Abstract
Toxin–antitoxin systems (TAs) are ubiquitous among bacteria and play a crucial role in the dissemination and evolution of antibiotic resistance, such as maintaining multi-resistant plasmids and inducing persistence formation. Generally, activities of the toxins are neutralised by their conjugate antitoxins. In contrast, antitoxins are more liable to degrade under specific conditions such as stress, and free active toxins interfere with essential cellular processes including replication, translation and cell-wall synthesis. TAs have also been shown to be responsible for plasmid maintenance, stress management, bacterial persistence and biofilm formation. We discuss here the recent findings of these multifaceted TAs (type I–VI) and in particular examine the role of TAs in augmenting the dissemination and maintenance of multi-drug resistance in bacteria.
Keywords: toxin–antitoxins, addictive systems, antimicrobial resistance, persistence
As antimicrobial resistance continues to escalate in Gram-negative bacteria, understanding the role of toxin–antitoxin systems in plasmid maintenance and inducing persistence becomes increasingly important.
INTRODUCTION
Antibiotic resistance has been highlighted as one of the most pressing concern of 21th century. The rapid spread of ‘superbugs’, including Enterobacteriaceae with NDM-1 (New Delhi Metallo-beta-lactamase-1), KPC-2 (Klebsiella pneumoniae carbapenemase-2) and the most recent reported MCR-1 (mobile colistin resistance-1), has been described as a global crisis and an impending return to the pre-antibiotics era (Moellering 2010; Liu et al. 2015). To rationally combat antibiotic resistance, we require a better understanding of which factors influence the emergence and persistence of antibiotic resistant clones. Bacterial toxin–antitoxin systems (TAs), originally linked to plasmid maintenance systems (Ogura and Hiraga 1983), exert important activities in the context of bacterial resistance and persistence formation (Wen et al. 2014; Harms, Maisonneuve and Gerdes 2016; Patel 2016). TAs are small modules consisting of a stable toxin and its unstable cognate antitoxin. Antitoxins are more labile than toxins and readily degraded under stress conditions; this allow the toxins to exert their detrimental effects, promoting plasmid maintenance, slow growth and dormancy, which is rather linked with chromosomally encoded TAs (Page and Peti 2016). TAs are not essential for normal cell growth but are nonetheless widely present on bacterial plasmids and chromosomes. It has been hypothesised that TAs play a central role that is advantageous for cell survival in their natural habitat, such as switching into a dormant, drug-resistance state to withstand high levels of antibiotic stress (Page and Peti 2016). The toxins inhibit cell growth by targeting a variety of important cellular processes, including DNA replication, transcription and cell-wall synthesis, which are similar to antibiotic activities (Davies and Davies 2010; Chan, Balsa and Espinosa 2015). Because of their ubiquity and crucial intracellular targets, the study of bacterial toxins will help us understand their role in the dissemination and evolution of bacterial antibiotic resistance. In this review, we will provide a synopsis of TAs and in particular examine the role of type II TAs in augmenting the dissemination and maintenance of multidrug resistance in Gram-negative bacteria.
TAs CLASSIFICATION
TAs are small genetic modules found on bacterial mobile genetic scaffolds like plasmids, as well as on bacterial chromosomes. The TA loci encode two-component systems that consist of a stable toxin whose overexpression either kills the bacterial cell or negates cell growth, and an unstable cognate antitoxin. As a result, when a plasmid encoding the TA is lost from a cell, the toxin is released from the existing TA complex and kills plasmid-free cells. In essence, this is an elegant model for perpetuating plasmid maintenance in bacterial population (Gerdes, Rasmussen and Molin 1986). This unique system is also called post-segregational killing. The first TA (ccdAB) identified was carried on a F-plasmid and was shown to play an important role in plasmid maintenance by coupling host cell division to plasmid proliferation (Ogura and Hiraga 1983). Since this initial discovery, a number of different TAs have been identified that are encoded on bacterial genomes. Based on their proteomic nature of their corresponding antitoxin, TAs are currently divided into six distinct classes (Table 1).
Table 1.
The intracellular targets of TAs.
| Targets of toxin | TA groups | Examples | Toxin | Antitoxin | Cellular process inhibited |
|---|---|---|---|---|---|
| Inner cell membranes | type I,V | hok-sok, tisB-istR, ghoTS | TisB, Hok, GhoT | TisA, Sok, GhoS | Cell membranes damage |
| Replication by DNA gyrase | type II | ccdAB, parDE | CcdB, ParE | CcdA, ParD | DNA replication |
| tRNAfMet | type II | vapBC | VapC | VapB | Translation |
| Ribosome-independent mRNA interferase | type II | mazEF | MazF | MazE | Translation |
| Ribosome-dependent mRNA interferase | type II | relBE, higBA | RelB, HigB | RelE, HigA | Translation |
| GltX:tRNA | type II | hipBA | HipA | HipB | Translation |
| Elongation factor EF-Tu | type II | phd-doc | Doc | Phd | Translation |
| Peptidoglycan precursors: UNAG | type II | ω-ε-ζ, pezTA | ζ, PezT | ε, PezA | Cell wall synthesis |
| Biofilm formation | type II,V | mqsRA, ghoST | MqsR, GhoT | MqsA, GhoS | Biofilm formation |
| mRNAs | type III | toxIN,cptTN, tenpIN | ToxN,CptN, TenpN | ToxI,CptI, TenpI | Growth arrest |
| Cytoskeletal protein MreB and FtsA | type IV | yeeU-cbtA | CbtA | YeeU | Cell morphology |
| Beta sliding clamp, protein DnaN | type VI | socAB | SocB | SocA | DNA elongation |
Type I TAs
All type I toxins are small hydrophobic proteins of approximately 60 amino acid and their gene expression is regulated by an antisense RNA transcribed from the toxin gene but in reverse orientation (Gerdes and Wagner 2007). Type I TAs are arranged either as overlapping, convergent transcribed genes pairs or as divergently transcribed gene pairs located apart. In the first case, the antitoxin is a cis-encoded antisense RNA (e.g. hok-sok, bsrG-SR4); in the latter case, it is a trans-encoded sRNA (e.g. tisB-IstR1, shoB-ohsC) (Brantl 2012). The first and best studied type I system is hok-sok (host killing, hok, and suppressor of killing, sok), which was first discovered on plasmid R1 from Escherichia coli (Gerdes, Rasmussen and Molin 1986; Thisted and Gerdes 1992). Later, other type I TAs were found in E. coli such as ldr-rdl, tisB-istR1, ibs-sib, shoB-ohsC and symER (Fozo 2012; Kawano 2012; Wagner and Unoson 2012).
All the toxins of type I TAs have an identical secondary structure consisting of one α-helical structure and are predicted to be localised in the inner membrane, and thus to induce pores into the bacterial cell membranes, resulting in inhibition of ATP synthesis (Fozo, Hemm and Storz 2008). Consequently, replication, transcription and translation maybe inhibited, leading to cellular death (Unoson and Wagner 2008). For instance, TisB produces clusters of narrow anion-selective pores in lipid bilayers that significantly disturbs the cytoplasmic membrane (Wagner and Unoson 2012). Many toxins are not bacteriocidal, but interfere with phage propagation, modulate the cell membrane or prevent mature particle formation, and in some cases, only overexpression of toxin genes shows a toxic effect (Yamaguchi and Inouye 2011).
