LTR_retriever is an accurate and sensitive program that identifies LTR retrotransposons and generates nonredundant exemplars from DNA sequences for whole-genome annotation and evolutionary studies.
Abstract
Long terminal repeat retrotransposons (LTR-RTs) are prevalent in plant genomes. The identification of LTR-RTs is critical for achieving high-quality gene annotation. Based on the well-conserved structure, multiple programs were developed for the de novo identification of LTR-RTs; however, these programs are associated with low specificity and high false discovery rates. Here, we report LTR_retriever, a multithreading-empowered Perl program that identifies LTR-RTs and generates high-quality LTR libraries from genomic sequences. LTR_retriever demonstrated significant improvements by achieving high levels of sensitivity (91%), specificity (97%), accuracy (96%), and precision (90%) in rice (Oryza sativa). LTR_retriever is also compatible with long sequencing reads. With 40k self-corrected PacBio reads equivalent to 4.5× genome coverage in Arabidopsis (Arabidopsis thaliana), the constructed LTR library showed excellent sensitivity and specificity. In addition to canonical LTR-RTs with 5′-TG…CA-3′ termini, LTR_retriever also identifies noncanonical LTR-RTs (non-TGCA), which have been largely ignored in genome-wide studies. We identified seven types of noncanonical LTRs from 42 out of 50 plant genomes. The majority of noncanonical LTRs are Copia elements, with which the LTR is four times shorter than that of other Copia elements, which may be a result of their target specificity. Strikingly, non-TGCA Copia elements are often located in genic regions and preferentially insert nearby or within genes, indicating their impact on the evolution of genes and their potential as mutagenesis tools.
Transposable elements (TEs) are ubiquitous interspersed repeats in most sequenced eukaryote genomes (Wessler, 2006). According to their transposition schemes, TEs are categorized into two classes. Class I TEs (retrotransposons) use RNA intermediates with a copy-and-paste transposition mechanism (Kumar and Bennetzen, 1999; Wicker et al., 2007). Class II TEs (DNA transposons) use DNA intermediates with a cut-and-paste mechanism (Feschotte and Pritham, 2007; Wicker et al., 2007). Depending on the presence of long terminal repeats (LTRs), class I TEs are further classified as LTR retrotransposons (LTR-RTs) and non-LTR-RTs, including short interspersed nuclear elements (SINEs) and long interspersed nuclear elements (LINEs) (Han, 2010). For simplicity, TEs other than LTR-RT, including both non-LTR-RTs and DNA transposons, are called non-LTR in this study. In plants, LTR-RTs contribute significantly to genome size expansion due to their high copy number and large size (Rensing et al., 2008; Schnable et al., 2009; Nystedt et al., 2013; Ming et al., 2015). For example, retrotransposons contribute to approximately 75% of the size of the maize (Zea mays) genome (Schnable et al., 2009). In Oryza australiensis, a wild relative of rice (Oryza sativa), the amplification of three families of LTR-RTs is attributed to the genome size doubling within the last 3 million years (MY; Piegu et al., 2006). The amplification and elimination of LTR-RTs has shaped genome landscapes, such as genome organization (Ammiraju et al., 2007, 2010) and epigenetic status of the insertion sites (Fedoroff, 2012), thereby affecting the expression of adjacent genes (Hollister and Gaut, 2009; Hollister et al., 2011; Vonholdt et al., 2012; Makarevitch et al., 2015).
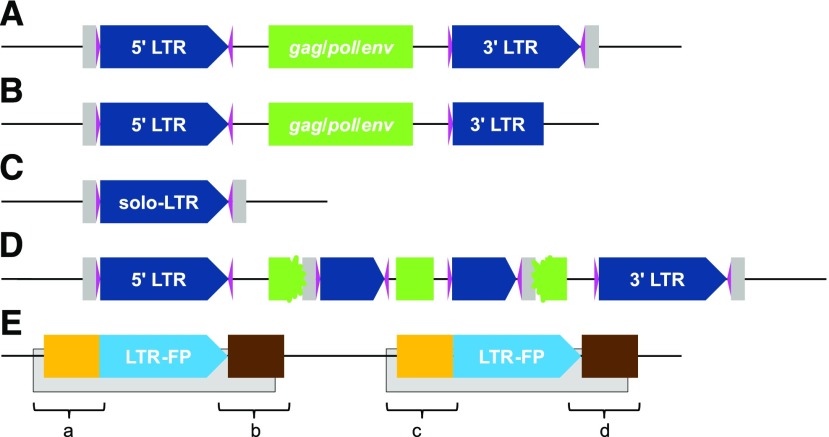
An intact LTR-RT carries a pair of LTRs that usually span 85 to 5,000 bp at both termini (Fig. 1A). In plants, LTRs are typically flanked by 2-bp palindromic motifs (Fig. 1A), commonly 5′-TG…CA-3′ (Zhao et al., 2016), with some rare exceptions. For instance, the first active TE detected in rice, the Tos17 LTR element, has a 5′-TG…GA-3′ motif (Hirochika et al., 1996). The sequence between the 5′ and 3′ LTRs is defined as the internal region and usually ranges from 1,000 to 15,000 bp (Supplemental Fig. S1). To confer transposition activities, the internal region of most autonomous LTR elements should contain a primer-binding site, a polypurine tract, a gag gene (i.e. encoding structural proteins for reverse transcription), and a pol gene (i.e. functioning as protease, reverse transcriptase, and integrase; Havecker et al., 2004). Depending on the order of protein domains in the pol gene, intact LTR-RTs can be categorized further into two superfamilies called Gypsy and Copia (Kumar and Bennetzen, 1999). If the internal region does not contain any open reading frames (e.g. reverse transcriptase genes), the belonging LTR-RT is unable to transpose independently, and it relies on the transposition-related proteins from other autonomous LTR-RTs (Havecker et al., 2004; Jiang, 2016). There are two groups of noncoding LTR-RTs: terminal-repeat retrotransposon in miniature (TRIM; Havecker et al., 2004; Gao et al., 2012) and large retrotransposon derivatives (Havecker et al., 2004). These noncoding LTR-RTs are distinguished by their average length: TRIMs are less than 1 kb and large retrotransposon derivatives are 5.5 to 9 kb (Havecker et al., 2004; Jiang, 2016).
Figure 1.
The structure of LTR-RTs, their derivatives, and false positives. A, The structure of an intact LTR-RT with LTR (navy pentagons), a pair of dinucleotide palindromic motifs flanking each LTR (magenta triangles), the internal region including protein-coding sequences for gag, pol, and env (green boxes), and a 5-bp target site duplication (TSD) flanking the element (gray boxes). B, A truncated LTR-RT with missing structural components. C, A solo LTR. D, A nested LTR-RT with another LTR-RT inserted into its coding region. E, A false LTR-RT detected due to two adjacent non-LTRs (gray boxes). The counterfeit also features a direct repeat (blue pentagons) but usually has extended sequence similarity on one or both sides of the LTR (orange and brown boxes). Regions a to d are extracted and analyzed by LTR_retriever.
The insertion of an LTR-RT is accompanied by the duplication of a small piece of sequence immediately flanking the element, which is called the TSD (4–6 bp in length; Fig. 1A). There are many mechanisms that can introduce mutations to a newly transposed LTR-RT. Due to the sequence similarity between the long direct repeat of an LTR-RT, intraelement recombination can occur, leading to the elimination of the internal region and the formation of a solo LTR (Fig. 1C). The number of solo LTRs indicates the frequency and efficiency of LTR removal in a genome (Tian et al., 2009). Compared with genes, LTR elements are prone to mutations, including deletions, resulting in truncated LTR-RTs (Fig. 1B). Truncated LTR-RTs could also be the product of illegitimate recombination, which generates deletions and translocations (Tian et al., 2009; Zhao et al., 2016). LTR-RTs often insert into other LTR-RTs, generating nested LTR-RTs (Fig. 1D; SanMiguel et al., 1998; Tian et al., 2009; Levy et al., 2010). Given these mutation mechanisms, intact elements only contribute a small fraction of all LTR-RT-related sequences in a genome. If the required structural components are altered (i.e. mutated, truncated, and nest inserted by other TEs; Fig. 1), the LTR element becomes nonautonomous and is difficult to identify using structural information.
Although the structure of the LTR-RT is conserved among species, their nucleotide sequences are not conserved except among closely related species. Particularly, substantial sequence diversity is observed within the LTR region. Therefore, LTR-RTs are usually not adequately identified based on sequence homology. Due to the lack of nucleotide sequence similarity among species, constructing a species-specific LTR library (i.e. exemplars) is essential for the identification of all LTR-RT-related sequences in a newly sequenced genome.
