Abstract
A recurrent de novo missense variant within the C-terminal Sin3-like domain of ZSWIM6 was previously reported to cause acromelic frontonasal dysostosis (AFND), an autosomal-dominant severe frontonasal and limb malformation syndrome, associated with neurocognitive and motor delay, via a proposed gain-of-function effect. We present detailed phenotypic information on seven unrelated individuals with a recurrent de novo nonsense variant (c.2737C>T [p.Arg913Ter]) in the penultimate exon of ZSWIM6 who have severe-profound intellectual disability and additional central and peripheral nervous system symptoms but an absence of frontonasal or limb malformations. We show that the c.2737C>T variant does not trigger nonsense-mediated decay of the ZSWIM6 mRNA in affected individual-derived cells. This finding supports the existence of a truncated ZSWIM6 protein lacking the Sin3-like domain, which could have a dominant-negative effect. This study builds support for a key role for ZSWIM6 in neuronal development and function, in addition to its putative roles in limb and craniofacial development, and provides a striking example of different variants in the same gene leading to distinct phenotypes.
Keywords: ZSWIM6, intellectual disability, autism, epilepsy, genomics, nonsense-mediated decay, exome sequencing, recurrent, de novo, ubiquitination
Main Text
Advances in high-throughput DNA sequencing combined with databases that allow the sharing of clinical and genotypic information among clinicians and researchers, such as those in the Matchmaker Exchange hub, have changed the landscape of research collaborations leading to accelerated disease variant discovery and validation for neurocognitive disorders during recent years.1, 2 This has led to an explosion in the diagnostic rate for neurocognitive disorders,1 with increasing appreciation of the complexity of clinical presentations of variants even in the same gene.2 In a multicenter study, we discovered a recurrent protein-truncating variant in ZSWIM6 (MIM: 615951) in a cohort of affected individuals with overlapping neurocognitive phenotypes. A recurrent de novo nonsynonymous ZSWIM6 variant (GenBank: NM_020928.1 [ZSWIM6]; c.3487C>T [p.Arg1163Trp]) had previously been reported to cause the rare Mendelian disorder acromelic frontonasal dysostosis (AFND)3, 4 characterized by severe frontonasal dysplasia, tibial hemimelia, pre-axial polydactyly, brain malformations, and severe neurocognitive and motor delay.3, 4 The p.Arg1163Trp variant was postulated to perturb the function of the highly conserved Sin3-like domain at the C terminus of the protein, with three-dimensional modeling suggesting disruption of an interaction surface.4 A gain-of-function mechanism was proposed, based on the observation that affected individuals with chromosomal deletions spanning ZSWIM65 lack the craniofacial and limb abnormalities seen in AFND.4
By sharing variants identified from exome or genome sequencing with Matchmaker Exchange6 or via contact with individual diagnostic laboratories, we identified seven unrelated individuals (four female, three male) with a recurrent single-nucleotide variant in ZSWIM6 that introduced a premature termination codon (PTC) in exon 13 (chr5[GRCh37]: g60837744C>T; GenBank: NM_020928.1; c.2737C>T [p.Arg913Ter]) (Figure 1A). All individuals had a severe-profound intellectual disability (ID)/developmental delay (DD) and additional neurological features but lacked the frontonasal or limb malformations reported in individuals with the p.Arg1163Trp variant. An overview of the clinical data is provided in Table 1, and further clinical details are in Table S3.
Figure 1.
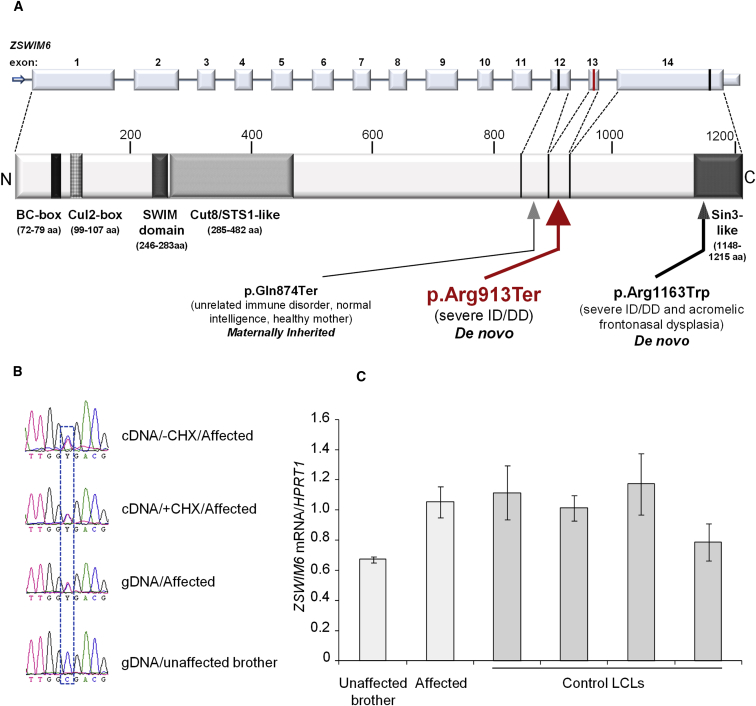
ZSWIM6 Domain Structure and Variants and Molecular Data on mRNA Encoding the p.Arg913Ter Variant
(A) Diagram showing predicted ZSWIM6 protein domains (lower) relative to exonic structure (upper) and positions of the p.Arg913Ter and p.Gln874Ter variants reported here (in red and black, respectively) and the previously reported p.Arg1163Trp recurrent missense variant located within the C-terminal Sin3-like domain (information adapted from Tischfield et al.7). For clarity, the correspondence between the exon boundaries and the protein sequence is depicted only for the final three exons. Introns not drawn to scale.
