Abstract
Copy-number variants (CNVs) of chromosome 15q13.3 manifest clinically as neuropsychiatric disorders with variable expressivity. CHRNA7, encoding for the α7 nicotinic acetylcholine receptor (nAChR), has been suggested as a candidate gene for the phenotypes observed. Here, we used induced pluripotent stem cells (iPSCs) and neural progenitor cells (NPCs) derived from individuals with heterozygous 15q13.3 deletions and heterozygous 15q13.3 duplications to investigate the CHRNA7-dependent molecular consequences of the respective CNVs. Unexpectedly, both deletions and duplications lead to decreased α7 nAChR-associated calcium flux. For deletions, this decrease in α7 nAChR-dependent calcium flux is expected due to haploinsufficiency of CHRNA7. For duplications, we found that increased expression of CHRNA7 mRNA is associated with higher expression of nAChR-specific and resident ER chaperones, indicating increased ER stress. This is likely a consequence of inefficient chaperoning and accumulation of α7 subunits in the ER, as opposed to being incorporated into functional α7 nAChRs at the cell membrane. Here, we showed that α7 nAChR-dependent calcium signal cascades are downregulated in both 15q13.3 deletion and duplication NPCs. While it may seem surprising that genomic changes in opposite direction have consequences on downstream pathways that are in similar direction, it aligns with clinical data, which suggest that both individuals with deletions and duplications of 15q13.3 manifest neuropsychiatric disease and cognitive deficits.
Keywords: iPSCs, NPCs, CHRNA7, 15q13.3 CNVs, neurodevelopmental disorders, neuropsychiatric disorders
Introduction
Copy-number variation at 15q13.3 has proven to be a major contributor to neuropsychiatric disease. Among individuals carrying recurrent 15q13.3 copy-number variants (CNVs), a wide range of neuropsychiatric phenotypes have been reported, including intellectual disability/developmental delay (ID/DD), epilepsy, autism spectrum disorder (ASD [MIM: 209850]), and schizophrenia (MIM: 181500).1, 2 The most severe of these phenotypes are observed in rare individuals with homozygous deletions, resulting in neonatal encephalopathy.3, 4, 5, 6 The highly penetrant heterozygous deletions (15q13.3 microdeletion syndrome [MIM: 612001]) result in moderate ID/DD, seizures, and ASD.7 Duplications at 15q13.3 are the most common CNVs contributing to neuropsychiatric disease. In a recent genotype-to-phenotype study of a cohort of individuals with 15q13.3 gains, we found that they tend to exhibit a milder, yet similar, phenotype in comparison to deletion probands, consisting of borderline ID, ASD, and attention deficit-hyperactivity disorder (ADHD [MIM: 143465]). This is in line with associations that have previously been reported, with 15q13.3 duplications found in 1.25% of probands with ADHD.8, 9 However, gains are also present at high frequency in control populations (0.6%),10 leading to open questions about the true pathogenicity of these CNVs.
While the full range of phenotypes for both 15q13.3 deletions and duplications has not been fully established, both our and others’ previously published work supports the idea that 15q13.3 duplication probands have a milder neurobehavioral phenotype than those with deletions, although with incomplete penetrance and considerable variability.
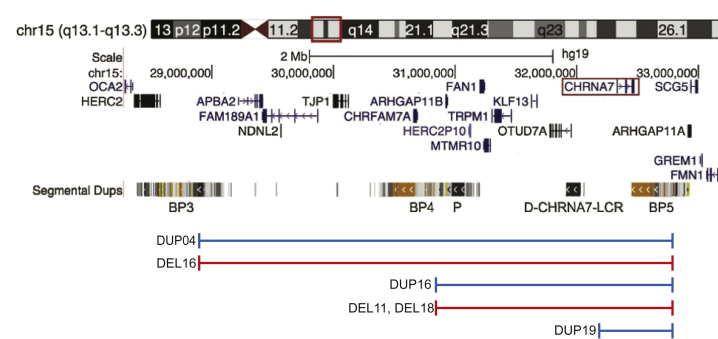
Due to an enrichment of low copy repeat (LCR) elements clustering into breakpoints (BPs) along the chromosome, 15q13.3 is one of the least stable regions in the human genome and thus is incredibly vulnerable to structural variation mediated by nonallelic homologous recombination (NAHR). While CNVs in this region vary in size depending on what BPs are used, the majority of deletions are mediated by BP4 and BP5 and contain six genes: FAN1 (MIM: 613534), MTMR10, TRPM1 (MIM: 603576), KLF13 (MIM: 605328), OTUD7A (MIM: 612024), and CHRNA7 (MIM: 118511), as well as one micro-RNA: hsa-mir-211 (Figure 1). The most frequent of duplications occur between the distal-CHRNA7-LCR (D-CHRNA7-LCR) and BP5, encompassing CHRNA7 and possibly the first noncoding exon of OTUD7A. While mechanistically it should be included, limitations in the resolution of clinical CMA analysis have left it unclear whether part of OTUD7A is encompassed by these small CNVs, and what the consequence, if any, would be on its expression.1, 2, 11 Furthermore, due to the 5′ end of CHRNA7 spanning BP5, it has not been confirmed whether the gene is fully duplicated, which may be relevant for its mode of pathogenesis.
Figure 1.
15q13.3 Region and CNVs in Probands
Region from breakpoint (BP) 3 to BP5 shown, including BP4 and the proximal (P) and distal (D-CHRNA7-LCR) CHRNA7-LCRs. Red/blue regions indicate recurrent CNVs observed. Probands’ CNVs in the study are shown underneath their appropriate recurrent CNV in blue (duplications) or red (deletions), labeled with their identifier. CHRNA7 is highlighted by a red box. Adapted from UCSC Genome Browser.
Small deletions and duplications mediated by the D-CHRNA7-LCR and BP5 highlight CHRNA7 as a candidate gene for the neuropsychiatric phenotypes observed.12 CHRNA7 encodes for the α7 nicotinic acetylcholine receptor (nAChR) subunit and is highly expressed in the brain, particularly in the hippocampus.13 The gene has been implicated in neuronal functions, including learning, memory, and attention.1, 13 α7 nAChRs are located pre-, post-, and extra-synaptically and are important for mediating fast signal transduction at synapses. When stimulated by agonists, these channels open and allow flux of Na+, K+, and Ca2+, with higher Ca2+ permeability than other nAChRs.14, 15 The influx of calcium can activate secondary messengers, depolarize the membrane, and activate voltage-gated ion channels to increase calcium flux and stimulate calcium-induced calcium release (CICR) from internal stores.16 This results in downstream calcium signaling effectors being activated, which are involved in a multitude of cellular processes.
When considering the human population, CHRNA7 is a strong candidate gene for many of the phenotypes observed in individuals with 15q13.3 CNVs. The majority of the probands carrying 15q13.3 CNVs do not carry additional CNVs, suggesting that a gene or genes in the region are responsible for their phenotypes.1 Additionally, multiple cohorts of individuals with neuropsychiatric disorders have had positive responses to treatment with α7 nAChR agonists and positive allosteric modulators (PAMs). In individuals with schizophrenia, α7 nAChR targeting agonists and PAMs have been utilized with clinically relevant results, including a reduction of negative symptoms and improvements in cognition. Similarly, although in much smaller cohorts, α7 nAChR agonists and PAMs have been utilized in ASD, with reported improvements in social behavior.17, 18, 19 Broad nAChR agonist galantamine, an FDA-approved drug, has been used in the treatment of Alzheimer disease (MIM: 104300), which also has the α7 nAChR implicated in its pathogenesis.15 This is likely impacting α7 nAChRs in particular, as it has also been used successfully in a proband carrying a 15q13.3 deletion diagnosed with aggressive behaviors and schizophrenia.20 Overall, this evidence strongly supports that CHRNA7 is playing a significant role in the phenotypes observed in probands, both with 15q13.3 CNVs and possibly in a broader population of individuals with neuropsychiatric phenotypes.
While the human data support the notion of CHRNA7 as a potential candidate gene for 15q13.3 CNV phenotypes, animal models of Chrna7 loss of function have provided little evidence. For deletions, Chrna7 knockout mice have been found to exhibit few of the human behavioral phenotypes.21, 22 On a functional level, decreased hippocampal inhibitory function and alterations in GABAA receptors have been noted, although these do not result in measurable behavioral changes.23 This suggests that there may be compensatory mechanisms in the mouse that do not occur in humans, although this has yet to be determined. Furthermore, it has been shown that RIC3, a chaperone required for biogenesis and trafficking of α7 nAChRs, may function differently in mouse versus humans.24 Knockout mice of the typical BP4/BP5 deletions have been shown to recapitulate some phenotypes observed in individuals with 15q13.3 losses, including epilepsy, autism, and schizophrenia-like behaviors, suggesting that other genes in the region could be contributing to the phenotypes observed in 15q13.3 microdeletion syndrome.25 While clinical CMA data have highlighted the phenotypes associated with 15q13.3 duplications, no models of these CNVs have been reported.
Induced pluripotent stem cells (iPSCs) have proven to be an extremely valuable resource for studying human disease. Many neuropsychiatric disorders, including Rett syndrome (MIM: 312750), Timothy syndrome (MIM: 601005), bipolar disorder, schizophrenia, and idiopathic autism, have been modeled using iPSCs, neural progenitor cells (NPCs), and terminally differentiated neurons.26, 27, 28, 29, 30 Recently, the expression and functionality of nAChRs have been explored in human iPSC-derived neurons.31, 32 In these studies, it was concluded that the α7 nAChR was the major functional nAChR subtype in iPSC-derived neuronal cells. In order to complement animal models of CHRNA7-dependent disease, with all their species-dependent limitations especially when it comes to modeling human behavior and psychiatric disease, we decided to develop human iPSC lines from both individuals with 15q13.3 deletions and 15q13.3 duplications. These iPSCs, along with the induced neuronal cell lines derived from them, represent an exciting model to study the molecular consequences of CHRNA7 copy-number variation, and may represent a tool for developing therapeutics for the affected individuals down the line.
