ABSTRACT
Purple prairie clover (PPC; Dalea purpurea Vent.) containing 84.5 g/kg dry matter (DM) of condensed tannin (CT) was ensiled without (control) or with polyethylene glycol (PEG) for 76 days, followed by 14 days of aerobic exposure. Changes in fermentation characteristics were determined, and the composition of bacterial and fungal communities were assessed using metagenomic sequencing. The addition of polyethylene glycol (PEG) that deactivated CT at ensiling increased (P < 0.05 to ∼0.001) soluble N, nonprotein N, lactic acid, total volatile fatty acids, ammonia N, deoxynivalenol (DON), and ochratoxin A (OTA) but decreased (P < 0.001) pH and water-soluble carbohydrates. The concentrations of DON and OTA increased (P < 0.001) for both silages, with the extent of increase being greater for control than for PEG-treated silage during aerobic exposure. The PEG-treated silage exhibited higher (P < 0.01 to ∼0.001) copy numbers of total bacteria, Lactobacillus, yeasts, and fungi than the control. The addition of PEG decreased (P < 0.01) bacterial diversity during both ensiling and aerobic exposure, whereas it increased (P < 0.05) fungal diversity during aerobic exposure. The addition of PEG at ensiling increased (P < 0.05) the abundances of Lactobacillus and Pediococcus species but decreased (P < 0.01) the abundances of Lactococcus and Leuconostoc species. Filamentous fungi were found in the microbiome at ensiling and after aerobic exposure, whereas Bacillus spp. were the dominate bacteria after aerobic exposure. In conclusion, CT decreased protein degradation and improved the aerobic stability of silage. These desirable outcomes likely reflect the ability of PPC CT to inhibit those microorganisms involved in lowering silage quality and in the production of mycotoxins.
IMPORTANCE The present study reports the effects of condensed tannins on the complex microbial communities involved in ensiling and aerobic exposure of purple prairie clover. This study documents the ability of condensed tannins to lower mycotoxin production and the associated microbiome. Taxonomic bacterial community profiles were dominated by Lactobacillales after fermentation, with a notable increase in Bacillus spp. as a result of aerobic exposure. It is interesting to observe that condensed tannins decreased bacterial diversity during both ensiling and aerobic exposure but increased fungal diversity during aerobic exposure only. The present study indicates that the effects of condensed tannins on microbial communities lead to reduced lactic acid and total volatile fatty acid production, proteolysis, and mycotoxin concentration in the terminal silage and improved aerobic stability. Condensed tannins could be used as an additive to control unfavorable microbial development and maybe enhanced feed safety.
KEYWORDS: bacteria, condensed tannins, fungi, purple prairie clover, silage
INTRODUCTION
Ensiling is a worldwide conservation method for green fodder because it allows high-quality forage to be conserved even under less-than-favorable weather conditions. However, conserving forage as silage has its challenges, as the quality of silage is highly dependent on the activities and types of microorganisms that contribute to the ensiling process. Therefore, understanding the microbial community involved in ensiling is central to ensuring the effective conservation of silage. Microbes involved in ensiling have been extensively studied using both culture-dependent methods and culture-independent molecular methods (1, 2). During ensiling, bacteria, such as Lactobacillus and Lactococcus strains, are known to enter into the viable-but-unculturable state (3). Hence, microbial communities being described solely on the basis of culture overlooks the contribution that these bacteria may make to the ensiling process. The recent development of high-throughput sequencing of phylogenetic gene markers for bacteria, yeasts, and fungi makes it possible to identify unculturable members of microbial communities that are associated with ensiling. Although such studies have been reported for grass and whole-crop barley silage (2, 4), they have not been undertaken for legume forages containing condensed tannins (CT). Extensive proteolysis occurs during ensiling that converts almost all plant protein to nonprotein N (NPN), thus lowering the protein nutritive value of silage (1, 5). Therefore, methods that reduce protein degradation during ensiling could improve the nutritional value of silage.
Condensed tannins are a group of naturally occurring phenolic plant secondary compounds that possess various biological activities, including protein precipitation and antimicrobial activity (1, 6). Both concentration and structure affect the biological activity of CT. It has been found that CT in temperate forage at lower than 40 g/kg dry matter (DM) generally have positive effects on ruminant performance, mainly due to the improved protein utilization via reducing extensive protein degradation in the rumen and increasing amino acid absorption in the small intestine (7–9). In contrast, CT concentrations higher than 50 g/kg DM usually depressed feed intake and reduced nutrient, especially protein, digestion (10, 11). It has also been reported that the addition of tannins decreased protein degradation during ensiling (12–14) and that ensiling CT-containing species, such as sainfoin (Onobrychis viciifolia cv. Common) and birdsfoot trefoil (Lotus corniculatus), reduce the degree of proteolysis compared to legumes that do not contain CT (1). However, the mechanisms whereby CT alter the silage microbiome to reduce proteolysis and improve silage quality during ensiling are not fully understood.
The objective of this study was to assess the effects of CT on fermentation and the bacterial and fungal microbiomes during ensiling and upon aerobic exposure.
RESULTS
Characteristics of terminal and aerobically exposed silage.
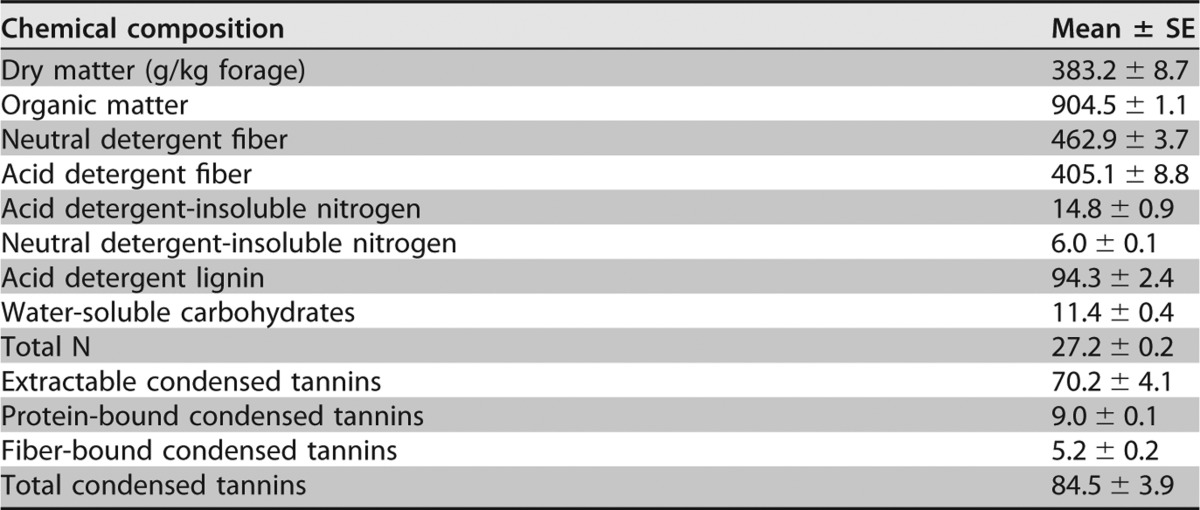
Whole-plant purple prairie clover (PPC; Dalea purpurea Vent.) contained 84.5 g CT/kg DM at ensiling, the majority of which was extractable CT (70.2 g/kg DM; Table 1). After 76-day ensiling, extractable CT decreased to about 27.4 g/kg silage DM (Table 2). The addition of polyethylene glycol (PEG) at ensiling increased (P < 0.05 to ∼0.001) soluble N (SN), NPN, lactic acid, total volatile fatty acids (VFA), ammonia N, acetic acid, and propionic acid but decreased (P < 0.001) the contents of acid detergent-insoluble nitrogen (ADIN) and water-soluble carbohydrates (WSC). Silage ensiled with PEG also had higher (P < 0.01) concentrations of deoxynivalenol (DON) and ochratoxin A (OTA) than with the control silage.
TABLE 1.
Chemical compositions of fresh whole-plant purple prairie clover harvested at full flower stage for silagea
n = 3. Composition measures are in grams per kilogram of DM, unless otherwise stated.
TABLE 2.
Chemical composition, fermentation products, and mycotoxin concentration of purple prairie clover ensiled without or with polyethylene glycol in terminal silage and after 7 and 14 days of aerobic exposure
| Item | Terminal silage (n = 5) |
Aerobic exposure silage (n = 4) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 0 |
SEM | P | Day 7 |
SEM | P | Day 14 |
SEM | P | ||||
| Control | PPC-p | Control | PPC-p | Control | PPC-p | |||||||
| Chemical composition (g/kg DM) | ||||||||||||
| Dry matter (g/kg forage) | 305.94 | 311.76 | 4.77 | 0.375 | 387.51 | 388.96 | 5.10 | 0.848 | 376.59 | 389.44 | 3.80 | 0.054 |
| Organic matter | 911.34 | 910.25 | 1.40 | 0.599 | 912.56 | 913.64 | 0.70 | 0.383 | 902.89 | 901.93 | 2.94 | 0.826 |
| Neutral detergent fiber | 437.43 | 428.28 | 13.55 | 0.637 | 437.11 | 438.40 | 4.65 | 0.850 | 527.00 | 509.77 | 20.08 | 0.545 |
| Acid detergent fiber | 357.21 | 366.93 | 8.27 | 0.430 | 369.04 | 380.78 | 5.76 | 0.199 | 442.74 | 420.33 | 13.06 | 0.292 |
| Acid detergent-insoluble nitrogen | 9.03 | 7.76 | 0.13 | <0.001 | 8.85 | 6.32 | 0.12 | <0.001 | 11.26 | 10.26 | 1.36 | 0.658 |
| Neutral detergent-insoluble nitrogen | 7.45 | 8.21 | 0.44 | 0.270 | 11.92 | 10.30 | 0.60 | 0.095 | 13.06 | 13.32 | 1.06 | 0.878 |
| Acid detergent lignin | 70.56 | 82.09 | 9.17 | 0.400 | 103.07 | 98.62 | 6.29 | 0.622 | 148.78 | 132.49 | 8.60 | 0.252 |
| Water-soluble carbohydrates | 2.40 | 0.30 | 0.12 | <0.001 | 0.69 | 0.47 | 0.06 | 0.034 | 0.51 | 0.42 | 0.09 | 0.482 |
| Total N | 28.16 | 28.24 | 0.06 | 0.457 | 28.62 | 28.32 | 0.18 | 0.363 | 27.82 | 28.45 | 0.83 | 0.569 |
| Soluble N | 0.29 | 0.45 | 0.02 | <0.001 | 0.26 | 0.45 | 0.02 | <0.001 | 0.22 | 0.42 | 0.02 | <0.001 |
| Nonprotein N | 0.35 | 0.50 | 0.02 | <0.001 | 0.24 | 0.38 | 0.01 | <0.001 | 0.16 | 0.35 | 0.02 | <0.001 |
| Extractable condensed tannins | 27.4 ± 1.5a | NDb | 4.0 ± 0.5a | ND | ND | ND | ||||||
| Fermentation products (g/kg DM) | ||||||||||||
| pH | 5.02 | 4.61 | 0.01 | <0.001 | 4.98 | 4.66 | 0.02 | <0.001 | 8.81 | 8.96 | 0.18 | 0.586 |
| Lactic acid | 40.99 | 91.04 | 5.36 | <0.001 | 27.13 | 63.48 | 2.57 | <0.001 | 1.92 | 3.14 | 0.40 | 0.161 |
| Total VFA | 15.59 | 26.84 | 1.70 | 0.002 | 22.38 | 36.12 | 2.55 | 0.009 | 1.85 | 1.60 | 0.46 | 0.730 |
| Acetic acid | 14.82 | 25.38 | 2.68 | 0.022 | 21.78 | 33.93 | 2.58 | 0.016 | 1.14 | 0.88 | 0.32 | 0.635 |
| Propionic acid | 0.17 | 0.97 | 0.06 | <0.001 | 0.17 | 1.59 | 0.12 | <0.001 | 0.05 | 0.10 | 0.02 | 0.184 |
| Butyric acid | 0.38 | 0.36 | 0.01 | 0.408 | 0.33 | 0.41 | 0.05 | 0.291 | 0.29 | 0.34 | 0.01 | 0.087 |
| Minor VFAc | 0.28 | 0.17 | 0.04 | 0.101 | 0.12 | 0.16 | 0.01 | 0.087 | 0.26 | 0.30 | 0.04 | 0.539 |
| Succinic acid | 5.54 | 2.60 | 0.39 | <0.001 | 3.72 | 1.84 | 0.32 | 0.006 | 1.20 | 0.66 | 0.48 | 0.453 |
| Lactic acid:acetic acid ratio | 2.49 | 3.62 | 0.18 | 0.003 | 1.28 | 1.80 | 0.17 | 0.067 | 1.09 | 3.60 | 0.20 | 0.001 |
| Ammonia N | 2.70 | 3.74 | 0.14 | <0.001 | 2.10 | 3.26 | 0.18 | 0.004 | 2.13 | 4.22 | 0.25 | 0.001 |
| Mycotoxin concn (μg/kg DM) | ||||||||||||
| Deoxynivalenol | 108.6 | 190.9 | 3.93 | 0.004 | 150.2 | 186.4 | 2.31 | 0.008 | 474.7 | 679.4 | 10.92 | 0.006 |
| Ochratoxin A | 12.0 | 44.0 | 5.29 | 0.003 | 13.67 | 39.51 | 3.34 | 0.009 | 25.60 | 71.64 | 1.68 | 0.003 |
Values = mean ± standard error.