(B) The mRNA encoding p.Arg913Ter is not degraded by nonsense-mediated decay (NMD) in LCLs. Sequence chromatograms showing normal and p.Arg913Ter-encoding reverse-transcribed mRNAs (cDNA) from LCLs (3 × 106) cultured in the presence or absence of 100 μg/mL NMD inhibitor cycloheximide (CHX) for 6 hr and genomic DNA (gDNA) of the affected individual and his unaffected brother. The cDNA of affected individual 1 and genomic DNA of affected individual 1 and his unaffected brother were PCR amplified using hZSWIM6_2633F and hZSWIM6_2871R (cDNA); and hZSWIM6_gDNA_F and hZSWIM6_gDNA_R (gDNA: Table S2) and subjected to Sanger sequencing. Presence of the variant mRNA in the affected individual’s LCLs cultured without CHX indicates absence of NMD. Normal/variant nucleotides are boxed.
(C) ZSWIM6 expression levels are not reduced in affected individual 1 relative to his unaffected brother and is consistent with four unrelated male control subjects. Total RNA from affected individual 1, his unaffected brother, and four unrelated normal male LCL pellets was extracted using a RNeasy Mini Kit (QIAGEN) and RNase-Free DNase set (QIAGEN) according to the manufacturers’ protocols. The cDNA was synthesized from total RNA using Super Script RT III Reverse Transcription kit (Life Technologies) following the manufacturer’s protocol. ZSWIM6 expression was determined by real-time quantitative PCR from cDNA using SYBR Green reagent (Bio-Rad Laboratories) and hZSWIM6_2633F and hZSWIM6_2871R primers (Table S2) and normalized to HPRT1 expression assayed alongside using HPRT1-F and HPRT1-R primers (Table S2). Each sample was assayed twice independently in triplicate. Error bars indicate standard deviation. The significance was calculated by Student’s t test.
Table 1.
Comparison of Clinical Features of Affected Individuals with the p.Arg913Ter or p.Arg1163Trp ZSWIM6 Variants
| ZSWIM6 Variant/Affected Individual | p.Arg913Ter Affected Individual 1 | p.Arg913Ter Affected Individual 2 | p.Arg913Ter Affected Individual 3 | p.Arg913Ter Affected Individual 4 | p.Arg913Ter Affected Individual 5 | p.Arg913Ter Affected Individual 6 | p.Arg913Ter Affected Individual 7 | Summary p.Arg913Ter Cohort (n = 7) | Summary Postnatal Non-mosaicap.Arg1163Trp Cohort (n = 6) |
|---|---|---|---|---|---|---|---|---|---|
| Method of testing and confirmation | WGS of proband, SS to confirm and segregate in family | Trio WES, SS confirmation | WES of proband, SS to confirm and segregate in family | WES of proband, SS to confirm and segregate in family | WES of proband, SS to confirm and segregate in family | trio WGS, SS to confirm and segregate in family | trio WES, SS confirmation | – | – |
| Inheritance, if known; parental ages at birth of child | not maternal;b paternal 32, maternal 32 | DN; paternal 31, maternal 28 | DN; paternal 34,maternal 25 | DN | DN; paternal 33, maternal 32 | DN; paternal 32, maternal 31 | DN; paternal 30, maternal 29 | 6/7 DN; 1/7 unknown | 4/6 DN; 1 NT; 1/6 inherited from mosaic parent |
| Evidence of mosaicism? (wild-type: mutant reads) | no (32:22) | no (75:62) | no (121/257) | no (99/191) | no (69/165) | no (50:50) | no (24:25) | – | – |
| Age (year) | 16 | 7 | 4 | 5 | 3 | 29 | 29 | 3–29 years | unknown |
| Gender | M | F | F | F | F | M | M | 3/7 male (43%) | 4/6 male (67%) |
| Level of ID | severe-profound | severe | severe | severe | severe | profound | severe | 7/7 severe-profound (100%) | 6/6 severe (100%) |
| OFC | normo-cephaly | progressive microcephaly (−2 SD) | progressive microcephaly | normocephaly | progressive microcephaly | progressive macro-cephaly to 90–97th centile | normo-cephaly | progressive microcephaly 3/7 (43%); progressive macrocephaly 1/7 (14%) | NR |
| Infantile hypotonia/delayed GM milestones? | + | + | + | + | + | + | + | 7/7 (100%) | 6/6 (100%) |
| ASD? | + | − | + | + | + | − | + | 5/7 (71%) | NR |
| Communication | non verbal | non verbal; limited comprehension | gestures; non verbal | few words; PECS | babble and one word (no); starting to use PECS | vocalizes, one sign | short sentences, articulation difficulties | 6/7 non-verbal or only few words (86%) | 2/2 non-verbal (100%) |
| Ambulation | non ambulant (wheelchair); previously ambulant at 2.5 yr with wide unsteady gait | ambulant (from 3 year) with wide-based ataxic gait | ambulant with wide unsteady gait (from 2 year) | ambulant with wide unsteady gait, stroller used for distances | starting to cruise | ambulant with wide-based gait; wheelchair for distances | ambulant with broad-based unsteady gait | 5/7 ambulant (71%) with wide-based gait | 2/2 non ambulant (100%) |
| Temperament/behavior | happy, affectionate, interested in family and TV; hyperactivity | happy; bursts of laughter; hyperactivity | happy; hyperactivity; repetitive behaviors; pica | interactive and sociable | no behavioral concerns | happy, affectionate; loves music and water play | happy; hyperactivity and attention deficit; history of pica | 4/7 hyperactivity (57%) | NR |
| Epilepsy | + (GTC and focal dyscognitive) | – | (under investigation for starting episodes and unusual movements) | – | + (IS: controlled with medication) | + (infrequent GTC and absence from age 9) | + possible absence seizures (no medication) | 4/7 seizure/possible seizure disorder (57%) | 1/6 (17%) |
| Progressive spasticity | + | − | − | + | − | + | − | 3/7 (43%) | NR |
