Abstract
Complex signaling networks between the chloroplast and the nucleus mediate the emergence of the seedling into the light and the establishment of photosynthesis.
Light is the main source of energy for life on Earth, and plants and algae are able to convert light energy, through photosynthesis, into chemical energy that can be used by all organisms. The photosynthetic reactions are housed in the chloroplasts, but the chloroplasts also are the site for synthesis of essential compounds like fatty acids, vitamins, amino acids, and tetrapyrroles. Given their essential role, the correct formation and function of chloroplasts is vital for the growth and development of plants and algae, and hence for almost all organisms. Chloroplasts evolved from an endosymbiotic event where a photosynthetic prokaryotic organism was acquired by a proeukaryotic cell. With time, the photosynthetic prokaryote lost or transferred most of its genes to the host genome. As a result, plastid protein complexes, such as the photosynthetic complexes, are encoded by genes of both the nuclear and plastid genomes. This division of genetic information requires a precise coordination between the two genomes to achieve proper plastid development and function. Plastid development and gene expression are under nuclear control, in what is referred to as anterograde control. However, there also is a signaling system originating in the plastids, so-called retrograde signals, transmitting information about the developmental and functional state of the plastids to the nucleus to regulate nuclear gene expression. Retrograde signaling is a complex network of signals that can be divided into “biogenic control,” referring to signals generated by the plastid as it develops from a proplastid or etioplast into a chloroplast, and “operational control” signals, including those generated from a mature chloroplast in response to environmental perturbations (Chan et al., 2016).
FROM THE PLASTID SIGNAL TO THE COMPLEX NETWORK OF PLASTID-TO-NUCLEUS SIGNALING
The original idea of a single plastid signal has evolved over the years, and we now know that the retrograde signaling system is a complex network of signals and pathways, most of which are still unknown. To date, four major signals/pathways have been described: (1) signals related to the tetrapyrrole biosynthesis pathway (TBP); (2) signals triggered by plastid gene expression (PGE); (3) reactive oxygen species (ROS) and changes to the activity of the photosynthetic electron transport (PET) chain; and (4) signals deriving from disturbed plastid metabolism, such as accumulation of 3′-phosphoadenosine 5′-phosphate, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate (MEcPP), or carotenoid derivatives like β-cyclocitral or apocarotenoids (Estavillo et al., 2011; Ramel et al., 2012; Xiao et al., 2012; Avendaño-Vázquez et al., 2014). The signals derived from plastid metabolism are mostly related to stress responses and operational control of the chloroplast. Those pathways have recently been extensively reviewed, and will not be covered in this review (Bobik and Burch-Smith, 2015; Chi et al., 2015; de Souza et al., 2017). Our focus here is on biogenic control and the retrograde signals linked to the early light response and to chloroplast development.
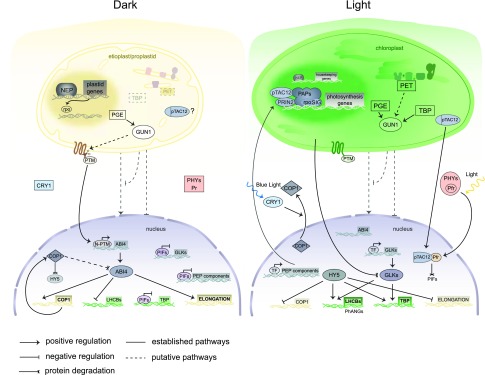
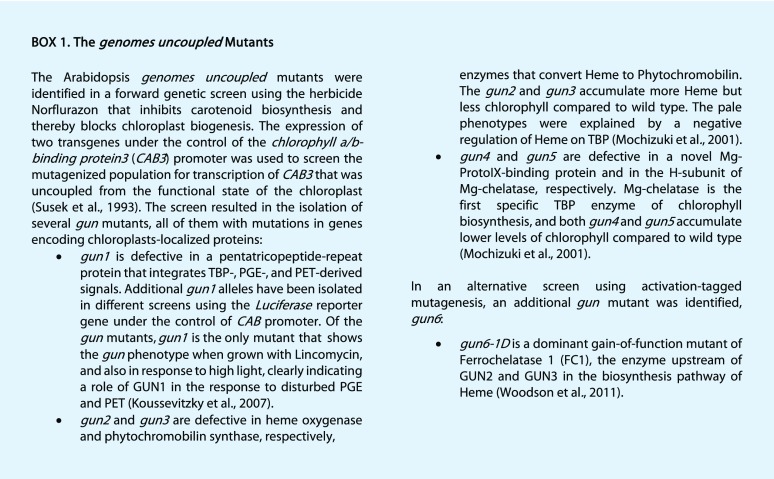
The biogenic signals can be linked to the TBP, PGE, and changes to the PET activity (Fig. 1). Chlorophyll and the other major tetrapyrroles, like heme, siroheme, and phytochromobilin, derive from a common biosynthetic pathway (TBP) located in the plastids. Arabidopsis (Arabidopsis thaliana) mutants with impaired communication between the chloroplast and the nucleus, referred to as the gun (for genomes uncoupled) mutants, were isolated from forward genetic screens (Susek et al., 1993; Mochizuki et al., 2001; Woodson et al., 2011). GUN2 to GUN6 all encode components closely associated with tetrapyrrole biosynthesis, and the respective mutants have changes to the flux through TBP (Box 1). The phenotype of the gun mutants has been linked to oxidative stress causing perturbations of flux through the TBP resulting in accumulation of both ROS and specific metabolites (for review, see Larkin, 2016). In addition, the tetrapyrrole heme has been proposed as a plastid signal positively regulating the expression of Photosynthesis Associated Nuclear Genes (PhANGs) during chloroplast development (Woodson et al., 2011). During chloroplast development, precise coordination between the synthesis of the light-absorbing pigments and the expression of the nuclear-encoded chlorophyll-binding proteins is required to correctly assemble the antennae-reaction center super complexes of PSII and PSI. Thus, regulation of PhANG expression is linked to tetrapyrrole biosynthesis, and it has been suggested that PhANG expression is controlled by a balance between light-signaling pathways and a plastid signal triggered by impaired flux through chlorophyll biosynthesis, Mg-ProtoIX and/or heme (Woodson et al., 2011; Barajas-López et al., 2013). Furthermore, 1O2, a known operational control signal, also has been proposed to play a role during chloroplast development by repressing PhANGs in response to moderate increases of chlorophyll precursors (Page et al., 2017b).
