Abstract
Genome-wide association studies have identified hundreds of genetic variants associated with blood pressure (BP), but sequence variation accounts for a small fraction of the phenotypic variance. Epigenetic changes may alter the expression of genes involved in BP regulation and explain part of the missing heritability. We therefore conducted a two-stage meta-analysis of the cross-sectional associations of systolic and diastolic BP with blood-derived genome-wide DNA methylation measured on the Infinium HumanMethylation450 BeadChip in 17,010 individuals of European, African American, and Hispanic ancestry. Of 31 discovery-stage cytosine-phosphate-guanine (CpG) dinucleotides, 13 replicated after Bonferroni correction (discovery: N = 9,828, p < 1.0 × 10−7; replication: N = 7,182, p < 1.6 × 10−3). The replicated methylation sites are heritable (h2 > 30%) and independent of known BP genetic variants, explaining an additional 1.4% and 2.0% of the interindividual variation in systolic and diastolic BP, respectively. Bidirectional Mendelian randomization among up to 4,513 individuals of European ancestry from 4 cohorts suggested that methylation at cg08035323 (TAF1B-YWHAQ) influences BP, while BP influences methylation at cg00533891 (ZMIZ1), cg00574958 (CPT1A), and cg02711608 (SLC1A5). Gene expression analyses further identified six genes (TSPAN2, SLC7A11, UNC93B1, CPT1A, PTMS, and LPCAT3) with evidence of triangular associations between methylation, gene expression, and BP. Additional integrative Mendelian randomization analyses of gene expression and DNA methylation suggested that the expression of TSPAN2 is a putative mediator of association between DNA methylation at cg23999170 and BP. These findings suggest that heritable DNA methylation plays a role in regulating BP independently of previously known genetic variants.
Keywords: blood pressure, DNA methylation, epigenome-wide association study, gene expression, sequence variation, Mendelian randomization
Introduction
Elevated blood pressure (BP) confers a higher risk of heart disease, stroke, diabetes, dementia, renal failure, and pregnancy-related complications and is a leading risk factor for death worldwide.1 BP is a highly heritable trait2 and recent genetic studies have revealed part of its complex genetic architecture,3, 4, 5, 6, 7, 8, 9, 10, 11 yet the genetic variants identified to date account for only a small fraction of its phenotypic variance.3, 6, 8, 12 Complex phenotypes, such as BP, often result from the interplay between genetic and environmental influences. DNA methylation, the covalent binding of a methyl group to the 5′ carbon of cytosine-phosphate-guanine (CpG) dinucleotide sequences in the genome, plays a critical role in the regulation of gene expression and may reflect a link between genes, environment, and complex phenotypes such as BP. Evidence is beginning to emerge that epigenetic modifications in genes relevant to BP may account for part of its regulation.13 Variation in DNA methylation may thus explain additional phenotypic variation in BP and provide new clues to the biological processes influencing its regulation.
We conducted genome-wide DNA methylation meta-analyses for systolic and diastolic BP with a discovery phase and independent replication among 17,010 individuals of European (EA), African American (AA), and Hispanic ancestries in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. DNA methylation was measured in peripheral blood samples. We further sought to identify transcriptional changes for the replicated CpG sites and used Mendelian randomization techniques to explore the causal relationship between DNA methylation and BP. We report that the effect of DNA methylation on BP is likely independent of previously known genetic variants, representing new insights into the biological mechanisms underlying BP regulation.
Material and Methods
Study Populations
The discovery and replication studies were conducted in the framework of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium, which comprises multiple population-based cohort studies.14 Cohorts participating at the discovery stage included 9,828 individuals of EA and AA ancestries in the Atherosclerosis Risk in Communities (ARIC) study, Cardiovascular Health Study (CHS), Framingham Heart Study (FHS), Genetic Epidemiology Network of Arteriopathy (GENOA) study, Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) study, Lothian Birth Cohort 1936 (LBC1936), Normative Aging Study (NAS), Rotterdam Study (RS), and TwinsUK registry. Cohorts participating at the replication stage consisted of 7,182 additional individuals of EA, AA, and Hispanic ancestries in the Amish Complex Disease Research Studies (Amish), ARIC, the Multi-Ethnic Study of Atherosclerosis (MESA), RS, adults in the Saguenay Youth Study (SYS), and the Women’s Health Initiative (WHI). Details for each cohort are provided in the Supplemental Data. All studies obtained written informed consent from participants and were approved by local institutional review boards and ethics committees.
Blood Pressure Measurements
Epigenome-wide association studies (EWASs) were conducted for systolic and diastolic BP, in mmHg. In each cohort, BP was measured in a sitting position after a period of rest and an average of sequential readings was used as the phenotype for each analysis. For most cohorts, BP was measured concurrently at the time of tissue collection for DNA methylation profiling, or in as close proximity as available for TwinsUK (0.8 years) and SYS adults (3.1 years). To adjust for the use of antihypertensive medication, we used the standard adjustment of adding 15 mmHg and 10 mmHg to measured systolic and diastolic BPs, respectively, when the use of any antihypertensive medications were self-reported.
DNA Methylation Profiling
DNA methylation was measured on the Infinium HumanMethylation450 (450k) BeadChip (Illumina) in all cohorts using whole-blood samples, excepting that GOLDN measured DNA methylation in CD4+ T cells. To correct the beta value distributions of the two types of probes on the 450k array, each cohort normalized methylation beta values using BMIQ,15 DASEN,16 ComBat,17 SWAN,18 or quantile normalization. LBC1936 did not normalize methylation beta values prior to analyses.
Cohort-Level Association Analyses
In each cohort, race-stratified linear mixed effect models were used to estimate associations adjusting for age, sex (in samples including men and women), blood cell counts, body mass index, smoking (current/former/never), and ancestry, as well as fixed and/or random effects for technical covariates to control for batch effects. Surrogate variables were calculated and adjusted in the modeling for ARIC AAs and FHS due to batch effects not controlled by other modeling techniques. The Amish, FHS, GOLDN, and TwinsUK accounted for sample relatedness in all analyses. Study-specific modeling details can be found in the Supplemental Data.
Epigenome-wide Meta-Analyses
Effect estimates from all cohorts were combined using inverse variance fixed effects meta-analysis using GWAMA.19 We assessed heterogeneity of effect estimates between strata of races, sexes, and methylation tissue source among discovery cohorts using a 1 degree of freedom chi-square test for effect differences between strata; no heterogeneous effects were observed so all cohorts were included in a single meta-analysis. Meta-analyses were conducted separately for the discovery and replication cohorts to identify probes associated with BP. Statistical significance was Bonferroni corrected for the epigenome-wide discovery meta-analysis (p < 1.0 × 10−7) and the number of discovery CpG sites sought for replication in the second meta-analysis. An overall meta-analysis was additionally performed to combine effect estimates across all cohorts. Significant CpG sites were annotated using information provided by Illumina, including chromosome, position (GRCh37/hg19), UCSC gene names, relationship to CpG islands, location in gene enhancer regions, and DNase I hypersensitivity sites (DHS). To assess the impact of antihypertensive medication use on our top findings, we additionally performed an overall meta-analysis among all individuals reporting no use of antihypertensive medications. For the top findings in the discovery meta-analysis, we compared effect and standard error estimates to those estimated in the non-medicated meta-analysis.
Percent Variance Explained
Percent variance explained was calculated in the ARIC AA and EA samples included in discovery and replication meta-analyses, as well as validated in a sample from the FHS Third Generation not included in the meta-analysis (N = 1,516). Methylation profile scores for BP were calculated as the weighted sum of CpG sites significant for either BP trait in the replication and overall meta-analyses, with weights coming from the magnitude and direction of effects in the overall meta-analysis. Selection of CpGs from meta-analyses including the prediction samples could overestimate percent variance explained, so additional meta-analyses were conducted excluding the ARIC samples to identify CpGs for their respective methylation profile scores. The probe sets based on exclusion of the ARIC samples and the probes identified in the primary replication and overall meta-analyses were used to generate methylation profile scores in the FHS sample. Race- and cohort-stratified linear regression models were used to estimate the percent of age-, sex-, and BMI-adjusted systolic and diastolic BP variances explained by each methylation profile score; ARIC models were additionally adjusted for visit and study site, and ARIC AA and FHS models included surrogate variables. Percent variance explained by the methylation profile scores is reported as the adjusted R2 from each model and compared to models without methylation profile scores (covariate-only models). We additionally assessed genetic risk scores derived using effect estimates from the UK Biobank for 146 previously reported independent variants (r2 < 0.2) and 115 validated novel variants11 among the FHS Third Generation sample with available genetic data (N = 1,421).
Heritability
The narrow-sense heritability estimate of a DNA methylation trait (β score) (denoted as ) was the proportion of the additive polygenic genetic variance of the total phenotypic variance of a DNA methylation trait: , where denotes the additive polygenic genetic variance and denotes the total phenotypic variance of a DNA methylation trait. Heritability estimation for all DNA methylation traits was performed using the FHS-Offspring participants (N = 2,377).
Functional Tissue and Gene Set Enrichment Analyses
Functional DNA elements regulated by methylation may be tissue specific, so the set of replicated CpGs was used to identify tissue- and cell type-specific signals using experimentally derived Functional element Overlap analysis of ReGions from EWAS (eFORGE).20 After pruning results for CpG sites within 1 kb (2 probes removed), we matched the top 11 EWAS signals for overlap with DNase I hypersensitive sites using data from ENCODE and Roadmap Epigenomics. 1,000 matched sets were used with the 450k array as the background set. FDR correction was applied to the results.
Gene Set Enrichment Analysis (GSEA)21 was conducted on the results of the overall meta-analyses for systolic and diastolic BP. For each gene annotated to DNA methylation measured on the 450k array, a composite ranking for BP was generated based on the CpG site with the minimum p value for either trait. All gene ontology biological process categories (c5.bp.v5.1) were assessed for enrichment at FDR Q < 0.05.
