Abstract
Cellular responses to low-oxygen stress and to respiratory inhibitors share common mitochondrial energy signaling pathways.
Cells of complex organisms typically rely on mitochondria for energy provision. The amount of energy required to sustain cellular activity can vary strongly depending on external conditions. Vice versa, constraints on respiratory activity due to metabolic status or stress insult require mitochondrial signaling to coordinate cellular physiology with the function of the organelle. In this update, we review recent insights into plant mitochondrial energy signaling in the light of their significance to stress acclimation. First, we focus on the characteristic adjustments of the nuclear transcriptome that occur after pharmacological inhibition of the mitochondrial electron transport chain as the output of mitochondrial retrograde signaling. Second, we discuss the proteins that have recently been identified as regulators of the transcript responses and the emerging picture of their action as a signaling network. We then pose the question of how well our current models of inducing mitochondrial dysfunction relate to conditions that plants face naturally. We reason that low-oxygen stress shows striking similarities with electron transport inhibitors with respect to their impact on mitochondrial energy physiology upstream, as well as the cellular transcriptomic response. Finally, we highlight and discuss changes in mitochondrial physiology that are common to both stimuli as candidates for upstream signals. The aim of this update is to better define the physiological context in which mitochondrial signaling operates to provide new directions for future research.
RESPONSES TO MITOCHONDRIAL ENERGY SIGNALING AT THE TRANSCRIPT LEVEL
The impact of mitochondrial-related stimuli on gene expression has been intensively studied using transcript abundance as a proxy, which has provided a rapidly growing number of genome-scale datasets in the recent years. The signals triggering organelle-related transcriptional changes are largely classified into anterograde and retrograde. While anterograde signaling describes the nuclear control of the mitochondrion, retrograde signals originate from the organelle and induce nuclear transcriptional reprogramming, often referred to as feedback or backward flow of information.
Anterograde signals include programs of plant growth and development. As an example, the abundances of at least 45 nuclear-encoded mitochondrial proteins in photosynthetic Arabidopsis (Arabidopsis thaliana) tissue were shown to fluctuate with the diurnal cycle, probably to adjust organelle function between day and night. Individual tricarboxylic acid (TCA) cycle enzymes were more abundant and enzymatically more active early after the transition from light to dark (Lee et al., 2010). Most of the underlying genes were later shown to possess specific site II elements in their promoter region that bind to the TEOSINTE BRANCHED 1, CYCLOIDEA, PCF1 (TCP) family of transcription factors (Giraud et al., 2010), which are conserved among plants and control a wide range of developmental programs (Martín-Trillo and Cubas, 2010). Diurnal regulation of TCA cycle transcripts has also been demonstrated in unicellular photosynthetic diatoms (Chauton et al., 2013; Smith et al., 2016; Matthijs et al., 2017), green algae (Zones et al., 2015), and prokaryotic cyanobacteria (Stöckel et al., 2008; Welkie et al., 2014), suggesting conservation across photosynthetic organisms. The synthesis rates of mitochondrially encoded transcripts also show diurnal patterns, with lowest rates of transcription observed in Arabidopsis at the end of the day and early in the dark (Okada and Brennicke, 2006). Intriguingly, the steady-state levels of the mitochondrial transcripts remained constant during day and night time, despite fluctuating mRNA synthesis rates. How mitochondrial transcript levels maintained stable is not known, but diurnal changes in RNA decay rates appear to play a role (Okada and Brennicke, 2006). In line with the stability of mitochondrial transcript levels, polysome loading (the number of ribosomes actively translating the same mRNA molecule) of mitochondrial transcripts remained stable and at a relatively high level throughout day and night (Pal et al., 2013). This is in contrast to plastid polysome loading, which is much higher during the light period. Notably, more rapid changes of mitochondrial physiology caused by chemical perturbation of the electron transport chain were found to impact on mitochondrial transcript abundance (Zubo et al., 2014).
Retrograde signals have been widely studied in model systems that perturb mitochondrial function genetically or chemically to monitor the transcriptional response in the nucleus. The control of yeast citrate synthase was the initial model that opened up the field of retrograde signaling. Mitochondrial citrate synthase (CS1) is part of the TCA cycle in the mitochondrial matrix that supplies yeast cells with energy and intermediates required for growth. At CS1 loss (genetic perturbation) or blockage of the electron transport chain through antimycin A (AA; chemical perturbation), the transcript of peroxisomal citrate synthase (CIT2) is induced, which has been interpreted as a mechanism to maintain the provision of biosynthetic intermediates required for cell growth in the absence of a functional TCA cycle (Liao et al., 1991). The authors later found that the up-regulation of CIT2 strictly depends on the transcription factor RTG1 and introduced the concept of retrograde communication (Liao and Butow, 1993).
AA treatment has become an intensively used model of respiratory inhibition and induction of a mitochondrial retrograde response (MRR) also in plants (Yu et al., 2001; Schwarzländer et al., 2012a; Umbach et al., 2012; De Clercq et al., 2013; Ng et al., 2013a; Van Aken et al., 2016a). AA inhibits the mitochondrial electron transport chain (mtETC) at the N site of complex III and leads to an increased rate of superoxide generation (Cadenas et al., 1977; Murphy, 2009). AA treatment of Arabidopsis tissues results in a characteristic transcriptomic response that includes the regulation of chaperones and heat-shock proteins, transporters, as well as the up-regulation of ALTERNATIVE OXIDASE1a (AOX1a) and other components of the alternative respiratory chain, such as NAD(P)H DEHYDROGENASE B2 (NDB2). Alternative oxidases (AOXs) and NAD(P)H dehydrogenases (NDHs) represent an alternative pathway of the ETC that does not commonly exist in animals and can decrease or even fully bypass the production of ATP and reactive oxygen species (ROS) through the classical oxidoreductases of the cyanide-sensitive pathway (Schertl and Braun, 2014). In line with the function of their proteins, AOX and NDH genes are frequently coexpressed, for instance at inhibition of ETC complex I by rotenone or the ATP synthase (complex V) by oligomycin (Clifton et al., 2005, 2006). Like AA, these inhibitors stop electron flow down the respiratory chain, albeit at different steps with different physiological outcomes. The concerted action of transcriptionally elevated alternative electron transport components has been interpreted as an inducible safety valve to counterbalance ROS production independent of adenosine phosphorylation status (Vanlerberghe and McIntosh, 1992; Maxwell et al., 1999; Cvetkovska and Vanlerberghe, 2013). In the third section of this article, we will discuss how AA treatment compares with more physiological stimuli that induce the plant MRR. A general transcriptional overlap with different stress insults has clearly been indicated by multiple studies (Clifton et al., 2005; Van Aken et al., 2009; Umbach et al., 2012; Van Aken and Whelan, 2012). Overlaps with the transcriptomic fingerprints of mutants with functionally impaired mitochondria provide further support (Van Aken et al., 2007; Kühn et al., 2009), although mutants generally show far less differentially expressed genes than AA treatment. This may be explained by acclimation of mutants over their development to adopt a different steady state, while the AA treatment necessitates acute reprogramming (Van Aken and Pogson, 2017). Ease of the treatment and the apparent representative nature of the response for mitochondrial stress established AA treatment as the current standard in investigating the mitochondrial retrograde response as measured by specific marker transcripts (AOX1a, but also transcriptionally more strongly induced markers such as UGT74E2 and UP-REGULATED BY OXIDATIVE STRESS (Ho et al., 2008; Tognetti et al., 2010; De Clercq et al., 2013)).
Not only inhibitors of the mtETC have been used to induce retrograde regulation in plants. Monofluoroacetate (MFA), an inhibitor of TCA cycle enzyme aconitase, also triggers a pronounced transcriptomic response that includes AOX1a and other marker transcripts and partly overlaps with the response to AA inhibition (Vanlerberghe and McLntosh, 1996; Umbach et al., 2012; Ng et al., 2013b; Van Aken et al., 2016b). Aconitase inhibition leads to a rearrangement of fluxes through the cellular metabolic network, and this is likely to include an accumulation of citrate, a candidate retrograde signaling molecule (Buffa and Peters, 1949). Indeed, addition of citrate led to AOX induction in soybean and tobacco (Vanlerberghe and McLntosh, 1996; Djajanegara et al., 2002; Gray et al., 2004). In Arabidopsis, induction of AOX1a by citrate treatments could not be confirmed, and the changes observed were either modest or absent (Ho et al., 2008; Finkemeier et al., 2013). Overall, the transcriptomic effects of externally supplied citrate in Arabidopsis are mild (Finkemeier et al., 2013) compared to those observed in tobacco and soybean (Djajanegara et al., 2002; Gray et al., 2004). In contrast, MFA leads to a very pronounced transcriptional effect in Arabidopsis, which peaks several hours later but is even stronger and longer-lasting than AA (Umbach et al., 2012; Van Aken et al., 2016b). This may indicate that the strong effects caused by MFA are not coupled to citrate accumulation directly but to more indirect metabolic and physiological rearrangements that are currently not fully understood. Citrate supplementation appears not to result in any pronounced changes in cellular ROS production (Djajanegara et al., 2002; Gray et al., 2004), and also MFA did not trigger a ROS response, as estimated using the unspecific probe H2DCF-DA (see fourth section) in Arabidopsis leaves (Umbach et al., 2012). Addressing the mechanisms by which citrate, MFA, and ROS production cause different or overlapping responses will enhance our understanding of the mechanisms behind the mitochondrial retrograde response.
PROTEINS REGULATING NUCLEAR TRANSCRIPTION IN MITOCHONDRIAL SIGNALING
The detailed characterization of the mitochondrial retrograde response at transcriptional level has allowed the identification of upstream regulatory factors in recent years. Three studies conducted meta-analyses of upregulated transcripts at several conditions of mitochondrial dysfunction such as chemical treatments with AA, oligomycin, and rotenone or genetic modulations (Schwarzländer et al., 2012a; Van Aken and Whelan, 2012; De Clercq et al., 2013). Thirty transcripts were consistently induced, and a majority of those shared a cis-regulatory motif. These genes were defined as MITOCHONDRIAL DYSFUNCTION STIMULON (De Clercq et al., 2013), and their promotors were found to be targeted by five transcription factors of the NAC (NO APICAL MERISTEM/ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR/CUP-SHAPED COTYLEDON) family (De Clercq et al., 2013), including ANAC013 and ANAC017/REGULATORS OF ALTERNATIVE OXIDASE 1a (RAO2). ANAC017 is an ER-membrane protein that is likely activated through endoproteolysis, supposedly through rhomboid protease activity. Its cleavage releases the N terminus of ANAC017 into the cytosol from where it translocates to the nucleus (Ng et al., 2013b). Also, ANAC013 and other Arabidopsis NACs have been shown to localize to the ER (De Clercq et al., 2013; Liang et al., 2015), and several, including ANAC017, contain well-defined nuclear localization sequences in the N-terminal region (Olsen et al., 2005; Ng et al., 2013b). ER localization, stress-induced proteolytic activation and translocation into the nucleus have been experimentally documented for a number of membrane-bound NACs from Arabidopsis (Kim et al., 2008; Yoon et al., 2008; Seo et al., 2010) and soybean (Li et al., 2016), suggestive of a more general signaling principle.
