Abstract
Using trio whole-exome sequencing, we have identified de novo heterozygous pathogenic variants in GRIA4 in five unrelated individuals with intellectual disability and other symptoms. GRIA4 encodes an AMPA receptor subunit known as GluR4, which is found on excitatory glutamatergic synapses and is important for learning and memory. Four of the variants are located in the highly conserved SYTANLAAF motif in the transmembrane protein M3, and the fifth is in an extra-cellular domain. Molecular modeling of the altered protein showed that three of the variants in the SYTANLAAF motif orient toward the center of the pore region and most likely lead to disturbance of the gating mechanism. The fourth variant in the SYTANLAAF motif most likely results in reduced permeability. The variant in the extracellular domain potentially interferes with the binding between the monomers. On the basis of clinical information and genetic results, and the fact that other subunits of the AMPA receptor have already been associated with neurodevelopmental disorders, we suggest that pathogenic de novo variants in GRIA4 lead to intellectual disability with or without seizures, gait abnormalities, problems of social behavior, and other variable features.
Keywords: GRIA4, de novo, exome sequencing, intellectual disability, speech delay, GluR4, AMPA receptor, seizures
Main Text
Intellectual disability (ID) has a prevalence of about 1%1 and is characterized by substantial limitations in both intellectual functioning and adaptive behavior starting before the age of 18 years (ICD-10 2016, World Health Organization). Recent studies have shown that de novo variants are a frequent cause of neurodevelopmental disorders.2
At five centers in Germany, Denmark, and the United States, we clinically examined five unrelated individuals with neurodevelopmental disorders, primarily ID. In addition, four of the five affected individuals had seizures or abnormal electroencephalography (EEG). Further symptoms were muscular hypertonia followed by spasticity in later age, abnormal brain MRI, and different behavioral disorders (Table 1 and Supplemental Note: Case Reports).
Table 1.
Clinical Features of Individuals with Predicted Deleterious Variants in GRIA4
|
Proband |
|||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Age at last examination | 15 years | 21 years | 4 years | 4 years | 4 years |
| Sex | male | male | male | male | female |
| HGVS DNA reference |
c.1915A>T | c.1921A>G | c.1928C>G | c.1931C>T | c.2090G>C |
| Variant | p.Thr639Ser | p.Asn641Asp | p.Ala643Gly | p.Ala644Val | p.Arg697Pro |
| Prenatal period | unremarkable pregnancy after IVF | unremarkable | maternal pre-eclampsia | unremarkable | unremarkable |
| Gestational week at birth | 40 | 40 | 34 | 39 | 40 |
| Birth parameters | weight 3,755 g (62P), length 53 cm (59P), OFC not known |
weight 3,320 g (25P), length and OFC not known | weight 3,033 g (95P), length 47 cm (61P), OFC 34 cm (86P) | weight 3,450 g (47P), length 53 cm (68P), OFC 37 cm (90P) |
weight 4,150 g (95P), length 53 cm (72P), OFC 36 cm (80P) |
| Neonatal period | stiffness (stiff baby syndrome), irritability, excessive startle reflex | irritability, stiffness | hypertonia, nystagmus, increased startle reflex | unremarkable | unremarkable |
| Congenital anomalies | none | none | none | none | none |
| Postnatal growth | unremarkable | failure to thrive, short stature, microcephaly | unremarkable | unremarkable | unremarkable |
| Developmental delay | mild to moderate | severe | severe | moderate to severe | mild to moderate |
| Speech | speech with dysarthria | non-verbal communication | non-verbal communication | non-verbal communication | single words and a few two-word sentences |
| Brain MRI | unremarkable | bilateral symmetric extensive atrophy of frontal lobes, mild frontal ventriculomegaly, thin corpus callosum | optic nerve hypoplasia | unremarkable | unremarkable |
| Social behavior | interaction with adults, reduced attention span, tension with irritability and anxiety | social smile, interaction with caregivers | occasional response to voice | 3 years: hyperactivity, reduced attention span, aggressive behavior, reduced interaction with other children; 4 years: non-verbal communication, ability to focus and play | lack of distance toward adults, reduced interaction with other children, strong searching for physical contact, mood changes with aggressive behavior and attention deficits |
| Muscle tone | hyperekplexia with exaggerated head-retraction reflex, stiffness, and hypertonia | severe spastic quadriplegia and hypertonia with contractures | spasticity | mild muscular hypotonia (neonatal) | unremarkable |
| Walking abilities | difficulties when walking in a straight line, stiff gait, ability to run | inability to walk | supported walking | clumsy gait | yes |
| Seizures | no, but severe contraction burst in relation to trauma | intractable generalized seizures, onset at 5 weeks | seizure-like episodes, onset at 14 months | febrile seizures at 13 months | no |
| Craniofacial dysmorphism | large ears | prognathism, midface retrusion, short philtrum, large ears | large ears | no | no |
| EEG | unremarkable | diffuse cerebral disturbance without electrographic correlates to the seizures | generalized slowing, no epileptiform discharges | generalized spikes and waves during sleep | unremarkable |
| Additional features or notes | sleeping problems in childhood | bilateral hiatal hernias, gastresophageal reflux, feeding difficulties, apneas, recurrent respiratory infections in first year of life, strabismus, choreiform movements | – | stereotypic hand movements | hyporeflexia, simian crease on both hands |
Abbreviations are as follows: IVF, in vitro fertilization; OFC, occipitofrontal circumference; MRI, magnetic resonance imaging; and P (e.g., 62P), percentile.
