Abstract
The present study aimed to estimate the molecular prevalence of Theileria annulata and Trypanosoma evansi infection in cattle in Northern Tunisia. A total number of 96 cattle from five farms were evaluated. T. annulata and T. evansi prevalences were 61% [56/66] and 10% [7/13], respectively, at a confidence interval (CI) of 95%, while co‐infection was present in 6% [4/8] of the tested animals at a CI of 95%. There was a significant correlation between age and the prevalence of T. annulata infection, whereas, there was no significant association shown with the age of cattle and T. evansi infection. Sequence and phylogenetic analyses showed that the T. annulata Tams1 gene and T. evansi ITS1 rDNA gene were highly conserved with 97.1–100% and 98.3–100% sequence identity, respectively.
Keywords: Prevalence, Theileria annulata, Trypanosoma evansi, PCR, Tunisia, Phylogenetic analysis
Introduction
Tropical theileriosis (Theileria annulata infection) and surra (Trypanosma evansi infection) are two major vector‐borne haemoprotozoan infections in cattle. Both of the diseases cause high financial losses as a consequence of severe clinical illness in infected cattle but also of carrier state (Gharbi et al. 2006, 2011). In Tunisia, tropical theileriosis is one of the most frequent summer diseases of cattle with annual declared clinical cases of more than 2500 (Bahri et al. 1995). However, in Tunisia, T. evansi infection prevalence in cattle has never been studied.
Theileria annulata is enzootic in sub‐humid and semi‐arid Tunisian bioclimatic zones (Darghouth et al. 1996). It is a major constraint to the development of cattle population, particularly for exoticdairy breeds (Darghouth 1991).
Trypanosoma evansi is the most widely distributed pathogenic salivarian trypanosome in animals, it is the causative agent of surra (Woo 1977).The infection is transmitted mechanically by haematophagous arthropods. This parasite is cosmopolitan, affects a wide range of hosts (including humans). In India, four cases of human infection by T. evansi were documented with one death case (Desquesnes et al. 2013).
Trypanosoma evansi is especially pathogenic in camels and horses (Sumbria et al. 2014), it causes, in domestic animals, high morbidity, an decreased milk and meat yield. This parasite also induces immunosuppression in infected animals (Singla et al. 2009; Desquesnes et al. 2013). Since 2008, surra has been recognized as a notifiable disease, but cattle infection prevalence remains unknown in Tunisian cattle (OIE, 2008).
The aim of our study was to estimate the infection prevalence of cattle in Tunisian enzootic region for tropical theileriosis by T. annulata and T. evansi. The genetic characterization of the isolates and their phylogenetic relationships with those sequences available in GenBank was also performed.
Materials and methods
Study region and animals
The present study was carried out in El Hessiene locality (Kalaat El Andalous, Ariana Gouvernorate, Northern Tunisia) (Fig. 1); it is a sub‐humid region with an annual average rainfall of 454 mm. The mean minimal and maximal temperatures are 25.9 and 10.7°C in August and January, respectively (climate‐data.org). The study was carried out in extensive cattle herds during April 2014 (spring season) and January 2015 (winter season). Five cattle herds were visited and a total of 96 Holstein cattle of both sexes, aged between 2 months and 15 years were used for Theleria annulata and Trypanosoma evansi testing. Blood samples were collected in EDTA tubes from the jugular vein of each animal.
Figure 1.
