Abstract
Deeper sequencing and improved bioinformatics in conjunction with single-cell and metagenomic approaches continue to illuminate undercharacterized environmental microbial communities. This has propelled the ‘who is there, and what might they be doing’ paradigm to the uncultivated and has already radically changed the topology of the tree of life and provided key insights into the microbial contribution to biogeochemistry. While characterization of ‘who’ based on marker genes can describe a large fraction of the community, answering ‘what are they doing’ remains the elusive pinnacle for microbiology. Function-driven single-cell genomics provides a solution by using a function-based screen to subsample complex microbial communities in a targeted manner for the isolation and genome sequencing of single cells. This enables single-cell sequencing to be focused on cells with specific phenotypic or metabolic characteristics of interest. Recovered genomes are conclusively implicated for both encoding and exhibiting the feature of interest, improving downstream annotation and revealing activity levels within that environment. This emerging approach has already improved our understanding of microbial community functioning and facilitated the experimental analysis of uncharacterized gene product space. Here we provide a comprehensive review of strategies that have been applied for function-driven single-cell genomics and the future directions we envision.
Keywords: function-driven sequencing, single-cell genomics, microbial dark matter, single-cell activity
This review highlights the use of function-driven single-cell genomics for the targeted recovery of single-cell genomes from uncultivated or uncharacterized microbes implicated with a specific function or phenotype.
INTRODUCTION
Metagenomics has evolved through the need to better characterize our world by studying the composition and coding potential of complex microbial communities where a majority of the diversity is not yet cultivated. Facilitated by next-generation sequencing platforms, metagenomics can provide incredible sequence output from a minimally disturbed complex environmental sample, making it the gold standard for recovering DNA sequences from samples of interest. Despite the magnitude of the output, the logistics of metagenome assembly are complicated and many community features can be lost within the sequence data. Furthermore, annotating assembled sequences depends on homology-based computational tools to infer function with varying degrees of ‘homology creep’ (Woyke and Jarett 2015), and provides no information regarding the activity of that organism within the environment unless coupled with experimental approaches like metatranscriptomics, proteomics or stable isotope probing. Even when an ‘all of the above’ approach is taken, the final result is the average of a bulk population, masking the large variability in functional activity due to spatial microenvironments or phenotypic noise known to drive divergent functions even in clonal populations (Ackermann 2013). Thus, the strength of metagenomics for its indiscriminate capture of all DNA from a heterogeneous sample is also its pitfall in that the complex output can confound the recovery of complete genomes, obscure genomic population heterogeneities (Engel, Stepanauskas and Moran 2014), overlook rare organisms (Ainsworth et al.2015), miss ecological relationships, blend spatially divergent populations and diminish perspective for the interpretation of recovered sequences of unknown function.
Single-cell genomics offers a complimentary approach to metagenomics by capturing and amplifying DNA from a single isolated cell, as opposed to many cells in metagenomics, and is routinely capable of producing partial (Zhang et al.2006), and even complete (Woyke et al.2010), non-composite genomes. This approach thus addresses some of the caveats of metagenomics at the sacrifice of throughput. Although technical limitations and inherent biases currently exist for single-cell approaches (Lasken 2012; Gawad, Koh and Quake 2016), within microbiology single-cell genomics has demonstrated itself as a powerful tool by generating genomes from rare (Martijn et al.2015), symbiotic (Siegl et al.2011) and previously uncharacterized microbial lineages (Marcy et al.2007; Blainey et al.2011; Rinke et al.2013). In addition, auxiliary DNA from viruses, organelles, plasmids, transformed DNA and symbionts is also captured and sequenced along with the host DNA due to its colocalization with the cell (Stepanauskas 2015). Thus, associations between these genetic elements and the host organism of interest are maintained with high resolution, allowing characterization of subtle ecological interactions (Yoon et al.2011; Roux et al.2014). When coupled with microscopy, single-cell genomics enables spatiotemporal characterization of microscale environments, improving the resolution at which our understanding of microbial ecology takes place (Landry et al.2013). Single-cell genomics has identified a novel genetic code (Campbell et al.2013), uncovered unexplored protein sequence space relevant for biotechnology and human medicine (Wilson et al.2014), and facilitated the expansion of novel branches in the bacterial and archaeal tree of life and improved phylogenetic read anchoring for metagenomic data sets (Rinke et al.2013). With these efforts, pipelines for the single-cell isolation and sequencing of environmental microbes have optimized their efficiency and throughput (Hutchison et al.2005; Woyke et al.2011; Rinke et al.2014), thereby advancing single-cell genomics methods to the next level.
The major outstanding limitation in metagenomic and single-cell approaches is the overreliance on using predicted protein function as a proxy for functional activity in the environment. These approaches lack the ability to discern which recovered populations are active, potentially overestimating the importance of abundant organisms while overlooking significant ecological contributions of lower abundance organisms (Martinez-Garcia et al.2012). This caveat becomes even more pronounced when trying to study ‘microbial dark matter’ i.e., microbes that lack characterized cultured representatives (Rinke et al.2013; Kamke et al.2014; Brown et al.2015) and whose genes often lack functional prediction (Youssef et al.2011; Kantor et al.2013; McLean et al.2013). Thus, although both metagenomic and single-cell genomic approaches can recover novel genes, neither approach currently provides insight into the function of the gene or activity level of the organism.
Identifying the function of a gene has traditionally relied on classical genetic knockout/complementation studies, and more recently high-throughput relative fitness studies (Wetmore et al.2015). Because these approaches require compatible genetic toolkits and cultivable organisms, they are unsuitable for studying the function of unknown genes from the uncultivated majority. Function-based screens of metagenomic DNA sequences have previously utilized clone libraries to heterologously express environment-derived DNA fragments in a gain-of-function approach (Daniel 2005). Escherichia coli, the workhorse of heterologous screening approaches, was computationally determined to be able to transcribe ∼40% of genes from well-known cultivated lineages of microbes with the typical expression library approach (Gabor, Alkema and Janssen 2004). Considering downstream incompatibilities such as codon bias (Tuller et al.2010), strategic rare codon utilization (Komar et al.1999), required metabolite pools, post-translational modification, accessory proteins, apoprotein activation, secretion and even the availability of a compatible read-out assay, it becomes apparent that only a small fraction of functional space is accessible through this approach. Furthermore, when introducing more divergent DNA sequences, such as those from candidate phyla, the success rate for accessing the vast functional diversity that exists with current tools is expected to rapidly diminish. Thus, the majority of unknown functions from uncultivated organisms remain obscured within their native expression hosts.
