Abstract
Significant events have taken place shaping the recent industrialization of physiologically based pharmacokinetic in vitro–in vivo extrapolation (PBPK‐IVIVE) use in drug development. Due to our knowledge gaps about drug‐independent systems parameters, there are limitations in the use of purely IVIVE‐based (bottom‐up) approaches. This has encouraged combining the classical data analysis (top‐down) with PBPK‐IVIVE‐linked models in order to optimize model parameters by taking advantage of observed clinical data. This concept, when initiated after clinical observations, can be viewed as “reverse translation,” since it refers back to available systems information preclinical data before trying to describe the observations. This review demonstrates the advantages of such strategies in filling knowledge gaps and discusses the perceived hurdles in widening applications. It is paramount that no clinical data are assessed on their own, but in conjunction with other studies for that drug in different populations and/or other similar drugs in the same population.
Broader use of drug‐independent “system” information is a concept that distinguishes quantitative systems pharmacology (QSP) from classical descriptive models of observed data using purely statistical/mathematical models. However, building QSP models requires a series of drug‐dependent parameters that are usually, but not exclusively, measured in vitro or in species other than human. Translation of these values within QSP models is associated with uncertainties related not only to the gaps in system parameters, but also the accuracy and translatability (scaling) of the drug parameters. Conversely, the majority of system parameters, particularly those related to physiology and anatomy, as opposed to biology, are mostly derived directly or indirectly from human studies (e.g., transit time through various segments of the gastrointestinal tract, tissue blood flows, renal glomerular filtration rate, functional turnover rate of enzymes). In the absence of wide experience in forward translational in vitro–in vivo extrapolation (IVIVE) approaches, qualification for the overarching model can be obtained by verifying the specific use examples through reverse translation. This involves fitting the models to observed data and optimizing the drug or system parameters for which prior confidence is not high. Needless to say, optimizing the system parameters is only valid as long as observations from several independent drugs can be described simultaneously with such optimized values. Physiologically based pharmacokinetic (PBPK) models are a branch of QSP models. They share the common principles with QSP regarding the separation of the systems data from the drug data. PBPK models rely heavily, but not exclusively, on the IVIVE process and the data generated from in vitro systems. Like QSP models, whenever there are information gaps they resort to combining observed clinical data from human studies and even preclinical animal studies. The distinction between PBPK and other QSP models, which sometimes becomes vague when talking about local kinetics in tissues, is the fact that PBPK models focus on how the body handles the drugs rather than the more holistic view of QSP that defines the way drugs affects the body.
Over the last 6 years and since the publication of reviews on scientific rationale1 and regulatory benefits of using PBPK‐IVIVE linked models2 in this journal, applications of PBPK models have increased momentum. Industrialization of PBPK‐IVIVE use led to the publication of draft guidance documents by the European Medicines Agency (EMA)3 and the US Food and Drug Administration (FDA)4 last year. These guidance documents acknowledge the limitations in the use of a purely IVIVE‐driven (bottom‐up) approach while emphasizing the added benefits of PBPK‐IVIVE models in “extrapolation” to conditions that have not yet been studied. These benefits are not commonly associated with the classical data analysis of clinical studies (top‐down approach). Hence, combining the two approaches with the purpose of optimizing model parameters of PBPK‐IVIVE models using some observed clinical data is becoming more popular.
These so‐called “middle‐out” models, which are also known as hybrid multilevel models, take the advantages and strength of two other approaches. Therefore, they are not just restricted to explaining the observed data but they intend to go backwards (in explaining the clinical observations) in order to go forwards beyond the perimeters of the initial clinical study using the prior in vitro and system information. This provides the necessary “qualification” for the model to be used with confidence for “Pre”‐dictions. This reverse translation approach can begin with the clinical data (if the purely bottom‐up models are not built) but goes backwards to use that data to create models that were not previously developed, but can now be readily created, providing immense benefit in understanding those clinical study results. Even when the bottom‐up model exists, using the clinical data to revisit some aspects of the system or drug data is beneficial. The schematics shown in Figure 1 a try to capture the separated bottom‐up and top‐down modeling in contrast with a combined approach in Figure 1 b, regardless of the starting point in the loops between clinical and preclinical studies. It should be noted that the boundary between bottom‐up and middle‐out as well as the distinction between top‐down and middle‐out gets blurry sometimes. For instance, top‐down models may also infer mechanistic meanings for the fitted parameters under investigating a clinical observation involving drug interactions by fitting functional inhibitory constants against a given enzyme and probe substrate. However, the frequency of using “external” data/information is less frequent in these models as opposed to middle‐out approach. Similarly, it is rare that a bottom‐up model does not use any clinical data in building the model and there are always some elements in the model that are derived from some clinical data.
Figure 1.