Figure 1.
Model of retrograde signaling during chloroplast biogenesis. Chloroplasts develop from etioplasts or proplastids in response to light. In the dark and in the etioplasts/proplastids, the plastid-encoded genes are mainly transcribed by NEP, and PET and TBP are not functional (dashed line boxes). During the greening process, activation of PEP plays a major role. PEP activity requires the rpo core components, SIGMA factors (SIG), PEP-associated proteins (PAPs, like pTAC12), and other proteins located in the nucleoid, like PRIN2. PRIN2 is involved in the light regulation of PEP activity. The disruption of PET, TBP, or PGE generates a signal that is transduced by GUN1. It is to date unknown if the retrograde signal is of a positive nature that is interrupted in the case of chloroplast disruption (for example, by GUN1) or if the disrupted chloroplasts emit a negative signal (that can be mediated by GUN1). In response to chloroplast signals, PTM is cleaved, and the N-PTM form is translocated to the nucleus where it activates ABI4 expression, inhibiting LHCB expression and promoting the expression of COP1 and genes involved in hypocotyl elongation. In the dark, HY5 is degraded by COP1, which also degrades ABI4. The PIFs repress a set of PhANGs, the GLKs, and genes encoding the nuclear-encoded PEP components. In the light, the light perception by the photoreceptors CRY1 and PHYs causes exclusion of COP1 from the nucleus and degradation of the PIFs. The chloroplast-localized pTAC12 is an essential part of the PEP complex, but also is processed and translocated to the nucleus. The nuclear pTAC12 interacts with the active form of PHYs (Pfr) and contributes to PIF degradation. The exclusion of COP1 allows for accumulation of HY5. Degradation of PIFs releases the transcription of the GLKs and other sets of genes, like the genes encoding the PEP component. HY5 and GLKs induce PhANG (LHCBs and genes coding TBP enzymes, among others) expression, and repress COP1 and the elongation-related genes. Figure by Daria Chrobok.
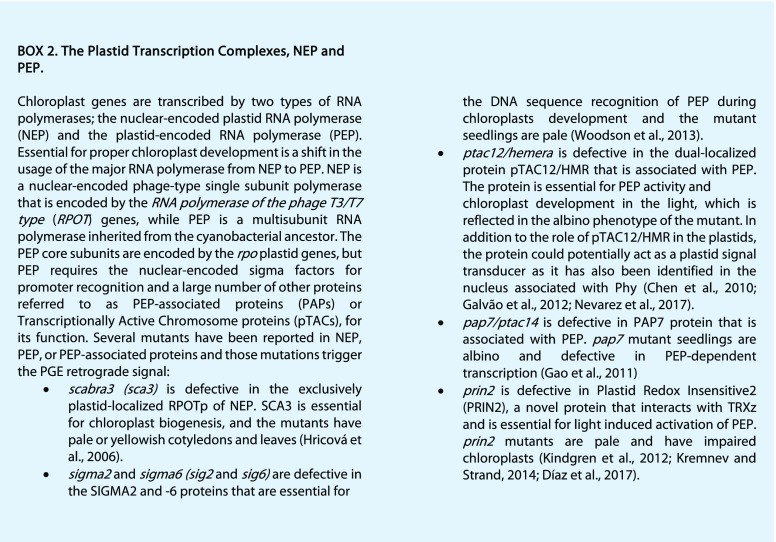
The status of plastid transcription and translation as a trigger for a retrograde signal has been demonstrated by repression of PhANG expression following chemical treatments such as the plastid translational inhibitor lincomycin or rifampicin, which selectively inhibits the plastid-encoded RNA polymerase (PEP; Woodson et al., 2013; Chan et al., 2016). The repression of the PhANG expression phenotype also was observed in mutants affected in the plastid transcriptional machinery, including: the RpoTp/SCA3 protein of the nuclear-encoded RNA polymerase (NEP); the sigma factors SIG2 and SIG6 essential for PEP activity; and other proteins required for full expression of the PEP-dependent genes like Polymerase-Associated Protein7 (PAP7), Plastid Redox Insensitive2 (PRIN2), or Yellow Seedlings1 (Hricová et al., 2006; Gao et al., 2011; Kindgren et al., 2012; Woodson et al., 2013; Box 2). The exact nature of the signal generated by PGE is unknown, and different but overlapping pathways have been suggested (Woodson et al., 2013). One of the proposed signaling pathways involves PEP-transcribed tRNAGlu, the starting and limiting substrate of TBP, thus linking PGE to TBP-mediated signaling with the regulation of PhANG expression during chloroplast biogenesis (Woodson et al., 2013).
In the mature chloroplast, ROS generated by photosynthesis and changes to the redox state of PET act as indicators of environmental fluctuations, and generate signals in the context of operational control (Chan et al., 2016). However, there also are reports where an imbalance of PET is perceived in the frame of biogenic control during the initial stages of plastid development. IMMUTANS (IM) encodes a plastid terminal oxidase (PTOX) required for chloroplast biogenesis, both in seedlings and in adult leaves. IM is a component of a redox pathway that desaturates phytoene, where electrons are transferred from phytoene to plastoquinone (PQ) via phytoene desaturase and from PQ to oxygen via IM. Thus, in the ptox mutant, this pathway is inhibited and carotene biosynthesis is blocked at the phytoene desaturase step. The lack of protective carotenoids results in photooxidation of plastid components, which halts plastid development (Kambakam et al., 2016; Pogorelko et al., 2016). Transcriptomic analyses of etiolated and de-etiolating im seedlings revealed altered expression of nuclear transcription factors that control PhANG expression (Kambakam et al., 2016). The retrograde signal or signals triggered by the imbalance of PET during chloroplast development could potentially coordinate chlorophyll with carotenoid biosynthesis during the early light response.
