Distinctive developmental stages lead to de novo root organogenesis in leaves guide genetically dissection of the primary developmental pathways.
Abstract
Body regeneration through formation of new organs is a major question in developmental biology. We investigated de novo root formation using whole leaves of Arabidopsis (Arabidopsis thaliana). Our results show that local cytokinin biosynthesis and auxin biosynthesis in the leaf blade followed by auxin long-distance transport to the petiole leads to proliferation of J0121-marked xylem-associated tissues and others through signaling of INDOLE-3-ACETIC ACID INDUCIBLE28 (IAA28), CRANE (IAA18), WOODEN LEG, and ARABIDOPSIS RESPONSE REGULATORS1 (ARR1), ARR10, and ARR12. Vasculature proliferation also involves the cell cycle regulator KIP-RELATED PROTEIN2 and ABERRANT LATERAL ROOT FORMATION4, resulting in a mass of cells with rooting competence that resembles callus formation. Endogenous callus formation precedes specification of postembryonic root founder cells, from which roots are initiated through the activity of SHORT-ROOT, PLETHORA1 (PLT1), and PLT2. Primordia initiation is blocked in shr plt1 plt2 mutant. Stem cell regulators SCHIZORIZA, JACKDAW, BLUEJAY, and SCARECROW also participate in root initiation and are required to pattern the new organ, as mutants show disorganized and reduced number of layers and tissue initials resulting in reduced rooting. Our work provides an organ regeneration model through de novo root formation, stating key stages and the primary pathways involved.
Plants have striking regeneration capacities, and can produce new organs from postembryonic tissues (Hartmann et al., 2010; Chen et al., 2014; Liu et al., 2014) as well as reconstitute damaged organs upon wounding (Xu et al., 2006; Heyman et al., 2013; Perianez-Rodriguez et al., 2014; Melnyk et al., 2015; Efroni et al., 2016). Intriguingly, root regeneration upon stem cell damage recruits embryonic pathways (Hayashi et al., 2006; Efroni et al., 2016), whereas in contrast, postembryonic formation of whole new organs, such as lateral roots, appears to use specific postembryonic pathways (Lavenus et al., 2013).
Cross talk between auxin and cytokinin signaling is required for many aspects of plant development and regeneration (El-Showk et al., 2013), although how their synergistic interaction is implemented at the molecular level has not been clarified (Skoog and Miller, 1957; Chandler and Werr, 2015). Exogenous in vitro supplementation of these two hormones results in continuous cell proliferation, to form a characteristic structure termed “callus”. Callus emerges as a common regenerative mechanism for almost all plant organs through in vitro culture (Atta et al., 2009; Sugimoto et al., 2010). There is increasing evidence that callus formation requires hormone-mediated activation of a lateral and meristematic root development program in pericycle-like cells defined by expression of the J0121 marker (Sugimoto et al., 2010). Accordingly, many regulators of lateral root development, such as AUXIN RESPONSE FACTOR7 (ARF7), ARF19, LATERAL ORGAN BOUNDARIES DOMAIN16 (LBD16), LBD17, LBD18, and LBD29, are required for hormone-induced callus formation (for review, see Ikeuchi et al., 2013).
Many species can regenerate new organs from explants (e.g. roots from leaves) without exogenous supplementation of hormones (Bellini et al., 2014). Making roots de novo requires generating the different tissues and cell types of the new organ. All roots have the same tissues, although the number of layers and cells types of these may vary (Kuroha et al., 2006; Lucas et al., 2011). Tissues are continuously formed by asymmetric division of initial cells, which are stem cells, followed by proliferative divisions of their daughter meristematic cells. Stem cell activity is maintained by a quiescent center (QC; van den Berg et al., 1997; Drisch and Stahl, 2015) and auxin activity (Della Rovere et al., 2013). Auxin accumulation in the QC area triggers a dose-dependent and slow response that activates PLETHORA (PLT) factors. PLT proteins form a gradient in the root meristem, which is required to position the QC, maintain stem cell activity, and trigger proliferation of meristematic cells (Aida et al., 2004; Mähönen et al., 2014). Position and activity of the QC also requires radial information delivered by the mobile factor SHORT-ROOT and its downstream target SCARECROW (Sabatini et al., 2003; Levesque et al., 2006; Moubayidin et al., 2016). In addition, WUSCHEL-RELATED HOMEOBOX5 (WOX5) is confined by auxin signaling into the QC and represses differentiation of the stem cell niche, primarily from the QC (Sarkar et al., 2007; Forzani et al., 2014; Pi et al., 2015; Zhang et al., 2015). Tissue formation in the primary root meristem also requires lineage-specific factors that function as cell fate determinants and as tissue endogenous signaling factors to incorporate positional information into patterning (Moreno-Risueno et al., 2015). However, little is known about how tissues are formed de novo.
