Abstract
Telomerase is the essential reverse transcriptase required for linear chromosome maintenance in most eukaryotes. Telomerase supplements the tandem array of simple-sequence repeats at chromosome ends to compensate for the DNA erosion inherent in genome replication. The template for telomerase reverse transcriptase is within the RNA subunit of the ribonucleoprotein complex, which in cells contains additional telomerase holoenzyme proteins that assemble the active ribonucleoprotein and promote its function at telomeres. Telomerase is distinct among polymerases in its reiterative reuse of an internal template. The template is precisely defined, processively copied, and regenerated by release of single-stranded product DNA. New specificities of nucleic acid handling that underlie the catalytic cycle of repeat synthesis derive from both active site specialization and new motif elaborations in protein and RNA subunits. Studies of telomerase provide unique insights into cellular requirements for genome stability, tissue renewal, and tumorigenesis as well as new perspectives on dynamic ribonucleoprotein machines.
Keywords: telomere, reverse transcriptase, ribonucleoprotein biogenesis, DNA replication
INTRODUCTION
Chromosome Ends Have Unique Function and Composition
Nearly a century ago, Müller (1) and McClintock (2) independently discovered that natural chromosome ends in fruit flies and maize are resistant to fusion, in contrast with ends generated by DNA breakage. The word telomere was used to describe chromosome ends sealed against rearrangement or, in modern parlance, with “end protection.” At the DNA level, the first clue to an explanation for eukaryotic chromosome end protection was revealed in the tandem telomeric repeats of (T2G4)n • (C4A2)n, shown to cap a highly amplified chromosome of the single-celled ciliated protozoan Tetrahymena thermophila (3).
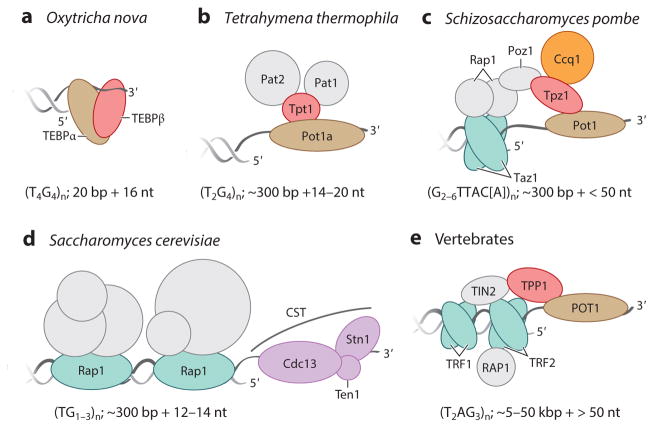
In general, telomeric repeats have one guanosine-rich strand (G-strand) and one cytosine-rich strand (C-strand) synthesized as the leading and lagging strands, respectively (4). Across organisms, tracts of telomeric repeats vary in length from a few to a few thousand repeats and have a G-strand 3′ overhang that varies from a few to more than a hundred nucleotides. At least in vertebrate cells, the relatively long G-strand overhang can invade the adjacent duplex to produce a lasso-like structure known as a telomeric loop (t-loop) (5). Telomere-bound proteins organized by DNA interaction (Figure 1) suppress DNA degradation and repair reactions that would be aberrant for authentic chromosome termini (6).
Figure 1.
Telomere structures for end protection. The telomeric repeat DNA structure and telomere-associated proteins important for chromosome end protection are illustrated for (a,b) two ciliates, (c,d ) two yeasts, and (e) vertebrate cells. Typical telomeric repeat tract lengths in each organism are indicated as base pairs of duplex and nucleotides of 3′ overhang, highlighting the evolutionary divergence of overall telomere and overhang length. Telomere proteins in direct contact with double-stranded DNA are colored blue-green. Proteins other than in CST that are in direct contact with single-stranded DNA are colored brown. Proteins that are orthologous are similarly colored in both this figure and Figure 4 using a shared coloring scheme. Other subunits colored in gray are indirectly bound to DNA. Saccharomyces cerevisiae proteins with functions other than end protection are not labeled; these proteins contribute to telomere length regulation and heterochromatin assembly. Abbreviations: bp, base pair; Cdc, cell division cycle; Ccq, coiled-coil protein quantitatively enriched; CST, heterotrimer containing Stn1, Ten1, and variable third subunit; nt, nucleotide; Pat, Pot1-associated Tetrahymena; Pot, protection of telomeres; Poz, Pot1-associated protein; Rap1, repressor/activator protein 1; Stn, suppressor of Cdc13; Taz, telomere-associated in Schizosaccharomyces pombe; TEBP, telomere end–binding protein; Ten1, telomeric pathways in association with Stn1, number 1; TIN2, TRF1-interacting nuclear protein 2; TPP1, vertebrate shelterin protein designation for proteins, initially named TINT1/PTOP/PIP1; Tpt, TPP1/Tpz1 in Tetrahymena thermophila; Tpz, TPP1 homolog in S. pombe; TRF, telomeric repeat binding factor.
Telomere complexes include numerous proteins that are telomere specific. In ciliates such as Oxytricha nova, with an extremely short, fixed-length telomere sequence, the telomere end–binding protein (TEBP) heterodimer buries the single-stranded 3′ overhang (Figure 1a) (7). In T. thermophila, the fission yeast Schizosaccharomyces pombe, and vertebrate cells (Figure 1b,c,e), the 3′ overhang is protected by a complex of the single-stranded telomeric-repeat DNA-binding protein protection of telomeres 1 (Pot1/POT1) and its heterodimer partner Tpt1/Tpz1/TPP1 (8). Fission yeast and vertebrate cells physically link POT1–TPP1 to protein(s) that bind the double-stranded telomeric repeat tract (Figure 1c,e). Altogether the complex bridging single-stranded and double-stranded repeats is termed shelterin (9). The budding yeast Saccharomyces cerevisiae has an atypical telomeric single-stranded DNA-binding protein, cell division cycle protein 13 (Cdc13), in a heterotrimeric complex, designated CST, with end-capping function (Figure 1d ) (10, 11). Telomeric chromatin assembled by the S. cerevisiae double-stranded telomeric-repeat DNA-binding repressor/activator protein 1 (Rap1) mediates telomere length regulation without physical connection to Cdc13 (12).
A Telomere-Devoted Reverse Transcriptase Supplements the Central Dogma
Terminal sequence loss is inherent to each round of genome replication by DNA-dependent DNA polymerases. Evolution has crafted a diversity of mechanisms to counteract this erosion (13). In early eukaryotes, end maintenance could have relied on telomere extension by an ancestral reverse transcriptase (RT) encoded within self-splicing introns (14, 15). Across extant eukaryotes, the nearly ubiquitous solution to the end-replication problem is a specialized RT, telomerase (16). Unlike retroviral RTs, telomerase functions as a ribonucleoprotein (RNP) complex, using a region within its integral RNA subunit as the template for telomeric repeat synthesis (17). Telomerase activity was first detected in T. thermophila cell extract (18) and then in other ciliates (19, 20). More than a decade later telomerase was detected in cells other than ciliates, first in the immortalized human cancer cell line HeLa (21) and eventually in a wide range of human cancers (22). This moved the study of telomerase from the realm of ciliates to general eukaryotic biology.
It was initially counterintuitive that human cell lines established from normal primary tissue lacked telomerase activity. From the relatively few surveys of normal human cells and tissues, telomerase activity appears widespread only in early human development (23). Consistent with this conclusion, human embryonic stem cells have robust telomerase activity (24). Some adult human somatic cells have detectable telomerase activity, but most of these nonetheless experience telomere shortening with proliferation (24, 25). Several human diseases arise from insufficient telomerase activity, including syndromes of bone marrow failure and pulmonary fibrosis (26–28). Also, aberrant reactivation of telomerase is a requirement of most human tumorigenesis (29). Together the consequences of insufficient or unrestrained telomerase function underscore the importance of ongoing studies to elucidate the developmental regulation of human telomerase activity using a combination of genome engineering and differentiation of human pluripotent stem cells (30).
Ciliate, yeast, and vertebrate telomerases are the focus of this review, owing to their relative depth of characterization. Telomerases in plants, trypanosomes, and additional fungal and metazoan species have recently been described elsewhere (31, 32).
THE TELOMERASE CATALYTIC CORE
An active telomerase minimal RNP can be assembled in a heterologous cell extract by expression or addition of two telomerase catalytic core subunits (33): telomerase RNA (TER) and telomerase reverse transcriptase (TERT). TER and its internal template were discovered by direct sequencing of an RNA coenriched with T. thermophila telomerase activity (17). TERT and its active site motifs were revealed by sequence comparison between ever-shortening telomeres protein 2 (Est2) from S. cerevisiae, identified as genetically essential for telomere length maintenance (34), and a telomerase protein from the particularly telomere-rich ciliate Euplotes aediculatus (35). In addition to the active site motifs shared across RTs, the TERT RT domain has two additional TERT-specific motifs: an Insertion in Fingers Domain (IFD) and motif 3 (Figure 2a) (36, 37). These and the T-motif in the TERT-specific high-affinity RNA–binding domain (TRBD) enable phylogenetically broad identification and comparison of TERTs (38).
