Abstract
Zellweger spectrum disorders (ZSDs) are autosomal-recessive disorders that are caused by defects in peroxisome biogenesis due to bi-allelic mutations in any of 13 different PEX genes. Here, we identified seven unrelated individuals affected with an apparent dominant ZSD in whom a heterozygous mutant PEX6 allele (c.2578C>T [p.Arg860Trp]) was overrepresented due to allelic expression imbalance (AEI). We demonstrated that AEI of PEX6 is a common phenomenon and is correlated with heterozygosity for a frequent variant in the 3′ untranslated region (UTR) of the mutant allele, which disrupts the most distal of two polyadenylation sites. Asymptomatic parents, who were heterozygous for PEX c.2578C>T, did not show AEI and were homozygous for the 3′ UTR variant. Overexpression models confirmed that the overrepresentation of the pathogenic PEX6 c.2578T variant compared to wild-type PEX6 c.2578C results in a peroxisome biogenesis defect and thus constitutes the cause of disease in the affected individuals. AEI promoting the overrepresentation of a mutant allele might also play a role in other autosomal-recessive disorders, in which only one heterozygous pathogenic variant is identified.
Keywords: peroxisome, peroxisome biogenesis disorder, peroxisomal disorder, metabolic, dominant-negative, PEX1, PEX6
Introduction
Peroxisomes are essential organelles involved in multiple metabolic pathways, such as the β-oxidation of branched-chain fatty acids and very long-chain fatty acids (VLCFAs), and the synthesis of plasmalogens and primary bile acids.1 Defects in genes encoding peroxisomal proteins result in various peroxisomal diseases, including the autosomal-recessive Zellweger spectrum disorders (ZSDs). ZSDs are multisystemic diseases and clinical symptoms can be mild to early lethal, ranging from mild neurosensory deficits to profound neonatal hypotonia and liver dysfunction.2 They are caused by bi-allelic mutations in any of 13 different PEX genes, which encode proteins involved in peroxisome biogenesis, including the import of peroxisomal proteins.1 Of these, PEX1 (∼60% [MIM: 602136]) and PEX6 (∼15% [MIM: 601498]) are most commonly defective.3 PEX1 and PEX6 both encode AAA+ ATPases, which form hetero-hexameric double-ring complexes composed of equal number of subunits.4, 5 They are anchored to the cytosolic face of the peroxisomal membrane by the interaction of PEX6 with the peroxisomal membrane protein PEX26. PEX1-PEX6 complexes facilitate the export of the peroxisomal matrix protein receptor PEX5 back into the cytosol after PEX5 has delivered its cargo to the peroxisome (Figure 1A).5 Since PEX5 export is crucial for peroxisomal matrix protein import,6 defects in PEX1 or PEX6 can prevent this import and consequently affect peroxisome-dependent metabolic pathways. This results in characteristic accumulations or shortages of metabolites, the degradation or synthesis of which depend on these pathways, such as the accumulation of VLCFAs.2
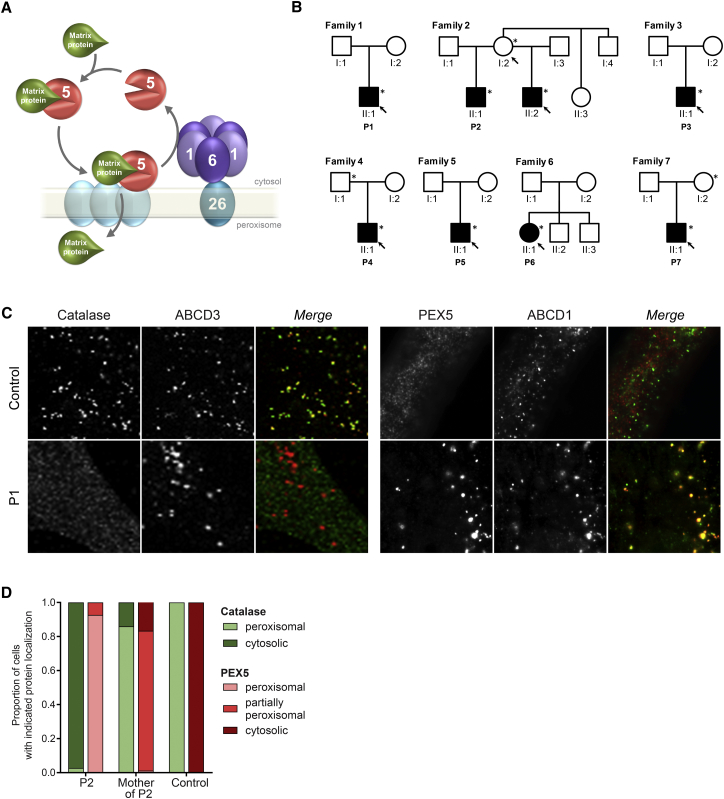
Figure 1.
PEX6 Defect in Individuals with ZSD
(A) Schematic presentation of the role of the PEX1-PEX6 complex in the import of peroxisomal matrix proteins into peroxisomes. The peroxisomal protein receptor PEX5 binds matrix proteins, such as catalase, in the cytosol and transports these to the peroxisome, where the receptor docks at PEX proteins in the peroxisomal membrane and releases the matrix proteins into the matrix. After this, PEX5 is exported back into the cytosol through the action of the hexameric PEX1-PEX6 complex, which is anchored to the peroxisome via interaction of PEX6 with PEX26.
(B) Pedigrees of seven affected families with the PEX6 c.2578C>T variant (only relevant members depicted). Filled symbols represent individuals suffering from a ZSD, asterisks indicate individuals heterozygous for the PEX6 c.2578C>T variant, and arrows indicate family members from whom primary skin fibroblasts were available for functional studies.
(C and D) Immunofluorescence microscopy assays to determine the subcellular localization of peroxisomal matrix protein catalase (green) and receptor PEX5 (red) together with peroxisomal membrane proteins ABCD3 (red) or ABCD1 (green), respectively.
(C) Representative microscopy images of cells of individual P1 and a control individual. In control cells catalase is peroxisomal (left) and PEX5 cytosolic (right), but in the cells of the affected individuals catalase is mislocalized to the cytosol and PEX5 to peroxisomes, demonstrating a severe defect in the import of the peroxisomal matrix proteins and the export of PEX5 in the cells of affected individuals (see Figure S1 for images of cells from other individuals).
(D) Relative subcellular localization of catalase and PEX5 in cells of individual P2 and his asymptomatic mother, demonstrating only a mild defect in cells of the asymptomatic mother of individual P2 (see also Figures S1 and S6 for additional images).
We report seven unrelated individuals and one half-brother with an apparent dominant ZSD, in whom we identified only one single heterozygous pathogenic variant in PEX6, the relative expression of which was increased due to allelic expression imbalance (AEI). The AEI-induced overrepresentation of the mutated PEX6 protein impairs the function of the PEX1-PEX6 complex, which results in a defective import of peroxisomal proteins and thus causes the clinical manifestations in the affected individuals. AEI-induced overrepresentation of a mutant allele might also play a role in other autosomal-recessive disorders, in which only one heterozygous pathogenic variant is identified.
Material and Methods
Clinical Information
All subjects or their legal representatives provided written informed consent for this study. The procedures followed were in accordance with the ethical standards of the responsible committees on human experimentation (institutional and national). An overview of the main clinical features of the eight affected individuals (seven unrelated individuals and one half-brother) is presented in Table 1. All individuals showed multiple symptoms characteristic for a ZSD. Most individuals had visual impairment and/or sensorineural hearing loss. Liver dysfunction and adrenal insufficiency, both typical for ZSD,2 were also frequently noted. All affected individuals showed neurological involvement, including profound hypotonia, gait abnormalities, developmental delay, neuropathy, and white matter abnormalities on brain magnetic resonance imaging (MRI). The clinical courses were progressive without a clear episode of rapid deterioration occurring. Individuals P4, P6, P2, and P2’s half-brother died between 8 and 20 years of age, whereas individuals P1, P3, P5, and P7 were still alive at the time of this study.