Due to the increasing recovery and accumulation of sequences of unknown function, a context-driven approach to investigate the roles of these genes within their native hosts and environment is required. This will facilitate improved protein annotation, interpretation of microbial networks and understanding of microbial influences on biogeochemistry (Hicks and Prather 2014; Woyke and Jarett 2015). Thus, an increased focus on characterizing the functional roles and activity levels of uncultivated organisms in conjunction with downstream genomic sequencing has motivated the development of methods to ascertain both functional trait and/or phenotype, and corresponding coding potential from these organisms at the resolution of a single cell. Here we present the approaches and limitations of pioneering studies focused on developing a suite of methods for the function-driven single-cell identification, isolation and sequencing of uncultivated environmental microbes. Though this review focuses on these approaches from a largely bacterial and archaeal perspective, many of the same strategies can be adopted to small environmental eukaryotic cells, and some are being applied in human cells to address human health issues (Gao et al.2004; Yu et al.2011).
FUNCTION-DRIVEN APPROACHES
Label-free
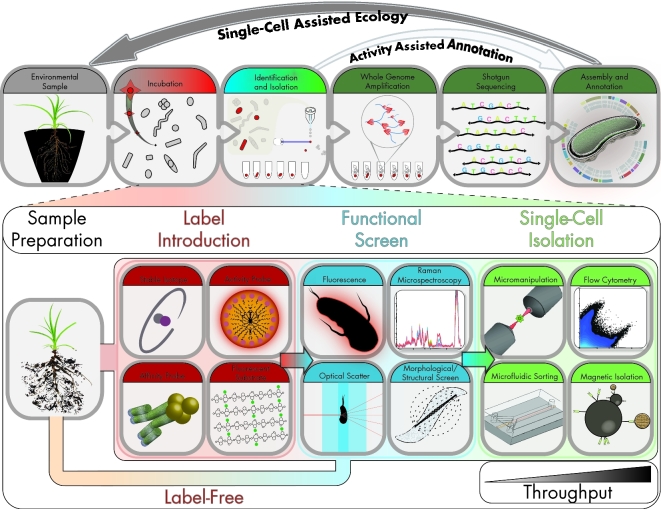
The earliest ‘targeted’ single-cell screens took advantage of previously characterized environments where distinguishing morphological traits were used as identifying selection criteria. Marcy et al. first applied this approach in 2007 for single-cell genomics from members of the uncultivated TM7 phylum (Marcy et al.2007). To isolate their target, rod-shaped cells from the human mouth were sorted and enriched in a microfluidic chip for single-cell genomics. A similar strategy was applied in 2010 by Woyke et al. to isolate Candidatus Sulcia muelleri, an obligate symbiont of the green sharpshooter, through micromanipulation, producing the first complete genome by single-cell sequencing approaches (Woyke et al.2010). Other morphological features relating directly to function, including photosynthetic pigments (Rodrigue et al.2009), polyhydroxybutyrate production (Tyo, Zhou and Stephanopoulos 2006) and magnetotaxis (Kolinko et al.2012), have also been used as selection criteria for identifying and isolating single cells in true function-driven screening approaches (Fig. 1). However, since a majority of activities of interest cannot be selected by a unique morphological trait, label-based screens provide an alternate route to visualize additional functions.
Figure 1.
Flow diagram of function-driven single-cell genomic pipeline. Top panel shows general workflow highlighting how function screen can aid in annotation and ecology of organisms recovered from original sample. Bottom panel highlights specific identification and isolation strategies. Throughput for flow cytometry and magnetic isolation is generally larger than micromanipulation and microfluidic systems.
Label-based approaches
Recent innovations in function-driven single-cell genomics have successfully exploited the association, metabolism or integration of labeled substrates to implicate functional activity in specific microbial guilds within environmental communities. These studies have utilized fluorescently tagged (Martinez-Garcia et al.2012; Geva-Zatorsky et al.2015) and isotopically labeled (Huang et al.2009; Li et al.2012) substrates (Fig. 1) to identify and recover individual active microbes that respond to, and interact with, these substrates.
Fluorescent substrate
The fluorescent substrate single amplified genome analysis (FS-SAGA) method was developed by Martinez-Garcia et al. (2012) to identify populations active in the degradation of the complex carbohydrate laminarin in aquatic systems. Though bacterial degradation of this abundant substrate had been demonstrated in aquatic environments (Arnosti 2011), the identity of the responsible population remained obscure (Alderkamp, van Rijssel and Bolhuis 2007). To conclusively identify this population, Martinez-Garcia et al. spiked fluorescently labeled laminarin into freshly recovered aquatic samples. The samples were then screened by flow cytometry to recover cells that became increasingly fluorescent (strategy outlined in Fig. 1). The captured cell population was largely dominated by a few Verrucomicrobia taxa, which had low overall relative abundances (<1%) within the starting population as determined by culture-independent methods. Single-cell genomes generated from this population were highly enriched in a number of glycoside hydrolase families, suggesting the accuracy of the FS-SAGA method for identifying this highly active, yet relatively rare, population and implicating the activity of these specific hydrolases with laminarin degradation.
Bioorthogonal tagging
A caveat of fluorescently labeled substrates is that the modification required to attach fluorophores may abolish native recognition, transport and metabolism of that substrate. One strategy to circumvent the complication of bulky fluorophore attachment has been the utilization of non-canonical substrates that instead contain only a small bioorthogonal tag. A bioorthogonal reaction, such as the commonly used azide-alkyne ‘click’ reaction, is advantageous since it readily proceeds within biological systems without cross-reaction (Baskin et al.2007). During incubation, these non-canonical substrates are integrated into the biomass of metabolically active, but potentially non-replicative, cells allowing incorporation into cellular structures before a subsequent introduction of a ‘click’ compatible fluorescent marker (Speers, Adam and Cravatt 2003) (Fig. 2, no. 7). This has been successfully demonstrated with non-canonical amino acids, nucleotides, lipids and sugar analogs modified to contain either an azide or alkyne group for ‘click’ chemistry compatibility (Dieterich et al.2007; Salic and Mitchison 2008; Neef and Schultz 2009; Paredes and Das 2011; Hatzenpichler et al.2014; Samo et al.2014; Geva-Zatorsky et al.2015), though the enzymatic promiscuity that permits integration of these analogues may not be universal among all bacteria.
Figure 2.
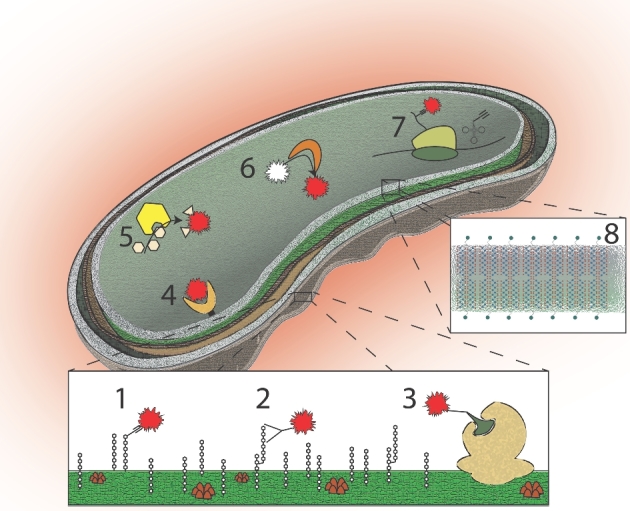
Labeling targets for function-based single-cell genomics exploiting activity-based profiling and incorporation approaches. (1) Extracellular integration, (2) extracellular recognition, (3) extracellular enzyme affinity, (4) intracellular receptor, (5) intracellular product, (6) intracellular enzymatic reaction, (7) intracellular integration, (8) isotope integration.