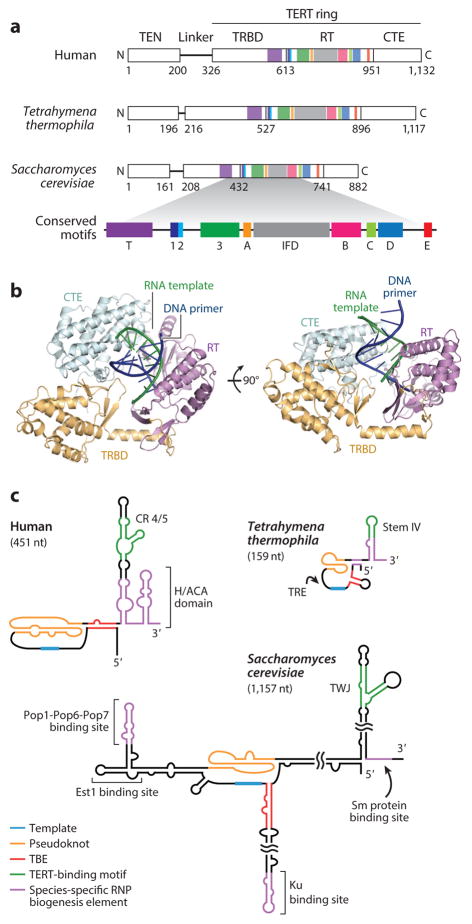
Figure 2.
The telomerase catalytic core. (a) Schematic of human, Tetrahymena thermophila, and Saccharomyces cerevisiae TERTs. TERT is composed of the TEN domain, linker, and TERT ring containing TRBD, RT domain, and CTE/thumb domains. Domain boundaries are labeled according to the TERT amino acid sequence. The TERT T-motif and motifs 1, 2, 3, A, IFD, B, C, D, and E are conserved across evolution (38). (b) The 2.7 Å –resolution structure of Tribolium castaneum TERT ring with DNA and RNA base paired as a primer–template duplex in the active site (40). The illustration was rendered from Protein Data Bank 3KYL with RNA in green and DNA in dark blue. Each TERT domain was given a different color. Tribolium TERT lacks a TEN domain. (c) TERs share conserved functional motifs including the template, pseudoknot, TBE, and template-distal TERT-binding motif corresponding to CR 4/5 in hTR, stem IV loop in T. thermophila TER, and the TWJ in S. cerevisiae TLC1. Species-specific TER binding sites for proteins involved in RNP biogenesis are indicated in purple. Abbreviations: CR, conserved region; CTE, C-terminal extension; Est1, ever-shorter telomeres protein 1; H/ACA, hairpin-H box-hairpin-ACA motif; hTR, human TER; IFD, insertion in fingers domain; Ku, dimeric protein complex that binds to DNA double-stranded break ends; nt, nucleotide; Pop, processing of precursor RNAs; RNP, ribonucleoprotein; RT, reverse transcriptase; Sm, proteins identified by Sm serotype antibodies from patients with autoimmune disease; TBE, template boundary element; TEN, TERT N-terminal; TER, telomerase RNA; TERT, telomerase reverse transcriptase; TLC1, S. cerevisiae TER identified as telomerase component 1; TRBD, TERT-specific high-affinity RNA-binding domain; TRE, template recognition element; TWJ, three-way junction.
TERT has four domains: the TERT N-terminal (TEN) domain, the TRBD, the RT domain, and the C-terminal extension (CTE) analogous to a polymerase thumb domain (Figure 2a). Intramolecular interaction of the TRBD and CTE constrains the contiguous TRBD–RT–CTE into a ring shape (Figure 2b), in contrast to the horseshoe shape of conventional RTs (39). The active site is central to the ring, within a cavity that holds an RNA–DNA duplex (40). A linker region of variable length and no obvious contribution to RNP assembly or catalytic activity connects the TEN domain to the TERT ring (38, 41, 42). TERTs from a few organisms lack a TEN domain altogether (38), S. cerevisiae telomerase can function without a TEN domain in vivo (43), and human telomerase without a TEN domain still catalyzes single-repeat synthesis in vitro (44).
In contrast to TERTs, TERs exhibit remarkable divergence in sequence and secondary structure (32, 45). Shared TER motifs are embedded within divergent motifs for telomerase holoenzyme protein interactions (Figure 2c). All TERs contain a template for repeat synthesis, albeit of different length and fidelity of copying. Other shared TER motifs include a pseudoknot (PK) with characteristic base triples and a stem-loop/stem-junction element distant from the template/PK that contributes to TERT binding (Figure 2c). The PK and template-distal stem-loop/stem-junction element are essential or strongly stimulatory for the activity of many telomerase enzymes, but studies of TERs in early branching eukaryotes suggest that the PK is not ancestral and that the distal TERT-binding element can be deleted in some cases without major loss of catalytic activity (32, 46). In S. cerevisiae TER, identified as telomerase component 1 (TLC1), the evolutionarily minimized stem-junction element is not essential for telomerase biochemical or biological function (47). Direct roles of PK and template-distal stem-loop/stem-junction elements remain to be established.
High-resolution structures of TERT domains and TER motifs (7, 48, 49), and recently of TERT domains bound to TER motifs (50, 51), complement the medium-resolution structures of entire telomerase holoenzyme complexes determined by electron microscopy (52). These structures and models from them serve as guides for ongoing pursuit of an atomic-resolution understanding of telomerase structure and mechanism. Progress has been hampered by the inability to combine purified TERT and TER to efficiently reconstitute active RNP. Furthermore, cellular overexpression generates surprisingly heterogeneous RNP complexes (42). Robust assembly and selective isolation of RNPs with functional folding of TERT and TER are future challenges.
THE TELOMERASE CATALYTIC CYCLE OF REPEAT SYNTHESIS
Catalytic Cycle Specialization
Like many polymerases, telomerase catalyzes nucleotide addition to a primer 3′ hydroxyl group, forming a product–template duplex. Accordingly, telomerase and other polymerases share a metal-dependent chemistry of nucleotide addition (53). Beyond these parallels, telomerase possesses unique properties of nucleic acid handling. Accurate telomeric repeat synthesis depends on strict boundaries of template copying within TER. Also, telomerases from most species studied have the exceptional ability to extend a primer by processive addition of repeats (54). Repeat addition processivity (RAP) obliges dissociation of the product–template duplex without product dissociation from the enzyme. The template-dissociated single-stranded DNA must maintain template-independent interactions while the template repositions for base pairing of its 3′ end, rather than the 5′ end, with the product. These coordinated nucleic acid handling events, discussed in more detail below, occur as part of the full catalytic cycle of repeat synthesis (Figure 3).
Figure 3.
The telomerase catalytic cycle. The telomerase catalytic cycle begins with base pairing of DNA primer (blue) to the template 3′ end, while the template 5′ end loops out (top left). After binding the duplex and dNTP, the active site closes to form the elongation-competent conformation (step ❶). Elongation proceeds until the template 5′ boundary, which is typically defined by a steric barrier, is reached (steps ❷ and ❸). As the template 5′ region is reeled into the active site, the template 3′ region and flanking RNA are displaced. If only 5–7 base pairs are stabilized by the telomerase active site, the initial duplex displaced by new repeat synthesis may fray in a manner that affects the conformation and positioning of the template 3′ end and flanking RNA (step ❹). This fraying could occur during or after repeat synthesis; it is shown here as occurring after repeat synthesis for illustration simplicity. We suggest that the displaced template 3′ end and flanking RNA favor a substantial active site opening necessary for strand separation (step ❺). DNA previously base paired to the template could retain and/or form additional protein contacts with an SRS of TERT, which holds the newly synthesized repeat of DNA while the template translocates (step ❻). After template translocation, default placement of the template 3′ end near the DNA 3′ end would promote the formation of a short duplex (step ❼). If this short duplex is captured into the TERT ring central cavity by the conformational changes necessary to restore a functional active site, another round of repeat synthesis begins. Alternatively, product release could occur prior to reestablishment of the active site. Abbreviations: dNTP, deoxynucleotide triphosphate; SRS, single-stranded DNA retention surface; TER, telomerase RNA; TERT, telomerase reverse transcriptase.
Defining an internal telomerase RNA region for primer base pairing and extension
For accurate repeat synthesis, the active site must copy only a restricted region of TER as template. For T. thermophila telomerase, a unique template position is the default starting point for elongation of nontelomeric primers with no template base pairing (55). The precision of default template positioning in ciliate telomerases can account for how the same permutation of telomeric repeat is added to each nontelomeric end of fragmented chromosomes (56). The ciliate TER template recognition element (TRE), located immediately 3′ of the template (Figure 2c), contributes to preferential positioning of the template 3′ end, rather than the template 5′ end, in the active site. Remarkably, RNA oligonucleotides containing only the wild-type TRE and template sequence are accurately positioned and copied by a recombinant T. thermophila telomerase RNP without a template (57). In the T. thermophila telomerase holoenzyme structure determined by cryoEM, the TRE threads between the CTE and the TEN domain positioned above the active site cavity of the TERT ring (58). Consistent with a TEN domain contribution to default template positioning, TRE mutations and the TEN domain mutation L14A inhibit copying of template 3′ positions but not template 5′ positions (41). Also, assays of T. thermophila TERT L14A recombinant enzyme by single-molecule fluorescence resonance energy transfer (FRET) suggest that the L14A substitution decreases the stability of DNA positioning at the template 3′ end but not at the template 5′ end (59).