FROM THE CHLOROPLAST TO THE NUCLEUS: TRANSDUCERS OF PLASTID SIGNALS
The different signals originating in the plastids need to be transduced to the nucleus and connected to transcriptional regulators that control the transcription of PhANGs. GUN1 seems to act as a central hub in the chloroplast, integrating different retrograde signals. Similar to GUN5, GUN1 plays a role in the tetrapyrrole-mediated pathway (Koussevitzky et al., 2007), and genetic analyses revealed that GUN1 also is implicated in the PGE-triggered retrograde signal, as supported by the gun1 mutation reverting the altered PhANG expression phenotype caused by mutations in SIG2, SIG6, and PRIN2 (Kindgren et al., 2012; Woodson et al., 2013). GUN1 is a nuclear-encoded, plastid-localized protein with homology to pentatricopeptide repeat (PPR) domain-containing proteins, but the exact molecular role(s) of GUN1 remains unknown. PPR proteins are involved in RNA processing, and initially a fragment of GUN1 including the PPR domain was shown to bind DNA. However, no interaction was detected between GUN1 and nucleic acids when the entire GUN1 protein was used (Koussevitzky et al., 2007; Tadini et al., 2016). Proteomic analyses have identified a large number of GUN1-interacting proteins representing a wide range of functions, including ribosomal proteins and factors involved in ribosome biogenesis, enzymes in tetrapyrrole biosynthesis, and protein chaperones involved in protein import to the chloroplast, assembly of proteins, and protein degradation (Tadini et al., 2016). However, the identified interactions were weak, suggesting that GUN1 could transiently associate with different protein complexes (Tadini et al., 2016). In addition, given the diversity of proteins identified, the proteomic efforts have so far generated few clues about the true function(s) of GUN1. Possibly, in vivo interaction studies would generate more relevant results.
A plant homeodomain transcription factor with transmembrane domains located to the chloroplast envelope, named PTM, was identified as a potential transduction component of the GUN1-mediated signaling pathway (Sun et al., 2011). Following treatments with lincomycin or norflurazon, a truncated form of PTM (N-PTM) accumulated in the nucleus in a Ser protease-dependent manner. Furthermore, the cleaved N-PTM form was shown to directly control the expression of transcription factors, such as Abscisic Acid Insensitive4 (ABI4), in the nucleus (Sun et al., 2011). The gun phenotype of the ptm mutant was recently challenged after analysis of three ptm mutant alleles (Page et al., 2017a), and alternative pathways for the transduction of the plastid signal to the nucleus were suggested. An alternative for the control of ABI4 activity in response to a retrograde signal is a calcium-dependent, three-component MAPK system. Perturbations of chloroplast function, following norflurazon and/or lincomycin treatments, cause transient increases in cytosolic Ca2+ that activates the MAPK cascade involving the calcium-binding protein 14-3-3ω. The activated MPK3 or MPK6 phosphorylates and activates ABI4 in the nucleus (Guo et al., 2016). In addition, an as yet unknown mechanism transduces retrograde signals to control the protein levels of the nuclear transcription factor Golden2-like1 (GLK1), in a GUN1-independent way (Tokumaru et al., 2017).
Overall, very few transcriptional regulators have been assigned a role in retrograde signaling during the process of chloroplast development. ABI4 acts downstream of GUN1 and the calcium signal, and in response to chloroplast dysfunction, the GUN1-PTM module activates the transcription of ABI4, whereas the Ca2+-activated MPK3/MPK6 phosphorylates ABI4, which will then bind the promoters of PhANGs and thus repress their transcription (Koussevitzky et al., 2007; Sun et al., 2011; Guo et al., 2016). The GLK1 and GLK2 transcription factors are major players in chloroplast biogenesis, integrating retrograde and light signals (Waters et al., 2009; Martín et al., 2016). GLK1 and GLK2 promote chloroplast development by directly activating the expression of nuclear genes encoding proteins involved in chlorophyll biosynthesis, light harvesting, and electron transport (Waters et al., 2009). The important role of the GLKs is reinforced by the pale glk1glk2 double mutant with partially developed chloroplasts and a, albeit weak, gun phenotype (Fitter et al., 2002; Waters et al., 2009). Although genetic analyses indicated that GLK1 and GLK2 are redundant, recent work has revealed specificity that is regulated at transcriptional and posttranscriptional levels (Powell et al., 2012; Kobayashi et al., 2013; Wang et al., 2013; Martín et al., 2016; Tokumaru et al., 2017).
INTERACTION OF PLASTID AND LIGHT SIGNALS DURING CHLOROPLAST DEVELOPMENT
Chloroplast biogenesis is a complex process, and the mechanisms involved differ not only between different plant species but also between different organs within the individual plant. Numerous studies have revealed the significance of proper chloroplast development during all stages of plant growth and the necessity for coordination between growth and chloroplast development (Pogson et al., 2015; Chan et al., 2016). Much of our current knowledge of the molecular mechanisms controlling chloroplast biogenesis comes from studies using the angiosperm model species Arabidopsis. Angiosperms germinate in darkness (e.g. covered by soil) and develop following a dark-adapted program named skotomorphogenesis, which is characterized by long hypocotyls and etiolated cotyledons that contain a form of plastid referred to as etioplasts. Following light exposure, the developmental program changes into photomorphogenesis, hypocotyl elongation is inhibited, and the cotyledons turn green and develop functional chloroplasts (Pogson et al., 2015). In adult leaves, mature chloroplasts develop from proplastids in the stem cells of the apical meristem. Proplastids start to differentiate and form the first extended thylakoid membranes in specific layers of the meristem and in the leaf primordia. Developmental gradients can be observed from the base to the tip (most matured chloroplasts) and from the margin to the midrib within a given leaf in Arabidopsis (Pogson et al., 2015; Gügel and Soll, 2017). However, the spatial (base-to-tip) developmental gradient is more easily detected in monocotyledonous leaves (Pogson et al., 2015).