Hatzenpichler et al. (2016) applied a bioorthogonal non-canonical amino acid tagging (BONCAT) approach to investigate the activity and microbial ecology of slow-growing methane-oxidizing/sulfur-reducing archaea/bacteria consortia with intra-aggregate resolution. Methane seep enrichment cultures were incubated, with and without methane stimulation, in the presence of L-homopropargylglycine (HPG). This alkyne-containing non-canonical amino acid is a structural analog for L-methionine that is readily taken up and integrated into nascent proteins within active cells. Following incorporation of HPG into cellular proteins, click-based fluorophore labeling marked ∼ 25% of aggregates as translationally active (compared to ∼2% in samples incubated without methane). The community identity of each individual aggregate was then determined by sorting the fluorescent aggregates with flow cytometery, followed by lysis, genome amplification with Phi29 polymerase (as detailed in Fig. 1), and amplification of the 16S rRNA gene with ‘universal’ primers. This approach allowed mapping the taxonomic identities of six distinct archaeal lineages (ANME-1a, 1b, 2a, 2b, 2c and 3) in individual aggregates with their preferred syntrophic sulfate-reducing partners.
Substrate-independent isotope
To avoid complications of modified substrates entirely, isotopically labeled substrates can be introduced instead. However, because of the cost and logistical challenge associated with synthesizing substrates from stable isotopes, Berry et al. (2015) developed a substrate-independent strategy using deuterated water for activity-based identification of individual cells following perturbation of a community with any native substrate. As microbes grow in the presence of heavy water (D2O), deuterium is actively integrated within their biomass through the synthesis of new lipids and proteins. The replacement of the C-Hx bond with the C-Dx bond causes a spectral shift due to the change in vibrational energy when measured by Raman microspectroscopy (Wei et al.2013). Thus, the emergence of the C-Dx peak in the Raman spectrum serves as a proxy for the extent of biosynthetic activity, thereby unambiguously labeling all active organisms in a substrate-independent manner (Kopf et al.2015). This strategy enables a targeted approach when obtaining labeled substrates of interest is prohibitive, or where environmentally relevant substrates are unknown. Berry et al. (2015) coupled the labeling of both pure culture and complex environmental microbial cultures with D2O and employed single-cell Raman microspectroscopy detection within capillary tubes (Fig. 1) to screen and sort individual cells. Following benchmarking of the method with cultures of model organisms from diverse phyla, the approach was applied to a mouse cecal microbiome stimulated with a range of carbohydrates. Responses from randomly screened cells following incubation ranged from 9% (no substrate addition) to 41% (mucin) and 81% (glucose) labeling, suggesting that different substrates successfully activated different subpopulations. To validate the observed result, fluorescent in situ hybridization (FISH) probes targeting two known mucin degraders served as positive controls for activity and correctly identified their targets as active under expected conditions. This study demonstrates the power of the substrate-independent isotope approach by making nearly any substrate and environment amenable to probing.
Activity-based probes
Though largely yet to be applied, there are many function-based probes compatible with application in single-cell genomics that originate from, and elaborate on, the field of activity-based profiling (ABP). ABP historically focused on the identification of a specific protein family activity from within a bulk protein pool isolated from a single organism (Barglow and Cravatt 2007). The most extensive developments in ABP have been employing family-specific probes for the identification of differential protein activity levels between healthy and diseased states (Jessani and Cravatt 2004). These probes have chiefly targeted large protein families including serine (Liu, Patricelli and Cravatt 1999), cysteine (Kato et al.2005) and metallohydrolases (Saghatelian et al.2004), and have resulted in numerous insights in proteome function and annotation (Adam et al. 2004). Strategies for improving the design and activity of these probes have been extensively reviewed (Cravatt, Wright and Kozarich 2008) and include methods for improving reactivity (Walvoort et al.2012), quantification (Okerberg et al.2005), labeled fraction recovery (Chan et al.2004), identification of participating proteins and active site residues (Speers and Cravatt 2005), and effectiveness of target competitive inhibitors (Evans et al.2005).
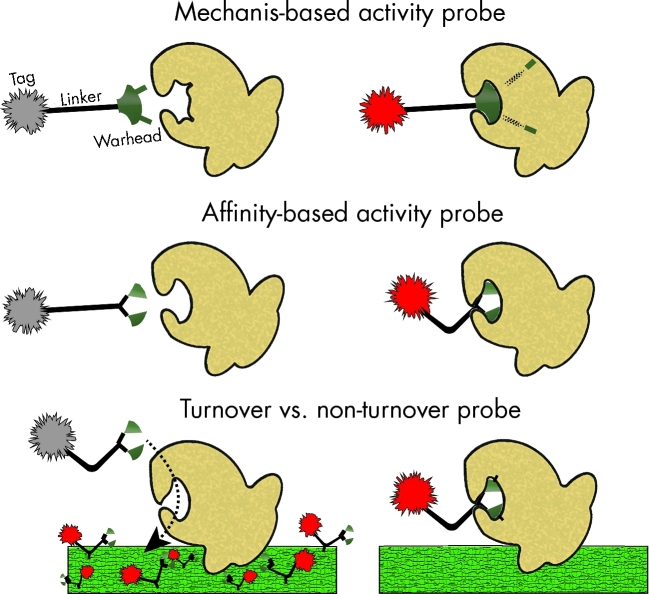
Activity-based probes are typically designed using a modular three-component strategy: a warhead for target specificity, a tag for detection, and a polyfunctional linker for joining the warhead and tag that can also aid in quenching and/or covalent attachment following activation (Sadler and Wright 2015) (Fig. 3). Two primary classes of functional probes, mechanism and affinity based, have been developed and successfully demonstrated (Chauvigne-Hines et al.2012). Mechanism-based probes rely on the direct enzymatic transformation of the probe by a specific cellular enzyme or metabolite to become activated. These probes often mark their target with high specificity by forming a covalent bond with a key active residue in the enzyme, functioning as an irreversible inhibitor (Walvoort et al.2012). This approach is advantageous when looking to characterize only active protein forms from zymogens (Adam et al. 2004). Affinity probes alternatively do not require mechanistic activation by the target, but physically associate with a receptor or molecule of interest for retention or tag activation (Chan et al.2004). An example of this strategy is the nanoparticle-based quorum-sensing probe that relies only on the affinity between the homoserine lactone warhead and the intracellular quorum-sensing receptor (Mukherji et al.2013) (Table 1). In this example, however, no strategy for tag activation or covalent attachment exists upon receptor binding, likely reducing the sensitivity of the probe screen when advancing to environmental samples.
Figure 3.
Activity-based probe profiling strategies.
Table 1.
