The LOFSEP transcription factors OsMADS1, OsMADS5, and OsMADS34 regulate rice spikelet morphogenesis, form higher order complexes, and promote the expression of other floral homeotic genes.
Abstract
SEPALLATA (SEP)-like genes, which encode a subfamily of MADS-box transcription factors, are essential for specifying floral organ and meristem identity in angiosperms. Rice (Oryza sativa) has five SEP-like genes with partial redundancy and overlapping expression domains, yet their functions and evolutionary conservation are only partially known. Here, we describe the biological role of one of the SEP genes of rice, OsMADS5, in redundantly controlling spikelet morphogenesis. OsMADS5 belongs to the conserved LOFSEP subgroup along with OsMADS1 and OsMADS34. OsMADS5 was expressed strongly across a broad range of reproductive stages and tissues. No obvious phenotype was observed in the osmads5 single mutants when compared with the wild type, which was largely due to the functional redundancy among the three LOFSEP genes. Genetic and molecular analyses demonstrated that OsMADS1, OsMADS5, and OsMADS34 together regulate floral meristem determinacy and specify the identities of spikelet organs by positively regulating the other MADS-box floral homeotic genes. Experiments conducted in yeast also suggested that OsMADS1, OsMADS5, and OsMADS34 form protein-protein interactions with other MADS-box floral homeotic members, which seems to be a typical, conserved feature of plant SEP proteins.
At present there are an estimated 352,000 species of angiosperms on Earth (The Plant List 2013), which have evolved a variety of inflorescences and flower morphologies (Theissen and Melzer, 2007). The monocot family of grasses (Poaceae) represents nearly one-fifth of all monocotyledons (Linder and Rudall, 2005) and includes many economically significant crops such as rice (Oryza sativa) and maize (Zea mays). Grasses have evolved a surprisingly diversified inflorescence structure and arrangement of spikelets and flowers, which not only differ from eudicot plants but also other monocots. The rice inflorescence meristem is determinate, because it arrests after producing a programmed number of primary and secondary branches, and panicle branching is an important agronomic trait for crop improvement. In all grasses, the basic inflorescence structural unit is the spikelet (Ciaffi et al., 2011; Zhang et al., 2013; Zhang and Yuan, 2014), which in rice contains only one floret and, arranged in a distichous pattern at its base, two highly reduced leaf-like rudimentary glumes and two depressed sterile lemmas (also called empty glumes) above the rudimentary glumes (Kellogg, 2001; Itoh et al., 2005; Yoshida and Nagato, 2011). In addition, rice florets have a bilateral symmetry with five types of floral organs with characteristic numbers, that is one bract-like lemma and one bract-like palea arranged in the adjacent outer whorls, the latter of which is hypothesized to be the first true floral whorl of grasses, two small fleshy organs called lodicules in the second whorl, six stamens in the third whorl, and one pistil in the fourth innermost whorl (Prasad et al., 2001; Prasad and Vijayraghavan, 2003; Zhang and Wilson, 2009; Ciaffi et al., 2011).
The classic ABC model of flower development in angiosperms was formulated more than 25 years ago (Coen and Meyerowitz, 1991). This model, which was based on the observation of mutants with defects in floral organ development, expounds how floral meristem (FM) determinacy and floral organ identity are genetically specified, and shows that the relatively distantly related eudicot flowering plants Arabidopsis (Arabidopsis thaliana L. Heynh.) and snapdragon (Antirrhinum majus) use conserved mechanisms in floral pattern regulation. In Arabidopsis, sepals form in floral whorl 1 because of expression of only the so-called class A genes (APETALA1 [AP1] and AP2), whereas class A and class B (AP3 and PISTILLATA) together direct petal development in whorl 2, B and C (AGAMOUS [AG]) together specify stamens in whorl 3, and class C genes expressed alone determine FM termination and carpel formation in the fourth and last whorl. More recently two gene classes, D and E, have been added to form an extended ABCDE model. The D-class gene (SEEDSTICK [STK]) controls the development of ovules (Pinyopich et al., 2003), while four E-class genes SEPALLATA1/2/3/4 (SEP1/2/3/4; formerly known as AGL2/3/4/9) specify the identities of all four whorls of floral organs and ovules and determine FM identity and determinacy (Pelaz et al., 2000; Pelaz et al., 2001; Favaro et al., 2003; Ditta et al., 2004). Except for AP2, all class A, B, C, D, and E genes encode MIKC-type MADS-box transcription factors.
These MADS-box proteins fall into different subfamilies whose functions are broadly conserved in angiosperms; thus, the ABCDE model can largely explain floral development in monocots such as grasses. Rice has four AP1/FRUITFULL (FUL)-like genes, OsMADS14, OsMADS15, OsMADS18, and OsMADS20, as putative class A genes (Kyozuka et al., 2000; Pelucchi et al., 2002; Fornara et al., 2004; Kater et al., 2006; Preston and Kellogg, 2006; Wu et al., 2017). The analysis of rice floral homeotic mutants for the AP3-like gene OsMADS16/SUPERWOMAN1 and the PI-like genes OsMADS2 and OsMADS4 showed that the function of B-class genes is conserved from grasses to eudicots (Kang et al., 1998; Nagasawa et al., 2003; Prasad and Vijayraghavan, 2003; Yamaguchi et al., 2006; Yadav et al., 2007; Yoshida et al., 2007; Yao et al., 2008; Yun et al., 2013). In rice, there are two AG orthologous genes, OsMADS3 and OsMADS58, and two STK orthologs, OsMADS13 and OsMADS21, which function as class C and class D genes, respectively (Lopez-Dee et al., 1999; Kyozuka and Shimamoto, 2002; Yamaguchi et al., 2006; Dreni et al., 2007; Dreni et al., 2011; Hu et al., 2011). Grasses have diverse SEP-like genes divided into five conserved lineages, which are represented in rice by OsMADS1, OsMADS5, OsMADS7 (allelic to OsMADS45), OsMADS8 (allelic to OsMADS24), and OsMADS34 (Greco et al., 1997; Kang et al., 1997; Malcomber and Kellogg, 2005; Zahn et al., 2005; Arora et al., 2007; Ciaffi et al., 2011). All the SEP-like genes of angiosperms form two major clades, the LOFSEP clade (also called SEP1/2/4 clade or AGL2/3/4 clade) comprising the rice genes OsMADS1, OsMADS5, and OsMADS34, and the SEP3 clade (also called AGL9 clade) including the two rice genes OsMADS7(45) and OsMADS8(24) (Malcomber and Kellogg, 2005; Zahn et al., 2005; Arora et al., 2007). The rice LOFSEP gene OsMADS1/LHS1 (LEAFY HULL STERILE1) was the first E-class gene to be partially characterized in grasses. OsMADS1 controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. The mutation of OsMADS1 causes elongated leafy palea and lemmas, an open hull, two pairs of leafy palea- or lemma-like lodicules, the decrease of stamen number, the increase of carpel number, and an additional floret arising from the same rachilla (Jeon et al., 2000; Prasad et al., 2001, 2005; Agrawal et al., 2005; Hu et al., 2015). No obvious defect in either panicles or vegetative organs was observed in the loss-of-function mutation of the LOFSEP gene OsMADS5, except for the lodicules remaining more tightly attached to the lemma and palea upon spikelet dissection (Agrawal et al., 2005). The LOFSEP gene OsMADS34 (PANICLE PHYTOMER2 [PAP2]) is required for rice inflorescence and spikelet development (Gao et al., 2010; Kobayashi et al., 2010; Lin et al., 2014). The osmads34-1 mutant displayed altered inflorescence morphology with increased primary branch number and decreased secondary branch number. Moreover, the osmads34-1 mutant had fewer spikelets and altered spikelet morphology, with elongated sterile lemmas acquiring lemma/palea-like features (Gao et al., 2010). OsMADS34/PAP2 is also involved in the transition from vegetative to reproductive growth through the specification of inflorescence meristem identity. Indeed, OsMADS34 and three AP1/FUL-like genes, OsMADS14, OsMADS15, and OsMADS18, coordinately act in the meristem to specify inflorescence meristem, downstream of the florigen signal (Kobayashi et al., 2012). As for the members of the SEP3 clade, knockdown of both OsMADS7(45) and OsMADS8(24) showed severe phenotypes including late flowering, homeotic changes of lodicules, conversion of stamens and carpels into palea/lemma-like organs, and a loss of floral determinacy (Cui et al., 2010). Therefore, all of the above SEP-like floral homeotic genes in rice show both functional conservation (E function) and diversification. In addition, AGL6 is an ancient subfamily of MADS-box genes, sister to the SEPs and comprising the rice genes OsMADS6/MOSAIC FLORAL ORGANS1 and OsMADS17 (Zahn et al., 2005; Ohmori et al., 2009). The characterization of rice osmads6 mutant alleles indicated that the AGL6-like genes contribute to the E-function in floral development, like the SEP genes (Ohmori et al., 2009; Li et al., 2010, 2011; Zhang et al., 2010), and this has been observed in other angiosperms (Rijpkema et al., 2009; Thompson et al., 2009; Dreni and Zhang, 2016).
Although the class E floral homeotic genes in rice have already been studied to define their role in molecular mechanisms regulating flower development, the presence of the five SEP genes has been an obstacle in elucidating their global functions, as rice is a much less amenable model plant in which to construct higher order mutants, compared to Arabidopsis. Therefore, the function of OsMADS5 and the genetic interactions among SEP-like genes are still largely unknown. In an effort to address this question, Cui et al. (2010) took an RNA interference (RNAi) knockdown approach targeting all rice SEP genes except OsMADS34, which led to the homeotic transformation of all floral organs except the lemma into leaf-like organs. However, the specific functions of the rice LOFSEP and SEP3 clades are still not clear, although there is evidence of more functional divergence than in Arabidopsis, since the inhibition of just OsMADS1 or OsMADS34, alone or of both these genes together, is sufficient to cause severe phenotypes (Jeon et al., 2000; Prasad et al., 2001, 2005; Agrawal et al., 2005; Cui et al., 2010; Gao et al., 2010; Kobayashi et al., 2010). To date, OsMADS5 remains the less studied SEP-like gene in rice, where only one mutant allele has been generated, which suggested only a minor role for this gene in spikelet development (Agrawal et al., 2005).
In this study, we characterized the genetic interaction of the LOFSEP members OsMADS1, OsMADS5, and OsMADS34 and explored their molecular mechanisms in controlling rice spikelet morphogenesis. To address this question, we used the stable mutant alleles osmads1-z (Gao et al., 2010; Hu et al., 2015) and osmads34-1 (Gao et al., 2010) and we identified and characterized the new osmads5-3 mutant allele. We constructed all possible double mutant combinations, namely osmads1-z osmads34-1 (Gao et al., 2010), osmads1-z osmads5-3, and osmads5-3 osmads34-1, and the triple mutant osmads1-z osmads5-3 osmads34-1, to obtain a detailed comparison of their spikelet phenotypes. Altogether, our data reveal that OsMADS5 is a key player in rice spikelet morphogenesis together with OsMADS1 and OsMADS34. Moreover, genetic interaction analysis of the LOFSEP members OsMADS1, OsMADS5, and OsMADS34 reveals their redundant role in regulating the identity of sterile lemma, lemma and palea, and also of all the inner floral organs, where their latter function is very likely to be dependent on the transcriptional activation of B-, C-, AGL6-, and SEP3-like genes. These new insights help to better understand the core genetic mechanisms underpinning floral development in rice and its conserved and diverged aspects across the evolution of angiosperms.
RESULTS
Identification of the OsMADS5 T-DNA Insertion Mutant
We searched the rice T-DNA insertion library database (Rice Mutant Database, RMD) and identified one insertion line for the OsMADS5 gene. The T-DNA was inserted in the seventh exon (Supplemental Fig. S1A). Because two Tos17 insertion alleles were previously reported (Agrawal et al., 2005), we named this T-DNA mutant osmads5-3. Three pairs of primers for quantitative reverse transcription PCR (qRT-PCR) were used to quantify OsMADS5 transcripts, revealing that the insertion caused a significant reduction of transcript when primer sets upstream of the T-DNA were used, but no transcript could be detected using the primer pair downstream of the T-DNA. It is therefore highly likely that most of the C terminus from the translated protein product was not present in osmads5-3 plants (Supplemental Fig. S1, B–D). The osmads5-3 line was subsequently backcrossed with its wild-type parent Zhonghua11 (Oryza sativa L. ssp. japonica) three times.
