor (Ratcliff et al.2015), required for septum destruction after cytokinesis. The comparative analysis of Z. bailii genes homologous to S. cerevisiae genes involved in daughter cell separation as well as other genes involved in cell budding or morphogenesis and whose deletion led to abnormal budding pattern or the formation of cell aggregates was performed in both IST302 and CLIB 213T strains (Table 7). No gene duplications or eliminations were detected, but a closer look into the protein sequences of the homolog of S. cerevisiae Ace2 transcription factor, ZBIST_2164 and BN860_01596g_h from Z. bailii IST302 and CLIB 213T, respectively, allowed the identification of five amino acid substitutions, of which three involving serine/threonine residues. The pairwise alignment of Z. bailii IST302 and CLIB 213T Ace2 homologs is available in Supplementary file 4 (Supporting Information). It was recently reported that non-synonymous mutations located or adjacent to Ace2 zinc finger-binding domain in the C-terminal region of the protein are responsible for the S. cerevisiae ‘snowflake’ phenotype (Ratcliff et al.2015). Although the comparison of Ace2 homologs from IST302 and CLIB 213T did not identify amino acid substitutions in the C-terminal, one cannot discard the hypothesis that the identified mutations might also be involved in the aggregation phenotype shown by CLIB 213T. Nevertheless, other proteins can also have a role in the aggregation phenotype. For example, Z. bailii IST302 and CLIB 213T homologs of Cbk1 and Cts1, which in S. cerevisiae encode, respectively, a serine/threonine kinase involved in the regulation of Ace2 localization and activity and an endochitinase required for cell separation after mitosis whose transcriptional activation is mediated by Ace2, also show differences in their amino acid sequences (Table 7, Supplementary file 4).
Figure 7.
Microscopic observation of the morphology of the colonies (A) and cells (B) of Z. bailii IST302, Z. bailii CLIB 213T and ISA1307 hybrid strain. (A) Colonies of Z. bailii IST302, Z. bailii CLIB 213T and ISA1307 hybrid strain cells plated onto YPD plates and grown for 5 days were observed using a stereomicroscope and photographed. (B) Cells of Z. bailii IST302, Z. bailii CLIB 213T and the hybrid strain ISA1307 grown in YPD were observed on a Zeiss® Axioplan microscope (×400 magnification) and photographed.
Table 7.
Zygosaccharomyces bailii homologs of S. cerevisiae genes involved in cell aggregation.
| S. cerevisiae gene | Phenotype of S. cerevisiae null mutant (reference) | Z. bailii IST302 | Z. bailii CLIB 213T | IST302 and CLIB 213T protein sequence alignment identity (%) |
|---|---|---|---|---|
| ACE2/SWI5 | Abnormal budding pattern (Voth et al.2005) | ZBIST_2164 | BN860_01596g_h | 99.3 |
| CBK1 | Abnormal budding pattern (Voth et al.2005) | ZBIST_0511 | BN860_14928g1_1 | 97.3 |
| Cells form aggregates and display cell separation defects (Nelson et al.2003) | ||||
| Cells form large aggregates that do not easily dissociate with a micro-manipulator or EDTA treatment but separate upon the expression of chitinase (CTS1) (Racki et al.2000) | ||||
| BUD4 | Abnormal budding pattern (Voth et al.2005) | ZBIST_1783 | BN860_04500g_e | 99.2 |
| MOB2 | Cells form aggregates and display cell separation defects (Nelson et al.2003) | ZBIST_4670 | BN860_00694g_d | 99.6 |
| TAO3 | Cells form aggregates and display cell separation defects (Nelson et al.2003) | ZBIST_4175 | BN860_01046g_h1 | 99.5 |
| KIC1 | Cells form aggregates and display cell separation defects (Nelson et al.2003) | ZBIST_1895 | BN860_02850g1_1 | 99.5 |
| SOG2 | Cells form aggregates and display cell separation defects (Nelson et al.2003) | ZBIST_1556 | BN860_05226g_b | 99.8 |
| HYM1 | Cells form aggregates and display cell separation defects (Nelson et al.2003) | ZBIST_3061 | BN860_02454g_d | 100 |
| Clumps of cells, possibly due to failed septum removal (Bidlingmaier et al.2001) | ||||
| CTS1 | Cell separation blocked (Kuranda and Robbins 1991) | ZBIST_3250 | BN860_01904g_j | 98.2 |
In conclusion, this study provides relevant information on the genome sequence and annotation of Z. bailii IST302 that, contrasting with the reference strain Z. bailii CLIB 213T, does not form cell aggregates, thus being amenable to physiological studies. This haploid strain is easy to transform (Palma et al.2015) and therefore eventually suitable to be engineered exploring genome editing tools. For these reasons, Z. bailii IST302 is proposed as an attractive strain to be used for enlightening the mechanisms underlying its remarkable tolerance to stress induced by acetic acid and other weak acid food preservatives or for the exploitation of its potential use as microbial cell factory. The availability of Z. bailii IST302 genome sequence and annotation is an essential step towards the achievement of these important objectives.
