Abstract
A series of dimeric isoxazolyl-1,4-dihydropyridines (IDHPs) were prepared by click chemistry and examined for their ability to bind the multi-drug resistance transporter (MDR-1), a member of the ATP-binding cassette superfamily (ABC). Eight compounds in the present study exhibited single digit micromolar binding to this efflux transporter. One monomeric IDHP m-Br-1c, possessed submicromolar binding of 510 nM at MDR-1. Three of the dimeric IDHPs possessed <1.5 μM activity, and 4b and 4c were observed to have superior binding selectivity compared to their corresponding monomers verses the voltage gated calcium channel (VGCC). The dimer with the best combination of activity and selectivity for MDR-1 was analog 4c containing an m-Br phenyl moiety in the 3-position of the isoxazole, and a tether with five ethyleneoxy units, referred to herein as Isoxaquidar. Two important controls, mono-triazole 5 and pyridine 6, also were examined, indicating that the triazole – incorporated as part of the click assembly as a spacer – contributes to MDR-1 binding. Compounds were also assayed at the allosteric site of the mGluR5 receptor, as a GPCR 7TM control, indicating that the p-Br IDHPs 4d, 4e and 4f with tethers of from n = 2 to 5 ethylenedioxy units, had sub-micromolar affinities with 4d being the most efficacious at 193 nM at mGluR5. The results are interpreted using a docking study using a human ABC as our current working hypothesis, and suggest that the distinct SARs emerging for these three divergent classes of biomolecular targets may be tunable, and amenable to the development of further selectivity.
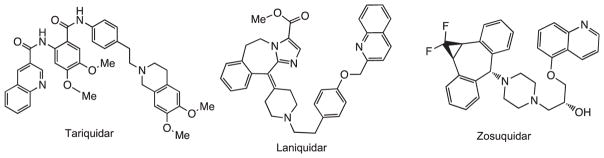
Molecules which reverse the effect of the multidrug resistance transporter (MDR-1, also known as P-glycoprotein or P-gp1–5), exemplified by Tariquidar,6 Laniquidar7 and Zosuquidar8(Fig. 1) are the subject of clinical trials as adjuvants to cancer chemotherapy. Dihydropyridines (DHPs) have been under intense study for many years for their ability to inhibit the multidrug transporter.9,10 More recently we have reported that 4-isoxazolyl-1,4-dihydropyridines (IDHPs) have significant binding at this target,11–13 and a unique structure activity relationship (SAR) which diverges from their voltage gated calcium channel (VGCC) activity. In our initial study, we found that branching at the C(5) of the isoxazole gave rise to three of the four most efficacious IDHPs at MDR-1.11 In a follow-up study, attachment of a fluorophore also enhanced activity. This suggested additional binding opportunities in proximity to the putative nucleotide binding domain (NBD) of IDHPs (vide infra).12,13
Fig. 1.
Selective MDR-1 inhibitors Tariquidar (XR9576), Laniquidar, and Zosuquidar have been investigated, alone and in combination with other chemotherapeutic agents, in clinical trials.
There have been previous indications that dimeric ligands enhance MDR-1 binding. The groups of Andrus14 and Chmielewski and Hrycyna15–17 established that there was a relationship between tether type, length and binding affinity. In contrast, during his study of dimeric tethered dihydropyridines at the VGCC, Triggle found that monomeric analogs lacking a second 1,4-DHP nucleus or possessing a second inactive pyridine were no more active,18 leading to the conclusion that such dimers likely do not bridge adjacent DHP receptor motifs at the VGCC. We reasoned we could use these observations, together with other known VGCC SAR, to design greater selectivity for MDR-1 into the IDHPs.
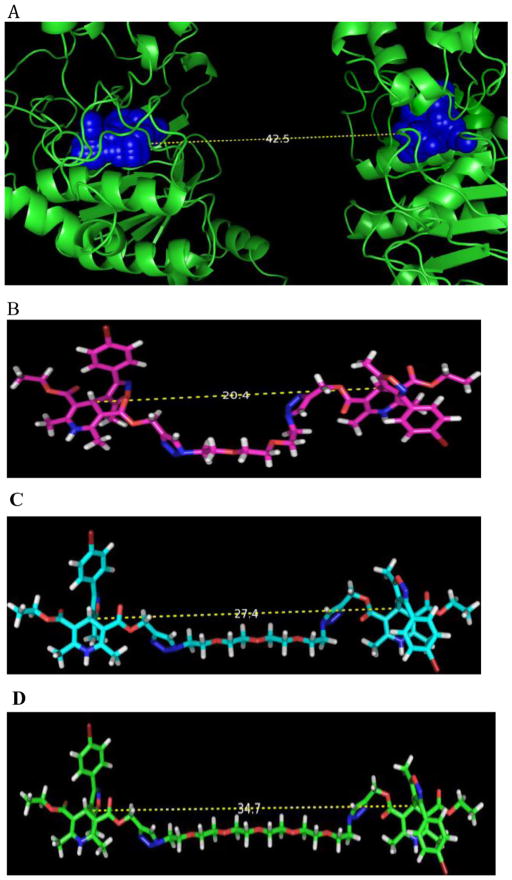
Our hypothesis is that a bivalent inhibitor could bind more tightly to MDR1 and thus be a more effective inhibitor than the corresponding monomer. Taking into account previous research the tether’s length is critical, however, the MDR undergoes significant conformational reorganization during the ATP driven efflux. In the apo state used in our previous homology model12,13 the NBDs reside over 40 Å apart using the mouse P-gp data (pdb accession number 3G5U) (Fig. 2, below), while in the ATP bound state exemplified by Sav1866 (pdb accession number 2ONJ), that distance is reported as 4 Å. In our previous study of IDHPs tethered to fluorophores,12 we used hydrocarbon tethers, and we were concerned that such groups could suffer hydrophobic collapse during interaction with the target, given that such folded conformers were evidenced in the NMR. We reasoned that oligo ethyleneoxy linkers (short PEGs), would be more likely to adopt an extended conformation able to intercept the ABC at a stage intermediate to the apo and outward facing ATP bound conformation. We then sought to synthesize such small molecules to test our hypothesis.
Fig. 2.
The SYBYL calculated distance between the Q-site predicted binding sites residing proximal to the nucleotide binding domains in the non-ATP bound state (apo) of MDR-1, based on the Mus musculus P-gp coordinates.12,13 B, C, D, calculated distances for n = 2, 3 and 5, tethered dimers, respectively, in energy minimized conformations.
1. Chemistry
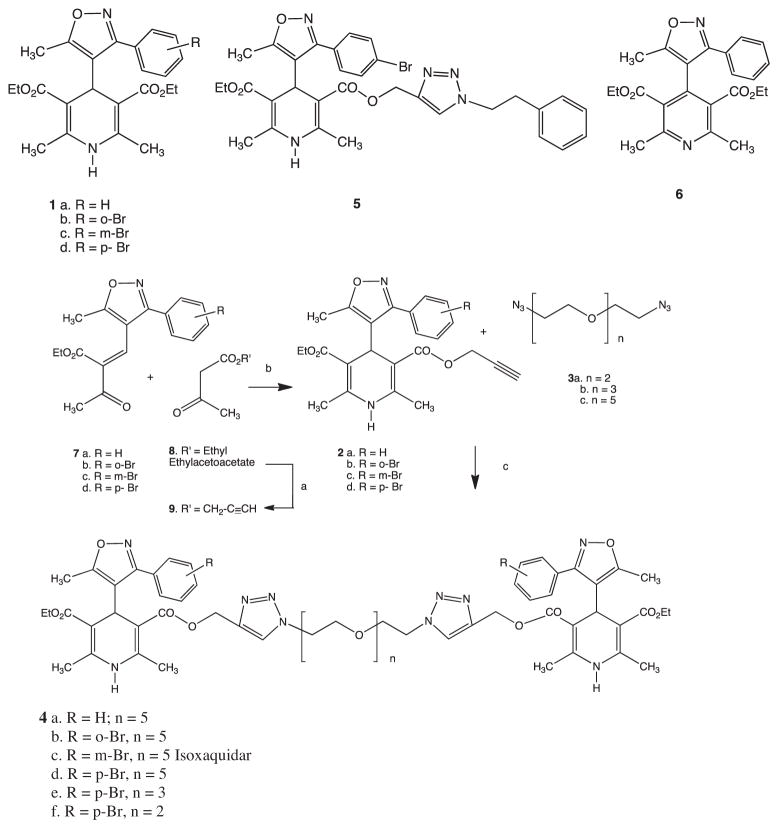
The monomeric IDHP 1a was prepared as previously described,19,20 The bromo IDHPs 1b–1d were prepared in analogous fashion from the corresponding commercial aryl aldehydes, briefly, by oxime formation, followed by oxidation to the oximidoyl chloride, nitrile oxide cycloaddition to the isoxazole ester, lithium aluminum hydride reduction to the 4-carbinol (carefully temperature controlled to avoid aryl debromination), chromium (VI) oxidation to the 4-carbaldehyde, and completed by Hantzsch pyridine synthesis.21 The full experimental details and characterization for all intermediates are detailed in the Supplementry Dt. The dimers were prepared by click chemistry22,23 by 1,3-dipolar cycloaddition of alkynyl-IDHPs 2 to bis-azide PEGs 3, to produce the Dimeric IDHPs 4 (Scheme 1). We observed that mono-incorporation of the alkyne moiety onto IDHPs was best accomplished by a modified two-component Hantzsch, adding the Knoevenagel adducts 7, to alkynyl keto esters 9 to produce the click ready IDHP-alkynes 2. The alkyne was incorporated onto ethylacetoacetate 8, using the excellent method of Anand using boric acid trans-esterification to produce 9.24
Scheme 1.