Table 1.
Clinical Characteristics of Affected Individuals and Parents Heterozygous for the PEX6 c.2578C>T (p.Arg860Trp) Variant
| P1a | P2 | Half-Brother of P2 | Mother of P2 | P3b | P4c | Father of P4 | P5 | P6 | P7 | Mother of P7 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subject Characteristics | |||||||||||
| Gender | male | male | male | female | male | male | male | male | female | male | female |
| Country of origin | the Netherlands | USA | USA | USA | Spain | Sweden | Sweden | Australia | USA | the Netherlands | the Netherlands |
| Year of birth | 2009 | 1984 | 1980 | 1963 | 2009 | 2005 | 1979 | 2010 | 1984 | 2015 | 1993 |
| Age at diagnosis | 4 yo | at birth | 4 yo | N/A | 1 yo | 6 yo | N/A | 3 yo | 2 yo | 1 yo | N/A |
| Age at death | N/A | 8 yo | 11 yo | N/A | N/A | 10 yo | N/A | N/A | 20 yo | N/A | N/A |
| Neurological | |||||||||||
| Hypotonia | + | + | + | − | + | − | − | + | + | + | − |
| Gait abnormalities | + | + | + | − | + | + | − | + | + | − | − |
| Developmental delay | + | + | + | − | + | + | − | + | + | − | − |
| Intellectual disability | + | ND | + | − | + | + | − | − | + | − | − |
| Neuropathy | + | + | + | ND | + | + | − | ND | ND | − | − |
| White matter abnormalities | + | ND | + | ND | + | + | − | + | ND | + | ND |
| Sensory | |||||||||||
| Vision | − | + | + | ND | + | ND | − | − | + | ND | − |
| Hearing | + | ND | ND | ND | + | ND | − | + | ND | − | − |
| Other | |||||||||||
| Hepatomegaly / liver dysfunction | − | + | + | ND | + | ND | − | + | + | + | − |
| Adrenal insufficiency | − | ND | ND | ND | + | ND | − | − | ND | − | − |
+, defect present; −, no abnormalities; yo, years old; N/A, not applicable; ND, no data.
Individual P1 additionally displays recurrent calcium oxalate kidney stones from age 7 onwards.
Individual P3 additionally displays cardiac malformation (atrial septal defect, multi-perforated foramen ovale type).
Individual P4 was diagnosed with partial trisomy 18 (SNP array 250k, 18p 0-3423586, hg18, spanning 18p11.32 and parts of 18p11.31).
Biochemical Analysis of Peroxisomal Parameters
We measured very long-chain fatty acid (VLCFA) concentrations and ratios in plasma of family 2 by using capillary gas chromatography of fatty acid methyl esters as previously described.7 VLCFA, phytanic, and pristanic acid concentrations in plasma of family 4 were measured as follows. The fatty acids were extracted and methylated in one step using HCl and methanol, before being separated in a gas chromatograph and detected by mass spectrometry using electron ionization and quantified using internal calibrators D3-Phytanic acid, C19:0, and C27:0. Plasma phytanic acid methyl ester of individual P1 and P6 was quantified by capillary gas chromatographym8 and plasma pipecolic acid was measured by isotope dilution mass-fragmentometry of the t-butyl-dimethylsilyl derivative of pipecolic acid.9 VLCFA concentrations and ratios in plasma of family 7 were measured by using UPLC-tandem MS in ESI-negative mode with Waters Micromass Quattro Micro API-tandem MS. VLCFAs in plasma were hydrolyzed with acetonitrile and HCl, extracted with hexane, and concentrated and separated on UPLC with Waters Acquity BEH C18 1.7 μm 2.1 × 50 mm column in mobile phase of acetic acid (no fragmentation of metabolites with similar mass of mother and daughter components). Quantification was done using the stable-isotope dilution technique; deuterated analogs of metabolites (D3-Pristanic acid, D3-Phytanic acid, D4-Docosanoic acid, D4-Tetracosanoic acid, D4-Hexacosanoic acid) were used as internal standards. Pipecolic acid in plasma of family 7 was measured by using ion-pair-UPLC and detected with a Quattro Premier-LC-MSMS in the multiple reaction monitoring mode (MRM) in the ESI-positive mode (internal standard: D3-L-leucine).10 We measured VLCFA concentrations and assessed the processing of thiolase in cultured primary skin fibroblasts as previously described.11, 12
Genetic Analysis
To perform whole-exome sequencing, DNA was isolated from skin fibroblasts of family 2 and from blood samples of individual P3 using established protocols. Whole-exome sequencing of individuals from family 2 was performed using SureSelect Human All Exon 51 Mb V5 capture kit (Agilent) on a HiSeq 2500 (Rapid Flow Cell V1; Illumina). Whole-exome sequencing of individual P3 was performed using the SeqCap EZ Human Exome Kit v3.0 (Roche Nimblegen) with 100-bp paired-end read sequences generated on a HiSeq2000 (Illumina) in the Centro Nacional de Análisis Genómico in Barcelona (CNAG). Sequences were aligned to hg19 by Burrows-Wheeler Aligner (BWA mem) and single variants and insertions/deletions (indels) were identified through the GATK best practices for Germline SNP & Indel discovery in whole-exome sequencing.
Peroxisome gene panel sequencing was performed with DNA of individuals P3, P4, and P7 using IonTorrent (Thermo Fisher Scientific; included genes: ABCD1, ABCD2, ABCD3, ACBD5, ACOX1, ACOX2, AGPS, AMACR, GNPAT, HSD17B4, PEX1, PEX2, PEX3, PEX5, PEX6, PEX7, PEX10, PEX11A, PEX11B, PEX11G, PEX12, PEX13, PEX14, PEX16, PEX19, PEX26, PHYH, SCP2), or with DNA of individual P5 using a 75 PEX and related peroxisome gene panel using the Nimblegen SeqCap EZ choice library (Roche) and a paired-end protocol on a MiSeq (Illumina). Sequences were aligned to hg19 with BWA and variations were detected with GATK.13
For Sanger sequencing of PEX6, genomic DNA was isolated from primary skin fibroblasts using the NucleoSpin Tissue genomic DNA purification kit (Macherey-Nagel). The majority of forward and reverse primers used for sequencing PEX6 exons, 3′ UTR, and potential promoter regions listed in Table S1 were tagged with a −21M13 (5′-TGTAAAACGACGGCCAGT-3′) sequence or M13rev (5′-CAGGAAACAGCTATGACC-3′) sequence, respectively. PCR fragments were sequenced using “−21M13,” “M13rev” primers, and/or the respective PEX6 primers, by means of BigDye Terminator v1.1 Cycle Sequencing Kits (Applied Biosystems) and analyzed on an Applied Biosystems 3130x1 or 3730x1 DNA analyzer (Applied Biosystems), following the manufacturer’s protocol. Sequence reads (electropherograms) were analyzed using CodonCode Aligner software package (CodonCode Corporation) and compared to PEX6 reference sequence GenBank: NM_000287.3 (GRCh38, hg38).
For mRNA sequence analysis, we isolated RNA from primary skin fibroblasts of all affected individuals and control individuals using trizol (Invitrogen) extraction and reverse transcribed it into cDNA using the QuantiTect Reverse Transcription Kit (QIAGEN) before Sanger sequencing. For mRNA stability assays, we incubated primary fibroblasts with 1 μM actinomycin D (ActD) for the indicated time or 20 μM emetine for 8 hr, before harvesting the cells for RNA isolation.
