Abstract
Daily injections of insulin provide lifesaving benefits to millions of diabetics. But currently available prandial insulins are suboptimal: The onset of action is delayed by slow dissociation of the insulin hexamer in the subcutaneous space, and insulin forms amyloid fibrils upon storage in solution. Here we show, through the use of non-canonical amino acid mutagenesis, that replacement of the proline residue at position 28 of the insulin B-chain (ProB28) by (4S)-hydroxyproline (Hzp) yields an active form of insulin that dissociates more rapidly, and fibrillates more slowly, than the wild-type protein. Crystal structures of dimeric and hexameric insulin preparations suggest that a hydrogen bond between the hydroxyl group of Hzp and a backbone amide carbonyl positioned across the dimer interface may be responsible for the altered behavior. The effects of hydroxylation are stereospecific; replacement of ProB28 by (4R)-hydroxyproline (Hyp) causes little change in the rates of fibrillation and hexamer disassociation. These results demonstrate a new approach that fuses the concepts of medicinal chemistry and protein design, and paves the way to further engineering of insulin and other therapeutic proteins.
Blood glucose levels are tightly controlled in mammals through a sensitive regulatory system mediated by insulin, a 51-amino acid endocrine hormone composed of two disulfide-linked polypeptide chains (designated A and B). Upon binding to its receptor, insulin initiates a signaling cascade that accelerates glucose uptake and glycogen production. In diabetic patients, this system malfunctions, and glucose levels must be controlled through subcutaneous injections of insulin1. The C-terminus of the B-chain is important in mediating dimerization of the hormone2–3, and the flexibility of the B-chain C-terminus is believed to contribute to aggregation through formation of amyloid fibrils4–6. Pharmaceutical formulations of insulin are stabilized with respect to fibrillation by addition of zinc and phenolic preservatives, which drive assembly of the R6 hexamer (Fig. 1a)7–9. The R6 form of insulin is inactive and dissociates slowly to its active monomeric form after subcutaneous injection; the lag time for dissociation delays the onset of action10. Mutation of ProB28 yields rapid-acting insulins (RAIs) by disrupting contacts that are critical for dimer formation8, but replacement of Pro through conventional mutagenesis also increases the flexibility and perturbs the trajectory of the protein backbone (Fig. 1B). We sought a means to disrupt the dimer interface without releasing the conformational constraints characteristic of proline by using non-canonical amino acid (ncAA) mutagenesis11–13. Specifically, we introduced hydroxyl groups at the 4-position of ProB28 (Fig. 1B, C) by replacing Pro with Hzp or Hyp. In addition to introducing a polar functional group and the capacity for hydrogen-bonding (including transannular hydrogen bonding), hydroxylation at the 4-position is known to alter the endo/exo preference of the pyrrolidine ring and the cis/trans equilibrium of the backbone amide bond14–16.
Figure. 1. Hydroxyinsulins retain activity.

(A) Schematic of hexamer disassembly (adapted from mechanism previously described 8). Phenolic ligand (Ph), zinc ion (Zn2+), insulin monomer (triangle). Darker shading indicates the R-state of the hexamer. (B) Structures of the B-chain C-termini of wild-type insulin (ProI) and RAI Aspart (AspI). (C) Chemical structures of L-proline (1), (2S,4S)-hydroxyproline (Hzp; 2), (2S,4R)-hydroxyproline (Hyp; 3). (D) Reduction of blood glucose following subcutaneous injection of 35 µg/kg insulins into streptozotocin-induced diabetic mice. Glucose levels were measured post-injection via tail vein sampling. ProI, AspI, HzpI, HypI or vector were formulated as described 22. Error bars denote one standard deviation (n = 3).