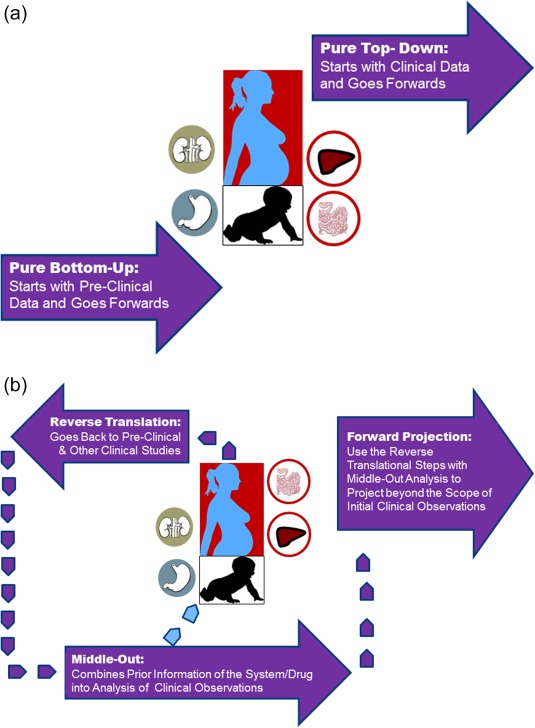
(a,b) The schematics of the differences between purely bottom‐up and pure top‐down approaches (a), vs. the combined models (b). For combined approach, in recent years we have witnessed many PBPK‐IVIVE approaches that are built based on preclinical data and then complemented by a designed study to qualify the model (Index Study, as it is called in the latest FDA Guidance). However, if for any reason such models are not built, it is still possible to do a “reverse translation” by taking observed clinical data and building a model that takes into account all prior knowledge about that drug and the systems information. In reality, the loop between clinical observations and preclinical data might be reiterated several times before a projection is made for cases which are not studied for a variety of ethical and practical reasons such as conducting drug–drug interaction studies in neonates, renal or hepatic impairment, pregnancy, the elderly, and so on; even the vulnerability of these patients could be very different from those of healthy volunteer populations.
Combining these models is not a seamless process and is fraught with issues, as reviewed by Tsamandouras et al. in 2014.5 If viewed purely from a mathematical point of view, these “middle‐out” approaches suffer from structural identifiability issues. Although it is argued by Agoram6 that this is not the right perspective when employing QSP models. Nonetheless, the absence of a unique correspondence between parameter values and the observed output is concerning for the researcher who wants to quantify the physiological process only based on a single observation set rather than considering a matrix of evidence from various sets (see later sections on fitting models to several datasets from varying drugs, or the same drug in various populations or conditions). It should be noted that even structurally identifiable models may suffer from numerical nonidentifiabilities. This occurs when clinical observations are made in a space that makes the model outcome insensitive to changes made to certain model parameter values (particularly in the face of noise and variations). Finally, estimating parameter values without considering the correlation between parameters can be an issue. This can also affect global sensitivity analysis (which is promoted by recent EMA PBPK draft guidance,3 although the importance of intercorrelations are ignored in all suggested global sensitivity approaches). The importance of such intercorrelations are exemplified by some recent research reports on this matter (Doki et al.,7 Liu et al.8).
QSP models, including PBPK, are now used to simulate and make inferences for conditions that are not tested (or cannot be tested easily) clinically (for example, see recent review by Yoshida et al.9 in this journal). In many cases, these models require inputs beyond those provided by in vitro or in silico experiments. Parameter estimation in these complex models enables reverse translation of observed clinical data, when available and relevant. The advantage over simpler empirical compartmental models offered by such a process is the ability to incorporate prior knowledge of the system while verifying the model predictions beyond the clinical dataset used to optimize the input values. An overview of selected examples for a “middle‐out” approach are provided below, followed by a critical analysis of common objections made to wider applications. The purpose of these examples and the critique that follows is to demonstrate that “bottom‐up” and “top‐down” modeling strategies, which until recently were considered separate functions, are now being incorporated into each other more and more, and borrowing strength from each other.
CASE EXAMPLES
Case 1
Purpose: Increasing ability to predict drug disposition in renal impairment.
Specific question to be addressed: Does renal impairment affect active secretion as much as glomerular filtration of drugs?
Scotcher et al.10 recently tried to delineate the role of various factors in renal disposition of digoxin by a reverse translational modeling that involved application of a physiologically based kidney model to clinically observed data (both on the plasma level of drug as well as the urinary excretion rate) from renal impairment populations. The submodel of kidney within PBPK (MechhKiM11) contained many active processes and went beyond the classical perfusion‐limited drug distribution into organs. However, this was challenged for an inadequacy of fully bottom‐up IVIVE approaches to recover some observed data (retro‐diction). The clinical observations for the modeling exercise came from divergent patient groups (healthy and renal impairment patients). Also, the observations were made both at the systemic circulation level (plasma drug concentrations vs. time) as well as in urine (rate of elimination). Those, which were coming from different studies, were assembled for assessment of altered renal secretion vs. glomerular filtration. Hence, the study combined the in vitro knowledge of drug affinity and handling by organic anion transporting peptide 4C1 (OATP4C1) and P‐glycoprotein (P‐gp) to build components of the model on tubular secretion and investigate the impact of age and renal impairment (moderate to severe) on renal drug disposition. The observed reduction in digoxin renal excretion clearance (CLR) in subjects with moderately impaired renal function relative to healthy was not consistent with models that assumed changes only in the glomerular filtration rate (GFR). Two hypotheses were tested based on reduction in either proximal tubule cell number or the OATP4C1 abundance and they successfully predicted a 59% decrease in renal clearance when these changes were proportional to reduction in GFR. However, the predicted proximal tubule concentration of the drug was only affected by changes in the transporter expression. Hence, the model not only provided evidence for reduction to active secretion (in line with GFR), as it also suggested as possible differentiation of the plausible causes based on consequences that might be associated with varying tubular concentrations in two scenarios. The implications of conditions that are not studied (“Pre”‐diction) were discussed such as those involving complex cases like transporter‐mediated drug–drug interactions (DDIs) in renal impairment patients.