The vertebrate telomerase active site relies on primer–template base pairing rather than template-flanking RNA motifs for template positioning. Indeed, human telomerase can elongate an annealed primer–template duplex containing only the remaining template region as single-stranded RNA (60). For telomerases in general, less base pairing is required to copy the template 3′ end than to copy the template 5′ end (61). This favors primer alignment in register with the beginning rather than the end of the template, which is a critical feature of internal template use. Stabilization of the template 3′-end duplex in the active site requires the TEN domain, accounting for the crucial contribution of the TEN domain to RAP (44, 62). Perhaps because S. cerevisiae telomerase has low RAP, the TEN domain is not genetically essential for its function (43).
Accurate template use also obliges DNA synthesis to halt at the template 5′ end (Figure 3, step ❸). The 5′ edge of the copied template is generally determined by spacing from an adjacent high-affinity RNA–protein and/or an RNA–RNA interaction (32). Curiously, in human telomerase, this typical template 5′ boundary determination mechanism makes less of a contribution than the sequence of the template–product duplex (63, 64). The structural elaboration(s) that give the vertebrate telomerase active site its sequence-specific duplex recognition remain to be defined.
Melting the base pairs between product and template
Telomerase dissociation from an elongated chromosome end and recycling of the internal template require unpairing the product–template duplex (Figure 3, step ❺). Models for the thermodynamic driver(s) of duplex melting include template stretching and compression against protein-anchored template-flanking motifs (65), duplex release from the active site (60), product DNA hairpin formation (66, 67), or product DNA interaction with protein (64, 68); below we extend this list with another model (see Models for Conformational Change Across the Catalytic Cycle). Establishing the change in state that is the rate-limiting barrier to product–template unpairing requires more knowledge of the telomerase catalytic cycle. At present, even the length of the product–template duplex when synthesis reaches the template 5′ boundary is unresolved. Chemical modification-protection assays of S. cerevisiae TLC1 using telomerase paused at different template positions revealed that some duplex unpaired as synthesis proceeded across the TLC1 template (69). At the two template positions tested, duplex length was 7 base pairs (69). Because the S. cerevisiae enzyme copied the TLC1 template with no RAP, the number of product–template base pairs dissociated during a catalytic cycle of processive repeat synthesis remains to be established.
Binding of template-dissociated single-stranded DNA and the realigned product–template duplex
RAP requires a mechanism of single-stranded DNA retention while base pairing of a product 3′ end shifts from the template 5′ end to the template 3′ end (Figure 3, steps ❺, ❻, ❼, and ❶). Early hypotheses envisioned an anchor site, independent from the active site, bound to single-stranded DNA (70, 71). In the T. thermophila telomerase holoenzyme, the telomeric repeat binding subunit 1 (Teb1) with high-affinity single-stranded DNA binding (see Telomerase Action at Telomeres) provides an envisioned type of anchor site in vitro, contributing to the exceptionally high in vitro RAP of this holoenzyme (72). However, because minimal telomerase RNPs have RAP, a crucial RAP-determining DNA interaction must occur only within a complex of TERT and TER. TERT interaction with single-stranded DNA immediately adjacent to the template duplex stimulates primer use, but primer single-stranded tails are not necessary for RAP of a minimal RNP (68, 73). Instead, the RAP-determining single-stranded DNA interaction appears to be made by DNA previously base paired with the template.
The first proposed single-stranded DNA-binding site on TERT was a groove on the T. thermophila TEN domain (74). Consistent with single-stranded DNA binding by the TEN domain, the TEN domain confers RAP to a TERT ring RNP (44, 62). However, recent biochemical and single-molecule FRET studies lend support to an alternate model of TEN domain function: active site capture of the realigned product–template duplex (59, 62). Structural studies of T. thermophila telomerase holoenzyme suggest that single-stranded DNA threads away from the putative DNA-binding site on the TEN domain (58). The DNA path could instead traverse an area assigned to the N- and C-terminal regions of the TEN domain that were not well ordered in the TEN domain crystal structure (74), consistent with active enzyme cross-linking to only the C terminus of the TEN domain (75). Because the cross-linked TEN domain residue does not contribute to enzyme activity (75), the RAP-determining DNA interaction is likely to occur elsewhere.
Models for Conformational Change Across the Catalytic Cycle
Studies of other polymerases revealed that binding of a nucleotide triphosphate suitable for incorporation triggers a subtle closure of the active site cavity to adopt the synthesis conformation (76). Telomerase could likewise undergo a conformational change upon binding nucleotide triphosphate that closes the active site to its synthesis conformation (Figure 3, step ❶). As synthesis proceeds from the template 3′ to 5′ end, each nucleotide addition would toggle the telomerase active site to and from the fully closed conformation. More challenging to envision are the conformational changes required for processive repeat synthesis. Template unpairing and repositioning for next-repeat synthesis require large-scale conformational changes relative to nucleotide addition (Figure 3, steps ❺ to ❼). Recently determined structures of the translesion DNA polymerase Pol ν revealed that the primer strand can be displaced from template pairing to form an extruded loop, which occupies a pocket created by thumb domain rotation relative to the rest of the polymerase (77). Modeling of a parallel conformational change in the Tribolium castaneum TERT ring suggests that the telomerase active site cavity could open by CTE rotation to accommodate a similar product DNA hairpin (67). DNA hairpin–induced slippage of the telomerase product 3′ end relative to the template C-tract would account for synthesis of homopolymeric G-ladders in ciliate telomerase activity assays with deoxyguanosine triphosphate as the only nucleotide substrate (78).
We propose a working model for the telomerase catalytic cycle hinging on several evolutionary innovations of nucleic acid recognition. Default template positioning places the template 3′ end near a bound primer 3′ end, with the unused template 5′ region “looped out” of the active site cavity, though not necessarily as a structured loop (Figure 3, 5′ template loop depicted in step ❼). Default positioning depends in part on template-flanking RNA structure and RNA–protein interaction; however, in addition, a template 5′ loop could be favored by other RNA–protein or RNA–RNA interactions. Duplex formation and stabilization in the active site initiate repeat synthesis (Figure 3, steps ❼ and ❶). As repeat synthesis progresses, the template 5′ loop reels in to the active site while the template 3′ end is extruded from the active site cavity (Figure 3, steps ❷ and ❸). A template 3′ loop formed by complete repeat synthesis (Figure 3, loop formation depicted in step ❹) could then make an RNA–protein or RNA–RNA interaction that favors rate-limiting conformational transition to a fully open active site (Figure 3, step ❺). Substantial protein remodeling may be required to dissociate the product–template duplex and promote template translocation to default positioning (Figure 3, steps ❺ and ❻). Active site opening to release the template 5′ end could also occur without participation of the displaced template 3′ region, perhaps with a difference in rate or progression to next-repeat synthesis.
In the current working model for telomerases with RAP (Figure 3), DNA unpaired from the template remains bound to TERT at a single-stranded DNA retention site (SRS). We envision the SRS to retain some contacts between DNA and TERT that were formed during repeat synthesis. Single-stranded DNA could thread passively across the TERT surface that mediates DNA protection from exonuclease digestion (62). Template translocation would bring the template 3′ end into closer proximity with the DNA 3′ end (Figure 3, step ❻), favoring base pairing (Figure 3, step ❼). Duplex formation could be a cause or consequence of reversing the structural change that released the template 5′ end from the active site (Figure 3, step ❺).
Overall, this working model differs from previous models in the prediction of RAP-influencing interactions made by single-stranded RNA that are dynamic with the catalytic cycle, supplementing the static RNA–RNA and RNA–protein contacts that govern template boundaries. Unlike previous models, because at least some SRS–DNA contacts are established prior to product dissociation from the template, there is also no requirement for DNA base recognition.
Repeat Addition Processivity in Telomere Maintenance
RAP assays in vitro do not recapitulate the complexity of telomeric repeat synthesis in cells. Thus, it remains uncertain how many repeats are synthesized with each telomerase engagement of a chromosome end in vivo. Does inherent RAP matter? Partial inhibition of human telomerase RAP by a chemical inhibitor (79, 80) or disease-associated TERT mutation (81, 82) leads to telomere shortening, suggesting that RAP contributes to telomere length maintenance. RAP in cells is challenging to characterize, even with simplifying assumptions such as no trimming of the G-strand extension. Infrequent RAP of S. cerevisiae telomerase was detected by sequencing the degenerate repeats added in one cell division cycle to shortened telomeres (83). Human telomerase RAP in cells is proposed to be relatively uniform, estimated as five to ten repeats per telomere elongation event based on density gradient centrifugation after telomere synthesis with labeled nucleotides (84). This length of repeat synthesis is likely to reflect competition between telomerase, other single-stranded telomeric repeat DNA-binding proteins, and replication protein A (RPA), the general single-stranded DNA-binding heterotrimer (see Telomerase Action at Telomeres). Direct assays of telomerase RAP in vivo would lead to better understanding of telomere length regulation.