Methylation Quantitative Trait Loci
To determine methylation levels at CpG sites that may be influenced by nearby DNA sequence, methylation quantitative trait loci (meQTL) analyses were performed for the 13 replicated BP CpGs in EA individuals from ARIC (N = 948), FHS (N = 2,357), and RS (N = 731) and AA individuals from ARIC (N = 2,173) and GENOA (N = 422). Residuals were obtained from regressing inverse-normal transformed methylation beta values on the first ten methylation principal components (PCs) and up to the first ten genetic PCs. The residuals were then regressed on 1000 Genomes Phase I imputed SNPs within 50 kb of the probe (CpG position ± 25 kb, GRCh37/hg19). SNPs with low imputation quality (r2 < 0.3), low frequency variants (MAF < 0.05), and SNPs present in only one cohort were removed from analyses. Results for each probe were combined using race-stratified p value-based meta-analysis weighted by sample size and direction of effects using METAL.22 Significant meQTLs were determined using a Bonferroni correction for all meQTLs tested in each race (EA: 0.05/1,447 = 3.5 × 10−5; AA: 0.05/1,952 = 2.6 × 10−5). To maximize statistical power for identifying meQTLs associated with BP, we then searched the largest genome-wide association studies (GWASs) for BP in each race for suggestive association of meQTL regions with BP.
To assess the association of SNPs reported by Kato et al.23 whose association may be mediated by DNA methylation, we additionally performed meQTL analyses for 35 sentinel SNPs and additional GWAS loci in high linkage disequilibrium (LD) with these regions.3, 4, 5, 23, 24, 25, 26, 27, 28, 29, 30 We assessed the association of DNA methylation within 1 Mb (CpG position ± 500 kb) of GWAS SNPs among ARIC EAs (N = 790) using the previously described methodology. SNPs associated with methylation after Bonferroni correction for the 28 meQTLs reported by Kato et al.23 (p < 0.0018) were then assessed for association with BP before and after adjustment for methylation at the CpG site. We additionally assessed the association of these CpG sites with BP in our overall meta-analysis.
Bidirectional Mendelian Randomization
To assess the directional association of DNA methylation and BP, we conducted bidirectional Mendelian randomization (MR) using 1000 Genomes imputed SNPs among EA individuals in ARIC, FHS, RS, and WHI-EMPC (N = 4,513). Forward MR was used to identify replicated CpG sites which may have an effect on BP. Instrumental variables (IVs) for DNA methylation were drawn from the meQTLs estimated among EAs and pruned for independence (r2 < 0.2). Forward MR was conducted for the six sentinel CpG sites with at least three independent meQTLs, which is the minimum number of IVs needed to perform multi-instrument MR. Reverse MR was used to identify DNA methylation at the 11 sentinel CpG sites that may be caused by BP. The 29 independent loci reported as associated with BP by the International Consortium for Blood Pressure (ICBP) were selected as IVs. The SNP rs805303 was not imputed in 1000 Genomes and rs805301 was used as a proxy when available (r2 = 1.0 in HapMap).
Each cohort estimated the associations of IVs with systolic BP, diastolic BP, and DNA methylation at the respective CpG sites. Cohort-level effect estimates for each IV were combined using inverse variance-weighted meta-analyses in METAL.22 For each CpG in forward and reverse MR, causation was formally tested based on the inverse variance-weighted effects across all IV-BP and IV-CpG estimates using the R package MendelianRandomization.31 Tests for causation with p value < 0.05 were considered significant. To ensure the validity of the inverse-variance weighted approach, the IVs were assessed for pleiotropy using the MR-Egger test. Inverse-variance weighted MR is invalid in the presence of pleiotropic effects of IVs, so Egger regression estimates of causality were assessed only when pleiotropy was indicated at a particular CpG site.
Associations of DNA Methylation and Gene Expression
Association tests of BP-associated CpGs with transcripts that were located within ±1 Mb distance of the corresponding CpGs were performed in 2,216 FHS-Offspring samples and 730 RS samples whose DNA methylation and gene expression data were both available. In FHS, linear mixed effect regression models were used with DNA methylation β scores as the dependent variable, gene expression as independent variables, age, sex, and technical covariates as fixed effects, and family structure as a random effect. In RS, we first created residuals for both DNA methylation and mRNA expression after regressing out age, sex, blood cell counts (fixed effect), and technical covariates (random effect). We then examined the association between the residuals of DNA methylation (independent variable) and mRNA expression (dependent variable) using a linear regression model. Estimates of the gene expression-methylation associations in RS and FHS were combined using sample size weighted fixed effects meta-analysis based on p values and direction of effects using GWAMA.19
Associations of Gene Expression and BP
Differential gene expression analysis of the transcripts assessed for association with DNA methylation were performed for systolic BP, diastolic BP, and hypertension in 3,679 FHS Offspring and 3rd-Gen participants who were not receiving anti-hypertensive treatments. Hypertension was defined as systolic BP ≥ 140 mmHg or diastolic BP ≥ 90 mmHg. See details in Huan et al.32
Two-Step Mendelian Randomization for Relationship of DNA Methylation, Gene Expression, and BP
To identify gene transcription that functionally mediates the relationship of DNA methylation and BP, we performed a two-step MR technique for genes with expression associated with both DNA methylation and BP (FDR Q < 0.05). The first step was to establish a directional relationship between DNA methylation and gene transcription. IVs for DNA methylation were drawn from estimated meQTLs pruned to be independent (r2 < 0.2). Using whole-blood eQTLs estimated in the Genotype-Tissue Expression (GTEx) project, we verified the association of each IV with the implicated gene expression. In the second step, IVs for each implicated gene were selected from the GTEx whole-blood dataset in order to establish a directional relationship between gene expression and BP. The top eQTL also present in the ICBP results was selected as the IV for each gene and assessed for association with systolic and diastolic BP in ICBP published GWAS results. Genes with p < 0.05 at both steps were considered to mediate a directional relationship of the respective CpG and BP; correction for multiple testing is not used because strong associations of IVs with an outcome would violate the assumptions of Mendelian randomization.
Results
Cohort Characteristics
Characteristics of the 14 studies participating in discovery and replication meta-analyses are presented in Table 1. Each cohort included middle-aged and older adults with a wide range of BP values. Mean systolic BP ranged from 116 mmHg in GOLDN to 152 mmHg among CHS AAs. Mean diastolic BP ranged from 68 mmHg in GOLDN to 89 mmHg in the RS replication sample. Prevalence of antihypertensive medication use varied with cohort age and health, with no use among the Amish to more than 62% among the CHS AA sample.
Table 1.
Characteristics of the Discovery and Replication Cohorts
| Cohort | Race | n | Cohort Type | Tissue | Normalization |
Age, years |
SBP, mmHg |
DBP, mmHg |
HTN |
AHT |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | % | % | ||||||
| Discovery (N = 9,828) | |||||||||||||
| ARIC | AA | 2,743 | unrelated | blood | BMIQ | 56.6 | 5.9 | 135.0 | 23.4 | 80.2 | 12.4 | 65.5 | 48.9 |
| CHS | AA | 196 | unrelated | blood | SWAN | 73.0 | 5.4 | 151.5 | 23.9 | 83.2 | 12.5 | 78.6 | 62.2 |
| CHS | EA | 189 | unrelated | blood | SWAN | 76.0 | 5.1 | 142.8 | 23.9 | 76.7 | 10.9 | 65.6 | 49.2 |
| FHS | EA | 2,645 | family | blood | DASEN | 66.4 | 8.9 | 128.6 | 17.2 | 73.4 | 10.0 | 59.0 | 49.0 |
| GENOA | AA | 239 | unrelated | blood | SWAN | 60.1 | 8.4 | 146.1 | 25.6 | 82.5 | 12.4 | 72.0 | 58.2 |
| GOLDN | EA | 822 | family | CD4+ T cells | ComBat | 48.8 | 15.9 | 115.7 | 16.3 | 68.4 | 9.4 | 25.7 | 21.0 |
| LBC1936 | EA | 903 | unrelated | blood | – | 69.5 | 0.8 | 149.4 | 19.0 | 81.3 | 10.1 | 40.7 | 43.0 |
| NAS | EA | 674 | unrelated | blood | BMIQ | 72.5 | 6.8 | 139.5 | 18.9 | 81.9 | 10.3 | 71.0 | 58.2 |
| RS-III | EA | 727 | unrelated | blood | DASEN | 59.7 | 8.2 | 138.9 | 22.0 | 85.8 | 12.5 | 53.2 | 30.1 |
| TwinsUK | EA | 690 | twins | blood | BMIQ | 58.4 | 9.3 | 126.0 | 16.6 | 77.2 | 9.8 | 25.7 | 21.5 |
| Replication (N = 7,182) | |||||||||||||
| Amish | EA | 192 | family | blood | quantile | 46.3 | 13.6 | 117.8 | 12.7 | 72.4 | 8.1 | 2.0 | 0.0 |
| ARIC | EA | 1,058 | unrelated | blood | BMIQ | 59.8 | 5.4 | 121.2 | 20.5 | 70.1 | 11.1 | 29.7 | 17.6 |
| MESA | AA | 236 | unrelated | blood | quantile | 60.6 | 9.2 | 127.5 | 19.6 | 73.3 | 9.5 | 55.2 | 48.0 |
| MESA | EA | 566 | unrelated | blood | quantile | 60.8 | 9.6 | 121.0 | 18.5 | 70.1 | 9.6 | 36.8 | 31.7 |
| MESA | HL | 381 | unrelated | blood | quantile | 59.0 | 9.5 | 122.6 | 18.4 | 72.0 | 9.3 | 37.8 | 31.3 |
| RS-III | EA | 711 | unrelated | blood | DASEN | 67.5 | 6.0 | 151.3 | 24.0 | 88.7 | 13.0 | 71.5 | 43.3 |
| SYS adults | EA | 111 | unrelated | blood | SWAN | 47.2 | 4.9 | 131.5 | 15.3 | 79.5 | 8.4 | 29.7 | 8.1 |
| WHI-BAA23 | AA | 666 | unrelated | blood | ComBat | 62.8 | 6.7 | 140.9 | 21.1 | 83.3 | 10.9 | 65.0 | 54.7 |
| WHI-BAA23 | EA | 965 | unrelated | blood | ComBat | 68.4 | 6.2 | 136.5 | 21.1 | 78.2 | 11.1 | 48.5 | 34.6 |
| WHI-BAA23 | HL | 333 | unrelated | blood | ComBat | 62.3 | 6.8 | 133.3 | 20.7 | 78.6 | 10.8 | 47.3 | 35.2 |
| WHI-EPMC | AA | 556 | unrelated | blood | BMIQ | 62.8 | 7.0 | 131.5 | 18.1 | 77.4 | 9.6 | 60.4 | 55.2 |
| WHI-EMPC | EA | 1,092 | unrelated | blood | BMIQ | 64.7 | 7.1 | 127.5 | 17.7 | 74.5 | 9.4 | 42.9 | 30.5 |
| WHI-EMPC | HL | 315 | unrelated | blood | BMIQ | 61.6 | 6.2 | 127.2 | 18.2 | 74.8 | 9.5 | 41.9 | 29.5 |
Hypertension is defined as systolic BP ≥ 140 mmHg or diastolic BP ≥ 90 mmHg or the use of antihypertensive treatment. Antihypertensive treatment is defined as the self-reported use of any antihypertensive medication. WHI-EMPC normalized DNA methylation data using BMIQ and plate-adjusted using ComBat. The discovery and replication samples from RS-III do not include overlapping or related individuals. Abbreviations: AA, African American; AHT, antihypertensive treatment; BMIQ, Beta Mixture Quantile dilation; ComBat, combatting batch effects when COMbining BATches of microarray data; DASEN, background-adjusted (D) between-array (S) without dye bias correction (N); DBP, diastolic blood pressure; EA, European ancestry; HL, Hispanic/Latino; HTN, hypertension; SBP, systolic blood pressure; SD, standard deviation; SWAN, Subset-quantile Within Array Normalization.