Conclusions beyond the intended purpose and identified gaps: None of the above would be possible without combining the PBPK‐IVIVE with analysis of clinically observed data in less complicated patient groups. In addition, the observation of two alternative models, describing the same set of observations equally well, highlighted that data where the two models differ from each other (renal tubular concentrations) would be necessary to distinguish between the rival models. However, since such data are not practical to collect, indirect observations related to epidemiologic evidence of nephrotoxic effects could be added into future analysis.
Case 2
Purpose: Increasing ability to predict drug disposition in renal impairment.
Specific question to be addressed: Is nonrenal elimination (e.g., metabolism in liver) affected by the renal impairment?
It is almost two decades since simultaneous modeling of urinary drug: metabolite ratios and urinary recoveries of metabolites for three different probe drug substrates of CYP2D6 indicated the possibility of reduced enzyme activity in renal impairment patients (Rostami‐Hodjegan et al.12). Such parallel impairment in enzyme activity with renal function were included in some of the PBPK‐IVIVE models that demonstrated applicability in nonrenally eliminated drugs such as paroxetine.13 Although it was claimed that these models may help with defining safe and effective dosage regimens in patients with renal impairment, particularly when there is a void in availability of clinical data for severe renal impairment,14 the wider application required a reverse translational study involving many more drugs. Yoshida et al.15 carried out such an analysis in 2016. The drugs that were subject of the analysis were selected based on clinical DDI and pharmacogenetic studies. This reverse translation study confirmed and provided confidence in the earlier notion on chronic kidney disease affecting the pharmacokinetics of nonrenally eliminated drugs for pathways involving CYP2D6. Nonetheless, the attempts by the investigators to determine the similar effects on another drug‐metabolizing enzyme, CYP3A4, was inconclusive—mainly due to lack of information on changes happening to unbound drug in patients with chronic kidney disease.
Conclusions beyond the intended purpose and identified gaps: Despite the ability to conclude a longstanding open question on the issue of changes to CYP2D6 in renal impairment, the reverse translation attempt on other enzymes was not conclusive. However, this highlighted the significance of obtaining information on unbound drug in these patients, not just for understanding the kinetic changes for that drug but for a higher benefit of understanding the interplay between kidney and liver by determining an interfering element (i.e., changes in drug plasma protein binding). Ongoing studies on other enzymes and transporters are considering these gaps in going forward.
Case 3
Purpose: Increasing ability to predict changes to drug disposition in pregnancy.
Specific question to be addressed: Does known induction in CYP3A in liver during pregnancy occur in parallel in the gut wall?
The capacity of drug‐eliminating organs change during pregnancy. While most of these are known to increase,16 some also may go down (e.g., CYP1A2‐related metabolism17). However, since most drugs are taken orally and as gut wall metabolism may play a significant role in determining overall bioavailability of drugs metabolized by enzymes such as the CYP3A family, it is important to know the direction and magnitude of possible changes in gut wall metabolism in addition to what happens in the liver. This issue was not tackled until Ke et al.18 analyzed three different sets of clinical observations with a view to discern the site of CYP3A induction. Simultaneous modeling of the three drugs was a reverse translational attempt to provide confidence in future predictions, knowing the existing gap of knowledge regarding what happens to the gut wall during pregnancy. The selection of three different drugs with varying degrees of gut and liver metabolism enabled the investigators to explore the site of CYP3A induction (i.e., liver, intestine, or both). The model accounted for gestational age‐dependent changes in maternal physiological function and hepatic CYP3A activity. The model successfully predicted midazolam, nifedipine, and indinavir disposition during pregnancy when it assumed that CYP3A induction is most likely hepatic and not intestinal. The model provided more confidence in applications to other drugs (when there is contribution from CYP3A).
Conclusions beyond the intended purpose and identified gaps: The exercise was an indication that relative changes in enzyme levels in gut are less than those in liver. However, the ability to quantitatively describe the changes would have been stronger if the data were simultaneously modeled with a formal fitting process as opposed to sensitivity analysis performed in the study. When both oral and intravenous (i.v.) data were available for a given drug in pregnancy, these could be added to the analysis (as in the case of indinavir, but not for midazolam or nifedipine). Thus, further refinement of systems information can be obtained by a larger modeling exercise with additional data from other drugs and other routes of administration.
Case 4
Purpose: Obtain an indication of CYP3A turnover in human liver.
Specific question to be addressed: What value of CYP3A turnover is associated with the best predictability of mechanism‐based inhibition?
