Abstract
HER2 transmembrane receptor is an important target in immunotherapy treatment of breast and gastroesophageal cancer. Molecular imaging of HER2 expression may provide essential prognostic and predictive information concerning disseminated cancer and aid in selection of an optimal therapy. Radiolabeled low molecular weight peptide ligands are particularly attractive as probes for molecular imaging, since they reach and bind to the target and clear from non-target organs and blood stream faster than bulky antibodies. In this study, we evaluated a potential HER2-imaging probe, an A9 nonapeptide, derived from the trastuzumab-Fab portion. Its cellular uptake was investigated by mass spectrometry analysis of the cytoplasmic cellular extracts. Moreover, based on in-silico modeling, DTPA chelator was conjugated to N-terminus of A9. 111In-labeled A9 demonstrated nanomolar affinity to HER2-expressing BT474 cells and favorable biodistribution profile in NMRI mice. This study suggests that the peptide A9 represents a good lead candidate for development of molecular probe, to be used for imaging purposes and for the delivery of cytotoxic agents.
Introduction
HER2 is a transmembrane protein, belonging to the ErbB or epidermal growth factor receptor (EGFR) family1. Structurally it is featured by an N-terminal extracellular ligand binding portion (ECD), a single alpha-helix transmembrane segment (TM), and an intracellular protein tyrosine kinase. Overexpression of HER2 is associated with a wide number of cancers, including lung, breast, and ovarian, as well as adenocarcinomas of colon and salivary gland2,3. In particular, HER2 receptor is found to be overexpressed in about 30% of primary breast cancers4,5, mainly because of its gene amplification. HER2 has a great tendency to dimerize with other ErbB family receptors, which results in activation of the HER signaling pathways6. Moreover, it has been found that among all ErbB receptor dimers, the heterodimers containing HER2 receptors have the highest mitogenic potential7–15.
HER2 has been extensively investigated, in order to develop new HER2-specific cancer therapies16,17. Several studies have focused on immunotherapy of HER2-positive tumors due to its transmembrane accessibility. Trastuzumab (Herceptin) is a humanized IgG1 monoclonal antibody that recognizes an epitope in the extracellular domain (ECD) of HER218,19. It was approved in 1998 by FDA (Food and Drug Administration) for therapy of metastatic breast cancer in combination with cytotoxic chemotherapy20. Several strategies have been used to increase the effectiveness of therapy of HER2-positive tumors. For instance, HER2-specific antibodies, their fragments and affibody molecules have been conjugated to cytotoxic molecules21–24.
It is essential to select patients with HER2-expressing metastases for HER2-targeting therapy, as this antigen has sufficient expression level only in a fraction of the tumors. A biopsy of primary breast cancer is not informative in this case as discordance in HER2 expression between primary tumor and metastases might be up to 18%25. Therefore, radionuclide molecular imaging using radiolabeled anti-HER2 antibodies was suggested, which is a non-invasive approach for determination of HER2 expression in the disseminated breast cancer20,26–30. However, intact antibodies are not considered ideal probes for radionuclide molecular imaging due to their long blood residence time, which results in low tumor/blood (T/B) ratios limiting imaging contrast31. Moreover, intact IgGs often have a high liver uptake, mediated by interaction of the Fc portion with hepatocyte receptors, which reduces the imaging sensitivity for detection of liver metastases.
Antibody fragments, such as Fab or F(ab’)2 have been considered better candidates for tumor imaging. In fact, their relatively small size results in higher extravasation and tissue penetration rates as well as improved blood clearance. Recently, radiolabeled anti-HER2 Fab fragments32–34, and novel recombinant antibody forms (scFv, diabodies, minibodies)35–37, as well as HER2-binding affibody38–40 have been successfully used for imaging purposes.
In this regard, peptide molecules, characterized by low molecular weight, can represent the most appropriate HER2 targeting tracers, since they exhibit superior ability to diffuse across tissues, improving tumor exposure to the drug. Moreover, the peptides are easily produced and the chemistry for their radiolabeling is easier and more flexible compared with monoclonal antibodies and their fragments.
In this context, we have recently developed and studied several trastuzumab (Fab)-derived peptides capable to bind HER241. In particular, the A9 peptide was found to be particularly interesting, since it showed a dissociation constant in nanomolar range for the receptor model HER2-DIVMP41–43. Therefore, it was studied as a promising candidate for development of an HER2-specific imaging probe. In this context, the ability of this peptide to be internalized into the tumor cell by receptor-mediated endocytosis has been investigated, since it is considered a crucial step in the process of in vivo receptor targeting with radiolabeled peptides44–46.
As main goal, we evaluated site-specific conjugation of DTPA chelator to A9, labelling of DTPA-A9 conjugate with radionuclide 111In, evaluation of 111In-DTPA-A9 binding to living HER2-expressing cancer cells in vitro, its stability in vitro and in vivo and biodistribution of 111In-DTPA-A9 in vivo.
Results and Discussion
Cellular uptake of A9 peptide
The internalization of radiolabeled peptide ligands, upon receptor binding, represents an important event for in vivo tumor targeting, since it results in accumulation of radioactive in the tumor tissues. Therefore, it can considerably improve the sensitivity of diagnostic procedures, as well as the efficacy of receptor-mediated radiotherapy.
To this aim, the cytoplasmic localization of A9 peptide was investigated by mass spectrometry analysis in HER2-positive human breast cancer BT474 cells. In particular, A9 peptide was bound to a biotin moiety at its N-terminus, in order to be easily extracted from cytoplasmic cell lysate by using streptavidin-coated magnetic beads. Then, the mixture obtained was analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
As shown in Fig. 1 (schematic view of the employed protocol), BT474 cells were treated with biotinylated-A9 for 3 hours, then washed with a PBS buffer in presence of 0.5% Trypsin-Ethylenediaminetetraacetic acid, in order to be collected. The obtained cell pellets were incubated with a digitonin solution to permeabilize the plasma membranes and recover the intracellular fractions47. Subsequently, a purification step was performed by incubating the soluble intracellular fraction with the streptavidin-coated magnetic beads, which strongly catch any biotinylated compounds. After filtration, the unbound proteins were removed and the beads were washed with a PBS buffer, before the treatment with a solution of EDTA in formamide at high temperature, which allowed the elution of biotinylated species. The final step consisted of a LC-MS/MS analysis of the collected biotinylated molecules (see Methods section for details). The same procedure was carried out on untreated BT474 cells.
Figure 1.
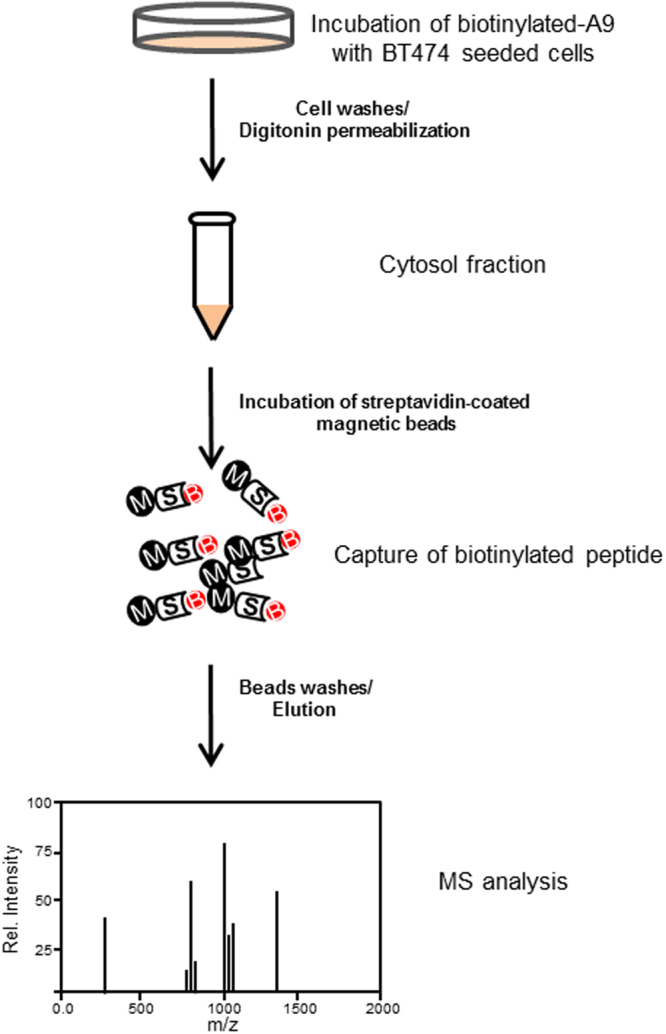
Schematic diagram of affinity tag in an in vitro pull-down assay and mass spectrometry analysis.