To assess the expression imbalance of two alleles, the height of the peaks of heterozygous variants in the electropherograms were measured and the peak height ratio of the mutant peak “mutant/(wild-type+mutant)” calculated, as described previously.14
Cell Culturing and Transfections
We used primary skin fibroblasts from seven affected individuals and the mother of individual P2, as well as primary skin fibroblast cell lines that are completely deficient of PEX6 (homozygous for c.402delC [p.Gly135Aspfs∗23]15). Cells were cultured in Dulbecco’s modified Eagle’s medium with L-glutamine (Bio-Whittaker), supplemented with 10% fetal bovine serum (Bio-Whittaker), 25 mM HEPES buffer (BioWhittaker), 100 U/mL penicillin, 100 μg/mL streptomycin (LifeTechnologies), and 250 ng/mL Fungizone (LifeTechnologies) in a humidified atmosphere of 5% CO2 at 37°C or 40°C. Transfection of fibroblasts for immunofluorescence and functional assays was performed using the AMAXA NHDF nucleofector kit (Lonza) following the manufacturer’s instructions (program U23). Transfection of HEK-FlpIn cells was performed in 96-well or 6-well plates using the jetPRIME DNA transfection kit (Polyplus transfection) according to the manufacturer’s instructions. 24 hr after transfection, the medium was changed and the cells were imaged 48–72 hr after transfection.
Genetic Complementation and Functional Assays
We performed genetic complementation of fibroblasts by transfection of the cells from the affected individuals with PEX cDNA, as described in Ebberink et al.3
To test the functional consequences of the PEX6 variants on peroxisomal protein import, we co-transfected plasmids containing the different PEX6 variants or pcDNA3 vectors without insert in various ratios (1 versus 3:1 versus 1:1 versus 1:3 versus 0) together with the peroxisomal matrix marker pGFP-SKL16 into skin fibroblasts deficient for PEX6 (6-well plates, 2 μg DNA per transfection in total). The PEX6 variant constructs pcDNA3-PEX6 c.821T17 and pcDNA3-PEX6 c.2578T were generated by site-directed mutagenesis of the mammalian expression vector pcDNA3 containing full-length PEX6 cDNA using the QuikChange Site-Directed Mutagenesis Kit from QIAGEN (Hilden) following the manufacturer’s instructions. We analyzed the subcellular localization of the fluorescent signal 2 to 3 days after transfection using the fluorescence microscope Zeiss Axio Observer A1. Cells with restored peroxisomal protein import displaying a punctate GFP signal and cells displaying cytosolic GFP signal were quantified by determining the GFP signal in 100–300 cells in three to seven independent experiments.
To test the ability of the PEX6 variants to localize the PEX1/6 complex to peroxisomes, we transfected pcDNA3-PEX1, pcDNA3-PEX6 and/or pcDNA3-PEX6 c.2578T into skin fibroblasts deficient for PEX6 before performing immunofluorescence assays.
To test the effect of miRNA inhibitors, which were predicted to target the longer PEX6 c.∗1_462 mRNA by TargetScan and microRNA.org, on the PEX6 allelic expression, we co-transfected skin fibroblasts derived from the affected individuals with GFP-SKL and miRIDIAN microRNA Human hsa-miR-150-5p (“miR150”) and hsa-miR-33a-5p (“miR33”) Hairpin Inhibitor or Hairpin Inhibitor Negative Control #1 (50-70 nM, Dharmacon). We determined the subcellular localization of GFP signal by fluorescence microscopy and quantified the number of peroxisome-positive fibroblasts after anonymizing samples for unbiased assessment after 2 to 7 days. Additionally, we isolated the RNA from the fibroblasts and performed quantitative RT-PCR to measure the mRNA levels of PEX6, long PEX6 c.∗1_462, and miR33 and miR150 target genes CPT1a and p53 2 to 4 days after transfection.
Quantitative RT-PCR
We isolated RNA from primary fibroblasts and reverse transcribed it to cDNA for quantitative RT-PCRs. We measured the mRNA levels of target genes in duplicates using the SensiFAST SYBR No-ROX Kit (Bioline) and the Lightcycler 480, Instrument II (Roche). The N0 values of target genes were normalized to the geometric mean of the reference genes Nono and hsH3F3A, which were determined most stable in expression and thus best suited for normalization out of ten assessed reference genes (determined with Normfinder algorithm18).
Immunofluorescence Imaging
We analyzed the peroxisomal phenotype in skin fibroblasts by confocal immunofluorescence microscopy. The cells were cultured on glass slides to a confluency of 50%–80%. For fixation, we treated the cells with 2% paraformaldehyde (Merck) in PBS for 20 minutes at room temperature and permeabilized with 0.5% Triton X-100 (Bio Rad) for 5 min. We used primary antibodies against the peroxisomal matrix protein catalase (monoclonal Mab 17E10, own production), the peroxisomal membrane protein ABCD3 (PMP70, rabbit, #71-8300, Thermo Fisher Scientific), PEX1 (mouse, # 611719, BD Biosciences), and PEX5 (rabbit, gift from G. Dodt) and as secondary antibodies either biotinylated α-mouse antibodies (E 433; Dako) and streptavidin-FITC (F 422; Dako) or Alexa Fluor 555 goat anti-rabbit (Invitrogen). The slides were fixed on mounting medium Vectashield H1000 (Brunschwig). Images were taken using the fluorescence microscope Zeiss Axio Observer A1 (100× magnification) or the Leica TCS-SP8 filter-free Spectral Confocal Microscope (63× magnification) and processed with the Leica Application Suite AF Lite software version 2.6.3 (Leica). If required, brightness and contrast of whole images were adjusted using the Adobe Photoshop CS6 software (Adobe Systems).
Immunoblotting
We performed immunoblot analyses using whole-cell lysates, of which proteins were separated by SDS polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane using a semidry blotting process. Subsequently, we used primary antibodies against PEX6 (HPA025924, Sigma-Aldrich), peroxisomal thiolase (3-ketoacyl-CoA thiolase, HPA007244, Atlas antibodies), α-tubulin (T6199, Sigma-Alderich), or β-actin (A5441, Sigma-Aldrich) and secondary antibodies IRDye 800CW Goat anti-Rabbit IgG (LI-COR Biosciences) or IRDye 680RD Goat anti-Mouse IgG (H+L; LI-COR Biosciences) with the Odyssey Imaging System (LI-COR Biosciences).
NanoLuc Assay
We constructed NanoLuc expression vectors by cloning the PEX6 3′ UTR sequence c.∗1_∗650 wild-type or with the deletion c.∗442_445delTAAA (GeneScript) into the pNL3.2[NlucP/minP] vector (Promega). Correct cloning was confirmed by Sanger sequencing of the vectors. We mutated the proximal polyadenylation signal by introducing a c.∗308A>C change, creating the vector pNluc_PEX6 c.∗308C, by site-directed mutagenesis of the pNluc-wild-type vector using the QuikChange II Site-Directed Mutagenesis Kit (Agilent). We integrated the NanoLuc-PEX6 3′ UTR constructs into HEK-FlpIn cells and confirmed the stable NanoLuc expression by quantitative RT-PCR. Additionally, we confirmed that Nanoluc-PEX6 c.∗308C was transcribed as NanoLuc mRNA with the longer PEX6 c.∗1_∗462 3′ UTR and that Nanoluc-PEX6 c.∗442_445delTAAA was transcribed as NanoLuc mRNA with the shorter PEX6 c.∗1_∗326 3′ UTR using the rapid amplification of cDNA ends (RACE) method. We transfected 50 nM miRIDIAN microRNA Mimic Negative Control #1 or miRIDIAN microRNA Human hsa-miR-33a-5p, hsa-miR-33b-5p, or hsa-miR-150-5p Mimics (Dharmacon) into the HEK-FlpIn-Nanoluc-PEX6 c.∗308C cells and HEK-FlpIn-Nanoluc-PEX6 c.∗442_445delTAAA cells. 24 hr after transfection, we used the Nano-Glo Dual-Luciferase Reporter Assay System (Promega) to measure luminescence at three subsequent time points. To validate miRNA efficiency, we confirmed the downregulated expression of miRNA target genes CPT1a or cMyb, respectively, using quantitative RT-PCR.