CELLULAR RIBONUCLEOPROTEIN ASSEMBLY AND ACTION AT TELOMERES
In cells, telomerase assembly and function rely on a plethora of factors beyond TERT and TER, including telomerase holoenzyme proteins that stably associate with the active RNP (Figure 4) and chaperones for folding and trafficking that participate transiently. Despite broad conservation of activity-critical TERT domains and TER motifs that co-fold to form the enzyme catalytic core, entirely different pathways of telomerase biogenesis and regulation have evolved in ciliates, fungi, and vertebrates (32, 45). Even within fungi, a remarkable extent of divergence has occurred in the telomerase RNP assembly and telomere recruitment pathways (85, 86). In this section, we focus on telomerase cellular requirements and regulations that can be compared across species.
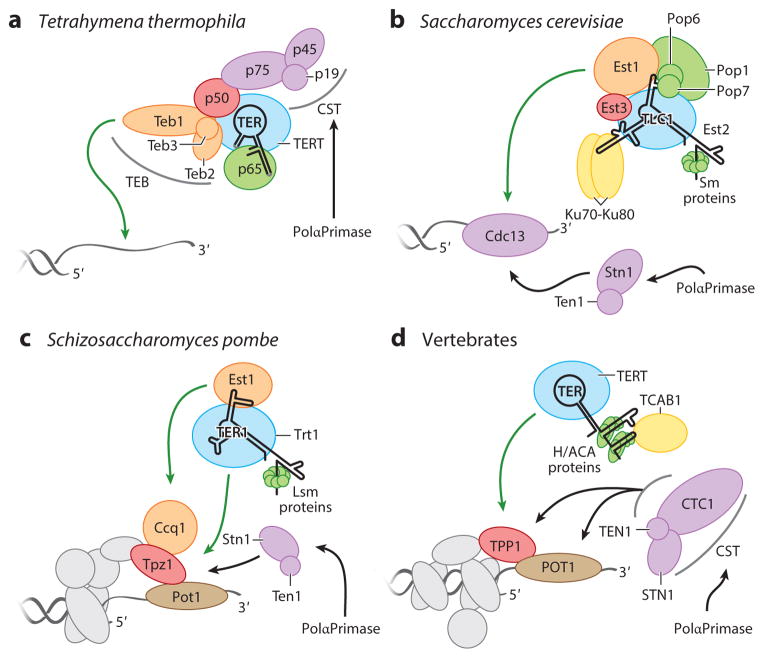
Figure 4.
Telomerase holoenzymes are illustrated for (a) Tetrahymena thermophila, (b) Saccharomyces cerevisiae, (c) Schizosaccharomyces pombe, and (d ) vertebrates. TERs are labeled TER, TLC1, or TER1; they are greatly simplified in secondary structure representation. TERT proteins are shaded blue and labeled TERT, Est2, or Trt1. Other colors group proteins with generally related biological functions. Green arrows indicate interactions important for G-strand synthesis, and black arrows indicate interactions thought to be important for C-strand synthesis. Abbreviations: Ccq1, coiled-coil protein quantitatively enriched 1; Cdc13, cell division cycle protein 13; CST, heterotrimer containing Stn1, Ten1, and variable third subunit; Est, ever-shorter telomeres; H/ACA, hairpin-H box-hairpin-ACA motif; Ku, dimeric protein complex that binds to DNA double-strand break ends; Lsm, like Sm; nt, nucleotide; PolαPrimase, polymerase α primase; p19-p45-p75, Tetrahymena telomerase CST with subunits of 19, 45, and 75 kDa; p65, Tetrahymena telomerase subunit of 65 kDa; Pop, processing of precursor RNAs; Sm proteins, proteins identified by Sm serotype antibodies from patients with autoimmune disease; Stn1, suppressor of cdc thirteen 1; TCAB1, telomerase and Cajal body protein 1; Teb1, telomeric repeat binding subunit 1; TEB, Teb1 heterotrimer complex; Ten1, telomeric pathways in association with Stn1, number 1; TER, telomerase RNA; TERT, telomerase reverse transcriptase; TLC1, S. cerevisiae TER identified as telomerase component 1; TPP1, vertebrate shelterin protein designation for proteins initially named TINT1/PTOP/PIP1; Tpz1, TPP1 homolog in S. pombe; Trt1, TERT of S. pombe.
Cellular Telomerase Ribonucleoproteins
TER folding and RNP assembly follow multistep pathways (45, 87). A biological complex of TER, RNA binding protein(s) essential for TER accumulation, and TERT can be described as a telomerase RNP catalytic core, to distinguish it from the recombinant minimal RNP of TERT and TER alone. The T. thermophila telomerase RNP catalytic core was the first to be defined and comprises TERT, TER, and the La domain and RRM domain RNA-binding protein p65 (Figure 4a) (88, 89). Roles of p65 include protection of the RNA polymerase III TER transcript ends from exonuclease access and bending of stem IV (Figure 2c) to promote TERT interaction with flanking TER motifs (90, 91).
Other TERs are transcripts of RNA polymerase II, typically bearing the 5′ trimethylguanosine (TMG) cap that stabilizes many small nuclear RNAs. The TER precursor 3′ end is determined in S. cerevisiae by the Nrd1/Nab3/Sen1 transcription termination pathway (92), in most other fungi by intentionally aborted splicing (85, 86, 93), and in human cells by transcription-coupled cleavage and polyadenylation machinery (94). Proteins protect a mature TER 3′ end by blocking exonuclease access. In most fungi, Sm proteins assemble on the TER 3′ end (Figure 4b) (32). In S. pombe, the like Sm (Lsm) proteins replace the Sm complex after initial RNP biogenesis (Figure 4c) (95). Also, some fungi have a TER 3′-end determination mechanism apparently independent of Sm or Lsm proteins that remains to be elucidated (32). In S. cerevisiae, in addition to Sm proteins, active RNP assembly relies on the processing of precursor RNAs (Pop) proteins Pop1, Pop6, and Pop7 and their RNA binding sites (Figure 4b), shared with ribonuclease P and the related ribonuclease for mitochondrial RNA processing (RNase MRP) (96).
In vertebrate cells, TER 3′-end formation is determined by assembly of a mature H/ACA RNP containing two subunits each of dyskerin, nonhistone chromatin protein 2 (NHP2), NOP10, and glycine/arginine-rich domain protein 1 (GAR1) on a two-hairpin H/ACA motif (Figure 4d ) (97, 98). Disease-associated mutations in the four human H/ACA proteins that cotranscriptionally assemble with nascent H/ACA RNA transcripts [dyskerin, NHP2, NOP10, and nuclear assembly factor 1 (NAF1)] (99) reduce levels of human TER (hTR) much more than the levels of other H/ACA small nucleolar and small Cajal body RNAs (28, 100). The selective impact of these mutations is proposed to arise from sensitization of the hTR precursor transcript to rapid degradation, balanced against H/ACA RNP assembly and productive hTR 3′-end processing by the polyadenosine-specific ribonuclease (PARN) (94, 101, 102). Accordingly, human PARN mutations impose premature tissue renewal failures that result from telomerase insufficiency (103, 104).
Telomerase Trafficking
Proteins that traffic telomerase subunits and RNPs within a cell play important roles in active RNP biogenesis and action at telomeres. Stepwise trafficking of S. cerevisiae TLC1 (Figure 5a) and hTR (Figure 5b) are the most thoroughly investigated (45, 87, 105, 106), although even in these pathways many uncertainties remain. In S. cerevisiae (87, 105), TLC1 assembles Sm proteins coincident with processing to the mature RNA 3′ end and with the 5′ TMG cap addition by trimethylguanosine synthase 1 (Tgs1) in nucleoli. The TMG-capped Sm RNP is exported to the cytoplasm by chromosome region maintenance 1 protein (Crm1), where it assembles with Est2 and other holoenzyme proteins, including Est1. Recent studies revealed that direct TLC1 interaction by Est1 and Est2 depends on assembly of Pop1, Pop6, and Pop7 with TLC1 adjacent to the Est1 binding site (96), implying that Pop1, Pop6, and Pop7 bind TLC1 either prior to nuclear export or in the cytoplasm. Est3 may be the final holoenzyme subunit to join the complex, regulated by progression of the cell cycle (107). Nuclear import of telomerase holoenzyme is mediated by mRNA transport defective protein 10 (Mtr10) and karyopherin protein 122 (Kap122), enhanced by Ku–TLC1 interaction (87). Once in the nucleoplasm, telomerase can be recruited to telomeres (105, 108). Quantitative imaging of TLC1 in cells suggests that each TLC1–monomer RNP diffuses independently through the nucleoplasm until encountering a replicated telomere (109, 110). S. cerevisiae telomerase complexes appear to cluster at telomeres (109), but how this finding relates to requirements for telomere elongation is not known.