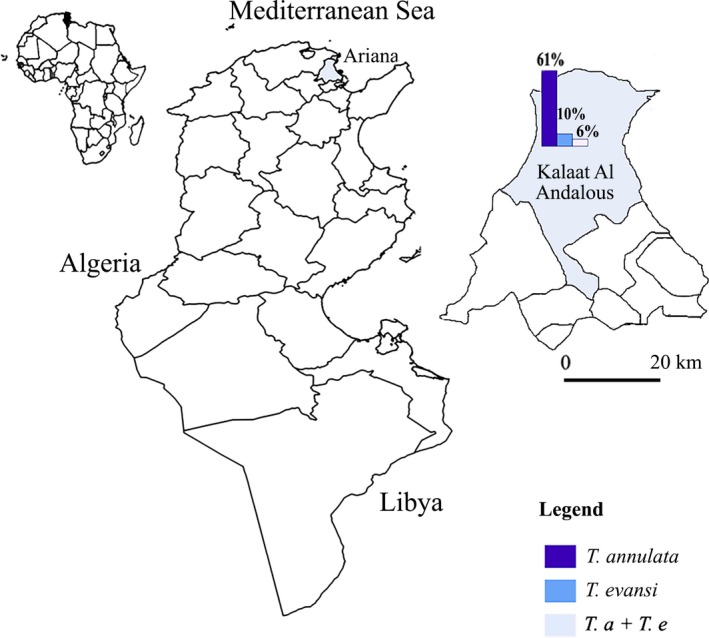
Theileria annulata and Trypanosoma evansi infection prevalence in cattle in Ariana (Northern Tunisia) estimated by PCR.
DNA extraction
The DNA was extracted from whole blood using a Bio Basic DNA Kit (Rapid Blood Genomic DNA Extraction Kit, Markham Ontario, Canada). Briefly, 300 μL of blood was mixed with 600 μL of lysis buffer and centrifuged at 4000 g. The supernatant was collected and 20 μL of proteinase K was added. PR buffer (60 μL) was added to the mixture, then, incubated at −20°C for 20–30 min and centrifugation at 12 000g. The supernatant was mixed with isopropanol twice volume followed by centrifugation; finally, the pellet was washed in 75% ethanol. The microtubes were centrifuged at 12 000 g for 5 min. The pellet was resuspended in 100 μL of Tris EDTA buffer and stored at −20°C until used.
Trypanosoma evansi DNA amplification
The PCR for T. evansi DNA detection was performed using the protocol described by Rjeibi et al. (2015). Trypanosoma evansi PCR was performed with a set of primers that amplifies a 480 bp region of T. evansi ITS1 rDNA gene (Njiru et al. 2005). The forward and reverse primers were: ITS1CF (5′‐CCGGAAGTTCACCGATATTG‐3′) and ITS1 BR (5′‐TGCTGCGTTCTTCAACGAA‐3′), respectively. PCR was performed in 25 μL volume consisting of 2.5 μL of 10× PCR buffer (50 mmol/L Tris‐HCl; pH 8.5; 50 mmol/L NaCl), 0.2 mmol/L of each dNTP, 2 mmol/L MgCl2, 0.2 μmol/L of each primer, 0.5 U Taq polymerase (Vivantis, Chino, California) and 3 μL of the DNA template.
In each PCR run, negative and positive (T. evansi DNA) controls were added. Amplification was performed in an automatic Swift MaxPro thermal cycler (SWT‐MXP). Thermocycling started with an initial denaturation for 5 min at 94°C, followed by 35 cycles (94°C for 40 s, 58°C for 40 s, and 72°C for 90 s) and a final extension at 72°C for 5 min.
Theileria annulata DNA amplification
Theileria annulata PCR amplifying a 721 bp fragment was performed with a set of specific primers of the gene encoding Tams1 of T. annulata. The forward primer was N516 (5′‐GTAACCTTTAAAAACGT‐3′) and the reverse primer was N517 (5′‐GTTACGAACATGGGTTT‐3′) (d'Oliveira et al. 1995). The PCR was carried out in 25 μL volume for each reaction consisting of 2.5 μL PCR buffer, 2 μL of the extracted DNA template, 0.4 mmol/L of each dNTP, 0.5 μmol/L of each primer, 3 mmol/L MgCl2 (25 mmol/L), and 0.05 U/μL of Taq DNA Polymerase (Vivantis). The reactions were performed in an automatic Swift MaxPro thermal cycler (SWT‐MXP), with initial denaturation at 94°C during 5 min, followed by 30 cycles (94°C, 55°C and 72°C for 1 min each) and a final extension at 72°C for 10 min.