Recently, a hormone-free method to study de novo root organogenesis in excised leaf blades has been described (Chen et al., 2014). YUCCA-mediated auxin biosynthesis was shown to be ubiquitously enhanced in the leaf mesophyll and indirectly contribute to auxin accumulation near the excision site to trigger localized auxin signaling in the vasculature (Liu et al., 2014; Chen et al., 2016). Formation of new roots involves formation of competent cells through auxin-induced expression of WOX11 transcription factor, which has been defined as a first step for cell fate transition during de novo organ regeneration (Liu et al., 2014). WOX11 and its homolog WOX12 can in addition promote callus formation and up-regulate the callus formation factors LBD16 and LDB29 (Fan et al., 2012; Liu et al., 2014), suggesting that de novo root formation might share similar regulatory mechanisms with callus formation. Subsequently in leaf blade rooting, WOX11 and WOX12 activate WOX5 and WOX7 factors, which are expressed in dividing cells forming root primordia, whereas WOX11/12 expression quickly decreases in dividing cells (Liu et al., 2014; Hu and Xu, 2016). Activation and maintenance of WOX5/7 expression also requires auxin signaling in an unknown pathway different from WOX11/12. Mutants in these WOX factors reduce the number of roots regenerated per leaf blades and affect rooting rate of leaf blades. As a considerably high percentage of leaf blades still root in these mutants, additional regulation must exist.
We have performed an extensive study to further understanding root regeneration from aerial organs. Whole leaves of many species can regenerate entire functional plants in hormone-free medium, and thus we used whole leaves with petioles of Arabidopsis (Arabidopsis thaliana) instead of excised leaf blades. We identified four developmental stages: 1) proliferation of some xylem-associated tissues, forming an endogenous callus; 2) specification of root founder cells within the callus; 3) root primordia initiation from founder cells and patterning; and 4) root meristem activation and emergence. We have also characterized a number of factors regulating these developmental stages. Some auxin and cytokinin signaling factors appear as critical for endogenous callus initiation and formation whereas some stem cell regulators control initiation and patterning of newly formed organs. These results define key stages and regulators required for leaf rooting establishing a developmental framework for de novo organ formation in plants.
RESULTS
Vasculature-Associated Cell Proliferation Is Required for De Novo Organ Regeneration in Arabidopsis
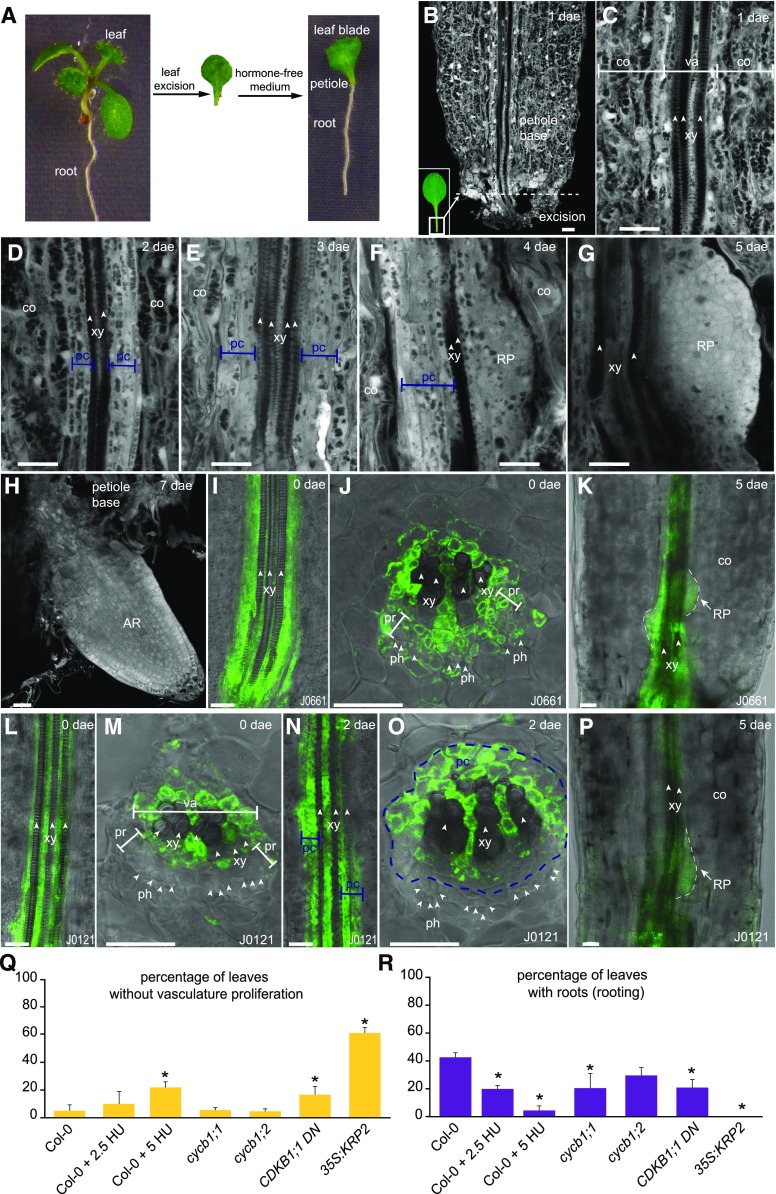