ND, not detectable.
Minor VFA is isobutyric acid + valeric acid + isovaleric acid + caproic acid.
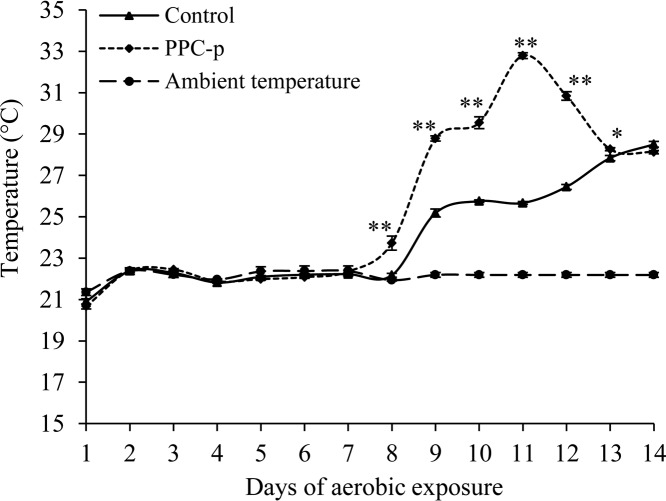
The temperature of aerobically exposed PEG-treated silage started to increase on day 7, whereas that of the control silage did not increase until on day 8 (Fig. 1). The magnitude of the temperature increase was also greater (P < 0.01) for PEG-treated (0 to ∼11°C) than for control (0 to ∼6°C) silages. The pH after 7-day aerobic exposure was similar to day 0 silage but increased (P < 0.001) after 14 days of aerobic exposure (Table 2).
FIG 1.
Temperature of purple prairie clover ensiled without (control) or with polyethylene glycol (PPC-p) in terminal silage (day 0) and after 7 and 14 days of aerobic exposure. *, P < 0.05; **, P < 0.01.
The increases in temperature and pH of silage during aerobic exposure were accompanied by a decrease in lactic acid concentration (Table 2). After 7 days of aerobic exposure, PEG-treated silage had higher (P < 0.05 to ∼0.001) concentrations of SN, NPN, ammonia N, total VFA, lactic acid, acetic acid, and propionic acid than control silage. However, this trend was observed for SN, NPN, and ammonia N after 14 days of aerobic exposure. On the contrary, the concentrations of DON and OTA increased (P < 0.001) during aerobic exposure, with concentrations of DON and OTA being greater (P < 0.01) in PEG-treated silage than in control silage.
Microbial rRNA copy numbers in terminal and aerobically exposed silage.
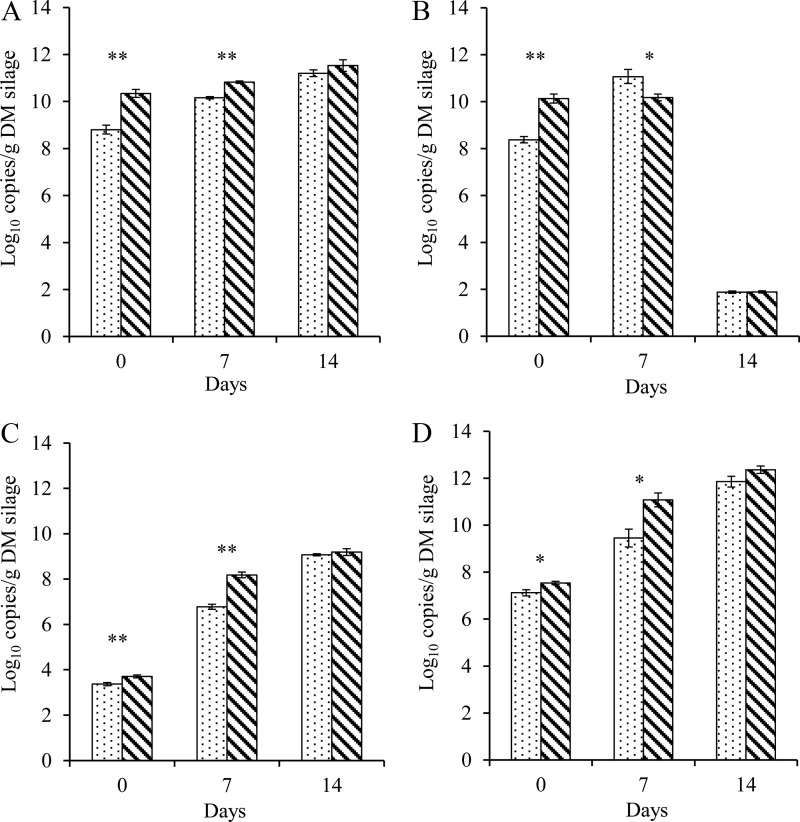
After a 76-day ensiling, PEG-treated silage exhibited higher (P < 0.05 to ∼0.01) rRNA copy numbers of total bacteria, Lactobacillus spp., yeasts, and fungi than the control silage (Fig. 2). The rRNA copies associated with the majority of microbes increased (P < 0.001) during aerobic exposure, with the exception of copy numbers of Lactobacillus, which declined (P < 0.001) from day 7 to day 14 of aerobic exposure (Fig. 2B). The PEG-treated silage had higher (P < 0.05 to ∼0.001) copy numbers of total bacteria, yeasts, and fungi but lower (P < 0.05) copy numbers of Lactobacillus spp. than the control silage. The two silages had similar copy numbers of total bacteria, Lactobacillus spp., yeasts, and fungi after 14 days of aerobic exposure.
FIG 2.
Copy numbers of 16S rRNA gene of total bacteria (A) and Lactobacillus (B), 26S rRNA gene of yeasts (C), and 18S rRNA gene of fungi (D). Purple prairie clover was ensiled without (control) or with polyethylene glycol (PPC-p) in terminal silage (day 0) and after 7 and 14 days of aerobic exposure. *, P < 0.05; **, P < 0.01.
Bacterial and fungal microbiome in terminal and aerobically exposed silage.
Across all samples, a total of 6,295,205 and 5,033,203 reads with average lengths of 450 and 380 bp were obtained for bacteria and fungi, respectively. After bioinformatics analysis, a total of 4,273,668 bacterial and 3,455,929 fungal sequences were classified. Rarefaction curves produced from bacterial and fungal sequences for the number of microbial genome operational taxonomic units (OTUs) are shown in Fig. S1A and B in the supplemental material, respectively. Rarefaction curves indicated that reasonable coverages of the bacterial and fungal communities were achieved, although the sequencing depth was insufficient to fully describe the diversity of these populations, as a plateau in the rarefaction curves was not achieved.
Bacterial microbiome.
More than 96% of the bacterial amplicon sequences were assigned at the phylum level. Within the 10 different phyla identified, over 92% of the sequences were classified within the phyla Firmicutes and Proteobacteria (Fig. S2). As many as 225 different bacterial genera were further identified, representing 58.9% of total sequences, with the 15 most relatively abundant genera shown in Table 3. Overall, most of reads assigned to the Firmicutes were related to the order Lactobacillales, with Lactobacillus, Lactococcus, Leuconostoc, and Pediococcus being the most predominant genera after 7 days of aerobic exposure. After 14 days of aerobic exposure, Bacillus, Lysinibacillus, Oceanobacillus, Paenibacillus, and Sporosarcina of the order Bacillales became more prominent than other genera.
TABLE 3.
Abundances of the 15 most relatively abundant bacterial genera in purple prairie clover ensiled without or with polyethylene glycol in terminal silage and after 7 and 14 days of aerobic exposure
| Phylum | Order | Genus | Terminal silage |
Aerobic exposure silage |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 0 abundance (%) |
SEM (%) | P | Day 7 abundance (%) |
SEM (%) | P | Day 14 abundance (%) |
SEM (%) | P | ||||||
| Control | PPC-p | Control | PPC-p | Control | PPC-p | |||||||||
| Firmicutes | Lactobacillales | Lactobacillus | 22.59 | 29.93 | 0.973 | <0.001 | 40.21 | 32.69 | 1.332 | 0.007 | 7.16 | 5.50 | 2.217 | 0.602 |
| Lactobacillales | Lactococcus | 15.67 | 0.57 | 0.617 | <0.001 | 14.06 | 0.62 | 1.001 | <0.001 | 0.45 | 0.01 | 0.109 | 0.046 | |
| Lactobacillales | Leuconostoc | 4.18 | 0.48 | 0.116 | <0.001 | 4.56 | 0.68 | 0.212 | <0.001 | 0.26 | 0.03 | 0.048 | 0.028 | |
| Lactobacillales | Pediococcus | 1.89 | 16.99 | 0.792 | <0.001 | 4.45 | 18.18 | 1.723 | 0.003 | 0.45 | 0.92 | 0.111 | 0.024 | |
| Lactobacillales | Weissella | 0.35 | 0.28 | 0.059 | 0.425 | 0.22 | 0.31 | 0.038 | 0.154 | 0.02 | 0.02 | 0.019 | 0.487 | |
| Bacillales | Bacillus | 0.04 | 0.02 | 0.009 | 0.096 | 0.03 | 0.04 | 0.012 | 0.810 | 41.17 | 38.16 | 2.546 | 0.450 | |
| Bacillales | Lysinibacillus | 0.003 | 0.002 | 0 | 0.328 | 0.003 | 0.002 | 0 | 0.316 | 2.44 | 4.63 | 0.490 | 0.141 | |
| Bacillales | Oceanobacillus | 0.001 | 0.001 | 0 | 0.946 | 0.002 | 0.003 | 0 | 0.632 | 0.67 | 2.21 | 0.181 | 0.002 | |
| Bacillales | Paenibacillus | 0.008 | 0 | 0.003 | 0.118 | 0.003 | 0.001 | 0 | 0.251 | 3.41 | 4.54 | 0.563 | 0.122 | |
| Bacillales | Sporosarcina | 0.002 | 0 | 0.001 | 0.214 | 0.002 | 0.002 | 0 | 0.287 | 1.91 | 2.80 | 0.213 | 0.001 | |
| Proteobacteria | Enterobacteriales | Erwinia | 2.49 | 1.01 | 0.303 | 0.009 | 2.07 | 1.05 | 0.254 | 0.030 | 0.02 | 0.01 | 0.002 | 0.112 |
| Sphingomonadales | Sphingomonas | 1.64 | 0.13 | 0.086 | <0.001 | 0.35 | 0.10 | 0.059 | 0.022 | 0.01 | 0.02 | 0.012 | 0.746 | |
| Rhizobiales | Methylobacterium | 1.32 | 0.12 | 0.059 | <0.001 | 0.26 | 0.11 | 0.034 | 0.021 | 0.02 | 0.02 | 0.015 | 0.782 | |
| Pseudomonadales | Pseudomonas | 0.41 | 0.02 | 0.059 | 0.001 | 0.06 | 0.01 | 0.010 | 0.012 | 0.004 | 0.001 | 0.003 | 0.495 | |
| Rhizobiales | Agrobacterium | 0.30 | 0.03 | 0.035 | <0.001 | 0.06 | 0.04 | 0.010 | 0.143 | 0.002 | 0.006 | 0.003 | 0.505 | |
Fungal microbiome.
