Abstract
A breast-risk score, published in 2016, was developed in white-American women using 92 genetic variants (GRS92), modifiable and non-modifiable risk factors. With the aim of validating the score in the Spanish population, 1,732 breast cancer cases and 1,910 controls were studied. The GRS92, modifiable and non-modifiable risk factor scores were estimated via logistic regression. SNPs without available genotyping were simulated as in the aforementioned 2016 study. The full model score was obtained by combining GRS92, modifiable and non-modifiable risk factor scores. Score performances were tested via the area under the ROC curve (AUROC), net reclassification index (NRI) and integrated discrimination improvement (IDI). Compared with non-modifiable and modifiable factor scores, GRS92 had higher discrimination power (AUROC: 0.6195, 0.5885 and 0.5214, respectively). Adding the non-modifiable factor score to GRS92 improved patient classification by 23.6% (NRI = 0.236), while the modifiable factor score only improved it by 7.2%. The full model AUROC reached 0.6244. A simulation study showed the ability of the full model for identifying women at high risk for breast cancer. In conclusion, a model combining genetic and risk factors can be used for stratifying women by their breast cancer risk, which can be applied to individualizing genetic counseling and screening recommendations.
Subject terms: Cancer epidemiology, Breast cancer
Introduction
Breast cancer is the most frequent type of cancer in women worldwide and a main cause of cancer death in developed countries1. Epidemiological research has led to the identification of several risk factors (age at menarche, parity, age at first full-term pregnancy, age at menopause), most of them associated with estrogen production2,3. A number of risk factors are related with lifestyle (tobacco smoking, alcohol consumption, overweight or obesity), although their importance seems to be smaller than the estrogen/reproductive life-associated risk factors4. Known risk factors can only explain about 40% of breast cancer risk.
A few highly penetrant genetic variants, like those in BRCA1 and BRCA2 genes, have been proved to increase breast cancer risk5,6; although their low prevalence hardly allows them to explain around 5% cases of breast cancer7, carrying any of these variants puts women in such a high risk that screening practices have been adapted in carrier women and even oophorectomy or early mastectomy can be considered in some cases in the absence of breast cancer diagnosis8. With the advent of next generation sequencing techniques, an increasing number of low-penetrant genetic variants are being identified as related to breast cancer7 and some polygenic tests have been marketed intending to identify women at high risk of breast cancer, although their clinical relevance is uncertain.
In this way, Maas et al. presented a breast cancer risk model among white women in the United States, which included modifiable and non-modifiable risk factors, as well as a genetic risk score with information from 92 genetic variants9. The goal of this article is to validate Maas et al.’s model in a Spanish case-control study.
Methods
Study design and population
The Multi Case-Control (MCC-Spain) study is a population-based case-control study of common tumors in Spain and has been described elsewhere. It has been carried out in 23 hospitals and primary care centers in 12 Spanish provinces and assesses five types of cancer (colorectal, breast, stomach, prostate and chronic lymphocytic leukemia) using the same series of population controls for all cases10. Cases and controls were recruited between September 1st, 2008 and December 31st, 2013.
We included 1,732 incident cases of breast cancer in women and 1,910 controls in ten participating centers (Asturias, Barcelona, Cantabria, Girona, Gipuzkoa, Huelva, León, Madrid, Navarra and Valencia). Cases were aged 20–85 and with a new pathology-confirmed diagnosis of breast cancer living in the catchment area of each hospital at least 6 months prior to the diagnosis. Controls were women without history of breast cancer living in the same catchment area as cases; they were randomly selected from the rosters of General Practitioners at the Primary Health Centers. Controls were frequency-matched to cases by 5-year age groups and study area. Response rates were 72% among cases and 52% among controls.
The study was carried out according to Spanish laws on biomedical research. All procedures were performed with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments.
The Ethics Committees of the participating hospitals (Ethical Committee of Clinical Research of Barcelona, Cantabria, Girona, Gipuzkoa, Huelva, León, Principado de Asturias, Madrid, Navarra and Valencia) approved the study protocols, and participants provided written informed consent at the time of enrollment.
