Abstract
Antibiotic resistance and its wider implications present us with a growing healthcare crisis. Recent research points to the environment as an important component for the transmission of resistant bacteria and in the emergence of resistant pathogens. However, a deeper understanding of the evolutionary and ecological processes that lead to clinical appearance of resistance genes is still lacking, as is knowledge of environmental dispersal barriers. This calls for better models of how resistance genes evolve, are mobilized, transferred and disseminated in the environment. Here, we attempt to define the ecological and evolutionary environmental factors that contribute to resistance development and transmission. Although mobilization of resistance genes likely occurs continuously, the great majority of such genetic events do not lead to the establishment of novel resistance factors in bacterial populations, unless there is a selection pressure for maintaining them or their fitness costs are negligible. To enable preventative measures it is therefore critical to investigate under what conditions and to what extent environmental selection for resistance takes place. In addition, understanding dispersal barriers is not only key to evaluate risks, but also to prevent resistant pathogens, as well as novel resistance genes, from reaching humans.
Keywords: antimicrobial resistance, dissemination, fitness costs, horizontal gene transfer, human health risks, microbial ecology
This review defines which ecological and environmental factors are important for the development of antibiotic resistance in human pathogens, and suggests some possible mitigation strategies to delay and reduce increased resistance.
INTRODUCTION
Antibiotic resistance accounts for hundreds of thousands of deaths annually (Review on Antimicrobial Resistance 2014), and its projected increase has made the WHO recognize it as a major global health threat (WHO 2014). Conventionally, the struggle against antibiotic resistance development has mainly taken place in clinical, community, and in more recent years also agricultural settings—aiming to reduce transmission and prevent selection of resistant bacteria during antibiotic treatment. Over the past years, the role of the environment as an important source and dissemination route of resistance has been increasingly recognized (Martinez 2008; Wright 2010; Ashbolt et al. 2013; Finley et al. 2013; Pruden et al. 2013; Bengtsson-Palme et al. 2014b; Bondarczuk, Markowicz and Piotrowska-Seget 2015), but our understanding of its contribution is still limited. The lack of knowledge of how, and under which circumstances, the environment facilitates resistance development makes mitigation of the emergence and dissemination of mobile resistance factors problematic (Berendonk et al. 2015). Several authors have highlighted the need to take on a holistic perspective on antibiotic resistance, including humans, animals and the external environment—a so-called one-health approach (Collignon 2013, 2015; So et al. 2015). Increased knowledge of the environmental factors that drive resistance may ultimately allow us to build models for how resistance emerges and is disseminated (Hiltunen, Virta and Laine 2017). Although such models would be descriptive at first, as most of their parameters remain unknown, and thus lack predictive power, they still would have value as indicators of the most urgent knowledge gaps to fill in order to develop mitigation strategies.
This paper aims to conceptualize and define the factors that influence the emergence, mobilization, dissemination and maintenance of antibiotic resistance genes in the environment. We have tried to accommodate both ecological and evolutionary aspects, but without any attempt to fully cover the growing literature on the environmental dimensions of antibiotic resistance (Fig. 1). In order to define those factors, we must first spell out some basic definitions for which ambiguous meanings exist in the literature. In this paper, we follow the operational definition of resistance by Martinez, Coque and Baquero (2015), which postulates that a strain is resistant against an antibiotic if its minimal inhibitory concentration (MIC) is higher than for the corresponding parental wild-type strain. Accordingly, we define a gene as a ‘resistance gene’ (or ‘resistance factor’) when its presence allows a bacterium to withstand a higher antibiotic concentration or when its absence increases susceptibility of the antibiotic (Martinez, Baquero and Andersson 2007), a definition that also includes many non-mobile chromosomal resistance genes. We furthermore define ‘novel’ (or ‘new’) resistance genes as genes that have not previously been described to have a resistance function, regardless of if they appear in pathogens or not, and regardless of if they appear on the bacterial chromosome or on a mobile genetic element. It is complicated to define ecological emergence (de Haan 2006), and depending on the viewpoint several definitions of when resistance genes emerge are possible (Baquero et al. 2015). In this paper, we will consider the ‘emergence’ of a resistance determinant as the event where it first appears in a context where it provides operational resistance. Further, we consider a gene to have undergone ‘mobilization’ when it appears on a mobile genetic element, such as a plasmid, transposon or integron. As a consequence of these definitions, most emergence and mobilization events will in this view remain undetected—at least before they result in a clinical problem, which many will probably never do.
Figure 1.
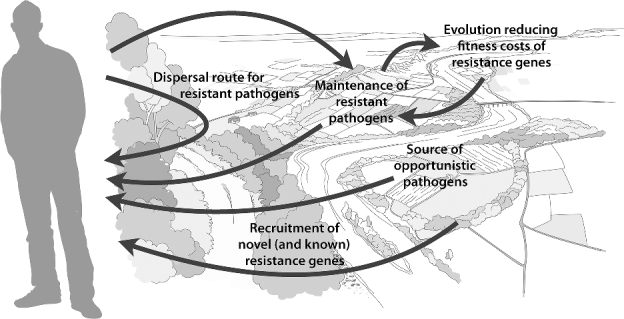
An overview of the main roles of the external environment in antibiotic resistance development and dissemination. Note that none of these processes requires a selection pressure for resistance to operate, although such a selection pressure would facilitate both maintenance and recruitment of resistance genes. Antibiotic exposure may select for resistant bacteria during dispersal if those bacteria are able to grow also in the external environment, which is the case for many opportunistic pathogens. Furthermore, the environment also serves as a source of opportunistic pathogens that are already resistant, or may acquire resistance genes from other human-associated bacteria in or on the human body and then cause resistant infections at a later stage. While both known and novel resistance genes may be recruited from the environmental resistome, the most severe long-term health consequences of such acquisitions are likely when genes not currently present in pathogens are added to their resistance repertoire.
THE ORIGINS OF RESISTANCE GENES
Novel antibiotic resistance factors could potentially emerge anywhere, at any time. The astounding number of bacterial cells on Earth, estimated to around 1030—a thousand billion billion billions (Kallmeyer et al. 2012), provide an immense genetic variability, and opportunities for mutations, rearrangements and horizontal gene transfer. Thus, new resistance factors likely appear regularly, although we never detect the vast majority of these events. As will be outlined below, there are nevertheless several reasons to why pathogens are not flooded by novel resistance genes. For a start, resistance factors are generally associated with some fitness cost. This cost may be particularly large for genes providing novel resistance functions for a bacterium, as their expression may not be sufficiently fine-tuned and their products may interfere with other cellular functions. Thus, novel resistance genes will be selected against unless there is a relatively strong selection pressure to maintain them. Furthermore, even if such a resistance factor would have a low or negligible fitness cost, it would still be rare, and may therefore not become permanently established in the bacterial population unless there is a positive selection pressure for it (Martinez 2011). This selection pressure may be weak, but unless it is present the only way by which a novel resistance factor would be retained is through genetic drift (Baquero et al. 1998).
MOBILIZATION OF RESISTANCE FACTORS
Similar to the emergence of novel resistance factors, resistance genes could be mobilized anywhere but need—unless their fitness costs are negligible—a selection pressure to be kept on a mobile genetic element until they have evolved to present smaller fitness costs to the host. The subsequent question therefore becomes: where are selection pressures strong enough to promote mobilization of chromosomal resistance genes and maintenance of already mobile resistance genes? Resistance factors that have recently been mobilized onto a mobile genetic element are likely to often be associated with increased fitness costs, associated with the burden of keeping multiple copies of the same gene and the difficulty to maintain expression control of the gene on a mobile element. This means that environments allowing sustained longevity of a resistance gene regardless of its cost would be of particular importance for the mobilization of resistance determinants. It is then reasonable to assume that once a mobile resistance gene has gained a foothold in a bacterial community, it can subsequently evolve towards diminished fitness cost (Salyers and Amábile-Cuevas 1997). Resistance genes with considerably lower fitness costs may of course be recruited to mobile genetic elements, and sub-inhibitory concentrations of an antibiotic could then suffice to select for carriage of them. It is important to consider that resistance genes have evolved in a context of competition and interaction between different species, some of which may use antibiotics as warfare agents. Thus, both known and novel resistance determinants can be selected for naturally, if they confer a competitive advantage against antibiotics producers, or allow host bacteria to survive higher concentrations of an antibiotic that they themselves produce. Such natural selection processes contribute to preserving an environmental pool of resistance genes, but only indirectly to further establishment of resistance genes in pathogens, as they are unlikely to drive mobilization of resistance factors.
An oft-neglected aspect of resistance gene mobilization is that a resistance gene can also be mobilized onto a non-transferrable mobile genetic element, such as an integron (Gillings 2017a). The cost-benefit tradeoffs for such mobilization are poorly investigated, but it can be imagined that on the positive side for the bacterium, this provides a mechanism for gene duplication and subsequent neofunctionalization. This may increase the expression levels of a resistance gene or contribute to fine-tuning of its expression, both of which could be beneficial under antibiotic selection. On the negative side, transposition of a gene may result in disruption of other genes or loss of expression control, both of which could reduce the overall fitness of the bacterium. The extent to which mobilization into chromosomal mobile elements contribute to the development of transferrable antibiotic resistance is virtually unknown, but it could potentially be an important step towards resistance recruitment onto plasmids as integrons often act in tandem with plasmids and transposons that are transferrable between cells (Stokes and Gillings 2011). Importantly, while integrons function as effective gene relocation devices that can capture a wide range of resistance genes, a selection pressure for resistance is still the main force upholding such relocations in bacterial genomes.