Synthesis of dimeric-IDHPs (4a–f) and relevant controls. a. B(OH)3, prop-2-yne-1-ol, toluene reflux. b. Aqueous Ammonia, EtOH, reflux. c. Cu(I)Br, 10 mol%.
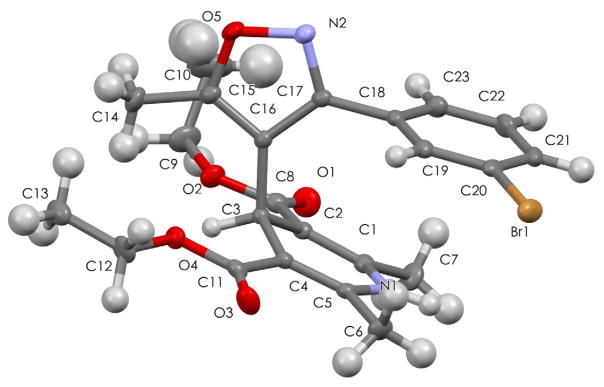
2. Single crystal X-ray diffractometry
The conformations of DHPs are important to their structure activity relationships, and IDHPs have particularly pronounced divergent topologies dependent on rotation about the bond joining the heterocyclic rings.13 In the VGCC arena, several parameters have been found to correlate with biological activity: the conformation of the esters at C-3 and C-5 of the DHP, the sum of the six intra-ring torsion angle in the flattened boat conformation, and the forward presentation of the C-4 aryl group of the DHP.25 We had earlier postulated that the heterocyclic ring juncture could well adapt divergent conformations dependent on the biomolecular target. We have found single crystal X-ray diffractometry (Sc-Xrd) useful to help inform our computational studies on rotational barriers and drug-receptor modeling, and Sc-Xrd of 1c was performed as previously described (Table 1).26 In the event, IDHP monomer 1c shows that both esters adopt the syn-conformation in the solid state, the C-3m-Br phenyl ring is oriented above the DHP ring – so that the O of the isoxazole is exo-. The sum of the six intra-ring torsion angles σ |τ | is 66.33° and thus 1c adopts a very shallow boat conformer. The forward presentation of the 4-isoxazolyl ring is reflected by the average magnitude of the C(2)-C(3)-C(4)-C(7) and C(6)-C(5)-C(4)-C(7) torsion angles, designated σ |ρ | 106.25°. These two variables are not independent, and the biological activity at the VGCC has been found to correlate with both parameters.
Table 2.
Summary of biological evaluation of the Dimer-IDHPs (4) and relevant controls.
| Structure | R | n | PDSP # | VGCC (IC50, nM) | mGluR5 (nM) | MDR-1 (% vs. Cyclosporin)b | MDR-1 (IC50, nM) |
|---|---|---|---|---|---|---|---|
| 1a | H | 0 | 21618 | 13.9a | ND | 48.9 | ND |
| 1b | o-Br | 0 | 27215 | 909 | 1524 | − 12 | >10,000 |
| 1c | m-Br | 0 | 27216 | 208 | >10,000 | 101.9 | 510 |
| 1d | p-Br | 0 | 27217 | 36 | >10,000 | 58.4 | 3096 |
| 4a | H | 5 | 27218 | >10,000 | >10,000 | 20.4 | >10,000 |
| 4b | o-Br | 5 | 27219 | >10,000 | 4858 | 17.7 | 2862 |
| 4c | m-Br | 5 | 27220 | 3766 | 8638 | 150.8 | 1296 |
| 4d | p-Br | 5 | 27221 | 2706 | 193 | 172.6 | 1394 |
| 4e | p-Br | 3 | 27222 | 516 | 546 | 11.6 | 3231 |
| 4f | p-Br | 2 | 27223 | 603 | 933 | 41.9 | 1449 |
| 5 | p-Br | 0 | 27224 | 127 | 1972 | 107.6 | 1643 |
| 6 | H | 0 | 27225 | >10,000 | >10,000 | 36.9 | >10,000 |
3. Important intermolecular interactions
There is a plausible Halogen bond in the Sc-Xrd, as defined by the Br1 to O3′ distance of 3.363 Å and halogen to nucleophile angle of 143.1°, which are within the tolerances for a moderate strength halogen bond according to the criteria suggested by Auffinger,27 and Wilcken.28
Tables containing the Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2), Atomic displacement parameters (Å2), and Geometric parameters (Å, °) are given in the Supplementry Dt.
4. MDR-1 Assay
Screening of IDHPs was performed by the Psychoactive Drug Screening Program (PDSP) of NIMH. The PDSP protocol utilizes live Caco-2 cells, which are derived from human colonic epithelium cells which express MDR-1.29 The assay is based on the passive diffusion of calcein acetoxymethyl ester (Calcein-AM), which is hydrolyzed inside the cell to calcein, which is both fluorescent and negatively charged, and therefore trapped inside the cell. MDR-1 can transport non-fluorescent Calcein-AM from cells, but not the hydrolysis product. The assay measures the increase in calcein fluorescence as a function of time using a FlexStation II fluorimeter (Molecular Devices) in 384 well plates in which cells were preincubated with IDHPs (30 μM–3 pM) for 30 min, upon which time calcein-AM was added to a final concentration of 150 nM. Fluorescence is monitored over 4 min, and each assay was performed in triplicate, with a cyclosporin control. The value from untreated cells is 0% and the slope of the fluorescence is normalized taking the value for cyclosporin as 100%.30 The results are shown in Table 2.
Table 1.
Crystal data, data collection and refinement parameters for IDHP monomer 1c.
| Crystal data | |
| C23H25BrN2O5 | |
| Mr = 489.36 | Dx = 1.459 Mg m− 3 |
| Monoclinic, C2/c | Melting point: ? K |
| a = 17.6211 (9) Å | radiation, λ = 0.71073 Å |
| b = 15.3041 (8) Å | Cell parameters from 323 reflections |
| c = 17.9370 (9) Å | θ = 3.4–28.4° |
| β = 112.8770 (6)° | μ = 1.88 mm− 1 |
| V = 4456.7 (4) Å3 | T = 100 K |
| Z = 8 | Prism, clear pale yellow |
| F(000) = 2016 | 0.52 × 0.51 × 0.24 mm |
| Data collection | |
| Bruker SMART BREEZE CCD diffractometer | 4567 independent reflections |
| Radiation source: Mo Kα | 4179 reflections with I > 2σ (I) Rint = 0.021 |
| Detector resolution: 8.3333 pixels mm− 1 | θ max = 26.4°, θ min = 1.8° |
| Absorption correction: numerical SADABS V2012/1 (Bruker AXS Inc.) | h = − 2221 |
| Tmin = 0.41, Tmax = 0.66 | k = − 1919 |
| 24,782 measured reflections | l = − 2222 |
| Refinement | |
| Refinement on F2 | |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ (F2)] = 0.035 | H-atom parameters constrained |
| wR(F2) = 0.098 | where |
| S = 1.07 | (Δ /σ )max = 0.002 |
| 4567 reflections | Δ ρ max = 1.46 e Å− 3 |
| 285 parameters | Δ ρ min = − 0.45 e Å− 3 |
| 80 restraints | Extinction correction: none |
5. Results and discussion
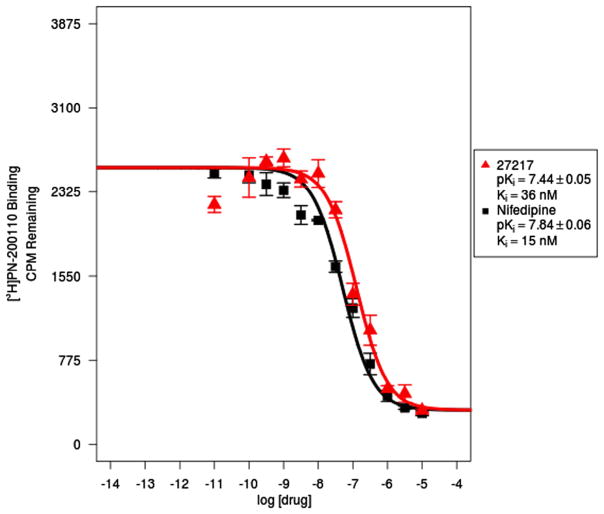
As expected the new monomeric IDHPs 1b, 1c and 1d exhibit robust calcium antagonist activity, with the relationship of activity to halogen substitution at the 3-aryl-isoxazole position p- > m- > o-as previously observed for the chloro-phenyl series,21 however, the range is more pronounced for the Br series, with the p-Br 1d with an IC50 of 36 nM (Fig. 4), over 25 times more potent than the o-Br 1b. However, to place this in perspective, the bromo-substituted phenyl IDHPs monomers including click control 5, were all inferior at the VGCC to the unsubstituted 3-isoxazole phenyl lead 1a (IC50 13.9 nM),31 and previously reported branched chiral32 and lipophilic IDHPs,33 which were in the single digit nanomolar range.
Fig. 4.
Binding isotherms of nifedipine and IDHP monomer PDSP 27217 (1d) at the L-type Voltage gated calcium Channel (VGCC).