SV40 Immortalized Cell Model with Stable PEX6 Overexpression
We generated PEX6 expression vectors containing wild-type PEX6 cDNA (pMono-hygro-PEX6) or mutant PEX6 c.2578T cDNA (pMono-neo-PEX6_c.2578T) by cloning wild-type PEX6 c.2578C or mutant PEX6 c.2578T from pcDNA3 vectors into pMono-hygro-mcs or pMono-neo-mcs (InvivoGen), using restriction enzymes with compatible ends (i.e., BspEI/XbaI and AgeI/AvrII, respectively) and then transfected them into SV40 immortalized human control fibroblasts. After selection with the respective antibiotics (100–200 μg/mL geneticin, 150 μg/mL hygromycin B), we confirmed the overexpression of PEX6 mRNA by immunoblot analysis, quantitative RT-PCR (see primers in Table S1) and—in case of pMono-neo-PEX6 c.2578T—by Sanger sequencing.
Statistical Methods
All values in figures are presented as the mean ± SD. Results were analyzed with Mann-Whitney U tests or unpaired t tests, in case of mean allele peak ratios, with one-sample t tests with a theoretical mean of 0.5. A p value less than 0.05 was considered statistically significant. Statistical analyses were carried out in the Graphpad Prism 6 software.
Results
Individuals with ZSD Display Same Heterozygous PEX6 Mutation
The seven unrelated individuals (Figure 1B) were diagnosed with a ZSD on the basis of their clinical symptoms (Table 1), aberrant peroxisomal metabolite levels in blood and fibroblasts (Tables 2 and S2), and an import defect of peroxisomal matrix proteins in fibroblasts (Figures 1C and S1).
Table 2.
Peroxisomal Parameters in Primary Skin Fibroblasts Derived from Affected Individuals and Parents Heterozygous for the PEX6 c.2578C>T (p.Arg860Trp) Variant
|
Individual |
P1 |
P2 |
Mother of P2 |
P3 |
P4 |
P5 |
P6 |
P7 |
Reference Range | |
|---|---|---|---|---|---|---|---|---|---|---|
| Age at sampling | 3 yo | 1 yo | 21 yo | 1 yo | 8 yo | ND | ND | 1 yo | ||
| VLCFAs concentration (in μmol/g protein) | C22:0 | 3.13 | 4.2 | 5.31 | 1.19 | 3.4 | 4.02 | 2.88 | 2.19 | 2.46–6.59 |
| C24:0 | 7.56 | 11.05 | 11.81 | 6.06 | 8.18 | 10.46 | 8.91 | 8.29 | 6.37–13.87 | |
| C26:0 | 0.95 | 1.19 | 0.41 | 1.39 | 0.61 | 1.51 | 1.18 | 1.35 | 0.16–0.41 | |
| VLCFA ratios | C24:0/C22:0 | 2.42 | 2.63 | 2.22 | 3.17 | 2.4 | 2.6 | 3.1 | 3.78 | 1.68–2.92 |
| C26:0/C22:0 | 0.3 | 0.28 | 0.08 | 0.73 | 0.18 | 0.38 | 0.41 | 0.62 | 0.03–0.1 | |
| Thiolase immunoblota | 44 kDa | +/− | +/− | − | +/− | +/− | +/− | +/− | − | − |
| 41 kDa | +/− | +/− | ++ | +/− | +/− | +/− | +/− | +/− | ++ | |
| Catalase import deficiencyb | [percentage of deficient cells] | >90% | >90% | <20% | >90% | >90% | >90% | >90% | 90% | 0% |
Abbreviations are as follows: VLCFAs, very long-chain fatty acids; yo, years old; ND, no data.
For assessment of peroxisomal processing of thiolase
Compare Figure S1
By means of functional genetic complementation of cultured primary skin fibroblasts3 (of individuals P1, P3, and P4) or fused cell complementation19 (of individual P6), we identified a defective PEX6 gene as the cause of the ZSD in four affected individuals. When we subsequently Sanger sequenced PEX6 (GenBank: NM_000287.3) to identify the disease-causing mutations, we detected only the heterozygous variant c.2578C>T (p.Arg860Trp) (rs61753230) in all four individuals, but no second potentially pathogenic variant (Table 3). The same single heterozygous PEX6 c.2578C>T variant was also independently identified by Sanger sequencing in individual P2 and his half-brother and in individuals P5 and P7. Peroxisome-specific gene-panel sequencing in four individuals (P3, P4, P5, and P7) and whole-exome sequencing in two individuals (P2 and P3) did not identify additional pathogenic variants in other genes encoding peroxisomal proteins that could explain the phenotype of the individuals.
Table 3.
Genomic PEX6 Variants in Affected Individuals and Parents Heterozygous for the PEX6 c.2578C>T (p.Arg860Trp) Variant
|
PEX6 Polymorphism |
Alleles in Individuals |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| refSNP Cluster ID | DNA Sequence Changea | Amino Acid Change | Minor Allele Frequency (1000 Genomes) | P1 | P2 | Mother of P2 | P3 | P4 | Father of P4 | Mother of P4 | P5 | P6 | P7 | Father of P7 | Mother of P7 |
| rs9462859 | c.1_55C>T | – | A = 0.334 / 1672 | T/C | T/C | T | T/C | T/C | T | T/C | T/C | T/C | T/C | C | T |
| c.279G>A | p.Arg93Arg | G | G | G/A | G | G | G | G | G | G | G | G | G | ||
| rs9462858 | c.399G>T | p.Val133Val | A = 0.332/1662 | T/G | T/G | T | T/G | T/G | T | T/G | T/G | T/G | T/G | G | T |
| rs61753220 | c.853C>G | p.Pro285Ala | C = 0.01/3b | C | C | C | C | C | C | C | C | C/G | C | C | C |
| rs9986447 | c.883−3T>C | – | G = 0.29/1451 | C/T | C/T | C | C/T | C/T | C | C/T | C/T | C/T | C/T | T | C |
| rs61753230 | c.2578C>T | p.Arg860Trp | C/T | C/T | C/T | C/T | C/T | C/T | C | C/T | C/T | C/T | C | C/T | |
| rs1129186 | c.2814G>A | p.Glu938Glu | T = 0.494/2472 | A/G | A/G | A | A/G | A/G | A | A/G | A/G | A/G | A/G | G | A |
| rs1129187 | c.2816C>A | p.Pro939Gln | T = 0.332/1660 | A/C | A/C | A | A/C | A/C | A | A/C | A/C | A/C | A/C | C | A |
| rs144286892 | c.∗442_445 delTAAA | – | T = 0.353/1766 | TAAA / delTAAA | TAAA / delTAAA | delTAAA / delTAAA | TAAA / delTAAA | TAAA / delTAAA | delTAAA / delTAAA | TAAA / delTAAA | TAAA / delTAAA | TAAA / delTAAA | TAAA / delTAAA | TAAA / TAAA | delTAAA / delTAAA |
n/a, not assessed.
Nucleotide numbering based on GenBank: NM_000287.3, GRCh38/hG38 (on reverse DNA strand)
Clinical significance (ClinVar) of polymorphism c.853C>G (p.Pro285Ala): likely benign/uncertain significance
PEX6 Mutation Impairs Peroxisome Biogenesis, but Not PEX6 Protein Localization
The mutated arginine-860 residue constitutes the arginine finger 2 of PEX6, which is a highly conserved residue in AAA+ ATPases and located in the second region of homology (SRH) of the ATP-binding D2 domain in close vicinity to bound ATP (Figure S2).20, 21 The PEX6 c.2578C>T variant is absent in genome databases of healthy populations or affected individuals, including ExAC and ClinVar, but was reported once as a single heterozygous variant in an individual diagnosed with ZSD.22 The variant is predicted as deleterious by SIFT23 (score 0), disease causing by MutationTaster24 (score 1.0), and probably damaging by PolyPhen-225 (score 1.0). We confirmed the predicted pathogenicity of the variant by expressing PEX6 p.Arg860Trp in PEX6-deficient skin fibroblasts, which, in contrast to wild-type PEX6, did not restore peroxisomal matrix protein import (Figure S3).
