ABSTRACT
Campylobacter jejuni is a microaerophilic bacterium and is believed to persist in a biofilm to antagonize environmental stress. This study investigated the influence of environmental conditions on the formation of C. jejuni biofilm. We report an extracellular DNA (eDNA)-mediated mechanism of biofilm formation in response to aerobic and starvation stress. The eDNA was determined to represent a major form of constitutional material of C. jejuni biofilms and to be closely associated with bacterial lysis. Deletion mutation of the stress response genes spoT and recA enhanced the aerobic influence by stimulating lysis and increasing eDNA release. Flagella were also involved in biofilm formation but mainly contributed to attachment rather than induction of lysis. The addition of genomic DNA from either Campylobacter or Salmonella resulted in a concentration-dependent stimulation effect on biofilm formation, but the effect was not due to forming a precoating DNA layer. Enzymatic degradation of DNA by DNase I disrupted C. jejuni biofilm. In a dual-species biofilm, eDNA allocated Campylobacter and Salmonella at distinct spatial locations that protect Campylobacter from oxygen stress. Our findings demonstrated an essential role and multiple functions of eDNA in biofilm formation of C. jejuni, including facilitating initial attachment, establishing and maintaining biofilm, and allocating bacterial cells.
IMPORTANCE Campylobacter jejuni is a major cause of foodborne illness worldwide. In the natural environment, the growth of C. jejuni is greatly inhibited by various forms of environmental stress, such as aerobic stress and starvation stress. Biofilm formation can facilitate the distribution of C. jejuni by enabling the survival of this fragile microorganism under unfavorable conditions. However, the mechanism of C. jejuni biofilm formation in response to environmental stress has been investigated only partially. The significance of our research is in identifying extracellular DNA released by bacterial lysis as a major form of constitution material that mediates the formation of C. jejuni biofilm in response to environmental stress, which enhances our understanding of the formation mechanism of C. jejuni biofilm. This knowledge can aid the development of intervention strategies to limit the distribution of C. jejuni.
KEYWORDS: Campylobacter jejuni, biofilms, extracellular DNA, stress response, environmental stress
INTRODUCTION
Biofilms are structured bacterial communities encased in a self-produced extracellular polymeric matrix. The matrix is composed mainly of proteins, polysaccharides, lipids, and nucleic acids. As one of the well-recognized bacterial survival modes, biofilms exhibit various physiological responses to environmental stress and inherent tolerance of the host defense system (1). Bacterial cells encased in a biofilm demonstrate significantly elevated tolerance of most of the conventional disinfectants and antimicrobial agents (2). Such survival behavior represents a potential risk because over 60% of chronic bacterial infections are associated with biofilm formation (1).
Campylobacter jejuni is recognized as one of the leading causes of bacterial foodborne illness worldwide. Raw meat, untreated water, unpasteurized milk, and animals (e.g., birds) are the major reservoirs of C. jejuni (3). The paradox associated with this fastidious bacterium is that C. jejuni is vulnerable to environmental stress and yet the incidence of Campylobacter infections is high and has progressively increased. For example, the reported cases of Campylobacter infection in the United States increased by 9% from 2006 to 2015 (4). It is suggested that biofilm may contribute to the survival and distribution of C. jejuni in the natural environment (5).
C. jejuni is capable of forming biofilms on different substrates (6, 7) as well as residing in a well-developed multispecies biofilm (8). C. jejuni cells in the biofilm can survive aerobic stress, and their viability can persist up to more than twice as long as that of planktonic cells (9). It is therefore reasonable to speculate that biofilm is the protection vehicle where C. jejuni resides for survival under different environmental conditions. This may also explain the high prevalence of Campylobacter-associated foodborne diseases. Recent studies suggest that environmental stresses, such as high oxygen levels and relatively low temperatures, can stimulate C. jejuni biofilm formation to a relatively high level (6, 10). Molecular investigation has identified several critical genetic factors, such as Campylobacter planktonic growth regulator cprRS, stringent response regulator spoT, and global regulator csrA, that can affect C. jejuni biofilm formation (6, 11–13). However, there is still a knowledge gap with respect to correlation of these genetic factors with the formation of C. jejuni biofilm in response to environmental stress.
DNA presents in abundance in the environment as a consequence of lysis of dead organisms or via active secretion from the living organisms (14). Bacteria can utilize that free DNA as an important supply of nutrients or integrate the free DNA into the genome for acquiring resistance capacity (15–17). In a previous study, Pseudomonas aeruginosa progressively released DNA into the environment during biofilm formation (18). Our recent study showed that the addition of meat juice with a high level of DNA content (e.g., chicken juice or pork juice) not only supported the growth of C. jejuni cells but also led to an increase of biofilm formation (7). Therefore, we speculate that eDNA might mediate biofilm formation in response to different environments. The aim of this study was to characterize the role of eDNA during the formation of C. jejuni biofilm under two common environmental stress conditions, namely, aerobic stress and starvation stress.
RESULTS
Biofilm formation under different environmental conditions.
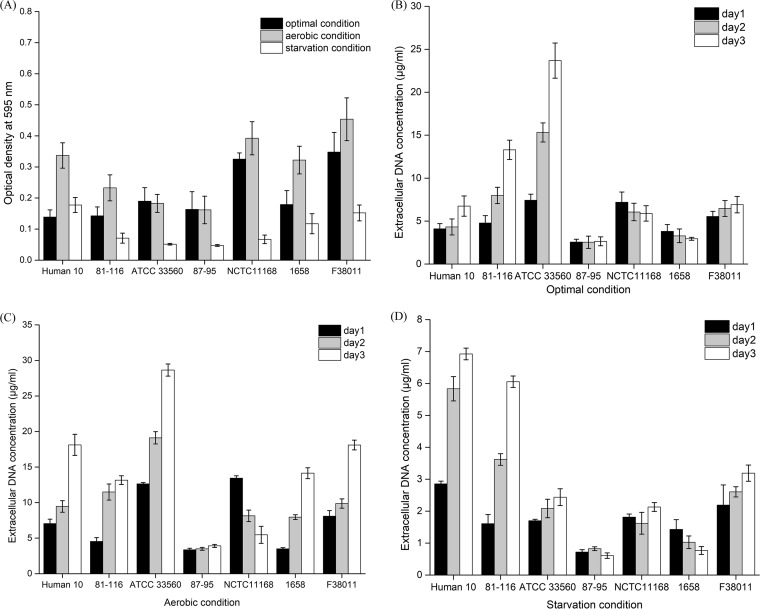
Biofilm formation of a wide range of C. jejuni strains was tested under different environmental conditions (i.e., optimal, aerobic, and starvation conditions). Under the optimal condition, most of the C. jejuni wild-type strains, including 3 clinical isolates (C. jejuni human 10, C. jejuni 1658, and C. jejuni 87-95) and 4 reference strains (C. jejuni 81-116, C. jejuni ATCC 33560, C. jejuni NCTC 11168, and C. jejuni F38011), were able to form relatively intensive biofilms (Fig. 1A). Among these wild-type strains, C. jejuni F38011 formed the highest level of biofilm, which was approximately 2.5 times higher than the lowest level of biofilm formed by C. jejuni human 10. The biofilm formation of most C. jejuni wild-type strains (i.e., C. jejuni human 10, 81-116, NCTC11168, 1658, and F38011) was significantly (P < 0.05) stimulated under the aerobic condition compared to the results seen under the optimal condition. The effects of stimulation on the biofilm formation ranged from an increase of 142% for C. jejuni human 10 to 20% for C. jejuni NCTC 11168. The stimulation effect on the biofilm formation by C. jejuni F38011 was an increase of 30%. In contrast, biofilm formation of C. jejuni ATCC 33560 and C. jejuni 87-95 under the aerobic condition was at the same level as that seen under the optimal condition. The starvation condition significantly (P < 0.05) inhibited biofilm formation for almost all C. jejuni wild-type strains except C. jejuni human 10. The inhibition effect on biofilm formation ranged from a 79% decrease for C. jejuni NCTC 11168 to a 34% decrease for C. jejuni 1658. The inhibition effect on the biofilm formation by C. jejuni F38011 was a decrease of 56%. C. jejuni F38011 was used as the representative strain for the following study due to its intense biofilm formation and remarkable response to different environmental conditions.
FIG 1.
Biofilm formation and release of extracellular DNA (eDNA) by wild-type C. jejuni strains (i.e., human 10, 81-116, ATCC 33560, 87-95, NCTC 11168, 1658, and F38011) under optimal, aerobic, and starvation conditions. (A) The level of biofilm formation was evaluated using crystal violet staining. The stained biofilm was released using 95% ethanol and determined by monitoring the value of OD595. (B to D) The concentration of eDNA during biofilm formation under optimal conditions (B), aerobic conditions (C), and starvation conditions (D) over 3 days was quantified using SYBR green I dye on the basis of a standard curve generated using a series of 10-fold dilutions of Lambda DNA from 80 μg/ml to 0.156 μg/ml.