Expression Pattern of OsMADS5
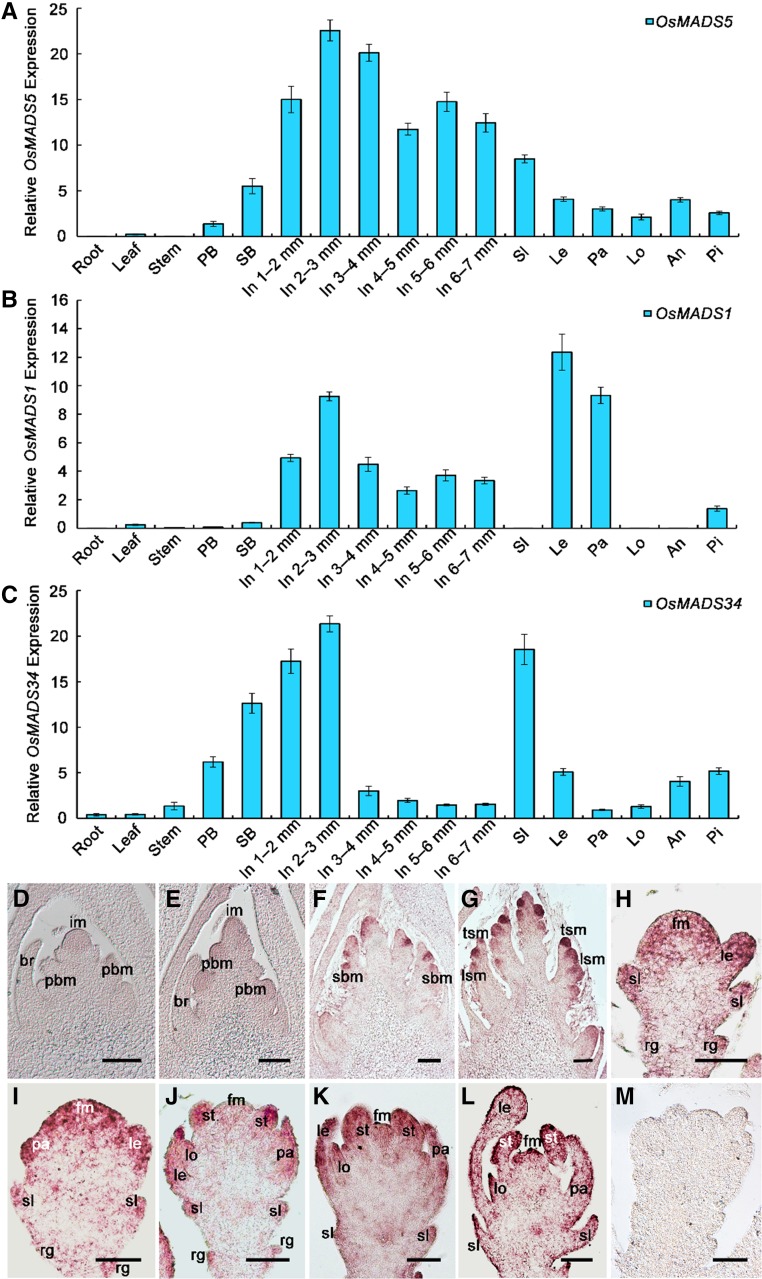
Based on previous studies in rice, the developmental courses of inflorescence and spikelet development have been divided into nine and eight stages, respectively (Ikeda et al., 2004). qRT-PCR analysis revealed a broad expression pattern of OsMADS5 in wild-type reproductive tissues. This included young inflorescences from 0.3 to 7 mm, which span the inflorescence and spikelet development stages from In3 to In5 and from Sp1 to Sp8, respectively, but also the mature sterile lemmas, lemmas, paleas, lodicules, anthers, and pistils at stage In8 to In9 when FM activity is terminated. No transcripts of OsMADS5 were detected in vegetative tissues like roots or leaves (Fig. 1A). Subsequently, RNA in situ hybridization was performed to study the spatial distribution of OsMADS5 mRNA, where faint signals indicated that OsMADS5 mRNA accumulation was first detected at comparatively low levels in the rachis meristem and primary branch primordia during early inflorescence development (Fig. 1, D and E). Subsequently, the OsMADS5 mRNA signal strengthened and was specific in the secondary branch meristem and spikelet meristem (SM) at stage In5 (Fig. 1F) and at the terminal and lateral SMs (Fig. 1G), persisting in the floret meristem during spikelet organogenesis. In spikelet and floral organs, OsMADS5 was expressed at the tips of the rudimentary glumes and sterile lemmas as well as in the lemma primordia at stage Sp3 (Fig. 1H), then in lemma and palea primordia at stage Sp4, and finally in the primordia of lodicules, stamens, and carpels from stage Sp6 to Sp8 (Fig. 1, I–L).
Figure 1.
Expression pattern analysis of rice LOFSEP genes. A to C, qRT-PCR detection of OsMADS5 (A), OsMADS1 (B), and OsMADS34 (C) transcripts. Total RNA was isolated from wild-type roots, leaves, stems, sterile lemmas, lemmas, paleas, lodicules, anthers, and pistils at stage In8 to In9 and also from 0.3- to 7-mm inflorescences. The inflorescence length around 0.3- to 0.6-mm covers the stages of In3 and In4 when the primary branches form and elongate, while inflorescence length 0.6 to 0.9 mm corresponds to stage In5 when the primary branch meristem produces secondary branches, but the SM has not yet formed. The results are presented as mean ± sd. Error bars indicate the SD for three biological replications. D to M, In situ hybridization analysis of OsMADS5 expression in the wild type at stage In3 (D), In4 (E), In5 (F), early In6 (G), Sp3 (H), Sp4 (I), Sp6 (J), Sp7 (K), and Sp8 (L), and at stage Sp6 with sense control (M). an, anther; br, bract; fm, floral meristem; im, florescence meristem; In, inflorescence; le, lemma; lo, lodicule; lsm, lateral spikelet meristem; pa, palea; PB, primary branch; pbm, primary branch meristem; pi, pistil; rg, rudimentary glume; SB, secondary branch; sbm, secondary branch meristem; sl, sterile lemma; tsm, terminal spikelet meristem. Bars = 100 µm (D–G) and 50 µm (H–M).
OsMADS5 and OsMADS34 Redundantly Regulate the Identity of Outer Spikelet Organs and Meristem Determinacy
A wild-type spikelet consists of two rudimentary glumes, two normal sterile lemmas, one lemma, one palea, two lodicules, six stamens, and one pistil (Fig. 2, A–D). Compared with the wild type, no obvious abnormal phenotype was detected in osmads5-3 (Fig. 2, E–H), and the abnormal attachment of lodicules to lemma and palea previously reported for another osmads5 mutant (Agrawal et al., 2005) was not observed. Since our T-DNA allele might not cause a complete loss of function of OsMADS5, we recently analyzed mutant lines created by the CRISPR/Cas9 system, which contained the mutations in the first or the second exon of OsMADS5, respectively (Supplemental Fig. S1A). Again, these lines did not show any visible floral phenotype compared with the wild type (Supplemental Fig. S5; Supplemental Tables S6 and S7), suggesting that the putative function of OsMADS5 might be completely hidden by genetic redundancy. Indeed, dramatically stronger phenotypes were scored for the double and triple mutants described below, carrying the homozygous T-DNA insertion in the OsMADS5 gene.
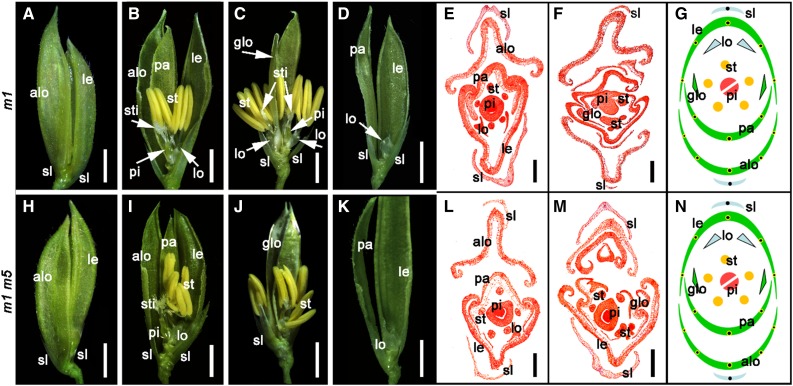
Figure 2.
Phenotypes of wild type, osmads5-3, osmads34-1, and osmads5-3 osmads34-1 mutants. A, E, I, and M, Spikelets of wild type (A), osmads5-3 (E), osmads34-1 (I), and osmads5-3 osmads34-1 (M) at stage In9. B, F, J, and N, Lemma and palea were removed in wild type (B) and osmads5-3 osmads34-1 (N) and half the lemma and palea were removed in osmads5-3 (F) and osmads34-1 (J) to show the inner floral organs of A, E, I, and M, respectively. C, G, K, and O, Transverse section of wild type (C), osmads5-3 (G), osmads34-1 (K), and osmads5-3 osmads34-1 (O) at stage In9 showing the identity change of sterile lemma and defects of inner floral organs in the mutants. Black stars mark the number of vascular bundles. D, H, L, and P, Diagrammatic representation of wild-type (D), osmads5-3 (H), osmads34-1 (L), and osmads5-3 osmads34-1 (P) spikelets. Q to Z, SEM analysis of the abaxial epidermis of wild-type sterile lemma (Q), lemma (R), palea (T), marginal region of palea (mrp) (U), lodicule (V), leaf blade (W), adaxial surface of wild type lemma (S), abaxial epidermis of lemma/palea-like sterile lemma of osmads34-1 (X), and osmads5-3 osmads34-1 (Y), and the abnormal elongated lodicule of osmads5-3 osmads34-1 with an mrp-like structure (Z). Black arrowheads indicate the stomata. elo, elongated lodicule; le, lemma; lo, lodicule; lsl, lemma/palea-like sterile lemma; m5, osmads5-3; m34, osmads34-1; pa, palea; pi, pistil; sl, sterile lemma; st, stamen; sti, stigma. Bars = 2 mm (A, B, E, F, I, J, M, and N), 200 µm (C, G, K, and O), 50 µm (R, S, T, U, W, and Y), and 10 µm (Q, V, X, Z).
To investigate the genetic interaction between OsMADS5 and OsMADS34, we crossed osmads5-3 with the previously identified mutant osmads34-1 (Gao et al., 2010) to generate the double mutant. Consistent with previous reports, the defects observed in osmads34-1 were limited to the conversion of sterile lemmas into elongated lemma/palea-like structures (Fig. 2, I–L), also consistent with the phenotype of CRISPR lines targeting the second exon of OsMADS34 (Supplemental Figs. S1E and S5; Supplemental Tables S6 and S7). The osmads5-3 osmads34-1 double mutant displayed a similar phenotype to the osmads34-1 single mutant, except the lemma/palea-like structures were even longer and, in addition, new abnormalities appeared in the lemma, palea, and inner whorls of floral organs, such as elongated lodicules and a change in the number of stamens and pistils (Fig. 2, M–P).
These differences were quantified, showing that osmads5-3 osmads34-1 displayed significantly longer lemma/palea-like sterile lemmas (12.01 ± 0.64 mm, n = 100) compared to osmads34-1 (9.01 ± 0.32 mm, n = 100), to osmads5-3 (3.12 ± 0.09 mm, n = 100), and to the wild type (3.12 ± 0.08 mm, n = 100; Supplemental Figs. S2 and S4B). There was also a modest increase in vascular bundle numbers per sterile lemma (5.27 ± 0.45, n = 30) compared with osmads34-1 (5.00 ± 0.00, n = 30), osmads5-3 (1.00 ± 0.00, n = 30), and the wild type (1.00 ± 0.00, n = 30), respectively (Supplemental Table S5).
