ABSTRACT
Salmonella enterica serovar Typhimurium is the only organism demonstrated to utilize fructose-asparagine (F-Asn) as a source of carbon and nitrogen. In this report, we first used a bioinformatics approach to identify other microorganisms that encode homologs of the Salmonella F-Asn utilization enzymes FraB (deglycase), FraD (kinase), and FraE (asparaginase). These candidate organisms were then tested with up to four different methods to confirm their ability to utilize F-Asn. The easiest and most broadly applicable method utilized a biological toxicity assay, which is based on the observation that F-Asn is toxic to a Salmonella fraB mutant. Candidate organisms were grown in a rich medium containing F-Asn, and depletion of F-Asn from the medium was inferred by the growth of a Salmonella fraB mutant in that same medium. For select organisms, the toxicity assay was cross-validated by direct mass spectrometry-aided measurement of F-Asn in the spent-culture media and through demonstration of FraB and FraD enzyme activity in cellular extracts. For prototrophs, F-Asn utilization was additionally confirmed by growth in a minimal medium containing F-Asn as the sole carbon source. Collectively, these studies established that Clostridium bolteae, Clostridium acetobutylicum, and Clostridium clostridioforme can utilize F-Asn, but Clostridium difficile cannot; Klebsiella oxytoca and some Klebsiella pneumoniae subspecies can utilize F-Asn; and some Citrobacter rodentium and Citrobacter freundii strains can also utilize F-Asn. Within Salmonella enterica, the host-adapted serovars Typhi and Paratyphi A have lost the ability to utilize F-Asn.
IMPORTANCE Fructose-asparagine (F-Asn) is a precursor to acrylamide that is found in human foods, and it is also a nutrient source for Salmonella enterica, a foodborne pathogen. Here, we determined that among the normal intestinal microbiota, there are species of Clostridium that encode the enzymes required for F-Asn utilization. Using complementary experimental approaches, we have confirmed that three members of Clostridium, two members of Klebsiella, and two members of Citrobacter can indeed utilize F-Asn. The Clostridium spp. likely compete with Salmonella for F-Asn in the gut and contribute to competitive exclusion. FraB, one of the enzymes in the F-Asn utilization pathway, is a potential drug target because inhibition of this enzyme leads to the accumulation of a toxic metabolite that inhibits the growth of Salmonella species. This study identifies the potential off-target organisms that need to be considered when developing therapeutics directed at FraB.
KEYWORDS: Salmonella, typhoid, pathovar, fructosamines, fructose-asparagine, Amadori products, Clostridium, Klebsiella, Citrobacter, FraB, phylogeny
INTRODUCTION
Salmonella enterica is a major contributor to human morbidity and mortality (1–8). There are over 2,500 serovars of S. enterica (9). Some of these serovars, notably S. enterica Typhimurium and Enteritidis, cause inflammatory diarrhea in humans. Other serovars, including S. enterica serovars Typhi and Paratyphi A, are less likely to cause diarrhea and instead cause either a severe systemic disease called typhoid or the less severe paratyphoid fever. These typhoidal serovars (also called extraintestinal serovars) have a more restricted host range than the nontyphoidal serovars and appear to be undergoing genome reduction as they become specialized to particular hosts and lose genes that contribute to metabolism during gastrointestinal inflammation (10–15). Such streamlining includes genes for the utilization of specific nutrient sources and for anaerobic metabolism (10). One of the loci that is either mutated or deleted in some typhoidal serovars is the fra locus, which encodes the enzymes necessary to use fructose-asparagine (F-Asn) as a carbon and nitrogen source (16). Prior to the discovery of F-Asn utilization by Salmonella, F-Asn was not known to be a nutrient source for any organism (16).
The proposed pathway of F-Asn utilization in Salmonella spp. requires a periplasmic asparaginase, FraE, which converts F-Asn to fructose-aspartate (F-Asp) (17). F-Asp is transported by FraA, a Dcu-type transporter, into the cytoplasm, where the FraD kinase phosphorylates F-Asp to form 6-phospho-F-Asp (6-P-F-Asp), which is then cleaved by the FraB deglycase to form glucose-6-phosphate (glucose-6-P) and aspartate (18). Interestingly, F-Asn is toxic to a fraB mutant due to the accumulation of 6-P-F-Asp (19). The mechanism of toxicity is unknown, but it engenders at least a 1,000-fold fitness defect in a fraB mutant in several mouse models (16, 19). We have determined that F-Asn is present in mouse chow and several human foods and that its abundance in the mouse intestinal tract is determined by the microbiota (34). In conventional mice with a normal microbiota, F-Asn is found at much lower concentrations than in germfree mice, presumably due to consumption by certain members of the microbiota. The fitness defects of a Salmonella fraB mutant correlate with the availability of F-Asn in these mouse models, consistent with F-Asn being toxic to the fraB mutant.
Here, we sought to determine which members of the normal microbiota can utilize F-Asn. Using an integrated approach encompassing bioinformatics, microbial toxicity assays, growth on minimal media containing F-Asn, and biochemical measurements, we have determined that some Clostridium, Klebsiella, and Citrobacter species can utilize F-Asn as a nutrient source. We also confirmed that the typhoidal serovars of Salmonella, Typhi and Paratyphi A, have lost the ability to utilize F-Asn.
RESULTS AND DISCUSSION
Strategy.
We first employed bioinformatics methods to determine which microorganisms have the genetic potential to utilize F-Asn as a nutrient. We then attempted to grow those members in minimal medium containing F-Asn as the sole nutrient source; however, this strategy proved to be unviable for many organisms due to our inability to mimic the complex growth requirements of the organisms involved. Instead, we grew selected members of the normal microbiota in rich medium supplemented with F-Asn and then sought to determine whether or not the F-Asn had been removed from the medium. To accomplish this goal, we devised a toxicity assay in which we determined if the spent-culture supernatant was toxic to a Salmonella fraB mutant. Toxicity, if any, indicates that the culture supernatant contains F-Asn. We cross-validated the inferences from the toxicity assay through mass spectrometry (MS)-aided measurements of F-Asn in some of the spent-culture supernatants and by biochemical assays of FraB and FraD enzyme activity in cellular extracts. Additionally, we tested some organisms for growth in minimal medium containing F-Asn as the sole carbon source. Using these methods, we have determined that some Clostridium, Klebsiella, and Citrobacter species can utilize F-Asn as a nutrient source and that some Salmonella serovars have lost this ability.
Bioinformatics reveals potential F-Asn utilizers within the gastrointestinal microbiome.