We expressed modified proinsulins (PIs) in the proline-auxotrophic E. coli strain CAG18515 in M9 minimal media supplemented with Hyp or Hzp. The extent of replacement of Pro by either Hyp or Hzp was approximately 90%17–18 as determined by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS; fig. S1). Denatured PIs were purified by Ni-NTA affinity chromatography in yields of 32 mg/L for Hzp-PI and 29 mg/L for Hyp-PI (table S1) from the inclusion body fraction. The PIs were refolded and cleaved with trypsin and carboxypeptidase B19. The resulting mature insulins were purified by reversed phase HPLC18–19, and proper proteolytic processing of each variant was verified by MALDI-MS (table S1). Wild-type insulin (ProI) and RAI Aspart (AspI, in which ProB28 is replaced by aspartic acid) were produced similarly. All of the variants caused similar reductions in blood glucose upon subcutaneous injection into diabetic mice (Fig. 1D)2, 20–22. RAIs cannot be distinguished from ProI in rodent models23.
In the absence of Zn2+ and phenolic preservatives, insulins dimerize with a dissociation constant (KD) of approximately 10 µM. In contrast, KD for RAIs is typically >500 µM, and it is believed that destabilization of the dimer interface causes the accelerated onset of insulin action after subcutaneous injection20, 24–25. Monomeric forms of insulin give rise to characteristic circular dichroism (CD) spectra with distinct minima at 208 and 222 nm26–27 (e.g., AspI; Fig. 2A). Dimerization causes a loss of negative ellipticity at 208 nm27 (e.g., ProI; Fig. 2A). At a concentration of 60 µM, Hypl appears to be monomeric (with a CD spectrum nearly identical to that of AspI; Fig. 2A) while the spectrum of HzpI suggests a dimeric insulin (Fig. 2A). Sedimentation velocity (SV) and sedimentation equilibrium (SE) experiments were consistent with the results of the CD analysis (fig. S3). SE data were fitted to a model of monomer-dimer-hexamer self-association (SEDPHAT) 28–29, and yielded monomer-dimer dissociation constants (KD) of >200 µM and 25 µM for HypI and HzpI, respectively.
Figure. 2. Hydroxylation at ProB28 modulates insulin dimerization, dissociation kinetics, and stability.
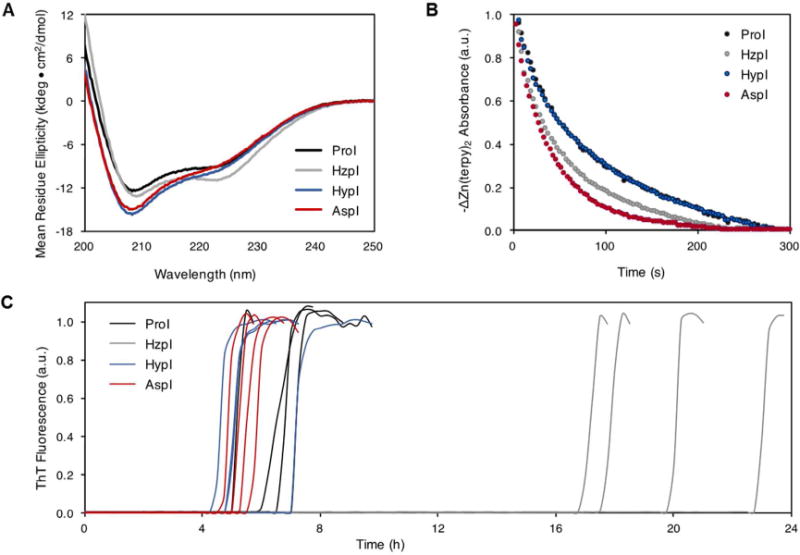
(A) Far UV CD spectra collected on 60 µM insulins in 10 mM phosphate buffer, pH 8.0 at 25°C. (B) Insulin hexamer dissociation following sequestration of Zn2+ by terpyridine. Zn2+-(terpy) signal was monitored at 334 nm and fitted to a mono-exponential decay. HzpI and HypI contain 10% ProI. The curves for HypI and ProI are indistinguishable in this plot. (C) Representative fibrillation curves for 60 µM insulins (37°C, 960 RPM; n=4). Insulin fibrils were detected by the rise in Thioflavin T (ThT) fluorescence that accompanies binding to fibrillar aggregates.