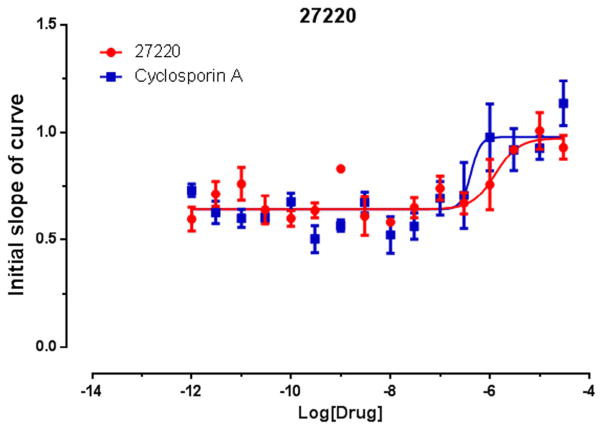
The effect of dimerization on both binding and selectivity is dramatic. For the monomeric 1b compared to dimer 4b, MDR-1 activity appears to be improved, where in contrast VGCC is essentially abolished for the dimer. The relationship for substitution at the isoxazole 3-aryl is maintained in both monomers and dimers, with m-Br > p-Br > o-Br > H, however, the m-Br 4c (Fig. 5) and p-Br 4d dimers have significantly more binding potency at MDR-1.
Fig. 5.
IC50 of dimeric IDHP PDSP 27220 (4c). verses cyclosporine A at the multidrug resistance transporter (MDR-1).
The isoxazolyl-pyridine 6 was envisioned as a control, as usually oxidation of DHPs abolished VGCC activity.13 This was found to be the case, as 6 was essentially bereft of activity at both the VGCC and mGluR5, however a small amount of binding at the MDR-1 was found in the one dose transport inhibition assay (shown in the Supplementry Dt).
G-Protein Coupled receptors are a significant class of receptors, with a large representation among drugs in general medical practice. 34 Furthermore, 7TM receptors have been reported which bind DHPs.35 Therefore, we deemed that screening at a representative 7TM receptor, exemplified by the allosteric site of mGluR5, represented an important control. To our surprise, while the monomeric IDHPs showed only modest binding at best, three dimeric IDHPs exhibited sub-micromolar binding, most notably IDHP 4d at 193 nM, with selectivity for mGluR5 over the VGCC and MDR-1. The putative 7TM allosteric binding site spans a large “big dipper” shaped area,36 and could plausibly accommodate both IDHPs, if the PEG tether oriented outside of the lipophilic cleft in the extracellular space. We are in the process of modeling 7TMs with IDHP analogs, and further experimental and computational studies will be reported at a later date.
5.1. Docking and a new working hypothesis
Our previous ABCB1 homology model12,13 was based on the mouse P-gp structure,37 which has a significantly different topology in its apo conformation than the recent human structure reported by Carpenter’s group (vide infra).38 Our calculation of the NBD distance in our previous human homology model based on the M. musculus P-gp template was 42.5 Å, and the C. elegans distance is 41 Å,2,38 which means that for our dimeric structures to be effective they would have to intercept the efflux pump in a conformation intermediate at some point between apo and Sav1866 nucleotide bound open-to-outside conformers. Calculations with SYBYL 8 and the Molecular Mechanics forcefield using our mouse homology model gave highest scoring solutions that were unsatisfying for the IDHP dimers, wherein the PEG tether collapsed inside the ATP binding site, and both IDHP moieties were bound to the edge of the same NBD (Supplementry Dt).
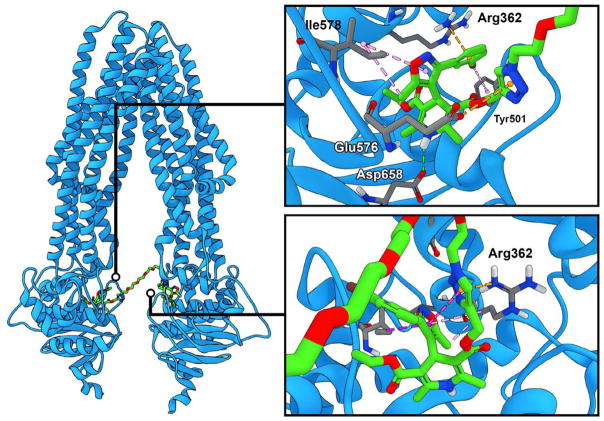
In contrast, for the human ABC structure the distance observed between NBDs in the apo state is 17 Å,38 and the tethers in our synthetic dimers are sufficiently long to bridge the NBD to NBD interface in the human apo state, therefore we examined docking to Carpenter’s ABC coordinates, PDB accession number 3ZDQ. The protein structure was typed with the CHARMM forcefield39 and energy minimized with the smart minimizer protocol within Discovery Studio40 using the Generalized-Born with simple switching implicit solvent model to a root mean square gradient (RMS) convergence <0.001 kcal/mol prior to use in the docking studies. Docking was performed using the flexible docking protocol,41 which allows for flexibility in both the ligand and the binding site residues. The final ranking of the docked poses was performed via consensus scoring, combining the predicted binding energy with the Jain,42 PLP2,43 and Ludi344 scoring functions. In general, the poses were much more reasonable using basic chemical intuition than our previous Sybyl results. We initially examined the docking of a DHP with a reactive azide incorporated that has been used for photo affinity labeling,13 which previously had experimentally established that DHPs bind near the NBD. The best scored docking poses place the azide moiety of azidopine proximal to the NBD, which we found encouraging. We then examined the docking of the synthesized IDHP dimers, and almost all of the dimer poses bridged the NBDs (Fig. 3). The highest scoring pose was obtained for the RS-diastereomer of Isoxaquidar 4c using the -CDocker scoring, and RR-4c had the highest calculated binding energy. In the highest scored poses all of the salient functional groups of both DHP moieties of the dimer are brought into interactions with critical moving parts of the ABC. The IDHP interacts with critical residues in the ATP binding region, and channel helix 2, also referred to as the sliding helix. In the interaction with the left NBD (as illustrated in Fig. 6 for the RS-4c), the isoxazole interacts with Ile578 by lipophilic interaction with the C(5) methyl group, and a π-alkyl interaction with the isoxazole ring, and the C(3) phenyl has a π-cation interaction with Arg362, which also bridges through a hydrogen bond with the C-3 ester carbonyl, and the m-Br interacts with Tyr 501. Discovery Studio assigns this as a lipophilic interaction, although the geometry suggests the potential for a halogen bonding interaction (op cit). This array of multiple interactions would prevent the sliding helix from performing its critical conformational change which is necessary for the efflux of xenobiotics.
Fig. 3.
ORTEP of monomeric IDHP 1c, the unit cell reveals a plausible halogen bond (vide infra).
Fig. 6.
Left, Docking of RS-IDHP dimer 4c Isoxaquidar (carbons green, nitrogens blue, and oxygens red) at the human ABCB10 transporter (blue). The dimer 4c spans the interface between the NBDs. Top right. Close up view of dimer 4c interacting with the left NBD, The isoxazole moiety locks Ile578 by interacting with the C5 methyl and the isoxazole ring, Arg362 with the 3-Ph, while the m-Bromo binds the Tyr501. Bottom right. Originally incorporated as a spacer, the triazole appears to contribute interactions with Arg362.
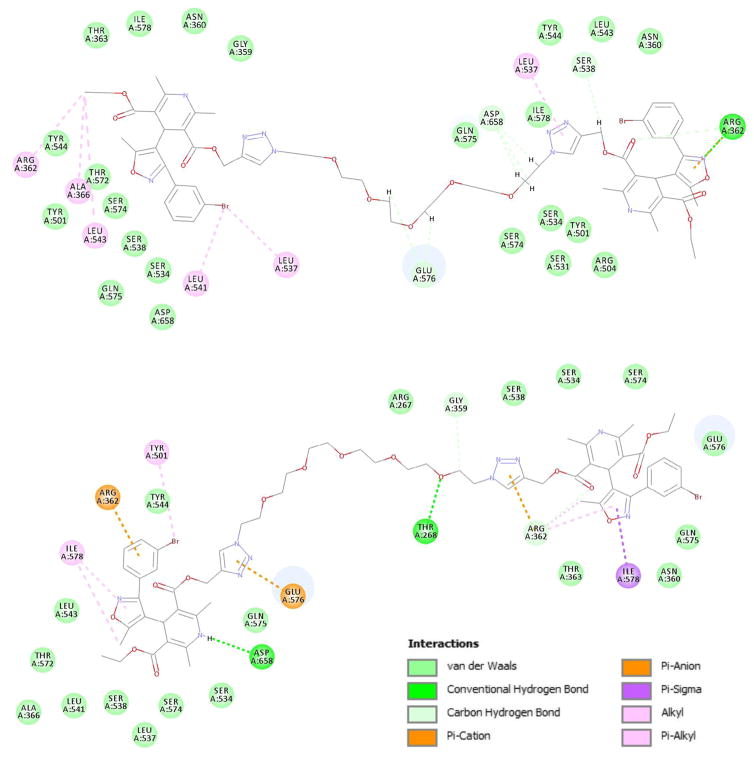
Similar significant interactions are observed for RR-4c, namely DHP N-H hydrogen bonding with Asp658, and surprisingly since the click incorporated triazole was envisioned as a spacer, there is a predicted π-alkyl interaction with Leu537, and an interaction of the polyethylene linker with Glu576. The full complement of interactions are tabulated schematically in the LigPlot diagram below (Fig. 7). More detailed discussion of the binding interactions are illustrated in the Supplementry Dt.
Fig. 7.
Top. Ligplot schematic representation of RR-IDHP dimer 4c interactions at the MDR-1. Bottom Ligplot of RS-IDHP dimer 4c.
5.2. Limitations of the computational model
Chang has noted that a flexible linker region connecting the two halves of the MDR efflux pump is disordered and does not diffract, perhaps owing to its conformational mobility.44 Interactions with this “missing linker” between the NBDs could provide an important avenue for improving efficacy and selectivity of inhibitors.
