Abstract
Aim:
This study aims to ascertain the advantages of Atomic Force Microscopy (AFM) in the morphologic study of microorganisms and their interactions within the subgingival biofilm in patients with gingivitis and periodontitis.
Settings and Design:
Conducted a study on twenty patients, ten patients with severe periodontitis with probing the pocket depth of ≥8 mm, with a clinical attachment loss (CAL) of ≥6 mm CAL and ten patients with gingivitis: ≥5 mm pocket depth, and no attachment loss, was selected for the study.
Materials and Methods:
Bacterial biofilms were collected and slide preparation done. Morphological study was done using AFM. AFM consists of a cantilever-mounted tip, a piezoelectric scanner, a photodetector diode, a laser diode, and a feedback control. The laser beam is reflected from back of the cantilever into the quadrant of the photodetector. AFM works on the principle of interaction between the tip and the sample which causes the cantilever to deflect, thereby changing the position of laser onto the photodetector. Methodology used for studying the bacteria through AFM includes the following: (1) Probe type: Platinum coated silicon nitrate tip. (2) Probe force: 0.11 N/m. (3) Probe geometry: Triangular shaped tip. (4) Probe frequency: 22 KHz. (5) Probe immobilization: Used in Contact mode. AFM Solver Pro-M (NT-MDT) equipped with ETALON probe was used to take images in Nova software.
Results:
The investigation showed various morphological features, such as shape, size, and secretory product-like vesicles of the bacterial species involved in gingivitis and periodontitis. More bacterial surface details were studied by reproducing a three-dimensional reconstruction using AFM.
Conclusions:
The morphological variations of bacteria of different sizes, and shapes, cell wall structures, secretory product-like vesicles flagellated and filamentous microorganisms, polymorphonuclear leukocytes, and bacterial coaggregation analysis were done by AFM. Results of the present study conclude that AFM is a quite a reliable method for studying morphology of bacterial species involving periodontal diseases and is also used to study microbial interactions in biofilm.
Keywords: Atomic Force Microscopy, biofilm, gingivitis, periodontitis
INTRODUCTION
The human oral cavity is assembled with a peculiar microbiological environment which is organized as a solid, nonshedding area abutting with epithelial cells and tissues of the periodontium allowing the expansion and spread of microbial colonies called “Biofilm” that renders natural accumulation of microorganisms on liquid-solid and liquid-air interfaces.[1,2,3,4] Periodontitis is a multifactorial entity caused by bacteria and their pernicious products encompassed in the dental plaque biofilm. Most natural biofilms contain multiple species and are termed “microbial communities.”[5,6] Evidence establishes that the aggregated organisms are not merely passive neighbors but rather are involved in a wide range of physical, metabolic, and molecular interactions.[7]
Culturing techniques have been the primary method of identifying and studying putative periodontal pathogens. However, the method of cultivating microorganisms is hampered by technical problems.[8] According to Staley and Konopka, <1% of the organisms can grow under laboratory conditions, suggesting that the complexity and diversity of microbial communities based on cultivation strategies are severely biased.[9] Fortunately, a number of molecular-based assay has helped overcome this limitation by allowing to obtain clearer picture of microbial communities in terms of their structural complexity and genetic diversity.