Data collection
Participants were interviewed face-to-face by trained interviewers with a comprehensive epidemiological questionnaire that assessed socio-demographic information, personal and family history of cancer, anthropometric data, alcohol consumption, smoking habits, reproductive and medical history, and family history. Participant’s weight was recorded by self-report, as estimated one year before diagnosis for cases and for controls. Body mass index (BMI) was estimated from self-reported weight and height one year before the diagnosis for cases and one year prior to the interview for controls. Blood samples were obtained following the study protocol. Elapsed time between breast cancer diagnosis and interview was 117 days on average.
Genotyping
The genotyping was performed for 1,138 cases and 1,239 controls using the Infinium Human Exome BeadChip (Illumina, San Diego, USA) that includes >200,000 coding markers plus 5,000 additional custom SNPs selected from previous GWAS or in genes of interest.
Genetic score
In order to construct the genetic score, we consider two SNP sets. The first set included 24 SNPs identified from Breast and Prostate Cancer Cohort Consortium (BPC3 study) (Supplementary Table 1)11–13. 17 of them were included in the performed genotyping; the remaining 7 (rs10069690, rs10941679, rs17530068, rs1250003, rs10483813, rs6504950 and rs2284378) were imputed using SNPSTATs14,15. The 24-SNP genetic score (GRS24) was estimated by the addition of the beta coefficients as obtained in BPC3 study13. Barrdahl et al.13 considered rs10483813 as surrogate for rs999737 as they are in strong linkage disequilibrium; therefore, we excluded rs10483813 from GRS24 in order to do not double the weight of this locus. As genotyping was not available for 594 cases and 671 controls, we simulated GRS24 in them using the method suggested by Chatterjee et al.16 as explained in the following paragraph.
The second set of SNPs included 68 identified in Breast Cancer Association Consortium (BCAC) and Collaborative Oncological Gene-Environment Study (COGS) (Supplementary Table 1). They were not included in the genotyping, so the 68-SNP genetic score (GRS68) was simulated with the Chatterjee et al. method16, as performed by Maas et al.9 when developing the score we are trying to validate. In brief, GRS68 was obtained using the reported beta coefficients (=log odds ratios) and conditional on case-control status and family history of breast cancer as:
| 1 |
where , , subindex i refers to the participants included in the study and k refers to the SNPs included in the model, are the estimates of the log odds ratios, and fk are the risk allele frequencies. and fk were obtained from the BCAC17,18.
Finally, a 92-SNP genetic score (GRS92) was obtained by adding GRS24 plus GRS68. Supplementary Table 2 displays R2 as a measure of linkage disequilibrium between the SNPs located in the same chromosome, according to the European population in the 1000 Genome Project and were obtained from https://analysistools.nci.nih.gov/LDlink/?tab=home.
Modifiable risk factor score
The modifiable risk factor score (MRFS) included BMI, menopausal hormone therapy, level of alcohol consumption, and smoking status. Alcohol use and BMI were categorized as in Maas et al.9 (Supplementary Table 3). In order to build MRFS, we carried out a multivariate logistic regression analysis including the above indicated modifiable risk factors. Then, MRFS was obtained by adding the estimated beta coefficients of the above indicated risk factors, every beta being adjusted for each other factor.
Non-modifiable risk factor score
The non-modifiable risk factor score (NMRFS) included family history, age at first birth, parity, age at menarche, height, menopausal status, and age at menopause. According to Maas et al.9, age at first birth and parity were considered “non-modifiable” as it is unlikely that women modify these factors because of their breast cancer risk. Age at menarche, age at first birth, height and age at menopause were categorized as in Maas et al.9 (Supplementary Table 3). NMRFS was built in the same way that MRFS.
Full model
The full model (FM) was obtained as the addition of GRS92 + MRFS + NMRFS.
Statistical analysis
The independence among the scores was tested using the Pearson correlation coefficient. Differences in scoring between cases and controls were tested with the Student-t test. In order to analyze the association among scores and breast cancer, scores were categorized into deciles; then, a logistic regression model was constructed for each score, with decile 1 as reference. ROC curves and their area under the curve (AUROC) were obtained for each logistic model.