WRKY-type transcription factors (WRKY15/WRKY40/WRKY63; Vanderauwera et al., 2012; Van Aken et al., 2013) and ABI4 (ABSCISIC ACID INSENSITIVE4; Giraud et al., 2009) have been identified as additional downstream regulators in mitochondrial retrograde signaling. Notably, ANAC017, WRKY40 and ABI4 are also involved in retrograde signaling of the chloroplast (Koussevitzky et al., 2007; Shang et al., 2010; Van Aken et al., 2013; 2016a). Converging retrograde signaling cascades may allow for synchronization of the downstream response in both endosymbiotic energy organelles, but the level at which the integration occurs remains elusive (Finkemeier and Schwarzländer, 2017). ABI4 in the retrograde response of the chloroplast has recently been shown to be activated through phosphorylation by a MAP kinase cascade in a Ca2+-dependent manner, suggesting potential molecular entry points of signal integration (Guo et al., 2016). ANAC017 appears also crucial for the transcriptional response to methyl viologen (MV; paraquat) that produces large amounts of superoxide using electrons from PSI under illuminated conditions and eventually leads to cell death (Van Aken and Van Breusegem, 2015; Van Aken et al., 2016a). Importantly, several MRR target genes (including AOX1a) are not induced by MV. Vice versa, MV induces additional transcripts (including HSP17.6) that are unaffected by AA treatment. ANAC017 appeared almost solely responsible for HSP17.6 induction by MV, indicating that ANAC017 can relay signals originating from either chloroplasts or mitochondria yet regulate the expression of separate gene sets. This suggests that ANAC017 operates in concert with other factors that define its specificity and have not yet been identified. ABI4 further regulates plant developmental processes, such as seed germination (Finkelstein et al., 1998), flower development (Foyer et al., 2012; Shu et al., 2016), and lateral root formation (Shkolnik-Inbar and Bar-Zvi, 2010). It remains unclear if and how those programs are linked or even integrated with retrograde regulation. Similarly, WRKYs take part in developmental programs and in a variety of abiotic and biotic stresses (Rushton et al., 2010). WRKY40 was found to be associated with plastids by binding the envelope-spanning Mg-Chelatase H subunit and may be involved in seed development (Shang et al., 2010). WRKY40 and WRKY63 were further found to act in leaf senescence of plants carrying a mutated mitochondrial AAA-protease FILAMENTATION TEMPERATURE SENSITIVE H4 (FTSH4) (Zhang et al., 2017b). As both mutation in FTSH4 (Zhang et al., 2014) and seed development (El-Maarouf-Bouteau and Bailly, 2008) have been associated with increased rates of mitochondrial reactive oxygen species (mtROS) production, the signal leading to developmental WRKY40/63 activation may be of mitochondrial (as opposed to nuclear) origin and provide a route to integrate mitochondrial signaling into regulation of plant growth and development.
A study using genetic mutants with dysfunctional mitochondria suggests that ANAC017 has a special role among the transcription factors identified so far in mitochondrial signaling. While loss of the PROHIBITIN3 scaffolding complex in the inner mitochondrial membrane (Van Aken et al., 2007) or of the dual-targeted RNA polymerase RPOTmp (Kühn et al., 2009) independently and constitutively induced marker transcripts of the mitochondrial retrograde response, their induction was drastically decreased in the absence ANAC017 (Van Aken et al., 2016b). A similar pattern was observed when mitochondrial dysfunction was induced by AA. The AOX1a transcript was unresponsive in the anac017 null mutant but regulated similar to wild type in the absence of other NACs, ABI4, and WRKY40/63 (Van Aken et al., 2016a). A central role for ANAC017 is further supported by a pronounced reduction of primary root growth induced by AA, rotenone, or MV in anac017. This effect was absent in lines lacking individual ANAC013/053/078 or WRKY40/63 transcripts, demonstrating the functional significance of ANAC017 for acclimation of the plant to mitochondrial dysfunction. Acclimation was even improved beyond wild-type capacity by overexpression of ANAC017, which led to longer primary roots in the presence of the inhibitors. Particularly high expression as compared to the other transcription factors of the NAC family is characteristic for ANAC017. This may provide an explanation for the phenotypes associated with its loss, implying that functional backup may be available for less abundant family members (Van Aken et al., 2016a).
Recently, Zhang et al. (2017a) identified MYB DOMAIN PROTEIN29/RAO7 as an additional transcription factor that has a negative, albeit indirect, regulatory impact on downstream genes related to mitochondrial stress, including AOX1a, but notably also WRKY40 and ANAC053. Since WRKY40 in turn regulates expression of ABI4 (Liu et al., 2012b) it appears that the transcription factors in retrograde signaling form expression networks. Being part of such a network, MYB DOMAIN PROTEIN29 acts as a positive regulator of plant growth, development, and energy metabolism and appears to balance needs under stressed and nonstressed conditions.
The CYCLIN-DEPENDENT KINASE E1 (CDKE1/RAO1) was identified in a screen for AOX1a regulators in response to AA and proposed to be a hub of retrograde signaling from mitochondria as well as chloroplasts (Ng et al., 2013a; Blanco et al., 2014). CDKE1 is predicted to be a subunit of the Arabidopsis mediator complex, which acts as a scaffold that bridges DNA-bound transcription factors to RNA polymerase II, linking signaling input to transcription (Mathur et al., 2011; Poss et al., 2013). Nuclear-localized CDKE1 interacts with the SNF1-Related Kinase1 (SnRK1) catalytic subunit KIN10, which integrates transcription networks of plant stress signaling, sugar metabolism, and developmental programs (Baena-González et al., 2007; Cho et al., 2012). KIN10 is conserved across plants and has been shown to interact with several transcription factors (Kleinow et al., 2009; Tsai and Gazzarrini, 2012; Zhai et al., 2017). The integrative role of KIN10 is further discussed by Wurzinger et al. (2018) as part of this Focus Issue. In the mitochondrial retrograde response, CDKE1 has been proposed to act independently of ABI4 (Blanco et al., 2014). In principle, the mediator complex would provide a mechanism for CDKE1 to operate upstream of different transcription factors, but systematic experimental validation is currently lacking.
LOW-OXYGEN STRESS: LINKING OUR INSIGHTS FROM MODELS OF MITOCHONDRIAL ENERGY SIGNALING TO THE NATURAL CONTEXT
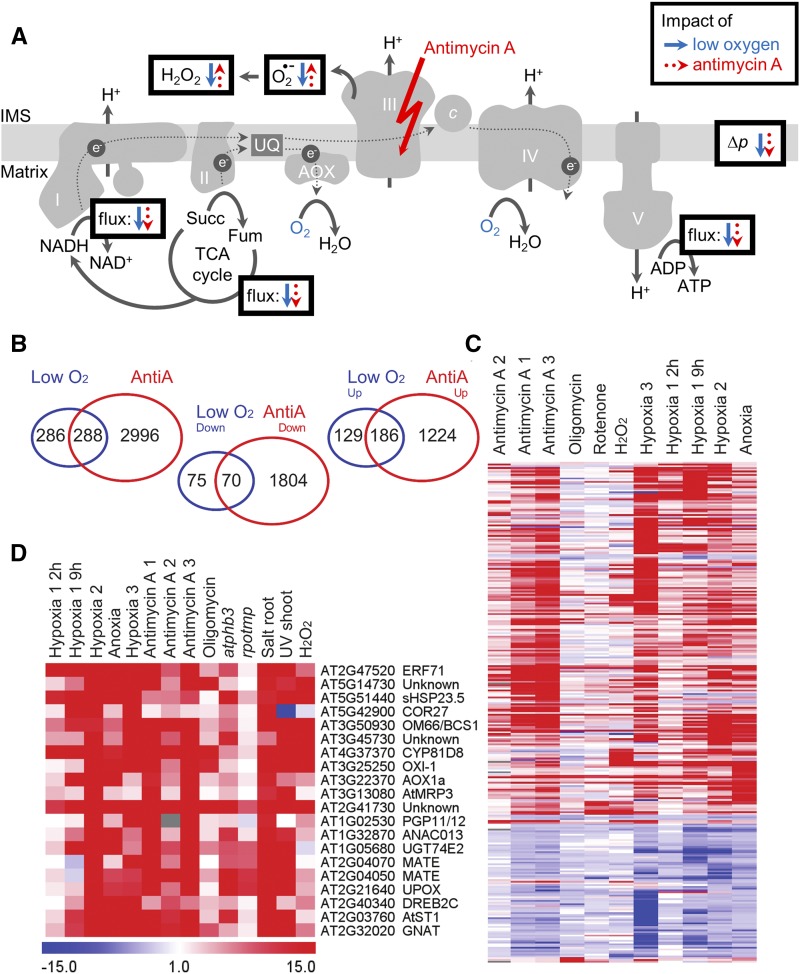
The progress that has been made in measuring the transcriptomic output and identifying regulatory proteins of mitochondrial energy signaling prompts the question of under what natural conditions their action is relevant for the plant. This deserves a closer look given that many of our insights rely on ETC inhibitors, like AA, as model stimuli. Only natural conditions that involve a rapid transition (minutes to hours) and require an acute response need considering because the inhibitors are typically applied in an abrupt fashion (e.g. sudden spraying rather than long-term supplementation of a medium). AA has been widely used with the rationale of triggering an increase in the production of ROS by the mitochondria and mitochondrial dysfunction in a sense that has not been further defined. Hence, natural conditions that lead to an increase in mtROS production may also activate the identified signaling responses. Indeed, several biotic and abiotic stresses have been associated with mtROS production (Rhoads et al., 2006; Yao and Greenberg, 2006; Chang et al., 2012; Cvetkovska and Vanlerberghe, 2013) as well as with the induction of AOX1a and other retrograde marker transcripts (Van Aken and Whelan, 2012). Yet, the link between mtROS and induction of signaling remains correlative, which is partly due to the difficulty to trigger increased mtROS production without causing gross perturbation of respiratory physiology. In addition to mtROS, mitochondrial physiology also shows changes in several other stimuli that represent equally good candidates as upstream signals. Complex III inhibition by AA enables the prediction of several changes in mitochondrial energy physiology (Fig. 1A): block of the cyanide-sensitive pathway (while AOX remaining active), drop in oxygen consumption rate, drop in proton pumping rate (with likely impact on membrane potential and pH gradient and acidification of the mitochondrial matrix affecting supercomplex composition; Ramírez-Aguilar et al., 2011), drop in ATP synthase activity, drop in adenosine energy charge, increase of superoxide release rate into the intermembrane space, increase in reduction of the ubiquinone pool, decrease of oxidation rate of matrix NADH, decrease of the rate of matrix catabolic pathways (including the TCA cycle) with impact on extramitochondrial pathways via modified rates of transport across the mitochondrial membranes. Drought and acute ozone exposure have both been shown to inhibit mitochondrial respiration by strongly decreasing the capacity of the classical electron transport pathway, while inducing the alternative pathway (Ederli et al., 2006; Dahal and Vanlerberghe, 2017). In that respect, both stresses are likely to include the response that is triggered by AA-induced signaling. Yet their primary impact on the mitochondria is mechanistically poorly defined and probably involves a combination of several changes. More specific inhibition of the electron transport with remarkable mechanistic resemblance to that of AA can be predicted for oxygen limitation (Fig. 1A). Lack of oxygen as the final mitochondrial electron acceptor of the respiratory chain may be conceptualized as a reasonably specific block of electron transport at the terminal oxidase (complex IV, AOX). Given that the mitochondrion is by far the main consumer of oxygen in the cell, the expected impact on mitochondrial energy physiology is likely to be particularly similar to that of AA. Otto Warburg made a similar assumption 60 years ago when studying the metabolism of animal cancer cells. “From the standpoint of energy”, he wrote, “no matter whether oxygen is withdrawn from the cell or whether the oxygen is prevented from reacting by a poison, the result is the same in both cases—namely, impairment of respiration from lack of energy” (Warburg, 1956). Although this consideration has not been made for plant mitochondrial energy signaling, it is indeed well supported by similarities of the transcriptomic response between anoxia/hypoxia and AA (Fig. 1, B and C): half of about 600 genes consistently differently regulated under anoxic or hypoxic conditions overlapped with the genomic response to AA. The majority of the MITOCHONDRIAL DYSFUNCTIN STIMULON (see above; De Clercq et al., 2013) is part of that overlap (Fig. 1D), suggesting that NAC-dependent signaling makes a substantial contribution to the response to low-oxygen stress.
