ABSTRACT
For the past 150 years, bacteria have been investigated primarily in liquid batch cultures. Contrary to most expectations, these cultures are not homogeneous mixtures of single-cell bacteria, because free-floating bacterial aggregates eventually develop in most liquid batch cultures. These aggregates share characteristics with biofilms, such as increased antibiotic tolerance. We investigated how aggregates develop and what influences this development in liquid batch cultures of Pseudomonas aeruginosa. We focused on how the method of inoculation affected aggregation by assessing aggregate frequency and size using confocal laser scanning microscopy. Several traditional methods of initiating an overnight bacterial culture, i.e., inoculation directly from frozen cultures, inoculation using agar-grown cells, or inoculation using cells grown in liquid cultures, were investigated. We discovered a direct link between the inoculation method and the size and frequency of biofilm aggregates in liquid batch cultures, with inoculation directly from a plate resulting in the most numerous and largest aggregates. These large aggregates had an overall impact on the cultures' subsequent tolerance toward tobramycin, indicating that the inoculation method has a profound impact on antibiotic tolerance. We also observed a mechanism whereby preformed aggregates recruited single cells from the surrounding culture in a “snowball effect,” building up aggregated biomass in the culture. This recruitment was found to rely heavily on the exopolysaccharide Psl. Additionally, we found that both Escherichia coli and Staphylococcus aureus produced aggregates in liquid batch cultures. Our results stress the importance of inoculation consistency throughout experiments and the substantial impact aggregate development in liquid batch cultures may have on the outcomes of microbiological experiments.
IMPORTANCE Pure liquid cultures are fundamental to the field of microbiological research. These cultures are normally thought of as homogeneous mixtures of single-cell bacteria; the present study shows that this is not always true. Bacteria may aggregate in these liquid cultures. The aggregation can be induced by the method chosen for inoculation. The presence of aggregates can significantly change the outcomes of experiments by altering the phenotype of the cultures. The study found a mechanism whereby preformed aggregates are able to recruit surrounding single cells in a form of snowball effect, creating more and larger aggregates in the cultures. Once formed, these aggregates are hard to remove. Aggregates in liquid cultures may be an immense unseen challenge for microbiologists.
KEYWORDS: aggregation, Pseudomonas aeruginosa, biofilms
INTRODUCTION
Since the days of Robert Koch and Walter Hesse, cultivation in pure liquid cultures has been the mainstay for the study of bacteria (1). When working with a bacterial liquid batch culture, it is often assumed that the culture consists of a phenotypically homogeneous population of single bacterial cells. This assumption is based on the fact that liquid cultures are generally thought to be initiated from clones of a single bacterial cell (1–3). With the traditional technique of streaking frozen stocks or liquid cultures onto a solid medium, researchers are able to select for colonies originating from, in theory, a single bacterium of a particular genotype or morphology and to examine cultures for contamination (3, 4). Subsequently, colonies may be selected and used to inoculate liquid cultures to study bacteria in a free-floating (planktonic) state (1, 4, 5).
Most undergraduate biology students have been taught the growth pattern of bacterial liquid batch cultures described by distinct growth phases (6–9). When cells are inoculated into fresh culture medium, they initially enter the lag phase. During the lag phase, cells adjust to the fresh medium by synthesizing proteins that prepare the cells for growth (9–14). This phase varies in length, depending on the growth state of the inoculated cells, and the entire population of the culture consists of the inoculum. When the cells have adjusted to the new environment, they start to increase in number through exponential growth (14–16). During this exponential phase, the rapid growth results in a rapid decrease in the proportion of the initial inoculated population. Following exponential growth, the growth rate decreases and eventually the population enters the stationary phase. In the stationary phase, cells either do not divide or divide at the same rate as they die, thereby maintaining a stable population number (17). Ultimately, the population enters a “death phase” in which the population numbers decrease (18). A basic assumption of batch bacterial growth is that the decrease in the proportion of the inoculated population eliminates all residues of the phenotypic state of the initial inoculum.
Recently, there has been an increased focus on investigating the degree of homogeneity of liquid batch cultures. Schleheck and colleagues showed how nonattached aggregates of the opportunistic pathogen Pseudomonas aeruginosa form spontaneously in liquid batch cultures (19). After 6 h of growth, 90% of the biomass of the cultures consisted of aggregated bacteria in spheres with diameters of 5 to 600 μm. With overexpression of the exopolysaccharide Psl, P. aeruginosa exhibited increased aggregation in liquid cultures (20). For P. aeruginosa, these studies have clinical significance because nonattached biofilm aggregates are proposed to play a central role in the bacterial life history, both in the environment and during infection (21–25). Nonattached aggregates present the same phenotypic characteristics as biofilms attached to a surface, including enhanced antibiotic tolerance and resilience toward the immune response, compared to single cells (25). Thus, the frequency of nonattached aggregates in liquid laboratory cultures has significant effects not only on population-level phenotypes such as antibiotic tolerance but also on the reproducibility of experiments within and between laboratories (25–27).
The ability to conduct in vitro microbiological experiments consistently has always been a fundamental aspect of microbiological research. Among many other benefits, the assumption that bacterial liquid batch cultures are relatively homogeneous has spurred the use of microbes as experimental systems in many other scientific fields, including physics, engineering, and evolution. However, differences in practices among laboratories may often lead to different results based on the same protocol. We have investigated how the method used to inoculate batch cultures influences the development of nonattached biofilm aggregates and subsequently the experimental outcomes. We discovered that the aggregated biomass of a liquid P. aeruginosa culture depends on the method of inoculation, with direct inoculation using agar-grown colonies leading to the highest level of aggregate biomass. The level of aggregation had a profound heritable effect on the phenotype of subsequent bacterial populations, including antibiotic tolerance. The data presented here emphasize the importance of consistency in experimental workflow to control the effects of unintended nonattached aggregates in experiments. Variations in the method of inoculation, intentional or not, may have downstream effects that influence the experimental conclusions.
RESULTS
Growth and aggregation in cultures inoculated by various methods.
To examine the impact of inoculation method on aggregate growth and abundance in liquid culture, four different inoculation methods were evaluated after 4 and 18 h of growth, reinoculated, and then evaluated again after an additional 4 and 18 h (Fig. 1). Cultures inoculated with a lysogeny broth (LB) overnight culture from a single colony (method 1) or directly from a single colony from a plate (method 2) or from a LB overnight culture started from frozen stock (method 3) grew with comparable growth kinetics (P = 0.49). The culture inoculated directly from frozen stock (method 4) displayed an increased lag phase, compared to other cultures, over the first 7 h (P = 0.003) (see Fig. S2 in the supplemental material). After 24 h of growth, all four cultures had reached similar cell densities (P = 0.75).
FIG 1.