Furthermore, Discovery Studio assigns the stereochemistry to the IDHP dimers, and the highest calculated -CDocker Energy was predicted for RR-4c, while the highest binding energy was predicted for specifically the RS-diastereomer of 4c., as well as the highest weighted average for both scoring algorithms, assuming a statistical distribution of RR-/RS-/SS of 1:2:1. In our hands, no separation of the diastereomers was observed during column or thin layer chromatography or HPLC–MS, likely owing to the distance and conformational flexibility of the tether between the DHP moieties of the dimers. Our recently reported organocatalytic asymmetric synthesis of chiral IDHPs45 could plausibly allow us to prepare optically pure dimers, and this will be the subject of future efforts.
6. Conclusion
The 4-isoxazolyl-dihydropyridines (IDHPs) have complex polypharmacology analogous to their 4-aryl analogs, which is often observed for such so-called privileged scaffolds. Several IDHP dimers described here have single digit micromolar binding to MDR-1, which is in the approximate range for cyclosporin (800–900 nM), with dimer 4c (Isoxaquidar) exhibiting selectivity for MDR-1 over both the VGCC and mGluR5. As a control for 7TM binding, it was observed that the p-Br dimer 4d exhibited 193 nM binding, with selectivity for mGluR5 over the VGCC and MDR-1, and further studies on 7TM binding will be the subject of future reports from our laboratories. The position of the bromine substitution exerts a profound effect on binding affinity, as well as selectivity, the question remains whether the critical contribution to selectivity results mainly from lipophilic character or relates directly to halogen bonding. However, the observation reported here that IDHPs exhibit selectivity between these three classes of biomolecular targets suggest further SAR development could produce improved selectivity. Such studies are part of an ongoing effort in our group, and our progress will be reported in due course.
7. Experimental
All reactions sensitive to air or moisture were run under an inert atmosphere of dry argon. Chemicals were obtained from commercial sources and purified as needed. Purification of all new compounds was performed using flash chromatography and/or preparative TLC. Melting points were determined in open capillary tubes using a Melt-Temp instrument and are uncorrected. NMR spectra were obtained using either a Bruker Avance III 400 MHz or a Varian NMR systems 500 MHz spectrometer. NMR samples were run in and referenced to CDCl3, unless stated otherwise. High-resolution mass spectrometry (HRMS) data were obtained using a Waters/Micromass ESI/LCT-TOF instrument. Single crystal X-ray diffractometry was performed on a Bruker SMART BREEZE CCD Diffractometer, data collection and refinement was performed as previously described. The biological screening of IDHPs was performed by the Psychoactive Drug Screening Program (PDSP) of NIMH. The known IDHP 1a was prepared as previously described.19 The preparation of the isoxazole-4-carbaldehydes required for the Hantzsch synthesis of the monomeric IDHPs 1b–d, IDHP alkynes 2, PEG-diazides 3, Knoevenagel products 7, and 3-Oxo-butyric acid prop-2-ynyl ester 9 were prepared by standard methods which are detailed in the Supplementry Dt.
7.1. General synthesis of 4-[3-(p-bromo-phenyl)-5-methyl-isoxazol-4-yl]-2,6-dimethyl-1,4-dihydro-pyridine-3,5-dicarboxylic acid diethyl ester (1d)
An oven dried 100 ml round bottom was charged with 0.107 g (0.405 mmol) of 3-(p-Bromo-phenyl)-5-methyl-isoxazole-4-carbaldehyde, 0.11 g (0.84 mmol) of ethyl acetoacetate, 50 ml of benzene and a magnetic stir bar. The solution was allowed to stir at room temperature. 0.10 ml of 30% aqueous ammonia was measured out along with 0.90 ml of methanol, the mixture was then added to the round bottom. The round bottom was then fitted with an oven dried condenser and the solution was brought to reflux (70 °C). The reaction process was monitored via TLC. Once complete the solution was then cooled to room temperature and the excess solvent was removed via rotovap. The product was then purified via recrystallization from ethyl acetate and hexanes to give (1d) as yellow crystals (0.084 g, 0.0411 mmol, 42.92%).
1H NMR: (CDCl3) δ 7.55 (d, J = 8.4 Hz, 2H), 7.37 (d, J = 8.7 Hz, 2H), 5.06 (s, 1H), 5.02 (bs, 1H), 3.52 (s, 6H), 2.43 (s, 3H).
13C NMR: (CDCl3) δ 167.27, 166.26, 162.71, 145.31, 143.76, 143.52, 131.24, 130.72, 130.01, 122.88, 119.46, 101.14, 59.85, 51.31, 29.29, 29.16, 19.35, 14.46, 11.46.
MS: m/z = 489.15 (M+, 100% rel. intensity), 530.36 ([M+]+CH3-CN, 70).
TLC: SiO2; Hexane:EtOAc 4:1 Rf = 0.17.
7.1.1. 2,6-Dimethyl-4-(5-methyl-3-phenyl-isoxazol-4-yl)-1,4-dihydro-pyridine-3,5-dicarboxylic acid diethyl ester (1a)
The spectral data was in accord with that previously reported.19
1H NMR: (CDCl3) δ 7.57 (d 2H J = 7.62 Hz), 7.30 (d 2H J = 7.62 Hz), 7.23 (s 1H), 4.98 (s 1H), 4.09 (q, J = 6.85 Hz, 2H), 4.01 (q, J = 4.89 Hz, 2H), 2.49 (s, 3H), 1.91 (s, 6H), 1.18 (t, J = 7.09 Hz, 6H).
13C NMR: (CDCl3) δ 171.23, 167.44, 166.32, 165. 81, 163.92, 147.32, 130.25, 128.63, 125.77, 121.32, 101.12, 59.81, 31.65, 20.87, 14.51, 12.34.
TLC: SiO2; Hexane:EtOAc 4:1 Rf = 0.17.
7.1.2. 4-[3-(o-Bromo-phenyl)-5-methyl-isoxazol-4-yl]-2,6-dimethyl-1,4-dihydro-pyridine-3,5-dicarboxylic acid diethyl ester (1b)
1H NMR: (CDCl3) δ 7.60 (d, J = 7.91 Hz, 1H), 7.31 (d, J = 7.40 Hz, 1H), 7.24 (d, J = 5.90 Hz, 1H), 7.18 (d, J = 7.53 Hz, 1H), 4.95 (s, 1H), 4.15 (q, J = 4.64 Hz, 2H), 4.13 (q, J = 4.64 Hz, 2H), 2.58 (s, 3H), 1.96 (s, 6H), 1.25 (t, J = 7.15 Hz, 6H).
13C NMR: (CDCl3) δ 167.33, 165.60, 162.95, 144.54, 132.46, 132.25, 130.00, 126.24, 125.01, 118.27, 100.47, 59.79, 29.24, 19.61, 14.61, 11.54.
HRMS: calculated for C23H26BrN2O5 489.1023 found 489.1025.
MS: m/z = 489 (M+, 100).
TLC: SiO2; Hexane:EtOAc 4:1 Rf = 0.07.
7.1.3. 4-[3-(m-Bromo-phenyl)-5-methyl-isoxazol-4-yl]-2,6-dimethyl-1,4-dihydro-pyridine-3,5-dicarboxylic acid diethyl ester (1c)
1H NMR: (CDCl3) δ 7.59 (s, 1H), 7.56 (s, 1H), 7.55 (s, 1H), 7.33 (s, 1H), 5.05 (s, 1H), 4.16 (q, J = 3.76 Hz, 2H), 4.09 (q, J = 3.76 Hz, 2H), 2.54 (s, 3H), 2.06 (s, 6H), 1.25 (t, J = 7.03 Hz, 6H).
13C NMR: (CDCl3) δ 167.27, 166.09, 162.48, 144.08, 133.18, 132.50, 129.21, 128.44, 121.64, 119.41, 101.00, 59.87, 29.41, 19.51, 14.51, 11.50.
HRMS: calculated for C23H26BrN2O5 489.1028 found 489.1025.
MS: m/z = 489 (M+, 100) 530 ([M+2]+CH3CN, 20).
TLC: SiO2; Hexane: EtOAc 4:1 Rf = 0.26.
7.2. General Synthesis of 4-[3-(p-Bromo-phenyl)-5-methyl-isoxazol-4-yl]-2,6-dimethyl-1,4-dihydro-pyridine-3,5-dicarboxylic acid 3-ethyl ester 5-prop-2-ynyl ester, 2d, Scheme 1
An oven dried 100 ml round bottom was charged with 0.82 g (2.17 mmol) of ethyl (2Z)-2-[(5-methyl-3-p-Bromophenyl-1,2-oxazol-4-yl)methylidene]-3-oxobutanoate, 0.305 g (2.17 mmol) of prop-2-yn-1-yl 3-oxobutanoate, 50 ml of absolute ethanol and a magnetic stir bar the vessel was sealed and the solution was allowed to stirrer at room temperature. In a 10 ml graduated cylinder 0.47 ml of 30% ammonia was measured out along with 0.53 ml of absolute ethanol, the mixture was then added to the 100 ml round bound bottom containing the reaction solution. The 100 ml round bottom was then placed on a heating mantel and fitted with a oven dried condenser. The solution was then refluxed (78 °C) for 12 h during which time the solution turned yellow. The solution was then cooled to room temperature and the excess solvent was removed via rotovap. The solution was then purified via recrystallized from ethyl acetate and hexanes to give off yellow crystals. (0.355 g, 0.713 mmol, 33%).