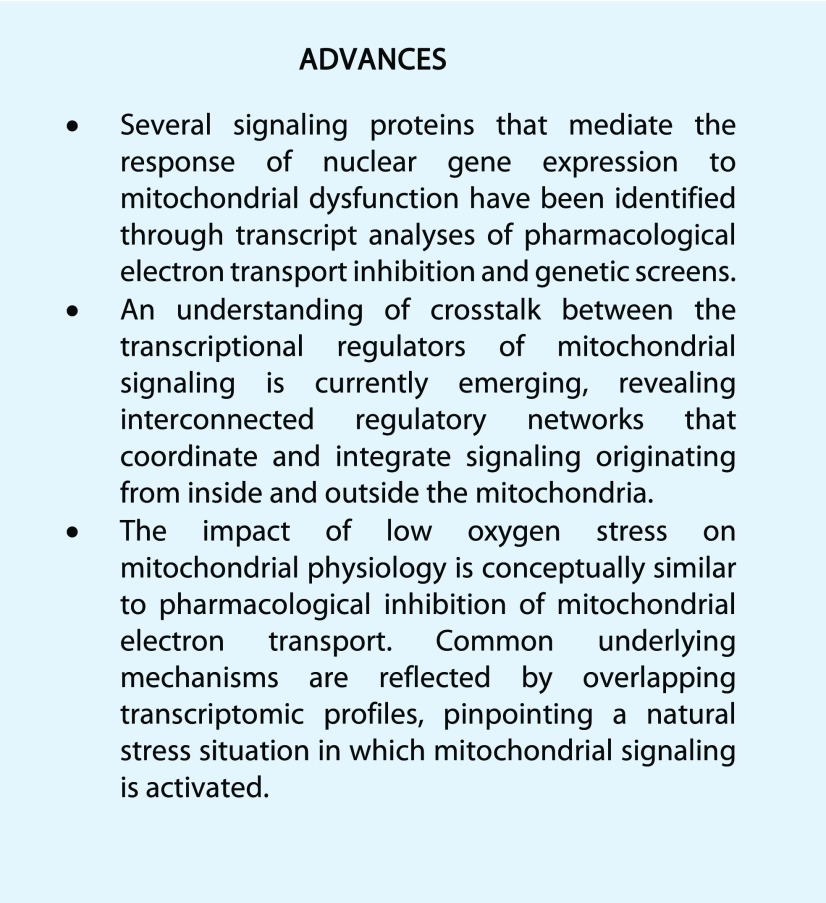
Figure 1.
Effects of low-oxygen and antimycin A treatments on mitochondrial energy physiology and the nuclear transcriptome. A, Central respiration-associated processes at the inner mitochondrial membrane are outlined. Parameters highlighted in boxes are further discussed in the text as candidate signaling molecules in mitochondrial energy signaling. Arrows in solid blue (low oxygen) and dotted red (antimycin A) indicate increases and decreases. The decrease in superoxide (O2•-) under low-oxygen stress represents a theoretical consideration; the transient increase in superoxide production has been proposed and is discussed in the text. B, To define the low-oxygen response at the transcript level, publicly available datasets (GSE accessions GSE9719 [hypoxia #1], GSE21504 [hypoxia #2], GSE16222 [anoxia], and GSE14420 [hypoxia #3]) were analyzed, and 574 genes were identified as commonly differentially regulated by low oxygen (using Cyber-T; Kayala and Baldi, 2012); 2-fold change, cum.ppde.P > 0.95). The antimycin A response was analyzed analogously using GSE accessions GSE41136 (antimycin A #1) and GSE36011 (antimycin A #2), identifying 3,284 genes differently regulated by AA. The Venn diagram shows the overall overlaps between the extracted genes, as well as the gene transcripts that were commonly up- or down-regulated. C, Normalized expression values for all 288 genes overlapping between low-oxygen and antimycin A treatment after hierarchical clustering (Pearson correlation and average linkage). For comparison, expression values from Arabidopsis cell culture responses to oligomycin (complex V inhibitor), rotenone (complex I inhibitor), and H2O2 are shown (GES3709). Expression values are fold changes normalized to the respective control experiments (set to 1). D, Normalized expression values for common differentially expressed genes at AA treatment (as defined in B) and in atphb3 mutant plants lacking the abundant PROHIBITIN3 scaffolding complex in the inner mitochondrial membrane (Van Aken et al., 2007). Expression values for salt (E-GEOD-5623) and UV (E-GEOD-5626) treatment are also shown.
Differences in the transcriptomic profiles may be accounted for by mechanistic differences between the consequences of AA and anoxia/hypoxia in mitochondrial energy physiology: low-oxygen stress will block additional oxygen-dependent pathways in the cell (e.g. cytosolic oxygenases, ER oxidoreductins that drive oxidative protein folding) that are not inhibited by AA. The concept of signal integration from various cellular compartments in low-oxygen signaling is reviewed by Schmidt et al. (2018) as part of this Focus Issue. In the mitochondria, lack of oxygen will lead to efficient inhibition of electron transport flux without the backup by AOX, as in the case of AA. Indeed, the direct impact of a gradual decline in oxygen on AOX activity is expected before that on complex IV, due to a higher Km of AOX for oxygen (Gupta et al., 2009). A comparable block of ETC flux at both terminal oxidases may be achieved pharmacologically by combined treatments of e.g. AA and the AOX inhibitor n-propyl gallate or salicylhydroxamic acid. Likewise, low-oxygen stress treatments of plant tissues can be varied to obtain different physiological outcomes. Typically, experiments are performed in the dark and involve a rapid transition to hypoxic or anoxic conditions, a scenario that naturally equals flash flooding at night. When the water retreats, reoxygenation can occur rapidly or may also occur internally once oxygen evolution by photosynthesis starts in the morning. Those scenarios show parallels to the ischemia-reperfusion situation that has been extensively studied in mammalian cells, predominately cardiac myocytes (Granger and Kvietys, 2015). In mammals, it appears that the severity of the stress impact is determined less by the hypoxic period itself, but rather by the restart of respiration that can involve excessive mtROS production at complex I through reverse electron transport leading to cell death (Chouchani et al., 2016). In analogy, tight regulation of the restart of respiration is likely to be critical to limit damage in plant cells, requiring specific mitochondrial signals. Mitochondrial signaling at the restart of mtETC after inhibition has not been studied, however, since removal of inhibitors such as AA is not feasible experimentally. Hence, low-oxygen stress may be considered a particularly relevant natural stress to study mitochondrial energy signaling. Its mode of biochemical action is well defined and comparable to that of the widely used ETC inhibitors, while carrying physiological meaning. In the following section, we will discuss physiological parameters and molecules that are affected when mtETC flux is inhibited, either in response to low oxygen or AA. In the absence of knowledge of any endogenous mitochondrial energy sensors, those are strong candidates to act upstream in mitochondrial signaling as primary stimuli or second messengers.
WHICH MOLECULES QUALIFY AS UPSTREAM MITOCHONDRIAL SIGNALS?
Reactive Oxygen Species
A candidate signal favored in several studies of mitochondrial energy signaling is an elevated rate of mtROS production. Although changes in plant mitochondrial ROS production have rarely been shown with any certainty and specificity in planta (Huang et al., 2016), ROS have been generally recognized for their signaling properties in a wide range of biological situations. Those include plant mitochondrial signaling based on indirect evidence (Ng et al., 2014; Huang et al., 2016), although a concept of where and by what mechanism different ROS may interact with a specific player downstream in the cascade is lacking. On the one hand, individual species, e.g. superoxide, the hydroxyl radical and hydrogen peroxide, differ vastly in their chemical properties, which may allow for distinct and specific cellular signals in principle (Schwarzländer and Finkemeier, 2013). On the other hand, their rapid interconversion may hamper exploitation of their distinct properties (Møller and Sweetlove, 2010).