HORIZONTAL TRANSFER OF RESISTANCE FACTORS
Horizontal gene transfer is central for the spread of novel (and known) resistance genes, as it allows resistance to expand beyond specific clones. This way, gene transfer makes resistance genes available to a much larger part of the bacterial community in a particular environment, often beyond species boundaries (Martinez 2011). As for the mobilization of resistance factors, transfer of genes between bacteria can in theory occur anywhere. However, for resistance genes to be horizontally transferred from environmental to pathogenic bacteria they need to, at least temporarily, share the same habitat (Matte-Tailliez et al. 2002; Wiedenbeck and Cohan 2011). Furthermore, horizontal gene transfer is much more likely to occur between phylogenetically closely related bacteria (Philippot et al. 2010; Smillie et al. 2011). Finally, transfer of genetic material between bacterial cells is induced by stressors such as antibiotics (Beaber, Hochhut and Waldor 2004; Hastings, Rosenberg and Slack 2004; Maiques et al. 2006; Jutkina et al. 2016), and potentially also metals and biocides (Seier-Petersen et al. 2014; Zhang et al. 2017). Subsequently, antibiotic selection also contributes to establishment of transferred resistance genes in their new host. Thus, resistance transfer to pathogens could be expected to be relatively frequent between human-associated bacteria (Salyers, Gupta and Wang 2004; Porse et al. 2017), particularly during treatment with antibiotics. In contrast, transfer of resistance genes to pathogens from environmental bacteria, which occupy other habitats and are often less phylogenetically related, would likely be less common, although environmental stressors may induce horizontal gene transfer to and from (opportunistic) human pathogens in environmental settings. This means that once a resistance factor has entered a human pathogen, it is more likely to further spread between commensals and pathogens than being transferred again into another pathogen from environmental bacteria, and in any case the consequences/contribution from the latter type of event would be expected to be small (Bengtsson-Palme and Larsson 2015). Moreover, avoiding transfer of resistance between pathogens (as well as evolutionary close commensals) is likely impossible, since they share habitats, often are phylogenetically related, and carriage of mobile resistance factors generally seem to be associated with low fitness costs in pathogens (Salyers and Amábile-Cuevas 1997; Andersson and Hughes 2010). Somewhat surprisingly, the human microbiome harbors a fairly large number of resistance genes that have not (yet) been transferred to human pathogens as far as we know (Sommer, Dantas and Church 2009; Sommer, Church and Dantas 2010). The reasons for this are unknown, but one can speculate that strong barriers to transfer are at play. For example, many of the resistance genes in the cultivable bacteria of the human gut derive from aerobic organisms and are identical to resistance genes in human pathogens. On the contrary, the uncultivable portion of the human microbiome, which mostly corresponds to anaerobic organisms, contained much higher fractions of previously undiscovered genes, conferring resistance to aminoglycosides, amphenicols, beta-lactams and tetracyclines (Sommer, Dantas and Church 2009). Differences in oxygen preferences may thus be one barrier to resistance transfer, but the carriers of those genes may also be evolutionary divergent from most human pathogens, the transfer of their resistance genes may be complicated by that they have not been mobilized onto a suitable mobile genetic element, or expression of those genes in a new host may impose too high fitness costs (Martinez 2011).
The vast majority of existing resistance factors, including those not yet described, are unlikely to be encountered in pathogens and human commensals, but would instead be expected to be present in environmental bacteria (Allen et al. 2010). Bacteria not typically associated with the human microbiome may have the opportunity to interact with human-associated species in various settings. One possibility is that environmental bacteria can be transiently present in the human microbiome, after e.g. interaction with wild animals, intake of raw food, or drinking contaminated water (Allen et al. 2010; De Boeck et al. 2012; Ghaly et al. 2017). The impact of these exposure scenarios is uncertain as the timeframes for interaction are limited, and there is virtually no knowledge of the factors triggering transfer of resistance genes in environmental bacteria under this type of stress. That said, there are also other settings where human bacteria can interface with animal-associated and environmental ones. A key consideration in these contexts is the length of the dispersal route from those milieus back into the human population (Baquero, Alvarez-Ortega and Martinez 2009). A pathogen (or commensal) that acquires a novel resistance factor from an environmental bacterium, but is eradicated before it can return to a human host, never causes any clinical resistance problems. Only those that eventually make it back to their hosts may become real human health threats. Sewage treatment plants (STPs) provide an obvious setting that offers interaction opportunities for a range of different bacterial species, and also may present sufficient conditions for resistance selection (Rizzo et al. 2013). Other milieus that could serve as breeding grounds for resistance transfer can be found in agriculture—particularly among livestock (Allen 2014; Bengtsson-Palme 2017), water bodies (Baquero, Martinez and Cantón 2008; Lupo, Coyne and Berendonk 2012), and the food supply chain (Rolain 2013; Bengtsson-Palme 2017). All these environments have in common that the exposure routes to humans after a potential transfer event may be relatively short (see the discussion on dissemination below). Finally, transfer of resistance factors from human pathogens to environmental bacteria is possible, enabling human-associated bacteria to use environmental bacterial populations as reservoirs for resistance genes that can later be re-recruited into the human-associated resistome (Baquero, Martinez and Cantón 2008; Martinez 2008). There is evidence for transfer of resistance genes between diverse organisms such as Clostridium perfringens, Streptococcus pneumoniae, Enterococcus faecalis and strains of Bacteroides (Shoemaker et al. 2001), and hence such transfer across divergent taxa is not unimaginable. Opportunistic pathogens usually thriving in soil, such as Burkholderia cepacia, Ochrobactrum intermedium and Stenotrophomonas maltophilia (Berg, Eberl and Hartmann 2005; Johnning et al. 2013), may be intermediary organisms that can act as recipients of resistance genes from human-associated bacteria and which could transfer those genes back at a later point, or even infect humans themselves. Although this process is not easy to quantify, resistance genes that circulate among pathogens are (as previously pointed out) nearly impossible to eliminate. Resistance factors can thus ‘re-emerge’ without being recruited from the environmental, non-pathogenic, bacteria again. The environment as a storage or reservoir of resistance genes commonly encountered in pathogens is, therefore, in our point of view likely of lesser concern than the environment as a source for novel resistance factors into pathogens or its role in the dissemination of resistant pathogens (Fig. 1). Furthermore, measures to prevent re-recruitment of resistance genes from the environment could overlap with mitigation strategies to avoid spread of novel resistance factors into pathogens, although which environments are associated with highest risks will likely differ for the two scenarios.
DISSEMINATION OF RESISTANT BACTERIA
The main route of exposure for humans to resistant pathogens is from other people, either in clinics or through the community setting. Typical dispersal routes here are through body contact or indirect contact transmission, aerosols, and food prepared by persons carrying the pathogen (Livermore 2000). These are also the typical transmission routes for infectious bacteria in general, and interventions preventing circulation of resistant pathogens among humans are essentially the same as those applied to prevent the spread of any bacterial pathogen (Rao 1998; Lipsitch, Bergstrom and Levin 2000; Livermore 2000; Levin, Baquero and Johnsen 2014). Importantly, proper hygiene routines constitute the principal dispersal barrier for resistant pathogens, and the significance of sanitation for preventing the spread of resistant bacteria between humans cannot be overstated (Mattner et al. 2012).
Apart from transmission between humans, environmental dissemination routes for resistant bacteria have also been pointed out as potentially important for the spread of antibiotic resistance (Allen et al. 2010; Finley et al. 2013; Pruden et al. 2013; Levin, Baquero and Johnsen 2014; Huijbers et al. 2015). Again, environments facilitating dissemination of resistant bacteria also enable spread of non-resistant human pathogens, and generally also opportunistic pathogens. Thus, sewage, wastewater treatment plants, water bodies and travel, but also air-borne aerosols, dust, and food colonized by bacteria, are important vectors enabling bacterial transmission between hosts through the environment (Fernando, Collignon and Bell 2010; Molton et al. 2013; Rolain 2013; Pruden 2014; Angelin, Evengård and Palmgren 2015; Barberán et al. 2015; Bengtsson-Palme et al. 2015; McEachran et al. 2015; Pal et al. 2016; Bengtsson-Palme 2017). STPs generally discharge their effluent (which has repeatedly been shown to contain resistance genes; see e.g. Bengtsson-Palme et al. 2016) into water bodies. Water contaminated by STP effluents is often used for irrigation of farmland, for recreational swimming and as drinking water supply (after further treatment). Domestic animals often drink such surface water untreated and may subsequently spread resistant bacteria to humans. However, for the dissemination of resistant bacteria, untreated sewage released into water bodies poses a considerably larger risk than STP effluents, as STPs often reduce the relative abundance of the vast majority of resistance genes, and also lower the total bacterial abundance from ten- up to thousand-fold (Bengtsson-Palme et al. 2016; Karkman et al. 2016).