Similarly, more than 98% of the fungal sequences were classified into 35 fungal phyla, with Ascomycota and Basidiomycota being the most abundant, accounting for 67.6% of the total sequences (data not shown) and more than 94% of the total phyla (Fig. S3). Of the two fungal phyla, the majority of reads mapped to the Ascomycota. Fungal sequences were classified into 142 different genera, accounting for 68.7% of the total sequences, with the 15 most relatively abundant genera shown in Table 4.
TABLE 4.
Abundances of the 15 most relatively abundant fungal genera in purple prairie clover ensiled without or with polyethylene glycol in terminal silage and after 7 and 14 days of aerobic exposure
| Phylum | Order | Genus | Species | Terminal silage |
Aerobic exposure silage |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 0 abundance (%) |
SEM (%) | P | Day 7 abundance (%) |
SEM (%) | P | Day 14 abundance (%) |
SEM (%) | P | |||||||
| Control | PPC-p | Control | PPC-p | Control | PPC-p | ||||||||||
| Ascomycota | Saccharomycetales | Candida | Candida sp. LPP-Y12 | 3.25 | 2.78 | 0.905 | 0.731 | 0.54 | 0.39 | 0.079 | 0.235 | 0.38 | 0.94 | 0.093 | 0.013 |
| Ascomycota | Capnodiales | Cladosporium | Cladosporium sp. 8_20_21 | 3.17 | 2.36 | 1.179 | 0.603 | 0.29 | 0.03 | 0.165 | 0.296 | 0.03 | 0.01 | 0.009 | 0.198 |
| Ascomycota | Glomerellales | Colletotrichum | Colletotrichum ledebouriae | 0.01 | 5.80 | 0.555 | 0.018 | 2.99 | 15.02 | 0.985 | 0.002 | 0.25 | 14.47 | 0.621 | <0.001 |
| Ascomycota | Incertae sedis | Xylogone | Xylogone sphaerospora | 0.01 | 0.39 | 0.09 | 0.040 | 0.95 | 0.08 | 0.33 | 0.101 | 11.11 | 0.08 | 0.52 | <0.001 |
| Ascomycota | Lichinales | Euopsis | Euopsis pulvinata | 0.96 | 2.90 | 1.280 | 0.256 | 0.54 | 0.02 | 0.116 | 0.020 | 0.01 | 0.02 | 0.011 | 0.504 |
| Ascomycota | Saccharomycetales | Galactomyces | Galactomyces sp. E234 | 0.03 | 0.19 | 0.030 | 0.020 | 0.10 | 0.25 | 0.080 | 0.220 | 0.05 | 0.28 | 0.107 | 0.174 |
| Ascomycota | Eurotiales | Aspergillus | aff. Aspergillus sp. | 0.13 | 0.62 | 0.148 | 0.053 | 0.24 | 0.03 | 0.036 | 0.054 | 0.05 | 0.03 | 0.013 | 0.316 |
| Ascomycota | Saccharomycetales | Wickerhamomyces | Wickerhamomyces cf. pijperi HMD-2015 | 0.05 | 0.22 | 0.060 | 0.072 | 0.34 | 0.21 | 0.151 | 0.591 | 0.04 | 0.06 | 0.004 | 0.039 |
| Ascomycota | Eurotiales | Penicillium | Penicillium wollemiicola | 0.02 | 0.12 | 0.02 | <0.001 | 0.04 | 0.16 | 0.02 | 0.002 | 0.13 | 0.24 | 0.02 | 0.003 |
| Ascomycota | Hypocreales | Fusarium | Fusarium sp. ELM149 | 0.16 | 0.23 | 0.01 | <0.001 | 0.18 | 0.30 | 0.03 | 0.019 | 0.26 | 0.34 | 0.04 | 0.144 |
| Basidiomycota | Tremellales | Hannaella | Hannaella sp. | 0.31 | 0.49 | 0.068 | 0.146 | 0.16 | 0.01 | 0.014 | 0.004 | 0.01 | 0.01 | 0.001 | 0.285 |
| Basidiomycota | Polyporales | Pterula | Pterula epiphylla | 0.41 | 2.03 | 0.604 | 0.093 | 0.29 | 0.03 | 0.018 | 0.002 | 0.03 | 0.06 | 0.008 | 0.038 |
| Basidiomycota | Tremellales | Tremella | Tremella dysenterica | 2.87 | 8.88 | 1.910 | 0.063 | 5.51 | 0.04 | 0.126 | <0.001 | 0.10 | 0.02 | 0.021 | 0.038 |
| Basidiomycota | Sporidiobolales | Rhodotorula | Rhodotorula sp. DMKU-RE22 | 1.74 | 2.26 | 0.608 | 0.581 | 1.13 | 0.05 | 0.118 | <0.001 | 0.04 | 0.03 | 0.026 | 0.707 |
| Basidiomycota | Tremellales | Cryptococcus | Cryptococcus sp. 5 AP-2015 | 0.47 | 1.15 | 0.140 | 0.016 | 0.46 | 0.01 | 0.112 | 0.031 | 0.03 | 0.03 | 0.014 | 0.955 |
Effects of CT on bacterial microbiome in terminal and aerobically exposed silage. (i) Terminal silage.
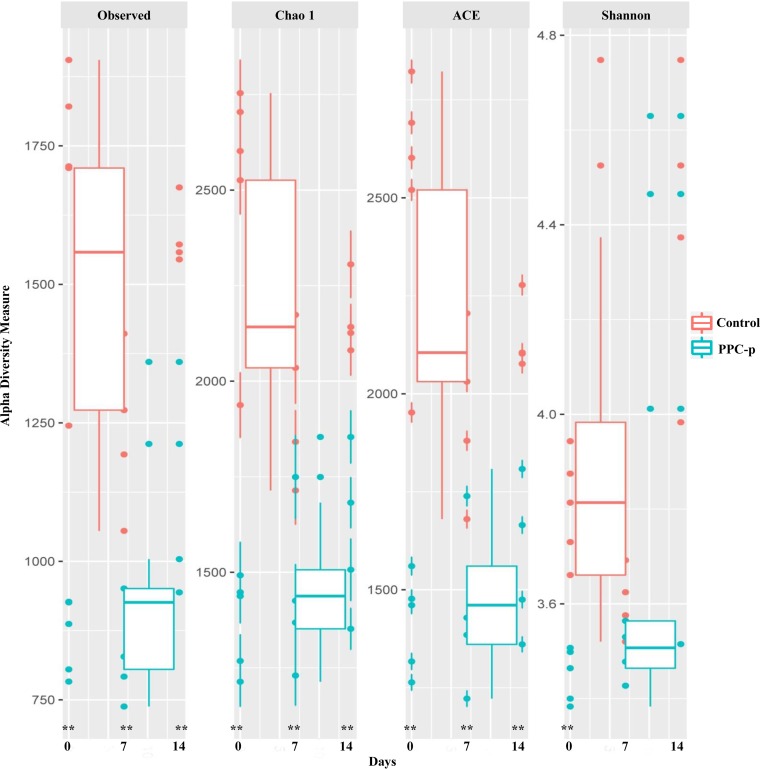
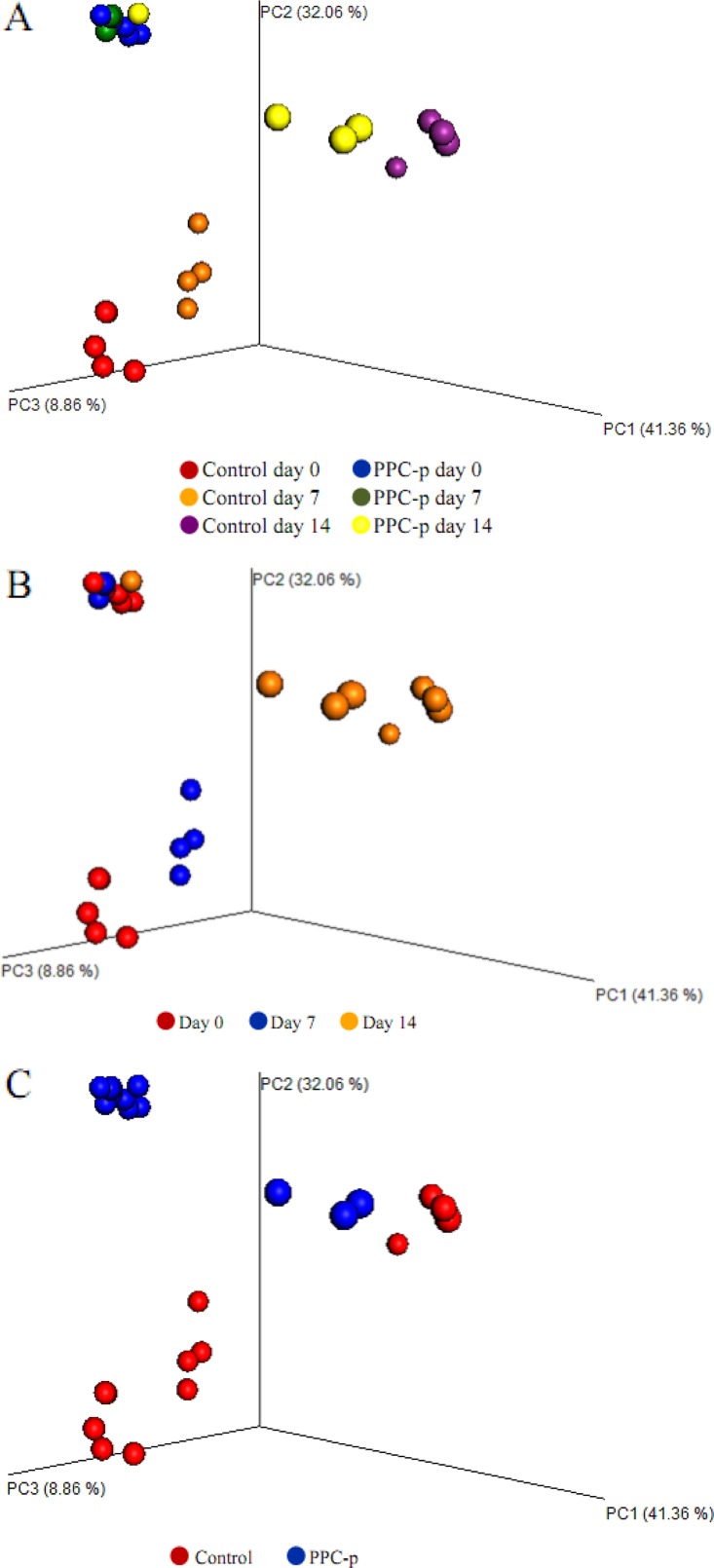
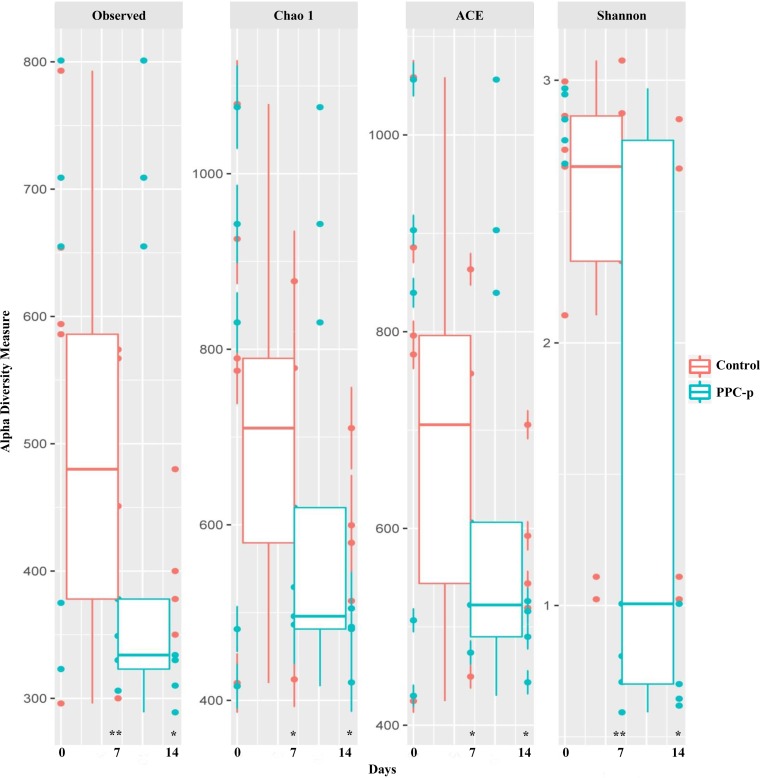
Upon opening, PEG-treated silage exhibited a greater (P < 0.001) abundance of Firmicutes but a lower (P < 0.01) abundance of Proteobacteria than the control silage (Fig. S2). Moreover, PEG-treated silage had a greater (P < 0.001) abundance of Lactobacillus and Pediococcus spp., with a lower (P < 0.01) abundance of Lactococcus, Leuconostoc, Erwinia, Sphingomonas, Methylobacterium, Pseudomonas, and Agrobacterium spp. than those of control silage (Table 3). The addition of PEG at ensiling decreased (P < 0.01) the observed number of operational taxonomic units (OTUs) and the Chao 1, abundance-based coverage estimator (ACE), and Shannon bacterial diversity indices in terminal silage (Fig. 3). In addition, when the structure of bacterial communities was assessed using weighted UniFrac distance, a clear distinction between bacterial communities in the PEG-treated and control silages on day 0 was noted, as the plots cluster separately (Fig. 4A; P < 0.001; R = 0.355). Moreover, the Pearson distance analysis showed that bacterial populations in terminal silage differed (P < 0.01) between PEG-treated and control silages (Fig. S4). The higher abundance of Lactococcus spp. in control silage, along with the greater abundances of Lactobacillus and Pediococcus spp. in PEG-treated silages corresponded to the bacterial genus analysis in Table 3.