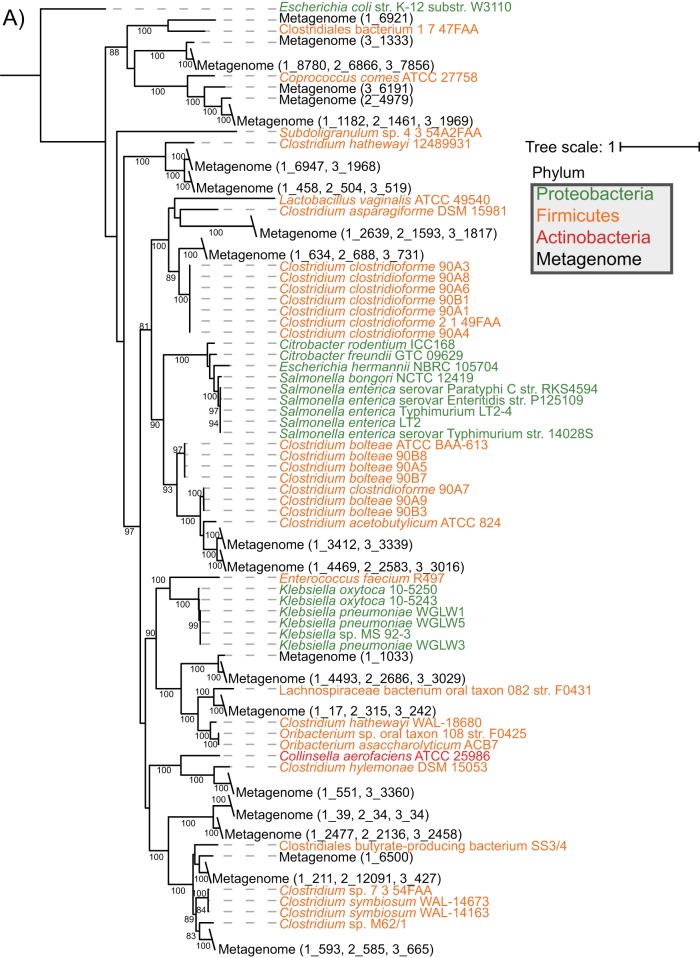
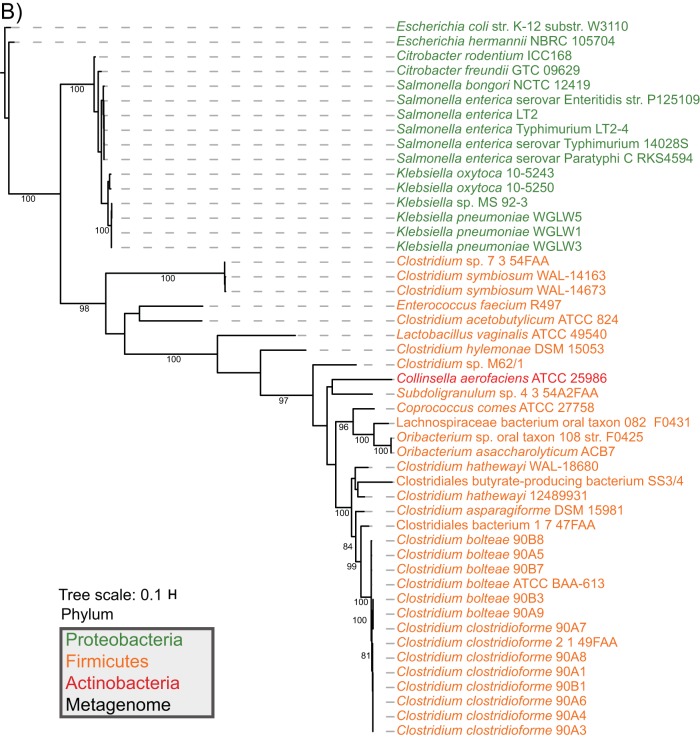
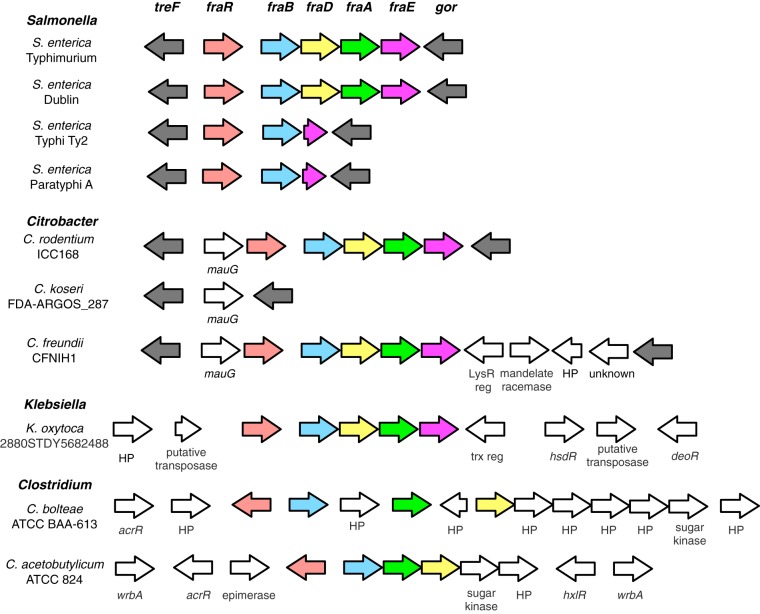
To firmly establish the presence of fraBDE genes with a function similar to those characterized in Salmonella enterica, we used S. Typhimurium strain LT2 amino acid sequences to identify homologs in the Human Microbiome Reference Genomes Database (HMRGD) (20). From this genomic database containing 1,322 genomes, we recovered 49 genomes from the Proteobacteria, Firmicutes, and Actinobacteria that contain homologs with high overall sequence similarity to proteins from strain LT2 (see Table S1 in the supplemental material). FraB and FraD had a similar phylogeny; therefore, we concatenated these two proteins (i.e., FraBD) to increase resolution in the final analyses (Fig. 1). The phylogenetic distribution of the FraBD concatenated protein is not consistent with the evolutionary history of the organisms involved. Instead, a comparison of the protein and organism phylogenies suggests horizontal gene transfer between Firmicutes and Proteobacteria (Fig. 1). In contrast, the FraE topology is consistent with vertical descent, with the FraE monophyletic groups mirroring the organism's phylogenetic affiliation at the phylum level (Fig. 1). Consistent with the different evolutionary histories, we note that outside the Salmonella genus, fraE is often not colocalized with fraB and fraD (Fig. 2).