The collected samples were then characterized by using the simple, sensitive and efficient analytical technique LC-MS/MS, in multiple-reaction monitoring (MRM) mode. The MRM strategy provides a unique fragment ion that can be selectively monitored, also in a complex mixture48. In order to investigate the fragmentation pattern for biotinylated-A9, pilot LC-MS/MS-MRM experiments were also performed on the synthetic peptide. They allowed us to identify the diagnostic fragment, which is essential for developing a high-sensitivity MRM assay. Therefore, the selection of the most intense ion, among all individual fragment ions, could be performed (see Supplementary Information Fig. S1). In particular, the fragment ion 477.1 m/z was selected as a diagnostic one of the precursor 615.3. On the basis of these data, SRM (selected transition monitoring) mode experiments at m/z 615.0–477.2 on our samples were also performed.
In Fig. 2a is shown the MS/MS profiles of the samples obtained for BT474 cells incubated with the biotinylated A9, in which the SRM peak is easily detectable (Fig. 2b; see also Supplementary Information Fig. S2). On the other hand, the same SRM peak was not detected in sample collected for untreated BT474 cells, which were used as a negative control (see Supplementary Information Fig. S3).
Figure 2.
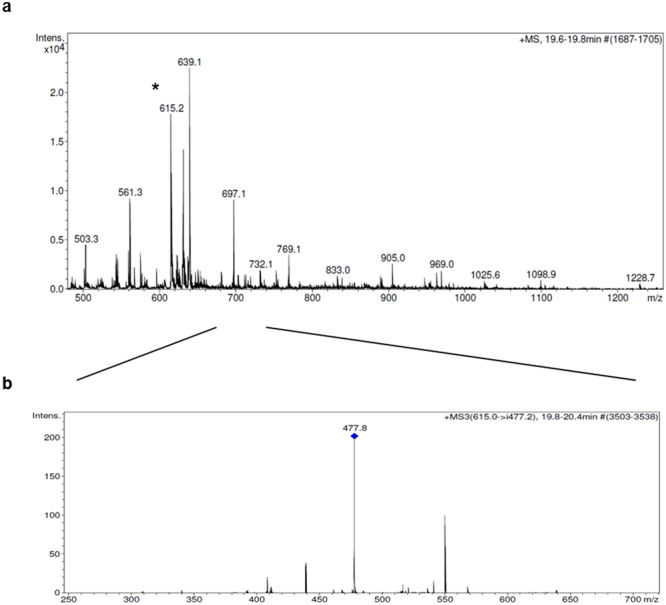
(a) MS/MS spectra of the [M + 2 H]2+ precursor of biotinylated A9 at 615.2 m/z (*). (b) SRM plot of the selected transition at m/z 615.0–477.2.
All these results suggest a putative internalization of A9 peptide inside the cells.
Design and synthesis of the conjugate A9-DTPA
The final goal of our studies is to develop new targeting agents, which are smaller than antibodies and affibody molecules, and possess a high specificity and affinity for HER2 receptor. They could be used in the non-invasive imaging of HER2-expression level in metastases, thus allowing stratification of patients who would most likely respond to targeted anti-HER2 therapies. This should prevent over- and under treatment and improve efficacy of breast cancer therapy. Therefore, we evaluated the possibility to develop a peptide A9-derivative bearing a chelating agent able to coordinate radioactive metals for applications in cancer diagnosis by nuclear medicine techniques.
We synthesized the A9 amidated at the C-terminus and coupled with an acyclic chelator at the N-terminus (Fig. 3). These peptide modifications represent a good strategy for stabilizing the peptide molecule and protect it against the exopeptidases. The acyclic chelator DTPA is routinely used to provide in vitro and in vivo stable complexes of the radioactive isotope 111In. The issue of the metal complex stability is crucial for the radiolabeling of peptide molecule: even though many of the chelating agents are both thermodynamically stable and kinetically inert, a complexation of a metal by a chelator is always a reversible process. In addition, some blood proteins, e.g. transferrin, possess metal-complexing properties, which lead to transchelation of a radionuclide from an imaging agent to these proteins49. This results in longer retention of a radionuclide in circulation and increases background during imaging. On the other hand, the required stability depends on the residence time of a radiolabeled probe in the circulation. For short peptides, which clear rapidly from blood via kidneys, the exposure to transferrin is short and no measurable transchelation takes place. For example, the heptadentate DTPA provides an adequate in vivo stability of binding of 111In to octapeptide octreotide, which has a rapid blood clearance50, and 111In-DTPA-octreotide (OctreoScan) is routinely used in clinics for imaging of neuroendocrine tumors51. Taking all these aspects in consideration, we synthesized the conjugate reported in Fig. 3.
Figure 3.
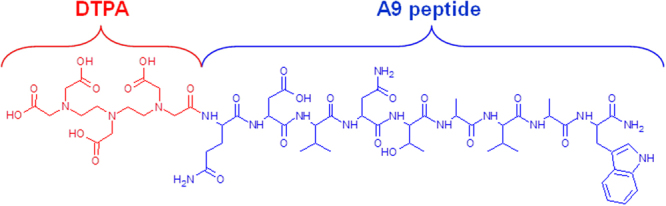
Chemical structure of DTPA-A9 conjugate.
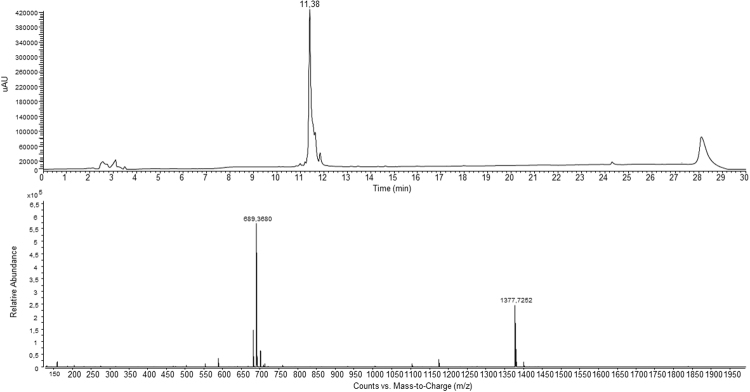
In more details, the peptide synthesis was performed manually in solid phase using the standard Fmoc-protocol. Rink Amide resin (Merck Millipore) was employed as solid support to release the peptide with the C-terminal amide. The heptadentate DTPA chelator was introduced by using its free carboxylic function to the peptide N-terminus. The final compound was obtained in good yield (60%) and with high purity grade (>98%) after RP-HPLC purification and it was fully characterized for its identity by mass spectrometry (Fig. 4).
Figure 4.
LC-MS profile of the purified DTPA-A9 conjugate.
Molecular modeling study
In a recent work, we have studied the putative complex between A9 ligand and the receptor model HER2-DIVMP by employing a computational approach; the identified binding site was further validated by mutagenesis, as well as fluorescence studies52.
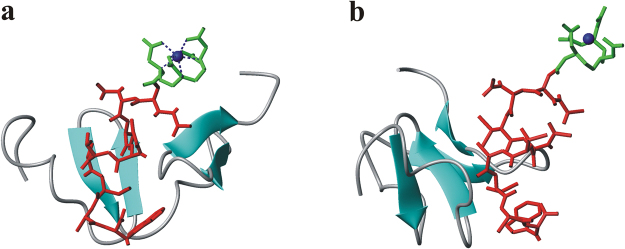
To investigate the chelating agent site of linkage, which does not interfere on the peptide-receptor interaction, we have modelled the 3D structure of the complex between HER2-DIVMP and 111In-DTPA-A9, starting from the previously performed molecular dynamics (MD) study52. In details, the structure of 111In-DTPA was retrieved from the CSD (The Cambridge Structural Database)53 (Fig. 5), whereas a representative conformation of the complex A9/HER2-DIVMP was obtained by our previous Molecular Dynamics study52.
Figure 5.