For analyzing whether a score adds or not to the predictive ability of another score, we estimated the net reclassification improvement (NRI), the integrated discrimination improvement (IMI) and the improvement in the AUROC. The goal for a score is to classify breast cancer cases and controls adequately; when two scores -say GRS24 and GRS92- are compared, GRS92 would classify some cases better than GRS24 and some others worse; likewise, GRS92 would classify some controls better and some others worse. NRI19 is the net sum of classifying cases and controls better using GRS92 rather than GRS24:
| 2 |
It is noteworthy that NRI can only be applied when both scores are nested one in each other, as it is the case between GRS24 and GRS92, because GRS92 equals GRS24 plus GRS68. In this article, we use the continuous version of NRI20.
While NRI measures the improvement in classification, IMI estimates the improvement in predicted probability. A better score is expected to predict higher probabilities in cases and lower probabilities in controls than a worse score. Therefore, IMI is defined as19:
| 3 |
Finally, in order to explore the ability of the scores to identify women at high risk of breast cancer, we simulated the expected breast cancer incidence rate according to their scoring in GRS92, NMRFS, MRFS and full model. For instance, the simulation for GRS92 was built as follows: First, a logistic regression model was estimated using GRS92 as regressor and breast cancer as event, obtaining a β coefficient (=log (OR)). Second, incidence rates by 5-year age groups were obtained from Globocan1,21; these rates were attributed to patients with the average GRS92. Lastly, incidence rates for each patients were simulated by multiplying the average age-specific incidence rate times exp[β*(GRS92 – average GRS92)]. Similar simulations were performed for NMRFS, MRFS and full model. All statistical analyses were performed with the software Stata 14/SE (Stata Co., College Station, TX, US).
Results
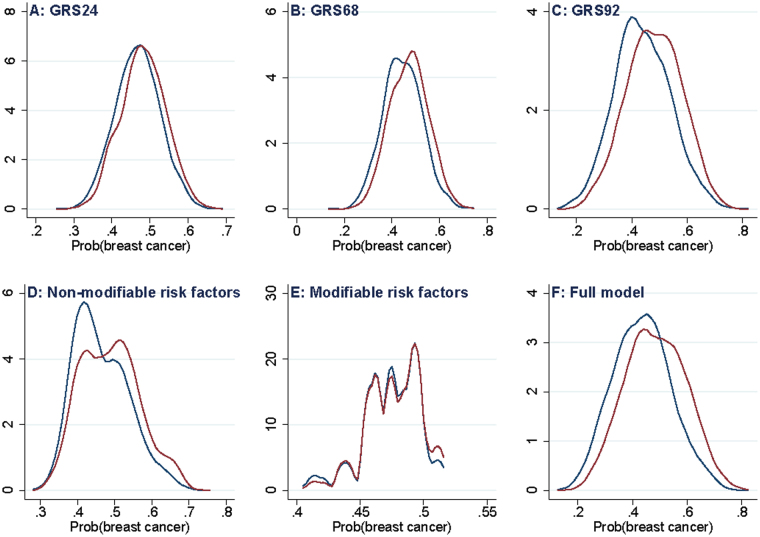
1,732 breast cancer cases and 1,910 controls were included in the analysis; their main characteristics are reported in Supplementary Table 4. Compared to controls, cases were 2.6 younger in average, they have about twice the probability of having a first degree relative affected of breast cancer, their age at menarche was a bit earlier and their age at menopause was later, and they have less children. There were no differences in height, BMI, alcohol consumption and age at first delivery. The distributions of genetic risk scores with 24 [GRS24], 68 [GRS68] and 92 [GRS92] SNPs; modifiable risk factor score [MRFS]; nonmodifiable risk factor score [NMRFS], and full model [FM] are reported in Fig. 1. All scores show a high degree of overlapping between cases and controls, being MRFS the less discriminant score. BC cases scored higher than controls in each risk score (Supplementary Table 5). GRS92, MRFS and NMRFS were not linearly dependent from each other as their Pearson correlation coefficient was <0.06 (Supplementary Table 6).
Figure 1.
Kernel density plots on distribution of genetic and risk factor scores in breast cancer cases (red line) and controls (blue line).