FIG 1.
Maximum likelihood trees of concatenated FraBD (A) and FraE (B) amino acid sequences that were recovered from a BLAST search of HMRGD isolates and CBA/J mouse fecal metagenomes are shown. All putative homologs found with BLAST search of the HMRGD and mouse metagenomes can be found in File S1 in the supplemental material. Bootstrap values are based on 100 resamplings, with values of >80 shown. The metagenome and scaffold associated with each FraBD from mouse metagenomes (black text) are reported with the metagenome number (1 to 3), followed by an underscore and scaffold number. E. coli K-12 strain W3110 was used as an outgroup that does not contain fra genes.
FIG 2.
Synteny map of the fra locus from select bacterial species. Putative homologs are indicated by the same color.
Additionally, we recovered 21 unique FraBD proteins from 3 CBA/J mouse metagenomes (Fig. 1), an indication of the scope of F-Asn utilization in the gut. Outside of human- and mouse-associated microbial genomes and metagenomes, we also found the genomic potential for F-Asn utilization in publically available soil and rhizosphere metagenomes (NCBI BioProject numbers PRJNA330336 and PRJNA336894, respectively).
Some Clostridium isolates can utilize F-Asn as a nutrient source.
Because some Clostridium species encode homologs of FraB, FraD, and FraE, we tested the hypothesis that these organisms can indeed utilize F-Asn as a carbon source. We initially attempted to grow the Clostridium isolates in a minimal medium with F-Asn as the sole carbon source. This approach was not successful, presumably due to a strict requirement for rich medium to support their growth. We then tested the ability of the Clostridium isolates to deplete F-Asn from a rich medium. For these studies, we used tryptic soy broth (TSB) containing 5% sheep blood supplemented with 5 mM F-Asn. After 48 h of anaerobic growth at 37°C, the spent-culture supernatant was tested for the presence of F-Asn using two different methods, a toxicity assay and MS. In the toxicity assay we tested the spent-culture supernatant for toxicity to a Salmonella fraB mutant, with toxicity indicating the presence of F-Asn (19). For this measurement, we added an equal mixture of wild-type and fraB mutant Salmonella cells to the spent-culture supernatant that was obtained after growing an organism of interest. After overnight growth at 37°C, the Salmonella cells were plated on LB-Kan and LB-Cam to enumerate the fraB mutant and wild type, respectively. If F-Asn was present, the fraB mutant would be recovered in lower numbers than the wild type.
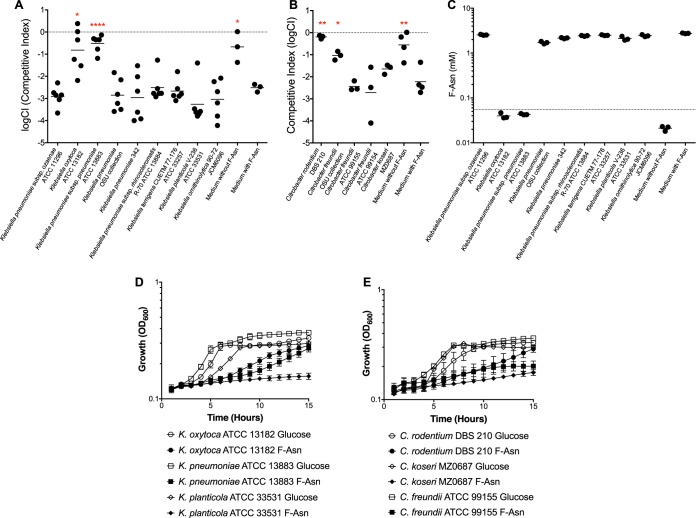
Based on the phylogenetic trees shown in Fig. 1, we tested Clostridium bolteae, C. acetobutylicum, C. clostridioforme, and C. difficile. All but C. difficile were predicted to utilize F-Asn. Indeed, growth of the Salmonella fraB mutant was not inhibited in the culture supernatants of C. bolteae, C. clostridioforme, or C. acetobutylicum, a finding that suggests that these three species can deplete F-Asn from the medium. In contrast, the Salmonella fraB mutant was inhibited in the spent-culture supernatants of C. difficile, an observation that suggests the failure of C. difficile to deplete F-Asn from the medium (Fig. 3A). We verified the presence or absence of F-Asn in the culture supernatants by removing a sample prior to the addition of Salmonella bacteria and measuring the concentration of F-Asn by mass spectrometry. The concentrations of F-Asn in the culture supernatants correlate with the inhibition of the Salmonella fraB mutant (Fig. 3B).
FIG 3.
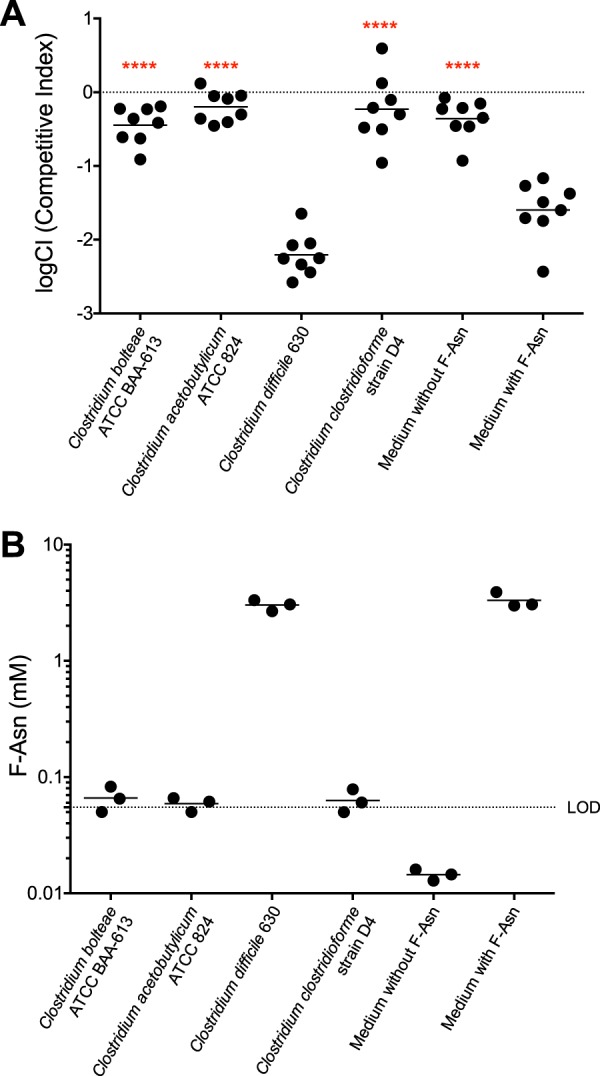
(A) A Salmonella toxicity assay used to determine if Clostridium species can deplete F-Asn from growth medium. The log competitive index (logCI) is plotted, with a horizontal bar indicating the mean of the results from three independent experiments that include a total of eight technical replicates. Competitions in which the result was closer to neutral than the “medium with F-Asn” competition were tested for statistical significance. Asterisks represent differences in an unpaired t test (parametric) compared to medium with F-Asn (****, P < 0.0001). (B) MS measurements to determine if Clostridium species can remove F-Asn from growth medium. The horizontal bar in the MS data indicates the mean value of three measurements; the dotted line represents the limit of detection (LOD) of 0.055 mM.