We found that excised whole leaves of Arabidopsis can root without hormone supplementation, similarly to leaf blades as previously described (Chen et al., 2014) and at similar percentages (Supplemental Fig. S1, A and B). Because some species can regenerate entire functional plants from whole leaves without the aid of external hormones, we performed our studies using whole leaves. As de novo formed roots emerged from the petiole base of whole leaves (Fig. 1A), petioles were microscopically observed (Fig. 1B). All petioles showed the same morphological changes during de novo organ regeneration. Although asynchrony was observed in the regeneration process, by 10 d after excision (dae) most leaves (85% to 100%) had regenerated at least one root. At 2 dae, cells adjacent to xylem started to proliferate, forming stratified layers from 3 dae onwards (Fig. 1, C to E) that pushed away xylem conducts and displaced the collenchyma. Vasculature proliferation and subsequent formation of primordia caused the proximal petiole to thicken (Supplemental Fig. S1C). First primordia were visible at 4 dae, and located at external layers of proliferating vasculature (Fig. 1F). At 5 dae, root primordia showed a layered pattern (Fig. 1G). Eventually, newly formed roots with well-organized meristems emerged through petiole tissues from 7 dae onwards (Fig. 1H).
Figure 1.
Developmental stages during rooting of Arabidopsis leaves. A, Whole leaf rooting procedure. B to H, Confocal microscopy pictures of petioles showing (C to E) proliferation of vasculature cells, (F and G) initiation and formation of primordia, and (H) de novo root emergence. I to K, J0661-GFP line in sections of petioles at (I and J) excision time or (K) 5 dae. L to P, J0121-GFP line in sections of petioles at (L and M) excision time, (N and O) 2 dae, and (P) 5 dae. J, M, and O, Transversal sections of petioles at the base. Scale bar: 25 μm. Q and R, Leaves of wild type and cell-cycle mutants, or upon treatment with HU, were assessed for (Q) vasculature proliferation and (R) rooting at 7 dae. Asterisks: statistically significant (P value < 0.05) by GLM and LSD posthoc test. AR, adventitious roots; co, collenchyma; pc, proliferating cells; ph, phloem; pr, procambium; RP, root primordium; va, vasculature; xy, xylem.
Pericycle-like cells (those expressing the J0121 reporter) have been associated to regenerative and morphogenic processes as the source of reprogrammable cells (Sugimoto et al., 2010; Chen et al., 2014). Sections of petioles at the time of excision revealed that the root-pericycle line J0661-GFP marks cells around xylem and procambium cells (Fig. 1, I to J), whereas the J0121-GFP line (Fig. 1, L and M) was restricted to a layer around xylem vessels, being excluded from procambium. Number of cells marked with J0661 and J0121increased quickly during first days of regeneration (Fig. 1, K, and N to P). We observed that all proliferating cells were marked with J0661-GFP whereas some proliferating cells in the J0121 line did not have the GFP (Fig. 1, K, and N to P), indicating that cell proliferation associated to the J0661 reporter. Although it cannot be ruled out that J0661-GFP is activated in proliferating cells, it is possible that xylem and procambium proliferate as part of the reprogramming process. In addition, we observed that primordia at early stages of development were marked with J0121-GFP (Fig. 1P), establishing an association between de novo primordia formation and J0121 identity, similarly to other developmental or regenerative processes such as callus or lateral root formation (Dubrovsky el al., 2006; Sugimoto et al., 2010).