Limiting the spread of human-associated bacteria—resistant or not—requires an understanding of the environmental dispersal barriers that exist. Contrary to the case of clinical- and community-transmitted bacteria, identifying relevant barriers to dispersal is considerably harder in the environment. We may here adopt a metacommunity ecology perspective and consider the human and/or animal hosts of primary pathogens as habitable patches, while most other external environments would serve as a dispersal matrix (Leibold et al. 2004). Metacommunity theory suggests that if patches are of equivalent quality in terms of allowing for a species to thrive and compete, the distance between patches and the dispersal capability of species determine their relative success (Bengtsson 2009). Thus, the quality of the dispersal matrix (i.e. how well it mediates movement between patches) and the ability to survive between hosts are fundamental properties for pathogens to spread between humans through the environment. Some understanding of how different pathogens survive in the external environment can be gained from epidemiology, although this is not a particularly well-studied subject outside of a few select model bacteria. Importantly, the dispersal barriers are largely species specific, as different bacteria have very different requirements for survival outside of their preferred habitat. For example, many gut bacteria are, in principle, obligate anaerobes and therefore show poor survival, and consequently also limited dispersal ability, outside of the human body. Since survival rather than growth is critical in the dissemination of human-associated bacteria (resistant or not) through the environment, the advantage of carrying resistance genes in the presence of low concentrations of antibiotics is likely to be small or negligible for species which only use the environment as a dispersal matrix. On the other hand, for those bacteria that utilize the environment as an alternative habitat—or even their main habitat—and therefore can readily grow there, eventual antibiotic exposure would be much more likely to contribute to the selection of resistance factors during environmental dissemination. The latter category includes opportunistic and emerging pathogens, such as Pseudomonas putida, Stenotrophomonas maltophilia and Bacillus cereus (Berg, Eberl and Hartmann 2005). Importantly, this means that the factors influencing selection for resistant strains during dispersal are specific both to the mechanism of the antibiotic and to the species in question. Another example of species-specific factors important for these dispersal processes is the ability to form inactive dormant stages, such as the highly resilient spores formed by some pathogenic bacteria, of which Bacillus anthracis is a good example (Leggett et al. 2012). Dormant life stages could vastly help bacteria survive in the dispersal matrix, almost regardless of matrix quality, and re-spawn once in a suitable host (Lennon and Jones 2011; Shade et al. 2012).
The dispersal routes for bacteria through the environment have not completely evaded investigation, however. Research on microbial source tracking, usually aiming at identifying the sources and health risks associated with e.g. leaks of untreated sewage, have generated some knowledge regarding the persistence and re-infection potential of human gut bacteria in the environment (Harwood et al. 2014). Furthermore, it is known that physical forces, such as wind and water movement, can move bacteria over large distances (Allen et al. 2010). Many bacteria have been isolated from air, including species of the Micrococcus, Staphylococcus, Bacillus and Aeromonas genera (Górny and Dutkiewicz 2002; Tsai and Macher 2005). Recently, air has also received attention as an underinvestigated potential route for dispersal of resistant bacteria (McEachran et al. 2015; Pal et al. 2016; Gat et al. 2017). However, due to the lack of nutrients in the air environment, the ability to grow in the presence of antibiotics becomes an almost negligible factor compared to survival and persistence in this dispersal scenario. Thus, air transfer would enable the spread of both resistant and non-resistant bacteria alike, and although horizontal transfer of resistance between bacteria in aerosols and on dust particles remain a possibility, such events are less likely to be permanently established in the recipient genomes unless they are subsequently exposed to antibiotics in an environment that better permits growth.
Wild birds and animals living close to humans are also known to harbor bacteria carrying resistance genes, and may contribute to spreading those genes across large areas (Baquero, Martinez and Cantón 2008; Bonnedahl et al. 2009; Stedt et al. 2015). In addition, global food trade has been shown to ship resistant pathogenic bacteria around the world, contributing for example to the German Shiga-toxin-producing Escherichia coli (O104:H4) outbreak in 2011 (Buchholz et al. 2011; Rasko et al. 2011). Still, much remains to be understood in terms of dispersal limitations, environmental survival, competitiveness versus environmental species and strains, resistance selection and alternative habitats for human-associated bacteria in the environment (Hiltunen, Virta and Laine 2017). Even less is known about how resistance-carrying, non-pathogenic environmental bacteria disperse and interact with human-associated bacteria. In this process, opportunistic pathogens with the environment as their chief habitat may play a very important role in mediating resistance from environmental bacteria to the human microbiome. The dissemination routes from environments that present a selection pressure for initial emergence, mobilization and maintenance of resistance genes, all the way to humans and/or animals are poorly understood. These routes need to be delineated, along with the factors determining survival of environmental bacteria in various dispersal matrices. This also calls for efforts to monitor the presence of pathogens and resistance genes in a variety of environmental settings.
EVOLUTIONARY PROCESSES INFLUENCING ENVIRONMENTAL ANTIBIOTIC RESISTANCE
For the long-term maintenance of antibiotic resistance genes in bacterial communities, two parallel evolutionary forces are at play: selection promoting resistance phenotypes, and selection leading to reduction of the fitness costs associated with carrying resistance genes (Andersson and Hughes 2010; Baquero, Coque and de la Cruz 2011; Hernando-Amado et al. 2017). As discussed earlier, gain and establishment of resistance genes in a bacterial population are largely dependent on a direct antibiotic selection pressure (Martinez 2011). The selective forces towards maintenance of resistance genes do not only include direct antibiotic selection pressure, however. Even in the absence of a direct selection pressure from an antibiotic, mobile resistance genes may be favored by co-selection by other substances present, such as metals and biocides (Baker-Austin et al. 2006; Wales and Davies 2015), as the resistance determinants for some of these compounds can be co-localized to the same mobile genetic elements as antibiotic resistance genes (Pal et al. 2015). The implications of co-selection of antibiotic resistance by metals were recently thoroughly reviewed by Pal et al. (2017). Similarly, exposure to other stressors than antibiotics may select for increased expression of genes encoding efflux pumps, which in turn may render bacteria less susceptible to antibiotics as well. Although this may not immediately contribute to the spread of resistance factors, it could contribute to the development of more efficient resistance genes by enabling bacteria to withstand low-level antibiotic exposure for longer times, enabling a recently acquired resistance gene to evolve to be more efficient and less costly within the new host.
In addition, resistance genes may be maintained because they confer advantages to the cell even in the absence of a selection pressure, in essence allowing bacteria to perform intrinsic functions more efficiently when they carry the resistance gene (Enne et al. 2004). However, carriage and maintenance of resistance genes usually come with a cost in terms of reduced fitness, although the cost is sometimes small. This cost is (apart from random reduction by genetic drift) the sole factor that acts to reduce the frequency of resistance genes in bacterial populations (Andersson and Hughes 2010). Random losses of resistance genes from bacterial cells happen all the time, but seldom result in complete elimination of the gene from the community, which means that once a selection pressure for resistance re-emerges (such as during antibiotic treatment), resistance development of bacterial populations previously subjected to resistance selection can be quick (Levin et al. 1997). Selection pressure acting specifically against the carriage of resistance genes is therefore crucial for complete eradication of resistance factors from a community.
The fitness costs associated with carrying and expressing genes providing antibiotic resistance are largely dictated by the nature of the resistance mechanism. Bacteria typically become resistant to antibiotics via (i) upregulation of efflux pumps exporting the substance from the cell, (ii) modifications to the cell wall or outer membrane, reducing permeability for the antibiotic substance, (iii) expression of degradation enzymes that can render the substance harmless, (iv) protection of the molecular target of the antibiotic, or (v) alternative means to perform inhibited functions (Walsh 2003; Arzanlou, Chai and Venter 2017). Resistance mechanisms associated with efflux pumps and cell wall modifications are often caused by mutations in chromosomal DNA, although many efflux pumps are transferrable between bacteria on plasmids. Degradation enzymes, target protection proteins, and enzymes allowing utilization of alternative enzymatic pathways are more likely to be transmissible on mobile genetic elements as they add functions to their carrier rather than modify existing ones. In general, fitness costs associated with the latter three mechanisms are primarily associated with the cost of carrying the resistance plasmid and expressing its genes, while costs of mutations are related to decreased growth rate due to changes in essential genes and/or altered resource usage. In both cases, compensatory mechanisms, including additional mutations, can reduce fitness costs over time (Andersson 2003; Hernando-Amado et al. 2017). Under antibiotic selection, evolution of a bacterial population towards mutation-mediated resistance depends on both the population size and the mutation rate (Perron et al. 2015). Certain mutations have little or no fitness cost, but there seems to be a tendency that those also confer lower degree of resistance than more costly mutations, at least for some antibiotics (Melnyk, Wong and Kassen 2015). Would the same also be true for resistance genes? The relationships between fitness costs and degree of resistance are still not elucidated for mobile resistance genes. Recent meta-analysis of fitness costs associated with different types of resistance factors suggests that while the costs associated with resistance phenotypes vary substantially, the overall trend is that maintenance of resistance plasmids infers a smaller cost relative to resistance caused by chromosomal mutations, and the fitness reduction by carrying a resistance plasmid seems to be relatively small (Vogwill and MacLean 2015). That said, there are substantial fitness costs associated with the initial uptake of horizontally transferred genes (Baltrus 2013), and if resistance genes are selectively moved from an acquired plasmid to the chromosome, fitness costs are reduced compared to retaining the entire source plasmids (Gullberg et al. 2014). Furthermore, both plasmids and hosts seem to compensate for the initial fitness costs within a comparably small number of generations through plasmid domestication (Bouma and Lenski 1988; San Millan et al. 2015; Vogwill and MacLean 2015; Hernando-Amado et al. 2017). Thus, the majority of horizontally transferred resistance genes may actually present little cost to their hosts. If that is the case, the advantage of losing a mobile resistance gene would be small for the individual cell, essentially reducing the gene loss mechanism to that of stochasticity. Random losses, however, will often not be sufficient to fully eradicate resistance genes from a population before a selection pressure favoring their presence reemerges (Levin et al. 1997).