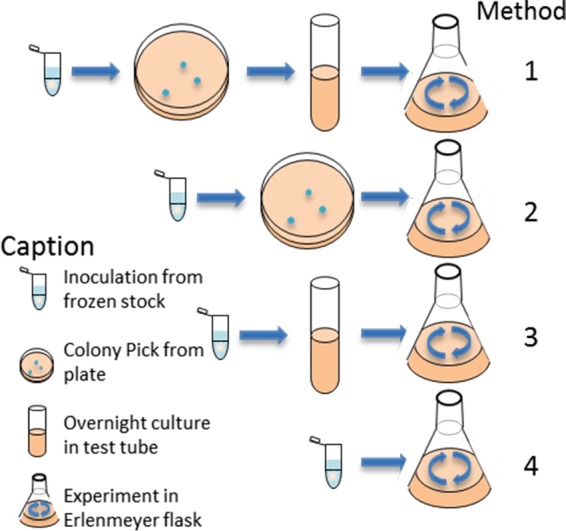
Schematic drawing of the four ways of inoculating liquid batch cultures. Method 1, frozen stock to a plate, a colony picked and added to LB for overnight culture in a test tube, and then inoculation for the experiment; method 2, frozen stock to a plate and then direct inoculation of a colony; method 3, frozen stock to LB for overnight culture in a test tube and then inoculation; method 4, direct inoculation from frozen stock.
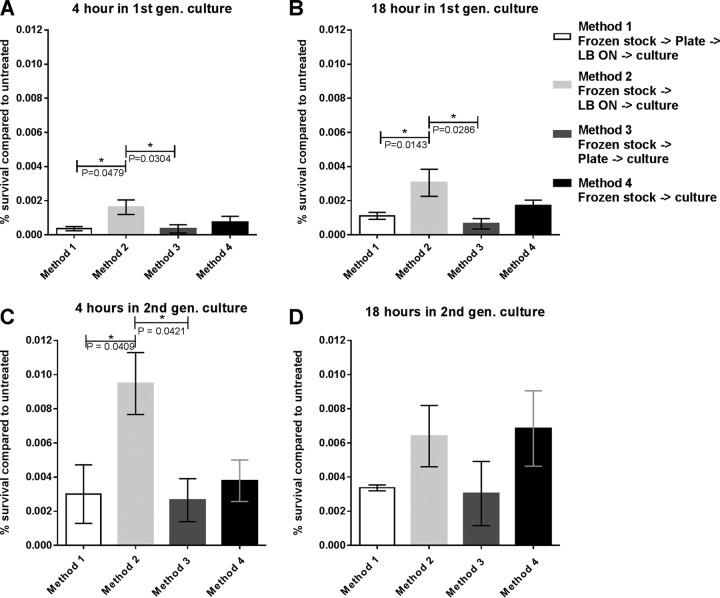
Aggregation was evaluated by confocal laser scanning microscopy (CLSM) after 4 and 18 h in first-generation cultures (defined as the cultures that were initially inoculated by the various methods) (Fig. 2). Significantly more aggregated biomass was observed after 4 h in the cultures inoculated from a single colony from a plate (method 2), compared to cultures from a single colony grown in LB overnight before inoculation (method 1) (P = 0.02) and cultures inoculated from frozen stock grown in LB overnight before inoculation (method 3) (P = 0.04). Cultures inoculated directly from the frozen stock were highly variable in the abundance of aggregation but exhibited significantly more aggregation than that found for methods 1 and 3 (P = 0.001 and P = 0.02, respectively) (Fig. 3A). After 18 h of growth, cultures inoculated from a plate (method 2) showed more aggregates than all other methods tested (Fig. 3B).
FIG 2.
Representative images of first-generation P. aeruginosa liquid cultures after 18 h of growth when inoculated by the four test methods. (A) Method 1. (B) Method 2. (C) Method 3. (D) Method 4. Collected z-stack Easy 3D images are shown. Magnification, ×630.
FIG 3.
Fractions of total P. aeruginosa biomass in aggregates for cultures inoculated by the four different methods. (A) Aggregation in cultures after 4 h in first-generation cultures. (B) Aggregation in cultures after 18 h in first-generation cultures. (C) Aggregation in cultures after 4 h in second-generation cultures. (D) Aggregation in cultures after 18 h in second-generation cultures. Aggregates were defined as cell clusters with biomass containing a minimum volume of 250 μm3. Biomass was normalized to the OD600 values of the cultures. Bars represent means with standard errors of the means (SEM). ON, overnight.
First-generation cultures were diluted to an optical density at 600 nm (OD600) of 0.005 in fresh medium and grown for an additional 4 h to create the second-generation culture. After 4 h, the second-generation cultures originally inoculated from a plate (method 2) contained significantly more aggregated biomass than did cultures inoculated with method 1 (P = 0.02) but not significantly more than cultures inoculated with method 3 or 4 (P = 0.14 and P = 0.59, respectively) (Fig. 3C). After 18 h of second-generation growth, cultures inoculated with method 2 had significantly more aggregated biomass than did cultures inoculated with methods 1 and 3 (P = 0.002 and P = 0.002, respectively). Cultures inoculated with method 4 displayed the same tendency to aggregate more than those inoculated with methods 1 and 3 (P = 0.02 and P = 0.02, respectively) (Fig. 3D). Thus, inoculation directly from an agar plate resulted in more aggregation in liquid cultures than did inoculation from a liquid LB overnight culture.
Tolerance toward tobramycin.
To test whether the method of inoculation altered antibiotic tolerance, cultures were exposed for 24 h to tobramycin at a concentration 10 times higher than the standard MIC determined by Etest for P. aeruginosa (25, 28). After 4 and 18 h of growth in both first- and second-generation cultures, samples were challenged with 10 μg ml−1 tobramycin for 24 h. Cultures treated after 4 h of growth in first-generation cultures, inoculated from plates (method 2), had significant higher survival rates than did cultures inoculated from LB overnight cultures (method 1 or 3) (P = 0.048 and P = 0.03, respectively). No increased survival in cultures initiated from a frozen stock (method 4), compared to those initiated from LB overnight cultures (method 1 or 3), was observed (Fig. 4A). After 18 h, the same relationship existed; plate-inoculated (method 2) cultures had significantly higher survival rates than did LB-overnight-culture-inoculated (methods 1 and 3) cultures (P = 0.014 and P = 0.029, respectively). Cultures inoculated from frozen stock inoculated (method 4) cultures had the same survival as seen in cultures inoculated from LB (method 1 and 3) (Fig. 4B).
FIG 4.
Survival of P. aeruginosa cultures treated with 10 μg ml−1 tobramycin for 24 h. (A) Survival in samples taken after 4 h in first-generation cultures. (B) Survival in samples taken after 18 h in first-generation cultures. (C) Survival in samples taken after 4 h in second-generation cultures. (D) Survival in samples taken after 18 h in second-generation cultures. The percent survival was based on CFU in treated versus untreated cultures. Bars represent means with SEM. ON, overnight.