1H NMR: (CDCl3) δ 7.52 (d, J = 7.83 Hz, 2H), 7.29 (d, J = 8.31 Hz, 2H), 5.01 (s, 1H), 4.98 (s, 2H) 4.56 (q, J = 9.78 Hz, 2H), 4.09 (qq, J = 6.36 Hz, 2H), 4.00 (qq, J = 6.85 Hz, 2H), 2.50 (s, 3H), 2.47 (s, 3H), 2.05 (s 6H), 1.21 (t, J = 7.03 Hz, 3H).
13C NMR: (CDCl3) δ 167.28, 166.21, 162.70, 145.35, 143.78, 131.23, 130.72, 122.84, 119.47, 101.62, 101.12, 74.68, 59.85, 51.31, 29.69, 29.16, 19.35, 14.46, 11.46.
MS: m/z = 499.35 (M+, 100% rel. intensity), 540.86 ([M+]+CH3-CN, 60).
TLC: SiO2; Hexane:EtOAc 4:1 Rf = 0.12.
7.2.1. 2,6-Dimethyl-4-(5-methyl-3-phenyl-isoxazol-4-yl)-1,4-dihydro-pyridine-3,5-dicarboxylic acid 3-ethyl ester 5-prop-2-ynyl ester, 2a
1H NMR: (CDCl3) δ 7.57 (d, J = 8.72 Hz, 2H), 7.30 (d, J = 8.72 Hz, 2H), 7.22 (s, 1H), 5.37 (s, 1H), 5.23 (s, 1H), 4.98 (s, 1H), 4.55 (q, J = 6.36 Hz, 2H), 4.08 (q, J = 6.85 Hz, 2H), 2.54 (s, 3H), 2.08 (s, 1H), 1.92 (s, 6H), 1.18 (t, J = 7.09 Hz, 3H).
13C NMR: (CDCl3) δ 171.18, 167.44, 166.41, 165.79, 163.81, 146.01, 144.39, 130.76, 129.52, 127.55, 119.43, 101.43, 74.59, 60.40, 29.20, 19.15, 14.47, 11.52.
TLC: SiO2; Hexane:EtOAc 4:1 Rf = 0.18.
7.2.2. 4-[3-(o-Bromo-phenyl)-5-methyl-isoxazol-4-yl]-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylic acid 3-ethyl ester 5-prop-2-ynyl ester, 2b
1H NMR: (CDCl3) δ 7.58 (d, J = 8.66 Hz, 1H), 7.31 (d, J = 6.53 Hz, 1H), 7.24 (d, J = 7.65 Hz, 1H), 7.18 (d, J = 7.53 Hz, 1H), 4.95 (s, 1H), 4.67 (d, J = 3.04 Hz, 2H), 4.15 (qq, J = 7.15 Hz, 2H), 2.58 (s, 3H), 2.05 (s, 1H), 1.96 (s, 6H), 1.25 (t, J = 7.15 Hz, 3H).
13C NMR: (CDCl3) δ 167.33, 165.50, 162.95, 144.54, 132.46, 132.25, 132.11, 130.00, 126.24, 125.01, 118.27, 100.47, 100.47, 59.79, 29.24, 19.61, 14.61, 11.54.
TLC: SiO2; Hexane:EtOAc 4:1 Rf = 0.12.
7.2.3. 4-[3-(m-Bromo-phenyl)-5-methyl-isoxazol-4-yl]-2,6-dimethyl-1,4-dihydro-pyridine-3,5-dicarboxylic acid 3-ethyl ester 5-prop-2-ynyl ester, 2c
1H NMR: (CDCl3) δ 7.59 (s, 1H), 7.56 (s, 1H), 7.55 (s, 1H), 7.33 (s, 1H), 5.05 (s, 1H), 4.65 (d, J = 2.51 Hz, 2H), 4.16 (qq, J = 7.15 Hz, 2H), 2.54 (s, 3H), 2.08 (s, 1H), 2.06 (s, 6H), 1.25 (t, J = 7.03 Hz, 3H).
13C NMR: (CDCl3) δ 167.27, 166.09, 162.48, 144.08, 133.18, 132.50, 131.51, 129.21, 128.44, 121.64, 119.41, 101.00, 59.87, 29.41, 19.51, 14.51, 11.50.
TLC: SiO2; Hexane:EtOAc 4:1 Rf = 0.15.
7.3. General synthesis of 3-[1-(17-{4-[({4-[3-(p-Bromophenyl)-5-methyl-1,2-oxazol-4-yl]-5-(ethoxycarbonyl)-2,6-dimethyl-1,4-dihydropyridin-3-yl}carbonyloxy)methyl]-1H-1,2,3-triazol-1-yl}-3,6,9,12,15-pentaoxaheptadecan-1-yl)-1H-1,2,3-triazol-4-yl]methyl 5-ethyl 4-[3-(p-Bromophenyl)-5-methyl-1,2-oxazol-4-yl]-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate, 4d
An oven dried 100 ml round bottom was charged with 0.164 g (0.328 mmol) of 4-[3-(4-Bromo-phenyl)-5-methyl-isoxazol-4-yl]-2,6-dimethyl-1,4-dihydro-pyridine-3,5-dicarboxylic acid 3-ethyl ester 5-prop-2-ynyl ester, .034 g of 1-Azido-2-[2-(2-{2-[2-(2-azido-ethoxy)-ethoxy]-ethoxy}-ethoxy)-ethoxy]-ethane 3c, 0.010 g of Copper(I) bromide, 0.010 g of anhydrous Copper sulfate and a magnetic stir bar. Freshly distilled THF (50 ml) was then added and the solution was allowed to stir at room temperature. Sodium ascorbate (0.003 g) was then added and the solution was allowed to stir, and the reaction progress was monitored by TLC, once the reaction was completed excess solvent was removed via rotovap. The solution was then purified via a silica column. The purification yielded 4d as a yellow oil. (0.0067 g, 0.005 mmol, 15.34%).
1H NMR: (CDCl3) δ 7.65 (s, 2H), 7.49 (d, J = 8.28 Hz, 4H), 7.22 (d, J = 8.28 Hz, 4H), 4.99 (s, 2H), 4.52 (q, J = 4.52 Hz, 4H), 4.13 (q, J = 7.28 Hz, 2H), 4.07 (q, J = 6.78 Hz, 2H), 3.89 (t, J = 5.52 Hz, 3H), 3.75 (t, J = 6.02 Hz, 4H), 3.65 (m, 16H), 2.33 (s, 6H), 2.05 (s, 9H), 1.19 (t, J = 7.03 Hz, 6H).
13C NMR: (CDCl3) δ 168.74, 166.68, 159.89, 156.71, 135.10, 131.91, 131.22, 130.77, 129.04, 128.17, 127.45, 124.90, 124.24, 109.43, 71.36, 70.59, 69.35, 69.303, 61.72, 58.48, 42.76, 37.76, 31.94, 29.71, 23.16, 14.13, 13.63, 11.25.
HRMS: calculated for C60H71Br2N10O15 1329.3467 found 1329.3462.
MS: m/z = 1327 (M (79Br) 100% rel. intensity), 1328 (M, 62), 1329 (M+1, 97), 1330 ([M+1](81Br), 60) 1331 ([M+1]+(81Br), 60), 1332 ([M+1]+2(81Br), 40).
IR: cm− 1 3014.92, 2962.68, 1746.26, 1492.10, 1369.40, 1216.41, 753.73.
TLC: SiO2; EtOAc:MeOH:CH2Cl2 (4:1:2) Rf = 0.56.
Then run a column in 4:1:3:2 (EtOAc:MeOH:Hexanes:CH2Cl2) Rf = 0.23.
7.3.1. 3-(1-{17-[4-({[5-(Ethoxycarbonyl)-2,6-dimethyl-4-(5-methyl-3-phenyl-1,2-oxazol-4-yl)-1,4-dihydropyridin-3-yl]carbonyloxy} methyl)-1H-1,2,3-triazol-1-yl]-3,6,9,12,15-pentaoxaheptadecan-1-yl}-1H-1,2,3-triazol-4-yl)methyl 5-ethyl 2,6-dimethyl-4-(5-methyl-3-phenyl-1,2-oxazol-4-yl)-1,4-dihydropyridine-3,5-dicarboxylate (4a)
1H NMR: (CDCl3) δ 7.64 (s, 2H), 7.52 (d, J = 8.28 Hz, 4H), 7.25 (d, J = 8.28 Hz, 4H), 4.98 (s, 2H), 4.54 (q, J = 4.77 Hz, 4H), 4.11 (q, J = 7.03 Hz, 2H), 4.04 (q, J = 7.03 Hz, 2H), 3.87 (t, J = 5.27 Hz, 3H), 3.75 (t, J = 5.77 Hz, 4H), 3.65 (m, 16H), 2.36 (s, 6H), 2.03 (d, J = 5.27 Hz, 9H), 1.19 (t, J = 7.03 Hz, 6H).
13C NMR: (CDCl3) δ 171.15, 168.55, 167.21, 169.96, 151.03, 144.05, 130.90, 129.78, 129.44, 129.30, 128.68, 128.40, 127.51, 106.88, 101.17, 88.97, 71.28, 70.60, 70.51, 69.93, 69.37, 68.94, 66.48, 63.51, 61.56, 59.56, 55.57, 51.58, 50.65, 42.73, 37.64, 35.87, 29.57, 29.20, 23.50, 22.58, 21.50, 18.84, 15.10, 14.40, 14.02, 12.41, 11.06.
HRMS: calculated for C60H71N10O15 1171.5100 found 1171.5134.
MS: m/z = 1169 ([M–2] 100% rel. intensity), 1170 ([M–1] 70), 1171 ([M] 40), 1172 ([M+] 10).