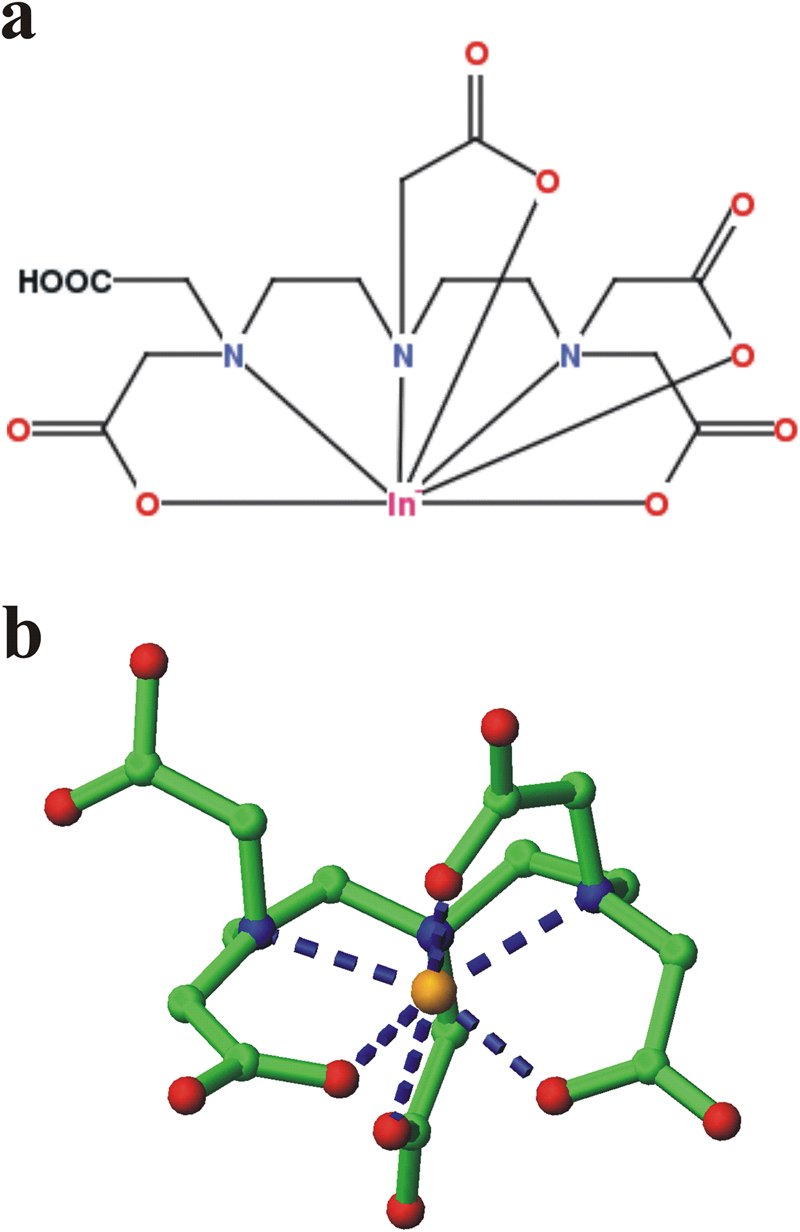
2D diagram (a) and 3D image (b) of 111In-DTPA complex from Cambridge Structural Database (CSD) (code XIKVES)54. (b) Atoms are displayed in ball-and-sticks mode. Indium atom is in orange.
Subsequently, the model complex between HER2-DIVMP and 111In-DTPA-A9 was built through modelling and energy minimization using Insight II package (see Methods section)54, it is shown in Fig. 6. The model indicates that the presence of 111In-DTPA is compatible with the structure suggested by our previous MD study52, since it is directly bounded to the N-terminus of A9, which is not involved into interaction with HER2-DIVMP receptor. A scrambled variant of A9 (SA9) has been designed and synthesized for specificity test. The sequence of scrambled-A9 (WAVQNTDAV) was selected by using a free of charge software.
Figure 6.
(a) Model complex between HER2-DIVMP and 111In-DTPA-A9. (b) Side-view of the model complex, rotated of 90° along z-axis. HER2-DIVMP receptor is shown in cartoon representation. 111In-DTPA-A9 is displayed in sticks (A9: red sticks, DTPA: green sticks).
Radiolabeling of 111In-DTPA-A9
Both DTPA-A9 and scrambled DTPA-SA9, which was used in specificity test, were labelled with 111In in 0.2 M ammonium acetate, pH 5.5, at room temperature. A blank experiment (without adding DTPA-A9) was performed to facilitate interpretation of chromatograms. Radio-HPLC chromatograms are presented in Fig. 7.
Figure 7.
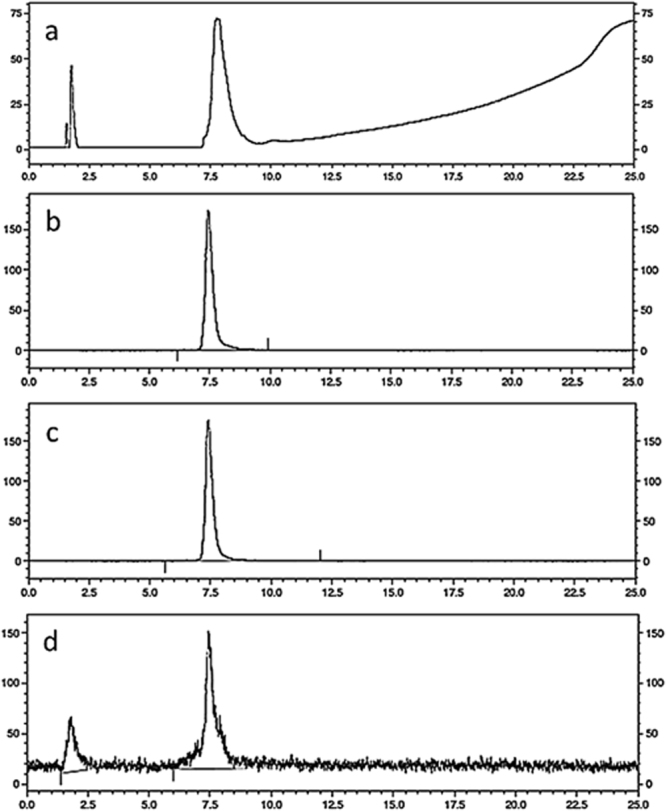
HPLC UV (a) and radio-chromatograms (b) of a freshly labeled 111In-DTPA-A9. (c) Radio-chromatogram of murine blood plasma after 60 min incubation with 111In-DTPA-A9 (d). Radio-chromatogram of murine blood 15 min after injection 111In-DTPA-A9.
As expected, non-conjugated 111In was eluted at 2.1 min, immediately after dead volume. The retention time of 111In-DTPA-A9 was 7.5 min (Fig. 7a,b) and the retention time of 111In-DTPA-SA9 was 7.3 min. Labeling was efficient at room temperature. The analytical radiochemical yield of both111In-DTPA-A9 and 111In-DTPA-SA9 labelling was over 98%. The specific radioactivity of 1 MBq/µg was achieved.
There was no measurable release of radioactivity after 15 or 60 min (Fig. 7c) of 111In-DTPA-A9 incubation with murine blood plasma, which demonstrates its stability against blood-borne peptidases.
In vitro evaluation of kinetics of 111In-DTPA-A9 binding to HER2-expressing cells
The kinetics of 111In-DTPA-A9 binding to and dissociation from HER2-expressing BT474 cells in vitro was measured using LigandTracer. The InteractionMap is presented in Fig. 8a. The best fitting of the binding curves was obtained using a 1:2 interaction model. This indicates that the binding of 111In-DTPA-A9 to HER2 on living cells is mediated by two binding sites, one strong, with affinity of 4.9 nM, and one weaker, with affinity 103 nM. The presence of two binding sites with different affinities on living cells is intriguing but not unique. Similar phenomenon has been found earlier for binding of 125I-labeled anti-HER2 antibody trastuzumab and 111In-labeled anti-HER2 affibody molecules to different HER2-expressing cells55. One possible explanation for two affinities of 111In-DTPA-A9 binding to HER2 might be homo- and heterodimerization of receptors on cancer cells. The dimer formation may slightly change the conformation of receptor, resulting in the change the interaction with a ligand. Similar effect has been demonstrated for other member of HER family, EGFR, by Björkelund and co-workers56.
Figure 8.