Type II TAs
Type II TAs have been most extensively studied among all TAs, and the huge number of type II TAs varies greatly from different bacterial species, even among the same species. Hitherto, 12 subgroups of type II TAs have been identified based on toxin amino sequence homology (Leplae et al. 2011), including mazEF (Aizenman, Engelberg-Kulka and Glaser 1996), relEB (Takagi et al. 2005), yefM-yoeB (Kamada and Hanaoka 2005), ω-ε-ζ (Zielenkiewicz and Ceglowski 2005) and mqsRA (Gonzalez Barrios et al. 2005; Brown et al. 2009). In type II systems, the antitoxin is a small, unstable protein that sequesters the toxin through protein complex formation. The expression of the two genes is regulated at the level of transcription by the TA complex that involves binding palindromic sequence at the promoter region. Therefore, as the concentration of the TA complex in the cell is reduced as a result of antitoxin degradation, the TA operon expression is suppressed to produce more toxin and antitoxin, and thus the type II system is also termed the ‘addiction module’ (Yarmolinsky 1995). In most cases, the antitoxin genes are located upstream of their cognate toxin genes so that the antitoxins appear to have an advantage for their production over their cognate toxins. Conversely, there are many exceptions of this genetic arrangement, such as higBA in which the toxin genes higB is located upstream of its cognate antitoxin genes, higB (Yamaguchi, Park and Inouye 2009; Christensen-Dalsgaard, Jørgensen and Gerdes 2010).
Type III TAs
The toxINPa was first identified on a plasmid from Erwinia carotovora subspecies atrosepticum (Pectobacterium atrosepticum) as an example of the novel type III protein–RNA TAs (Fineran et al. 2009). The toxINPa locus consists of a toxin ToxNPa inhibiting bacterial growth and RNA antisense ToxIPa counteracting the toxicity. The arrangements of type III TAs are unusual, as a toxin gene is preceded by a short palindromic repeat, which separates the toxin from its small RNA antitoxin, composed of several repeats of short nucleotide sequences. The short palindromic repeat acts as a transcriptional terminator, regulating the relative levels of antitoxin and toxin transcript. For example, toxINBt located on pAW63 from Bacillus thuringiensis encodes a toxic protein ToxNBt and a antitoxin ToxIBt containing 2.9 tandem repeats of a 34-nucleotide sequence (Short, Monson and Salmond 2015; Goeders et al. 2016). Currently, type III TAs are divided into three subgroups sharing the same genetic organisation, namely toxIN, cptIN (for Coprococcus type III inhibitor-toxiN) and tenpIN (for type III ENdogenous to Photorhabdus inhibitor-toxiN) (Blower et al. 2012; Goeders et al. 2016). Though these subgroups were identified by shared identity with ToxN, their cognates diverge between and within the subtypes, such as the number of repeats and the length of repeats (Blower et al. 2012; Goeders et al. 2016). All type III toxins discovered so far serve as endoRNase that cleave mRNAs in adenine-rich regions, whose activities inhibit by forming RNA pseudoknot–toxin complex.
Type IV TAs
The yeeU-cbtA has been proposed for the new type IV TAs in which the protein antitoxin interferes with binding of the toxin to its target rather than inhibiting the toxin via direct TA binding (Masuda et al. 2012). Unlike most toxins targeting the macromolecular biosynthesis, CbtA is the first toxin of the TAs that affects cellular morphology (Tan, Awano and Inouye 2011). CbtA binds and inhibits the polymerisation of bacterial cytoskeletal proteins, MreB and FtsZ. The antitoxin, YeeU, suppresses the CbtA toxicity by stabilising the CbtA target proteins rather than by directly interacting with CbtA to suppress its toxicity (Masuda et al. 2012). Specifically, YeeU directly binds to both MreB and FtsZ and enhances the bundling of their filaments in vitro. Notably, this is a unique feature of the yeeU-cbtA system, distinguishing it from all the other TAs in that CbtA and YeeU does not form a complex. Nevertheless, YeeU is able to neutralise CbtA toxicity. Thus, the yeeU-cbtA constitutes a new type of TA.
Type V TAs
The ghoTS is a new type of TA, where GhoS (ghost cell suppressor) is the first known antitoxin to neutralise the toxicity of GhoT ghost cell toxin, by specifically cleaving its mRNA (Wang et al. 2012). Compared to the high overlapping catalytic sites of CRISPR-associated-2 protein SSO1404 structures, Wang et al. (2012) suggested that the antitoxin, GhoS, is a sequence-specific endoRNase that cleaves ghoT transcription and thereby prevents GhoT translation. GhoT is a membrane-damaging protein, and its production can lyse the cell membrane and change its morphologies. Ultimately, this causes the formation of ghost cells, a group of dead or dying cells in which cell outline is still visible but the cytoplasmic area is transparent (Wang et al. 2012). GhoT has also been shown to contribute to biofilm formation—after the deletion of its repressor GhoS, the formation of biofilm and cell motility increased by approximately 6- and 2-fold, respectively (Wang et al. 2012).
Type VI TAs
In contrast to typical TAs, in which toxicity of the toxin is neutralised by the antitoxin, socB is unique and constitutively controlled by the protease CIpXP, while its cognate socA acts as a proteolytic adaptor, promoting the degradation of SocB by CIpXP. SocB identified in Caulobacter crescentus has been proposed to inhibit DNA replication through direct interaction with DnaN, a ring-shaped protein that encodes a central component for DNA elongation (Markovski and Wickner 2013). This interaction disrupts the association of DnaN and Pol III and other DnaN-binding proteins, resulting in the collapse of the DNA replication forks. It also has been shown that this DNA damage can cause the accumulation of SocB, suggesting that it may play a regulatory role in the induction of the RecA-mediated SOS response (Aakre et al. 2013; Markovski and Wickner 2013; Page and Peti 2016). Therefore, the socA-socB system may play important roles in promoting Caulobacter adaptation to varying environmental conditions by preventing DNA replication.
THE CELLULAR TARGETS OF TAs
In last decade, an increasing number of cellular targets for TAs have been elucidated, and most of them are involved in many essential bacterial processes such as DNA replication, RNA transcription and protein translational modification as shown in Table 1 and Fig. 1. Interestingly, TAs share many cellular targets with that of the antibiotics. For instance, zeta toxin phosphorylates the essential nucleotide sugar UDP-N-acetylglucosamine (UNAG), and leads to the inhibition of cell-wall synthesis, much like the activity of penicillin, that inhibits the formation of peptidoglycan across-links in the bacterial cell wall or glycopeptides that bind cell-wall precursors (Kohanski, Dwyer and Collins 2010). Another example are DNA gyrases that can induce and relax supercoils during DNA replication yet are the target of toxins CcdB and ParE, as well as quinolone antibiotics that disrupt DNA replication by binding to DNA-gyrase complexes (Kohanski, Dwyer and Collins 2010). Due to their remarkable similarity in cellular targets between TAs and antibiotics, TAs may provide novel insights into the discovery and development of new antimicrobials.