IR: cm− 1 2972.26, 2831.20, 1734.06, 1571.92, 1357.96, 1225.00, 727.32.
TLC: SiO2; EtOAc:MeOH:CH2Cl2 (4:1:2) Rf = 0.52.
7.3.2. 3-[1-(17-{4-[({4-[3-(o-Bromophenyl)-5-methyl-1,2-oxazol-4-yl]-5-(ethoxycarbonyl)-2,6-dimethyl-1,4-dihydropyridin-3-yl}carbonyloxy)methyl]-1H-1,2,3-triazol-1-yl}-3,6,9,12,15-pentaoxaheptadecan-1-yl)-1H-1,2,3-triazol-4-yl]methyl 5-ethyl 4-[3-(o-bromophenyl)-5-methyl-1,2-oxazol-4-yl]-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate (4b)
1H NMR: (CDCl3) δ 7.59 (d, J = 6.78 Hz, 2H), 7.58 (d, J = 6.78 Hz, 2H), 7.31 (d, J = 7.03 Hz, 2H), 7.23 (d, 2H J = 7.53 Hz), 7.15 (d, J = 7.03 Hz, 2H), 4.92 (s, 2H), 4.54 (q, J = 5.52 Hz, 4H), 4.10 (q, J = 8.78 Hz, 2H), 3.97 (t, J = 5.77 Hz, 3H), 3.81 (t, J = 6.27 Hz, 4H), 3.66 (m, 16H), 2.36 (s, 6H), 1.96 (s, 9H), 1.23 (t, J = 7.03 Hz, 6H).
13C NMR: (CDCl3) δ 167.24, 132.21, 131.98, 130.28, 126.40, 100.36, 71.28, 70.56, 69.96, 69.47, 68.96, 65.56, 61.91, 59.69, 50.64, 47.62, 42.81, 37.70, 32.16, 30.48, 29.63, 23.41, 19.16, 15.14, 14.56, 11.28.
HRMS: calculated for C60H71Br2N10O15 1329.3467 found 1329.3467.
MS: m/z = 1327 (M (79Br) 35% rel. intensity), 1328 (M, 30), 1329 (M+1, 97), 1330 ([M+1](81Br), 50) 1331 ([M+1]+(81Br), 100), 1332 ([M+1]+2(81Br), 40).
IR: cm− 1 2917.91, 2850.74, 1731.34, 1496.26, 1261.19, 1164.17, 733.12.
TLC: SiO2; EtOAc:MeOH:CH2Cl2 (4:1:2) Rf = 0.48.
7.3.3. Isoxaquidar. 3-[1-(17-{4-[({4-[3-(m-bromophenyl)-5-methyl-1,2-oxazol-4-yl]-5-(ethoxycarbonyl)-2,6-dimethyl-1,4-dihydropyridin-3-yl}carbonyloxy)methyl]-1H-1,2,3-triazol-1-yl}-3,6,9,12,15-pentaoxaheptadecan-1-yl)-1H-1,2,3-triazol-4-yl]methyl 5-ethyl 4-[3-(m-bromophenyl)-5-methyl-1,2-oxazol-4-yl]-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate (4c)
1H NMR: (CDCl3) δ 7.73 (s, 2H), 7.64 (d, J = 5.52 Hz, 2H), 7.51 (s, 2H), 7.25 (s, 2H), 7.18 (d, J = 10.54 Hz, 2H), 4.98 (s, 2H), 4.53 (q, J = 4.02 Hz, 4H), 4.38 (q, J = 4.27 Hz, 2H), 4.09 (q, J = 10.79 Hz, 2H), 3.86 (t, J = 5.02 Hz, 3H), 3.76 (t, J = 5.02 Hz, 4H), 3.62 (m, 16H), 2.35 (s, 6H), 2.01 (s, 9H), 1.19 (t, J = 7.28 Hz, 6H).
13C NMR: (CDCl3) δ 167.18, 166.31, 162.49, 145.12, 144.12, 133.04, 132.73, 132.40, 131.54, 130.44, 13.026, 129.28, 128.39, 126.18, 124.96, 121.62, 199.30, 101.11, 100.33, 71.35, 70.62, 69.39, 69.01, 66.64, 59.85, 58.44, 56.92, 50.69, 43.38, 42.79, 41.44, 37.75, 29.41, 23.17, 23.09, 19.46, 19.29, 14.48, 13.62, 11.30.
HRMS: calculated for C60H71Br2N10O15 1329.3467 found 1329.3450.
MS: m/z = 1327 (M (79Br) 35% rel. intensity), 1328 (M, 20), 1329 (M+1, 100), 1330 ([M+1](81Br), 60) 1331 ([M+1]+(81Br), 100), 1332 ([M+1]+2(81Br), 50).
IR: cm− 1 3007.46, 2970.14, 1738.80, 1432.83, 1227.61, 1216.41, 761.51.
TLC: SiO2; EtOAc:MeOH:CH2Cl2 (4:1:2) Rf = 0.52.
7.3.4. 3-[1-(2-{2-[2-(2-{4-[({4-[3-(p-Bromophenyl)-5-methyl-1,2-oxazol-4-yl]-5-(ethoxycarbonyl)-2,6-dimethyl-1,4-dihydropyridin-3-yl}carbonyloxy)methyl]-1H-1,2,3-triazol-1-yl}ethoxy)ethoxy]ethoxy} ethyl)-1H-1,2,3-triazol-4-yl]methyl 5-ethyl 4-[3-(p-Bromophenyl)-5-methyl-1,2-oxazol-4-yl]-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate (4e)
1H NMR: (CDCl3) δ 7.60 (d, J = 5.52, 2H), 7.50 (t, J = 8.28 Hz, 4H), 7.26 (t, J = 8.28 Hz, 4H), 5.17 (m, 4H), 4.99 (s, 2H), 4.51 (q, J = 5.52 Hz, 4H), 4.07 (q, J = 6.78 Hz, 2H), 3.94 (q, J = 6.78 Hz, 2H), 3.85 (t, J = 5.27 Hz, 4H), 3.62 (m, 8H), 2.34 (s, 6H), 2.01 (d, J = 5.02 Hz, 9H), 1.15 (t, J = 7.28 Hz, 6H).
13C NMR: (CDCl3) δ 167.21, 162.62, 145.12, 143.89, 131.90, 131.12, 130.78, 129.92, 129.02, 122.88, 119.54, 101.29, 100.39, 72.46, 70.67, 70.06, 69.34, 61.72, 59.81, 50.67, 42.84, 29.69, 19.03, 14.45, 11.30.
HRMS: calculated for C56H63 N10O13Br2 1241.2943 found 1241.2950.
MS: m/z = 1239 ([M(79Br)] 60% rel. intensity), 1240 ([M] 40), 1241 ([M+1] 100), 1242 ([M+1](81Br) 60), 1243 ([M+2] 70), 1244 ([M+3] 50).
IR: cm− 1 2917.91, 2850.74, 1731.31, 1496.26, 1261.19, 1164.17, 697.76.
TLC: SiO2; EtOAc:MeOH:CH2Cl2 (4:1:2) Rf = 0.52.
7.3.5. 3-(1-{2-[2-(2-{4-[({4-[3-(4-Bromophenyl)-5-methyl-1,2-oxazol-4-yl]-5-(ethoxycarbonyl)-2,6-dimethyl-1,4-dihydropyridin-3-yl}carbonyloxy)methyl]-1H-1,2,3-triazol-1-yl}ethoxy)ethoxy]ethyl}-1H-1,2,3-triazol-4-yl)methyl 5-ethyl 4-[3-(4-bromophenyl)-5-methyl-1,2-oxazol-4-yl]-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate (4f)
1H NMR: (CDCl3) δ 7.61 (d, J = 7.53 Hz 2H), 7.49 (d, J = 8.28 Hz 4H), 7.25 (d 4H J = 8.53 Hz), 5.17 (m 4H), 4.98 (s 2H), 4.51 (q 4H J = 5.27 Hz), 4.06 (q 2H J = 6.83 Hz), 3.88 (q 2H J = 5.02 Hz), 3.72 (t 4H J = 6.02 Hz), 3.64 (d 4H J = 6.27 Hz), 2.33 (s 6H), 2.01 (s 9H), 1.15 (t 6H J = 7.28 Hz).
13C NMR: (CDCl3) δ 167.25, 143.96, 131.97, 131.18, 130.81, 129.89, 129.03, 122.98, 101.28, 71.30, 70.74, 70.47, 70.32, 69.15, 61.75, 59.83, 53.51, 50.67, 42.92, 29.70, 29.36, 19.04, 15.16, 14.46, 13.64, 11.36.
HRMS: calculated for C54H59 N10O12Br2 1197.2633 found 1197.2681.
MS: m/z = 1195 ([M(79Br)] 80% rel. intensity), 1196 ([M] 60), 1197 ([M+1] 100), 1198 ([M+1](81Br) 80), 1199 ([M+2] 100), 1200 ([M+3] 60).
IR: cm− 1 3014.92, 2962.68, 1746.26, 1429.10, 1369.40, 1216.41, 753.73.
TLC: SiO2; EtOAc:MeOH:CH2Cl2 (4:1:2) Rf = 0.5.
