Abstract
The phytohormone auxin, indole-3-acetic acid (IAA), regulates nearly all aspects of plant growth and development. Despite substantial progress in our understanding of auxin biology, delineating specific auxin response remains as a major challenge. Auxin regulates transcriptional response via its receptors, TIR1/AFB F-box proteins. Here we report an engineered, orthogonal auxin-TIR1 receptor pair, developed through a bump-and-hole strategy, that triggers auxin signaling without interfering with endogenous auxin or TIR1/AFBs. A synthetic, convex IAA (cvxIAA) hijacked the downstream auxin signaling in vivo both at the transcriptomic level and in specific developmental contexts, only in the presence of a complementary, concave TIR1 (ccvTIR1) receptor. Harnessing the cvxIAA-ccvTIR1 system, we provide conclusive evidence for the role of TIR1-mediated pathway in auxin-induced seedling acid growth. The cvxIAA-ccvTIR1 system serves as a powerful tool for solving outstanding questions in auxin biology and for precise manipulation of auxin-mediated processes as a controllable switch.
Introduction
Auxin mediates virtually every aspect of plant growth and development1,2. Substantial progress has been made in our understanding of auxin biology. Yet, multiple layers of auxin regulation at the levels of biosynthesis, catabolism, conjugation, transport and signaling as well as complex redundancies and severe developmental defects of higher-order mutants impose major challenges in specifically delineating auxin response of interest1,2. The auxin receptor TIR1 is a subunit of SKP1-Cullin-F-box (SCF) ubiquitin ligase complex3–5. Binding of auxin to TIR1 stabilizes its association with AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) repressor proteins6. Subsequent, proteasome-mediated degradation of AUX/IAA proteins by the SCFTIR1 complex releases auxin response factors (ARFs) to initiate gene expression7. There are six TIR1-related AUXIN SIGNALING F-BOX (AFB) proteins and 29 AUX/IAA proteins in Arabidopsis, and their combinatorial associations likely provide further complexity in auxin perception8.
To overcome these limitations, we employed the bump-and-hole strategy, a collaborative approach between chemistry and biology that enables the engineering of shape-complementary synthetic ligand and receptor pairs. It was first developed for delineating the function of protein kinases via engineering the kinases to specifically interact with synthetic ATP analogues9 or modified kinase inhibitors10. More recently, the bump-and-hole approach was successfully applied for identification of the individual roles of BET bromodomains11. We developed an orthogonal auxin-TIR1 pair, that can trigger auxin response, both at transcriptomic level and specific developmental context, without interfering with the endogenous auxin or TIR1/AFB-mediated signaling. The synthetic system further enabled us to decipher the role of TIR1-mediated signaling in acid growth, an auxin-mediated rapid cell elongation. Our work highlights the power of synthetic chemistry approach in bypassing the complexity of genetic redundancies and feedback regulation in order to understand and manipulate hormone signaling in plants.
Results
Bump-and-hole approach for orthogonal auxin-TIR1 pair
For a chemical control of auxin perception and signaling without interference with the endogenous counterparts, we took a bump-and-hole strategy and rationally designed a synthetic cvxIAA-ccvTIR1 pair based on the crystal structure of the TIR1-IAA-AUX/IAA complex6 (Fig. 1a–d). The auxin binding pocket of TIR1 consists of a floor made of TIR1 residues and the side walls made of two phenylalanine residues (F79 and F82) as well as the solenoid inner surface of the LRR domain as reported previously6 (Fig. 1a). Our strategy for cvxIAA-ccvTIR1 pair is to carve the wall of TIR1 and, reciprocally, introduce a bump to the indole ring of IAA to fill the cavity. We precluded modifications at the LRR polypeptide backbone, as it will likely influence AUX/IAA docking or structural integrity of the SCFTIR1 complex. In contrast, a substitution of F79 or F82 on the loop domain should not affect overall structure of the SCFTIR1 complex. A removal of a phenyl residue from F79 or F82 will create an open cavity, which is predicted to compromise stable binding of IAA (Fig. 1b). Having extra aryl residues in IAA, either at the fifth position or fourth position, will be predicted to introduce a molecular collision with the exposed phenyl residue corresponding at F79 (Fig. 1c) or F82, respectively.
Fig. 1. Engineering cvxIAA-ccvTIR1 pair by a bump-and-hole approach.
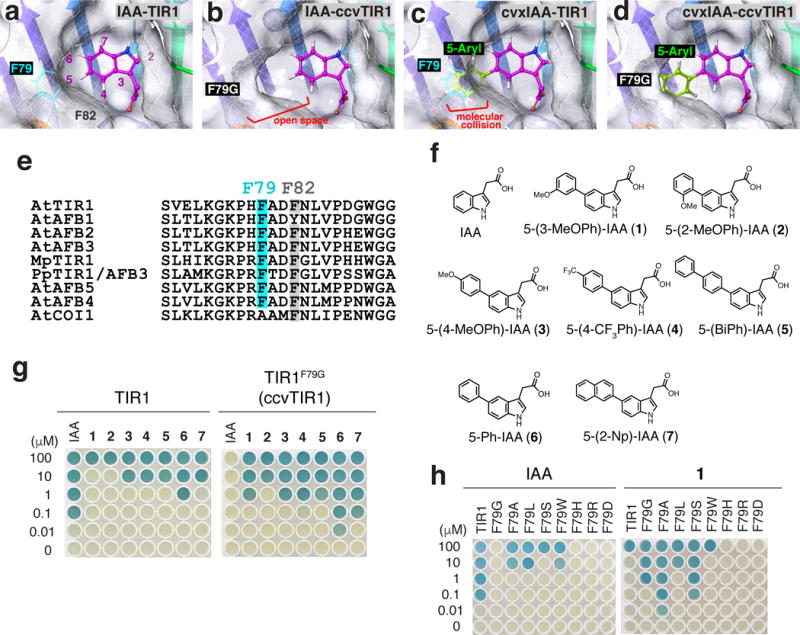
(a-d) TIR1 and ccvTIR1 auxin binding pocket modeled from the published X-ray crystal structure. (a) IAA and TIR1. (b) IAA and ccvTIR1. (c) cvxIAA and TIR1. (d) cvxIAA and ccvTIR1. The 5-aryl moiety of the cvxIAA would replace the lost phenyl moiety due to F79G substitution. (e) Amino-acid alignments of TIR1 paralogs and orthologs. The F79 (cyan) and F82 (gray) residues. (f) Chemical structures of IAA and 5-aryl-IAAs. (g) Yeast two-hybrid (Y2H) screening for cvxIAA. Association of the LexA-fused TIR1 and TIR1F79G with the activation-domain-fused IAA3DI+DII was tested in the presence of IAA or 5-aryl-IAAs. (h) Y2H screening for ccvTIR1. Association of the LexA-fused mutant TIR1 and the activation-domain-fused IAA3DI+DII tested in the presence of serial dilutions of IAA or cvxIAA.
Amino-acid sequence comparison reveals that F79 is evolutionarily conserved among TIR1 orthologs and paralog, including the ancestral TIR1 from basal land plant species, Marchantia polymorpha12 and Pyscomitrella patens13, but not in Arabidopsis COI1, the receptor for an unrelated phytohormone, jasmonic acid14 (Fig. 1e). On the other hand, F82 is conserved both in TIR1/AFBs and COI1, therefore may serve a structural role or a role unrelated to auxin perception (Fig. 1e). Thus, F79 of TIR1 emerged the primary engineering target site to create ccvTIR1.