This study was approved by the ethics committees of the University of Leipzig (402/16-ek) and the University Medical Center of Hamburg-Eppendorf (PV3802). Informed consent was obtained from all examined individuals or their guardians. Otherwise, testing was done as part of routine clinical care, and therefore institutional ethics approval was not required. All families provided informed consent for clinical testing and publication.
We performed exome sequencing for all five individuals as a trio analysis including DNA samples of both biological parents. DNA from the proband and the parents was subjected to exome capture by NimbleGen SeqCap EZ MedExome (Roche), NimbleGen SeqCap EZ VCR (Roche), SureSelect Human All Exon 50Mb V5 (Agilent), or the Nextera Rapid Capture Exome Kit (Illumina). Sequencing was performed on an Illumina NextSeq 500, NextSeq 550, HiSeq 2000, or HiSeq 2500. For proband 1, raw reads were aligned with the Burrows-Wheeler Aligner (BWA-MEM) v.0.7.15,3 and the Genome Analysis Toolkit Best Practices pipeline v.3.8-0 was used for variant calling.4 Annotation and filtering of variants was performed with VarSeq 1.4.6 (Golden Helix) for proband 1 and his parents. For the other four probands, bioinformatic preparation of the data and data annotation and interpretation were performed with house-made pipelines as previously reported (see Supplemental Note: Exome Sequencing).5, 6, 7 Variants of interest were confirmed by dideoxy sequencing, and cosegregation analysis was performed in all informative, available family members. This revealed de novo missense variants in GRIA4(MIM: 138246) in the five probands: c.1915A>T (p.Thr639Ser), c.1921A>G (p.Asn641Asp), c.1928C>G (p.Ala643Gly), c.1931C>T (p.Ala644Val), and c.2090G>C (p.Arg697Pro) (see Table S1). Further protein-changing, de novo variants were not identified in any of the families (see also Table S2). None of the variants in GRIA4 have been observed in gnomAD8 (accessed October 2017) or the 1000 Genomes Project.9 Most of the in silico programs predicted the variants to be pathogenic, however inconstantly. In addition, GRIA4 is intolerant of loss-of-function variants (pLI = 0.99) and missense variants (Z score = 3.16).8
GRIA4 encodes GluR4, a subunit of the alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor.10, 11 These glutamate-dependent receptors consist of the four subunits GluR1, GluR2, GluR3, and GluR412 and have an important function in synaptic transmission, activity-dependent synaptic plasticity, and the control of network activity.13 AMPA receptors belong to the group of glutamate-gated cationic ion channels and are important for various neurologic functions, such as synaptic communication11 and long-term potentiation.14 AMPA receptors (encoded by GRIA1 [MIM: 138248], GRIA2 [MIM: 138247], GRIA3 [MIM: 305915], and GRIA4) share vast similarities with other ionotropic glutamate receptors, comprising kainate receptors (encoded by GRIK1 [MIM: 138245], GRIK2 [MIM: 138244], GRIK3 [MIM: 138243], GRIK4 [MIM: 600282], and GRIK5 [MIM: 600283]), NMDA receptors (encoded by GRIN1 [MIM: 138249], GRIN2A [MIM: 138253], GRIN2B [MIM: 138252], GRIN2C [MIM: 138254], GRIN2D [MIM: 602717], GRIN3A [MIM: 606650], and GRIN3B [MIM: 606651]), and δ receptors (encoded by GRID1 [MIM: 610659] and GRID2 [MIM: 602368]).12, 15 Pathogenic variants in several of these genes have been associated with developmental delay, ID, different epilepsy disorders (including epileptic encephalopathy), and ataxia.16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35