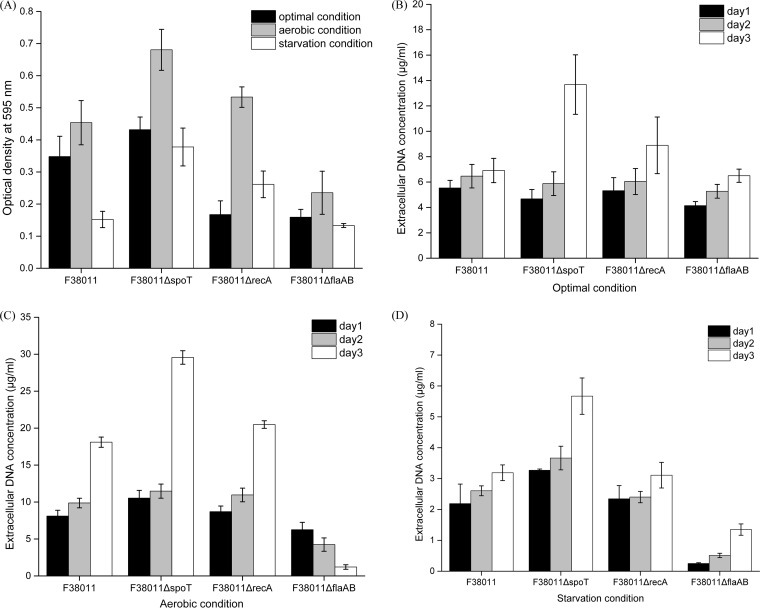
Stress response deficiency mutants (mutation of the stringent response regulator spoT gene and mutation of the DNA repair system recA gene) and a motility deficiency mutant (mutation of flagellin protein flaAB genes) were generated using C. jejuni F38011 as the parental strain, and their corresponding biofilm formation was tested under different environmental conditions (Fig. 2A). Under the optimal condition, the biofilm formation of recA and flaAB deletion mutants was significantly (P < 0.05) lower than that of their parental counterpart by 52% and 55%, respectively, whereas the biofilm formation of the spoT deletion mutant was significantly (P < 0.05) higher than that of its parental counterpart by 24%. These deletion mutants demonstrated similar responses to the environmental stress. Thus, the biofilm formation of the deletion mutants was stimulated under the aerobic condition and impaired under the starvation condition. Compared to the wild-type strain, the levels of biofilm formation of spoT and recA deletion mutants under the aerobic condition were significantly (P < 0.05) higher by 50% and 17%, respectively, whereas the biofilm formation of the flaAB deletion mutant was significantly (P < 0.05) lower by 48%. Under the starvation condition, the biofilm formation of spoT and recA deletion mutants was significantly (P < 0.05) higher by 149% and 72%, respectively, whereas the biofilm formation of flaAB deletion mutant was lower by 12%. The biofilm formation of the complementary strains (i.e., the spoT, recA, and flaAB strains) was also tested under the optimal condition, and their biofilm formation was shown to have been restored to the same level as that seen with their wild-type counterpart (see Fig. S8 in the supplemental material). We believed that the complementation restored the capacity of biofilm formation of C. jejuni F38011 mutants. These complementary strains are believed to have the same response as their wild-type counterpart to environmental stress in terms of biofilm formation.
FIG 2.
Biofilm formation and release of extracellular DNA (eDNA) by wild-type C. jejuni F38011 and the corresponding spoT, recA, and flaAB deletion mutants under optimal, aerobic, and starvation conditions. (A) The level of biofilm formation was evaluated using crystal violet staining. The stained biofilm was released using 95% ethanol and determined by monitoring the value of OD595. (B to D) The concentration of eDNA during biofilm formation under optimal conditions (B), aerobic conditions (C), and starvation conditions (D) over 3 days was quantified using SYBR green I dye on the basis of a standard curve generated using a series of 10-fold dilutions of Lambda DNA from 80 μg/ml to 0.156 μg/ml.
Determination of chemical compositions of C. jejuni biofilms in a microfluidic “lab-on-a-chip” device.
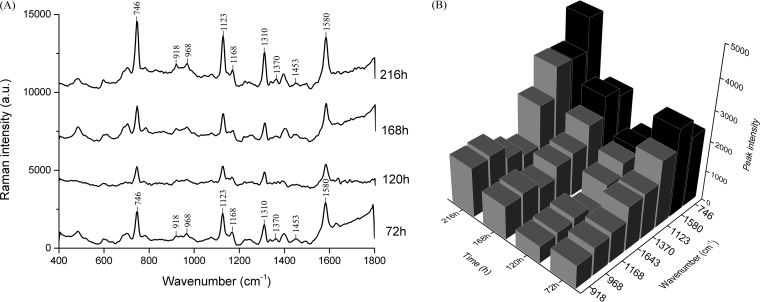
A microfluidic “lab-on-a-chip” device coupled with confocal micro-Raman spectroscopy was applied to characterize the chemical composition of the biofilm over a long period of cultivation (up to 216 h) in a continuous and nondestructive manner. The Raman spectral pattern of the C. jejuni F38011 biofilm was different from that of the microfluidic substrate (glass and polydimethylsiloxane [PDMS]). The microfluidic substrate generated intensive Raman scattering signals at 485, 610, 703, 785, 860, 1,250, and 1,402 cm−1 (Fig. S2). After the initial attachment, the C. jejuni F38011 biofilm progressively accumulated and covered the substrate. Meanwhile, the intensity of Raman peaks derived from the substrate substantially decreased. Only the substrate peaks at 485, 610, 703, and 1,402 cm−1 could still be detected at low levels. In contrast, Raman peaks derived from the biofilm appeared at different wavenumbers and demonstrated an intensity higher than that seen with the substrate.
The characteristic Raman spectrum of the C. jejuni F38011 biofilm showed prominent peaks at 746, 918, 968, 1,123, 1,168, 1,310, and 1,580 cm−1 after 72 h of cultivation in the microfluidic chip (Fig. 3A). Peaks at 746 and 1,580 cm−1 were assigned to thymine ring and pyrimidine ring structures of nucleic acids, while peaks at 918, 968, 1,123, 1,168, and 1,310 cm−1 were derived from proline ring, lipid representative band, C—N stretching vibration, C=C vibration, and the CH3/CH2 twisting or bending mode of lipids, respectively (see Table S3 in the supplemental material). From 72 h to 168 h, the intensity of the prominent Raman peaks (i.e., 746, 918, 968, 1,123, 1,168, 1,310, and 1,580 cm−1) significantly (P < 0.05) declined by ∼30% at 120 h and then rebounded at 168 h. Up to 216 h, the intensity of these peaks reached the highest levels, which was significantly (P < 0.05) higher by 15% than that seen at 72 h (Fig. 3B). According to Raman band assignment, nucleic acids, lipids, proteins, and polysaccharides were the major compositions of the C. jejuni F38011 biofilm. The amounts of these components changed along with the development of the biofilm (i.e., dispersion and regrowth), which was reflected by the change in the intensity of Raman peaks from biofilms. The Raman peaks assigned to nucleic acids demonstrated a significantly (P < 0.05) higher intensity than those derived from proteins, polysaccharides, and lipids, indicating that nucleic acids were the major components in a developed monospecies C. jejuni biofilm (Fig. 3B).
FIG 3.
Confocal micro-Raman spectroscopy monitors the development of C. jejuni F38011 biofilm in the microfluidic “lab-on-a-chip” platform. C. jejuni F38011 biofilm was cultivated in a microfluidic device, and the chemical component was determined at 72 h, 120 h, 168 h, and 216 h using confocal micro-Raman spectroscopy coupled with a 532-nm-wavelength laser. (A) Prominent Raman peaks during biofilm formation. (B) Variations in the intensity of the corresponding Raman peaks (746, 918, 968, 1,123, 1,168, 1,370, 1,580, and 1,643 cm−1) over time. The Raman peaks derived from nucleic acid components (746 and 1,580 cm−1) are shown in black, and the Raman peaks derived from other components (i.e., proteins, lipids, and polysaccharides) are shown in gray.
Accumulation of eDNA occurs together with biofilm development.
Since eDNA was identified as the major component of monospecies C. jejuni biofilm, we hypothesized that the release of eDNA might be closely associated with the development of C. jejuni biofilm. Accumulation of eDNA during biofilm formation was quantified using specific fluorescent double-stranded DNA stain SYBR green I. The released eDNA could be detected from all C. jejuni strains, including wild-type and mutant strains, since the first day of biofilm formation (Fig. 1B, C, and D and 2B, C, and D). However, the concentrations and accumulation patterns of eDNA varied among different strains and were influenced by the environmental condition as well.
Under the optimal condition, C. jejuni 81-116 and ATCC 33560 produced the largest amounts of eDNA among all wild-type strains and their eDNA concentrations reached 13.3 and 23.7 μg/ml in a 3-day developed biofilm, respectively (Fig. 1B). The C. jejuni F38011 spoT deletion mutant produced the most eDNA among all the mutant strains, and its eDNA concentration reached 13.7 μg/ml in a 3-day developed biofilm (Fig. 2B). During the biofilm formation of C. jejuni 81-116 and ATCC 33560, eDNA progressively accumulated at from ∼4 μg/ml to ∼23 μg/ml, while the eDNA concentration in the biofilms of C. jejuni human 10, 87-95, NCTC 11168, 1658, and F38011 remained relatively constant at below 10 μg/ml (Fig. 1B). For C. jejuni F38011 mutant strains, spoT and recA deletion mutants released amounts of eDNA similar to those seen with their wild-type counterparts at day 1 and day 2 of biofilm formation, but the level of released DNA was significantly increased at day 3. Specifically, the concentration of eDNA reached 13.7 μg/ml for the spoT deletion mutant and 8.89 μg/ml for the recA deletion mutant at day 3. In comparison, the flaAB deletion mutant shared an eDNA release pattern similar to that seen with its wild-type counterpart (Fig. 2B).