FIG 3.
Alpha diversity of bacterial communities in purple prairie clover ensiled without (control) or with polyethylene glycol (PPC-p) in terminal silage (day 0) and after 7 and 14 days of aerobic exposure. Each panel represents one alpha diversity measure, as follows: Observed, total number of OTUs; Chao 1 and ACE, richness estimators to estimate the total number of OTUs present in a community; Shannon, microbial index of diversity. **, P < 0.01.
FIG 4.
Principal-coordinate analysis (PCoA) plots of weighted UniFrac distances for the bacterial microbiota of treatments × days (A), days (B), and treatments in purple prairie clover ensiled without (control) or with polyethylene glycol (PPC-p) (C). PC1, principal-coordinate analysis 1; PC2, principal-coordinate analysis 2.
(ii) Aerobically exposed silage.
The taxonomic profile of the bacterial microbiome indicated that PEG-treated silage was more abundant (P < 0.001) in Firmicutes after 7- and 14-day aerobic exposures but lower (P < 0.001) in Proteobacteria after 7 days than control silage (Fig. S2). Similar to the PEG-treated terminal silage, PEG-treated silage aerobically exposed for 7 days was less abundant (P < 0.05) in Lactococcus, Leuconostoc, Erwinia, Sphingomonas, Methylobacterium, and Pseudomonas spp. than control silage but possessed a greater abundance (P = 0.003) of Pediococcus (Table 3). Unexpectedly, the abundance of Lactobacillus declined (P = 0.007) with the addition of PEG. After 14 days of aerobic exposure, the dominant bacterial genera (i.e., Bacillus, Lysinibacillus, Paenibacillus, and Sporosarcina) did not differ (P = 0.05) between control and PEG-treated silages. In addition, the abundances of Lactococcus and Leuconostoc spp. in PEG-treated silage at day 14 remained lower (P < 0.05) than in the control silage, whereas Pediococcus, Oceanobacillus, and Sporosarcina spp. were greater (P < 0.05) in the PEG-treated silage than the control silage. Noticeably, the abundance of the majority bacteria was decreased (P < 0.001) over 14 days of aerobic exposure.
The key diversity indices of bacteria showed that Chao 1 richness was lower (P < 0.05) in PEG-treated silage than in control silage after 7 and 14 days of aerobic exposure (Fig. 3), and the same trend was also observed for number of OTUs and ACE index. The bacterial communities assessed using the weighted UniFrac distances appeared to be influenced by time (Fig. 4B) and PEG treatment (Fig. 4C) after aerobic exposure (P < 0.001). A clear separation between control and PEG treatments was observed after 7 days of aerobic exposure, although the bacterial sequences obtained after 14 days of aerobic exposure tended to cluster together (Fig. 4A and B; P > 0.05). In addition, the heatmap of bacterial taxon abundance showed that the control silage had bacterial populations that were obviously distinct from those of the PEG-treated silage either at day 7 or day 14 of aerobic exposure (Fig. S4). In addition, most of the classified bacterial species were assigned to the genus Lactobacillus on day 7 and Bacillus on day 14, which was consistent with observations in the analysis of the relative abundances of bacterial genera.
Effects of CT on fungal microbiome in terminal and aerobically exposed silage. (i) Terminal silage.
The abundances of Basidiomycota (35% versus 30%) and Ascomycota (64% versus 67%) did not differ significantly (P > 0.05) between the control and PEG-treated silages (Fig. S3). The PEG-treated silage had higher (P < 0.05 to ∼0.001) abundances of Colletotrichum, Xylogone, Galactomyces, Penicillium, Fusarium, and Cryptococcus spp., with a tendency (P = 0.053 to ∼0.093) also to contain higher abundances of Aspergillus, Wickerhamomyces, Pterula, and Tremella spp. than control silage (Table 4).
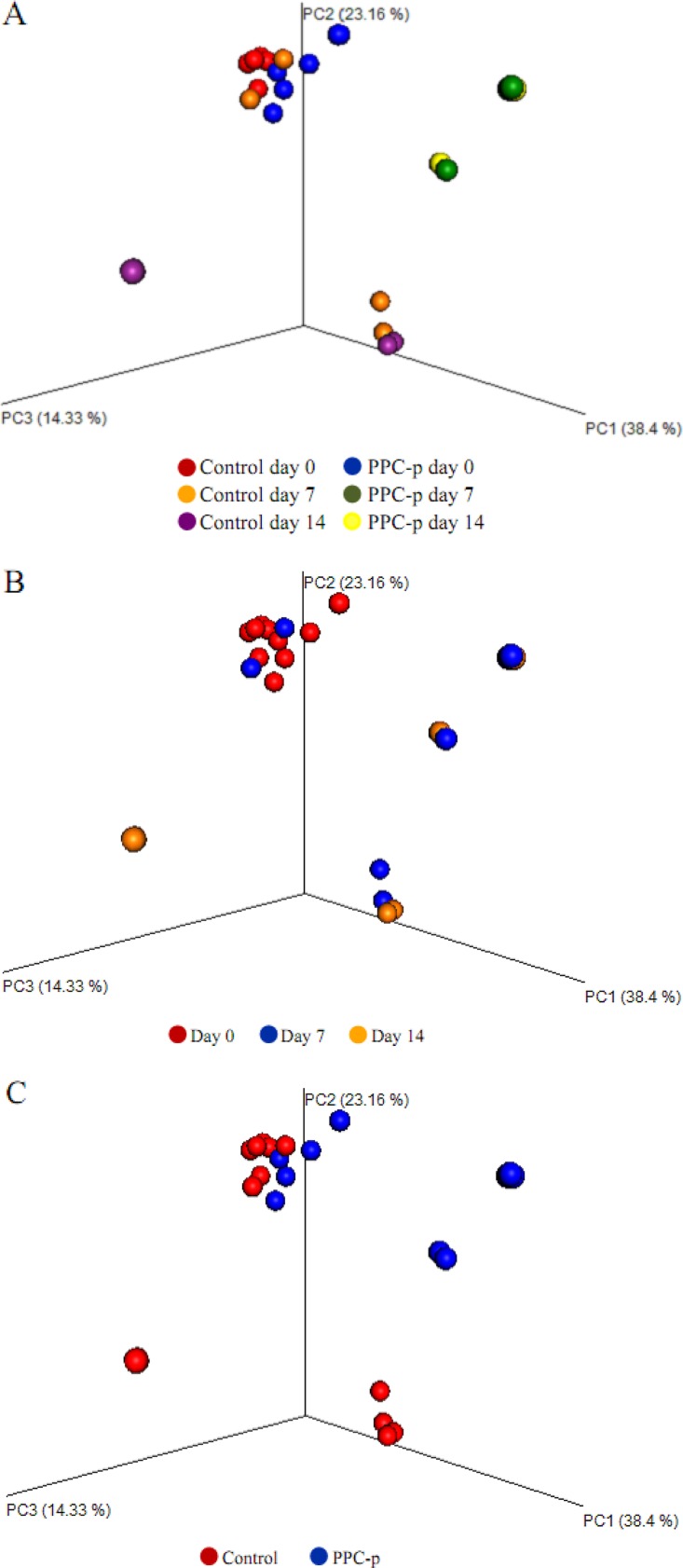
The addition of PEG at ensiling did not affect the diversity of fungal communities in terminal silage (Fig. 5). In addition, Bray-Curtis distance metric assessment identified no significant (P > 0.05) distinction in fungal communities between the control and PEG-treated silages (Fig. 6A), with no distinguishing difference in the fungal compositions within silages, as indicated by the heatmap based on Pearson distance (Fig. S5).
FIG 5.
Alpha diversity of fungal communities in purple prairie clover ensiled without (control) or with polyethylene glycol (PPC-p) in terminal silage (day 0) and after 7 and 14 days of aerobic exposure. Each panel represents one alpha diversity measure, as follows: Observed, total number of OTUs; Chao 1 and ACE, richness estimators to estimate the total number of OTUs present in a community; Shannon, microbial index of diversity. *, P < 0.05; **, P < 0.01.
FIG 6.
PCoA plots of Bray-Curtis distances for the fungal microbiota of treatments × days (A), days (B), and treatments in purple prairie clover ensiled without (control) or with polyethylene glycol (PPC-p) (C). PC3, principal-coordinate analysis 3.
(ii) Aerobically exposed silage.
The PEG-treated silage had a lower (P < 0.001) abundance of Basidiomycota (37% versus 4%) but a higher (P < 0.001) abundance of Ascomycota (60% versus 96%) than the control silage after 7 days of aerobic exposure (Fig. S3). The abundances of Colletotrichum, Penicillium, and Fusarium spp. were higher (P < 0.05 to ∼0.001) after aerobic exposure, whereas the abundances of Candida, Wickerhamomyces, Penicillium, and Pterula spp. were lower (P < 0.05) after 14 days of aerobic exposure for PEG-treated silage than for control silage (Table 4). In contrast, PEG-treated silage contained lower (P < 0.05 to ∼0.001) abundances of Euopsis, Hannaella, Pterula, Tremella, Rhodotorula, and Cryptococcus spp. after 7 days and lower (P < 0.01 to ∼0.001) abundances of Xylogone and Tremella spp. after 14 days of aerobic exposure compared to those in the control silage.
Similar to the bacterial diversity, the total observed number of OTUs and the key diversity measures showed that Chao 1 richness, ACE, and Shannon indices for fungi were all lower (P < 0.05) in PEG-treated silage than in control silage after aerobic exposure (Fig. 5). Moreover, the addition of PEG had a greater impact on diversity than the duration of aerobic exposure, as the principal-coordinate analysis (PCoA) plots were not clustered separately (P > 0.05) between days 7 and 14 of aerobic exposure (Fig. 6B).
The decline in the abundances of majority fungal species, as shown by heatmap, also are reflected by a similar trend shown in Table 4. The composition of fungal communities in control formed a cluster either at day 7 or day 14 of aerobic exposure, while most of those PEG-treated individual samples fell into individual clades that were distinct from one another in the tree (Fig. S5).