One of the mechanisms involved in metabolic DDIs involves permanent deactivation of the enzyme (tight binding or irreversible binding of the drug or metabolic product) as opposed to the more common competition between the drugs to engage with the enzyme. While the latter (competitive inhibition) is the most prevalent metabolic interaction, the former, so‐called mechanism‐based inhibition (MBI), has a significant and lasting impact on handling any substrates for that enzyme, as it removes the enzyme from the available pool and reduces the capacity until more enzymes are synthesized.19 Therefore, the turnover rate of the enzyme is a key factor in determining the magnitude and duration of the inhibitory effects. However, the data obtained on the turnover values are sparse at best and have employed various indirect measures involving both in vitro and in vivo assessment.19 Some of the values for specific enzymes have been adequately consistent (e.g., CYP1A2) despite the methodologies being different, while some others (e.g., CYP3A) have been very different. Rowland‐Yeo et al. in 201120 used a reverse‐translation approach to obtain the most optimally defined turnover number for CYP3A based on analyzing a series of drug interaction studies by incorporating some in vitro data into their analysis. The study involved prediction of time‐dependent metabolic DDIs related to CYP3A observed in 29 clinical studies. The most predictive model outcome, considering all studies together, contained a turnover of 2 days, although the authors emphasized that this is the best value conditioned upon current methodologies in obtaining the inactivation constants for the inhibitor.
Conclusions beyond the intended purpose and identified gaps: The studies investigating the biologic turnover of enzymes involved the incorporation of [3H]‐leucine into the enzyme by preincubation and chasing the radioactivity in specific enzymes over time. Although these have generated turnover values using human hepatocytes (e.g., 51 h and 44 h for turnover half‐life for CYP1A2 and CYP3A4, respectively21, 22) they determined using material from only one individual.19 Hence, the translatability of values might be questioned. Gathering all seemingly unrelated clinical studies (such as DDI with different drugs) could produce knowledge that went beyond the specific case: the system parameter values. Of course, this has not resolved all issues but opened new frontiers for research into differences in turnover of enzyme in different tissues (gut vs. liver) or the turnover of enzyme within a cell that itself turns over and sometimes more rapidly that the enzyme (e.g., gut23).
Case 5
Purpose: Define ontogeny of CYP3A in liver from birth to adulthood.
Specific question to be addressed: What function describes the change in activity/abundance of CYP3A per gram of tissue and after accounting for allometrical changes in liver size?
It is now well recognized that in addition to allometric changes in the size of organs during a human lifetime, the content and activity of the given content of the tissue, including enzymes and transporters that handle disposition of drugs, change as well. It has been shown that some functions are nonexistent or at a negligible level at birth, while others are comparable to that of adults. Understanding the changes that occur with age regarding abundance and activity of enzymes and transporters (and even receptors and other drug targets), after correcting for the size differences, helps to account for variation in handling of the drugs and their anticipated effects, particularly at a younger age. However, despite progress in new methodologies in measuring abundance of proteins (for review, see Al‐Feteisi et al.)24 the availability of tissue at different age groups, as well as the possible disconnect between activity and abundance due to cofactors and their ontogeny, has a major effect. This means that building confidence in predictive value, beyond a specific drug, may require reverse translation of observed data from several sources and its combination with known parameters defining other biological and physiological phenomena. Salem et al. in 201425 employed such an approach by deconvolution of observed clearance values from a series of independent clinical studies. This was to address the reported underprediction of clearance values for some substrates when purely bottom‐up in vitro values were used to assign ontogeny functions. The new models developed by the reverse translation was validated based on improved predictions of the systemic clearances of a drug that had not been used in derivation of the ontogeny function (alfentanil). In addition to improving the predictions, the study also highlighted the importance of considering potential confounding factors (e.g., disease) that may affect the physiological conditions of the patient.
Conclusions beyond the intended purpose and identified gaps: The sparsity of data and biological samples from neonates and younger children makes this group particularly relevant to activities of QSP, PBPK, and reverse translation. Efforts in gathering various clinical data and analyzing them alongside available information is a big task in this area, but with rewarding results that informs many other drugs that have not been part of the study.
PERCEIVED HURDLES TO CONDUCTING REVERSE TRANSLATION AND INCORPORATING PRIOR SYSTEMS AND DRUG DATA
As demonstrated by the examples above, all QSP model building, including PBPK‐IVIVE, go through cycles that are defined by the integration of available experimental data and existing biological knowledge at a given time (Figure 1 b). This is regardless of the starting point being the preclinical data (which is a recent phenomenon with the advent of PBPK‐IVIVE models) or clinical observations (which is common in the setups where PBPK‐IVIVE modeling is not industrialized or their capacity is limited and not carried out for all candidates until they reach the later stages of clinical studies). These, in turn, generate hypotheses and make predictions that sometimes can be tested prospectively but sometimes require retrospective analysis of the field data or anecdotal evidence. Conducting DDIs in renal impairment or frail elderly patients or in very young pediatrics are among such examples where there is adequate system information to hypothesize a variation in susceptibility to interaction beyond the healthy volunteers, but conducting actual studies might be very challenging both on practical and ethical grounds. So the issue of “Pre”‐diction and giving guidance on management in such situations is greatly beneficial to the most vulnerable patients as opposed to leaving a void in the label.