Subunits GluR1–GluR4 of the AMPA receptor can form both homo- and heteromers but are usually heteromeric.12, 36 The relative combination ratios of the four subunits varies in different types of neurons,36 and the conductance properties of the receptors are dependent on the assembly of their subunits.37 GRIA1 encodes GluR1 and is seen as an important element in associative memory formation.38 GRIA1 has been associated with ID and neurodevelopmental disorders due to a recurrent de novo missense variant, c.1906G>A (p.Ala636Thr).24, 28 GRIA2 encodes GluR2. It has been shown that receptors lacking GluR2 have a higher Ca2+ permeability and channel conductance (reviewed by Isaac et al.39). A deletion comprising exons 1 and 2 of GRIA2 has been described as a possible cause of ID (and gait abnormalities and abnormal behavior),40 and an in-frame deletion in GRIA2 has been described in a person with ID.2 Pathogenic variants in GRIA3 have been associated with X-linked mental retardation 94 (MIM: 300699). Different variants are described in the literature in persons with ID, seizures, autistic features, short stature, and behavioral problems.16, 27, 29, 30 GRIA4, the main subject of this study, has not yet been reported in association with any disorder. In rat brains, the subunit GluR4, encoded by Gria4, is found ubiquitously in the CNS, but its amount is relatively high in CA1 pyramidal cells and the dentate gyrus of the hippocampus, layers III and IV of the cerebral cortex, and the granule cells of the cerebellum.41 In the hippocampus, GluR4 was more abundant only in the first postnatal week.42 Sagata et al.11 produced Gria4−/− mice, which showed 10% lower weight than wild-type mice, normal anxiety-like behavior in a novel environment, muscle weakness, and more faults in the training period for the hippocampus-dependent Barnes circular maze test, which proofs spatial reference memory. In addition, these knockout mice showed normal spontaneous locomotor activity, and the authors suggest that their social behavior was improved. The authors concluded that the Gria4−/− mice were impaired solely in the acquisition of spatial reference memory and not the retention of spatial reference memory.11 However, Beyer et al.36 demonstrated that mice with a genetic deficiency of GRIA4, either resulting in reduced function or its complete absence, showed highly frequent spike-wave discharges on EEG, which are associated with absence epilepsy. In addition, some of the mice homozygous for the knockout allele developed a mild cerebellar ataxia.36 Notably, in both studies,11, 36 the modification of GRIA4 was toward loss of function. However, the identified variants in the present report are heterozygous and do not cause loss of function per se.
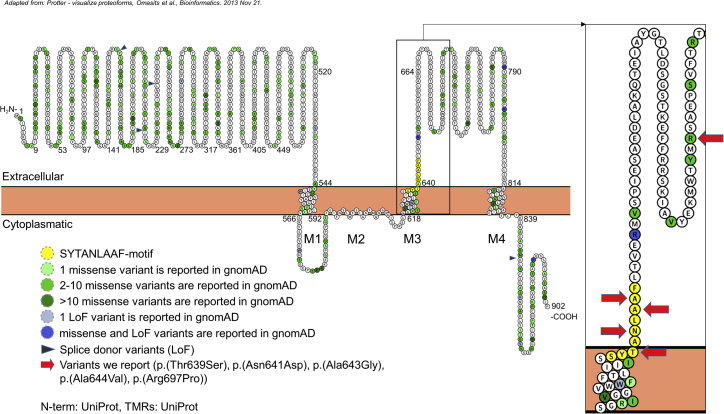
The variants p.Thr639Ser, p.Asn641Asp, p.Ala643Gly, and p.Ala644Val in probands 1, 2, 3, and 4, respectively, are located in the SYTANLAAF motif. AMPA and other glutamate receptors have three transmembrane domains (M1, M3, and M4) plus the re-entrant membrane loop M243 (Figure 1). The M2 loop and the M3 segment represent the major pore-lining domains in GluRs.44 The channel transition between symmetries possibly occurs at the M3 segment.45 This region contains the highly conserved SYTANLAAF motif,46 which has the strictest amino acid conservation among all members of the ionotropic glutamate receptor family.47 It is known that the region moves in response to activation and that a distinction among channel types is the degree of exposure.48, 49 Changes in the SYTANLAAF motif at Thr639 and Ala64346, 50 are associated with constitutively open channels.51 Several other studies have also proved that other changes in the SYTANLAAF motif lead to a permanently open channel (e.g., Chang and Kuo46) in the sense of gain of function.52, 53, 54 Human de novo missense variants affecting this motif have already been described as disease causing in several genes encoding ionotrophic glutamate receptors: GRIN1, GRIN2A, and GRID2.18, 24, 26, 55 Also, a recurrent de novo variant in GRIA1 affects this motif in a person with ID24, 28 (see Table S3). In addition, Li et al. reported a functionally proven gain-of-function de novo variant in GRIN2D, c.1999G>A (p.Val667Ile), affecting the encoded protein very close to the SYTANLAAF motif in two unrelated persons with epileptic encephalopathy.22
Figure 1.