In angiosperms, chloroplast biogenesis is dependent on light. Exposure to light leads to extensive transcriptional reprogramming, involving up to one-third of the nuclear-encoded genes that are either induced or repressed in response to light signals (Ma et al., 2001; Jiao et al., 2005; Dubreuil et al., 2018). A significant number of the induced genes encode PhANGs. Light and plastid signals regulate expression of the same group of photosynthesis-related genes, and it has been reported that light and retrograde signals also are mediated by cis-elements found in close proximity (Koussevitzky et al., 2007; Chi et al., 2013). Recent studies revealed that nuclear genes encoding photosynthesis-associated genes, such as genes encoding light-harvesting antenna proteins and TBP enzymes, are enriched in G-box elements in their promoters and could be regulated directly by ELON-GATED HYPOCOTYL5 (HY5) and indirectly by GLKs, which are central regulators of light and chloroplast development signaling, respectively (Lee et al., 2007; Waters et al., 2009). This suggests a close interaction between these two signaling pathways that we have recently begun to untangle. However, these two pathways also can act independently, as retrograde signals from the plastid also were shown to regulate PhANG expression in the dark (Sullivan and Gray, 1999; Larkin, 2014). Interestingly, this specific feature of plastid signaling is observed in many gymnosperms where the seedlings green in the dark, and recent work from pine suggested that activation of gymnosperm PhANGs in the dark may be regulated by a plastid-to-nucleus signal (Hills et al., 2015).
The first regulatory step leading to the development of functional chloroplasts is the perception of light by a set of photoreceptors, phytochromes (PHY) and cryptochromes (CRY), which undergo conformational changes to interact with downstream proteins and initiate intracellular signaling pathways (Jiao et al., 2007; Waters and Langdale, 2009). Genetic data revealed that the blue light receptor CRY1 is involved in chloroplast development during blue light induction and that the transcription factor HY5 also is involved in this pathway (Ruckle et al., 2007; Fig. 1). The red and far-red light receptors, the PHYs, also regulate HY5 upon light activation, but PHY signaling mainly proceeds through the Phytochrome-Interacting Proteins (PIFs), which are transcriptional repressors of chloroplast biogenesis (Jiao et al., 2007). A recent quantitative mathematical model, validated in vivo, established a direct link between light input via PHYB-PIF3 and the initiation of chloroplast development. The light-activated form of PHYB degrades PIF3, which represses the nuclear-encoded components of the plastid transcriptional machinery required for transcription of the plastid-encoded photosynthesis genes (Dubreuil et al., 2017; Fig. 1). pTAC12/HMR, one of the components associated with the PEP, was shown to be dually targeted to the plastid and the nucleus. In the nucleus, pTAC12/HMR interacts with the photoactivated PHYs, and PHY-pTAC12/HMR participates in the degradation of PIFs in the light, thereby promoting photomorphogenesis (Chen et al., 2010; Galvão et al., 2012; Fig. 1). The newly synthetized pTAC12/HMR has a plastid transit peptide and is transported to the plastids where it is processed for the subsequent nuclear localization. However, the mechanisms and regulation of this translocation from the plastids to the nucleus is unknown (Nevarez et al., 2017). Detailed investigations of the timing of pTAC12/HMR localization during early light response and chloroplast development could confirm the exciting proposition that pTAC12/HMR might be involved in the coordination of photosynthetic gene expression in both the nucleus and the chloroplast in response to light.
Two molecular pathways for the interaction of light and retrograde signals have recently been described during the transition from skotomorphogenesis to photomorphogenesis in Arabidopsis seedlings (Fig. 1). One of the pathways involves the GUN1-PTM-ABI4 retrograde signaling module, which in this model antagonizes the COP1-HY5 light signal. During de-etiolation of seedlings, ABI4 and HY5 integrate both signals by directly controlling the expression of a set of genes involved in hypocotyl elongation and chloroplast development (Xu et al., 2016). Moreover, the proposed pathway involves HY5 or ABI4 degradation by COP1. Conversely, HY5 and ABI4 both regulate the expression of COP1: ABI4 stimulates and HY5 inhibits expression (Fig. 1). Thus, the balance between these three components provides the means to fine-tune the response to light (Xu et al., 2016).
The second pathway describes the convergence of PHY-PIF-dependent light signal and the GUN1-dependent retrograde signal. The interaction point is downstream of the PIFs and antagonistically regulates the transcriptional photomorphogenic network (Martín et al., 2016). This pathway is independent of ABI4; instead, the GUN1- and the PIF-dependent signals converge on GLK1, repressing or inducing its expression, respectively. This transcriptional control over GLK1 is both ABI4- and HY5-independent, and the existing evidence suggests that GLK1 is a direct target of PIF-mediated repression in the dark (Oh et al., 2012; Leivar and Monte, 2014; Martín et al., 2016). In the model that has been proposed, PIFs directly inhibit GLK1 expression in the dark to support skotomorphogenesis. In the light, GLK1 expression is released and allows photomorphogenesis to proceed. When the plastid is dysfunctional or damaged, a GUN1-mediated retrograde signal represses GLK1 and attenuates photomorphogenesis (Martín et al., 2016; Fig. 1).
Clearly, there is a close interaction between light and plastid signals, but the timing of these two signals during the normal progression of chloroplast biogenesis has been unclear. Early research provided strong indications that a plastid retrograde signal is required for full expression of the nuclear-encoded photosynthetic genes (Sullivan and Gray, 1999). Blocking PEP-driven plastid transcription with rifampicin or using mutants of various PEP components blocked PhANG expression in a light-independent way (Woodson et al., 2013). However, a recent comparative transcriptomic analysis of seedlings grown in light or dark, and the albino pap7 mutant, revealed that altered PGE and blocked plastid development does not affect global PhANG expression (Grübler et al., 2017). On the other hand, a new model where the light response has been investigated in an Arabidopsis cell culture with very high temporal resolution of the chloroplast developmental process suggests that the light signal precedes a plastid signal (Dubreuil et al., 2018). Two phases were clearly observed in the expression profile of the PhANGs in the cell culture. The first phase is dependent on light and triggers changes that will initiate chloroplast development, and more importantly initiates expression of the PEP components. The second phase is dependent on the activation of the chloroplast as the second phase of PhANG induction was absent when chloroplast development was blocked (Dubreuil et al., 2018).