In the second-generation culture, similar results were observed after 4 h of growth. The culture from plates (method 2) had an increased survival compared to the cultures from LB overnight (method 1 and 3) (P = 0.041 and P = 0.042, respectively). However, frozen stock inoculated (method 4) cultures did not show an increased survival compared to LB overnight inoculated (method 1 and 3) (Fig. 4C). After 18 h of growth in second-generation cultures, there was no significant difference in survival for any inoculation method (Fig. 4D).
Break up of aggregates by sonication.
To test whether the disruption of preformed aggregates before inoculation could remove the differences in aggregation seen in the cultures, the inoculum was sonicated just prior to inoculation, according to the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) guidelines for biofilm diagnostics (29). The sonication did not break up the preformed aggregates found in the inoculum from agar-grown colonies (method 2) to any significant degree (P = 0.54) (Fig. S3). The inoculated cultures grew for 18 h and were evaluated again. After 18 h, the cultures inoculated from a plate (method 2) had large visible aggregates observed with CLSM, as seen in cultures inoculated with nonsonicated inocula. The cultures inoculated from frozen stock (method 4) had some degree of aggregation as well. No preformed aggregates could be found in inocula from LB overnight cultures (methods 1 and 3) (Fig. S4). These data indicate that sonication appears unable to disrupt preformed aggregates.
Mixed-culture aggregates.
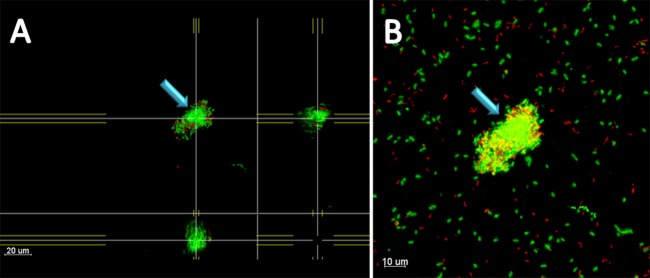
To elucidate the mechanism of formation of aggregates in the second-generation cultures, a PAO1 green fluorescent protein (GFP)-expressing first-generation culture containing aggregates was diluted and mixed 1:1 with a PAO1 mCherry-expressing culture of primarily planktonic single cells. With CLSM, we were able to observe aggregates formed in the second-generation culture after 4 h of growth. In several aggregates, a clear core consisting exclusively of GFP-marked cells surrounded by a layer of mixed GFP- and mCherry-marked cells was visible; no cores consisting of mCherry-marked cells were observed (Fig. 5). This finding suggests that aggregates were formed either during the inoculation process or in the first-generation culture and that, when transferred to the second-generation culture, aggregates recruited planktonic cells from the surrounding culture.
FIG 5.
CLSM presentation of P. aeruginosa aggregates in a second-generation culture after 4 h of growth. The first-generation culture was inoculated only with GFP-expressing P. aeruginosa according to method 2. At reinoculation into a second-generation culture, the first-generation culture was diluted and mixed 1:1 with a primarily single-cell culture of P. aeruginosa expressing mCherry. The turquoise arrows mark a central core of GFP-expressing cells. This core is surrounded by mixed GFP- and mCherry-expressing cells. (A) Top-down and side views. (B) Compiled Easy 3D image of all slides in a z-stack. Magnification, ×630.
Involvement of biofilm matrix components in aggregate formation.
To investigate the mechanism underlying the observed recruitment of single cells around aggregates, we tested the ability of several P. aeruginosa strains with different deficiencies in extracellular matrix production to aggregate in liquid culture. A PAO1 strain deficient in production of the exopolysaccharide Psl showed a significant decrease in aggregation, compared to wild-type PAO1 and a PAO1 strain deficient in production of the exopolysaccharide Pel (P = 0.0105 and P = 0.0049, respectively). Deletion of the surface adhesin CdrA, which has been shown to associate with the Psl exopolysaccharide (30), did not affect the ability to aggregate, compared to the wild-type strain. A decrease in aggregation similar to the PAO1 psl mutant was also observed for P. aeruginosa strain PA14, a strain not able to produce Psl (31), indicating that the phenotype was not specific to strain PAO1 (P = 0.0094) (Fig. 6).
FIG 6.
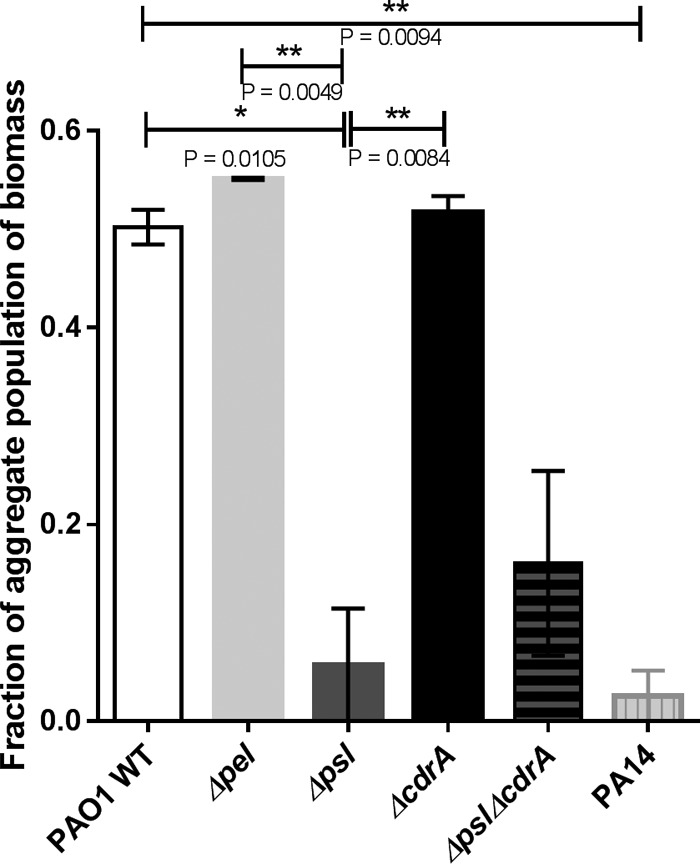
Fraction of P. aeruginosa biomass present as aggregates for strains with various deficiencies in extracellular matrix production. Cultures were inoculated according to method 2 and examined after 18 h of growth in a first-generation culture. The strains tested were PAO1 wild type (WT), PAO1 Δpel, PAO1 Δpsl, PAO1 ΔcdrA, PAO1 Δpsl ΔcdrA, and PA14 wild type. Bars represent means with SEM.
To further investigate the involvement of Psl exopolysaccharide in the aggregate recruitment of single cells in liquid cultures, a strain with inducible Psl production and Pel deficiency (Δpel PBAD-psl) was created. Aggregation of psl-induced or noninduced cultures was similar after 4 h of growth; after 18 h of growth, however, there was significantly more aggregated biomass in the cultures in which Psl production was induced, compared to the uninduced cultures (P = 0.037). Using these induced and uninduced cultures as inocula showed that the aggregate phenotype was conserved in the second-generation cultures (P = 0.0039), indicating that the aggregate phenotype was heritable at least over a few generations (Fig. 7).