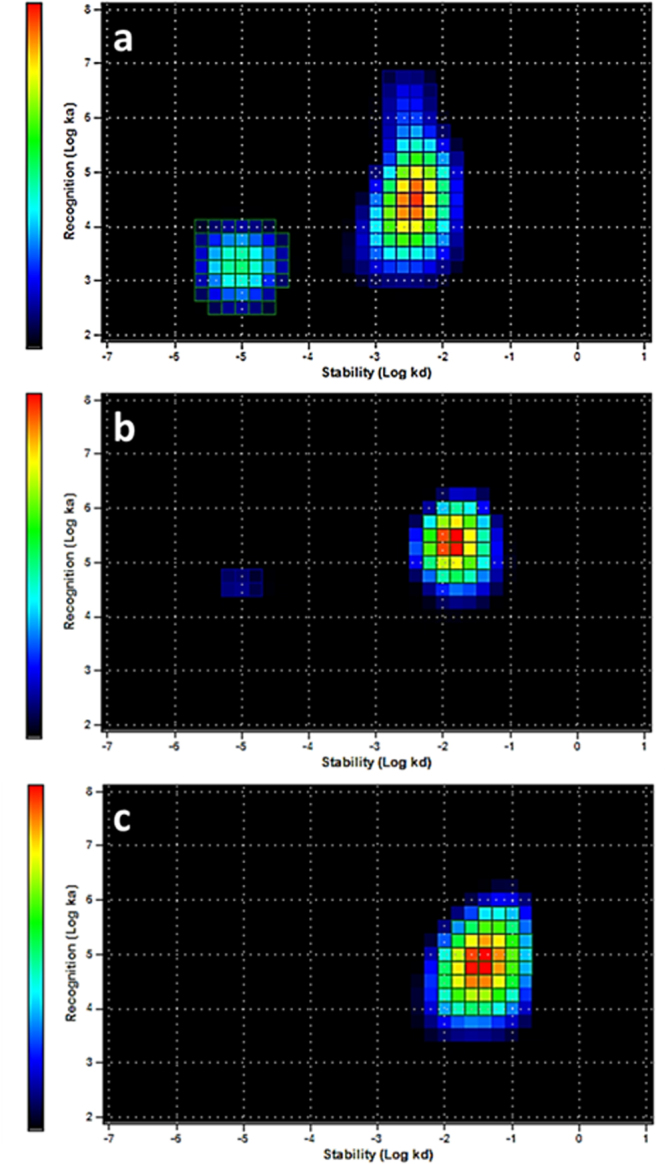
InteractionMaps of 111In-DTPA-A9 interaction with HER2-expressing BT474 cells (a). Concentrations of 111In-DTPA-A9 were 4, 12 and 36 nM. Fitting binding LigandTracer curves to 1:2 interaction models suggest presence of two interactions, one stronger (4.9 nM) and one weaker but predominant (103 nM); (b) 111In-DTPA-A9 interaction with HER2-expressing BT474 cells in the presence of large excess of trastuzumab. Concentrations of 111In-DTPA-A9 and timing were the same as above; (c) 111In-DTPA-S-A9 interaction with HER2-expressing BT474 cells. Concentrations of 111In-DTPA-S-A9 (4, 12 and 36 nM) and timing were the same as for 111In-DTPA-A9.
Further elucidation of this phenomenon was obtained by performing LigandTracer experiment in the presence of a large excess of trastuzumab (Fig. 8b). In these conditions, the high affinity interaction was nearly completely suppressed, upon traztuzumab displacement, while low affinity interaction remained. This suggested that this low affinity interaction might be not HER2-specific.
To further clarify this issue, an additional experiment was performed. Specifically, a scrambled conjugate (111In-DTPA-SA9), i.e. having the same amino acids of A9 but in a different order, variant of DTPA-A9, was created. As it shown in Fig. 8c, the InteractionMap, evaluating the binding of 111In-DTPA-SA9 to Her2-positive BT474 cell, revealed only the low affinity interaction (103 nM).
Taken our results together, the InteractionMap measurements suggest that there are two interactions of 111In-DTPA-A9 with HER2-expressing cell. One is HER2-specific, as it can be suppressed by trastuzumab and does not appear in the case of the scrambled variant. Another is non-HER2 specific and might be characteristic for peptides containing the same amino acids as A9. Accordingly, the hypothesis concerning homo- or hetero- dimerization as a cause of low affinity binding should be excluded.
Our previous studies have demonstrated that affinity in a single digit nanomolar range is required for efficient targeting of tumors with high HER2 expression using affibody molecules57. A possible way to increase the affinity of A9-based probe to HER2 is a di- or multimerization. Even dimeric form of A9 is small enough to provide both efficient extravasation and rapid renal excretion of an unbound tracer. However, this might require re-modelling and, possibly, designing of a branched linker between N-termini of monomeric units, permitting conjugation of a chelator.
In vivo studies
Specific high affinity binding to molecular target is a necessary precondition for development of an imaging probe, but it is not sufficient. Off-target interaction of the probe with blood and normal tissue might result in undesirable accumulation of an imaging probe in normal organs. Such accumulation would reduce an imaging contrast leading to reduced sensitivity of in vivo diagnostics. Particularly, a weak non-HER2-specific interaction found in the InteractionMap experiments might be a reason for undesirable accumulation in normal tissues. Therefore, we measured biodistribution of 111In-DTPA-A9 in normal mice at 1 h after injection. Data concerning the biodistribution of 111In-DTPA-A9 in BALB/c nu/nu mice 1 h post injection is presented in Table 1. 111In-DTPA-A9 showed rapid clearance from blood and normal organs (Table 1). The low (approximately 1% of injected activity) radioactivity in the gastrointestinal tract with content indicates minor role of hepatobiliary excretion in the elimination of the radiolabeled peptide. More apparent was renal excretion with subsequent tubular reabsorption, as kidney uptake of 111In-DTPA-A9 was more than 7% ID/g (2% ID/sample).
Table 1.
Data are presented as average ID%/g and ID%/sample values for 3 animals ± standard deviation. (Data for intestines with content and carcass are presented only as % ID/sample).
| organ | % ID/g | % ID/organ (sample) |
|---|---|---|
| blood | 0.24 ± 0.04 | |
| lung | 0.5 ± 0.3 | 0.06 ± 0.03 |
| liver | 0.44 ± 0.04 | 0.47 ± 0.03 |
| spleen | 0.22 ± 0.01 | 0.02 ± 0.01 |
| stomach | 0.27 ± 0.07 | 0.05 ± 0.06 |
| pancreas | 0.13 ± 0.03 | 0.01 ± 0.00 |
| brain | 0.03 ± 0.03 | 0.00 ± 0.01 |
| kidney | 7.0 ± 0.6 | 2.00 ± 0.1 |
| muscle | 0.01 ± 0.01 | 0.04 ± 0.01 |
| bone | 0.16 ± 0.13 | 0.01 ± 0.01 |
| GI tract (with content) | ------------ | 0.83 ± 0.48 |
| carcass | ------------- | 3.47 ± 1.01 |
However, this value was much lower than renal uptake of 111In-labeled trastuzumab Fab fragments (53.0 ± 15.94%ID/g at 24 h after injection)33, minibodies (16–30%ID/g at 2 h after injection)34 or affibody molecules (200–250%ID/g at 4 h after injection)58 in murine models. The clearance from blood was rapid, suggesting that the label was stable, and no transchelation to transferrin took place. The radioactivity accumulation was low in liver, lung and bones, i.e. in the major metastatic sites for breast cancer, the main target of anti-HER2 therapies. Thus, the in vivo data suggest that 111In-DTPA-A9 has favorable biodistribution profile, and the low affinity unspecific interaction does not cause any undesirable accumulation in healthy organs and tissues
It has to be noted that although incubation of A9 in vitro with blood serum demonstrated high stability during one hour (see Supplementary Information Fig. S4 and Fig. 7c), peptidases anchored in blood vessels might degrade peptides in vivo59–61. Therefore, we evaluated stability of 111In-DTPA-A9 in vivo using the method developed by Nock and co-workers59. Our experiment demonstrated that 92% of blood-borne activity was associated with DTPA-A9 at 5 min after injection and ca. 70% at 15 min after injection (Fig. 7d). For comparison, only 5% of activity was bound to mingastrin analogue DOTA-MG11 at 5 min after injection, and the rest was associated with the peptide degradation products. In the case of 111In-CP04, over 70% of 111In in blood was bound to peptide at 5 min after injection61. This was considered as a good stability, and 111In-CP04 is currently prepared for clinical study for imaging of medullary thyroid carcinoma. Apparently, 111In-DTPA-A9 has sufficient in vivo stability.