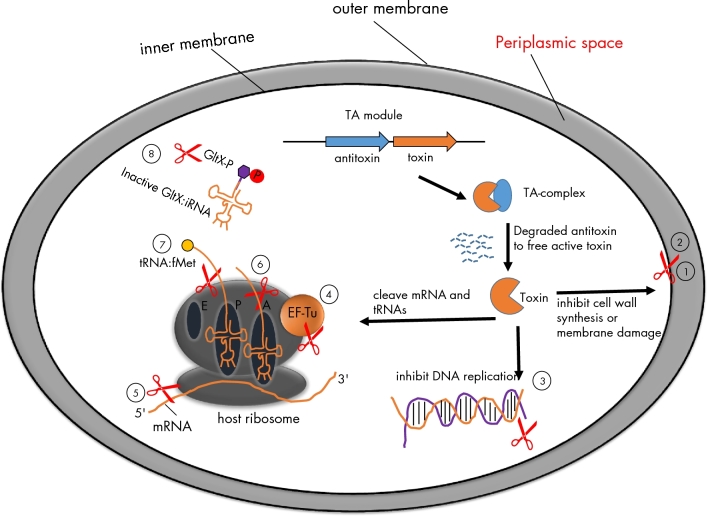
Figure 1.
The intracellular targets of TAloci. TA loci usually encode two genes: one is a stable toxin and the other one is an unstable antitoxin. The antitoxins sequester the toxins but are subjected to proteolytic degradation by cellular proteases (Lon or ClpXP) under stress condition. Consequently, free active toxins alter cellular processes including DNA replication, translation or cell-wall synthesis, which ultimately results in slow growth or the formation of highly drug-tolerant persisters. TAs examples for the cellular targets are given below. (1) Zeta toxin inhibits cell-wall synthesis by specific phosphorylation of peptidoglycan precursor UNAG. (2) TisB, HokB and GhoT: the products of TisB and HokB can decrease the level of membrane potential motive force (pmf) and ATP by inserting into cytoplasmic membrane, while protein GhoT can lyse cell membrane and change cell morphologies. (3) CcdB and ParE inhibit DNA replication by poison DNA gyrase. (4) Doc inhibits translation by phosphoralation of elongation factor Tu (EF-Tu). (5–7) MazF, RelB and VapC inhibit translation by cleavage of mRNAs like single-stranded mRNA, A-site on ribosome and initiator tRNAfMet, respectively. (8) HipA inhibits translation by phosphoration of GltX. tRNA:fMet indicates initiator tRNA at P site carried formyl methionine; ‘p’ indicates phosphorylation.
Targeting cell-wall synthesis: Zeta toxin
The epsilon zetas were originally discovered as plasmid maintenance modules on a 29-kb low-copy plasmid, pSM19035, isolated from Streptococcus pyogenes (Zielenkiewicz and Ceglowski 2005). pSM19035 stability is conferred by two regions (segA and segB), and their corresponding products, SegA and SegB, control the plasmid partitioning (Ceglowski et al. 1993; Ana et al. 2000). The segB gene complex consists of four genes (δ and ω-ε-ζ), ensuring a ‘better-than-random’ plasmid segregation. The gene δ shares a significant homology to ATPases involved in active plasmid partitioning, but stabilisation function is dependent on the ω-ε-ζ operon. Therefore, among TAs, the organisation of the ω-ε-ζ operon is unique. The first three-component operon with the ε and ζ genes encodes an antitoxin and toxin, respectively, and the transcription of this operon is controlled by the additional gene ω (Ana et al. 2000; Zielenkiewicz and Ceglowski 2005). It has been show that the product of ω binds to the promoter of the entire operon as a dimer, and in the absence of ω repression, the intensity of transcription from ω is increased about 40-fold (Ana et al. 2000). Plasmid-encoded epsilon-zeta TAs enhance plasmid maintenance by killing the plasmid-free daughter cells (Zielenkiewicz and Ceglowski 2005), whereas chromosomally encoded epsilon-zeta TA family (pezAT for pneumococcal epsilon-zeta) identified from S. pneumoniae kills bacteria through the inhibition of cell-wall synthesis. More recently, Mutschler and Meinhart (2011) showed that toxin PezT inhibits the bacteria cell-wall synthesis by phosphorylating the UNAG into a toxic module UNAG-3-phosphate (UNAG-3P). UNAG-3P accumulates and competitively inhibits MurA, which is the essential enzyme for peptidoglycan synthesis (Barreteau et al. 2008), thereby freeing PezT toxin that poisons bacteria through inhibition of the cell-wall formation, causing the cells to lyse (Mutschler and Meinhart 2011; Mutschler et al. 2011).
The zeta toxin systems have been identified as highly abundant modules in multi-resistance plasmids and chromosomes of various Gram-positive pathogens, including S. pyogenes (Zielenkiewicz and Ceglowski 2005) and S. pneumoniae (Khoo et al. 2007). It has long been thought that epsilon-zeta systems only can be found in Gram-positive bacteria; however, a novel zeta homolog has been first identified from the Gram-negative bacterium Escherichia coli (Rocker and Meinhart 2015). This zeta toxin, designated EzeT for E. coli zeta toxin, is located in 3.4-kb islet, consisting of two domains featuring EzeT toxin and epsilon-like antitoxin within a single polypeptide chain. Similar to the toxin PzeT, the C-terminal domain of EzeT containing all catalytic motifs of UNAG kinases is capable of phosphorylating UNAG in vitro (Mutschler et al. 2011; Rocker and Meinhart 2015). In contrast to conventional type II TAs, N-terminal domain of EzeT contains an antitoxin; thus, EzeT is an authentic zeta-like UNAG kinase and is also the first auto-inhibited TA system, since it can be inhibited by its own N-terminal cis-acting antitoxin domain (Rocker and Meinhart 2015).
Targeting tRNAs: VapC and HipA
PIN (N-terminus of the pilin biogenesis protein PiIT) domains are small protein domains consisting of 130 amino acid in length, and serve as ribonuclease enzymes that cleave single-stranded RNA in a sequence-dependent manner (Arcus et al. 2011). The TA module vapBC (virulence-associated protein) is associated with PIN-domain proteins. The vapBC (at that time called vagCD) locus derived from virulence plasmid of Salmonella Dublin strain G19 was proposed to prevent plasmid loss under nutrient-limiting conditions (Pullinger and Lax 1992). VapC is the PIN-domain ribonuclease, co-expressed with cognate inhibitor VapB that forms a novel PIN-domain–inhibitor complex (Bunker et al. 2008; Arcus et al. 2011). vapBC are surprisingly abundant; for example, the genome of Mycobacterium tuberculosis encodes 47 different vapBC homologs (Ahidjo et al. 2011). The transcription of vapBC operon is regulated by the DNA promoter, via the N-terminal ribbon-helix-helix domain of antitoxin VapB. The proteolytic degradation of the more labile VapB by Lon protease results in the accumulated level of VapC toxin. Once activated, VapC inhibits mRNA transcription presumably by site-specific cleavage of tRNAfMet, which plays a crucial role in the protein synthesis of bacteria (Bunker et al. 2008; Winther and Gerdes 2012). HipA function acts in similar manner to VapC, but has different binding sites. In contrast to phosphorylate EF-Tu, free HipA inactivates GltX by phosphorylation at its ATP-binding site Ser239, and thus GltX is unable to charge tRNA with glutamate (tRNAGlu). Consequently, this induces amino acid starvation and the invariable activation of RelA to more (p)ppGpp synthesis. Thus, high accumulated levels of (p)ppGpp trigger a stringent response that inhibits the global translation process such as protein synthesis (Kaspy et al. 2013; Germain et al. 2015).