The composition of bacterial biofilm consists of polysaccharides, DNA, protein, peptidoglycans, phospholipids, and lipids that play a major role in adhesion of bacteria to tooth surfaces.[10] Most microbial surfaces are structurally very complex emphasizing a well-defined cell envelope, which consists of a plasma membrane, cell wall, often an outer polysaccharide layer and various surface appendages.[11] Over many years, microbiologists and surface scientists used different methods to characterize microbial cell surfaces.[12] However, the evolution of nanoscience and nanotechnology in the 21st century has impacted human life to a great extent.[13,14] One such example was the invention of Atomic Force Microscopy (AFM) by Binning in 1986, an advanced technology for observing the force interaction between the tip and the surface being observed in the AFM.[15,16] The great advantage of AFM is its ability to image the surface topography of individual cells under physiological conditions with nanometric lateral resolution.[15,17] In addition, it also provides information on physical properties of the sample being tested and observed.[13,18,19]
Traditional methods such as cell counting, bacterial labeling, light microscopic analysis, flow chamber or quantifying cell removal from the surfaces, and ultrastructural techniques like electron microscopy have failed to provide satisfactory information on the native surfaces being observed.[20,21,22] In recent years, AFM has risen to be an essential aid by substantiating its high resolution in both air and fluid environment, and microstructural analysis of microbiological agents, by facilitating simple preparation of the sample. The capability of AFM to collect quantitative and qualitative analysis of dentine and collagen at the nanoscale are used by researchers to prove it essential and effective in prevention and management of periodontal diseases and development of collagen-dependent materials, such as bone, cartilage, tendons, skin, collagen-based materials in tissue engineering and biomedical device coating.[23]
Multispecies communities like dental plaque can also produce polymicrobial infections in which microorganisms interact in a synergistic fashion leading to pathogenesis. Periodontal diseases represent one of the best documented polymicrobial infections.[24]
Bacteria have discrete cell body shapes ranging from sphere (cocci) to rods (bacilli) of various curvatures and helices; more so, to peculiar shapes such as stars. Bacteria produce a variety of appendages such as pilli or flagella which reveal diversity in overall shape, length, and width as well as placement with respect to the cell body. Bacteria can change their morphology during their life cycle in response to environmental conditions. Mutants with altered colonization or virulence properties have an altered shape. Hence, cell shape of bacteria itself may be a virulent factor modified by a change in the environment in its vicinity.[25]
Virulence is a pathogen's or microbe's ability to infect or damage host. The description of different possible interactions between biofilm residents implies that the net effect of these interactions may influence localization or colonization of certain organisms within the biofilm structure. A better understanding of bacterial interactions using AFM will elucidate the role of different virulence factors involved in a mixed infection like periodontitis. The surface components of bacterial cells are major determinants of virulence for many pathogens. In general, pathogenic bacteria produce more vesicles than their nonpathogenic counterparts, and vesicles are commonly observed near colonizing bacteria. Bacterial vesicles are implicated in many aspects of periodontal diseases including colonization, inflammation, and disseminates sequel.
Ryoma Nakao et al.[26] in their study by electron microscopy revealed the presence of small vesicles on the outer membrane of Gram-negative bacteria, such as Porphyromonas gingivalis. The outer membrane vesicles of P. gingivalis contain virulence factors, such as lipopolysaccharides and gingipain. The gingipain laden outer membrane vesicles may contribute to tissue destruction in periodontal disease functioning as a vehicle for the antigens and active proteases.
Azari et al.[27] in their study through electron micrograph have revealed rugosa appearance on the bacterial surface of periodontal pathogen, such as Aggregatibacter actinomycetemcomitans and propensity of Gram-negative bacterium to secrete vesicles containing virulence factors such as leukotoxins. Hence, the electron microscopic studies have revealed that the observation of the outer membrane of the Gram-negative bacteria, especially the vesicles dictates the virulence of such bacteria.
Till date, studies to delineate the morphological characteristics of bacteria are scarce; therefore, an attempt is made through this study to describe the morphology of bacterial species in patients with gingivitis and periodontitis using AFM.
MATERIALS AND METHODS
The present study conducted on 20 patients (10 chronic periodontitis cases and 10 gingivitis cases) at the Department of Periodontics, after obtaining an ethical committee clearance from the institution.
Sample selection
Bacterial biofilm samples were collected from ten patients with probing pocket depth of ≥8 mm in at least six teeth in the oral cavity, detected using UNC probe and clinical attachment loss ≥6 mm of diagnosed as chronic generalized severe periodontitis, and from 10 patients with gingivitis with a probing pocket depth ranging from ≥5 mm, diagnosed as chronic generalized gingivitis.[28] Informed consent was obtained from patients participating in the study after explaining the procedure of study. Patients with any systemic pathologic conditions, patients on any steroids or antibiotics or any medications like analgesic drugs over previous 6 months were excluded in the investigations.