Mitochondria produce superoxide as the primary species at complex I (NADH dehydrogenase) and complex III (cytochrome bc1 complex) through one-electron reduction of molecular oxygen. In animal and plant tissues, typically between 1% and 2% of consumed oxygen is considered to drive ROS production and, intuitively, the rate of ROS production is positively correlated with the availability of oxygen (Chance et al., 1979; Turrens et al., 1982; Puntarulo et al., 1988). At first glance, and paradoxically, rapid transition to low-oxygen conditions has also been found to elevate ROS production in cultured human cells (Chandel et al., 1998). The authors used the fluorescent dye 2′,7′-dichlorofluorescin (DCF), which indicated pronounced ROS production when cells were shifted from normoxic (21% O2) to hypoxic (2% O2) conditions, which they attributed to superoxide production at complex III using mETC inhibitors. While controversies persist, mtROS are now widely considered a potential early trigger in the mammalian transcriptional response to low oxygen (Bailey-Serres and Chang, 2005; Waypa et al., 2016). Whether, however, superoxide itself has a role as trigger remains challenging to answer. Superoxide is efficiently converted to H2O2 by superoxide dismutase (SOD) and most, if not all, probes that have been used in the relevant ROS quantitation experiments show poor specificity for an individual species. Despite this limitation, Chang et al. (2012) used H2DCF-DA in Arabidopsis seedlings to monitor ROS production when plants were deprived of oxygen. Intriguingly, they observed an increase in DCF fluorescence that colocalized with mitochondria, suggesting low-oxygen-induced ROS production in or near mitochondria. The interpretation of localized ROS production is, however, complicated by the fact that DCF, like a number of other fluorescent dyes, tends to accumulate in mitochondria (Rashid and Horobin, 1991; Huang et al., 2016). Similar to the observations of Chandel et al. (1998), AA treatment mimicked this effect, while myxothiazol, a chemically distinct complex III inhibitor that does not elevate superoxide production at the N site, did not. Increased production of superoxide at mitochondrial complex III in response to low oxygen thus appears conserved in animals and plants. In a plant that is subjected to submergence, this elevation is likely to occur in a transient fashion; however, as ROS formation from oxygen becomes chemically implausible when oxygen availability is strongly reduced and even impossible under bona fide anoxia, i.e. the complete absence of oxygen (Sasidharan et al., 2017). A sustained increase in ROS production as observed in different plant species (Yan et al., 1996; Liu et al., 2012a; Vergara et al., 2012) is unlikely to originate from the mitochondria and more likely the result of respiratory burst NADPH oxidase activity at the plasma membrane. Alternative mechanisms of increasing in steady-state ROS levels, e.g. due to progressing failure of NADPH-dependent ROS detoxification systems or as a result of other metabolic changes, such as the drop in adenylate energy charge and acidification of the cytosol (Blokhina et al., 2003), have rarely been considered. Li et al. (2013) proposed that superoxide in rice triggers a mitochondrial retrograde response. They induced superoxide production through MV treatment and found that the induction of OsAOX1a as a retrograde marker was much more pronounced than following H2O2 treatment. The induction through MV was independent of light, suggesting that superoxide formation at complex I of mitochondria and not chloroplast was responsible for OsAOX1a elevation. Further, the overexpression of SOD in mitochondria, but neither in the chloroplasts nor cytosol, and application of TEMPOL (an SOD mimetic) dampened the transcriptional response to drought, cold, and salinity. The effect of mtSOD was, however, not tested in response to direct MRR triggers such as AA. In Arabidopsis, the transcriptional landscape in a SOD knockout mutant did not obviously overlap with AA-responsive genes such as AOX1a, questioning a conserved function throughout plants (Schwarzländer et al., 2012a).
Hydrogen peroxide (H2O2) has a comparatively long half-life in the cell with 1 ms as a very rough estimate (Dat et al., 2000). As such, steady-state concentrations of H2O2 are about three orders of magnitude higher than those of superoxide. Aquaporin-facilitated diffusion of uncharged H2O2 across membranes (Bienert and Chaumont, 2014) may add to its role as a cellular signaling molecule. Recently, H2O2 has been proposed as a mobile signal in the chloroplast retrograde response, where it originates from the organelle, moves to the nucleus, and might directly act on transcription factors (Exposito-Rodriguez et al., 2017). Exogenous application of H2O2 in cell suspensions of Arabidopsis and Chlamydomonas has been shown to induce mitochondrial retrograde marker transcripts such as AOX1a and NDB2 (Vanlerberghe and McLntosh, 1996; Ho et al., 2008; Blaby et al., 2015). A large overlap exists also between the transcriptional footprints of H2O2 and AA in Arabidopsis (Ng et al., 2013b; Huang et al., 2016), making H2O2 a strong candidate for transmitting plant MRR signals. While the cascade from H2O2 production to transcriptional reprogramming via the nucleus requires further clarification, organelle function can also be more directly adjusted by posttranslational modifications. Mitochondrial citrate synthase, for instance, can be directly inactivated by H2O2 and is activated through interaction with and reduction by thioredoxins (Schmidtmann et al., 2014). Redox regulation was also demonstrated for other plant TCA cycle enzymes (Daloso et al., 2015) and previously for the AOX (Vanlerberghe et al., 1995). While oxidation of the enzyme thiols directly by H2O2 is kinetically unlikely in vivo, H2O2 may selectively react with the high-reactivity thiols of the mitochondrial thiol peroxidase system (e.g. Prx IIF), transferring the oxidation via the thioredoxin system to the different enzyme targets. Thiol-switching of mitochondrial enzymes could act as a direct signal to readjust organellar function on site even before and not necessarily linked directly to transcriptional programs.
Nitric Oxide
The plant hypoxic response is centrally regulated through the transcription factor RELATED TO APETALA (RAP) 2.12 that interacts with a hypoxia responsive promoter element conserved across plant species (Gasch et al., 2016). It controls mitochondrial respiration and TCA cycle enzymes, offering a genetic handle to adjust mitochondrial function when oxygen becomes limiting (Paul et al., 2016). RAP2.12 undergoes proteasome-mediated proteolysis through the N-end rule pathway under normoxic conditions and is protected from degradation under hypoxia (Gibbs et al., 2011; Licausi et al., 2011). Mitochondria could provide stability to RAP2.12 through nitric oxide (NO) production. NO production by the plant mtETC is elevated under hypoxia (reviewed by Hebelstrup and Møller, 2015). Alber et al. (2017) recently proposed that NO generation happens at complex III, while AOX dampens NO generation rate. Gibbs et al. (2014) further showed that NO acts in the stabilization of RAP2.12, and this requires cytosolic nitrate reductase activity in vivo. As nitrate reductase in the cytosol could theoretically fuel mitochondrial NO production (via nitrite), this finding does not rule out that mitochondrially produced NO acts in RAP2.12-mediated hypoxic signaling. Isolated pea root mitochondria were found to generate NO under hypoxic conditions when nitrite is present, which had stabilizing impact on mitochondrial function (Gupta et al., 2017). A key protective mechanism of NO was recently identified in mouse, where a specific Cys of complex I subunit ND3 is S-nitrosated to prevent reverse electron flow and excess mtROS production at reoxygenation (Chouchani et al., 2013). This Cys is highly conserved, suggesting a similar mechanism to operate also in plants (Braun et al., 2014). A nitrate reductase mutant was also used by Gupta et al. (2012) to suggest that NO inhibits activity of the TCA cycle enzyme aconitase. This block resulted in citrate accumulation as a potential retrograde and NO-mediated energy signal in hypoxic plants. Aconitase activity is further decreased through superoxide and H2O2 and may be considered an integration point of plant oxidative stress (Sweetlove et al., 2002).
Posttranslational Modification of Proteins
More direct regulation could be achieved through protein succinylation. In animals and plants, succinate accumulates under hypoxia (Rocha et al., 2010; Sweetlove et al., 2010; Narsai et al., 2011; Chouchani et al., 2014; António et al., 2016). In animals, succinate can indirectly stabilize the 1α subunit of the transcriptional regulator hypoxia-inducible factor, and succinylation, which relies on succinyl-CoA as substrate, regulates TCA cycle and glycolytic enzymes (Mills and O’Neill, 2014). Interestingly, proteins active in the TCA cycle and glycolysis have also been found succinylated in tomato (Solanum lycopersicum), wheat (Triticum durum), rice (Oryza sativa), and Brachypodium, suggesting a conserved role of succinate and succinyl-CoA in the regulation of metabolic processes (He et al., 2016; Jin and Wu, 2016; Zhen et al., 2016; Zhang et al., 2017c). Other posttranslational modifications such as protein phosphorylation (e.g. through SnRK1, as discussed by Wurzinger et al. [2018] in this Focus Issue) and protein Lys acetylation also critically rely on substrates that can be derived from mitochondrial energy metabolism, i.e. ATP and acetyl-CoA. Both classes have been discussed as signals in the retrograde response (Hartl and Finkemeier, 2012), but their contributions are currently largely unknown (Ng et al., 2013a; Blanco et al., 2014).
ATP
Lack of oxygen as the terminal electron acceptor in oxidative phosphorylation results in a rapid drop of MgATP concentration in the cytosol, which is otherwise comparatively well buffered (De Col et al., 2017). As alternative ATP sources activated under hypoxic conditions are comparatively inefficient, this ultimately results in an energy crisis of the cell (van Dongen and Licausi, 2015). Notably, the adenylate energy state is already changed very early at the onset of hypoxia, when oxygen concentrations are still considerably above the Km of cytochrome c oxidase (complex IV) for oxygen, suggesting that in principle oxidative phosphorylation at the mETC could be maintained even under relatively strong hypoxic conditions. The kinetic properties of complex IV, however, have been shown to directly depend on molecular oxygen, and in vivo flux through the cytochrome c oxidase is expected to gradually decrease at oxygen concentrations above a theoretically achievable Km (Chandel et al., 1996). Adenylate changes at the onset of low-oxygen stress have been additionally explained by proactive regulation that rely on proteins other than complex IV as oxygen sensors, and recent studies have helped to gain mechanistic insights (Geigenberger, 2003; Weits et al., 2014; António et al., 2016). In the retrograde response of budding yeast (Saccharomyces cerevisiae), ATP can act as a signaling trigger to adjust metabolic processes according to energy availability. The transcription factors RTG1 and RTG3, responsible for transcriptomic reprogramming, require activation through RTG2. RTG2 has an N-terminal ATP-binding domain, and ATP-binding in yeast releases RTG2 from another protein, MKS1P, that is a cytosolic dephosphorylase and acts on RTG3 to make the RTG1/RTG3 complex translocate to the nucleus for genomic control (Zhang et al., 2013). In Arabidopsis, inhibition of the ATP synthase through oligomycin transcriptionally activates retrograde markers (e.g. AOX1a and NDB2) and overlaps markedly with the transcript responses to different abiotic and biotic stresses (Geisler et al., 2012; Fig. 1, C and D). Whether ATP and/or the ATD/ADP/AMP ratios are directly involved in activating those responses is, however, not known.
Phosphoadenosine Phosphate
PAP (3′-phosphoadenosine 5′-phosphate) has been proposed as an active signal in the retrograde response of chloroplast and mitochondria. PAP is dephosphorylated by SAL1 to form AMP and SAL1 is a dual-localized protein that resides in both chloroplasts and mitochondria (Estavillo et al., 2011). In yeast, PAP inhibits nuclear 5′ to 3′ exoribonucleases (XRNs), thereby modulating RNA catabolism and transcript abundances of XRN targets. Accordingly, Arabidopsis sal1 mutants that accumulate PAP show transcriptional changes largely overlapping with xrn2 xrn3 double-mutant plants (Gregory et al., 2008; Estavillo et al., 2011). A significant number of these transcripts is also changed under specific AOX1a-inducing conditions including high light, drought, and ABA treatment, pointing to a function in retrograde stress signaling. The idea of PAP acting as a direct signal in this response is supported by the fact that loss of SAL1 in both plastids and mitochondria can be compensated by artificial expression of SAL1 in the nucleus (Estavillo et al., 2011). Furthermore, there is significant overlap between the transcriptional signatures of PAP-related mutants sal1 and xrn2 xrn3 with MRR-inducing conditions such as AA, atphb3, and rpotmp mutants (Van Aken and Whelan, 2012; Van Aken and Pogson, 2017). As many of these commonly regulated genes are ANAC017 dependent, it was recently suggested that the PAP and ANAC017 pathways are mechanistically linked (Van Aken and Pogson, 2017). At least part of this common pathway appears capable of suppressing cell death and lesion formation, potentially by suppressing, e.g., extrusion of toxic compounds such as chlorophyll degradation products (Bruggeman et al., 2016; Van Aken and Pogson, 2017).