The association between GRS92 and breast cancer is displayed in Table 1, where the odds ratio increased decile by decile; patients in the 10th decile had 3.8 the odds of breast cancer than patients in the first decile and double the odds than patients around the median of GRS92 distribution (i.e.: patients in 5th or 6th deciles). The area under the ROC curve (AUROC) was 0.6195; GRS24 and GRS68 -the two components of GRS92- had smaller prediction ability, with AUROC being equal to 0.5676 and 0.5936, respectively (Supplementary Figure 1).
Table 1.
Relationship among breast cancer and different scores. Odds ratios per decile of each score.
| Decile | GRS92 | NMRFS | MRFS | Full model |
|---|---|---|---|---|
| 1 | 1 (ref.) | 1 (ref.) | 1 (ref.) | 1 (ref.) |
| 2 | 1.27 (0.90–1.78) | 0.90 (0.67–1.22) | 1.03 (0.79–1.34) | 1.82 (1.29–2.56) |
| 3 | 1.22 (0.87–1.71) | 1.18 (0.88–1.58) | 1.05 (0.76–1.46) | 1.54 (1.09–2.17) |
| 4 | 1.79 (1.28–2.49) | 1.02 (0.75–1.37) | 1.16 (0.88–1.53) | 2.56 (1.83–3.60) |
| 5 | 2.00 (1.43–2.78) | 1.36 (1.01–1.83) | 0.83 (0.64–1.08) | 2.12 (1.51–2.98) |
| 6 | 2.17 (1.56–3.03) | 1.63 (1.21–2.20) | 1.33 (0.96–1.84) | 2.08 (1.48–2.92) |
| 7 | 2.35 (1.69–3.27) | 1.59 (1.18–2.14) | 1.11 (0.85–1.45) | 2.34 (1.67–3.29) |
| 8 | 2.54 (1.82–3.54) | 2.00 (1.48–2.70) | 1.06 (0.78–1.44) | 3.28 (2.34–4.60) |
| 9 | 3.82 (2.74–5.35) | 2.03 (1.50–2.73) | 1.14 (0.87–1.49) | 4.45 (3.16–6.26) |
| 10 | 3.80 (2.72–5.32) | 2.43 (1.79–3.29) | 1.46 (1.08–1.97) | 5.70 (4.02–8.07) |
GRS92: Genetic Risk Score with 92 SNPs.
NMRFS: Non-modifiable Risk Factor Score.
MRFS: Modifiable Risk Factor Score.
Nonmodifiable risk factors show a dose-response relationship with breast cancer from the fifth decile on; the first four deciles, however, do not exhibit any increase in risk (Table 1). The AUROC was 0.5885 (Supplementary Figure 1).
Modifiable risk factor score had little association with breast cancer (Table 1); only patients in the 10th decile had an odds ratio around 1.5 respect to patients in the first decile. Its ROC curve was hardly over the diagonal, displaying small discrimination power (AUROC = 0.5214) (Supplementary Figure 1).
The improvement in risk prediction when adding more component scores from GRS24 on was measured with three indicators: net reclassification index (NRI) (equation (2)), integrated discrimination improvement (IDI) (equation (3)) and improvement in AUROC, whose results are displayed in Table 2. Adding GRS68 to GRS24 (i.e.: constructing GRS92) improved patient classification by 28.2% (NRI = 0.282) and the difference in the probabilities of suffering breast cancer between cases and controls improved by 2.7% (IDI = 0.027). Adding NMRFS up to GRS92 still improved patient classification by 23.6%, in spite of moderate improvements in IDI (0.015) and AUROC (0.0141). Adding MRFS to GRS92, however, only scored 7.2% in NRI with marginal enhancements in IDI and AUROC. Being low the betterment of MRFS over GRS92, most of it seems to be redundant with NMRFS as adding MRFS to NMRFS hardly improved patient classification (NRI = 0.017), discrimination (IDI = 0.000) and AUROC (−0.0013).
Table 2.