DISCUSSION
Effect of CT on ensiling properties.
It is generally recognized that legumes are difficult to ensile due to their low WSC content and high buffering capacity, which limits lactic acid production and hinders the rapid pH decline. The chemical composition and production of fermentation products (lactic acid, VFA, etc.) during ensiling in both control and PEG-treated silages were within the normal range measured in legume silages (15, 16), which indicated that PPC was ensiled well and that CT in PPC had no effect on overall silage quality. Extensive proteolysis during ensiling is recognized to reduce the nutritive value of plant protein, and it appeared that the proteolysis was decreased by the presence of CT in silage. Research has shown that other CT-containing forages, such as sainfoin and birdsfoot trefoil, undergo less proteolysis during ensiling, with transformation of their plant protein N into NPN being inhibited compared to that in forages that lack CT (17, 18). Even the direct addition of tannic acid (13) or isolated tannins (i.e., mimosa, myrabolam, quebracho, and chestnut sources) reduces proteolysis during ensiling (6, 14), likely as a result of their ability to precipitate proteins and inhibit proteolytic microorganisms (1, 19). The higher lactic acid and VFA concentrations, along with the lower WSC content and pH, in PEG-treated than in control silages indicated that CT broadly decreased the microbial activity during ensiling. In the present study, this possibility is supported by both real-time PCR and metagenomics sequencing data, which identified an increased population and abundance of lactate-producing bacteria (LAB) in PEG-treated than in control silages. Anti-LAB activity of CT during ensiling was also reported in other studies (1, 19). It was suggested that CT have ability to precipitate bacterial enzymes and bind cell membranes (20), thereby inducing changes in bacterial morphology, and to decrease the activity of bacterial enzymes (21) which led to a reduced growth of bacteria. Condensed tannins were also reported to negatively affect the growth of LAB, mainly by decreasing lactic acid production, and this effect may be due to a modification of the permeability of the bacterial cell membrane (22).
An interesting finding of this study is that control silage contained less DON and OTA than PEG-treated silage. Ochratoxin A is produced almost exclusively by Penicillium spp. (23), and DON is mainly produced by Fusarium species (24). It is generally regarded that there is little opportunity for mold/fungal growth under the proper ensiling conditions. However, there are some other commonly detected filamentous fungi during ensiling, such as Penicillium spp. (24–26) and Fusarium spp. (26–28), that are able to tolerate organic acids, carbon dioxide, and the low availability of oxygen, and their spores are always present even in healthy-looking silages (29, 30). Purple prairie clovers for both control and PEG treatment at ensiling were from the same source, and the fermentation products indicated that both silages had undergone normal ensiling fermentation. Therefore, it is difficult to explain the reason why CT reduced the production of these mycotoxins during the ensiling. Nevertheless, the lower DON and OTA concentrations were consistent with the lower copy numbers of fungi, along with the decreased abundances of Penicillium and Fusarium species in control than in PEG-treated silages in this study. This suggests that CT might have inhibited the production of these mycotoxins via inhibiting the activity of the respective mold species with an unknown mechanism during ensiling. Further research is still needed. Our study documents the ability of CT to lower mycotoxin production during ensiling. Bargiacchi et al. (E. Bargiacchi, P. Bellotti, P. Pinelli, G. Costa, S. Miele, A. Romani, P. Zambelli, and A. Scardigli A, U.S. patent application EP20150154069) also reported that chestnut tannin (a hydrolysable tannin) decreased mycotoxins in raw food materials, such as oils, oilseeds, and cereals.
Effect of CT on fermentation of silage during aerobic exposure.
Once minisilos were opened, the growth of aerobic microbes resulted in increases in both temperature and pH, a response that has been well established upon aerobic exposure of silage. The delayed increase in temperature for control silage compared to that in PEG-treated silage after aerobic exposure indicates that biologically active CT improved aerobic stability. Exposure to oxygen is known to trigger the growth of undesirable aerobic microorganisms (usually yeasts and molds) and the synthesis of toxins (31, 32). Therefore, the higher temperature in aerobically exposed PEG-treated than control silages likely reflects the ability of CT to inhibit the activity of aerobic microbes. This is consistent with the real-time PCR results that showed that the majority of microorganisms were present at a higher density in aerobically exposed PEG-treated than control silages. This is also supported by the higher NPN, SN, and ammonia N in aerobically exposed PEG-treated than in control silages. It is interesting to note that the concentrations of DON and OTA did not increase over the first 7 days of aerobic exposure in PEG-treated silages and increased only slightly for control silages. This is in spite of real-time PCR results documenting a significant increase in yeast and mold populations during this time. In contrast, DON and OTA concentrations markedly increased after 14 days of aerobic exposure even though the increase in yeast and mold populations was similar to that which occurred from day 0 to day 7. This may indicate that mycotoxins in silage are mainly produced by the aerobic microbes associated with spoilage. Nevertheless, the higher DON and OTA concentrations at each sampling date (day 7 or day 14) in PEG-treated than in control silages suggests that PPC CT inhibited the activity of spoilage microbes producing these mycotoxins during aerobic exposure.
Effects of CT on bacterial microbiome in terminal and aerobically exposed silage.
Although several studies have reported bacterial community changes in grass and legume silages (4, 33, 34), there is no information available about the effects of CT on the bacterial and fungal microbiome in terminal or aerobically exposed silage. In this study, higher numbers of rRNA copies associated with total bacteria, Lactobacillus, yeasts, and fungi were observed in PEG-treated than in control silages, indicating that CT in PPC likely reduce the total number of these microorganisms during ensiling. Similarly, Salawu (19) documented that CT that extracted from Calliandra calothyrsus inhibited sheep gut bacteria and rumen fungal activities using traditional culturing technology. For bacterial communities identified in terminal PPC silages, the phyla Firmicutes and Proteobacteria were the most abundant, which were also observed to be the predominant phyla in grass (4) and alfalfa (34) silages. Among the Firmicutes, the majority of sequencing reads were associated with Lactobacillus, Pediococcus, and Lactococcus spp. of homofermentative LAB. Sequences associated with Lactobacillus and Pediococcus of the homofermentative LAB have also previously been found in alfalfa silage (33). The higher population of Lactobacillus spp., as determined by real-time PCR, and higher relative abundances of Lactobacillus and Pediococcus spp. in PEG-treated than in control silages suggest that PPC CT inhibited the growth of these LAB during ensiling. This hypothesis was supported by the higher concentration of lactic acid and lower pH observed in PEG-treated than in control silages. In contrast, the lower abundance of Leuconostoc spp. in PEG-treated than in control silages suggested that CT may have promoted the growth of this genus in silage. Vivas et al. (35) also reported that oak wood tannins increased the presence of Leuconostoc strains in culture media. Unlike Lactobacillus and Pediococcus, which are homofermentative, all species within Leuconostoc are heterofermentative. It is regarded that heterofermentation is advantageous over homofermentation during ensiling in terms of improved aerobic stability of silage due to the production of acetic acid by heterofermentation. This could partially explain the enhanced aerobic stability by CT observed in this study. Condensed tannins are polyphenolics with antimicrobial activity mostly toward Gram-positive bacteria through ionophoric-like action (21, 36). In addition, previous studies have demonstrated that CT in PPC strongly inhibit Escherichia coli and E. coli O157:H7 activity (37, 38). Although all LAB are Gram-positive bacteria, the observations that LAB in different genera responded differently to CT indicate that the effects of CT on LAB might be species dependent. Tabasco et al. (39) also observed that the growth of different LAB (i.e., Lactobacillus species) responded to grape seed flavan-3-ol differently.
The greater bacterial diversity and richness indices in control silage, combined with the significant separation between control and PEG-treated silages in the microbiota structure, as demonstrated by the weighted UniFrac distances, suggest that PPC CT increased bacterial diversity in terminal silage. Zhang (40) also reported that green tea tannins increased the ruminal bacterial diversity of both sheep and cashmere goats. It has been found that bacterial population structure but not the overall levels of bacteria diversity changed significantly during ensiling (34). Therefore, it seems that the increased bacterial diversity in terminal silage by CT is likely due to the inhibitory effect of CT that inhibited some bacteria, such as LAB, resulting in niches within the silage that could be occupied by other genera, such as Erwinia, Sphingomonas, and Methylobacterium.
Upon aerobic exposure, the structure of bacterial community in silages changed substantially over time. A significant increase in Firmicutes, with a corresponding decline in the abundance of Proteobacteria, was observed after 7 and 14 days of aerobic exposure in both silages. McGarvey et al. (34) also reported a similar shift in phyla in aerobically exposed alfalfa silage. Although the populations of most microorganisms in both silages increased upon aerobic exposure, as reflected by the real-time PCR results, the extent of the increase was greater for PEG-treated than for control silages at day 7 of the aerobic exposure, indicating that the slower growth of bacteria in control than in PEG-treated silages after aerobic exposure is either from the carryover inhibitory effect of CT on bacterial growth during ensiling or PPC CT in exposed silage still exhibit antibacterial effect or both. The greater extent of the increase of Lactobacillus in control than in PEG-treated silages at the first 7-day aerobic exposure suggested that species in this genus likely proliferated faster than those in PEG-treated silage at the initial aerobic exposure, which may indicate that the action of PPC CT at the concentration used in this study on these species during ensiling was bacteriostatic, which might have been reversed under aerobic conditions. Further research is needed to confirm that Lactobacillus spp. can convert lactic acid to acetic acid under the aerobic conditions (41), and this partially explained the significant increase but significant decrease in acetic acid concentration in 7-day and 14-day aerobically exposed silage compared to terminal silage in this study.
Bacillus spp. are considered undesirable in silage, as they can produce butyric acid, which can promote the growth of less acid-tolerant spoilage microorganisms and result in reduced silage intake (2, 42). After 14 days of aerobic exposure, Bacillus was observed to be the dominant genus in both silages, followed by Paenibacillus, Lysinibacillus, and Oceanobacillus, which are belong to the same order, Bacillales. The significant increase in abundance of Bacillales from day 7 to day 14 of aerobic exposure was concomitant with the deterioration of silage after 7 days of aerobic exposure. Others have also found that Bacillales contribute to the deterioration of corn (42, 43) and small-grain silages (2) during aerobic exposure. The notable growth of Bacillus as a result of aerobic exposure is probably owed to the presence of oxygen, because these strains are aerobic bacteria. In addition, the similar extent of the increase in genus Bacillus abundance between the two silages during days 7 to 14 of aerobic exposure is likely due to the low extractable CT concentration in the exposed silage, because only 4 g was extractable of CT/kg DM at day 7, and none was detected at day 14 of the control samples.
Another undesirable group of bacteria, Clostridia, which tend to thrive in wet silage, can result in excessive protein degradation, DM losses, and the production of toxins. Kung (44) reported that wilting forage with above 30 to ∼35% DM can reduce the incidence of Clostridia. The relative abundance of genus Clostridium across the whole period of aerobic exposure was less than 0.01% (data not shown) in this study, indicating that the addition of water by the treatment did not promote the overgrowth of this organism.
Effects of CT on fungal microbiome in terminal and aerobically exposed silage.
Little information is available with regard to the nature of fungal communities in legume silage, although the presence and growth of those mycotoxigenic fungi have been studied in grass and cereal silages (2, 45). This study showed that the copy numbers associated with the total fungal populations were higher in PEG-treated than in control silages in both terminal and 7-day aerobically exposed silages. Another study using culture-dependent methods showed that CT can inhibit the growth and enzyme activity of fungi (46). Fungal growth during ensiling and aerobic exposure leads to a loss of nutrients and reductions in the palatability and feed value of silage (47). Therefore, the decreased fungal populations by CT during ensiling and aerobic exposure would improve the feeding value of silage.
Candida, Tremellales, Cladosporium, Cryptococcus, Colletotrichum, and Rhodotorula spp. were the predominant fungi within the microbiome of terminal PPC silages. Similar fungal communities have been reported in alfalfa (16), barley (48), corn (49), and small-grain silages (2). The sequencing results also showed that the abundances of mycotoxigenic fungi, such as those from the genera Penicillium and Fusarium, were significantly lower in control than terminal and aerobically exposed PEG-treated silages. In addition, lower concentrations of DON and OTA in control than those in PEG-treated silages were observed in this study. These phenotypic and molecular results suggest that PPC CT potentially inhibit the growth of mycotoxigenic fungi and prevent the development of mycotoxin during ensiling and aerobic exposure. As mycotoxin in feed is currently a big problem worldwide, PPC CT could be used as a natural additive for the prevention of mycotoxin in silage production.