Mitochondrial Membrane Potential
In yeast, the mitochondrial membrane potential has been proposed to be central in triggering the mitochondrial retrograde response. Miceli et al. (2012) modulated the membrane potential of Saccharomyces cerevisiae strain rho0 lacking mitochondrial DNA by expressing either a mutated F1-ATPase that results in an elevated membrane potential or by deleting COX4 to reduce the membrane potential. Hyperactivity of the F1-ATPase suppressed translocation of RTG3 into the nucleus and thus dampened the MRR. Reduction of the membrane potential, conversely, elevated the response. This was independent of increasing concentrations of a free radical scavenger, indicating that indeed the membrane potential and not ROS formation downstream of that, triggered up-regulation of retrograde marker genes such as CIT2. The interplay between ROS formation, the mitochondrial membrane potential, and ATP production complicates making statements about cause and effect in mitochondrial retrograde signaling, also in plants. This was nicely illustrated by Krause and Durner (2004), who treated Arabidopsis cells in suspension with a bacterial elicitor. The activated defense response decreased the mitochondrial membrane potential and total ATP levels and led to an accumulation of H2O2 and, ultimately, AOX1a transcript. A better understanding of the sequence of events, which we can similarly assume to happen in hypoxia, is still required.
Calcium Ions
Ca2+ is involved in the mammalian mitochondrial retrograde response (Butow and Avadhani, 2004), where Ca2+ concentrations are linked to ATP production (Griffiths and Rutter, 2009). In both mammals and plants, a uniporter complex is involved in mitochondrial Ca2+ import (Kamer and Mootha, 2015; Wagner et al., 2015; Teardo et al., 2017), which makes use of the mitochondrial membrane potential. Ca2+ transients in the mitochondrial matrix have been shown to occur in response to various abiotic stresses (Logan and Knight, 2003; Loro et al., 2012) that also activate an MRR. Treatment of tobacco cells with Ca2+ channel blockers (ruthenium red and LaCl3) suppressed the induction of AOX capacity at AA treatment, suggesting a contribution of Ca2+ to mitochondrial stress signaling (Vanlerberghe et al., 2002). Ruthenium red also partially suppressed AOX1 induction in rice and Arabidopsis at more general stress such as cold, drought, salinity, and hypoxia (Tsuji et al., 2000; Vanderauwera et al., 2012; Li et al., 2013). Conversely, addition of cyclopiazonic acid (an ER calcium channel inhibitor that results in increased cytosolic Ca2+ concentration in animal cells) intensified AOX1a gene expression induction by salt (Vanderauwera et al., 2012). Hypoxia has been suggested to drive Ca2+ extrusion from mitochondrial stores in cultured maize cells (Subbaiah et al., 1998), but supporting experimental data are scarce. Conceptually, the existence of such Ca2+ stores in the matrix under nonpathological conditions requires further scrutiny, considering a minor gradient in free Ca2+ concentration (about 100 nm in the cytosol versus 200 nm in the matrix at baseline; Wagner et al., 2015). Considering the highly negative potential across the inner mitochondrial membrane, this strongly favors uptake rather than release of Ca2+. A breakdown of membrane potential due to sustained hypoxia may allow for Ca2+ release and mobilize the considerable total amounts of bound Ca2+ in the matrix. Even if such a scenario could be empirically confirmed, the small volume of mitochondria, as compared to the vacuole or the extracellular space, appear not optimal for mitochondria to act as stores for cytosolic Ca2+ signaling. Ca2+ release from mitochondria may rather represent a hallmark of cellular energy crisis and loss of homeostasis than contribute to a specific signal. Yet, the cytosolic increase in Ca2+ that has been consistently observed at hypoxia appears to be conserved throughout plants and chemical suppression hampers transcriptional induction of hypoxia-responsive genes such as ALCOHOL DEHYDROGENASE (Subbaiah et al., 1994; Sedbrook et al., 1996; Yemelyanov et al., 2011). The mechanism by which Ca2+ fluxes modulate nuclear transcription is currently unknown.
Abscisic Acid
Levels of the phytohormone abscisic acid (ABA) are typically elevated under various stress conditions, including hypoxia, and induce AOX1a and other typical transcripts of the mitochondrial retrograde response (Giraud et al., 2009; Van Aken et al., 2009). ABA, although not ultimately synthesized in the plant organelles, has been proposed to act as an integrator of retrograde signals. A mitochondrial protein (RRL; RETARDED ROOT GROWTH-LIKE) has been suggested to mediate between ABA and the transcription factor ABI4. The overexpression of RRL markedly increased the transcriptional up-regulation of ABI4 and AOX1a in response to ABA, while both transcripts were not ABA inducible in an rrl mutant (Yao et al., 2015). The link between ABA and ABI4 in that scenario might further involve WRKY40 that represses ABI4 in the absence of ABA (Shang et al., 2010). ABA also suppresses biosynthesis of ethylene through ABI4-mediated transcriptional changes (Dong et al., 2016) and the interplay between ABA, ethylene, and also auxin is supposed to adjust plant growth under hypoxic conditions (Xu et al., 2013).
Auxin
Auxin, in contrast to ABA, represses AOX1a. Mutants compromised in auxin transport show elevated levels of AOX1a transcript at stress treatment. Conversely, treating plants with auxin represses the induction of AOX1a, e.g. upon AA-induced mitochondrial dysfunction (Ivanova et al., 2014; Kerchev et al., 2014). He et al. (2012) characterized a mitochondrial DEXH RNA helicase (ABO6) that is necessary for proper splicing of multiple mitochondrial complex I subunits. Upon ABO6 loss, mitochondria were proposed to produce ROS at elevated levels that were further modulated by exogenous application of ABA and auxin. The authors proposed that both hormones act as mitochondrial signals through ROS formation. Those conclusions will require future reassessment because the employed ROS-detection methodology based on the cpYFP sensor protein has since proven inappropriate (Schwarzländer et al., 2014; Demaurex and Schwarzländer, 2016). A potential link between mitochondrial ROS and auxin is independently supported by Arabidopsis plants lacking the mitochondrial FTSH4 AAA-protease that is required for the assembly and/or stability of complex I and, even more, complex V. Similar to abo6 mutants, auxin accumulation or responsiveness was reduced in these plants, and external application of auxin could revert the phenotypic symptoms of ftsh4 mutant plants (Zhang et al., 2014). Interestingly, ftsh4 plants also accumulated salicylic acid, suggesting plant mitochondrial signaling is linked to hormonal regulation at several levels. It appears that the constant balancing act between fast growth during optimal conditions and reduced growth/stress response during challenging conditions may be achieved by antagonistic relationships between for instance auxin (growth promoting) and MRR (stress-response promoting; Ivanova et al., 2014; Kerchev et al., 2014).
CONCLUSION
The understanding of mitochondrial signaling has seen major advances in recent years. More and more protein players of mitochondrial dysfunction signaling are being identified, and a complex signaling network is emerging that integrates and coordinates a wide range of seemingly distinct cellular function (Ng et al., 2014). Yet, little is known about the natural circumstances under which mitochondrial signaling operates to the advantage of the plant. Comparative transcriptomics provide first insights and hint to a role of mitochondrial energy signaling in different stress scenarios and hypoxia in particular. Yet, the complex signaling network that separates primary stimuli from gene expression response can hamper unambiguous deduction of a natural condition from a transcriptome response. Hence, the upstream stimuli in response to a naturally occurring transition deserve focused investigation. The capture of those changes at the right time and in the correct place, including the mitochondria, has been limiting, which is why the primary events of mitochondrial signaling remain unknown, similarly to those of most stress-signaling pathways (Zhu, 2016). The usage of genetically encoded biosensors has the potential to overcome several critical hurdles and deliver information from the upstream end of signaling, similarly to what transcript analysis achieves at the downstream end. Sensors for some of the promising candidates of mitochondrial signaling stimuli have been created or adapted for use in plants and an understanding of the cell-compartment-specific in vivo dynamics of several of the molecules discussed here has moved into reach, including H2O2, ATP, and Ca2+ (Costa et al., 2010; Loro et al., 2012, 2016; Bonza et al., 2013; De Col et al., 2017). A next major step forward is likely to come from measuring the dynamics of a large array of chemical species in concert. Multiplexing approaches will resolve the order of events and help to dissect cause from coincidence as well as the interconnectivity of the physiological network that feeds into the signaling network through specific messengers. Recent in vivo sensing approaches have unearthed first links between membrane potential, pH, Ca2+, ABA, and GA, although more than two or three parameters have rarely been assessed at a time (Schwarzländer et al., 2012b; Rizza et al., 2017; Waadt et al., 2017). Breaking those limitations and driving sensor multiplexing toward the omics scale of small molecular signal candidates would enable us to get direct and specific fingerprints of any transition, may it be a stress stimulus or the effect of a specific inhibitor. This would not only elucidate upstream triggers of signaling but also discriminate different scenarios of mitochondrial signaling. Eventually, this opens the door to reliably match different mitochondrial energy signaling modes with the natural conditions they have evolved for in the plant.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through the Emmy-Noether programme (SCHW1719/1-1), the Research Training Group GRK2064 and a grant (SCHW1719/5-1) as part of the package PAK918. O.V.A. was supported by Crafoord Foundation (20170862) and the Australian Research Council (DP160103573).
Articles can be viewed without a subscription.