FIG 7.
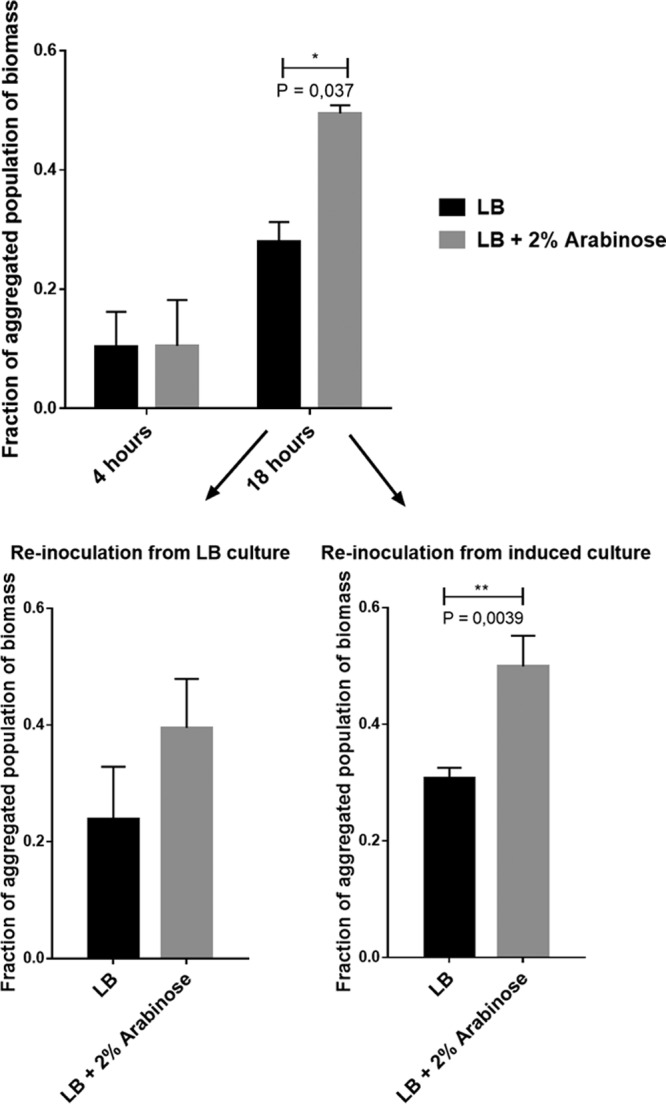
Fraction of aggregated biomass of the PAO1 Δpel PBAD-psl strain in cultures inoculated according to method 2, into LB with or without 2% arabinose (to induce psl). After 24 h of growth, cultures were reinoculated into fresh LB medium with or without 2% arabinose, creating four second-generation cultures. (Top) First-generation cultures sampled after 4 and 18 h. (Bottom left) Reinoculated cultures from the original noninduced (no arabinose) cultures. (Bottom right) Reinoculated cultures from the original induced (2% arabinose) cultures. Bars represent means with SEM.
To test the involvement of extracellular DNA (eDNA) in recruitment of single cells and in the structural integrity of aggregates, cultures inoculated directly from a plate were treated with DNase. After 4 h of treatment, no difference in the degree of aggregation between DNase-treated and untreated cultures could be observed (P = 0.38). After 18 h, the cultures treated with DNase had significantly (P = 0.0019) less aggregated biomass, compared to untreated cultures (Fig. S5).
Stability of the aggregated population in continuous culture.
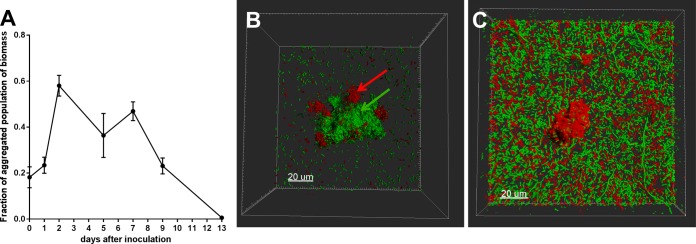
How long is the aggregate phenotype able to persist? To answer this question, a long-term chemostat experiment was performed with a 1:1 mixture of GFP- and mCherry-marked cells and a dilution rate of 0.2 per hour (bacterial doubling time of 5 h). Both the GFP- and mCherry marked populations were inoculated using agar-grown colonies (method 2), in an effort to induce maximal initial aggregation. For the entire duration of the experiment (13 days), aggregates were present to a degree comparable to that of an 18-h-old batch culture. However, the amount of aggregated population decreased toward the end of the 13 days. This may be a result of the constant exchange of medium, with slow dilution of the aggregate population because the formation of aggregates was not able to keep up with dilution (Fig. 8A). By visual examination of the aggregates being produced in the culture, a clear pattern emerged. Most aggregates had clear zones of monocolored bacteria in the first 2 to 3 days (Fig. 8B) but, as the experiment progressed, aggregates began to appear more homogeneous and mixed between GFP- and mCherry-tagged strains (Fig. 8C). Thus, aggregates remained a stable part of the population for least 7 to 9 days after inoculation.
FIG 8.
Findings from chemostat cultures grown for 13 days. (A) Fraction of aggregated biomass in samples taken on day 0, 1, 2, 5, 7, 9, and 13 postinoculation. Bars represent means with SEM. (B) Three-dimensional projection of an aggregate in chemostat culture after 2 days of growth. Red and green arrows indicate monocolored sections of the aggregate. (C) Three-dimensional projection of an aggregate in chemostat culture after 9 days of growth. The aggregate consists of a mixed population of red and green cells both originally inoculated directly from a plate, in an effort to distinguish between original aggregates (monocolor) and aggregates generated in chemostat culture (mixed colors). Magnification, ×630.
Aggregation of other common pathogenic bacterial species in liquid culture.
To test whether the observed link between the method of inoculation and the degree of aggregation is a general phenomenon, Escherichia coli and Staphylococcus aureus were tested in the aggregation assay. Inoculation from agar-grown colonies and LB overnight cultures (methods 2 and 3, respectively) were employed as two extremes of aggregate induction, according to the results from P. aeruginosa experiments. E. coli showed increased aggregation, compared to PAO1, with a mean aggregated population fraction of approximately 0.7 after 4 h in first-generation cultures and approximately 0.2 in 18-h-old first-generation cultures. No significant correlation between the method of inoculation and the degree of aggregation was observed for E. coli. S. aureus exhibited the same link between inoculation directly from a plate and increased aggregation during early growth (4 h) as seen for P. aeruginosa cultures (P < 0.0001) (Fig. 9). Thus, aggregation in liquid cultures is not limited to P. aeruginosa.
FIG 9.