Here, we explored gene expression, α7 nAChR-dependent calcium flux, and consequences of altered calcium signaling in iPSCs and NPCs derived from 15q13.3 CNV probands. With the limitations of clinical CMA for this region of the genome, we first answered questions of gene expression of CHRNA7 and OTUD7A for duplication NPCs. We then determined that these proband-derived cells have α7 nAChR-dependent phenotypes, with both CNVs resulting in decreased α7 nAChR-dependent calcium flux, known to be an extremely efficient way to increase calcium concentrations in the cell. This decreased α7 nAChR-dependent calcium flux has similar consequences for both 15q13.3 deletions and duplications, resulting in downregulation of downstream effectors. In NPCs with 15q13.3 deletions, this is likely due to haploinsufficiency of CHRNA7. We propose a mechanism for the pathogenicity of CHRNA7 duplications of increased α7 protein levels in the ER, resulting in ER stress and impaired chaperoning of α7 subunits to the membrane. While the decreased α7 nAChR functionality in 15q13.3 duplication cells is surprising based on genomics and mRNA expression, these similar molecular phenotypes are in line with the clinical traits observed in the respective probands.
Material and Methods
Establishment of Cohort of CHRNA7 CNV Probands
Probands were recruited based on known 15q13.3 CNVs identified by clinical CMA testing, as previously described.7, 10 Studies recruiting the probands were approved by the Institutional Review Board at Baylor College of Medicine, and probands consented to use of their cells in research. A total of six probands (three with 15q13.3 deletion and three with 15q13.3 duplication) and three control siblings of both sexes were biopsied (probands aged 7–14, controls aged 4–7, Figure 1). Five of the six individuals carrying 15q13.3 CNVs had previously been phenotyped by a team of specialists at Texas Children’s Hospital, and the final duplication individual was reportedly asymptomatic (Table 1).7, 10 Secondary CNVs of uncertain clinical significance were detected in one control subject (CONTROL2) and her 15q13.3 duplication sibling (DUP16), as well as in one deletion proband (DEL18) (Table 1).7, 10 Genotypes of all individuals in this study were confirmed using multiplex ligation-dependent probe amplification (MLPA) as previously described.7
Table 1.
Clinical Characteristics of Probands
| Identifier | Sex | Age at Biopsy | 15q13.3 CNV | ID/DD | ASD | ADHD |
|---|---|---|---|---|---|---|
| DUP04a | M | 9 years | BP3/BP5 duplication | yes | no | yes |
| DUP16a,b | F | 13 years | BP4/BP5 duplication | borderline | yes | yes |
| DUP19c | F | 7 years | D-CHRNA7-LCR/BP5 duplication | no | no | no |
| DEL11d | M | 13 years | BP4/BP5 deletion | borderline | yes | no |
| DEL16d,e | M | 14 years | BP3/BP5 deletion | yes | yes | no |
| DEL18d | M | 15 years | BP4/BP5 deletioni | yes | no | no |
| CONTROL01f | F | 4 years | copy neutral | no | no | no |
| CONTROL02g | M | 7 years | copy neutral | no | no | no |
| CONTROL03e,h | M | 7 years | copy neutral | yes | no | no |
Generation of Human Induced Pluripotent Stem Cells (iPSCs)
15q13.3 deletion, 15q13.3 duplication, and control fibroblasts were derived from individuals’ biopsies by the IDDRC Tissue Culture Facility Core. Fibroblasts were reprogrammed into iPSCs by the Baylor College of Medicine Human Stem Cell Core (HSCC) using the CytoTune-iPS 2.0 Sendai Reprogramming Kit (Life Technologies), following the manufacturer’s protocol. Following transduction, cells were grown in high-glucose DMEM+10%FBS+1× MEM non-essential amino acids (Life Technologies) for 5 days, in TeSR-E7 (StemCell) for 8 days, and in TeSR-E8 (StemCell) until day 21 when colonies were manually picked. Lower passaged colonies were picked manually, while later passaged colonies were passaged using ReLeSR (StemCell) or 0.5 mM EDTA. After reprogramming, iPSCs were maintained in mTeSR1 (StemCell) on Matrigel (Corning) or Cultrex (Trevigen). All cells were incubated at 37°C with 95% humidity and 5% CO2.
Neural Progenitor Cell (NPC) Differentiation
To differentiate iPSCs into NPCs, a dual SMAD inhibition protocol based on previous publications was used.33, 34 Generally one clone was differentiated, with two clones for Control1 and DUP04. iPSCs were dissociated using Accutase (Sigma) and 2 million cells per well were plated into an Aggrewell plate (StemCell) in Neural Induction Medium (DMEM/F-12 and Neurobasal media [Life Techonologies], 2% B27-supplement minus vitamin A [Life Technologies], 1% N2-supplement [Thermo Fisher], 2 mM Glutamax [Life Techonologies]). 10 μM Y-27632 (Selleckchem) was used during initial 24 hr of culturing in Aggrewells to promote cell survival. On the following day media was changed and 10 μM SB-431542 (Selleckchem) was added to initiate dual SMAD inhibition. Starting at day 3, 4 μM dorsomorphin (Santa Cruz Technologies) was added. On day 5, embryoid bodies were collected, sieved through a 37 μm reversible strainer (StemCell), and transferred to Matrigel- or Cultrex (Trevigen)-coated plates in Neural Proliferation Medium (DMEM/F-12 and Neurobasal media, 1% B27-supplement minus vitamin A, 0.5% N2-supplement, 20 ng bFGF/mL [Peprotech], and 20 ng EGF/mL [Peprotech]). Both SMAD inhibitors were present in media until day 9 with addition of 10 μM cyclopamine (CCP) (VWR) starting from day 6. Media was changed daily until rosettes of neural progenitors were collected at day 12 by incubation for 1 hr with Neural Rosette Selection Reagent (StemCell) at 37°C. Dislodged rosettes were incubated for 1 hr in plates coated with 0.2% porcine gelatin (Sigma). The floating fraction was collected and transferred into a non-coated plate and incubated in suspension overnight. On the following day, floating spheres of NPCs were plated onto Matrigel- or Cultrex-coated 6-well plates and propagated until confluent. Cells were passaged using Accutase and split into media containing 10 μM Y-27632 to support single-cell survival. All cultures were maintained in presence of 1% penicillin-streptomycin (Thermo Fisher).
iPSC and NPC Characterization
Two or three iPSC clones were characterized for each individual by karyotyping at an early passage post reprogramming, flow cytometry for cell-specific iPSCs markers OCT4 and SSEA-4, immunofluorescence at a later passage, and by qPCR twice for appropriate markers (Figures 2, S1, and S5). Flow cytometry was performed by the Baylor College of Medicine Human Stem Cell Core and the Cytometry and Cell Sorting Core. Cells were trypsinized and centrifuged at 200 × g for 5 min. Cells were resuspended in FACS buffer (DBPS without Ca2+ or Mg2+, 2% FBS, 0.1% sodium azide) and then stained for cell surface antigens SSEA-4-APC (R&D FAB1435A) and CD-29-Alexa 488 (AbD Serotec MCA2298A488) as a negative control for 30 minutes at 4°C. Cells were then fixed for 30 min in 2% paraformaldehyde (PFA) at room temperature followed by permeabilization in permeabilization buffer (DBPS without Ca2+ or Mg2+, 0.1% BSA, 0.1% Saponin) for 30 min at room temperature. Cells were then stained for nuclear antigen OCT4-PE (BD 560186) for 1 hr at room temperature. Finally, cells were stained with DAPI and run through FACS on same day.
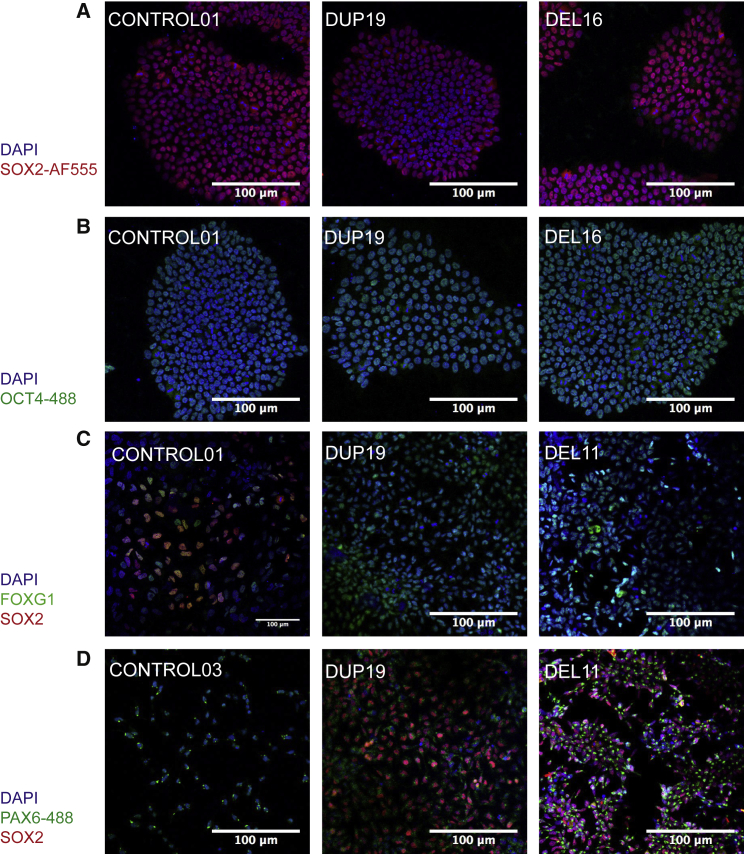
Figure 2.
Characterization of iPSCs and NPCs
(A and B) iPSCs stain positive for SOX2 (A) and OCT4-AF488 (B), both pluripotency markers. Figure S1 shows additional characterization of iPSCs using flow cytometry and karyotypes.
(C and D) NPCs stain positive for neuronal markers FOXG1 (C), SOX2 (see D for duplications and deletions), and PAX6 (D).