Supplementary Material
Supplementary data are available at FEMSYR online.
Acknowledgments
We thank Dr Cláudia L. da Silva, Ricardo Pereira and António Soure from iBB for helpful suggestions regarding flow cytometry data acquisition and interpretation.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSYR online.
FUNDING
This work was partially supported by Institute for Bioengineering and Biosciences - iBB [BBI14]. Funding received by iBB from Fundação para a Ciência e a Tecnologia (FCT) [UID/BIO/04565/2013] and from Programa Operacional Regional de Lisboa 2020 [Project N. 007317] is also acknowledged. MP is the recipient of a post-doctoral fellowship [SFRH/BPD/73306/2010] awarded by FCT.
Conflict of interest. None declared.
REFERENCES
- Alarco A-M, Balan I, Talibi D et al. . AP1-mediated Multidrug Resistance in Saccharomyces cerevisiae requires FLR1 Encoding a transporter of the major facilitator superfamily. J Biol Chem 1997;272:19304–13. [DOI] [PubMed] [Google Scholar]
- Arneborg N, Jespersen L, Jakobsen M. Individual cells of Saccharomyces cerevisiae and Zygosaccharomyces bailii exhibit different short-term intracellular pH responses to acetic acid. Arch Microbiol 2000;174:125–8. [DOI] [PubMed] [Google Scholar]
- Bidlingmaier S, Weiss EL, Seidel C et al. . The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol Cell Biol 2001;21:2449–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branduardi P, Dato L, Porro D. Molecular tools and protocols for engineering the acid-tolerant yeast Zygosaccharomyces bailii as a potential cell factory. Methods Mol Biol 2014;1152:63–85. [DOI] [PubMed] [Google Scholar]
- Branduardi P, Valli M, Brambilla L et al. . The yeast Zygosaccharomyces bailii: a new host for heterologous protein production, secretion and for metabolic engineering applications. FEMS Yeast Res 2004;4:493–504. [DOI] [PubMed] [Google Scholar]
- Brôco N, Tenreiro S, Viegas CA et al. . FLR1 gene (ORF YBR008c) is required for benomyl and methotrexate resistance in Saccharomyces cerevisiae and its benomyl-induced expression is dependent on Pdr3 transcriptional regulator. Yeast 1999;15:1595–608. [DOI] [PubMed] [Google Scholar]
- Butler G, Kenny C, Fagan A et al. . Evolution of the MAT locus and its Ho endonuclease in yeast species. P Natl Acad Sci USA 2004;101:1632–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrito TR, Teixeira MC, Singh A et al. . The yeast ABC transporter Pdr18 (ORF YNR070w) controls plasma membrane sterol composition, playing a role in multidrug resistance. Biochem J 2011;440:195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang TDT, De Maeseneire SL, Zhang BY et al. . Monitoring the intracellular pH of Zygosaccharomyces bailii by green fluorescent protein. Int J Food Microbiol 2012;156:290–5. [DOI] [PubMed] [Google Scholar]
- Dato L, Branduardi P, Passolunghi S et al. . Advances in molecular tools for the use of Zygosaccharomyces bailii as host for biotechnological productions and construction of the first auxotrophic mutant. FEMS Yeast Res 2010;10:894–908. [DOI] [PubMed] [Google Scholar]
- Dato L, Sauer M, Passolunghi S et al. . Investigating the multibudded and binucleate phenotype of the yeast Zygosaccharomyces bailii growing on minimal medium. FEMS Yeast Res 2008;8:906–15. [DOI] [PubMed] [Google Scholar]
- Dias PJ, Sá-Correia I. The drug:H+ antiporters of family 2 (DHA2), siderophore transporters (ARN) and glutathione:H+ antiporters (GEX) have a common evolutionary origin in hemiascomycete yeasts. BMC Genomics 2013;14:901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E, Muller H, Therizols P et al. . Comparative genomics in hemiascomycete yeasts: evolution of sex, silencing, and subtelomeres. Mol Biol Evol 2005;22:856–73. [DOI] [PubMed] [Google Scholar]
- Fernandes AR, Mira NP, Vargas RC et al. . Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem Bioph Res Co 2005;337:95–103. [DOI] [PubMed] [Google Scholar]
- Ferreira MM, Loureiro-Dias MC, Loureiro V. Weak acid inhibition of fermentation by Zygosaccharomyces bailii and Saccharomyces cerevisiae. Int J Food Microbiol 1997;36:145–53. [DOI] [PubMed] [Google Scholar]