The bioassay and MS measurements together confirmed that Clostridium bolteae, Clostridium clostridioforme, and Clostridium acetobutylicum have the genes for F-Asn utilization in their genome and that these species can deplete F-Asn from their growth media, or at least alter it, such that it is no longer toxic to a Salmonella fraB mutant and no longer detectable at the F-Asn m/z value. In contrast, Clostridium difficile does not encode the F-Asn utilization enzymes in its genome and is unable to remove F-Asn from its growth medium.
Some Klebsiella and Citrobacter isolates can utilize F-Asn as a nutrient source.
Some Klebsiella and Citrobacter species are predicted to encode FraB, FraD, and FraE (Citrobacter rodentium, C. freundii, Klebsiella pneumoniae, and K. oxytoca) (Fig. 1 and 2). However, it should be noted that only some isolates of each of these species contain the fra genes, while other isolates do not. Whether or not any of these organisms can truly utilize F-Asn is unknown. As with the Clostridium experiments described in Materials and Methods, we used the Salmonella fraB mutant-based toxicity assay to determine if a variety of Klebsiella and Citrobacter isolates can deplete F-Asn from a rich growth medium. The only two differences in the assay were that (i) sheep blood was not added to the TSB, and (ii) the isolates were grown aerobically for 16 to 18 h, while the Clostridium isolates were grown anaerobically for 48 h. Based on the Salmonella toxicity assay, our K. oxytoca isolate, one of three K. pneumoniae isolates, a C. rodentium isolate, and a C. freundii isolate did remove F-Asn from their growth media (Fig. 4A and B). Of the two K. pneumoniae isolates that did not remove F-Asn, one is a plant isolate (21, 22), and the other is from an in-house collection and is of unknown origin. Several species or subspecies of Klebsiella and Citrobacter that were not predicted to encode F-Asn utilization enzymes were unable to deplete F-Asn from their growth medium.
FIG 4.
Identification of Klebsiella and Citrobacter species that can utilize F-Asn. The Salmonella toxicity assay was used to determine which Klebsiella (A) or Citrobacter (B) strains could deplete F-Asn from their medium. The log competitive index (logCI) is plotted, with a horizontal bar indicating the mean of independent replicates. Competitions in which the result was closer to neutral than the “medium with F-Asn” competition were tested for statistical significance. Asterisks indicate difference from medium with F-Asn (unpaired t test, parametric: *, P < 0.05; **, P < 0.01; ****, P < 0.0001). (C) MS measurements to determine if Klebsiella species can deplete F-Asn from growth medium. The horizontal bar in the MS data indicates mean value of three independent measurements; the dotted line represents the limit of detection (LOD) of 0.055 mM. Growth at 37°C was tested in minimal medium containing either glucose (open symbols) or F-Asn (closed symbols) as the sole carbon source for Klebsiella (D) or Citrobacter (E) strains. Growth curves represent two independent experiments with six total replicates for each point. Error bars indicate the standard deviation. OD600, optical density at 600 nm.
MS was used to verify the presence or absence of F-Asn in the Klebsiella spent-culture supernatants (Fig. 4C). As with the Clostridium spent-culture supernatants, the MS results correlated well with the toxicity assay results. Although less likely, we considered the possibility that species that are depleting F-Asn from their growth medium are merely modifying F-Asn to a form that is either not toxic to a Salmonella fraB mutant or that is not detectable by MS. To address this possibility, we tested the cellular extracts of two Klebsiella strains for FraB and FraD enzyme activities, one which was expected to have them (K. oxytoca) and one which was not (K. planticola). Indeed, the cellular extracts of K. oxytoca contained FraB and FraD activity, while the extracts of K. planticola did not. Furthermore, the enzyme activities were induced ∼18-fold or more in the presence of F-Asn that was included in the growth medium (Table 1).
TABLE 1.
FraB and FraD enzymatic activities in Klebsiella cellular extracts
| Klebsiella strain reference | Klebsiella strain and growth condition | Sp act (× 104 U/mg)a |
|
|---|---|---|---|
| FraD | FraB | ||
| ATCC 13182 | K. oxytoca (−F-Asn) | 3.2 ± 2.5 | 1.6 ± 1.6 |
| K. oxytoca (+F-Asn) | 58.4 ± 2.4 | 46.8 ± 1.5 | |
| ATCC 33531 | K. planticola (−F-Asn) | ND | ND |
| K. planticola (+F-Asn) | ND | ND | |
The specific activities were calculated from two independent measurements of three different biological replicates and are expressed as the mean values and the associated standard error. ND, activity was not detected.
Finally, we attempted to grow all of the Klebsiella and Citrobacter isolates in minimal medium with F-Asn as the sole source of carbon. Minimal medium with glucose was used as a control. K. pneumoniae subsp. rhinoscleromatis was the only organism that failed to grow on glucose and must be an auxotroph. The remaining Klebsiella strains gave unambiguous results. Two strains were able to grow on F-Asn while the remainder could not, and these results correlated with the strains that could deplete F-Asn from their growth medium according to the Salmonella toxicity assay and MS measurements (Fig. 4D). The results with Citrobacter spp. were not as straightforward to interpret. All strains grew well with glucose but grew poorly, or not at all, with F-Asn (Fig. 4E); it is possible that these results are influenced by differences in the rates of uptake and utilization, which remain uncharacterized. However, the strains that grew at all were the same strains that were able to deplete F-Asn from their growth medium.
Some Salmonella serovars have lost the ability to utilize F-Asn as a nutrient source.