Targeting DNA gyrase: CcdB and ParE
The ccd (couple cell division) locus is adjacent to the origin of replication of the F plasmid and promotes the stable maintenance of F plasmids by coupling host cell division to plasmid proliferation (Ogura and Hiraga 1983). The target of toxin ccdB, DNA gyrase, is a ubiquitous bacterial enzyme essential for negative supercoiling of DNA during DNA replication and transcription (Dao-Thi et al. 2005; Nöllmann, Crisona and Arimondo 2007). Gyrase is known to consist of two subunits (GyrA and GyrB), GyrA contains a catalytic domain for DNA binding and cleavage, and GyrB contains the ATPase domain. Quinolones are able to inhibit the topoisomerase ligase domain by forming a DNA-topoisomerase-quinolone complex to block DNA and RNA polymerases (Wentzell and Maxwell 2000). The bacterial toxins ccdB and parE present similar properties to those of quinolones, but interact at a different site to DNA gyrase (Jiang et al. 2002; Dao-Thi et al. 2005). Under normal growth conditions, the antitoxin CcdA inhibits CcdA toxic activity by forming a tight CcdA2–(CcdB)2 complex. Once the bacterium loses its plasmid, unstable CcdA degrades and CcdB and GyrA form a symmetric complex. After CcdB-GyrA binding, ATP is hydrolysed and the supercoiled DNA is released resulting in blocking bacterial transcription and immediate cell death (Critchlow et al. 1997; Dao-Thi et al. 2005). More recently, an additional role for CcdB has been that of a persistence factor (Tripathi et al. 2012). When faced with antibiotic or heat stress, the increased levels of the ATP-dependent protease Lon (Kuroda et al. 2001), responsible for the rapid turnover of unstable proteins, degrade the antitoxin CcdA, freeing toxin CcdB. Free active toxin CcdB causes DNA damage through forming a GyrA-CcdB cleavage complex, which triggers the RecA-mediated SOS response. Ultimately, multidrug-tolerant persister cells are formed.
Targeting membrane potential: HokB and TisB
tisB/istR-1 is the first TA locus involved in the SOS response. The locus encodes two small RNA molecules: one is an antisense RNA, istR-1, that inhibits TisB toxicity, and the toxin, TisB, under the control of Lex (Vogel et al. 2004; Darfeuille et al. 2007) and is localised on the inner membrane (Unoson and Wagner 2008) (Fig. 2). The induction of tisB results in membrane damage that entails a rapid decrease in DNA replication, RNA transcription and protein synthesis (Unoson and Wagner 2008). HokB is similar to TisB in that both are small proteins, and exert toxicity by damaging the inner membrane. The hokB-sokB locus derived from chromosome of E. coli K-12 codes for three genes: sokB, mokB and hokB (Pedersen and Gerdes 1999). The sokB is a small antisense RNA that controls the translation of the mokB reading frame. hokB translation is under the control of mokB, thereby sokB can also suppress hokB toxicity. Recent studies have shown that HokB acts as a potential persistence factor (Verstraeten et al. 2015) and its accumulated leads to a loss of membrane electrochemical potential, ultimately resulting in persistence.
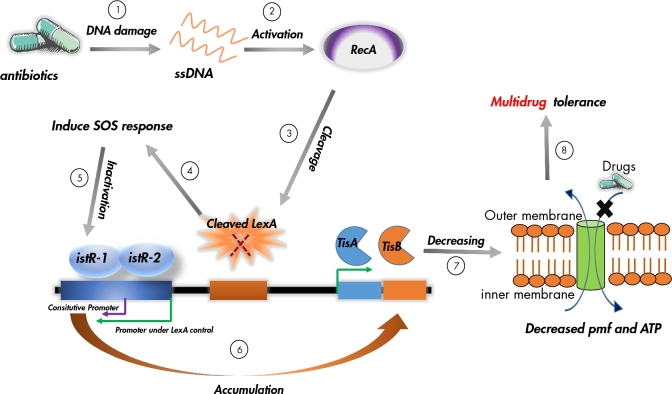
Figure 2.
Model of the TisB toxin induced SOS response and persistence formation. (1) Antibiotics (like ciprofloxacin) kill bacteria by damaging their DNA; (2) the SOS response gene recA is activated by the accumulation of single-stranded DNA (ssDNA). (3) The induced RecA interacts with the LexA repressor, leading to facilitate the LexA autocleavage. (4) Once the degradation of LexA repressor, the SOS genes are induced to repair DNA damage. (5) Concurrently, the SOS induction results in cleavage of the istR-1 pool. (6) The expression of tisB is activated by degrading the level of antitoxin IstR-1, this causes membrane damage and the loss of membrane proton motive force (pmf) and ATP level (7); as a result, drugs were drive to out of the cells, leading to persister formation (8). The green and purple arrowheads representing the promoters under LexA control and istR-1 constitutive promoter, respectively.
Targeting ribosomes: Doc, MazF and RelE
The toxin doc (death on cure) and its conjugate antidote, phd (prevent host death), are derived from the bacteriophage P1 and play a major role in plasmid stability persistence, programmed cell death and stress response (Lehnherr et al. 1993; Gazit and Sauer 1999). Doc has been showed to be a representative member of the Fic protein subfamily, which is ubiquitous in bacteria and involved in crucial functions (such as bacterial pathogenesis) (Garcia-Pino et al. 2008; Harms, Maisonneuve and Gerdes 2016). Fic proteins have a central conserved HXFX(D/E)N(K/G)R motif that is present in Doc structures. Phd dimers are subject to cleavage by CIpXP protease, an ATP-dependent protease of E. coli (Lehnherr and Yarmolinsky 1995). It has been shown that mRNA is significantly stabilised upon Doc induction, suggesting that Doc does not cleave mRNA. In fact, Doc toxicity has been proposed to act in a similar manner to hygromycin B (HygB), an aminoglycoside antibiotic (Liu et al. 2008). After degradation of Phd by CIpXP protease, the free Doc binds on the 30S ribosomal subunit that includes the HygB-binding site and phosphorylates the conserved threonine (Thr382) of the elongation factor EF-Tu. Subsequently, Doc-bound EF-Tu is unable to bind to aminoacylated tRNAs which leads to an accumulation of stalled ribosomes, blocking protein synthesis, and thus a dormant state is formed (Liu et al. 2008; Castro-Roa et al. 2013). The MazF and RelE proteins are also RNases, which inhibit translation by the cleavage of mRNA. Purified MazF is a sequence-specific (ACA) endoribonuclease, which only cleaves single-stranded mRNA at VUUV' sites independently of the ribosomes, by a mechanism very similar to that of E. coli RelE (Christensen et al. 2003; Zhang et al. 2003; Donegan and Cheung 2009). In the context of the stringent response, antitoxin RelB is degraded by ATP-dependent protease Lon, which leads to activate RelE. The activated RelE induces a global inhibition of translation by cleavage of the mRNA at the ribosome A-site, with the consequence of the tRNA degradation (Christensen and Gerdes 2003; Pedersen et al. 2003). Consequently, this activates RelE to trigger a stringent response, creating high-tolerant persisters.