Potential strategies for single-cell identification and targets of interest. Numbering corresponds to labeling targets in Figure 2.
| Target | Example |
|---|---|
| Labeling strategies | |
| (1) Extracellular integration | Oligosaccharide labeling (Geva-Zatorsky et al.2015) |
| (2) Extracellular recognition | DNA aptamer (Chang et al.2013), phage binding domain (Kretzer et al.2007) |
| (3) Extracellular enzyme affinity | Fluorescent substrate (Martinez-Garcia et al.2012), glycoside hydrolases (Chauvigne-Hines et al.2012), phytase (Berry and Harich 2013), penicillin binding proteins (Kocaoglu and Carlson 2013), β-lactamases (Bottcher and Sieber 2012; Shao and Xing 2012), showdomycin (Bottcher and Sieber 2010) |
| (4) Intracellular receptor | Quorum sensing (Mukherji et al.2013) |
| (5) Intracellular product | Polyhydroxybutyrate granules (Tyo, Zhou and Stephanopoulos 2006), hypochlorous acid production (Sun et al.2014), ROS production (Gomes, Fernandes and Lima 2005) |
| (6) Intracellular enzymatic reaction | Non-ribosomal peptide synthetases (Konno et al.2015), polyketide synthases (Meier et al.2009), nucleophilic RNAs (McDonald et al.2014), adenylation (Duckworth et al.2012), glyceraldehyde-3-phosphate dehydrogenase (Kaschani et al.2012), glutathione reductase (Lou et al.2014), sulfatase (Park, Rhee and Hong 2012), phosphatase (Kim et al.2011), aldehyde dehydrogenase (Adam, Cravatt and Sorensen 2001), palmitoyl actytransferases (Zheng et al.2013), β-glucosidases (Kallemeijn et al.2012; Walvoort et al.2012), agmatine deiminase (Marchenko et al.2015), monoamine oxidase (Li et al.2014), N-acetylgalactosaminidase (Kalidasan et al.2013), alkaline phosphatase (Kim et al.2011), metalloproteases (Sieber et al.2006), cysteine proteases (Kato et al.2005), serine hydrolase (Liu, Patricelli and Cravatt 1999) |
| (7) Intracellular integration | BONCAT (Dieterich et al.2007), degenerate tRNA synthatases (Ngo et al.2009), RNA labeling (Paredes and Das 2011) |
| (8) Isotope integration | 2H2O (Berry et al.2015), 13CO2 (Li et al.2012), [13C] naphthalene (Huang et al.2009) |
| Characteristics | |
| General properties | Magnetotaxis (Kolinko et al.2012), cell morphology (Marcy et al.2007) |
| Cellular activity | Electron transport chain (Kaprelyants and Kell 1993), live/dead (Nocker, Cheung and Camper 2006), reductase (Lahtinen et al.2008) |
| Intracellular ions | pH (Han and Burgess 2010), [Zn2+] (Walkup et al.2000) |
Similar to how probing enzyme targets from a bulk pool is a powerful approach as it can greatly reduce the complexity of results when screening the proteome, activity-based probes can analogously be applied to a complex microbial community to return a narrowed result of what microorganisms in the given environment are actively performing an activity of interest (Fig. 2). Translating this technology from its previous proteomic application to single-cell thus shifts the focus from a family of proteins within a bulk pool as the fundamental unit of activity to the functional profile of an individual cell within a community as a fundamental unit. Though applications in activity-based profiling of environmental microbes are just emerging (Sadler and Wright 2015), we envision that by coupling this approach with cell sorting and single-cell genomics, activity-based probe selection offers a strategy to parse a complex community, identify individual organisms participating in a given function and relate this activity, with the context of their genome, to the community at large. In addition to surveying individual cellular activities with a specific functional probe, screens can be multiplexed with other cellular function or status signals using a suite of probes developed for cellular conditions and activity levels (Sieracki, Cucci and Nicinski 1999; Kalyuzhnaya, Lidstrom and Chistoserdova 2008; Chan, Dodani and Chang 2012) (Table 1).
Magnetic capture
Beyond magnetic enrichment of magnetotatic bacteria as a function-driven screen (Kolinko et al.2016), magnetic capture using functionalized magnetic beads presents itself as a high-throughput function-driven selection screen with the ability to enrich all members with a given extracellular marker from an entire sample (Fig. 2, no. 2). The ability to recover cells using aptamer (Chang et al.2013) or phage binding domains (Kretzer et al.2007) has already demonstrated that this approach could readily be applied for function-driven single-cell genomics.
CHALLENGES
Beyond increasing the array of compatible functional approaches, one of the primary obstacles to improving the application of function-driven single-cell screens within environmental samples is ensuring a sufficiently strong probe signal beyond that of background noise e.g. natural phycobiliproteins can produce higher fluorescence than organic fluorophores (Chiu 2014). Due to the limited number of targets for tagging within a cell (e.g. receptors or enzyme active sites for a specific function of interest), ensuring conclusive detection of a positive result beyond background noise, whether fluorescent or Raman, is paramount to the effectiveness of the single-cell approach. Though general strategies for improving probe design have been widely studied and extensively reviewed (Chan, Dodani and Chang 2012; Vendrell et al.2012), coupling the addition of these probe improvements with the specificity required for function-driven screens remains largely a case-by-case approach. Here we outline some of the general strategies that have been successfully used to improve the detection of function-driven activities.
Activity-based probes
Turn on signal
Minimizing avoidable background signals from non-specific associations of fluorescent probes have resulted in the design of ‘turn-on’ probes that display improved signal intensity only following structural or conformational activation (Fig. 3). For fluorescent probes, this can be achieved with quenching modules that absorb the fluorescent reporter signal (Li et al.2014), fluorogenic precursor probes (Weissleder et al.1999; Kim et al.2011), photoinduced electron transfer (de Silva, Moody and Wright 2009), structural conformation (Mello and Finney 2001) and a variety of other strategies (Chan, Dodani and Chang 2012). In each case, a covalent or conformational modification to the probe through a specific activity of the cell results in an increase in detectable signal, localizing the activated probe fluorescence to within functionally active cells.
Turnover vs non-turnover
Two primary types of mechanism-based probes have been described with regard to activation: turnover and non-turnover. Non-turnover probes are essentially irreversible inhibitors that occupy the enzymatic active site following activation. These probes often function through high-affinity covalent bonding to active site residues that result in a structural change for probe-signal activation (Kallemeijn et al.2012). Although this probe strategy ensures the activated fluorophore is retained inside the cell, this approach imposes a maximum signal for each cell due to the finite number of targets. In addition, regardless of enzyme kcat or km (the parameters that would largely determine relative cellular activity levels), if the concentration of target enzyme is present at low levels within the cell, detecting a discernable signal above background could be prevented. This limitation thereby prevents a relative comparison in true activity levels between different organisms as measured by the fluorescent signal in the function-driven screen. Turnover probes, alternatively, do not result in inactivation of the target enzyme following modification and thereby allow repeated activation of multiple probe molecules by a single enzyme (Kalidasan et al.2013). This strategy improves the maximum theoretical signal attainable by a given cell by maintaining a functional pool of enzymatic activity independent of probe activation. In addition, kcat and km could influence the rate and concentration to which the activated probe can accumulate, allowing semiquantitative insights into relative activity levels between cells.