Fractions of aggregated biomass for cultures of E. coli and S. aureus inoculated using method 2 or method 3. Cultures were evaluated for aggregation after 4 and 18 h of growth. (Left) E. coli. (Right) S. aureus. Bars represent means with SEM. ON, overnight.
To assess whether the recruitment of single cells to aggregates can occur between different species, tagged P. aeruginosa and E. coli were mixed as described above. Preaggregated E. coli cells were mixed with predominantly single-cell P. aeruginosa. After 18 h of combined growth, several aggregates with a monocolored center of E. coli could be found with a mixture of P. aeruginosa and E. coli surrounding the center (Fig. 10).
FIG 10.

Two examples of three-dimensional projections of multispecies aggregates. GFP-tagged E. coli cells from aggregate-rich cultures were mixed with predominately single-cell P. aeruginosa cultures. After 18 h of growth, the mixed cultures contained large proportions of aggregates with a central core of green GFP-tagged E. coli (marked with blue arrows) surrounded by a mixture of green E. coli cells and red mCherry-tagged P. aeruginosa cells.
DISCUSSION
Microbiology has seen a resurgence as evolutionary biologists, physicists, and engineers have adopted microbial model systems for testing theories and producing and testing products. The traditional microbiology viewpoint is that test-tube-grown populations are relatively homogeneous and distinct from biofilm-grown bacteria. The result is that phenotypic differences, including enhanced tolerance toward antibiotics and host defenses, are frequently assumed to discriminate test-tube-grown planktonic cells from biofilms (32–34). Here we show that the method of inoculation of planktonic cultures has a profound effect on the phenotypes observed, including antibiotic tolerance.
The antibiotic tolerance of planktonic cultures depends on bacterial aggregation, which varies based on the method of inoculation. Indeed, the increased survival rates seen for P. aeruginosa cultures inoculated directly from colonies on plates are in accordance with the findings that these cultures had more aggregated biomass and that tobramycin, an antibiotic known for killing actively growing cells (35), failed to kill parts of nonattached aggregates (25). Therefore, it is likely that the increased tobramycin tolerance of cultures inoculated using method 2 is a result of cells being protected either due to reduced metabolic activity in parts of the larger aggregated population or as a result of Psl providing a shielding effect against tobramycin (36, 37). This hidden difference in antibiotic tolerance among macroscopically identical cultures could have major effects on the outcomes of various studies; for example, this could be the case in susceptibility testing of antimicrobial agents, where aggregation in cultures may induce various fractions of tolerant cells.
Interestingly, the greater prevalence of aggregated biomass in second-generation cultures originally inoculated from a colony or directly from frozen stock was maintained even after ∼1,000-fold dilution in fresh medium. This indicates that cultures first seeded with aggregates inherit the frequency of aggregation of the parent culture and maintain it over several generations. We propose that the maintenance of aggregate frequency in second-generation cultures is mediated by recruitment of planktonic cells to preformed aggregates, which results in larger aggregates that eventually seed smaller aggregates that again recruit planktonic cells, resulting in a snowball effect of aggregate formation. Our proposal is supported by the fact that we were able to find a core of the ancestral aggregate within aggregates produced in second-generation cultures, indicating that these core parts were inherited from the first-generation culture and recruited cells. The fact that our chemostat cultures maintained high aggregate frequencies for 7 days (33 generations) indicates that a failure to pay attention to the possible introduction of preformed aggregates may have important repercussions for both short- and long-term experiments.
Psl has been shown repeatedly to be crucial for the formation of surface-attached P. aeruginosa biofilms (30, 38, 39). Our results indicate a similar dependency in the formation of nonattached aggregates in the liquid phase. The Psl-associated adhesin CdrA was not required for aggregation, indicating that Psl does not need this adhesin for aggregation. In contrast to Psl, the exopolysaccharide Pel, which has been the focus of great interest for its involvement in P. aeruginosa biofilms (40, 41), was not involved in aggregate formation in our system. Thus, the exopolysaccharide Psl seems to be a central mediator for the recruitment of single cells around preformed aggregates introduced into a liquid culture. eDNA was reported previously to be critical for the integrity of biofilm aggregates (25). With removal of eDNA from cultures by DNase supplementation, we did not see any change in aggregation in the early hours of growth (4 h), compared to untreated cultures. In contrast to this, all aggregates were removed from the cultures in the late stages of growth (18 h). This finding suggests that eDNA is involved as an adhesive that maintains the cohesiveness of the aggregate structure, as well as Psl, but may not be as essential for the recruitment of single cells in young cultures.
The ability of E. coli to aggregate in liquid cultures was remarkable but was not affected by the method of inoculation. It is worth noting that, even with the method found to generate the least aggregation for P. aeruginosa, E. coli cultures could still contain close to 70% of the biomass in an aggregated state. This raises the question of when, if ever, an E. coli culture is a completely homogeneous mixture of single cells. One possible mediator of this high propensity to aggregate could be curli, which has been shown to play a major role in the ability of E. coli to form aggregates and to adhere to surfaces (42, 43). Although curli expression in most E. coli strains is recognized to reach a maximum during growth below 30°C, the K-12 strain that was used in this study produces curli at 37°C (43). Interestingly, we found that preformed E. coli aggregates were able to recruit a mixture of E. coli and P. aeruginosa single cells, creating multispecies aggregates. This finding reveals a completely new area of possible interactions between species of aggregated bacteria and leads to ideas of multispecies aggregated biofilms.
S. aureus aggregation after 4 h of growth was greater when cultures were inoculated from a plate than when they were inoculated from a LB overnight culture, as found with P. aeruginosa. In contrast to what was observed with P. aeruginosa, however, almost no aggregates could be found in 18-h-old S. aureus cultures. The dissolution of aggregates in older S. aureus cultures might be explained by the ability of S. aureus to secrete proteases in the late stationary phase, which enables cells to disperse from an aggregated state to a single-cell state. The secretion of protease K, for example, is regulated by the agr quorum-sensing (QS) system in S. aureus (44). Despite this possible late-stationary-phase dispersal, our results indicate effects of the method of inoculation on S. aureus in early-exponential-phase cultures similar to those seen for P. aeruginosa. Interestingly, Haaber et al. described previously how S. aureus aggregates recruit single cells from the surrounding liquid phase during exponential-phase and late-exponential-phase growth, in a manner similar to that we observed with P. aeruginosa (45). Therefore, our results from studies of E. coli and S. aureus indicate that aggregation in liquid culture is not limited to P. aeruginosa.