Targeting bacterial biofilm formation: MqsR
Bacterial biofilms are communities in which cells aggregate on a solid surface and are further enveloped in an exopolysaccharide matrix (Mah and O’Toole 2001; Stewart and Costerton 2001). It has been shown that biofilms are closely linked to antibiotic resistance and that a biofilm can form slimy layers that surround the bacteria and act as a barrier to antimicrobial agents, decreasing the penetration of antibiotics to the bacterium's surface (Davies 2003). When cells are embedded in a biofilm, their MIC has been shown to increase from 6.25 to >400 μg/ml depending on the antimicrobial agent (Evans and Holmes 1987). Besides failure of antibiotic diffusion, some studies have demonstrated that biofilm-associated multidrug-resistant Pseudomonas aeruginosa cells can cause slow growth, lipopolysaccharide modification and antibiotic degradation, ultimately accompanied by an increase in antibiotic resistance (de la Fuente-Núñez et al. 2013). The first TAs shown to be involved in biofilm formation was mqsRA(motility quorum-sensing regulator), a typical type II TAs in which the toxicity of protein MqsR is neutralised by its conjugate antitoxin MqsA (Gonzalez Barrios et al. 2005; Brown et al. 2009; Wang and Wood 2011). Gonzalez Barrios et al. (2005) demonstrated that toxin MqsR is significantly stimulated by biofilm formation and enhanced cell motility. It has been suggested that MqsR is an RNase and prevents translation by cleaving RNAs (Brown et al. 2009). In addition, antitoxin MqsA has been linked to the regulation of the general stress responses, such as oxidative stress (Wang et al. 2011). Wang et al. confirmed that MqsA represses the stress regulator, RpoS, leading to the decreased concentration of messenger 3,5-cyclic diguanylic acid and thus decreasing biofilm formation. However, upon stress, for example, oxidative stress, MsqA is unstable and is rapidly degraded by Lon and ClpXP protease, causing the accumulation of RpoS. As a result, the stringent response is triggered, and the bacterial state is switched from high motility (planktonic) to sessile (biofilm) state.
BIOLOGICAL ROLE OF TAs IN ANTIMICROBIAL RESISTANCE
Initially, TAs were identified on plasmids and used to be considered as selfish genes with little or no physiological benefit to the host cells. Because if a plasmid encoding the TAs is absent in the daughter cell, the stable toxin is released by rapidly degrading antitoxin to kill plasmid-free cells, in order to increase plasmid maintenance in host cells. Since their discovery, the role of TAs has been debated over decades. Hitherto, mounting evidence has testified that TAs are far more than selfish loci and that they play key roles in promoting cell survival. In particular, in response to antibiotic stress, toxins can be activated by stress-induced protease like CIpXP and Lon. This phenomenon results in slow cellular growth in which the bacterium can now effectively tolerate antibiotic challenge.
THE MAINTENANCE OF MULTIDRUG RESISTANCE PLASMIDS
Conjugative plasmids identified as reservoirs for resistance genes are one of the most effective physical forums to develop and disseminate the antibiotic resistance genes among bacteria (Carattoli 2013; Mathers, Peirano and Pitout 2015). In many cases, plasmids can carry genes that are highly beneficial to the host bacteria by enabling them to persist in unfavourable environments, e.g. protection against potentially lethal antibiotics. Therefore, plasmids serve as effective DNA shuttles for antibiotic resistance genes that are, in part, linked to the clinical failure of antibiotics treatments. However, because plasmids are extrachromosomal, mobile genetic elements presented in host cells, plasmids impose a metabolic burden to the host cells, which are prone to elimination from bacterial genome in the absence of selective pressure (Zielenkiewicz and Ceglowski 2001). The stable inheritance of plasmids is achieved by activating the plasmid-specified partitioning proteins into dividing cells and selective killing of the cells that failed to acquire a plasmid (Hayes 2003). As discussed above, TAs, like hok-sok and ccdAB, are responsible for the plasmids stabilisation; thus, TAs also have been viewed as ‘addiction modules’ (Engelberg-Kulka et al. 2006). Beside plasmids, TAs appear to play a stabilising role in genomic islands, for instance, SXT, an integrative and conjugative element that mediates tolerance to multiple antibiotics in Vibrio cholera (Wozniak and Waldor 2009). One novel TA pair (designated mosAT) within SXT has been identified to promote SXT stability. Ectopic expression of mosT causes growth inhibition and MosA can neutralise the toxic effect of overexpressed MosT. Similar to plasmid-borne toxins, when SXT is vulnerable to loss, MosT expression is activated to minimise the SXT-free cells. Therefore, the activity of mosAT may contribute to the maintenance of SXT in bacterial populations (Wozniak and Waldor 2009).
BACTERIAL STRESS RESPONSE
The SOS response is important for bacterial survival under stress conditions that can trigger disruption of the DNA replication fork and result in the accumulation of single-stranded DNA. Both RecA and LexA proteins have an important role in the SOS response as regulators (Yamaguchi and Inouye 2011). RecA, activated by single-strand DNA, is involved in the inactivation of the repressor LexA. Normally, LexA binds to a specific sequence in the DNA (the SOS box) and represses the expression of genes involved in DNA repair, mutagenesis and cell growth arrest. The SOS response is an important factor for persister formation in response to the fluoroquinolone antibiotic, ciprofloxacin, which can cause DNA damage (Dorr, Lewis and Vulic 2009; Lewis 2010). The first TA locus, tisAB-istR-1, is involved in the SOS response to DNA damage (Vogel et al. 2004). This locus encodes a toxic gene tisAB and two small RNAs, IstR-1 and IstR-2, as shown in Fig. 2. TisAB is under LexA control and thus activated by the SOS response, but only TisB is responsible to the toxicity (Vogel et al. 2004). The transcription of istR-2 is also SOS regulated and not involved in the TisB control, whereas the antitoxin IstR-1 binds with the LexA-independent promoter and inhibits TisB expression by inducing RNase III-dependent cleavage of tisB mRNA (Vogel et al. 2004; Darfeuille et al. 2007). In the absence of an SOS response, istR-1 is constitutively transcribed to inactive the toxicity of TisAB by inducing RNase III-dependent cleavage of tisB mRNA (Vogel et al. 2004). When DNA damage is caused by ciprofloxacin, it activates the RecA protein leading to LexA repressor cleavage, and then the SOS response is induced. The antitoxin IstR-1 that controls the Lex promoter is almost complete cleaved, while the toxin TisB gradually accumulates and rapidly binds to the cytoplasmic membrane, leading to membrane damage, and the proton motive force (pmf) and ATP levels are decreased. This causes the rates of DNA, RNA and protein synthesis to decrease, and the intake of drugs to the cells is blocked. As a result, growth slows down and a multidrug-resistant persister is formed (Vogel et al. 2004; Darfeuille et al. 2007; Unoson and Wagner 2008; Dorr, Vulic and Lewis 2010) (Fig. 2).