7.4. General synthesis of 4-[3-(p-Bromo-phenyl)-5-methyl-isoxazol-4-yl]-2,6-dimethyl-1,4-dihydro-pyridine 3,5-dicarboxylic acid 3-ethyl ester 5-(1-phenethyl-1H-[1,2,3]triazol-4-ylmethyl) ester, 5
An oven dried 100 mL round bottom was charged with 0.038 g (0.259 mmol) of (2-Azido-ethyl)-benzene and a magnetic stir bar. The sample was then taken up in freshly distilled THF and the solution was allowed to stir at room temperature. Once the (2-Azidoethyl)-benzene was completely dissolved 0.129 g (0.259 mmol) of 4-[3-(p-Bromo-phenyl)-5-methyl-isoxazol-4-yl]-2,6-dimethyl-1,4-dihydro-pyridine-3,5-dicarboxylic acid 3-ethyl ester 5-prop-2-ynyl ester 2d was added in portions. The round bottom was then capped with a rubber septum and the solution was allowed to stir. The solution was allowed to stir until all reagents where dissolved, 0.011 g of Copper(I)-bromide was then added and the solution was allowed to stir at room temperature for 48 Hr. Reaction progress was monitored via TLC, once the compete excess solvent was removed via rotovap. The solution was then purified via silica column yielding 5 as a yellow oil (0.0527 g, 0.0816 mmol, 32%). TLC: SiO2; EtOAc:MeOH:CH2Cl2 (4:1:2) Rf = 0.55.
1H NMR: (CDCl3) δ 7.51 (d, J = 8.53 Hz, 2H), 7.30 (d, J = 7.53 Hz, 2H), 7.26 (s, 1H), 7.24 (d, J = 9.29 Hz, 2H), 7.12 (d, J = 8.28 Hz, 2H), 5.15 (q, J = 12.30 Hz, 2H), 5.03 (s, 1H), 4.99 (s, 1H), 4.56 (t, J = 7.53 Hz, 2H), 4.14 (q, J = 7.03 Hz, 2H), 4.07 (t, J = 7.03 Hz, 2H), 3.20 (t, J = 7.28 Hz, 2H), 2.35 (s, 3H), 2.02 (d, J = 5.27 Hz, 6H), 1.27 (t, J = 7.28 Hz, 3H).
13C NMR: (CDCl3) δ 167.14, 167.04, 166.55, 144.59, 143.54, 136.89, 131.20, 130.79, 129.93, 128.85, 128.69, 127.13, 122.89, 101.52, 100.64, 60.43, 59.89, 56.88, 51.68, 45.91, 36.63, 26.39, 21.08, 19.46, 19.26, 14.45, 19.26, 14.45, 14.22, 11.33, 8.67.
HRMS: calculated for C23H32BrN5O5 646.1685 found 646.1665.
MS: m/z = 645 (M (79Br), 20% rel. intensity), 646 (M+, 80), 647 ([M+1]+, 35), 648 ([M+1]+(81Br), 78).
IR: cm− 1 2992.36, 1734.06, 1365.56, 1255.39, 1278.18, 1221.20, 761.51, 746.32.
7.4.1. Synthesis of 2,6-dimethyl-4-(5-methyl-3-phenyl-isoxazol-4-yl)-pyridine-3,5-dicarboxylic acid diethyl ester, 6
The 2,6-Dimethyl-4-(5-methyl-3-phenyl-isoxazol-4-yl)-1,4-dihydro-pyridine-3,5-dicarboxylic acid diethyl ester (1a) was placed in a 50 mL oven dried round bottom along with a magnetic stir bar and 0.085 g of deionized water. The solution was then allowed to stir at room temperature while, 0.227 g of HNO3 and 0.0246 g of H2SO4 was added. The solution was then heated to 110 °C and white foam was observed, after 10 min. at 110 °C the solution turned a light yellow. The solution was then allowed to cool to room temperature. The solution was then taken up in ice water and LiOH was added slowly until the solution was neutral. The solution was then added to a separatory funnel and extracted with CHCl3 and Brine (×3). The solution was then dried over Na2SO4 and filtered; the excess solvent was then removed via rotovap yielding 6 as a pale yellow oil that solidified on standing, and was then crystallized with CH2Cl2 and Hexanes. 6, (0.100 g, 0.245 mmol, 98%). TLC: SiO2; Hexanes:EtOAc (4:1) Rf = 0.31. 1H NMR: (CDCl3) 7.46 (d, J = 7.28 Hz, 2H), 7.34 (d, J = 7.11 Hz, 2H), 7.29 (s, 1H), 4.03 (q, J = 9.56 Hz, 2H), 2.59 (s, 3H), 2.25 (s, 3H), 0.97 (t, J = 7.01 Hz, 3H).
13C NMR: (CDCl3) δ 167.18, 165.91, 159.70, 155.29, 134.52, 128.66, 127.56, 126.56, 109.18, 60.63, 22.09, 12.57, 10.38.
HRMS: calculated for C23H24N2O5 408.1685 found 408.1692.
MS: m/z = 409 (M+1, 100), 410 (M+2, 40).
IR: cm− 1 3007.46, 1737.86, 1441.54, 1365.56, 1259.19, 1217.60, 746.32.
7.5. General synthesis of 2-acetyl-3-[3-(p-Bromo-phenyl)-5-methylisoxazol-4-yl]-acrylic acid ethyl ester, 7d
An oven dried 50 mL round bottom was charged with 0.95 g of 5-methyl-3-p-Bromophenyl-1,2-oxazole-4-carbaldehyde, 0.799 g of Ethyl Acetoacetate, 4 mL of Benzene where added at room temperature. The solution was allowed to stir, and 0.1325 g of glacial acidic acid and 0.054 g of piperidine was added. The round bottom was then fitted with a dean stark trap and a condenser. The solution was then allowed to reflux for three hours; with the reaction progress monitored my TLC. The solution was then cooled to room temperature and the excess solvent was removed via rotovap. The solution was then purified via a silica column. The purification yielded yellow oil. (0.761 g, 2.53 mmol, 52%).
1H NMR: (CDCl3) δ 7.54 (d, J = 9.78 Hz, 2H), 7.40 (d, J = 8.56 Hz, 2H), 7.33 (s, 1H), 4.13 (q, J = 7.09 Hz, 2H), 2.35 (s, 3H), 2.19 (s, 3H), 1.20 (t, J = 7.09 Hz, 3H).
13C NMR: (CDCl3) δ 194.12, 169.17, 165.80, 160.44, 137.70, 132.11, 129.50, 131.45, 127.45, 124.46, 109.55, 61.63, 13.75, 12.19.
MS: m/z = 377.03 (M+, 50% rel. intensity) 418.45 ([M+]+CH3CN, 100).
TLC: SiO2; Hexane:EtOAc 1:1 Rf = 0.39.
7.5.1. 2-Acetyl-3-(5-methyl-3-phenyl-isoxazol-4-yl)-acrylic acid ethyl ester, 7a
1H NMR: (CDCl3) δ 7.57 (d, J = 12.72 Hz, 2H), 7.44 (d, J = 12.72 Hz, 2H), 7.25 (s, 1H), 3.99 (q, J = 7.52 Hz, 2H), 2.40 (s, 3H), 2.37 (s, 3H), 1.14 (t, J = 6.05 Hz, 3H).
13C NMR: (CDCl3) δ 194.36, 168.96, 166.03, 161.50, 137.26, 132.67, 128.93, 128.05, 109.77, 61.60, 27.82, 13.79, 12.31.
TLC: SiO2; Hexane:EtOAc 1:1 Rf = 0.39.
7.5.2. 2-Acetyl-3-[3-(o-bromo-phenyl)-5-methyl-isoxazol-4-yl]-acrylic acid ethyl ester, 7b
1H NMR: (CDCl3) δ 7.65 (s, 1H), 7.54 (s, 1H), 7.36 (s, 1H) 7.24 (s, 1H), 7.10 (s, 1H), 4.17 (q, J = 6.57 Hz, 2H), 2.37 (s, 3H), 2.15 (s, 3H), 1.17 (t, J = 6.98 Hz, 3H).
13C NMR: (CDCl3) δ 194.36, 168.96, 166.03, 161.50, 138.26, 132.30, 130.71, 128.93, 128.05, 109.77, 61.60, 27.82, 13.79, 12.13.
TLC: SiO2; Hexane:EtOAc 1:1 Rf = 0.41.
7.5.3. 2-Acetyl-3-[3-(3-bromo-phenyl)-5-methyl-isoxazol-4-yl]-acrylic acid ethyl ester, 7c
1H NMR: (CDCl3) δ 7.65 (s, 1H), 7.43 (s, 1H), 7.35 (s, 1H), 7.30 (s, 1H), 7.68 (s, 1H), 4.15 (q, J = 6.84 Hz, 2H), 2.34 (s, 3H), 2.17 (s, 3H), 1.16 (t, J = 5.42 Hz, 3H).
13C NMR: (CDCl3) δ 194.24, 169.12, 165.83, 162.78, 148.65, 137.68, 132.11, 130.41, 131.96, 124.93, 127.45, 110.15, 59.67, 24.65, 14.04, 12.18.
TLC: SiO2; Hexane:EtOAc 1:1 Rf = 0.30.
7.6. Synthesis of 3-oxo-butyric acid prop-2-ynyl ester, 9
An oven dried round bottom (250 mL) was charged with a magnetic stir bar, 4.64 g of Ethyl Acetoacetate, 2.00 g of propargyl alcohol, 0.22 g of boric acid, and 40 mL of toluene added at room temperature. The round bottom was then fitted with a Dean Stark trap, a condenser and a drying tube. The solution was then heated to reflux and left for 144 h. The reaction yielded a light red solution. The solution was then purified via a silica column. The purification compound yielded yellow oil. (2.868 g, 20.46 mmol, 57%).