In conclusion, the ability of A9 peptide to specifically bind the HER2-positive cell plasma membrane with a subsequent internalization was demonstrated. The conjugation of DTPA to N-terminus of A9 via amide bond enables stable labeling with 111In. The 111In-DTPA-A9 has nanomolar affinity to HER2-expressing cells. The biodistribution data suggest that 111In-DTPA-A9 clear rapidly from blood and does not bind to any noticeable extent to any tissues. Thus, 111In-DTPA-A9 is a promising starting point for development of HER2-imaging probes and also for optimization of ligands useful for the targeting of HER2-positive tumor tissue.
Methods
Materials
Fmoc-protected amino acids, N-hydroxybenzotriazole (HOBT), O-Benzotriazole-N,N,N’,N’-tetramethyl-uroniumhexafluoro- phosphate (HBTU) and Benzotriazolyloxy-tris[pyrrolidino]-phosphonium hexafluorophosphate (PyBOP) were purchased from Calbiochem-Novabiochem (Laufelfingen, Switzerland), piperidine and diisopropylethylamine (DIPEA) were purchased from Fluka (Milwaukee, WI, USA), Rink Amide MBHA resins were purchased from Sigma-Aldrich (St Louis, MO, USA). DTPA(tBu)4 was purchased from CheMatech (Dijon, France).
Other solvents were purchased from Sigma-Aldrich or Fluka (Milwaukee, WI, USA) and were used without further purification, unless otherwise stated.
Analytical RP-HPLC runs were carried out on a HP Agilent Series 1100 system using a C18 column, 250*4.6 mm ID (Phenomenex, Torrance, CA, USA) at a flow rate of 1.0 mL min−1 or on a C18 monolithic column 50*2 mm ID (Phenomenex, Torrance, CA, USA), operating at 0.6 mL min−1. Preparative RP-HPLC was carried out on a Shimadzu 8 A chromatograph coupled with an UV detector, using a C18 column, 22*250 mm (Phenomenex Torrance, CA, USA) at a flow rate of 20 mL min−1. For all the RP-HPLC procedures the solvent system used was: H2O 0.1% TFA (A) and CH3CN 0.1% TFA (B). Separations were achieved applying a linear gradient of B from 20% to 80% in 20 min and monitoring at 210 and 280 nm.
LC–MS data were obtained using a Finnigan Surveyor MSQ single quadrupole or LCQ DECA XP MAX electrospray ionization mass spectrometers, coupled with a Surveyor HPLC (ThermoFisher), operating in the full scan positive mode, between 200 and 2000 m/z.
Buffers, including 0.1 M phosphate buffered saline (PBS), pH 7.5, 0.2 M ammonium acetate, pH 5.5, and 0.2 M citric acid were prepared using common methods from chemicals supplied by Merck (Darmstadt, Germany). High-quality Milli-Q© water (resistance higher than 18 MΩ cm) was used for preparing the solutions. Buffers, which were used for labeling, were purified from metal contamination using Chelex 100 resin (Bio-Rad Laboratories, Richmond, USA). [111In]-indium chloride was purchased from Covidien (Hazelwood, US) as a solution in 0.05 M hydrochloric acid. The radiochemical purity of the labelled peptide was analysed using HPLC system from Beckman. Ketalar [ketamine] (50 mg/mL, Pfizer, NY, USA), Rompun [xylazin] (20 mg/mL, Bayer, Leverkusen, Germany), and Heparin (5000 IE/mL, Leo Pharma, Copenhagen, Denmark) were obtained commercially.
The radioactivity was measured using an automated gamma-counter with a 3-inch NaI(Tl) detector (1480 WIZARD, Wallac OY, Turku, Finland). The data were corrected for background.
Peptide synthesis
The synthesis of A9 and scrambled A9 (SA9) peptides were carried out manually by solid phase method using the standard Fmoc-protecting group strategy. Appropriate Fmoc-amino acid derivatives were employed and a Rink Amide MBHA resin (0.7 mmol g−1 substitution; 50 µmol scale) was used as solid support, it releases peptides amidated at C-terminus upon acid treatment. All Fmoc-amino acids were activated by in situ PyBop/HOBt//DIPEA activation procedure. Amino acid coupling steps were monitored by Kaiser test after 60 min coupling cycles. Fmoc-deprotection was performed with 20% piperidine in DMF for 5 + 10 min.
After removal of the final N-terminal Fmoc-protecting group, the coupling step with the chelating agent was performed using 2 equiv of DTPA(tBu)4 and PyBOP and 4 equiv of DIPEA in DMF. The coupling time was increased to 2 h and completion of the reaction was checked by Kaiser test.
The conjugate cleavage from the solid support and the simultaneous deprotection of all side chains were performed by suspending the fully protected compound-resins in TFA/H2O/TIS/EDT/tioanisole (95:2:1:1:1) for 3 h. The peptide conjugate was isolated by precipitation into cold diethyl ether and centrifuged to form a pellet.
The final product was purified by RP-HPLC and analyzed by mass spectrometry.
DTPA-A9, DTPA-SA9: [M + H]+ calculated 1377.45 m/z, found 1377.7.
For the internalization studies, Biotin was linked to the N-terminus of A9 peptide.
Biotinylated-A9 peptide: [M + H]+ calculated 1228.4 m/z, found 1228.2. For details see Supplementary Information Figs S5 and S6.
Internalization studies of A9 in BT474 cells
BT474 cells were seeded at a density of 106 cells/well in 6-well-plates for incubation. After 24 h of preincubation, medium was replaced with fresh medium containing the biotinylated-A9 peptide (at a concentration of 200 μg/mL), and the cells were further incubated for 3 h. The cells were then washed twice with ice cold phosphate buffered saline (PBS; Sigma-Aldrich) before adding ice cold 0.5% Trypsin-Ethylenediaminetetraacetic acid (EDTA; Sigma-Aldrich). Cell pellets and culture media fractions were collected by centrifugation for 10 min at 5,000 rpm and 4 °C. For the subsequent estimation of cellular uptake, BT474 cell pellets were permeabilized with 40 μg/mL digitonin solution (Sigma-Aldrich), which contains 10 mM Hepes, 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT, complete protease inhibitors tablet (Roche) at pH 7.4, for 15 min in an ice bath47. The supernatants, which represent the intracellular fraction, were collected by centrifugation for 20 min at 16000 rpm and at 4 °C. The Samples obtained were incubated with streptavidin-coated magnetic beads (ThermoFisher Scientific) for 12 h at 4 °C, under continuous agitation, in order to catch the biotinylated-peptide. The streptavidin-bound compounds, after several washes with PBS buffer (pH7.4), were eluted with formamide 95% EDTA 0.5 mM for 2 min at 95 °C, as reported by the manufacturer, and stored at −80 °C until next mass spectrometry analysis. Intracellular fractions of untreated BT474 cells, were used as negative control. Protein amounts were determined with the Pierce BCA kit (Perbio Science, Bonn, Germany).
MS analyses were performed using a ESI Ion Trap HCT ETD II HC Ultra PTM discovery system Bruker mass spectrometer coupled with an HPLC System Alliance e2695 separation module fitted out with a 2998 PDA detector (Waters, Milan, Italy). Chromatographic separations of samples (50 μL injected) were performed on an Alliance HT WATERS 2795 system, equipped with a PDA WATERS detector 2996 using a C18 Waters xBridge (3 mm, 4.6 × 5.0 mm) column. The optimized mobile phase consisted of water containing 0.1% TFA (solvent A) and CH3CN containing 0.1% TFA (solvent B). The elution was carried out by using a linear gradient from 1% to 70% of solvent B over 20 min, from 70% to 95% of solvent B over 1 min, following an isocratic step at 95% B for 5 min, and a re-equilibration step at 2% B for 4 min was always performed before injections. The total analysis time was thereby kept at 30 minutes.
The ESI source operating parameters were set as follows: positive ionization mode; capillary voltage 3.5 kV; source temperature 150 °C; desolvation temperature 600 °C; desolvation nitrogen gas flow rate 1000 L/h; cone gas 50 L/h; sampling cone voltage 40 V. The acquisitions were made in MRM/SRM (multiple reaction monitoring/selected reaction monitoring)48 mode with a dwell time of 63 ms and a collision energy of 25 V. For the SRM mode the following transitions was monitored: biotinylated-A9 at m/z 615.0–477.2.