For all PCR runs, electrophoresis was performed in 1% agarose gel with ethidium bromide and visualized under UV light.
DNA sequencing and phylogenetic analyses
Six PCR products obtained with primers ITS1CF/ITS1BR and N516/N517 amplifying T. evansi and T. annulata, respectively were purified with the unincorporated terminators with the BigDye® Terminator v3.1 Cycle Sequencing Kits (Applied Biosystems) according to manufacturer's instructions. The PCR products were sequenced in both directions, using the same primers as for PCR. Sequencing reactions were performed in the DNA Engine Tetrad 2 Peltier Thermal Cycler (BIO‐RAD) using the ABI BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), following the protocols supplied by the manufacturer. Chromatograms were evaluated with Chromas Pro software (version 2.4.3; Technelysium Pty Ltd 2014). The MEGA 6.1 software programme was used to perform multiple sequence alignments (Tamura et al. 2013). The sequences were compared with the GenBank database by a nucleotide sequence homology search carried out at the network server of the National Centre for Biotechnology Information (NCBI) using BLAST (http://blast.ncbi.nlm.nih.gov). Phylogenetic tree was constructed by using the neighbour‐joining method. The two alignments were analysed by Maximum Composite Likelihood. The outgroup that are known to fall outside of the group of interest (the ingroup) is the way to root tree.
Statistical analysis
The observed prevalence was estimated as follows:
Age was categorized in three groups: less than 2 years, between 2 and 5 years and superior or equal to 6 years. Age and gender were tested for significant association with the infections status of animals to T. annulata and T. evansi, respectively, using chi‐square test. This one was performed with Epi Info 6 software at the threshold value of 0.05 (Schwartz 1993).
To assess the significant effect of each factors (age and gender) while simultaneously controlling for the effect of the other factors an analysis with logistic regression was carried out based on prevalence status to T. annulata and T. evansi of the animals as a binary outcome. A backward stepwise method was used for significant (P < 0.05) variables in the univariate analysis. The logistic regression was performed using SPSS, version 21 software.
Results and discussion
Molecular prevalence of T. annulata and T. evansi in Tunisian cattle
Overall T. annulata infection prevalence was estimated at 61 [56; 66] CI at 95%. The highest infection prevalence was observed in cattle aged of ≥ 6 years 92 [85; 99] CI at 95%, while the lowest prevalence infection was observed in the animals aged of less than 2 years 34 [26; 42] CI at 95% (P = 0.00001) (Table 1). According to logistic regression results, animals aged more than 6 years were 20 times at greater risk of positivity compared to animals between 2 and 5 and five times more likely to be positive compared to animals less than 2 years. The infection prevalence of T. annulata was higher than cattle in Turkey (39%; 99/252) (Aktas et al. 2006), in Egypt (9.56%) (Elsify et al. 2015) than those estimated by blood smear in Faisalabad, Pakistan (10.8%; 28/260) (Saleem et al. 2014) and India (Tuli et al. 2015). This high prevalence is related to the abundance of the vector tick (Hyalomma scupense) in this region (Gharbi et al. 2014). Moreover, in our survey, studied cattle are reared in extensive system. This breeding system is a determining risk factor for tropical theileriosis infection explained by the endophilic character of the tick vector in North Africa (Gharbi et al. 2012). A positive association between the age of the animals and the infection prevalence was detected (P < 0.005) (Fig. 2). The logistic regression model showed that only age was associated to T. annulata infection. The age was shown to be positively associated with multiplicity of T. annulata infection (Weir et al. 2011). The same trend was reported by Flach et al. (1995). This could be because of: (1) the multiple re‐infections of the animals, (2) the low tick‐attractiveness of calves or (3) colostral antibodies persistence (Gharbi et al., 2014). However, there was no statistically significant difference of T. annulata infection prevalence according to gender.
Table 1.