Improvement in risk prediction when adding more component scores.
| Base score | Enhanced score | Net Reclassification Improvement | Integrated Discrimination Improvement | Improvement in AUROC | p value* |
|---|---|---|---|---|---|
| GRS24 | GRS24 + GRS68 = GRS92 | 0.282 | 0.027 | 0.0484 | <0.001 |
| GRS92 | GRS92 + NMRFS | 0.236 | 0.015 | 0.0141 | 0.01 |
| GRS92 | GRS92 + MRFS | 0.072 | 0.003 | 0.0024 | 0.37 |
| NMRFS | NMRFS + MRFS = RFS | 0.017 | 0.000 | −0.0013 | 0.18 |
*p value for the improvement in AUROC.
GRS24: Genetic Risk Score with 24 SNPs.
GRS68: Genetic Risk Score with 68 SNPs.
GRS92: Genetic Risk Score with 92 SNPs.
NMRFS: Non-modifiable Risk Factor Score.
MRFS: Modifiable Risk Factor Score.
RFS: Risk Factor Score.
The full model, thus, mainly reflects the combination of the genetic score GRS92 and the non-modifiable risk score (NMRFS). Its relationship with breast cancer stepped up by each decile (Table 1); the AUROC reached 0.6244 (Supplementary Figure 1).
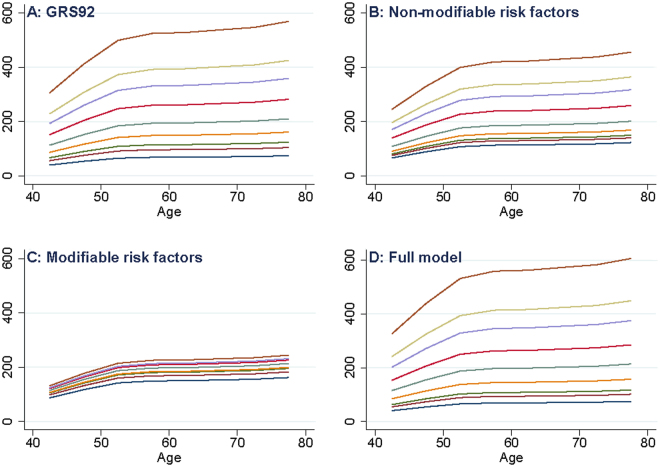
Figure 2 exhibits the expected breast cancer incidence rates by age, according to GRS92, NMRFS, MRFS and full model. Lines represent percentiles 1, 5, 10, 25, 50, 75, 90, 95 and 99 of each score distribution. Figure 2A–D are represented in the same Y scale in order to easily identify which models are more able to classify patients according to their risk: the more separated the lines are, the higher the model ability to classify patients. GRS92 (Fig. 2A), NMRFS (Fig. 2B) and especially full model (Fig. 2D) allowed for identifying patients in high risk of breast cancer.
Figure 2.
Projected breast-cancer incidence rate by age, according to the percentiles of the scores. (A) Genetic score; (B) Non-modifiable risk factors; (C) Modifiable risk factors; (D) Full model. In each graphic, lines from bottom to top represent percentiles 1, 5, 10, 25, 50, 75, 90, 95 and 99.
Discussion
In this case-control study, we have found that a model for breast cancer risk stratification developed in white women in the United States9 similarly performs in the Spanish population.
Relative relevance of GRS, NMRFS, MRFS
According to our results, modifiable risk factors do not discriminate well between breast cases and controls, which questions our ability to implement breast cancer primary prevention policies. Of note, several modifiable factors (namely, alcohol consumption, BMI and tobacco smoking) have been identified as risk factors for many other frequent diseases (e.g.: cardiovascular diseases, lung cancer and other cancers); thus, general recommendations for moderating alcohol consumption, avoiding tobacco smoking and maintaining BMI in its optimal range are considered among the most important health promotion recommendations, despite we do not observe that these factors can discriminate between breast cases and controls.