In conclusion, we have discovered a connection between the method of inoculation of bacterial liquid cultures and the degree of aggregation within those cultures. This variation in P. aeruginosa aggregation was associated with altered tolerance toward the antibiotic tobramycin. These differences in aggregation and tobramycin tolerance were inherited by next-generation cultures when they were initiated from dilutions of the first-generation cultures. Our results stress the importance of consistency throughout experimental procedures beginning with inoculation, and they indicate that differences in inoculation procedures can have phenotypic consequences for >30 generations of bacterial growth and thereby can affect the outcomes of experiments.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa strains used in this study were of the PAO1 strain background, from the University of Washington (Seattle, WA) (46). The strain P. aeruginosa PAO1 ΔpelA PBAD-psl was created from P. aeruginosa PAO1 PBAD-psl (47) by introduction of the pelA allelic exchange vector pMPELA (20), essentially as described in reference 48. The E. coli strain used was K-12 MG1655 (ATCC 47076). S. aureus strain RN4220, constitutively expressing GFP from pAH13, was used (49). To enable visualization with CLSM, the wild-type PAO1 strain was made to express GFP or mCherry by Tn7 transformation (50). Other strains were stained using Syto9 (Molecular Probes, USA).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Pseudomonas aeruginosa | ||
| PAO1 | Wild-type strain | 53 |
| PAO1 Δpel | Pel-deficient knockout mutant | 30 |
| PAO1 Δpsl | Psl-deficient knockout mutant | 54 |
| PAO1 ΔcdrA | CdrA-deficient knockout mutant | 30 |
| PAO1 Δpsl ΔcdrA | Psl- and CdrA-deficient knockout mutant | 30 |
| PAO1 ΔpelA PBAD-psl | Pel-deficient knockout mutant with arabinose-inducible expression of psl | This study |
| PA14 | Wild-type strain | |
| Escherichia coli MG1655 | Wild-type strain derived from E. coli K-12 | 55 |
| Staphylococcus aureus AH13 | Strain derived from S. aureus RN4220, with constitutive expression of gfp | 49 |
| Plasmids | ||
| pMPELA | pelA allelic exchange vector | 20 |
| pAH13 | Plasmid for GFP expression in S. aureus | 49 |
Growth conditions.
All growth experiments were performed at 37°C. Experiments were performed in LB (BD, USA), and LB agar (2% [wt/vol]) plates were used for routine cultures. Frozen stocks (−80°C) consisted of 80% overnight cultures in LB and 20% glycerol (Sigma, USA). Liquid cultures were shaken at 180 rpm.
Growth experiments and aggregate evaluation.
Erlenmeyer flasks were inoculated in four ways (Fig. 1), as follows. In method 1, frozen stock was streaked on LB plates and left to grow overnight. Subsequently, one colony was inoculated into LB, grown overnight, and used for inoculation of the Erlenmeyer flask. In method 2, frozen stock was streaked on LB plates and left to grow overnight. One colony was picked, suspended in 1 ml LB, and used for inoculation of the Erlenmeyer flask. In method 3, LB was inoculated directly from frozen stock, left to grow overnight, and used for inoculation of the Erlenmeyer flask. In method 4, frozen stock was directly suspended in 1 ml of LB before inoculation of the Erlenmeyer flask.
All four types of P. aeruginosa inocula were added to obtain a final OD600 of 0.005 in the Erlenmeyer flask. Each 250-ml Erlenmeyer flask contained 100 ml LB medium. The four cultures were monitored by OD600 measurements with a UV-1800 spectrophotometer (Shimadzu, Japan) every 30 min during the first 7 h of growth. At 4 and 18 h, samples were taken for microscopic evaluation. These cultures are referred to as first-generation cultures. Time points were chosen to represent exponential-phase cultures (4 h) and stationary-phase cultures (18 h). For microscopic evaluation, 50 μl of culture was applied to a counting chamber (μ-slide VI Flat; Ibidi, Germany). Chambers were scanned in z-stacks in a tile scan (5 by 5 fields of view; 674.56 μm by 674.56 μm by 40 μm) with a Zeiss Imager Z2 microscope with a LSM 880 CLSM system and the accompanying Zen 2010 version 6.0 software (Zeiss, Germany). Imaris image analysis software (Bitplane, Switzerland) was used to measure the size and frequency of aggregated biomass, using the Measuring Pro package. Dense collections of cells with a volume of at least 250 μm3 were considered to be aggregates. This threshold was chosen to enable sufficient discrimination from several single cells connected on the surface and dense three-dimensional structural aggregates (see Fig. S1 in the supplemental material). The average volume of a single bacterium measured in this way is ∼4 μm3; therefore, 250-μm3 aggregates contain ∼62 cells.
First-generation cultures were reinoculated into fresh medium after 24 h of growth in Erlenmeyer flasks. The 24-h-old cultures were diluted into fresh LB to a final OD600 of 0.005 (∼1,000-fold dilution) and grown for an additional 18 h, with samples taken after 4 and 18 h for microscopic evaluation of aggregate formation. These cultures are referred to as second-generation cultures.
Aggregation of strains deficient in Psl, Pel, and CdrA expression.
PAO1 strains deficient in production of the exopolysaccharides Psl (Δpsl) and Pel (Δpel) and the surface adhesin CdrA (ΔcdrA), as well as the Psl-deficient P. aeruginosa strain PA14, were tested. Based on results from previous experiments, method 2 (frozen stock then agar plate then inoculation) was chosen for inoculation, to achieve the greatest possible initial aggregated population. Strains were grown for 24 h in first-generation cultures and reinoculated into fresh medium in second-generation cultures before being evaluated for aggregation after 18 h of growth.
A PAO1 Pel-deficient strain with inducible Psl production (Δpel PBAD-psl) was also tested for aggregation. Cultures inoculated from plates, according to method 2, were grown in LB with or without 2% arabinose. After 24 h, the cultures were reinoculated into fresh LB either with or without 2% arabinose. Samples were evaluated for aggregation at 4 and 18 h of growth in both the first- and second-generation cultures.
To assess eDNA involvement in aggregation, DNase (Pulmozyme [dornase alpha]; Genentech, USA) was added at 0 h after inoculation of cultures directly from a plate, according to method 2. DNase was diluted to a final concentration of 90 U ml−1, which was reported previously to disrupt P. aeruginosa biofilm aggregates (25).
E. coli and S. aureus aggregation.
For evaluation of the ability of E. coli and S. aureus to aggregate in liquid batch cultures, Erlenmeyer flasks were inoculated according to methods 2 and 3, directly from agar plates or LB overnight cultures. Aggregation was evaluated in samples taken at 4 and 18 h from first-generation cultures, as described above.
Disruption of aggregates in inocula by ultrasonication.
To disrupt preformed aggregates before introduction into cultures, inocula were treated with sonication (ultrasound) immediately prior to inoculation. Inocula were created based on the four previously described methods and subsequently were treated in a Branson 2510 ultrasonication bath (Branson, USA) with a program consisting of 5 min of degassing followed by 5 min of ultrasonication at 42 kHz (±6%) and 100 W. The level of aggregation was evaluated by microscopy in the inoculum before and after ultrasonication treatment, as well as in the resulting culture after 18 h of growth.
Antibiotic treatment of inoculated cultures.