We next studied mutants defective in cell cycle progression, at the G1/S transition, such as the KIP-RELATED PROTEIN2 (KRP2) overexpressor, and at the G2/M transition, such as cyclinB1;1 (cycb1;1) and cycb1;2 mutants and a dominant negative form of the CDKB1;1 kinase (CDKB1;1 DN161). Percentage of petioles showing vasculature-associated proliferation was reduced in Pro35S:KRP2 and CDKB1;1 DN161 lines (Fig. 1Q), whereas only size of proliferating mass of cells was reduced in the rest of the lines. In addition, Pro35S:KRP2 blocked de novo organ regeneration, whereas cycb1;1 and CDKB1;1 DN161 showed a significant reduction in the number of petioles regenerating roots (Fig. 1R). We also chemically inactivated the G1/S transition by incubating leaves with either 2.5 or 5 mm hydroxyurea (HU). We observed a significant decrease in rate of petioles showing vascular-associated proliferation and subsequent new root formation by the HU treatment (Fig. 1, Q and R; Supplemental Fig. S2, A and B). HU treatment did not associate with increased cell death around the vasculature near the leaf excision site upon Trypan Blue staining (Supplemental Fig. S2C). Taken together, these results indicate that cell division activation of vasculature cells is the first and required stage for de novo organogenesis during rooting of leaves.
Cytokinin Biosynthesis and Response during De Novo Root Regeneration
As we had found an association between de novo root regeneration and J0661 and J0121 identities, and callus originates from J0121-marked cells after hormonal induction (Sugimoto et al., 2010), we hypothesized that vasculature proliferation required for leaf rooting could be a type of callus. Cytokinin and auxin signaling are required for callus formation and regeneration, and thus we tested if these two hormones were involved in the developmental pathway leading to de novo organ regeneration from leaves.
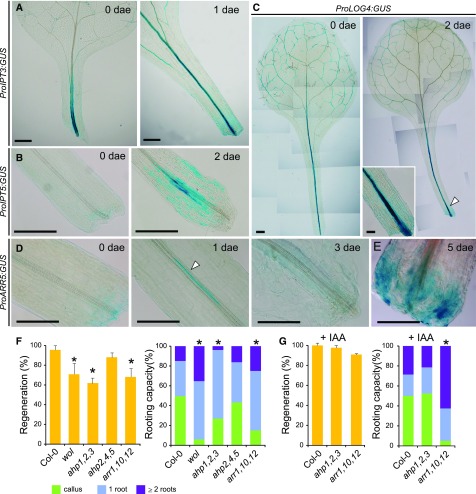
First, we investigated cytokinin biosynthesis and signaling (Zürcher and Müller, 2016). We found enriched expression of ProIPT3:GUS in petioles right after excision (Fig. 2A). ProIPT5:GUS was highly expressed in vascular-associated cells in the petiole base at 2 dae (Fig. 2B), whereas ProLOG4:GUS, which was originally expressed in leaf vasculature at 0 dae, increased expression at the petiole base over time (Fig. 2C, arrowhead). Cytokinin signaling, as reported by ProARR5:GUS (D’Agostino et al., 2000), was restricted to a subset of vascular-associated cells near the petiole base at 2 dae, which associated with proliferation of vasculature, to decrease at later time points (Fig. 2D) and it did not show expression during de novo primordia formation. Consistent with ARABIDOPSIS RESPONSE REGULATORS5 (ARR5) reporting primary cytokinin response during petiole vasculature proliferation (D’Agostino et al., 2000), incubation of leaf explants with synthetic cytokinin 6-benzylaminopurine (6-BAP) increased ProARR5:GUS expression in the petiole vasculature and basal region (Fig. 2E).
Figure 2.