Yeasts, such as Candida spp., found in this study belong to the order Saccharomycetales, which have been shown to be associated with whole-crop barley (48) and corn silages (50). Besides Candida, the present study also identified some other uncultured yeast species, including those of Galactomyces and Wickerhamomyces, members of the Saccharomycetales, which dominated the fungal microbiome of silage after aerobic exposure. The higher population of yeast in terminal and 7-day aerobically exposed PEG-treated than in control silages suggests that the CT inhibited the growth of some yeasts. However, similar to those of bacteria and other fungi, the differential abundances of yeast species within the microbiome of the two silages suggested that effects of CT on yeast were also species dependent.
In contrast to these measures for the bacterial community, the diversity and richness of the fungal community were similar between the two terminal silages. This suggested that PPC CT decreased the fungal population without affecting diversity. Winder et al. (51) also reported that high levels of CT (up to 110 g/kg DM) in poplar leaves had no impact on the composition of soil fungal communities. On the contrary, it was observed that fungal communities in 7- and 14-day aerobically exposed silage have greater diversity in control than in PEG-treated silages. Furthermore, the PCoA plots between treatments were separated, suggesting that PPC CT affected silage microbiota structure during aerobic exposure. The mechanism by which CT increases fungal diversity during aerobic exposure of silage needs further study.
Conclusion.
Condensed tannins inhibited the growth of total bacteria, LAB, yeast, and fungi during ensiling and aerobic exposure of silage, with the effect appearing to be species dependent, as demonstrated by the differences in their relative abundances within the microbiome. Condensed tannins increased bacterial diversity during ensiling and aerobic exposure, whereas they increased fungal diversity during aerobic exposure. The effects of CT on the microbial communities reduced lactic acid and VFA production, proteolysis, and mycotoxin concentration in terminal silage and improved aerobic stability.
MATERIALS AND METHODS
Forage and ensiling.
Whole-plant PPC was harvested in July 2015 at full flower from 3 irrigated Swinton silt loam soil plots at the Lethbridge Research and Development Centre, Alberta, Canada. Purple prairie clover is a native legume in the prairie region of North America and contains high levels of CT (up to 94 g/kg DM) that exhibit significant antimicrobial activity (37, 38). The plants were manually cut 5.0 cm above the ground using a pair of scissors, wilted in the field to approximately 30% DM, and chopped to a theoretical length of 1.0 cm using a paper cutter (X-Acto 26612; Westerville, OH, USA). Subsamples of harvested fresh forage from each plot were immediately stored at −40°C for chemical analysis. Equal portions of chopped PPC from of the three plots were then combined into a single lot for ensiling.
Chopped PPC was divided into 2 equal lots, with each being evenly spread on two separate plastic sheets. A chopped sample was immediately analyzed for CT, as described below. To inactivate the biological activity of CT, one lot was sprayed with a solution of 333 g/liter PEG (Sigma, molecular weight [MW], 6,000; PPC-p) at the rate of 420 ml/kg DM of PPC to achieve a CT-to-PEG ratio of 1:2 (52). Polyethylene glycol specifically binds with CT with H-bonds to form PEG-CT complexes without affecting other compounds in the plant and therefore specifically inactivates CT activity (53). The other lot was sprayed with an equivalent amount (280 ml/kg DM) of deionized water (control). Comparisons between PEG-treated (PPC-p) and control PPC were assumed to reflect the effects of CT. Once sprayed, PPC forage in each plastic sheet was manually tumbled for approximately 3 min to ensure uniform mixing. Forage in each sheet was then packed (≈2.5 kg) into polyvinyl chloride (PVC) laboratory minisilos (diameter, 10 cm; height, 35.5 cm) and compacted with a hydraulic press to achieve a packing density of approximately 890 kg (fresh weight)/m3.
Silos were sealed at both ends with rubber lids and metal bands, with one lid fitted with 7.0-cm-long rubber tubing as a vent. Five silos were prepared for each treatment and stored indoors at 22°C. Silos were opened after 76 days for an evaluation of silage quality and evaluation of aerobic stability. Upon opening, silage within 5.0 cm at both ends was discarded, and the remaining material was thoroughly mixed by hand. Subsamples of silage were either stored at −40°C for chemical analysis or at −80°C for microbial characterization. The remaining silage was used to assess aerobic stability, as outlined below.
Determination of aerobic stability.
Silage (400 g) from each of 4 randomly selected minisilos of each treatment was separately transferred into four 4-liter insulated containers (diameter, 13.5 cm; height, 31.0 cm). The containers were covered with 4 layers of cheesecloth and stored at 20°C for 14 days. Two Dallas Thermochron iButtons (Embedded Data Systems, Lawrenceburg, KY, USA) were embedded in the lower and midlayers of the silage in each container to monitor temperature, and two additional iButtons were placed outside the containers to record ambient temperature. The temperature was recorded at 15-min intervals for 14 days. The contents within each container were thoroughly mixed by hand and subsampled after 7- and 14-day aerobic exposures for chemical analysis and microbial profiling. Aerobic stability was defined as the period of time between initial aerobic exposure, and the temperature of the aerobically exposed silage exceeded baseline ambient temperature by 2°C (54).
Chemical analysis.
Silage samples were freeze-dried and ground to pass through a 1.0-mm screen using a Thomas Wiley cutting mill (Arthur H. Thomas Co., Philadelphia, PA, USA). The samples were subjected to analysis of DM and organic matter (OM) using an AOAC method (55), neutral detergent fiber (NDF), and acid detergent fiber (ADF) using an Ankom 200 system (Ankom Technology Corp., Fairport, NY, USA), with sodium sulfite and α-amylase added for NDF analysis. Nitrogen in NDF (NDIN) and ADF (ADIN) residues were determined with N measured by flash combustion, as described below. Samples were ball grounded in a planetary micromill (Retsch, Inc., Newtown, PA, USA) for measurement of total nitrogen (TN) by flash combustion analysis using an NA1500 nitrogen analyzer (Carlo Erba Instruments, Milan, Italy). Acid detergent lignin was measured using a method of the AOAC (55). Commercial enzyme-linked immunosorbent assay (ELISA) kits (Sigma-Aldrich, MO, USA) were used to determine DON (catalog no. SE120009) and OTA (catalog no. SE120014) concentrations, according to the manufacturer's instructions. The concentrations of extractable, protein-bound, and fiber-bound CT were determined using the method of Terrill et al. (56), with CT purified from whole-PPC plants as a standard.
For analysis of VFA, lactic acid, ammonia N, NPN, SN, and WSC content, 15 g of subsamples from fermented silage at each sampling day was immediately combined with 135 g of deionized H2O and blended in a homogenizer (Osterizer; Sunbeam, Fontana, CA, USA) for 30 s. The homogenate was strained through four layers of cheesecloth, and the supernatant was sampled and analyzed for WSC content, as described by Zahiroddini et al. (57), for VFA and lactic acid content using a 5890A gas liquid chromatograph (Phenomenex, Torrance, CA, USA), as described by Wang et al. (58), and for ammonia N content by the phenol-hypochlorite method (59). Both SN and NPN were analyzed by combustion analysis, as described above. The pH of the extract was measured using a portable pH meter (Orion; Thermo Electron Corp., Kent, WA, USA).
DNA extraction.
Samples stored at −80°C were lyophilized, ground through a 1.0-mm screen, and further ball ground using a planetary micromill (Resch, Inc., Newtown, PA, USA), and DNA was extracted according to the protocol described by Healey et al. (60). This extraction method has been successfully used for the reliable isolation of high-molecular-weight genomic DNA from tannin-containing plant materials (61, 62). All samples were further purified through a QIAamp DNA stool minikit column (Qiagen Sciences, Germantown, MD, USA) and eluted in nuclease-free water. The DNA concentration was quantified using a NanoDrop 3300 (NanoDrop Technologies, Inc., Wilmington, DE, USA). High-molecular-weight samples of DNA (50 μl) at a minimal concentration of 20 ng/μl were used for sequencing.
Primer design and standard curve generation for real-time PCR.
The primers used for real-time PCR are described in Table 5. The purified PCR products were cloned into One Shot DH5α Escherichia coli competent cells through TOPO TA vector (Life Technologies, Carlsbad, CA, USA), according to the manufacturer's instructions. Plasmids were extracted using the QIAprep Spin miniprep kit (Qiagen Science, Germantown, MD, USA), and DNA concentration was determined using a NanoDrop 3300. The copy numbers of rRNA genes were obtained based on equation 1 of Whelan et al. (63):
| (1) |
TABLE 5.
Primers and real-time PCR conditions
| PCR target (gene) | Primer name | Primer sequence (5′ to 3′) | Amplicon size (bp) | PCR cycling conditions |
|---|---|---|---|---|
| Total bacterial population (16S) | 16S F | TCCTACGGGAGGCAGCAGT | 466 | 3 min at 95°C, followed by 35 cycles of 15 s at 95°C, 1 min at 60°C, and 1 min at 72°C |
| 16S R | GGACTACCAGGGTATCTAATCCTGTT | |||
| Lactobacillus (16S) | UF-lac | TTTAYGCGGAACAYYTRGGKGT | 450 | 2 min at 95°C, followed by 40 cycles of 60 s at 95°C, 90 s at 55°C, 90 s at 57°C, and 120 s at 72°C |
| UR-lac | CCAAACATCACVCCRACTT | |||
| Total fungal population (18S) | FR1 | AICCATTCAATCGGTAIT | 390 | 5 min at 98°C, followed by 40 cycles of 15 s at 98°C, 30 s at 50°C, and 30 s at 72°C |
| FF390 | CGATAACGAACGAGACCT | |||
| Yeast population (26S) | TQ 26S 1B | TCAGGATAGCAGAAGCTCGT | 332 | 5 min at 98°C, followed by 40 cycles of 15 s at 98°C, 30 s at 64°C, and 30 s at 72°C |
| TQ 26S 2C | GTTCATTCGGCCGGTGAGTT |
Plasmid DNA was serially diluted 10-fold from 108 copies to 1 copy per μl and run in duplicate in a real-time PCR assay. Amplification efficiency (E) was estimated by using the slope of the standard curve and using equation 2 of Rasmussen (64):
| (2) |
Microbial quantification by real-time PCR.
For bacteria, amplifications were performed in a final volume of 20 μl containing 1 to 10 ng of DNA template, 0.3 μM each respective primer, and 10 μl of SsoAdvanced Universal SYBR Green supermix (Bio-Rad Laboratories, Inc., Hercules, CA, USA). For fungi, gene quantification (20 μl) consisted of 1 to 10 ng of DNA template, 0.3 μM each respective primer, 0.25 μl of bovine serum albumin (BSA; New England BioLabs, Inc., Ipswich, MA, USA), and 10 μl of iQ SYBR Green supermix (Bio-Rad Laboratories, Inc.). All amplifications were carried out in optical-grade 96-well plates on an Applied Biosystems 7500 Fast real-time PCR (RT-PCR) machine (Applied Biosystems, Foster City, CA, USA), as described in Table 5. Minor populations were quantified, and the low copy numbers were analyzed only when the coefficient of the standard curve regression was higher than 0.98 and the run efficiency was higher than 90%. The rRNA gene copy numbers quantified by real-time PCR were expressed as log10 copies/DM silage.
Microbiome characterization by metagenomic sequencing.