The computational identification of LTR-RTs based on structural features has been implemented multiple times. Such methods are usually used jointly to maximize power in genome annotation projects. However, inconsistent results are often obtained from these tools (Hoen et al., 2015), which could be due to the differences in defining the LTR structure in the program and the different implementation of these methods. LTR_STRUC was one of the earliest developments of genome-wide LTR identification programs (McCarthy and McDonald, 2003), but its scalability and computational potency are limited by the Windows platform, since most genome annotation pipelines are Linux based. LTR_finder (Xu and Wang, 2007) and LTRharvest (Ellinghaus et al., 2008) are, by far, the most sensitive programs in finding LTRs. Nevertheless, these programs suffer from reporting large numbers of false positives (Lerat, 2010). MGEScan-LTR is another early development of LTR-searching programs (Rho et al., 2007). Its recent update on the Web-based platform allows wider usage (Lee et al., 2016), but it is still associated with the issue of false identifications. As the most sizeable content of plant genomes, the assembly of LTR-RTs in plant genomes is typically compromised due to the collapse of short reads from such regions. Fragmented and misassembled repetitive sequences could lead to further error propagation in downstream genome annotation. Unfortunately, most of the current programs are not well adapted to the nature of draft genomes.
In this study, we introduce LTR_retriever, a novel tool for the identification of LTR-RTs. This package efficiently removes false positives from initial software predictions. We benchmarked the performance of LTR_retriever with existing programs using the well-assembled and well-annotated rice genome (International Rice Genome Sequencing Project, 2005) as well as other high-quality monocot and dicot model genomes, such as maize (Jiao et al., 2017), sacred lotus (Nelumbo nucifera; Ming et al., 2013), and Arabidopsis (Arabidopsis thaliana; Arabidopsis Genome Initiative, 2000). Our results indicated that LTR_retriever achieved very high specificity, accuracy, and precision without significantly sacrificing sensitivity, hence significantly outperforming existing methods. In addition, we implemented a module to accurately search for noncanonical LTR-RTs that featured non-TGCA motifs in LTR regions. A search in 50 published genomes identified seven types of noncanonical LTR-RTs, which are mainly Copia elements with substantially shorter length compared with regular Copia elements. Further characterizations show that noncanonical LTR-RTs are less abundant in the genomes but inserted preferentially into genic regions. Finally, we demonstrated the feasibility of making high-quality LTR libraries from self-corrected PacBio reads.
NEW APPROACHES
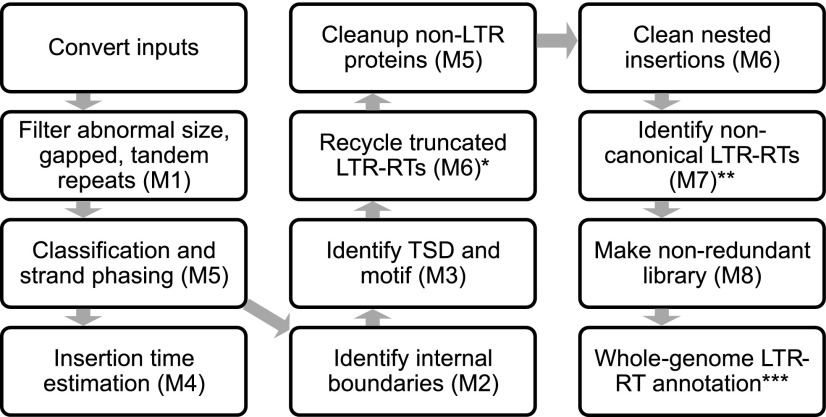
The de novo prediction of LTR-RTs can produce large amounts of false positives. To detect and filter out non-LTR sequences and obtain high-quality LTR-RT exemplars (representative LTR-RT sequences), we developed eight modules with adjustable parameters in LTR_retriever (Fig. 2). A detailed description of each individual module can be found in Supplemental Methods S1.
Figure 2.
Workflow of LTR_retriever. Modules 1 to 8 are indicated in parentheses. *, Optional; supply the -notrunc parameter to deactivate this step. **, Optional; require -nonTGCA [extra_input_file] to activate this module. ***, Optional; supply the -noanno parameter to deactivate this step.
RESULTS
The recovery of LTR elements based on structural features has been implemented in multiple packages. However, high levels of false positives are a key issue. It is possible to reduce false positives by defining more stringent parameters, such as high LTR similarity, intermediate LTR length, and TGCA motif (Fig. 3; Supplemental Table S1). Unfortunately, the level of false negatives becomes high when more stringent parameters are applied (Fig. 3; Supplemental Table S1). The tradeoff between sensitivity and specificity cannot be minimized by merely adjusting parameters of existing tools (Fig. 3; Supplemental Table S1). To establish efficient filters, it is essential to understand the fundamental differences between true LTR elements and false positives. In this study, we employed four statistical metrics (sensitivity, specificity, accuracy, and precision) to evaluate the performance of LTR-RT recovery programs (see “Materials and Methods”).
Figure 3.
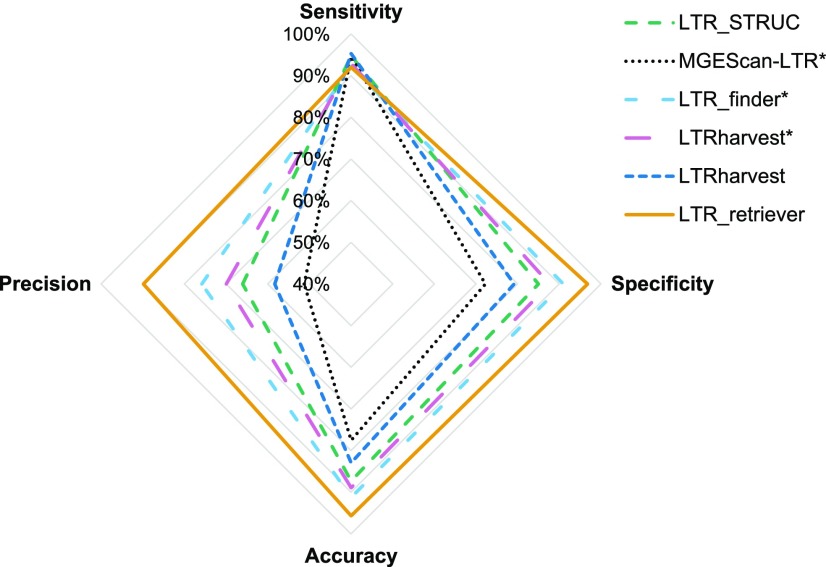
Comparison of the performance of LTR-RT recovery programs on the rice genome. LTR libraries of the rice genome were constructed using LTR_STRUC, MGEScan-LTR, LTR_finder, LTRharvest, and LTR_retriever and then were used to identify LTR sequences in the genome using RepeatMasker. Identified candidate sequences were compared with whole-genome LTR sequences recognized by the manually curated standard library. The genomic size (bp) of true positive, false positive, true negative, and false negative were used to calculate sensitivity, specificity, accuracy, and precision. *, The analysis used optimized parameters (see “Materials and Methods”), while the remainder were in default parameters. The output of optimized LTRharvest was used as input for LTR_retriever. Parameters of LTR identity (-similar), alignment seed length (-seed), and TSD search range (-vic) in LTRharvest were optimized based on the sensitivity and FDR of LTR-RT recovery in rice and further applied to other search programs.
Features of LTR False Positives and Solutions
In genome-assembling practices, one of the most difficult tasks is to assemble highly repetitive regions. Even in the best-assembled genomes, there are still gaps to be filled. In assemblies of nonoverlapping scaffolds, sequence space (gaps) is added manually based on their inferred order. For a piece of sequence with gaps, it is not uncommon that genome assemblers mistakenly join two similar sequences that belong to different TEs from the same family. Under these situations, the ambiguous sequence replaced by gaps is much less reliable than continuous sequence.