Liquid P. aeruginosa batch cultures inoculated according to the four methods described were grown for 24 h, with constant shaking (first-generation cultures). The cultures were then reinoculated into second-generation cultures and grown for 18 h. At 4 and 18 h, two 15-ml aliquots of the cultures (both first- and second-generation cultures) were transferred to two new Erlenmeyer flasks. One flask was treated with 10 μg ml−1 tobramycin (Sigma), and one was left untreated as a control. The cultures were grown for an additional 24 h. One milliliter of culture was washed three times, degassed for 5 min, and sonicated before serial dilutions and plating to obtain CFU counts.
Recruitment and mixed cultures.
Cultures of a GFP-producing P. aeruginosa strain were inoculated according to method 2 and grown for 24 h. Concurrently, overnight cultures of an mCherry-marked strain were inoculated according to method 3. The mCherry-producing overnight culture was diluted to an OD600 of 0.005, regrown in LB to an OD600 of 0.1, diluted, and regrown again for 3 h in fresh medium, to keep the formation of aggregates to a minimum. The first-generation GFP-producing culture and the primarily single-cell mCherry-producing culture were diluted to an OD600 of 0.005 and mixed in a 1:1 ratio. The mixed cultures were grown for 4 h. Samples were subsequently taken for examination with CLSM.
Interspecies aggregation was tested by mixing, in a 1:1 ratio, a primarily single-cell mCherry-tagged P. aeruginosa culture with an E. coli culture tagged with GFP and inoculated according to method 2. Subsequent incubation samples were taken for examination with CLSM.
Aggregation in chemostat-grown cells.
A chemostat was constructed based on the design of Whiteley et al. (51). A medium based on a trace metal minimal medium with 10% (vol/vol) A10 phosphate buffer (52) and 10% (vol/vol) LB was used. This medium resulted in an OD450 of ∼0.5. The chemostat contained 200 ml of culture, with a flow rate of 40 ml h−1, resulting in a dilution rate of 0.2. The culture was aerated by continuously bubbling atmospheric air through it. The culture was continuously mixed with a magnetic stirrer set at ∼200 rpm. The chemostat was inoculated with a 1:1 ratio of GFP- and mCherry-tagged cells taken directly from agar plates according to method 2. The culture was adjusted to a final starting OD450 of 0.005. The chemostat was operated for 13 days, with samples being evaluated for aggregation at day 0, 1, 2, 5, 7, 9, and 13.
Statistical analysis.
Statistical significance was evaluated by one-way analysis of variance (ANOVA) with multiple comparisons for quantitative parametric data. Quantification of the area under the curve was used for the evaluation of growth patterns. P values of <0.05 were considered significant. All tests were performed in GraphPad Prism 6 (GraphPad Software, USA). All quantitative experiments were performed with at least biological triplicates. Antibiotic treatment was evaluated using three technical replicates for each biological sample.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Human Frontiers Science Program (grant RGY0081/201 to T.B.), the Lundbeck Foundation (T.B.), the Danish Council for Independent Research (grant DFF-1323-00177 to T.T.-N.). the National Institutes of Health (grant R01GM116547-01A1 to M.W.), and the Cystic Fibrosis Foundation (grant WHITEL16G0 to M.W.).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02264-17.
REFERENCES
- 1.Koch R. 1881. Methods for the study of pathogenic organisms. Mitt Kaiserlichen Gesundbeitsamte 1:1–48. [Google Scholar]
- 2.Jeanson S, Floury J, Gagnaire V, Lortal S, Thierry A. 2015. Bacterial colonies in solid media and foods: a review on their growth and interactions with the micro-environment. Front Microbiol 6:1284. doi: 10.3389/fmicb.2015.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austin B. 2017. The value of cultures to modern microbiology. Antonie Van Leeuwenhoek 110:1247–1256. doi: 10.1007/s10482-017-0840-8. [DOI] [PubMed] [Google Scholar]
- 4.Chick H. 1905. The biological limitations of the method of pure culture. New Phytol 4:120–124. doi: 10.1111/j.1469-8137.1905.tb05889.x. [DOI] [Google Scholar]
- 5.Houpikian P, Raoult D. 2002. Traditional and molecular techniques for the study of emerging bacterial diseases: one laboratory's perspective. Emerg Infect Dis 8:122–131. doi: 10.3201/eid0802.010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madigan MT, Martinko JM. 2006. Brock biology of microorganisms, 11th ed Pearson Prentice Hall, Upper Saddle River, NJ. [Google Scholar]
- 7.Willey JM, Sherwood L, Woolverton CJ. 2013. Prescott's microbiology, 9th ed McGraw-Hill Science Engineering, New York, NY. [Google Scholar]
- 8.Tille P. 2015. Bailey & Scott's diagnostic microbiology, 13th ed Mosby Elsevier Health Science, St. Louis, MO. [Google Scholar]
- 9.Holborow EJ. 1965. Topley & Wilson's principles of bacteriology and immunity, 5th edn. Immunology 8:529. [Google Scholar]
- 10.Sherman JM, Albus WR. 1924. The function of lag in bacterial cultures. J Bacteriol 9:303–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chesney AM. 1916. The latent period in the growth of bacteria. J Exp Med 24:387–418. doi: 10.1084/jem.24.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller M. 1895. Ueber den Einfluss von Fiebertemperaturen auf die Wachsthumsgeschwindigkeit und die Virulenz des Typhusbacillus. Z Hyg Infektionskr 20:245–280. [Google Scholar]
- 13.Wittmann HG. 1982. Structure and evolution of ribosomes. Proc R Soc Lond B Biol Sci 216:117–135. doi: 10.1098/rspb.1982.0065. [DOI] [PubMed] [Google Scholar]
- 14.Kjeldgaard NO, Maaloe O, Schaechter M. 1958. The transition between different physiological states during balanced growth of Salmonella typhimurium. J Gen Microbiol 19:607–616. doi: 10.1099/00221287-19-3-607. [DOI] [PubMed] [Google Scholar]
- 15.Sutherland JP, Bayliss AJ, Braxton DS. 1995. Predictive modelling of growth of Escherichia coli O157:H7: the effects of temperature, pH and sodium chloride. Int J Food Microbiol 25:29–49. doi: 10.1016/0168-1605(94)00082-H. [DOI] [PubMed] [Google Scholar]
- 16.Gibson AM, Bratchell N, Roberts TA. 1987. The effect of sodium chloride and temperature on the rate and extent of growth of Clostridium botulinum type A in pasteurized pork slurry. J Appl Bacteriol 62:479–490. doi: 10.1111/j.1365-2672.1987.tb02680.x. [DOI] [PubMed] [Google Scholar]
- 17.Fujikawa H, Kai A, Morozumi S. 2003. A new logistic model for bacterial growth. Shokuhin Eiseigaku Zasshi 44:155–160. doi: 10.3358/shokueishi.44.155. [DOI] [PubMed] [Google Scholar]