Identification of Epigenome-wide CpG Sites Associated with Blood Pressure
In the discovery stage, we conducted genome-wide associations of DNA methylation with systolic and diastolic BP in nine cohort studies (N = 9,828). Multiethnic meta-analyses identified methylation at 31 CpG sites associated with BP after Bonferroni correction for the number of DNA methylation CpG sites measured on the Illumina 450K array (p < 1.0 × 10−7; Table S1, Figures S1 and S2). Replication of the 31 discovery CpG sites was sought in multiethnic meta-analyses of an additional six cohort studies (N = 7,182). Methylation at 13 of the 31 discovery CpG sites was associated with BP at p < 0.0016 in the replication meta-analysis (0.05/31; Table 2). A schematic of the overall study design, including subsequent integrative analyses, is found in Figure S3.
Table 2.
Results of Discovery, Replication, and Overall Meta-analyses for CpG Sites Replicated for Association with BP
| CpG site | Chr | Position | UCSC Gene |
Systolic BP |
Diastolic BP |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Discovery |
Replication |
Overall |
Discovery |
Replication |
Overall |
||||||||||
| Coeff | p Value | Coeff | p Value | Coeff | p Value | Coeff | p Value | Coeff | p Value | Coeff | p Value | ||||
| cg23999170 | 1 | 115628111 | TSPAN2 | −0.0001 | 2.7 × 10−6 | −0.0001 | 1.6 × 10−5 | −0.0001 | 1.5 × 10−10 | −0.0002 | 6.4 × 10−8 | −0.0002 | 3.4 × 10−7 | −0.0002 | 1.9 × 10−13 |
| cg16246545 | 1 | 120255941 | PHGDH | −0.0002 | 2.4 × 10−10 | −0.0002 | 3.3 × 10−14 | −0.0002 | 1.2 × 10−22 | −0.0002 | 2.2 × 10−4 | −0.0003 | 4.3 × 10−7 | −0.0002 | 1.1 × 10−9 |
| cg14476101 | 1 | 120255992 | PHGDH | −0.0003 | 1.5 × 10−16 | −0.0004 | 7.0 × 10−21 | −0.0003 | 2.7 × 10−34 | −0.0004 | 6.0 × 10−11 | −0.0005 | 1.9 × 10−12 | −0.0004 | 2.1 × 10−21 |
| cg19693031 | 1 | 145441552 | TXNIP | −0.0002 | 7.7 × 10−13 | −0.0003 | 3.8 × 10−19 | −0.0002 | 3.1 × 10−29 | −0.0002 | 6.0 × 10−7 | −0.0004 | 7.5 × 10−10 | −0.0003 | 1.8 × 10−14 |
| cg08035323 | 2 | 9843525 | – | −0.0001 | 4.2 × 10−5 | −0.0001 | 4.1 × 10−3 | −0.0001 | 9.6 × 10−7 | −0.0003 | 1.4 × 10−8 | −0.0002 | 2.6 × 10−4 | −0.0003 | 2.6 × 10−11 |
| cg06690548 | 4 | 139162808 | SLC7A11 | −0.0001 | 3.4 × 10−16 | −0.0002 | 8.3 × 10−20 | −0.0002 | 1.6 × 10−32 | −0.0002 | 5.5 × 10−14 | −0.0003 | 9.9 × 10−14 | −0.0003 | 7.9 × 10−26 |
| cg18120259 | 6 | 43894639 | LOC100132354 | −0.0001 | 1.5 × 10−8 | −0.0002 | 9.4 × 10−15 | −0.0002 | 2.2 × 10−21 | −0.0002 | 1.9 × 10−5 | −0.0003 | 6.9 × 10−10 | −0.0002 | 8.9 × 10−14 |
| cg00533891 | 10 | 80919242 | ZMIZ1 | −0.0001 | 2.4 × 10−7 | −0.0001 | 3.7 × 10−3 | −0.0001 | 5.5 × 10−9 | −0.0003 | 4.4 × 10−9 | −0.0002 | 8.9 × 10−4 | −0.0002 | 2.0 × 10−11 |
| cg17061862 | 11 | 9590431 | – | −0.0001 | 6.9 × 10−5 | −0.0002 | 6.6 × 10−9 | −0.0001 | 9.4 × 10−12 | −0.0003 | 5.1 × 10−8 | −0.0003 | 1.2 × 10−6 | −0.0003 | 4.3 × 10−13 |
| cg00574958 | 11 | 68607622 | CPT1A | −0.0001 | 1.9 × 10−8 | −4.8 × 10−5 | 1.4 × 10−6 | −0.0001 | 1.2 × 10−13 | −0.0001 | 5.9 × 10−7 | −0.0001 | 2.5 × 10−4 | −0.0001 | 3.0 × 10−10 |
| cg10601624 | 12 | 6404377 | – | −0.0001 | 6.6 × 10−8 | −0.0001 | 1.6 × 10−10 | −0.0001 | 2.4 × 10−16 | −0.0001 | 3.5 × 10−7 | −0.0002 | 1.7 × 10−7 | −0.0002 | 4.3 × 10−13 |
| cg22304262 | 19 | 47287778 | SLC1A5 | −0.0001 | 5.4 × 10−10 | −0.0001 | 8.7 × 10−9 | −0.0001 | 1.4 × 10−17 | −0.0002 | 6.0 × 10−7 | −0.0002 | 4.9 × 10−5 | −0.0002 | 9.6 × 10−11 |
| cg02711608 | 19 | 47287964 | SLC1A5 | −0.0001 | 3.0 × 10−11 | −0.0001 | 1.1 × 10−11 | −0.0001 | 2.0 × 10−21 | −0.0002 | 3.2 × 10−5 | −0.0002 | 3.0 × 10−6 | −0.0002 | 4.3 × 10−10 |
Position is Hg19. Coefficients give the percent change in DNA methylation for every 1 mmHg change in blood pressure. Abbreviations: BP, blood pressure; Chr, chromosome; Coeff, coefficient; CpG, cytosine-phosphate-guanine; UCSC, University of California Santa Cruz.
The top two CpG sites for both systolic and diastolic BP were at the PHGDH locus, cg14476101 (systolic BP: coefficient = 0.03% decrease in DNA methylation per 1 mmHg increase in BP, p = 2.7 × 10−34; diastolic BP: coefficient = 0.04% decrease in DNA methylation per 1 mmHg increase in BP, p = 2.1 × 10−21), and the SLC7A11 locus, cg06690548 (systolic BP: coefficient = 0.02% decrease in DNA methylation per 1 mmHg increase in BP, p = 1.6 × 10−32; diastolic BP: coefficient = 0.03% decrease in DNA methylation per 1 mmHg increase in BP, p = 7.9 × 10−26). cg14476101 is located on chromosome 1p12 in the first intron of PHGDH, which encodes a phosphoglycerate dehydrogenase that catalyzes the rate-limiting step of serine biosynthesis. Located on chromosome 4q28.3, cg06690548 is in the first intron of SLC7A11, which encodes a sodium-independent cysteine/glutamate antiporter. All replicated CpG sites demonstrated associations of decreased DNA methylation with increases in BP (Table S1 and Figure S4). None of the replicated CpG sites cross-hybridize with sequence variation on the sex chromosomes, and one CpG, SLC1A5 cg02711608, is polymorphic.33 An additional CpG site in SLC1A5, cg22304262, was also associated with BP and not polymorphic, so we did not exclude cg02711608 from our results. Narrow-sense heritability estimates of the 13 replicated CpG sites are moderate to high (h2 = 30%–100%) relative to all epigenome-wide probes (average h2 = 12%; Table 3). Of the 13 replicated CpG sites, 4 are in DNase I hypersensitivity sites and enhancer regions (Table S2). In PHGDH and SLC1A5, we identified two nearby CpG sites in each gene associated with BP. We regard cg14476101 as the sentinel CpG site in PHGDH and cg02711608 as the sentinel CpG site in SLC1A5 due to the strength of association p value with BP. Methylation levels at the two CpG sites in PHGDH were strongly correlated (AA and EA ρ = 0.85), whereas the two CpG sites in SLC1A5 were only modestly correlated (AA: ρ = 0.24, EA: ρ = 0.37; Figure S5). Heterogeneity (Cochran’s Q) that may be attributable to cell type or race was observed in the discovery panel for SLC7A11 cg06690548 (Table S3); however, estimates in the replication panel for this CpG site were homogeneous with the same direction of effect and similar magnitude of association p value as in the discovery meta-analyses (Table 2). All other reported CpG sites showed homogeneous effects in discovery and replication meta-analyses.