Retaining signal
Permeable probes are required to pass through intact membranes to reach intracellular targets of interest in physiologically active cells. Following activation, however, turnover mechanism-based and affinity probes often remain free to diffuse out of the cell into the bulk environment, diminishing the intensity of the signal within the cell and increasing background. While non-turnover probes are retained in the cell through their mechanistic bonding to the enzyme active site, if activated turnover probes could be retained inside the cell through non-specific covalent bonding to cell biomass instead of the enzymatic site, this approach would allow for both the covalent retention of the probe within a cell of interest while allowing the target enzyme to continue activating probes (Kalidasan et al.2013). Alternatively, activated probes that become unable to diffuse through the membrane due to changes in charge or solubility are other strategies that could be designed for retaining signal. Using classical probe design approaches, those structures that can integrate modular components to address all the above design strategies including turn-on signal, enzyme active site turnover, and a strategy for intracellular probe retention present a route for best chances for detection of signal above noise in a function-driven detection screen (Kalidasan et al.2013).
Other challenges
Modern fluorophores
Many of the fluorescent markers conjugated to functional probes have remained classic organic dyes (e.g. fluorescein, rhodamine) although advances in materials science have produced new fluorescent molecules with improved properties. Examples include semiconductor quantum dots (Qdots) (Gao et al.2004) and π-conjugated polymer dots (Pdots) (Wu et al.2008). This limitation for function-driven single-cell genomics from classical dyes stems from their relatively low absorptivity and poor photostability (Chiu 2014). Recent advances in dot fluorophore technology have improved the absorptivity and quantum efficiency of applied probes and have even demonstrated compatibility with click chemistry labeling (Wu et al.2010).
Surface-enhanced Raman detection
Raman microspectroscopy is a very powerful tool as it provides an entire cell ‘fingerprint’ by measuring inelastic scattering of photons from abundant bonds within a single cell. However, Raman vibrations are relatively rare events in cells, with only 1 in 106–8 photons causing a detectable vibration, and often require measurements on the order of minutes per cell (Jarvis and Goodacre 2008). In addition, due to the complexity of simultaneously probing all vibrational energies within the cell, specific signals from within the spectrum can become convoluted. The addition of nanometer-scale Ag/Au particles or colloids to a culture of interest results in a surface-enhanced Raman spectroscopy (SERS) phenomenon, whereby the sensitivity and acquisition time are improved in isotopic measurements on single cells (Kubryk et al.2015). SERS not only enables improved throughput of the approach, but also potentially reduces the amount of isotope incorporation necessary for detection, minimizing any negative impacts of high levels of isotopically labeled substrates. Current limitations with the SERS approach within function-driven single-cell screens include heterogeneity of nanoparticle binding (Kubryk et al.2015), and any potential toxicity due to nanoparticle association with cells (Rai, Yadav and Gade 2009).
Sample activity
Unlike traditional metagenomics or single-cell sampling methods where samples can be taken and immediately preserved through freezing or fixation as instantaneous snapshots, function-driven methods require both viability and activity to be maintained following sampling. Because of this, and in the interest of maintaining an as close to in situ observation as possible, minimal disturbance to the physical and chemical properties of the sample is necessary. In addition, delays in sample screening perturb the natural abundances and activities of the microbial community within the sample, displacing events from the true in situ activity. These limitations logistically restrict the extent of samples that can be screened to those that can be quickly recovered with minimal disturbance from the target environment to the lab.
Disaggregation of cells
As the name implies, single-cell genomics requires the ability to manipulate individual cells, a task which is not always trivial depending on sample type. Vortexing, sonication, aspiration, centrifugation and even grinding are all approaches that have been used to liberate single cells from difficult sample environments (Rinke et al.2014). The optimal method for liberating cells while still maintaining activity necessary for downstream screening will vary by sample type and likely require a case-by-case approach.
Sample fixation
Fixation is often a critical step for the preparation of samples where membrane permeabilization is required for introduction of a probe or label. For function-driven single-cell genomics this process can become problematic, as fixation often interferes with downstream whole genome amplification. Formaldehyde is a common fixation agent for the stabilization and inactivation of cells through global crosslinking. However, crosslinking of genomic DNA with formaldehyde has been shown to prevent amplification (Ben-Ezra et al.1991), alter nucleotide sequences and degrade DNA (Williams et al.1999), making it incompatible with single-cell genomics. While ethanol, a precipitating fixative, has been found to produce fixed cells that yield sufficient amplification product for single-cell sequencing, it comes at the cost of reduced genome recovery (Clingenpeel et al.2014). A range of alternative fixation agents and protocols with varying degrees of previously reported DNA amplification success could be screened with Phi29 for compatibility with whole genome amplification (Srinivasan, Sedmak and Jewell 2002).
Cell lysis
Lysing an isolated cell under mild enough conditions to maintain the integrity of the genomic DNA prior to whole genome amplification can be challenging due to the varying degree of difficulty lysing structurally diverse cells. Alkaline lysis, lysozyme treatment, heat, freeze-thaw and detergents have been used with varying success of subsequent amplification by method and cell type (Rinke et al.2014). Developing efficient cell lysis methods that routinely liberate DNA from a wide range of microbial cells will broaden potential targets of single-cell approaches.
FUTURE
While many compatible tools and methodological strategies outlined here for probing the function of the uncultivated majority exist, only a handful of remarkable studies to date have applied these tools for the characterization and recovery of single-cell genomes from the environment. These findings, such as the high levels of functional activity from low abundance keystone organisms and the resolution of taxonomic partnerships within diverse methane-oxidizing aggregates, have contributed to the fundamental understanding of microscale ecological dynamics. Continuing to apply existing tools and developing new approaches for characterizing the in situ function of uncultivated microbes in the environment will continue to expand our understanding of the functional role of ‘microbial dark matter’. Moving forward, one primary pitfall that remains with this strategy is that linking the activity with a particular coding sequence can be easy to apply if the encoded function is homologous to known and previously characterized genes. If recovered genes involved in the targeted function screen are so divergent that they can only be assigned as hypothetical, attributing the screened activity to specific sequences in the genome becomes obfuscated. However, compiling recovered genes of unknown function implicated with the screened activity and coupling them with an optimized DNA synthesis and expression approach could allow the targeted discovery of truly novel gene function from uncultivated organisms.
SUMMARY
Function-driven single-cell genomic approaches offer a unique route to directly study the activities of uncultivated organisms, at single-cell resolution, in an in situ style approach without any prior knowledge of cellular activity. The ability to screen all members of a microbial community, independent of our ability to cultivate them, and implicate individual organisms as actively involved with a specific functional activity or exhibiting a certain morphological phenotype provides valuable information regarding their ecological role within a given ecosystem. Being able to then couple the genomic sequence of the identified organisms back to the screened activity or trait allows for the characterization of potentially unknown and unstudied genes and pathways in their native hosts. This represents a critical strategy to advance our understanding of microbial community functioning and move beyond purely sequence-based predictions.