Prevalence of Theileria annulata and Trypanosoma evansi infections and different parameters based on PCR in studied cattle
| Parameter | Theileria annulata | Trypanosoma evansi | Co‐infection (T. annulata and T. evansi) | ||||
|---|---|---|---|---|---|---|---|
| +ive/examined (% ± SEb) | OR [95% CIc] | +ive/examined (% ± SEb) | OR [95% CIc] | +ive/examined (% ± SEb) | OR [95% CIc] | ||
| Gender | Female | 46/69 (66 ± 0.06) | 2.15 [0.79; 5.88] | 7/69 (10 ± 0.04) | 0.90 [0.19; 4.85] | 5/69 (7 ± 0.03) | 2.03 [0.21; 48.22] |
| Male | 13/27 (48 ± 0.1) | 3/27 (11 ± 0.06) | 1/27 (3 ± 0.04) | ||||
| Age group | <2 years | 12/35 (34 ± 0.08) |
2.1 [0.08; 0.74]a
5.3 [0.01; 0.27]a |
5/35 (11 ± 0.06) | 1.88 [0.10; 13.12] | 1/35 (2 ± 0.03) |
3.09 [0.00; 1.44]a
1.03 [0.01; 3.35] |
| 2–5 years | 25/37 (67 ± 0.08) | 3/37 (8 ± 0.05) | 3/37 (8 ± 0.05) | ||||
| ≥6 years | 22/24 (92 ± 0.06) | 2/24 (8 ± 0.06) | 1.83 [0.20; 10.04] | 2/24 (8 ± 0.06) | |||
| Overall | 59/96 (61 ± 0.05) | 10/96 (10 ± 0.03) | 6/96 (6 ± 0.02) | ||||
OR, Odds Ratio; NA, not applicable.
0.001 ≤ P < 0.05.
Standard Error.
95% Confidence Interval.
Figure 2.
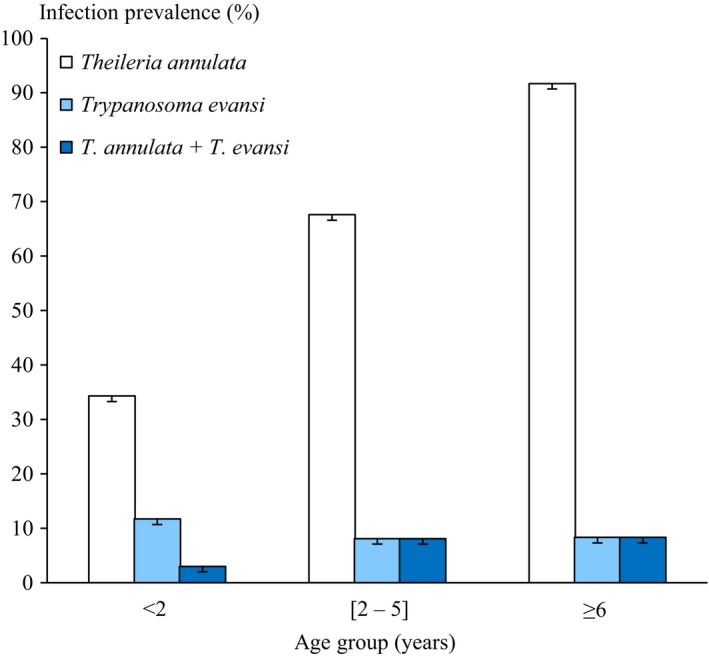
Age ‐ prevalence relation of Theileria annulata and Trypanosoma evansi infection.
A total 10 samples were positive to T. evansi, giving an infection prevalence of 10 [7; 13] CI at 95% (Table 1). The distribution of arthropod‐borne diseases is associated with the presence and distribution of its vector host (Baticados et al. 2011). In this context, the low prevalence of surra can be due to the scarcity of vector arthropod, the low parasitaemia in cattle and the low susceptibility of this species to T. evansi. There were no statistically significant differences in age and gender and infection prevalence. Trypanosma evansi has a large host range, but it is particularly pathogenic in camels and horses (Desquesnes et al. 2013).