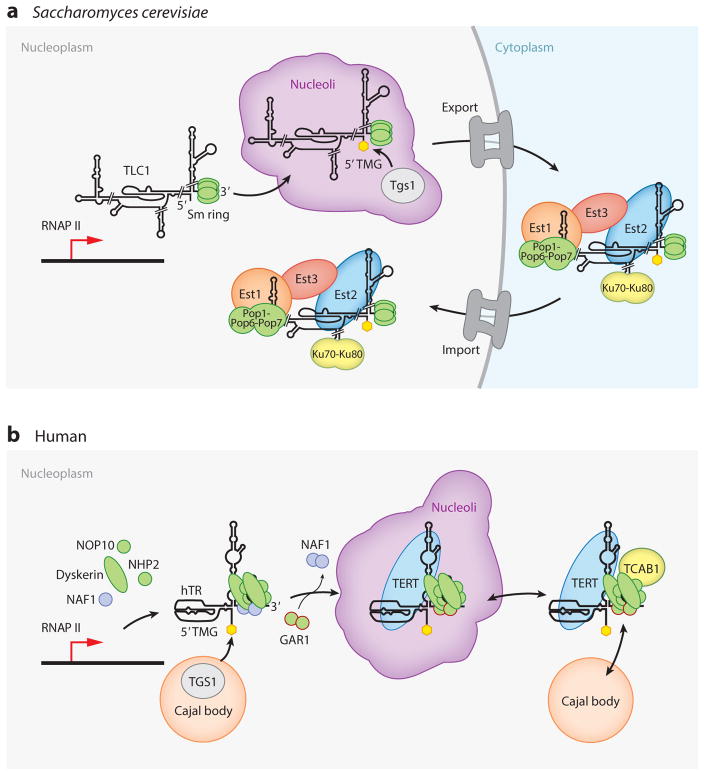
Figure 5.
Pathways of telomerase biogenesis. Cellular trafficking for biogenesis of (a) Saccharomyces cerevisiae and (b) human telomerase holoenzymes is illustrated using a light gray background for nuclear localization and a light blue background for cytoplasmic localization. TERT proteins are labeled TERT or Est2 and are shaded blue. Other colors group proteins with generally related biological functions. The 5′ TMG cap added to TLC1 and hTR is shown as a yellow hexagon. Individual steps of trafficking are described in the text. Biogenesis pathways are illustrated to finish with the nucleoplasmic localization of telomerase holoenzyme that precedes its binding to telomeres. Abbreviations: Est, ever-shorter telomeres; GAR1, glycine/arginine-rich domain protein 1; hTR, human telomerase RNA; Ku, dimeric protein complex that binds to DNA double-stranded break ends; NAF1, nuclear assembly factor 1; NHP2, nonhistone chromatin protein 2; Pop, processing of precursor RNAs; RNAP II, RNA polymerase II; Sm ring, heteroheptamer of Sm proteins; TCAB1, telomerase and Cajal body protein 1; TERT, telomerase reverse transcriptase; Tgs1, trimethylguanosine synthase 1; TLC1, S. cerevisiae TER identified as telomerase component 1; TMG, trimethylguanosine.
Human telomerase RNP biogenesis involves trafficking between Cajal bodies, nucleoli, and nucleoplasm (Figure 5b). The nascent hTR transcript binds two sets of core H/ACA proteins (dyskerin, NOP10, and NHP2), scaffolded together by NAF1 (101). Construction of the NAF1-orchestrated RNA-binding platform requires numerous protein chaperones, including some with the exclusive function of trafficking H/ACA proteins (99). The hTR H/ACA RNP released from the site of transcription is thought to rapidly transit Cajal bodies to acquire a 5′ TMG cap (111). Also, the RNA helicase DEAH box protein 36 (DHX36) resolves guanosine quadruplex structures formed in the hTR 5′ leader (112). At some point, NAF1 is exchanged for the mature H/ACA RNP subunit GAR1. A fraction of hTR binds TERT to form catalytically active RNP, and the hTR 3′ stem loop CAB-box motif (Figure 2c) binds Telomerase and Cajal body protein 1 (TCAB1). Because active RNPs appear to assemble irreversibly (113) and have a long cellular half-life (114), only ~100 active RNP assembly events may occur in each cell cycle. The location within the nucleus where TERT assembles with hTR is unknown. Active RNP assembly in nucleoli is supported by studies of hTR without H/ACA proteins: Assembly of this nucleoplasmic hTR into active RNP requires TERT overexpression, whereas assembly of hTR localized to the nucleolus by loss of TCAB1 does not (115). Alternately, active RNP assembly could occur in Cajal bodies or the nucleoplasm.
TCAB1 binds active and inactive telomerase RNPs and shifts their distribution from nucleoli to the nucleoplasm and Cajal bodies (Figure 5b) (116, 117). TCAB1 gene disruption decreases the length at which telomeres are maintained in human embryonic stem cells (115). Decreased telomere length in stem cells, and thus reduced proliferative capacity of cell lineages differentiated from them, would explain why mutations in TCAB1 cause diseases of premature telomere shortening (28, 118). Because genetic disruption of Coilin protein and thus Cajal bodies does not affect active RNP assembly or telomere length in human cancer cells or embryonic stem cells (115, 119), telomere access by telomerase can occur from the nucleoplasm. This conclusion is consistent with the biological function of active RNPs containing hTR without an H/ACA motif, which is exclusively nucleoplasmic (115). Live cell imaging of tagged TERT suggests that TERT encounters telomeres in the nucleoplasm (120). Trafficking of the small fraction of cellular TERT that is assembled into active RNP remains to be determined, for example, using the approach developed to track TLC1 in S. cerevisiae (109).
Telomerase Action at Telomeres
The low endogenous level of telomerase presents a technical barrier to determining requirements for its action at telomeres. Chromatin immunopurification assays of S. cerevisiae and S. pombe telomerase subunits, telomere proteins, and DNA replication factors across a multitude of genetic backgrounds have given the most detailed understanding of requirements for telomerase recruitment and activation at telomeres (12, 121). In human cells, most assays of telomerase recruitment to telomeres have relied on enzyme overexpression, which can undermine the cell cycle restriction of telomerase–telomere interaction and telomerase regulation by telomere length feedback (122).
Telomerase action at telomeres requires one protein domain that is structurally conserved across evolution: the oligosaccharide/oligonucleotide-binding (OB) fold of T. thermophila p50, S. cerevisiae Est3, S. pombe Tpz1, and vertebrate TPP1 (Figure 4a–d). The telomerase holoenzyme subunits p50 and Est3 give the RNP catalytic core a conformation active for telomere synthesis (123–125). The telomere proteins TPP1 and Tpz1 are major contributors to telomerase recruitment as well as subsequent telomerase activation for telomere elongation (122, 126). Each of the four proteins has an OB fold, TEL-patch surface that biochemical, genetic, and/or structural studies indicate to be an interaction site for the TERT TEN domain (58, 122, 125, 126). Structural or functional evidence suggests a second interaction of the p50 and TPP1 OB fold domains with the TERT IFD (58, 127), which future studies will test for conservation in other species.
Other than a poorly defined activating role of the p50/Est3/Tpz1/TPP1 OB fold, no obvious commonality exists among mechanisms that direct telomerase action to its cellular substrates in different organisms. Two general strategies for telomere-specific recruitment of telomerase can be considered. First is recruitment to single-stranded telomeric repeat DNA, exemplified by T. thermophila and S. cerevisiae telomerases (Figure 4a,b). The second strategy is recruitment to telomere-bound shelterin complexes nucleated on double-stranded telomeric repeats (Figure 4c,d ). With more depth of study, a regulatory role for kinases such as ataxia-telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3 related (ATR) could emerge as a shared principle of telomerase–telomere interaction (128, 129).
T. thermophila telomerase recruitment to telomeres is mediated by the holoenzyme protein Teb1 (Figure 4a), which has high affinity and specificity for binding to telomeric-repeat single-stranded DNA (89, 130). Teb1 is paralogous to the RPA large subunit, Rpa1 (89). Like Rpa1, tandem OB fold domains of Teb1 bind an ~15-nt length of single-stranded DNA (131). Also like Rpa1, Teb1 assembles with the RPA middle subunit (Teb2/Rpa2) and RPA small subunit (Teb3/Rpa3) to form a TEB heterotrimer (58, 132). However, unlike the case for RPA, in the TEB complex only the Teb1 subunit contributes to DNA binding (132).
S. cerevisiae telomerase holoenzyme recruitment to telomeres occurs via Cdc13, another high-affinity, telomere-specific, single-stranded DNA-binding protein (Figure 4b) (7, 11). Cdc13 alternatively interacts with Stn1–Ten1 to form the telomere-capping CST complex (133) or recruits telomerase to chromosome ends through the holoenzyme subunit Est1 (12, 134). The Ku heterodimer has been proposed to play a role in telomerase recruitment to telomeres, which may result from Ku enhancement of holoenzyme nuclear import and/or retention (135).
In S. pombe, TER1-bound Est1 binds shelterin-assembled coiled-coil protein quantitatively enriched 1 (Ccq1) and TERT binds Tpz1 (Figure 4c), but this initial interaction network rearranges to dissociate Est1–Ccq1 from the activated, elongation-competent Tpz1-associated telomerase holoenzyme (121, 126). In vertebrate cells (Figure 4d ), only TPP1–TERT interaction is known to be essential (122, 135, 136). Because a streamlined active human telomerase without H/ACA proteins can maintain stable telomere length homeostasis (115), none of the known human telomerase holoenzyme proteins other than TERT are necessary for telomerase action at telomeres.