Structure of GRIA4 and Variant Location
In addition, we show the allele count of genetic variants reported by gnomAD8 for the canonical transcript ENST00000282499 (GenBank: NM_000829.3). At position Arg697, where we describe the variant p.Arg697Pro in proband 5 in this report, another variant (p.Arg697Gln) is reported in gnomAD.8 In the main text, we describe the most likely mild impact of this change on the function of the protein. The figure was made with the help of Protter.64
The alanine residues at positions 643 and 644 orient toward the center of the pore region and most likely inhibit closing of the channel either by trapping the channel in the open state or allowing leakage, thus resulting in constitutive opening of the channel.56 Like p.Ala643Gly, the change from threonine to serine at position 639 removes a methyl group that partially occludes the closed channel, most likely resulting in a partially open or leaky channel in the closed state. As Table S4 shows, variants at Ala643 and Ala644 introduce moderate steric perturbations. However, they significantly affect the function of the channel by disrupting the interactions between monomers in the membrane and disrupting the gating and transport of ions through the channel. In addition, the variant c.1931C>T (p.Ala644Val) in proband 4 is at the same position in the motif as the pathogenic variants in GRIA1 (p.Ala636Thr)24, 28 and GRID2 (p.Ala654Asp and p.Ala654Thr),26 as well as in the functionally well-studied Lurcher mouse model. The Lurcher variant (SYTANLAAF to SYTANLATF in the GluRδ2 receptor) in one subunit of a heteromeric glutamate receptor is functionally dominant, leads to spontaneous channel activity, and slows channel kinetics, even though the changes in gating kinetics are increased in homomeric Lc-mutated forms.52
Conversely, for the known variants p.Asn641Lys and p.Asn641Cys, variants at the equivalent position in the paralog GRIA2 (p.Asn619Lys and p.Asn619Cys) are associated with significant loss of permeability.57 Our molecular modeling showed that residue Asn641 orients toward the adjacent monomers, interacts with the backbone atoms of the nearby loops, and thus stabilizes the open and closed states and most likely mediates the transition between states. Loss of this functionality either inhibits the transition or allows the channel to become trapped in an inactivated state.
The fifth variant in this study is c.2090G>C, leading to the protein change p.Arg697Pro, which is located outside the SYTANLAAF motif. Pathogenic variants leading to protein changes outside of the SYTANLAAF sequence have already been described in association with ID, different epilepsy disorders, or ataxia in the glutamate receptor genes GRIA3, GRIN1, GRIN2A, GRIN2B, GRIN2D, GRID2, and GRIK2.16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 29, 30, 31, 32, 33, 34, 55 In the homologous segments of the protein sequence around our variant p.Arg697Pro, pathogenic variants (e.g., the two variants in GluR3, p.Met706Thr [c.2117T>C] and p.Gly721Arg [c.2161G>A]) have also been identified.16, 27 The region around Arg697 lies at the interface between monomers in the tetramer within the ligand binding domain (Figure 2), which is involved in agonist and antagonist binding and activation of the transmembrane domain.58 The residue itself is partially exposed and does not appear to engage in strong interactions with residues on adjacent monomers. It should be noted that between the homology models of the bound (PDB: 3KG245) and apo (PDB: 4U2P59) states, Arg697 moves by more than 10 Å (see Figure 2). Additionally, the conformation for the ligand-bound state places Arg697 near repulsive side chains on Lys787 and Lys792, whereas in the apo state it interacts favorably with Glu444 of the adjacent monomers, thereby stabilizing the apo state. The variant identified in this study, p.Arg697Pro, leads to the insertion of a helix breaker in the middle of an α helix at the interface between monomers. So, p.Arg697Pro leads to the disruption of the α helix and potentially to local unwinding, which in turn interferes with binding between monomers. Additionally, the loss of the charged sidechain disturbs the equilibrium between the two states. This is not the case for variant p.Arg697Gln, which has an allele frequency of 0.00002 in gnomAD8 (see Figure 1). The destabilization energy of p.Arg697Gln is small (0.17 kcal), most likely indicating that the change does not strongly disrupt protein function. Variant p.Arg697Gln has a more subtle effect and could interfere at an intermediate stage in monomer folding or tetramerization and most likely affects functionality more mildly such that it might be imperceptible from baseline variations.
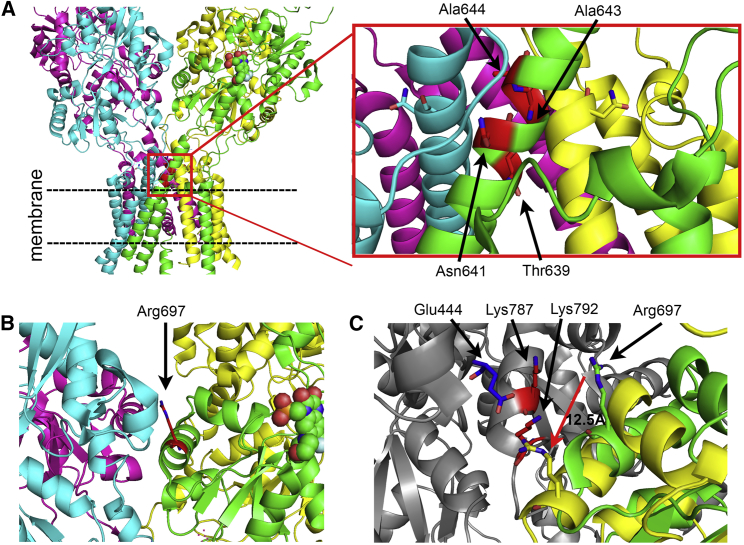
Figure 2.