The release of eDNA was stimulated in a well-developed (3-day) biofilm under the aerobic condition (Fig. 1C and 2C). For C. jejuni wild-type strains, the level of release of eDNA in C. jejuni human 10, ATCC 33560, F38011, and 1658 was significantly higher (P < 0.05) than that seen under the optimal condition and their eDNA concentrations reached 18.1, 28.7, 18.1, and 14.1 μg/ml, respectively. In contrast, the eDNA concentration of C. jejuni 87-95 was not influenced by the aerobic condition and remained constant at the same low level as that seen under the optimal condition (∼3.5 μg/ml) during biofilm formation. In addition, C. jejuni NCTC 11168 demonstrated a distinct eDNA release pattern in that its eDNA concentration progressively decreased from 13.4 μg/ml at day 1 to 5.5 μg/ml at day 3. For the mutant strains, both the C. jejuni F38011 spoT and recA deletion mutants produced a significantly (P < 0.05) larger amount of eDNA under the aerobic condition than under the optimal condition and their concentrations reached 29.6 and 20.5 μg/ml, respectively, in a 3-day developed biofilm. In contrast, the C. jejuni F38011 flaAB deletion mutant produced less eDNA under the aerobic condition than under the optimal condition.
The starvation condition significantly (P < 0.01) inhibited the release of eDNA for all C. jejuni strains, including the wild-type strains and the deletion mutants, such that the eDNA concentration was less than 8 μg/ml (Fig. 1D and 2D).
Taking the results together, a synchronous relationship between eDNA accumulation and biofilm formation was observed. Massive accumulation of eDNA usually occurred along with a high level of C. jejuni biofilm formation. In addition, a threshold eDNA concentration ranging from 10 to 20 μg/ml was seen to be associated with a high level of biofilm formation.
Source of eDNA during biofilm formation.
Bacterial lysis can release DNA into the environment. Therefore, we speculated that bacterial lysis is responsible for the accumulation of DNA during biofilm formation. The autolysis capacity of C. jejuni F38011 was tested using a 0.02% Triton X-100 autolysis solution, and the rate of the reduction of optical density at 600 nm (OD600) was recorded over time (Fig. S4). The autolysis capacity of Salmonella was used as the reference. Salmonella enterica serovar Typhimurium SL1344 could persist in a Triton X-100 solution for over 90 min, and only 10% of the total population was lysed. In contrast, the C. jejuni F38011 wild-type strain was extremely vulnerable to Triton X-100-induced autolysis pressure such that over 55% of the C. jejuni population was lysed within 70 min. The autolysis capacity of C. jejuni F38011 mutants (i.e., the spoT, recA, and flaAB mutant strains) was at a level similar to that of the wild-type counterpart.
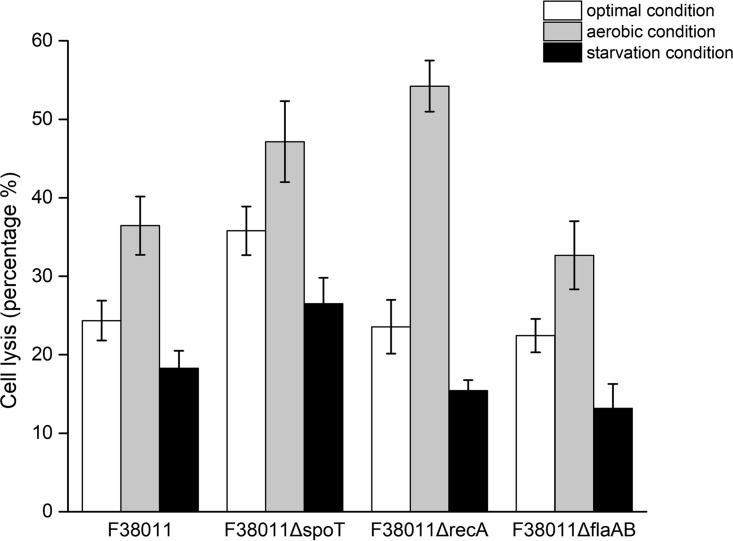
The release of genomic DNA was an important indicator of bacterial lysis. Using a DNA gel electrophoresis assay, the DNA collected from C. jejuni F38011 biofilm showed the same length as the genomic DNA extracted from C. jejuni F38011 (Fig. S5). We further evaluated the relative rates of lysis of C. jejuni cells under various conditions using quantitative PCR (qPCR) (Fig. 4). Under the optimal condition, the C. jejuni F38011 wild-type strains and the recA and flaAB deletion mutants shared similar lysis levels such that approximately 20% to 25% of the total population was lysed in a 3-day biofilm. Further, the spoT deletion mutant showed high lysis levels such that 35.8% of the population was lysed in a 3-day biofilm. Compared to the optimal condition, the aerobic condition significantly (P < 0.05) stimulated bacterial lysis. Among these strains, the lysis levels of the recA deletion mutant showed the greatest increase (from 23.6% [optimal condition] to 54.2% [aerobic condition]). Under the starvation condition, bacterial lysis was significantly (P < 0.05) inhibited. Although the spoT deletion mutant maintained a relatively high lysis level that accounted for 26.5% of the total population, the lysis levels were still lower than that seen under the optimal condition, which was 35.7%. Other C. jejuni F38011 strains (i.e., the wild-type strain and the recA and flaAB mutant strains), which accounted for ∼15% of the total population, shared similarly low levels of lysis.
FIG 4.
Lysis level of C. jejuni cells was stimulated by aerobic conditions and inhibited by starvation conditions during biofilm formation. The presence of genomic DNA is an indicator of bacterial lysis. After 3 days of biofilm cultivation, the genomic DNA in the supernatant and biofilm of the C. jejuni wild-type strain and spoT, recA, and flaAB deletion mutant strains was purified. The relative levels of content of genomic DNA in the supernatant and biofilm were individually determined via real-time quantitative PCR (qPCR) using the rpoA housekeeping gene. The lysis level was calculated by dividing the value corresponding to the genomic DNA content in the supernatant by the sum of the values corresponding to the genomic DNA content in the supernatant and in the biofilm.
Interestingly, the flaAB deletion mutant had a lysis level similar to that of its parental counterpart but released less eDNA. We therefore speculated that there exists a flaA and flaB gene-dependent mechanism to regulate the release of eDNA. Accordingly, the expression profiles of flaA and flaB were determined using qPCR and arbitrary fold change cutoff values were set at more than 2 (Fig. S6). Under the aerobic condition, both the flaA and flaB genes were significantly (>2-fold and P < 0.05) upregulated among the C. jejuni F38011 wild-type (2.8-fold for flaA and 2.2-fold for flaB) and spoT (3.2-fold for flaA and 4.1-fold for flaB) and recA (5.3-fold for flaA and 3.0-fold for flaB) deletion mutant strains, but the upregulation could be detected only at the first day of biofilm formation. However, the expression profiles of both the flaA and flaB genes were at the same levels among the wild-type and spoT and recA deletion mutant strains under the starvation condition as under the optimal condition, indicating that the flaA and flaB genes might play a role under the oxidative stress condition rather than the starvation stress condition.
Addition of DNA stimulates biofilm formation.
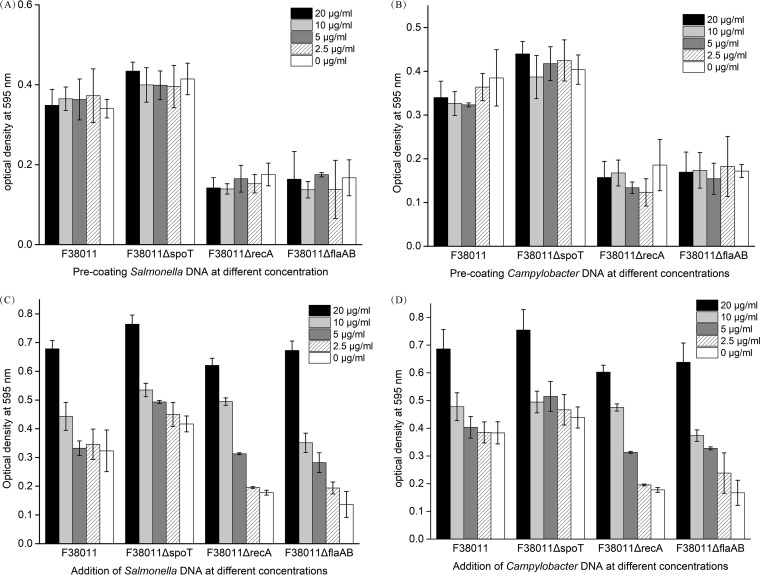
We identified a close correlation between the release of eDNA and biofilm formation (Fig. 1 and 2), but the consequence was not clear. A DNA addition assay was conducted to investigate whether eDNA was the inducing factor or a by-product of biofilm formation. In the natural environment, bacterial cells usually inhabit in a multispecies bacterial community. Therefore, DNA released by other bacterial species may also contribute to biofilm formation. In the current study, genomic DNA of C. jejuni F38011 and genomic DNA of S. Typhimurium SL 1344 were supplemented either by direct addition into the bacterial culture or by forming a precoating layer for the subsequent biofilm formation in the 96-well plate. Direct addition of DNA demonstrated a concentration-dependent stimulation effect on C. jejuni biofilm formation, and this effect could be triggered not only by C. jejuni DNA but also by Salmonella DNA (Fig. 5C and D). For the C. jejuni F38011 wild-type strain, the stimulation effect on biofilm formation was triggered when the concentration of the added DNA reached 10 μg/ml. For the spoT deletion mutant, the stimulation effect was observed when the concentration of the added DNA reached 20 μg/ml. For the recA deletion mutant, the stimulation effect started at 5 μg/ml and addition of 10 μg/ml DNA was found to restore the biofilm formation to the level seen with the wild-type strain. For the flaAB deletion mutant, the stimulation effect started at the concentration of 2.5 μg/ml and addition of 10 μg/ml DNA restored biofilm formation to the level seen with the wild-type strain. When the concentration of the added DNA reached 20 μg/ml, biofilm formation of the C. jejuni F38011 wild-type and mutant strains was promoted to similarly high levels, indicating that 20 μg/ml was close to the saturation concentration of DNA that could maximize the biofilm formation in the environment.