For immunofluorescence, iPSCs or NPCs were plated on Matrigel- or Cultrex-coated glass coverslips. Cells were rinsed twice with PBS and fixed with 3.7%–4% PFA for 15 min at room temperature. Cells were then permeabilized in 0.5% Triton-100X in PBS for 10 minutes at room temperature and washed two times with PBS. Fixed cells were blocked for 1 hr in 1% BSA + 22.25 mg/mL glycine in PBST. iPSCs were incubated overnight at 4°C with antibodies for pluripotency markers (1:100 goat anti-SOX2 [Santa Cruz, SC-17320] and 1:60 mouse anti-OCT4 [Santa Cruz, SC-5279]). NPCs were incubated overnight at 4°C in in neural progenitor markers (1:200 rabbit anti-SOX2 [Abcam, ab97959], 1:50 rabbit anti-PAX6 [Abcam, ab5790], 1:200 rabbit anti-PAX6 [BioLegend, 901301], and 1:200 FOXG1 anti-rabbit [Abgent, AP9793c]). Appropriate secondary antibodies (1:200 AlexaFluor-488, -555, or -647) were incubated with cells for 1 hr at room temperature in the dark. Cells were imaged on a Zeiss 880 or Zeiss 710 confocal microscope and analyzed using ImageJ.
RNA Isolation and qPCR
RNA was isolated from confluent iPSCs and NPCs in 6-well plates using miRNeasy Mini Kit (QIAGEN) following manufacturer’s protocol. cDNA was synthesized using M-MLV Reverse Transcriptase (Invitrogen) according to the manufacturer’s directions. Quantitative PCR (qPCR) was performed using the primers in Table S1, designed manually using DNASTAR SeqBuilder or used in previous publications,35, 36 with the following cycle: 95°C for 10 minutes; 40 cycles 95°C for 10 s, 60°C for 20 s, and 72°C for 15 s; followed by 95°C for 10 s and then 25°C for 30 s. All qPCR products were between 95 and 150 basepairs. qPCR was run using the CFX96 Real Time System (Bio-Rad) using SYBR green. Of note, CHRNA7 qPCR primers had no overlap with the fusion gene, CHRFAM7A (MIM: 609756) (Figure S2, Table S1). For calculation of significance, only biological replicates were considered. However, error bars in Figure 3, Figure 4D, and Figure 5 take biological and technical replicates into consideration.
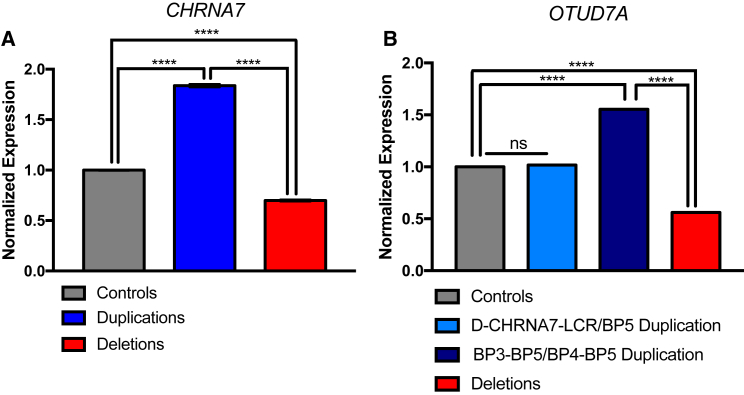
Figure 3.
15q13.3 CNV NPCs Have Correlative Gene Expression by qPCR
Control NPCs are shown in gray, duplication NPCs are in blue, and deletion NPCs are in red. All groups were compared using a one-way ANOVA.
(A) CHRNA7 is fully duplicated for all sizes of 15q13.3 gains (n = 3 clones, 3 individuals).
(B) The expression of OTUD7A in small D-CHRNA7-LCR/BP5 gains (n = 1 clone, 1 individual) is not altered as compared to controls (n = 3 clones, 3 individuals), while its expression is appropriately up- or downregulated in larger duplications (n = 2 clones, 2 individuals) and deletions (n = 3 clones, 3 individuals), respectively.
∗∗∗∗p < 0.0001; ns, not significant; BP, breakpoint. Error bars indicate standard error.
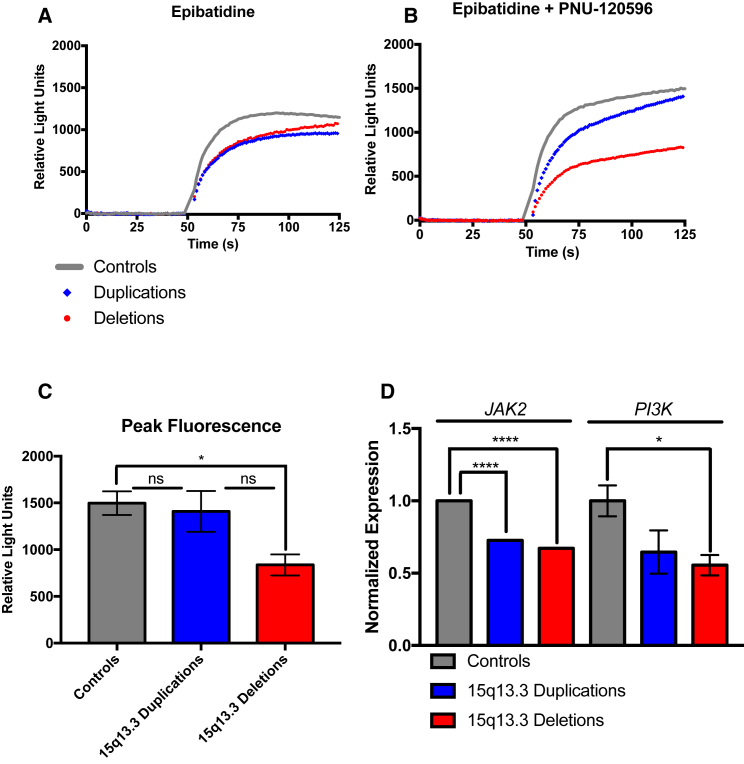
Figure 4.
15q13.3 Deletion and Duplication NPCs Have Decreased α7 nAChR-Dependent Calcium Flux
(A and B) Controls are shown with a gray solid line, duplications are shown as blue diamonds, and deletions are shown as red circles. Error bars are not shown for simplicity in the figure but are shown in Figure S4.
(A) With application of 1 μM epibatidine, an orthosteric nAChR agonist, NPCs have a response that is decreased for both 15q13.3 deletion and duplication NPCs (two-way ANOVA p < 0.0001; s: seconds).
(B) With co-application of 1 μM epibatidine and 3 μM PNU-120596, an α7 nAChR-specific type II positive allosteric modulator (PAM), decreased α7 nAChR-dependent calcium flux is decreased for both 15q13.3 deletion and duplication NPCs (two-way ANOVA p < 0.0001).
(C) Peak fluorescence, as measured by normalized relative light units, for each genotype, with co-application of 1 μM epibatidine and 3 μM PNU-120596. Deletions have significantly decreased peak fluorescence, while duplications have trending decreased peak fluorescence, indicating less calcium flux. Data are of three replicates for each condition for each individual (one NPC line each) in two experiments, and averages of each genotype (controls n = 3, duplications n = 3, deletions n = 3); p < 0.05; ns, not significant. Data are normalized to baseline values.
(D) 15q13.3 deletion and duplication NPCs have decreased α7 nAChR-dependent calcium signaling via the JAK2-PI3K pathway, as determined by qPCR (p < 0.05, ∗∗∗∗p < 0.0001, one-way ANOVA).
Error bars indicate standard error.
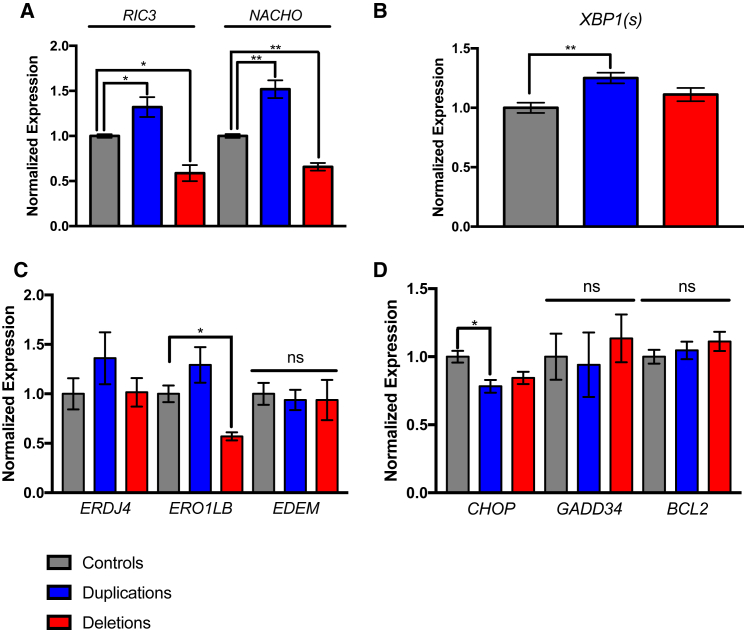
Figure 5.
15q13.3 Duplications Have Increased Expression of nAChR-Specific and Resident ER Chaperones
All groups were compared using a one-way ANVOA.
(A) RIC3, a nAChR-specific ER chaperone involved in folding, assembly, and trafficking of nAChRs, is upregulated in duplication NPCs (n = 3 clones, 3 individuals). The gene is downregulated in deletion NPCs (n = 3 clones, 3 individuals), which may be due to fewer nAChR subunits. NACHO, a nAChR-specific ER chaperone involved in folding, assembly, trafficking, and cell surface expression, is upregulated in duplication NPCs (n = 3 clones, 3 individuals) and downregulated in deletion NPCs (n = 3 clones, 3 individuals).
(B) ER stress marker XBP1(s), the spliced mRNA of XBP1, is increased in duplication NPCs (n = 3 clones, 3 individuals) and is unchanged in deletion NPCs (3 clones, 3 individuals).
(C) Downstream targets of XBP1 have differential changes in expression. ERDJ4, an ER-resident chaperone important in folding misfolded or unfolded proteins, as well as those that fold slowly, is upregulated by 26.5%, although not significantly due to small sample size, in duplication NPCs (n = 3 clones, 3 individuals). ERO1LB, an ER-resident chaperone and ERAD protein, is similarly upregulated in duplication NPCs (22.5%) (n = 3 clones, 3 individuals) and downregulated in deletion NPCs (n = 3 individuals, 3 clones). EDEM, an ER-associated degradation factor, has no changes in expression in either 15q13.3 duplication or deletion lines (n = 3 clones, 3 individuals for each), suggesting that the ER stress is mediated.