To study the functional consequences of the PEX6 p.Arg860Trp variant, we assessed PEX6 protein levels in cell lysates of the affected individuals by immunoblot analysis, but this did not show marked changes when compared to control fibroblasts (Figure S4). Moreover, immunofluorescence microscopy showed that the PEX1-PEX6 complexes in fibroblasts of the affected individuals were correctly localized at peroxisomes (Figure S5). This observation corroborates our finding that expression of PEX6 p.Arg860Trp in PEX6-deficient cells also resulted in correct peroxisomal localization of PEX1-PEX6 complexes (Figure S5). PEX5, on the other hand, which is predominantly cytosolic in control cells, was mainly localized at peroxisomal membranes in cells of the affected individuals (Figure 1C). These observations indicate that PEX6 p.Arg860Trp results in defects in PEX5 export and peroxisome biogenesis in cells of the affected individuals, although it is correctly localized (Figure 1C).
Asymptomatic Parents with PEX6 Mutation Show Only Very Mild Peroxisomal Defects
We next Sanger sequenced PEX6 of the parents of the affected individuals, which revealed that individuals P1 and P3 obtained the variant de novo (biological parenthood confirmed). Individual P2 and his half-brother inherited the variant from their mother, individual P4 from his father, and individual P7 from his mother (Table 3). For the other individuals, parental DNA was not available.
All three parents who were heterozygous for the PEX6 c.2578C>T variant showed no clinical symptoms or any signs that could be related to a peroxisomal defect (Table 1) and, in contrast to the affected individuals, only minor biochemical abnormalities in blood and fibroblasts (Tables 2 and S1). Furthermore, we observed that PEX5 was only partly mislocalized to peroxisomes in fibroblasts of the mother of individual P2, who was the only parent heterozygous for the PEX6 c.2578C>T variant of whom cells were available for analysis (Figures 1D and S6). This slight PEX5 export defect corresponded to a very mild import defect of the peroxisomal matrix protein catalase in these cells (Figures 1D and S1). Based on these observations we considered a classical dominant-negative disease mechanism in the affected individuals (due to mere heterozygosity of the pathogenic PEX6 c.2578C>T variant) unlikely.
Affected and Control Individuals with a Heterozygous 3′ UTR Variant Show Allelic Expression Imbalance of PEX6
Interestingly, when we analyzed PEX6 mRNA expression by Sanger sequencing cDNA derived from the affected individuals’ fibroblasts, we noted that the levels of the mutant PEX6 c.2578T cDNA were consistently three to five times higher than those of wild-type PEX6 (c.2578C) cDNA (Figures 2A and 2B). In contrast, the cDNA levels of both PEX6 alleles were equal in the mother of individual P2. When we Sanger sequenced the PEX6 cDNAs of 22 different fibroblast cell lines derived from unrelated control individuals, we noted AEI of PEX6 in over 40% of the cell lines. This indicates that the AEI is not specific for the cells of the affected individuals but a common feature of PEX6.
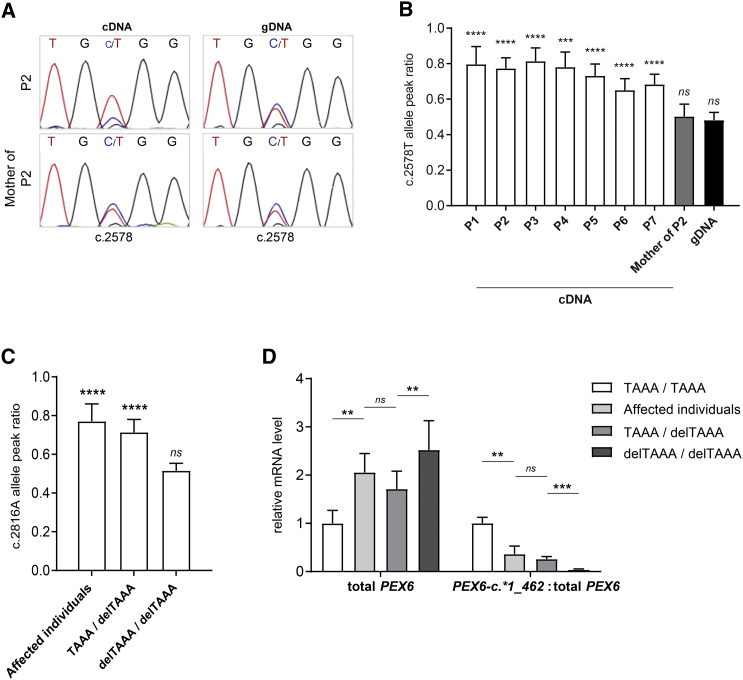
Figure 2.
AEI of PEX6 in Affected Individuals and Control Individuals with the Heterozygous 3′ UTR Variant PEX6 c.∗442_445delTAAA
(A and B) Allelic expression of PEX6 mRNA in cells of affected individuals and the mother of affected individual 2. PEX6 cDNA and genomic DNA were Sanger sequenced and the peak height of both PEX6 alleles were used to determine the relative abundance of each allele (see representative sequence reads in A). Mean c.2578T allele peak ratios in the different cell lines (B), expressed as the ratio of the mutant allele peak height (c.2578T) over the total peak height (c.2578T+c.2578C) as described in Moncini et al.,14 depicted as mean with standard deviation of 6–19 analyzed electropherograms per cell line (or gDNA group) using 2–3 different primer sets. While the PEX6 allele ratio is in balance in genomic DNA of the affected individuals and genomic DNA and cDNA of the mother of individual P2 (i.e., equal peak heights), the mutant allele ratio in cDNA of all affected individuals is significantly increased, demonstrating the AEI of PEX6.
(C and D) PEX6 mRNA expression in fibroblasts of affected individuals and fibroblasts of 22 unrelated control individuals with different combinations of the PEX6 c.∗442_445delTAAA allele (homozygous for the allele without the deletion [“TAAA / TAAA”], homozygous for the PEX6 c.∗442_445delTAAA allele [“delTAAA / delTAAA”] or compound heterozygous [“TAAA / delTAAA”]).
(C) Mean allele peak ratios for the PEX6 c.∗442_445delTAAA allele in cDNA of cells of the affected individuals and control cells heterozygous and homozygous for this allele, confirming that the AEI of PEX6 occurs only in the heterozygous cells. Shown are the peak ratios of the c.2816A variant, which is a common polymorphism (MAF 0.39 [ExAc] / 0.33 [1000 Genomes]) that is in cis with PEX6 c.∗442_445delTAAA (see also Table 3). The peak ratios were determined as described in (B) and Moncini et al.14 and are presented as mean with standard deviation based on analysis of 5–10 electropherograms from 4–7 different cell lines per allele combination.
(D) Results of quantitative RT-PCR experiments, demonstrating an increased level of total PEX6 mRNA but no expression of longer PEX6 c.∗1_462 mRNA in cells homozygous for the PEX6 c.∗442_445delTAAA. Depicted are the PEX6 mRNA levels normalized to reference gene expression or as ratio PEX6 c.∗1_462 over total PEX6, with “TAAA / TAAA” values set as 1, as mean with standard deviation determined in 4–13 cell lines per allele combination.
Statistical analyses for (B) and (C) were performed using one-sample t tests with a theoretical mean of 0.5, and for (D) using Mann-Whitney U tests (∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗ p ≤ 0.0001, ns not significant).