FUNDING
The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under Contract No. DE-AC02-05CH11231. This work was supported under the LBNL Microbes to Biomes LDRD entitled ‘Tackling microbial-mediated plant carbon decomposition using “function-driven” genomics’.
Conflict of interest. None declared
REFERENCES
- Ackermann M. Microbial individuality in the natural environment. ISME J 2013;7:465–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam GC, Burbaum J, Kozarich JW et al. . Mapping enzyme active sites in complex proteomes. J Am Chem Soc 2004;126:1363–8. [DOI] [PubMed] [Google Scholar]
- Adam GC, Cravatt BF, Sorensen EJ. Profiling the specific reactivity of the proteome with non-directed activity-based probes. Chem Biol 2001;8:81–95. [DOI] [PubMed] [Google Scholar]
- Ainsworth TD, Krause L, Bridge T et al. . The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J 2015;9:2261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderkamp AC, van Rijssel M, Bolhuis H. Characterization of marine bacteria and the activity of their enzyme systems involved in degradation of the algal storage glucan laminarin. FEMS Microbiol Ecol 2007;59:108–17. [DOI] [PubMed] [Google Scholar]
- Arnosti C. Microbial Extracellular Enzymes and the Marine Carbon Cycle. Annu Rev Mar Sci 2011;3:401–25. [DOI] [PubMed] [Google Scholar]
- Barglow KT, Cravatt BF. Activity-based protein profiling for the functional annotation of enzymes. Nat Methods 2007;4:822–7. [DOI] [PubMed] [Google Scholar]
- Baskin JM, Prescher JA, Laughlin ST et al. . Copper-free click chemistry for dynamic in vivo imaging. P Natl Acad Sci USA 2007;104:16793–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ezra J, Johnson DA, Rossi J et al. . Effect of fixation on the amplification of nucleic acids from paraffin-embedded material by the polymerase chain reaction. J Histochem Cytochem 1991;39:351–4. [DOI] [PubMed] [Google Scholar]
- Berry D, Mader E, Lee TK et al. . Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. P Natl Acad Sci USA 2015;112:E194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DF, Harich K. Tetrachlorofluorescein TInsP(5) as a substrate analog probe for measuring phytase activity in surface water: proof of concept. J Environ Qual 2013;42:56–64. [DOI] [PubMed] [Google Scholar]
- Blainey PC, Mosier AC, Potanina A et al. . Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 2011;6:e16626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher T, Sieber SA. Showdomycin as a versatile chemical tool for the detection of pathogenesis-associated enzymes in bacteria. J Am Chem Soc 2010;132:6964–72. [DOI] [PubMed] [Google Scholar]
- Bottcher T, Sieber SA. beta-Lactams and beta-lactones as activity-based probes in chemical biology. Medchemcomm 2012;3:408–17. [Google Scholar]
- Brown CT, Hug LA, Thomas BC et al. . Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 2015;523:208–11. [DOI] [PubMed] [Google Scholar]
- Campbell JH, O’Donoghue P, Campbell AG et al. . UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. P Natl Acad Sci USA 2013;110:5540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EWS, Chattopadhaya S, Panicker RC et al. . Developing photoactive affinity probes for proteomic profiling: Hydroxamate-based probes for metalloproteases. J Am Chem Soc 2004;126:14435–46. [DOI] [PubMed] [Google Scholar]
- Chan J, Dodani SC, Chang CJ. Reaction-based small-molecule fluorescent probes for chemoselective bioimaging. Nat Chem 2012;4:973–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Yang CY, Sun RL et al. . Rapid single cell detection of Staphylococcus aureus by aptamer- conjugated gold nanoparticles. Sci Rep-UK 2013;3:1863–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvigne-Hines LM, Anderson LN, Weaver HM et al. . Suite of activity-based probes for cellulose-degrading enzymes. J Am Chem Soc 2012;134:20521–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu DT. Highly fluorescent semiconducting polymer dots for biology and medicine. Abstr Pap Am Chem S 2014;248:3086–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clingenpeel S, Schwientek P, Hugenholtz P et al. . Effects of sample treatments on genome recovery via single-cell genomics. ISME J 2014;8:2546–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem 2008;77:383–414. [DOI] [PubMed] [Google Scholar]
- Daniel R. The metagenomics of soil. Nat Rev Microbiol 2005;3:470–8. [DOI] [PubMed] [Google Scholar]
- de Silva AP, Moody TS, Wright GD. Fluorescent PET (Photoinduced Electron Transfer) sensors as potent analytical tools. Analyst 2009;134:2385–93. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Lee JJ, Link AJ et al. . Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc 2007;2:532–40. [DOI] [PubMed] [Google Scholar]
- Duckworth BP, Wilson DJ, Nelson KM et al. . Development of a selective activity-based probe for adenylating enzymes: profiling MbtA involved in siderophore biosynthesis from Mycobacterium tuberculosis. ACS Chem Biol 2012;7:1653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P, Stepanauskas R, Moran NA. Hidden diversity in honey bee gut symbionts detected by single-cell genomics. PLoS Genet 2014;10:e1004596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Saghatelian A, Sorensen EJ et al. . Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nat Biotechnol 2005;23:1303–7. [DOI] [PubMed] [Google Scholar]
- Gabor EM, Alkema WBL, Janssen DB. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ Microbiol 2004;6:879–86. [DOI] [PubMed] [Google Scholar]
- Gao X, Cui Y, Levenson RM et al. . In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol 2004;22:969–76. [DOI] [PubMed] [Google Scholar]
- Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet 2016;17:175–88. [DOI] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Alvarez D, Hudak JE et al. . In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat Med 2015;21:1091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes A, Fernandes E, Lima JLFC. Fluorescence probes used for detection of reactive oxygen species. J Biochem Bioph Meth 2005;65:45–80. [DOI] [PubMed] [Google Scholar]
- Han J, Burgess K. Fluorescent indicators for intracellular pH. Chem Rev 2010;110:2709–28. [DOI] [PubMed] [Google Scholar]
- Hatzenpichler R, Connon SA, Goudeau D et al. . Visualizing in situ translational activity for identifying and sorting slow-growing archaeal-bacterial consortia. P Natl Acad Sci USA 2016;113:E4069–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenpichler R, Scheller S, Tavormina PL et al. . In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry. Environ Microbiol 2014;16:2568–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks MA, Prather KL. Bioprospecting in the genomic age. Adv Appl Microbiol 2014;87:111–46. [DOI] [PubMed] [Google Scholar]
- Huang WE, Ferguson A, Singer AC et al. . Resolving genetic functions within microbial populations: in situ analyses using rRNA and mRNA stable isotope probing coupled with single-cell Raman-fluorescence in situ hybridization. Appl Environ Microb 2009;75:234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CA, Smith HO, Pfannkoch C et al. . Cell-free cloning using phi 29 DNA polymerase. P Natl Acad Sci USA 2005;102:17332–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis RM, Goodacre R. Characterisation and identification of bacteria using SERS. Chem Soc Rev 2008;37:931–6. [DOI] [PubMed] [Google Scholar]