- 18.Steinhaus EA, Birkeland JM. 1939. Studies on the life and death of bacteria. I. The senescent phase in aging cultures and the probable mechanisms involved. J Bacteriol 38:249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schleheck D, Barraud N, Klebensberger J, Webb JS, McDougald D, Rice SA, Kjelleberg S. 2009. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PLoS One 4:e5513. doi: 10.1371/journal.pone.0005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD, Wozniak DJ, Harwood CS, Parsek MR. 2009. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol 191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner M, Loy A, Nogueira R, Purkhold U, Lee N, Daims H. 2002. Microbial community composition and function in wastewater treatment plants. Antonie Van Leeuwenhoek 81:665–680. doi: 10.1023/A:1020586312170. [DOI] [PubMed] [Google Scholar]
- 22.Alldredge AL, Cole JJ, Caron DA. 1986. Production of heterotrophic bacteria inhabiting macroscopic organic aggregates (marine snow) from surface waters. Limnol Oceanogr 31:68–78. doi: 10.4319/lo.1986.31.1.0068. [DOI] [Google Scholar]
- 23.Bjarnsholt T, Alhede M, Alhede M, Eickhardt-Sørensen SR, Moser C, Kühl M, Jensen PØ, Høiby N. 2013. The in vivo biofilm. Trends Microbiol 21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Kragh KN, Alhede M, Jensen PØ, Moser C, Scheike T, Jacobsen CS, Seier Poulsen S, Eickhardt-Sørensen SR, Trøstrup H, Christoffersen L, Hougen H-P, Rickelt LF, Kühl M, Høiby N, Bjarnsholt T. 2014. Polymorphonuclear leukocytes restrict the growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect Immun 82:4477–4486. doi: 10.1128/IAI.01969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alhede M, Kragh KN, Qvortrup K, Allesen-Holm M, van Gennip M, Christensen LD, Jensen PØ, Nielsen AK, Parsek M, Wozniak D, Molin S, Tolker-Nielsen T, Høiby N, Givskov M, Bjarnsholt T, Hoiby N, Givskov M, Bjarnsholt T. 2011. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS One 6:e27943. doi: 10.1371/journal.pone.0027943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Folsom JP, Richards L, Pitts B, Roe F, Ehrlich GD, Parker A, Mazurie A, Stewart PS. 2010. Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol 10:294. doi: 10.1186/1471-2180-10-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 28.Bjarnsholt T, Jensen PØ, Burmølle M, Hentzer M, Haagensen JAJ, Hougen HP, Calum H, Madsen KG, Moser C, Molin S, Høiby N, Givskov M. 2005. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151:373–383. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- 29.Hoiby N, Bjarnsholt T, Moser C, Bassi GL, Coenye T, Donelli G, Hall-Stoodley L, Hola V, Imbert C, Kirketerp-Moller K, Lebeaux D, Oliver A, Ullmann AJ, Williams C. 2015. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect 21(Suppl 1):S1–S25. doi: 10.1016/j.cmi.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 30.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol 51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 32.Costerton JW. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 33.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. 1987. Bacterial biofilms in nature and disease. Annu Rev Microbiol 41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 34.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuomanen E, Durack DT, Tomasz A. 1986. Antibiotic tolerance among clinical isolates of bacteria. Antimicrob Agents Chemother 30:521–527. doi: 10.1128/AAC.30.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Billings N, Ramirez Millan M, Caldara M, Rusconi R, Tarasova Y, Stocker R, Ribbeck K. 2013. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog 9:e1003526. doi: 10.1371/journal.ppat.1003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goltermann L, Tolker-Nielsen T. 2017. Importance of the exopolysaccharide matrix in antimicrobial tolerance of Pseudomonas aeruginosa aggregates. Antimicrob Agents Chemother 61:e02696-16. doi: 10.1128/AAC.02696-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kundukad B, Seviour T, Liang Y, Rice SA, Kjelleberg S, Doyle PS. 2016. Mechanical properties of the superficial biofilm layer determine the architecture of biofilms. Soft Matter 12:5718–5726. doi: 10.1039/C6SM00687F. [DOI] [PubMed] [Google Scholar]
- 39.Wang S, Liu X, Liu H, Zhang L, Guo Y, Yu S, Wozniak DJ, Ma LZ. 2015. The exopolysaccharide Psl-eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Environ Microbiol Rep 7:330–340. doi: 10.1111/1758-2229.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooley BJ, Thatcher TW, Hashmi SM, L'her G, Le HH, Hurwitz DA, Provenzano D, Touhami A, Gordon VD. 2013. The extracellular polysaccharide Pel makes the attachment of P. aeruginosa to surfaces symmetric and short-ranged. Soft Matter 9:3871–3876. doi: 10.1039/c3sm27638d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, Secor PR, Tseng BS, Scian M, Filloux A, Wozniak DJ, Howell PL, Parsek MR. 2015. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci U S A 112:11353–11358. doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. 2005. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol Immunol 49:875–884. doi: 10.1111/j.1348-0421.2005.tb03678.x. [DOI] [PubMed] [Google Scholar]
- 44.Boles BR, Horswill AR. 2008. agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog 4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haaber J, Cohn MT, Frees D, Andersen TJ, Ingmer H. 2012. Planktonic aggregates of Staphylococcus aureus protect against common antibiotics. PLoS One 7:e41075. doi: 10.1371/journal.pone.0041075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. 2006. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol 188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rybtke MT, Borlee BR, Murakami K, Irie Y, Hentzer M, Nielsen TE, Givskov M, Parsek MR, Tolker-Nielsen T. 2012. Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl Environ Microbiol 78:5060–5069. doi: 10.1128/AEM.00414-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malone CL, Boles BR, Lauderdale KJ, Thoendel M, Kavanaugh JS, Horswill AR. 2009. Fluorescent reporters for Staphylococcus aureus. J Microbiol Methods 77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hutchison JB, Rodesney CA, Kaushik KS, Le HH, Hurwitz DA, Irie Y, Gordon VD. 2014. Single-cell control of initial spatial structure in biofilm development using laser trapping. Langmuir 30:4522–4530. doi: 10.1021/la500128y. [DOI] [PubMed] [Google Scholar]
- 51.Whiteley M, Brown E, Mclean RJC. 1997. An inexpensive chemostat apparatus for the study of microbial biofilms. J Microbiol Methods 30:125–132. doi: 10.1016/S0167-7012(97)00054-7. [DOI] [Google Scholar]
- 52.Clark DJ, Maaløe O. 1967. DNA replication and the division cycle in Escherichia coli. J Mol Biol 23:99–112. doi: 10.1016/S0022-2836(67)80070-6. [DOI] [Google Scholar]
- 53.Holloway BW. 1995. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol 13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 54.Kirisits MJ, Prost L, Starkey M, Parsek MR. 2005. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 23:4809–4821. doi: 10.1128/AEM.71.8.4809-4821.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tatum EL, Lederberg J. 1947. Gene recombination in the bacterium Escherichia coli. J Bacteriol 53:673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.