References
- Alber NA, Sivanesan H, Vanlerberghe GC (2017) The occurrence and control of nitric oxide generation by the plant mitochondrial electron transport chain. Plant Cell Environ 40: 1074–1085 [DOI] [PubMed] [Google Scholar]
- António C, Päpke C, Rocha M, Diab H, Limami AM, Obata T, Fernie AR, van Dongen JT (2016) Regulation of primary metabolism in response to low oxygen availability as revealed by carbon and nitrogen isotope redistribution. Plant Physiol 170: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Chang R (2005) Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann Bot 96: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert GP, Chaumont F (2014) Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim Biophys Acta 1840: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Blaby IK, Blaby-Haas CE, Pérez-Pérez ME, Schmollinger S, Fitz-Gibbon S, Lemaire SD, Merchant SS (2015) Genome-wide analysis on Chlamydomonas reinhardtii reveals the impact of hydrogen peroxide on protein stress responses and overlap with other stress transcriptomes. Plant J 84: 974–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco NE, Guinea-Díaz M, Whelan J, Strand Å (2014) Interaction between plastid and mitochondrial retrograde signalling pathways during changes to plastid redox status. Philos Trans R Soc Lond B Biol Sci 369: 20130231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhina O, Virolainen E, Fagerstedt KV (2003) Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Bot 91: 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonza MC, Loro G, Behera S, Wong A, Kudla J, Costa A (2013) Analyses of Ca2+ accumulation and dynamics in the endoplasmic reticulum of Arabidopsis root cells using a genetically encoded Cameleon sensor. Plant Physiol 163: 1230–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun HP, Binder S, Brennicke A, Eubel H, Fernie AR, Finkemeier I, Klodmann J, König AC, Kühn K, Meyer E, et al. (2014) The life of plant mitochondrial complex I. Mitochondrion 19: 295–313 [DOI] [PubMed] [Google Scholar]
- Bruggeman Q, Mazubert C, Prunier F, Lugan R, Chan KX, Phua SY, Pogson BJ, Krieger-Liszkay A, Delarue M, Benhamed M, et al. (2016) Chloroplast activity and 3′phosphadenosine 5′phosphate signaling regulate programmed cell death in Arabidopsis. Plant Physiol 170: 1745–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffa P, Peters RA (1949) The in vivo formation of citrate induced by fluoroacetate and its significance. J Physiol 110: 488–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG (2004) Mitochondrial signaling: The retrograde response. Mol Cell 14: 1–15 [DOI] [PubMed] [Google Scholar]
- Cadenas E, Boveris A, Ragan CI, Stoppani AO (1977) Production of superoxide radicals and hydrogen peroxide by NADH-ubiquinone reductase and ubiquinol-cytochrome c reductase from beef-heart mitochondria. Arch Biochem Biophys 180: 248–257 [DOI] [PubMed] [Google Scholar]
- Chance B, Sies H, Boveris A (1979) Hydroperoxide metabolism in mammalian organs. Physiol Rev 59: 527–605 [DOI] [PubMed] [Google Scholar]
- Chandel NS, Budinger GR, Schumacker PT (1996) Molecular oxygen modulates cytochrome c oxidase function. J Biol Chem 271: 18672–18677 [DOI] [PubMed] [Google Scholar]
- Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT (1998) Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci USA 95: 11715–11720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R, Jang CJ, Branco-Price C, Nghiem P, Bailey-Serres J (2012) Transient MPK6 activation in response to oxygen deprivation and reoxygenation is mediated by mitochondria and aids seedling survival in Arabidopsis. Plant Mol Biol 78: 109–122 [DOI] [PubMed] [Google Scholar]
- Chauton MS, Winge P, Brembu T, Vadstein O, Bones AM (2013) Gene regulation of carbon fixation, storage, and utilization in the diatom Phaeodactylum tricornutum acclimated to light/dark cycles. Plant Physiol 161: 1034–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Hong JW, Kim EC, Yoo SD (2012) Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol 158: 1955–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Methner C, Nadtochiy SM, Logan A, Pell VR, Ding S, James AM, Cochemé HM, Reinhold J, Lilley KS, et al. (2013) Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat Med 19: 753–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Pell VR, Gaude E, Aksentijević D, Sundier SY, Robb EL, Logan A, Nadtochiy SM, Ord ENJ, Smith AC, et al. (2014) Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 515: 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Pell VR, James AM, Work LM, Saeb-Parsy K, Frezza C, Krieg T, Murphy MP (2016) A unifying mechanism for mitochondrial superoxide production during ischemia-reperfusion injury. Cell Metab 23: 254–263 [DOI] [PubMed] [Google Scholar]
- Clifton R, Lister R, Parker KL, Sappl PG, Elhafez D, Millar AH, Day DA, Whelan J (2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol Biol 58: 193–212 [DOI] [PubMed] [Google Scholar]
- Clifton R, Millar AH, Whelan J (2006) Alternative oxidases in Arabidopsis: A comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim Biophys Acta 1757: 730–741 [DOI] [PubMed] [Google Scholar]
- Costa A, Drago I, Behera S, Zottini M, Pizzo P, Schroeder JI, Pozzan T, Lo Schiavo F (2010) H2O2 in plant peroxisomes: An in vivo analysis uncovers a Ca(2+)-dependent scavenging system. Plant J 62: 760–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovska M, Vanlerberghe GC (2013) Alternative oxidase impacts the plant response to biotic stress by influencing the mitochondrial generation of reactive oxygen species. Plant Cell Environ 36: 721–732 [DOI] [PubMed] [Google Scholar]
- Dahal K, Vanlerberghe GC (2017) Alternative oxidase respiration maintains both mitochondrial and chloroplast function during drought. New Phytol 213: 560–571 [DOI] [PubMed] [Google Scholar]
- Daloso DM, Müller K, Obata T, Florian A, Tohge T, Bottcher A, Riondet C, Bariat L, Carrari F, Nunes-Nesi A, et al. (2015) Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria. Proc Natl Acad Sci USA 112: E1392–E1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat J, Vandenabeele S, Vranová E, Van Montagu M, Inzé D, Van Breusegem F (2000) Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci 57: 779–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq I, Vermeirssen V, Van Aken O, Vandepoele K, Murcha MW, Law SR, Inzé A, Ng S, Ivanova A, Rombaut D, et al. (2013) The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25: 3472–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Col V, Fuchs P, Nietzel T, Elsässer M, Voon CP, Candeo A, Seeliger I, Fricker MD, Grefen C, Møller IM, et al. (2017) ATP sensing in living plant cells reveals tissue gradients and stress dynamics of energy physiology. eLife 6: e26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaurex N, Schwarzländer M (2016) Mitochondrial flashes: Dump superoxide and dance with protons now. Antioxid Redox Signal 25: 550–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djajanegara I, Finnegan PM, Mathieu C, McCabe T, Whelan J, Day DA (2002) Regulation of alternative oxidase gene expression in soybean. Plant Mol Biol 50: 735–742 [DOI] [PubMed] [Google Scholar]
- Dong Z, Yu Y, Li S, Wang J, Tang S, Huang R (2016) Abscisic acid antagonizes ethylene production through the ABI4-mediated transcriptional repression of ACS4 and ACS8 in Arabidopsis. Mol Plant 9: 126–135 [DOI] [PubMed] [Google Scholar]
- Ederli L, Morettini R, Borgogni A, Wasternack C, Miersch O, Reale L, Ferranti F, Tosti N, Pasqualini S (2006) Interaction between nitric oxide and ethylene in the induction of alternative oxidase in ozone-treated tobacco plants. Plant Physiol 142: 595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarouf-Bouteau H, Bailly C (2008) Oxidative signaling in seed germination and dormancy. Plant Signal Behav 3: 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Rodriguez M, Laissue PP, Yvon-Durocher G, Smirnoff N, Mullineaux PM (2017) Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat Commun 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I, König AC, Heard W, Nunes-Nesi A, Pham PA, Leister D, Fernie AR, Sweetlove LJ (2013) Transcriptomic analysis of the role of carboxylic acids in metabolite signaling in Arabidopsis leaves. Plant Physiol 162: 239–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I, Schwarzländer M (2017) Mitochondrial regulation in the photosynthetic cell: principles and concepts. In Logan D.C., ed, Annual Plant Reviews: Plant Mitochondria, Ed 2, Vol 50 Wiley, Hoboken, NJ, pp 185–226 [Google Scholar]
- Foyer CH, Kerchev PI, Hancock RD (2012) The ABA-INSENSITIVE-4 (ABI4) transcription factor links redox, hormone and sugar signaling pathways. Plant Signal Behav 7: 276–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch P, Fundinger M, Müller JT, Lee T, Bailey-Serres J, Mustroph A (2016) Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28: 160–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P. (2003) Response of plant metabolism to too little oxygen. Curr Opin Plant Biol 6: 247–256 [DOI] [PubMed] [Google Scholar]
- Geisler DA, Päpke C, Obata T, Nunes-Nesi A, Matthes A, Schneitz K, Maximova E, Araújo WL, Fernie AR, Persson S (2012) Downregulation of the δ-subunit reduces mitochondrial ATP synthase levels, alters respiration, and restricts growth and gametophyte development in Arabidopsis. Plant Cell 24: 2792–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, et al. (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Md Isa N, Movahedi M, Lozano-Juste J, Mendiondo GM, Berckhan S, Marín-de la Rosa N, Vicente Conde J, Sousa Correia C, Pearce SP, et al. (2014) Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol Cell 53: 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Ng S, Carrie C, Duncan O, Low J, Lee CP, Van Aken O, Millar AH, Murcha M, Whelan J (2010) TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22: 3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Van Aken O, Ho LH, Whelan J (2009) The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol 150: 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger DN, Kvietys PR (2015) Reperfusion injury and reactive oxygen species: The evolution of a concept. Redox Biol 6: 524–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray GR, Maxwell DP, Villarimo AR, McIntosh L (2004) Mitochondria/nuclear signaling of alternative oxidase gene expression occurs through distinct pathways involving organic acids and reactive oxygen species. Plant Cell Rep 23: 497–503 [DOI] [PubMed] [Google Scholar]
- Gregory BD, O’Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR (2008) A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Griffiths EJ, Rutter GA (2009) Mitochondrial calcium as a key regulator of mitochondrial ATP production in mammalian cells. Biochim Biophys Acta 1787: 1324–1333 [DOI] [PubMed] [Google Scholar]
- Guo H, Feng P, Chi W, Sun X, Xu X, Li Y, Ren D, Lu C, David Rochaix J, Leister D, et al. (2016) Plastid-nucleus communication involves calcium-modulated MAPK signalling. Nat Commun 7: 12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KJ, Lee CP, Ratcliffe RG (2017) Nitrite protects mitochondrial structure and function under hypoxia. Plant Cell Physiol 58: 175–183 [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Shah JK, Brotman Y, Jahnke K, Willmitzer L, Kaiser WM, Bauwe H, Igamberdiev AU (2012) Inhibition of aconitase by nitric oxide leads to induction of the alternative oxidase and to a shift of metabolism towards biosynthesis of amino acids. J Exp Bot 63: 1773–1784 [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Zabalza A, van Dongen JT (2009) Regulation of respiration when the oxygen availability changes. Physiol Plant 137: 383–391 [DOI] [PubMed] [Google Scholar]