The removal of phenyl moiety of F79 should result in the formation of cavity around the 5-position of IAA. This cavity could be refilled by introducing aryl groups at the 5-position of IAA (Fig. 1d). We therefore synthesized a series of 5-aryl-IAAs: 5-(3-methoxyphenyl)indole-3-acetic acid (5-(3-MeOPh)-IAA, 1), 5-(2-methoxyphenyl)indole-3-acetic acid (5-(2-MeOPh)-IAA, 2), 5-(4-methoxyphenyl)indole-3-acetic acid (5-(4-MeOPh)-IAA, 3), 5-(4-trifluoromethylphenyl)indole-3-acetic acid (5-(4-CF3Ph)-IAA, 4), 5-(biphenyl-4-yl)-indole-3-acetic acid (5-(BiPh)-IAA, 5), 5-phenyl-indole-3-acetic acid (5-Ph-IAA, 6), and 5-(2-naphthyl)indole-3-acetic acid (5-(2-Np)-IAA, 7) (Fig. 1f, Supplementary Note).
To screen for the best cvxIAA-ccvTIR1 pair, we performed yeast two-hybrid (Y2H) assays, which can detect the IAA-dependent association of TIR1 and IAA3 at concentrations as low as 40 nM (Fig. 1g, Supplementary Fig. 1a). In contrast, seven 5-aryl-IAAs exhibited reduced efficacy in promoting TIR1-IAA association: among them 5-(3-MeOPh)-IAA (1) and 5-(2-MeOPh)-IAA (2) did not promote TIR1-IAA3 association at 10 μM, well above physiologically relevant concentrations (Fig. 1g). Next, site-directed mutagenesis of TIR1 was conducted to substitute F79 with eight different amino acids (Fig. 1h). Among them, TIR1F79G exhibited the most preferred characteristics: IAA, even at 100 μM, failed to elicit TIR1F79G-IAA3 interaction (Fig. 1h). Likewise, active synthetic auxin analogs, 1-naphthaleneacetic acid (1-NAA, 8) and 2,4-dichlorophenoxyacetic acid (2,4-D, 9), both failed to elicit TIR1F79G-IAA3 interaction even at 100 μM (Supplementary Fig. 1b). In contrast, 5-(3-MeOPh)-IAA (1) successfully induced TIR1F79G-IAA3 interaction at concentration ten times lower than 5-(2-MeOPh)-IAA (2) (Fig. 1g). Competition assays further showed that IAA is unable to displace 5-(3-MeOPh)-IAA-mediated TIR1F79G-IAA3 association (Supplementary Fig. 1c). Unlike F79, site-directed mutagenesis at F82 (TIR1F82A, TIR1F82G, and TIR1F82S), which we predicted to create a cavity for 4-aryl-IAA (4-phenyl-indole-3-acetic acid, 10), abolished any Y2H interactions tested (Supplementary Fig. 1d, e). Based on these results, 5-(3-MeOPh)-IAA (1) and TIR1F79G were chosen for further studies as cvxIAA and ccvTIR1, respectively.
Biochemical property of engineered cvxIAA-ccvTIR1 pair
To test the biochemical specificity of cvxIAA-mediated ccvTIR1-AUX/IAA association, pull down assays were performed using in vitro translated TIR1-FLAG and ccvTIR1-FLAG proteins. The biotinylated DII peptide from IAA7 pulled down TIR1-FLAG protein in the presence of IAA or 1-NAA, but not cvxIAA (Fig. 2a). Conversely, ccvTIR1-FLAG protein was pulled down by the DII peptide only in the presence of cvxIAA, but neither IAA nor 1-NAA (Fig. 2a). The mutant DII peptide (mDII), which lacks the ability to associate with TIR18, was unable to pull down either TIR1 or ccvTIR1 (Fig 2a). We next performed a quantitative analysis of in vitro cvxIAA-triggered ccvTIR1-IAA7DII association. The binding affinity of cvxIAA-triggered ccvTIR1-IAA7DII based on the dose-response curves (BD50 = 3.98 ± 0.60 μM) is highly comparable to that of IAA-triggered TIR1-IAA7DII (BD50 = 11.54 ± 1.23 μM) (Fig. 2b, c; Supplementary Fig. 2). Due to lower sensitivity of the densitometry-based detection method, these BD50 values are higher than the previously-reported Kd values using [3H]-IAA3,5. The important point here, however, is that both cvxIAA-ccvTIR1 and IAA-TIR1 pairs recruit IAA7DII with a similar, if not identical, binding property, therefore providing a biochemical support to justify the further use of our cvxIAA-ccvTIR1 system.
Fig. 2. cvxIAA, but not active auxins, promotes association of ccvTIR1 and IAA7DII peptide.
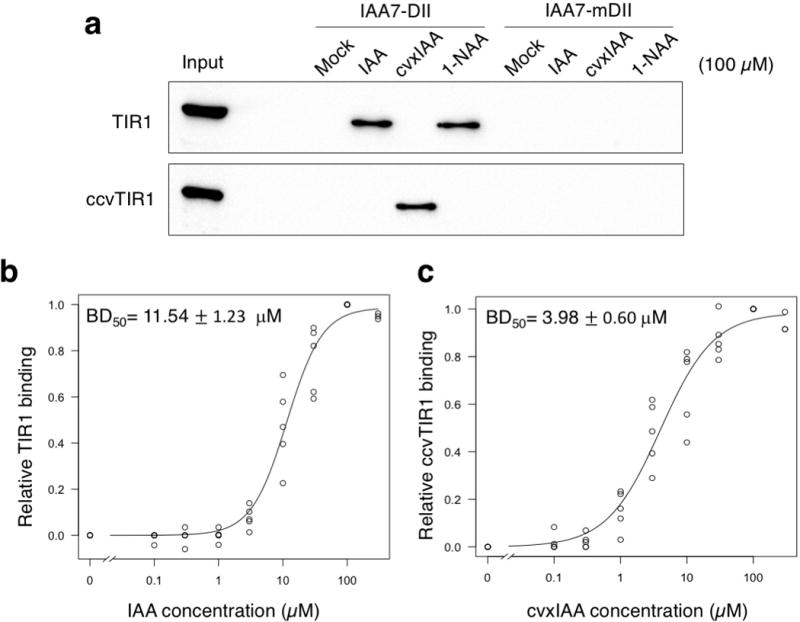
(a) In vitro pull-down assays of TIR1 and ccvTIR1 co-incubated with the biotinylated IAA7 DII or mDII peptide in the presence or absence of 100 μM IAA, 1-NAA, and cvxIAA. Number of experimental repeats: n = 5 for IAA7DII pull downs; n = 3 for IAA7mDII pull downs. For uncropped blot data, see Supplementary Fig. 11. (b) Fitted dose-response binding curve of TIR1 to IAA7DII peptide in the function of IAA concentration. (c) Fitted dose-response binding curve of ccvTIR1 to IAA7DII peptide in the function of cvxIAA concentration. For b and c, BD50 is defined as a concentration of IAA or cvxIAA required for the 50 % pull down of TIR or ccvTIR1 bound with IAA7DII. For mean values and standard deviation, see Supplementary Fig. 2b and Supplementary Table 1.