- Jessani N, Cravatt BF. The development and application of methods for activity-based protein profiling. Curr Opin Chem Biol 2004;8:54–9. [DOI] [PubMed] [Google Scholar]
- Kalidasan K, Su Y, Wu XY et al. . Fluorescence-activated cell sorting and directed evolution of alpha-N-acetylgalactosaminidases using a quenched activity-based probe (qABP). Chem Commun 2013;49:7237–9. [DOI] [PubMed] [Google Scholar]
- Kallemeijn WW, Li KY, Witte MD et al. . Novel activity-based probes for broad-spectrum profiling of retaining beta-exoglucosidases in situ and in vivo. Angew Chem Int Edit 2012;51:12529–33. [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Real-time detection of actively metabolizing microbes by redox sensing as applied to methylotroph populations in Lake Washington. ISME J 2008;2:696–706. [DOI] [PubMed] [Google Scholar]
- Kamke J, Rinke C, Schwientek P et al. . The candidate phylum Poribacteria by single-cell genomics: new insights into phylogeny, cell-compartmentation, eukaryote-like repeat proteins, and other genomic features. PLoS One 2014;9:e87353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor RS, Wrighton KC, Handley KM et al. . Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. Mbio 2013;4:e00708–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaprelyants AS, Kell DB. The use of 5-cyano-2,3-ditolyl tetrazolium chloride and flow-cytometry for the visualization of respiratory activity in individual cells of Micrococcus luteus. J Microbiol Meth 1993;17:115–22. [Google Scholar]
- Kaschani F, Clerc J, Krahn D et al. . Identification of a selective, activity-based probe for glyceraldehyde 3-phosphate dehydrogenases. Angew Chem Int Edit 2012;51:5230–3. [DOI] [PubMed] [Google Scholar]
- Kato D, Boatright KM, Berger AB et al. . Activity-based probes that target diverse cysteine protease families. Nat Chem Biol 2005;1:33–8. [DOI] [PubMed] [Google Scholar]
- Kim TI, Kim H, Choi Y et al. . A fluorescent turn-on probe for the detection of alkaline phosphatase activity in living cells. Chem Commun 2011;47:9825–7. [DOI] [PubMed] [Google Scholar]
- Kocaoglu O, Carlson EE.. Penicillin-Binding Protein Imaging Probes. Curr Protoc Chem Biol 2013;5:239–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolinko S, Jogler C, Katzmann E et al. . Single-cell analysis reveals a novel uncultivated magnetotactic bacterium within the candidate division OP3. Environ Microbiol 2012;14:1709–21. [DOI] [PubMed] [Google Scholar]
- Kolinko S, Richter M, Glockner FO et al. . Single-cell genomics of uncultivated deep-branching magnetotactic bacteria reveals a conserved set of magnetosome genes. Environ Microbiol 2016;18:21–37. [DOI] [PubMed] [Google Scholar]
- Komar AA, Lesnik T, Reiss C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett 1999;462:387–91. [DOI] [PubMed] [Google Scholar]
- Konno S, Ishikawa F, Suzuki T et al. . Active site-directed proteomic probes for adenylation domains in nonribosomal peptide synthetases. Chem Commun 2015;51:2262–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf SH, McGlynn SE, Green-Saxena A et al. . Heavy water and (15) N labelling with NanoSIMS analysis reveals growth rate-dependent metabolic heterogeneity in chemostats. Environ Microbiol 2015;17:2542–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzer JW, Lehmann R, Schmelcher M et al. . Use of high-affinity cell wall-binding domains of bacteriophage endolysins for immobilization and separation of bacterial cells. Appl Environ Microb 2007;73:1992–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubryk P, Kolschbach JS, Marozava S et al. . Exploring the potential of stable isotope (resonance) Raman microspectroscopy and surface-enhanced Raman scattering for the analysis of microorganisms at single cell level. Anal Chem 2015;87:6622–30. [DOI] [PubMed] [Google Scholar]
- Lahtinen SJ, Ahokoski H, Reinikainen JP et al. . Degradation of 16S rRNA and attributes of viability of viable but nonculturable probiotic bacteria. Lett Appl Microbiol 2008;46:693–8. [DOI] [PubMed] [Google Scholar]
- Landry ZC, Giovanonni SJ, Quake SR et al. . Optofluidic cell selection from complex microbial communities for single-genome analysis. Methods Enzymol 2013;531:61–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken RS. Genomic sequencing of uncultured microorganisms from single cells. Nat Rev Microbiol 2012;10:631–40. [DOI] [PubMed] [Google Scholar]
- Li MQ, Canniffe DP, Jackson PJ et al. . Rapid resonance Raman microspectroscopy to probe carbon dioxide fixation by single cells in microbial communities. ISME J 2012;6:875–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XF, Yu JJ, Zhu Q et al. . Visualization of monoamine oxidases in living cells using ‘Turn- ON’ fluorescence resonance energy transfer probes. Analyst 2014;139:6092–5. [DOI] [PubMed] [Google Scholar]
- Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. P Natl Acad Sci USA 1999;96:14694–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou XD, Hong YN, Chen SJ et al. . A selective glutathione probe based on AIE fluorogen and its application in enzymatic activity assay. Sci Rep-Uk 2014;4:4272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald RI, Guilinger JP, Mukherji S et al. . Electrophilic activity-based RNA probes reveal a self-alkylating RNA for RNA labeling. Nat Chem Biol 2014;10:1049–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JS, Lombardo MJ, Badger JH et al. . Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. P Natl Acad Sci USA 2013;110:E2390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchenko M, Thomson A, Ellis TN et al. . Development of a clickable activity-based protein profiling (ABPP) probe for agmatine deiminases. Bioorgan Med Chem 2015;23:2159–67. [DOI] [PubMed] [Google Scholar]
- Marcy Y, Ouverney C, Bik EM et al. . Dissecting biological "dark matter" with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. P Natl Acad Sci USA 2007;104:11889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martijn J, Schulz F, Zaremba-Niedzwiedzka K et al. . Single-cell genomics of a rare environmental alphaproteobacterium provides unique insights into Rickettsiaceae evolution. ISME J 2015;9:2372–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia M, Brazel DM, Swan BK et al. . Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS One 2012;7:e35314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JL, Niessen S, Hoover HS et al. . An orthogonal active site identification system (OASIS) for proteomic profiling of natural product biosynthesis. ACS Chem Biol 2009;4:948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello JV, Finney NS. Dual-signaling fluorescent chemosensors based on conformational restriction and induced charge transfer. Angew Chem Int Ed Engl 2001;40:1536–8. [DOI] [PubMed] [Google Scholar]
- Mukherji R, Samanta A, Illathvalappil R et al. . Selective imaging of quorum sensing receptors in bacteria using fluorescent Au nanocluster probes surface functionalized with signal molecules. Acs Appl Mater Inter 2013;5:13076–81. [DOI] [PubMed] [Google Scholar]
- Neef AB, Schultz C. Selective fluorescence labeling of lipids in living cells. Angew Chem Int Ed Engl 2009;48:1498–500. [DOI] [PubMed] [Google Scholar]