The purified DNA samples were analyzed by Génome Québec (McGill University, Génome Québec Innovation Centre, Quebec, Canada) using Illumina MiSeq (San Francisco, CA, USA) sequencing technology. For bacteria, the V3-V4 region of 16S was targeted using universal primers 347F-CS1 (ACACTGACGACATGGTTCTACAGGAGGCAGCAGTRRGGAAT) and 803R-CS2 (TACGGTAGCAGAGACTTGGTCTCTACCRGGGTATCTAATCC) (2). Primers nu-SSU-0817 (TTAGCAT-GGAATAATRRAATAGGA) and nu-SS-1196 (TCTGGACCTGGTGAGTTTCC), originally designed by Borneman and Hartin (65), were used to target a 400-bp genomic region of the V4 and total V5 variable domains of the short subunit (SSU) ribosomal DNA (rDNA) gene from all four major phyla of fungi. The purified amplicons were paired-end sequenced with Illumina MiSeq (San Francisco, CA, USA) by Génome Québec, according to the manufacturer's recommendations.
The V3-V4 and V4-V5 regions of the Illumina data sets were initially demultiplexed using MiSeq Reporter version 2.0. The demultiplexed paired-end reads were submitted and processed using the fully automated open-source systems of the European Bioinformatics Institute (EBI) (https://www.ebi.ac.uk/metagenomics/pipelines/2.0). Briefly, overlapping reads were first merged into single contigs via SeqPrep, and low-quality ends and sequences were trimmed (>10% undetermined nucleotides) via Trimmomatic and filtered for length via Biopython. The processed reads were then computed for identification of rRNA-encoding sequences using rRNA Selector. For taxonomic classifications, 16S rRNA reads were annotated via QIIME version 1.9.1 using the Greengenes reference database (default closed-reference OTU-picking protocol with Greengenes 13.8 reference with reverse-strand matching enabled). The 18S rRNA operational taxonomic units (OTUs) were picked from the reads using a closed-reference OTU-picking protocol against the SILVA 119 database (Eukarya only) (66), at 99% identity, via the QIIME software package (version 1.9.1). The BIOM files (see http://biom-format.org/) containing phylogenetic classification information provided by EBI metagenomics were used to construct rarefaction curves using the QIIME software (version 1.9.1).
Calculations and statistical analysis.
PPC-p data were calibrated by the amount of PEG present in this treatment. For an estimation of alpha diversity, the data set was rarefied to the same level (67), and the observed OTU richness, Chao 1 (68), and ACE (69) richness estimators, and Shannon (70) diversity indices were calculated and further plotted using the phyloseq package (71). The bacterial community structure (beta diversity) of each treatment and sampling time was evaluated using the weighted UniFrac distances (72) and visualized as principal-coordinate analysis (PCoA) plots using EMPeror (73). The fungal community structure was assessed using Bray-Curtis distances and PCoA plots. Analysis of similarity (ANOSIM) with 999 permutations was used to compare the weighted UniFrac and Bray-Curtis distances.
All data were analyzed by analysis of variance as a completely randomized design using the PROC MIXED procedure of SAS (74). For the minisilo study, treatment was considered a fixed effect, and individual silos (n = 5) were considered random factors. The repeated-measures model was used for analysis of data obtained from aerobic stability study, which included treatment, time (aerobic exposure time), and the treatment × time interaction. When time or time × treatment interaction effects were significant (i.e., P < 0.05), the means of the treatments were compared on each sampling date. Differences among treatments were determined using LSMEANS with the PDIFF function and declared significant at a P value of <0.05. A tendency of significant differences was described at P values between 0.05 and 0.1.
Accession number(s).
All sequencing data generated in the study have been submitted to the EBI (accession numbers ERR1413924 to ERR1413975) under project accession number ERP015646.
Supplementary Material
ACKNOWLEDGMENTS
This study was partially supported by the AAFC-Beef Science Cluster and the Alberta Livestock and Meat Agency. K.P. acknowledges the China Scholarship Council for awarding a scholarship to conduct this research at the Lethbridge Research and Development Centre (LRDC) of Agriculture and Agri-Food Canada.
Y.W., T.A.M., S.A., and S.W. conceived and designed the experiments. K.P., L.J., Q.H., and Z.X. performed the experiments. K.P., L.J., Y.D.N., H.D., and H.E.Y. analyzed the data. Y.W. and T.A.M. contributed reagents, materials, and analysis tools. K.P., Y.W., and T.A.M. wrote the paper.
We declare no competing conflicts of interest.
Footnotes
This article is contribution 387-17038 from the Lethbridge Research and Development Centre.
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02274-17.
REFERENCES
- 1.Wang Y, Barbieri LR, Berg BP, McAllister TA. 2007. Effects of mixing sainfoin with alfalfa on ensiling, ruminal fermentation and total tract digestion of silage. Anim Feed Sci Technol 135:296–314. doi: 10.1016/j.anifeedsci.2006.07.002. [DOI] [Google Scholar]
- 2.Duniere L, Xu S, Long J, Elekwachi C, Wang Y, Turkington K, Forster R, McAllister TA. 2017. Bacterial and fungal core microbiomes associated with small grain silages during ensiling and aerobic spoilage. BMC Microbiol 17:50. doi: 10.1186/s12866-017-0947-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliver JD. 2005. The viable but nonculturable state in bacteria. J Microbiol 43:93–100. [PubMed] [Google Scholar]
- 4.Eikmeyer FG, Köfinger P, Poschenel A, Jünemann S, Zakrzewski M, Heinl S, Mayrhuber E, Grabherr R, Puhler A, Schluter A. 2013. Metagenome analyses reveal the influence of the inoculant Lactobacillus buchneri CD034 on the microbial community involved in grass ensiling. J Biotechnol 167:334–343. doi: 10.1016/j.jbiotec.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Muck RE. 1987. Dry matter level effects on alfalfa silage quality I. nitrogen transformations. Trans ASAE 30:7–14. doi: 10.13031/2013.30393. [DOI] [Google Scholar]
- 6.Salawu MB, Acamovic T, Stewart CS, Hvelplund T, Weisbjerg MR. 1999. The use of tannins as silage additives: effects on silage composition and mobile bag disappearance of dry matter and protein. Anim Feed Sci Technol 82:243–259. doi: 10.1016/S0377-8401(99)00105-4. [DOI] [Google Scholar]
- 7.Wang Y, Douglas GB, Waghorn GC, Barry TN, Foote AG. 1996. Effect of condensed tannins in Lotus corniculatus upon lactation performance in ewes. J Agric Sci 126:353–362. doi: 10.1017/S0021859600074918. [DOI] [Google Scholar]
- 8.Waghorn GC, Shelton ID. 1997. Effect of condensed tannins in Lotus corniculatus on the nutritive value of pasture for sheep. J Agric Sci 128:365–372. doi: 10.1017/S0021859697004218. [DOI] [Google Scholar]
- 9.Min BR, Fernandez JM, Barry TN, McNabb WC, Kemp PD. 2001. The effect of condensed tannins in Lotus corniculatus upon reproductive efficiency and wool production in ewes during autumn. Anim Feed Sci Technol 92:185–202. doi: 10.1016/S0377-8401(01)00258-9. [DOI] [Google Scholar]
- 10.Min BR, Barry TN, Attwood GT, McNabb WC. 2003. The effect of condensed tannins on the nutrition and health of ruminants fed fresh temperate forages: a review. Anim Feed Sci Technol 106:3–19. doi: 10.1016/S0377-8401(03)00041-5. [DOI] [Google Scholar]
- 11.Pritchard DA, Stocks DC, O'Sullivan BM, Martin PR, Hurwood IS, O'Rourke PK. 1988. The effect of polyethylene glycol (PEG) on wool growth and liveweight of sheep consuming a mulga (Acacia aneura) diet. Proc Aust Soc Anim Prod 11:290–293. [Google Scholar]
- 12.Jones BA, Muck RE, Hatfleld RD. 1995. Red clover extracts inhibit legume proteolysis. J Sci Food Agric 67:329–333. doi: 10.1002/jsfa.2740670309. [DOI] [Google Scholar]
- 13.Santos GT, Oliveira RL, Petit HV, Cecato U, Zeoula LM, Rigolon LP, Damasceno UC, Branco AF, Bett V. 2000. Short communication: effect of tannic acid on composition and ruminal degradability of bermudagrass and alfalfa silages. J Dairy Sci 83:2016–2020. doi: 10.3168/jds.S0022-0302(00)75080-6. [DOI] [PubMed] [Google Scholar]
- 14.Tabacco E, Borreani G, Crovetto GM, Galassi G, Colombo D, Cavallarin L. 2006. Effect of chestnut tannin on fermentation quality, proteolysis, and protein rumen degradability of alfalfa silage. J Dairy Sci 89:4736–4746. doi: 10.3168/jds.S0022-0302(06)72523-1. [DOI] [PubMed] [Google Scholar]
- 15.McAllister TA, Feniuk R, Mir Z, Mir P, Selinger LB, Cheng KJ. 1998. Inoculants for alfalfa silage: effects on aerobic stability, digestibility and the growth performance of feedlot steers. Livest Prod Sci 53:171–181. doi: 10.1016/S0301-6226(97)00150-4. [DOI] [Google Scholar]
- 16.Rossi F, Dellaglio F. 2007. Quality of silages from Italian farms as attested by number and identity of microbial indicators. J Appl Microbiol 103:1707–1715. doi: 10.1111/j.1365-2672.2007.03416.x. [DOI] [PubMed] [Google Scholar]
- 17.Rioux R, Dos Santos GT, Petit HV, Proulx JG. 1995. Effect of cultivars on in vitro and ruminal degradation of the nitrogen fraction in birdsfoot trefoil silage. J Dairy Sci 78:1766. doi: 10.3168/jds.S0022-0302(95)76802-3. [DOI] [PubMed] [Google Scholar]
- 18.Fraser MD, Fychan R, Jones R. 2000. Voluntary intake, digestibility and nitrogen utilization by sheep fed ensiled forage legumes. Grass Forage Sci 55:271–279. doi: 10.1046/j.1365-2494.2000.00225.x. [DOI] [Google Scholar]
- 19.Salawu MB. 1997. The nutritive value of the leguminous browse Calliandra calothyrus [sic] and the role of tannins in ruminant feeds. Ph.D. thesis University of Aberdeen, Scotland, United Kingdom. [Google Scholar]
- 20.Min BR, Attwood GT, McNabb WC, Molan AL, Barry TN. 2005. The effect of condensed tannins from Lotus corniculatus on the proteolytic activities and growth of rumen bacteria. Anim Feed Sci Technol 121:45–58. doi: 10.1016/j.anifeedsci.2005.02.007. [DOI] [Google Scholar]
- 21.Jones GA, McAllister TA, Muir AD, Cheng KJ. 1994. Effects of sainfoin (Onobrychis viciifolia Scop.) condensed tannins on growth and proteolysis by four strains of ruminal bacteria. Appl Environ Microbiol 60:1374–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scalbert A. 1991. Antimicrobial properties of tannins. Phytochemistry 30:3875–3883. doi: 10.1016/0031-9422(91)83426-L. [DOI] [Google Scholar]
- 23.Frisvad JC. 1989. The connection between the penicillia and aspergilli and mycotoxins with special emphasis on misidentified isolates. Arch Environ Contam Toxicol 18:452–467. doi: 10.1007/BF01062373. [DOI] [PubMed] [Google Scholar]
- 24.Cheli F, Campagnoli A, Dell'Orto V. 2013. Fungal populations and mycotoxins in silages: from occurrence to analysis. Anim Feed Sci Technol 183:1–16. doi: 10.1016/j.anifeedsci.2013.01.013. [DOI] [Google Scholar]
- 25.González Pereyra ML, Chiacchiera SM, Rosa CA, Sager R, Dalcero AM, Cavaglieri L. 2011. Comparative analysis of the mycobiota and mycotoxins contaminating corn trench silos and silo bags. J Sci Food Agric 91:1474–1481. doi: 10.1002/jsfa.4336. [DOI] [PubMed] [Google Scholar]