Table 3.
Narrow-Sense Heritability Estimated in the FHS for CpG Sites Replicated for Association with BP
| CpG Site | Chr | Position | Gene | CpG h2 | (95% CI) |
|---|---|---|---|---|---|
| cg23999170 | 1 | 115628111 | TSPAN2 | 0.45 | (0.39, 0.50) |
| cg16246545 | 1 | 120255941 | PHGDH | 0.47 | (0.41, 0.55) |
| cg14476101 | 1 | 120255992 | PHGDH | 0.53 | (0.43, 0.63) |
| cg19693031 | 1 | 145441552 | TXNIP | 0.55 | (0.47, 0.63) |
| cg08035323 | 2 | 9843525 | – | 0.65 | (0.57, 0.73) |
| cg06690548 | 4 | 139162808 | SLC7A11 | 0.35 | (0.27, 0.44) |
| cg18120259 | 6 | 43894639 | LOC100132354 | 0.32 | (0.26, 0.38) |
| cg00533891 | 10 | 80919242 | ZMIZ1 | 0.54 | (0.47, 0.63) |
| cg17061862 | 11 | 9590431 | – | 0.54 | (0.46, 0.62) |
| cg00574958 | 11 | 68607622 | CPT1A | 1.00 | (0.95, 1.05) |
| cg10601624 | 12 | 6404377 | – | 0.30 | (0.27, 0.34) |
| cg22304262 | 19 | 47287778 | SLC1A5 | 0.46 | (0.39, 0.52) |
| cg02711608 | 19 | 47287964 | SLC1A5 | 0.31 | (0.28, 0.35) |
Epigenome-wide average heritability is 0.12. Position is Hg19. Abbreviations: Chr, chromosome; CpG, cytosine-phosphate-guanine.
We additionally conducted an overall meta-analysis of the discovery and replication cohorts and identified 126 CpG sites associated with BP after Bonferroni correction (p < 1.0 × 10−7; Table S4). To assess the effects of antihypertensive medication use, we performed epigenome-wide meta-analyses among the 9,894 individuals reporting no concurrent use of antihypertensive medications in the discovery and replication samples. This combined sample free from antihypertensive medication use is of comparable size to the discovery meta-analysis. We did not identify a large difference in effect estimates among the discovery CpG sites that met our strict replication standards (Figure S6). Many replicated CpGs were also epigenome-wide significant in the non-medicated analysis and three CpG sites on chromosome 10p15.1 were identified that were not significant in the discovery stage (Table S5). These CpG sites map to the first intron of PFKFB3, which encodes a glycolytic enzyme.
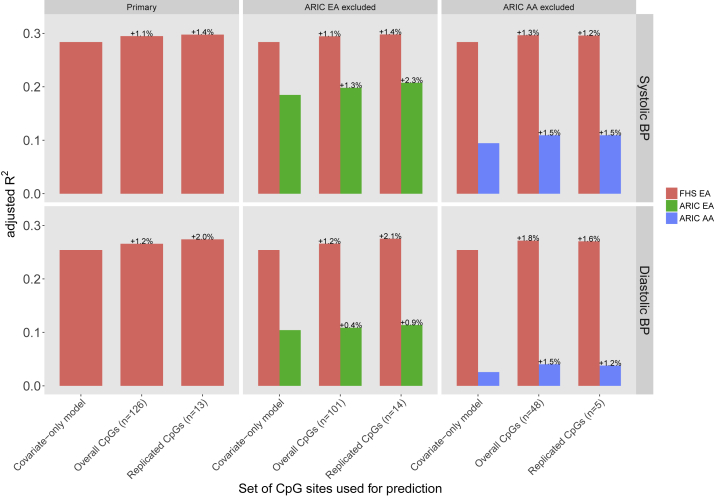
Percent Variance Explained
A methylation profile score based on the replicated CpG sites explained an additional 1.4% and 2.0% of the interindividual variation in systolic and diastolic BP, respectively, beyond the traditional BP covariates of age, sex, and BMI in an additional sample set from the FHS (N = 1,516, Third Generation Cohort) not included in the discovery or replication meta-analyses (Figure 1). Expanding the DNA methylation risk score to include the 126 CpG sites that were significant in the overall meta-analysis did not explain additional phenotypic variance in samples of either ancestry. Up to 261 BP-associated genetic variants explained minimal variance in the FHS Third Generation sample set (N = 1,421; PVE = 0.003%–0.1%). We elected to report only percent variances explained for methylation risk scores since our estimates are independent of the distally located known genetic loci.
Figure 1.
Percent Variance Explained by Traditional Covariates and Methylation Profile Scores for Systolic and Diastolic BP
The plot presents adjusted R2 values from covariate-adjusted models including a methylation profile risk score based on methylation CpG sites identified to be associated with BP in the overall and replication meta-analyses. The number of CpG sites included in the methylation profile scores is indicated as n. Percent variance explained for the CpG sites identified in the primary replication and overall meta-analyses was calculated among an independent sample from FHS. The two ARIC samples participating in the discovery and replication stages were excluded from meta-analyses used to identify CpGs for their respective methylation risk scores, which caused the sets of methylation sites to differ. Abbreviations: AA, African American; ARIC, Atherosclerosis Risk in Communities; BP, blood pressure; CpG, cytosine-phosphate-guanine; EA, European ancestry; FHS, Framingham Heart Study.
Functional Tissue and Gene Set Enrichment Analyses
Tissues enriched for DNase I hypersensitive sites in regions of the replicated CpGs include blood cells, vascular tissues, brain tissues, and cardiac tissues (Figure S7). Gene set enrichment analysis (GSEA) was conducted for intragenic CpG sites identified in the overall meta-analyses for systolic and diastolic BP. DNA methylation associated with either BP trait mapped to genes involved in the transport of neutral amino acids (FDR Q = 0.01; Figure S8). The transport of neutral amino acids was also identified as significantly enriched in the individual meta-analyses for systolic and diastolic BP (FDR Q < 0.05). 43 biological processes reached FDR Q < 0.25 including brain development, hematopoietic or lymphoid organ development, and the transport of amino acids and amines.
Methylation Quantitative Trait Loci
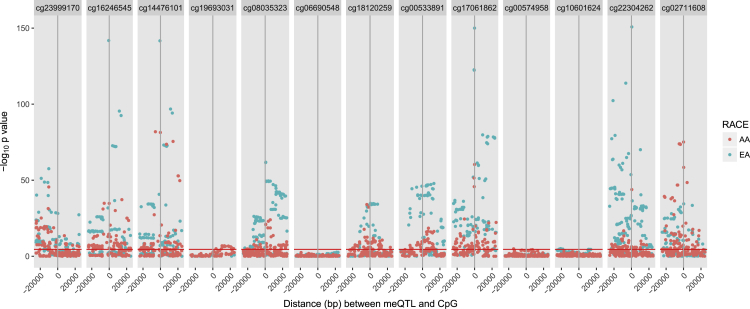
We assessed genetic determinants of DNA methylation at the 13 replicated CpG sites in 4,036 EA individuals and 2,595 AA individuals in ARIC, FHS, GENOA, and RS. Of the 13 CpG sites, 9 demonstrated substantial evidence for methylation quantitative trait loci (meQTLs) in both ancestries (EA p < 3.5 × 10−5, AA p < 2.6 × 10−5), with evidence for weak meQTLs at one additional CpG site in each ancestry (Figure 2). We confirmed our estimated EA meQTLs in an independent EA dataset published by ARIES34 and found almost all estimated meQTLs were significant or in linkage disequilibrium (r2 > 0.2 or D’ = 1) with ARIES meQTLs. We assessed the association of EA meQTLs with BP in 1000 Genomes analysis by the International Consortium for Blood Pressure (ICBP)3 that is yet to be published. Seven of the ten CpGs demonstrated nominal association with systolic or diastolic BP (0.05 > p > 1.0 × 10−3; Table S6). The strongest association with both systolic and diastolic BP was observed at rs561931 for PHGDH cg14476101 and cg16246545 (systolic p = 0.007; diastolic p = 0.01). Though phenotypic association of exposure SNPs can serve as an indication of causality, we chose to formally test causality using multi-instrument Mendelian randomization, as follows, due to the complex genetic architecture of both DNA methylation and BP.
Figure 2.
Distribution of Unpruned 1000 Genomes Imputed SNPs Assessed for Association with Methylation Relative to the CpG Location (±25 kb)
SNP position relative to the replicated methylation CpG position (X = 0) is plotted against –log10 of the p value for meQTL meta-analysis in each race. SNPs above the red line are significant after Bonferroni correction for multiple testing (p < 3.0 × 10−5). Abbreviations: AA, African American; bp, base pair; CpG, cytosine-phosphate-guanine; EA, European ancestry; meQTL, methylation quantitative trait locus; SNP, single-nucleotide polymorphism.