FIG 5.
Addition of genomic DNA extracted either from Campylobacter or Salmonella had a concentration-dependent stimulation effect on biofilm formation of C. jejuni F38011; the precoating layer formed by DNA extracted either from Campylobacter or Salmonella did not contribute to the development of biofilm formation of C. jejuni F38011. Genomic DNA of C. jejuni F38011 or S. Typhimurium SL1344 was separately extracted and added for biofilm formation. (A and B) To form a precoating layer, 200 μl of DNA of Salmonella (A) or C. jejuni (B) at different concentrations was added into each well of the 96-well plate and maintained for 4 h. The unbounded DNA was washed out before the addition of C. jejuni F38011 culture. (C and D) To directly add DNA for biofilm formation, DNA of Salmonella (C) or C. jejuni (D) was mixed with C. jejuni F38011 culture to reach a certain final concentration and 200 μl of this mixed culture was added into the 96-well plate. The plate was then cultivated in a microaerobic environment at 37°C for up to 72 h.
In contrast, formation of a precoated layer did not enhance biofilm formation regardless of the DNA concentration (Fig. 5A and B). The biofilm formation levels of the C. jejuni F38011 wild-type strain and the spoT, recA, and flaAB mutants on a precoated DNA layer were the same as those seen on the untreated substrate. Thus, eDNA can be regarded as a leading factor for biofilm formation and eDNA derived from Salmonella can also stimulate the formation of monospecies C. jejuni biofilm. Although the initial attachment was the prerequisite for biofilm formation, the precoated layer of DNA had little influence on biofilm development.
DNase I treatment prevents biofilm formation and disrupts biofilm structure.
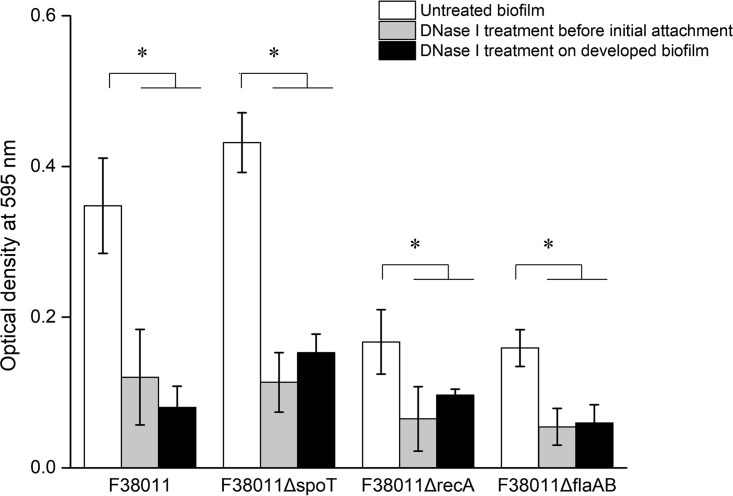
DNase I treatment was performed to further elucidate the role of eDNA in biofilm formation. The treatment was conducted at two distinct stages, namely, the initial attachment stage and the maturation stage (Fig. 6). Treatment at the initial attachment stage significantly (P < 0.05) prevented biofilm formation. Compared to the results seen with the untreated group, DNase I treatment reduced the biofilm formation levels of the C. jejuni F38011 wild-type strain (65%), spoT deletion mutant (74%), recA deletion mutant (61%), and flaAB deletion mutant (66%). DNase I treatment at the maturation stage significantly (P < 0.05) disrupted the well-developed biofilm. The treatment reduced the biofilm biomass of the C. jejuni F38011 wild-type strain (77%), spoT deletion mutant (64%), recA deletion mutant (43%), and flaAB deletion mutant (63%).
FIG 6.
DNase I treatment before bacterial initial attachment prevented C. jejuni biofilm formation and DNase I treatment of the well-developed C. jejuni biofilm disrupted biofilm structure, leading to the reduction of biomass. To treat biofilm at the initial attachment stage, DNase I was mixed with C. jejuni culture to reach a final concentration of 2 U/ml and then added into the 96-well plate for biofilm formation. To treat the well-developed biofilm in the 96-well plate, 200 μl of DNase I solution (2 U/ml) was added into a well with a 3-day cultivated C. jejuni biofilm. The treatment was maintained for 15 min at room temperature. The reduction level of biofilm was evaluated using the aforementioned crystal violet staining assay. Asterisks denote significant difference (P < 0.05).
Atomic force microscopy (AFM) was applied to further investigate the effect of DNase I treatment on C. jejuni F38011 biofilm. The topographic images of the treated and untreated biofilms were collected in contact scanning mode. Trace and retrace images were well matched (data not shown), indicating that the AFM tips in the contact mode did not scratch the biofilm surfaces and that the data corresponding to the morphological properties derived from AFM characterization were highly reliable. The topographic image of C. jejuni F38011 biofilm demonstrated a compact structure and smooth surface. On the top surface of the biofilm, protruding cell-shaped patterns with a size of 1.5 × 0.2 μm2 were observed. These cell-shaped patterns were squeezed and highly organized (Fig. S7A and B). DNase I treatment resulted in morphological variations where the top surface of the biofilm was altered from smooth to rough and from compact to loose. Cell-shaped patterns could still be observed, but the shape was concave after the treatment (Fig. S7C and D).
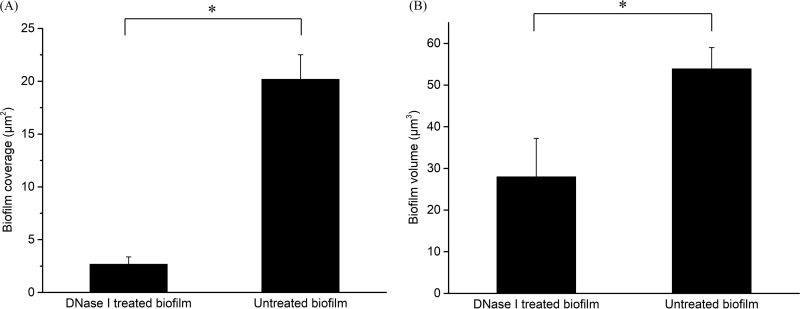
The volume and coverage area of the biofilms were also determined on the basis of a three-dimensional (3D) reconstruction of biofilm topographic images. As shown in Fig. 7, DNase I treatment significantly (P < 0.05) disrupted the biofilm such that its volume was reduced from 53.93 μm3 to 28 μm3 and the coverage area of the biofilm was reduced from 20.19 μm2 to 2.68 μm2. Taken together, these findings demonstrated the role of eDNA, which not only facilitated the initial attachment but also maintained the integrity of the biofilm structure, in biofilm formation.
FIG 7.
DNase I treatment reduced the coverage and volume of C. jejuni biofilm. The well-developed (3-day) C. jejuni F38011 biofilm on a nitrocellulose membrane was treated with DNase I solution (2 U/ml) for 15 min and then air-dried for the analysis of atomic force microscopy. (A) Coverage reduction caused by DNase I treatment. (B) Volume reduction caused by DNase I treatment. Asterisks denote significant difference (P < 0.05).
DNA allocates C. jejuni and Salmonella cells in a dual-species biofilm.
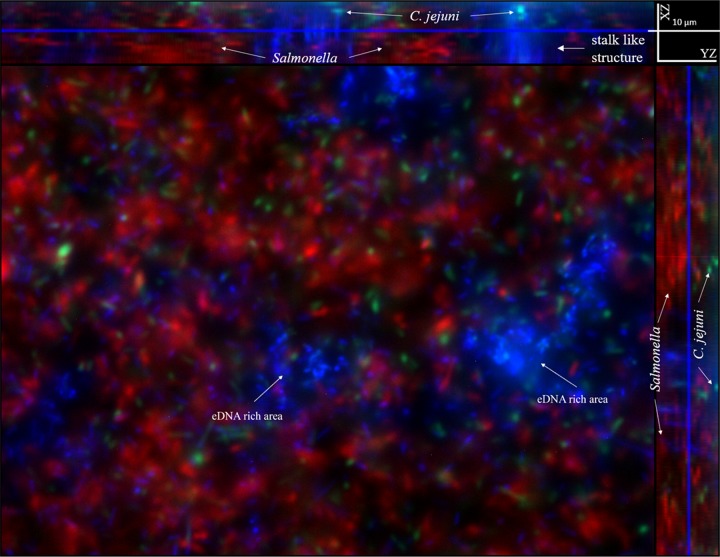
Multispecies biofilm is the dominant type in the natural environment (19). In the current study, Campylobacter-Salmonella biofilm was used as a representative model to investigate the role of eDNA in a dual-species biofilm. A green fluorescent protein (GFP)-tagged C. jejuni F38011 strain was mixed with a red fluorescent protein (RFP)-tagged S. Typhimurium SL 1344 strain for biofilm formation using the microfluidic “lab-on-a-chip” platform. After a 3-day cultivation, the biofilm was stained using DAPI (4′,6-diamidino-2-phenylindole), followed by imaging (Fig. 8). Campylobacter was seen to coexist with Salmonella and to form a dual-species biofilm. Within this dual-species biofilm, C. jejuni cells were mainly located at the bottom layer whereas S. Typhimurium cells were mainly located at the top layer. A large amount of eDNA (shown in blue) was identified in this dual-species biofilm and mainly assembled as eDNA-rich areas, although discretely distributed eDNA could still be observed.