In vivo auxin transcriptional response by cvxIAA-ccvTIR1
To address whether our designed cvxIAA-ccvTIR1 system operates in vivo, we first expressed ccvTIR1 driven by the constitutive promoter (35S::ccvTIR1) into Arabidopsis plants that carry the auxin output marker DR5::GFP15 (hereafter referred as “wild type”). Both 35S::ccvTIR1 and control 35S::TIR1 transgenes are fused with a FLAG epitope-tag for specific detection of transgene products. First, we performed an immunoblot analysis to confirm the accumulation of ccvTIR1 proteins. As shown in Supplementary Fig. 3, both 35S::TIR1 and 35S::ccvTIR1 line with similar transcript levels accumulated similar amounts of TIR1 and ccvTIR1 proteins, respectively. Thus, the F79G substitution, which serves as basis for our bump-and-hole strategy, does not significantly affect the stability of TIR1 protein.
We subsequently characterized the 35S::ccvTIR1 seedlings. In the absence of cvxIAA treatment, 35S::ccvTIR1 roots exhibited DR5::GFP patterns at the root tip, just like wild-type roots carrying the same marker, therefore ccvTIR1 appears inert by itself (Fig. 3a; Supplementary Fig. 4). Treatments with cvxIAA conferred strong GFP signals throughout the roots in a dosage-dependent manner only in the seedlings carrying 35S::ccvTIR1, but not 35S::TIR1 or untransformed controls (Fig. 3a; Supplementary Fig. 4), indicating that cvxIAA triggers auxin transcriptional outputs via ccvTIR1 in plants. When wild-type Arabidopsis seedlings were treated with 0.1 or 0.3 μM cvxIAA on agar media, which are the concentrations used in the experiments with liquid culture, their roots showed normal gravitropism (Supplementary Fig. 5a, b), which is in sharp contrast to the treatment of an auxin transport inhibitor, 1-naphthylphthalamic acid (NPA). Likewise, unlike the NPA treatment, the cvxIAA treatment did not influence a characteristic DR5::GFP pattern with highest signal at the root tip16 (Supplementary Fig. 5c). These results indicate that cvxIAA does not clearly affect the endogenous auxin transport or response when treated at a physiological concentration. However, a statistical analysis suggests that at 0.3 μM, cvxIAA may have a subtle effect on root gravitropism (p=0.045, Welch’s 2-sample T-test, unpaired, Supplementary Fig. 5b). Therefore, the possibility that cvxIAA disrupts endogenous auxin transport at higher concentrations, such as submillimolar levels, cannot be excluded.
Fig. 3. cvxIAA triggers global auxin-induced gene expression only in the seedlings expressing the engineered ccvTIR1.
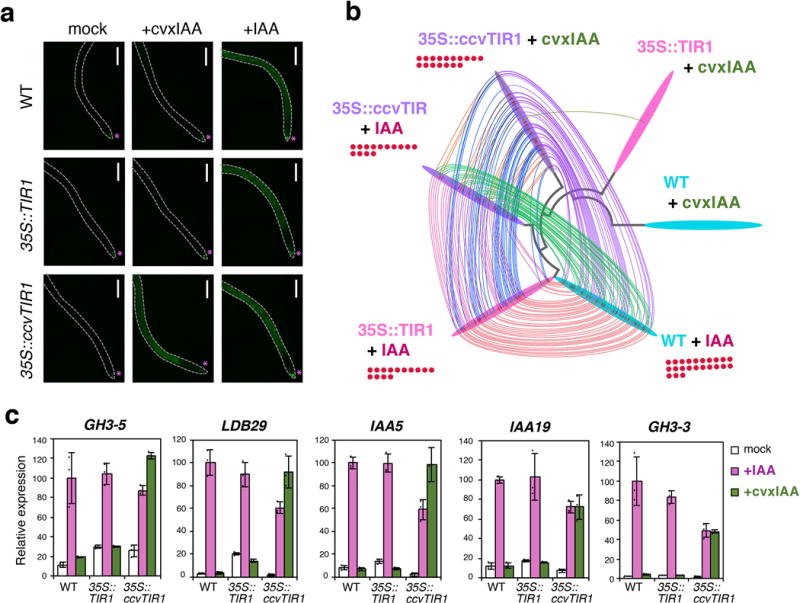
(a) Auxin output marker (DR5::GFP) in response to IAA or cvxIAA treatment. Shown are the fluorescent micrographs of 4-day-old roots from Arabidopsis DR5::GFP seedlings (wild-type (WT), 35S::TIR1, and 35S::ccvTIR1) treated with mock, 1 μM cvxIAA or 1 μM IAA for 24 hours. Asterisks, auxin response peak to the endogenous auxin. Dotted line, root outline. Scale bars, 300 μm. (b) Bobbin Dendrogram. The RNA-seq profiles of genes induced by 0.1 μM IAA or cvxIAA (Log2FC>1, qVal < 0.01) in wild-type (cyan), 35S::TIR1 (pink), and 35S::ccvTIR1 (purple) seedlings over respective mock control (3 biological replicates). Each genotype/treatment is placed at each node of the dendrogram based on the relatedness of their global gene expression profiles. Among the induced genes, those shared by the other samples/treatments are connected with colored threads. Red dots, genes classified with GO Category ‘auxin’. (c) qRT-PCR analysis. Shown are relative expression levels of five known auxin induced genes from seedlings (wild-type (WT), 35S::TIR1, and 35S::ccvTIR1) treated with mock, 0.1 μM cvxIAA (green) or IAA (magenta) for 3 hours. Bars, mean values. Error bars, standard deviation. Dots, the exact data points of individual samples. Experiments were performed three times. The normalized mean value of IAA-treated WT was set at 100.
To investigate the specificity of cvxIAA-induced transcriptional response at the genome-wide level, we furthermore performed RNA-seq profiling (Fig. 3b; Supplementary Dataset 1). For this purpose, wild-type, 35S::TIR1, and 35S::ccvTIR1 seedlings were mock treated, treated with IAA or cvxIAA for three hours and subjected to transcriptome analysis. To most effectively visualize the similarities of differentially-expressed genes (DEGs) in multi-parameter transcriptomic analyses, we developed a software visualization package, Bobbin Dendrogram Plot Generator (https://github.com/takaho/bobinplot), whereby each sample is connected by a node representing the phylogenetic relationships based on the DEGs. Commonly upregulated genes between each pairwise sample are connected by the lines. As shown in Fig. 3b, IAA-treated wild-type, 35S::TIR1 and 35S::ccvTIR1 seedlings showed very similar DEG profiles with cvxIAA-treated 35S::ccvTIR1. Importantly, auxin-specific genes are neither induced nor repressed by cvxIAA in wild-type and 35S::TIR1 seedlings (log2FC >1.0, qVal <0.01 and log2FC <-1.0, qVal <0.01), indicating that cvxIAA is highly selective and does not trigger false-positive auxin transcriptional response via 35S::TIR1 or endogenous TIR1/AFB proteins (Fig. 3b, Supplementary Dataset 1). Traditional Pearson correlation analyses also highlighted the resemblance of the genome-wide transcriptional response by cvxIAA-ccvTIR1 and IAA-TIR1 (Supplementary Fig. 6). qRT-PCR analyses of the representative auxin-inducible genes further validated their specific induction by cvxIAA only in the presence of ccvTIR1 (Fig. 3c). Together, we conclude that the cvxIAA-ccvTIR1 pair is capable of hijacking the auxin transcriptional response in Arabidopsis.
cvxIAA-ccvTIR1 triggers auxin-mediated development
To examine whether cvxIAA-ccvTIR1 promotes auxin-specific developmental processes, we first carried out auxin-induced root growth inhibition assays (Supplementary Fig. 7). Wild-type, 35S::TIR1, and 35S::ccvTIR1 seedlings all exhibited dose-dependent root growth inhibition by the application of natural auxin IAA, resulting in 80-90 % inhibition by 0.3 μM IAA. The findings that 35S::ccvTIR1 seedlings respond normally to the IAA treatment suggests that the introduced ccvTIR1 does not interfere with the auxin perception by endogenous TIR1/AFBs. By contrast, cvxIAA had no effects on the elongation of wild-type and 35S::TIR1 roots even at 0.3 μM. At this concentration, the root growth of 35S::ccvTIR1 seedlings was inhibited by 60-70 % (Supplementary Fig. 7).