- 26.Keller LAM, González Pereyra ML, Keller KM, Alonso VA, Oliveira AA, Almeida TX, Barbosa TS, Nunes LMT, Cavaglieri LR, Rosa CAR. 2013. Fungal and mycotoxins contamination in corn silage: monitoring risk before and after fermentation. J Stored Prod Res 52:42–47. doi: 10.1016/j.jspr.2012.09.001. [DOI] [Google Scholar]
- 27.Garon D, Richard E, Sage L, Bouchart V, Pottier D, Lebailly P. 2006. Mycoflora and multimycotoxin detection in corn silage: experimental study. J Agric Food Chem 54:3479–3484. doi: 10.1021/jf060179i. [DOI] [PubMed] [Google Scholar]
- 28.González Pereyra ML, Alonso VA, Sager R, Morlaco MB, Magnoli CE, Astoreca AL, Rosa CAR, Chiacchiera SM, Dalcero AM, Cavaglieri LR. 2008. Fungi and selected mycotoxins from pre- and post-fermented corn silage. J Appl Microbiol 104:1034–1041. doi: 10.1111/j.1365-2672.2007.03634.x. [DOI] [PubMed] [Google Scholar]
- 29.Mansfield MA, Kuldau GA. 2007. Microbiological and molecular determination of mycobiota in fresh and ensiled maize silage. Mycologia 99:269–278. [DOI] [PubMed] [Google Scholar]
- 30.Storm IDM, Sørensen JL, Rasmussen RR, Nielsen KF, Thrane U. 2008. Mycotoxins in silage. Stewart Postharvest Rev 4:1–12. doi: 10.2212/spr.2008.6.4. [DOI] [Google Scholar]
- 31.Danner H, Holzer M, Mayrhuber E, Braun R. 2003. Acetic acid increases stability of silage under aerobic conditions. Appl Environ Microbiol 69:562–567. doi: 10.1128/AEM.69.1.562-567.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filya I. 2004. Nutritive value and aerobic stability of whole crop maize silage harvested at four stages of maturity. Anim Feed Sci Technol 116:141–150. doi: 10.1016/j.anifeedsci.2004.06.003. [DOI] [Google Scholar]
- 33.Stevenson DM, Muck RE, Shinners KJ, Weimer PJ. 2006. Use of real time PCR to determine population profiles of individual species of lactic acid bacteria in alfalfa silage and stored corn stover. Appl Microbiol Biotechnol 71:329–338. doi: 10.1007/s00253-005-0170-z. [DOI] [PubMed] [Google Scholar]
- 34.McGarvey JA, Franco RB, Palumbo JD, Hnasko R, Stanker L, Mitloehner FM. 2013. Bacterial population dynamics during the ensiling of Medicago sativa (alfalfa) and subsequent exposure to air. J Appl Microbiol 114:1661–1670. doi: 10.1111/jam.12179. [DOI] [PubMed] [Google Scholar]
- 35.Vivas N, Augustin M, Lonvaud-Funel A. 2000. Influence of oak wood and grape tannins on the lactic acid bacterium Oenococcus oeni (Leuconostoc oenos, 8413). J Sci Food Agric 80:1675–1678. doi:. [DOI] [Google Scholar]
- 36.Min BR, Attwood GT, Reilly K, Sun W, Peters JS, Barry TN, McNabb WC. 2002. Lotus corniculatus condensed tannins decrease in vivo populations of proteolytic bacteria and affect nitrogen metabolism in the rumen of sheep. Can J Microbiol 48:1–6. doi: 10.1139/w01-128. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Jin L, Ominski KH, He M, Xu Z, Krause DO, Acharya SN, Wittenberg KM, Liu XL, Stanford K, McAllister TA. 2013. Screening of condensed tannins from Canadian prairie forages for anti–Escherichia coli O157:H7 with an emphasis on purple prairie clover (Dalea purpurea Vent). J Food Prot 76:560–567. doi: 10.4315/0362-028X.JFP-12-259. [DOI] [PubMed] [Google Scholar]
- 38.Liu XL, Hao YQ, Jin L, Xu ZJ, McAllister TA, Wang Y. 2013. Anti-Escherichia coli O157:H7 properties of purple prairie clover and sainfoin condensed tannins. Molecules 18:2183–2199. doi: 10.3390/molecules18022183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabasco R, Sánchez-Patán F, Monagas M, Bartolomé B, Moreno-Arribas MV, Peláez C, Requena T. 2011. Effect of grape polyphenols on lactic acid bacteria and bifidobacteria growth: resistance and metabolism. Food Microbiol 28:1345–1352. doi: 10.1016/j.fm.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 40.Zhang MM. 2015. Effects of tannin supplemented in diet on the quantity and diversity of ruminal bacteria in sheep and cashmere goats. M.Sc. thesis Inner Mongolia Agricultural University, Hohhot, China. [Google Scholar]
- 41.Oude Elferink SJWH, Krooneman J, Gottschal JC, Spoelstra SF, Faber F, Driehuis F. 2001. Anaerobic conversion of lactic acid to acetic acid and 1, 2-propanediol by lactobacillus buchneri. Appl Environ Microbiol 67:125. doi: 10.1128/AEM.67.1.125-132.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vissers MMM, Driehuis F, Te Giffel MC, De Jong P, Lankveld JMG. 2007. Concentrations of butyric acid bacteria spores in silage and relationships with aerobic deterioration. J Dairy Sci 90:928–936. doi: 10.3168/jds.S0022-0302(07)71576-X. [DOI] [PubMed] [Google Scholar]
- 43.Borreani G, Dolci P, Tabacco E, Cocolin L. 2013. Aerobic deterioration stimulates outgrowth of spore-forming paenibacillus in corn silage stored under oxygen-barrier or polyethylene films. J Dairy Sci 96:5206–5216. doi: 10.3168/jds.2013-6649. [DOI] [PubMed] [Google Scholar]
- 44.Kung L. 2008. Silage fermentation end products and microbial populations: their relationships to silage quality and animal productivity. Proc 2008 Ann Conf Am Assoc Bovine Practitioners, 25 to 27 September 2008, Charlotte, NC. [Google Scholar]
- 45.Alonso VA, Pereyra CM, Keller LAM, Dalcero AM, Rosa CAR, Chiacchiera SM, Cavaglieri LR. 2013. Fungi and mycotoxins in silage: an overview. J Appl Microbiol 115:637–643. doi: 10.1111/jam.12178. [DOI] [PubMed] [Google Scholar]
- 46.McAllister TA, Bae HD, Yanke LJ, Cheng KJ, Muir A. 1994. Effect of condensed tannins from birdsfoot trefoil on endoglucanase activity and the digestion of cellulose filter paper by ruminal fungi. Can J Microbiol 40:298–305. doi: 10.1139/m94-048. [DOI] [PubMed] [Google Scholar]
- 47.O'Brien M, O'Kiely P, Forristal PD, Fuller HT. 2007. Visible fungal growth on baled grass silage during the winter feeding season in Ireland and silage characteristics associated with the occurrence of fungi. Anim Feed Sci Technol 139:234–256. doi: 10.1016/j.anifeedsci.2007.01.010. [DOI] [Google Scholar]
- 48.Inglis GD, Yanke LJ, Kawchuk LM, McAllister TA. 1999. The influence of bacterial inoculants on the microbial ecology of aerobic spoilage of barley silage. Can J Microbiol 45:77–87. doi: 10.1139/w98-207. [DOI] [PubMed] [Google Scholar]
- 49.May LA, Smiley B, Schmidt MG. 2001. Comparative denaturing gradient gel electrophoresis analysis of fungal communities associated with whole plant corn silage. Can J Microbiol 47:829. doi: 10.1139/w01-086. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Nishino N. 2011. Effects of inoculation of Lactobacillus rhamnosus and Lactobacillus buchneri on fermentation, aerobic stability and microbial communities in whole crop corn silage. Grassland Sci 57:184–191. doi: 10.1111/j.1744-697X.2011.00226.x. [DOI] [Google Scholar]
- 51.Winder RS, Lamarche J, Constabel CP, Hamelin RC. 2012. The effects of high-tannin leaf litter from transgenic poplars on microbial communities in microcosm soils. Front Microbiol 4:290. doi: 10.3389/fmicb.2013.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Makkar HP, Blummel M, Becker K. 1995. Formation of complexes between polyvinyl pyrrolidones or polyethylene glycols and tannins, and their implication in gas production and true digestibility in in vitro techniques. Br J Nutr 73:897–913. doi: 10.1079/BJN19950095. [DOI] [PubMed] [Google Scholar]
- 53.Jones WT, Mangan JL. 1977. Complexes of the condensed tannins of sainfoin (Onobrychis viciifolia scop.) with fraction 1 leaf protein and with submaxillary mucoprotein, and their reversal by polyethylene glycol and pH. J Sci Food Agric 28:126–136. doi: 10.1002/jsfa.2740280204. [DOI] [Google Scholar]
- 54.Teller RS, Schmidt RJ, Whitlow LW, Kung L Jr. 2012. Effect of physical damage to ears of corn before harvest and treatment with various additives on the concentration of mycotoxins, silage fermentation, and aerobic stability of corn silage. J Dairy Sci 95:1428–1436. doi: 10.3168/jds.2011-4610. [DOI] [PubMed] [Google Scholar]
- 55.AOAC. 1999. Official methods of analysis of the association of official agricultural chemists, 16th ed, 5th revision AOAC International, Gaithersburg, MD. [Google Scholar]
- 56.Terrill TH, Rowan AM, Douglas GB, Barry TN. 1992. Determination of extractable and bound condensed tannin concentrations in forage plants, protein concentrate meals and cereal grains. J Sci Food Agric 58:321–329. doi: 10.1002/jsfa.2740580306. [DOI] [Google Scholar]
- 57.Zahiroddini H, Baah J, Absalom W, McAllister TA. 2004. Effect of an inoculant and hydrolytic enzymes on fermentation and nutritive value of whole crop barley silage. Anim Feed Sci Technol 117:317–330. doi: 10.1016/j.anifeedsci.2004.08.013. [DOI] [Google Scholar]
- 58.Wang Y, Berg BP, Barbieri LR, Veira DM, McAllister TA. 2006. Comparison of alfalfa and mixed alfalfa-sainfoin pastures for grazing cattle: effects on incidence of bloat, ruminal fermentation, and feed intake. Can J Anim Sci 86:383–392. doi: 10.4141/A06-009. [DOI] [Google Scholar]
- 59.Weatherburn MW. 1967. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem 39:971–974. doi: 10.1021/ac60252a045. [DOI] [Google Scholar]
- 60.Healey A, Furtado A, Cooper T, Henry RJ. 2014. Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 10:21. doi: 10.1186/1746-4811-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krause DO, Smith WJ, McSweeney CS. 2001. Extraction of microbial DNA from rumen contents containing plant tannins. Biotechniques 31:294–298. [DOI] [PubMed] [Google Scholar]
- 62.Toader VA, Moldovan IC, Sofletea N, Abrudan IV, Curtu AL. 2009. DNA isolation and amplification in oak species (Quercus spp.). Bulletin of the Transilvania University of Brasov, Series II-Forestry, Wood Industry, Agricultural Food Engineering. 2:45–50. [Google Scholar]
- 63.Whelan JA, Russell NB, Whelan MA. 2003. A method for the absolute quantification of cDNA using real-time PCR. J Immunol Methods 278:261–269. doi: 10.1016/S0022-1759(03)00223-0. [DOI] [PubMed] [Google Scholar]
- 64.Rasmussen R. 2001. Quantification on the LightCycler, p 21–34. In Meuer S, Wittwer C, Nakagawara K (ed), Rapid cycle real-time PCR: methods and applications. Springer, Berlin, Germany. [Google Scholar]
- 65.Borneman J, Hartin RJ. 2000. PCR primers that amplify fungal rRNA genes from environmental samples. Appl Environ Microbiol 66:4356–4360. doi: 10.1128/AEM.66.10.4356-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lemos LN, Fulthorpe RR, Triplett EW, Roesch LF. 2011. Rethinking microbial diversity analysis in the high throughput sequencing era. J Microbiol Methods 86:42–51. doi: 10.1016/j.mimet.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 68.Chao A. 1984. Nonparametric estimation of the number of classes in a population. Scand J Stat 11:265–270. [Google Scholar]
- 69.Chao A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 783–791. [PubMed] [Google Scholar]
- 70.Shannon CE. 1948. A note on the concept of entropy. Bell Syst Tech J 27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 71.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. 2013. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2:1. doi: 10.1186/2047-217X-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.SAS Institute, Inc. 2012. SAS OnlineDoc 9.1.3. SAS Institute, Inc., Cary, NC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.