A total number of six animals were co‐infected by both parasites 6 [3.5; 8.5] CI at 95% which was lower than cattle in India (11.25%) (Sudan et al. 2015). The highest rate co‐infection was observed in 2 to 5 years age group 8 [3; 13] CI at 95%. However, animals aged of less than 2 years showed the lowest infection rate (Fig. 2). While, no significant difference was observed in gender infection prevalence.
Additional studies are needed to further understand the importance of co‐infection and the presence of any interaction between the two pathogens. Veterinarians should consider the possibility of co‐infection and the presence of surra in Northern Tunisia.
Phylogenetic analysis
Theileria annulata phylogeny
T. annulata Tams1 gene (GenBank Accession Number: KU145624) was identified in this study. Nucleotide sequence identity data demonstrated that the Tunisian T. annulata strain share 100% sequence identity with Mauritania and Egypt (AF214819 and AB917290, respectively), in which they were found in a single group (group1) for the considered gene. Our isolate has 98.2 and 97.1% nucleotide homology with Bahrain (AF214795) and Turkey (U22888), respectively (group 2). The Tams1 sequence from this work shared 98‐97.1% identity with other Tunisian isolates (group 3). It was related to sequences from India and Iraq (AF214844 and GU130194, group 4) (95.9 and 94.8% sequence identity, respectively). Furthermore, sequences from Spain, Iran, Italy and Sudan were classified in a single group (group 5) (Fig. 3).
Figure 3.
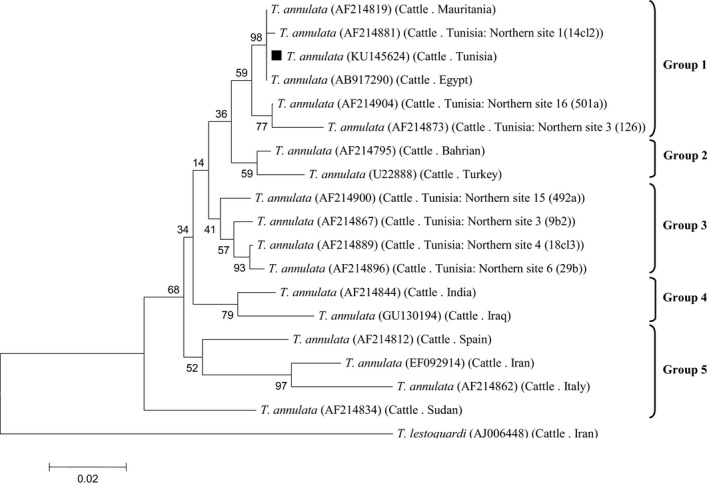
Phylogenetic relationships based on the partial nucleotide sequences (450 pb) of Tams1 gene for merozoite‐piroplasm surface antigen of T. annulata with other variants from T. annulata stains available in GenBank. The host, the country of origin and the GenBank accession numbers are given between parentheses. Sequence obtained in the present study is indicated with a black square.
The Tunisian Tams1 gene sequences were relatively diverse (97.1–100% identity values), dispersing themselves across several groups in the phylogenetic tree containing sequences from other countries. Knowing that, the Tams1 gene of T. annulata is antigenically diverse (Dickson & Shiels 1993). Moreover, T. annulata may generate novel antigenic Tams1 types by differential glycosylation (Katzer et al. 1998). This leads to high diversity levels allowing the parasite to evade the host's immune responses. However, identical Tams1 sequences were established in widely separate regions Habibi (2013). In this study, a close relationship was detected between Tunisian, Mauritanian and Egyptian.