To find an explanation for this AEI of PEX6, we compared the coding sequences and 1,000 bp 5′ of the start and 500 bp 3′ of the stop codon of PEX6 of all affected individuals to those of the parents, who were heterozygous for the PEX6 c.2578C>T variant (mothers of individuals P1 and P7, father of individual P4). This revealed several common variants that were heterozygous in the affected individuals but homozygous in the parents, and thus could be responsible for the AEI (Table 3). Of these, we found the c.1_55C>T (rs9462859), c.399G>T (rs9462858), c.883−3T>C (rs9986447), c.2814G>A (rs1129186), and c.2816C>A variants (rs1129187) also heterozygous in some of the control cells that did not display AEI of PEX6, and consequently excluded these as candidates (data not shown). Heterozygosity for the c.∗442_445delTAAA variant in the 3′ UTR region of PEX6 (rs144286892), however, always correlated with AEI of PEX6 (Figure 2C). Long-range PCR and clone sequencing confirmed that the PEX6 c.∗442_445delTAAA variant was allelic to the PEX6 c.2578T mutation (data not shown).
The PEX6 c.∗442_445delTAAA variant is a frequently occurring polymorphism with a minor allele frequency of 0.35 (1000 Genomes). According to the APASdb database, it disrupts the most distal of the two known polyadenylation signals of PEX6, which are located at c.∗305_310 and c.∗440_445. Usage of the two polyadenylation signals results in PEX6 mRNA with a shorter (PEX6 c∗1_∗326) or longer (PEX6 c.∗1_∗462 mRNA) 3′ UTR, respectively. Quantitative analysis of PEX6 mRNA expression in fibroblast cell lines with different PEX6 allele combinations indeed confirmed that the PEX6 c.∗442_445delTAAA allele does not produce the longer PEX6 c.∗1_462 mRNA (Figure 2D). Nevertheless, the total levels of PEX6 mRNA expressed by the PEX6 c.∗442_445delTAAA allele were consistently higher than those expressed by the allele without the deletion (Figure 2D). This expression level difference of the two alleles explains why AEI of PEX6 occurs in all cell lines heterozygous for PEX6 c.∗442_445delTAAA, but not in the cell lines homozygous for any of the PEX6 alleles (Figure 2C).
AEI Is Not Caused by Differences in Stability of Longer or Shorter PEX6 mRNA
To determine whether the relatively increased levels of the shorter PEX6 c.∗1_326 mRNA, which is expressed by the PEX6 c.∗442_445delTAAA allele, may be due to a decreased stability of the longer PEX6 c.∗1_462 mRNA, we treated cells of the affected individuals with the nonsense-mediated mRNA decay-inhibitor emetine or the transcriptional inhibitor actinomycin D. We did not observe increased degradation nor decreased stability of the longer PEX6 c.∗1_462 mRNA (Figure S7). Furthermore, we could not find indications that the stability of the longer PEX6 c.∗1_462 mRNA is negatively regulated by the microRNAs hsa-miR-33-5p and hsa-miR-150-5p, both of which are predicted to target sequences present in the extended 3′ UTR of the longer PEX6 c.∗1_462 mRNA by TargetScan and microRNA.org (Figures S8 and S9). These results indicate that differential stability of the two PEX6 mRNA isoforms is not the underlying reason for the AEI.
Overrepresentation of Mutant PEX6 Impairs Wild-Type PEX6 Function and Causes Peroxisome Biogenesis Defect
We hypothesized that the peroxisome biogenesis defect in the affected individuals is caused by the AEI-induced overrepresentation of the pathogenic PEX6 p.Arg860Trp variant. To confirm this, we overexpressed PEX6 p.Arg860Trp in SV40-immortalized control fibroblasts that express endogenous PEX6 (Figure 3A). This resulted in an accumulation of PEX5 at the peroxisomal membranes and a concomitant defect of catalase import into peroxisomes (Figures 3B and S10). Moreover, when we co-expressed PEX6 p.Arg860Trp and wild-type PEX6 in different ratios in PEX6-deficient fibroblasts, we observed a significant negative correlation between the restoration of peroxisomal matrix protein import and the ratio of PEX6 p.Arg860Trp over wild-type PEX6. This negative effect on peroxisomal import was far more pronounced than in cells co-expressing wild-type PEX6 with the previously characterized recessive variant, PEX6 p.Pro274Leu17 (Figure 3C). These results indicate that the PEX6 p.Arg860Trp variant has a negative effect on PEX1-PEX6 complex function but results in a disease phenotype only when PEX6 p.Arg860Trp is at least two to three times more abundant than wild-type PEX6.
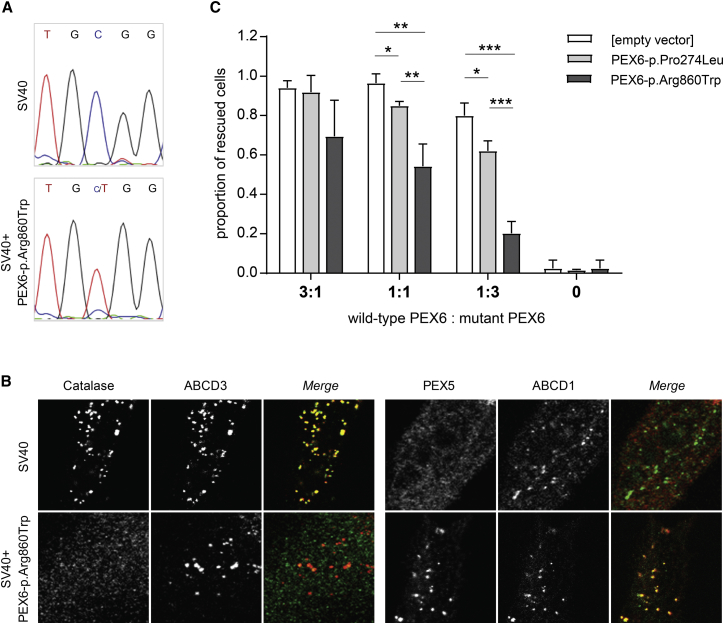
Figure 3.
Overexpression Models of Dominant-Negative PEX6-p.Arg860Trp in Presence of Wild-Type PEX6
(A and B) Peroxisome biogenesis inSV40 immortalized control cells overexpressing PEX p.Arg860Trp.
(A) Representative sequence reads obtained by Sanger sequencing of PEX6 cDNA in SV40 cells and SV40 cells over-expressing PEX6 c.2578C>T. The mutant PEX6 c.2578T allele displays a similar overrepresentation when compared to the endogenous wild-type PEX6 allele as observed in cells of the affected individuals (compare Figure 2A).
(B) Imunofluorescence microscopy images of cells, which were double labeled to determine the subcellular localizations of catalase (green) and ABCD3 (red), or PEX5 (red) and ABCD1 (green), demonstrating a defect in peroxisomal catalase import and PEX5 export in SV40 cells over-expressing PEX6 p.Arg860Trp (compare also Figure S10).
(C) Co-expression of mutant and wild-type PEX6 in PEX6-deficient cells. PEX6-deficient cells were co-transfected with wild-type PEX6 cDNA and PEX6 variants PEX6 c.2578T (p.Arg860Trp) (dark grey) or PEX6 c.821T (p.Pro274Leu) (light grey) in different ratios, together with the peroxisomal matrix protein marker GFP-SKL, whereupon the degree of GFP-SKL import into peroxisomes was determined. GFP-SKL import after transfection with wild-type PEX6 only was set as 1. Increased relative expression of PEX6-p.Arg860Trp results in a decreased peroxisomal import of GFP-SKL, which was far more pronounced than observed with PEX6 p.Pro274Leu.
Data are depicted as mean with standard deviation of three independent experiments counting 180–300 cells per condition, and statistical analyses were performed using unpaired t tests (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, while an absence of a p value indicates non-significance).