One may ask the question why these models are not built and used more commonly then? The list below is by no means comprehensive in gathering perceived views on hurdles for a wider use of reverse‐translation and forward projections from PBPK‐IVIVE (or any other QSP) models; however, they indicate some misconceptions associated with these perceived hurdles.
These models are data hungry!
Typically, those who bring this up as a hurdle do not distinguish between two separate sets of data that are required in these models, namely, system information vs. drug data. Systems information, although a massive task to gather, curate, and integrate, needs only be done “once.” The repeated use of the models, therefore, does not require as extensive data as the objectors have in mind. The number of drug‐related data to inform these models is limited. In fact, most of the data are gathered during the drug development procedure anyway, with a key difference being they are currently viewed in isolation as opposed being part of an integrated model.
Building these models is time‐consuming!
Again, if the intended use was for one drug and one occasion, the time spent in building these models could not be justified. However, repeated use for many drugs and extension to applications to areas not yet explored clinically saves lots of time and effort in conducting unnecessary studies with uncertain outcomes later on.
Building these models requires expertise!
Most of the required experts are in fact not associated with the modeling aspect by the deep field information on system attributes. These are inherently available in companies who develop a drug in each particular field. There are also many initiatives to identify the generic (precompetitive) aspects of the models and build them via a consortia approach up to the level where propriety aspects kick in. The mechanistic nature of these models make them much easier and intuitive to communicate compared to classical (empirical) models, which are not perceived by field experts (nonmodelers) as a reasonable integration of what they already know. Therefore, these models provide the opportunity for closer collaborations between various expertise rather than relying (unduly) on modelers only.
The models require specialist software!
This might be true, however many alternatives always exist. Expecting all modeling and simulation endeavors to remain as an “individual task” on the back of a piece of paper, or as a piece of a file in a commonly available tool, is the same as asking for an individually packed high‐performance liquid chromatography (HPLC) column for laboratory chemical analysis or manual preparation of all aspects of gene sequencing! Time moves on and many aspects of what was originally considered a complex operation becomes automated, while the experts move to a new frontier. It is often ignored that the largest investment in shifting to the use of these models is not the cost of software and modeling team, but the creation of infrastructure that is aligned with the philosophy of “integration” rather than “compartmentalization.” Of course, if there are elements in models that are common (at a precompetitive level), then getting a global agreement rather than reinventing and adding variations becomes important to enable comparisons and consistency.
These models make many assumptions!
There is a complete misconception stemming from the mere fact of “declaring all assumptions transparently.” Representation of a complex system in any simple formalism requires many more assumptions than a more complex model representing the same biological entity. However, since the simpler models rarely list all of the (many, many) assumptions they make, to a naive bystanding observer it may look like they have fewer assumptions and they can take comfort in even “not making any assumption”! No complex biological phenomenon can be described in a simple form unless many assumptions are made. Clarity in declaring assumptions provides a good opportunity to check the validity or degree of confidence in each assumption and put them to the test. Complex models make fewer assumptions than simpler models, as they take known facts and incorporate them into the models rather than making assumptions about them. Also, when they make assumptions, they declare them transparently.
Not all aspects are transparent!
This is in contradiction to the previous item; however, what is usually meant by it is not transparency but accessibility to modelers who wish to modify the components. The complex nature of these models means tracing the changes made (and their consequences), which is not an easy task at all. Hence, these models require a gatekeeper who ensures that the changes are properly documented. Some modelers find this frustrating, as they may wish to have access to all elements and this frustration is manifested in calling the models a “black box” while, as described above, in relation to declaring assumptions, these models are anything but a black box. In trying to separate the issue of access ability and transparency, I have recently used the term “glass box.” The latter conveys the message that all components are visible (unlike black box), but that at the same time all aspects within the box are outside the interference by every user and only certain gatekeepers have access to open the box and change them. One should realize that without such a quality control system, assessment of the models, considering their complexity and traceability of any changes, becomes an impossible task. Nonetheless, it is crucial that the details related to all elements of these large‐scale models are documented and ideally published through the peer review process. Those modelers who struggle to comprehend all various components of the large‐scale models have to realize that perhaps one single modeler may never adequately cover the vast number of submodels in these multiscale models unless they go through all the relevant publications. This is not an easy task, and even with the simplest PBPK‐IVIVE models there are now hundreds of publications each dealing with only a very narrow area of the model.
The effort does not match the added value!
This could have been argued several years ago when there was not abundant evidence on the use and acceptability of these models to accelerate the drug development and improve the informative nature of the drug labels.9 Nonetheless, it is difficult to measure and attach a value to making “right” (most optimal) decisions considering that the great majority of the use of reverse translation occurs during the internal decisions for drug development. It is hard to imagine a system that does not integrate available information (in a formal way) that will produce a better outcome than a formalization of integrated knowledge. Hence, the argument is whether the time and effort justifies the improvement. This will remain an open question, however, as the drug regulatory authorities start to accept the outcome of qualified models as an alternative to leaving gaps in drug labeling or even avoiding certain studies (which are deemed to be predictable based on the models; see Wagner et al.26 on DDI for instance). Indications of financial value as well as health impact (to include guidance on dosage for groups which were not previously served; see Jadhav et al.14) are starting to emerge and encourage the use of models further.