Given that most antibiotics in use are derived from natural compounds produced by microorganisms in the environment, the presence of genes conferring resistance to those compounds across a range of habitats is not surprising (Allen et al. 2010; Pal et al. 2016). Most likely, however, resistance genes did not evolve as a means to fight the high concentrations of antibiotics used in therapy, since such high concentrations are unlikely to be encountered in environments with no or little anthropogenic impact (Kümmerer 2009a,b). Many antibiotics instead seem to primarily function as pigments, toxins, and effectors in microbial communities (Demain 1998), or be involved in microbial signaling (Linares et al. 2006). A curious property of antibiotics is that, at low concentrations, many of them seem to upregulate efflux pumps, escalate mutation rates and mobilize DNA (Aminov 2009; Blázquez et al. 2012). The exact reasons for this remain poorly understood, but it has been hypothesized that higher mutation rates enable quicker niche adaption (Aminov 2009). Likewise, it seems reasonable that upregulation of efflux pumps would also contribute to better adaptability to changing environments. Thus, a signaling role for antibiotics as secondary metabolites may be that when resources in the habitat begin to decay, the release of antibiotics initiates generation of genetic variability, some of which may be favorable in the search for new suitable niches and habitats. In essence, this would ensure more efficient utilization of resources. In this case, resistance genes may have evolved to balance these needs, or to protect bacteria against such signaling schemes of other species. This implies that it might be advantageous to carry resistance genes regardless of anthropogenic antibiotic selection. Similarly, resistance mutations may also have functions aside of providing antibiotic resistance. For example, the D87G substitution in the target gene for fluoroquinolones—gyrA—seem to increase tolerance also to general stress (Webber et al. 2013), which may explain why these substitutions in this position are common among environmental Escherichia species, also in environments with a low level of anthropogenic impact (Johnning et al. 2015). The almost ubiquitous presence of resistance genes in a vast range of environments (Allen et al. 2009; Sommer, Dantas and Church 2009; Lang et al. 2010; D’Costa et al. 2011; Martiny et al. 2011; Forsberg et al. 2012; Segawa et al. 2012; Munck et al. 2015; Pal et al. 2016) further supports that this is the case, and that the cost associated with carrying resistance genes is almost negligible unless the niche is extremely resource-poor, with genome streamlining as a result (Yooseph et al. 2010; Giovannoni, Cameron Thrash and Temperton 2014; Bengtsson-Palme et al. 2014a).
THE ECOLOGY OF ANTIBIOTIC RESISTANCE DEVELOPMENT
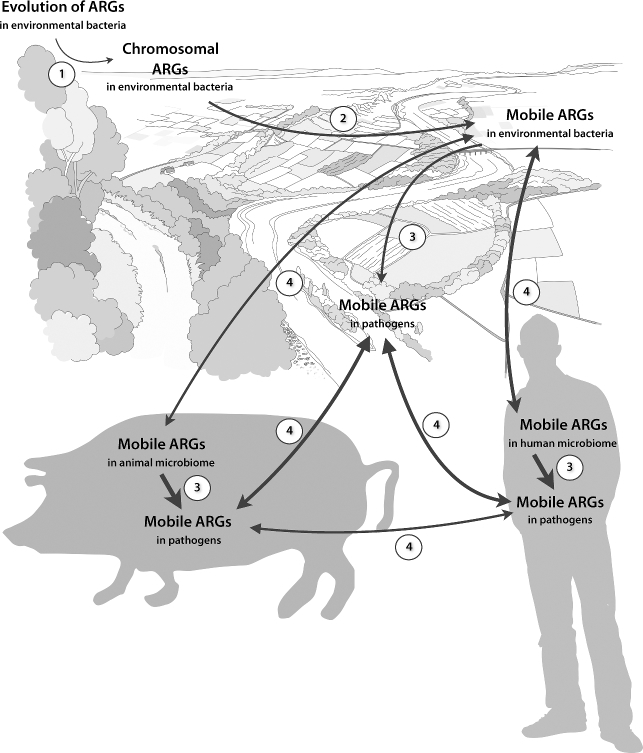
Based on the above reasoning, we propose that four steps are central on the route to clinically important antibiotic resistance: emergence of novel resistance factors, mobilization, transfer to human pathogens, and dissemination. Notably, these steps need not to happen in this particular order; transfer to human pathogens may occur before or after dissemination to the human microbiome, and certain steps in the process may repeat (Fig. 2). The arsenal of resistance factors currently present in pathogens and opportunistic pathogens constitute a set of genes that are at the later stages of this route. A crucial factor for a resistance gene to reach human pathogens is that it is maintained throughout all these steps. Resistance genes with high fitness costs are more likely to be lost in the absence of a selection pressure, particularly if located on a mobile genetic element. Furthermore, a scenario with a constant selection pressure by antibiotics—from the environmental emergence of a resistance gene to its transfer to a human pathogen—seems improbable, although one could argue that there are places in the world where this may be possible. Taken together, it seems reasonable that successfully maintained resistance genes have either evolved towards low fitness cost in a mobile context (a sort of evolutionary rescue (Gonzalez et al. 2013) on the individual gene level), or were associated with low fitness costs from the beginning. Since losses of resistance genes are likely as long as they bestow their carrier with a significant fitness cost, recently mobilized genes that do not provide an obvious fitness advantage are undoubtedly sorted out early from mobile genetic elements such as plasmids (Baquero, Tedim and Coque 2013). This highlights the importance of environments in which resistance genes provide a strong selective advantage, for example milieus subjected to industrial pollution with antibiotics (Larsson 2014a,b). Since these environments would also present bacteria with conditions that favor increased mutation frequency, one consequence may be that resistance genes could be present in several slightly different variants, all selected for detoxification efficiency, of which only those with a low fitness cost are maintained when the selection pressure is removed (for example, after dispersal of the carrier to a non-polluted environment). Given how long the dispersal route from initial mobilization to human pathogens generally is for a novel resistance gene, it is not surprising that mobile resistance factors found in pathogens today are terribly hard to eliminate from bacterial populations (Levin et al. 1997; Andersson 2003; Jernberg et al. 2007; Löfmark et al. 2008) and seem to present little fitness cost to their carriers (Enne et al. 2004; Andersson and Hughes 2010; Gullberg et al. 2014; Vogwill and MacLean 2015). This suggests that once a resistance gene is widely spread among human pathogens (or even among human commensals), part of the game is lost and we are restricted to manage its spread. Mitigation of the spread of resistance factors to human pathogens should therefore ideally take place before they get a foothold in the human microbiome. Thus, detection of resistance determinants in the environment that are not yet widespread among clinical bacteria should be an important component in risk assessment and management of antibiotic resistance (Bengtsson-Palme and Larsson 2015).
Figure 2.
The role of the environment in the recruitment of antibiotic resistance genes (ARGs) to human pathogens includes four major steps: (1) emergence of novel resistance factors in the environment, (2) mobilization onto mobile genetic elements, (3) transfer of ARGs to human pathogens, and (4) dissemination of ARGs into the human microbiome. The width of each arrow roughly corresponds to the assumed frequency of each event, although this is of course largely speculative. Many events are likely more frequent when antibiotic selection pressure is stronger or recurrent. Still, due to the much larger numbers of environmental bacteria than human- and animal-associated, environmental emergence and mobilization of ARGs is probably common on a global scale. Note that the focus of this figure is on the processes involving the external environment.
WHICH ENVIRONMENTS POSE THE MOST PERTINENT RISKS TO HUMAN HEALTH?
Ultimately, the most urgent reason to study antibiotic resistance in the environment is to gain further insights into health risks for humans and domestic animals that often are dependent on effective antibiotics. This knowledge can then ideally be used to design interventions that could prevent or delay the recruitment of resistance factors to pathogens from environmental bacteria and reduce environmental dissemination of resistant pathogens. To identify suitable mitigation strategies, we first need to define what environments and scenarios constitute the most severe risks. This, however, is not completely straightforward. Some have argued that the most severe risk scenarios involve known resistance genes that have previously been reported to reside on mobile genetic elements hosted by human bacterial pathogens (Martinez, Coque and Baquero 2015). This is a valid argument when such genes are encountered in the human microbiome, but while they are clearly of importance, finding them in environmental bacterial communities is not necessarily indicative of a high-risk situation. Well-known resistance genes present on mobile genetic elements easily spread with human feces, and detection of them in the external environment may be an indication of human fecal contamination (Pruden et al. 2006; Pruden, Arabi and Storteboom 2012; Bengtsson-Palme, Larsson and Kristiansson 2017). Risks associated with human fecal pollution should not be neglected, but are primarily related to the dissemination of already resistant bacteria. Furthermore, these genes are already circulating among pathogens and, as argued earlier, transfer of them between pathogens within the human microbiome is expected to be vastly more frequent than transfer of the same genes from environmental bacteria. Thus, in terms of future treatment outcomes, the clinical consequences of recruitment of resistance genes already present among pathogens from environmental sources are likely to be minor (Bengtsson-Palme and Larsson 2015).