PERSISTER CELLS
TAs can also contribute to bacteria persistence formation (Lewis 2010; Maisonneuve and Gerdes 2014; Page and Peti 2016). Persistence is observed when a small subpopulation of cells survive antibiotic treatment that has efficiently killed off the rest of the population. In contrast to resistance, persistence is a form of antimicrobial tolerance that is not link with genetic mutation or DNA acquisition, but rather with a spontaneous switch of a dormant, non-dividing state. Therefore, persisters are able to survive in the presence of antibiotics even if they are genetically not programmed to become resistant. More importantly; however, rather than causing cell death, some toxins convert cells into a dormant or a semidormant state that is resistance to antibiotics, and then revive them when environmental conditions become more conducive for growth (Hayes 2003). TAs have been shown to play a major role in persister formation in many model systems. An example of TAs mediating persister states involves the intracellular metabolite, guanosine tetraphosphate and pentaphosphate [(p)ppGpp], the main regulator of the stringent response (Amato, Orman and Brynildsen 2013; Maisonneuve, Castro-Camargo and Gerdes 2013). In Escherichia coli, (p)ppGpp was discovered as a alarmone to alter cellular transcription globally by interacting with RNA polymerase activity directly, in response to nutrient starvation or other stress (Dalebroux and Swanson 2012). As a consequence, bacteria can survive even faced with limiting nutrients, suggesting that the coupling accumulation of (p)ppGpp level may induce growth arrest, drug tolerance and the formation of persisters. It has been proposed that high levels of (p)ppGpp trigger persistence by activation of the TA loci, resulting in translation inhibition and growth arrest (Korch, Henderson and Hill 2003; Maisonneuve, Castro-Camargo and Gerdes 2013; Schumacher et al. 2015; Harms, Maisonneuve and Gerdes 2016). Contrary to previous reports, there is growing evidence to suggest that EF-Tu is not the target of HipA during the inactivation of translation, but HipA-mediated persistence depends stochastically on the (p)ppGpp-TA pathway (Germain et al. 2013; Kaspy et al. 2013; Maisonneuve, Castro-Camargo and Gerdes 2013; Wen et al. 2014). Most likely, the current molecular model explaining HipA-mediated persistence is shown in Fig. 3 (Korch, Henderson and Hill 2003; Germain et al. 2013; Kaspy et al. 2013; Maisonneuve, Castro-Camargo and Gerdes 2013; Germain et al. 2015). When faced with particular stresses, bacteria rapidly swift transcription profile to trigger the nucleotide alarmone (p)ppGpp synthesis, which involved in catalytic activity of SpoT and RelA, the two (p)ppGpp synthetases of E. coli (Dalebroux and Swanson 2012). The resulting increased (p)ppGpp levels accumulate inorganic polyphosphate (PolyP) through inhibition of exopolyphosphatase (PPX), a phosphatase enzyme that degrades PolyP. The accumulation of PolyP combines with Lon protease preferentially to cleave the antitoxin HipB, resulting in an excess of toxin HipA. In return, free active toxin HipA inactivates GltX by phosphorylation of its ATP-binding site Ser239, with the consequence of uncharged tRNA accumulation in the cell. Consequently, the amino acid starvation triggers the activation of RelA to more (p)ppGpp synthesis. Thereby, the high level of (p)ppGpp accumulation induces a stringent response that inhibits the synthesis of DNA, RNAs, ribosomal proteins and membrane components, promoting cells entry into dormant state. Conversely, a recent study showed that the activation of yefM-yoeB (Christensen et al. 2004), a well-characterised type II TAs, is not dependent on the level inorganic PolyP and (p)ppGpp (Ramisetty et al. 2016), and further suggested that the pathways of TAs-mediated persistence formation may be far more complicated than previously known.
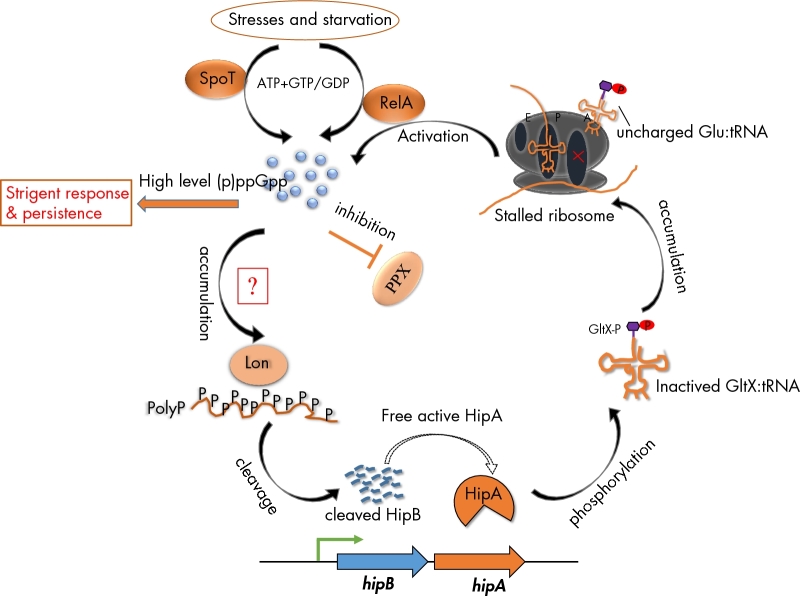
Figure 3.
(p)ppGpp-hipA mediated persister pathway. In response to particular stresses, SpoT and RelA are activated to synthesise the nucleotide alarmone (p)ppGpp, The increased (p)ppGpp levels lead to the accumulation of inoganic polyphosphate (PolyP) through inhibition of exopolyphosphatase (PPX), that the cellular enzyme to degrades PolyP. The accumulated PolyP combines with Lon protease preferentially to cleave the antitoxin HipB, resulting in an excess of toxin HipA. In return, free active toxin HipA inactivates GltX by phosphorylation of its ATP-binding site Ser239, with the consequence of uncharged tRNA with glutamate (tRNAGlu) accumulation in the cell. Uncharged tRNAGlu loads at empty ribosomal sites and triggers the activation of RelA to more (p)ppGpp synthesis, promoting cells entry into dormant state. Note that SpoT and RelA are bifunctional synthetase-hydrolase enzyme, if the stresses have been removed, they can hydrolase (p)ppGpp and bring cells to normal growth (Dalebroux and Swanson 2012). The red box labelled with ‘?’ indicates that the link between stringent response-associated genes (including ppGpp, Lon, PolyP) and TAs has been exploring in some TAs, such as relBE, mazEF and yefM-yeoB. It has been proved that the activation of toxin MazF and YoeB is dependent on the Lon-mediated degradation of their cognates, antitoxins, but not on the accumulation of PolyP and ppGpp (Christensen et al. 2001, 2003; Ramisetty et al. 2016).
CONCLUSION
In the last decade, antimicrobial resistance in Gram-negative pathogens has outpaced the production of novel and even new drugs entering the market place providing an increasing void that is unlikely to be bridged. The drivers and maintenance of antimicrobial resistance was hitherto thought to be antimicrobials themselves; however, increasingly we are becoming aware that antimicrobial resistance is as much to do with genetic maintenance systems, e.g. TAs, as it is to do with the presence of the drug. TAs are remarkable systems that parasitise the bacteria and hold it hostage. TAs are also extremely varied and are a testimony to the dexterity and plasticity of genetic systems to adapt and evolve. Although yet to be fully established, TAs are becoming increasingly numerous and more associated with antimicrobial genes present on the same plasmid thereby providing maintenance of the antimicrobial resistance in the absence of the drug. Worryingly, the SOS induction triggered by drugs such as fluoroquinolones activates TA systems such as TisAB via LexA. The fact that fluoroquinolones are widespread and poorly degraded implies an ever-present pressure on certain TA systems to further be mobilised throughout bacterial populations.
FUNDING
QY is funded by a CSC scholarship and TRW funded by HEFC. TRW and QY were also supported by MRC grant DETER-XDR-CHINA (MR/P007295/1).