Other modeling types can be applied
Most of the other types of models (classical data analysis using empirical models) will continue to be used, but in many cases these models are starting to include some elements of physiology and biology and the distinction between the empirical and mechanistic models are slowly fading. Realization that “extrapolation” beyond the initial dataset is not possible and “interpolation” within the parameter space of observed clinical studies will not satisfy the needs of the healthcare community to address permutation of conditions that are not previously studied, has brought the two communities of modelers closer to each other. Some communication barriers still exist when the two groups refer to the same impression and word but they mean different things. Examples of these are discussed in the final part of this review in relation to the use of word “pre‐diction.”
Confidence in the output is not high!
The frequency of use will bring this confidence over time. However, it is worth noting that the alternatives involve less rigorous and subjective gathering of various pieces of information in an informal process that can be named an “in cerebro” modeling, as opposed to formal model‐based integration under “in silico” modeling. Confidence in areas where many examples of tested and qualified cases exist (such as CYP‐related DDI) is high, particularly when the complexity prevents a more confident “in cerebro” judgment to be made. Some other areas are slowly gaining such a level of confidence (such as ADME in young pediatrics or special subpopulations), while many gaps are identified in other areas that define the research directions. So it can be argued that this will just be a matter of time, and unless the models are tried we will not find out about their performance.
WHAT IS THE WORD? QUALIFICATION VS. VERIFICATION VS. VALIDATION AND MIDDLE‐OUT VS. REVERSE TRANSLATION
Following the publication of the draft guidance by the EMA on PBPK models, substantial discussions have been taking place at various events regarding the qualification of PBPK models. These are useful discussions, which have implications beyond PBPK. The latter, as described in the introduction, is a subdivision within the larger family of QSP models; therefore, any conclusions reached with regard to PBPK may equally apply to all QSP modeling too. In every‐day conversation, we use “qualification” frequently by referring to someone or some organization that are permitted to conduct certain procedures because through some means they have passed what is required for such qualifications (take a General Practitioner Physician (family doctor), a certified lawyer, a pharmacist, an accountant or accountancy firm, etc.). The mere fact that a person or an organization is “qualified” does not guarantee that what they do would be correct, flawless, or optimal (best in class); it only legislates for the fact that they have passed through several tests and they are distinguished from others (who have not passed the test and hence not qualified/certified). The qualification process gives comfort (by a huge margin) in using the individual or organization for the intended purpose. Nonetheless, knowing that even within the qualified group there exists some variations; we seek advice and take recommendations from those who have used the person or firm based on prior performance, even though this performance may not exactly replicate what we want to get from the service. Taking recommendations and looking into past (postqualification) performance might be seen as “verification.” Both qualification and verification, albeit at different levels of expectations (importance of the decision), are indicators of “predictive power.” However, “validation,” with respect to prediction, is almost useless, as it is always after the event. Of course, validation can be used as an added verification for a subsequent expectation from the model and increases the trust in the model and likelihood of its predictive value. Setting standards for qualification are absolutely necessary; however, one should realize that, like professional qualification, the standards change with time. What was expected from a qualified pharmacist half a century ago is totally different from what is expected now. Hence, revising these standards and upgrading them with the minimum level of added knowledge and skill are essential too (in the same way that checking capabilities through continuous educational credits is set for a profession). In the context of expectations from the model after qualification (mainly predictive power), understanding alternatives is important and a matter of a wealth of evidence on verified cases they have handled.
Another item that is worth mentioning is the relationship between the middle‐out approach and reverse translation. If we start with forward translation, the issue may become clearer. When the in vitro drug data and prior knowledge of a system is put together to make a projection, we are conducting forward translation that contains a bottom‐up model. On our way and as we gather clinical data, we may conduct middle‐out modeling to verify assumptions and obtain optimal values (of system or drug). However, if we have not done such projections and landed in a set of clinical observations that cannot be described (easily), then we may wish to go back (reverse) and gather all the peripheral information that we had missed and might be of relevance to our observations until we make sense of what we have observed (translation). This will include, again, middle‐out modeling. Thus, reverse (or forward) translation is the philosophy of the activity, while the middle‐out approach is a tool. While the middle‐out approach is commonly used in the forward translation for verification and individual cases, they can be used in a wider context for modeling series of studies on various drugs, or the same drug in various conditions, or clinical studies of system with no drugs, all simultaneously.
EPILOGUE
The new FDA commissioner, Scott Gottlieb, on his blog posted on July 7, 2017,27 outlined his views on how the FDA is going to capitalize on advances in science for the benefit of consumers. He highlighted, as an example, investing in and expanding on innovations in the use of in silico tools for improving drug development and making regulation more efficient. This matches with the approved PDUFA reauthorization performance goals and procedures for fiscal years 2018 through 2022,28 where enhancing benefit–risk assessment in regulatory decision‐making gets a specific mention and the role of modeling is explicitly defined. Of particular interest is the emphasis on developing a regulatory science and review expertise and capacity for “Model‐Informed Drug Development” (MIDD) and the encouragement for defining best practices in conducting:
-
1
(1) physiologically based pharmacokinetic modeling;
-
2
(2) design analysis and inferences from dose–exposure–response studies;
-
3
(3) disease progression model development, including natural history and trial simulation;
-
4
(4) immunogenicity and correlates of protection for evaluating.