Slide preparation
After meticulous removal of the supragingival plaque, a sterile curette was used to collect the plaque biofilm from the gingival sulcus of gingivitis and deep pocket areas of periodontitis patients. Anaerobic protocol was not followed as the samples were collected from the deepest sites of periodontal pockets favoring the presence of anaerobic environment in itself, harboring anaerobic organisms such as spirochetes and filamentous bacteria.
Using a sterile syringe of gauge 10 mm, the plaque samples were dispersed into a 200 μl solution of sterile distilled water. To get turbid solutions without any visible bacterial clumping or aggregation, a gentle movement of suction was obtained. Of the 200 μl of solution containing bacterial aggregate of plaque biofilm, 20 μl was carefully micropipetted and then smeared onto sterile glass slides of 1 cm × 1 cm diameter. It was then allowed to dry and stored at room temperature till the microscopic examination to be performed within 2 days of smear preparation. Two slides from each sample were prepared from the included patients.
AFM solver Pro-M (NT-MDT, Moscow-Russia) in contact mode and equipped with ETALON HA_NC/15 probes were used to take images in Nova software system [Figure 1a and b]. AFM consists of a cantilever-mounted tip, a piezoelectric scanner, a photodetector diode, a laser diode, and a feedback control. The laser beam is reflected from back of the cantilever onto the quadrant of the photodetector. AFM works on the principle of interaction between the tip and the sample which causes the cantilever to deflect, thereby changing the position of the laser onto the photodetector.
Figure 1.

(a and b) Atomic Force Microscopy Solver Pro-M equipped with ETALON HA_NC/15 probes
The methodology used for studying bacteria through AFM includes the following: Triangular-shaped Platinum coated Silicon Nitrate tip, with a Probe force of 0.11N/m with a probe frequency of 22 KHz.[29]
It is important to note that since AFM works on the principle of interaction between the tip and the sample, which causes the cantilever to deflect, thereby changing the position of laser onto the photodetector; it does not require post smear slide preparation, like fixation/staining of the sample as done in other microscopic techniques. There are various protocols to visualize surface morphology and topography of samples, like:
Pretreatment such as air-drying and chemical fixation. However, it causes denaturation of the surface molecules
Immobilizing the samples mechanically in the agar gel. Agar gel is used as a soft, deformable immobilization matrix, which allows visualization of growth process
Immobilizing the cells mechanically in a porous membrane. Although this approach is simple and direct, its limitation is that it essentially works only for spherical cells (i.e., some bacteria, fungal spores, yeasts, etc.) and not for rod-like cells.[21]
RESULTS
Biofilm samples of chronic gingivitis patients
The three-dimensional reconstruction of bacterial species using AFM images was useful in getting more information regarding various morphology of bacteria, such as different size, shape, and morphological features in detail. AFM images showed predominance of coccoids and straight rod bacterial species from the biofilms obtained from gingivitis patients [Figure 2a]. Owing to the slide preparation, coccoidal bacteria appeared flattened [Figure 2b]. A classical image of a divided diplococcus can be seen represented three-dimensionally [Figure 3a and b].
Figure 2.
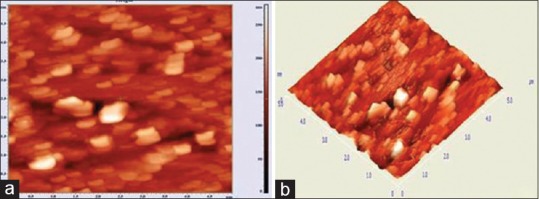
(a) Atomic Force Microscopy image showed predominance of coccoids and straight rods bacterial species from the biofilm obtained from gingivitis patients; (b) Flattened coccoidal bacteria
Figure 3.