Discussion
We identified AEI-induced overrepresentation of a pathogenic PEX6 c.2578C>T allele as the cause of disease in seven unrelated individuals and one half-brother with an apparent dominant ZSD. The fact that the same PEX6 c.2578C>T mutation occurred in seven unrelated families, among which two individuals in which the mutation arose de novo, may be related to the fact that c.2578C is part of a CpG site, which is prone to mutations.26 De novo occurrence of PEX mutations have been reported previously.27 Four of the affected individuals inherited the PEX6 c.2578C>T allele from one of the parents, who in all cases were asymptomatic and did not show AEI of PEX6.
We showed that the PEX6 p.Arg860Trp change has a dominant-negative effect on the function of the PEX1-PEX6 complex in peroxisomal matrix protein import. However, as observed in the parents, heterozygosity for the PEX6 c.2578C>T allele does not result in a disease phenotype when this allele is equally expressed as the wild-type allele, but only when overrepresented due to AEI. The AEI is correlated with heterozygosity for a common PEX6 c.∗442_445delTAAA deletion in the 3′ UTR of PEX6. This deletion eliminates the most distal of two known polyadenylation sites of PEX6, resulting in the exclusive and increased expression of the PEX6 mRNA with a shortened 3′ UTR (PEX6 c.∗1_326). mRNAs with shorter 3′ UTRs have been described previously to be higher expressed than their counterpart with a longer 3′ UTR, which often has been attributed to differences in their mRNA stability.28, 29 We have not been able to elucidate the cause for the observed AEI of PEX6, but excluded enhanced degradation and decreased stability of the longer mRNA. This AEI might thus possibly be caused by differences in gene transcription or mRNA processing of the longer and shorter PEX6 isoform.
The arginine at position 860 constitutes the arginine finger 2 in the D2 ATPase domain of PEX6, which is a highly conserved domain in AAA+ ATPases (Figure 2).20, 21 Previous studies have shown that ATP binding in this domain is essential for the physical interaction of PEX6 with both PEX1 and the peroxisomal membrane anchor PEX26 (Figure 1A).5 The PEX5 export by the PEX1-PEX6 complex requires hydrolysis of the bound ATP.4, 5 Our finding that the PEX1-PEX6 p.Arg860Trp complex still localizes to peroxisomes but is incapable of PEX5 export implies that the mutation does not affect ATP binding in D2 but prevents ATP hydrolysis. In line with this, mutations of arginine finger 2 in the two double-ring AAA+ ATPases NSF and p97 have also been reported to abrogate ATP hydrolysis without affecting complex formation and to have a dominant-negative effect on complex function.30, 31
Our study reports a dominant presentation of an autosomal-recessive disorder due to AEI promoting the overrepresentation of a pathogenic allele. In autosomal-dominant disorders and several forms of cancer, AEI of various genes has been implicated as risk factors or as modifiers of disease severity.14, 32, 33, 34, 35 Given the high prevalence and genome-wide occurrence of AEI,36, 37, 38 this disease mechanism should be considered as a possible underlying cause of other autosomal-recessive disorders, in which only single heterozygous mutations are found. Thus, our findings bear great relevance for the interpretation of whole-exome and genome sequence data in clinical diagnostic laboratories, where allelic expression levels are not functionally verified and heterozygous variants inherited from asymptomatic parents are usually filtered out or considered non-pathogenic. Finally, similar as described for autosomal-dominant disorders, AEI may modulate the phenotypic variability in compound heterozygous individuals with autosomal-recessive disorders.
Acknowledgements
We thank the subjects and their families for their contributions to this study.
We thank Petra Mooijer, Connie Dekker, and Janet Koster for expert technical assistance and Rob Ofman and Lodewijk IJlst for scientific discussions. We acknowledge Gabi Dodt for providing the antibodies against PEX5 and the Coriell Institute for providing primary fibroblasts cell lines derived from members of family 2. We thank Wendy Mitchell at Children’s Hospital of Los Angeles and Michael Nigro at Children’s Hospital of Michigan, Detroit, for providing clinical information of the individuals of families 2 and 6. We acknowledge Sebastien Levesque at University of Sherbrooke, Quebec, for genetic testing of individual P5 on PEX sequencing panel and Avihu Boneh at University of Melbourne, Australia, for providing clinical data of individual P5 and genetic testing of the mother of individual P5. We thank Laia Grau and Juan José Martinez for excellent technical support and Stéphane Fourcade for scientific discussion. Special thanks to the ALE-ELA España for logistics and support of subjects.
Work at the Laboratory Genetic Metabolic Diseases in Amsterdam was supported by Marie Curie Initial Training Networks action (FP7-2012-PERFUME-316723 to K.D.F. and H.R.W.), at Kennedy Krieger Institute by the grant “Intellectual and Developmental Disabilities Research Centers 2013” Type: (1 U54 HD079123-01A1) NICHD, and at the Neurometabolic Diseases Laboratory by grants from the Autonomous Government of Catalonia (SGR 2014SGR1430), the Spanish Center for Biomedical Research on Rare Diseases (CIBERER, ER15PR02ACCI14), the Foundation Marató de TV3 (2014-0830), and the Hesperia Foundation.
Published: December 7, 2017
Footnotes
Supplemental Data include ten figures and two tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.11.007.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
APASdb, http://genome.bucm.edu.cn
Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/
ExAC Browser, http://exac.broadinstitute.org/
miRNA.org, http://www.microRNA.org
OMIM, http://www.omim.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
TargetScan 6.2, http://www.targetscan.org/
UniProt, http://www.uniprot.org/
Supplemental Data
References
- 1.Braverman N.E., D’Agostino M.D., Maclean G.E. Peroxisome biogenesis disorders: Biological, clinical and pathophysiological perspectives. Dev. Disabil. Res. Rev. 2013;17:187–196. doi: 10.1002/ddrr.1113. [DOI] [PubMed] [Google Scholar]
- 2.Klouwer F.C.C., Berendse K., Ferdinandusse S., Wanders R.J.A., Engelen M., Poll-The B.T. Zellweger spectrum disorders: clinical overview and management approach. Orphanet J. Rare Dis. 2015;10:151. doi: 10.1186/s13023-015-0368-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebberink M.S., Mooijer P.A.W., Gootjes J., Koster J., Wanders R.J.A., Waterham H.R. Genetic classification and mutational spectrum of more than 600 patients with a Zellweger syndrome spectrum disorder. Hum. Mutat. 2011;32:59–69. doi: 10.1002/humu.21388. [DOI] [PubMed] [Google Scholar]
- 4.Ciniawsky S., Grimm I., Saffian D., Girzalsky W., Erdmann R., Wendler P. Molecular snapshots of the Pex1/6 AAA+ complex in action. Nat. Commun. 2015;6:7331. doi: 10.1038/ncomms8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamura S., Yasutake S., Matsumoto N., Fujiki Y. Dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p. J. Biol. Chem. 2006;281:27693–27704. doi: 10.1074/jbc.M605159200. [DOI] [PubMed] [Google Scholar]
- 6.Bhogal M.S., Lanyon-Hogg T., Johnston K.A., Warriner S.L., Baker A. Covalent label transfer between peroxisomal importomer components reveals export-driven import interactions. J. Biol. Chem. 2016;291:2460–2468. doi: 10.1074/jbc.M115.686501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moser H., Moser A. Techniques in Diagnostic Human Biochemical Genetics: A Laboratory Manual. Wiley-Liss Inc; New York: 1991. Measurement of saturated very long chain fatty acids in plasma; pp. 177–191. [Google Scholar]