It is important to note that these elements are a continuum and are not separate from each other, while in the current development practice, each are handled in isolation rather than as a coherent and interwoven package. Part of the challenge in overcoming the barriers is defining a common language. For instance, while many population pharmacokinetic modelers are debating the various technical elements of so‐called “visual predictive checks” (VPC), for the PBPK modeling community there is nothing “Pre”‐dictive about these tests, since they are a mere reflection of consistency between the proposed model and the data from which these models were derived from. Similarly, while classical modelers will be horrified by a twofold difference between model outcome and observed data, PBPK modelers will see that as triumph! This follows since the first group do not realize that the observed data actually were not used to make the prediction (true meaning of the “Pre”‐diction, i.e., saying something about an event or result before it is known). Thus, reverse translation could be seen as an opportunity where the meaning of “predict” (saying or estimating that a specified thing will happen in the future; made known beforehand) can be commonly defined and a separation made between its purpose vs. “postdiction” (explanation after the fact) or “retrodiction” (making a “prediction” about the past events but without using the observed data involved in that event—through blinding, etc). It is true that reverse translation takes us backwards, but it is only with the view to integrate independent sources of information with the observed data and move forward faster and with confidence.
CONFLICT OF INTEREST/DISCLOSURE
The author holds shares in Certara, a company focusing on Model‐Informed Drug Development. The author has completed the Unified Competing Interest form at http://www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author) and declares no support from any organization for the submitted work.
ACKNOWLEDGMENTS
The research of the author is sponsored by various pharmaceutical companies contributing to CAPKR and Simcyp Consortia, UK and EU granting bodies. The assistance by Susan Burkhill and Ellen Leinfuss in preparing the article is appreciated.
References
- 1. Rostami‐Hodjegan, A . Physiologically based pharmacokinetics joined with in vitro‐in vivo extrapolation of ADME: a marriage under the arch of systems pharmacology. Clin. Pharmacol. Ther. 92, 50–61 (2012). [DOI] [PubMed] [Google Scholar]
- 2. Zhao, P . et al Applications of physiologically based pharmacokinetic (PBPK) modeling and simulation during regulatory review. Clin. Pharmacol. Ther. 89, 259–267 (2011). [DOI] [PubMed] [Google Scholar]
- 3. European Medicines Agency . EMA/CHMP/458101/2016 — Guideline on the qualification and reporting of physiologically based pharmacokinetic (PBPK) modeling and Simulation Draft. < http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2016/07/WC500211315.pdf > (2016). Accessed August 2017.
- 4. U.S. Food and Drug Administration . Physiologically Based Pharmacokinetic Analyses — Format and Content. Guidance for Industry. <https://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM531207.pdf> (2016). Accessed August 2017.
- 5. Tsamandouras, N. , Rostami‐Hodjegan, A. & Aarons, L. Combining the ‘bottom up’ and ‘top down’ approaches in pharmacokinetic modeling: fitting PBPK models to observed clinical data. Br. J. Clin. Pharmacol. 79, 48–55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Agoram, B. Evaluating systems pharmacology models is different from evaluating standard pharmacokinetic‐pharmacodynamic models. CPT Pharmacometrics Syst. Pharmacol. 3, e101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doki, K. , Darwich, A.S. , Achour, B. , Tornio, A. , Backman, J.T. & Rostami‐Hodjegan, A. Accounting for inter‐correlation between hepatic CYP enzymes when predicting drug‐drug interactions and their inter‐individual variability. Br. J. Clin. Pharmacol. (Under Review). [Google Scholar]
- 8. Liu, D. , Li, L. & Jamei M. Application of Global Sensitivity Analysis Methods to Determine the Most Influential Parameters of a Minimal PBPK Model of Quinidine. Poster presented at PAGE 2017. 2017 June 6–9; Budapest, Hungary.