Salmonella enterica contains over 2,500 serovars, which have a range of disease manifestations and host specificities. They can be broadly classified into two pathovars, gastrointestinal and extraintestinal (10). Members of the gastrointestinal pathovar (e.g., S. enterica serovars Typhimurium and Enteritidis) have a broad host range and tend to cause gastroenteritis. In contrast, members of the extraintestinal pathovar (e.g., S. enterica serovars Typhi and Paratyphi A) tend to have a narrow host range and cause systemic infections, and they are less likely to cause gastroenteritis. S. Typhi infects only humans and is less likely to cause diarrhea, but it causes a systemic disease called typhoid fever that can last for up to 6 weeks. In ∼3% of cases, the bacterium can establish a carrier state in which the host is asymptomatic but sheds bacteria in the feces for years (23, 24). S. Typhi and other members of the extraintestinal pathovar also appear to be undergoing genome degradation concomitant with loss of genes that extend their host range and genes involved in anaerobic metabolism that are beneficial during gastroenteritis (10–15). The fra locus is one of the loci that has been mutated in some members of the extraintestinal pathovar (10) (Fig. 2). These serovars would not grow in minimal medium, so we used the toxicity assay to test these serovars for their ability to remove F-Asn from their growth medium (Fig. 5). Their genetic content correlates with the toxicity assays, with S. Enteritidis able to utilize F-Asn, while S. Typhi and S. Paratyphi A cannot.
FIG 5.
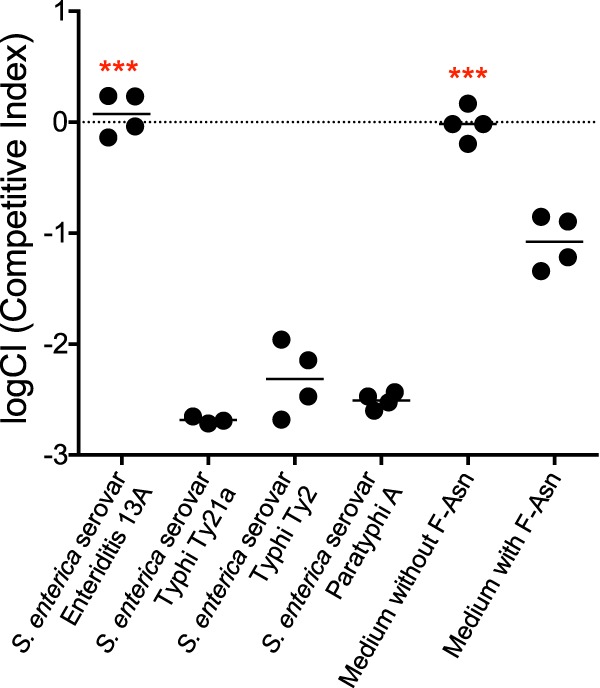
Some Salmonella serovars have lost the ability to utilize F-Asn. The log competitive index (logCI) is plotted, with a horizontal bar indicating the mean of the results from two independent experiments that include a total of four technical replicates. Competitions in which the result was closer to neutral than the “medium with F-Asn” competition were tested for statistical significance. Asterisks indicate difference from medium with F-Asn (unpaired t test, parametric: ***, P < 0.001).
Conclusion.
Although there are parallels in the catabolic routes for utilization of other Amadori products (e.g., fructose-lysine [25]), the presence of both FraB (deglycase) and FraD (kinase) homologs appears to be a reliable indicator of the ability of an organism to utilize F-Asn; in contrast, FraE (asparaginase) is not a good indicator. Many organisms encode one or more asparaginases that are difficult to distinguish from a bona fide fructose-asparaginase. Also, the FraE homolog is often not colocalized with FraB and FraD, although this observation could merely imply that the organism exploits a generic asparaginase and does not use a dedicated fructose-asparaginase (Fig. 2). In fact, we recently determined that the AnsB asparaginase of Salmonella can contribute to F-Asn utilization and contribute to growth of a fraE mutant in a medium containing F-Asn as the sole source of carbon and nitrogen (17). The F-Asn utilization system also includes a transporter of the Dcu type (FraA), which is present in all species that can utilize F-Asn and is colocalized with fraB and fraD (Fig. 2). Thus, FraA may be useful as an additional indicator of the ability of an organism to utilize F-Asn.
The toxicity assay is a rapid and facile method to determine if an organism can deplete F-Asn from its growth medium. The accuracy of the toxicity assay was rigorously cross-validated by mass spectrometric measurements of F-Asn in the spent media. However, it is still possible that organisms that encode the F-Asn utilization genes might not be able to utilize F-Asn but somehow still alter F-Asn such that it is no longer toxic to the Salmonella fraB mutant and no longer detectable by MS. To address this possibility, we chose a pair of organisms that either could or could not utilize F-Asn according to gene content, the toxicity assay, and MS (K. oxytoca and K. planticola) and tested these organisms for FraB and FraD enzyme activity. Consistent with the predictions, FraB and FraD activity was detected in K. oxytoca but not K. planticola. Importantly, this trend is mirrored in growth assays on minimal media, as K. oxytoca but not K. planticola is able to grow on F-Asn as the sole carbon source (Fig. 4D). Furthermore, these enzyme activities were induced by the presence of F-Asn in the medium (Table 1). Thus, we have five independent indications that K. oxytoca can utilize F-Asn (i.e., gene content, toxicity assay, MS of spent-culture supernatants, enzyme activities, and growth in minimal F-Asn medium). All of the other organisms in this study have at least two indications of their ability to utilize F-Asn. Gene content analysis and the toxicity assay are likely the most facile computational and experimental methods that can be used to investigate F-Asn utilization by any organism; additional cross-corroborating methods (MS, biochemical assays) will certainly help minimize false positives. Growth in minimal medium containing F-Asn is a quick and easy test but does not work for auxotrophs (e.g., all of the Clostridium species, some Salmonella serovars, and K. pneumoniae subsp. rhinoscleromatis, tested here). Interestingly, while Salmonella grows as well on F-Asn as it does on glucose (16, 19), Klebsiella and Citrobacter strains do not grow as well on F-Asn as they do on glucose. It is not clear why their metabolism does not allow them to grow as rapidly on F-Asn as they do on glucose. The presence of F-Asn utilization genes in Klebsiella and Citrobacter genomes is also not uniform. For example, some C. freundii genomes contain the fra genes, while others do not. Consistent with this, we have one C. freundii isolate that can utilize F-Asn and two isolates that cannot.