Tandem repeats are locally duplicated sequences of two or more bases such as centromere repeats and satellite sequences (Benson, 1999). Although it is possible that an LTR element carries small portions of tandem repeats, it becomes an LTR false positive when the majority sequence of an LTR-RT candidate consists of tandem repeats including low-complexity sequences. We deploy Module 1 in LTR_retriever to eliminate candidates that contain substantial amounts of gaps and tandem repeats (Fig. 2; Supplemental Methods S1). Module 1 also controls sequence length in consideration of both extremely long (15 kb) and short (100 bp) LTR-RTs. The broad range of length settings allows LTR_retriever to identify very short elements like TRIM or exceptionally long elements. The implementation of Module 1 allows LTR_retriever to exclude 4% to 19% of total candidates that are very likely false positives in the genomes of maize and rice, respectively. For example, of 6,159 LTR candidates from the rice genome, 1,162 (18.9%) of them were identified as false positives by Module 1.
Identifying the exact boundaries of an LTR candidate is critical for further structural analysis such as motifs and TSDs. Published methods have applied some schemes to define boundaries. In practice, we found that the external boundaries of an LTR candidate were defined quite precisely by these prediction methods. However, for the internal boundaries that define the start and end of the internal region, the predictions of existing methods are often incorrect. By manual inspections, we found that the percentage of inaccurate internal boundary could be as high as 30% in the rice genome. The misdefined internal boundary of an LTR candidate will result in an incorrect prediction of LTR structures, such as motif, primer-binding site, and polypurine tract, which is likely to fail in the next filtering steps. Thus, we developed Module 2 for correction of the internal boundaries of raw LTR predictions (Fig. 2; Supplemental Methods S1), which could recover an extra 27% of high-quality LTR candidates in the rice genome.
LTR-RT features LTRs flanking each side of the internal region. To exhaustively search for LTR candidates from genomic sequences, most published tools start with finding sequence alignments that are close to each other. This approach can effectively identify LTR elements featured with a pair of LTRs as well as finding non-LTR TE pairs that are similar to each other (Fig. 1). Such non-LTR TE fragments could be contributed by tandem repeats, DNA TEs, SINEs, LINEs, solo LTRs from the same LTR-RT family, or other repetitive sequences, including tandemly located gene families. Excluding such LTR-like false positives is challenging. Moreover, considering that some TEs prefer to insert into other TE sequences, TE clusters are found frequently (SanMiguel et al., 1998; Bergman et al., 2006). The dense distribution of TEs creates a significant amount of false LTRs in de novo predictions. With close inspection, we found that, in most cases, the intraelement sequence similarity of such false positives extended beyond the predicted boundary of the direct repeat (Fig. 1E). In contrast, for a true LTR-RT, the sequence alignment terminates at the boundary of the LTR region. This represents an important structural feature that could distinguish LTR-RTs and its false positives. Another distinctive feature between true LTRs and such false positives is the existence of TSDs. In an LTR-RT, TSDs flanking the element are identical (Fig. 1A). However, in an LTR false positive, sequences at each end have different origins (Fig. 1E). For 4- to 6-bp random sequences, the probability of one being identical to the other is only 0.0002 to 0.0039; meanwhile, a TSD could become unrecognizable due to mutation, depending on the age of the element. This indicates that the detection of TSD could effectively reduce the number of false positives but also may exclude some true positives. Nevertheless, the loss of true positives would not influence the sensitivity unless this LTR-RT is a single-copy element in the genome. To utilize the structural difference between LTR-RT and false positives, Module 3 was developed (Fig. 2; Supplemental Methods S1) to exclude elements with extended alignment beyond LTR regions and those without a TSD immediately flanking the termini of LTRs. Benefiting from the accurate boundaries of candidate elements corrected by Module 2, this module could effectively identify most of the false positives, which could account for approximately half (54%) of total LTR candidates. For example, 2,711 out of 4,997 candidates were further removed by Module 3 in the rice genome.
Module 3 also allows fine-grained adjustment of the internal and external element boundaries by jointly searching TSDs and motifs. As LTR-RTs are represented predominantly by 5-bp TSD and the 5′-TG…CA-3′ motif, searching for such a sequence structure at the termini of direct repeats is prioritized. If the canonical motif is absent, the seven noncanonical motifs (Supplemental Table S2) are searched instead. This function allows LTR_retriever flexibility while accurately characterizing the terminal structure of an LTR candidate. In rice, about 99% of recognized LTR-RTs carry the canonical 5′-TG…CA-3′ motif immediately flanked by 5-bp TSDs, while less than 1% of LTR-RTs have noncanonical motifs with 5-bp TSDs. In other cases, LTR candidates were found carrying the canonical motif with TSDs less than 5 bp, which could be due to interelement recombination or mutation. For example, in the maize genome, LTR-RTs with TSD length of 3 and 4 bp have 108 and 483 occurrences out of 43,226 intact LTR-RTs, respectively.
Similar to retroviruses, direct repeats of a newly inserted LTR-RT are identical to each other. Based on the neutral theory (Vonholdt et al., 2012), Module 4 was developed for the estimation of insertion time of each intact LTR-RT (Fig. 2; Supplemental Methods S1). We applied the Jukes-Cantor model for the estimation of divergence time in noncoding sequences (Jukes and Cantor, 1969). In the rice genome, more than 98% of intact LTR-RTs are inserted within 2.5 MY, given the rice mutation rate of 1.3 × 10−8 mutations per site per year (Ma and Bennetzen, 2004; Supplemental Fig. S2).
In the internal region of an autonomous LTR element, coding sequences like gag, pol, and env are usually found (Fig. 1A; Ellinghaus et al., 2008), which also could help to discriminate LTR-RTs and non-LTRs efficiently. In Module 5, we applied the profile hidden Markov model (pHMM) to identify conserved protein domains that occur in LTR-RT candidate sequences (Fig. 2; Supplemental Methods S1). A total of 102 TE-related pHMMs were identified using the rice TE library, with 55 non-LTR profiles and 47 LTR-RT profiles, which include 30 Gypsy profiles, nine Copia profiles, and eight profiles with ambiguous LTR-RT superfamily classifications (unknown). In rice and Arabidopsis, 79% and 54% of intact LTR-RTs could be classified as either Copia or Gypsy using Module 5, respectively. Furthermore, the direction of LTR-RT could be phased using the profile-match information. Eventually, 65% and 90% of LTR-RTs in rice and Arabidopsis could be phased to either the positive strand or the negative strand, respectively. Since the superfamily classification and strand phasing are dependent on the structure of coding regions, the varying efficiency may imply the structural variation of LTR-RTs in these genomes. A BLAST-based search for non-LTR transposase and plant coding proteins in LTR-RT candidates also is implemented in Module 5 for the further exclusion of non-LTR contaminations. About 1% to 4% of the candidate sequences in the genomes of rice and Arabidopsis were recognized as non-LTR originated and were further eliminated.
After screening and adjustment of LTR candidates using Module 1 to Module 5, the retained candidates are structurally intact LTR-RTs. However, since the screening criteria are very stringent, some true LTR-RTs could be excluded. Through manual inspection, we found that some LTR-RT candidates passed all the screening criteria but have only minor deletions at either the 5′ or 3′ terminus, resulting in failure in the identification of terminal structures. Such candidates are categorized as truncated LTR-RTs, whose intact LTR region and internal region will be retained if there is no highly similar copy in the intact LTR element pool. Module 6 was designed to retain sequence information from truncated LTR-RTs, which contributes about 10% of the sensitivity increment of LTR_retriever (Fig. 2; Supplemental Methods S1). For example, 48 nonredundant LTR sequences were recovered from 590 truncated LTR-RTs in the rice genome.
LTR-RT tends to insert into other LTR-RTs, creating nested insertions. To exclude nested insertions from the LTR exemplars, we developed a function in Module 6, which utilizes all newly identified LTR regions to search for homologous sequences in identified internal regions. This search could recognize and remove LTR-RTs that are nested in intact LTR-RTs. Using this method, about 8% of LTR-RT internal regions in rice and 67.7% in maize are identified as nested with other LTR elements. By removing such nested insertions, the library size can be reduced significantly without sacrifice of sensitivity. More importantly, it avoids the misannotation of LTR sequences as internal regions. Module 6 also can be used to remove nested insertions by other TE elements (i.e. miniature inverted TEs or MITEs). Users can use a curated contaminant source (i.e. a MITE library) to BLAST against the LTR-RT library, then the purger.pl script in the package can be used to remove the entire sequence that is heavily contaminated (default, 70% or greater coverage) or only purge the aligned part of the sequence (optional). MITEs are most abundant in plant genomes in terms of copy number, and MITE libraries can be generated through programs such as MITE-Hunter (Han and Wessler, 2010) and detectMITE (Ye et al., 2016).