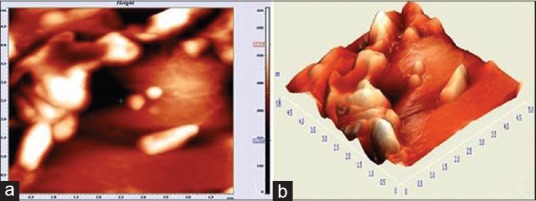
Three-dimensional image of diplococcus
A salient feature observed under AFM is the visualization of bacterial secretory products like vesicles, which could probably have been released from the neighboring bacterial aggregate. A small vesicle measuring about 4 μm in length and 3.2 μm in width was seen. These vesicles were typically found to have an icosahedral morphology. They not only represent the secretory product of the bacteria but also throw light on various virulence factors that could be associated with the bacterium [Figure 4a and b].
Figure 4.
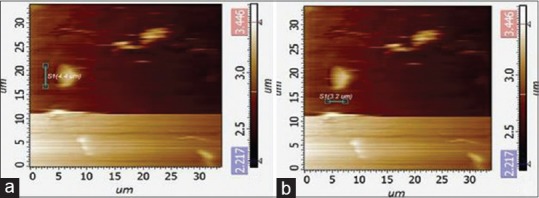
Three-dimensional reconstructions, the icosahedral form of the bacterial vesicles
Biofilm samples of chronic periodontitis patients
Unlike seen in the images obtained from chronic gingivitis patients, the bacterial biofilm images obtained from the chronic periodontitis group showed predominance of spirochetes, flagellated forms and filamentous organisms, and less representation of coccoid and straight rod species. Different features and sizes of the spirochetes were analyzed. Three categories of spirochetes were observed-small, with a length of 10 μm and width of 0.1to 0.2 μm; “intermediate” sized with 12–14 μm in length and 0.2–0.3 μm in width and large spirochetes with 0.5 μm width and up to 15–20 μm in length [Figure 5a and b]. A knob-like structure is present along the length of the spirochete. Three-dimensional image of the spirochete has been described in Figure 5c and d.
Figure 5.
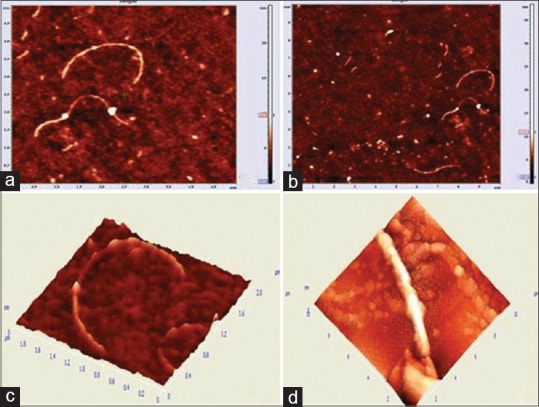
(a and b) Spirochetes were observed; (c) A knob-like structure attached with the spirochete; (d) Three-dimensional image of the spirochete
AFM image showed a round cell compatible with polymorphonuclear leukocytes (PMN) measuring about 15 μm in size and its morphology in detail. The usually found inflammatory cells in the oral biofilm include leukocytes especially PMN and macrophages owing to the inflammatory conditions in gingivitis and periodontitis [Figure 6a and b]. Various bacteria with different morphologies have been found to be present in contact with the leukocytes. Fusiform bacteria attached to the cell membrane represent phagocytized bacterium. A complex biofilm from a periodontitis patient representing filamentous bacteria and spirochetes is found to survive in conglomeration with rod-shaped bacterium and other cells.
Figure 6.