- Ngo JT, Champion JA, Mahdavi A et al. . Cell-selective metabolic labeling of proteins. Nat Chem Biol 2009;5:715–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocker A, Cheung CY, Camper AK. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbiol Meth 2006;67:310–20. [DOI] [PubMed] [Google Scholar]
- Okerberg ES, Wu JY, Zhang BH et al. . High-resolution functional proteomics by active-site peptide profiling. P Natl Acad Sci USA 2005;102:4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes E, Das SR. Click chemistry for rapid labeling and ligation of RNA. ChemBioChem 2011;12:125–31. [DOI] [PubMed] [Google Scholar]
- Park HJ, Rhee HW, Hong JI. Activity-based fluorescent probes for monitoring sulfatase activity. Bioorg Med Chem Lett 2012;22:4939–41. [DOI] [PubMed] [Google Scholar]
- Rai M, Yadav A, Gade A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol Adv 2009;27:76–83. [DOI] [PubMed] [Google Scholar]
- Rinke C, Lee J, Nath N et al. . Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nat Protoc 2014;9:1038–48. [DOI] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A et al. . Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013;499:431–7. [DOI] [PubMed] [Google Scholar]
- Rodrigue S, Malmstrom RR, Berlin AM et al. . Whole genome amplification and de novo assembly of single bacterial cells. PLoS One 2009;4:e6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S, Hawley AK, Torres Beltran M et al. . Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta-genomics. eLife 2014;3:e03125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler NC, Wright AT. Activity-based protein profiling of microbes. Curr Opin Chem Biol 2015;24:139–44. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Jessani N, Joseph A et al. . Activity-based probes for the proteomic profiling of metalloproteases. P Natl Acad Sci USA 2004;101:10000–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. P Natl Acad Sci USA 2008;105:2415–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samo TJ, Smriga S, Malfatti F et al. . Broad distribution and high proportion of protein synthesis active marine bacteria revealed by click chemistry at the single cell level. Front Mar Sci 2014;1:48. [Google Scholar]
- Shao Q, Xing B. Enzyme responsive luminescent ruthenium(II) cephalosporin probe for intracellular imaging and photoinactivation of antibiotics resistant bacteria. Chem Commun 2012;48:1739–41. [DOI] [PubMed] [Google Scholar]
- Sieber SA, Niessen S, Hoover HS et al. . Proteomic profiling of metalloprotease activities with cocktails of active-site probes. Nat Chem Biol 2006;2:274–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegl A, Kamke J, Hochmuth T et al. . Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J 2011;5:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieracki ME, Cucci TL, Nicinski J. Flow cytometric analysis of 5-cyano-2,3-ditolyl tetrazolium chloride activity of marine bacterioplankton in dilution cultures. Appl Environ Microb 1999;65:2409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(I)-catalyzed azide-alkyne [3+2] cycloaddition. J Am Chem Soc 2003;125:4686–7. [DOI] [PubMed] [Google Scholar]
- Speers AE, Cravatt BF. A tandem orthogonal proteolysis strategy for high-content chemical proteomics. J Am Chem Soc 2005;127:10018–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol 2002;161:1961–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanauskas R. Wiretapping into microbial interactions by single cell genomics. Front Microbiol 2015;6:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun MT, Yu H, Zhu HJ et al. . Oxidative cleavage-based near-infrared fluorescent probe for hypochlorous acid detection and myeloperoxidase activity evaluation. Anal Chem 2014;86:671–7. [DOI] [PubMed] [Google Scholar]
- Tuller T, Waldman YY, Kupiec M et al. . Translation efficiency is determined by both codon bias and folding energy. P Natl Acad Sci USA 2010;107:3645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyo KE, Zhou H, Stephanopoulos GN. High-throughput screen for poly-3-hydroxybutyrate in Escherichia coli and Synechocystis sp strain PCC6803. Appl Environ Microb 2006;72:3412–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrell M, Zhai DT, Er JC et al. . Combinatorial strategies in fluorescent probe development. Chem Rev 2012;112:4391–420. [DOI] [PubMed] [Google Scholar]
- Walkup GK, Burdette SC, Lippard SJ et al. . A new cell-permeable fluorescent probe for Zn2+. J Am Chem Soc 2000;122:5644–5. [Google Scholar]
- Walvoort MT, Kallemeijn WW, Willems LI et al. . Tuning the leaving group in 2-deoxy-2-fluoroglucoside results in improved activity-based retaining beta-glucosidase probes. Chem Commun 2012;48:10386–8. [DOI] [PubMed] [Google Scholar]
- Wei L, Yu Y, Shen Y et al. . Vibrational imaging of newly synthesized proteins in live cells by stimulated Raman scattering microscopy. P Natl Acad Sci USA 2013;110:11226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R, Tung CH, Mahmood U et al. . In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat Biotechnol 1999;17:375–8. [DOI] [PubMed] [Google Scholar]
- Wetmore KM, Price MN, Waters RJ et al. . Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. Mbio 2015;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Ponten F, Moberg C et al. . A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol 1999;155:1467–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MC, Mori T, Ruckert C et al. . An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 2014;506:58–62. [DOI] [PubMed] [Google Scholar]
- Woyke T, Jarett J. Function-driven single-cell genomics. Microb Biotechnol 2015;8:38–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, Sczyrba A, Lee J et al. . Decontamination of MDA reagents for single cell whole genome amplification. PLoS One 2011;6:e26161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, Tighe D, Mavromatis K et al. . One bacterial cell, one complete genome. PLoS One 2010;5:e10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Bull B, Szymanski C et al. . Multicolor conjugated polymer dots for biological fluorescence imaging. ACS nano 2008;2:2415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Jin Y, Schneider T et al. . Ultrabright and bioorthogonal labeling of cellular targets using semiconducting polymer dots and click chemistry. Angew Chem Int Ed Engl 2010;49:9436–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Price DC, Stepanauskas R et al. . Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 2011;332:714–7. [DOI] [PubMed] [Google Scholar]
- Youssef NH, Blainey PC, Quake SR et al. . Partial genome assembly for a candidate division OP11 single cell from an anoxic spring (Zodletone Spring, Oklahoma). Appl Environ Microb 2011;77:7804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Stott S, Toner M et al. . Circulating tumor cells: approaches to isolation and characterization. J Cell Biol 2011;192:373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Martiny AC, Reppas NB et al. . Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol 2006;24:680–6. [DOI] [PubMed] [Google Scholar]
- Zheng BH, DeRan M, Li XY et al. . 2-Bromopalmitate analogues as activity-based probes to explore palmitoyl acyltransferases. J Am Chem Soc 2013;135:7082–5. [DOI] [PubMed] [Google Scholar]