The nature of the plastid signal, whether it is a negative or a positive signal, is another open question, and different hypotheses have been proposed over the years (Pfannschmidt, 2010; Terry and Smith, 2013). However, recent experimental work goes some way to clarifying this question. Supporting the early study by Sullivan and Gray (1999), the work of Martín et al. (2016) demonstrated that the inhibitory effect of lincomycin blocks the photomorphogenic responses early and rapidly, indicating that the retrograde signal is already present in the proplastids/etioplasts or is developed rapidly in response to defective chloroplasts to prevent normal development in light. The results from the experiments by Martín et al. (2016) can be interpreted in two ways: (1) a positive signal from intact plastids that is necessary for GLK1 expression is interrupted by plastid dysfunction in a GUN1-dependent manner; or (2) functional chloroplasts do not emit signals, whereas dysfunctional chloroplasts generate a GUN1-dependent negative signal that represses GLK1 expression. Support for the positive nature of the plastid signal comes from analyses of LHCB expression in pine, where LHCBs are expressed in the dark and thus PhANG expression was shown to be independent of light but dependent on plastid-to-nucleus signals (Hills et al., 2015). These authors proposed an evolutionary model in which angiosperms recruited light-signaling repressors to suppress the response of PhANGs to a positive plastid signal also present in the dark (Hills et al., 2015). Furthermore, modeling of expression data from the plastid and the nuclear genomes strongly suggests that the plastid signal required for full PhANG induction is positive in nature (Dubreuil et al., 2018). In addition, it was recently shown using the prin2 mutant that LHCB expression was directly correlated with the recovery of PEP activity in the prin2 mutant complemented with different PRIN2 variants. It was further suggested that a positive signal is generated by PEP activity in the plastids that stimulates LHCB expression in the nucleus (Díaz et al., 2018). On the other hand, the transcriptomic results using pap7 seedlings have identified different sets of genes regulated by either healthy chloroplast or arrested plastids, respectively, suggesting the existence of both positive and negative signals (Grübler et al., 2017).
HIGHER-ORDER RESPONSES IN THE NUCLEUS TO RETROGRADE SIGNALS
Light has been shown to induce changes in splicing activity (Petrillo et al., 2014; Shikata et al., 2014; Hernando et al., 2017), and the splicing pattern of specific splicing factors involved in dark-to-light shifts is regulated in response to the redox status of PET. Proper splicing of these factors was proposed to play a role in the adaptation of the photosynthetic machinery to changes in the light conditions (Petrillo et al., 2014). However, more experimental work is required to identify the components that regulate splicing in response to retrograde signals, the genes affected, and how those changes affect plant development and photosynthetic activity.
Light has been shown to induce changes to nuclear size and architecture that correlate with transcriptional changes (Bourbousse et al., 2015; Perrella and Kaiserli, 2016). A specific light-regulated locus, CAB, was shown to relocate to the nuclear periphery just before transcriptional induction in a PHYA, PHYB, PIF, COP1, and DET1-dependent manner (Feng et al., 2014). A change to the nuclear position was also observed for the photosynthesis-related genes RBCS1A, GUN5, and PC, supporting the model of repositioning of loci to the nuclear periphery as a mechanism to activate gene expression in response to light (Feng et al., 2014). So far, neither the repositioning of the gene loci nor the changes to nuclear architecture have been linked to retrograde signals, but due to the close relationship between light and plastid signals, a detailed analysis of a potential contribution of plastid signals to those interesting features would be worth pursuing.
In addition, the proportion of heterochromatin was shown to change in light-exposed cotyledons in a CRY1/CRY2-DET1/COP1-HY5-dependent manner (Bourbousse et al., 2015), and RNA-seq, in combination with DNase I hypersensitive site sequencing, revealed that the level of chromatin condensation in darkness is correlated with blocked expression of light- and photosynthesis-related genes. The genes affected by changes in chromatin condensation are enriched in plastid signaling-related genes and their promoters frequently contain the GLK1 binding elements (Liu et al., 2017), but at this point no clear link between retrograde signal(s) and chromatin changes has been established. Currently, much research attention is directed toward the connection between metabolism and chromatin dynamics in animal systems. Thus, exploring the dynamics of global chromatin organization during chloroplast development is a critical future direction for plant sciences.
THE IMPACT OF RETROGRADE SIGNALS ON WHOLE-PLANT PHYSIOLOGY AND DEVELOPMENT
Chloroplast function and integrity are important not only for seedling development during the transition to photomorphogenesis, but also for leaf and plant development throughout the entire plant life cycle. The importance of chloroplast activity during seed and embryo development is reflected in the large number of essential proteins localized to the chloroplast, many of them involved in PGE. The complete loss of SIG5 or RUG2/BSM results in embryo-lethal phenotypes (Hsu et al., 2010; Babiychuk et al., 2011; seedgene.org). The significant role of PEP activity and therefore PGE-triggered retrograde signaling during embryo development was further supported by the embryo pigment-defective phenotype of mutants in PEP components, like pTAC3 or MurE, or with altered PEP activity, like PRIN2 (Garcia et al., 2008; Kremnev and Strand, 2014; seedgene.org). Similarly, the complete loss of essential components of the plastid ribosomal complex, like ClpP proteins or Plastid Ribosomal Protein S5 (Scabra1), also results in embryo lethality (Pogson and Albrecht, 2011; Mateo-Bonmatí et al., 2015). Furthermore, the NEP-defective sca3 mutant demonstrated reticulate patterns with perturbed mesophyll cell differentiation (Hricová et al., 2006). In addition, several anu (for angulata) mutants with abnormal leaf development are affected in genes encoding chloroplast proteins (Casanova-Sáez et al., 2014, Muñoz-Nortes et al., 2017). ANU7 is involved in the regulation of PGE and PhANG expression and interacts genetically with GUN1, indicating a role for retrograde signals in the development of leaf lamina and mesophyll cells (Muñoz-Nortes et al., 2017). PGE was specifically shown to regulate leaf abaxial-adaxial pattern in a GUN1-dependent manner, affecting the expression of specific genes that control asymmetric abaxial-adaxial differentiation (Tameshige et al., 2013). Thus, mutants with impaired plastid development have revealed the close relationship between plastid integrity and leaf development (Aluru et al., 2006; Casanova-Sáez et al., 2014; Lundquist et al., 2014; Van Dingenen et al., 2016; Muñoz-Nortes et al., 2017).