The results presented here expand our understanding of the diversity of microbes that can utilize F-Asn. Because F-Asn is toxic to a fraB mutant due to the accumulation of 6-P-F-Asp, FraB is a promising drug target (18, 19). A combination of F-Asn and a FraB inhibitor should be able to inhibit Salmonella growth in vivo. Here, we have determined that this therapeutic approach would have the advantage of being fairly specific to Salmonella and would not destroy large numbers of species within the gastrointestinal tract. Species-specific drugs are becoming important as we better recognize the undue consequences of broad-spectrum antibiotics. The Klebsiella species that can utilize F-Asn are unlikely to be present in a patient undergoing therapy for a Salmonella infection; even if they were present, their elimination by a FraB inhibitor is unlikely to pose ill effects. However, adverse consequences are possible from depleting Clostridium species, which are key and essential players of gut homeostasis. If developed, the effects of FraB inhibitors on the microbiota would need to be determined. Additionally, it may be possible to develop inhibitors that specifically target Salmonella FraB and not the homologs from Clostridium and other organisms.
MATERIALS AND METHODS
Bacterial strains and growth.
Salmonella, Klebsiella, and Citrobacter strains were routinely grown aerobically in LB broth (Fisher BioReagents) or on agar plates with the addition of 1.5% (wt/vol) agar (Fisher BioReagents), except for toxicity assays that were performed in tryptic soy broth (TSB; Becton Dickinson). Chloramphenicol (Cam) or kanamycin (Kan) was added at 30 or 50 μg/ml, respectively. Clostridium strains were routinely grown anaerobically on tryptic soy agar with 5% (vol/vol) sheep blood (TSA IITM plates; Becton Dickinson) or in TSB supplemented with 5% (vol/vol) defibrinated sheep blood (Remel). Anaerobic conditions utilized a Bactron 1 anaerobic chamber containing 90% N2, 5% CO2, and 5% H2 (Shel Lab). Media for the growth of anaerobes were degassed in the anaerobic chamber for 24 h prior to use. For growth studies using minimal medium, we used the recipe described by Bender et al. (26) supplemented with 5 mM NH4Cl and either 5 mM glucose or 5 mM F-Asn as the sole carbon source. Growth assays at 37°C were performed in a 96-well clear-bottom plate, and the optical density at 600 nm was recorded every hour over a 15-h period using a SpectraMax M5 microplate reader (Molecular Devices). Data were analyzed using the SoftMax Pro 6.1 software.
Synthesis of fructose-aspartate, fructose-asparagine, and [13C]fructose-asparagine.
Fructose-aspartate and fructose-asparagine were synthesized as described previously (27). 13C-labeled fructose-asparagine was made in a manner similar to the unlabeled counterpart, except for the use of glucose uniformly labeled with 13C (Cambridge Isotope Laboratories).
Fructose-asparagine toxicity assay.
Clostridium strains were subcultured 1:50 into 5 ml TSB supplemented with 5% (vol/vol) sheep blood and 5 mM F-Asn, and then they were grown standing at 37°C for 48 h in an anaerobic chamber. Salmonella and Klebsiella strains were subcultured 1:50 into 5 ml TSB (without sheep blood) for 16 h in a roller drum within a 37°C incubator. This spent culture was then tested for the presence of F-Asn. First, 50 μl was removed for enumeration of CFU by dilution plating (to confirm that the organism of interest did indeed grow); then, another 0.5-ml aliquot was withdrawn for mass spectrometry. The remainder of the spent-culture supernatant was then used for a competition experiment between wild-type and fraB mutant Salmonella strains (JLD1214 and HMB206, respectively); 107 CFU of each strain was mixed together, added to the culture supernatant, and incubated standing at 37°C for 24 h in an anaerobic chamber (for Clostridium strains) or rolling at 37°C for 16 h in a roller drum within a 37°C incubator (for Salmonella and Klebsiella strains). The wild-type and mutant strains were enumerated by plating on LB plus Cam and LB plus Kan. The log competitive index (logCI) was calculated by log([CFU of mutant recovered/CFU of WT recovered]/[CFU mutant input/CFU WT input]). If the mutant is defective compared to the wild type, it should have a logCI of <0. Because Klebsiella pneumoniae 342 was resistant to chloramphenicol and Citrobacter rodentium DBS 210 was resistant to kanamycin, their culture supernatants were clarified by centrifugation at 5,000 × g for 10 min at 4°C and filter sterilized prior to the addition of the wild-type and the fraB mutant Salmonella competition strains.
Identifying Fra homologs.
We used BLASTP with FraB (accession no. WP_010989080.1), FraD (accession no. WP_000028429), and FraE (accession no. WP_000702755.1) of Salmonella enterica serovar Typhimurium strain LT2 (accession no. NC_003197.1) as queries and an E value of >1e−20 (for FraB and FraD) or >1e−10 (for FraE) (20) to identify Salmonella homologs from the Human Microbiome Reference Genomes Database (HMRGD). Using the same search criteria and E value cutoffs, we also mined metagenomes from 3 CBA/J mouse metagenomes (NCBI BioProject PRJNA348350) and non-host-associated systems in the Integrated Microbial Genomes-Joint Genome Institute (IMG-JGI) database (July 2017) (28). The recovered homologs were linked to their corresponding genome and taxonomic information (Table S1).
Phylogenetic analyses.
FraB, FraD, and FraE proteins were included in phylogenetic analyses only if the isolate genome encoded all three. Individual Fra proteins were aligned using MUSCLE (version 3.8.31) and then manually curated to remove end gaps and ambiguously aligned regions (29). Phylogenetic trees were first constructed individually, prior to concatenation of the curated FraB and FraD alignments, while the FraE phylogenetic analysis remained separate. In the concatenated FraBD phylogenetic analyses, we also included FraB and FraD from CBA/J mouse metagenomes if they were located on the same scaffold. The concatenated FraBD alignments and individual FraE alignments were run through ProtPipeliner, a python script developed in-house for the generation of phylogenetic trees (https://github.com/rwolfe45/Protpipeliner). Briefly, alignments were first curated with low-level editing by gBlocks, followed by evolutionary model selection by ProtTest (30, 31). A maximum likelihood phylogenetic tree for the concatenated FraBD alignment and the FraE alignment was conducted using RAxML version 6.0.2 under the LG model of evolution, with 100 bootstrap replicates (32). Phylogenetic trees were visualized in iTOL (33).
Measurement of F-Asn concentration in culture media.