- 8.Moser H., Moser A. Techniques in Diagnostic Human Biochemical Genetics: A Laboratory Manual. Wiley-Liss Inc; New York: 1991. Measurement of phytanic acid levels; pp. 193–203. [Google Scholar]
- 9.Kelley R.I. Techniques in Diagnostic Biochemical Genetics: A Laboratory Manual. Wiley-Liss; New York: 1991. Quantification of pipecolic acid in plasma and urine by isotope-dilution gas chromatography/ mass spectrometry. [Google Scholar]
- 10.Waterval W.A.H., Scheijen J.L.J.M., Ortmans-Ploemen M.M.J.C., Habets-van der Poel C.D., Bierau J. Quantitative UPLC-MS/MS analysis of underivatised amino acids in body fluids is a reliable tool for the diagnosis and follow-up of patients with inborn errors of metabolism. Clin. Chim. Acta. 2009;407:36–42. doi: 10.1016/j.cca.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 11.Vreken P., van Lint A.E.M., Bootsma A.H., Overmars H., Wanders R.J.A., van Gennip A.H. Rapid stable isotope dilution analysis of very-long-chain fatty acids, pristanic acid and phytanic acid using gas chromatography-electron impact mass spectrometry. J. Chromatogr. B Biomed. Sci. Appl. 1998;713:281–287. doi: 10.1016/s0378-4347(98)00186-8. [DOI] [PubMed] [Google Scholar]
- 12.Wanders R.J., Dekker C., Ofman R., Schutgens R.B., Mooijer P. Immunoblot analysis of peroxisomal proteins in liver and fibroblasts from patients. J. Inherit. Metab. Dis. 1995;18(Suppl 1):101–112. doi: 10.1007/BF00711433. [DOI] [PubMed] [Google Scholar]
- 13.Levesque S., Morin C., Guay S.-P., Villeneuve J., Marquis P., Yik W.Y., Jiralerspong S., Bouchard L., Steinberg S., Hacia J.G. A founder mutation in the PEX6 gene is responsible for increased incidence of Zellweger syndrome in a French Canadian population. BMC Med. Genet. 2012;13:72. doi: 10.1186/1471-2350-13-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moncini S., Bonati M.T., Morella I., Ferrari L., Brambilla R., Riva P. Differential allelic expression of SOS1 and hyperexpression of the activating SOS1 c.755C variant in a Noonan syndrome family. Eur. J. Hum. Genet. 2015;23:1531–1537. doi: 10.1038/ejhg.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebberink M.S., Kofster J., Wanders R.J.A., Waterham H.R. Spectrum of PEX6 mutations in Zellweger syndrome spectrum patients. Hum. Mutat. 2010;31:E1058–E1070. doi: 10.1002/humu.21153. [DOI] [PubMed] [Google Scholar]
- 16.Waterham H.R., Koster J., van Roermund C.W.T., Mooyer P.A.W., Wanders R.J.A., Leonard J.V. A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 17.Ratbi I., Falkenberg K.D., Sommen M., Al-Sheqaih N., Guaoua S., Vandeweyer G., Urquhart J.E., Chandler K.E., Williams S.G., Roberts N.A. Heimler syndrome is caused by hypomorphic mutations in the peroxisome-biogenesis genes PEX1 and PEX6. Am. J. Hum. Genet. 2015;97:535–545. doi: 10.1016/j.ajhg.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen C.L., Jensen J.L., Ørntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 19.Moser A.B., Rasmussen M., Naidu S., Watkins P.A., McGuinness M., Hajra A.K., Chen G., Raymond G., Liu A., Gordon D. Phenotype of patients with peroxisomal disorders subdivided into sixteen complementation groups. J. Pediatr. 1995;127:13–22. doi: 10.1016/s0022-3476(95)70250-4. [DOI] [PubMed] [Google Scholar]
- 20.Ogura T., Whiteheart S.W., Wilkinson A.J. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 2004;146:106–112. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Wendler P., Ciniawsky S., Kock M., Kube S. Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta. 2012;1823:2–14. doi: 10.1016/j.bbamcr.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Yik W.Y., Steinberg S.J., Moser A.B., Moser H.W., Hacia J.G. Identification of novel mutations and sequence variation in the Zellweger syndrome spectrum of peroxisome biogenesis disorders. Hum. Mutat. 2009;30:E467–E480. doi: 10.1002/humu.20932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 24.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 25.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Youssoufian H., Kazazian H.H., Jr., Phillips D.G., Aronis S., Tsiftis G., Brown V.A., Antonarakis S.E. Recurrent mutations in haemophilia A give evidence for CpG mutation hotspots. Nature. 1986;324:380–382. doi: 10.1038/324380a0. [DOI] [PubMed] [Google Scholar]
- 27.Steinberg S.J., Snowden A., Braverman N.E., Chen L., Watkins P.A., Clayton P.T., Setchell K.D.R., Heubi J.E., Raymond G.V., Moser A.B., Moser H.W. A PEX10 defect in a patient with no detectable defect in peroxisome assembly or metabolism in cultured fibroblasts. J. Inherit. Metab. Dis. 2009;32:109–119. doi: 10.1007/s10545-008-0969-8. [DOI] [PubMed] [Google Scholar]
- 28.Mayr C., Bartel D.P. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W., Park J.Y., Zheng D., Hoque M., Yehia G., Tian B. Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol. 2016;14:6. doi: 10.1186/s12915-016-0229-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q., Song C., Irizarry L., Dai R., Zhang X., Li C.C.H. Multifunctional roles of the conserved Arg residues in the second region of homology of p97/valosin-containing protein. J. Biol. Chem. 2005;280:40515–40523. doi: 10.1074/jbc.M509636200. [DOI] [PubMed] [Google Scholar]
- 31.Zhao C., Matveeva E.A., Ren Q., Whiteheart S.W. Dissecting the N-ethylmaleimide-sensitive factor: required elements of the N and D1 domains. J. Biol. Chem. 2010;285:761–772. doi: 10.1074/jbc.M109.056739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butt C., Zheng H., Randell E., Robb D., Parfrey P., Xie Y.G. Combined carrier status of prothrombin 20210A and factor XIII-A Leu34 alleles as a strong risk factor for myocardial infarction: evidence of a gene-gene interaction. Blood. 2003;101:3037–3041. doi: 10.1182/blood-2002-09-2888. [DOI] [PubMed] [Google Scholar]
- 33.Amin A.S., Giudicessi J.R., Tijsen A.J., Spanjaart A.M., Reckman Y.J., Klemens C.A., Tanck M.W., Kapplinger J.D., Hofman N., Sinner M.F. Variants in the 3′ untranslated region of the KCNQ1-encoded Kv7.1 potassium channel modify disease severity in patients with type 1 long QT syndrome in an allele-specific manner. Eur. Heart J. 2012;33:714–723. doi: 10.1093/eurheartj/ehr473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broderick P., Carvajal-Carmona L., Pittman A.M., Webb E., Howarth K., Rowan A., Lubbe S., Spain S., Sullivan K., Fielding S., CORGI Consortium A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat. Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 35.Rhee J.-K., Lee S., Park W.-Y., Kim Y.-H., Kim T.-M. Allelic imbalance of somatic mutations in cancer genomes and transcriptomes. Sci. Rep. 2017;7:1653. doi: 10.1038/s41598-017-01966-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson A.D., Zhang Y., Papp A.C., Pinsonneault J.K., Lim J.E., Saffen D., Dai Z., Wang D., Sadée W. Polymorphisms affecting gene transcription and mRNA processing in pharmacogenetic candidate genes: detection through allelic expression imbalance in human target tissues. Pharmacogenet. Genomics. 2008;18:781–791. doi: 10.1097/FPC.0b013e3283050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pastinen T., Sladek R., Gurd S., Sammak A., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brändström H. A survey of genetic and epigenetic variation affecting human gene expression. Physiol. Genomics. 2004;16:184–193. doi: 10.1152/physiolgenomics.00163.2003. [DOI] [PubMed] [Google Scholar]
- 38.Lo H.S.S., Wang Z., Hu Y., Yang H.H.H., Gere S., Buetow K.H.H., Lee M.P.P. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.