The lemma/palea length of osmads5-3 osmads34-1 was 8.87 ± 0.24 mm (n = 100), which was significantly longer than the wild type, and each of the osmads5-3 and osmads34-1 single mutant (Supplemental Figs. S2 and S4A). About 59.56% (n = 273) of osmads5-3 osmads34-1 spikelets exhibited abnormal elongated lodicules in the second whorl (Supplemental Table S1). The stamen number in osmads5-3 osmads34-1 fluctuated, but the average number of stamens per spikelet was 5.89 ± 0.87 (n = 273; Supplemental Figure S4, C–F; Supplemental Table S1). In addition, the average number of pistils and stigmas in osmads5-3 osmads34-1 was 1.05 ± 0.26 (n = 273) and 2.55 ± 0.83 (n = 273) per spikelet, respectively, displaying a statistically significant increase compared with the wild type and with the two single mutants osmads5-3 and osmads34-1 (Supplemental Fig. S4, C–F; Supplemental Table S1).
Scanning electron microscopy (SEM) analysis was performed to examine abnormalities in osmads5-3 osmads34-1 in detail. For osmads5-3 osmads34-1, the variation in the number of stamen primordia was obvious at Sp6 and the sterile lemmas were larger at Sp8, compared to the wild type and the two single mutants osmads5-3 and osmads34-1 (Supplemental Fig. S3, A–D). Unlike the sterile lemmas of the wild type with a smooth epidermal surface (Fig. 2Q), the abaxial (outer) epidermal surface of osmads5-3 osmads34-1 and osmads34-1 displayed a pattern with regularly arrayed bulges (Fig. 2, X and Y), which showed obvious similarities with the abaxial epidermal surface of wild-type lemma/palea (Fig. 2, R and T), suggesting a homeotic transformation of each sterile lemma into a lemma/palea-like structure. In the second whorl of osmads5-3 osmads34-1 spikelets, the abaxial epidermal surface in most of the elongated lodicules exhibited a normal cellular morphology compared with the wild type, while occasionally displaying conversion to a structure resembling the marginal region of the palea (mrp) (Fig. 2, U, V and Z).
Taken together, these phenotypes reveal redundant regulatory roles of OsMADS5 and OsMADS34 not only in the outer bracts, that is the sterile lemmas, lemma, and palea, but also in the three innermost whorls of floral organs. Thus, OsMADS5 and OsMADS34 redundantly specify the identity of the sterile lemmas, lemma/palea, and lodicules as well as control the number of stamen and carpel primordia through the maintenance of meristem determinacy.
Osmads5-3 Partially Rescues the Defects of osmads1-z in the Second Whorl
To investigate the genetic interaction between OsMADS5 and OsMADS1, we generated a double mutant by crossing osmads5-3 with the previously identified mutant osmads1-z (Gao et al., 2010). Based on the previous analysis of OsMADS1 mutants (Jeon et al., 2000; Agrawal et al., 2005; Prasad et al., 2005; Chen et al., 2006; Gao et al., 2010; Khanday et al., 2013) and on their inner organ defects, we grouped the various phenotypes of osmads1-z into five types in our study (Supplemental Table S2): Type I (77.54%, n = 187) was classified as a weak phenotype with an additional lemma/palea-like organ, normal lodicules on the lemma side and/or an extra pair of glume/mrp-like organs in the second whorl, a decreasing number of stamens, and an increasing number of carpels and stigmas (Fig. 3, A, B and E; Supplemental Fig. S3E). Type II (1.60%, n = 187) was represented by twin florets closely packed together (Fig. 3, C and F); type III (6.95%, n = 187) had normal lodicules inside the lemma and palea but lacked any stamens and pistil (Fig. 3D); type IV (13.37%, n = 187) displayed a complete loss of the inner three whorls of floral organs, and finally type V (0.53%, n = 187) had an additional new spikelet borne on an elongated pedicel (Supplemental Fig. S3E2). The phenotypes of the osmads1-z osmads5-3 double mutant were very similar to the single osmads1-z, so they were also classified into five types (Fig. 3, H–M; Supplemental Fig. S3F). About 74.33% (n = 187) and 9.63% (n = 187) of the spikelets of osmads1-z osmads5-3 exhibited the type I and type III phenotypes, respectively (Supplemental Table S2). In the second whorl, the total number of organs per spikelet (1.94 ± 0.65, n = 143) in osmads1-z osmads5-3 did not change significantly compared with osmads1-z (1.99 ± 0.70, n = 149), whereas the ratio of normal lodicules to extra glume/mrp-like organs appeared remarkably altered (Supplemental Table S3). In the second whorl of osmads1-z, approximately 34.22% of the spikelets examined displayed normal lodicules, and the average number per spikelet of these lodicules was reduced to 0.81 ± 0.96 (n = 149), whereas 65.78% of the spikelets showed the abnormal extra glume-like organs at an average number of 1.19 ± 0.92 (n = 149). While in osmads1-z osmads5-3, a reduced percentage (only 37.76%) of the spikelets presented the extra glume-like organs with the significantly decreased average number of 0.60 ± 1.10 (n = 143), and the remaining 62.24% of the spikelets showed normal lodicules with the significantly increased average number of 1.27 ± 0.91 (n = 143; Supplemental Table S3). These results suggested that simultaneous mutation of both OsMADS5 and OsMADS1 can partially rescue the second whorl defects of osmads1-z, yet no significant differences were noticed in the other spikelet organs between osmads1-z osmads5-3 and osmads1-z.
Figure 3.
Phenotypes of osmads1-z and osmads1-z osmads5-3 mutants. A and H, Spikelets of osmads1-z (A) and osmads1-z osmads5-3 (H) at stage In9. B to D and I to K, The whole lemma and palea were removed in osmads1-z (C) and osmads1-z osmads5-3 (J) spikelets with type II phenotype; half the lemma, palea, and additional lemma/palea-like organs were removed in osmads1-z (B) and osmads1-z osmads5-3 (I) spikelets with type I phenotype; and half the lemma and palea were removed in osmads1-z (D) and osmads1-z osmads5-3 (K) spikelets with type III phenotype to show the inner floral organs at stage In9. E, F, L, and M, Transverse section of osmads1-z with type I phenotype (E) and type II phenotype (F) and osmads1-z osmads5-3 with type I phenotype (L) and type II phenotype (M) at stage In9 showing the defects in spikelets. G and N, Diagrammatic representations of osmads1-z (G) and osmads1-z osmads5-3 (N) spikelets. alo, additional lemma/palea-like organ; glo, extra glume-like second whorl organ; le, lemma; lo, lodicule; m1, osmads1-z; m5, osmads5-3; pa, palea; pi, pistil; sl, sterile lemma; st, stamen; sti, stigma. Bars = 2 mm (A–D and H–K) and 200 µm (E, F, M, and N).
OsMADS5, OsMADS1, and OsMADS34 Together Determine the Spikelet Identity
To investigate the reproductive roles of the three LOFSEP-like members OsMADS5, OsMADS1, and OsMADS34, we further generated the osmads1-z osmads5-3 osmads34-1 triple mutant and reanalyzed the osmads1-z osmads34-1 double mutant that was identified previously (Gao et al., 2010) as a control, because among the double mutant combinations it was the most similar to the triple mutant. OsMADS1 is not expressed in the sterile lemmas of wild type, but its expression has been reported in the lemma/palea-like sterile lemmas of osmads34-z mutants (Ren et al., 2016). Consistently, the osmads1-z osmads34-1 spikelet displayed abnormally elongated and leafy sterile lemmas and lemma/palea. It also showed both extra leaf/glume/mrp-like organs and normal lodicules in the second whorl, plus a decreased number of stamens and increased number of pistils compared with the wild type and each single mutant of osmads1-z and osmads34-1 (Fig. 4, A and C). The osmads1-z osmads5-3 osmads34-1 triple mutant displayed more severe defects of the spikelet organs, including the significantly elongated leafy sterile lemmas (15.61 ± 1.41 mm, n = 100) and lemma/palea (38.08 ± 5.46 mm, n = 100) compared to those of osmads1-z osmads34-1 at an average length of 9.08 ± 0.31 mm (n = 100) and 17.90 ± 2.17 mm (n = 100), respectively (Fig. 4B; Supplemental Figs. S2 and S4, A and B). Moreover, for osmads1-z osmads5-3 osmads34-1, the average number of vascular bundles per sterile lemma (6.75 ± 0.85, n = 20) and per lemma/palea (13.06 ± 1.26, n = 20) were also notably increased compared to osmads1-z osmads34-1 with an average number of 6.00 ± 0.73 (n = 20) and 9.75 ± 1.64 (n = 20), respectively (Fig. 4, I–N; Supplemental Table S5). These differences suggest that a more severe homeotic transformation of the sterile lemma and lemma/palea into leaves occurred in the triple mutant. A further reduction of organs occurred in the inner three whorls of osmads1-z osmads5-3 osmads34-1, including a reduced number of elongated glume-like organs arranged in the second whorl, and the sharply reduced number of stamens, pistils, and stigmas (1.51 ± 0.80 1.28 ± 0.93 and 2.50 ± 1.67, respectively, n = 103) in comparison with osmads1-z osmads34-1 (Fig. 4, C–F; Supplemental Fig. S4, C–F; Supplemental Table S4). Also visible was the growth of undifferentiated tissues or a cluster of additional FMs defective in determinacy (Fig. 4, G, M, and N) instead of the determinate FMs of osmads1-z osmads34-1. In addition, the total number of leaf-like lemmas/paleas per spikelet was significantly increased in osmads1-z osmads5-3 osmads34-1 (3.54 ± 0.50, n = 103) compared with osmads1-z osmads34-1 (2.00 ± 0.00, n = 102) and the wild type (2.00 ± 0.00, n = 200), resulting in an increased total number of outer spikelet organs including the sterile lemmas and the lemma/paleas (5.86 ± 0.67, n = 103, occasionally up to 11 in the most severely affected spikelets; Fig. 4H; Supplemental Table S4). This suggested the homeotic transformation of floral organs of the inner three whorls into outer whorl/leaf-like organs in osmads1-z osmads5-3 osmads34-1, or, alternatively, a delay in the switch of SM into FM leading to a consequent reiteration of outer whorls.
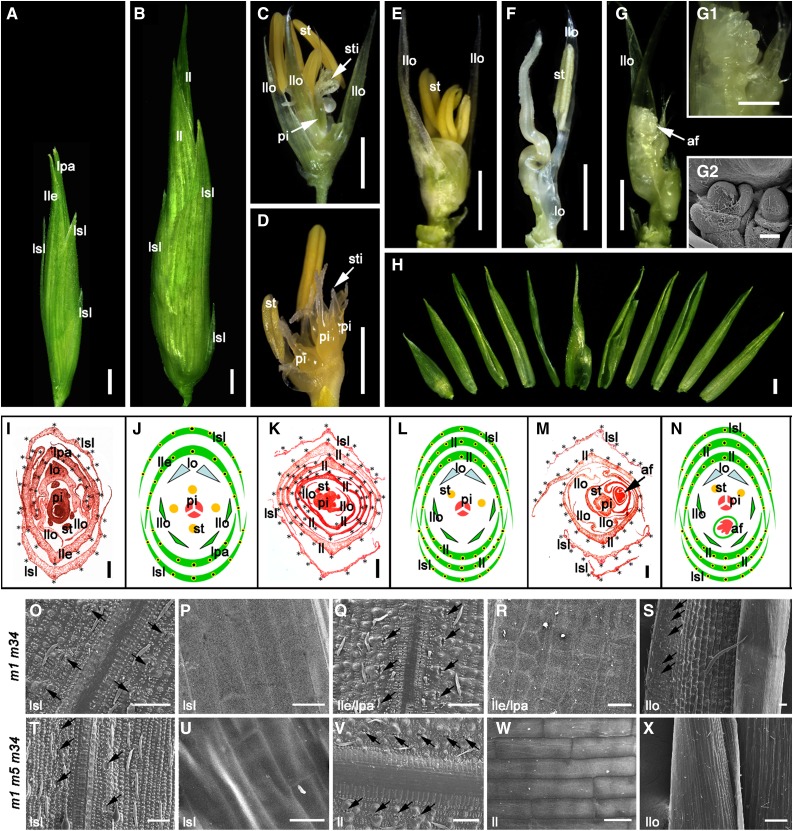
Figure 4.