Trypanosoma evansi phylogeny
A T. evansi ITS1 rDNA genotype named KU145623 (GenBank Accession Number) was identified in this study. The BLAST comparison of the partial sequences of ITS1 rDNA gene revealed 100% homology with the ITS1 sequence from the T. evansi strain collected from a dog in Tunisia, Sousse (KJ741365), 99.8% homology with T. evansi evansi from cattle in Thailand (AY912277) and 98.8% homology with isolates from water Buffalo in Thailand and Philippines (AY912275, HQ593643). In China, 99 and 99.3% homology from mule and buffalo (FJ712712 and FJ712711, respectively), 99.3% homology with isolates from Iranian camels (JN896754) and 98.8% homology with isolates from Philippine buffalos (HQ593643). T. evansi and T. brucei (AJ673390) share a high similarity (97%) in their ITS1 rDNA gene sequences (Fig. 4).
Figure 4.
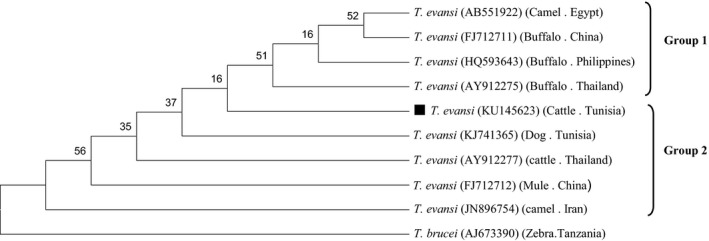
Phylogenetic relationships based on the partial nucleotide sequences (400 pb) of ITS1 rDNA of Trypanosoma with other variants from T. evansi stains available in GenBank. The host, the country of origin and the GenBank accession numbers are given in parentheses. Sequence obtained in the present study is indicated with a black square.
Phylogenetic analyses of T. evansi revealed neither host nor geographic specificity. In this context, the introduction of infected animals is a risk factor for spread of the disease. Furthermore, Pourjafar et al. (2013) showed that T. evansi probably related to the capacity for rapid adaptation to different host species and environments. We recommend undertaking additional studies to further understand the importance of co‐infection between the two pathogens.
Source of funding
The work was funded by the laboratory of “Laboratoire d’épidémiologie des infections enzootiques des herbivores en Tunisie: application a la lutte” (Ministère de l'enseignement superieur et de la recherche scientifique, Tunisia) and the Deutsche Forschungsgemeinschaft project ‘Molecular epidemiology network for promotion and support of delivery of live vaccines against Theileria parva and Theileria annulata infection in eastern and northern Africa’ (DFG SE 862/2‐1).
Conflicts of interest
The authors declare no conflicts of interest in relation to this work.
Ethical statement
This study was conducted in accordance with relevant national and international guidelines on handling animals, taking care to respect animal welfare.
Contribution
S Sallemi and M Gharbi conceived and designed the experiments. S Sallemi, M Rouatbi, S Amairia, M Khamassi Khbou and M Ben Said performed the experiments. S Sallemi and M Ben Said involved in the collection of samples. MR Rjeibi performed the phylogenetic analysis. M Gharbi, MR Rjeibi and S Sallemi wrote the manuscript.
Acknowledgement
The study was supported financially by Laboratoire d’épidémiologie d'infections enzootiques des herbivores en Tunisie (Ministère de l'enseignement supérieur, de la recherché scientifique et des technologies de l'information et de la communication) and the Deutsche Forschungsgemeinschaft project ‘Molecular epidemiology network for promotion and support of delivery of live vaccines against Theileria parva and Theileria annulata infection in eastern and northern Africa’ (DFG SE 862/2‐1). This work was financed by IRESA, programme de recherche multidisciplinaire et interinstitutionnelle: Étude des contraintes relatives à la filière lait bovin dans la région de Bizerte: vers un nouveau bassin laitier ?
The authors thank Mr Mohamed Jedidi, Mr Limam Sassi and Mr Tawfik Lahmer for their support, as well as the farmers who agreed to let us handle their animals.