Coordination of G-Strand and C-Strand Synthesis
Telomere maintenance involves not only G-strand synthesis but also complementary C-strand synthesis and 3′ overhang processing. Synthesis of the final C-strand Okazaki fragment has unique requirements, including mechanisms for recruiting DNA polymerase α primase (PolαPrimase) after replication fork disassembly and for removing the terminal RNA primer without 5′-flap displacement by an upstream Okazaki fragment. Although the mechanisms that ensure complete C-strand synthesis remain mostly a mystery, one conserved player in the process has been identified: an RPA-like single-stranded DNA-binding heterotrimer designated CST. The CST subunits Stn1 (suppressor of Cdc13 protein 1) and Ten1 (telomeric pathways in association with Stn1 protein 1) are paralogous to RPA middle and small subunits, respectively, and are largely conserved in domain structure across evolution. In contrast, the CST subunits T. thermophila p75, S. cerevisiae Cdc13, and vertebrate CTC1 paralogous to Rpa1 are evolutionarily divergent (10, 11). In S. pombe, no ortholog of the CST large subunit has been identified. CST complexes share single-stranded DNA-binding activity that is either selective or preferential for G-strand telomeric DNA, yet human CST can bind stably to a length of nontelomeric DNA about twice that necessary for G-strand binding (10, 137, 138). CST physically interacts with PolαPrimase and can functionally stimulate one or more substeps of its composite RNA–DNA primer synthesis (10, 11, 139).
The most direct coupling of G-strand and C-strand synthesis machineries occurs in ciliates, as first demonstrated by their co-purification from extracts of Euplotes crassus (140). Structural studies of T. thermophila telomerase revealed that CST is brought to telomeres as a part of the telomerase holoenzyme (Figure 4a) (58, 138). The p75–p45–p19 CST heterotrimer rotates relative to the RNP catalytic core as a subcomplex hinged on p50 (123). Overexpression of p19 defective for holoenzyme interaction uncouples G-strand and C-strand synthesis, as detected by an elongated G-strand overhang (138).
In S. cerevisiae, the Cdc13 subunit of CST binds with high affinity to telomeric repeats and alternatively assembles with telomerase or Stn1–Ten1 (Figure 4b), controlled by posttranslational modifications of Cdc13 (11). In S. pombe, Tpz1 posttranslational modification is involved in Stn1–Ten1 recruitment to telomeres (Figure 4c) (141, 142). Human or mouse CST association with telomeres involves TPP1 and/or POT1 (Figure 4d ) (137, 143). Because completion of C-strand synthesis is delayed for several hours after S phase in human cells (144), telomerase and PolαPrimase interactions with CST would be temporally uncoupled. Nonetheless, human CST restricts the amount of telomere synthesis by telomerase (137). Handoffs of G-strand binding between the telomerase active site, CST, and PolαPrimase may be more direct in T. thermophila (58, 138), providing a future opportunity to investigate their biochemical coordination.
Additional Considerations for Telomerase Function and Regulation
Much remains to be understood about how higher-order telomere structural organization influences telomerase recruitment and end engagement. For example, it is unknown whether only terminus-proximal TPP1 or Tpz1 recruits telomerase or whether telomerase interactions occur along the entire shelterin array. Furthermore, after telomere recruitment, the active site of telomerase must capture the extreme 3′ chromosome terminus. This process could be accelerated by RPA-like sliding of telomerase-adjacent POT1–TPP1 (145), or T. thermophila TEB, along single-stranded repeats. However, POT1–DNA interaction appears unnecessary for robust telomere elongation (146). Also, although telomerase activity is stimulated in vitro by addition of excess TPP1–POT1 (147) or by holoenzyme assembly of Teb1 (89), the biological significance of these in vitro activity assay observations remains to be investigated in cells.
Endogenous telomerase action at telomeres is restricted to the cell cycle S phase (108, 122, 148). This control of telomerase action does not derive from regulated assembly of the RNP catalytic core, because telomerase activity is present in cell extracts at all stages of the cell cycle in all examined model systems. Holoenzyme assembly is regulated to some extent: S. cerevisiae assembles, disassembles, and degrades holoenzyme subunits with some cell cycle regulation (107, 149), and human telomerase dissociates TCAB1 in the cell cycle M phase (113). However, subunit overexpression studies indicate that these regulations do not account for S phase restriction of telomerase action. Additional layers of regulation derive from telomere chromatin dynamics, changes in DNA structure, and posttranslational modifications (4, 9, 122, 128, 135, 150). The process of replication, or the replication fork itself, could recruit telomerase (148, 151). In the next decade, discovering the mechanisms for telomerase collaboration and competition with other telomere synthesis, processing, and end protection machineries will be a key direction of research.
Acknowledgments
The Collins laboratory acknowledges National Institutes of Health grants GM054198 and HL079585 for funding. We thank Collins laboratory members and Dirk Hockemeyer for discussions and comments on the manuscript.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Müller HJ. The remaking of chromosomes. Collect Net. 1938;13:181–98. [Google Scholar]
- 2.McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–82. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- 4.Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–38. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 5.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–14. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 6.de Lange T. How shelterin solves the telomere end-protection problem. Cold Spring Harb Symp Quant Biol. 2010;75:167–77. doi: 10.1101/sqb.2010.75.017. [DOI] [PubMed] [Google Scholar]
- 7.Lewis KA, Wuttke DS. Telomerase and telomere-associated proteins: structural insights into mechanism and evolution. Structure. 2012;20:28–39. doi: 10.1016/j.str.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumann P, Price C. Pot1 and telomere maintenance. FEBS Lett. 2010;584:3779–84. doi: 10.1016/j.febslet.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmutz I, de Lange T. Shelterin. Curr Biol. 2016;26:R397–99. doi: 10.1016/j.cub.2016.01.056. [DOI] [PubMed] [Google Scholar]
- 10.Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE. Evolution of CST function in telomere maintenance. Cell Cycle. 2010;9:3157–65. doi: 10.4161/cc.9.16.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lloyd NR, Dickey TH, Hom RA, Wuttke DS. Tying up the ends: plasticity in the recognition of single-stranded DNA at telomeres. Biochemistry. 2016;55:5326–40. doi: 10.1021/acs.biochem.6b00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wellinger RJ, Zakian VA. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics. 2012;191:1073–105. doi: 10.1534/genetics.111.137851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biessmann H, Mason JM. Telomere maintenance without telomerase. Chromosoma. 1997;106:63–69. doi: 10.1007/s004120050225. [DOI] [PubMed] [Google Scholar]
- 14.de Lange T. A loopy view of telomere evolution. Front Genet. 2015;6:321. doi: 10.3389/fgene.2015.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambowitz AM, Belfort M. Mobile bacterial group II introns at the crux of eukaryotic evolution. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0050-2014. MDNA3–0050–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–38. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 17.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–37. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 18.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–13. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 19.Zahler AM, Prescott DM. Telomere terminal transferase activity in the hypotrichous ciliate Oxytricha nova and a model for replication of the ends of linear DNA molecules. Nucleic Acids Res. 1988;16:6953–72. doi: 10.1093/nar/16.14.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shippen-Lentz D, Blackburn EH. Telomere terminal transferase activity from Euplotes crassus adds large numbers of TTTTGGGG repeats onto telomeric primers. Mol Cell Biol. 1989;9:2761–64. doi: 10.1128/mcb.9.6.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–29. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 22.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–15. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 23.Aubert G. Telomere dynamics and aging. Prog Mol Biol Transl Sci. 2014;125:89–111. doi: 10.1016/B978-0-12-397898-1.00004-9. [DOI] [PubMed] [Google Scholar]
- 24.Hiyama E, Hiyama K. Telomere and telomerase in stem cells. Br J Cancer. 2007;96:1020–24. doi: 10.1038/sj.bjc.6603671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins K, Mitchell JR. Telomerase in the human organism. Oncogene. 2002;21:564–79. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- 26.Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012;13:693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holohan B, Wright WE, Shay JW. Telomeropathies: an emerging spectrum disorder. J Cell Biol. 2014;205:289–99. doi: 10.1083/jcb.201401012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanley SE, Armanios M. The short and long telomere syndromes: paired paradigms for molecular medicine. Curr Opin Genet Dev. 2015;33:1–9. doi: 10.1016/j.gde.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shay JW. Role of telomeres and telomerase in aging and cancer. Cancer Discov. 2016;6:584–93. doi: 10.1158/2159-8290.CD-16-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hockemeyer D, Jaenisch R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell. 2016;18:573–86. doi: 10.1016/j.stem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson AD, Shippen DE. Evolution of TERT-interacting lncRNAs: expanding the regulatory landscape of telomerase. Front Genet. 2015;6:277. doi: 10.3389/fgene.2015.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Podlevsky JD, Chen JJ. Evolutionary perspectives of telomerase RNA structure and function. RNA Biol. 2016;13:720–32. doi: 10.1080/15476286.2016.1205768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 34.Lendvay TS, Morris DK, Sah J, Balasubramamian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–67. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 36.Lue NF, Lin YC, Mian IS. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol Cell Biol. 2003;23:8440–49. doi: 10.1128/MCB.23.23.8440-8449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie M, Podlevsky JD, Qi X, Bley CJ, Chen JJ. A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res. 2010;38:1982–96. doi: 10.1093/nar/gkp1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ. The telomerase database. Nucleic Acids Res. 2008;36:D339–43. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–37. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–18. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 41.Eckert B, Collins K. Roles of telomerase reverse transcriptase N-terminal domain in assembly and activity of Tetrahymena telomerase holoenzyme. J Biol Chem. 2012;287:12805–14. doi: 10.1074/jbc.M112.339853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu RA, Dagdas YS, Yilmaz ST, Yildiz A, Collins K. Single-molecule imaging of telomerase reverse transcriptase in human telomerase holoenzyme and minimal RNP complexes. eLife. 2015;4:e08363. doi: 10.7554/eLife.08363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman KL, Cech TR. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 1999;13:2863–74. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robart AR, Collins K. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol Cell. 2011;42:308–18. doi: 10.1016/j.molcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. RNA. 2012;18:1747–59. doi: 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Podlevsky JD, Li Y, Chen JJ. The functional requirement of two structural domains within telomerase RNA emerged early in eukaryotes. Nucleic Acids Res. 2016;44(20):9891–901. doi: 10.1093/nar/gkw605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zappulla DC, Goodrich K, Cech TR. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol. 2005;12:1072–77. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]