Phenotypes of osmads1-z osmads34-1 and osmads1-z osmads5-3 osmads34-1 mutants. A and B, Spikelets of osmads1-z osmads34-1 (A) and osmads1-z osmads5-3 osmads34-1 (B) at stage In9. C to G, Elongated leaf-like sterile lemma, lemma and palea were removed in osmads1-z osmads34-1 (C) and osmads1-z osmads5-3 osmads34-1 (D–G) to show the inner floral organs at stage In9. G1, A close-up view of the additional floral meristems marked by white arrowheads in G. G2, SEM analysis of the additional floral meristems marked by white arrowheads in G. H, All the leaf-like sterile lemmas, lemmas and paleas which were dissected from the same spikelet shown in G to show the increased number of outer spikelet organs in osmads1-z osmads5-3 osmads34-1. I, K, and M, Transverse section of osmads1-z osmads34-1 (I) and osmads1-z osmads5-3 osmads34-1 (K and M) at stage In9. Black stars mark the number of vascular bundles. J, L, and N, Diagrammatic representations of osmads1-z osmads34-1 (J) and osmads1-z osmads5-3 osmads34-1 (L and M) spikelets. O to S, SEM analysis of the abaxial epidermis of osmads1-z osmads34-1 leaf-like sterile lemma (O), leaf-like lemma/palea (Q), and a mixed epidermal pattern of extra second whorl organs consisting of leaf-like (left side), lemma/palea-like (middle), and a marginal region of the palea-like (mrp-like) structures (right side) (S), and the inner/adaxial surface of leaf-like sterile lemma (P) and lemma/palea (R). T to X, SEM analysis of the abaxial epidermis of osmads1-z osmads5-3 osmads34-1 leaf-like sterile lemma (T), leaf-like lemma/palea (V), mrp-like extra lodicules (X), and the adaxial surface of leaf-like sterile lemma (U) and lemma/palea (W). Black arrowheads indicate the stomata. af, additional floral meristems; ll, leaf-like lemma/palea; lle, leaf-like lemma; llo, leaf/glume/mrp-like second whorl organ; lpa, leaf-like palea; lsl, leaf-like sterile lemma; m1, osmads1-z; m5, osmads5-3; m34, osmads34-1; pi, pistil; st, stamen; sti, stigma. Bars = 2 mm (A–H), 1 mm (G1), 200 µm (I, K, and M), 50 µm (G2, O, Q, S, T, V, and X), and 20 µm (P, R, U, and W).
SEM analysis was used to gain a better understanding of the identity of the aberrant spikelet organs observed in the mutants. During spikelet development, osmads1-z osmads5-3 osmads34-1 sterile lemmas and lemma/palea were already abnormally enlarged at Sp6 and Sp8, as seen in osmads1-z osmads34-1 (Supplemental Fig. S3, G2–G4, and H3–H4), but more defects were detected inside the triple mutant spikelet, such as double FMs (Supplemental Fig. S3H2). Because sterile lemmas and lemma/palea had a leafy appearance in the mutants, we compared them with mature leaves. In wild-type leaf blades, the abaxial (lower) epidermal surface showed small bulges and several ordered lines of stomata along both sides of each vein (Fig. 2W). For osmads1-z osmads5-3 osmads34 and osmads1-z osmads34-1, the corresponding abaxial (outer) epidermal surface of the elongated sterile lemmas and lemma/palea exhibited similar patterns to wild-type leaf blades, except that only one line of stomata was found along both sides of each vein (Fig. 4, O, Q, T, and V), while the adaxial (inner) surface displayed smooth rectangular cells reminiscent of the lemma and palea adaxial surface rather than the adaxial (upper) leaf blade surface (Figs. 2S and 4, P, R, U, and W). These results indicated a partial homeotic transformation of the sterile lemma and lemma/palea to leaf-like structures in the double and triple mutant, especially on the adaxial side. The outer surface of the elongated glume-like second whorl organs was also examined, and a mixed epidermal pattern consisting of leaf-like, lemma/palea-like, and mrp-like structures was observed in osmads1-z osmads34-1 (Fig. 4S). However, only mrp-like cells were observed in the corresponding ectopic organs of the triple mutant (Fig. 4X), revealing that the mutation of OsMADS5 reduced the range of second whorl defects in osmads1-z osmads34-1 double mutants. The results above indicated that the three LOFSEP-like members OsMADS5, OsMADS1, and OsMADS34 together regulate the identity of sterile lemma, lemma, palea, and lodicule and the determinacy of FM and determine the correct initiation of stamen and pistil.
LOFSEP Genes Positively Regulate the Expression of Floral Homeotic Genes
To further study the regulatory function of rice LOFSEP members, qRT-PCR was performed to measure transcript levels of most of the other MADS-box genes known to be involved in flower development. Young inflorescences from 2 mm to 7 mm long were examined in the lofsep mutants and the wild type, covering five developmental time points across spikelet development, from Sp4 to Sp8.
The transcripts of two AP1/FUL-like genes OsMADS14 and OsMADS15 were marginally increased in lofsep mutants, in particular in the triple mutant, compared with their parental varieties Zhonghua11 and 9522 (Fig. 5, A and B).
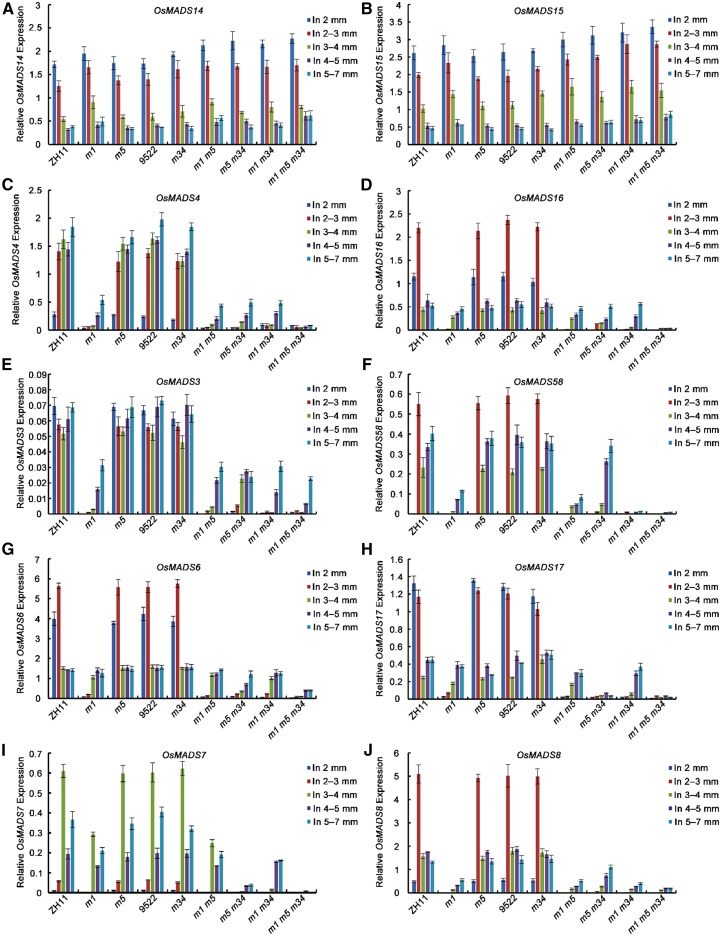
Figure 5.
Expression analysis of some floral homeotic genes in the young panicles of lofsep mutants and the wild-type backgrounds. A to J, Expression levels of class A (OsMADS14 and OsMADS15) (A and B), class B (OsMADS4 and OsMADS16) (C and D), class C (OsMADS3 and OsMADS58) (E and F), AGL6-like (OsMADS6 and OsMADS17) (G and H), and SEP3-like (OsMADS7 and OsMADS8) (I and J) genes were detected by qRT-PCR. Total RNA was isolated from 2- to 7-mm young inflorescences of the mutants of osmads1-z, osmads5-3, osmads34-1, osmads1-z osmads5-3, osmads5-3 osmads34-1, osmads1-z osmads34-1, and osmads1-z osmads5-3 osmads34-1 and also the wild-type parents Zhonghua11 and 9522. The results are presented as mean ± sd. The error bars indicate the SD for three biological replications. In, inflorescence; m1, osmads1-z; m5, osmads5-3; m34, osmads34-1; ZH11, Zhonghua 11.
The expression levels of other floral homeotic genes, that is the two SEP3-like genes OsMADS7(45) and OsMADS8(24), the PI-like gene OsMADS4, the AP3-like gene OsMADS16, and the two AG lineage genes OsMADS3 and OsMADS58 were significantly decreased in osmads1-z, especially in the early stages, but did not change significantly in the osmads5-3 and osmads34-1 single mutants. Similarly, compared to osmads1-z, the expression level of these homeotic genes did not show obvious changes in osmads1-z osmads5-3 or osmads1-z osmads34-1, except for OsMADS58 at extremely low levels in the latter. But in the osmads5-3 osmads34-1 double mutant, all genes tested were strongly suppressed across all developmental stages examined (Fig. 5, C–F, I and J). The AGL6-like genes OsMADS6 and OsMADS17 were down-regulated significantly only at the earliest developmental stages in osmads1-z and all the double mutants, except for OsMADS17, which was strongly down-regulated in osmads5-3 osmads34-1 from stage Sp4 to Sp8 (Fig. 5, G and H). In the osmads1-z osmads5-3 osmads34-1 triple mutant, SEP3-, PI-, AP3-, AG-, and the AGL6-like genes were strongly suppressed, except for OsMADS3 in 4 to 5-mm and 5 to 7-mm panicles (spikelet stages Sp7 and Sp8; Fig. 5, C–J).
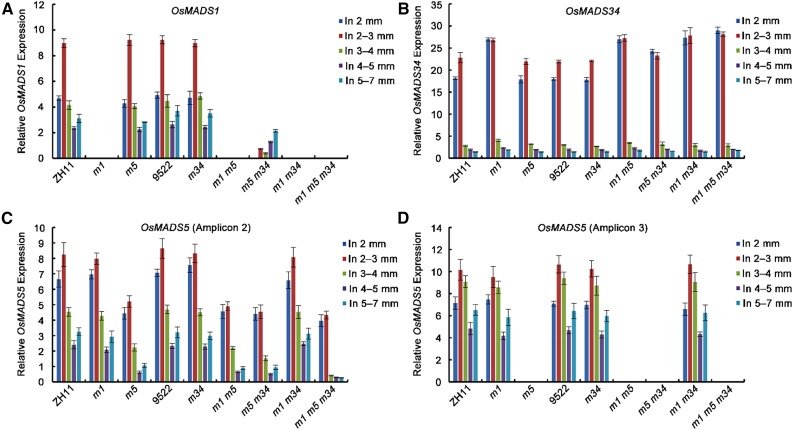
Subsequently, the expression of the LOFSEP genes themselves was examined in their mutant backgrounds. According to our previous work, no notable change in the amount of OsMADS34 transcript was detected in the osmads34-1 frame-shift mutant, which is caused by a change in the 3′ acceptor splice site in the fifth intron of the gene (Gao et al., 2010). In contrast, OsMADS34 was moderately up-regulated in osmads1-z, consistently with previous data (Khanday et al., 2013), and also in osmads1-z osmads5-3, osmads1-z osmads34-1, and osmads1-z osmads5-3 osmads34-1 (Fig. 6B). The OsMADS1 transcript was not detectable in any mutant background that included osmads1-z, confirming that osmads1-z is a full knock-out mutant. Interestingly, OsMADS1 was down-regulated in the osmads5-3 osmads34-1 double mutant, suggesting that OsMADS5 and OsMADS34 may act redundantly as positive regulators of OsMADS1 (Fig. 6A). Two pairs of primers were used to detect the OsMADS5 transcripts in the lofsep mutants, either upstream (Fig. 6C) or downstream (Fig. 6D) of the T-DNA insertion site in osmads5-3, respectively. The OsMADS5 transcript was decreased when amplified using the upstream primer set (Fig. 6C) and was depleted using the downstream primer set (Fig. 6D) in all of the mutant combinations involving osmads5-3 but was not altered in osmads1-z and osmads34-1 single or double mutants.