(D) ER stress factors downstream of PERK dimerization and phosphorylation CHOP, GADD34, and BCL2, known to lead to cell death with changes in expression, are unchanged in both duplication and deletion NPCs (3 clones, 3 individuals for both), supporting that the ER stress is mediated in 15q13.3 duplication cells.
∗p < 0.05, ∗∗p < 0.01, ns, not significant. Error bars indicate standard error.
Measurement of Intracellular Calcium using FLIPR
5 × 104 NPCs, one clone per individual, were plated in an optical bottom 384-well plate coated with Cultrex (Trevigin). Once confluent after 2 days, changes in calcium flux were measured in response to an orthosteric nAChR agonist agonist (epibatidine), α7 nAChR-specific PAM (PNU-120596) positive allosteric modulator (PAM) in the presence of agonist, and an α7 nAChR-specific antagonist (methyllcaconitine, MLA) in the presence of PAM and agonist with the Fluo-8 No Wash Calcium Assay kit (Abcam) according to kit instructions. Cells were transferred to the FLIPRTetra (Molecular Devices) after incubation in Fluo-8 for 1 hr. Drug dilutions (2×, epibatidine [Tocris], PNU-120596 [Tocris], and MLA [Tocris]) were prepared in a separate 384-well compound plate and were added in 25 μL volumes by automated pipetting. Intracellular calcium levels were recorded before and after drug dispensing. Data were exported and analyzed by two-way ANOVA using Microsoft Excel and GraphPad Prism and are normalized to the baseline relative light units. Peak fluorescence values were determined by averaging the peak normalized values, typically at the last time point, of each group.
Measurement of Apoptosis and Cell Viability
Apoptosis and cell viability were measured for 15q13.3 deletion and duplication NPCS, normalized to controls, using the ApoTox-Glo Asssay (Promega) according to manufacturer’s instructions. Luminescence and fluorescence were measured using a BioTek 2 Synergy Multi-Mode plate reader (BioTek).
Results
Generation and Characterization of 15q13.3 CNV Proband-Derived iPSCs and NPCs
In order to elucidate the molecular pathogenesis of 15q13.3 CNVs, we utilized proband-derived iPSCs and differentiated them into cortical-like NPCs. Three iPSC clones were generated per individual from fibroblasts of three individuals with 15q13.3 deletions, three with 15q13.3 duplications, and three sibling control subjects. Three iPSC clones were used unless one was found to have an abnormal karyotype. Two 15q13.3 deletions spanned BP4/BP5 and one spanned BP3/BP5, while duplications spanned BP3/BP5, BP4/BP5, or D-CHRNA7-LCR/BP5 (Figure 1). Phenotypes of probands are outlined in Table 1.7, 10 All iPSC lines expressed pluripotency markers OCT4 and SSEA4 by flow cytometry (Figure S1). Additionally, all lines showed SOX2 and OCT4 at the protein level as determined by immunofluorescence (Figures 2A and 2B). Post reprogramming, iPSC clones were also karyotyped to ensure no additional chromosomal abnormalities were introduced during the reprogramming process.
All control, deletion, and duplication lines were successfully differentiated into NPCs. NPC markers SOX2, FOXG1, and PAX6 were present at the protein level, as determined by immunofluorescence, and NESTIN (MIM: 600915) was expressed, as determined by qPCR (Figures 2C, 2D, and S5).
Assessment of Potentially Dosage-Sensitive 15q13.3 Genes CHRNA7 and OTUD7A
By qPCR, 15q13.3 microdeletion iPSCs (data not shown) and NPCs, with one clone per individual pooled with the appropriate genotype, showed significantly decreased expression of CHRNA7 compared to pooled controls (p < 0.0001 for both deletion sizes, Figure 3A). For duplications, it has been unclear whether CHRNA7 is fully duplicated, because part of the gene is located in an LCR element. mRNA expression of CHRNA7 in 15q13.3 duplication iPSCs (data not shown) and NPCs was significantly increased compared to controls, suggesting that CHRNA7 is fully duplicated in 15q13.3 duplication iPSCs and NPCs (p < 0.0001 for all duplication sizes pooled and individually). qPCR primers did not overlap with the fusion gene, CHRFAM7A, and are specific for CHRNA7 (Figure S2).
While CMA analysis and the genomic structure of the LCRs utilized suggests that the first noncoding exon of OTUD7A is included in D-CHRNA7-LCR/BP5 CNVs, it has been unclear whether this aberration would affect OTUD7A gene expression. This is of particular importance for duplications, as a duplicated first exon could be capable of disrupting expression. Utilizing qPCR, we determined that in D-CHRNA7-LCR/BP5 duplication iPSCs (data not shown) and NPCs, gene expression of OTUD7A resembles that of control cells, but is appropriately up- or downregulated in pooled larger duplications and pooled deletions (Figure 3B).
α7 nAChR-Dependent Calcium Flux Is Decreased in Both 15q13.3 Deletion and Duplication Proband-Derived NPCs
The α7 nAChR is unique among nAChRs in that it has a very high permeability to calcium, being one of the most efficient ways to increase cytoplasmic calcium concentrations. Utilizing the FLIPRTetra High-Throughput Cellular Screening System (Molecular Devices), intracellular calcium levels in response to α7 nAChR-signaling of control, deletion, and duplication NPCs were measured in response to epibatidine, an orthosteric agonist of nAChRs, PNU-120596, an α7 nAChR-specific type II positive allosteric modulator (PAM), and methyllycaconitine (MLA), an α7 nAChR-specific antagonist. Both 15q13.3 deletion and duplication NPCs had decreased α7 nAChR-dependent calcium flux. With application of 1 μM epibatidine, 15q13.3 deletion and duplication NPCs had significantly reduced calcium flux compared to controls (interaction of two-way ANOVA p < 0.0001) (Figures 4A and S4). Agonist alone is unable to detect α7 nAChR-dependent calcium flux accurately, as the channels open and desensitize within milliseconds. PAMs allow for the ion channel to remain open for a longer period of time, allowing for measurement of α7 nAChR-specific signal. With co-application of 1 μM epibaditine and 3 μM PNU-120596, the response was significantly lower for deletion NPCs, by 46%, and duplication NPCs, by 17%, compared to control NPCs, as determined by two-way ANOVA (p < 0.0001) (Figure 4B). Of note, duplication NPCs’ calcium flux is between control and deletion NPCs, which is remarkably similar to what has been observed with clinical phenotypes. Peak fluorescence, indicative of peak calcium levels, was significantly decreased for 15q13.3 deletions (p = 0.0458, ANOVA) (Figure 4C) as compared to controls, with 15q13.3 duplications having an intermediate value not significantly different from either controls or deletions. Time to reach peak fluorescence from stimulation was not significantly different between groups, indicating the channel kinetics are normal, as can be appreciated by the similar shape in the calcium flux curves (data not shown). In response to co-application of 1 μM epibatidine, 3 μM PNU-120596, and 10 μM MLA, this response was decreased for each group, indicating that this is an α7 nAChR-specific phenomenon (Figure S3).
Gene Expression of Chaperones Involved in Folding, Assembly, and Trafficking α7 nAChRs Is Increased in 15q13.3 Duplication NPCs
From gene to fully functional receptor, α7 nAChRs must be folded and assembled in the ER, followed by trafficking of fully assembled receptors to the membrane. In order to explore a mechanism explaining decreased α7 nAChR-specific calcium flux in duplication NPCs, we looked to the folding, assembly, and trafficking of the receptors. Two chaperones have been identified to be necessary for α7 nAChR assembly in the ER and transport to the membrane: RIC3 (MIM: 610509) and NACHO.24, 37, 38, 39 By qPCR, both of these chaperones had significantly increased mRNA expression in 15q13.3 duplication NPCs (RIC3 p = 0.0456, NACHO p = 0.0066; Figure 5) and decreased expression in deletion NPCs (RIC3 p = 0.0106, NACHO p = 0.0019).
Expression of a Subset of ER Stress Markers Is Increased in 15q13.3 Duplication NPCs
Since two nAChR-specific ER chaperones had altered gene expression, we explored other ER stress markers. Activation of three pathways is implicated in ER stress and the unfolded protein response: IREα splicing of XBP1 (MIM: 194355), PERK activation, and ATF6 translocation. Spliced XBP1 (XBP1(s)) had significantly increased expression in 15q13.3 duplication NPCs (p = 0.0052). Two known ER stress markers transcribed by XBP1(s) had increased expression, although not significantly, in duplication NPCs ERDJ4 (also known as DNAJB9 [MIM: 602634], p = 0.3055) and ERO1LB ([MIM: 615437], p = 0.2158). ERDJ4 had no change in deletion NPCs, but a decrease in expression of ERO1LB (p = 0.0104) was observed. Of note, CHOP and GADD34, known to be downstream of PERK dimerization and autophosphorylation and to lead to apoptosis, did not have increased expression in 15q13.3 CNV NPCs. BCL2 (MIM: 151430), affected downstream of CHOP during ER stress-induced apoptosis, did not have altered expression in either deletion or duplication NPCs. Additionally, neither duplication or deletion NPCs showed increased apoptosis or decreased viability, as compared to control NPCs (Figure S7).
15q13.3 CNV NPCs Have Decreased α7 nAChR-Dependent Calcium Effectors Downstream Signaling
The α7 nAChR has many important consequences in neuronal cells, including modulation of neuronal excitability and neurotransmitter release as well as calcium flux directly impacting multiple cellular signaling pathways. We explored the JAK2-PI3K pathway and found that multiple components had decreased mRNA expression in both 15q13.3 deletion and duplication lines (Figure 4D). This pathway, with JAK2 an early target of calcium flux through the α7 nAChR, has been implicated in anti-apoptotic effects, anti-inflammation effects, and neuroprotection.40, 41, 42 Notably, each of these downstream effectors alterations followed the same trend of 15q13.3 duplications being intermediate between controls and 15q13.3 deletions.