- 48.Mason M, Schuller A, Skordalakes E. Telomerase structure function. Curr Opin Struct Biol. 2011;21:92–100. doi: 10.1016/j.sbi.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Q, Kim NK, Feigon J. Architecture of human telomerase RNA. PNAS. 2011;108:20325–32. doi: 10.1073/pnas.1100279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang J, Brown AF, Wu J, Xue J, Bley CJ, et al. Structural basis for protein-RNA recognition in telomerase. Nat Struct Mol Biol. 2014;21:507–12. doi: 10.1038/nsmb.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jansson LI, Akiyama BM, Ooms A, Lu C, Rubin SM, Stone MD. Structural basis of template-boundary definition in Tetrahymena telomerase. Nat Struct Mol Biol. 2015;22:883–88. doi: 10.1038/nsmb.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feigon J, Chan H, Jiang J. Integrative structural biology of Tetrahymena telomerase: insights into catalytic mechanism and interaction at telomeres. FEBS J. 2016;283:2044–50. doi: 10.1111/febs.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–98. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 54.Collins K. Single-stranded DNA repeat synthesis by telomerase. Curr Opin Chem Biol. 2011;15:643–48. doi: 10.1016/j.cbpa.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, Blackburn EH. De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J. 1997;16:866–79. doi: 10.1093/emboj/16.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melek M, Shippen DE. Chromosome healing: spontaneous and programmed de novo telomere formation by telomerase. BioEssays. 1996;18:301–8. doi: 10.1002/bies.950180408. [DOI] [PubMed] [Google Scholar]
- 57.Miller MC, Collins K. Telomerase recognizes its template by using an adjacent RNA motif. PNAS. 2002;99:6585–90. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang J, Chan H, Cash DD, Miracco EJ, Ogorzalek Loo RR, et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science. 2015;350:aab4070. doi: 10.1126/science.aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akiyama BM, Parks JW, Stone MD. The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA-DNA hybrids. Nucleic Acids Res. 2015;43:5537–49. doi: 10.1093/nar/gkv406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi X, Xie M, Brown AF, Bley CJ, Podlevsky JD, Chen JJ. RNA/DNA hybrid binding affinity determines telomerase template-translocation efficiency. EMBO J. 2012;31:150–61. doi: 10.1038/emboj.2011.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H, Gilley D, Blackburn EH. A novel specificity for the primer-template pairing requirement in Tetrahymena telomerase. EMBO J. 1998;17:1152–60. doi: 10.1093/emboj/17.4.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu RA, Collins K. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J. 2014;33:921–35. doi: 10.1002/embj.201387205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown AF, Podlevsky JD, Qi X, Chen Y, Xie M, Chen JJ. A self-regulating template in human telomerase. PNAS. 2014;111:11311–16. doi: 10.1073/pnas.1402531111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu RA, Collins K. Sequence specificity of human telomerase. PNAS. 2014;111:11234–35. doi: 10.1073/pnas.1411276111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berman AJ, Akiyama BM, Stone MD, Cech TR. The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat Struct Mol Biol. 2011;18:1371–75. doi: 10.1038/nsmb.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shippen-Lentz D, Blackburn EH. Functional evidence for an RNA template in telomerase. Science. 1990;247:546–52. doi: 10.1126/science.1689074. [DOI] [PubMed] [Google Scholar]
- 67.Yang W, Lee YS. A DNA-hairpin model for repeat-addition processivity in telomere synthesis. Nat Struct Mol Biol. 2015;22:844–47. doi: 10.1038/nsmb.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hardy CD, Schultz CS, Collins K. Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J Biol Chem. 2001;276:4863–71. doi: 10.1074/jbc.M005158200. [DOI] [PubMed] [Google Scholar]
- 69.Förstemann K, Lingner J. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMBO Rep. 2005;6:361–66. doi: 10.1038/sj.embor.7400374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morin GB. Recognition of a chromosome truncation site associated with α-thalassaemia by human telomerase. Nature. 1991;353:454–56. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 71.Collins K, Greider CW. Nucleolytic cleavage and non-processive elongation catalyzed by Tetrahymena telomerase. Genes Dev. 1993;7:1364–76. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- 72.Greider CW. Telomerase is processive. Mol Cell Biol. 1991;11:4572–80. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baran N, Haviv Y, Paul B, Manor H. Studies on the minimal lengths required for DNA primers to be extended by the Tetrahymena telomerase: implications for primer positioning by the enzyme. Nucleic Acids Res. 2002;30:5570–78. doi: 10.1093/nar/gkf676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–25. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 75.Romi E, Baran N, Gantman M, Shmoish M, Min B, et al. High-resolution physical and functional mapping of the template adjacent DNA binding site in catalytically active telomerase. PNAS. 2007;104:8791–96. doi: 10.1073/pnas.0703157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doublie S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Structure. 1999;7:R31–35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 77.Lee YS, Gao Y, Yang W. How a homolog of high-fidelity replicases conducts mutagenic DNA synthesis. Nat Struct Mol Biol. 2015;22:298–303. doi: 10.1038/nsmb.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Collins K. Ciliate telomerase biochemistry. Annu Rev Biochem. 1999;68:187–218. doi: 10.1146/annurev.biochem.68.1.187. [DOI] [PubMed] [Google Scholar]
- 79.Damm K, Hemmann U, Garin-Chesa P, Hauel N, Kauffmann I, et al. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 2001;20:6958–68. doi: 10.1093/emboj/20.24.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pascolo E, Wenz C, Lingner J, Hauel N, Priepke H, et al. Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. J Biol Chem. 2002;277:15566–72. doi: 10.1074/jbc.M201266200. [DOI] [PubMed] [Google Scholar]
- 81.Robart AR, Collins K. Investigation of human telomerase holoenzyme assembly, activity, and processivity using disease-linked subunit variants. J Biol Chem. 2010;285:4375–86. doi: 10.1074/jbc.M109.088575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zaug AJ, Crary SM, Jesse Fioravanti M, Campbell K, Cech TR. Many disease-associated variants of hTERT retain high telomerase enzymatic activity. Nucleic Acids Res. 2013;41:8969–78. doi: 10.1093/nar/gkt653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang M, Arneric M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–94. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao Y, Abreu E, Kim J, Stadler G, Eskiocak U, et al. Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol Cell. 2011;42:297–307. doi: 10.1016/j.molcel.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kannan R, Helston RM, Dannebaum RO, Baumann P. Diverse mechanisms for spliceosome-mediated 3′ end processing of telomerase RNA. Nat Commun. 2015;6:6104. doi: 10.1038/ncomms7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qi X, Rand DP, Podlevsky JD, Li Y, Mosig A, et al. Prevalent and distinct spliceosomal 3′-end processing mechanisms for fungal telomerase RNA. Nat Commun. 2015;6:6105. doi: 10.1038/ncomms7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gallardo F, Chartrand P. Telomerase biogenesis: the long road before getting to the end. RNA Biol. 2008;5:212–15. doi: 10.4161/rna.7115. [DOI] [PubMed] [Google Scholar]
- 88.Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 2004;18:1107–18. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Min B, Collins K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell. 2009;36:609–19. doi: 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stone MS, Mihalusova M, O’Connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–61. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singh M, Wang Z, Koo BK, Patel A, Cascio D, et al. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol Cell. 2012;47:16–26. doi: 10.1016/j.molcel.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Noël JF, Larose S, Abou Elela S, Wellinger RJ. Budding yeast telomerase RNA transcription termination is dictated by the Nrd1/Nab3 non-coding RNA termination pathway. Nucleic Acids Res. 2012;40:5625–36. doi: 10.1093/nar/gks200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Box JA, Bunch JT, Tang W, Baumann P. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature. 2008;456:910–14. doi: 10.1038/nature07584. [DOI] [PubMed] [Google Scholar]