The risk landscape can essentially be partitioned into three main components: (i) the risks for mobilization and subsequently permanent establishment of (novel) resistance determinants, (ii) the risks for recruitment of resistance genes not carried by human pathogens through horizontal gene transfer, and (iii) the risks associated with dissemination of resistant bacteria (pathogens or not) through the environment to the human population (Table 1). Antibiotic selection may influence all these components, particularly in the former two cases, while dispersal barriers are most critical for the latter one (Fig. 2). That said, selection could play a major role in the dissemination of resistant bacteria which have the environment as their primary habitat, including opportunistic pathogens. Although mobilization of resistance genes could happen anywhere, stronger selection pressures are directly related to higher risks for their persistent establishment in bacterial populations, as the fitness costs for carrying recently emerged mobile resistance determinants probably are high. Recently mobilized resistance genes that do not confer a fitness advantage are likely to quickly be selected against and lost in microbial communities. This identifies environments in which antibiotics concentrations are clearly above established minimal selective concentrations (MSCs), or even the MICs for many bacteria, as particular high-risk environments in early resistance emergence and mobilization. This includes the human and animal microbiome during antibiotics treatment, intensive aquaculture assisted by antibiotics (Cabello 2006), as well as environments polluted with high levels of antibiotics from industrial sources (Larsson 2014b). The conditions bacteria face within treatment plants treating wastewater from antibiotics production are largely unexplored, but are also likely to be extensively selective, resulting in very limited diversity of bacteria present in such settings (Marathe et al. 2016). Furthermore, as sub-lethal concentrations of antibiotics also can select for resistance (Gullberg et al. 2011, 2014; Lundström et al. 2016), attention has to be paid to raw sewage, agricultural settings and STPs, where concentrations of antibiotics around the predicted MSCs have been determined (Michael et al. 2013; Bengtsson-Palme and Larsson 2016a). Furthermore, mobilization and transfer of resistance factors also increase during antibiotics exposure (Beaber, Hochhut and Waldor 2004; Hocquet et al. 2012), and similarly occur even at sub-inhibitory antibiotics concentrations (Dörr, Lewis and Vulić 2009; Jutkina et al. 2016, 2017). Thus, polluted environments once again pose a high risk, but sewage, for example, may also contain sufficient toxicant concentrations to induce horizontal gene transfer.
Table 1.
Human health risks associated with environmental antibiotic resistance and examples of risk environments and appropriate mitigations.
| Risk scenario | Environments of particular concern | Possible mitigations |
|---|---|---|
| Emergence and fixation of novel resistance genes | Human and animal microbiome | Reduce and optimize antibiotics usage |
| Intensive aquaculture | Restrict antibiotics usage | |
| Environments polluted by discharges from antibiotic manufacturing | Incentivize better control of discharges | |
| Mobilization and transfer of resistance genes to human pathogens | Environments polluted by discharges from antibiotic manufacturing | Incentivize better control of discharges |
| Sewage | Disinfection of treated sewage and sludge | |
| Human and animal microbiome | Reduce antibiotics usage, limit transmission of pathogens | |
| Dissemination of resistant bacteria | Human-to-human contacts, hospitals | Hygiene |
| Contact with environmental opportunistic pathogens, directly or through food | Hygiene, proper handling of fresh produce | |
| Animal husbandry | Reduce antibiotics usage, create transmission barriers between animals, humans and the external environment | |
| Poorly treated sewage | Improved infrastructure for sewage treatment |
For the transfer of resistance to human pathogens, the abundance of pathogenic bacteria that can act as recipients is crucial. This means that the human microbiome could potentially play a role in this process, and that human commensals may act as intermediary resistance reservoirs (Sommer, Church and Dantas 2010; Forslund et al. 2013). The role of human commensals in the transfer of resistance from environmental bacteria to pathogens is however not well investigated. Furthermore, transient environmental bacteria could potentially carry resistance genes into the human microbiome, where these genes could be transferred to human-associated bacteria, including (opportunistic) pathogens. However, for most transient bacteria, the interaction times with human-associated bacteria are likely to be short. In addition, animals may also serve as intermediate hosts for resistant bacteria, and contribute a breeding ground for resistance transfer to human pathogens (Allen et al. 2010). Risks associated with transfer of novel resistance genes to human pathogens in STPs appear somewhat smaller due to seemingly lacking selection pressures and lower proportions of human-associated bacteria in the effluent (Bengtsson-Palme et al. 2016), although such transfer is certainly not impossible. The dissemination routes for environmental bacteria carrying resistance genes are much less clear. Regardless, the most critical factor is whether there is a quick dispersal route to the human population (Baquero, Alvarez-Ortega and Martinez 2009). The shorter the ‘length’ of this route, the higher the risks associated with a particular environment.
Taken together, it is not clear how high-risk settings for human health associated with environmental antibiotic resistance should be defined. However, obvious scenarios where interventions could already be applied are environments with strong selection pressures from antibiotics (Table 1). Thus, limiting discharges of pharmaceutical waste from antibiotics production and reducing unnecessary use of antibiotics in humans, animals, and aquaculture are all important first steps towards mitigation of environmental antibiotic resistance development (Pruden et al. 2013; Bengtsson-Palme and Larsson 2016a,b; Review on Antimicrobial Resistance 2016). Second, identifying and reducing dispersal routes for resistant bacteria to the human microbiome should also be a high priority. For dissemination, targeting critical control points for resistance spread, such as STPs, would be of particular importance (Berendonk et al. 2015). For example, disinfection of treated effluent could be an efficient means of controlling the dispersal of resistant bacteria. However, building out any kind of modern treatment of sewage in developing countries would probably have larger effects on resistance dissemination and would thus be a strategy of even higher priority (Pruden et al. 2013; Graham et al. 2014; Kookana et al. 2014), as the resistance problem is global (Johnson and Woodford 2013; Molton et al. 2013; Bengtsson-Palme et al. 2015).
A FUTURE OF RESISTANT ‘SUPERBUGS’?
Apart from the obvious health hazards associated with increased prevalence of resistance genes among human pathogens, there are additional disturbing circumstances suggesting that the future holds a yet darker development than what we may currently appreciate. First of all, many resistance genes seem to be associated with small fitness costs for their hosts, and are readily transferred both between bacteria and between plasmids (Normark and Normark 2002). Resistance genes with low costs tend to be maintained, and can evolve in response to more efficient variants of the same antibiotic, as observed for cephalosporins and the TEM beta-lactamases (Baquero et al. 1998) as well as tigecycline and some tet tetracycline resistance genes (Linkevicius, Sandegren and Andersson 2016). Furthermore, resistance genes against several different antibiotics can accumulate on the same mobile genetic element. Given that several broad-spectrum antibiotic classes are used to treat the same bacterial strains, such co-localization is more likely than not to emerge over time. Once two genes have been co-localized on the same plasmid in this way, the evolutionary forces to separate them are generally small relative to the gain of keeping them on the plasmid in the face of eventual antibiotic exposure. Thus, we would expect to see an increase of bacteria with plasmid-borne multiresistance phenotypes, and that the rate of this increase would also increase with time. Indeed, this is what has been observed among clinical isolates (Livermore 2009; European Centre for Disease Prevention and Control 2013). Importantly, this increase would appear even if antibiotics usage did not surge. Troublingly, global antibiotics usage is also on the rise (Laxminarayan 2014), likely accelerating the multiresistance problem even further. The use of biocides and metals as antibacterials may also promote multidrug resistance, although to what extent is still uncertain (Baker-Austin et al. 2006; Pal et al. 2015; Wales and Davies 2015; Pal et al. 2017).
Multidrug resistance may not be the only problem we will face in the future though. Bacteria can generate genetic diversity through mutations, recombination and horizontal gene transfer. Each of these processes is under balancing selection, where the benefits of generating potentially adaptive genetic variants are weighted against the risk for fitness-reducing mutations (Gillings 2013). Antibiotic exposure has been shown to increase the mutation and recombination frequencies in bacteria, even at sub-inhibitory levels, through the SOS response (Beaber, Hochhut and Waldor 2004; López et al. 2007; Blázquez et al. 2012). Exposure of environmental bacteria to varying levels of antibiotics is therefore likely to generate variants with higher rates of genetic change, in addition to the selecting for resistance. Since populations of bacteria that have higher mutation rates are more likely to rapidly acquire beneficial mutations, and also more likely to quickly generate compensatory genetic changes, antibiotic exposure may select for fixation of bacterial populations with generally higher rates of genetic change (Gillings and Stokes 2012). In addition, antibiotics are often released into the environment together with bacteria carrying integrons and other mobile genetic elements (Gaze et al. 2011, 2013). Since integrase activity is also induced by antibiotics (Maiques et al. 2006), this may further increase bacterial evolvability, generating ever more complex mobile genetic elements (Gillings 2014, 2017b). If such reorganizations of resistance genes come together with a generally increased mutation rate, the net result would be an even quicker evolution towards mobile resistance genes with lower fitness costs. It is impossible to predict exactly what consequences this may have for bacterial pangenomes. Since integrons and other mobile genetic elements allow bacteria to adapt faster to new niches (Gillings 2014), genes mobilized in the future would likely not be restricted to conferring antibiotic resistance, but may also encompass genes that provide a fitness advantage in terms of adaption to changing environments. Thus, genes allowing bacteria to survive highly variable abiotic conditions, handle toxicants, utilize novel carbon sources, compete with other microbes, adhere to different types of surfaces, re-engineer their ecosystems, and allow formation of highly durable spores would be good candidates for future mobilization. From a human health perspective, it is easy to imagine that selection by antibiotics would favor strains with attributes that are beneficial for colonization and invasion of the human host. This could include mobilization of genes involved in virulence, transmission and pathogenicity (Gillings 2014), but also genes that increase competitive ability with human commensals. This paints a picture of a bleak future in which human pathogens would not only be non-treatable by most antibiotics, but also would become more aggressive and spread more easily between humans. We might already be witnessing a foretaste of what is to come, with hypervirulent Klebsiella pneumoniae resistant to all antibiotics tested recently appearing in Chinese hospitals (Gu et al. 2017). It is thus important to understand not only the risks for resistance transmission, but also the evolutionary consequences of antibiotic releases into the environment.