| Movement disorder? | paroxysmal hypertonicity; unusual tongue movements | stereotypical hand movements; midline tongue protrusion; ataxia | paroxysmal hypertonicity; unusual tongue movements; ataxia | body rocking; ataxia | stereotypical hand movements; ataxia | − | ataxia, tongue thrusting, head tics | 6/7 (86%) | NR |
| Ophthalmological features | impairment lateral gaze | strabismus | − | accommodative esotropia | right sided esotropia | − | right sided esotropia | 5/7 (71%) | variable: cataract, glaucoma, myopia, optic nerve hypoplasia |
| Brain MRI | cortical atrophy (13 year) | normal | normal | normal (7 mo) | normal | mild cortical atrophy (22 mo) but later MRI considered normal | normal | cortical atrophy 1/7 (14%) | interhemispheric lipoma 6/6 (100%); abnormal corpus callosum 3/6 (50%); other abnormalities variable |
| Additional neurological features | mixed peripheral neuropathy | – | truncal hypotonia, abnormally high pain threshold | truncal hypotonia | bilateral ankle pronation and toe pointing, torticollis | full neurological examination not possible | abnormally high pain threshold, self-sustained injuries | 5/7 (71%) | NR |
| Gastro-intestinal symptoms | failure to thrive and severe GERD requiring fundoplication; constipation; ulcerative colitis from mid-teens | infantile cow’s milk protein intolerance; recurrent diarrhea and vomiting from 18 mo improved on gluten-free diet | GERD | severe GERD requiring gastrostomy | GERD and failure to thrive | – | intermittent constipation | 6/7 (86%) significant GI symptoms | NR |
| Additional non- neurological features | marked equinovarus deformity of feet; thoracolumbar scoliosis, distinctive facial features | distinctive facial features | otitis media with effusions (requiring tympanostomy); distinctive facial features | premature eruption of teeth; distinctive facial features | distinctive facial features | facial asymmetry; premature eruption of teeth; puberty at 12; male pattern baldness by age 17; distinctive facial features | severe bilateral planovalgus; nocturnal eneuresis; distinctive facial features | – | 5/6 (83%) limb abnormalities; 2/4 males (50%) cryptorchidism; 1/6 (17%) scoliosis; 2/6 (33%) hypopituitarism; 6/6 hyperteloric (100%); 4/6 median facial cleft 67%); 4/6 cleft palate (67%) |
Abbreviations: AFO, ankle foot orthoses; ASD, autism or autism spectrum disorder; DN, de novo; WES, whole-exome sequencing; GM, gross motor; GERD, gastro-esophageal reflux disease; WGS, whole-genome sequencing; GTC, generalized tonic-clonic; ID, intellectual disability; IS, infantile spasms; mo, months; MRI, magnetic resonance imaging; ND, not done; NR, not relevant; NT, not tested; OFC, occipital frontal circumference; PECS, picture exchange communication system; SS, Sanger sequencing; yr, year.
Mildly affected mother with mosaicism and fetus terminated at 20 weeks excluded.
Not inherited from mother, not present in either of two unaffected brothers, father not available for testing.
Genetic studies were approved by local ethics committees and written informed consent was obtained for molecular genetic analysis, functional studies on affected individual-derived cells, and the publication of clinical and radiological data and photographs from participants or their legal guardians. All individuals were undiagnosed prior to exome/genome sequencing despite clinical genetic assessment and prior genetic screening including chromosomal array (details provided in supplementary clinical data, Table S3). Individuals 2–5 and 7 were screened by whole-exome sequencing (WES) using previously described methodology.8, 9, 10 Whole-genome sequencing (WGS) was utilized for individuals 1 and 6. DNA from individual 1 was sequenced on the Illumina HiSeqX platform at the Kinghorn Centre for Clinical Genomics, Sydney, obtaining ∼120 Gb data per sample, equivalent to >30× average coverage. Variant alignment, single nucleotide and short indel variant identification, annotation, and quality assessment was performed as previously described.8 Read alignment bam files were used as input to call structural variation including copy-number variation with ClinSV (unpublished data). Variants were annotated using the ENSEMBL gene set v7511 and previously reported structural variants from the Database of Genomic Variants.12 Candidate variant calls and the corresponding read-alignments were inspected using the Integrative Genomics Viewer. Individual 6 and his unaffected parents underwent WGS as part of the 100,000 Genomes Project. Genomic DNA was processed using a kit (TruSeq DNA PCR-Free Sample Preparation; Illumina) and sequenced using a high-throughput sequencing platform (HiSeq X Ten; Illumina), generating minimum coverage of 15 times for more than 97% of the callable autosomal genome. Readings were aligned to build GRCh37 of the human genome using the Isaac aligner (Illumina). Single-nucleotide variants and indels (insertions or deletions) were identified using Platypus software (v.0.8.1; Wellcome Trust Centre for Human Genetics) and annotated using Cellbase software. Variant filtering was performed using minor allele frequency in publicly available and in-house datasets, predicted protein effect, and familial segregation. Surviving variants were prioritized using the Intellectual Disability v.1.2 prespecified virtual gene panel from PanelApp, which includes ZSWIM6. Allelic state was required to match the curated mode of inheritance for variants in panel genes.