Previous studies of RAIs have shown that destabilization of the dimer interface correlates with accelerated dissociation of the hexamer and rapid onset of insulin action8, 13. Triggered dissociation of Zn2+-hexamers by addition of terpyridine30 revealed nearly identical rates of dissociation for HypI and ProI, (τ1/2 = 87.0 ± 10 s and 90.4 ± 4.2 s, respectively; Fig. 2B and fig. S4) while HzpI exhibited kinetics similar to those of AspI (τ1/2 = 53.6 ± 3.7 s and 42.7 ± 4.3 s, respectively; Fig. 2B and fig. S4). We found these results surprising – replacement of Pro by Hyp destabilizes the dimer but has essentially no effect on hexamer dissociation, while introduction of Hzp causes little change in dimer stability but a substantial increase in the rate of hexamer disassembly.
Each of the insulin variants was subjected to fibrillation lag time analysis (Fig. 2C)31. We found similar times to onset of fibrillation for HypI, ProI and AspI; in contrast, HzpI is markedly more resistant to aggregation, with a mean time to onset more than three-fold longer than that observed for ProI. The behavior of HzpI is especially striking, in that it combines fast hexamer dissociation with enhanced stability toward fibrillation.
Each subunit in the insulin hexamer adopts one of two conformational states (T or R), depending on the concentration of phenolic ligand (Fig. 1A)13. Pharmaceutical formulations are prepared in the more stable R6 form, whereas the T-state is observed in the absence of phenolic ligands, most commonly in the form of T2-dimers32. To elucidate the molecular origins of the dissociation and fibrillation behavior of HypI and HzpI, we examined crystal structures of both states.
Hydroxylation at ProB28 does not cause substantial perturbation of the overall insulin structure (Fig. 3, fig. S5). In comparison with ProI, the backbone RMSD values of HypI and HzpI are 0.31 Å (T2- HypI), 0.44 Å (T2- HypI), 0.38 Å (R6- HypI) and 0.69 Å (R6- HypI)33. The most notable feature of the HzpI structures is the proximity of the hydroxyl group of Hzp to the backbone carbonyl oxygen atom of GluB21′, which lies across the dimer interface (denoted by prime; Fig. 3B, E). The inter-oxygen distances (2.8 Å in the T2 structure, 2.7 Å in R6), are consistent with the formation of strong hydrogen bonds between the hydroxyl group of HzpB28 and the backbone carbonyl of GluB21′ in both structures. An analogous hydrogen bond has been observed in a structure (PDB ID: 1ZEH) of R6-AspI co-crystallized with m-cresol34; here the phenolic ligand serves as the hydrogen-bond donor (fig. S6). Although the significance of this hydrogen bond has not been discussed in the literature, we suggest that it may play an important role in determining the relative stabilities of the insulin species involved in dissociation and fibrillation. In contrast to the (4S)-hydroxyl group of Hzp, the (4R)-hydroxyl of HypB28 does not contact any crystallographically resolved hydrogen bond acceptor in the T2-structure (Fig. 3C), and appears to bond to an ordered water molecule in the R6-hexamer (Fig. 3F). The absence of new hydrogen-bonding interactions is consistent with the unaltered dissociation and fibrillation kinetics of HypI.
Figure. 3. Crystal structures of HzpI and HypI.