Next, we characterized whether the cvxIAA-ccvTIR1 pair can take control of well-studied auxin-induced lateral root (LR) development (Fig. 4)17. While 1-NAA increases LR density in a dose-dependent manner (Fig. 4a; Supplementary Fig. 8), cvxIAA has no effects on LR density in wild-type or 35S::TIR1 roots (Fig. 4a,b,d). By contrast, cvxIAA treatments conferred a significant increase in LR density if the seedlings carry ccvTIR1, reaching to ~4-fold increase at 1 μM (Fig. 4a). SOLITARY-ROOT (SLR)/IAA14 acts as a downstream negative regulator of auxin-dependent LR development, and its stabilized mutation within the DII motif in slr-1 completely blocks LR formation (Fig. 4c, e; Supplementary Fig. 8)18. cvxIAA treatment was unable to restore LR formation in 35S::ccvTIR1 slr-1 roots (Fig. 4c,e; Supplementary Fig. 8), consistent with the action of cvxIAA-ccvTIR1 upstream of IAA14. These results demonstrate that cvxIAA-ccvTIR1 pair can be used to evoke auxin-mediated developmental programs in vivo.
Fig. 4. cvxIAA inhibits root elongation and induces auxin-induced lateral root development only in the seedlings expressing the engineered ccvTIR1.
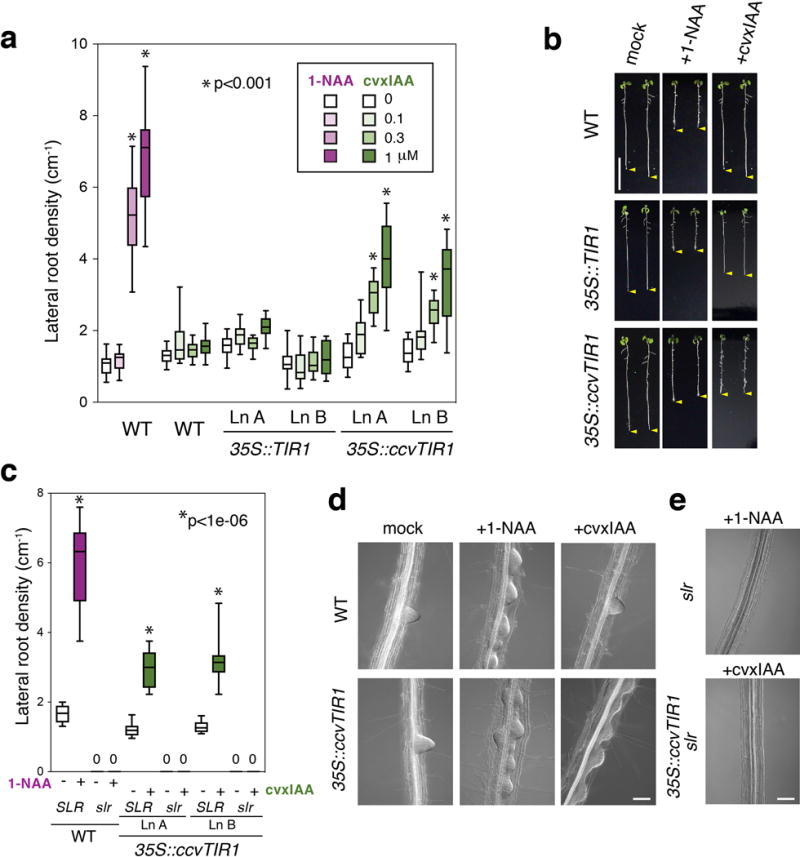
(a) Lateral root density of 8-day-old Arabidopsis seedlings from wild type (WT), two representative lines of 35S::TIR1 and 35S::ccvTIR1 treated with 1-NAA (magenta) or cvxIAA (green) for 3 days. Box-and-whisker plots show a median (centerline), upper/lower quartiles (box limits) and maximum/minimum (upper/lower whiskers). n=10. Experiments are performed three times. *p<0.001, Welch’s two sample t-test, unpaired. (b) 8-day-old Arabidopsis seedlings from wild type (WT), 35S::TIR1, and 35S::ccvTIR1 treated with 1 μM 1-NAA or cvxIAA for 3 days. Yellow arrowhead; the tip of roots. Images were taken under the same magnification. Scale bar, 1 cm. (c) slr is epistatic to the cvxIAA-induced lateral root formation in 35S::ccvTIR1 seedlings. Box-and-whisker plots show a median (centerline), upper/lower quartiles (box limits) and maximum/minimum (upper/lower whiskers). 0, no lateral roots detected; n=10. Experiments are performed three times. (d) DIC microscopy of representative roots from wild-type (WT) and 35S::ccvTIR1 seedlings either mock treated or treated with 1 μM 1-NAA or cvxIAA for 40 hrs. Images were taken under the same magnification. Scale bar, 100 μm. (E) DIC microscopy of representative roots of slr treated with 1 μM 1-NAA and 35S::ccvTIR1 slr treated with 1 μM cvxIAA. Scale bar, 100 μm.
TIR1-mediated pathway in promoting hypocotyl acid growth
The exact identity of the auxin receptor that evokes the auxin-mediated rapid seedling elongation, a phenomenon originally described by Charles Darwin and later termed ‘acid growth’, is not fully resolved. Harnessing the cvxIAA-ccvTIR1 system, we tackled the mechanism of auxin perception for acid growth, which occurs via auxin-induced rapid phosphorylation and activation of the plasma membrane H+-ATPase within minutes19, resulting in subsequent acidification of apoplasts and cell-wall loosening20. The phosphorylation occurs in the absence of TIR1 and AFB2, the major auxin receptors for seedling growth19, raising a question that yet another class of auxin receptor might be responsible for the acid growth21. Severe growth defects in higher-order tir1/afb mutants22 hinder finer dissection. Among the TIR1/AFB family members, AFB5 recognizes a synthetic auxin analog, picloram23. Making use of this, a recent report showed that picloram stimulates hypocotyl elongation after 50 min of the treatment24. This was substantially slower than the IAA-triggered response, which occurs in 10-20 min. While their pioneering work highlights TIR1/AFB as potential receptors for acid growth, direct role for the TIR1-mediated pathway in the faster auxin response, however, remains unclear.