Discussion
In this study, we established induced pluripotent stem cells (iPSCs) from individuals with 15q13.3 microdeletions and microduplications and differentiated them into cortical-like neural progenitor cells (NPCs), from which we were able to determine molecular consequences of CHRNA7 CNVs. Remarkably, we identified a mechanism of pathogenesis for both CHRNA7 deletions and duplications, both of which result in decreased α7 nAChR-dependent calcium flux, although due to different pathomechanisms (Figure 6).
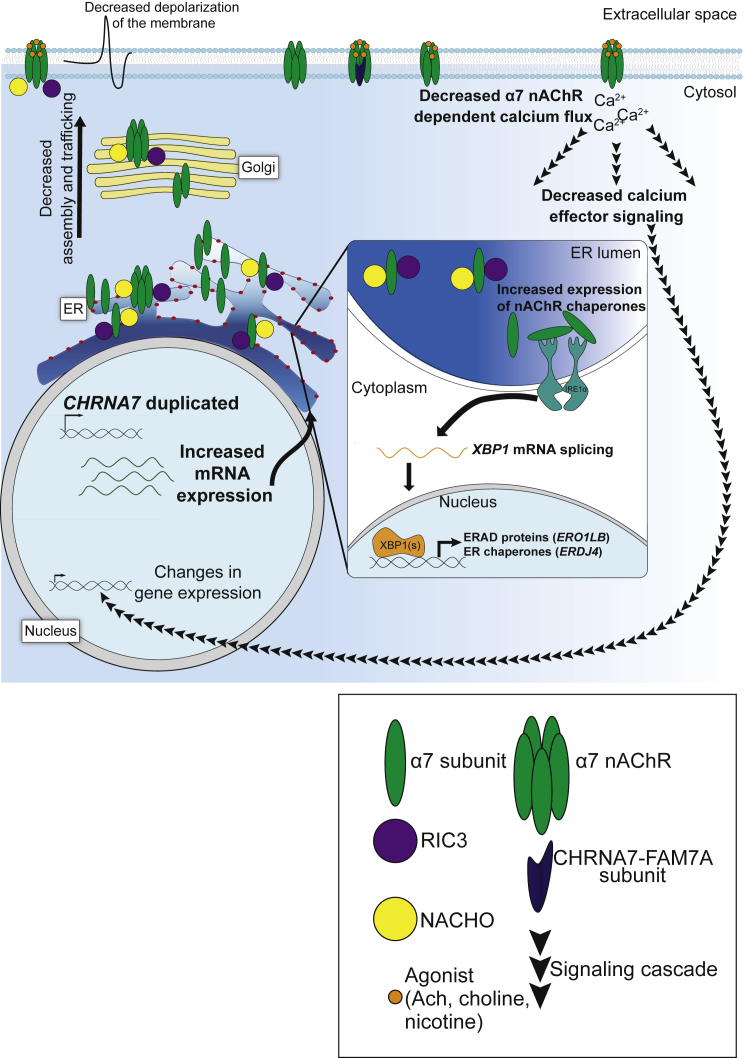
Figure 6.
Proposed Mechanism of Pathogenesis of 15q13.3 Duplications
We propose that CHRNA7 duplications result in increased gene expression. This results in increased α7 nAChR subunits (green) in the ER, which overwhelm the ER nAChR-specific chaperones RIC3 (purple) and NACHO (yellow). This causes adaptive ER stress, as indicated by increased ER stress marker expression of select pathways. A portion of these subunits are folded, assembled, and trafficked by RIC3 and NACHO, ending up at the cell membrane. However, there are fewer than in controls, and some of those at the membrane may have inappropriate stoichiometry, including incorporation of CHRFAM7A-encoded subunits (dark blue). This likely causes decreased depolarization at the cell membrane and does result in decreased α7 nAChR-dependent calcium flux. The decreased calcium flux downregulates calcium effectors, which likely has impacts on gene expression. These changes in calcium effectors and gene expression not only explain the phenotypes observed in probands, but may also contribute to the incomplete penetrance and variable expressivity of 15q13.3 CNVs.
15q13.3 microdeletions and duplications have been difficult to study for a variety of reasons, including the incomplete penetrance and variable expressivity of human phenotypes, the imperfect resolution of CMA analysis, especially in proximity of repetitive sequences, and the challenges of recapitulating phenotypes in murine models. Proband-derived cells provide an opportunity to study genes and mechanisms in a relevant human model. In the future, our 15q13.3 iPSCs and NPCs have potential to be a valuable tool in drug discovery, due to their ease of proliferating and use in high-throughput experiments.
15q13.3 Gene Expression Is Altered in Proband-Derived Cells
First, we explored gene expression of 15q13.3 genes in proband-derived iPSCs and NPCs (Figure 3). For deletions, we saw expression of both CHRNA7 and OTUD7A decreased by about 50%, as one would expect. Due to the repetitive nature of the genome at 15q13.3, it had yet to be determined whether CHRNA7 was fully duplicated in probands carrying 15q13.3 gains, as the 5′ end of the gene resides in an LCR element.11 With a partial duplication, the mechanism of pathogenesis would likely be different, so determination of the full duplication of CHRNA7 was key. Using qPCR primer sets with no overlap with the human fusion gene of CHRNA7, CHRFAM7A, we identified that in all three of our duplication proband iPSC and NPC lines, CHRNA7 was fully duplicated, further implicating the gene in neuropsychiatric phenotypes.
Additionally, in smaller duplications that span from the D-CHRNA7-LCR to BP5, it has been unclear whether the duplication of the first noncoding exon of OTUD7A alter its expression. It has been hypothesized that duplication of the first exon may disrupt the gene and decrease its expression in these probands, resulting in haploinsufficiency of the gene. While we had only one proband with a small D-CHRNA7-LCR/BP5 duplication, the iPSCs and NPCs generated from this individual had expression levels of OTUD7A similar to controls, while duplications that encompassed the entirety of OTUD7A had appropriately increased expression. This suggests that changes in CHRNA7 copy number are likely the major contributor to the pathogenesis these duplications. Recently, evidence has been found that OTUD7A loss-of-function mutations or deletions may contribute to neuropsychiatric phenotypes (J. Yin, personal communication), so it may be possible that loss OTUD7A contributes to phenotypes observed in 15q13.3 deletion probands. Unfortunately, due to the rarity of the probands (about 16% of all individuals carrying 15q13.3 deletions), we were unable to determine whether the expression of OTUD7A is altered in individuals with D-CHRNA7-LCR/BP5 deletions.
α7 nAChR-Dependent Calcium Flux Is Decreased in Both 15q13.3 Deletion and Duplication NPCs
As protein levels of α7 subunits cannot be determined reliably with many commercial antibodies, we chose to directly assess the α7 nAChR functionality in our NPC lines. Interestingly, we found that α7 nAChR-dependent calcium flux was decreased in both 15q13.3 deletion and duplication NPCs (Figure 4B). 15q13.3 deletion NPCs were expected to have decreased calcium flux due to haploinsufficiency of CHRNA7. For the duplication NPCs, the decreased calcium flux was surprising, considering the genomic and expression data. However, looking at the reported phenotypic manifestations in probands with 15q13.3 CNVs, the decreased calcium flux in both groups mirrors what has been observed in the clinic: deletions often having a similar but more severe phenotype than duplications.
In response to a non-selective nAChR agonist, epibatidine, we saw the same pattern of decreased calcium response for both 15q13.3 deletion and duplication NPCs (Figure 4A). Due to the activation of multiple nAChRs via epibatidine, the decrease in calcium flux following its application suggests that additional nAChRs cannot, or do not, fully compensate for the functional effects resulting from CHRNA7 copy-number variation. Other nAChRs, similar to what has been observed in iPSC-derived neurons, are expressed in our NPCs (Figure S6). However, particularly for duplications, it is likely that these are dysregulated or have altered function due to their chaperones shared with the α7 nAChR, which could vary between individuals. Furthermore, it is likely that alterations in α7-dependent calcium flux may alter gene expression of additional nAChRs. While some compensation from additional nAChRs, or even other Cys-loop receptors, may be occurring in 15q13.3 NPCs and/or neurons, it is unclear whether this would be consistent among probands or whether there would be significant impact.
With co-application of epibatidine and PNU-120596, a positive allosteric modulator of α7 nAChRs that reduces its desensitization kinetics, there was a considerably larger change in calcium flux as compared to epibatidine alone. 15q13.3 deletions had the lowest peak levels of calcium flux, and duplications had a non-significant decrease in α7 nAChR-dependent calcium flux compared to controls. This places 15q13.3 duplication NPCs’ α7 nAChR-dependent calcium flux at an intermediate between controls and 15q13.3 deletions, not significantly different from either. While somewhat speculative, this is remarkably similar to what is often observed in the clinic, with 15q13.3 deletion and duplication probands sharing phenotypes with differing severity.
Adaptive ER Stress Is Increased in 15q13.3 Duplication NPCs
Folding, assembly, and trafficking of nAChRs is a notoriously inefficient process, so any perturbation of the system would likely decrease the number of functional nAChRs at the membrane.43 Two chaperones have been found to be necessary for α7 nAChR assembly and trafficking: RIC3 and NACHO. RIC3, a Cys-loop receptor-specific ER chaperone, has previously been observed to have increased mRNA expression in individuals with bipolar disorder, with significantly higher levels in bipolar individuals manifesting psychosis.44 Similarly, our 15q13.3 duplication NPCs had increased expression of RIC3 (Figure 5A). Additionally, NACHO, a recently identified ER chaperone specific to nAChRs and important for their folding, assembly, trafficking, and cell surface expression, was also increased in 15q13.3 duplication NPCs. This suggested ER stress as a result of increased α7 subunits in the ER with inefficient and/or diminished processing.