Bidirectional Mendelian Randomization
DNA methylation can be the cause or consequence of complex phenotypes. To provide support for causal relationships between DNA methylation and BP, we conducted bidirectional Mendelian randomization among up to 4,513 EA individuals in ARIC, FHS, RS, and WHI-EMPC. We used inverse-variance weighted tests to assess both forward causal roles of DNA methylation on BP and reverse causation where BP influences DNA methylation. For the six sentinel CpG sites with multiple genetic determinants, we were able to test forward causality using independent meQTLs as the instrumental variables. The mean causal effect estimated across its seven independent meQTLs suggests that methylation at cg08035323 (TAF1B-YWHAQ) influences BP (causal effect estimate = 20.9 [11.1] change in systolic BP, p value = 0.009, and 15.1 [6.4] change in diastolic BP, p = 0.01, per one-percent change in DNA methylation; Table 4). There is also some evidence for reverse causation at cg08035323 (diastolic BP p = 0.02); however, the causal p values for both BP traits are smaller for, and thus favor, forward causation. We performed an additional Mendelian randomization using BP effect estimates from ICBP and confirmed a causal relationship of methylation at cg08035323 with BP (systolic BP p = 0.007; Table S7).
Table 4.
Bidirectional Mendelian Randomization Results Showing the Inverse-Variance Weighted Effects of Multiple SNPs Used as Instrumental Variables in the Association of DNA Methylation and BP
| CpG | Trait |
Forward Mendelian Randomization CpG → BP |
Reverse Mendelian Randomization BP → CpG |
||||||
|---|---|---|---|---|---|---|---|---|---|
| IV SNPs, n | Mean estimate | (SE) | p Value | IV SNPs, n | Mean Estimate | (SE) | p Value | ||
| cg00533891 | SBP | 6 | −10.3 | (13.5) | – | 29 | −0.0008 | (0.0004) | 0.0388 |
| cg00533891 | DBP | 6 | −14.9 | (7.8) | 0.0405 | 29 | −0.0020 | (0.0006) | 0.0013 |
| cg00574958 | SBP | – | – | – | – | 29 | 0.0001 | (0.0001) | 0.2301 |
| cg00574958 | DBP | – | – | – | – | 29 | 0.0001 | (0.0002) | – |
| cg02711608 | SBP | 3 | −31.1 | (30.2) | 0.2953 | 29 | −0.0006 | (0.0002) | 0.0204 |
| cg02711608 | DBP | 3 | −30.2 | (17.3) | – | 29 | −0.0008 | (0.0004) | 0.0495 |
| cg06690548 | SBP | – | – | – | – | 29 | −0.0004 | (0.0003) | 0.2267 |
| cg06690548 | DBP | – | – | – | – | 29 | −0.0002 | (0.0006) | 0.7724 |
| cg08035323 | SBP | 7 | 20.9 | (11.1) | 0.0091 | 29 | −0.0004 | (0.0003) | 0.2206 |
| cg08035323 | DBP | 7 | 15.1 | (6.4) | 0.0111 | 29 | −0.0012 | (0.0006) | 0.0226 |
| cg10601624 | SBP | – | – | – | – | 29 | −0.0004 | (0.0002) | 0.1069 |
| cg10601624 | DBP | – | – | – | – | 29 | −0.0010 | (0.0003) | – |
| cg14476101 | SBP | 7 | −2.5 | (14.3) | 0.8669 | 29 | −0.0001 | (0.0005) | 0.7977 |
| cg14476101 | DBP | 7 | 1 | (5.6) | 0.8623 | 29 | −0.0002 | (0.0008) | 0.7757 |
| cg17061862 | SBP | 10 | −8.6 | (10.2) | 0.4224 | 29 | 0.0002 | (0.0004) | 0.6574 |
| cg17061862 | DBP | 10 | 4.8 | (5.1) | 0.1112 | 29 | 0.0000 | (0.0006) | 0.9844 |
| cg18120259 | SBP | – | – | – | – | 29 | 0.0001 | (0.0003) | 0.8084 |
| cg18120259 | DBP | – | – | – | – | 29 | −0.0002 | (0.0005) | 0.6968 |
| cg19693031 | SBP | – | – | – | – | 29 | 0.0003 | (0.0004) | 0.3889 |
| cg19693031 | DBP | – | – | – | – | 29 | 0.0006 | (0.0006) | 0.3509 |
| cg23999170 | SBP | 5 | 5.9 | (18.4) | 0.7547 | 29 | 0.0004 | (0.0003) | 0.1954 |
| cg23999170 | DBP | 5 | −1.2 | (10.6) | 0.9151 | 29 | 0.0003 | (0.0005) | 0.6080 |
Causal mean effect and standard error estimates for the 11 sentinel CpGs are shown and causal p values have been omitted when IVs showed significant pleiotropic effects (p < 0.05). Mean forward causal estimates are in percent DNA methylation change per mmHg increase in BP; mean reverse causal estimates are mmHg change in BP per 0.01 percent change in DNA methylation. Abbreviations: BP, blood pressure; CpG, cytosine-phosphate-guanine; DBP, diastolic blood pressure; IV, instrumental variable; Pos, position; SBP, systolic blood pressure; SE, standard error; SNP, single-nucleotide polymorphism.
We assessed reverse causation for 11 sentinel CpG sites using 29 independent GWAS loci as instrumental variables to estimate the mean causal effect of BP on DNA methylation. In the absence of pleiotropic effects, inverse-variance weighted tests suggest that DNA methylation at cg00533891 (systolic BP p = 0.04, diastolic BP p = 0.001) and SLC1A5 cg02711608 (systolic BP p = 0.02, diastolic BP p = 0.0495) is influenced by BP (Table 4). Reverse causation at both cg00533891 and SLC1A5 cg02711608 is also supported by the lower-powered Egger test for causality (Table S8). Additionally, tests for causality of the second CpG in SLC1A5, cg22304262, support reverse causality at this locus (diastolic BP p = 0.04; Table S8). The significant reverse causal effect estimates are consistent in magnitude and direction with those estimated by our EWAS. We additionally identified significant pleiotropic effects of the instrumental variables with methylation at cg10601624 and diastolic BP (p = 0.02; Table S8). Pleiotropy overpowers the inverse-variance weighted test, and we did not identify a causal effect at cg10601624 using Egger regression (p = 0.9; Table S8). There was a significant test result for forward causation at cg00533891; however, there also was evidence of pleiotropic effects among the forward instrumental variables and both the inverse-variance weighted and Egger tests favored reverse causality (Table S8). We also identified an effect of diastolic BP on DNA methylation at cg00574958 using Egger regression (p = 0.04) in the presence of pleiotropic instrumental variables (Table S8). Using Mendelian randomization, we demonstrate that complex phenotypes can have an effect on DNA methylation among top EWAS signals and that forward causality can be assessed when instrumental variables are available.
Gene Expression Associations with Replicated CpG Sites and Blood Pressure Traits
In whole-blood gene expression analyses, 4 of the 13 replicated CpG sites were found to have one or more cis-located genes (TSPAN2, SLC7A11, UNC93B1, CPT1A, PTMS, and LPCAT3) where transcription levels are associated with both CpG methylation (FHS and RS, N = 2,946) and systolic BP, diastolic BP, or hypertension (FHS, N = 3,679; Tables 5 and S9). The direction of effects for all six gene transcripts was consistent with the negative associations of BP with DNA methylation at each CpG (Tables 2 and 5). The mRNA expression of TSPAN2 showed the strongest associations with both CpG methylation and BP among all transcripts tested. Methylation at cg23999170, located in the first intron of TSPAN2, was strongly associated with decreased expression of TSPAN2 in blood (p = 8.6 × 10−14) and expression levels were associated with systolic BP (p = 5.0 × 10−29), diastolic BP (p = 1.3 × 10−16), and hypertension (p = 2.4 × 10−10).
Table 5.
Genes in a cis-Region (±1 Mb) of Replicated CpG Sites (1) Associated with DNA Methylation in Meta-analyses of FHS and RS at FDR Q Value < 0.05, and (2) Associated with BP Traits with at Least One FDR Q Value < 0.05
| CpG Site | Chr | Gene |
Gene Expression: DNA Methylation |
Gene Expression: Blood Pressure Traits |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
FHS |
RS |
Meta-Analysis |
||||||||||
| Coeff | p Value | Coeff | p Value | Z-Score | FDR Q | Trait | Coeff | t Test | FDR Q | |||
| cg23999170 | 1 | TSPAN2 | −1.38 | 2.7 × 10−14 | −1.92 | 0.0062 | −7.32 | 2.8 × 10−12 | SBP | 0.0048 | 11.36 | 5.0 × 10−29 |
| DBP | 0.0054 | 8.43 | 1.3 × 10−16 | |||||||||
| HTN | 0.1161 | 6.49 | 2.4 × 10−10 | |||||||||
| −1.38 | 2.7 × 10−14 | −2.72 | 0.0005 | −7.86 | 8.6 × 10−14 | SBP | 0.0048 | 11.36 | 5.0 × 10−29 | |||
| DBP | 0.0054 | 8.43 | 1.3 × 10−16 | |||||||||
| HTN | 0.1161 | 6.49 | 2.4 × 10−10 | |||||||||
| cg06690548 | 4 | SLC7A11 | −0.62 | 2.8 × 10−14 | NA | NA | −7.61 | 2.2 × 10−13 | SBP | 0.0003 | 1.00 | 0.3173 |
| DBP | 0.0002 | 0.32 | 0.7471 | |||||||||
| HTN | 0.0304 | 2.12 | 0.0338 | |||||||||
| cg00574958 | 11 | UNC93B1 | 0.46 | 0.1375 | 2.84 | 0.0130 | 2.81 | 0.0376 | SBP | −0.0006 | −2.52 | 0.0472 |
| DBP | −0.0008 | −2.06 | 0.0790 | |||||||||
| HTN | −0.0184 | −1.69 | 0.2399 | |||||||||
| CPT1A | −2.95 | 1.4 × 10−13 | −2.36 | 0.0003 | −7.79 | 2.1 × 10−13 | SBP | 0.0007 | 2.03 | 0.0846 | ||
| DBP | 0.0014 | 2.65 | 0.0324 | |||||||||
| HTN | 0.0225 | 1.56 | 0.2399 | |||||||||
| cg10601624 | 12 | PTMS | −0.78 | 0.0002 | −4.50 | 0.0020 | −4.83 | 2.8 × 10−5 | SBP | 0.0009 | 2.40 | 0.0807 |
| DBP | 0.0015 | 2.67 | 0.0381 | |||||||||
| HTN | 0.0170 | 1.10 | 0.8100 | |||||||||
| LPCAT3 | 0.58 | 0.0012 | 1.18 | 0.2404 | 3.13 | 0.0134 | SBP | −0.0010 | −3.40 | 0.0069 | ||
| DBP | −0.0012 | −2.73 | 0.0381 | |||||||||
| HTN | −0.0302 | −2.48 | 0.1321 | |||||||||
Abbreviations: Chr, chromosome; Coeff, coefficient; CpG, cytosine-phosphate-guanine; DBP diastolic blood pressure; FDR, false discovery rate; FHS, Framingham Heart Study; HTN, hypertension; RS, Rotterdam Study; SBP, systolic blood pressure.