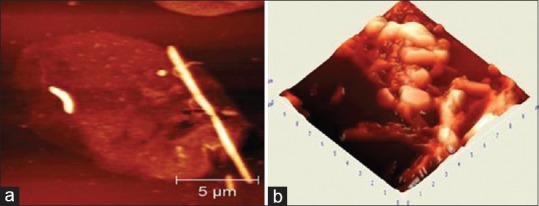
The oral biofilm showing polymorphonuclear leukocytes and macrophages owing to the inflammatory conditions in gingivitis (a); and periodontitis (b)
DISCUSSION
Many indigenous bacteria and microbes residing in various environmental niches in distinct communities at different anatomic sites can be detected in the human body.[30] These microbial communities which are also present in the oral cavity pose an important implication for pathosis. One of the most common inflammatory conditions of the oral cavity is chronic periodontitis which is associated with microbial colonies that are different from those related to health.[31] The importance of understanding and identifying microbial structural features goes a long way as it helps the clinician identify the concerned bacteria with specific pathological entity. This way it helps the patient in a prognostic approach, based on which further cultural and genomic studies may be considered to confirm the identity of bacteria. The oral cavity lodges more than 600 microbial species, out of which about 100 species contribute to the saprophytic microbiota of an individual. Although different sites of oral cavity are colonized by various microbial species, they can significantly vary from site to site. Oral cavity and healthy intestinal tract have been recorded with individual diversities of bacterium in various studies. In the oral cavity, through a process of sequential and specific adhesive interactions, a favorable community for bacterial survival and growth is created in the supragingival and subgingival plaque.
The aim of the present study is to three dimensionally view and reconstruct the bacterial morphology including bacterial cell wall and vesicles with the help of the novel microscopic technique, AFM. As one of the landmark discoveries in 1986 by Binning, AFM opened a new spectrum of opportunities for examining microbial cell surface, behavior of bacteriophage and also molecular interactions of the surface of bacterial cell.[32] The property of three-dimensional acquisition of data, detection of adherent mechanical properties at molecular level, the feasibility of understanding and studying surface characteristics of the bacterium in different environment without pretreatment protocol such as metal coating, staining, fixing or microtoming (that could alter the slide properties) were added advantages of AFM over existing microscopic techniques such as Scanning Electron Microscopy. These advantages have increased the biological applications of AFM in ascertaining host-pathogen association.[33]
Knowledge of research on periodontal bacterial species could also be increased during this technique of AFM. Periodontitis is a multifactorial disease involving the hard and soft tissue, preceded by colonization of bacterial species and elicited by inflammatory and immune responses. The effectiveness of protective functioning of the host tissue has been altered due to the presence of local tissue elements and the bacterial breakdown products, produced in the host response mechanisms.[34] Amro et al.[35] in their study have described the structural basis for permeability of the outer membrane of Escherichia coli using high-resolution AFM revealing patches of lipopolysaccharides.
The purpose of the present study was to disclose the advantages of AFM in the study of bacterial morphology and their interaction within the biofilm in patients with gingivitis and periodontitis. This study has revealed different morphological features of bacteria such as wall structures, different size and shapes, secretory products of bacteria like vesicles evaluated using AFM, in accordance with the study by Germano et al.[6] Analysis of the nanoscopic analysis of the bacterial morphology attained in this preliminary study was also seen conforming to the study by Azari et al., which emphasized on the ultrastructural analysis of the rugose cell envelope of a member of the pasteurellaceae family. On three-dimensional reconstruction, icosahedral form of bacterial vesicles was observed at high magnification [Figure 4a and b], suggesting compatibility with the head of the bacteriophage structure. Kazuya Iwai et al. in their study on examination of salivary microvesicles under AFM have shown that the size salivary microvesicle ranges from (47.89 ± 12.3 nm). Since the images of observed vesicles under AFM were seen ranging much higher diameter, and since they were present around clumps of bacterial co-aggregation, it was presumed that these could be of bacterial and not of salivary or GCF origin. Further, more extensive studies such as density-based fractionation and centrifugation, western blot and reverse polymerization chain techniques have to be taken up to exactly trace the origin of these extracellular vesicles.[36]
This technique facilitates the possibility of visualization of nanoparticles and bimolecular and submolecular structures at higher resolution and also facilitates the measurement of forces between the probe tip and the sample examined, within a spatial resolution of <1 nm. The local physical, chemical, and mechanical properties of the bacterial surfaces such as adhesion, viscosity, charge density, and elasticity could also be measured using AFM.[37] A few limitations of AFM when compared to other scanning microscopy techniques are its single scan image size of 150 μm × 150 μm. Its relative slow scan time can affect the accuracy of the sample by causing thermal drift.[38]
The tremendous potential of using AFM in molecular analysis is yet to be explored in depth and the adjunctive use of AFM with various other characterization techniques such as confocal microscopy and Raman spectroscopy can interpret the dynamic cellular processes.[39] This study has opened a gate in better understanding the bacterial features by the three-dimensional investigations. However, in future, more studies are required to correlate morphological features of biofilm to severity of periodontal diseases and thus help in addressing patient-directed treatment protocol.