Construction of a Nonredundant LTR Library
Construction of the repeat library with nonredundant, high-quality TE sequences is critical for RepeatMasker-based TE and gene annotations, with the size of the repeat library being one of the limiting factors for speed. The required time for whole-genome TE annotations using RepeatMasker is highly correlated to the size of the TE libraries. Since the identified LTR-RTs are redundant, it would significantly speed up whole-genome LTR-RT annotation if the redundancy were eliminated. To reduce redundancy, we developed Module 8 with an 80-90-100 rule compared with the commonly used 80-80-80 rule (Wicker, et al., 2007) to retain sensitivity. In brief, identified LTR-RTs are separated into LTR regions and internal regions for clustering by BLASTclust or CD-HIT with at least 80% sequence identity at the DNA level covering at least 90% of the longest sequence and minimum entry length of 100 bp. Due to the reduced redundancy and exclusion of nested insertions (Module 6), the LTR-RT sequence size was reduced to 10% to 30% of its original size for genomes of rice and maize. For example, the library size of the rice genome decreased from 20 to 5.9 Mb after the redundancy was removed. Accordingly, whole-genome LTR-RT annotation could be accelerated ∼4-fold with similar sensitivity compared with a nonredundant LTR library.
Comparison of Performance with Other LTR Identification Tools
To compare the performance between LTR_retriever and other existing methods, we employed the rice genome as a reference. The rice genome is one of the best sequenced and assembled genomes (International Rice Genome Sequencing Project, 2005). To set a standard for our comparison study, we manually curated representative LTR elements obtained from the rice genome (cv Nipponbare) and generated a compact repeat library that contains 897 sequences with the size of 2.34 Mb. The 897 sequences represent 508 nonredundant LTR elements (Supplemental Methods S1; Supplemental Sequences S1). Using this library, LTR-RT contributes 23.5% of the assembled genome (374 Mb). This number is slightly higher than the two highest estimates from previous studies (20.6% and 22%; Ma et al., 2004; Chaparro et al., 2007), suggesting that the current identification of LTR retrotransposon in cv Nipponbare is close to saturation and that the library is reasonably comprehensive. As a result, this library is used as a reference library for subsequent benchmarking of program performance. The accurate annotation of LTR-RTs in the rice genome allows us to summarize the sequence length of true positive, true negative, false positive, and false negative of a de novo LTR-RT prediction and annotation, hence allowing the evaluation of different methods.
The sensitivity of all existing LTR discovery tools was reported to be very high (Xu and Wang, 2007; Ellinghaus et al., 2008; You et al., 2015); however, systematic evaluation of specificity using the whole-genome sequence length is not available. Specificity describes the proportion of true negative (i.e. non-LTR sequences) being correctly ruled out, which is as important as sensitivity for evaluation of a diagnostic test (Zhu et al., 2010). To better describe the performance of these methods, precision and accuracy are also calculated (Fawcett, 2006). Precision, or positive predictive value, is the proportion of true positives (i.e. LTR sequences) among all positive results revealed by the test. The precision is an indication of false discovery rate (FDR), with the equation FDR = 1 − precision. Accuracy is the proportion of true predictions, which controls systemic errors and random errors (see “Materials and Methods”).
For comparison, we chose four of the most widely used LTR-searching methods, LTR_STRUC (McCarthy and McDonald, 2003), MGEScan-LTR (Rho et al., 2007), LTR_finder (Xu and Wang, 2007), and LTRharvest (Ellinghaus et al., 2008), for performance benchmarks. As LTRharvest is the most flexible program, with more than 20 modifiable parameters, we optimized some of the most influential parameters, including LTR identity (-similar), alignment seed length (-seed), and TSD search range (-vic). The sensitivity and FDR of LTR-RT were used to evaluate the performance of different parameter combinations. The LTR identity of 90%, alignment seed length of 20 bp, and TSD search range of 5 bp achieved the best balance between sensitivity (93%) and FDR (30%; Fig. 3). The optimized parameters were also applied to the parameter settings of LTR_finder and MGEScan-LTR. LTR_retriever can utilize multiple input sources, including the results from LTR_finder, LTRharvest, and MGEScan-LTR. We used input from a single program or inputs from two or more programs in LTR_retriever for comparisons.
As expected, the sensitivities of the most published methods are very high, ranging from 92.2% to 95.3% (Fig. 3; Supplemental Table S1). However, the specificities of these methods are not as high, ranging from 72.3% to 87.7% (Fig. 3; Supplemental Table S1), with the exception of LTR_finder using optimized parameters (91%). The specificity of 72.3% indicates that 27.7% of non-LTR genomic sequences were falsely recognized as LTR-RT sequences. The optimized parameters in LTRharvest led to an improvement of the specificity from 79.2% to 87.7% (Supplemental Table S1). The optimized LTR_finder had the best balance, with sensitivity and specificity both reaching the level of 90%; however, its precision is only 75.8% (Fig. 3; Supplemental Table S1). Although LTR_finder has the highest precision among the published methods, the precision of 75.8% indicates that 24.2% of LTR-RT-related sequences identified in the genome were falsely reported as LTR-RT. The accuracy of existing methods ranges from 77.5% to 91.3%, showing variations in true prediction rate.
We tested LTR_retriever using the optimized LTRharvest results as input. As a stringent filter, LTR_retriever achieved specificity and accuracy of 96.8% and 95.5%, respectively, greatly outperforming existing methods (Fig. 3; Supplemental Table S1). The precision also increased from the original 69.9% to 89.9%, indicating that the FDR dropped to one-third and is among the lowest of all methods (Fig. 3; Supplemental Table S1). Strikingly, the sensitivity of LTR_retriever remained as high as 91.1% compared with the original 93%, meaning that we only sacrificed less than 2% of sensitivity to achieve the observed performance improvements (Fig. 3; Supplemental Table S1). Other input sources, such as those from LTR_finder and MGEScan-LTR, also were tested and showed excellent performance (Supplemental Table S1). Upon combination of two or more input sources, the sensitivity is increased to 94.5%, which is equivalent to the highest level that was achieved by the existing methods, providing a workaround to achieve comprehensive and high-quality predictions (Supplemental Table S1). By excluding the majority of false positives, the final library size was reduced substantially, from the largest 44.4 Mb by MGEScan-LTR to the final 4.4 Mb by LTR_retriever (Supplemental Table S1). The reduced library size significantly reduced the annotation time using RepeatMasker. Based on our experience, we recommend using a reduced library for whole-genome annotation when the original library size is larger than 10 Mb.
Benchmarking on Other Genomes
LTR_retriever was developed based on the rice genome, which has demonstrated the highest specificity, accuracy, and precision among its counterparts with the same level of sensitivity. To test whether the excellent performance of LTR_retriever can be reproduced with other genomes, we chose four other genomes with variable amounts of LTR elements, including two maize genomes (cv B73 and cv Mo17; Xin et al., 2013, Jiao et al., 2017), Arabidopsis (Arabidopsis Genome Initiative, 2000), and sacred lotus (Ming et al., 2013). All these genomic sequences are associated with reasonable repeat libraries, so that the performance of LTR_retriever could be evaluated by comparisons between the respective standard annotations and LTR_retriever-generated libraries.
For all the genomes we tested, LTR_retriever demonstrated very sensitive and accurate performance in retrieving LTR-RTs. Most metrics reached the level of 90% (Table I). For Arabidopsis, we obtained very high specificity and accuracy, 98.9% and 98.4%, respectively, indicating nearly perfect prediction by LTR_retriever. For the ancient eudicot sacred lotus, the four metrics ranged from 81.2% to 91.3%. The maize genome is known to be highly repetitive, and we used both the reference cv B73 (v4) and the cv Mo17 genomes to evaluate the performance of LTR_retriever. With LTR-RTs comprising ∼75% of the 2.1-Gb genome, LTR_retriever could identify 91.1% and 95.7% of LTR-RTs with specificities of 90.6% and 95.7% in the genome assemblies of cv B73 and cv Mo17, respectively. Due to the high LTR-RT content and the nearly perfect performance of LTR_retriever, the precisions reached 96.6% (FDR = 3.4%) and 98.7% (FDR = 1.3%), respectively. It is known that the structure of the maize genome is very complex due to intensive nested TE insertions (SanMiguel et al., 1996); LTR_retriever is able to overcome complex structures and recover most LTR-RTs from the genome.