In all case subjects, the c.2737C>T variant was confirmed by Sanger sequencing using standard methodology, and segregation analysis was consistent with the variant being de novo for individuals 2 to 7. For individual 1 the variant was not present in his unaffected mother or either of his two unaffected brothers, while his unaffected father’s DNA was unavailable for testing. Wild-type: variant read counts (Table 1) and analysis of the dideoxy sequencing traces in forward and reverse sequences was not supportive of mosaicism in any family. For individuals 1 and 3–5, in whom only the proband has WES/WGS, no other potentially causative variants were found to match the curated mode of inheritance after segregation studies. For individuals 2, 6, and 7 where a trio sequencing approach was undertaken, the other identified de novo variants and the rationale for considering them unlikely to be contributory to the individual’s phenotype is described in Table S1.
All affected individuals had a severe-to-profound ID and required early developmental interventions or a special school. The two oldest individuals (individuals 6 and 7) live in fully supported independent accommodation. A particular deficit in verbal communication was noted for all individuals. Five of the affected individuals (71%) met diagnostic criteria for autism or autism spectrum disorder: this diagnosis was based on features of repetitive movements and limited play and communication. All affected individuals were noted to be content and socially responsive, showing affection to and interest in their families. Hyperactivity was noted in four of the seven (57%).
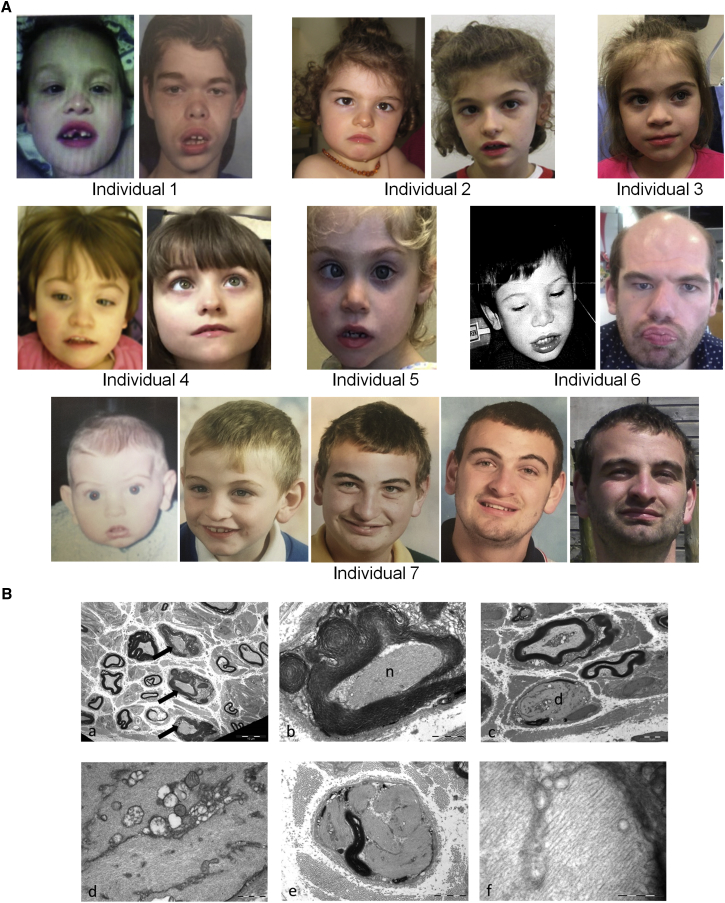
Additional neurological features were described in all affected individuals. Low truncal tone, delayed motor milestones in the first year of life, and delayed onset of walking (age range to achieve independent ambulation: 2 to 5 years) was universal. When ambulant, all had a wide-based, unsteady gait. Progressive neurological features were described: individuals 1, 4, and 6 had progressive spasticity, individual 1 lost independent ambulation in mid-childhood, and individuals 4 and 6 require a wheelchair or stroller for mobility outside of the home. In addition, individual 1 has a progressive neuropathy and in later childhood lost strength and reflexes in his distal lower limbs. No sequence or structural candidate variants in known neuropathy genes were identified on genome analysis for this individual (Table S3). Nerve conduction studies for individual 1 demonstrated mixed axonal and demyelinating features, and light and electron microscopy of a sural nerve biopsy (Figure 2B) showed a reduction in myelinated fiber density, reduction in the thickness of myelination in many myelinated axons, and evidence of abnormal accumulation of neurofilaments (Figure 2B). These findings are reminiscent of those seen in disorders of ubiquitination such as giant axonal neuropathy 1 (GAN1 [MIM: 25685021]) and also in certain toxic neuropathies such as those secondary to exposure to n-Hexane and acrylamide. This similarity in pathology between a known disorder of ubiquitination and ZSWIM6 p.Arg913Ter-related neuropathy is intriguing given a postulated role of ZSWIM6 in the ubiquitin pathway.7 Individuals 3 and 7 have low sensitivity to pain, leading to self-sustained injuries. No other individuals in the cohort have had nerve conduction or nerve biopsy studies and detailed neurological assessments to fully screen for neuropathies were not possible in the majority of individuals due to the severity of their neurocognitive disabilities or young age. Unusual movements have also been described in six of the seven (86%) affected individuals: four have paroxysmal tongue movements (tongue thrusting or tremor), two have stereotypical hand movements, one has head tics, and two have episodes of whole-body hypertonicity, which was not consistent with a seizure. Four (57%) had strabismus and individual 1 had limitation of the extremes of gaze, although none had a confirmed visual or hearing impairment. Three of the individuals developed a seizure disorder which could be controlled on medication, while individual 7 had a suspected seizure disorder that has never required antiepileptic medication. Brain MRI, performed in all individuals, was reported as normal in five; mild cortical atrophy was reported only for individual 1 (Figure S4).