Tetrameric Structure of GRIA4 with Variants and Allosteric Movements
The structure of GRIA4 was built off the homologous structure of GRIA2 for Rattus norvegicus in the complex with an agonist (PDB: 3KG245). The homology model of Gria4 was built off the homology modeling module of ROSETTA,60 where the sequence alignments were generated with TCOFFEE.61, 62 The energy calculations of both and the destabilization of internal structure were calculated with the FoldX program.63 The images were generated with PyMOL (PyMOL Molecular Graphics System, v.1.8, Schrödinger).
(A) GRIA4 tetramer with residues in the SYTANLAAF motif of M3. The residue positions of missense variants are indicated by red sticks.
(B) GRIA4 tetramer with a variant in the ligand binding domain. The residue position of wild-type Arg697 is indicated by red sticks.
(C) Arg697 moves by more than 10 Å from the ligand-bound (green; PDB: 3KG210) to the apo (yellow; PDB: 4U2P62) state. Arg697 interacts with residues on the adjacent monomer (gray), including the repulsive Lys787 and Lys792 (red sticks) in the ligand-bound state and the attractive Glu444 (blue sticks) in the apo state.
The importance of GRIA4 in neuronal function, the positions of the identified variants and their predicted effects on the protein, and the previously published data on the other subunits of the AMPA receptor and other glutamate receptors let us consider the de novo variants in GRIA4 in this study as causative for the phenotypes of our probands. The data that we have let us assume that the pathogenic effect is due to a dominant functional effect and not loss of function. This is in line with other pathogenic variants in glutamate receptors. In addition, the positions of our variants and the executed molecular modeling suggest some degree of genotype-phenotype correlation: a mild phenotype of the girl with the p.Arg697Pro variant outside the SYTANLAAF-motif and a more severe phenotype for variants within the motif. Also, within the motif, the variant at position 639 (which has a lesser effect on the structural destabilization of GRIA4) is in an individual with milder symptoms. However, this still requires further analysis and the identification of additional affected individuals. In conclusion, we suggest de novo, heterozygous pathogenic variants in GRIA4 as causative for ID with or without seizures and gait abnormalities.
Conflicts of Interest
A.C., D.N.S., and K.L.H. are employed by and receive a salary from Ambry Genetics. A.F. is a consultant to Ambry Genetics. J.S.C. is a consultant to Invitae. Exome sequencing is a commercially available test.
Published: December 7, 2017
Footnotes
Supplemental Data include two Supplemental Notes and four tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.11.004.
Accession Numbers
The accession numbers for the GRIA4 variants reported in this paper are ClinVar: SCV000611121, SCV000611122, SCV000611123, SCV000611124, SCV000611125, and SCV000611126.
Web Resources
Genome Aggregation Database (gnomAD), http://gnomad.broadinstitute.org/
MutationTaster, http://www.mutationtaster.org/
OMIM, http://omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Protter, http://wlab.ethz.ch/protter/
REVEL: Rare Exome Variant Ensemble Learner, https://sites.google.com/site/revelgenomics/
RSCB Protein Data Bank, https://www.rcsb.org/pdb/home/home.do
UCSC Genome Browser, https://genome.ucsc.edu/
Varvis, https://www.limbus-medtec.com/
wANNOVAR, http://wannovar.wglab.org/
Supplemental Data
References
- 1.Maulik P.K., Mascarenhas M.N., Mathers C.D., Dua T., Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res. Dev. Disabil. 2011;32:419–436. doi: 10.1016/j.ridd.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 2.McRae J.F., Clayton S., Fitzgerald T.W., Kaplanis J., Prigmore E., Rajan D., Sifrim A., Aitken S., Akawi N., Alvi M., Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li, H. (2013). Aligning sequence reads, clone sequences and assembly contains with BWA-MEM. arXiv, https://arxiv.org/abs/1303.3997.