- 94.Nguyen D, Grenier St-Sauveur V, Bergeron D, Dupuis-Sandoval F, Scott MS, Bachand F. A polyadenylation-dependent 3′ end maturation pathway is required for the synthesis of the human telomerase RNA. Cell Rep. 2015;13:2244–57. doi: 10.1016/j.celrep.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 95.Tang W, Kannan R, Blanchette M, Baumann P. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature. 2012;484:260–64. doi: 10.1038/nature10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lemieux B, Laterreur N, Perederina A, Noel JF, Dubois ML, et al. Active yeast telomerase shares subunits with ribonucleoproteins RNase P and RNase MRP. Cell. 2016;165:1171–81. doi: 10.1016/j.cell.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999;19:567–76. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase in vivo and in vitro. Mol Cell. 2000;6:361–71. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 99.Massenet S, Bertrand E, Verheggen C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2016;2016:1–13. doi: 10.1080/15476286.2016.1243646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stanley SE, Gable DL, Wagner CL, Carlile TM, Hanumanthu VS, et al. Loss-of-function mutations in the RNA biogenesis factor NAF1 predispose to pulmonary fibrosis–emphysema. Sci Transl Med. 2016;8:351ra107. doi: 10.1126/scitranslmed.aaf7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Egan ED, Collins K. An enhanced H/ACA RNP assembly mechanism for human telomerase RNA. Mol Cell Biol. 2012;32:2428–39. doi: 10.1128/MCB.00286-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tseng CK, Wang HF, Burns AM, Schroeder MR, Gaspari M, Baumann P. Human telomerase RNA processing and quality control. Cell Rep. 2015;13:2232–43. doi: 10.1016/j.celrep.2015.10.075. [DOI] [PubMed] [Google Scholar]
- 103.Stuart BD, Choi J, Zaidi S, Xing C, Holohan B, et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat Genet. 2015;47:512–17. doi: 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tummala H, Walne A, Collopy L, Cardoso S, de la Fuente J, et al. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J Clin Invest. 2015;125:2151–60. doi: 10.1172/JCI78963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Londono-Vallejo JA, Wellinger RJ. Telomeres and telomerase dance to the rhythm of the cell cycle. Trends Biochem Sci. 2012;37:391–99. doi: 10.1016/j.tibs.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 106.Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015;29:1095–105. doi: 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tucey TM, Lundblad V. Regulated assembly and disassembly of the yeast telomerase quaternary complex. Genes Dev. 2014;28:2077–89. doi: 10.1101/gad.246256.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gallardo F, Laterreur N, Wellinger RJ, Chartrand P. Telomerase caught in the act: united we stand, divided we fall. RNA Biol. 2012;9:1139–43. doi: 10.4161/rna.21498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gallardo F, Laterreur N, Cusanelli E, Ouenzar F, Querido E, et al. Live cell imaging of telomerase RNA dynamics reveals cell cycle-dependent clustering of telomerase at elongating telomeres. Mol Cell. 2011;44:819–27. doi: 10.1016/j.molcel.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 110.Bajon E, Laterreur N, Wellinger RJ. A single templating RNA in yeast telomerase. Cell Rep. 2015;12:441–48. doi: 10.1016/j.celrep.2015.06.045. [DOI] [PubMed] [Google Scholar]
- 111.Fu D, Collins K. Human telomerase and Cajal body ribonucleoproteins share a unique specificity of Sm protein association. Genes Dev. 2006;20:531–36. doi: 10.1101/gad.1390306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sexton AN, Collins K. The 5′ guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Mol Cell Biol. 2011;31:736–43. doi: 10.1128/MCB.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vogan JM, Collins K. Dynamics of human telomerase holoenzyme assembly and subunit exchange across the cell cycle. J Biol Chem. 2015;290:21320–35. doi: 10.1074/jbc.M115.659359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yi X, Tesmer VM, Savre-Train I, Shay JW, Wright WE. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol Cell Biol. 1999;19:3989–97. doi: 10.1128/mcb.19.6.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vogan JM, Zhang X, Youmans DT, Regalado SG, Johnson JZ, et al. Minimized human telomerase maintains telomeres and resolves endogenous roles of H/ACA proteins, TCAB1, and Cajal bodies. eLife. 2016;5:e18221. doi: 10.7554/eLife.18221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, et al. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–48. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, et al. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev. 2011;25:11–16. doi: 10.1101/gad.2006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen Y, Deng Z, Jiang S, Hu Q, Liu H, et al. Human cells lacking coilin and Cajal bodies are proficient in telomerase assembly, trafficking and telomere maintenance. Nucleic Acids Res. 2015;43:385–95. doi: 10.1093/nar/gku1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schmidt JC, Zaug AJ, Cech TR. Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell. 2016;166:1188–97. doi: 10.1016/j.cell.2016.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Moser BA, Nakamura TM. Protection and replication of telomeres in fission yeast. Biochem Cell Biol. 2009;87:747–58. doi: 10.1139/o09-037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hockemeyer D, Collins K. Control of human telomerase action at telomeres. Nat Struct Mol Biol. 2015;22:848–52. doi: 10.1038/nsmb.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, et al. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013;496:187–92. doi: 10.1038/nature12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hong K, Upton H, Miracco EJ, Jiang J, Zhou ZH, et al. Tetrahymena telomerase holoenzyme assembly, activation, and inhibition by domains of the p50 central hub. Mol Cell Biol. 2013;33:3962–71. doi: 10.1128/MCB.00792-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rao T, Lubin JW, Armstrong GS, Tucey TM, Lundblad V, Wuttke DS. Structure of Est3 reveals a bimodal surface with differential roles in telomere replication. PNAS. 2014;111:214–18. doi: 10.1073/pnas.1316453111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hu X, Liu J, Jun HI, Kim JK, Qiao F. Multi-step coordination of telomerase recruitment in fission yeast through two coupled telomere-telomerase interfaces. eLife. 2016;5:e15470. doi: 10.7554/eLife.15470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chu TW, D’Souza Y, Autexier C. The insertion in fingers domain in human telomerase can mediate enzyme processivity and telomerase recruitment to telomeres in a TPP1-dependent manner. Mol Cell Biol. 2016;36:210–22. doi: 10.1128/MCB.00746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tong AS, Stern JL, Sfeir A, Kartawinata M, de Lange T, et al. ATM and ATR signaling regulate the recruitment of human telomerase to telomeres. Cell Rep. 2015;13:1633–46. doi: 10.1016/j.celrep.2015.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee SS, Bohrson C, Pike AM, Wheelan SJ, Greider CW. ATM kinase is required for telomere elongation in mouse and human cells. Cell Rep. 2015;13:1623–32. doi: 10.1016/j.celrep.2015.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Upton HE, Hong K, Collins K. Direct single-stranded DNA binding by Teb1 mediates the recruitment of Tetrahymena thermophila telomerase to telomeres. Mol Cell Biol. 2014;34:4200–12. doi: 10.1128/MCB.01030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Min B, Collins K. Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J Biol Chem. 2010;285:16434–43. doi: 10.1074/jbc.M110.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Upton HE, Chan H, Feigon J, Collins K. Shared subunits of Tetrahymena telomerase holoenzyme and Replication Protein A have different functions in different cellular complexes. J Biol Chem. 2017;292:217–28. doi: 10.1074/jbc.M116.763664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–14. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 134.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–20. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 135.Nandakumar J, Cech TR. Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol. 2013;14:69–82. doi: 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sexton AN, Regalado SG, Lai CS, Cost GJ, O’Neil CM, et al. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev. 2014;28:1885–99. doi: 10.1101/gad.246819.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen LY, Redon S, Lingner J. The human CST complex is a terminator of telomerase activity. Nature. 2012;488:540–44. doi: 10.1038/nature11269. [DOI] [PubMed] [Google Scholar]
- 138.Wan B, Tang T, Upton H, Shuai J, Zhou Y, et al. The Tetrahymena telomerase p75–p45–p19 subcomplex is a unique CST complex. Nat Struct Mol Biol. 2015;22:1023–26. doi: 10.1038/nsmb.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lue NF, Chan J, Wright WE, Hurwitz J. The CDC13-STN1-TEN1 complex stimulates Pol α activity by promoting RNA priming and primase-to-polymerase switch. Nat Commun. 2014;5:5762. doi: 10.1038/ncomms6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ray S, Karamysheva Z, Wang L, Shippen DE, Price CM. Interactions between telomerase and primase physically link the telomere and chromosome replication machinery. Mol Cell Biol. 2002;22:5859–68. doi: 10.1128/MCB.22.16.5859-5868.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Miyagawa K, Low RS, Santosa V, Tsuji H, Moser BA, et al. SUMOylation regulates telomere length by targeting the shelterin subunit Tpz1(Tpp1) to modulate shelterin-Stn1 interaction in fission yeast. PNAS. 2014;111:5950–55. doi: 10.1073/pnas.1401359111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Garg M, Gurung RL, Mansoubi S, Ahmed JO, Dave A, et al. Tpz1TPP1 SUMOylation reveals evolutionary conservation of SUMO-dependent Stn1 telomere association. EMBO Rep. 2014;15:871–77. doi: 10.15252/embr.201438919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu P, Takai H, de Lange T. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell. 2012;150:39–52. doi: 10.1016/j.cell.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dai X, Huang C, Bhusari A, Sampathi S, Schubert K, Chai W. Molecular steps of G-overhang generation at human telomeres and its function in chromosome end protection. EMBO J. 2010;29:2788–801. doi: 10.1038/emboj.2010.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hwang H, Buncher N, Opresko PL, Myong S. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure. 2012;20:1872–80. doi: 10.1016/j.str.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–18. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 147.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–10. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 148.Greider CW. Regulating telomere length from the inside out: the replication fork model. Genes Dev. 2016;30:1483–91. doi: 10.1101/gad.280578.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lin KW, Zakian VA. 21st century genetics: mass spectrometry of yeast telomerase. Cold Spring Harb Symp Quant Biol. 2015;80:111–16. doi: 10.1101/sqb.2015.80.027656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Martinez P, Blasco MA. Replicating through telomeres: a means to an end. Trends Biochem Sci. 2015;40:504–15. doi: 10.1016/j.tibs.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 151.Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009;28:810–20. doi: 10.1038/emboj.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]