CONCLUDING REMARKS
In this paper, we have attempted to define and formalize relevant ecological and environmental factors for antibiotic resistance development and transmission. We propose that although emergence of novel resistance factors and mobilization of existing ones probably happen continuously, only few of these determinants are selected for and permanently established among bacterial populations. As a consequence, those that do make it to pathogenic species are likely to be evolved towards conveying very little fitness cost to their hosts, and will thus be hard to eliminate from pathogen populations. Successful mitigation strategies for environmental resistance development are therefore in principle limited to (i) avoiding creation of settings that select for, mobilize and allow persistence of resistance genes in bacterial communities, (ii) reducing dispersal routes for resistant bacteria to the human microbiome, and (iii) limiting the selection pressure for resistant pathogens (i.e. prudent use of antibiotics for humans and animals). The diversity of resistance genes present in the environment suggests that there are still many more resistance genes available for pathogens to recruit. Resistance genes common among the bacterial populations in the human microbiome are not likely to be eradicated, even in the absence of antibiotics selection. Thus, the mobile resistance genes that are already circulating among human pathogens may easily re-emerge during antibiotics treatment. Recruitment of additional novel resistance genes into pathogens, on the other hand, has the potential to cause devastating consequences for human health, as mobile resistance genes against new antibiotics, or more efficient resistance mechanisms against the ones that already face resistance, would further reduce treatment options.
FUNDING
This work was supported by the Swedish Research Council (VR) [grant numbers 2013-8633 and 2015-02492 to DGJL, grant numbers 2011-4744 and 2012-5975 to EK]; the Swedish Research Council for Environment, Agriculture and Spatial Planning (FORMAS) [grant numbers 2012-86 and 2015-750 to DGJL, grant number 2016-786 to JBP]; the Swedish Foundation for Strategic Environmental Research (MISTRA) [grant number 2004-147]; Life Science Area of Advance at Chalmers University of Technology; the Wallenberg Foundation; and the Centre for Antibiotic Resistance Research at University of Gothenburg (CARe).
Conflict of interest. None declared.
REFERENCES
- Allen HK. Antibiotic resistance gene discovery in food-producing animals. Curr Opin Microbiol 2014;19C:25–9. [DOI] [PubMed] [Google Scholar]
- Allen HK, Donato J, Wang HH et al. . Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 2010;8:251–9. [DOI] [PubMed] [Google Scholar]
- Allen HK, Moe LA, Rodbumrer J et al. . Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J 2009;3:243–51. [DOI] [PubMed] [Google Scholar]
- Aminov RI. The role of antibiotics and antibiotic resistance in nature. Environ Microbiol 2009;11:2970–88. [DOI] [PubMed] [Google Scholar]
- Andersson DI. Persistence of antibiotic resistant bacteria. Curr Opin Microbiol 2003;6:452–6. [DOI] [PubMed] [Google Scholar]
- Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 2010;8:260–71. [DOI] [PubMed] [Google Scholar]
- Angelin M, Evengård B, Palmgren H. Illness and risk behaviour in health care students studying abroad. Med Educ 2015;49:684–91. [DOI] [PubMed] [Google Scholar]
- Arzanlou M, Chai WC, Venter H. Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays Biochem 2017;61:49–59. [DOI] [PubMed] [Google Scholar]
- Ashbolt NJ, Amézquita A, Backhaus T et al. . Human Health Risk Assessment (HHRA) for Environmental Development and Transfer of Antibiotic Resistance. Environ Health Perspect 2013;121:993–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Austin C, Wright MS, Stepanauskas R et al. . Co-selection of antibiotic and metal resistance. Trends Microbiol 2006;14:176–82. [DOI] [PubMed] [Google Scholar]
- Baltrus DA. Exploring the costs of horizontal gene transfer. Trends Ecol Evol 2013;28:489–95. [DOI] [PubMed] [Google Scholar]
- Baquero F, Alvarez-Ortega C, Martinez JL. Ecology and evolution of antibiotic resistance. Environ Microbiol Rep 2009;1:469–76. [DOI] [PubMed] [Google Scholar]
- Baquero F, Coque TM, de la Cruz F. Ecology and evolution as targets: the need for novel eco-evo drugs and strategies to fight antibiotic resistance. Antimicrob Agents Ch 2011;55:3649–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero F, Lanza VF, Cantón R et al. . Public health evolutionary biology of antimicrobial resistance: priorities for intervention. Evol Appl 2015;8:223–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero F, Martinez JL, Cantón R. Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol 2008;19:260–5. [DOI] [PubMed] [Google Scholar]
- Baquero F, Negri MC, Morosini MI et al. . Antibiotic-selective environments. Clin Infect Dis 1998;27(Suppl 1):S5–11. [DOI] [PubMed] [Google Scholar]
- Baquero F, Tedim AP, Coque TM. Antibiotic resistance shaping multi-level population biology of bacteria. Front Microbiol 2013;4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberán A, Ladau J, Leff JW et al. . Continental-scale distributions of dust-associated bacteria and fungi. P Natl Acad Sci USA 2015;112:5756–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 2004;427:72–4. [DOI] [PubMed] [Google Scholar]
- Bengtsson J. Applied (meta)community ecology: diversity and ecosystem services at the intersection of local and regional processes. In: Verhoef HA, Morin PJ (eds). Community Ecology: Processes, Models, and Applications. Oxford: Oxford University Press, 2009, 115–30. [Google Scholar]
- Bengtsson-Palme J, Alm Rosenblad M, Molin M et al. . Metagenomics reveals that detoxification systems are underrepresented in marine bacterial communities. BMC Genomics 2014a;15:749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Angelin M, Huss M et al. . The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrob Agents Ch 2015;59:6551–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Boulund F, Fick J et al. . Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Front Microbiol 2014b;5:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Hammarén R, Pal C et al. . Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci Total Environ 2016;572:697–712. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Larsson DGJ, Kristiansson E. Using metagenomics to investigate human and environmental resistomes. J Antimicrob Chemoth 2017, DOI: 10.1093/jac/dkx199. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Larsson DGJ. Antibiotic resistance genes in the environment: prioritizing risks. Nat Rev Microbiol 2015;13:396. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Larsson DGJ. Concentrations of antibiotics predicted to select for resistant bacteria: proposed limits for environmental regulation. Environ Int 2016a;86:140–9. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Larsson DGJ. Time to regulate antibiotic pollution. Med Maker 2016b;0416:17–8. [Google Scholar]
- Bengtsson-Palme J. Antibiotic resistance in the food supply chain: where can sequencing and metagenomics aid risk assessment? Curr Opin Food Sci 2017;14:66–71. [Google Scholar]
- Berendonk TU, Manaia CM, Merlin C et al. . Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol 2015;13:310–7. [DOI] [PubMed] [Google Scholar]
- Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ Microbiol 2005;7:1673–85. [DOI] [PubMed] [Google Scholar]
- Blázquez J, Couce A, Rodríguez-Beltrán J et al. . Antimicrobials as promoters of genetic variation. Curr Opin Microbiol 2012;15:561–9. [DOI] [PubMed] [Google Scholar]
- Bondarczuk K, Markowicz A, Piotrowska-Seget Z. The urgent need for risk assessment on the antibiotic resistance spread via sewage sludge land application. Environ Int 2015;87:49–55. [DOI] [PubMed] [Google Scholar]
- Bonnedahl J, Drobni M, Gauthier-Clerc M et al. . Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS One 2009;4:e5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma JE, Lenski RE. Evolution of a bacteria/plasmid association. Nature 1988;335:351–2. [DOI] [PubMed] [Google Scholar]
- Buchholz U, Bernard H, Werber D et al. . German outbreak of Escherichia coli O104:H4 associated with sprouts. N Engl J Med 2011;365:1763–70. [DOI] [PubMed] [Google Scholar]
- Cabello FC. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ Microbiol 2006;8:1137–44. [DOI] [PubMed] [Google Scholar]
- Collignon P. Ban routine use of critically important antibiotics in food animals. BMJ 2013;347:f4976. [DOI] [PubMed] [Google Scholar]
- Collignon P. Antibiotic resistance: are we all doomed? Intern Med J 2015;45:1109–15. [DOI] [PubMed] [Google Scholar]
- D’Costa VM, King CE, Kalan L et al. . Antibiotic resistance is ancient. Nature 2011;477:457–61. [DOI] [PubMed] [Google Scholar]
- De Boeck H, Miwanda B, Lunguya-Metila O et al. . ESBL-positive Enterobacteria isolates in drinking water. Emerg Infect Dis 2012;18:1019–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan J. How emergence arises. Ecol Complex 2006;3:293–301. [Google Scholar]
- Demain AL. Induction of microbial secondary metabolism. Int Microbiol 1998;1:259–64. [PubMed] [Google Scholar]
- Dörr T, Lewis K, Vulić M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet 2009;5:e1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enne VI, Bennett PM, Livermore DM et al. . Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J Antimicrob Chemoth 2004;53:958–63. [DOI] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control Antimicrobial Resistance Surveillance in Europe 2012. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). Stockholm: European Centre for Disease Prevention and Control, 2013, DOI: 10.2900/93403. [Google Scholar]