CONCLUSIONS
The present study concludes that AFM is a more reliable method to identify species by studying the morphology of different bacteria with three-dimensional reconstructive images, which is also used to study various microbial interactions in biofilm. Further studies should be conducted to delineate the role of AFM in the observation of the bacterial interaction as well as host-microbial interaction and their role in the pathogenicity.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Socransky SS, Haffajee AD. Evidence of bacterial etiology: A historical perspective. Periodontol 2000. 1994;5:7–25. doi: 10.1111/j.1600-0757.1994.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 2.Costerton JW, Geesey GG, Cheng KJ. How bacteria stick. Sci Am. 1978;238:86–95. doi: 10.1038/scientificamerican0178-86. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW, Lewandowski Z. The biofilm lifestyle. Adv Dent Res. 1997;11:192–5. [Google Scholar]
- 4.Harper-Owen R, Dymock D, Booth V, Weightman AJ, Wade WG. Detection of unculturable bacteria in periodontal health and disease by PCR. J Clin Microbiol. 1999;37:1469–73. doi: 10.1128/jcm.37.5.1469-1473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–20. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 6.Germano F, Bramanti E, Arcuri C, Cecchetti F, Cicciù M. Atomic force microscopy of bacteria from periodontal subgingival biofilm: Preliminary study results. Eur J Dent. 2013;7:152–8. doi: 10.4103/1305-7456.110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsh PD. Dental plaque: Biological significance of a biofilm and community life-style. J Clin Periodontol. 2005;32(Suppl 6):7–15. doi: 10.1111/j.1600-051X.2005.00790.x. [DOI] [PubMed] [Google Scholar]
- 8.Marija IK, Nataπa B, Greta SS. Diagnostic methods for evaluation of microbial flora in periodontitis. Acta Stomatol Croat. 2001;35:137–40. [Google Scholar]
- 9.Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–46. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 10.Hu Y, Jens U, Zhang J. Kgs. Lyngby: DTU Chemistry; 2012. Bacterial Biofilms Investigated by Atomic Force Microscopy and Electrochemistry. [Google Scholar]
- 11.Klis FM, Boorsma A, De Groot PW. Cell wall construction in saccharomyces cerevisiae. Yeast. 2006;23:185–202. doi: 10.1002/yea.1349. [DOI] [PubMed] [Google Scholar]
- 12.Dufrêne YF. Atomic force microscopy in microbiology: New structural and functional insights into the microbial cell surface. MBio. 2014;5:e01363–14. doi: 10.1128/mBio.01363-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dufrêne YF. Application of atomic force microscopy to microbial surfaces: From reconstituted cell surface layers to living cells. Micron. 2001;32:153–65. doi: 10.1016/s0968-4328(99)00106-7. [DOI] [PubMed] [Google Scholar]
- 14.Dufrêne YF. Direct characterization of the physicochemical properties of fungal spores using functionalized AFM probes. Biophys J. 2000;78:3286–91. doi: 10.1016/S0006-3495(00)76864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binnig G, Quate CF, Gerber C. Atomic force microscopy. Phys Rev Lett. 1986;56:930–3. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 16.Dorobantu LS, Gray MR. Application of atomic force microscopy in bacterial research. Scanning. 2010;32:74–96. doi: 10.1002/sca.20177. [DOI] [PubMed] [Google Scholar]
- 17.Morris VJ, Kirby AR, Gunning AP. Atomic Force Microscopy for Biologists. 1st ed. London, UK: Imperial College Press; 1999. [Google Scholar]
- 18.Scheuring S, Dufrêne YF. Atomic force microscopy: Probing the spatial organization, interactions and elasticity of microbial cell envelopes at molecular resolution. Mol Microbiol. 2010;75:1327–36. doi: 10.1111/j.1365-2958.2010.07064.x. [DOI] [PubMed] [Google Scholar]