To unambiguously address this question, we generated tir1 afb2 mutant seedlings expressing the engineered ccvTIR1 driven by the endogenous TIR1 promoter. Unlike TIR1pro::TIR1, TIR1pro::ccvTIR1 failed to rescue the reduced auxin response of tir1 afb2 in root elongation assays (Supplementary Fig. 9). Thus, ccvTIR1 does not complement tir1. TIR1pro::ccvTIR1 tir1 afb2 seedlings responded sensitively to low concentrations (30-100 nM) of cvxIAA and inhibited root elongation, indicating that TIR1pro::ccvTIR1 can substitute for TIR1 only in the presence of cvxIAA (Supplementary Fig. 9). Consistent with the previous report19 TIR1pro::ccvTIR1 tir1 afb2 hypocotyl segments responded to IAA, suggesting that receptors other than TIR1 and AFB2, which could be remaining AFB proteins or yet unidentified auxin receptors, are able to trigger auxin-mediated rapid elongation (Fig. 5a). Strikingly, upon treatment with cvxIAA (1 μM), hypocotyl segments only from the seedlings containing TIR1pro::ccvTIR1 exhibited significant, rapid elongation within 20 min (p= 8.01 e-07, Welch two sample T-test, unpaired)(Fig. 5b). The elongation kinetics are similar to that of IAA (1 μM)-treated wild type (p= 0.525, Welch two sample T-test, unpaired)(Fig. 5b). The results demonstrate that the TIR1-mediated pathway is sufficient to trigger the rapid hypocotyl elongation at full extent. Time-course analysis shows that the kinetics of hypocotyl elongation by cvxIAA-ccvTIR1 mirrors that by IAA-TIR1 (Fig. 5b).
Fig. 5. Hypocotyl acid growth mediated by the synthetic cvxIAA and engineered ccvTIR1 pair.
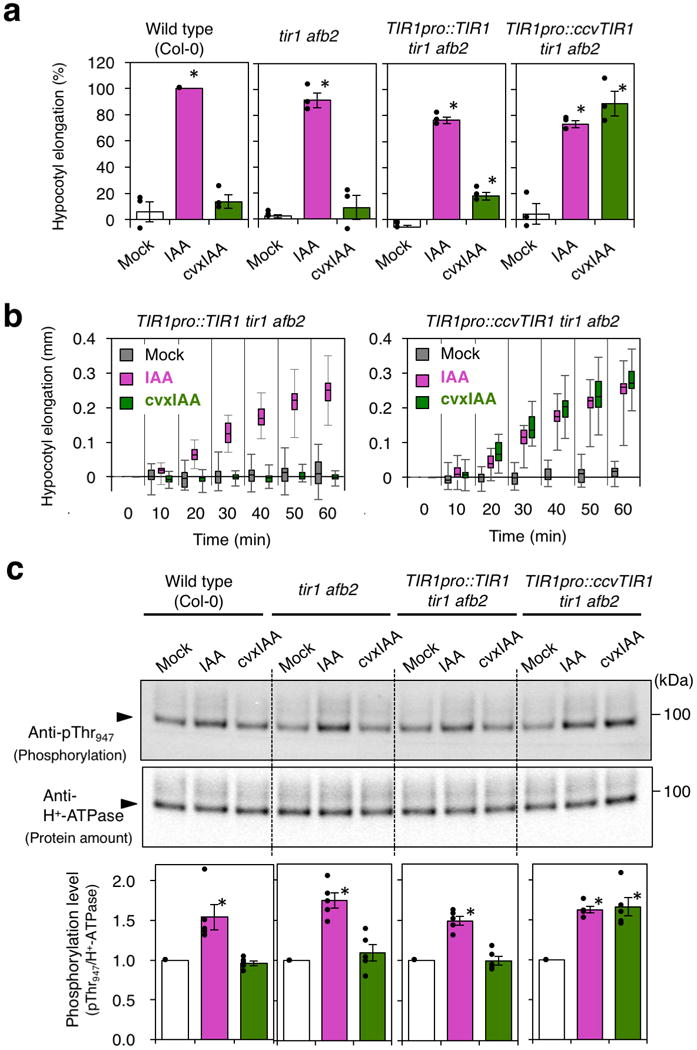
(a) Relative elongation of hypocotyl segments from 3-day-old Arabidopsis etiolated seedlings from wild-type, tir1 afb2, TIR1pro::TIR1 tir1 afb2, and TIR1pro::ccvTIR1 tir1 afb2 that are mock treated (white), treated with 1 μM IAA (magenta) or 1 μM cvxIAA (green) for 30 min. The three independent experiments were performed. Bars, mean. Error bars, standard error. Dots, the exact data points of individual samples. *p<0.01, Student t-test. (b) Elongation of the hypocotyl segments of 3-day-old Arabidopsis etiolated seedlings from TIR1pro::TIR1 tir1 afb2 and TIR1pro::ccvTIR1 tir1 afb2 that are mock treated (gray), treated with 1 μM IAA (magenta) or 1 μM cvxIAA (green). Box-and-whisker plots show a median (centerline), upper/lower quartiles (box limits) and maximum/minimum (upper/lower whiskers). n=15. Experiments were performed three times. (c) Phosphorylation of H+-ATPase. Hypocotyl segments of 3-day-old Arabidopsis etiolated seedlings from wild-type, tir1 afb2, TIR1pro::TIR1 tir1 afb2, and TIR1pro::ccvTIR1 tir1 afb2 were mock treated (white), treated with 1 μM IAA (magenta) or 1 μM cvxIAA (green) for 30 min and subjected to immune blot analysis using anti-pThr947 H+-ATPase antibody (top) as well as anti H+-ATPase antibody (middle). The five independent experiments were performed. The pT947 signals were normalized against the total H+-ATPase signals (bottom). Bars, mean. Error bars, standard error. Dots, the exact data points of individual samples. *p<0.01, Student t-test. For uncropped blot data, see Supplementary Fig. 12.
Next, to test whether the cvxIAA-ccvTIR1-mediated rapid hypocotyl elongation occurs via the activation of H+-ATPase in vivo, we conducted an immuno-detection of the AHA2 T947 phosphorylation (p-T947) from hypocotyls of etiolated seedlings. IAA treatment resulted in approx. 1.5-fold increase in p-T947 signals in all genotypes tested. Strikingly, cvxIAA treatment induced ~1.5-fold increase in p-T947 only in the TIR1pro::ccvTIR1 tir1 afb2 seedlings (Fig. 5c). Furthermore, only in the presence of ccvTIR1, cvxIAA treatment elicited rapid induction of SMALL AUXIN UP RNA19 (SAUR19), which promotes auxin-mediated cell elongation by counteracting protein phosphatases that dephosphorylate p-T94725,26 (Supplementary Fig. 10). Combined, this study convincingly demonstrates the role for TIR1-mediated pathway in the rapid activation of H+-ATPase and elongation growth, the bases of the acid growth theory27. Our finding accords with the very recent report showing that the overexpression of SAUR19 is sufficient for the acid growth28. In conclusion, our results corroborate the earlier report24 and further demonstrate that TIR1/AFB-mediated pathway can fully explain the kinetics of rapid auxin-induced acid growth.