- Hartl M, Finkemeier I (2012) Plant mitochondrial retrograde signaling: Post-translational modifications enter the stage. Front Plant Sci 3: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Duan Y, Hua D, Fan G, Wang L, Liu Y, Chen Z, Han L, Qu L-J, Gong Z (2012) DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell 24: 1815–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, Wang Q, Li M, Damaris RN, Yi X, Cheng Z, Yang P (2016) Global proteome analyses of lysine acetylation and succinylation reveal the widespread involvement of both modification in metabolism in the embryo of germinating rice seed. J Proteome Res 15: 879–890 [DOI] [PubMed] [Google Scholar]
- Hebelstrup KH, Møller I (2015) Mitochondrial signaling in plants under hypoxia: Use of reactive oxygen species (ROS) and reactive nitrogen species (RNS). In Gupta K, Igamberdiev A, eds, Reactive Oxygen and Nitrogen Species Signaling and Communication in Plants. Vol 23 Springer, Cham, Switzerland, pp 63–77 [Google Scholar]
- Ho LH, Giraud E, Uggalla V, Lister R, Clifton R, Glen A, Thirkettle-Watts D, Van Aken O, Whelan J (2008) Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol 147: 1858–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Van Aken O, Schwarzländer M, Belt K, Millar AH (2016) The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants. Plant Physiol 171: 1551–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A, Law SR, Narsai R, Duncan O, Lee JH, Zhang B, Van Aken O, Radomiljac JD, van der Merwe M, Yi K, et al. (2014) A functional antagonistic relationship between auxin and mitochondrial retrograde signaling regulates alternative oxidase1a expression in Arabidopsis. Plant Physiol 165: 1233–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Wu F (2016) Proteome-wide identification of lysine succinylation in the proteins of tomato (Solanum lycopersicum). PLoS One 11: e0147586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamer KJ, Mootha VK (2015) The molecular era of the mitochondrial calcium uniporter. Nat Rev Mol Cell Biol 16: 545–553 [DOI] [PubMed] [Google Scholar]
- Kayala MA, Baldi P (2012) Cyber-T web server: Differential analysis of high-throughput data. Nucleic Acids Res 40: W553–W559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchev PI, De Clercq I, Denecker J, Mühlenbock P, Kumpf R, Nguyen L, Audenaert D, Dejonghe W, Van Breusegem F (2014) Mitochondrial perturbation negatively affects auxin signaling. Mol Plant 7: 1138–1150 [DOI] [PubMed] [Google Scholar]
- Kim SG, Lee AK, Yoon HK, Park CM (2008) A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J 55: 77–88 [DOI] [PubMed] [Google Scholar]
- Kleinow T, Himbert S, Krenz B, Jeske H, Koncz C (2009) NAC domain transcription factor ATAF1 interacts with SNF1-related kinases and silencing of its subfamily causes severe developmental defects in Arabidopsis. Plant Sci 177: 360–370 [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Krause M, Durner J (2004) Harpin inactivates mitochondria in Arabidopsis suspension cells. Mol Plant Microbe Interact 17: 131–139 [DOI] [PubMed] [Google Scholar]
- Kühn K, Richter U, Meyer EH, Delannoy E, de Longevialle AF, O’Toole N, Börner T, Millar AH, Small ID, Whelan J (2009) Phage-type RNA polymerase RPOTmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell 21: 2762–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CP, Eubel H, Millar AH (2010) Diurnal changes in mitochondrial function reveal daily optimization of light and dark respiratory metabolism in Arabidopsis. Mol Cell Proteomics 9: 2125–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CR, Liang DD, Li J, Duan YB, Li H, Yang YC, Qin RY, Li L, Wei PC, Yang JB (2013) Unravelling mitochondrial retrograde regulation in the abiotic stress induction of rice ALTERNATIVE OXIDASE 1 genes. Plant Cell Environ 36: 775–788 [DOI] [PubMed] [Google Scholar]
- Li S, Wang N, Ji D, Xue Z, Yu Y, Jiang Y, Liu J, Liu Z, Xiang F (2016) Evolutionary and functional analysis of membrane-bound NAC transcription factor genes in soybean. Plant Physiol 172: 1804–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Li H, Zhou F, Li H, Liu J, Hao Y, Wang Y, Zhao H, Han S (2015) Subcellular distribution of NTL transcription factors in Arabidopsis thaliana. Traffic 16: 1062–1074 [DOI] [PubMed] [Google Scholar]
- Liao X, Butow RA (1993) RTG1 and RTG2: Two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 72: 61–71 [DOI] [PubMed] [Google Scholar]
- Liao XS, Small WC, Srere PA, Butow RA (1991) Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol Cell Biol 11: 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LA, Perata P, van Dongen JT (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479: 419–422 [DOI] [PubMed] [Google Scholar]
- Liu P, Sun F, Gao R, Dong H (2012a) RAP2.6L overexpression delays waterlogging induced premature senescence by increasing stomatal closure more than antioxidant enzyme activity. Plant Mol Biol 79: 609–622 [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Yan L, Wu Z, Mei C, Lu K, Yu YT, Liang S, Zhang XF, Wang XF, Zhang DP (2012b) Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J Exp Bot 63: 6371–6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC, Knight MR (2003) Mitochondrial and cytosolic calcium dynamics are differentially regulated in plants. Plant Physiol 133: 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loro G, Drago I, Pozzan T, Schiavo FL, Zottini M, Costa A (2012) Targeting of Cameleons to various subcellular compartments reveals a strict cytoplasmic/mitochondrial Ca2+ handling relationship in plant cells. Plant J 71: 1–13 [DOI] [PubMed] [Google Scholar]
- Loro G, Wagner S, Doccula FG, Behera S, Weinl S, Kudla J, Schwarzländer M, Costa A, Zottini M (2016) Chloroplast-specific in vivo Ca2+ imaging using Yellow Cameleon fluorescent protein sensors reveals organelle-autonomous Ca2+ signatures in the Stroma. Plant Physiol 171: 2317–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P (2010) TCP genes: A family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Mathur S, Vyas S, Kapoor S, Tyagi AK (2011) The Mediator complex in plants: Structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol 157: 1609–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijs M, Fabris M, Obata T, Foubert I, Franco-Zorrilla JM, Solano R, Fernie AR, Vyverman W, Goossens A (2017) The transcription factor bZIP14 regulates the TCA cycle in the diatom Phaeodactylum tricornutum. EMBO J 36: 1559–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miceli MV, Jiang JC, Tiwari A, Rodriguez-Quiñones JF, Jazwinski SM (2012) Loss of mitochondrial membrane potential triggers the retrograde response extending yeast replicative lifespan. Front Genet 2: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E, O’Neill LA (2014) Succinate: A metabolic signal in inflammation. Trends Cell Biol 24: 313–320 [DOI] [PubMed] [Google Scholar]
- Møller IM, Sweetlove LJ (2010) ROS signalling—specificity is required. Trends Plant Sci 15: 370–374 [DOI] [PubMed] [Google Scholar]
- Murphy MP. (2009) How mitochondria produce reactive oxygen species. Biochem J 417: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Rocha M, Geigenberger P, Whelan J, van Dongen JT (2011) Comparative analysis between plant species of transcriptional and metabolic responses to hypoxia. New Phytol 190: 472–487 [DOI] [PubMed] [Google Scholar]
- Ng S, De Clercq I, Van Aken O, Law SR, Ivanova A, Willems P, Giraud E, Van Breusegem F, Whelan J (2014) Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Mol Plant 7: 1075–1093 [DOI] [PubMed] [Google Scholar]
- Ng S, Giraud E, Duncan O, Law SR, Wang Y, Xu L, Narsai R, Carrie C, Walker H, Day DA, et al. (2013a) Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J Biol Chem 288: 3449–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I, Wang Y, Carrie C, Xu L, Kmiec B, et al. (2013b) A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25: 3450–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, Brennicke A (2006) Transcript levels in plant mitochondria show a tight homeostasis during day and night. Mol Genet Genomics 276: 71–78 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Pal SK, Liput M, Piques M, Ishihara H, Obata T, Martins MC, Sulpice R, van Dongen JT, Fernie AR, Yadav UP, et al. (2013) Diurnal changes of polysome loading track sucrose content in the rosette of wild-type arabidopsis and the starchless pgm mutant. Plant Physiol 162: 1246–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MV, Iyer S, Amerhauser C, Lehmann M, van Dongen JT, Geigenberger P (2016) Oxygen sensing via the ethylene response transcription factor RAP2.12 affects plant metabolism and performance under both normoxia and hypoxia. Plant Physiol 172: 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss ZC, Ebmeier CC, Taatjes DJ (2013) The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol 48: 575–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntarulo S, Sánchez RA, Boveris A (1988) Hydrogen peroxide metabolism in soybean embryonic axes at the onset of germination. Plant Physiol 86: 626–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Aguilar SJ, Keuthe M, Rocha M, Fedyaev VV, Kramp K, Gupta KJ, Rasmusson AG, Schulze WX, van Dongen JT (2011) The composition of plant mitochondrial supercomplexes changes with oxygen availability. J Biol Chem 286: 43045–43053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid F, Horobin RW (1991) Accumulation of fluorescent non-cationic probes in mitochondria of cultured cells: observations, a proposed mechanism, and some implications. J Microsc 163: 233–241 [DOI] [PubMed] [Google Scholar]
- Rhoads DM, Umbach AL, Subbaiah CC, Siedow JN (2006) Mitochondrial reactive oxygen species. Contribution to oxidative stress and interorganellar signaling. Plant Physiol 141: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizza A, Walia A, Lanquar V, Frommer WB, Jones AM (2017) In vivo gibberellin gradients visualized in rapidly elongating tissues. Nat Plants 3: 803–813 [DOI] [PubMed] [Google Scholar]
- Rocha M, Licausi F, Araújo WL, Nunes-Nesi A, Sodek L, Fernie AR, van Dongen JT (2010) Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol 152: 1501–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Sasidharan R, Bailey-Serres J, Ashikari M, Atwell BJ, Colmer TD, Fagerstedt K, Fukao T, Geigenberger P, Hebelstrup KH, Hill RD, et al. (2017) Community recommendations on terminology and procedures used in flooding and low-oxygen stress research. New Phytol 214: 1403–1407 [DOI] [PubMed] [Google Scholar]
- Schertl P, Braun HP (2014) Respiratory electron transfer pathways in plant mitochondria. Front Plant Sci 5: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Weits DA, Feulner CFJ, van Dongen JT (2018) Oxygen sensing and integrative stress signaling in plants. Plant Phys (in press) [DOI] [PMC free article] [PubMed]
- Schmidtmann E, König AC, Orwat A, Leister D, Hartl M, Finkemeier I (2014) Redox regulation of Arabidopsis mitochondrial citrate synthase. Mol Plant 7: 156–169 [DOI] [PubMed] [Google Scholar]
- Schwarzländer M, Finkemeier I (2013) Mitochondrial energy and redox signaling in plants. Antioxid Redox Signal 18: 2122–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzländer M, König AC, Sweetlove LJ, Finkemeier I (2012a) The impact of impaired mitochondrial function on retrograde signalling: a meta-analysis of transcriptomic responses. J Exp Bot 63: 1735–1750 [DOI] [PubMed] [Google Scholar]