1H NMR: (CDCl3) δ 4.68 (d, J = 2.45 Hz, 2H), 3.46 (s 2H), 2.49 (t, J = 2.45 Hz, 1H), 2.22 (s, 3H).
13C NMR: (CDCl3) δ 200.03, 166.27, 104.98, 89.06, 52.60, 49.56, 30.08.
MS: m/z = 140.047 (M+, 100% rel. intensity) 142.07 (M+2, 90), 183.78 ([M+2]+CH3CN, 60).
TLC: SiO2; Hexane:EtOAc 4:1 Rf = 0.18.
Supplementary Material
Acknowledgments
The authors thank NIH for grants NS038444 (NN), P20RR015583 (NN, SS), and CBSD CoBRE grant P20GM103546 (NN). These studies were also funded by the ALSAM Foundation Skaggs Scholars Program Grant (DSB, NN). Computational modeling studies were conducted at the University of Colorado Computational Chemistry and Biology Core Facility, which is funded in part by NIH/NCATS UL1 TR001082. We thank Dr. Mike Braden of the Core Facility for Molecular Computation (UM) for helpful discussions during computer modeling.
MDR1 data was generously provided by the National Institute of Mental Health’s Psychoactive Drug Screening Program (NIMH PDSP), Contract # HHSN-271-2008-00025-C (NIMH PDSP). The NIMH PDSP is directed by Bryan L. Roth MD, PhD at the University of North Carolina at Chapel Hill and Project Officer Jamie Driscoll at NIMH, Bethesda MD, USA. We also thank Dr. X.P. Huang of PDSP for helpful suggestions with the manuscript.
A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bmc.2017.04.008.
References
- 1.Beis K. Biochem Soc Trans. 2015;43:889–893. doi: 10.1042/BST20150047. [DOI] [PubMed] [Google Scholar]
- 2.ter Beek J, Guskov A, Slotboom DJ. J Gen Physiol. 2014;143:419–435. doi: 10.1085/jgp.201411164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hitchcock SA. J Med Chem. 2012;55:4877–4895. doi: 10.1021/jm201136z. [DOI] [PubMed] [Google Scholar]
- 4.Gillet J-P, Gottesman MM. Chapter 4 in Multi-Drug Resistance in Cancer. In: Zhou J, editor. Methods in Molecular Biology. Vol. 596. N.Y: Humana Press; 2010. [Google Scholar]
- 5.Gottesman MM, Fojo T, Bates SE. Nat Rev Cancer. 2002:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 6.Robey RW, Shukla S, Finley EM, et al. [last accessed February 14, 2017];Biochem Pharmacol. 2008 3:1302–1312. doi: 10.1016/j.bcp.2007.12.001. Tariquidar (XR9576), Laniquidar, Zosuquidar were listed as the subject of 15 clinical trials on clinicaltrials.gov during the writing of this manuscript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross DD. Best Pract Res Clin Haematol. 2004;17:641–651. doi: 10.1016/j.beha.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Cripe LD, Uno H, Paietta EM, et al. Blood. 2010;116:4077–4085. doi: 10.1182/blood-2010-04-277269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zarrin A, Mehdipour AR, Miri R. Chem Biol Drug Des. 2010;76:369–381. doi: 10.1111/j.1747-0285.2010.01025.x. [DOI] [PubMed] [Google Scholar]
- 10.Shah A, Bariwal J, Molnár J, Kawase M, Motohashi N. Advanced dihydropyridines as novel multidrug resistance modifiers and reversing agents. Top Heterocycl Chem. 2008;15:201–252. [Google Scholar]
- 11.Hulubei V, Meikrantz SB, Quincy DA, et al. 4-Isoxazolyl-1,4-dihydropyridines exhibit binding at the multidrug-resistance transporter. Bioorg Med Chem. 2012;20:6613–6620. doi: 10.1016/j.bmc.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szabon-Watola Monika, Ulatowski Sarah V, George Kathleen M, Hayes Christina D, Steiger Scott. Fluorescent probes of the isoxazole-dihydropyridine scaffold: MDR-1 binding and homology model. Bioorg Med Chem. 2014;24:117–121. doi: 10.1016/j.bmcl.2013.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steiger S, Natale NR. Future Med Chem. 2014;6:923–943. doi: 10.4155/fmc.14.46. [DOI] [PubMed] [Google Scholar]
- 14.Sauna ZE, Andrus MB, Turner TM, Ambudkar SV. Biochemistry. 2004;43:2262–2271. doi: 10.1021/bi035965k. [DOI] [PubMed] [Google Scholar]
- 15.Namanja HA, Emmert D, Davis DA, et al. J Am Chem Soc. 2011;134:2976–2980. doi: 10.1021/ja206867t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pires MM, Emmert D, Hrycyna CA, Chmielewski J. Mol Pharmacol. 2009;75:92–100. doi: 10.1124/mol.108.050492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pires MM, Hrycyna CA, Chmielewski J. Biochemistry. 2006;45:11695–11702. doi: 10.1021/bi0608109. [DOI] [PubMed] [Google Scholar]
- 18.Joslyn AF, Luchkowski E, Triggle DJ. Dimeric 1,4-dihydropyridines as calcium channel antagonists. J Med Chem. 1988;31:1489–1492. doi: 10.1021/jm00403a002. [DOI] [PubMed] [Google Scholar]
- 19.Natale NR, Quincy DA. Synth Commun. 1983;13:817–822. [Google Scholar]
- 20.Schauer CK, Anderson OP, Quincy DA, Natale NR. Acta Crystallogr C, Cryst Struct Commun. 1986;C42:884–886. [Google Scholar]
- 21.Zamponi G, Stotz SC, Staples RJ, et al. J Med Chem. 2003;46:87–96. doi: 10.1021/jm020354w. [DOI] [PubMed] [Google Scholar]
- 22.Hansen TV, Wu P, Folkin VV. J Org Chem. 2005;70:7761–7764. doi: 10.1021/jo050163b. [DOI] [PubMed] [Google Scholar]
- 23.Himo F, Lovell T, Hilgraf R, et al. J Am Chem Soc. 2005;127:210–216. doi: 10.1021/ja0471525. [DOI] [PubMed] [Google Scholar]
- 24.Kondaiah GCM, Reddy LA, Babu KS, et al. Tetrahedron Lett. 2008;49:106–109. [Google Scholar]
- 25.Triggle DJ, Langs DA, Janis RA. Med Res Rev. 1989;9:123–180. doi: 10.1002/med.2610090203. [DOI] [PubMed] [Google Scholar]
- 26.Steiger SA, Monacelli AJ, Li C, Hunting JL, Natale NR. Acta Crystallogr. 2014;E70:o791–o792. doi: 10.1107/S1600536814013294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auffinger P, Hays FA, Westhof E, Ho PS. Proc Natl Acad Sci USA. 2004;101:16789–16794. doi: 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilcken R, Zimmermann MO, Lange A, Joerger AC, Boeckler FM. J Med Chem. 2013;56:1363–1388. doi: 10.1021/jm3012068. [DOI] [PubMed] [Google Scholar]
- 29.Tiberghien F, Loor F. Anticancer Drugs. 1996;7:568–578. doi: 10.1097/00001813-199607000-00012.(b) https://pdspdb.unc.edu/pdspWeb/?site=mdr.
- 30.Hamilton G, Cosentini EP, Teleky B, et al. Anticancer Res. 1993;13:2059–2063. [PubMed] [Google Scholar]
- 31.Natale NR, Triggle DJ, Palmer RB, Lefler BJ, Edwards WD. J Med Chem. 1990;33:2255–2259. doi: 10.1021/jm00170a032. [DOI] [PubMed] [Google Scholar]
- 32.Mirzaei YR, Simpson BM, Triggle DJ, Natale NR. J Org Chem. 1992;57:6271–6279. [Google Scholar]
- 33.Natale NR, Rogers ME, Staples R, Triggle DJ, Rutledge A. J Med Chem. 1999;42:3087–3093. doi: 10.1021/jm980439q. [DOI] [PubMed] [Google Scholar]
- 34.Conn PJ, Lindsley CW, Meiler J, Niswender CM. Nat Rev Drug Discov. 2014;13:692–708. doi: 10.1038/nrd4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang J, Li A, Jang S, et al. J Med Chem. 1999;42:3055–3065. doi: 10.1021/jm980688e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W, Chun E, Thompson AA, et al. Science. 2012;337:222. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aller SG, Yu J, Ward A, et al. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shintre CA, Pike ACW, Li Q, et al. Proc Natl Acad Sci USA. 2013;110:9710–9715. doi: 10.1073/pnas.1217042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, et al. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koska J, Spassov VZ, Maynard AJ, et al. J Chem Inf Model. 2008;48:1965–1973. doi: 10.1021/ci800081s. [DOI] [PubMed] [Google Scholar]
- 41.Jain AN. J Comput Aided Mol Des. 1996;10:427–440. doi: 10.1007/BF00124474. [DOI] [PubMed] [Google Scholar]
- 42.Parrill L, Reddy MR, Gehlhaar DKB, Bouzida D, Rejto P. In: Rational drug design: novel methodology and practical applications. Parrill L, Reddy MR, editors. Washington, DC: [Google Scholar]
- 43.Bohm HJ. J Comput Aided Mol Des. 1994;8:623–632. doi: 10.1007/BF00123669. [DOI] [PubMed] [Google Scholar]
- 44.Ward AB, Szewczyk P, Grimard V, et al. Proc Natl Acad Sci USA. 2013;110:13386–13391. doi: 10.1073/pnas.1309275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steiger SA, Li C, Campana CF, Natale NR. Tetrahedron Lett. 2016;57:423–425. doi: 10.1016/j.tetlet.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.