Discussion
Auxin is regulated at biosynthesis, catabolism, conjugation, transport, signaling, and intricate feedbacks among them, and this complication hampers specific delineation of auxin responses of interest1. Using the artificial, engineered cvxIAA-ccvTIR1 system designed and validated here, we successfully demonstrated the conclusive role of TIR1-mediated pathway that is sufficient for fully triggering rapid seedling elongation, known as acid growth. Our work highlights the power of a synthetic-chemistry for breaking redundancies at the level of hormone perception to uncover the specific contributions of each receptor.
A bump-and-hole approach is a simple, clean way to generating orthogonal ligand-receptor pairs as well as increasing agonist selectivity11. Here, we carved an aryl group from F79, replacing phenylalanine by glycine, and complementarily introduced an additional aryl group to the 5-position of IAA to generate an orthogonal cvxIAA-ccvTIR1 pair (Figs. 1 and 2). Since no modification was introduced at the AUX/IAA binding interface, these complementary substitutions between the ligand and the receptor likely impose a minimal influence on recruitment of the AUX/IAA proteins. In support of this, the cvxIAA-ccvTIR1 pair exhibits binding (BD50) with the IAA7DII peptide is nearly identical to that of IAA-TIR1 (Fig. 2). It has been shown that AUX/IAA proteins determine the affinity of the TIR1-AUX/IAA co-receptor complex to auxin, from 10 nM to > 1μM, depending on the AUX/IAA proteins8. Extended biochemical analyses of cvxIAA-ccvTIR1 pair with different AUX/IAAs will reveal whether this is the case for ccvTIR1 to perceive cvxIAA.
The F79 residue is conserved among TIR1/AFB families, including those ancestral auxin receptors in moss and liverworts, but not in the JA receptor COI1 (Fig. 1e). The corresponding F-to-G substitution therefore can be introduced into TIR1/AFB orthologs as well as paralogs to delineate their specific downstream response in a native promoter context or in a cell/tissue-type-specific manner. For example, as reported previously19, the auxin-mediated acid growth occurs in tir1/afb2 (also see Fig. 5), indicating that in addition to TIR1 other AFBs also participate in this process. The F-to-G substitution may be introduced into AFBs to generate ccvAFBs that can be systematically tested for their roles in auxin-mediated acid growth or address their specific roles during plant growth and development.
A synthetic, auxin-inducible degradation (AID) system has been successfully applied for controllable protein depletion in yeast, mammalian culture cells, and intact C. elegans29–32. Here, a target protein of interest will be fused with the DII degron motif from AUX/IAA and subjected to the SCFTIR1-complex mediated proteasome degradation upon auxin treatment. Notably, a recent adaptation of CRISPR-Cas9 technology further enabled auxin-degron tagging in human cell lines for effective AID33. Taking advantage of the absence of a signal interference, cvxIAA-ccvTIR1 and IAA-TIR1 systems could be simultaneously used in these heterologous systems as a dual-switch for functional studies. Finally, previous mutagenesis studies on TIR1 have identified the residues influencing the affinity for AUX/IAA proteins as well as those untethering TIR1 from the SCF complex34,35. Incorporation of additional properties to the cvxIAA-ccvTIR1 system may enable further mechanistic understanding of auxin signaling and controllability of auxin-mediated plant growth and development for potential crop improvement36.
Methods
Plasmids
The plasmids generated in this study and the primers used for plasmid construction are listed in Supplementary Table 2 and Supplementary Table 3, respectively.
Plant materials
The Arabidopsis thaliana Columbia (Col) accession was used as wild type. solitary root mutant (slr-1) harboring the dominant allele of IAA14 was described previously18. The engineered TIR1 constructs (Supplementary Table 2) were transformed into DR5::GFP37 or tir1-1 afb2-3 double mutant38,39 by floral dipping using Agrobacterium tumefaciens strain GV3101. Over 20 independent transgenic lines were isolated for each construct, and those showing clear monogenic inheritance were subjected to detailed phenotypic characterizations. Some of the transgenes were transferred from the original lines to the slr-1 background by genetic cross.
Plant culture and treatment
For root growth assays and gene expression analyses, seeds were sterilized, stored in the dark at 4 °C for a few days, transferred to 0.5 × Murashige and Skoog (MS) liquid medium, and grown on a shaker set at 140 rpm under continuous white light at 22 °C. For root growth assays, after 1-day incubation, various concentrations of auxin or its analogs were added to growth media, and root length was measured after additional 1-week incubation. For DR5::GFP observation, 3-day-old seedlings were transferred to 0.5 × MS liquid medium containing various concentrations of the compounds, and after 24 hours, GFP fluorescence at root tip regions was observed using Zeiss SteREO Discovery V20. For lateral root induction assays, 5-day-old seedlings grown on plates were transferred to 0.5 × MS plates containing various concentrations of the compounds indicated in the corresponding figures. Roots were cleared by chloral hydrate after 40-hour incubation with the compounds, and lateral root primordia were observed using Zeiss Axio Imager 2. The number of emerging lateral roots was counted after 3-day incubation with the compounds.
RNA extraction, qRT-PCR and RNA-seq analysis
7-day-old seedlings in liquid media were treated with 0.1 μM IAA or cvxIAA, and total RNAs were extracted and purified after 3 hours using RNeasy Plant Mini Kit (QIAGEN). Each RNA sample was prepared from a pool of 30 whole seedlings. Three independent RNA samples for each condition were used for the following analyses. For qRT-PCR, reverse transcription was carried out using ReverTra Ace (TOYOBO) according to the manufacturer’s instructions. Quantitative PCR was performed using SYBER FAST qPCR Kit (KAPA) and LightCycler 96 (Roche). The primers used for qPCR are listed in Supplementary Table 3. For RNA-seq analysis, after RNA integrity was confirmed by running the RNA samples on Agilent RNA 6000 Nano Chip (Agilent Technologies), 0.5 μg of the samples were used for library preparation using Illumina TruSeq Stranded mRNA LT Sample kit. The resulting barcoded libraries were pooled and sequenced on Illumina NextSeq500 sequencing platform, and 75-bp single-end reads were obtained. All primary sequence read data for the RNA-seq analysis have been submitted to the DNA Data bank of Japan (DDBJ: http://www.ddbj.nig.ac.jp) with the accession number DRA005718.
The qRT-PCR analysis in the hypocotyl segments was performed according to the previous report19. Briefly, the hypocotyl segments excised from 3-day-old etiolated seedlings were treated with 0.1 μM IAA or cvxIAA for 30 min and total RNAs were extracted and purified using the NucleoSpin RNA Plant Kit (TaKaRa). Each RNA sample was prepared from a pool of 50 hypocotyl segments. Reverse transcription was performed using PrimeScript First Strand cDNA Synthesis Kit (TaKaRa) and qRT-PCR was carried out with primer-pairs specific for the corresponding genes (Supplementary Table 3), Power SYBR Green PCR Master Mix, and StepOne Real-Time PCR system (Applied Biosystems). For characterization of transgenic seedlings, qRT-PCR was performed with primer-pairs specific for nucleotide sequence of FLAG-TIR1 (Supplementary Table 3).