Table I. Performance of LTR_retriever on model plant genomes.
| Parameter | Genomes |
||||
|---|---|---|---|---|---|
| Rice cv Nipponbare | Sacred Lotus | Maize cv B73 Version 4 | Maize cv Mo17 | Arabidopsisa | |
| Library size (Mb)b | 5.92 | 2.75 | 35.97 | 2.57 | 1.21 |
| Standard library masking | 23.53% | 28.70% | 75.40% | 77.44% | 6.98% |
| Fraction masked | 25.30% | 29.61% | 70.08% | 75.05% | 7.43% |
| Run time (-t 20)c | 42 min | 2.08 h | 94.88 h | 24.8 h | 10 min |
| Sensitivity | 94.48% | 89.35% | 91.10% | 95.65% | 91.17% |
| Specificity | 95.99% | 91.26% | 90.58% | 95.66% | 98.92% |
| Accuracy | 95.64% | 90.70% | 90.97% | 95.65% | 98.38% |
| Precision | 87.90% | 81.18% | 96.61% | 98.69% | 86.33% |
The redundancy of the Arabidopsis library is not reduced, since it is already very compact. bLTR-RT libraries were generated by LTR_retriever using both LTRharvest and LTR_finder inputs. cUsing 20 threads to run the program.
Direct LTR Library Construction from PacBio Reads
The recent development of long-read sequencing technologies has provided a solution for resolving highly repetitive regions in de novo genome-sequencing projects (VanBuren et al., 2015). The PacBio single-molecule, real-time sequencing technology produces long reads with an average length of 10 to 15 kb. Empirically, more than 95% of LTR-RTs range from 1 to 15 kb (Supplemental Fig. S1). Thus, theoretically, the long-read sequencing technology may allow us to identify intact LTR elements directly from the reads.
It is known that the current PacBio RS II platform has an average sequencing error rate of 15%. In our experience, most LTR-RT insertions are structurally detectable if inserted 4 MY ago or younger (Supplemental Fig. S2), which is equivalent to 89.6% of identity between two LTR regions. When mutations/sequencing errors accumulated, the fine structure, such as TSD and terminal motifs, could be mutated and the LTR element would be beyond the detection limit. Thus, the sequencing error rate of 15% would make the actual LTR element undetectable. We tested LTR_retriever using raw PacBio reads, and no confident intact LTR element was recovered. However, LTR_retriever performed excellently using self-corrected PacBio reads that have an error rate of ∼2%.
To test the efficiency of LTR_retriever, we used 20 thousand (k) self-corrected PacBio reads from Arabidopsis Landsberg erecta (Ler-0) as an initial input (see “Materials and Methods”), and with 20k reads as an increment until 180k. The Arabidopsis repeat library from Repbase was used to calculate sensitivity, specificity, accuracy, and precision. The LTR library constructed from the Arabidopsis Ler-0 genome was used as the control to compare with the quality of LTR libraries constructed from PacBio reads. As more reads were used, the prediction of intact LTR-RTs increased linearly (Fig. 4A). However, the sizes of the LTR libraries constructed from these candidates are not increased at the same rate (Fig. 4A), and the sensitivity exceeds the library developed from the genome sequence after 40k reads input and is saturated at 93% after 120k reads being used (Fig. 4B). Since the average length of these reads is 14.6 kb and the Arabidopsis Ler-0 genome was assembled as ∼131 Mb, the sample of 40k and 200k reads is equivalent to 4.5- and 13.4-fold genome coverage, respectively. Moreover, despite the number of reads being used, the average specificity, accuracy, and precision were 99.5%, 98.8%, and 94%, respectively, indicating that very high-quality LTR libraries could be constructed from PacBio reads. Furthermore, the masking potentials (the percentage of the genome that could be masked) of PacBio LTR libraries surpass the standard library level after using 40k or more reads (Supplemental Fig. S3), indicating that it is sufficient to construct a comprehensive library using as little as 4.5× PacBio self-corrected reads. To summarize, LTR_retriever shows high sensitivity, specificity, accuracy, and precision to construct LTR libraries directly from self-corrected PacBio reads prior to genome assembly.
Figure 4.
Direct library construction using self-corrected PacBio reads. A, Identification of intact LTR elements and construction of libraries using the Arabidopsis Ler-0 genome and 20k to 180k self-corrected PacBio reads. B, The performance of custom LTR libraries compared with that from the Arabidopsis reference (Columbia-0) genome.
Identification of LTR-RTs with Noncanonical Motifs
LTR-RT features dinucleotide motifs flanking the direct repeat regions (Fig. 1). The most common motif is the palindromic 5′-TG…CA-3′ motif. However, during manual curation of LTR-RTs, we discovered many LTRs with non-TGCA motifs (A.A. Ferguson and N. Jiang, unpublished data). These noncanonical motifs can be nonpalindromic: for example, Tos17, a rice LTR-RT that can be activated by tissue culture, has noncanonical motifs of 5′-TG…GA-3′ (Hirochika et al., 1996); AtRE1 in Arabidopsis has 5′-TA…TA-3′ motifs (Kuwahara et al., 2000); and TARE1, intensively amplified in the tomato (Solanum lycopersicum) genome, has 5′-TA…CA-3′ motifs (Yin et al., 2013). In addition, three copies of Gypsy-like elements with 5′-TG…CT-3′ motifs were annotated in the soybean (Glycine max) genome (Du et al., 2010).
To recover LTR elements with certain terminal motifs, LTRharvest enables the -motif parameter, allowing users to specify the motif to be discovered, which requires prior motif knowledge. When users apply the default setting (no motif specified), the number of LTR-RT candidates can be 2 to 4 times more than the result with -motif TGCA specified. The significant increase of predicted candidates does not necessarily indicate a large number of non-TGCA LTR recovered. With annotations and further curations, we found that 99% of the additional candidates are false positives in the rice genome.
To identify non-TGCA LTR-RT with high confidence, we developed Module 7 as an optional add-on to LTR_retriever (Supplemental Methods S1). The sacred lotus genome carries many noncanonical LTR elements. We tested the performance of LTR_retriever in identifying such elements using the manually curated noncanonical LTR-RTs from this genome (Supplemental Methods S1). Our results showed that LTR_retriever could identify high-quality noncanonical LTR-RTs, with a sensitivity of 75.6% and a precision of 87.8% (FDR = 12.2%). And the specificity and accuracy were 99.1% and 97.1%, respectively, indicating that the identified noncanonical LTR-RTs are highly accurate.
Noncanonical LTR-RTs Are Widespread in Plants and Insert Preferentially in Genic Regions
To characterize non-TGCA LTR-RTs, we searched through 50 publicly available plant genomes. A total of 870 high-confidence non-TGCA LTR-RTs were found from 42 of these genomes (see “Materials and Methods”). Further categorization of non-TGCA LTR-RTs identified seven types of high-confidence noncanonical motifs, including three (TACT, TGTA, and TCCA) that were not reported previously (Supplemental Table S2). Further classification of open reading frames within these elements based on pHMM search indicated that 89% of classified non-TGCA LTR elements were the Copia type, while only 11% were the Gypsy type (Supplemental Table S2). We also identified 83,368 canonical LTR-RTs in these genomes, with a Gypsy:Copia ratio of 2.9:1 (Table II).
Table II. Average element size of different types of LTR-RTs in 50 sequenced plant genomes.
| Sample | Non-TGCA LTR-RT |
TGCA LTR-RT |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Count | Percentage | LTR | Internal Region | Total | Count | Percentage | LTR | Internal Region | Total | |
| bp | bp | |||||||||
| Copia | 255 | 29.2% | 272 | 4,435 | 4,979 | 14,854 | 17.8% | 911 | 5,765 | 7,588 |
| Gypsy | 34 | 3.9% | 1,115 | 5,044 | 7,273 | 42,667 | 51.2% | 1,288 | 7,352 | 9,928 |
| Unknown | 583 | 66.9% | 233 | 4,684 | 5,151 | 25,847 | 31.0% | 1,184 | 4,656 | 7,025 |
| All LTR | 872 | 100% | 279 | 4,625 | 5,184 | 83,368 | 100% | 1,189 | 6,234 | 8,611 |
For canonical LTR-RTs, the length of the LTR region in Gypsy elements is about 40% longer than Copia elements (Table II). However, in the case of noncanonical LTR-RTs, this size difference is intensified to 400%. This is due to the significant reduction of LTR length of noncanonical Copia elements, from an average size of 911 to 272 bp (Table II). The sizes of the internal region and the whole element of noncanonical Copia also are much shorter than those of Copia elements carrying the TGCA motif (Table II). These results suggest that shorter LTRs may have facilitated the amplification and survival of non-TGCA LTR-RTs.