FIG 8.
Spatial distribution of C. jejuni cells, Salmonella cells, and extracellular DNA (eDNA) within a Campylobacter-Salmonella dual-species biofilm formed in the microfluidic platform. Fluorescence microscopy was applied to determine the spatial distribution of eDNA and bacterial cells in a dual-species Campylobacter-Salmonella biofilm. After a 3-day cultivation, 30 nM DAPI solution was injected into the microfluidic device to stain eDNA in the biofilm. Images were collected at multiple channels: 405 nm (blue for DAPI signal), 488 nm (green for GFP signal), and 543 nm (red for RFP signal). Within this Campylobacter-Salmonella dual-species biofilm, C. jejuni cells were mainly located at the bottom whereas Salmonella cells were mainly located at the top layer. The eDNA mainly assembled and formed several eDNA-rich areas and maintained a spatial distance between C. jejuni and Salmonella in the biofilm.
The view from a spatial perspective demonstrated that an eDNA-rich area formed a stalk-like structure and occupied a large space. C. jejuni cells at the bottom layer were usually associated with eDNA-rich areas and covered by the stalk-like structure. In contrast, Salmonella cells did not directly interact with eDNA but were distributed throughout the eDNA-rich structure (see Video S1 in the supplemental material). Therefore, we speculated that eDNA was responsible for the positioning of different species of bacteria by maintaining a spatial distance between them.
DISCUSSION
C. jejuni is a fastidious microaerobic bacterium and is extremely vulnerable to environmental stress. However, this microbe is highly prevalent in the environment and is difficult to remove from the food chain. Campylobacteriosis is still the most frequently reported foodborne illness in Canada, with the reported cases outnumbering those of Salmonella, Listeria monocytogenes, and Shiga-toxigenic Escherichia coli infections (20). C. jejuni can antagonize stress by activating an internal stress response system. Previous studies demonstrated that the stringent response mediated global regulation of bacterial metabolism in reaction to stress, especially that represented by nutrient starvation. The spoT gene in C. jejuni 81-176 has been reported to be essential for stringent response. Deletion of this gene inhibited the expression of the genes related to redox balance, metabolism, energy production, and conversion pathways (11). Oxidative stress is a lethal challenge for microaerophilic bacteria because it can induce DNA damage and have a subsequent negative impact on the microbes (21, 22). A 38-kDa protein encoded by the recA gene was shown to mediate both DNA repair and homologous recombination, aiding C. jejuni in maintaining physiological activities when it encountered oxidative stress-induced DNA damage. Disruption of the recA gene impaired the viability of C. jejuni in a DNA damage-induced environment (23). However, bacterial (e.g., C. jejuni) cells can withstand only a limited level of stress. As a self-produced bacterial community, biofilm demonstrates unique physiological properties, such as tolerance of dehydration, persistence under harsh conditions, and resistance to antibiotic treatment, all of which provide remarkable protection to the encased bacterial cells (24). Apart from the inner stress response system, C. jejuni was also shown to survive by forming a biofilm or by residing in a mature biofilm for protection against the unfavorable condition. Our previous study showed that C. jejuni could survive an extended period of oxidative stress by forming a biofilm. After a 3-day exposure to atmosphere conditions, no viable C. jejuni cells could be detected from planktonic culture, but ∼5 log CFU/cm2 of C. jejuni cells in the biofilm was still culturable (9). Although biofilm formation of C. jejuni has been defined under different conditions, the correlation between the stress response system and biofilm formation of C. jejuni has been investigated only partially (6, 10). In the current study, we further investigated the influence of the stress response of C. jejuni on biofilm formation.
Biofilm formation and stress.
Most of the C. jejuni wild-type strains shared similar biofilm-forming capabilities in response to stress. In short, these strains produced more biofilm under the aerobic condition and less biofilm under the starvation condition (Fig. 1A). This observation was in agreement with a previous study that showed that biofilm formation of C. jejuni increased under oxygen-enriched conditions (10). In addition, another study also reported that the biofilm formation of C. jejuni under nutrient-limited conditions (e.g., Brucella or Bolton medium) was inhibited due to the slow growth of C. jejuni cells (6).
Biofilm formation of the C. jejuni F38011 spoT deletion mutant was increased by 26% compared to that seen with its parental counterpart under optimal conditions (Fig. 2A), which was consistent with a previous study that showed that mutation of the spoT gene resulted in an biofilm formation-increased phenotype in C. jejuni 81-176 (25). In contrast, the levels of biofilm formation of the recA deletion mutant and the flaAB deletion mutant were reduced by ∼52% and 55%, respectively, compared to those seen with their parental counterparts under the optimal condition. Our study demonstrated that deletion of recA inhibited C. jejuni biofilm formation. Flagella are known to be responsible for bacterial attachment onto the surface of a substrate. The loss of flagellar apparatus heavily impairs biofilm formation (6, 26–28). Consistent with those studies, the deletion of both flaA and flaB genes in C. jejuni F38011 resulted in a loss of motility as well in reduced biofilm formation (Fig. 2A; see also Fig. S3 in the supplemental material). Compared to a C. jejuni F38011 flagellum deletion mutant (i.e., flaAB) strain, C. jejuni F38011 stress response deletion mutants (i.e., spoT and recA) demonstrated a distinctive response to environmental stress in terms of biofilm formation. Compared to the wild-type strain results, the biofilm formation of the spoT and recA deletion mutants seen under the starvation condition significantly increased by 149% and 72%, respectively, whereas the change in biofilm formation of the flaAB deletion mutant was not significant. Therefore, we believe that the stress response is one of the critical factors affecting biofilm formation of C. jejuni and that the effect is greater than that of the flagellum under specific stress conditions.
eDNA and biofilms.
Stress response systems mainly regulate the physiological metabolism inside bacterial cells, whereas biofilm formation occurs outside bacterial cells. There is likely a factor that can mediate this inside-outside transition. DNA is present in abundance in the environment as a consequence of the presence of lysed dead organisms or of active secretion of the living organisms (14). Bacteria can utilize this free DNA as an important supply of nutrients or can integrate this free DNA into the genome for acquisition of resistant capacity (15, 16). Previous studies showed that bacteria progressively released DNA during biofilm formation. For example, the DNA content in a 3-day P. aeruginosa biofilm reached 20 to 25 μg/ml and the concentration of DNA was up to 220 μg/mg (eDNA/cellular DNA) when the biofilm cultivation was extended to 5 days (18, 29).
Our current study also identified the presence of eDNA during biofilm formation of C. jejuni. According to Raman spectroscopic analysis, the level of eDNA content in the biofilm was higher than the levels of other biofilm components (i.e., proteins, lipids, and polysaccharides). In addition, the content of eDNA would increase or decrease along with the growth or dispersion of the biofilm, respectively (Fig. 3A and B), demonstrating that eDNA was an important constitutional component of C. jejuni biofilm.
Bacterial lysis is regarded as the most important source for the release of eDNA (30, 31). As shown in Fig. S4 and S5, C. jejuni F38011 was vulnerable to the induced lysis pressure and the length of the released eDNA fragment was equal to the length of genomic DNA extracted from C. jejuni F38011. Taking the data together, there was a high possibility that bacterial lysis contributed to the release of eDNA during the formation of C. jejuni biofilm. However, the results of the autolysis assay could not fully explain the release of genomic DNA during the formation of biofilm of C. jejuni F38011 wild-type and deletion mutant strains in response to different stresses. Because Triton X-100 induced a much stronger lysis pressure than environmental stress, the autolysis assay results reflected only the tendency of bacterial lysis and not the precise proportion of the lysed C. jejuni cells during biofilm formation. Accordingly, qPCR was performed to evaluate the lysis level of C. jejuni F38011 biofilms under different environmental conditions (Fig. 4). Specifically, the aerobic condition significantly (P < 0.05) increased the lysis rate by over 32% and the starvation condition significantly (P < 0.05) decreased the lysis rate by over 25%. The recA deletion mutant was vulnerable to the aerobic condition, with 54% of the total population lysed in a 3-day biofilm. Aerobic conditions can generate oxidative pressure and induce DNA damage (22). The high rate of lysis of the recA deletion mutant was highly likely due to the lack of function of the DNA repair system.