Bioinformatics and Data Analysis
Sequences of RNA-seq experiments were mapped on TAIR 10 genome using STAR (version 2.5.1b, https://github.com/alexdobin/STAR) with default options, and the Illumina reads were mapped and the read counts were calculated using our in-house programs with the gene location GFF file distributed at TAIR (ftp://ftp.arabidopsis.org/Genes/TAIR10_genome_release/TAIR10_gff3/TAIR10_GFF3_genes.gff). Differentially expressed genes (DEGs) and their statistical significance were evaluated using DESeq2 R package (http://bioconductor.org/packages/release/bioc/html/DESeq2.html). Upregulated and downregulated genes in RNA-seq experiments were defined as log2(treated/control) more than 1 or less than 1, and q-values less than 0.01. The experiments were clustered using a hierarchal approach, in which the pairs having most score were clustered into a new virtual node until no pairs exist. For Person correlation analysis, genes classified into GO category for “auxin” (e.g. Response to Auxin, Auxin-activated Signaling Pathway, Auxin Polar Transport, Auxin Homeostasis) are extracted from DEGs based on the GO classification search (AmiGO2: http://amigo.geneontology.org/amigo/search/ontology?q=auxin). Scatter plots were generated using our in house phython scripts. DEGs and highly enriched GO categories are supplied as Supplementary Dataset 1.
The Bobbin Dendrogram Plot Generator was developed using Phython (requires ReportLab) for effective visualization of commonly up- or down-regulated genes among multiple-parameter samples (multiple biological samples with multiple treatments). Like molecular phylogenetic trees, each node represents the relatedness between pairwise samples. For each experiment, three biological replicates were clustered using a hierarchal approach, in which the pairs having most score were clustered into a new virtual node until no pairs exist. DEGs that are common between each pairwise combination is connected with lines. Denser the lines more similar the DEGs. The Bobbin Dendrogram Plot Generator is available at GitHub (https://github.com/takaho/bobinplot).
Yeast assays
EGY48 strain was transformed with pSH18-34 harboring a LexA-operon::LacZ reporter, a pGLex313-based plasmid for expression of LexA-DNA-binding-domain-fused constructs (TIR1 or a series of site-directed mutagenized ccvTIR1), and pJG4-5-based plasmid for expression of B42-transcriptional-activator-fused constructs (DI and DII domain of IAA3)40. Transformed strains were grown at 30 °C on agar plates composed of minimal SD base (Clontech, cat. 630411) and –His/–Trp/–Ura dropout supplement (Clontech, cat. 630424). Colonies were picked up and grown in liquid SD/–His/–Trp/–Ura medium for one night at 30 °C, and then medium was replaced with liquid medium composed of minimal SD/Gal/Raf base (Clontech, cat. 630420), –His/–Trp/–Ura dropout supplement, 50 mM Na-phosphate buffer (pH 7.0), 80 μg/ml X-gal (Wako) and various concentrations of compounds indicated in corresponding figures. After 3-day incubation at 30 °C, cultured media containing yeast were transferred to white 96-well plates (flat bottom) and observed. See Supplementary Table 2 for plasmid information.
Pull-down assays
Biotinyl-DII (biotinyl-(NH)-AKAQVVGWPPVRNYRKN) peptide and biotinyl-mDII (biotinyl-(NH)-AKAQVVEWSSGRNYRKN) peptide were reported previously8. C-terminally FLAG-tagged TIR1 or ccvTIR1 proteins were synthesized in vitro using a wheat germ extract system according to the manufacturer’s instructions (NUProtein, Nagoya, Japan). Templates for the protein synthesis were amplified using the primers listed in Supplementary Table 3. The biotinyl peptides were incubated with Dynabeads M-280 Streptavidin beads (Invitrogen) in binding buffer (50 mM Tris-HCl, pH 8.0, 200 mM NaCl, 10% glycerol, 0.1% Tween 20) at room temperature for 30 min and washed 10 times with binding buffer. The beads attached with the biotinyl peptides were incubated with the synthesized proteins in the binding buffer supplemented with various concentrations of compounds indicated in corresponding figures. The beads were washed 3 times with the same binding buffer supplemented with compounds. The proteins were extracted from the beads by SDS-PAGE sample buffer and separated by SDS-PAGE. Immunoblotting was performed using anti-FLAG antibody (Sigma, F3165), and immunoreactive bands were detected using SuperSignal WestPico Chemiluminescence reagent (Thermo Scientific) and Light-Capture cooled CCD camera system (ATTO). For a calculation of BD50 (= a concentration of IAA or cvxIAA required for the 50 % pull down of TIR or ccvTIR1 bound with IAA7DII), the pull down assays were performed five times for 0, 0.1, 0.3, 1, 3, 10, 30 and 100 μM IAA or cvxIAA, and 3 times for 300 μM IAA or cvxIAA. The R-package ‘drc’41 was used to fit the binding curve to the shifted log logistic distribution (the three-parameter log logistic function with limit to 0).
Immunodetection of TIR1 and ccvTIR1 proteins in Arabidopsis seedlings
Twenty seedlings (DR5::GFP, DR5::GFP 35S::FLAG-TIR1-WT, and DR5::GFP 35S::FLAG-TIR1-F79G) grown on 0.5x MS agar medium at 16h-light/8h-dark cycle for 5 days were harvested and frozen in liquid nitrogen. Protein extracts obtained from the seedlings were subjected to SDS-PAGE. The transferred membrane was first stained with Panceau-S (MP Biomedicals) to visualize total proteins, and subsequently subjected to immunoblot analysis using anti-FLAG (Sigma, F3165) and a control anti-H+-ATPase antibody raised against the catalytic domain of AHA242. Three biological replicates were performed.
Root gravitropism assays
DR5::GFP seedlings were grown on 0.5x MS agar medium at 16h light/8h dark for 5 days. The seedlings with a straight root grown along the agar surface were transferred onto 0.5x MS agar medium containing the indicated chemical substances and pre-incubated for 6 hours at the condition that roots oriented downward. Then, the seedlings were subjected to a gravistimulus by rotating the agar plates 90°. After 48 hours the images of the seedlings were captured by digital camera. The gravitropic angle of the roots was measured and the data were presented as a gravitropism diagram. The bar length in the diagram shows the percentage of roots seedlings assigned to the respective sector. The seedlings which were incubated with the indicated chemical substances were stained with propidium iodide and the GFP signal in their root tips was observed with confocal microscopy. Three independent experiments were performed.
Hypocotyl elongation assay and detection of H+-ATPase phosphorylation
Hypocotyl elongation assays were performed according to the previous report19. Briefly, seedlings were grown on 0.5 × MS plates in darkness for 3 days. Hypocotyl sections of 4 mm were excised using a razor blade and incubated in darkness on growth medium (10 mM KCl, 1 mM MES-KOH [pH 6.0], and 0.8% [w/v] agar) for 0.5-2 hours to deplete endogenous auxin. Chemical treatment was performed by transferring the hypocotyl sections to growth medium containing compounds indicated in corresponding figures. The length of hypocotyl sections was measured using ImageJ software. The average values were calculated from 15 hypocotyls. Experiments were repeated at least three times. The amount of plasma membrane H+-ATPase and the phosphorylation level of its penultimate Thr residue (T947) in the hypocotyl sections were determined as descried previously19. Briefly, 15 hypocotyl segments were ground in liquid nitrogen and solubilized with SDS-sample buffer. The proteins were separated by SDS-PAGE. Immunoblotting was performed using specific antibody against the catalytic domain of AHA2 and also that against phosphorylated T947 of AHA242. Immunoreactive bands were detected as described in the section above.