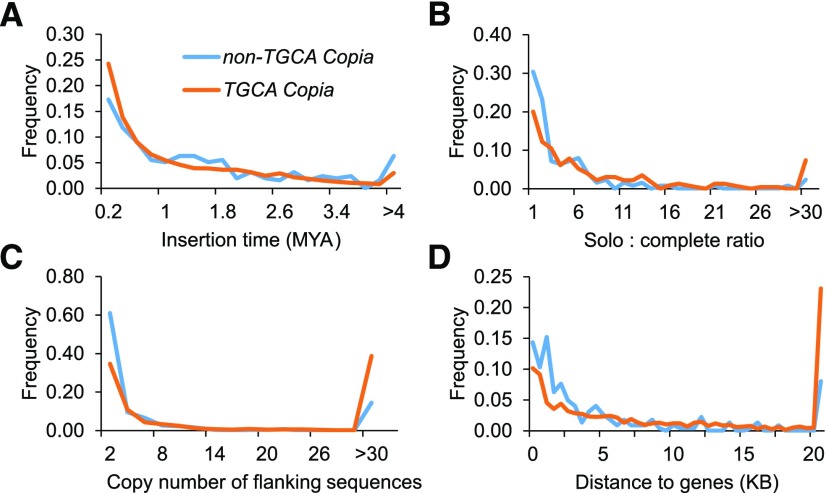
Compared with canonical Copia elements, fewer new insertions (5% less for elements younger than 0.2 MY) and more old elements (7% more of 1.2–1.8 MY elements; Fig. 5A) were observed for noncanonical Copia elements. Meanwhile, we found that elements with canonical motifs were more likely to form solo LTRs: 54% of the noncanonical Copia elements have very low (less than 3) solo-complete LTR ratios; only 32% of canonical Copia elements fall in this category (Fig. 5B). To characterize the insertion preference, we extracted 200-bp flanking sequences of each element and BLAST tested against the genome for the determination of copy numbers. The majority (70%) of the flanking sequences of noncanonical Copia elements have copy numbers less than five, while that of canonical Copia elements is 46% (Fig. 5C). Strikingly, 40% of non-TGCA Copia elements are located within 1 kb of protein-coding genes, which is 16% more frequent than canonical Copia elements (Fig. 5D). Taken together, our results show that noncanonical Copia elements prefer nonrepetitive genomic regions and are often inserted within or close to genes.
Figure 5.
Characterization of noncanonical Copia elements in plants. A, Non-TGCA Copia is older than canonical Copia. B, Non-TGCA Copia has a lower ratio of solo LTR to complete LTR, indicating ineffective exclusion for this type of LTR element. C, Non-TGCA Copia elements are associated predominantly with nonrepetitive flanking sequences. D, Non-TGCA Copia elements are located closer to genes than canonical Copia elements. Blue lines represent non-TGCA (noncanonical) Copia elements, and orange lines represent TGCA (canonical) Copia elements. All analyses were based on 50 plant genomes.
DISCUSSION
Technological advances have minimized the cost of sequencing a genome. The real bottleneck to establishing genomic resources of an organism is the annotation of its genomic sequence. As mentioned above, TEs, particularly LTR-RTs, are the largest component of most plant genomes. If TEs are left unmasked prior to gene annotation, they would seed numerous spurious sequence alignments, producing false evidence for gene identification. Even worse, the open reading frames of TEs look like bona fide genes to most gene-prediction software, corrupting the final annotations. As a result, the first step of genome annotation is to identify TEs and other repeats. Subsequently, these repeats are masked to facilitate gene annotation. As a result, the quality of a repeat library is not only important for the study of repeats but also critical for high-quality gene prediction.
In this study, we reported the development of LTR_retriever, a multithreading-empowered Perl program that can process LTR-RT candidates from LTR_finder, LTRharvest, and MGEScan-LTR and generate high-quality and compact LTR libraries for genome annotations or the study of TEs. We curated LTR elements identified from the rice genome and used the curated LTR library as the standard to test the performance of LTR_retriever in terms of sensitivity, specificity, accuracy, and precision. Benchmark tests on existing programs indicated very high sensitivities achieved; however, specificities and accuracies were not satisfactory, and the FDR could be as high as 49%, suggesting the necessity for improvement (Supplemental Table S1).
Since the annotation of TE sequences usually precedes the annotation of functional genes for a newly sequenced genome, the propagation of false positives in the construction of an LTR library will significantly increase the probability of misidentification of LTR sequences in the genome and further dampen the power of downstream annotations. For example, it is known that most DNA transposons target genic regions and avoid repetitive sequences (Feschotte and Pritham, 2007; Han et al., 2013). As a result, it is not uncommon that the sequence between two adjacent DNA transposons represents gene-coding regions or regulatory sequences. If the two DNA transposons are mistakenly annotated as the LTR of an individual LTR-RT, the intervening genes would be considered as the internal region of an LTR-RT and would be masked before gene annotation. In this scenario, the false positives could be extremely detrimental for downstream analyses. LTR_retriever effectively eliminates such false positives. By processing LTR-RT candidates using LTR_retriever, the specificity and accuracy reached 96.8% and 95.5%, respectively, and the FDR was reduced to 10%, which is among the lowest of all existing methods (Fig. 3; Supplemental Table S1). Strikingly, the sensitivity of LTR_retriever remained as high as 91.2%, meaning that we only sacrificed less than 2% of sensitivity to achieve all these performance improvements (Fig. 3; Supplemental Table S1). Further benchmark tests on two maize genomes, the sacred lotus genome, and the Arabidopsis genome also showed excellent performance (Table I), suggesting that LTR_retriever is compatible with both monocot and dicot genomes.
The majority of LTR-RTs we identified carried a palindromic dinucleotide motif flanking each direct repeat. The motif is well conserved and is usually 5′-TG…CA-3′. Despite the conservation, non-TGCA motifs also were found, but in a much lower frequency. LTR_retriever also demonstrated high performance in identifying such noncanonical LTR-RTs. A broad scan of 50 published plant genomes retrieved seven non-TGCA-type LTR-RTs, with the majority belonging to the Copia superfamily (Supplemental Table S2). For some, the abundance is not ignorable. It appears that, among the four terminal nucleotides (TGCA), only the first nucleotide (T) is invariable. Our systemic survey for the presence of noncanonical termini provides guidance for the future annotation of LTR elements.
Previous studies indicate that Gypsy and Copia elements are differentially located in plant genomes. The distribution of Copia elements is biased toward euchromatic chromosomal arms that are relatively close to genes, whereas Gypsy elements are more likely located in the gene-poor, heterochromatic or pericentromeric regions (Baucom et al., 2009; Bousios et al., 2012). Here, we demonstrate that the noncanonical Copia elements are even closer to genes than canonical Copia elements and insert preferentially into nonrepetitive sequences (Fig. 5). Apparently, there is a negative correlation between the distance to genes and element size, particularly the size of LTRs. As a result, the limited amplification and smaller size are likely the consequences of the target specificity of noncanonical LTR elements.
In Arabidopsis, TEs are separated into two classes based on their locations (Sigman and Slotkin, 2016). One class is present in large constitutive heterochromatic regions, and their CHH methylation is maintained by chromomethylase2; the other class is located near genes where CHH methylation is constantly targeted by RNA-directed DNA methylation. TEs in genic regions are subject to more stringent epigenetic control and demonstrate a higher level of CHH methylation compared with TEs in the nongenic region (Gent et al., 2013; Li et al., 2015). Moreover, TE insertions in genic regions are less likely to spread in the population, since some of them are deleterious. In addition, genic space in a genome is limited compared with the nongenic sequence space. The combined effect of epigenetic control, negative selection, and limited target sites is attributed to the low abundance of noncanonical LTR elements. Furthermore, selection against the insertion of large TEs would result in the relatively small size of both LTR and internal regions of these elements. To this notion, the Tos17 element in rice (with a 5′-TG…GA-3′ terminal motif) is an excellent example. The length of the Tos17 element is only 4.3 kb with an LTR of 138 bp, which is very small compared with other autonomous LTR elements (Table II). It inserts preferentially into genic regions and may amplify rapidly during tissue culture (Miyao et al., 2003). Nevertheless, there are only a few copies of Tos17 in natural populations of rice (Hirochika et al., 1996), suggesting the selective pressure against insertion of this element (Hirochika et al., 1996; Miyao et al., 2003). Because of its insertion preference, Tos17 has been applied as a tool for mutagenesis (Miyao et al., 2003). In our study, we identified 870 high-confidence noncanonical LTRs in 42 out of 50 plant genomes, which is likely an underestimate due to high stringency. These elements also prefer genic insertions, which could contain other Tos17-like active elements in these species. In conclusion, the annotation of noncanonical LTR elements is important not only due to their prevalent distribution but also for the potential application in functional studies in plants.