Computational Methods
The 3D structure of the complex between HER2-DIVMP and 111In-DTPA-A9 was modelled. The atomic coordinates of 111In-DTPA were retrieved from CSD (The Cambridge Structural Database)53, code XIKVES54, whereas a representative conformation of the complex A9/HER2-DIVMP was extracted from the most populated cluster of structures obtained by our previous Molecular Dynamics study52. Subsequently, the model complex between HER2-DIVMP and 111In-DTPA-A9 was built using the Builder module of Insight II package and was subjected to energy minimization by 1000 steps of CG. MOLMOL package was used for visual inspection and images production62.
Radiolabeling chemistry and HPLC analysis
Before labelling, freeze-dried DTPA-A9 or DTPA-SA9 was reconstituted in 0.2 M ammonium acetate, pH 5.5, to a concentration of 2 mg/ml. Aliquots containing 30 µg of conjugate were stored frozen at −20 °C. For labelling, an aliquot of a conjugate was mixed with a predetermined amount of 111In-chloride solution. The reaction mixtures were incubated at room temperature for 60 min. After 60 min incubation, the radioconjugate radiochemical yield was measured using HPLC. For a blank experiment, 111In-chloride was incubated with 0.2 M ammonium acetate, pH 5.5, without DTPA-A9 for 60 min, and the mixture was analyzed using radio-HPLC.
Analytic LC was performed using an HPLC system from Beckman consisting of a 126 pump, a 166 UV detector (set at wavelength of 220 nm), and a radiation detector coupled in series. The column used was a VydacRP C18 protein column (3 × 150 mm). The applied gradient elution had the following parameters, A: 0.1% TFA in H2O; B: 0.1% TFA in CH3CN, 1 mL/min flow rate using a linear gradient starting from 5% to 80% B in 20 min. Data acquisition and handling were performed using the Beckman System Gold or Hitachi Chromaster HPLC systems.
For stability test, murine blood plasma was prepared from whole blood by centrifuging for 10 min at 2000 g (4 °C). A sample of freshly labeled 111In-DTPA-A9 (10 µL) was mixed with the plasma (200 µL). Samples were incubated at 37 °C. At 15 or 60 min after incubation start, pre-chilled acetonitrile (200 µL) was added to the sample to precipitate blood proteins, the sample was centrifuged for 10 min (10000 g, 4 °C). The supernatant was collected, diluted with water (1:1) and analyzed with radio-HPLC as described above. Two experiments per data point were performed to check reproducibility.
Measurements of affinity of 111In-DTPA-A9 peptide binding to living HER2-expressing cells using LigandTracer Yellow
For cell studies, the HER2-expressing breast carcinoma cell line BT474 (from ATCC) was used. The cell line was cultured in RPMI medium (Flow Irvine, UK) supplemented with 10% fetal calf serum (Sigma, USA), 2 mM L-glutamine and PEST (penicillin 100 IU/mL and 100 μg/mL streptomycin), all from Biokrom Kg, Germany.
Cells from the human breast carcinoma cell line BT474 were seeded on a local area of a cell culture dish (NunclonTM, Size 100620, NUNC A/S, Roskilde, Denmark), as described by Björke and Andersson63. The binding of 111In-DTPA-A9 to living cells was monitored in real-time at room temperature using LigandTracer Yellow, using method established by Björke and Andersson63. In brief, the cell culture dish was placed on the sloping and rotating cell dish holder in LigandTracer Yellow, followed by a continuous monitoring of the radioactivity level along the rim of the dish. If the labelled peptide binds to the cells, an elevated signal will be recorded when the cells pass by the detector. This makes it possible to accurately detect binding events both during incubation and after wash. In order to cover the concentration span needed for proper affinity estimation entirely, three increasing concentrations 4, 12, and 36 nM (selected based on the fluorescence spectroscopy results). Each concentration was incubated long enough to approach steady state. Thereafter, the ligand solution was replaced with fresh medium and the dissociation rate was followed overnight. The data were corrected for decay of indium-111. Binding and dissociation curves were analyzed by InteractionMap software as described by Björkelund et al. to calculate affinity of 111In-DTPA-A9 binding to living HER2-expressing cells56.
Additionally, specificity of 111In-DTPA-A9 binding to HER2 expressing cells was evaluated using pre-saturation of receptors by trastuzumab and by using 111In-labeled scrambled DTPA-A9 (111In- DTPA-S-A9). The protocol for 111In- DTPA-S-A9 was exactly as for 111In-DTPA-A9. In the case of pre-saturation experiment, trastuzumab was added to cell dishes to the concentration of 400 nM and cells were incubated for 60 min immediately before affinity measurements, The medium was withdrawn and new medium was added. Thereafter, binding affinity of 111In-DTPA-A9 was measured using exactly the same protocol as described above. Two experiments per data point were performed to check reproducibility.
In vivo studies
The animal experiment was planned and performed in accordance with Swedish national legislation on laboratory animals’ protection and was approved by the Ethics Committee for Animal Research in Uppsala, Sweden.
Four BALB/C nu/nu mice were used for the biodistribution studies. At the time of experiment, the average animal weight was 20.5 ± 0.9 g.
Animals were injected intravenously (tail vein) with 3 µg (30 kBq) of 111In-DTPA-A9 in 100 µL PBS. Since DTPA-A9 peptide resulted stable to serum proteolytic degradation in a range of 90 minutes (for details see Supplementary Information Fig. S6), at 1 hour after injection, a mixture of Ketalar-Rompun (20 µL of solution per gram body weight; Ketalar: 10 mg/mL; Rompun: 1 mg/mL) was injected intraperitonealy, and the mice were euthanized by a heart puncture using a syringe, pre-washed with diluted heparin (5000 IE/mL). Blood as well as brain, stomach, pancreas, lung, liver, spleen, kidneys, samples of muscle and bone, gastrointestinal tract (with its content) and remaining carcass were collected in pre-weighed plastic vials. Organs and tissue samples were weighed and measured for radioactivity using an automatic gamma counter. The tissue uptake values were calculated as per cent injected activity per gram of tissue (% ID/g) and per whole sample (% ID/sample). The gastrointestinal tract and the carcass uptakes were calculated only as % ID per whole sample. The radioactivity in the gastrointestinal tract with its content was used as a measure of hepatobiliary excretion.
Additionally, in vivo stability to proteolytic enzymes anchored to the vasculature walls was assessed using to method described earlier61. Briefly, 10 MBq of 111In-DTPA-A9 were injected in NMRI mice via tail vein. At 5 or 15 min, animals were sacrificed, blood was withdrawn and transferred to a EDTA-containing vial. Each sample was centrifuged for 10 min (2000 g, 4 °C), and a supernatant was collected. A pre-chilled acetonitrile (200 µL) was added to the supernatant to precipitate blood proteins, the sample was centrifuged for 10 min (10000 g, 4 °C). The supernatant was collected, diluted with water (1:1) and analyzed with radio-HPLC as described above. Two experiments per data point were performed to check reproducibility63.
Electronic supplementary material
Acknowledgements
This research was financially supported by grants from the Swedish Cancer Society (grant CAN 2015/350) and the Swedish Research Council (grant 2015-02353).
Author Contributions
H.H. performed the nuclear medicine experiments; E.C. and N.D. performed the synthesis of the peptides; N.D. performed the cellular uptake experiments and stability experiments in vitro; E.L. performed the molecular modeling studies; A.O. performed experiments concerning in vivo stability of labeled peptides; J.B. performed InteractionMap evaluation of data obtained in LigandTracer expriments; V.D.A. and R.B. provided the cellular pools; M.S. contributed to the design of the research; V.T. designed the nuclear medicine experiments, preformed radiolabeling, in vitro stability test and Ligand Tracer experiments; and wrote the paper; S.D.L. designed the synthetic strategies and the research; and wrote the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Enrica Calce and Nunzianna Doti contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21283-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vladimir Tolmachev, Email: vladimir.tolmachev@igp.uu.se.
Stefania De Luca, Email: stefania.deluca@cnr.it.