Figure 2.
Facial and Neurological Features of Individuals with the p.Arg913Ter ZSWIM6 Variant
(A) Facial features of individuals 1 to 7. In early childhood frequent features include fine, arched eyebrows, short nose with depressed bridge and blunt tip, broad columella, thick everted lower lip vermillion, widely spaced teeth, downturned corners to mouth, mouth often held open, and esotropia. Individuals 1 and 6 were noted with age to develop prominent forehead and supraorbital ridges, thick eyebrows, and thicker everted lower lip vermillion, reminiscent of the facial appearance of older individuals with Coffin-Lowry syndrome. This was not the case for individual 7.
(B) Sural nerve biopsy from individual 1 demonstrating demyelination and abnormal accumulation of neurofilaments within numerous myelinated axons. (a) Low-power electron micrograph showing three fibers containing proliferated neurofilaments (arrows) among thinly myelinated fibers which demonstrate the normal density of axonal filaments. (b) Accumulated neurofilaments [n] in an axon with thick and folded myelin. (c) Demyelinated axon [d] packed with neurofilaments that displace other axonal organelles. (d) High magnification of (c) showing mitochondria and other organelles in septa between neurofilaments. (e) Demyelinated axon with proliferated neurofilaments in distinct whorled bundles. (f) High magnification of proliferated neurofilament bundle. The individual filaments measure 9–15 nm in diameter.
Many of the extra-neurocognitive features in the cohort are commonly seen in children with ID/DD due to other causes. None had structural congenital anomalies. Feeding difficulties, failure to thrive, or gastro-esophageal reflux were reported as significant for six of the seven, with two individuals requiring fundoplication/gastrostomy. Growth parameters were generally within the normal range; however, three individuals (2, 3, and 5) had progressive microcephaly whereas one (individual 6) had progressive macrocephaly. When assessed as a group, some similarity in facial features was apparent, particularly at younger ages (Figure 2A). None of the affected individuals with the p.Arg913Ter variant had the midline clefting, parietal foramina, or extreme hypertelorism characteristic of affected individuals with the p.Arg1163Trp variant.3, 4 Two of the three older male individuals (individuals 1 and 6) had progressive coarsening of their facial features, despite not being treated with an antiepileptic medication that could cause this, and in both individuals a differential diagnosis of Coffin-Lowry syndrome had been considered (based on facial and neurological features without the characteristic hand and foot features), but no pathogenic variants in RPS6KA3 had been identified. In view of this, we screened a cohort of 90 individuals with facial features suggestive of Coffin-Lowry syndrome, for whom screening had previously failed to detect a RPS6KA3 variant, for the recurrent c.2737C>T variant using a PCR and HphI restriction site digestion screen (see Figure S2). However, none of these 90 individuals carried the c.2737C>T variant. We also used the PCR and HphI restriction site digestion approach for screening this variant in an additional cohort of 672 individuals who were recruited from South Australia with a diagnosis of developmental delay, ID, or autism, and who had normal fragile X and chromosomal microarray, but no further individuals carrying the variant were identified.
The severe neurodevelopmental phenotype in individuals harboring the p.Arg913Ter variant is consistent with several lines of evidence showing neurodevelopmental roles for ZSWIM6. ZSWIM6 is expressed in a localized and developmentally regulated manner in the brain. In zebrafish larvae, zswim6 is relatively highly expressed in regions of the telencephalon, midbrain, hindbrain, and retina.4 In the embryonic mouse, Zswim6 is initially expressed in the ganglionic eminences and subsequently also in the cortical plate, developing amygdala, and portions of the thalamus and hypothalamus. Postnatally, telencephalic expression becomes more restricted to the striatum.7 A similar distribution has been noted during human brain development.7 Zswim6 knockout mice have abnormal neocortical and striatal development with reduced cortical and striatal volumes, and alterations in the number and structure of medium spiny neurons in the striatum.7 These morphological changes are accompanied by alterations in motor control and behavior, including impaired learning on the accelerating rotarod, hyperactivity, and an increase in stereotypical repetitive movements.7 Finally, genome-wide association (GWA) studies provide further support for a role for ZSWIM6 in brain development and function. ZSWIM6 has been reported as one of the top 15 candidate genes implicated most consistently across various GWA analyses with variation in educational attainment.13 ZSWIM6 was also included within a chromosomal region that reached genome-wide significance for schizophrenia.14, 15
ZSWIM6, at 5q12.1, has 14 exons and encodes a 133.5 kDa protein belonging to a recently described group of proteins characterized by a SWIM-type zinc finger domain16 (found in SWI2/SNF2 and MuDR proteins) (Figure 1A). SWIM domains are present in diverse archaeal, bacterial, and eukaryotic proteins, including bacterial ATPases of the SWI2/SNF2 family and vertebrate MEK kinase-1, and have putative roles in DNA and protein binding.16 In addition to the SWIM domain (246–283 aa), ZSWIM6 (GenBank: NP_065979.1) contains a BC-box (72–79 aa), Cul2-box (99–107 aa), a Cut8/STS1-like domain (285–482 aa), and a C-terminal Sin-3 like domain (1,148–1,215 aa)4 (Figure 1A). The c.2737C>T variant lies within the penultimate exon 13, 48 base pairs (bp) upstream of the last exon/exon junction (Figure 1A), suggesting that the ZSWIM6 PTC-encoding mRNA may escape nonsense-mediated decay (NMD). We therefore sought to confirm this in cells from the affected individuals. Lymphoblastoid cell lines (LCLs) from affected individual 1 and control subjects were generated using standard methods. We cultured LCLs from affected individual 1 in the presence or absence of cycloheximide, a translational inhibitor and suppressor of NMD. Sanger sequencing of ZSWIM6 cDNA revealed that the c.2737C>T transcript is not subjected to NMD (Figure 1B). This was consistent with real-time quantitative PCR data that showed no significant difference in ZSWIM6 expression between the affected individual and his unaffected brother and four unrelated male control LCLs (Figure 1C). Sequencing of cDNA generated from peripheral blood of individual 2 also suggested that the variant does not result in NMD (Figure S1). We could not detect the full-length (in LCLs of unaffected brother of individual 1) or truncated (in affected individual 1) ZSWIM6 protein using an anti-ZSWIM6 antibody raised against amino acids 634–696 (Sigma HPA035938). Because an alternative commercial antibody against the N-terminal region of ZSWIM6 protein is unavailable, we could not confirm the presence of a truncated ZSWIM6 protein, which we assume is translated from the mRNA encoding the PTC.