The recent development of single-molecule sequencing technology enables the assembly of low-complexity and repetitive regions. Many genome-sequencing projects have benefited from the PacBio single-molecule, real-time sequencing technique, which features 10- to 15-kb average read lengths (Ming et al., 2015; VanBuren et al., 2015). Given that the length of most LTR elements is less than 15 kb (Supplemental Fig. S1), it is possible to identify full-length LTRs from PacBio long reads. We applied LTR_retriever on self-corrected PacBio reads, which proved a successful strategy to identify LTR-RTs. For the Arabidopsis Ler-0 genome, 40,000 self-corrected reads covering approximately 4.5× of the genome were more than sufficient to generate an LTR library with higher quality compared with that generated from the assembled genome (Fig. 4). Although self-corrected reads still have an ∼2% sequencing error rate, the generated LTR library was proven highly sensitive and accurate (Fig. 4). The preidentified full-length LTRs may help to estimate LTR percentages of the new genome, study the evolution of LTR-RTs without performing the computationally intensive whole-genome assembly, and facilitate downstream de novo gene annotation.
In summary, we developed a package that takes genome sequences or corrected PacBio reads as input and generates high-quality, nonredundant libraries for LTR elements. It also provides information about the insertion time and location of intact LTR elements in the genome. This tool demonstrates significant improvements in specificity, accuracy, and precision while maintaining high sensitivity compared with existing methods. As a result, it will facilitate future genome assembly and annotation as well as enable rapid comparative studies of LTR-RT dynamics in multiple genomes.
MATERIALS AND METHODS
Implementation of LTR_retriever
LTR_retriever is a command line program developed based on Perl. The package supports multithreading, which was achieved using the Semaphore module in Perl, and multithreading requests are passed to dependent packages. LTR_retriever takes genomic sequences in the FASTA format as input. The program can handle fragmentized and gapped regions, which is a benefit when annotating draft genomes. LTR_retriever has been optimized for plant genomes; however, its parameters can be adjusted for the genomes of other organisms. The output of the program contains a set of high-quality, comprehensive but nonredundant LTR exemplars (library), which can be used to identify or mask LTR sequences using RepeatMasker. A redundant library is also available for further studies. Additionally, a summary table that includes LTR-RT coordinates, length, TSDs, motifs, insertion time, and LTR superfamilies is produced. The program also provides gff3 format output, which is convenient for downstream analysis.
Genomes and Sequences
The initial bacterial artificial chromosome sequences of rice (Oryza sativa ‘Nipponbare’) were downloaded from the Rice Genome Research Program (http://rgp.dna.affrc.go.jp) for our early efforts to construct the rice TE library. The rice reference genome cv Nipponbare release 7 was downloaded from the Michigan State University Rice Genome Annotation Project (http://rice.plantbiology.msu.edu; Kawahara et al., 2013). The sacred lotus (Nelumbo nucifera) genome was downloaded from the National Center for Biotechnology Information under the project identifier AQOG01. The Arabidopsis (Arabidopsis thaliana) reference genome Columbia version 10 was downloaded from TAIR (www.arabidopsis.org; Berardini et al., 2015). The maize (Zea mays) genome cv B73 version AGPv4 was downloaded from Ensembl Plants release 34. An additional of 46 plant genomes were downloaded from Phytozome version 11 (Goodstein et al., 2012; Supplemental Methods S1).
The Arabidopsis Ler-0 genome was sequenced and assembled by Pacific Biosciences using the PacBio RS II platform and P5-C3 chemistry. The assembly is about 131 Mb with a contig N50 (the shortest contig length at 50% of the genome) of 6.36 Mb (https://github.com/PacificBiosciences/DevNet). A total of 184,318 self-corrected reads were also downloaded, which is about 2.69 Gb, with an average read length of 14.6 kb and sequence error rate less than 2%, providing 20.58× coverage of the genome.
Standard LTR Libraries
In this study, LTR libraries from four genomes (rice, maize, Arabidopsis, and sacred lotus) were used to evaluate the performance of LTR_retriever as well as existing tools. The TE database of maize was downloaded from the Maize TE database (http://maizetedb.org). The Arabidopsis repeat library athrep.ref was downloaded from Repbase (Jurka, 2000). The LTR libraries for rice and sacred lotus were curated manually in the Jiang laboratory (Supplemental Methods S1; Supplemental Sequences S1 and S2).
Benchmark Programs and Parameters
LTR_STRUC (McCarthy and McDonald, 2003) was obtained from Vinay Mittal (vinaykmittal@gatech.edu) via personal communication. No parameter settings were available for LTR_STRUC. LTRharvest (Ellinghaus et al., 2008) is part of GenomeTools version 1.5.4 (Gremme et al., 2013). Parameters for running LTRharvest were empirically optimized with -minlenltr 100 -maxlenltr 7000 -mintsd 4 -maxtsd 6 -motif TGCA -motifmis 0 -similar 90 -vic 10 -seed 20. Optimization was focused on LTR identity (-similar; levels 95, 90, 85 [default], and 80), alignment seed length (-seed; levels 50, 40, 30 [default], 20, and 10), and TSD search range (-vic; levels 60 [default], 20, 15, 10, 9, 8, 7, 6, 5, 4, and 3) with combinations of different levels, which were evaluated based on whole-genome (rice) search sensitivity and FDR. The optimum parameter was determined in considering the reduction of FDR and the maintenance of sensitivity. Optimized parameters were also applied to MGEScan-LTR (Rho et al., 2007) and LTR_finder (Xu and Wang, 2007). The modified version of MGEScan-LTR was obtained from the DAWG-PAWS package (Estill and Bennetzen, 2009) and was run with parameter settings -min-mem=20 -mim-dist=1000 -max-dist=15000 -min-ltr=50 -max-ltr=7000 -min-orf=200. LTR_finder version 1.0.6 was run with parameter settings -D 15000 -d 1000 -L 7000 -l 100 -p 20 -M 0.9. To tolerate sequencing errors on corrected PacBio reads, the parameters -motif TGCA -motifmis 1 were used in related LTRharvest runs. To identify extra noncanonical LTR-RTs, no -motif parameter was specified for the maximum sensitivity.
Based on the annotation using the standard LTR library, the whole genome was categorized into four parts, which are true positive (TP; LTR was identified), false negative (FN; LTR was not identified), false positive (FP; non-LTR was identified as LTR), and true negative (TN; non-LTR was not identified as LTR). Four metrics were used to evaluate the performance of LTR_retriever and its counterparts, which are sensitivity, specificity, accuracy, and precision, defined as follows.
The sensitivity, specificity, accuracy, and precision of each test were calculated using genomic sequence lengths by custom Perl scripts.
Data Access
LTR_retriever is an open-source software available in the GitHub repository (https://github.com/oushujun/LTR_retriever). Manually curated LTR libraries for rice and sacred lotus are available as supplemental files.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Size distributions of full-length LTRs, internal regions, and LTR regions in the rice genome.
Supplemental Figure S2. Insertion time distributions of intact LTRs in the rice genome.
Supplemental Figure S3. Masking efficiency of LTR libraries derived from PacBio reads of the Arabidopsis Ler-0 genome.
Supplemental Table S1. Performance of LTR-RT recovery programs on the rice genome.
Supplemental Table S2. LTR-RTs with noncanonical motifs from 50 sequenced plant genomes.
Supplemental Methods S1. Detailed description of each Modules, characterization of Copia elements with a list of 46 plant genomes, and manual curation of LTR elements.
Supplemental Sequence S1. Jiang_rice6.9.lib_LTR: the manually curated nonredundant LTR library of rice.
Supplemental Sequence S2. Jiang_lotus3.3_LTR: the manually curated nonredundant LTR library of sacred lotus.
Acknowledgments
We thank Dr. Yi Liao (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for valuable discussions. We thank Stefan Cerbin and Drs. Cornelius Barry, Rebecca Grumet, Steve van Nocker, and Wayne Loescher for critical reading of the article.
Footnotes
This work was supported by the National Science Foundation (MCB-1121650 and IOS-1126998 to N.J.) and the United States Department of Agriculture National Institute of Food and Agriculture and AgBioResearch at Michigan State University (Hatch grant MICL02408 to N.J.).
Articles can be viewed without a subscription.