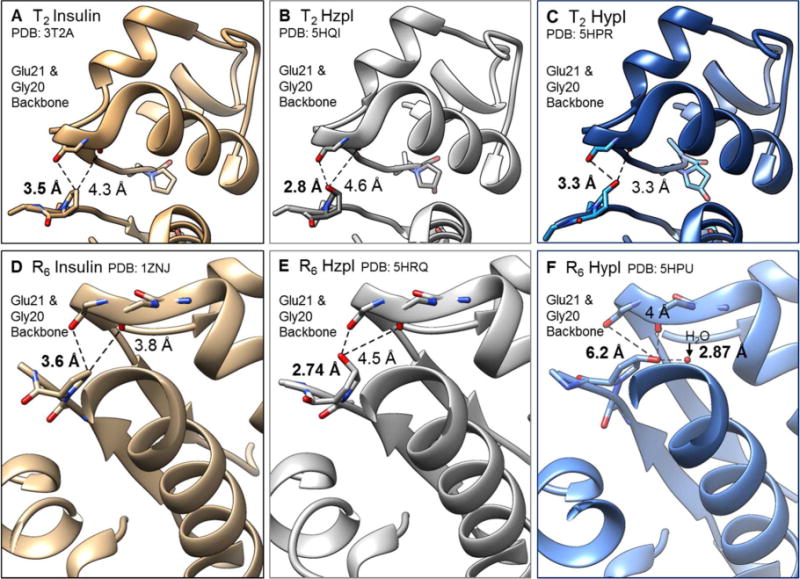
In tan (left), wt insulins from PDB (3T2A, 1ZNJ) highlighting the distance between the carbon atom at the 4th position of ProB28 and its closest neighbors, backbone carbonyl oxygen atoms of GlyB20’ and GluB21 in the T2 dimer (A) and R6 hexamer (D) forms. In grey (middle), HzpI in the T2 dimer (B) and R6 hexamer (E) forms. In blue (right), HypI in the T2 dimer (C) and R6 hexamer (F) forms.
Taken together, our results show that replacement of Pro by Hzp at position 28 of the insulin B-chain introduces a new hydrogen bond across the inter-subunit interface, accelerates hexamer dissociation and delays the onset of fibrillation (Table 1). We suggest that the hydrogen bond between Hzp and Glu21′ may stabilize the dimer relative to the hexamer, or perhaps reduce the energy of the transition state for the conformational change from the R-state to T-state, and thereby speed disassembly. The delayed onset of fibrillation may reflect changes in the structure and dynamics of the HzpI monomer or in the kinetics of fibril nucleation. Subtle conformational effects caused by 4S-substitution on the pyrrolidine ring may also contribute to the observed behavior.35 Whether or not these hypotheses are correct, the results described here demonstrate the power of ncAA mutagenesis to control functionally relevant biophysical properties of therapeutic proteins. We anticipate that this approach will find increasing application in the design of antibody-drug conjugates, bispecific antibodies, and other novel protein therapeutics.
Table 1.
Biophysical Characteristics of Insulin Variants
| Protein | Hexamer t½ (s) | Fibrillation lag time(h) | KD dimer (μM) |
|---|---|---|---|
| Prol | 90.4 ± 4.2 | 5.1 ± 1.5 | 9 μM |
| Hzpl* | 53.6 ± 3.7 | 19.6 ± 2.6 | ~25 μM |
| Hypl* | 87.0 ± 10 | 5.5 ± 1.2 | >200 μM |
| Aspl | 42.7 ± 4.3 | 5.3 ± 1.0 | >500 μM |
Errors are given as one standard deviation (n > 4).
10% Prol present.
Supplementary Material
Acknowledgments
We thank J. T. Kaiser, P. Nikolovski, S. Russi, S. Virgil, M. Shahgholi, A. Lakshmanan, and the scientific staff of Beamline 12-2 at the Stanford Synchrotron Radiation Laboratory for assistance. We thank W. Glenn, A. Madhavi, and T. Hoeg-Jensen for discussions.
Funding Sources
The work was supported by the Novo Nordisk Foundation. Fellowships from Amgen and from the Natural Sciences and Engineering Research Council of Canada (NSERC, PGS-D) provided partial support for S. A. L. and K. Y. F., respectively. J. K. B. C. acknowledges support of the Resnick Sustainability Institute (Caltech). Support by a grant to H.T.K. from the National Institutes of Health (R01DK099734) is also acknowledged.