We identified nominal triangular associations of gene expression levels with methylation at 11 of the replicated CpG sites (p < 0.05) and at least 1 BP trait (p < 0.05) and present estimates of association and correlation in Table S10. These genes include YWHAQ (cg08035323 and diastolic BP), PPIF (cg00533891 and diastolic BP), and GRLF1 (cg02711608/cg22304262 and diastolic BP).
Two-Step Mendelian Randomization for Genes Mediating the BP-DNA Methylation Association
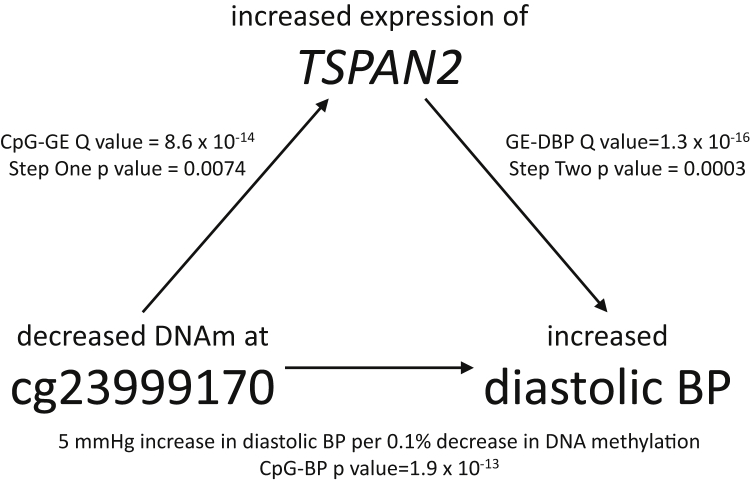
We used two-step Mendelian randomization to characterize causal mediation by gene transcripts significantly associated with methylation and BP. Using expression data available from the GTEx project and BP GWAS from ICBP, we first sought to establish a directional relationship from DNA methylation to gene expression, then a directional relationship from gene expression to BP. We showed that independent SNPs associated with methylation at cg23999170 are associated with expression of TSPAN2 and that an additional independent variant associated with TSPAN2 expression in blood is associated with BP (Table 6). The instrumental variables used for the exposures in each step achieved the associations needed to establish causality without showing strong evidence of association with the outcome that would invalidate the causal test (generally, 0.05 > p > 1 × 10−5). Using two-step Mendelian randomization, we demonstrated that TSPAN2 expression in whole blood is influenced by methylation at cg23999170 and that TSPAN2 expression affects diastolic BP (Figure 3). Taken together, the direction of causality and our epigenome-wide estimate of association suggest that diastolic BP may increase by 5 mmHg for every 0.1% decrease in DNA methylation at cg23999170.
Table 6.
Two-Step Mendelian Randomization Results for Genes with Transcription Significantly Associated with DNA Methylation and BP
| CpG site | Gene |
Step One: CpG → GE |
Step Two: GE → BP |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
IV: meQTLs |
GTEx eQTL |
IV: eQTLs |
ICBP SBP |
ICBP DBP |
|||||||
| SNP | Pos | p Value | p Value | SNP | Pos | p Value | p Value | p Value | |||
| cg23999170 | TSPAN2 | rs4240539 | 115,603,844 | 5.5 × 10−10 | 0.0074 | rs12143357 | 115,321,323 | 9.7 × 10−4 | 0.0986 | 0.0003 | |
| rs72697925 | 115,604,454 | 4.5 × 10−10 | 0.0254 | – | – | – | – | – | |||
| rs72697930 | 115,613,073 | 2.2 × 10−20 | 0.0512 | – | – | – | – | – | |||
| rs1286366 | 115,617,856 | 2.6 × 10−58 | 0.3148 | – | – | – | – | – | |||
| rs10858064 | 115,625,696 | 8.2 × 10−12 | 0.5198 | – | – | – | – | – | |||
| cg06690548 | SLC7A11 | NA | NA | NA | NA | rs17050398 | 139,350,368 | 3.6 × 10−3 | 0.0745 | 0.0852 | |
| cg00574958 | CPT1A | NA | NA | NA | NA | rs546233 | 69,311,449 | 1.7 × 10−3 | 0.6250 | 0.8880 | |
| UNC93B1 | NA | NA | NA | NA | rs1793252 | 67,740,366 | 1.3 × 10−4 | 0.2410 | 0.0747 | ||
| cg10601624 | LPCAT3 | rs984337 | 6,383,525 | 8.8 × 10−6 | 0.1149 | rs2110073 | 7,075,882 | 8.2 × 10−4 | 0.1920 | 0.2840 | |
| rs4764572 | 6,417,998 | 3.1 × 10−5 | 0.5567 | – | – | – | – | – | |||
| PTMS | rs984337 | 6,383,525 | 8.8 × 10−6 | 0.5104 | rs9668071 | 7,123,076 | 1.9 × 10−3 | 0.2590 | 0.6930 | ||
| rs4764572 | 6,417,998 | 3.1 × 10−5 | 0.5109 | – | – | – | – | – | |||
Position is Hg19. Abbreviations: BP, blood pressure; CpG, cytosine-phosphate-guanine; DBP diastolic blood pressure; eQTL, expression quantitative trait locus; GE, gene expression; GTEx, Genotype-Tissue Expression project; ICBP, International Consortium for Blood Pressure Genome-Wide Association Studies; IV, instrumental variable; meQTL, methylation quantitative trait locus; Pos, position; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
Figure 3.
Illustration of the Relationship of Methylation at cg23999170 with Diastolic BP, Mediated by Expression of TSPAN2
Methylation at cg23999170 was identified as associated with diastolic BP in discovery and replication meta-analyses of genome-wide DNA methylation (N = 17,010). Expression of TSPAN2 was associated with methylation at cg23999170 in meta-analyses of FHS and RS and diastolic BP in FHS. The direction of arrows in the diagram are inferred from significant two-step Mendelian randomization using data from the Genotype-Tissue Expression project and International Consortium for Blood Pressure, which suggests that methylation at cg23999170 influences BP through the expression of TSPAN2. The epigenome-wide association of DNA methylation and diastolic BP is interpreted given the evidence of causal direction and based on a 0.1% change in DNA methylation at cg23999170. Abbreviation: DBP, diastolic blood pressure.
Discussion
In a two-stage design of discovery and replication meta-analyses comprising 17,010 individuals, we identified DNA methylation at 13 CpG sites located in 8 intragenic regions and 3 intergenic regions significantly associated with systolic or diastolic BP. These CpGs are heritable, which suggests that DNA methylation that is the cause or consequence of BP may have a transgenerational effect. We identified substantial cis-located genetic variation associated with methylation at many of these sites in both EA and AA populations, and these regions have moderate genetic associations with BP but are independent of the top GWAS loci previously reported in either race. Through cis gene expression analyses, we identified four CpGs significantly associated with one or more genes that may functionally connect DNA methylation and BP. Mendelian randomization techniques characterized the direction of association for six CpG sites with BP, including the mediation of a causal relationship of cg23999170 with BP through expression of TSPAN2. Through the analysis of DNA methylation, we have identified genes that provide insight into the biological mechanisms underlying BP regulation and target genes possibly affected by BP-induced DNA methylation.
We identified expression of TSPAN2 to influence BP via DNA methylation. TSPAN2 encodes the tetraspanin 2 protein that is involved in signal transduction. TSPAN2 is highly expressed in vascular tissues and implicated in the contractile ability and differentiation of vascular smooth muscle cells.35 Sequence variation mapped to TSPAN2 has previously been associated with large artery atherosclerosis-related stroke36 and migraine37, 38 and TSPAN2 suppresses inflammation in the central nervous system.39 We additionally identified DNA methylation at cg08035323 to affect BP and the transcription of YWHAQ has a suggestive triangular relationship. YWHAQ encodes a 14-3-3 theta protein involved in signal transduction by binding to phosphoserine-containing proteins. YWHAQ has been implicated in phenotypes related to vascular response through transcriptional and DNA methylation changes in human preeclamptic placental tissues,40 DNA sequence variation associated with exercise heart rate response,41 and an effect on cardiomyocyte survival in animal models.42
We identified an effect of BP on DNA methylation at 4 of the 13 replicated CpG sites: ZMIZ1 cg00533891, CPT1A cg00574958, and SLC5A1 cg02711608/cg22304262. Previous epigenome-wide association studies have identified relationships of CPT1A cg00574958 and SLC5A1 cg02711608/cg22304262 with other metabolic phenotypes, particularly lipids and adiposity (Table S11). An effect of triglycerides on methylation at cg00574958 has previously been identified,43 which supports our hypothesis that an underlying cardiometabolic disease process related to BP and lipids alters DNA methylation within CPT1A. Transcriptional changes caused by DNA methylation at these four CpG sites could affect the risk of downstream phenotypes.