This case series indicates that the recurrent de novo nonsense variant c.2737C>T in ZSWIM6 should be considered causal of severe-profound autosomal-dominant ID associated with a wide-based ataxic gait, limited communication, happy disposition and, in some, significant gastrointestinal symptoms, repetitive behaviors on the autistic spectrum, distinctive facial features, spasticity, and strabismus. In two of the older individuals, an alternative diagnosis of Coffin-Lowry syndrome had been considered, and the constellation of happy disposition, ataxia, and seizures (in 3/7) and microcephaly (in 3/7) means that this condition could also be added to a growing list of differential diagnoses for Angelman syndrome.17 In one older individual, a debilitating progressive peripheral neuropathy is a striking feature, and we would recommend surveillance of affected individuals for this potential complication, to determine how frequently this is part of the clinical condition.
This report expands the phenotypes associated with pathogenic variants in ZSWIM6. Previously, the recurrent de novo p.Arg1163Trp variant located within the C-terminal Sin3-like domain was implicated in AFND.3, 4 Although the p.Arg913Ter cohort shares the severe-profound ID of affected individuals with AFND, all lack the characteristic facial clefting, significant hypertelorism, interhemispheric lipomas, and limb anomalies of the latter.
The molecular and cellular mechanisms explaining how the two ZSWIM6 variants cause distinct phenotypes remain to be determined. Hypothesizing that the recurrent c.2737C>T (p.Arg913Ter) variant is not a simple haploinsufficiency allele, we examined the frequency of ZSWIM6 loss-of-function (LoF) variants or chromosomal deletions in case and control populations. In the Database of Genomic Variants and the Copy Number Variation Morbidity map available through the UCSC genome browser, the only deletions affecting coding sequence correspond to a small, highly GC-rich region containing exon 1 and are therefore likely false positives (Figures S3A and S3B). The only LoF variants listed in the gnomAD database18 fall within a region of very poor coverage in a highly GC-rich and repetitive region of exon 1 (Figure S3C); all are indels and visual inspection suggests these are also artifactual. No individuals in our in-house cohorts, DECIPHER, or the literature have chromosomal deletions confined to ZSWIM6. The shortest deletion involving ZSWIM6 in DECIPHER (carried by affected individual 267601) is a 2.91 Mb region that also contains six other protein-coding genes including the OMIM morbid listed gene KIF2A, in which monoallelic missense variants have been reported to cause malformations of cortical development (MIM: 615411).19 The phenotype in this affected individual is mild ID in a healthy, sociable girl with short stature and microcephaly (D. Fitzpatrick, personal communication). ZSWIM6 is one of the 12 genes in the 2.63 Mb shortest region of overlap of microdeletions at 5q12.1, which are characterized by neurocognitive disorders.5 Given the number of genes in the above two intervals, the contribution of ZSWIM6 haploinsufficiency to the phenotype remains uncertain. Analysis of more than 10,000 whole exomes performed at the Institut Imagine indicated one further LoF variant in ZSWIM6, c.2620C>T (p.Gln874Ter) (Figure 1A), in a proband who died of an immunodeficiency phenotype caused by a homozygous recessive mutation in IFNGR2 (MIM: 147569; GenBank: NM_005534.3; c.679G>A [p.Gly227Arg]). Sanger sequencing indicated that this heterozygous ZSWIM6 variant was inherited from the proband’s mother; neither individual had significant neurological deficits. c.2620C>T, falling within exon 12, is predicted to result in NMD of its mRNA. Unfortunately, this could not be tested experimentally. Available data suggest that a heterozygous loss of ZSWIM6 does not cause ID. As the c.2737C>T ZSWIM6 variant mRNA escapes NMD, cellular consequences may be due to a dominant-negative effect caused by the presence of truncated ZSWIM6 protein. Several human diseases are caused by clustered C-terminal protein-truncating mutations.20, 21 The variant resulting in p.Arg913Ter falls on one of only three CpG dinucleotides in the penultimate two exons where a C to T transition could result in a premature stop codon. The most terminal of these sites, Arg1176, would also likely escape nonsense-mediated decay but has not been identified in any of these cohorts. p.Arg913Ter would result in ZSWIM6 lacking the C-terminal Sin3-like domain (Figure 1A). In comparison, the p.Arg1163Trp variant that causes AFND lies within the Sin3-like domain and is postulated to have a gain-of-function effect.4 The Sin3-like domain has similarity to the four paired amphipathic alpha-helix (PAH) motifs that are a prominent feature of the protein encoded by yeast Sin3 and its mammalian paralogs SIN3A and SIN3B22 that code for part of the key SIN3-HDAC-MeCP2 transcriptional co-repressor complex involved in gene regulation in neuronal progenitors.23 The biochemical function of the ZSWIM6 Sin3-like domain and the divergent molecular consequences of its truncation (p.Arg913Ter) or local surface alteration (p.Arg1163Trp) will be an important area for future investigations.