- 19.Dorobantu LS, Goss GG, Burrell RE. Atomic force microscopy: A nanoscopic view of microbial cell surfaces. Micron. 2012;43:1312–22. doi: 10.1016/j.micron.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Weisenhorn AL, Drake B, Prater CB, Gould SA, Hansma PK, Ohnesorge F, et al. Immobilized proteins in buffer imaged at molecular resolution by atomic force microscopy. Biophys J. 1990;58:1251–8. doi: 10.1016/S0006-3495(90)82465-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaboriaud F, Dufrêne YF. Atomic force microscopy of microbial cells: Application to nanomechanical properties, surface forces and molecular recognition forces. Colloids Surf B Biointerfaces. 2007;54:10–9. doi: 10.1016/j.colsurfb.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 22.Beveridge TJ, Graham LL. Surface layers of bacteria. Microbiol Rev. 1991;55:684–705. doi: 10.1128/mr.55.4.684-705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 24.Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007;71:653–70. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang DC, Blair KM, Salama NR. Staying in shape: The impact of cell shape on bacterial survival in diverse environments. Microbiol Mol Biol Rev. 2016;80:187–203. doi: 10.1128/MMBR.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho MH, Chen CH, Goodwin JS, Wang BY, Xie H. Functional advantages of Porphyromonas gingivalis vesicles. PLoS One. 2015;10:e0123448. doi: 10.1371/journal.pone.0123448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azari F, Nyland L, Yu C, Radermacher M, Mintz KP, Ruiz T, et al. Ultrastructural analysis of the rugose cell envelope of a member of the pasteurellaceae family. J Bacteriol. 2013;195:1680–8. doi: 10.1128/JB.02149-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 29.NT-MDT Brochure on Atomic Force Microscopy. [Last accessed on 2017 Sep 10]. Available from: http://www.ntmdt-tips.com .
- 30.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–8. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–74. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradley DE. Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev. 1967;31:230–314. doi: 10.1128/br.31.4.230-314.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubrovin EV, Voloshin AG, Kraevsky SV, Ignatyuk TE, Abramchuk SS, Yaminsky IV, et al. Atomic force microscopy investigation of phage infection of bacteria. Langmuir. 2008;24:13068–74. doi: 10.1021/la8022612. [DOI] [PubMed] [Google Scholar]
- 34.Dunne WM., Jr Bacterial adhesion: Seen any good biofilms lately? Clin Microbiol Rev. 2002;15:155–66. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amro NA, Kotra LP, Wadu-Mesthrige K, Bulychev A, Mobashery S, Liu G. High-Resolution Atomic Force Microscopy Studies of the Escherichia coli Outer Membrane: Structural Basis for Permeability. Langmuir. 2000;16:2789–96. [Google Scholar]
- 36.Iwai K, Minamisawa T, Suga K, Yajima Y, Shiba K. Isolation of human salivary extracellular vesicles by iodixanol density gradient ultracentrifugation and their characterizations. J Extracell Vesicles. 2016;5:30829. doi: 10.3402/jev.v5.30829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansma HG, Pietrasanta L. Atomic force microscopy and other scanning probe microscopies. Curr Opin Chem Biol. 1998;2:579–84. doi: 10.1016/s1367-5931(98)80086-0. [DOI] [PubMed] [Google Scholar]
- 38.Dufrêne YF. Atomic force microscopy, a powerful tool in microbiology. J Bacteriol. 2002;184:5205–13. doi: 10.1128/JB.184.19.5205-5213.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright CJ, Shah MK, Powell LC, Armstrong I. Application of AFM from microbial cell to biofilm. Scanning. 2010;32:134–49. doi: 10.1002/sca.20193. [DOI] [PubMed] [Google Scholar]