- 4.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:1–33. doi: 10.1002/0471250953.bi1110s43. 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farwell K.D., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Tippin Davis B., Baxter R.M., Zeng W., Mroske C., Parra M.C. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 6.Hempel M., Cremer K., Ockeloen C.W., Lichtenbelt K.D., Herkert J.C., Denecke J., Haack T.B., Zink A.M., Becker J., Wohlleber E. De Novo Mutations in CHAMP1 Cause Intellectual Disability with Severe Speech Impairment. Am. J. Hum. Genet. 2015;97:493–500. doi: 10.1016/j.ajhg.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trujillano D., Bertoli-Avella A.M., Kumar Kandaswamy K., Weiss M.E., Köster J., Marais A., Paknia O., Schröder R., Garcia-Aznar J.M., Werber M. Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur. J. Hum. Genet. 2017;25:176–182. doi: 10.1038/ejhg.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barad Z., Shevtsova O., Arbuthnott G.W., Leitch B. Selective loss of AMPA receptors at corticothalamic synapses in the epileptic stargazer mouse. Neuroscience. 2012;217:19–31. doi: 10.1016/j.neuroscience.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 11.Sagata N., Iwaki A., Aramaki T., Takao K., Kura S., Tsuzuki T., Kawakami R., Ito I., Kitamura T., Sugiyama H. Comprehensive behavioural study of GluR4 knockout mice: implication in cognitive function. Genes Brain Behav. 2010;9:899–909. doi: 10.1111/j.1601-183X.2010.00629.x. [DOI] [PubMed] [Google Scholar]
- 12.Traynelis S.F., Wollmuth L.P., McBain C.J., Menniti F.S., Vance K.M., Ogden K.K., Hansen K.B., Yuan H., Myers S.J., Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bettler B., Fakler B. Ionotropic AMPA-type glutamate and metabotropic GABAB receptors: determining cellular physiology by proteomes. Curr. Opin. Neurobiol. 2017;45:16–23. doi: 10.1016/j.conb.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Malinow R. AMPA receptor trafficking and long-term potentiation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003;358:707–714. doi: 10.1098/rstb.2002.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan H., Low C.-M., Moody O.A., Jenkins A., Traynelis S.F. Ionotropic GABA and Glutamate Receptor Mutations and Human Neurologic Diseases. Mol. Pharmacol. 2015;88:203–217. doi: 10.1124/mol.115.097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu H., Haas S.A., Chelly J., Van Esch H., Raynaud M., de Brouwer A.P.M., Weinert S., Froyen G., Frints S.G.M., Laumonnier F. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol. Psychiatry. 2016;21:133–148. doi: 10.1038/mp.2014.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zehavi Y., Mandel H., Zehavi A., Rashid M.A., Straussberg R., Jabur B., Shaag A., Elpeleg O., Spiegel R. De novo GRIN1 mutations: An emerging cause of severe early infantile encephalopathy. Eur. J. Med. Genet. 2017;60:317–320. doi: 10.1016/j.ejmg.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Ohba C., Shiina M., Tohyama J., Haginoya K., Lerman-Sagie T., Okamoto N., Blumkin L., Lev D., Mukaida S., Nozaki F. GRIN1 mutations cause encephalopathy with infantile-onset epilepsy, and hyperkinetic and stereotyped movement disorders. Epilepsia. 2015;56:841–848. doi: 10.1111/epi.12987. [DOI] [PubMed] [Google Scholar]
- 19.Lemke J.R., Geider K., Helbig K.L., Heyne H.O., Schütz H., Hentschel J., Courage C., Depienne C., Nava C., Heron D. Delineating the GRIN1 phenotypic spectrum: A distinct genetic NMDA receptor encephalopathy. Neurology. 2016;86:2171–2178. doi: 10.1212/WNL.0000000000002740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemke J.R., Hendrickx R., Geider K., Laube B., Schwake M., Harvey R.J., James V.M., Pepler A., Steiner I., Hörtnagel K. GRIN2B mutations in West syndrome and intellectual disability with focal epilepsy. Ann. Neurol. 2014;75:147–154. doi: 10.1002/ana.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemke J.R., Lal D., Reinthaler E.M., Steiner I., Nothnagel M., Alber M., Geider K., Laube B., Schwake M., Finsterwalder K. Mutations in GRIN2A cause idiopathic focal epilepsy with rolandic spikes. Nat. Genet. 2013;45:1067–1072. doi: 10.1038/ng.2728. [DOI] [PubMed] [Google Scholar]
- 22.Li D., Yuan H., Ortiz-Gonzalez X.R., Marsh E.D., Tian L., McCormick E.M., Kosobucki G.J., Chen W., Schulien A.J., Chiavacci R. GRIN2D Recurrent De Novo Dominant Mutation Causes a Severe Epileptic Encephalopathy Treatable with NMDA Receptor Channel Blockers. Am. J. Hum. Genet. 2016;99:802–816. doi: 10.1016/j.ajhg.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi Y., Tohyama J., Kato M., Akasaka N., Magara S., Kawashima H., Ohashi T., Shiraishi H., Nakashima M., Saitsu H., Matsumoto N. High prevalence of genetic alterations in early-onset epileptic encephalopathies associated with infantile movement disorders. Brain Dev. 2016;38:285–292. doi: 10.1016/j.braindev.