The ER has been proposed to be involved in neuropsychiatric disease, as many psychotropic medications likely involve the ER in their mechanism of action.45 For nAChRs in particular, the ER has been implicated in pathogenesis, as nicotine is known to mediate ER stress and upregulate (increase number of receptors at the membrane) nAChRs.46, 47, 48 ER stress in 15q13.3 duplication NPCs was confirmed by measuring expression of known ER stress markers in the 15q13.3 duplication cells. Increased expression of ER stress marker spliced XBP1 highlights the unfolded protein response pathway of inositol requiring enzyme 1α (IRE1α), which is bound by unfolded or misfolded proteins. We also identified increased expression of ERDJ4, an ER-resident chaperone and transcriptional target of spliced XBP1, that is known to bind to both unfolded and misfolded proteins as well as proteins that fold especially slowly. Furthermore, we found increased expression of ERO1LB, whose expression is also dependent on XBP1 splicing and is important in ER-associated degradation (ERAD). A second pathway in the unfolded protein response activated by PERK dimerization, resulting in CHOP expression and ultimately apoptosis, was not upregulated in these cells. Activation of a subset of ER stress pathways is thought to be indicative of adaptive ER stress and has been noted in disorders of protein accumulation.49 Altogether, this suggests that an increase in genomic copy number of CHRNA7 results in increased mRNA and protein product, which is retained in the ER due to insufficient chaperoning. Due to unreliable human α7 antibodies, we were unable to show changes on the protein level, but this conclusion has both functional and literature support.50
Protein accumulation in the ER, resulting in ER stress, has been reported in other neurodevelopmental disorders and is implicated in their pathogenesis. Pelizaeus-Merzbacher disease (PMD [MIM: 312080]), a dysmyelinating disorder of the central nervous system, is known to result in apoptotic pathways of the unfolded protein response being activated due to accumulation of misfolded protein in the ER, specifically the membrane protein proteolipid protein 1 (PLP1).51 Both missense mutations and duplications at PLP1 (MIM: 300401) result in PMD, with duplications having a less severe phenotype.20 This has been suggested to be the case due to differing molecular consequences, with missense mutations resulting in accumulation of mutant PLP1 in the ER and genomic duplications resulting in accumulation of PLP1 in endosomes or lysosomes, resulting in less PLP1 at the membrane. Deletions of PLP1, while showing a phenotype due to less PLP1 at the plasma membrane, have an even less severe phenotype than duplications, likely due to no trafficking consequences on the protein. In iPSCs, genomic duplications of PLP1 have been explored, as well as a partial duplication that disrupts the expression of PLP1 similar to deletions.52 In iPSC-derived oligodendrocytes, missense mutations in PLP1 have been shown to have a molecular phenotype of increased apoptosis due to increased ER stress.53 Broadly, this supports the notion of differing molecular consequences for different genetic changes of membrane proteins. However, unlike PMD iPSC-derived cells, it appears that the ER stress phenotype does not result in a cell death phenotype in our 15q13.3 duplication NPCs, but instead an adaptive ER stress phenotype that is allowing the cell viability to be the same as control cells.
It is possible that a similar range of cellular phenotypes may be seen with 15q13.3 CNVs, with deletions having a more severe phenotype due to decreased α7 nAChRs at the membrane and duplications having a less severe phenotype since they are able to still traffic some, but not all, α7 nAChRs to the membrane. Interestingly, in contrast to PLP deletions, although difficult to address due to the 99% homology with CHRFAM7A, no single-nucleotide variants or small indels have been reported in the coding region of CHRNA7.
Dysregulation of nAChR chaperones and/or nAChR trafficking has been implicated previously in neuropsychiatric disease, as well as shown to occur in vitro.45, 46, 47, 48 Similar to our model, a model of ER stress in response to nAChR dysfunction has been proposed for Alzheimer disease, a disease for which the α7 nAChR has also been implicated.54 In this model, chaperone dysfunction or ER stress results in increased assembly and trafficking of non-functional receptors or those with inappropriate stoichiometry, which may be less sensitive to ligands. Inappropriate stoichiometry of these receptors may include homomeric α7 nAChRs, although with the wrong number of subunits or inclusion of other nAChR subunits that are known to interact with α7 subunits, such as β2 subunits, but again in inappropriate numbers. Furthermore, these receptors may include CHRNA7-FAM7A subunits, encoded by CHRFAM7A, which has been proposed to have a dominant-negative effect on α7 nAChRs.15, 55 In overexpression studies, it is known that CHRNA7-FAM7A and α7 subunits interact in mammalian cells, although a dominant-negative effect in humans has yet to be determined.17 It is likely that a combination of decreased receptors, as well as increased dysfunctional receptors, contribute to the phenotypes seen in 15q13.3 duplication probands.
Downstream Signaling to α7 nAChR Activation Is Decreased in Both 15q13.3 Deletion and Duplication NPCs
Calcium signaling is extremely important in neuronal cells, with downstream effectors being activated and involved in many facets of cellular function. Beyond what we have explored here, α7 nAChR calcium flux is known to initiate calcium flux through voltage-dependent calcium channels (VDCCs) and cause calcium-induced calcium release (CICR) from the ER, each of which may have their own impact on the cell.56 Many pathways have been found to be active in response to α7 nAChR calcium influx, including the JAK2-PI3K pathway, which, among many downstream effects, is anti-apoptotic, anti-inflammatory, and involved in neuroprotection upon activation by the α7 nAChR. Here, we found that in both CNV groups JAK2 (MIM: 147796), directly activated by α7 nAChR-dependent calcium flux, had significantly decreased expression compared to controls, with its downstream target, PI3K, having decreased mRNA expression in both groups, but only significantly for deletions (Figure 4D). The wide variety of processes that may be affected by the changes in calcium flux in the cells of probands with 15q13.3 CNVs may contribute to the variable expressivity of phenotypes observed.
Potential Cellular Consequences
Here, we have identified an early cellular response to changes in copy number of CHRNA7. As an nAChR, there is a wide range of cellular phenotypes that could result from decreased α7 nAChR calcium flux, which we only began to explore here. The calcium influx from the α7 nAChR activates a range of secondary messengers, some or all of which could contribute to human phenotypes.56 Furthermore, many of these pathways likely impact gene expression and/or neuronal differentiation.18, 57 Future studies will need to address how neuronal differentiation may be impacted by CHRNA7 CNVs and whether this is a rescuable phenotype.
Clinical Implications
While 15q13.3 deletions have a prevalence similar to other genomic disorders (unpublished data), 15q13.3 duplications are the most common CNV contributing to neuropsychiatric disease, found in 1/123 individuals submitted for clinical chromosomal microarray analysis.10 Here, NPCs were utilized for their future potential in drug discovery, as these cells can be generated quickly from iPSCs and propagated easily for high-throughput experiments.58 Pharmacological compounds that target α7 nAChRs have been utilized in individuals with a range of neuropsychiatric phenotypes. Most relevant to 15q13.3 CNVs, the nAChR type I PAM and AChE inhibitor, galantamine, has been used in an individual with a 15q13.3 microdeletion, with some positive short- and long-term results observed, although there is definitely opportunity for improvement.59 Both type I and type II PAMs specific for the α7 nAChR have been in development, some tested in humans, some in primates, and many more in rodents.52, 53 These two drug classes have varying effects on α7 nAChRs, with type I PAMs not altering the normal kinetics of receptor desensitization and type II PAMs blocking desensitization. Not only does our study support the use of α7 nAChR targeting compounds, it also expands the pool of probands that would likely benefit from such therapies.
Interestingly, it has been observed that mood-stabilizing drugs, including lithium and valproate, increase expression of ER stress markers, but do not lead to apoptosis, indicative of adaptive ER stress.60, 61 Lithium treatment has also been shown in vitro to prevent cell death induced by drugs that increase ER stress, such as thapsigargin.59 This suggests that mitigating ER stress toward a nonapoptotic response could likely benefit individuals with a variety of neuropsychiatric disorders, especially those in which ER stress in implicated, such as those associated with 15q13.3 duplications. With increased ER stress marker expression but avoiding apoptosis, it may be possible to increase the number of chaperones in the ER, thus making it easier for the cell to assemble nAChRs. In Alzheimer disease models, drugs known to reduce ER stress, such as 4-phenylbutric acid (4-PBA), have been found to have positive effects on neuronal cells, which have increased Aβ secretion upon ER stress, but increased α/γ cleavage of APP with treatment reducing ER stress.62 Furthermore, 4-PBA has been tested in humans as a diabetes treatment, suggesting that it is a safe treatment option.63 Thus, there may be a two-pronged approach to treatment of 15q13.3 duplications: targeting the α7 nAChRs that do make it to the membrane, or targeting the process that causes them to be reduced.
Open Questions in 15q13.3 CNVs
While we have demonstrated a molecular pathology for 15q13.3 CNVs, there are still many unanswered questions. The first of these is the variable expressivity of phenotypes in 15q13.3 CNV probands. CHRFAM7A, the human-specific fusion gene of CHRNA7 and FAM7A shown to have a dominant-negative effect on α7 nAChR in some recombinant systems, likely plays a role in this variable expressivity. Our study provides the basis for using iPSC-derived neuronal cells to explore whether CHRFAM7A, or other potential modifiers, has an impact on α7 nAChR calcium flux. As CHRFAM7A is human specific, its exploration in a human model is necessary to understand its contribution to neuropsychiatric disease.
Other factors likely contribute to the variable expressivity as well. Recently, Rex et al.64 identified seven novel modulators of α7 nAChR calcium flux in a high-throughput screen. Six of these seven modulators are expressed in the brain and include proteins in the ER, proteins involved in transcription, and proteins involved in translation. Changes in these genes or their functions may contribute to the variable expressivity observed, as well as changes in other known modulators, including RIC3 and NACHO. RIC3 has been implicated in Parkinson disease, with loss-of-function mutations, showing that variants in the gene may be clinical relevant. Currently, probands with 15q13.3 CNVs are generally tested only by chromosomal microarray and not whole-genome or whole-exome sequencing, so additional variants that may contribute to phenotypes have yet to be uncovered.
Conclusions
Here, we generated 15q13.3 CNV iPSCs and NPCs, which may have clinical utility in the future for developing targeted therapeutics. We show that 15q13.3 CNVs both exhibit the same molecular pathogenesis, although to varying degrees and through differing mechanisms. This suggests that future studies into drug development for treatment with these CNVs should focus on both deletion and duplication probands.