Figure 6.
Expression analysis of LOFSEP genes in young panicles of lofsep mutants and wild-type backgrounds. A to D, Expression levels of LOFSEP genes OsMADS1 (A), OsMADS34 (B), and OsMADS5 (C and D) were detected by qRT-PCR. The second and third primer pairs shown in Supplemental Figure S1A were used to quantify OsMADS5 transcripts upstream (C) and downstream (D) of the T-DNA insertion site, which causes the osmads5-3 mutant, respectively. Total RNA was isolated from 2- to 7-mm young inflorescences from lofsep mutants and their wild-type backgrounds. The results are presented as mean ± sd. The error bars indicate the SD for three biological replications. In, inflorescence; m1, osmads1-z; m5, osmads5-3; m34, osmads34-1; ZH11, Zhonghua 11.
These results suggested that rice LOFSEP members act, mostly redundantly, as upstream activators of B- and C-class genes as well as of AGL6-like and SEP3-like genes. LOFSEP genes may regulate the expression of each other, in particular OsMADS5 and OsMADS34 seem to promote OsMADS1, which in turn could be involved in the fine tuning of OsMADS34 expression.
Physical Interactions of OsMADS5 and OsMADS34 with Other Floral Homeotic Proteins
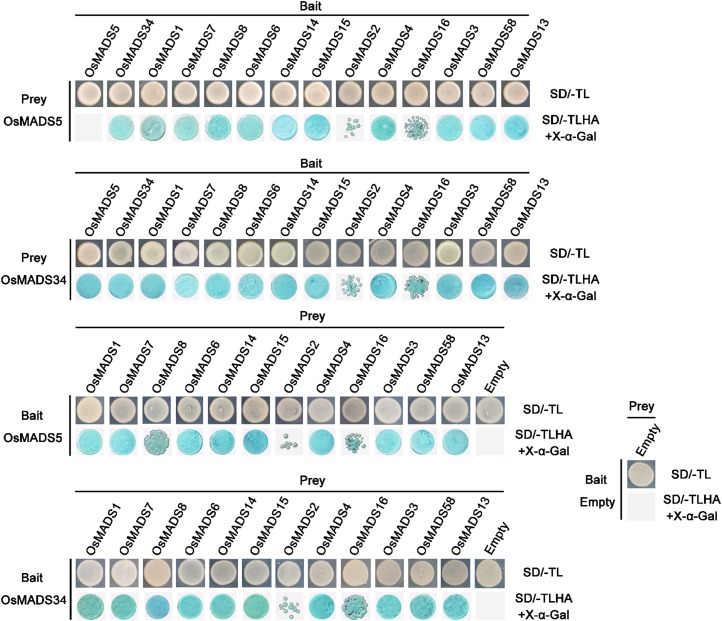
In Arabidopsis, the identity of the different floral organs, sepals, petals, stamens, and carpels is determined by four combinations of floral homeotic MADS-box proteins, as described by the “quartet model” of floral organ specification (Theissen and Saedler, 2001). Therefore, we performed yeast two-hybrid assay to investigate whether OsMADS5 and OsMADS34 can interact with other MADS-box proteins. We found that OsMADS34 is able to form homodimers, in contrast to OsMADS5, while all the three LOFSEP-like members could interact with each other to form heterodimers. Both OsMADS5 and OsMADS34 could form heterodimers with other A-, B-, C-, D-, and E-class as well as AGL6-like proteins including OsMADS7(45), OsMADS8(24), OsMADS6, OsMADS14, OsMADS15, OsMADS2, OsMADS4, OsMADS16, OsMADS3, OsMADS58, and OsMADS13, respectively, albeit the interaction with OsMADS2 and OsMADS16 appeared weak (Fig. 7). These results suggested that the two LOFSEP proteins OsMADS5 and OsMADS34 have the ability to form higher order complexes together with class A, B, C, D, E, or AGL6-like proteins to control multifarious transcriptional programs required for flower development.
Figure 7.
Interaction analysis of OsMADS5 and OsMADS34 with other floral homeotic MADS-box proteins. Yeast two-hybrid assay shows the interaction patterns of rice LOFSEP members OsMADS5 and OsMADS34 with class A (OsMADS14 and OsMADS15), class B (OsMADS2, OsMADS4 and OsMADS16), class C (OsMADS3 and OsMADS58), class D (OsMADS13), AGL6-like (OsMADS6), SEP3-like (OsMADS7 and OsMADS8), and LOFSEP-like (OsMADS1, OsMADS5, and OsMADS34) proteins, respectively. The transformants were grown on selective Minimal Synthetic Dropout (sd) media at high stringency (SD/-Ade/-His/-Leu/-Trp/X-α-Gal). Cotransformants with empty vectors pGADT7 and pGBKT7 were used as negative controls. A, adenine; H, His; L, Leu; T, Trp.
DISCUSSION
LOFSEP Genes Are Key Regulators of Rice Spikelet Organogenesis
In recent years, the function of SEP-like genes has gradually been characterized in rice. OsMADS7(45) and OsMADS8(24) are functionally redundant genes whose simultaneous silencing affects the development of only lodicules, stamens, and carpels in the innermost three whorls, consistent with their expression pattern (Cui et al., 2010).
OsMADS34 has been shown to be a key regulator of rice inflorescence architecture and of sterile lemma identity (Gao et al., 2010; Kobayashi et al., 2010, 2012). In our present study, the mutation of both OsMADS5 and OsMADS34 caused not only an increase in outer whorl defects greater than the mutation of OsMADS34 alone, but also affected the inner whorls. Thus, there was an increased number of organs in the second whorl that acquired a palea-like identity rather than resembling lodicules, a variable number of stamens, and an increased number of carpels in the innermost two whorls. The number of stigmas also increased, due to the extra carpels and also because most carpels had more than two stigmas. This mimics the defects in the three inner whorls caused by the double knockdown of OsMADS7(45) and OsMADS8(24) (Cui et al., 2010) and also resembles the mild phenotype of osmads1-z. These phenotypes correlate strongly with the sharp decrease and delay of the onset of expression of OsMADS7(45) and OsMADS8(24), along with B function, C function, and AGL6-like genes, during spikelet formation in the osmads1-z and osmads5-3 osmads34-1 plants (Fig. 5, C–J). The phenotypic defects of the osmads5-3 osmads34-1 double mutant reveal the functional redundancy between the two genes, as suggested by their sequence similarity and their highly overlapping expression patterns during spikelet organogenesis.
Our comparison of osmads1-z and osmads1-z osmads5-3 revealed similar phenotypes suggesting that OsMADS1 is mostly epistatic to OsMADS5. However, despite the total number of second whorl organs being similar in these two mutants, the ratio of extra glume-like organs and normal lodicules was nearly halved in the double mutant, suggesting that the mutation of OsMADS5 in the osmads1-z background could partially recover the defects of osmads1-z in the second whorl.
Compared with osmads1-z, osmads1-z osmads34-1 shows slightly enhanced defects of inner floral organs, suggesting that OsMADS34 is also involved in determining their identity redundantly with OsMADS1 (Gao et al., 2010). In our study, more severe defects were found in the osmads1-z osmads5-3 osmads34-1 triple mutant, compared to osmads1-z osmads34-1, including the enhanced elongation of leaf-like sterile lemmas and leaf-like lemma/paleas, the reduction in the number of elongated mrp-like second whorl organs, normal lodicules and reproductive organs, and defective meristem determinacy. Moreover, the total number of the leaf-like organs in the outer whorls was dramatically increased. This suggests either the homeotic transformation from the organs of the innermost three whorls to the leaf-like structures of the outer whorls, and/or the SM-to-FM transition is gravely affected in osmads1-z osmads5-3 osmads34-1. A previous study has shown that simultaneous knockdown of four SEP-like genes OsMADS1, OsMADS5, OsMADS7(45), and OsMADS8(24) led to homeotic transformation of the lodicule and reproductive organs into leaf-like organs, while the lemma and palea were marginally affected (Cui et al., 2010). Our osmads1-z osmads5-3 osmads34-1 triple mutant showed floral organs phenotypes very similar to those of the quadruple RNAi lines in the innermost three whorls, whereas the severe conversion to leaf-like organs in the outer whorls were not observed in the quadruple RNAi lines. In addition, the spikelet of osmads1-z osmads5-3 osmads34-1 exhibited an elongated, slender, and cylindrical pattern with little empty space inside, and the average length of the elongated leaf-like sterile lemmas and leaf-like lemma/paleas were 5.03 and 4.86 times that of wild type, and 2.92 and 2.28 times that of osmads1-z osmads34-1, respectively (Supplemental Figs. S2 and S4, A and B), causing the osmads1-z osmads5-3 osmads34-1 spikelets to become more like vegetative shoots. In summary, the SEP subfamily genes are functionally diversified in rice, with the SEP3-like genes showing restricted functions in the innermost three whorls, as in Arabidopsis and Petunia x hybrida hort. ex E.Vilm. (Mandel and Yanofsky, 1998; Pelaz et al., 2000; Ferrario et al., 2003; Vandenbussche et al., 2003; Rijpkema et al., 2009), while the function of LOFSEP genes starts earlier, in the specification of all spikelet and floral organs and also influencing meristem identity and determinacy as well.
Conservation and Diversification of SEPs in Arabidopsis and Rice
Phylogenetic analysis has revealed that the class E genes are clustered into an angiosperm-specific SEP subfamily that consists of two major, ubiquitous clades, called LOFSEP and SEP3, which probably arose via the whole genome duplication preceding the most recent common ancestor of angiosperms (Malcomber and Kellogg, 2005; Zahn et al., 2005). In Arabidopsis, SEP1, SEP2, and SEP4 (LOFSEP clade) are expressed throughout the FM at stage 2 and continue in the primordia of all floral organs: sepals, petals, stamens, carpels, and ovules, while the expression of SEP3 starts slightly later in late stage 2 floral primordia and continues in the inner three whorls rather than in sepals (Flanagan and Ma, 1994; Huang et al., 1995; Savidge et al., 1995; Mandel and Yanofsky, 1998; Ditta et al., 2004), suggesting that Arabidopsis SEP genes may encode functionally redundant proteins. In agreement with this, single SEP gene mutations produce only subtle or no phenotypes, while in a sep1 sep2 sep3 triple mutant the floral organs in the innermost three whorls turn into ectopic sepals and an even more striking phenotype occurs in sep1 sep2 sep3 sep4 quadruple mutant, where the floral organs of all four whorls are converted into leaf-like organs, indicating the central roles of the SEP genes in specifying FM and organ identity (Pelaz et al., 2000; Pelaz et al., 2001; Ditta et al., 2004). However, the sep1 sep2 sep4 (lofsep) triple mutant shows no significant perturbation of floral organ development, suggesting that SEP3 is significantly more critical for flower development than other SEPs (Ditta et al., 2004).
The expression patterns of rice SEP3-like genes OsMADS7(45) and OsMADS8(24) share much similarity with SEP3 in eudicots (Pelucchi et al., 2002; Cui et al., 2010), suggesting that they share similar conserved roles. Supportively, simultaneous knockdown of OsMADS7(45) and OsMADS8(24) in rice induces strong defects specifically in whorls 2, 3, and 4 (Cui et al., 2010). Despite in situ RNA hybridization results indicating an earlier expression of OsMADS8(24) in the inflorescence branch meristems (Cui et al., 2010), this finding seems in contrast with independent quantitative experiments (Kobayashi et al., 2010; Harrop et al., 2016).