The quantitative analysis of released eDNA revealed a synchronous relationship with biofilm formation in response to the environmental stress (Fig. 1 and 2). For example, the levels of eDNA release and biofilm formation increased simultaneously under the aerobic condition. However, C. jejuni ATCC 33560 was an exception. Although over 10 μg/ml of eDNA was detected from the biofilm of C. jejuni ATCC 33560 under the aerobic condition, its biofilm formation levels were significantly lower (P < 0.05) than those of other C. jejuni strains, such as C. jejuni F38011 and NCTC 11168, whose eDNA production was ∼10 μg/ml. According to a previous study, C. jejuni ATCC 33560 contains a single-nucleotide deletion that might lead to the nonfunction of CmeR (32). CmeR has been well characterized as a transcriptional repressor of the efflux pump of CmeABC (33) and is also involved in various forms of metabolic regulation, such as control of membrane transporters and biosynthesis of periplasmic capsule (34). Therefore, C. jejuni ATCC 33560 is more vulnerable and easily lysed under stress conditions, resulting in an increase of released eDNA. However, the heavy loss of viable cells reduces the initial attachment and subsequently impairs the biofilm formation. This might be responsible for the low levels of biofilm formation of C. jejuni ATCC 33560 seen under different stress conditions.
Mutations in the stress response system (spoT and recA) enhanced the impact of stress on eDNA secretion and biofilm formation, which was in agreement with the cell lysis results (Fig. 4). Although the deletion mutation of the genes corresponding to the flagellar apparatus (flaA and flaB) did not directly affect the stress response capacity of C. jejuni, both motility and chemotaxis can be impaired (35). Hence, the lysis level of flaAB deletion mutant was also increased due to the accumulation of redox pressure induced by the aerobic condition. We believe that the bacterial lysis was the consequence of the stress response, which eventually releases the DNA into the environment. The accumulation of eDNA then facilitated the development of biofilm as the constitutional material. To our surprise, not only the DNA from Campylobacter but also the DNA from Salmonella could mediate this process. DNase I treatment and the corresponding characterization further confirmed the role of eDNA in biofilm formation. Thus, eDNA was essential to the initial attachment because DNase I treatment at the initial attachment stage almost eliminated biofilm formation. In addition, DNase I treatment of the well-developed biofilm disrupted over 70% of the biomass, confirming that eDNA was the major constitutional component in a developed C. jejuni biofilm (Fig. 6). AFM analysis revealed that the degradation of eDNA in the biofilm disrupted the biofilm structure and resulted in an ∼87% reduction of biofilm coverage and ∼48% reduction of biofilm volume (Fig. 7 and S7). Taking the results together, eDNA was shown to be involved in biofilm formation by performing multiple functions. At the initial stage, eDNA facilitated bacterial initial attachment and established the basis for biofilm formation. At the developing stage, eDNA was included in the biofilm as the major constitutional material that built the biofilm structure. In a developed biofilm, eDNA was responsible for maintaining the biofilm structure.
Flagella and the release of eDNA.
Although the flaAB deletion mutant showed a lysis rate similar to that of its wild-type counterpart under the environmental stress condition, its levels of DNA release and biofilm formation were significantly (P < 0.05) lower (Fig. 2). Previous studies reported that both eDNA and flagella were responsible for bacterial attachment and biofilm formation. DNA could facilitate the attachment via an acid-base interaction and flagellum-mediated autoagglutination and immobilize bacterial cells on a surface (36–40). The deletion mutation of flagellum synthesis genes flaA and flaB resulted in a biofilm-repressed phenotype (6, 10). We believed that the low level of biofilm formation of the flaAB mutant was due to both a deficiency of eDNA and a lack of flagella, but this cannot explain why the flaAB deletion mutant released a small amount of eDNA. Interestingly, it was reported that expression of flagellin A (flaA) and flagellin B (flaB) was activated since biofilm formation and that their expression level was significantly higher in the biofilms than in the planktonic cells (27). Hence, we hypothesized that there might be a correlation between the synthesis of flagellin and eDNA release. However, the outcome of the qPCR assay did not completely support this hypothesis. Aerobic conditions upregulated the expression levels of flaA and flaB genes in C. jejuni F38011 wild-type and spoT and recA deletion mutant strains, but upregulation was shown only at the first day of biofilm formation. In contrast, the expression profiles of flaA and flaB under the starvation condition were maintained at levels similar to those seen under the optimal condition during biofilm formation. Taking the data together, although the upregulation of the flaA and flaB genes occurred along with the increase of eDNA release under the aerobic stress condition, the downregulation of the flaA and flaB genes was not observed under the starvation condition where eDNA release was inhibited. Therefore, the correlation between flagella and DNA release might be more specific to aerobic stress.
Spatial distribution in dual-species biofilms.
In the natural environment, Campylobacter bacteria are frequently isolated from environments where Salmonella bacteria also appear, such as poultry farm and sewage water (41, 42). Our previous study demonstrated that C. jejuni survived a longer period of time in a Salmonella-C. jejuni dual-species biofilm than in a monospecies C. jejuni biofilm under the aerobic condition (9). In the current study, we found that the distinct spatial distribution of bacterial cells might be responsible for this survival advantage in a dual-species biofilm and that eDNA played a role in maintaining spatial distributions by allocating different species of bacterial cells to different locations (Fig. 8; see also Video S1 in the supplemental material). In a well-developed dual-species biofilm, released eDNA was shown to assemble and form a stalk-like structure. At the bottom layer of the biofilm, eDNA tended to interact with C. jejuni cells and form a cover structure on top of C. jejuni cells. In contrast, the stalk-like eDNA structure tended to repel Salmonella cells and suspend Salmonella cells at the top layer of the biofilm. Therefore, C. jejuni was allocated at the bottom layer of biofilm away from the aerobic conditions and the top space was occupied by eDNA and Salmonella cells that further limited the penetration of oxygen. In the natural environment, the penetration of oxygen would be limited by the presence of this unique structure, which facilitates the survival of C. jejuni cells under the aerobic condition.
In this study, a comprehensive investigation of C. jejuni biofilm formation in response to the environmental stress was conducted. Oxidative stress and starvation stress could significantly influence the biofilm formation of a broad range of C. jejuni isolates, and a synchronous relationship was observed between biofilm formation and eDNA released from bacterial lysis. We propose that environmental stress may induce a high rate of bacterial lysis and that the DNA subsequently released from the dead bacterial cells promotes biofilm formation. This report reveals the essential role and multiple functions of eDNA in the biofilm formation of C. jejuni and provides insights into understanding their molecular mechanisms. This knowledge can aid in developing intervention strategies to limit the prevalence and distribution of C. jejuni in the environment.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
The bacterial strains and plasmids used in the current study are summarized in Table S1 in the supplemental material. Routine cultivation of C. jejuni was conducted either on Mueller-Hinton (MH) agar supplemented with 5% defibrinated sheep blood (MHB agar) or in MH broth with constant shaking at 37°C under microaerobic conditions (85% N2, 10% CO2, 5% O2). Salmonella and E. coli strains were cultivated in Luria-Bertani (LB) broth (BD Difco) at 37°C under aerobic conditions. When necessary, streptomycin, ampicillin, kanamycin, chloramphenicol, or tetracycline was supplemented into MHB agar or MH broth or LB broth at a final concentration of 100 μg/ml, 100 μg/ml, 50 μg/ml, 8 μg/ml, or 10 μg/ml, respectively. The details of the construction of the C. jejuni F38011 ΔspoT and ΔrecA mutant strains and the C. jejuni F38011 spoT and recA complementary strains are summarized in the Materials and Methods section of the supplemental material.
Biofilm formation either in a 96-well plate or on a nitrocellulose membrane under different environmental conditions.
To cultivate biofilm in a sterile polystyrene 96-well plate in an optimal growth environment, an overnight culture of C. jejuni was collected, washed, and diluted to an OD600 of 0.003 (∼107 CFU/ml) in MH broth. A total of 0.2 ml of the diluted bacterial culture was added into each well of a sterile polystyrene 96-well plate. The 96-well plate was incubated in a microaerobic environment (85% N2, 10% CO2, 5% O2) at 37°C for up to 72 h. To cultivate biofilm in 96-well plate under starvation conditions, C. jejuni overnight culture was diluted to an OD600 of 0.003 in phosphate-buffered saline (PBS) (pH = ∼7.0 to 7.2). A total of 200 μl of the diluted bacterial culture was added into each well of a 96-well plate and incubated under microaerobic conditions at 37°C for up to 72 h. To cultivate biofilm in a 96-well plate under aerobic conditions, a C. jejuni overnight culture was washed and diluted to an OD600 of 0.003 in MH broth. A total of 200 μl of the diluted bacterial culture was added into each well of a 96-well plate. The plate was incubated in an aerobic environment (79% N2, 21% O2) at 37°C for up to 72 h.
To cultivate biofilm on the nitrocellulose membrane (Sartorius Stedim-type filters) (0.45-mm pore size, 47-mm diameter), an overnight culture of C. jejuni was washed and diluted to an OD600 of 0.003 in MH broth. A total of 100 μl of the diluted bacterial culture was deposited onto the membrane in a surface area of ∼3 cm2. The membrane was incubated on MHB agar in a microaerobic environment at 37°C and aseptically transferred to a fresh MHB agar plate every 24 h for up to 72 h.
Crystal violet biofilm assay.
A crystal violet staining assay was applied to quantify the level of formation of biofilms developed in a 96-well plate (43). The details are summarized in the Materials and Methods section of the supplemental material.
Fabrication of microfluidic “lab-on-a-chip” platform for biofilm formation.