The classic variegated Arabidopsis mutant im is defective in PTOX, and the hypothesis for the formation of the white and green sectors is based on a threshold model where the variegation is dependent on the redox and excitation pressures during the early stages of chloroplast biogenesis when the thylakoid membranes are being formed. The green sectors in the im mutants emerge from plastids that managed to escape the overreduction of the membrane by the action of compensating factors that affect the excitation pressure threshold (Kambakam et al., 2016; Pogorelko et al., 2016). Using detailed kinematic and gene expression studies, it was shown that the leaf becomes photosynthetically active at the same time as it shifts from primary to secondary morphogenesis (Andriankaja et al., 2012). Thus, chloroplast differentiation is an important regulator of the simultaneous onset of cell expansion and photosynthesis. Furthermore, it was suggested that retrograde signaling through tetrapyrroles plays a role in the shift to cell expansion and activation of photosynthesis (Andriankaja et al., 2012).
Regulation of flowering time in response to plastid signals was indicated by the flowering phenotypes of mutants like crd (which overaccumulates MgProto-IX and MgProto-IX-ME), mutants with impaired NEP activity, and the sco1 mutant that is defective in the plastid elongation factor G (Baba et al., 2004; Albrecht et al., 2006; Barajas-López et al., 2013). A model has been proposed where chloroplasts act as stress sensors that transmit information to the nucleus to regulate flowering-related gene expression and thereby also the timing of the plant life cycle. MEcPP, a plastid metabolite that plays a role in a stress-induced retrograde signaling pathway, also regulates flowering time through the regulation of the transcription factor BBX19, which interacts with CO to regulate FT expression (Wang et al., 2014). In addition, the cleaved N-PTM transcription factor was shown to be involved in the repression of the flowering time regulator FLC by recruiting FVE and interacting with the promoter of FLC (Feng et al., 2016).
The GLK transcription factors have been shown to be implicated in fruit development, for example, through the pale siliques of the Arabidopsis glk2 mutant or the identification of GLK2 as the locus affected in the uniform ripening mutation that results in uniformly green tomatoes (Solanum lycopersicum; Waters et al., 2008; Powell et al., 2012; Nguyen et al., 2014). Similarly, a correlation was reported between CaGLK2 and variation in chlorophyll content and color in pepper (Capsicum annuum) fruit (Brand et al., 2014). However, a clear involvement of a retrograde signal controlling GLK2 expression and/or activity during fruit development remains to be demonstrated. A theoretical model referred to as “degradational control” has been proposed where the degradation of Rubisco during senescence, as a source of nitrogen, and its export out of the chloroplast constitute a potential retrograde signal (Pfannschmidt, 2010). Senescence is a highly controlled light- and/or age-dependent program that is accompanied by a transcriptional reprogramming. The light-dependent signaling pathway (PHYB-PIFs-GLKs) involved in senescence is equivalent to the light-signaling pathway regulating chloroplast biogenesis, but the outcome is repression rather than induction of the GLKs and PhANGs (Liebsch and Keech, 2016). However, as is the case for fruit development, the involvement and nature of a plastid retrograde signal in the senescence process has yet to be experimentally demonstrated.
CONCLUDING REMARKS
The field of plastid-to-nucleus signaling has been very dynamic over the last few years, and there have been several major breakthroughs leading to a much more advanced understanding of the mechanisms involved in the communication between the plastids and the nucleus. These recent findings, as highlighted in this review, have made it clear that chloroplast development is controlled by a delicate interplay between light and plastid signaling pathways. In addition, a number of key players involved in this interplay are now identified (Advances box). Critical future directions for this field of plant sciences include efforts to finally understand the mechanism of the elusive key signaling component GUN1, to identify cytosolic components acting as signal transducers from the chloroplast to the nucleus, and to explore the dynamics of global chromatin organization in response to retrograde signals and the establishment of photosynthesis (Outstanding Questions box).
Acknowledgments
We apologize to the authors who did not get their work discussed in this review due to space limitations.
Footnotes
This work was supported by grants from the Swedish Research Foundation, VR (Å.S.).
Articles can be viewed without a subscription.