The expression patterns of rice LOFSEP-like genes are diversified compared to their orthologs SEP1, SEP2, and SEP4 in Arabidopsis. In rice, the global expression profile of LOFSEP genes starts earlier than that of the SEP3 genes and is associated with the development of the outer spikelet organs. Rice sterile lemmas, lemma, and, eventually, the palea represent new bract-like organs surrounding the flower, which evolved in the monocot grass lineage, and LOFSEP genes were clearly neofunctionalized and recruited to regulate their identity. This effect might be indirect and dependent on the LOFSEP function in SM identity specification: as the SM identity is increasingly lost in lofsep mutant combinations, the glumes are gradually converted toward a default leaf identity. A similar mechanism has been proposed to explain the effect of A-class mutants on sepal identity in Arabidopsis, alternatively to the existence of a true and direct A function (reviewed by Causier et al., 2010). On the other hand, rice LOFSEP genes continue to be expressed during the formation and growth of glume primordia. OsMADS34 is expressed in the inflorescence meristem and branch meristems, and later in the primordia of rudimentary glumes and sterile lemmas. During later stages, OsMADS34 is expressed in the primordia of lemma, palea, stamens, carpels, and ovules but is absent from the lodicule primordia (Gao et al., 2010; Kobayashi et al., 2010; Lin et al., 2014), although in this study we detected its expression in mature lodicules (Fig. 1C). The expression of OsMADS5 starts in branch meristems (Kobayashi et al., 2010). In this study, we showed that OsMADS5 is increasingly expressed in the SM and subsequently in the FM. Notably, OsMADS5 was expressed in the primordia of all spikelet organs including the tip of rudimentary glumes and sterile lemmas, and the primordia of lemma, palea, lodicules, stamens, and carpel (Fig. 1, A and D–L). Furthermore, OsMADS5 and OsMADS34 showed an overlapping pattern of mRNA accumulation in all the mature spikelet organs, that is sterile lemmas, lemma, palea, lodicules, anthers, and pistil at stage In8 to In9 when the FM terminated (Fig. 1, A and C). In contrast, the transcript of OsMADS1 is not detected during panicle branching; its earliest expression is detected in the SM, but not in the rudimentary glumes and sterile lemmas, then is confined to the lemma and palea; within the floral whorls, it is weakly expressed only in the carpel but undetectable in lodicules or stamens (Chung et al., 1994; Prasad et al., 2001; Hu et al., 2015; Fig. 1B). Despite this, the loss of OsMADS1 function causes strong defects in the inner floral whorls (Jeon et al., 2000; Prasad et al., 2001; Agrawal et al., 2005), demonstrating a pivotal role of OsMADS1 in the establishment of the FM. In agreement with their expression maxima (Fig. 1, A–C), OsMADS34 is more important for the development of sterile lemmas, whereas OsMADS1 is required for lemma and palea development. OsMADS5 single mutants have no phenotype; however, all three genes share significant redundancy in the development of both glumes and floral organs, as revealed by double and triple mutants. In Arabidopsis, flower development is not significantly affected even in the lofsep triple mutant sep1 sep2 sep4, whereas we have found drastic effects in the corresponding mutant of rice. In this study, the sterile lemmas, lemmas, and paleas, as well as the inner floral organs of osmads1-z osmads5-3 osmads34-1, were converted into leaf-like structures mimicking the phenotype of sep1 sep2 sep3 sep4 of Arabidopsis. This demonstrates that there is a clear functional conservation of the SEP subfamily in the regulation of floral organ and meristem identity, but in rice we observe a higher degree of subfunctionalization.
Contrary to previous assumptions, we reveal that OsMADS5 actively participates together with its paralogs in spikelet and floral morphogenesis. This is a significant advance in understanding the basal genetic model of flower development in rice, because, previously, OsMADS5 was thought to play a minor role at best (Agrawal et al., 2005). Interestingly, besides its general redundancy with OsMADS1 and OsMADS34, OsMADS5 shows some antagonistic effect in the second whorl, because its mutation partially rescued the formation of extra glume-like organs in osmads1 and osmads1 osmads34-1 mutant backgrounds.
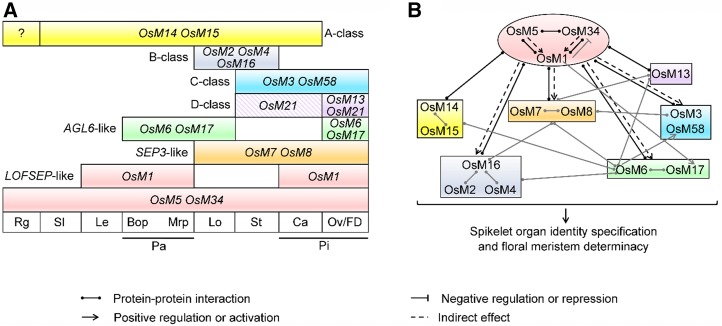
The main implication of the expression patterns of SEP genes is that, in rice, LOFSEP and SEP3 genes are coexpressed with all the other MADS-box genes involved in flower development. Here, the comprehensive comparison of the expression patterns of floral homeotic MADS-box genes enabled us to construct an overarching model of expression profiles during spikelet morphogenesis (Fig. 8A). This model has facilitated the integration of previously published data with the findings of this study.
Figure 8.
A working model summarizing the roles of homeotic MADS-box genes during rice spikelet development. A, The expression domains of A-, B-, C-, D-, and E-class and AGL6-like genes in different spikelet organs. During the spikelet development, the expression of the FUL1-like gene OsMADS14 is found in the SM; later restricted to the sterile lemmas, lemma, and palea (Pelucchi et al., 2002; Preston and Kellogg, 2006, 2007); and the FUL2-like gene OsMADS15 is also expressed in the SM and continues in the sterile lemmas, lemma, palea, and lodicules (Kyozuka et al., 2000; Preston and Kellogg, 2006, 2007). The PI/GLO-like genes OsMADS2 and OsMADS4 and the AP3/DEF-like gene OsMADS16 share a common conserved expression domain in the lodicule and stamen primordia (Nagasawa et al., 2003; Yadav et al., 2007; Yun et al., 2013). In general, the AG lineage genes OsMADS3 and OsMADS58 exhibit a very similar expression profile in the stamen, carpel, and ovule primordia (Dreni et al., 2011). The expression of the AGL11 lineage gene OsMADS13 is specific in the ovule primordium (Lopez-Dee et al., 1999; Dreni et al., 2011), and yet the other AGL11 lineage gene OsMADS21 is expressed in the developing ovule integument and very weakly in stamens and carpels (Arora et al., 2007; Dreni et al., 2007; Dreni et al., 2011). The two AGL6-like genes OsMADS6 and OsMADS17 show a largely overlapping expression patterns in the FM and, later, in palea, lodicule, and ovule (Favaro et al., 2002; Pelucchi et al., 2002; Ohmori et al., 2009; Li et al., 2010). B, Genetic and physical interactions of MADS-box factors in regulating rice spikelet morphogenesis. Black arrows indicate the interactions presented in this study and gray arrows show the interactions reported in previous works (see manuscript text for references). Bop, body of palea; Ca, carpel; FD, floral meristem determinacy; Le, lemma; Lo, lodicule; Mrp, marginal region of palea; OsM, OsMADS; Ov, ovule; Pa, palea; Pi, pistil; Rg, rudimentary glume; Sl, sterile lemma; St, stamen.
LOFSEP-Like Members of Rice Are Upstream Activators of Class B, C, D, E, and AGL6-Like Genes
Another key difference in rice is that the expression of SEP3-like genes seems to be activated by LOFSEPs, which is not observed in Arabidopsis. In osmads1-z osmads5-3 osmads34-1, the expression of SEP3-like genes, like one of the AGL6-like, B, and C genes, is dramatically delayed and reduced during floral organogenesis from the stages of lemma primordium, FM, and palea primordium initiation (Sp3/Sp4) to the formation of floral organs and the termination of FM (Fig. 5, C–J). Yet this apparent function of LOFSEP genes as activators may not be absolute, as also in the triple mutant the conversion of the SM into FM is retarded not abolished, and defective or reduced floral organs still develop after several whorls of leaf-like bracts. However, because osmads5-3 and omads34-1 may not be knock-out mutations, this assumption needs to be verified with more alleles in the future.
In Arabidopsis, the B and C genes do not strictly depend on SEP to be activated in the floral primordium (Pelaz et al., 2000), although SEP proteins probably bind to their promoters to ensure proper expression levels (Kaufmann et al., 2009). SEP3 itself does not appear to act downstream of LOFSEP genes in Arabidopsis, as in the sep1 sep2 sep4 triple (lofsep) mutant is still expressed and is sufficient to provide the E function (Ditta et al., 2004). Therefore, our analysis suggests that the rice LOFSEP genes may regulate floral organ identity and meristem determinacy through the initial activation of all the other floral homeotic MADS-box genes. The hypothesis that these genes are direct targets of LOFSEP proteins is a likely scenario but challenging to test because of the difficulty in developing efficient antibodies for chromatin immunoprecipitation, although using this method we have previously shown that OsMADS17, one of the rice AGL6-like members, is a direct downstream target of OsMADS1 (Hu et al., 2015). Regarding the relationship among the LOFSEP-like members, OsMADS1 is down-regulated in the osmads5-3 osmads34-1 double mutant, but the expression level of OsMADS34 is increased in osmads1-z, suggesting that OsMADS5 and OsMADS34 may together promote OsMADS1 during spikelet morphogenesis and that OsMADS1 may antagonize OsMADS34. This is consistent with previous reports that OsMADS1 promotes and ensures the SM to FM transition by reducing the expression of the SM identity factors such as OsMADS34 in young spikelets, probably by direct targeting, and contributes to the maintenance of FM identity and its determinate development (Jeon et al., 2000; Agrawal et al., 2005; Prasad et al., 2005; Chen et al., 2006; Gao et al., 2010; Khanday et al., 2013; Xu et al., 2017). However, we show that the SM to FM transition is further delayed in osmads1-z osmads5-3 osmads34-1, suggesting that OsMADS34 may function either as an SM or an FM identity gene, perhaps depending on the genetic context and presence of stage specific interaction partners. In addition, OsMADS34 may repress OsMADS1 in developing sterile lemmas, based on recent reports (Ren et al., 2016).
LOFSEP Proteins Participate in Rice Floral Quartets
The SEP proteins appear to act as the “glue” mediating the assembly of ABCD floral homeotic proteins into alternative MADS-box complexes specifying different floral whorls in eudicots (Theissen, 2001; Theissen and Saedler, 2001; Malcomber and Kellogg, 2005; Immink et al., 2009). The SEP proteins of both rice and Arabidopsis either form homodimers or heterodimers to interact with A-, B-, C-, and D-class and AGL6-like proteins (Honma and Goto, 2001; Favaro et al., 2002, 2003; Cooper et al., 2003; Lee et al., 2003; de Folter et al., 2005; Kater et al., 2006; Cui et al., 2010). For example, to date OsMADS1 has been shown to interact with OsMADS14 and OsMADS15 (A-class), OsMADS16 (B-class), OsMADS3 and OsMADS58 (C-class), OsMADS13 (D-class), OsMADS6 (AGL6-like), OsMADS7, and OsMADS8 (SEP3-like), and to form homodimers weakly (Moon et al., 1999b; Lim et al., 2000; Cui et al., 2010; Hu et al., 2015). Similarly, OsMADS34 can form homodimers, or heterodimers with OsMADS14, OsMADS15, and OsMADS18, OsMADS58, OsMADS6, and OsMADS7 (Kobayashi et al., 2012; Lin et al., 2014). In this study, we reveal that OsMADS5 and OsMADS34 have a similar ability to interact with candidate class A, B, C, D, E, and AGL6-like floral homeotic proteins. Previously, no direct interactions were detected when using the K domain of OsMADS16 or the KC domain of OsMADS4 as bait and the KC domain of OsMADS5 as prey (Moon et al., 1999a; Lee et al., 2003). Here, we observed heterodimerization using the extended IKC domain of OsMADS4 and OsMADS16, and the full-length protein of OsMADS5. These results confirmed that I and K domains are essential to mediate protein interactions while the C domain plays a crucial role in the formation of higher order complexes (Moon et al., 1999a; Causier et al., 2010; Lin et al., 2014).