- Fortuna M, Sousa MJ, Côrte-Real M et al. . Cell cycle analysis of yeasts. In: Robinson JP, Darzynkiewicz Z, Dean P et al. (eds). Current Protocols in Cytometry. Vol Chapter 11. New York, USA: John Wiley & Sons, Inc., 2000, 11.13.1–11.13.9. [DOI] [PubMed] [Google Scholar]
- Galeote V, Bigey F, Devillers H et al. . Genome sequence of the food spoilage yeast Zygosaccharomyces bailii CLIB 213T. Genome Announc 2013;1:e00606–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro JF, Mira NP, Sá-Correia I. Adaptive response to acetic acid in the highly resistant yeast species Zygosaccharomyces bailii revealed by quantitative proteomics. Proteomics 2012;12:2303–18. [DOI] [PubMed] [Google Scholar]
- Hellborg L, Piskur J. Complex nature of the genome in a wine spoilage yeast, Dekkera bruxellensis. Eukaryot Cell 2009;8:1739–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CF. Multiple molecular mechanisms for multidrug resistance transporters. Nature 2007;446:749–57. [DOI] [PubMed] [Google Scholar]
- Hitchcock CA, Dickinson K, Brown SB et al. . Interaction of azole antifungal antibiotics with cytochrome P-450-dependent 14 alpha-sterol demethylase purified from Candida albicans. Biochem J 1990;266:475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holyoak CD, Bracey D, Piper PW et al. . The Saccharomyces cerevisiae weak-acid-inducible ABC transporter Pdr12 transports fluorescein and preservative anions from the cytosol by an energy-dependent mechanism. J Bacteriol 1999;181:4644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques N, Sacerdot C, Derkaoui M et al. . Population polymorphism of nuclear mitochondrial DNA insertions reveals widespread diploidy associated with loss of heterozygosity in Debaryomyces hansenii. Eukaryot Cell 2010;9:449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SA, Stratford M. Zygosaccharomyces. In: Kurtzman CP, Fell JW, Boekhout T (eds). The Yeasts, a Taxonomic Study. 5th edn London: Elsevier, 2011, 937–47. [Google Scholar]
- Kalathenos P, Sutherland JP, Roberts TA. Resistance of some wine spoilage yeasts to combinations of ethanol and acids present in wine. J Appl Bacteriol 1995;78:245–50. [Google Scholar]
- Kuranda MJ, Robbins PW. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem 1991;266:19758–67. [PubMed] [Google Scholar]
- Li R, Zhu H, Ruan J et al. . De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 2010;20:265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L, Genheden S, Eriksson LA et al. . Sphingolipids contribute to acetic acid resistance in Zygosaccharomyces bailii. Biotechnol Bioeng 2016;113:744–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg L, Santos AX, Riezman H et al. . Lipidomic profiling of Saccharomyces cerevisiae and Zygosaccharomyces bailii reveals critical changes in lipid composition in response to acetic acid stress. PLoS One 2013;8:e73936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro V, Malfeito-Ferreira M. Spoilage yeasts in the wine industry. Int J Food Microbiol 2003;86:23–50. [DOI] [PubMed] [Google Scholar]
- McKay SJ, Vergara IA, Stajich JE. Using the Generic Synteny Browser (GBrowse_syn). Curr Protoc Bioinformatics 2010;9:9.1–9.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L, Lydall D. Pulsed-field gel electrophoresis of budding yeast chromosomes. Methods Mol Biol 2006;313:65–73. [DOI] [PubMed] [Google Scholar]
- Martorell P, Stratford M, Steels H et al. . Physiological characterization of spoilage strains of Zygosaccharomyces bailii and Zygosaccharomyces rouxii isolated from high sugar environments. Int J Food Microbiol 2007;114:234–42. [DOI] [PubMed] [Google Scholar]
- Mira NP, Munsterkotter M, Dias-Valada F et al. . The genome sequence of the highly acetic acid-tolerant Zygosaccharomyces bailii-derived interspecies hybrid strain ISA1307, isolated from a sparkling wine plant. DNA Res 2014;21:299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira NP, Teixeira MC, Sá-Correia I. Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: a genome-wide view. OMICS 2010;14:525–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Piper P. Targeted gene deletion in Zygosaccharomyces bailii. Yeast 2001;18:173–86. [DOI] [PubMed] [Google Scholar]
- Muller H, Thierry A, Coppée J-Y et al. . Genomic polymorphism in the population of Candida glabrata: Gene copy-number variation and chromosomal translocations. Fungal Genet Biol 2009;46:264–76. [DOI] [PubMed] [Google Scholar]