- Schwarzländer M, Logan DC, Johnston IG, Jones NS, Meyer AJ, Fricker MD, Sweetlove LJ (2012b) Pulsing of membrane potential in individual mitochondria: A stress-induced mechanism to regulate respiratory bioenergetics in Arabidopsis. Plant Cell 24: 1188–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzländer M, Wagner S, Ermakova YG, Belousov VV, Radi R, Beckman JS, Buettner GR, Demaurex N, Duchen MR, Forman HJ, et al. (2014) The ‘mitoflash’ probe cpYFP does not respond to superoxide. Nature 514: E12–E14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Kronebusch PJ, Borisy GG, Trewavas AJ, Masson PH (1996) Transgenic AEQUORIN reveals organ-specific cytosolic Ca2+ responses to anoxia and Arabidopsis thaliana seedlings. Plant Physiol 111: 243–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, Kim J, Park CM (2010) Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J 61: 661–671 [DOI] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, et al. (2010) The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Bar-Zvi D (2010) ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 22: 3560–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Chen Q, Wu Y, Liu R, Zhang H, Wang S, Tang S, Yang W, Xie Q (2016) ABSCISIC ACID-INSENSITIVE 4 negatively regulates flowering through directly promoting Arabidopsis FLOWERING LOCUS C transcription. J Exp Bot 67: 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SR, Gillard JT, Kustka AB, McCrow JP, Badger JH, Zheng H, New AM, Dupont CL, Obata T, Fernie AR, et al. (2016) Transcriptional orchestration of the global cellular response of a model pennate diatom to Diel light cycling under iron limitation. PLoS Genet 12: e1006490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckel J, Welsh EA, Liberton M, Kunnvakkam R, Aurora R, Pakrasi HB (2008) Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proc Natl Acad Sci USA 105: 6156–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Bush DS, Sachs MM (1994) Elevation of cytosolic calcium precedes anoxic gene expression in maize suspension-cultured cells. Plant Cell 6: 1747–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Bush DS, Sachs MM (1998) Mitochondrial contribution to the anoxic Ca2+ signal in maize suspension-cultured cells. Plant Physiol 118: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Beard KF, Nunes-Nesi A, Fernie AR, Ratcliffe RG (2010) Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci 15: 462–470 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32: 891–904 [DOI] [PubMed] [Google Scholar]
- Teardo E, Carraretto L, Wagner S, Formentin E, Behera S, De Bortoli S, Larosa V, Fuchs P, Lo Schiavo F, Raffaello A, et al. (2017) Physiological characterization of a plant mitochondrial calcium uniporter in vitro and in vivo. Plant Physiol 173: 1355–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti VB, Van Aken O, Morreel K, Vandenbroucke K, van de Cotte B, De Clercq I, Chiwocha S, Fenske R, Prinsen E, Boerjan W, et al. (2010) Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22: 2660–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AY, Gazzarrini S (2012) AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J 69: 809–821 [DOI] [PubMed] [Google Scholar]
- Tsuji H, Nakazono M, Saisho D, Tsutsumi N, Hirai A (2000) Transcript levels of the nuclear-encoded respiratory genes in rice decrease by oxygen deprivation: Evidence for involvement of calcium in expression of the alternative oxidase 1a gene. FEBS Lett 471: 201–204 [DOI] [PubMed] [Google Scholar]
- Turrens JF, Freeman BA, Levitt JG, Crapo JD (1982) The effect of hyperoxia on superoxide production by lung submitochondrial particles. Arch Biochem Biophys 217: 401–410 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Zarkovic J, Yu J, Ruckle ME, McIntosh L, Hock JJ, Bingham S, White SJ, George RM, Subbaiah CC, et al. (2012) Comparison of intact Arabidopsis thaliana leaf transcript profiles during treatment with inhibitors of mitochondrial electron transport and TCA cycle. PLoS One 7: e44339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, De Clercq I, Ivanova A, Law SR, Van Breusegem F, Millar AH, Whelan J (2016a) Mitochondrial and chloroplast stress responses are modulated in distinct touch and chemical inhibition phases. Plant Physiol 171: 2150–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Ford E, Lister R, Huang S, Millar AH (2016b) Retrograde signalling caused by heritable mitochondrial dysfunction is partially mediated by ANAC017 and improves plant performance. Plant J 88: 542–558 [DOI] [PubMed] [Google Scholar]
- Van Aken O, Pecenková T, van de Cotte B, De Rycke R, Eeckhout D, Fromm H, De Jaeger G, Witters E, Beemster GT, Inzé D, et al. (2007) Mitochondrial type-I prohibitins of Arabidopsis thaliana are required for supporting proficient meristem development. Plant J 52: 850–864 [DOI] [PubMed] [Google Scholar]
- Van Aken O, Pogson BJ (2017) Convergence of mitochondrial and chloroplastic ANAC017/PAP-dependent retrograde signalling pathways and suppression of programmed cell death. Cell Death Differ 24: 955–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Van Breusegem F (2015) Licensed to kill: Mitochondria, chloroplasts, and cell death. Trends Plant Sci 20: 754–766 [DOI] [PubMed] [Google Scholar]
- Van Aken O, Whelan J (2012) Comparison of transcriptional changes to chloroplast and mitochondrial perturbations reveals common and specific responses in Arabidopsis. Front Plant Sci 3: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Carrie C, Uggalla V, Paynter E, Giraud E, Whelan J (2009) Defining the mitochondrial stress response in Arabidopsis thaliana. Mol Plant 2: 1310–1324 [DOI] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Law S, Narsai R, Whelan J (2013) AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol 162: 254–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Licausi F (2015) Oxygen sensing and signaling. Annu Rev Plant Biol 66: 345–367 [DOI] [PubMed] [Google Scholar]
- Vanderauwera S, Vandenbroucke K, Inzé A, van de Cotte B, Mühlenbock P, De Rycke R, Naouar N, Van Gaever T, Van Montagu MC, Van Breusegem F (2012) AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 109: 20113–20118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Day DA, Wiskich JT, Vanlerberghe AE, McIntosh L (1995) Alternative oxidase activity in tobacco leaf mitochondria (dependence on tricarboxylic acid cycle-mediated redox regulation and pyruvate activation). Plant Physiol 109: 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L (1992) Coordinate regulation of cytochrome and alternative pathway respiration in tobacco. Plant Physiol 100: 1846–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, McLntosh L (1996) Signals regulating the expression of the nuclear gene encoding alternative oxidase of plant mitochondria. Plant Physiol 111: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Robson CA, Yip JY (2002) Induction of mitochondrial alternative oxidase in response to a cell signal pathway down-regulating the cytochrome pathway prevents programmed cell death. Plant Physiol 129: 1829–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara R, Parada F, Rubio S, Pérez FJ (2012) Hypoxia induces H2O2 production and activates antioxidant defence system in grapevine buds through mediation of H2O2 and ethylene. J Exp Bot 63: 4123–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Krebs M, Kudla J, Schumacher K (2017) Multiparameter imaging of calcium and abscisic acid and high-resolution quantitative calcium measurements using R-GECO1-mTurquoise in Arabidopsis. New Phytol 216: 303–320 [DOI] [PubMed] [Google Scholar]
- Wagner S, Behera S, De Bortoli S, Logan DC, Fuchs P, Carraretto L, Teardo E, Cendron L, Nietzel T, Füßl M, et al. (2015) The EF-hand Ca2+-binding protein MICU choreographs mitochondrial Ca2+ dynamics in Arabidopsis. Plant Cell 27: 3190–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. (1956) On the origin of cancer cells. Science 123: 309–314 [DOI] [PubMed] [Google Scholar]
- Waypa GB, Smith KA, Schumacker PT (2016) O2 sensing, mitochondria and ROS signaling: The fog is lifting. Mol Aspects Med 47-48: 76–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weits DA, Giuntoli B, Kosmacz M, Parlanti S, Hubberten HM, Riegler H, Hoefgen R, Perata P, van Dongen JT, Licausi F (2014) Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat Commun 5: 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welkie D, Zhang X, Markillie ML, Taylor R, Orr G, Jacobs J, Bhide K, Thimmapuram J, Gritsenko M, Mitchell H, et al. (2014) Transcriptomic and proteomic dynamics in the metabolism of a diazotrophic cyanobacterium, Cyanothece sp. PCC 7822 during a diurnal light-dark cycle. BMC Genomics 15: 1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzinger B, Nukarinen E, Nägele T, Weckwerth W, Teige M (2018) The SnRK1 kinase as 1 central mediator of energy signalling between different organelles. Plant Physiol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Jia L, Shi W, Liang J, Zhou F, Li Q, Zhang J (2013) Abscisic acid accumulation modulates auxin transport in the root tip to enhance proton secretion for maintaining root growth under moderate water stress. New Phytol 197: 139–150 [DOI] [PubMed] [Google Scholar]
- Yan B, Dai Q, Liu X, Huang S, Wang Z (1996) Flooding-induced membrane damage, lipid oxidation and activated oxygen generation in corn leaves. Plant Soil 179: 261–268 [Google Scholar]
- Yao N, Greenberg JT (2006) Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 18: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Li J, Liu J, Liu K (2015) An Arabidopsis mitochondria-localized RRL protein mediates abscisic acid signal transduction through mitochondrial retrograde regulation involving ABI4. J Exp Bot 66: 6431–6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yemelyanov VV, Shishova MF, Chirkova TV, Lindberg SM (2011) Anoxia-induced elevation of cytosolic Ca2+ concentration depends on different Ca2+ sources in rice and wheat protoplasts. Planta 234: 271–280 [DOI] [PubMed] [Google Scholar]
- Yoon HK, Kim SG, Kim SY, Park CM (2008) Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol Cells 25: 438–445 [PubMed] [Google Scholar]
- Yu J, Nickels R, McIntosh L (2001) A genome approach to mitochondrial-nuclear communication in Arabidopsis. Plant Physiol Biochem 39: 345–353 [Google Scholar]
- Zhai Z, Liu H, Shanklin J (2017) Phosphorylation of WRINKLED1 by KIN10 results in its proteasomal degradation, providing a link between energy homeostasis and lipid biosynthesis. Plant Cell 29: 871–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ivanova A, Vandepoele K, Radomiljac J, Van de Velde J, Berkowitz O, Willems P, Xu Y, Ng S, Van Aken O, et al. (2017a) The transcription factor MYB29 is a regulator of ALTERNATIVE OXIDASE1a. Plant Physiol 173: 1824–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Li C, Wang R, Chen Y, Shu S, Huang R, Zhang D, Li J, Xiao S, Yao N, et al. (2017b) The Arabidopsis mitochondrial protease FtSH4 Is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol 173: 2294–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Pracheil T, Thornton J, Liu Z (2013) Adenosine triphosphate (ATP) is a candidate signaling molecule in the mitochondria-to-nucleus retrograde response pathway. Genes (Basel) 4: 86–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang G, Song L, Mu P, Wang S, Liang W, Lin Q (2017c) Global analysis of protein lysine succinylation profiles in common wheat. BMC Genomics 18: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wu J, Yuan D, Zhang D, Huang Z, Xiao L, Yang C (2014) Perturbation of auxin homeostasis caused by mitochondrial FtSH4 gene-mediated peroxidase accumulation regulates arabidopsis architecture. Mol Plant 7: 856–873 [DOI] [PubMed] [Google Scholar]
- Zhen S, Deng X, Wang J, Zhu G, Cao H, Yuan L, Yan Y (2016) First comprehensive proteome analyses of lysine acetylation and succinylation in seedling leaves of Brachypodium distachyon L. Sci Rep 6: 31576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2016) Abiotic Stress Signaling and Responses in Plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zones JM, Blaby IK, Merchant SS, Umen JG (2015) High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27: 2743–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubo YO, Potapova TV, Yamburenko MV, Tarasenko VI, Konstantinov YM, Borner T (2014) Inhibition of the electron transport strongly affects transcription and transcript levels in Arabidopsis mitochondria. Mitochondrion 19: 222–230 [DOI] [PubMed] [Google Scholar]