Abbreviations
- Hzp
(4S)-hydroxyproline
- Hyp
(4R)-hydroxyproline
- RAI
Rapid Acting Insulin
- ncAA
non-canonical amino acid
- PI
proinsulin
- HypI
Hyp-Insulin
- HzpI
Hzp-Insulin
- ProI
Wild-type human insulin
- AspI
Insulin Aspart
- CD
circular dichroism
- SV
sedimentation velocity
- SE
sedimentation equilibrium
Footnotes
Supporting Information
The following files are available free of charge via the internet at http://pubs.acs.org. Experimental procedures and methods, supporting figures and tables, additional references.
References
- 1.Zaykov AN, Mayer JP, DiMarchi RD. Nat Rev Drug Discov. 2016;15:425–439. doi: 10.1038/nrd.2015.36. [DOI] [PubMed] [Google Scholar]
- 2.Ciszak E, Beals JM, Frank BH, Baker JC, Carter ND, Smith GD. Structure. 1995;3:615–22. doi: 10.1016/s0969-2126(01)00195-2. [DOI] [PubMed] [Google Scholar]
- 3.Menting JG, Whittaker J, Margetts MB, Whittaker LJ, Kong GKW, Smith BJ, Watson CJ, Žáková L, Kletvíková E, Jiráček J, Chan SJ, Steiner DF, Dodson GG, Brzozowski AM, Weiss MA, Ward CW, Lawrence MC. Nature. 2013;493:241–5. doi: 10.1038/nature11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang K, Maiti NC, Phillips NB, Carey PR, Weiss MA. Biochemistry. 2006;45:10278–10293. doi: 10.1021/bi060879g. [DOI] [PubMed] [Google Scholar]
- 5.Menting JG, Yang Y, Chan SJ, Phillips NB, Smith BJ, Whittaker J, Wickramasinghe NP, Whittaker LJ, Pandyarajan V, Wan Z-l, Yadav SP, Carroll JM, Strokes N, Roberts CT, Ismail-Beigi F, Milewski W, Steiner DF, Chauhan VS, Ward CW, Weiss MA, Lawrence MC. Proc Natl Acad Sci U S A. 2014;111:E3395–404. doi: 10.1073/pnas.1412897111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanova MI, Sievers SA, Sawaya MR, Wall JS, Eisenberg D. Proc Natl Acad Sci U S A. 2009;106:18990–18995. doi: 10.1073/pnas.0910080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman JS. J Am Osteopath Assoc. 2009;109:26–36. [PubMed] [Google Scholar]
- 8.Birnbaum DT, Kilcomons MA, DeFelippis MR, Beals JM. Pharm Res. 1997;14:25–36. doi: 10.1023/a:1012095115151. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter MC, Wilcox DE. Biochemistry. 2014;53:1296–301. doi: 10.1021/bi4016567. [DOI] [PubMed] [Google Scholar]
- 10.Bakaysa DL, Radziuk J, Havel HA, Brader ML, Li S, Dodd SW, Beals JM, Pekar AH, Brems DN. Protein Sci. 1996;5:2521–2531. doi: 10.1002/pro.5560051215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandey AK, Naduthambi D, Thomas KM, Zondlo NJ. J Am Chem Soc. 2013;135:4333–4363. doi: 10.1021/ja3109664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu CC, Schultz PG. Annu Rev Biochem. 2010;79:413–44. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 13.Pandyarajan V, Weiss MA. Curr Diab Rep. 2012;12:697–704. doi: 10.1007/s11892-012-0318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shoulders MD, Kotch FW, Choudhary A, Guzei IA, Raines RT. J Am Chem Soc. 2010;132:10857–10865. doi: 10.1021/ja103082y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuemin M, Nagel YA, Schweizer S, Monnard FW, Ochsenfeld C, Wennemers H. Angew Chem, Int Ed. 2010;49:6324–6327. doi: 10.1002/anie.201001851. [DOI] [PubMed] [Google Scholar]