- Fernando GA, Collignon PJ, Bell JM. A risk for returned travellers: the “post-antibiotic era”. Med J Aust 2010;193:59. [DOI] [PubMed] [Google Scholar]
- Finley RL, Collignon P, Larsson DGJ et al. . The scourge of antibiotic resistance: the important role of the environment. Clin Infect Dis 2013;57:704–10. [DOI] [PubMed] [Google Scholar]
- Forsberg KJ, Reyes A, Wang B et al. . The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012;337:1107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K, Sunagawa S, Kultima JR et al. . Country-specific antibiotic use practices impact the human gut resistome. Genome Res 2013;23:1163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gat D, Mazar Y, Cytryn E et al. . Origin-dependent variations in the atmospheric microbiome community in eastern Mediterranean dust storms. Environ Sci Technol 2017;51:6709–18. [DOI] [PubMed] [Google Scholar]
- Gaze WH, Krone SM, Larsson DGJ et al. . Influence of humans on evolution and mobilization of environmental antibiotic resistome. Emerg Infect Dis 2013;19, DOI: 10.3201/eid1907.120871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaze WH, Zhang L, Abdouslam NA et al. . Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J 2011;5:1253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaly TM, Chow L, Asher AJ et al. . Evolution of class 1 integrons: mobilization and dispersal via food-borne bacteria. PLoS One 2017;12:e0179169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings MR. Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Front Microbiol 2013;4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings MR. Integrons: past, present, and future. Microbiol Mol Biol Rev 2014;78:257–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings MR. Lateral gene transfer, bacterial genome evolution, and the Anthropocene. Ann NY Acad Sci 2017a;1389:20–36. [DOI] [PubMed] [Google Scholar]
- Gillings MR. Class 1 integrons as invasive species. Curr Opin Microbiol 2017b;38:10–5. [DOI] [PubMed] [Google Scholar]
- Gillings MR, Stokes HW. Are humans increasing bacterial evolvability? Trends Ecol Evol 2012;27:346–52. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Cameron Thrash J, Temperton B. Implications of streamlining theory for microbial ecology. ISME J 2014;8:1553–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Ronce O, Ferriere R et al. . Evolutionary rescue: an emerging focus at the intersection between ecology and evolution. Philos T Roy Soc B 2013;368:20120404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górny RL, Dutkiewicz J. Bacterial and fungal aerosols in indoor environment in Central and Eastern European countries. Ann Agric Environ Med 2002;9:17–23. [PubMed] [Google Scholar]
- Graham DW, Collignon P, Davies J et al. . Underappreciated role of regionally poor water quality on globally increasing antibiotic resistance. Environ Sci Technol 2014;48:11746–7. [DOI] [PubMed] [Google Scholar]
- Gu D, Dong N, Zheng Z et al. . A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 2017, DOI: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- Gullberg E, Albrecht LM, Karlsson C et al. . Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. MBio 2014;5:e01918–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullberg E, Cao S, Berg OG et al. . Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 2011;7:e1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood VJ, Staley C, Badgley BD et al. . Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 2014;38:1–40. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Rosenberg SM, Slack A. Antibiotic-induced lateral transfer of antibiotic resistance. Trends Microbiol 2004;12:401–4. [DOI] [PubMed] [Google Scholar]
- Hernando-Amado S, Sanz-García F, Blanco P et al. . Fitness costs associated with the acquisition of antibiotic resistance. Essays Biochem 2017;61:37–48. [DOI] [PubMed] [Google Scholar]
- Hiltunen T, Virta M, Laine A-L.. Antibiotic resistance in the wild: an eco-evolutionary perspective. Philos T Roy Soc B 2017;372, DOI: 10.1098/rstb.2016.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocquet D, Llanes C, Thouverez M et al. . Evidence for induction of integron-based antibiotic resistance by the SOS response in a clinical setting. PLoS Pathog 2012;8:e1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers PMC, Blaak H, De Jong MC et al. . Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ Sci Technol 2015;49:11993–2004. [DOI] [PubMed] [Google Scholar]
- Jernberg C, Löfmark S, Edlund C et al. . Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 2007;1:56–66. [DOI] [PubMed] [Google Scholar]
- Johnning A, Kristiansson E, Fick J et al. . Resistance mutations in gyrA and parC are common in Escherichia communities of both fluoroquinolone-polluted and uncontaminated aquatic environments. Front Microbiol 2015;6:1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnning A, Moore ER, Svensson-Stadler L et al. . Acquired genetic mechanisms of a multiresistant bacterium isolated from a treatment plant receiving wastewater from antibiotic production. Appl Environ Microb 2013;79:7256–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AP, Woodford NN. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol 2013;62:499–513. [DOI] [PubMed] [Google Scholar]
- Jutkina J, Marathe NP, Flach C-F et al. . Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci Tot Environ 2017, DOI: 10.1016/j.scitotenv.2017.10.312. [DOI] [PubMed] [Google Scholar]
- Jutkina J, Rutgersson C, Flach C-F et al. . An assay for determining minimal concentrations of antibiotics that drive horizontal transfer of resistance. Sci Total Environ 2016;548-549:131–8. [DOI] [PubMed] [Google Scholar]
- Kallmeyer J, Pockalny R, Adhikari RR et al. . Global distribution of microbial abundance and biomass in subseafloor sediment. P Natl Acad Sci USA 2012;109:16213–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkman A, Johnson TA, Lyra C et al. . High-throughput quantification of antibiotic resistance genes from an urban wastewater treatment plant. FEMS Microbiol Ecol 2016, DOI: 10.1093/femsec/fiw014. [DOI] [PubMed] [Google Scholar]
- Kookana RS, Williams M, Boxall ABA et al. . Potential ecological footprints of active pharmaceutical ingredients: an examination of risk factors in low-, middle- and high-income countries. Philos T Roy Soc B 2014;369:20130586–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmerer K. Antibiotics in the aquatic environment–a review–part I. Chemosphere 2009a;75:417–34. [DOI] [PubMed] [Google Scholar]
- Kümmerer K. Antibiotics in the aquatic environment–a review–part II. Chemosphere 2009b;75:435–41. [DOI] [PubMed] [Google Scholar]
- Lang KS, Anderson JM, Schwarz S et al. . Novel florfenicol and chloramphenicol resistance gene discovered in Alaskan soil by using functional metagenomics. Appl Environ Microb 2010;76:5321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson DGJ. Antibiotics in the environment. Ups J Med Sci 2014a;119:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson DGJ. Pollution from drug manufacturing: review and perspectives. Philos T Roy Soc B 2014b;369, DOI: 10.1098/rstb.2013.0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxminarayan R. Antibiotic effectiveness: balancing conservation against innovation. Science 2014;345:1299–301. [DOI] [PubMed] [Google Scholar]
- Leggett MJ, McDonnell G, Denyer SP et al. . Bacterial spore structures and their protective role in biocide resistance. J Appl Microbiol 2012;113:485–98. [DOI] [PubMed] [Google Scholar]
- Leibold M, Holyoak M, Mouquet N et al. . The metacommunity concept: a framework for multi-scale community ecology. Ecol Lett 2004;7:601–13. [Google Scholar]
- Lennon JT, Jones SE. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 2011;9:119–30. [DOI] [PubMed] [Google Scholar]
- Levin BR, Baquero F, Johnsen PJ. A model-guided analysis and perspective on the evolution and epidemiology of antibiotic resistance and its future. Curr Opin Microbiol 2014;19C:83–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR, Lipsitch M, Perrot V et al. . The population genetics of antibiotic resistance. Clin Infect Dis 1997;24(Suppl 1):S9–16. [DOI] [PubMed] [Google Scholar]
- Linares JF, Gustafsson I, Baquero F et al. . Antibiotics as intermicrobial signaling agents instead of weapons. P Natl Acad Sci USA 2006;103:19484–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkevicius M, Sandegren L, Andersson DI. Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob Agents Ch 2016;60:789–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M, Bergstrom CT, Levin BR. The epidemiology of antibiotic resistance in hospitals: paradoxes and prescriptions. P Natl Acad Sci USA 2000;97:1938–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore DM. Epidemiology of antibiotic resistance. Intensive Care Med 2000;26(Suppl 1):S14–21. [DOI] [PubMed] [Google Scholar]
- Livermore DM. Has the era of untreatable infections arrived? J Antimicrob Chemoth 2009;64(Suppl 1):i29–36. [DOI] [PubMed] [Google Scholar]
- Löfmark S, Jernberg C, Billström H et al. . Restored fitness leads to long-term persistence of resistant Bacteroides strains in the human intestine. Anaerobe 2008;14:157–60. [DOI] [PubMed] [Google Scholar]
- López E, Elez M, Matic I et al. . Antibiotic-mediated recombination: ciprofloxacin stimulates SOS-independent recombination of divergent sequences in Escherichia coli. Mol Microbiol 2007;64:83–93. [DOI] [PubMed] [Google Scholar]