Conflict of interest. None declared.
REFERENCES
- Aakre CD, Phung TN, Huang D et al. . A bacterial toxin inhibits DNA replication elongation through a direct interaction with the beta sliding clamp. Mol Cell 2013;52:617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahidjo BA, Kuhnert D, McKenzie JL et al. . VapC toxins from Mycobacterium tuberculosis are ribonucleases that differentially inhibit growth and are neutralized by cognate VapB antitoxins. PLoS One 2011;6:e21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal ‘addiction module’ regulated by guanosine [corrected] 3΄, 5΄-bispyrophosphate: a model for programmed bacterial cell death. P Natl Acad Sci USA 1996;93:6059–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato SM, Orman MA, Brynildsen MP. Metabolic control of persister formation in Escherichia coli. Mol Cell 2013;50:475–87. [DOI] [PubMed] [Google Scholar]
- Ana B, Ayora S, Sitkiewicz I et al. . Plasmid copy-number control and better-than-random segregation genes of pSM19035 share a common regulator. P Natl Acad Sci USA 2000;97:728–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcus VL, McKenzie JL, Robson J et al. . The PIN-domain ribonucleases and the prokaryotic VapBC toxin–antitoxin array. Protein Eng Des Sel 2011;24:33–40. [DOI] [PubMed] [Google Scholar]
- Barreteau H, Kovač A, Boniface A et al. . Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev 2008;32:168–207. [DOI] [PubMed] [Google Scholar]
- Blower TR, Short FL, Rao F et al. . Identification and classification of bacterial Type III toxin–antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res 2012;40:6158–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S. Bacterial type I toxin-antitoxin systems. RNA Biol 2012;9:1488–90. [DOI] [PubMed] [Google Scholar]
- Brown BL, Grigoriu S, Kim Y et al. . Three dimensional structure of the MqsR: MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog 2009;5:e1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker RD, McKenzie JL, Baker EN et al. . Crystal structure of PAE0151 from Pyrobaculum aerophilum, a PIN-domain (VapC) protein from a toxin-antitoxin operon. Proteins 2008;72:510–8. [DOI] [PubMed] [Google Scholar]
- Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol 2013;303:298–304. [DOI] [PubMed] [Google Scholar]
- Castro-Roa D, Garcia-Pino A, De Gieter S et al. . The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat Chem Biol 2013;9:811–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceglowski P, Boitsov A, Karamyan N et al. . Characterization of the effectors required for stable inheritance of Streptococcus pyogenes pSM19035-derived plasmids in Bacillus subtilis. Mol Gen Genet 1993;241:579–85. [DOI] [PubMed] [Google Scholar]
- Chan WT, Balsa D, Espinosa M. One cannot rule them all: are bacterial toxins-antitoxins druggable? FEMS Microbiol Rev 2015;39:522–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Gerdes K. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol 2003;48:1389–400. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Maenhaut-Michel G, Mine N et al. . Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol Microbiol 2004;51:1705–17. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Mikkelsen M, Pedersen K et al. . RelE, a global inhibitor of translation, is activated during nutritional stress. P Natl Acad Sci USA 2001;98:14328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Pedersen K, Hansen FG et al. . Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol 2003;332:809–19. [DOI] [PubMed] [Google Scholar]
- Christensen-Dalsgaard M, Jørgensen MG, Gerdes K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol Microbiol 2010;75:333–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchlow SE, O’Dea MH, Howells AJ et al. . The interaction of the F plasmid killer protein, CcdB, with DNA gyrase: induction of DNA cleavage and blocking of transcription. J Mol Biol 1997;273:826–39. [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Swanson MS. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 2012;10:203–12. [DOI] [PubMed] [Google Scholar]
- Dao-Thi MH, Van Melderen L, De Genst E et al. . Molecular basis of gyrase poisoning by the addiction toxin CcdB. J Mol Biol 2005;348:1091–102. [DOI] [PubMed] [Google Scholar]
- Darfeuille F, Unoson C, Vogel J et al. . An antisense RNA inhibits translation by competing with standby ribosomes. Mol Cell 2007;26:381–92. [DOI] [PubMed] [Google Scholar]
- Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2003;2:114–22. [DOI] [PubMed] [Google Scholar]
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol R 2010;74:417–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente-Núñez C, Reffuveille F, Fernández L et al. . Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr Opin Microbiol 2013;16:580–9. [DOI] [PubMed] [Google Scholar]
- Donegan NP, Cheung AL. Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J Bacteriol 2009;191:2795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr T, Lewis K, Vulic M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLos Genet 2009;5:e1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr T, Vulic M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 2010;8:e1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Amitai S, Kolodkin-Gal I et al. . Bacterial programmed cell death and multicellular behavior in bacteria. PLos Genet 2006;2:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RC, Holmes CJ. Effect of vancomycin hydrochloride on Staphylococcus epidermidis biofilm associated with silicone elastomer. Antimicrob Agents Ch 1987;31:889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran PC, Blower TR, Foulds IJ et al. . The phage abortive infection system, ToxIN, functions as a protein–RNA toxin–antitoxin pair. P Natl Acad Sci USA 2009;106:894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM. New type I toxin-antitoxin families from “wild” and laboratory strains of E. coli: Ibs-Sib, ShoB-OhsC and Zor-Orz. RNA Biol 2012;9:1504–12. [DOI] [PubMed] [Google Scholar]
- Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol R 2008;72:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Pino A, Christensen-Dalsgaard M, Wyns L et al. . Doc of prophage P1 is inhibited by its antitoxin partner Phd through fold complementation. J Biol Chem 2008;283:30821–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit E, Sauer RT. The Doc toxin and Phd antidote proteins of the bacteriophage P1 plasmid addiction system form a heterotrimeric complex. J Biol Chem 1999;274:16813–8. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Rasmussen PB, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. P Natl Acad Sci USA 1986;83:3116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Wagner EG. RNA antitoxins. Curr Opin Microbiol 2007;10:117–24. [DOI] [PubMed] [Google Scholar]
- Germain E, Castro-Roa D, Zenkin N et al. . Molecular mechanism of bacterial persistence by HipA. Mol Cell 2013;52:248–54. [DOI] [PubMed] [Google Scholar]
- Germain E, Roghanian M, Gerdes K et al. . Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. P Natl Acad Sci USA 2015;112:5171–6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Goeders N, Chai R, Chen B et al. . Structure, evolution, and functions of bacterial type III toxin-antitoxin systems. Toxins (Basel) 2016;8:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Barrios AF, Zuo R, Hashimoto Y et al. . Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J Bacteriol 2005;188:305–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A, Maisonneuve E, Gerdes K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016;354:aaf4268. [DOI] [PubMed] [Google Scholar]
- Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 2003;301:1496–9. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Pogliano J, Helinski DR et al. . ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol Microbiol 2002;44:971–9. [DOI] [PubMed] [Google Scholar]
- Kamada K, Hanaoka F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol Cell 2005;19:497–509. [DOI] [PubMed] [Google Scholar]
- Kaspy I, Rotem E, Weiss N et al. . HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat Commun 2013;4:3001. [DOI] [PubMed] [Google Scholar]
- Kawano M. Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E. coli: hok/sok, ldr/rdl, symE/symR. RNA Biol 2012;9:1520–7. [DOI] [PubMed] [Google Scholar]