Consequences for screening
The US Preventive Services Task Force recommends biennial breast cancer screening with mammography for all women aged 50 to 74 “who are not at high risk of breast cancer because of a known underlying genetic mutation”22. GRS92, NMRFS and full model, however, could be useful in stratifying women according to their breast cancer risk. Women scoring in the 10th decile in GRS92 or full model could have double the risk of women with the average scoring and four- to six-fold the risk of women in the first decile, which makes it sensible to use these scores for individualizing the breast cancer screening recommendations. Genetic risk is already being the basis for changing screening periodicity; for instance, women carrying mutations in the gene BRCA1 have about 4-fold the risk of the average women, which supports performing screen mammography every six months23 and could even sustain recommendations for oophorectomy and/or mastectomy on an individual basis. In a similar way, a two-fold increase in risk over the average could lead to shorten the usual two-year interval for mammography in women scoring high in GRS92 or in the full model; alternatively, women scoring high in GRS92 or in the full model could benefit from initiating screening mammography before being 50; in this way, results from a simulation study24 suggest that women with two-fold to four-fold increased risk for breast cancer could be annually screened starting at age 40 with similar harm-to-benefit ratio than the biennial screening starting at age 50 for the average-risk women. Likewise, women in the first decile have about half the risk of the average women, which could induce to reconsider the balance of harms and benefits they have when being screened and, thus, to discuss the possibility of widening their mammography interval, delaying their starting age or even reconsidering the utility the screening program has for them.
Consequences for communicating risk
Genetic counseling has been available since testing for BRCA1 and BRCA2 was developed about 20 years ago25. New sequencing technologies make polygenic panel tests accessible26,27; applying them to risk assessment would allow genetic counseling to go further than a few highly-penetrant variants, which opens new ways for advising women before breast cancer can develop. Moreover, genetic scores on breast cancer and non-modifiable risk factors seem to be independent from each other; according to our results, stratifying breast cancer risk combining both genetic score (GRS92) and non-modifiable risk factors (NMRFS) improves patient classification by 28%. From a woman’s point of view, it is of no relevance whether her [possible] breast cancer would be caused by a sum of genetic variant or non-modifiable risk factors or a combination of both; in this way, it could be time for moving from genetic counseling towards risk counseling, irrespective of the factors related to that risk. Maas et al.’s model9, which we have here validated in a Spanish population, could be a well-founded instrument for doing it.
Limitations
Our study has some limitations. First, genotyping was available only for 65% of our patients; therefore, GRS24 had to be imputed in the remaining 35%. In order to validate this imputation, we carried out a sensitivity analysis on the GRS24 – breast cancer relationship in participants with and without genotyping data (Supplementary Table 7); OR by deciles were very similar in both groups, reinforcing the way genotype was imputed. Genotyping of the SNPs included in GRS68, however, was not available at all, so GRS68 was completely imputed as it actually was in the paper were the score was developed9. Second, when constructing GRS92, rs10483813 was excluded because of its almost perfect correlation with rs999737; apart from it, only two other SNPs (rs6678914 and rs4245739) displayed R2 with each other greater than 0.3 (Supplementary Table 2), involving some degree of linkage disequilibrium; however, we consider that their correlation was not high enough to exclude one of them in spite of admitting some redundancy degree. Third, a main concern in our results was the weak breast cancer - modifiable risk factors association; of note, reproductive and lifestyle factors have been measured by self-reporting; in this way, data are exposed to recall bias. As breast cancer participants were aware of their condition, we cannot rule out that their reports could be more biased that those of controls; this could be especially considered when reporting unhealthy lifestyles such as alcohol consumption or tobacco smoking, eventually leading to downward OR estimations. On the other hand, hormone replacement therapy has been used for less than 10% Spanish women28, which makes it difficult to find a relevant impact on breast cancer risk. Fourth, we limited our analysis to validate Maas et al.’s model in our case-control study; therefore, we did not go further than their article13 in exploring interactions between genetic and non-genetic risk factors, which -on the other hand- could be limited given the sample size our study has. Fifth, other risk factor models for BC have been published; for instance, Rosner and Colditz developed29 and refined30 a cumulative risk model based on the Nurses’ Health Study. Rosner – Coldtiz model, however, does not include genetic variants, which is a main point in Maas et al.’s score; therefore, we have not conducted a specific comparison between them.
In conclusion, when validating a breast cancer model, we have found that adding a non-modifiable risk factor score to a polygenic score can largely improve patient’s risk stratification. Its potential utilities would include risk-based screening programs or changes in genetic/risk counseling on breast cancer.