References
- Aktas M., Altay K. & Dumanli N. (2006) A molecular survey of bovine Theileria parasites among apparently healthy cattle and with a note on the distribution of ticks in eastern Turkey. Veterinary Parasitology 138, 179–185. [DOI] [PubMed] [Google Scholar]
- Bahri S., Kallel A. & Gouia A. (1995) La theilériose bovine en Tunisie, étude rétrospective sur 5 ans. Bull Epidémiol Inf. Vét Instit Rech Vét Tun 5, 1–3. [Google Scholar]
- Baticados W.N., Fernandez C.P. & Baticados A.M. (2011) Molecular detection of Trypanosoma evansi in cattle from Quirino Province, Philippines. Veterinarski Arhiv 81, 635–646. [Google Scholar]
- Climate data.org . Available at: http://fr.climate-data.org/ (Accessed June 2015).
- Darghouth M.A. (1991) Tropical theileriosis in Tunisia: status of the disease and research being undertaken at the ENMV Sidi Thabet Tunisia In: Proceeding of the Second EEC Workshop on Orientation and Coor‐Dination of Research on Tropical Theileriosis, 18–19. (eds Singh D.K. & Varshney B.C.), National Dairy Develop‐Ment Board, Anand: India. [Google Scholar]
- Darghouth M.A., Bouattour A., Ben Miled L., Kilani M. & Brown C.G.D. (1996) Epidemiology of tropical theileriosis (Theileria annulata infection of cattle) in an endemic region of Tunisia: characterization of endemicity states. Veterinary Parasitology 65, 199–211. [DOI] [PubMed] [Google Scholar]
- Desquesnes M., Holzmuller P., Lai D‐H., Dargantes A., Lun Z‐R., Jittaplapong S. (2013) Trypanosoma evansiand surra: a review and perspectives on origin, history, distribution, taxonomy, morphology, hosts, and pathogenic effects. Bio Med Research International 2013, 194176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson J. & Shiels B.R. (1993) Antigenic diversity of a major merozoite surface molecule in Theileria annulata . Molecular and Biochemical Parasitology 57, 55–64. [DOI] [PubMed] [Google Scholar]
- Elsify A., Sivakumar T., Nayel M., Salama A., Elkhtam A., Rizk M. et al (2015) An epidemiological survey of bovine Babesia and Theileria parasites in cattle, buffaloes, and sheep in Egypt. Parasitology International 64, 79–85. [DOI] [PubMed] [Google Scholar]
- Flach E.J., Ouhelli H., Waddington D., Oudich M. & Spooner R.L. (1995) Factors influencing the transmission and incidence of tropical theileriosis (Theileria annulata infection of cattle) in Morocco. Veterinary Parasitology 59, 177–188. https://doi.org/10.1016/0304-4017(94)00760-A. [DOI] [PubMed] [Google Scholar]
- Gharbi M., Sassi L., Dorchies P., Darghouth M. A. (2006) Infection of calves with Theileria annulata in Tunisia: Economic analysis and evaluation of the potential benefit of vaccination. Veterinary Parasitology 137, 231–241. [DOI] [PubMed] [Google Scholar]
- Gharbi M., Touay A., Khayeche M., Laarif J., Jedidi M., Sassi L. & Darghouth M.A. (2011) Ranking control options for tropical theileriosis in at‐risk dairy cattle in Tunisia, using benefit‐cost analysis. Revue Scientifique et Technique‐OIE 30, 763. [DOI] [PubMed] [Google Scholar]
- Gharbi M., Mhadhbi M. & Darghouth M.A. (2012) Diagnostic de la theilériose tropicale du bœuf (infection par Theileria annulata) en Afrique du Nord. Revue Méd. Vét. 163, 563–571. [Google Scholar]
- Gharbi M., Rjeibi M.R. & Darghouth M.A. (2014) Épidémiologie de la theilériose tropicale bovine (infection par Theileria annulata) en Tunisie: une synthèse. Revue D’Élevage et de Médecine Vétérinaire des Pays Tropicaux 67, 241–247. [Google Scholar]
- Habibi G. (2013) A comparative phylogenetic analysis of Theileria spp. by usingtwo 18S ribosomal RNA and Theileria annulata merozoite surface antigen gene sequences. Archives of Razi Institute 68, 47–52. [Google Scholar]
- Katzer F., McKellar S., Miled L., d'Oliveira C. & Shiels B. (1998) Selection for antigenic diversity of Tams1, the major merozoite antigen of Theileria annulata . Annals of the New York Academy of Sciences 849, 96–108. [DOI] [PubMed] [Google Scholar]
- Njiru Z.K., Constantine C.C., Guya S., Crowther J., Kiragu J.M., Thompson R.C.A. & Dávila A.M.R. (2005) The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitology Research 95, 186–192. https://doi.org/10.1007/s00436-004-1267-5. [DOI] [PubMed] [Google Scholar]
- OIE , 2008. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Online: www.oie.int.