Prior to the addition of the Salmonella competition strains for the F-Asn toxicity assay, 1-ml aliquots were removed from the cultures and centrifuged at 19,000 × g for 1 min, and the supernatant was stored at −80°C until use. For MS analysis, the supernatant was thawed on ice, and 100 μl was transferred to a 1.5-ml microcentrifuge tube, followed by the addition of 500 μl of chilled methanol and 490 μl of water spiked with 100 nmol [13C]F-Asn. After being vortexed and centrifuged at 14,800 × g for 1 h, the supernatant was transferred to a new 1.5-ml microcentrifuge tube, frozen, and lyophilized. Before MS analysis, these dried pellets were resuspended in 500 μl of water and further diluted 100-fold with 4:1 acetonitrile–0.1% (vol/vol) formic acid (liquid chromatography-MS [LC-MS] grade; Thermo Scientific) and then filtered using a 0.2-μm-pore-size polytetrafluoroethylene (PTFE) filter (Thermo Scientific). The flowthrough was further diluted 5-fold before analysis by LC-MS. A nanoACQUITY ultraperformance LC (UPLC) system (Waters, Milford, MA) with a UPLC M-class BEH 130 amide column (75 μm by 150 mm, 1.7 μm; Waters) was coupled to a triple quadrupole mass spectrometer (Waters Xevo TQ-S) for F-Asn quantification. Buffer A (0.1% formic acid in water with 10% acetonitrile) and buffer B (0.1% formic acid in acetonitrile) were used for elution, which started with 80% B for 6 min, followed by a gradient of 6 to 20 min, 80 to 50% B; 20 to 26 min, 50% B; 26 to 28 min, 50 to 80% B; and 28 to 35 min, 80% B (flow rate, 0.75 μl/min). The mass spectrometer was operated in positive ion nano-electrospray ionization mode (nano-ESI+), with a capillary voltage of 3.5 kV, source temperature of 70°C, cone voltage of 2 V, and source offset of 2 V. The gas flow rate for the collision cell was 0.15 ml/min. The transition m/z 295 → 211 of F-Asn with collision energy 13 eV was selected for quantitation, and the m/z 301 → 216 of [13C]F-Asn with collision energy 13 eV was used for normalization. Skyline-daily (version 3.5; MacCoss Lab, Department of Genome Sciences, University of Washington, Seattle, WA, USA) was used for calculating the peak area of transitions. The limit of detection (LOD) was calculated as 3.3 × (the standard deviation of the intercept/the slope), with the intercept and slope derived from a standard curve based on a series of F-Asn concentrations spiked into the growth medium.
Preparation of Klebsiella extracts.
Klebsiella oxytoca (ATCC 13182) and Klebsiella planticola (ATCC 33531) crude extracts were prepared using essentially the protocol described in reference 19. These strains were grown in 5 ml LB with or without 5 mM F-Asn for 16 h at 37°C with shaking. The cells were harvested by centrifugation (5,000 × g at 4°C), washed with 1.5 ml water, and resuspended in 300 μl of 25 mM HEPES (pH 7.5) and 0.1 mM phenylmethylsulfonyl fluoride. Cells were then lysed by sonication (50% output power for 60 s, with cycles of 2 s on and 5 s off in an ultrasonic processor; Cole-Parmer) and centrifuged at 13,000 × g for 20 min at 4°C to obtain the crude lysates, which were dialyzed against 25 mM HEPES (pH 7.5) at 4°C, with two changes over 60 min. These crude extracts were then used for enzyme and protein (Bradford) assays.
Enzyme assays.
While the assay protocols used mirrored those described in reference 19, there were some notable differences, including the use of a continuous rather than a discontinuous assay to determine the initial velocities. All assays were carried out at 37°C in a 20-μl total reaction volume. To measure the FraB deglycase activities in the crude extracts, a glucose-6-phosphate dehydrogenase (G6PD)-based coupled enzyme assay was employed. The NADPH produced by the G6PD was taken as a direct readout of the glucose-6-phosphate (G6P) produced by FraB. The 20-μl FraB reaction mixture contained 1 mM 6-P-F-Asp, 25 mM HEPES (pH 7.5), 5 mM MgCl2, 0.1 mM EGTA, and 0.5 mM NADP+. To measure FraD kinase activity, we exploited a G6PD–FraB-based coupled assay. The 20-μl reaction mixture for the kinase assay contained 1 mM fructose-aspartate (F-Asp), 25 mM HEPES (pH 7.5), 25 mM KCl, 1 mM MgCl2, 1 mM dithiothreitol, 1 mM ATP, 0.1 mM EGTA, 0.5 mM NADP+, and 0.3 μM recombinant FraB (Sengupta and Gopalan, unpublished data).
In the case of FraB, 12 μl of crude extract was added to the 6.5-μl reaction mixture and incubated at 37°C for 4 min; for FraD, we determined that better reproducibility resulted when the crude extract and reaction mixture were preincubated separately for 4 min at 37°C prior to the addition of G6PD. In both instances, the addition of 1.5 μl baker's yeast G6PD (100 U/ml; catalog no. G6378; Sigma), which had been preincubated separately at 37°C for 4 min, enabled continuous spectrophotometric readout. From the 20-μl reaction mixture that was assembled, 18 μl was immediately transferred to a 384-well microplate, and the absorbance at 340 nm was monitored in real time using a SpectraMax M5 (Molecular Devices) microplate reader (integration time, 1,000 ms; settle time, 300 ms). Linear regression analysis of NADPH generated as a function of time was used to calculate the initial velocity (0.97 ≤ r2 ≤ 0.998; Excel).
The blanks for the FraB and FraD assays included all the assay components except the corresponding substrates, thus accounting for residual G6P in the crude extract that was being tested. One unit of activity is defined as the amount of enzyme catalyzing the formation of 1 μmol NADPH per min; specific activities (units per milligram) were calculated using the protein amount determined by the Bradford assay. The mean and standard errors of the specific activities were calculated from a total of six measurements, with two independent assays using crude extracts prepared from three separate cultures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Valley Stewart, Vincent Young, Michael Bailey, Jean Petter, Michael McClelland, Eric Triplett, and Thaddeus Ezeji for strains. We thank Edward Behrman and Alex Bogard for synthesis of F-Asn, F-Asp, and 6-P-F-Asp.
This work was funded by grant number 1R01AI116119 from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01957-17.