Electronic supplementary material
Acknowledgements
The study was partially funded by the “Acción Transversal del Cancer” project, approved by the Spanish Council of Ministers on the 11th October 2007, by the Instituto de Salud Carlos III-FEDER (PI08/1770, PI08/0533, PI08/1359, PI09/00773-Cantabria, PI09/01286-León, PI09/01903-Valencia, PI09/02078-Huelva, PI09/01662-Granada, PI11/01403, PI11/01889-FEDER, PI11/00226, PI11/01810, PI11/02213, PI12/00488, PI12/00265, PI12/01270, PI12/00715, PI12/00150, PI14/01219), by the Fundación Marqués de Valdecilla (API 10/09), by the ICGC International Cancer Genome Consortium CLL (The ICGC CLL-Genome Project is funded by Spanish Ministerio de Economía y Competitividad (MINECO) through the Instituto de Salud Carlos III (ISCIII) and Red Temática de Investigación del Cáncer (RTICC) del ISCIII (RD12/0036/0036)), by the Junta de Castilla y León (LE22A10-2), by the Consejería de Salud of the Junta de Andalucía (2009-S0143), by the Conselleria de Sanitat of the Generalitat Valenciana (AP_061/10), by the Recercaixa (2010ACUP 00310), by the Regional Government of the Basque Country, by the European Commission grants FOOD-CT-2006-036224-HIWATE, by the Spanish Association Against Cancer (AECC) Scientific Foundation and by the Catalan Government DURSI grant 2009SGR1489.
Author Contributions
T.D.S., I.G.A., C.P., M.K., M.P. and J.L. contributed substantially to the conception, design and acquisition of data. T.D.S., I.G.A., C.P. and J.L. contributed to the analysis and interpretation of the data. T.D.S., I.G.A., P.R.C. and J.L. contributed to devising the draft of the article. The remaining authors (B.P.G., J.M.A., C.G.D., E.A., M.B., J.A.M., C.V., R.M.G., V.M., G.C.V., M.E., L.V.M., P.A., A.T., J.B.C., D.S.T., N.A. and P.F.N.) participated in the patients’ recruitment, acquisition of data and critical revision of the manuscript. All authors approved the final version to be published.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
4/1/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20832-0.
References
- 1.Ferlay J, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Golubnitschaja O, et al. Breast cancer epidemic in the early twenty-first century: evaluation of risk factors, cumulative questionnaires and recommendations for preventive measures. Tumour Biol. 2016;37(10):12941–12957. doi: 10.1007/s13277-016-5168-x. [DOI] [PubMed] [Google Scholar]
- 3.Lambertini M, et al. Reproductive behaviors and risk of developing breast cancer according to tumor subtype: A systematic review and meta-analysis of epidemiological studies. Cancer Treatment Reviews. 2016;49:65–76. doi: 10.1016/j.ctrv.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Hankinson S., Tamimi R., Hunter D. Breast Cancer in Textbook of CancerEpidemiology (eds Adami HO., Hunter D., Trichopoulos D.) 403–445 (Oxford University Press, 2008).