References
- Albrecht V, Ingenfeld A, Apel K (2006) Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: the impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol Biol 60: 507–518 [DOI] [PubMed] [Google Scholar]
- Aluru MR, Yu F, Fu A, Rodermel S (2006) Arabidopsis variegation mutants: new insights into chloroplast biogenesis. J Exp Bot 57: 1871–1881 [DOI] [PubMed] [Google Scholar]
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GTS, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Avendaño-Vázquez A-O, Cordoba E, Llamas E, San Román C, Nisar N, De la Torre S, Ramos-Vega M, Gutiérrez-Nava MD, Cazzonelli CI, Pogson BJ, León P (2014) An uncharacterized apocarotenoid-derived signal generated in ζ-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis. Plant Cell 26: 2524–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K, Schmidt J, Espinosa-Ruiz A, Villarejo A, Shiina T, Gardeström P, Sane AP, Bhalerao RP (2004) Organellar gene transcription and early seedling development are affected in the rpoT;2 mutant of Arabidopsis. Plant J 38: 38–48 [DOI] [PubMed] [Google Scholar]
- Babiychuk E, Vandepoele K, Wissing J, Garcia-Diaz M, De Rycke R, Akbari H, Joubès J, Beeckman T, Jänsch L, Frentzen M, et al. (2011) Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc Natl Acad Sci USA 108: 6674–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas-López J de D, Kremnev D, Shaikhali J, Piñas-Fernández A, Strand A (2013) PAPP5 is involved in the tetrapyrrole mediated plastid signalling during chloroplast development. PLoS One 8: e60305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik K, Burch-Smith TM (2015) Chloroplast signaling within, between and beyond cells. Front Plant Sci 6: 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbousse C, Mestiri I, Zabulon G, Bourge M, Formiggini F, Koini MA, Brown SC, Fransz P, Bowler C, Barneche F (2015) Light signaling controls nuclear architecture reorganization during seedling establishment. Proc Natl Acad Sci USA 112: E2836–E2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A, Borovsky Y, Hill T, Rahman KAA, Bellalou A, Van Deynze A, Paran I (2014) CaGLK2 regulates natural variation of chlorophyll content and fruit color in pepper fruit. Theor Appl Genet 127: 2139–2148 [DOI] [PubMed] [Google Scholar]
- Casanova-Sáez R, Mateo-Bonmatí E, Kangasjärvi S, Candela H, Micol JL (2014) Arabidopsis ANGULATA10 is required for thylakoid biogenesis and mesophyll development. J Exp Bot 65: 2391–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, Chory J (2010) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Feng P, Ma J, Zhang L (2015) Metabolites and chloroplast retrograde signaling. Curr Opin Plant Biol 25: 32–38 [DOI] [PubMed] [Google Scholar]
- Chi W, Sun X, Zhang L (2013) Intracellular signaling from plastid to nucleus. Annu Rev Plant Biol 64: 559–582 [DOI] [PubMed] [Google Scholar]
- de Souza A, Wang J-Z, Dehesh K (2017) Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu Rev Plant Biol 68: 85–108 [DOI] [PubMed] [Google Scholar]
- Díaz MG, Hernández-Verdeja T, Kremnev J, Crawford T, Dubreuil C, Strand Å (2018) Redox regulation of PEP activity during seedling establishment in Arabidopsis thaliana. Nat Commun 9: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil C, Ji Y, Strand Å, Grönlund A (2017) A quantitative model of the phytochrome-PIF light signalling initiating chloroplast development. Sci Rep 7: 13884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil C, Jin X, Barajas-López JD, Hewitt TC, Tanz SK, Dobrenel T, Schröder WP, Hanson J, Pesquet E, Grönlund A, et al. (2018) Establishment of photosynthesis through chloroplast development is controlled by two distinct regulatory phases. Plant Physiol 176: 1199–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C-M, Qiu Y, Van Buskirk EK, Yang EJ, Chen M (2014) Light-regulated gene repositioning in Arabidopsis. Nat Commun 5: 3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng P, Guo H, Chi W, Chai X, Sun X, Xu X, Ma J, Rochaix J-D, Leister D, Wang H, et al. (2016) Chloroplast retrograde signal regulates flowering. Proc Natl Acad Sci USA 113: 10708–10713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA (2002) GLK gene pairs regulate chloroplast development in diverse plant species. Plant J 31: 713–727 [DOI] [PubMed] [Google Scholar]
- Galvão RM, Li M, Kothadia SM, Haskel JD, Decker PV, Van Buskirk EK, Chen M (2012) Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes Dev 26: 1851–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z-P, Yu Q-B, Zhao T-T, Ma Q, Chen G-X, Yang Z-N (2011) A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol 157: 1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Myouga F, Takechi K, Sato H, Nabeshima K, Nagata N, Takio S, Shinozaki K, Takano H (2008) An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J 53: 924–934 [DOI] [PubMed] [Google Scholar]
- Grübler B, Merendino L, Twardziok SO, Mininno M, Allorent G, Chevalier F, Liebers M, Blanvillain R, Mayer KFX, Lerbs-Mache S, et al. (2017) Light and plastid signals regulate different sets of genes in the albino mutant pap7-1. Plant Physiol 175: 1203–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gügel IL, Soll J (2017) Chloroplast differentiation in the growing leaves of Arabidopsis thaliana. Protoplasma 254: 1857–1866 [DOI] [PubMed] [Google Scholar]
- Guo H, Feng P, Chi W, Sun X, Xu X, Li Y, Ren D, Lu C, David Rochaix J, Leister D, Zhang L (2016) Plastid-nucleus communication involves calcium-modulated MAPK signalling. Nat Commun 7: 12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando CE, Garcia C, Mateos JL (2017) Casting away the shadows: elucidating the role of light-mediated posttranscriptional control in plants. Photochem Photobiol 93: 656–665 [DOI] [PubMed] [Google Scholar]
- Hills AC, Khan S, López-Juez E (2015) Chloroplast biogenesis-associated nuclear genes: control by plastid signals evolved prior to their regulation as part of photomorphogenesis. Front Plant Sci 6: 1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hricová A, Quesada V, Micol JL (2006) The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol 141: 942–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S-C, Belmonte MF, Harada JJ, Inoue K (2010) Indispensable roles of plastids in Arabidopsis thaliana embryogenesis. Curr Genomics 11: 338–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Ma L, Strickland E, Deng XW (2005) Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17: 3239–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambakam S, Bhattacharjee U, Petrich J, Rodermel S (2016) PTOX mediates novel pathways of electron transport in etioplasts of Arabidopsis. Mol Plant 9: 1240–1259 [DOI] [PubMed] [Google Scholar]