Therefore, both LOFSEP and SEP3 proteins can interact with B, C, and AGL6-like proteins. Moreover, apart from the absence of OsMADS1 in the second and third whorls, LOFSEP genes are consistently expressed in all floral organs, demonstrated in this work and by independent data, indicating that rice LOFSEP proteins likely participate in the formation of the putative floral quartets or of functionally equivalent complexes (Fig. 8, A and B). However, because SEP3-like genes are dramatically down-regulated in lofsep mutants, genetic tests are much more complicated in rice than in Arabidopsis and will require further studies in the future.
In summary, the MADS-box floral quartet complexes and even higher order complexes with non-MADS transcription factors may also be formed in rice and other monocots, similar to those surmised in core eudicots (West et al., 1998; Egea-Cortines et al., 1999; West and Sharrocks, 1999; Honma and Goto, 2001; Theissen and Saedler, 2001). In yeast two-hybrid screenings using LOFSEP proteins as baits, the AP1/FUL-like proteins OsMADS14 and OsMADS15 emerged as the MADS-box factors with the highest affinity (Lim et al., 2000; Q. Meng, W. Liang, and D. Zhang, unpublished data). Their encoding genes are coexpressed with LOFSEP genes in both SMs and FMs. Interestingly, a recent analysis of these two transcription factors underlined their essential role in inflorescence development and spikelet development (Wu et al., 2017), with double mutants showing leafy-like bracts, papery lodicules, and other floral defects reminiscent of the osmads1-z osmads5-3 osmads34-1 phenotype. The combination of previous data and molecular models predicting SEP-AP1 complexes as activators of B and C function genes in Arabidopsis (Honma and Goto, 2001; de Folter et al., 2005; Gregis et al., 2009), and the higher order mutants between AP1, CAL, SEP1/2/4 as well as SEP4 overexpression, confirmed that the LOFSEP genes of Arabidopsis are indeed promoters of FM identity (Ditta et al., 2004). The results presented here suggest that the same mechanism has been only partially rewired during the evolution of grass reproductive organs.
MATERIALS AND METHODS
Plant Materials
The three rice single mutants osmads5-3, osmads1-z, and osmads34-1 were used in this study. Subsequently, three CRISPR/Cas9 lines osmads5(M)-CRISPR, osmads5(I)-CRISPR, and osmads34(I)-CRISPR were developed to confirm the mutant phenotypes. The parental cultivar of osmads5-3 and osmads1-z is Zhonghua11 (Oryza sativa L. ssp. japonica) while for osmads34-1, osmads5(M)-CRISPR, osmads5(I)-CRISPR, and osmads34(I)-CRISPR it is 9522 (Oryza sativa L. ssp. japonica). Both osmads1-z and osmads34-1 have been previously described by our group (Gao et al., 2010; Hu et al., 2015). Double and triple mutants were isolated by genotyping and phenotype observation. All plant materials used in this study were grown in the paddy field at Shanghai Jiao Tong University (at 31°2′3.55″ North latitude, 121°26′39.70″ East longitude) from May to September.
Genotyping of lofsep Mutant Plants
For genotyping the T-DNA line osmads5-3, the PCR primer set OsM5 GF and OsM5 GR was used to detect the OsMADS5 wild-type allele and OsM5 GF with NTLB5 for the osmads5-3 allele. For the CRISPR mutant lines osmads5(M)-CRISPR, osmads5(I)-CRISPR and osmads34(I)-CRISPR, the genotyping was performed by PCR amplification and sequencing using three primer sets OsM5-C G1F and OsM5-C G1R, OsM5-C G2F and OsM5-C G2R, and OsM34-C GF and OsM34-C GR, respectively. The genotyping of osmads1-z and osmads34-1 plants was described previously (Gao et al., 2010; Hu et al., 2015). All primers are listed in Supplemental Table S8.
Morphological Analysis and Microscopy Observation
Fresh spikelets at stage In9 were photographed with a Leica stereomicroscope (S8AP0). For cytological analysis, young spikelets of mutants and wild type were fixed in fresh FAA solution (50% ethanol, 5% acetic acid, and 3.7% formaldehyde) and dehydrated through a series of graded ethanol solutions (70% twice, 80%, 90% and 95% once each, and 100% twice). Samples for paraffin sectioning were infiltrated with Histo-Clear II, embedded in Paraplast Plus, sectioned into 6-μm-thick slices using a Leica rotary microtomes (RM2245), deparaffinized, and photographed under a Nikon microscope (Eclipse 80i). The samples for SEM were prepared and analyzed as described previously (Li et al., 2006).
cDNA Preparation and qRT-PCR Analysis
For the expression analysis of OsMADS1, OsMADS5, and OsMADS34, total RNA of three biological replicates was extracted using Trizol Reagent (Invitrogen) from young panicles between 0.3 to 7 mm long, thus covering broad inflorescence and spikelet developmental stages of In4, In5 and Sp1 to Sp8, and also from mature sterile lemmas, lemmas, paleas, lodicules, anthers, and pistils at stage In8 to In9.
To analyze the expression of floral homeotic genes in lofsep mutants, young inflorescences from 2 mm to 7 mm long were harvested from the lofsep mutants and from the wild type, covering five time-points across spikelet development. Indeed a 2-mm-long inflorescence in the wild-type cultivars Zhonghua11 or 9522 has mostly spikelet primordia at the Sp4 stage, with a few still at stage Sp3. When the inflorescence length becomes 2 to 3 mm, most of the spikelet primordia are at Sp5, with a small number at Sp4 or Sp6. Similarly, we sampled inflorescences at 3 to 4 mm, 4 to 5 mm, or 5 to 7 mm long, which mostly bear stage Sp6, Sp7, and Sp8 spikelet primordia, respectively.
The cDNA was synthesized using the PrimeScript RT reagent Kit with gDNA Eraser (Takara). The qRT-PCR analysis was performed using the CFX96 Real-Time PCR Detection System (Bio-Rad) with SuperReal PreMix Plus (SYBR Green, TIANGEN). Primers for OsMADS16, OsMADS58, OsMADS7, and OsMADS8 were described by (Cui et al., 2010); OsMADS4, OsMADS17, and OsMADS1 by (Hu et al., 2015); OsMADS3 by (Chen et al., 2006); OsMADS6 by (Ohmori et al., 2009); and OsMADS34 by (Gao et al., 2010). All primers are listed in Supplemental Table S8. Data were processed using the CFX Manager software and Excel, based on the relative quantitation delta-delta Ct method.
RNA In Situ Hybridization
Fresh young panicles dissected from wild type were fixed immediately in FAA solution, then dehydrated, infiltrated, embedded, and sectioned as described above in the paraffin section method. The digoxygenin-labeled gene-specific RNA probes of OsMADS5 were generated in vitro from a cDNA PCR amplicon comprising the C terminal of coding region and the 3′-UTR region of OsMADS5. Primers for in situ hybridization are listed in Supplemental Table S8. Digoxygenin-labeled RNA hybridization and hybridized probes detection were performed essentially according to (Dreni et al., 2007). Images were captured on a Nikon microscope (Eclipse 80i).
Yeast Two-Hybrid Assay
The full-length cDNAs of OsMADS5, OsMADS34, OsMADS1, OsMADS7, OsMADS8, OsMADS14, OsMADS15, and OsMADS58; the cDNA fragments encoding the IKC domains of OsMADS2, OsMADS4, OsMADS16, OsMADS3, OsMADS13; and the cDNA fragments encoding the MIK domains as well as the first 14 amino acid residues of C domains of OsMADS6 were amplified and cloned into both the activation-domain vector pGADT7 and the DNA-binding domain vector pGBKT7 (Clontech). Yeast two-hybrid assay was subsequently performed according to the MATCHMAKER GAL4 Two-Hybrid System 3 & Libraries User Manual (Clontech). Protein interactions were tested and no single constructs gave rise to detectable autonomous activation as indicated by yeast growth ability on selective Minimal Synthetic Dropout media with high stringency (Minimal Synthetic Dropout/-Ade/-His/-Leu/-Trp/X-α-Gal). The transformants containing empty plasmids of both pGADT7 and pGBKT7 were used as negative controls.
Accession Numbers
Sequence data from this article can be found in the Rice Genome Annotation Project Database under the following accession numbers: OsMADS5 (Os06g06750), OsMADS1 (Os03g11614), OsMADS34 (Os03g54170), OsMADS7 (Os08g41950), OsMADS8 (Os09g32948), OsMADS6 (Os02g45770), OsMADS17 (Os04g49150), OsMADS14 (Os03g54160), OsMADS15 (Os07g01820), OsMADS2 (Os01g66030), OsMADS4 (Os05g34940), OsMADS16 (Os06g49840), OsMADS3 (Os01g10504), OsMADS58 (Os05g11414), and OsMADS13 (Os12g10540).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The osmads5 and osmads34 mutant alleles.
Supplemental Figure S2. Spikelets of lofsep mutants and of the wild-type parental cultivars.
Supplemental Figure S3. SEM analysis of spikelet primordia from wild type and lofsep mutants.
Supplemental Figure S4. Statistics of spikelet organs of lofsep mutants and the wild type.
Supplemental Figure S5. Phenotypes of wild-type, osmads5, and osmads34 CRISPR lines.
Supplemental Table S1. The number and percentage of floral organs of the osmads5-3, osmads34-1, and osmads5-3 osmads34-1 mutants.
Supplemental Table S2. The classification and percentage of five types of phenotypes presented by osmads1-z and osmads1-z osmads5-3 mutants.
Supplemental Table S3. The number and percentage of spikelet organs of osmads1-z and osmads1-z osmads5-3 mutants.
Supplemental Table S4. The number of spikelet organs of osmads1-z osmads34-1 and osmads1-z osmads5-3 osmads34-1 mutants.
Supplemental Table S5. The number of vascular bundles of lofsep mutants.
Supplemental Table S6. The floral organ number of the CRISPR mutants of OsMADS5 and OsMADS34 compared to osmads5-3 and osmads34-1.
Supplemental Table S7. The number of vascular bundles of the CRISPR mutants of OsMADS5 and OsMADS34 compared to osmads5-3 and osmads34-1.
Supplemental Table S8. Primers used in this study.
Acknowledgments
We thank the staff of Rice Mutant Database for providing osmads5-3, Professor Hongwei Xue for providing osmads1-z, Zhijing Luo for rice cultivation and crosses, Changsong Yin and Jie Wang for technical assistance during in situ hybridization experiments, and Xiaonan Nie for the genotyping of osmads5-3.
Footnotes
This work was supported by the National Natural Science Foundation of China (NSFC) (31230051), the NSFC Research Fund for International Young Scientists (31550110198), the National Key Technologies Research and Development Program of China, Ministry of Science and Technology (grant no. 2016YFD 0100804; 2016YFE0101000), the H2020-MSCA-RISE-2015 project ExpoSEED (691109), the China Postdoctoral Science Foundation (2014M560328), the National Transgenic Major Program (grant no. 2016ZX08009003-003-007), the Innovative Research Team, Ministry of Education; the 111 Project (grant no. B14016), Australian Research Council (DP170103352), an Australia-China Science and Research Fund Joint Research Centre grant (ACSRF48187), and the National Key Technologies Research and Development Program of China, Ministry of Science and Technology (grant no. 2016YFD 0100804). L.D. is supported by a Marie Skłodowska-Curie Global Fellowship (H2020-MSCA-IF-2014_GF, proposal no. 661678 ‘RiceStyle’).
Articles can be viewed without a subscription.