References
- 1.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell. Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Niehans GA, Singleton TP, Dykoski D, Kiang DT. Stability of HER-2/neu Expression Over Time and at Multiple Metastatic Sites. J. Natl. Cancer Inst. 1993;85:1230–1235. doi: 10.1093/jnci/85.15.1230. [DOI] [PubMed] [Google Scholar]
- 3.Neve RM, Lane HA, Hynes NE. The role of overexpressed HER2 in transformation. Ann. Oncol. 2001;12(Suppl. 1):S9–S13. doi: 10.1093/annonc/12.suppl_1.S9. [DOI] [PubMed] [Google Scholar]
- 4.Slamon DJ, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 5.Paik S, et al. Pathologic findings from the national surgical adjuvant breast and bowel project: Prognostic significance of erbE-2 protein overexpression in primary breast cancer. J. Clin. Oncol. 1990;8:103–112. doi: 10.1200/JCO.1990.8.1.103. [DOI] [PubMed] [Google Scholar]
- 6.Garrett TP, et al. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol. Cell. 2003;11:495–505. doi: 10.1016/S1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 7.Engel RH, Kaklamani VG. HER2-positive breast cancer: Current and future treatment strategies. Drugs. 2007;67:1329–1341. doi: 10.2165/00003495-200767090-00006. [DOI] [PubMed] [Google Scholar]
- 8.Faltus T, et al. Silencing of the HER2/neu gene by siRNA inhibits proliferation and induces apoptosis in HER2/neu-overexpressing breast cancer cells. Neoplasia. 2004;6:786–795. doi: 10.1593/neo.04313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang G, Cai KQ, Thompson-Lanza JA, Bast RC, Jr., Liu J. Inhibition of breast and ovarian tumor growth through multiple signaling pathways by using retrovirus-mediated small interfering RNA against Her-2/neu gene expression. J. Biol. Chem. 2004;279:4339–4345. doi: 10.1074/jbc.M311153200. [DOI] [PubMed] [Google Scholar]
- 10.Pils D, et al. In ovarian cancer the prognostic influence of HER2/neu is not dependent on the CXCR4/SDF-1 signalling pathway. Br. J. Cancer. 2007;96:485–491. doi: 10.1038/sj.bjc.6603581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roh H, Pippin J, Drebin JA. Down-regulation of HER2/neu expression induces apoptosis in human cancer cells that overexpress HER2/neu. Cancer Res. 2000;60:560–565. [PubMed] [Google Scholar]
- 12.Tai W, Qin B, Cheng K. Inhibition of breast cancer cell growth and invasiveness by dual silencing of HER2 and VEGF. Mol. Pharmaceutics. 2010;7:543–556. doi: 10.1021/mp9002514. [DOI] [PubMed] [Google Scholar]
- 13.Bookman MA, Darcy KM, Clarke-Pearson D, Boothby RA, Horowitz IR. Evaluation of monoclonal humanized anti-HER2 antibody, trastuzumab, in patients with recurrent or refractory ovarian or primary peritoneal carcinoma with overexpression of HER2: a phase II trial of the Gynecologic Oncology Group. J. Clin. Oncol. 2003;21:283–290. doi: 10.1200/JCO.2003.10.104. [DOI] [PubMed] [Google Scholar]
- 14.Steffensen KD, et al. The prognostic importance of cyclooxygenase 2 and HER2 expression in epithelial ovarian cancer. Int. J. Gynecol. Cancer. 2007;17:798–807. doi: 10.1111/j.1525-1438.2006.00855.x. [DOI] [PubMed] [Google Scholar]
- 15.Hellstrom I, Goodman G, Pullman J, Yang Y, Hellström KE. Overexpression of HER2 in ovarian carcinomas. Cancer Res. 2001;61:2420–2423. [PubMed] [Google Scholar]
- 16.Calce E, Monfregola L, Saviano M, De Luca S. HER2-mediated anticancer drug delivery: strategies to prepare targeting ligands highly specific for the receptor. Curr. Med. Chem. 2015;22:1–14. doi: 10.2174/0929867322666150521091103. [DOI] [PubMed] [Google Scholar]
- 17.Monfregola, L. & De Luca, S. The role of HER2 in targeted cancer therapy and hypothesized future perspectives. in HER2 and Cancer: Mechanism, Testing and Targeted Therapy. (Ed. :Williams, S. I.; Rogers, C. E.) 101–122 (Nova Science Publishers, 2011).
- 18.Tan AR, Swain SM. Ongoing adjuvant trials with trastuzumab in breast cancer. Semin. Oncol. 2003;30:54–64. doi: 10.1053/j.seminoncol.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Leonard DS, et al. Anti-human epidermal growth factor receptor 2 monoclonal antibody therapy for breast cancer. Br. J. Surg. 2002;89:262–271. doi: 10.1046/j.0007-1323.2001.02022.x. [DOI] [PubMed] [Google Scholar]
- 20.Behr TM, Béhé M, Wörmann B. Trastuzumab and breast cancer. N. Engl. J. Med. 2001;345:995–996. doi: 10.1056/NEJM200109273451312. [DOI] [PubMed] [Google Scholar]
- 21.Burris HA, 3rd, Tibbitts J, Holden SN, Sliwkowski MX, Lewis Phillips GD. Trastuzumab emtansine (T-DM1): a novel agent for targeting HER2+ breast cancer. Clin. Breast Cancer. 2011;11:275–282. doi: 10.1016/j.clbc.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 22.Cao Y, et al. Design optimization and characterization of HER2/neu-targeted immunotoxins: comparative in vitro and in vivo efficacy studies. Oncogene. 2014;33:429–439. doi: 10.1038/onc.2012.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zielinski R, et al. J. HER2-affitoxin: a potent therapeutic agent for the treatment of HER2-overexpressing tumors. Clin Cancer Res. 2011;17:5071–5081. doi: 10.1158/1078-0432.CCR-10-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altai M, Liu H, Orlova A, Tolmachev V, Gräslund T. Influence of molecular design on biodistribution and targeting properties of an Affibody-fused HER2-recognising anticancer toxin. Int J. Oncol. 2016;49:1185–1194. doi: 10.3892/ijo.2016.3614. [DOI] [PubMed] [Google Scholar]
- 25.Houssami N, Macaskill P, Balleine RL, Bilous M, Pegram MD. HER2 discordance between primary breast cancer and its paired metastasis: tumor biology or test artefact? Insights through meta-analysis. Breast Cancer Res Treat. 2011;129:659–674. doi: 10.1007/s10549-011-1632-x. [DOI] [PubMed] [Google Scholar]
- 26.Blend MJ, Stastny JJ, Swanson SM, Brechbiel MW. Labeling anti- HER2/neu monoclonal antibodies with 111In and 90Y using a bifunctional DTPA chelating agent. Cancer Biother. Radiopharm. 2004;18:355–363. doi: 10.1089/108497803322285107. [DOI] [PubMed] [Google Scholar]
- 27.Garmestani K, Milenic DE, Plascjak PS, Brechbiel MW. A new and convenient method for purification of 86Y using a Sr(II) selective resin and comparison of biodistribution of 86Y and 111In labeled Herceptin. Nucl. Med. Biol. 2002;29:599–606. doi: 10.1016/S0969-8051(02)00322-0. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi H, et al. Rapid accumulation and internalization of radiolabeled herceptin in an inflammatory breast cancer xenograft with vasculogenic mimicry predicted by the contrast-enhanced dynamic MRI with the macromolecular contrast agent G6-(1B4M-Gd)256. Cancer Res. 2002;62:860–866. [PubMed] [Google Scholar]
- 29.Palm S, et al. Pharmacokinetics and biodistribution of 86Y-trastuzumab for 90Y dosimetry in an ovarian carcinoma model: correlative MicroPET and MRI. J. Nucl. Med. 2003;44:1148–1155. [PubMed] [Google Scholar]
- 30.Winberg KJ, Persson M, Malmstrӧm P-U, Sjӧberg S, Tolmachev V. Radiobromination of anti-HER2/neu/ErbB-2 monoclonal antibody using the p-isothiocyanatobenzene derivative of the [76Br]undecahydro-bromo-7,8-dicarba-nido-undecaborate(1-) ion. Nucl. Med. Biol. 2004;31:425–433. doi: 10.1016/j.nucmedbio.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Reilly RM, et al. Problems of delivery of monoclonal antibodies. Pharmaceutical and pharmacokinetic solutions. Clin Pharmacokinet. 1995;28:126–142. doi: 10.2165/00003088-199528020-00004. [DOI] [PubMed] [Google Scholar]