References
- Ammiraju JS, Zuccolo A, Yu Y, Song X, Piegu B, Chevalier F, Walling JG, Ma J, Talag J, Brar DS, et al. (2007) Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. Plant J 52: 342–351 [DOI] [PubMed] [Google Scholar]
- Ammiraju JSS, Fan C, Yu Y, Song X, Cranston KA, Pontaroli AC, Lu F, Sanyal A, Jiang N, Rambo T, et al. (2010) Spatio-temporal patterns of genome evolution in allotetraploid species of the genus Oryza. Plant J 63: 430–442 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, SanMiguel PJ, Bennetzen JL (2009) Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet 5: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E (2015) The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53: 474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman CM, Quesneville H, Anxolabéhère D, Ashburner M (2006) Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol 7: R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousios A, Kourmpetis YAI, Pavlidis P, Minga E, Tsaftaris A, Darzentas N (2012) The turbulent life of Sirevirus retrotransposons and the evolution of the maize genome: more than ten thousand elements tell the story. Plant J 69: 475–488 [DOI] [PubMed] [Google Scholar]
- Chaparro C, Guyot R, Zuccolo A, Piégu B, Panaud O (2007) RetrOryza: a database of the rice LTR-retrotransposons. Nucleic Acids Res 35: D66–D70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Tian Z, Bowen NJ, Schmutz J, Shoemaker RC, Ma J (2010) Bifurcation and enhancement of autonomous-nonautonomous retrotransposon partnership through LTR swapping in soybean. Plant Cell 22: 48–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U (2008) LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estill JC, Bennetzen JL (2009) The DAWGPAWS pipeline for the annotation of genes and transposable elements in plant genomes. Plant Methods 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett T. (2006) An introduction to ROC analysis. Pattern Recognit Lett 27: 861–874 [Google Scholar]
- Fedoroff NV. (2012) Presidential address. Transposable elements, epigenetics, and genome evolution. Science 338: 758–767 [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41: 331–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Chen J, Chen M, Meyers BC, Jackson S (2012) A highly conserved, small LTR retrotransposon that preferentially targets genes in grass genomes. PLoS ONE 7: e32010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Ellis NA, Guo L, Harkess AE, Yao Y, Zhang X, Dawe RK (2013) CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res 23: 628–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremme G, Steinbiss S, Kurtz S (2013) GenomeTools: a comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans Comput Biol Bioinform 10: 645–656 [DOI] [PubMed] [Google Scholar]
- Han JS. (2010) Non-long terminal repeat (non-LTR) retrotransposons: mechanisms, recent developments, and unanswered questions. Mob DNA 1: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Qin S, Wessler SR (2013) Comparison of class 2 transposable elements at superfamily resolution reveals conserved and distinct features in cereal grass genomes. BMC Genomics 14: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wessler SR (2010) MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res 38: e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Gao X, Voytas DF (2004) The diversity of LTR retrotransposons. Genome Biol 5: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M (1996) Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci USA 93: 7783–7788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoen DR, Hickey G, Bourque G, Casacuberta J, Cordaux R, Feschotte C, Fiston-Lavier AS, Hua-Van A, Hubley R, Kapusta A, et al. (2015) A call for benchmarking transposable element annotation methods. Mob DNA 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS (2009) Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res 19: 1419–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Smith LM, Guo YL, Ott F, Weigel D, Gaut BS (2011) Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc Natl Acad Sci USA 108: 2322–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jiang N. (2016) Plant transposable elements. eLS 1–7 [Google Scholar]
- Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin CS, et al. (2017) Improved maize reference genome with single-molecule technologies. Nature 546: 524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR (1969) Evolution of protein molecules. In HN Munro, ed, Mammalian Protein Metabolism. Academic Press, pp 21–132 [Google Scholar]
- Jurka J. (2000) Repbase update: a database and an electronic journal of repetitive elements. Trends Genet 16: 418–420 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33: 479–532 [DOI] [PubMed] [Google Scholar]
- Kuwahara A, Kato A, Komeda Y (2000) Isolation and characterization of copia-type retrotransposons in Arabidopsis thaliana. Gene 244: 127–136 [DOI] [PubMed] [Google Scholar]
- Lee H, Lee M, Mohammed Ismail W, Rho M, Fox GC, Oh S, Tang H (2016) MGEScan: a Galaxy-based system for identifying retrotransposons in genomes. Bioinformatics 32: 2502–2504 [DOI] [PubMed] [Google Scholar]
- Lerat E. (2010) Identifying repeats and transposable elements in sequenced genomes: how to find your way through the dense forest of programs. Heredity (Edinb) 104: 520–533 [DOI] [PubMed] [Google Scholar]
- Levy A, Schwartz S, Ast G (2010) Large-scale discovery of insertion hotspots and preferential integration sites of human transposed elements. Nucleic Acids Res 38: 1515–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Gent JI, Zynda G, Song J, Makarevitch I, Hirsch CD, Hirsch CN, Dawe RK, Madzima TF, McGinnis KM, et al. (2015) RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc Natl Acad Sci USA 112: 14728–14733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA 101: 12404–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14: 860–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I, Waters AJ, West PT, Stitzer M, Hirsch CN, Ross-Ibarra J, Springer NM (2015) Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet 11: e1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy EM, McDonald JF (2003) LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics 19: 362–367 [DOI] [PubMed] [Google Scholar]
- Ming R, VanBuren R, Liu Y, Yang M, Han Y, Li LT, Zhang Q, Kim MJ, Schatz MC, Campbell M, et al. (2013) Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol 14: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, VanBuren R, Wai CM, Tang H, Schatz MC, Bowers JE, Lyons E, Wang ML, Chen J, Biggers E, et al. (2015) The pineapple genome and the evolution of CAM photosynthesis. Nat Genet 47: 1435–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15: 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584 [DOI] [PubMed] [Google Scholar]
- Piegu B, Guyot R, Picault N, Roulin A, Sanyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, et al. (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16: 1262–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Rho M, Choi JH, Kim S, Lynch M, Tang H (2007) De novo identification of LTR retrotransposons in eukaryotic genomes. BMC Genomics 8: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nat Genet 20: 43–45 [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, et al. (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sigman MJ, Slotkin RK (2016) The first rule of plant transposable element silencing: location, location, location. Plant Cell 28: 304–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z, Rizzon C, Du J, Zhu L, Bennetzen JL, Jackson SA, Gaut BS, Ma J (2009) Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons? Genome Res 19: 2221–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R, Bryant D, Edger PP, Tang H, Burgess D, Challabathula D, Spittle K, Hall R, Gu J, Lyons E, et al. (2015) Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature 527: 508–511 [DOI] [PubMed] [Google Scholar]
- Vonholdt BM, Takuno S, Gaut BS (2012) Recent retrotransposon insertions are methylated and phylogenetically clustered in japonica rice (Oryza sativa spp. japonica). Mol Biol Evol 29: 3193–3203 [DOI] [PubMed] [Google Scholar]
- Wessler SR. (2006) Transposable elements and the evolution of eukaryotic genomes. Proc Natl Acad Sci USA 103: 17600–17601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8: 973–982 [DOI] [PubMed] [Google Scholar]
- Xin M, Yang R, Li G, Chen H, Laurie J, Ma C, Wang D, Yao Y, Larkins BA, Sun Q, et al. (2013) Dynamic expression of imprinted genes associates with maternally controlled nutrient allocation during maize endosperm development. Plant Cell 25: 3212–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wang H (2007) LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 35: W265–W268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Ji G, Liang C (2016) detectMITE: a novel approach to detect miniature inverted repeat transposable elements in genomes. Sci Rep 6: 19688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Liu J, Xu Y, Liu X, Zhang S, Ma J, Du J (2013) TARE1, a mutated Copia-like LTR retrotransposon followed by recent massive amplification in tomato. PLoS ONE 8: e68587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You FM, Cloutier S, Shan Y, Ragupathy R (2015) LTR Annotator: automated identification and annotation of LTR retrotransposons in plant genomes. Int J Biosci Biochem Bioinform 5: 165–174 [Google Scholar]
- Zhao D, Ferguson AA, Jiang N (2016) What makes up plant genomes: the vanishing line between transposable elements and genes. Biochim Biophys Acta 1859: 366–380 [DOI] [PubMed] [Google Scholar]
- Zhu W, Nancy Z, Ning W (2010) Sensitivity, specificity, accuracy, associated confidence interval and ROC analysis with practical SAS implementations. NorthEast SAS Users Group [Google Scholar]