References
- Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H (2005) Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol Biol 59: 125–135 [DOI] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Schwarz-Sommer Z, Davies B (2010) Floral organ identity: 20 years of ABCs. Semin Cell Dev Biol 21: 73–79 [DOI] [PubMed] [Google Scholar]
- Chen ZX, Wu JG, Ding WN, Chen HM, Wu P, Shi CH (2006) Morphogenesis and molecular basis on naked seed rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta 223: 882–890 [DOI] [PubMed] [Google Scholar]
- Chung YY, Kim SR, Finkel D, Yanofsky MF, An G (1994) Early flowering and reduced apical dominance result from ectopic expression of a rice MADS box gene. Plant Mol Biol 26: 657–665 [DOI] [PubMed] [Google Scholar]
- Ciaffi M, Paolacci AR, Tanzarella OA, Porceddu E (2011) Molecular aspects of flower development in grasses. Sex Plant Reprod 24: 247–282 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Cooper B, Clarke JD, Budworth P, Kreps J, Hutchison D, Park S, Guimil S, Dunn M, Luginbühl P, Ellero C, et al. (2003) A network of rice genes associated with stress response and seed development. Proc Natl Acad Sci USA 100: 4945–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z (2010) Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J 61: 767–781 [DOI] [PubMed] [Google Scholar]
- de Folter S, Immink RG, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. (2005) Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PB, An G, Colombo L, Kater MM (2007) The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J 52: 690–699 [DOI] [PubMed] [Google Scholar]
- Dreni L, Pilatone A, Yun D, Erreni S, Pajoro A, Caporali E, Zhang D, Kater MM (2011) Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23: 2850–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreni L, Zhang D (2016) Flower development: the evolutionary history and functions of the AGL6 subfamily MADS-box genes. J Exp Bot 67: 1625–1638 [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M, Saedler H, Sommer H (1999) Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R, Immink RG, Ferioli V, Bernasconi B, Byzova M, Angenent GC, Kater M, Colombo L (2002) Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Mol Genet Genomics 268: 152–159 [DOI] [PubMed] [Google Scholar]
- Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, Yanofsky MF, Kater MM, Colombo L (2003) MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15: 2603–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario S, Immink RG, Shchennikova A, Busscher-Lange J, Angenent GC (2003) The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 15: 914–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan CA, Ma H (1994) Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant Arabidopsis flowers. Plant Mol Biol 26: 581–595 [DOI] [PubMed] [Google Scholar]
- Fornara F, Parenicová L, Falasca G, Pelucchi N, Masiero S, Ciannamea S, Lopez-Dee Z, Altamura MM, Colombo L, Kater MM (2004) Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol 135: 2207–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Liang W, Yin C, Ji S, Wang H, Su X, Guo C, Kong H, Xue H, Zhang D (2010) The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol 153: 728–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco R, Stagi L, Colombo L, Angenent GC, Sari-Gorla M, Pè ME (1997) MADS box genes expressed in developing inflorescences of rice and sorghum. Mol Gen Genet 253: 615–623 [DOI] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Dorca-Fornell C, Kater MM (2009) The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J 60: 626–637 [DOI] [PubMed] [Google Scholar]
- Harrop TW, Ud Din I, Gregis V, Osnato M, Jouannic S, Adam H, Kater MM (2016) Gene expression profiling of reproductive meristem types in early rice inflorescences by laser microdissection. Plant J 86: 75–88 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Hu L, Liang W, Yin C, Cui X, Zong J, Wang X, Hu J, Zhang D (2011) Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23: 515–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Liang W, Yin C, Yang X, Ping B, Li A, Jia R, Chen M, Luo Z, Cai Q, et al. (2015) Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol Plant 8: 1366–1384 [DOI] [PubMed] [Google Scholar]
- Huang H, Tudor M, Weiss CA, Hu Y, Ma H (1995) The Arabidopsis MADS-box gene AGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol Biol 28: 549–567 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Sunohara H, Nagato Y (2004) Developmental course of inflorescence and spikelet in rice. Breed Sci 54: 147–156 [Google Scholar]
- Immink RG, Tonaco IA, de Folter S, Shchennikova A, van Dijk AD, Busscher-Lange J, Borst JW, Angenent GC (2009) SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biol 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Jeon JS, Jang S, Lee S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG, et al. (2000) leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Jang S, Chung JE, Cho YG, An G (1997) Characterization of two rice MADS box genes that control flowering time. Mol Cells 7: 559–566 [PubMed] [Google Scholar]
- Kang HG, Jeon JS, Lee S, An G (1998) Identification of class B and class C floral organ identity genes from rice plants. Plant Mol Biol 38: 1021–1029 [DOI] [PubMed] [Google Scholar]
- Kater MM, Dreni L, Colombo L (2006) Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J Exp Bot 57: 3433–3444 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC (2009) Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA. (2001) Evolutionary history of the grasses. Plant Physiol 125: 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanday I, Yadav SR, Vijayraghavan U (2013) Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways. Plant Physiol 161: 1970–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J (2010) PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol 51: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, Yoshida H, Nagamura Y, Kyozuka J (2012) Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24: 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J, Kobayashi T, Morita M, Shimamoto K (2000) Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiol 41: 710–718 [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Shimamoto K (2002) Ectopic expression of OsMADS3, a rice ortholog of AGAMOUS, caused a homeotic transformation of lodicules to stamens in transgenic rice plants. Plant Cell Physiol 43: 130–135 [DOI] [PubMed] [Google Scholar]
- Lee S, Jeon JS, An K, Moon YH, Lee S, Chung YY, An G (2003) Alteration of floral organ identity in rice through ectopic expression of OsMADS16. Planta 217: 904–911 [DOI] [PubMed] [Google Scholar]
- Li H, Liang W, Hu Y, Zhu L, Yin C, Xu J, Dreni L, Kater MM, Zhang D (2011) Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23: 2536–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liang W, Jia R, Yin C, Zong J, Kong H, Zhang D (2010) The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res 20: 299–313 [DOI] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Moon YH, An G, Jang SK (2000) Two rice MADS domain proteins interact with OsMADS1. Plant Mol Biol 44: 513–527 [DOI] [PubMed] [Google Scholar]
- Lin X, Wu F, Du X, Shi X, Liu Y, Liu S, Hu Y, Theißen G, Meng Z (2014) The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the ‘empty glumes’ of rice (Oryza sativa) spikelets are in fact rudimentary lemmas. New Phytol 202: 689–702 [DOI] [PubMed] [Google Scholar]
- Linder HP, Rudall PJ (2005) Evolutionary history of poales. Annu Rev Ecol Evol Syst 36: 107–124 [Google Scholar]
- Lopez-Dee ZP, Wittich P, Enrico Pè M, Rigola D, Del Buono I, Gorla MS, Kater MM, Colombo L (1999) OsMADS13, a novel rice MADS-box gene expressed during ovule development. Dev Genet 25: 237–244 [DOI] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA (2005) SEPALLATA gene diversification: brave new whorls. Trends Plant Sci 10: 427–435 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF (1998) The ArabidopsisAGL9 MADS box gene is expressed in young flower primordia. Sex Plant Reprod 11: 22–28 [Google Scholar]
- Moon YH, Jung JY, Kang HG, An G (1999. a) Identification of a rice APETALA3 homologue by yeast two-hybrid screening. Plant Mol Biol 40: 167–177 [DOI] [PubMed] [Google Scholar]
- Moon YH, Kang HG, Jung JY, Jeon JS, Sung SK, An G (1999. b) Determination of the motif responsible for interaction between the rice APETALA1/AGAMOUS-LIKE9 family proteins using a yeast two-hybrid system. Plant Physiol 120: 1193–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y (2003) SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718 [DOI] [PubMed] [Google Scholar]
- Ohmori S, Kimizu M, Sugita M, Miyao A, Hirochika H, Uchida E, Nagato Y, Yoshida H (2009) MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21: 3008–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Tapia-López R, Alvarez-Buylla ER, Yanofsky MF (2001) Conversion of leaves into petals in Arabidopsis. Curr Biol 11: 182–184 [DOI] [PubMed] [Google Scholar]
- Pelucchi N, Fornara F, Favalli C, Masiero S, Lago C, Pè EM, Colombo L, Kater MM (2002) Comparative analysis of rice MADS-box genes expressed during flower development. Sex Plant Reprod 15: 113–122 [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF (2003) Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Prasad K, Parameswaran S, Vijayraghavan U (2005) OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J 43: 915–928 [DOI] [PubMed] [Google Scholar]
- Prasad K, Sriram P, Kumar CS, Kushalappa K, Vijayraghavan U (2001) Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev Genes Evol 211: 281–290 [DOI] [PubMed] [Google Scholar]
- Prasad K, Vijayraghavan U (2003) Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ patterning. Genetics 165: 2301–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA (2006) Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae). Genetics 174: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA (2007) Conservation and divergence of APETALA1/FRUITFULL-like gene function in grasses: evidence from gene expression analyses. Plant J 52: 69–81 [DOI] [PubMed] [Google Scholar]
- Ren D, Rao Y, Leng Y, Li Z, Xu Q, Wu L, Qiu Z, Xue D, Zeng D, Hu J, et al. (2016) Regulatory role of OsMADS34 in the determination of glumes fate, grain yield, and quality in rice. Front Plant Sci 7: 1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema AS, Zethof J, Gerats T, Vandenbussche M (2009) The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. Plant J 60: 1–9 [DOI] [PubMed] [Google Scholar]
- Savidge B, Rounsley SD, Yanofsky MF (1995) Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell 7: 721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Plant List (2013) Version 1.1. http://www.theplantlist.org/ (January 2, 2017)
- Theissen G. (2001) Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol 4: 75–85 [DOI] [PubMed] [Google Scholar]
- Theissen G, Melzer R (2007) Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann Bot 100: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Saedler H (2001) Plant biology. Floral quartets. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Thompson BE, Bartling L, Whipple C, Hall DH, Sakai H, Schmidt R, Hake S (2009) bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 21: 2578–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M, Zethof J, Souer E, Koes R, Tornielli GB, Pezzotti M, Ferrario S, Angenent GC, Gerats T (2003) Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 15: 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AG, Causier BE, Davies B, Sharrocks AD (1998) DNA binding and dimerisation determinants of Antirrhinum majus MADS-box transcription factors. Nucleic Acids Res 26: 5277–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AG, Sharrocks AD (1999) MADS-box transcription factors adopt alternative mechanisms for bending DNA. J Mol Biol 286: 1311–1323 [DOI] [PubMed] [Google Scholar]
- Wu F, Shi X, Lin X, Liu Y, Chong K, Theißen G, Meng Z (2017) The ABCs of flower development: mutational analysis of AP1/FUL-like genes in rice provides evidence for a homeotic (A)-function in grasses. Plant J 89: 310–324 [DOI] [PubMed] [Google Scholar]
- Xu W, Tao J, Chen M, Dreni L, Luo Z, Hu Y, Liang W, Zhang D (2017) Interactions between FLORAL ORGAN NUMBER4 and floral homeotic genes in regulating rice flower development. J Exp Bot 68: 483–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav SR, Prasad K, Vijayraghavan U (2007) Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ. Genetics 176: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, Hirano HY (2006) Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao SG, Ohmori S, Kimizu M, Yoshida H (2008) Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development. Plant Cell Physiol 49: 853–857 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Itoh J, Ohmori S, Miyoshi K, Horigome A, Uchida E, Kimizu M, Matsumura Y, Kusaba M, Satoh H, et al. (2007) superwoman1-cleistogamy, a hopeful allele for gene containment in GM rice. Plant Biotechnol J 5: 835–846 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Nagato Y (2011) Flower development in rice. J Exp Bot 62: 4719–4730 [DOI] [PubMed] [Google Scholar]
- Yun D, Liang W, Dreni L, Yin C, Zhou Z, Kater MM, Zhang D (2013) OsMADS16 genetically interacts with OsMADS3 and OsMADS58 in specifying floral patterning in rice. Mol Plant 6: 743–756 [DOI] [PubMed] [Google Scholar]
- Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, Depamphilis CW, Ma H (2005) The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169: 2209–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wilson ZA (2009) Stamen specification and anther development in rice. Chin Sci Bull 54: 2342–2353 [Google Scholar]
- Zhang D, Yuan Z (2014) Molecular control of grass inflorescence development. Annu Rev Plant Biol 65: 553–578 [DOI] [PubMed] [Google Scholar]
- Zhang D, Yuan Z, An G, Dreni L, Hu J, Kater MM (2013) Panicle development. In Zhang Q, Wing RA, eds, Genetics and Genomics of Rice. Springer, New York, NY, pp 279–295 [Google Scholar]
- Zhang J, Nallamilli BR, Mujahid H, Peng Z (2010) OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa). Plant J 64: 604–617 [DOI] [PubMed] [Google Scholar]