Frequency data were available from the Baylor Genetics and DDD cohorts: the recurrent p.Arg913Ter variant was found in 3/6,100 case subjects with a neurological phenotype who had exome sequencing through Baylor Genetics, and 1/5,873 case subjects with pediatric-onset intellectual disability or global developmental delay who had exome sequencing through the DDD cohort, suggesting a frequency of 1.7–4.9 × 10−4 in individuals with neurocognitive/neurological disorders. The variant was not present in any of the 123,136 exomes and 15,496 genomes listed in the gnomAD database, which is depleted of individuals and their first-degree relatives with severe pediatric disease. Although this variant is rare, we postulate that it is likely to be present in more affected individuals who have already had diagnostic WES/WGS, as the lack of the distinctive AFND phenotype reported for the separate recurrent p.Arg1163Trp may have meant that the variant was reported as a variant of uncertain significance.
In summary, we have described a syndromic neurocognitive disorder due to a recurrent de novo ZSWIM6 p.Arg913Ter variant that is distinct from the multiple congenital anomaly disorder AFND, caused by the recurrent p.Arg1163Trp variant within the C-terminal Sin3-like domain. The distinct clinical phenotypes likely reflect differing molecular mechanisms: the p.Arg913Ter variant may result in a dominant-negative effect due to production of a truncated ZSWIM6 protein that lacks the Sin-3-like domain, whereas the p.Arg1163Trp variant may result in a gain of function. Such distinctive genotype-phenotype relationships for dominant conditions are still relatively rare in clinical genetics; examples include the disparate conditions caused by variants in RET (e.g., multiple endocrine neoplasia [MIM: 171300] and central hypoventilation syndrome [MIM: 209880]) and FGFR1 (Kallman syndrome [MIM: 147950] and Pfeiffer syndrome [MIM: 101600]), although this may be due to a bias in WES/WGS data interpretation. Our study further highlights genotype-phenotype complexity2 and serves as a cautionary tale to clinicians and pathologists analyzing high-throughput sequencing data not to discount the possibility that distinctive clinical phenotypes may result from different variants within the same Mendelian gene. Our study also highlights the benefits of platforms such as Matchmaker Exchange that can connect clinicians and genomicists and result in the rapid building of cohorts of affected individuals to delineate the natural history of recently described genetic conditions.2 The consistency of a severe ID phenotype across both p.Arg913Ter and p.Arg1163Trp cohorts builds on accumulating evidence that ZSWIM6 has critical roles in neuronal development and function and that this gene is a valid target for ongoing basic and clinical neuroscience research.
Acknowledgments
The authors thank the affected individuals and their families for participation in this study. We would like to thank the Greenwood Centre, South Carolina, USA, and in particular the Director of Molecular Studies Professor Charles Schwartz for arranging the sending of DNA of individuals with a Coffin-Lowry phenotype for investigation of the recurrent ZSWIM6 variant. J.G. was supported by NHMRC Program Grant 1091593, Senior Research Fellowship 1041920, and Channel 7 Children’s Research Foundation. This research was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the United Kingdom Department of Health). The 100,000 Genomes Project is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK, and the Medical Research Council have also funded research infrastructure. The DDD Study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute (grant number WT098051). The views expressed in this publication are those of the authors and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). We acknowledge the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network. C.T.G. and J.A. were supported by funding from the Agence Nationale de la Recherche (ANR-10-IAHU-01, CranioRespiro). C.B. received support from the Fondation Maladies Rares. A.T. was supported by the Medical Genetics Research Fellowship Program NIH/NIGMS, grant 5T32GM007526-40. The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing done at Baylor Genetics Laboratory.
Published: November 30, 2017
Footnotes
Supplemental Data include three figures, three tables, and Supplemental Subjects and Methods and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.10.009.
Contributor Information
Michael Field, Email: mike.field@health.nsw.gov.au.
Jozef Gecz, Email: jozef.gecz@adelaide.edu.au.
Accession Numbers
The accession number for the recurrent variant in ZSWIM6 reported in this study is ClinVar: SCV000588874.
Web Resources
Cellbase, https://github.com/opencb/cellbase
ClinSV, https://github.com/KCCG/ClinSV
Database of Genomic Variants (DGV), http://dgv.tcag.ca/dgv/app/home
DECIPHER, https://decipher.sanger.ac.uk/
Ensembl Genome Browser, http://www.ensembl.org/index.html
ExAC Browser, http://exac.broadinstitute.org/
GeneReviews, Kuhlenbaumer, G., Timmerman, V., and Bomont, P. (1993). Giant axonal neuropathy. https://www.ncbi.nlm.nih.gov/books/NBK1136/
gnomAD Browser, http://gnomad.broadinstitute.org/
Matchmaker Exchange, http://www.matchmakerexchange.org/
OMIM, http://www.omim.org/
PanelApp, https://panelapp.genomicsengland.co.uk/
Platypus software, http://www.well.ox.ac.uk/platypus
PROVEAN, http://provean.jcvi.org
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
Abbreviations: ABA, applied behavioral analysis; DD, developmental delay; DQ, developmental quotient; GDD, global developmental delay; ID, intellectual disability; mo, month; yr, year.