- Kindgren P, Kremnev D, Blanco NE, de Dios Barajas López J, Fernández AP, Tellgren-Roth C, Kleine T, Small I, Strand A (2012) The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J 70: 279–291 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Sasaki D, Noguchi K, Fujinuma D, Komatsu H, Kobayashi M, Sato M, Toyooka K, Sugimoto K, Niyogi KK, et al. (2013) Photosynthesis of root chloroplasts developed in Arabidopsis lines overexpressing GOLDEN2-LIKE transcription factors. Plant Cell Physiol 54: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Kremnev D, Strand A (2014) Plastid encoded RNA polymerase activity and expression of photosynthesis genes required for embryo and seed development in Arabidopsis. Front Plant Sci 5: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RM. (2014) Influence of plastids on light signalling and development. Philos Trans R Soc Lond B Biol Sci 369: 20130232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RM. (2016) Tetrapyrrole signaling in plants. Front Plant Sci 7: 1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebsch D, Keech O (2016) Dark-induced leaf senescence: new insights into a complex light-dependent regulatory pathway. New Phytol 212: 563–570 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang W, Zhang K, You Q, Yan H, Jiao Y, Jiang J, Xu W, Su Z (2017) Genome-wide mapping of DNase I hypersensitive sites reveals chromatin accessibility changes in Arabidopsis euchromatin and heterochromatin regions under extended darkness. Sci Rep 7: 4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist PK, Rosar C, Bräutigam A, Weber APM (2014) Plastid signals and the bundle sheath: mesophyll development in reticulate mutants. Mol Plant 7: 14–29 [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo-Bonmatí E, Casanova-Sáez R, Quesada V, Hricová A, Candela H, Micol JL (2015) Plastid control of abaxial-adaxial patterning. Sci Rep 5: 15975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Nortes T, Pérez-Pérez JM, Ponce MR, Candela H, Micol JL (2017) The ANGULATA7 gene encodes a DnaJ-like zinc finger-domain protein involved in chloroplast function and leaf development in Arabidopsis. Plant J 89: 870–884 [DOI] [PubMed] [Google Scholar]
- Nevarez PA, Qiu Y, Inoue H, Yoo CY, Benfey PN, Schnell DJ, Chen M (2017) Mechanism of dual targeting of the phytochrome signaling component HEMERA/pTAC12 to plastids and the nucleus. Plant Physiol 173: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen CV, Vrebalov JT, Gapper NE, Zheng Y, Zhong S, Fei Z, Giovannoni JJ (2014) Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell 26: 585–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu J-Y, Wang Z-Y (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MT, Kacprzak SM, Mochizuki N, Okamoto H, Smith AG, Terry MJ (2017a) Seedlings lacking the PTM protein do not show a genomes uncoupled (gun) mutant phenotype. Plant Physiol 174: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MT, McCormac AC, Smith AG, Terry MJ (2017b) Singlet oxygen initiates a plastid signal controlling photosynthetic gene expression. New Phytol 213: 1168–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrella G, Kaiserli E (2016) Light behind the curtain: photoregulation of nuclear architecture and chromatin dynamics in plants. New Phytol 212: 908–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo E, Godoy Herz MA, Fuchs A, Reifer D, Fuller J, Yanovsky MJ, Simpson C, Brown JWS, Barta A, Kalyna M, et al. (2014) A chloroplast retrograde signal regulates nuclear alternative splicing. Science 344: 427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T. (2010) Plastidial retrograde signalling--a true “plastid factor” or just metabolite signatures? Trends Plant Sci 15: 427–435 [DOI] [PubMed] [Google Scholar]
- Pogorelko GV, Kambakam S, Nolan T, Foudree A, Zabotina OA, Rodermel SR (2016) Impaired chloroplast biogenesis in immutans, an Arabidopsis variegation mutant, modifies developmental programming, cell wall composition and resistance to Pseudomonas syringae. PLoS One 11: e0150983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Albrecht V (2011) Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol 155: 1545–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Ganguly D, Albrecht-Borth V (2015) Insights into chloroplast biogenesis and development. Biochim Biophys Acta 1847: 1016–1024 [DOI] [PubMed] [Google Scholar]
- Powell ALT, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernández-Muñoz R, Vicente A, et al. (2012) Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336: 1711–1715 [DOI] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H, Hanada K, Ushijima T, Nakashima M, Suzuki Y, Matsushita T (2014) Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc Natl Acad Sci USA 111: 18781–18786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Gray JC (1999) Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in roots of the pea lip1 mutant. Plant Cell 11: 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang L (2011) A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat Commun 2: 477. [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Tadini L, Pesaresi P, Kleine T, Rossi F, Guljamow A, Sommer F, Mühlhaus T, Schroda M, Masiero S, Pribil M, et al. (2016) GUN1 controls accumulation of the Plastid Ribosomal Protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol 170: 1817–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameshige T, Fujita H, Watanabe K, Toyokura K, Kondo M, Tatematsu K, Matsumoto N, Tsugeki R, Kawaguchi M, Nishimura M, et al. (2013) Pattern dynamics in adaxial-abaxial specific gene expression are modulated by a plastid retrograde signal during Arabidopsis thaliana leaf development. PLoS Genet 9: e1003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MJ, Smith AG (2013) A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front Plant Sci 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumaru M, Adachi F, Toda M, Ito-Inaba Y, Yazu F, Hirosawa Y, Sakakibara Y, Suiko M, Kakizaki T, Inaba T (2017) Ubiquitin-proteasome dependent regulation of the GOLDEN2-LIKE 1 transcription factor in response to plastid signals. Plant Physiol 173: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dingenen J, Blomme J, Gonzalez N, Inzé D (2016) Plants grow with a little help from their organelle friends. J Exp Bot 67: 6267–6281 [DOI] [PubMed] [Google Scholar]
- Wang C-Q, Guthrie C, Sarmast MK, Dehesh K (2014) BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 26: 3589–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Fouracre J, Kelly S, Karki S, Gowik U, Aubry S, Shaw MK, Westhoff P, Slamet-Loedin IH, Quick WP, et al. (2013) Evolution of GOLDEN2-LIKE gene function in C(3) and C(4) plants. Planta 237: 481–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Langdale JA (2009) The making of a chloroplast. EMBO J 28: 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Moylan EC, Langdale JA (2008) GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J 56: 432–444 [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J 73: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J (2011) Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol 21: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EEK, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K (2012) Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149: 1525–1535 [DOI] [PubMed] [Google Scholar]
- Xu X, Chi W, Sun X, Feng P, Guo H, Li J, Lin R, Lu C, Wang H, Leister D, et al. (2016) Convergence of light and chloroplast signals for de-etiolation through ABI4-HY5 and COP1. Nat Plants 2: 16066. [DOI] [PubMed] [Google Scholar]