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 24.de Ligt J., Willemsen M.H., van Bon B.W., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 25.Swanger S.A., Chen W., Wells G., Burger P.B., Tankovic A., Bhattacharya S., Strong K.L., Hu C., Kusumoto H., Zhang J. Mechanistic Insight into NMDA Receptor Dysregulation by Rare Variants in the GluN2A and GluN2B Agonist Binding Domains. Am. J. Hum. Genet. 2016;99:1261–1280. doi: 10.1016/j.ajhg.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coutelier M., Burglen L., Mundwiller E., Abada-Bendib M., Rodriguez D., Chantot-Bastaraud S., Rougeot C., Cournelle M.-A., Milh M., Toutain A. GRID2 mutations span from congenital to mild adult-onset cerebellar ataxia. Neurology. 2015;84:1751–1759. doi: 10.1212/WNL.0000000000001524. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y., Arai A.C., Rumbaugh G., Srivastava A.K., Turner G., Hayashi T., Suzuki E., Jiang Y., Zhang L., Rodriguez J. Mutations in ionotropic AMPA receptor 3 alter channel properties and are associated with moderate cognitive impairment in humans. Proc. Natl. Acad. Sci. USA. 2007;104:18163–18168. doi: 10.1073/pnas.0708699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geisheker M.R., Heymann G., Wang T., Coe B.P., Turner T.N., Stessman H.A.F., Hoekzema K., Kvarnung M., Shaw M., Friend K. Hotspots of missense mutation identify neurodevelopmental disorder genes and functional domains. Nat. Neurosci. 2017;20:1043–1051. doi: 10.1038/nn.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen N.M., Conroy J., Shahwan A., Lynch B., Correa R.G., Pena S.D.J., McCreary D., Magalhães T.R., Ennis S., Lynch S.A., King M.D. Unexplained early onset epileptic encephalopathy: Exome screening and phenotype expansion. Epilepsia. 2016;57:e12–e17. doi: 10.1111/epi.13250. [DOI] [PubMed] [Google Scholar]
- 30.Philips A.K., Sirén A., Avela K., Somer M., Peippo M., Ahvenainen M., Doagu F., Arvio M., Kääriäinen H., Van Esch H. X-exome sequencing in Finnish families with intellectual disability--four novel mutations and two novel syndromic phenotypes. Orphanet J. Rare Dis. 2014;9:49. doi: 10.1186/1750-1172-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao K., Tankovic A., Zhang Y., Kusumoto H., Zhang J., Chen W., XiangWei W., Shaulsky G.H., Hu C., Traynelis S.F. A de novo loss-of-function GRIN2A mutation associated with childhood focal epilepsy and acquired epileptic aphasia. PLoS ONE. 2017;12:e0170818. doi: 10.1371/journal.pone.0170818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Platzer K., Yuan H., Schütz H., Winschel A., Chen W., Hu C., Kusumoto H., Heyne H.O., Helbig K.L., Tang S. GRIN2B encephalopathy: novel findings on phenotype, variant clustering, functional consequences and treatment aspects. J. Med. Genet. 2017;54:460–470. doi: 10.1136/jmedgenet-2016-104509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen A.S., Berkovic S.F., Cossette P., Delanty N., Dlugos D., Eichler E.E., Epstein M.P., Glauser T., Goldstein D.B., Han Y., Epi4K Consortium. Epilepsy Phenome/Genome Project De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Endele S., Rosenberger G., Geider K., Popp B., Tamer C., Stefanova I., Milh M., Kortüm F., Fritsch A., Pientka F.K. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat. Genet. 2010;42:1021–1026. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- 35.Fogel B.L., Lee H., Deignan J.L., Strom S.P., Kantarci S., Wang X., Quintero-Rivera F., Vilain E., Grody W.W., Perlman S. Exome sequencing in the clinical diagnosis of sporadic or familial cerebellar ataxia. JAMA Neurol. 2014;71:1237–1246. doi: 10.1001/jamaneurol.2014.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beyer B., Deleuze C., Letts V.A., Mahaffey C.L., Boumil R.M., Lew T.A., Huguenard J.R., Frankel W.N. Absence seizures in C3H/HeJ and knockout mice caused by mutation of the AMPA receptor subunit Gria4. Hum. Mol. Genet. 2008;17:1738–1749. doi: 10.1093/hmg/ddn064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuner T., Beck C., Sakmann B., Seeburg P.H. Channel-lining residues of the AMPA receptor M2 segment: structural environment of the Q/R site and identification of the selectivity filter. J. Neurosci. 2001;21:4162–4172. doi: 10.1523/JNEUROSCI.21-12-04162.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rumpel S., LeDoux J., Zador A., Malinow R. Postsynaptic receptor trafficking underlying a form of associative learning. Science. 2005;308:83–88. doi: 10.1126/science.1103944. [DOI] [PubMed] [Google Scholar]
- 39.Isaac J.T., Ashby M.C., McBain C.J. The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron. 2007;54:859–871. doi: 10.1016/j.neuron.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Hackmann K., Matko S., Gerlach E.-M., von der Hagen M., Klink B., Schrock E., Rump A., Di Donato N. Partial deletion of GLRB and GRIA2 in a patient with intellectual disability. Eur. J. Hum. Genet. 2013;21:112–114. doi: 10.1038/ejhg.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keinänen K., Wisden W., Sommer B., Werner P., Herb A., Verdoorn T.A., Sakmann B., Seeburg P.H. A family of AMPA-selective glutamate receptors. Science. 1990;249:556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- 42.Zhu J.J., Esteban J.A., Hayashi Y., Malinow R. Postnatal synaptic potentiation: delivery of GluR4-containing AMPA receptors by spontaneous activity. Nat. Neurosci. 2000;3:1098–1106. doi: 10.1038/80614. [DOI] [PubMed] [Google Scholar]