Supplementary Material
Acknowledgments
We thank Mark Estelle for inspiring us and providing tir1-1 afb2-3 seeds; Hidehiro Fukaki for slr-1 seeds; Yeast Genetics Resource Center and Yuichi Tsuchiya for Y2H vectors; Peptide/protein center at WPI-ITbM for biotinyl peptides; Jeffery Bode for his visionary comments during the conceptualization of the project; Jennifer Nemhauser and Robert Cleland for commenting on the manuscript. KUT dedicates this manuscript to Robert Cleland, who proposed the acid growth theory in 1970, for his continued mentorship at the Univ. Washington. This work was funded by MEXT/JSPS KAKENHI (JP26291057 JP16H01237 and JP17H06476 to KUT; JP16H01462, JP17H03695 and JP17KT0017 to NU; JP26440140 to KT; JP15H05956 to TK; JP17H06350 to SH; JP16H01472 and S1511023 to SK), Japan Science and Technology Agency (PRESTO, JPMJPR15Q9 to SH; ERATO, JPMJER1302 to KI), Howard Hughes Medical Institute (HHMI) and Gordon and Betty Moore Foundation (GBMF3035) to KUT. SH is a JST PRESTO investigator, KI is a JST ERATO investigator and KUT is an HHMI-GBMF Investigator and Univ. Washington Endowed Distinguished Professor of Biology.
Footnotes
Accession Number
All primary sequence reads for the RNA-seq analysis have been deposited to DDBJ (Accession Number: DRA005718).
Author Contributions
KUT conceived the project; NU, SH, KT and KUT designed research; NU and RI performed molecular cloning, Y2H assays, transgenic plants generation; NU, RI, and KT conducted qRT-PCR and phenotypic characterization; SH, HZ, RY, MY performed synthesis and NMR analyses of auxin analogs; KT performed biochemical analyses; SK performed NGS, and SK, NU and TAE performed bioinformatics; TAE developed Bobbin Dendrogram Plot Generator; MN and YT provided reagents; NU, SH, KT, TK, KI, and KUT analyzed data; KUT performed R-drc analysis; KUT, NU, KT, and SH wrote the manuscript; all authors read and approved the manuscript.
Competing financial interests
The cvxIAA-ccvTIR1 system reported here has been filed for US provisional patent (No. 62/468642) where NU, RI, KI, SH, and KUT appear as inventors.
References
- 1.Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teale WD, Paponov IA, Palme K. Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol. 2006;7:847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- 3.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 4.Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 1999;13:1678–1691. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 6.Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 7.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 8.Calderón-Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, Zheng N, Napier R, Kepinski S, Estelle M. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol. 2012;8:477–485. doi: 10.1038/nchembio.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah K, Liu Y, Deirmengian C, Shokat KM. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc Natl Acad Sci U S A. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 11.Baud MG, Lin-Shiao E, Cardote T, Tallant C, Pschibul A, Chan KH, Zengerle M, Garcia JR, Kwan TT, Ferguson FM, Ciulli A. Chemical biology. A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes. Science. 2014;346:638–641. doi: 10.1126/science.1249830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato H, Ishizaki K, Kouno M, Shirakawa M, Bowman JL, Nishihama R, Kohchi T. Auxin-mediated transcriptional system with a minimal set of components is critical for morphogenesis through the life cycle in Marchantia polymorpha. PLoS Genet. 2015;11:e1005084. doi: 10.1371/journal.pgen.1005084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prigge MJ, Lavy M, Ashton NW, Estelle M. Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr Biol. 2010;20:1907–1912. doi: 10.1016/j.cub.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 14.Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, He SY, Rizo J, Howe GA, Zheng N. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 16.Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 17.Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L. Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 2013;18:450–458. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi K, Hayashi K, Kinoshita T. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 2012;159:632–641. doi: 10.1104/pp.112.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hager A. Role of the plasma membrane H+-ATPase in auxin-induced elongation growth: historical and new aspects. J Plant Res. 2003;116:483–505. doi: 10.1007/s10265-003-0110-x. [DOI] [PubMed] [Google Scholar]
- 21.Schenck D, Christian M, Jones A, Lüthen H. Rapid auxin-induced cell expansion and gene expression: a four-decade-old question revisited. Plant Physiol. 2010;152:1183–1185. doi: 10.1104/pp.109.149591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci U S A. 2009;106:22540–22545. doi: 10.1073/pnas.0911967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh TA, Neal R, Merlo AO, Honma M, Hicks GR, Wolff K, Matsumura W, Davies JP. Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol. 2006;142:542–552. doi: 10.1104/pp.106.085969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fendrych M, Leung J, Friml J. TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. Elife. 2016;5 doi: 10.7554/eLife.19048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012;70:978–990. doi: 10.1111/j.1365-313X.2012.04946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell. 2014;26:2129–2142. doi: 10.1105/tpc.114.126037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rayle DL, Cleland R. Enhancement of wall loosening and elongation by acid solutions. Plant Physiol. 1970;46:250–253. doi: 10.1104/pp.46.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spartz AK, Lor VS, Ren H, Olszewski NE, Miller ND, Wu G, Spalding EP, Gray WM. Constitutive expression of Arabidopsis Small Auxin Up RNA19 (SAUR19) in tomato confers auxin-independent hypocotyl elongation. Plant Physiol. 2016 doi: 10.1104/pp.16.01514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 30.Holland AJ, Fachinetti D, Han JS, Cleveland DW. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc Natl Acad Sci U S A. 2012;109:E3350–3357. doi: 10.1073/pnas.1216880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Ward JD, Cheng Z, Dernburg AF. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C elegans. Development. 2015;142:4374–4384. doi: 10.1242/dev.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169:930–944e922. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Natsume T, Kiyomitsu T, Saga Y, Kanemaki MT. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 2016;15:210–218. doi: 10.1016/j.celrep.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Yu H, Moss BL, Jang SS, Prigge M, Klavins E, Nemhauser JL, Estelle M. Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol. 2013;162:295–303. doi: 10.1104/pp.113.215582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu H, Zhang Y, Moss BL, Bargmann BO, Wang R, Prigge M, Nemhauser JL, Estelle M. Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat Plants. 2015;1:14030. doi: 10.1038/nplants.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nemhauser JL, Torii KU. Plant synthetic biology for molecular engineering of signalling and development. Nat Plants. 2016;2:16010. doi: 10.1038/nplants.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 38.Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savaldi-Goldstein S, Baiga TJ, Pojer F, Dabi T, Butterfield C, Parry G, Santner A, Dharmasiri N, Tao Y, Estelle M, Noel JP, Chory J. New auxin analogs with growth-promoting effects in intact plants reveal a chemical strategy to improve hormone delivery. Proc Natl Acad Sci U S A. 2008;105:15190–15195. doi: 10.1073/pnas.0806324105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estojak J, Brent R, Golemis EA. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritz C, Baty F, Streibig JC, Gerhard D. Dose-Response Analysis Using R. PLoS One. 2015;10:e0146021. doi: 10.1371/journal.pone.0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayashi Y, Nakamura S, Takemiya A, Takahashi Y, Shimazaki K, Kinoshita T. Biochemical characterization of in vitro phosphorylation and dephosphorylation of the plasma membrane H+-ATPase. Plant Cell Physiol. 2010;51:1186–1196. doi: 10.1093/pcp/pcq078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