- 9. Yoshida, K. , Budha, N. & Jin, J.Y. Impact of physiologically based pharmacokinetic models on regulatory reviews and product labels: frequent utilization in the field of oncology. Clin. Pharmacol. Ther. 101, 597–602 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scotcher, D. , Jones, C.R. , Galetin, A. & Rostami‐Hodjegan, A. Delineating the role of various factors in renal disposition of digoxin through application of physiologically based kidney model to renal impairment populations. J. Pharmacol. Exp. Ther. 360, 484–495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neuhoff, S . et al Accounting for transporters in renal clearance: towards a mechanistic kidney model (Mech KiM) In: Transporters in Drug Development: Discovery, Optimization, Clinical Study and Regulation (eds. Sugiyama Y. &. Steffansen B.) (Springer, New York, 2013). [Google Scholar]
- 12. Rostami‐Hodjegan, A. , Kroemer, H.K. & Tucker, G.T. In‐vivo indices of enzyme activity: the effect of renal impairment on the assessment of CYP2D6 activity. Pharmacogenetics 9, 277–286 (1999). [DOI] [PubMed] [Google Scholar]
- 13. Rowland Yeo, K. , Aarabi, M. , Jamei, M. & Rostami‐Hodjegan, A. Modeling and predicting drug pharmacokinetics in patients with renal impairment. Expert Rev. Clin. Pharmacol. 4, 261–274 (2011). [DOI] [PubMed] [Google Scholar]
- 14. Jadhav, P.R. et al A proposal for scientific framework enabling specific population drug dosing recommendations. J. Clin. Pharmacol. 55, 1073–1078 (2015). [DOI] [PubMed] [Google Scholar]
- 15. Yoshida, K. et al Systematic and quantitative assessment of the effect of chronic kidney disease on CYP2D6 and CYP3A4/5. Clin. Pharmacol. Ther. 100, 75–87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abduljalil, K. , Furness, P. , Johnson, T.N. , Rostami‐Hodjegan, A. & Soltani, H. Anatomical, physiological and metabolic changes with gestational age during normal pregnancy: a database for parameters required in physiologically based pharmacokinetic modeling. Clin. Pharmacokinet. 51, 365–396 (2012). [DOI] [PubMed] [Google Scholar]
- 17. Gaohua, L. , Abduljalil, K. , Jamei, M. , Johnson, T.N. & Rostami‐Hodjegan, A. A pregnancy physiologically based pharmacokinetic (p‐PBPK) model for disposition of drugs metabolized by CYP1A2, CYP2D6 and CYP3A4. Br. J. Clin. Pharmacol. 74, 873–885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ke, A.B. , Nallani, S.C. , Zhao, P. , Rostami‐Hodjegan, A. & Unadkat, J.D. A PBPK model to predict disposition of CYP3A‐metabolized drugs in pregnant women: verification and discerning the site of CYP3A induction. CPT Pharmacometrics Syst. Pharmacol. 1, e3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang, J. et al Cytochrome p450 turnover: regulation of synthesis and degradation, methods for determining rates, and implications for the prediction of drug interactions. Curr. Drug Metab. 9, 384–394 (2008). [DOI] [PubMed] [Google Scholar]
- 20. Rowland Yeo, K. , Walsky, R.L. , Jamei, M. , Rostami‐Hodjegan, A. & Tucker, G.T. Prediction of time‐dependent CYP3A4 drug‐drug interactions by physiologically based pharmacokinetic modelling: impact of inactivation parameters and enzyme turnover. Eur. J. Pharm. Sci. 43, 160–173 (2011). [DOI] [PubMed] [Google Scholar]
- 21. Diaz, D. et al Omeprazole is an aryl hydrocarbon‐like inducer of human hepatic cytochrome P450. Gastroenterology 99, 737–747 (1990). [DOI] [PubMed] [Google Scholar]
- 22. Pichard, L. , Fabre, I. , Daujat, M. , Domergue, J. , Joyeux, H. & Maurel, P. Effect of corticosteroids on the expression of cytochromes P450 and on cyclosporin A oxidase activity in primary cultures of human hepatocytes. Mol. Pharmacol. 41, 1047–1055 (1992). [PubMed] [Google Scholar]
- 23. Darwich, A.S. , Aslam, U. , Ashcroft, D.M. & Rostami‐Hodjegan, A. Meta‐analysis of the turnover of intestinal epithelia in preclinical animal species and humans. Drug Metab. Dispos. 42, 2016–2022 (2014). [DOI] [PubMed] [Google Scholar]
- 24. Al Feteisi, H. , Achour, B. , Rostami‐Hodjegan, A. & Barber, J. Translational value of liquid chromatography coupled with tandem mass spectrometry‐based quantitative proteomics for in vitro‐in vivo extrapolation of drug metabolism and transport and considerations in selecting appropriate techniques. Expert Opin. Drug Metab. Toxicol. 11, 1357–1369 (2015). [DOI] [PubMed] [Google Scholar]
- 25. Salem, F. , Johnson, T.N. , Abduljalil, K. , Tucker, G.T. & Rostami‐Hodjegan, A. A re‐evaluation and validation of ontogeny functions for cytochrome P450 1A2 and 3A4 based on in vivo data. Clin. Pharmacokinet. 53, 625–636 (2014). [DOI] [PubMed] [Google Scholar]
- 26. Wagner, C. , Pan, Y. , Hsu, V. , Sinha, V. & Zhao, P. Predicting the effect of CYP3A inducers on the pharmacokinetics of substrate drugs using physiologically based pharmacokinetic (PBPK) modeling: an analysis of PBPK submissions to the US FDA. Clin. Pharmacokinet. 55, 475–483 (2016). [DOI] [PubMed] [Google Scholar]
- 27. Gottlieb, S. How FDA Plans to Help Consumers Capitalize on Advances in Science. < https://blogs.fda.gov/fdavoice/index.php/2017/07/how-fda-plans-to-help-consumers-capitalize-on-advances-in-science/>. Accessed July 2017.
- 28. U.S. Food and Drug Administration . PDUFA Reauthorization Performance Goals and Procedures Fiscal Years 2018 through 2022. <https://www.fda.gov/downloads/forindustry/userfees/prescriptiondruguserfee/ucm511438.pdf> (2016). Accessed August 2017.