- Nelson B, Kurischko C, Horecka J et al. . RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol Biol Cell 2003;14:3782–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma M, Dias PJ, Roque F et al. . The Zygosaccharomyces bailii transcription factor Haa1 is required for acetic acid and copper stress responses suggesting subfunctionalization of the ancestral bifunctional protein Haa1/Cup2. BMC Genomics 2017;18:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma M, Roque F, de C et al. . Search for genes responsible for the remarkably high acetic acid tolerance of a Zygosaccharomyces bailii-derived interspecies hybrid strain. BMC Genomics 2015;16:1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina C, Gonçalves P, Prista C et al. . Ffz1, a new transporter specific for fructose from Zygosaccharomyces bailii. Microbiology 2004;150:2429–33. [DOI] [PubMed] [Google Scholar]
- Piper P, Calderon CO, Hatzixanthis K et al. . Weak acid adaptation: the stress response that confers yeasts with resistance to organic acid food preservatives. Microbiology 2001;147:2635–42. [DOI] [PubMed] [Google Scholar]
- Piper P, Mahé Y, Thompson S et al. . The Pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J 1998;17:4257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt JI, Hocking AD. Yeasts. In: Fungi and Food Spoilage. Boston, MA: Springer, 2009, 357–82. [Google Scholar]
- Prasad R, Shah AH, Rawal MK. Antifungals: Mechanism of Action and Drug Resistance. Adv Exp Med Biol 2016;892:327–49. [DOI] [PubMed] [Google Scholar]
- Pribylova L, Straub M-L, Sychrova H et al. . Characterisation of Zygosaccharomyces rouxii centromeres and construction of first Z. rouxii centromeric vectors. Chromosome Res 2007;15:439–45. [DOI] [PubMed] [Google Scholar]
- Racki WJ, Bécam AM, Nasr F et al. . Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J 2000;19:4524–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff WC, Denison RF, Borrello M et al. . Experimental evolution of multicellularity. P Natl Acad Sci USA 2012;109:1595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff WC, Fankhauser JD, Rogers DW et al. . Origins of multicellular evolvability in snowflake yeast. Nat Commun 2015;6:6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues F, Ludovico P, Sousa MJ et al. . The spoilage yeast Zygosaccharomyces bailii forms mitotic spores: a screening method for haploidization. Appl Environ Microb 2003;69:649–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues F, Sousa MJ, Ludovico P et al. . The fate of acetic acid during glucose co-metabolism by the spoilage yeast Zygosaccharomyces bailii. PLoS One 2012;7:e52402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues F, Zeeman AM, Alves C et al. . Construction of a genomic library of the food spoilage yeast Zygosaccharomyces bailii and isolation of the beta-isopropylmalate dehydrogenase gene (ZbLEU2). FEMS Yeast Res 2001;1:67–71. [DOI] [PubMed] [Google Scholar]
- Rustchenko-Bulgac E. Variations of Candida albicans electrophoretic karyotypes. J Bacteriol 1991;173:6586–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá-Correia I, dos Santos SC, Teixeira MC et al. . Drug:H+ antiporters in chemical stress response in yeast. Trends Microbiol 2009;17:22–31. [DOI] [PubMed] [Google Scholar]
- Sá-Correia I, Guerreiro JF, Loureiro-Dias MC et al.Zygosaccharomyces. In: Batt CA, Tortorello ML (eds). Encyclopedia of Food Microbiology. 2nd edn Cambridge, Massachusetts: Elsevier Ltd/Academic Press, 2014, 849–55. [Google Scholar]
- Seret M-L, Diffels JF, Goffeau A et al. . Combined phylogeny and neighborhood analysis of the evolution of the ABC transporters conferring multiple drug resistance in hemiascomycete yeasts. BMC Genomics 2009;10:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares EV, Vroman A, Mortier J et al. . Carbohydrate carbon sources induce loss of flocculation of an ale-brewing yeast strain. J Appl Microbiol 2004;96:1117–23. [DOI] [PubMed] [Google Scholar]
- Solieri L, Cassanelli S, Croce MA et al. . Genome size and ploidy level: New insights for elucidating relationships in Zygosaccharomyces species. Fungal Genet Biol 2008;45:1582–90. [DOI] [PubMed] [Google Scholar]