A polydimethylsiloxane (PDMS)-based microfluidic device was fabricated using a soft lithographic technique (44). The details are summarized in the Materials and Methods section of the supplemental material. Overnight cultures of C. jejuni F38011 and Salmonella enterica serovar Typhimurium SL1344 were individually washed twice using PBS and diluted to an OD600 of 0.03 (∼108 CFU/ml) in MH broth. The C. jejuni F38011 culture was either individually introduced into the microfluidic chip to form a monospecies biofilm or mixed with S. Typhimurium SL1344 to form a dual-species biofilm. Bacterial culture in the microfluidic device was maintained for 2 h to allow initial bacterial attachment. Biofilm cultivation was conducted by continuously flowing MH broth at a rate of 0.0002 μl/min under aerobic conditions at room temperature.
Characterization of C. jejuni biofilm in a microfluidic platform using confocal micro-Raman spectroscopy.
A confocal micro-Raman spectroscopic system (Renishaw, Gloucestershire, United Kingdom) with a 532-nm-wavelength diode green laser was applied to characterize the chemical composition of the C. jejuni F38011 biofilm in the microfluidic device. A Raman laser was introduced into the cultivation chamber through the use of a 50× objective (Leica Biosystems, Wetzlar, Germany) and was focused onto the biofilm at a laser illumination power of 0.2 mW. Raman scattering signal was collected and dispersed by a diffraction grating and was then recorded using a 578-pixel-by-384-pixel charge-coupled-device (CCD) array detector. An integration time of 20 s was applied for spectral collection over a simultaneous Raman shift range of 400 to 1,800 cm−1. A Raman spectrometer was equipped with a 1,200-line/mm grating and controlled via WiRE software for spectral acquisition and processing (Renishaw, United Kingdom).
Quantification of the released eDNA.
The amount of eDNA released during biofilm formation was quantified using SYBR green I dye (Invitrogen) according to the manufacturer's protocol. Briefly, 200 μl of bacterial culture was removed from the 96-well plate and pelleted by centrifugation at 12,000 ×g for 2 min. The supernatant was collected for further analysis. SYBR green I dye was diluted 100 times using TE buffer (10 mM Tris HCl, 1 mM EDTA, pH 8) as the working solution. A total of 5 μl of the SYBR green I working solution was mixed with 95 μl of the collected supernatant in a well of a black 96-well plate (Greiner Bio-One), and the plate was incubated on an orbital shaker for 5 min. The fluorescence signal was recorded using a Tecan plate reader (Infinite 200 Pro; Tecan Life Sciences) with an excitation wavelength of 485 nm and an emission wavelength of 535 nm. The amount of eDNA was calculated via the use of a standard curve generated using Lambda DNA (Invitrogen) (using a series of 10 dilutions from 80 μg/ml to 0.156 μg/ml).
Gel electrophoresis of the released DNA.
Gel electrophoresis was performed to demonstrate the length of the released DNA fragment. After 3 days of biofilm cultivation, each bacterial culture in a 96-well plate was collected and centrifuged at 16,000 × g for 5 min. A total of 10 μl of the supernatant was mixed with 2 μl of DNA loading dye solution (FroggaBio), and then the mixture was loaded in a 1% agarose gel for electrophoresis. A 1-kb ladder (Invitrogen) was used as the reference. The DNA was stained using SYBR safe DNA gel stain (Life Technologies) according to the manufacturer's protocol, and the DNA band was visualized on a ChemiDoc XRS gel documentation system (Bio-Rad).
DNase I treatment of C. jejuni F38011 biofilm.
In order to determine the role of eDNA in biofilm formation, DNase I treatment was applied either at the initial bacterial attachment stage or to a well-developed biofilm. The treatment was conducted either in a 96-well plate or on a nitrocellulose membrane. To treat biofilm at the initial attachment stage, DNase I (Thermo Scientific DNase I) was diluted using DNase- and protease-free water and was added into the bacterial culture at a final concentration of 2 U/ml before biofilm formation. To treat the well-developed biofilm in a 96-well plate, the biofilm was removed and then 200 μl of DNase I solution (2 U/ml) was added. The treatment was maintained for 15 min at room temperature. The reduction of biofilm was evaluated using the aforementioned crystal violet assay. To treat the well-developed biofilm on a nitrocellulose membrane, the biofilm was immersed in DNase I solution (2 U/ml) for 15 min at room temperature and then washed with PBS. The treated biofilm was air-dried and then analyzed using atomic force microscopy.
Atomic force microscopy.
The morphological variation of the biofilm due to DNase I treatment was determined using a Cypher atomic force microscope (Bruker) (Innova high-resolution system) with TR400PB tip cantilevers (Bruker) (nominal spring constant: k = 0.02 N/m). The details are summarized in the Materials and Methods section of the supplemental material.
Addition of DNA for biofilm formation.
Genomic DNA of C. jejuni F38011 or S. Typhimurium SL1344 was individually extracted from the overnight bacterial culture using a Presto Mini gDNA Bacteria kit (FroggaBio) according to the manufacturer's protocol. Genomic DNA was quantified using a Nano Drop 2000 spectrophotometer (Thermo Scientific). Addition of DNA for biofilm formation was conducted in the following two manners. (i) To form a precoating layer, 200 μl of DNA solution was individually added at different concentrations (20, 10, 5, 2.5, and 0 μg/ml) into each well of the 96-well plate and maintained for 4 h. The unbounded DNA was then washed out using sterile PBS before bacterial inoculation. (ii) DNA was mixed with bacterial culture to reach a certain final concentration (20, 10, 5, 2.5, or 0 μg/ml), and 200 μl of this mixed culture was cultivated in a 96-well plate. The plate was then cultivated in a microaerobic environment at 37°C for up to 72 h.
Autolysis assay.
The autolysis assay was adapted from a previous study with further modifications (45). Briefly, autolysis buffer was prepared by diluting Triton X-100 with 0.05 M Tris-HCl to achieve a final concentration of 0.02% (vol/vol). Bacterial cells were harvested in the late exponential phase by centrifugation at 8,000 ×g for 5 min at 4°C, washed twice with chilled water, and resuspended in autolysis buffer to an OD600 of 0.3. The reduction of absorbance (OD600) was measured using a microplate reader (SpectraMax M2; Molecular Devices, Sunnyvale, CA, USA) every 3 min for a total of 90 min.
Motility test.
Motility of C. jejuni cells was assessed on the soft agar plates as described previously (27). Briefly, 5 μl of the overnight C. jejuni culture was spotted onto the Brucella media supplemented with 0.4% agar. The plate was then incubated under microaerobic conditions at 37°C for 2 days. The halo size of C. jejuni cells on the soft agar plate was measured. The results were compared to demonstrate the relative levels of motility among different C. jejuni strains.
Quantification of cell lysis.
Genomic DNA is an indicator of bacterial lysis. The cell lysis was quantified via real-time quantitative PCR (qPCR) as described in a previous study (46). Briefly, the C. jejuni F38011 wild-type strain and spoT, recA, and flaAB deletion mutant strains were individually inoculated into the 96-well plate for biofilm formation as described above. After 3 days of biofilm cultivation, the supernatant was collected. Biofilm was detached using 0.1% trypsin solution (Sigma), washed twice with PBS, and pelleted by centrifugation at 10,000 ×g for 2 min. The genomic DNA either in the supernatant or in the biofilm cells was purified using a PicoPure DNA extraction kit (Applied Biosystems). The qPCR was performed on an ABI Prism 7000 Fast instrument (Life Technologies) using a primer pair of a housekeeping gene, rpoA (47). The percentage of lysis was calculated by dividing the amount of genomic DNA in the supernatant by the sum of the amounts of genomic DNA in the supernatant and the encased cells in the biofilm.
Real-time qPCR analysis of gene expression.
The real-time qPCR was performed to plot the expression profiles of flaA and flaB in response to the aerobic and starvation conditions in the C. jejuni F38011 wild-type strain as well as the spoT and recA deletion mutant strains. The details are summarized in the Materials and Methods section of the supplemental material.
Fluorescence microscopy for biofilm structure analysis.
Fluorescence microscopy was applied to investigate the spatial distribution of eDNA and bacterial cells in a developed dual-species Campylobacter-Salmonella biofilm. The eDNA was stained using 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) according to the manufacturer's protocol. After a 3-day cultivation, the DAPI working solution was prepared as a 30 nM concentration in PBS and injected into the microfluidic device with a well-developed dual-species Campylobacter-Salmonella biofilm at a rate of 0.0002 μl/min. The microfluidic device was incubated at room temperature for 15 min and then rinsed with flowing PBS at a rate of 0.002 μl/min for 10 min. Images were collected using an Axiovert 200 microscope (Carl Zeiss) equipped with an Axiocam camera (Carl Zeiss) and the following multiple channels: 405 nm (blue for DAPI signal), 488 nm (green for GFP signal), and 543 nm (red for RFP signal). Analysis of the three-dimensional reconstruction and analysis of the spatial distribution were conducted using ImageJ software (National Institutes of Health, USA) and ZEN software (Zeiss, blue edition), respectively.
Statistical analysis.
All the experiments were performed in at least three biological replicates. Results were reported as the averages of replicates ± the standard deviations with significance (P < 0.05) determined by one-way analysis of variance (ANOVA).
Supplementary Material
ACKNOWLEDGMENTS
Financial support to X.L. in the form of a Discovery Grant from the National Sciences and Engineering Research Council of Canada (NSERC RGPIN-2014-05487) is gratefully acknowledged. J.F. received a 4-year Ph.D. fellowship from the China Scholarship Council.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02068-17.