- d'Oliveira C., dervan Weide M. , Habela M.A., Jacquiet P., Jongejan F. (1995) Detection of Theileria annulata in blood samples of carrier cattle by PCR. Journal of Clinical Microbiology 33, 2665–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourjafar M., Badiei K., Sharifiyazdi H., Chalmeh A., Naghib M., Babazadeh M. et al 2013. Genetic characterization and phylogenetic analysis of Trypanosoma evansi in Iranian dromedary camels. Parasitology Research 112, 899–903. [DOI] [PubMed] [Google Scholar]
- Rjeibi M.R., Ben Hamida T., Dalgatova Z., Mahjoub T., Rejeb A., Dridi W. & Gharbi M. (2015) First report of surra (Trypanosoma evansi infection) in a Tunisian dog. Parasite 22, https://doi.org/10.1051/parasite/2015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem M.I., Tariq A., Shazad A. & Mahfooz S.A. (2014) Clinical, epidemiological and therapeutic studies on bovine tropical theileriosis in Faisalabad, Pakistan. Iraqi J Vet Sci 28, 87–93. [Google Scholar]
- Schwartz D. (1993) Méthodes statistiques à l'usage des médecinset des biologistes, 3ème éd. Flammarion: Paris, France. [Google Scholar]
- Singla L.D., Juyal P.D. & Sharma N.S. (2009) Immune responses to haemorrhagic septicaemia (HS) vaccination in Trypanosoma evansi infected buffalo‐calves. Tropical Animal Health and Production. 42, 589–595. [DOI] [PubMed] [Google Scholar]
- Sudan V., Jaiswal A.K., Parashar R. & Shanker D. (2015) A duplex PCR‐based assay for simultaneous detection of Trypanosoma evansi and Theileria annulata infections in water buffaloes. Tropical Animal Health and Production 47, 915–919. https://doi.org/10.1007/s11250-015-0808-5. [DOI] [PubMed] [Google Scholar]
- Sumbria D., Singla L.D., Sharma A., Moudgil A.D. & Bal M.S. (2014) Equine trypanosomosis in central and western Punjab: prevalence, haemato‐biochemical response and associated risk factors. Acta Tropica 138, 44–50. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The National Center for Biotechnology Information . Available at: http://www.ncbi.nlm.nih.gov/ (Accessed October 2015).
- Tuli A., Singla L.D., Sharma A., Bal M.S., Filia G. & Kaur P. (2015) Molecular epidemiology, risk factors and haematochemical alterations induced by Theileria annulata in dairy animals of Punjab (India). Acta Parasitologica. 60, 378–390. [DOI] [PubMed] [Google Scholar]
- Weir W., Karagenç T., Gharbi M., Simuunza M., Aypak S., Aysul N. et al (2011) Population diversity and multiplicity of infection in Theileria annulata . International Journal for Parasitology 41, 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.T. 1977. Salivarian trypanosomes producing disease in livestock outside of sub‐Saharan Africa [Trypanosoma evansi, Trypanosoma equiperdum, Trypanosoma vivax viennei, trypanosomiasis, insect vectors, Tabanidae, Stomoxys]. Parasit Protozoa, 269–296. [Google Scholar]