- Khoo SK, Loll B, Chan WT et al. . Molecular and structural characterization of the PezAT chromosomal toxin-antitoxin system of the human pathogen Streptococcus pneumoniae. J Biol Chem 2007;282:19606–18. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 2010;8:423–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch SB, Henderson TA, Hill TM. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol 2003;50:1199–213. [DOI] [PubMed] [Google Scholar]
- Kuroda A, Nomura K, Ohtomo R et al. . Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 2001;293:705–8. [DOI] [PubMed] [Google Scholar]
- Lehnherr H, Maguin E, Jafri S et al. . Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol 1993;233:414–28. [DOI] [PubMed] [Google Scholar]
- Lehnherr H, Yarmolinsky MB. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. P Natl Acad Sci USA 1995;92:3274–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R, Geeraerts D, Hallez R et al. . Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res 2011;39:5513–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Persister cells. Annu Rev Microbiol 2010;64:357–72. [DOI] [PubMed] [Google Scholar]
- Liu M, Zhang Y, Inouye M et al. . Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit. P Natl Acad Sci USA 2008;105:5885–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-Y, Wang Y, Walsh TR et al. . Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2015;16:161–8. [DOI] [PubMed] [Google Scholar]
- Mah T-FC, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 2001;9:34–9. [DOI] [PubMed] [Google Scholar]
- Maisonneuve E, Castro-Camargo M, Gerdes K (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 2013;154:1140–50. [DOI] [PubMed] [Google Scholar]
- Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell 2014;157:539–48. [DOI] [PubMed] [Google Scholar]
- Markovski M, Wickner S. Preventing bacterial suicide: a novel toxin-antitoxin strategy. Mol Cell 2013;52:611–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, Tan Q, Awano N et al. . YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol Microbiol 2012;84:979–89. [DOI] [PubMed] [Google Scholar]
- Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 2015;28:565–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering RC., Jr NDM-1–a cause for worldwide concern. N Engl J Med 2010;363:2377–9. [DOI] [PubMed] [Google Scholar]
- Mutschler H, Gebhardt M, Shoeman RL et al. . A novel mechanism of programmed cell death in bacteria by toxin–antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol 2011;9:e1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutschler H, Meinhart A. Epsilon/zeta systems: their role in resistance, virulence, and their potential for antibiotic development. J Mol Med (Berl) 2011;89:1183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nöllmann M, Crisona NJ, Arimondo PB. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie 2007;89:490–9. [DOI] [PubMed] [Google Scholar]
- Ogura T, Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. P Natl Acad Sci USA 1983;80:4784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R, Peti W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol 2016;12:208–14. [DOI] [PubMed] [Google Scholar]
- Patel S. Drivers of bacterial genomes plasticity and roles they play in pathogen virulence, persistence and drug resistance. Infect Genet Evol 2016;45:151–64. [DOI] [PubMed] [Google Scholar]
- Pedersen K, Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol 1999;32:1090–102. [DOI] [PubMed] [Google Scholar]
- Pedersen K, Zavialov AV, Pavlov MY et al. . The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 2003;112:131–40. [DOI] [PubMed] [Google Scholar]
- Pullinger GD, Lax AJ. A Salmonella dublin virulence plasmid locus that affects bacterial growth under nutrient-limited conditions. Mol Microbiol 1992;6:1631–43. [DOI] [PubMed] [Google Scholar]
- Ramisetty BC, Ghosh D, Roy Chowdhury M et al. . What is the link between stringent response, endoribonuclease encoding type II toxin-antitoxin systems and persistence? Front Microbiol 2016;7:1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocker A, Meinhart A. A cis-acting antitoxin domain within the chromosomal toxin-antitoxin module EzeT of Escherichia coli quenches toxin activity. Mol Microbiol 2015;97:589–604. [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Balani P, Min J et al. . HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature 2015;524:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short FL, Monson RE, Salmond GP. A Type III protein-RNA toxin-antitoxin system from Bacillus thuringiensis promotes plasmid retention during spore development. RNA Biol 2015;12:933–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet 2001;358:135–8. [DOI] [PubMed] [Google Scholar]
- Takagi H, Kakuta Y, Okada T et al. . Crystal structure of archaeal toxin-antitoxin RelE–RelB complex with implications for toxin activity and antitoxin effects. Nat Struct Mol Biol 2005;12:327–31. [DOI] [PubMed] [Google Scholar]
- Tan Q, Awano N, Inouye M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol Microbiol 2011;79:109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisted T, Gerdes K. Mechanism of post-segregational killing by the hok/sok system of plasmid R1: Sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene. J Mol Biol 1992;223:41–54. [DOI] [PubMed] [Google Scholar]
- Tripathi A, Dewan PC, Barua B et al. . Additional role for the ccd operon of F-plasmid as a transmissible persistence factor. P Natl Acad Sci USA 2012;109:12497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoson C, Wagner EG. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol Microbiol 2008;70:258–70. [DOI] [PubMed] [Google Scholar]
- Verstraeten N, Knapen WJ, Kint CI et al. . Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell 2015;59:9–21. [DOI] [PubMed] [Google Scholar]
- Vogel J, Argaman L, Wagner EG et al. . The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr Biol 2004;14:2271–6. [DOI] [PubMed] [Google Scholar]
- Wagner EGH, Unoson C. The toxin-antitoxin system tisB-istR1: Expression, regulation, and biological role in persister phenotypes. RNA Biol 2012;9:1513–9. [DOI] [PubMed] [Google Scholar]
- Wang X, Kim Y, Hong SH et al. . Antitoxin MqsA helps mediate the bacterial general stress response. Nat Chem Biol 2011;7:359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lord DM, Cheng H-Y et al. . A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol 2012;8:855–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wood TK. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microb 2011;77:5577–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Behiels E, Devreese B. Toxin-Antitoxin systems: their role in persistence, biofilm formation, and pathogenicity. Pathog Dis 2014;70:240–9. [DOI] [PubMed] [Google Scholar]
- Wen Y, Behiels E, Felix J et al. . The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence. Nucleic Acids Res 2014;42:10,134–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell LM, Maxwell A. The complex of DNA gyrase and quinolone drugs on DNA forms a barrier to the T7 DNA polymerase replication complex. J Mol Biol 2000;304:779–91. [DOI] [PubMed] [Google Scholar]
- Winther KS, Gerdes K. Regulation of enteric vapBC transcription: induction by VapC toxin dimer-breaking. Nucleic Acids Res 2012;40:4347–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RA, Waldor MK. A toxin–antitoxin system promotes the maintenance of an integrative conjugative element. PLos Genet 2009;5:e1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Inouye M. Regulation of growth and death in Escherichia coli by toxin–antitoxin systems. Nat Rev Microbiol 2011;9:779–90. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Park J-H, Inouye M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J Biol Chem 2009;284:28,746–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmolinsky MB. Programmed cell death in bacterial populations. Science 1995;267:836. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hoeflich KP et al. . MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell 2003;12:913–23. [DOI] [PubMed] [Google Scholar]
- Zielenkiewicz U, Ceglowski P. Mechanisms of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochim Pol 2001;48:1003–23. [PubMed] [Google Scholar]
- Zielenkiewicz U, Ceglowski P. The toxin-antitoxin system of the streptococcal plasmid pSM19035. J Bacteriol 2005;187:6094–105. [DOI] [PMC free article] [PubMed] [Google Scholar]