Acknowledgments
This study was supported in part by IDDRC grant number 1U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. M.A.G. was supported by Award Number T32GM008307 from the National Institute of General Medical Sciences. The IDDRC Microscopy Core was used for this project. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. We would like to thank Emeline Crutcher, Ezequiel Sztainberg, and Ricky Lozoya for helping maintaining cells, as well as Dr. Frank Horrigan for training and use of the FLIPRTetra.
Published: November 9, 2017
Footnotes
Supplemental Data include seven figures and one table and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.09.024.
Supplemental Data
References
- 1.Gillentine M.A., Schaaf C.P. The human clinical phenotypes of altered CHRNA7 copy number. Biochem. Pharmacol. 2015;97:352–362. doi: 10.1016/j.bcp.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowther C., Costain G., Stavropoulos D.J., Melvin R., Silversides C.K., Andrade D.M., So J., Faghfoury H., Lionel A.C., Marshall C.R. Delineating the 15q13.3 microdeletion phenotype: a case series and comprehensive review of the literature. Genet. Med. 2015;17:149–157. doi: 10.1038/gim.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spielmann M., Reichelt G., Hertzberg C., Trimborn M., Mundlos S., Horn D., Klopocki E. Homozygous deletion of chromosome 15q13.3 including CHRNA7 causes severe mental retardation, seizures, muscular hypotonia, and the loss of KLF13 and TRPM1 potentially cause macrocytosis and congenital retinal dysfunction in siblings. Eur. J. Med. Genet. 2011;54:e441–e445. doi: 10.1016/j.ejmg.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Lepichon J.B., Bittel D.C., Graf W.D., Yu S. A 15q13.3 homozygous microdeletion associated with a severe neurodevelopmental disorder suggests putative functions of the TRPM1, CHRNA7, and other homozygously deleted genes. Am. J. Med. Genet. A. 2010;152A:1300–1304. doi: 10.1002/ajmg.a.33374. [DOI] [PubMed] [Google Scholar]
- 5.Masurel-Paulet A., Drumare I., Holder M., Cuisset J.M., Vallée L., Defoort S., Bourgois B., Pernes P., Cuvellier J.C., Huet F. Further delineation of eye manifestations in homozygous 15q13.3 microdeletions including TRPM1: a differential diagnosis of ceroid lipofuscinosis. Am. J. Med. Genet. A. 2014;164A:1537–1544. doi: 10.1002/ajmg.a.36471. [DOI] [PubMed] [Google Scholar]
- 6.Liao J., DeWard S.J., Madan-Khetarpal S., Surti U., Hu J. A small homozygous microdeletion of 15q13.3 including the CHRNA7 gene in a girl with a spectrum of severe neurodevelopmental features. Am. J. Med. Genet. A. 2011;155A:2795–2800. doi: 10.1002/ajmg.a.34237. [DOI] [PubMed] [Google Scholar]
- 7.Ziats M.N., Goin-Kochel R.P., Berry L.N., Ali M., Ge J., Guffey D., Rosenfeld J.A., Bader P., Gambello M.J., Wolf V. The complex behavioral phenotype of 15q13.3 microdeletion syndrome. Genet. Med. 2016;18:1111–1118. doi: 10.1038/gim.2016.9. [DOI] [PubMed] [Google Scholar]
- 8.Williams N.M., Zaharieva I., Martin A., Langley K., Mantripragada K., Fossdal R., Stefansson H., Stefansson K., Magnusson P., Gudmundsson O.O. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet. 2010;376:1401–1408. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams N.M., Franke B., Mick E., Anney R.J., Freitag C.M., Gill M., Thapar A., O’Donovan M.C., Owen M.J., Holmans P. Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13.3. Am. J. Psychiatry. 2012;169:195–204. doi: 10.1176/appi.ajp.2011.11060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillentine M.A., Berry L.N., Goin-Kochel R.P., Ali M.A., Ge J., Guffey D., Rosenfeld J.A., Hannig V., Bader P., Proud M. The cognitive and behavioral phenotypes of individuals with CHRNA7 duplications. J. Autism Dev. Disord. 2017;47:549–562. doi: 10.1007/s10803-016-2961-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szafranski P., Schaaf C.P., Person R.E., Gibson I.B., Xia Z., Mahadevan S., Wiszniewska J., Bacino C.A., Lalani S., Potocki L. Structures and molecular mechanisms for common 15q13.3 microduplications involving CHRNA7: benign or pathological? Hum. Mutat. 2010;31:840–850. doi: 10.1002/humu.21284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinawi M., Schaaf C.P., Bhatt S.S., Xia Z., Patel A., Cheung S.W., Lanpher B., Nagl S., Herding H.S., Nevinny-Stickel C. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat. Genet. 2009;41:1269–1271. doi: 10.1038/ng.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaaf C.P. Nicotinic acetylcholine receptors in human genetic disease. Genet. Med. 2014;16:649–656. doi: 10.1038/gim.2014.9. [DOI] [PubMed] [Google Scholar]
- 14.Albuquerque E.X., Pereira E.F., Alkondon M., Rogers S.W. Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol. Rev. 2009;89:73–120. doi: 10.1152/physrev.00015.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinkus M.L., Graw S., Freedman R., Ross R.G., Lester H.A., Leonard S. The human CHRNA7 and CHRFAM7A genes: A review of the genetics, regulation, and function. Neuropharmacology. 2015;96(Pt B):274–288. doi: 10.1016/j.neuropharm.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickinson J.A., Hanrott K.E., Mok M.H., Kew J.N., Wonnacott S. Differential coupling of alpha7 and non-alpha7 nicotinic acetylcholine receptors to calcium-induced calcium release and voltage-operated calcium channels in PC12 cells. J. Neurochem. 2007;100:1089–1096. doi: 10.1111/j.1471-4159.2006.04273.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Xiao C., Indersmitten T., Freedman R., Leonard S., Lester H. The duplicated alpha7 subunits assemble and form functional nicotinic receptors with the full-length alpha7. J. Biol. Chem. 2014;289 doi: 10.1074/jbc.M114.582858. 26451–26363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arredondo J., Nguyen V.T., Chernyavsky A.I., Bercovich D., Orr-Urtreger A., Kummer W., Lips K., Vetter D.E., Grando S.A. Central role of alpha7 nicotinic receptor in differentiation of the stratified squamous epithelium. J. Cell Biol. 2002;159:325–336. doi: 10.1083/jcb.200206096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freedman R. α7-nicotinic acetylcholine receptor agonists for cognitive enhancement in schizophrenia. Annu. Rev. Med. 2014;65:245–261. doi: 10.1146/annurev-med-092112-142937. [DOI] [PubMed] [Google Scholar]
- 20.Shimojima K., Inoue T., Imai Y., Arai Y., Komoike Y., Sugawara M., Fujita T., Ideguchi H., Yasumoto S., Kanno H. Reduced PLP1 expression in induced pluripotent stem cells derived from a Pelizaeus-Merzbacher disease patient with a partial PLP1 duplication. J. Hum. Genet. 2012;57:580–586. doi: 10.1038/jhg.2012.71. [DOI] [PubMed] [Google Scholar]
- 21.Yin J., Chen W., Yang H., Xue M., Schaaf C.P. Chrna7 deficient mice manifest no consistent neuropsychiatric and behavioral phenotypes. Sci. Rep. 2017;7:39941. doi: 10.1038/srep39941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paylor R., Nguyen M., Crawley J.N., Patrick J., Beaudet A., Orr-Urtreger A. Alpha7 nicotinic receptor subunits are not necessary for hippocampal-dependent learning or sensorimotor gating: a behavioral characterization of Acra7-deficient mice. Learn. Mem. 1998;5:302–316. [PMC free article] [PubMed] [Google Scholar]
- 23.Adams C., Yonchek J., Schulz K., Graw S., Stitzel J., Teschke P., Stevens K. Reduced Chrna7 expression in mice is associated with decreases in hippocampal markers of inhibitory function. Neurosience. 2012;207:274–282. doi: 10.1016/j.neuroscience.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-David Y., Mizrachi T., Kagan S., Krisher T., Cohen E., Brenner T., Treinin M. RIC-3 expression and splicing regulate nAChR functional expression. Mol. Brain. 2016;9:47. doi: 10.1186/s13041-016-0231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forsingdal A., Fejgin K., Nielsen V., Werge T., Nielsen J. 15q13.3 homozygous knockout mouse model display epilepsy-, autism- and schizophrenia-related phenotypes. Transl. Psychiatry. 2016;6:e860. doi: 10.1038/tp.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paşca S.P., Portmann T., Voineagu I., Yazawa M., Shcheglovitov A., Paşca A.M., Cord B., Palmer T.D., Chikahisa S., Nishino S. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat. Med. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchetto M.C., Carromeu C., Acab A., Yu D., Yeo G.W., Mu Y., Chen G., Gage F.H., Muotri A.R. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran N.N., Ladran I.G., Brennand K.J. Modeling schizophrenia using induced pluripotent stem cell-derived and fibroblast-induced neurons. Schizophr. Bull. 2013;39:4–10. doi: 10.1093/schbul/sbs127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stern S., Santos R., Marchetto M.C., Mendes A.P., Rouleau G.A., Biesmans S., Wang Q.W., Yao J., Charnay P., Bang A.G. Neurons derived from patients with bipolar disorder divide into intrinsically different sub-populations of neurons, predicting the patients’ responsiveness to lithium. Mol. Psychiatry. 2017 doi: 10.1038/mp.2016.260. Published online February 28, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X., Campanac E., Cheung H.-H.H., Ziats M.N., Canterel-Thouennon L., Raygada M., Baxendale V., Pang A.L., Yang L., Swedo S. Idiopathic autism: cellular and molecular phenotypes in pluripotent stem cell-derived neurons. Mol. Neurobiol. 2017;54:4507–4523. doi: 10.1007/s12035-016-9961-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatzidaki A., Fouillet A., Li J., Dage J., Millar N.S., Sher E., Ursu D. Pharmacological characterisation of nicotinic acetylcholine receptors expressed in human iPSC-derived neurons. PLoS ONE. 2015;10:e0125116. doi: 10.1371/journal.pone.0125116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dage J.L., Colvin E.M., Fouillet A., Langron E., Roell W.C., Li J., Mathur S.X., Mogg A.J., Schmitt M.G., Felder C.C. Pharmacological characterisation of ligand- and voltage-gated ion channels expressed in human iPSC-derived forebrain neurons. Psychopharmacology (Berl.) 2014;231:1105–1124. doi: 10.1007/s00213-013-3384-2. [DOI] [PubMed] [Google Scholar]