- 5.Walker LC, et al. Evaluation of copy-number variants as modifiers of breast and ovarian cancer risk for BRCA1 pathogenic variant carriers. European Journal of Human Genetics. 2017;25:432–438. doi: 10.1038/ejhg.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuchenbaecker KB, et al. Associations of common breast cancer susceptibility alleles with risk of breast cancer subtypes in BRCA1 and BRCA2 mutation carriers. Breast cancer research. 2014;16(6):3416. doi: 10.1186/s13058-014-0492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skol AD, Sasaki MM, Onel K. The genetics of breast cancer risk in the post-genome era: thoughts on study design to move past BRCA and towards clinical relevance. Breast Cancer Research. 2016;18(1):99. doi: 10.1186/s13058-016-0759-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, et al. Effectiveness of Prophylactic Surgeries in BRCA1 or BRCA2 Mutation Carriers: A Meta-analysis and Systematic Review. Clin Cancer Res. 2016;1;22(15):3971–81. doi: 10.1158/1078-0432.CCR-15-1465. [DOI] [PubMed] [Google Scholar]
- 9.Maas P, et al. Breast cancer risk from modifiable and nonmodifiable risk factors among white women in the United States. JAMA Oncol. 2016;2(10):1295–1302. doi: 10.1001/jamaoncol.2016.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castaño-Vinyals G, et al. Population-based multicase-control study in common tumors in Spain (MCC-Spain): rationale and study design. Gac Sanit. 2015;29(4):308–15. doi: 10.1016/j.gaceta.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Hunter DJ, et al. A candidate gene approach to searching for low-penetrance breast and prostate cancer genes. Nat Rev Cancer. 2005;5(12):977–85. doi: 10.1038/nrc1754. [DOI] [PubMed] [Google Scholar]
- 12.Joshi AD, et al. Additive interactions between susceptibility single-nucleotide polymorphisms identified in genome-wide association studies and breast cancer risk factors in the Breast and Prostate Cancer Cohort Consortium. Am J Epidemiol. 2014;180(10):1018–1027. doi: 10.1093/aje/kwu214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrdahl M, et al. Post-GWAS gene-environment interplay in breast cancer: results from the Breast and Prostate Cancer Cohort Consortium and a meta-analysis on 79,000 women. Hum Mol Genet. 2014;23(19):5260–5270. doi: 10.1093/hmg/ddu223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed E, et al. A guide to genome‐wide association analysis and post‐analytic interrogation. Statistics in medicine. 2015;34(28):3769–3792. doi: 10.1002/sim.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 16.Chatterjee N, et al. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nature Genet. 2013;45:400–405. doi: 10.1038/ng.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nature Genetics. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michailidou K, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nature Genetics. 2015;47:373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pencina MJ, D’Agostino RB, Sr., D’Agostino RBJ, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Statistics in Medicine. 2008;27:157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 20.Pencina MJ, D’Agostino RB, Sr., Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30(1):11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Globocan, http://globocan.irac.fr/Pages/online.aspx. Accessed Feb 20 (2017).
- 22.Siu AL. Screening for Breast Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med. 2016;164(4):279–96. doi: 10.7326/M15-2886. [DOI] [PubMed] [Google Scholar]
- 23.Wong EM, et al. Constitutional methylation of the BRCA1 promoter is specifically associated with BRCA1 mutation-associated pathology in early-onset breast cancer. Cancer Prevention Research. 2011;4(1):23–33. doi: 10.1158/1940-6207.CAPR-10-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandelblatt JS, et al. Collaborative Modeling of the Benefits and Harms Associated With Different U.S. Breast Cancer Screening Strategies. Ann Intern Med. 2016;164(4):215–25. doi: 10.7326/M15-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cini G, et al. Tracking of the origin of recurrent mutations of the BRCA1 and BRCA2 genes in the North-East of Italy and improved mutation analysis strategy. BMC medical genetics. 2016;17(1):11. doi: 10.1186/s12881-016-0274-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurian AW, Ford JM. Multigene panel testing in oncology practice: how should we respond? JAMA Oncol. 2015;1(3):277–8. doi: 10.1001/jamaoncol.2015.28. [DOI] [PubMed] [Google Scholar]
- 27.Kurian AW, et al. Genetic Testing and Counseling Among Patients with Newly Diagnosed Breast Cancer. JAMA. 2017;317(5):531–534. doi: 10.1001/jama.2016.16918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costas L, et al. Hormonal contraception and postmenopausal hormone therapy in Spain: time trends and patterns of use. Menopause. 2015;22:1138–46. doi: 10.1097/GME.0000000000000487. [DOI] [PubMed] [Google Scholar]
- 29.Rosner B, Colditz GA. Nurses’ Health Study: Log-incidence mathematical model of breast cancer incidence. JNCI. 1996;88:359–64. doi: 10.1093/jnci/88.6.359. [DOI] [PubMed] [Google Scholar]
- 30.Colditz GA, Rosner B. Cumulative risk of breast cancer to age 70 years according to risk factor status: Data from the Nurses’ Health Study. Am J Epidemiol. 2000;152:950–64. doi: 10.1093/aje/152.10.950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.