- Souciet J, Dujon B, Gaillardin C et al. . Comparative genomics of protoploid Saccharomycetaceae. Genome Res 2009;19:1696–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa MJ, Rodrigues F, Corte-Real M et al. . Mechanisms underlying the transport and intracellular metabolism of acetic acid in the presence of glucose in the yeast Zygosaccharomyces bailii. Microbiology 1998;144:665–70. [DOI] [PubMed] [Google Scholar]
- Sousa-Dias S, Goncalves T, Leyva JS et al. . Kinetics and regulation of fructose and glucose transport systems are responsible for fructophily in Zygosaccharomyces bailii. Microbiology 1996;142:1733–8. [Google Scholar]
- Stratford M. Yeast flocculation: Calcium specificity. Yeast 1989;5:487–96. [Google Scholar]
- Stratford M. Food and beverage spoilage yeasts. In: Querol A, Fleet G (eds). Yeasts in Food and Beverages. New York: Springer, 2006, 335–79. [Google Scholar]
- Stratford M, Steels H, Nebe-von-Caron G et al. . Extreme resistance to weak-acid preservatives in the spoilage yeast Zygosaccharomyces bailii. Int J Food Microbiol 2013;166:126–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh S-O, Gujjari P, Beres C et al. . Proposal of Zygosaccharomyces parabailii sp. nov. and Zygosaccharomyces pseudobailii sp. nov., novel species closely related to Zygosaccharomyces bailii. Int J Syst Evol Micr 2013;63:1922–9. [DOI] [PubMed] [Google Scholar]
- Teixeira MC, Dias PJ, Simões T et al. . Yeast adaptation to mancozeb involves the up-regulation of FLR1 under the coordinate control of Yap1, Rpn4, Pdr3, and Yrr1. Biochem Bioph Res Co 2008;367:249–55. [DOI] [PubMed] [Google Scholar]
- Teixeira MC, Godinho CP, Cabrito TR et al. . Increased expression of the yeast multidrug resistance ABC transporter Pdr18 leads to increased ethanol tolerance and ethanol production in high gravity alcoholic fermentation. Microb Cell Fact 2012;11:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MC, Sá-Correia I. Saccharomyces cerevisiae resistance to chlorinated phenoxyacetic acid herbicides involves Pdr1p-mediated transcriptional activation of TPO1 and PDR5 genes. Biochem Bioph Res Co 2002;292:530–7. [DOI] [PubMed] [Google Scholar]
- Tenreiro S, Nunes PA, Viegas CA et al. . AQR1 gene (ORF YNL065w) encodes a plasma membrane transporter of the major facilitator superfamily that confers resistance to short-chain monocarboxylic acids and quinidine in Saccharomyces cerevisiae. Biochem Bioph Res Co 2002;292:741–8. [DOI] [PubMed] [Google Scholar]
- Tenreiro S, Rosa PC, Viegas CA et al. . Expression of the AZR1 gene (ORF YGR224w), encoding a plasma membrane transporter of the major facilitator superfamily, is required for adaptation to acetic acid and resistance to azoles in Saccharomyces cerevisiae. Yeast 2000;16:1469–81. [DOI] [PubMed] [Google Scholar]
- Ter-Hovhannisyan V, Lomsadze A, Chernoff YO et al. . Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res 2008;18:1979–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DS, Davenport RR. Zygosaccharomyces bailii — a profile of characteristics and spoilage activities. Food Microbiol 1985;2:157–69. [Google Scholar]
- Voth WP, Olsen AE, Sbia M et al. . ACE2, CBK1, and BUD4 in budding and cell separation. Eukaryot Cell 2005;4:1018–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MC, Rattei T, Arnold R et al. . PEDANT covers all complete RefSeq genomes. Nucleic Acids Res 2009;37:D408–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe J, Uehara K, Mogi Y. Diversity of mating-type chromosome structures in the yeast Zygosaccharomyces rouxii caused by ectopic exchanges between MAT-like loci. PLoS One 2013;8:e62121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TC, Marr KA, Bowden RA. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev 1998;11:382–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Zavaljevski N, Desai V et al. . QuartetS: a fast and accurate algorithm for large-scale orthology detection. Nucleic Acids Res 2011;39:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YO, Sherlock G, Petrov DA. Whole genome analysis of 132 clinical Saccharomyces cerevisiae strains reveals extensive ploidy variation. G3 2016;6:2421–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at FEMSYR online.