A recent GWAS identified 28 of 35 sentinel SNPs to have methylation-mediated associations with BP using a Mendelian randomization technique.23 In our overall meta-analyses, we assessed the association of the 28 CpG sites reported by Kato et al.,23 but were not able to confirm the direct association of any of these CpG sites with BP (p > 1.0 × 10−5; Table S12). We further could not confirm the association of the sentinel SNPs with the CpG sites reported to mediate the associations with BP. However, we identified an association of rs12567136 (CLCN6) with methylation at an upstream CpG, cg20946054 (DRAXIN). This CpG site has a nominal association with BP (systolic BP p = 0.0318, diastolic BP p = 0.0015) and adjustment for methylation at this locus attenuated the association of rs12567136 with systolic BP but not diastolic BP (Table S13). Diastolic BP was the primary phenotype identified by Kato et al. Differences between our findings and those reported by Kato et al.23 may be due to differing linkage disequilibrium structure in the populations examined in the two studies.
The strength of this study is our use of multiple data sources and analytic techniques to characterize functional relationships of DNA methylation and BP. We used strict replication standards to identify 13 CpG sites associated with BP and characterized gene transcriptional changes that could underlie these associations. However, these CpG sites were identified through cross-sectional analyses and we were able to characterize the direction of association only for six CpG sites. Mendelian randomization requires exposure-predictive SNPs to be selected as instrumental variables. For three CpG sites, we estimated no significant cis-meQTLs (cg19693031, cg06690548, and cg00574958) and an additional two CpG sites had insufficient independent meQTLs to assess forward causation using multiple instrumental variables (cg18120259 and cg10601624), so we lacked the ability to perform causal tests for select CpG sites. We additionally a priori chose to examine gene expression 1 Mb up- and downstream of each CpG site in whole blood, so more distal regulatory effects, and effects in different tissues, could have been missed. However, we did identify at least one biologically plausible gene to have a nominal triangular association with both methylation and BP for 11 of the replicated CpG sites, so it may be that larger sample sizes will uncover the functional and causal relationships of DNA methylation and BP. Despite these limitations, we discovered heritable CpG sites associated with BP among 17,010 individuals and showed that these CpGs explain additional phenotypic variance in an independent sample in which up to 261 BP-associated genetic variants explained minimal trait variance. We characterized the methylation-BP relationship using gene expression analyses and Mendelian randomization techniques to understand both which genes may regulate BP and how BP may affect gene transcription.
In conclusion, our genome-wide analysis of DNA methylation has uncovered loci influencing BP variation independently of underlying genetic variation. We additionally characterized functional and causal relationships connecting methylation at these loci with BP. In particular, we have identified TSPAN2 as a candidate gene for BP that is regulated by heritable DNA methylation. TSPAN2 and other methylated regions point to vascular contractility and inflammatory processes that functionally and causally connect DNA methylation and BP, and, thus, may represent new targets for the treatment and prevention of hypertension. Additional studies are needed to provide further mechanistic insights into the environmental exposures and genetic variation that influence DNA methylation and lead to high blood pressure. Nonetheless, our findings suggest that information on DNA methylation is likely to yield additional insights into the pathobiology of complex traits.
Consortia
BIOS consortium members are as follows: Bastiaan T. Heijmans, Peter A.C. ’t Hoen, Joyce van Meurs, Aaron Isaacs, Rick Jansen, Lude Franke, Dorret I. Boomsma, René Pool, Jenny van Dongen, Jouke J. Hottenga, Marleen M.J. van Greevenbroek, Coen D.A. Stehouwer, Carla J.H. van der Kallen, Casper G. Schalkwijk, Cisca Wijmenga, Alexandra Zhernakova, Ettje F. Tigchelaar, P. Eline Slagboom, Marian Beekman, Joris Deelen, Diana van Heemst, Jan H. Veldink, Leonard H. van den Berg, Cornelia M. van Duijn, Albert Hofman, André G. Uitterlinden, P. Mila Jhamai, Michael Verbiest, H. Eka D. Suchiman, Marijn Verkerk, Ruud van der Breggen, Jeroen van Rooij, Nico Lakenberg, Hailiang Mei, Maarten van Iterson, Michiel van Galen, Jan Bot, Peter van ’t Hof, Patrick Deelen, Irene Nooren, Matthijs Moed, Martijn Vermaat, Dasha V. Zhernakova, René Luijk, Marc Jan Bonder, Freerk van Dijk, Wibowo Arindrarto, Szymon M. Kielbasa, Morris A. Swertz, and Erik W. van Zwet.
Acknowledgments
We thank all participants for contributing to this study. See Supplemental Data for detailed study acknowledgments and funding.
Published: November 30, 2017
Footnotes
Supplemental Data include 8 figures, 13 tables, and Supplemental Material and Methods, including funding acknowledgments and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.09.028.
Contributor Information
Melissa A. Richard, Email: melissa.a.lee@uth.tmc.edu.
Myriam Fornage, Email: myriam.fornage@uth.tmc.edu.
BIOS Consortium:
Bastiaan T. Heijmans, Peter A.C. ’t Hoen, Joyce van Meurs, Aaron Isaacs, Rick Jansen, Lude Franke, Dorret I. Boomsma, René Pool, Jenny van Dongen, Jouke J. Hottenga, Marleen M.J. van Greevenbroek, Coen D.A. Stehouwer, Carla J.H. van der Kallen, Casper G. Schalkwijk, Cisca Wijmenga, Alexandra Zhernakova, Ettje F. Tigchelaar, P. Eline Slagboom, Marian Beekman, Joris Deelen, Diana van Heemst, Jan H. Veldink, Leonard H. van den Berg, Cornelia M. van Duijn, Albert Hofman, André G. Uitterlinden, P. Mila Jhamai, Michael Verbiest, H. Eka D. Suchiman, Marijn Verkerk, Ruud van der Breggen, Jeroen van Rooij, Nico Lakenberg, Hailiang Mei, Maarten van Iterson, Michiel van Galen, Jan Bot, Peter van ’t Hof, Patrick Deelen, Irene Nooren, Matthijs Moed, Martijn Vermaat, Dasha V. Zhernakova, René Luijk, Marc Jan Bonder, Freerk van Dijk, Wibowo Arindrarto, Szymon M. Kielbasa, Morris A. Swertz, and Erik W. van Zwet
Supplemental Data
Position is Hg19. Coefficients give the percent change in DNA methylation for every 1-unit change in blood pressure. Abbreviations: BP, blood pressure; Chr, chromosome; Coeff, coefficient; CpG, cytosine-phosphate-guanine; UCSC, University of California Santa Cruz.
Start and stop positions are in Hg19. Abbreviations: Chr, chromosome; Coeff, coefficient; Corr, correlation coefficient for continuous traits; CpG, cytosine-phosphate-guanine; DBP, diastolic blood pressure; HTN, hypertension; SBP, systolic blood pressure; UCSC, University of California Santa Cruz.