- 16.Erdmann RS, Wennemers H. Angew Chem, Int Ed. 2011;50:6835–6838. doi: 10.1002/anie.201008118. [DOI] [PubMed] [Google Scholar]
- 17.Kim W, George A, Evans M, Conticello VP. Chembiochem. 2004;5:928–936. doi: 10.1002/cbic.200400052. [DOI] [PubMed] [Google Scholar]
- 18.Kemmler W, Peterson JD, Steiner DF. J Biol Chem. 1971;246:6786–6791. [PubMed] [Google Scholar]
- 19.Min C-K, Son Y-J, Kim C-K, Park S-J, Lee JW. J Biotechnol. 2011;151:350–356. doi: 10.1016/j.jbiotec.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 20.Brems DN, Alter LA, Beckage MJ, Chance RE, DiMarchi RD, Green LK, Long HB, Pekar AH, Shields JE, Frank BH. Protein Eng. 1992;5:527–533. doi: 10.1093/protein/5.6.527. [DOI] [PubMed] [Google Scholar]
- 21.Sakata N, Yoshimatsu G, Tsuchiya H, Egawa S, Unno M. Exp Diabetes Res. 2012;2012 doi: 10.1155/2012/256707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandyarajan V, Phillips NB, Cox GP, Yang Y, Whittaker J, Ismail-Beigi F, Weiss MA. J Biol Chem. 2014;289:23367–23381. doi: 10.1074/jbc.M114.588277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plum A, Agersø H, Andersen L. Drug Metab Dispos. 2000;28:155–160. [PubMed] [Google Scholar]
- 24.Attri AK, Fernández C, Minton AP. Biophys Chem. 2010;148:28–33. doi: 10.1016/j.bpc.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antolikova E, Zakova L, Turkenburg JP, Watson CJ, Hanclova I, Sanda M, Cooper A, Kraus T, Brzozowski AM, Jiracek J. J Biol Chem. 2011;286:36968–77. doi: 10.1074/jbc.M111.265249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawa SH, Tager HS, Steiner DF. Biochemistry. 2000;39:15826–15835. doi: 10.1021/bi001802+. [DOI] [PubMed] [Google Scholar]
- 27.Pocker Y, Biswas SB. Biochemistry. 1980;19:5043–5049. doi: 10.1021/bi00563a017. [DOI] [PubMed] [Google Scholar]
- 28.Zhao H, Schuck P. Acta Crystallogr, Sect D: Biol Crystallogr. 2015;71:3–14. doi: 10.1107/S1399004714010372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown PH, Schuck P. Biophys J. 2006;90:4651–61. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahuel-Clermont S, French CA, Kaarsholm NC, Dunn MF. Biochemistry. 1997;36:5837–5845. doi: 10.1021/bi963038q. [DOI] [PubMed] [Google Scholar]
- 31.Vinther TN, Norrman M, Strauss HM, Huus K, Schlein M, Pedersen TÅ, Kjeldsen T, Jensen KJ, Hubálek F. PLoS ONE. 2012;7:e30882. doi: 10.1371/journal.pone.0030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmieri LC, Favero-Retto MP, Lourenco D, Lima LM. Biophys Chem. 2013;173–174:1–7. doi: 10.1016/j.bpc.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Marshall H, Venkat M, Seng NS, Cahn J, Juers DH. Acta Crystallogr, Sect D: Biol Crystallogr. 2012;68:69–81. doi: 10.1107/S0907444911050360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith GD, Ciszak E, Magrum LA, Pangborn WA, Blessing RH. Acta Crystallogr, Sect D: Biol Crystallogr. 2000;56:1541–1548. doi: 10.1107/s0907444900012749. [DOI] [PubMed] [Google Scholar]
- 35.DeRider ML, Wilkens SJ, Waddell MJ, Bretscher LE, Weinhold F, Raines RT, Markley JL. J Am Chem Soc. 2002;124:2497–2505. doi: 10.1021/ja0166904. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