- 33.Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J.-E., O’Sullivan M.L., Sanchez C.A., Hwang M., Israel M.A., Brennand K., Deerinck T.J., Goldstein L.S., Gage F.H., Ellisman M.H., Ghosh A. Investigating synapse formation and function using human pluripotent stem cell-derived neurons. Proc. Natl. Acad. Sci. USA. 2011;108:3005–3010. doi: 10.1073/pnas.1007753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oslowski C., Urano F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 2011;490:71–92. doi: 10.1016/B978-0-12-385114-7.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Galen P., Kreso A., Mbong N., Kent D.G., Fitzmaurice T., Chambers J.E., Xie S., Laurenti E., Hermans K., Eppert K. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510:268–272. doi: 10.1038/nature13228. [DOI] [PubMed] [Google Scholar]
- 37.Walstab J., Hammer C., Lasitschka F., Möller D., Connolly C.N., Rappold G., Brüss M., Bönisch H., Niesler B. RIC-3 exclusively enhances the surface expression of human homomeric 5-hydroxytryptamine type 3A (5-HT3A) receptors despite direct interactions with 5-HT3A, -C, -D, and -E subunits. J. Biol. Chem. 2010;285:26956–26965. doi: 10.1074/jbc.M110.122838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Treinin M. RIC-3 and nicotinic acetylcholine receptors: biogenesis, properties, and diversity. Biotechnol. J. 2008;3:1539–1547. doi: 10.1002/biot.200800179. [DOI] [PubMed] [Google Scholar]
- 39.Gu S., Matta J.A., Lord B., Harrington A.W., Sutton S.W., Davini W.B., Bredt D.S. Brain α7 nicotinic acetylcholine receptor assembly requires NACHO. Neuron. 2016;89:948–955. doi: 10.1016/j.neuron.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 40.Marrero M.B., Bencherif M. Convergence of alpha 7 nicotinic acetylcholine receptor-activated pathways for anti-apoptosis and anti-inflammation: central role for JAK2 activation of STAT3 and NF-kappaB. Brain Res. 2009;1256:1–7. doi: 10.1016/j.brainres.2008.11.053. [DOI] [PubMed] [Google Scholar]
- 41.Parada E., Egea J., Romero A., del Barrio L., García A.G., López M.G. Poststress treatment with PNU282987 can rescue SH-SY5Y cells undergoing apoptosis via α7 nicotinic receptors linked to a Jak2/Akt/HO-1 signaling pathway. Free Radic. Biol. Med. 2010;49:1815–1821. doi: 10.1016/j.freeradbiomed.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 42.Shaw S., Bencherif M., Marrero M.B. Janus kinase 2, an early target of α 7 nicotinic acetylcholine receptor-mediated neuroprotection against Abeta-(1-42) amyloid. J. Biol. Chem. 2002;277:44920–44924. doi: 10.1074/jbc.M204610200. [DOI] [PubMed] [Google Scholar]
- 43.Millar N.S. Assembly and subunit diversity of nicotinic acetylcholine receptors. Biochem. Soc. Trans. 2003;31:869–874. doi: 10.1042/bst0310869. [DOI] [PubMed] [Google Scholar]
- 44.Severance E.G., Yolken R.H. Lack of RIC-3 congruence with beta2 subunit-containing nicotinic acetylcholine receptors in bipolar disorder. Neuroscience. 2007;148:454–460. doi: 10.1016/j.neuroscience.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 45.Lewis A.S., Picciotto M.R. High-affinity nicotinic acetylcholine receptor expression and trafficking abnormalities in psychiatric illness. Psychopharmacology (Berl.) 2013;229:477–485. doi: 10.1007/s00213-013-3126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srinivasan R., Henderson B.J., Lester H.A., Richards C.I. Pharmacological chaperoning of nAChRs: a therapeutic target for Parkinson’s disease. Pharmacol. Res. 2014;83:20–29. doi: 10.1016/j.phrs.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srinivasan R., Richards C.I., Xiao C., Rhee D., Pantoja R., Dougherty D.A., Miwa J.M., Lester H.A. Pharmacological chaperoning of nicotinic acetylcholine receptors reduces the endoplasmic reticulum stress response. Mol. Pharmacol. 2012;81:759–769. doi: 10.1124/mol.112.077792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srinivasan R., Pantoja R., Moss F.J., Mackey E.D., Son C.D., Miwa J., Lester H.A. Nicotine up-regulates alpha4beta2 nicotinic receptors and ER exit sites via stoichiometry-dependent chaperoning. J. Gen. Physiol. 2011;137:59–79. doi: 10.1085/jgp.201010532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rutkowski D.T., Kaufman R.J. That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem. Sci. 2007;32:469–476. doi: 10.1016/j.tibs.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Moser N., Mechawar N., Jones I., Gochberg-Sarver A., Orr-Urtreger A., Plomann M., Salas R., Molles B., Marubio L., Roth U. Evaluating the suitability of nicotinic acetylcholine receptor antibodies for standard immunodetection procedures. J. Neurochem. 2007;102:479–492. doi: 10.1111/j.1471-4159.2007.04498.x. [DOI] [PubMed] [Google Scholar]
- 51.Numasawa-Kuroiwa Y., Okada Y., Shibata S., Kishi N., Akamatsu W., Shoji M., Nakanishi A., Oyama M., Osaka H., Inoue K. Involvement of ER stress in dysmyelination of Pelizaeus-Merzbacher disease with PLP1 missense mutations shown by iPSC-derived oligodendrocytes. Stem Cell Reports. 2014;2:648–661. doi: 10.1016/j.stemcr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gee K.W., Olincy A., Kanner R., Johnson L., Hogenkamp D., Harris J., Tran M., Edmonds S.A., Sauer W., Yoshimura R. First in human trial of a type I positive allosteric modulator of alpha7-nicotinic acetylcholine receptors: Pharmacokinetics, safety, and evidence for neurocognitive effect of AVL-3288. J. Psychopharmacol. (Oxford) 2017;31:434–441. doi: 10.1177/0269881117691590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olincy A., Blakeley-Smith A., Johnson L., Kem W.R., Freedman R. Brief report: initial trial of alpha7-nicotinic receptor stimulation in two adult patients with autism spectrum disorder. J. Autism Dev. Disord. 2016;46:3812–3817. doi: 10.1007/s10803-016-2890-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sadigh-Eteghad S., Majdi A., Talebi M., Mahmoudi J., Babri S. Regulation of nicotinic acetylcholine receptors in Alzheimer's disease: a possible role of chaperones. Eur. J. Pharmacol. 2015;755:34–41. doi: 10.1016/j.ejphar.2015.02.047. [DOI] [PubMed] [Google Scholar]
- 55.Araud T., Graw S., Berger R., Lee M., Neveu E., Bertrand D., Leonard S. The chimeric gene CHRFAM7A, a partial duplication of the CHRNA7 gene, is a dominant negative regulator of α7∗nAChR function. Biochem. Pharmacol. 2011;82:904–914. doi: 10.1016/j.bcp.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen J.X., Yakel J.L. Nicotinic acetylcholine receptor-mediated calcium signaling in the nervous system. Acta Pharmacol. Sin. 2009;30:673–680. doi: 10.1038/aps.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Resende R.R., Adhikari A. Cholinergic receptor pathways involved in apoptosis, cell proliferation and neuronal differentiation. Cell Commun. Signal. 2009;7:20. doi: 10.1186/1478-811X-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lorenz C., Lesimple P., Bukowiecki R., Zink A., Inak G., Mlody B., Singh M., Semtner M., Mah N., Auré K. Human iPSC-derived neural progenitors are an effective drug discovery model for neurological mtDNA disorders. Cell Stem Cell. 2017;20:659–674.e9. doi: 10.1016/j.stem.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 59.Cubells J.F., Deoreo E.H., Harvey P.D., Garlow S.J., Garber K., Adam M.P., Martin C.L. Pharmaco-genetically guided treatment of recurrent rage outbursts in an adult male with 15q13.3 deletion syndrome. Am. J. Med. Genet. A. 2011;155A:805–810. doi: 10.1002/ajmg.a.33917. [DOI] [PubMed] [Google Scholar]
- 60.Shao L., Sun X., Xu L., Young L.T., Wang J.-F.F. Mood stabilizing drug lithium increases expression of endoplasmic reticulum stress proteins in primary cultured rat cerebral cortical cells. Life Sci. 2006;78:1317–1323. doi: 10.1016/j.lfs.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 61.Hiroi T., Wei H., Hough C., Leeds P., Chuang D.-M.M. Protracted lithium treatment protects against the ER stress elicited by thapsigargin in rat PC12 cells: roles of intracellular calcium, GRP78 and Bcl-2. Pharmacogenomics J. 2005;5:102–111. doi: 10.1038/sj.tpj.6500296. [DOI] [PubMed] [Google Scholar]
- 62.Wiley J.C., Meabon J.S., Frankowski H., Smith E.A., Schecterson L.C., Bothwell M., Ladiges W.C. Phenylbutyric acid rescues endoplasmic reticulum stress-induced suppression of APP proteolysis and prevents apoptosis in neuronal cells. PLoS ONE. 2010;5:e9135. doi: 10.1371/journal.pone.0009135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao C., Giacca A., Lewis G.F. Sodium phenylbutyrate, a drug with known capacity to reduce endoplasmic reticulum stress, partially alleviates lipid-induced insulin resistance and beta-cell dysfunction in humans. Diabetes. 2011;60:918–924. doi: 10.2337/db10-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rex E.B., Shukla N., Gu S., Bredt D., DiSepio D. A genome-wide arrayed cDNA screen to identify functional modulators of α7 nicotinic acetylcholine receptors. SLAS Discov. 2017;22:155–165. doi: 10.1177/1087057116676086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.