References
- 1.Campbell N.R., Khalsa T., Lackland D.T., Niebylski M.L., Nilsson P.M., Redburn K.A., Orias M., Zhang X.H., Burrell L., Horiuchi M., World Hypertension League Executive. International Society of Hypertension Executive. World Stroke Organization. International Diabetes Federation. International Council of Cardiovascular Prevention and Rehabilitation. International Society of Nephrology High blood pressure 2016: why prevention and control are urgent and important. The World Hypertension League, International Society of Hypertension, World Stroke Organization, International Diabetes Foundation, International Council of Cardiovascular Prevention and Rehabilitation, International Society of Nephrology. J. Clin. Hypertens. (Greenwich) 2016;18:714–717. doi: 10.1111/jch.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy D., DeStefano A.L., Larson M.G., O’Donnell C.J., Lifton R.P., Gavras H., Cupples L.A., Myers R.H. Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the framingham heart study. Hypertension. 2000;36:477–483. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- 3.Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Hwang S.-J., International Consortium for Blood Pressure Genome-Wide Association Studies. CARDIoGRAM consortium. CKDGen Consortium. KidneyGen Consortium. EchoGen consortium. CHARGE-HF consortium Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy D., Ehret G.B., Rice K., Verwoert G.C., Launer L.J., Dehghan A., Glazer N.L., Morrison A.C., Johnson A.D., Aspelund T. Genome-wide association study of blood pressure and hypertension. Nat. Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newton-Cheh C., Johnson T., Gateva V., Tobin M.D., Bochud M., Coin L., Najjar S.S., Zhao J.H., Heath S.C., Eyheramendy S., Wellcome Trust Case Control Consortium Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franceschini N., Fox E., Zhang Z., Edwards T.L., Nalls M.A., Sung Y.J., Tayo B.O., Sun Y.V., Gottesman O., Adeyemo A., Asian Genetic Epidemiology Network Consortium Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am. J. Hum. Genet. 2013;93:545–554. doi: 10.1016/j.ajhg.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox E.R., Young J.H., Li Y., Dreisbach A.W., Keating B.J., Musani S.K., Liu K., Morrison A.C., Ganesh S., Kutlar A., International Consortium for Blood Pressure Genome-wide Association Studies (ICBP-GWAS) Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum. Mol. Genet. 2011;20:2273–2284. doi: 10.1093/hmg/ddr092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehret G.B., Ferreira T., Chasman D.I., Jackson A.U., Schmidt E.M., Johnson T., Thorleifsson G., Luan J., Donnelly L.A., Kanoni S., CHARGE-EchoGen consortium. CHARGE-HF consortium. Wellcome Trust Case Control Consortium The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat. Genet. 2016;48:1171–1184. doi: 10.1038/ng.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Surendran P., Drenos F., Young R., Warren H., Cook J.P., Manning A.K., Grarup N., Sim X., Barnes D.R., Witkowska K., CHARGE-Heart Failure Consortium. EchoGen Consortium. METASTROKE Consortium. GIANT Consortium. EPIC-InterAct Consortium. Lifelines Cohort Study. Wellcome Trust Case Control Consortium. Understanding Society Scientific Group. EPIC-CVD Consortium. CHARGE+ Exome Chip Blood Pressure Consortium. T2D-GENES Consortium. GoT2DGenes Consortium. ExomeBP Consortium. CHD Exome+ Consortium Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat. Genet. 2016;48:1151–1161. doi: 10.1038/ng.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C., Kraja A.T., Smith J.A., Brody J.A., Franceschini N., Bis J.C., Rice K., Morrison A.C., Lu Y., Weiss S., CHD Exome+ Consortium. ExomeBP Consortium. GoT2DGenes Consortium. T2D-GENES Consortium. Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia. CKDGen Consortium Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat. Genet. 2016;48:1162–1170. doi: 10.1038/ng.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren H.R., Evangelou E., Cabrera C.P., Gao H., Ren M., Mifsud B., Ntalla I., Surendran P., Liu C., Cook J.P., International Consortium of Blood Pressure (ICBP) 1000G Analyses. BIOS Consortium. Lifelines Cohort Study. Understanding Society Scientific group. CHD Exome+ Consortium. ExomeBP Consortium. T2D-GENES Consortium. GoT2DGenes Consortium. Cohorts for Heart and Ageing Research in Genome Epidemiology (CHARGE) BP Exome Consortium. International Genomics of Blood Pressure (iGEN-BP) Consortium. UK Biobank CardioMetabolic Consortium BP working group Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat. Genet. 2017;49:403–415. doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niiranen T.J., Havulinna A.S., Langén V.L., Salomaa V., Jula A.M. Prediction of blood pressure and blood pressure change with a genetic risk score. J. Clin. Hypertens. (Greenwich) 2016;18:181–186. doi: 10.1111/jch.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowley A.W., Jr., Nadeau J.H., Baccarelli A., Berecek K., Fornage M., Gibbons G.H., Harrison D.G., Liang M., Nathanielsz P.W., O’Connor D.T. Report of the national heart, lung, and blood institute working group on epigenetics and hypertension. Hypertension. 2012;59:899–905. doi: 10.1161/HYPERTENSIONAHA.111.190116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Psaty B.M., O’Donnell C.J., Gudnason V., Lunetta K.L., Folsom A.R., Rotter J.I., Uitterlinden A.G., Harris T.B., Witteman J.C.M., Boerwinkle E., CHARGE Consortium Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teschendorff A.E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pidsley R., Y Wong C.C., Volta M., Lunnon K., Mill J., Schalkwyk L.C. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 18.Maksimovic J., Gordon L., Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mägi R., Morris A.P. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breeze C.E., Paul D.S., van Dongen J., Butcher L.M., Ambrose J.C., Barrett J.E., Lowe R., Rakyan V.K., Iotchkova V., Frontini M. eFORGE: a tool for identifying cell type-specific signal in epigenomic data. Cell Rep. 2016;17:2137–2150. doi: 10.1016/j.celrep.2016.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato N., Loh M., Takeuchi F., Verweij N., Wang X., Zhang W., Kelly T.N., Saleheen D., Lehne B., Leach I.M., BIOS-consortium. CARDIo GRAMplusCD. LifeLines Cohort Study. InterAct Consortium Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat. Genet. 2015;47:1282–1293. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Y., Tomlinson B., Chu T., Fang Y.J., Gui H., Tang C.S., Yip B.H., Cherny S.S., Hur Y.-M., Sham P.C. A genome-wide linkage and association scan reveals novel loci for hypertension and blood pressure traits. PLoS ONE. 2012;7:e31489. doi: 10.1371/journal.pone.0031489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato N., Takeuchi F., Tabara Y., Kelly T.N., Go M.J., Sim X., Tay W.T., Chen C.-H., Zhang Y., Yamamoto K. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat. Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy D., Larson M.G., Benjamin E.J., Newton-Cheh C., Wang T.J., Hwang S.-J., Vasan R.S., Mitchell G.F. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med. Genet. 2007;8(Suppl 1):S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Org E., Eyheramendy S., Juhanson P., Gieger C., Lichtner P., Klopp N., Veldre G., Döring A., Viigimaa M., Sõber S., KORA. BRIGHT Genome-wide scan identifies CDH13 as a novel susceptibility locus contributing to blood pressure determination in two European populations. Hum. Mol. Genet. 2009;18:2288–2296. doi: 10.1093/hmg/ddp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padmanabhan S., Melander O., Johnson T., Di Blasio A.M., Lee W.K., Gentilini D., Hastie C.E., Menni C., Monti M.C., Delles C., Global BPgen Consortium Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010;6:e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wain L.V., Verwoert G.C., O’Reilly P.F., Shi G., Johnson T., Johnson A.D., Bochud M., Rice K.M., Henneman P., Smith A.V., LifeLines Cohort Study. EchoGen consortium. AortaGen Consortium. CHARGE Consortium Heart Failure Working Group. KidneyGen consortium. CKDGen consortium. Cardiogenics consortium. CardioGram Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat. Genet. 2011;43:1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huan T., Meng Q., Saleh M.A., Norlander A.E., Joehanes R., Zhu J., Chen B.H., Zhang B., Johnson A.D., Ying S., International Consortium for Blood Pressure GWAS (ICBP) Integrative network analysis reveals molecular mechanisms of blood pressure regulation. Mol. Syst. Biol. 2015;11:799. doi: 10.15252/msb.20145399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y.A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaunt T.R., Shihab H.A., Hemani G., Min J.L., Woodward G., Lyttleton O., Zheng J., Duggirala A., McArdle W.L., Ho K. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016;17:61. doi: 10.1186/s13059-016-0926-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J., Wu W., Zhang W., Lu Y.W., Tou E., Ye J., Gao P., Jourd’heuil D., Singer H.A., Wu M., Long X. Selective expression of TSPAN2 in vascular smooth muscle is independently regulated by TGF-β1/SMAD and myocardin/serum response factor. FASEB J. 2017;31:2576–2591. doi: 10.1096/fj.201601021R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.NINDS Stroke Genetics Network (SiGN) International Stroke Genetics Consortium (ISGC) Loci associated with ischaemic stroke and its subtypes (SiGN): a genome-wide association study. Lancet Neurol. 2016;15:174–184. doi: 10.1016/S1474-4422(15)00338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esserlind A.-L., Christensen A.F., Le H., Kirchmann M., Hauge A.W., Toyserkani N.M., Hansen T., Grarup N., Werge T., Steinberg S. Replication and meta-analysis of common variants identifies a genome-wide significant locus in migraine. Eur. J. Neurol. 2013;20:765–772. doi: 10.1111/ene.12055. [DOI] [PubMed] [Google Scholar]
- 38.Anttila V., Winsvold B.S., Gormley P., Kurth T., Bettella F., McMahon G., Kallela M., Malik R., de Vries B., Terwindt G., North American Brain Expression Consortium. UK Brain Expression Consortium Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat. Genet. 2013;45:912–917. doi: 10.1038/ng.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Monasterio-Schrader P., Patzig J., Möbius W., Barrette B., Wagner T.L., Kusch K., Edgar J.M., Brophy P.J., Werner H.B. Uncoupling of neuroinflammation from axonal degeneration in mice lacking the myelin protein tetraspanin-2. Glia. 2013;61:1832–1847. doi: 10.1002/glia.22561. [DOI] [PubMed] [Google Scholar]
- 40.Liu H., Tang Y., Liu X., Zhou Q., Xiao X., Lan F., Li X., Hu R., Xiong Y., Peng T. 14-3-3 tau (YWHAQ) gene promoter hypermethylation in human placenta of preeclampsia. Placenta. 2014;35:981–988. doi: 10.1016/j.placenta.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Rankinen T., Sung Y.J., Sarzynski M.A., Rice T.K., Rao D.C., Bouchard C. Heritability of submaximal exercise heart rate response to exercise training is accounted for by nine SNPs. J. Appl. Physiol. 2012;112:892–897. doi: 10.1152/japplphysiol.01287.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau J.M.C., Jin X., Ren J., Avery J., DeBosch B.J., Treskov I., Lupu T.S., Kovacs A., Weinheimer C., Muslin A.J. The 14-3-3tau phosphoserine-binding protein is required for cardiomyocyte survival. Mol. Cell. Biol. 2007;27:1455–1466. doi: 10.1128/MCB.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dekkers K.F., van Iterson M., Slieker R.C., Moed M.H., Bonder M.J., van Galen M., Mei H., Zhernakova D.V., van den Berg L.H., Deelen J., BIOS Consortium Blood lipids influence DNA methylation in circulating cells. Genome Biol. 2016;17:138. doi: 10.1186/s13059-016-1000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Position is Hg19. Coefficients give the percent change in DNA methylation for every 1-unit change in blood pressure. Abbreviations: BP, blood pressure; Chr, chromosome; Coeff, coefficient; CpG, cytosine-phosphate-guanine; UCSC, University of California Santa Cruz.
Start and stop positions are in Hg19. Abbreviations: Chr, chromosome; Coeff, coefficient; Corr, correlation coefficient for continuous traits; CpG, cytosine-phosphate-guanine; DBP, diastolic blood pressure; HTN, hypertension; SBP, systolic blood pressure; UCSC, University of California Santa Cruz.