REFERENCES
- 1.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Fux C, Costerton J, Stewart P, Stoodley P. 2005. Survival strategies of infectious biofilms. Trends Microbiol 13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Allos BM. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis 32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2016. Food Safety Report 2015. https://www.cdc.gov/foodnet/pdfs/foodnet-mmwr-progress-508-final.pdf.
- 5.Buswell CM, Herlihy YM, Lawrence LM, McGuiggan JT, Marsh PD, Keevil CW, Leach SA. 1998. Extended survival and persistence of Campylobacter spp. in water and aquatic biofilms and their detection by immunofluorescent-antibody and-rRNA staining. Appl Environ Microbiol 64:733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reeser RJ, Medler RT, Billington SJ, Jost BH, Joens LA. 2007. Characterization of Campylobacter jejuni biofilms under defined growth conditions. Appl Environ Microbiol 73:1908–1913. doi: 10.1128/AEM.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Feng J, Ma L, de la Fuente Núñez C, Gölz G, Lu X. 2017. Effects of meat juice on biofilm formation of Campylobacter and Salmonella. Int J Food Microbiol 253:20–28. doi: 10.1016/j.ijfoodmicro.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Hanning I, Jarquin R, Slavik M. 2008. Campylobacter jejuni as a secondary colonizer of poultry biofilms. J Appl Microbiol 105:1199–1208. doi: 10.1111/j.1365-2672.2008.03853.x. [DOI] [PubMed] [Google Scholar]
- 9.Feng J, Lamour G, Xue R, Mirvakliki MN, Hatzikiriakos SG, Xu J, Li H, Wang S, Lu X. 2016. Chemical, physical and morphological properties of bacterial biofilms affect survival of encased Campylobacter jejuni F38011 under aerobic stress. Int J Food Microbiol 238:172–182. doi: 10.1016/j.ijfoodmicro.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Reuter M, Mallett A, Pearson BM, van Vliet AH. 2010. Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl Environ Microbiol 76:2122–2128. doi: 10.1128/AEM.01878-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaynor EC, Wells DH, MacKichan JK, Falkow S. 2005. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol Microbiol 56:8–27. doi: 10.1111/j.1365-2958.2005.04525.x. [DOI] [PubMed] [Google Scholar]
- 12.Fields JA, Thompson SA. 2008. Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J Bacteriol 190:3411–3416. doi: 10.1128/JB.01928-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svensson SL, Pryjma M, Gaynor EC. 2014. Flagella-mediated adhesion and extracellular DNA release contribute to biofilm formation and stress tolerance of Campylobacter jejuni. PLoS One 9:e106063. doi: 10.1371/journal.pone.0106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen KM, Johnsen PJ, Bensasson D, Daffonchio D. 2007. Release and persistence of extracellular DNA in the environment. Environ Biosafety Res 6:37–53. doi: 10.1051/ebr:2007031. [DOI] [PubMed] [Google Scholar]
- 15.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 16.Lorenz MG, Wackernagel W. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev 58:563–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avrain L, Vernozy-Rozand C, Kempf I. 2004. Evidence for natural horizontal transfer of tetO gene between Campylobacter jejuni strains in chickens. J Appl Microbiol 97:134–140. doi: 10.1111/j.1365-2672.2004.02306.x. [DOI] [PubMed] [Google Scholar]
- 18.Das T, Manefield M. 2012. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS One 7:e46718. doi: 10.1371/journal.pone.0046718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elias S, Banin E. 2012. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol Rev 36:990–1004. doi: 10.1111/j.1574-6976.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- 20.Public Health Agency of Canada. 2017. Reported cases from 1924 to 2015 in Canada—notifiable diseases on-line. http://diseases.canada.ca/notifiable/charts?c=plt.
- 21.Cabiscol E, Tamarit J, Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3:3–8. [PubMed] [Google Scholar]
- 22.Storz G, Imlayt JA. 1999. Oxidative stress. Curr Opin Microbiol 2:188–194. doi: 10.1016/S1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 23.Gaasbeek EJ, van der Wal FJ, van Putten JP, de Boer P, van der Graaf-van Bloois L, de Boer AG, Vermaning BJ, Wagenaar JA. 2009. Functional characterization of excision repair and RecA-dependent recombinational DNA repair in Campylobacter jejuni. J Bacteriol 191:3785–3793. doi: 10.1128/JB.01817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623. [DOI] [PubMed] [Google Scholar]
- 25.McLennan MK, Ringoir DD, Frirdich E, Svensson SL, Wells DH, Jarrell H, Szymanski CM, Gaynor EC. 2008. Campylobacter jejuni biofilms up-regulated in the absence of the stringent response utilize a calcofluor white-reactive polysaccharide. J Bacteriol 190:1097–1107. doi: 10.1128/JB.00516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 27.Kalmokoff M, Lanthier P, Tremblay T-L, Foss M, Lau PC, Sanders G, Austin J, Kelly J, Szymanski CM. 2006. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J Bacteriol 188:4312–4320. doi: 10.1128/JB.01975-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joshua GP, Guthrie-Irons C, Karlyshev A, Wren B. 2006. Biofilm formation in Campylobacter jejuni. Microbiology 152:387–396. doi: 10.1099/mic.0.28358-0. [DOI] [PubMed] [Google Scholar]
- 29.Steinberger R, Holden P. 2005. Extracellular DNA in single-and multiple-species unsaturated biofilms. Appl Environ Microbiol 71:5404–5410. doi: 10.1128/AEM.71.9.5404-5410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A 104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol 59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 32.Hyytiäinen H, Hänninen M-L. 2012. Quality control strain Campylobacter jejuni ATCC 33560 contains a frameshift mutation in the CmeR regulator. Antimicrob Agents Chemother 56:1148–1148. doi: 10.1128/AAC.06228-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin J, Akiba M, Sahin O, Zhang Q. 2005. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob Agents Chemother 49:1067–1075. doi: 10.1128/AAC.49.3.1067-1075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo B, Wang Y, Shi F, Barton Y-W, Plummer P, Reynolds DL, Nettleton D, Grinnage-Pulley T, Lin J, Zhang Q. 2008. CmeR functions as a pleiotropic regulator and is required for optimal colonization of Campylobacter jejuni in vivo. J Bacteriol 190:1879–1890. doi: 10.1128/JB.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lertsethtakarn P, Ottemann KM, Hendrixson DR. 2011. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol 65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das T, Sharma PK, Busscher HJ, van der Mei HC, Krom BP. 2010. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl Environ Microbiol 76:3405–3408. doi: 10.1128/AEM.03119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie A, Bryner J, Foley J. 1983. Role of DNA and bacteriophage in Campylobacter auto-agglutination. J Med Microbiol 16:333–340. doi: 10.1099/00222615-16-3-333. [DOI] [PubMed] [Google Scholar]
- 38.Brown HL, Hanman K, Reuter M, Betts RP, Van Vliet AH. 2015. Campylobacter jejuni biofilms contain extracellular DNA and are sensitive to DNase I treatment. Front Microbiol 6:699. doi: 10.3389/fmicb.2015.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lillard HS. 1986. Role of fimbriae and flagella in the attachment of Salmonella typhimurium to poultry skin. J Food Sci 51:54–56. doi: 10.1111/j.1365-2621.1986.tb10834.x. [DOI] [Google Scholar]
- 40.Brown HL, Reuter M, Hanman K, Betts RP, van Vliet AH. 2015. Prevention of biofilm formation and removal of existing biofilms by extracellular DNases of Campylobacter jejuni. PLoS One 10:e0121680. doi: 10.1371/journal.pone.0121680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Craven S, Stern N, Line E, Bailey J, Cox N, Fedorka-Cray P. 2000. Determination of the incidence of Salmonella spp., Campylobacter jejuni, and Clostridium perfringens in wild birds near broiler chicken houses by sampling intestinal droppings. Avian Dis 44:715–720. [PubMed] [Google Scholar]
- 42.Slader J, Domingue G, Jørgensen F, McAlpine K, Owen R, Bolton F, Humphrey T. 2002. Impact of transport crate reuse and of catching and processing on Campylobacter and Salmonella contamination of broiler chickens. Appl Environ Microbiol 68:713–719. doi: 10.1128/AEM.68.2.713-719.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng J, De La Fuente-Núñez C, Trimble MJ, Xu J, Hancock RE, Lu X. 2015. An in situ Raman spectroscopy-based microfluidic “lab-on-a-chip” platform for non-destructive and continuous characterization of Pseudomonas aeruginosa biofilms. Chem Commun (Camb) 51:8966–8969. doi: 10.1039/C5CC02744F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin D, Xia Y, Whitesides GM. 2010. Soft lithography for micro-and nanoscale patterning. Nat Protoc 5:491. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- 45.Kreth J, Vu H, Zhang Y, Herzberg MC. 2009. Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. J Bacteriol 191:6281–6291. doi: 10.1128/JB.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma Q, Wood TK. 2009. OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ Microbiol 11:2735–2746. doi: 10.1111/j.1462-2920.2009.02000.x. [DOI] [PubMed] [Google Scholar]
- 47.Ritz M, Garenaux A, Berge M, Federighi M. 2009. Determination of rpoA as the most suitable internal control to study stress response in C. jejuni by RT-qPCR and application to oxidative stress. J Microbiol Methods 76:196–200. doi: 10.1016/j.mimet.2008.10.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.