- Lundström SV, Östman M, Bengtsson-Palme J et al. . Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Sci Total Environ 2016;553:587–95. [DOI] [PubMed] [Google Scholar]
- Lupo A, Coyne S, Berendonk TU. Origin and evolution of antibiotic resistance: the common mechanisms of emergence and spread in water bodies. Front Microbiol 2012;3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiques E, Ubeda C, Campoy S et al. . beta-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J Bacteriol 2006;188:2726–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe NP, Shetty SA, Shouche YS et al. . Limited bacterial diversity within a treatment plant receiving antibiotic-containing waste from bulk drug production. PLoS One 2016;11:e0165914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JL. Antibiotics and antibiotic resistance genes in natural environments. Science 2008;321:365–7. [DOI] [PubMed] [Google Scholar]
- Martinez JL. Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Front Microbiol 2011;2:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JL, Baquero F, Andersson DI. Predicting antibiotic resistance. Nat Rev Microbiol 2007;5:958–65. [DOI] [PubMed] [Google Scholar]
- Martinez JL, Coque TM, Baquero F. What is a resistance gene? Ranking risk in resistomes. Nat Rev Microbiol 2015;13:116–23. [DOI] [PubMed] [Google Scholar]
- Martiny AC, Martiny JBH, Weihe C et al. . Functional metagenomics reveals previously unrecognized diversity of antibiotic resistance genes in gulls. Front Microbiol 2011;2:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matte-Tailliez O, Brochier C, Forterre P et al. . Archaeal phylogeny based on ribosomal proteins. Mol Biol Evol 2002;19:631–9. [DOI] [PubMed] [Google Scholar]
- Mattner F, Bange F-C, Meyer E et al. . Preventing the spread of multidrug-resistant gram-negative pathogens: recommendations of an expert panel of the German Society For Hygiene and Microbiology. Dtsch Arztebl Int 2012;109:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachran AD, Blackwell BR, Hanson JD et al. . Antibiotics, bacteria, and antibiotic resistance genes: aerial transport from cattle feed yards via particulate matter. Environ Health Perspect 2015;123:337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk AH, Wong A, Kassen R. The fitness costs of antibiotic resistance mutations. Evol Appl 2015;8:273–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael I, Rizzo L, McArdell CS et al. . Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: a review. Water Res 2013;47:957–95. [DOI] [PubMed] [Google Scholar]
- Molton JS, Tambyah PA, Ang BSP et al. . The global spread of healthcare-associated multidrug-resistant bacteria: a perspective from Asia. Clin Infect Dis 2013;56:1310–8. [DOI] [PubMed] [Google Scholar]
- Munck C, Albertsen M, Telke A et al. . Limited dissemination of the wastewater treatment plant core resistome. Nat Commun 2015;6:8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark BH, Normark S. Evolution and spread of antibiotic resistance. J Intern Med 2002;252:91–106. [DOI] [PubMed] [Google Scholar]
- Pal C, Asiani K, Arya S et al. . Metal resistance and its association with antibiotic resistance. Adv Microb Physiol 2017;70:261–313. [DOI] [PubMed] [Google Scholar]
- Pal C, Bengtsson-Palme J, Kristiansson E et al. . Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics 2015;16:964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Bengtsson-Palme J, Kristiansson E et al. . The structure and diversity of human, animal and environmental resistomes. Microbiome 2016;4:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron GG, Inglis RF, Pennings PS et al. . Fighting microbial drug resistance: a primer on the role of evolutionary biology in public health. Evol Appl 2015;8:211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L, Andersson SGE, Battin TJ et al. . The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol 2010;8:523–9. [DOI] [PubMed] [Google Scholar]
- Porse A, Gumpert H, Kubicek-Sutherland JZ et al. . Genome dynamics of Escherichia coli during antibiotic treatment: transfer, loss, and persistence of genetic elements in situ of the infant gut. Front Cell Infect Mi 2017;7:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruden A. Balancing water sustainability and public health goals in the face of growing concerns about antibiotic resistance. Environ Sci Technol 2014;48:5–14. [DOI] [PubMed] [Google Scholar]
- Pruden A, Arabi M, Storteboom HN. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ Sci Technol 2012;46:11541–9. [DOI] [PubMed] [Google Scholar]
- Pruden A, Larsson DGJ, Amézquita A et al. . Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ Health Perspect 2013;121:878–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruden A, Pei R, Storteboom HN et al. . Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ Sci Technol 2006;40:7445–50. [DOI] [PubMed] [Google Scholar]
- Rao GG. Risk factors for the spread of antibiotic-resistant bacteria. Drugs 1998;55:323–30. [DOI] [PubMed] [Google Scholar]
- Rasko DA, Webster DR, Sahl JW et al. . Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med 2011;365:709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Review on Antimicrobial Resistance In: O’Neill J. (ed). Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. London: Wellcome Trust & HM Government, 2014. [Google Scholar]
- Review on Antimicrobial Resistance In: O’Neill J. (ed). Tackling Drug-Resistant Infections Globally: Final Report and Recommendations London: Wellcome Trust & HM Government, 2016. [Google Scholar]
- Rizzo L, Manaia C, Merlin C et al. . Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 2013;447:345–60. [DOI] [PubMed] [Google Scholar]
- Rolain J-M. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front Microbiol 2013;4:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Amábile-Cuevas CF. Why are antibiotic resistance genes so resistant to elimination? Antimicrob Agents Ch 1997;41:2321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol 2004;12:412–6. [DOI] [PubMed] [Google Scholar]
- San Millan A, Santos-Lopez A, Ortega-Huedo R et al. . Small-plasmid-mediated antibiotic resistance is enhanced by increases in plasmid copy number and bacterial fitness. Antimicrob Agents Ch 2015;59:3335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa T, Takeuchi N, Rivera A et al. . Distribution of antibiotic resistance genes in glacier environments. Environ Microbiol Rep 2012;5:127–34. [DOI] [PubMed] [Google Scholar]
- Seier-Petersen MA, Jasni A, Aarestrup FM et al. . Effect of subinhibitory concentrations of four commonly used biocides on the conjugative transfer of Tn916 in Bacillus subtilis. J Antimicrob Chemoth 2014;69:343–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Peter H, Allison SD et al. . Fundamentals of microbial community resistance and resilience. Front Microbiol 2012;3:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker NB, Vlamakis H, Hayes K et al. . Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl Environ Microb 2001;67:561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie CS, Smith MB, Friedman J et al. . Ecology drives a global network of gene exchange connecting the human microbiome. Nature 2011;480:241–4. [DOI] [PubMed] [Google Scholar]
- So AD, Shah TA, Roach S et al. . An integrated systems approach is needed to ensure the sustainability of antibiotic effectiveness for both humans and animals. J Law Med Ethics 2015;43(Suppl 3):38–45. [DOI] [PubMed] [Google Scholar]
- Sommer MOA, Church GM, Dantas G. The human microbiome harbors a diverse reservoir of antibiotic resistance genes. Virulence 2010;1:299–303. [DOI] [PubMed] [Google Scholar]
- Sommer MOA, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 2009;325:1128–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedt J, Bonnedahl J, Hernandez J et al. . Carriage of CTX-M type extended spectrum β-lactamases (ESBLs) in gulls across Europe. Acta Vet Scand 2015;57:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes HW, Gillings MR. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol Rev 2011;35:790–819. [DOI] [PubMed] [Google Scholar]
- Tsai FC, Macher JM. Concentrations of airborne culturable bacteria in 100 large US office buildings from the BASE study. Indoor Air 2005;15(Suppl 9):71–81. [DOI] [PubMed] [Google Scholar]
- Vogwill T, MacLean RC. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol Appl 2015;8:284–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales A, Davies R. Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics 2015;4:567–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. Antibiotics: Actions, Origins, Resistance. Washington, DC: ASM Press, 2003. [Google Scholar]
- Webber MA, Ricci V, Whitehead R et al. . Clinically relevant mutant DNA gyrase alters supercoiling, changes the transcriptome, and confers multidrug resistance. MBio 2013;4:e00273–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Antimicrobial Resistance: Global Report on Surveillance 2014. Geneva, Switzerland: World Health Organization, 2014. [Google Scholar]
- Wiedenbeck J, Cohan FM. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev 2011;35:957–76. [DOI] [PubMed] [Google Scholar]
- Wright GD. Antibiotic resistance in the environment: a link to the clinic? Curr Opin Microbiol 2010;13:589–94. [DOI] [PubMed] [Google Scholar]
- Yooseph S, Nealson KH, Rusch DB et al. . Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature 2010;468:60–6. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gu AZ, He M et al. . Subinhibitory concentrations of disinfectants promote the horizontal transfer of multidrug resistance genes within and across genera. Environ Sci Technol 2017;51:570–80. [DOI] [PubMed] [Google Scholar]