- 43.Dingledine R., Borges K., Bowie D., Traynelis S.F. The glutamate receptor ion channels. Pharmacol. Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 44.Sobolevsky A.I., Yelshansky M.V., Wollmuth L.P. State-dependent changes in the electrostatic potential in the pore of a GluR channel. Biophys. J. 2005;88:235–242. doi: 10.1529/biophysj.104.049411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sobolevsky A.I., Rosconi M.P., Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang H.R., Kuo C.C. The activation gate and gating mechanism of the NMDA receptor. J. Neurosci. 2008;28:1546–1556. doi: 10.1523/JNEUROSCI.3485-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan H., Erreger K., Dravid S.M., Traynelis S.F. Conserved structural and functional control of N-methyl-D-aspartate receptor gating by transmembrane domain M3. J. Biol. Chem. 2005;280:29708–29716. doi: 10.1074/jbc.M414215200. [DOI] [PubMed] [Google Scholar]
- 48.Blanke M.L., van Dongen A.M.J. Activation Mechanism of the NMDA receptor. In: van Dongen A.M., editor. Biology of the NMDA Receptor. CRC Press/Taylor & Francis; 2009. [Google Scholar]
- 49.Sobolevsky A.I., Yelshansky M.V., Wollmuth L.P. Different gating mechanisms in glutamate receptor and K+ channels. J. Neurosci. 2003;23:7559–7568. doi: 10.1523/JNEUROSCI.23-20-07559.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wollmuth L.P., Sobolevsky A.I. Structure and gating of the glutamate receptor ion channel. Trends Neurosci. 2004;27:321–328. doi: 10.1016/j.tins.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 51.Zuo J., De Jager P.L., Takahashi K.A., Jiang W., Linden D.J., Heintz N. Neurodegeneration in Lurcher mice caused by mutation in delta2 glutamate receptor gene. Nature. 1997;388:769–773. doi: 10.1038/42009. [DOI] [PubMed] [Google Scholar]
- 52.Schwarz M.K., Pawlak V., Osten P., Mack V., Seeburg P.H., Köhr G. Dominance of the lurcher mutation in heteromeric kainate and AMPA receptor channels. Eur. J. Neurosci. 2001;14:861–868. doi: 10.1046/j.0953-816x.2001.01705.x. [DOI] [PubMed] [Google Scholar]
- 53.Lee J.H., Wei L., Deveau T.C., Gu X., Yu S.P. Expression of the NMDA receptor subunit GluN3A (NR3A) in the olfactory system and its regulatory role on olfaction in the adult mouse. Brain Struct. Funct. 2016;221:3259–3273. doi: 10.1007/s00429-015-1099-3. [DOI] [PubMed] [Google Scholar]
- 54.Jones K.S., VanDongen H.M., VanDongen A.M. The NMDA receptor M3 segment is a conserved transduction element coupling ligand binding to channel opening. J. Neurosci. 2002;22:2044–2053. doi: 10.1523/JNEUROSCI.22-06-02044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lesca G., Rudolf G., Bruneau N., Lozovaya N., Labalme A., Boutry-Kryza N., Salmi M., Tsintsadze T., Addis L., Motte J. GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat. Genet. 2013;45:1061–1066. doi: 10.1038/ng.2726. [DOI] [PubMed] [Google Scholar]
- 56.Jatzke C., Hernandez M., Wollmuth L.P. Extracellular vestibule determinants of Ca2+ influx in Ca2+-permeable AMPA receptor channels. J. Physiol. 2003;549:439–452. doi: 10.1113/jphysiol.2002.034413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perrais D., Coussen F., Mulle C. Atypical functional properties of GluK3-containing kainate receptors. J. Neurosci. 2009;29:15499–15510. doi: 10.1523/JNEUROSCI.2724-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stern-Bach Y., Bettler B., Hartley M., Sheppard P.O., O’Hara P.J., Heinemann S.F. Agonist selectivity of glutamate receptors is specified by two domains structurally related to bacterial amino acid-binding proteins. Neuron. 1994;13:1345–1357. doi: 10.1016/0896-6273(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 59.Dürr K.L., Chen L., Stein R.A., De Zorzi R., Folea I.M., Walz T., Mchaourab H.S., Gouaux E. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell. 2014;158:778–792. doi: 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Erb I., Notredame C. How should we measure proportionality on relative gene expression data? Theory Biosci. 2016;135:21–36. doi: 10.1007/s12064-015-0220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Catuara-Solarz S., Espinosa-Carrasco J., Erb I., Langohr K., Gonzalez J.R., Notredame C., Dierssen M. Combined Treatment With Environmental Enrichment and (-)-Epigallocatechin-3-Gallate Ameliorates Learning Deficits and Hippocampal Alterations in a Mouse Model of Down Syndrome. eNeuro. 2016;3 doi: 10.1523/ENEURO.0103-16.2016. ENEURO.0103-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Omasits U., Ahrens C.H., Müller S., Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30:884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.