- 32.Scollard DA, Chan C, Holloway CMB, Reilly RM. A kit to prepare 111In-DTPA-trastuzumab (Herceptin) Fab fragments injection under GMP conditions for imaging or radioimmunoguided surgery of HER2 positive breast cancer. Nucl. Med. Biol. 2011;38:129–136. doi: 10.1016/j.nucmedbio.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Tang Y, et al. Imaging of HER2/neu-positive BT-474 human breast cancer xenografts in athymic mice using 111In-trastuzumab (Herceptin) Fab fragments. Nucl. Med. Biol. 2005;32:51–58. doi: 10.1016/j.nucmedbio.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Tang Y, et al. Imaging of HER2/neu expression in BT-474 human breast cancer xenografts in athymic mice using 99mTc-HYNIC-trastuzumab (Herceptin) Fab fragments. Nucl. Med. Commun. 2005;26:427–432. doi: 10.1097/00006231-200505000-00006. [DOI] [PubMed] [Google Scholar]
- 35.Adams GP, et al. Increased affinity leads to improved selective tumor delivery of single-chain Fv antibodies. Cancer Res. 1998;58:485–490. [PubMed] [Google Scholar]
- 36.Robinson MK, et al. Quantitative immuno-positron emission tomography imaging of HER2-positive tumor xenografts with an iodine-124 labeled anti-HER2 diabody. Cancer Res. 2005;65:1471–1478. doi: 10.1158/0008-5472.CAN-04-2008. [DOI] [PubMed] [Google Scholar]
- 37.Olafsen T, et al. Optimizing radiolabeled engineered anti-p185HER2 antibody fragments for in vivo imaging. Cancer Res. 2005;65:5907–5916. doi: 10.1158/0008-5472.CAN-04-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tolmachev V. Imaging of HER2 overexpression in tumors for guiding therapy. Curr. Pharm. Design. 2008;14:2999–3019. doi: 10.2174/138161208786404290. [DOI] [PubMed] [Google Scholar]
- 39.Kramer-Marek G, Kiesewetter DO, Capala J. Changes in HER2 expression in breast cancer xenografts after therapy can be quantified using PET and 18F- labeled affibody molecules. J. Nucl. Med. 2009;50:1131–1139. doi: 10.2967/jnumed.108.057695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orlova A, Wållberg H, Stone-Elander S, Tolmachev V. On the selection of a tracer for PET imaging of HER2-expressing tumors: direct comparison of a 124I-labeled affibody molecule and trastuzumab in a murine xenograft model. J. Nucl. Med. 2009;50:417–425. doi: 10.2967/jnumed.108.057919. [DOI] [PubMed] [Google Scholar]
- 41.Calce E, Monfregola L, Sandomenico A, Saviano M, De Luca S. Fluorescence study for selecting specific ligands toward HER2 receptor: an example of receptor fragment approach. Eur. J. Med. Chem. 2013;61:116–121. doi: 10.1016/j.ejmech.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 42.Monfregola L, Vitale RM, Amodeo P, De Luca S. A SPR strategy for high-throughput ligand screenings based on synthetic peptides mimicking a selected subdomain of the target protein: A proof of concept on HER2 receptor. Bioorg. Med. Chem. 2009;17:7015–7020. doi: 10.1016/j.bmc.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Calce E, Sandomenico A, Saviano M, Ruvo M, De Luca S. Cysteine co-oxidation process driven by native peptide folding: an example on HER2 receptor model system. Amino Acids. 2014;46:1197–1206. doi: 10.1007/s00726-014-1681-7. [DOI] [PubMed] [Google Scholar]
- 44.Ginj M, et al. Radiolabeled somatostatin receptor antagonists are preferable to agonists for in vivo peptide receptor targeting of tumors. Proc. Natl. Acad. Sci. USA. 2006;103:16436–16441. doi: 10.1073/pnas.0607761103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalm SU, et al. Comparison of the Therapeutic Response to Treatment with a 177Lu-Labeled Somatostatin Receptor Agonist and Antagonist in Preclinical Models. J. Nucl. Med. 2016;57:260–265. doi: 10.2967/jnumed.115.167007. [DOI] [PubMed] [Google Scholar]
- 46.Reubi JC. Peptide receptors as molecular targets for cancer diagnosis and therapy. Endocr Rev. 2003;24:389–427. doi: 10.1210/er.2002-0007. [DOI] [PubMed] [Google Scholar]
- 47.Holden P, Horton WA. Crude subcellular fractionation of cultured mammalian cell lines. BMC Research Notes. 2009;2:243–253. doi: 10.1186/1756-0500-2-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russo R, et al. Ultra-high performance liquid chromatography tandem mass spectrometry for the detection of durum wheat contamination or adulteration. J Mass Spectrom. 2014;49:1239–46. doi: 10.1002/jms.3451. [DOI] [PubMed] [Google Scholar]
- 49.Hnatowich DJ. Label stability in serum of four radionuclides on DTPA-coupled antibodies–an evaluation. Int. J. Rad. Appl. Instrum. B. 1986;13:353–358. doi: 10.1016/0883-2897(86)90009-7. [DOI] [PubMed] [Google Scholar]
- 50.Bakker WH, et al. In vivo application of [111In-DTPA-D-Phe1]-octreotide for detection of somatostatin receptor-positive tumors in rats. Life Sci. 1991;49:1593–1601. doi: 10.1016/0024-3205(91)90053-E. [DOI] [PubMed] [Google Scholar]
- 51.van Essen M, Sundin A, Krenning EP, Kwekkeboom DJ. Neuroendocrine tumours: the role of imaging for diagnosis and therapy. Nat. Rev. Endocrinol. 2014;10:102–114. doi: 10.1038/nrendo.2013.246. [DOI] [PubMed] [Google Scholar]
- 52.Langella E, Calce E, Saviano M, De Luca S. Structural identification of HER2 receptor model binding pocket to optimize lead compounds: a combined experimental and computational approach. Mol. BioSyst. 2016;12:2159–2167. doi: 10.1039/C6MB00158K. [DOI] [PubMed] [Google Scholar]
- 53.Groom CR, Bruno IJ, Lightfoot MP, Ward SC. The Cambridge StructuralDatabase. Acta Cryst. 2016;B72:171–179. doi: 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, et al. Synthesis and structure determination of seven coordinate K[InIII(Hdtpa)]·3.5H2O. Chem. J. Chin. Univ., Chinese Edition. 2000;21:1468–1470. [Google Scholar]
- 55.Barta P, et al. Protein interactions with HER-family receptors can have different characteristics depending on the hosting cell line. Int. J. Oncol. 2012;40:1677–1682. doi: 10.3892/ijo.2011.1307. [DOI] [PubMed] [Google Scholar]
- 56.Björkelund H, Gedda L, Barta P, Malmqvist M, Andersson K. Gefitinib induces epidermal growth factor receptor dimers which alters the interaction characteristics with 125I-EGF. PLoS One. 2011;6:e24739. doi: 10.1371/journal.pone.0024739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tolmachev V, et al. Tumor targeting using affibody molecules: interplay of affinity, target expression level, and binding site composition. J. Nucl. Med. 2012;53:953–960. doi: 10.2967/jnumed.111.101527. [DOI] [PubMed] [Google Scholar]
- 58.Tran TA, Rosik D, Abrahmsén L, Sandström M. Design, synthesis and biological evaluation of a multifunctional HER2-specific affibody molecule for molecular imaging. Eur. J. Nucl. Med. Mol. Imaging. 2009;36:1864–1873. doi: 10.1007/s00259-009-1176-z. [DOI] [PubMed] [Google Scholar]
- 59.Nock BA, Maina T, Krenning EP, de Jong M. “To serve and protect”: enzyme inhibitors as radiopeptide escorts promote tumor targeting. J. Nucl. Med. 2014;55:121–127. doi: 10.2967/jnumed.113.129411. [DOI] [PubMed] [Google Scholar]
- 60.Chatalic KL, et al. In Vivo Stabilization of a Gastrin-Releasing Peptide Receptor Antagonist Enhances PET Imaging and Radionuclide Therapy of Prostate Cancer in Preclinical Studies. Theranostics. 2016;6:104–117. doi: 10.7150/thno.13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maina T, et al. Preclinical pharmacokinetics, biodistribution, radiation dosimetry and toxicity studies required for regulatory approval of a phase I clinical trial with (111)In-CP04 in medullary thyroid carcinoma patients. Eur. J. Pharm. Sci. 2016;91:236–242. doi: 10.1016/j.ejps.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14(51-5):29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 63.Björke H, Andersson K. Automated, high-resolution cellular retention and uptake studies in vitro. Appl. Radiat. Isot. 2006;64:901–905. doi: 10.1016/j.apradiso.2006.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.