REFERENCES
- 1.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 2.Pires SM, Fischer-Walker CL, Lanata CF, Devleesschauwer B, Hall AJ, Kirk MD, Duarte ASR, Black RE, Angulo FJ. 2015. Aetiology-specific estimates of the global and regional incidence and mortality of diarrhoeal diseases commonly transmitted through food. PLoS One 10:e0142927. doi: 10.1371/journal.pone.0142927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scallan E, Mahon BE, Hoekstra RM, Griffin PM. 2013. Estimates of illnesses, hospitalizations and deaths caused by major bacterial enteric pathogens in young children in the United States. Pediatr Infect Dis J 32:217–221. [DOI] [PubMed] [Google Scholar]
- 4.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crump JA, Mintz ED. 2010. Global trends in typhoid and paratyphoid fever. Clin Infect Dis 50:241–246. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon MA. 2011. Invasive nontyphoidal Salmonella disease. Curr Opin Infect Dis 24:484–489. doi: 10.1097/QCO.0b013e32834a9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease ‘Burden of Illness’ Studies. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 8.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Döpfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, BockemUhl J, Grimont PAD, Weill F-X. 2010. Supplement 2003-2007 (no. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol 161:26–29. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Nuccio S-P, Bäumler AJ. 2014. Comparative analysis of Salmonella genomes identifies a metabolic network for escalating growth in the inflamed gut. mBio 5:e00929-14. doi: 10.1128/mBio.00929-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, Feasey NA, Kingsley RA, Thomson NR, Keane JA, Weill F-X, Edwards DJ, Hawkey J, Harris SR, Mather AE, Cain AK, Hadfield J, Hart PJ, Thieu NTV, Klemm EJ, Glinos DA, Breiman RF, Watson CH, Kariuki S, Gordon MA, Heyderman RS, Okoro C, Jacobs J, Lunguya O, Edmunds WJ, Msefula C, Chabalgoity JA, Kama M, Jenkins K, Dutta S, Marks F, Campos J, Thompson C, Obaro S, MacLennan CA, Dolecek C, Keddy KH, Smith AM, Parry CM, Karkey A, Mulholland EK, Campbell JI, Dongol S, Basnyat B, Dufour M, Bandaranayake D, Naseri TT, Singh SP, et al. 2015. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet 47:632–639. doi: 10.1038/ng.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt KE, Thomson NR, Wain J, Langridge GC, Hasan R, Bhutta ZA, Quail MA, Norbertczak H, Walker D, Simmonds M, White B, Bason N, Mungall K, Dougan G, Parkhill J. 2009. Pseudogene accumulation in the evolutionary histories of Salmonella enterica serovars Paratyphi A and Typhi. BMC Genomics 10:36. doi: 10.1186/1471-2164-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill F-X, Goodhead I, Rance R, Baker S, Maskell DJ, Wain J, Dolecek C, Achtman M, Dougan G. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet 40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker S, Dougan G. 2007. The genome of Salmonella enterica serovar Typhi. Clin Infect Dis 45(Suppl 1):S29–S33. doi: 10.1086/518143. [DOI] [PubMed] [Google Scholar]
- 15.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Hague A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O'Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852. [DOI] [PubMed] [Google Scholar]
- 16.Ali MM, Newsom DL, Gonzalez JF, Sabag-Daigle A, Stahl C, Steidley B, Dubena J, Dyszel JL, Smith JN, Dieye Y, Arsenescu R, Boyaka PN, Krakowka S, Romeo T, Behrman EJ, White P, Ahmer BMM. 2014. Fructose-asparagine is a primary nutrient during growth of Salmonella in the inflamed intestine. PLoS Pathog 10:e1004209. doi: 10.1371/journal.ppat.1004209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabag-Daigle A, Sengupta A, Blunk HM, Biswas PK, Cron MC, Bogard A, Behrman EJ, Gopalan V, Ahmer BMM. 2017. Salmonella FraE, an asparaginase homolog, contributes to fructose-asparagine but not asparagine utilization. J Bacteriol 199:e00330-17. doi: 10.1128/JB.00330-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biswas PK, Behrman EJ, Gopalan V. 2017. Characterization of a Salmonella sugar kinase essential for the utilization of fructose-asparagine. Biochem Cell Biol 95:304–309. doi: 10.1139/bcb-2016-0138. [DOI] [PubMed] [Google Scholar]
- 19.Sabag-Daigle A, Blunk HM, Sengupta A, Wu J, Bogard AJ, Ali MM, Stahl C, Wysocki VH, Gopalan V, Behrman EJ, Ahmer BMM. 2016. A metabolic intermediate of the fructose-asparagine utilization pathway inhibits growth of a Salmonella fraB mutant. Sci Rep 6:28117. doi: 10.1038/srep28117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Dong Y, Iniguez AL, Ahmer BMM, Triplett EW. 2003. Kinetics and strain specificity of rhizosphere and endophytic colonization by enteric bacteria on seedlings of Medicago sativa and Medicago truncatula. Appl Environ Microbiol 69:1783–1790. doi: 10.1128/AEM.69.3.1783-1790.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iniguez AL, Dong Y, Carter HD, Ahmer BMM, Stone JM, Triplett EW. 2005. Regulation of enteric endophytic bacterial colonization by plant defenses. Mol Plant Microbe Interact 18:169–178. doi: 10.1094/MPMI-18-0169. [DOI] [PubMed] [Google Scholar]
- 23.Crawford RW, Rosales-Reyes R, Ramírez-Aguilar MDLL, Chapa-Azuela O, Alpuche-Aranda C, Gunn JS. 2010. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc Natl Acad Sci U S A 107:4353–4358. doi: 10.1073/pnas.1000862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunn JS, Marshall JM, Baker S, Dongol S, Charles RC, Ryan ET. 2014. Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol 22:648–655. doi: 10.1016/j.tim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiame E, Delpierre G, Collard F, Van Schaftingen E. 2002. Identification of a pathway for the utilization of the Amadori product fructoselysine in Escherichia coli. J Biol Chem 277:42523–42529. doi: 10.1074/jbc.M200863200. [DOI] [PubMed] [Google Scholar]
- 26.Bender RA, Janssen KA, Resnick AD, Blumenberg M, Foor F, Magasanik B. 1977. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J Bacteriol 129:1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen AL, Behrman EJ. 2016. Synthesis of 6-phosphofructose aspartic acid and some related Amadori compounds. Carbohydr Res 431:1–5. doi: 10.1016/j.carres.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 31.Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 32.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 33.Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 34.Wu J, Sabag-Daigle A, Metz TO, Deatherage Kaiser BL, Gopalan V, Behrman EJ, Wysocki VH, Ahmer BMM. 12 December 2007. Measurement of fructose-asparagine concentrations in human and animal foods. J Agric Food Chem. doi: 10.1021/acs.jafc.7b04237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.