References
- 1.Vissers L.E., Gilissen C., Veltman J.A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 2.Boycott K.M., Rath A., Chong J.X., Hartley T., Alkuraya F.S., Baynam G., Brookes A.J., Brudno M., Carracedo A., den Dunnen J.T. International cooperation to enable the diagnosis of all rare genetic diseases. Am. J. Hum. Genet. 2017;100:695–705. doi: 10.1016/j.ajhg.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Twigg S.R., Ousager L.B., Miller K.A., Zhou Y., Elalaoui S.C., Sefiani A., Bak G.S., Hove H., Hansen L.K., Fagerberg C.R. Acromelic frontonasal dysostosis and ZSWIM6 mutation: phenotypic spectrum and mosaicism. Clin. Genet. 2016;90:270–275. doi: 10.1111/cge.12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith J.D., Hing A.V., Clarke C.M., Johnson N.M., Perez F.A., Park S.S., Horst J.A., Mecham B., Maves L., Nickerson D.A., Cunningham M.L., University of Washington Center for Mendelian Genomics Exome sequencing identifies a recurrent de novo ZSWIM6 mutation associated with acromelic frontonasal dysostosis. Am. J. Hum. Genet. 2014;95:235–240. doi: 10.1016/j.ajhg.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaillard S., Andrieux J., Plessis G., Krepischi A.C., Lucas J., David V., Le Brun M., Bertola D.R., David A., Belaud-Rotureau M.A. 5q12.1 deletion: delineation of a phenotype including mental retardation and ocular defects. Am. J. Med. Genet. A. 2011;155A:725–731. doi: 10.1002/ajmg.a.33758. [DOI] [PubMed] [Google Scholar]
- 6.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., Brunner H.G., Buske O.J., Carey K., Doll C. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tischfield D.J., Saraswat D.K., Furash A., Fowler S.C., Fuccillo M.V., Anderson S.A. Loss of the neurodevelopmental gene Zswim6 alters striatal morphology and motor regulation. Neurobiol. Dis. 2017;103:174–183. doi: 10.1016/j.nbd.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poirier K., Hubert L., Viot G., Rio M., Billuart P., Besmond C., Bienvenu T. CSNK2B splice site mutations in patients cause intellectual disability with or without myoclonic epilepsy. Hum. Mutat. 2017;38:932–941. doi: 10.1002/humu.23270. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deciphering Developmental Disorders S., Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yates A., Akanni W., Amode M.R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L. Ensembl 2016. Nucleic Acids Res. 2016;44(D1):D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacDonald J.R., Ziman R., Yuen R.K., Feuk L., Scherer S.W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okbay A., Beauchamp J.P., Fontana M.A., Lee J.J., Pers T.H., Rietveld C.A., Turley P., Chen G.B., Emilsson V., Meddens S.F., LifeLines Cohort Study Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ripke S., O’Dushlaine C., Chambert K., Moran J.L., Kähler A.K., Akterin S., Bergen S.E., Collins A.L., Crowley J.J., Fromer M., Multicenter Genetic Studies of Schizophrenia Consortium. Psychosis Endophenotypes International Consortium. Wellcome Trust Case Control Consortium 2 Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova K.S., Aravind L., Koonin E.V. SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem. Sci. 2002;27:384–386. doi: 10.1016/s0968-0004(02)02140-0. [DOI] [PubMed] [Google Scholar]
- 17.Tan W.H., Bird L.M., Thibert R.L., Williams C.A. If not Angelman, what is it? A review of Angelman-like syndromes. Am. J. Med. Genet. A. 2014;164A:975–992. doi: 10.1002/ajmg.a.36416. [DOI] [PubMed] [Google Scholar]
- 18.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavallin M., Bijlsma E.K., El Morjani A., Moutton S., Peeters E.A., Maillard C., Pedespan J.M., Guerrot A.M., Drouin-Garaud V., Coubes C. Recurrent KIF2A mutations are responsible for classic lissencephaly. Neurogenetics. 2017;18:73–79. doi: 10.1007/s10048-016-0499-8. [DOI] [PubMed] [Google Scholar]
- 20.Jansen S., Geuer S., Pfundt R., Brough R., Ghongane P., Herkert J.C., Marco E.J., Willemsen M.H., Kleefstra T., Hannibal M., Deciphering Developmental Disorders Study De novo truncating mutations in the last and penultimate exons of PPM1D cause an intellectual disability syndrome. Am. J. Hum. Genet. 2017;100:650–658. doi: 10.1016/j.ajhg.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White J.J., Mazzeu J.F., Hoischen A., Bayram Y., Withers M., Gezdirici A., Kimonis V., Steehouwer M., Jhangiani S.N., Muzny D.M., Baylor-Hopkins Center for Mendelian Genomics DVL3 alleles resulting in a -1 frameshift of the last exon mediate autosomal-dominant Robinow syndrome. Am. J. Hum. Genet. 2016;98:553–561. doi: 10.1016/j.ajhg.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grzenda A., Lomberk G., Zhang J.S., Urrutia R. Sin3: master scaffold and transcriptional corepressor. Biochim. Biophys. Acta. 2009;1789:443–450. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witteveen J.S., Willemsen M.H., Dombroski T.C., van Bakel N.H., Nillesen W.M., van Hulten J.A., Jansen E.J., Verkaik D., Veenstra-Knol H.E., van Ravenswaaij-Arts C.M. Haploinsufficiency of MeCP2-interacting transcriptional co-repressor SIN3A causes mild intellectual disability by affecting the development of cortical integrity. Nat. Genet. 2016;48:877–887. doi: 10.1038/ng.3619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Abbreviations: ABA, applied behavioral analysis; DD, developmental delay; DQ, developmental quotient; GDD, global developmental delay; ID, intellectual disability; mo, month; yr, year.