Cytokinin acts locally during de novo root regeneration. A and C, Expression of (A) ProIPT3:GUS, (B) ProIPT5:GUS, (C) ProLOG4:GUS, and (D and E) ProARR5:GUS in petioles at indicated dae. E, Leaves were incubated with 5 μM 6-BAP. F, Mutants impaired in cytokinin signaling show defective regeneration (which accounts for petioles with vasculature proliferation, primordia, and roots; yellow barplots) and rooting capacity (multicolored barplots show frequencies of vasculature proliferation and of roots or primordia, indistinctly) at 7 dae. G, Cytokinin signaling mutant leaf explants incubated with 1 μM indole-3-acetic acid and measured for root regeneration responses as in (F). Scale bars: (A to C) 0.5 mm; (D and E) 0.25 mm. Asterisks: P value < 0.05 by GLM followed by LSD posthoc or χ2 test.
Next, we studied the ability of several cytokinin signaling mutant combinations in ARABIDOPSIS HIS KINASE4 (AHK4) and ARABIDOPSIS HIS PHOSPHOTRANSFER PROTEIN1 (AHP1) to AHP5 and ARR1, ARR10, and ARR12 genes in regulating both vasculature proliferation and de novo root regeneration. We quantified petioles regenerating as petioles showing vasculature proliferation/thickening, root primordia formation, or visible roots. Leaf petioles of wooden leg (wol, a dominant negative mutant in AHK4 receptor) and ahp1 ahp2 ahp3 and arr1 arr10 arr12 loss-of-function mutants displayed lower regeneration percentage at 7 dae (Fig. 2F). Interestingly, wol, ahp1 ahp2 ahp3, and arr1 arr10 arr12 mutants were also defective in hormone-induced callus formation from different tissue explants, such as leaves, cotyledons, and roots (Supplemental Fig. S3), indicating that specific cytokinin signaling is required for both callus formation and vasculature proliferation in leaf petioles. Despite cytokinin signaling being required for vasculature proliferation in petioles during rooting, for those leaf petioles of cytokinin signaling mutants that regenerated, we detected a higher number of roots (which we categorized by frequencies in numbers of roots and designated as “rooting capacity”; Fig. 2F). Higher auxin-to-cytokinin ratios have been shown to induce specification and growth of new root primordia (Müller and Sheen, 2008). Thus, we wondered if we could alter new primordia initiation by altering hormone ratios. Cytokinin treatment increased vascular proliferation in a concentration- and time-dependent manner (Supplemental Fig. S2, D and E). We observed that regeneration deficiencies of most cytokinin signaling mutants could be compensated by low levels of exogenous auxin (Fig. 2G) that also increased vasculature proliferation at the expense of reducing rooting capacity in the ahp1 ahp2 ahp3 mutant (Fig. 2G). Taken together, these results indicate a dual role for cytokinin first as a positive activator of vasculature cell division, and second as a negative regulator of root primordia initiation.
Specific Auxin Signaling Factors Regulate De Novo Root Regeneration
Next, we investigated auxin signaling during rooting of leaves using the DR5 reporter line (Ulmasov et al., 1997). ProDR5:GUS was expressed in vascular-associated cells at the proximal region of the petiole, as early as 12 h after excision (hae), to increase quickly to 1 dae, remaining high during proliferative stages and decreasing over time coincident with deceleration of vasculature growth (Fig. 3, A and B). We observed high localized expression of ProDR5:GUS in clusters of cells at the time of primordia initiation and formation. Consistent with a regulatory role of auxin in rooting, local INDOLE-3-ACETIC ACID INDUCIBLE (IAA) application significantly increased root formation, along with expansion of ProDR5:GUS expression domain (Fig. 3, A and C). In addition, the auxin-overproducing mutant superroot2 (sur2; Barlier et al., 2000) also showed increased number of de novo formed roots at 7 dae, similarly to auxin-treated petioles (Fig. 3C).
Figure 3.
Distinctive auxin signaling pathways regulate de novo root regeneration. A and B, Auxin response reported by (A) ProDR5:GUS expression in Arabidopsis leaf petioles and (B) quantification of GUS-stained area. (C) ProDR5:GUS expression and de novo-formed adventitious roots in leaves supplemented with 1 μM indole-3-acetic acid (left) and in the sur2-1 auxin overproduction mutant (right). D, Regeneration percentage and (E) rooting capacity at 7 d after excision of auxin signaling mutants (nph4-1 arf19-1, axr2-1, slr-1, crane-2, and iaa28-1) and auxin overproducer sur2-1. F, Vasculature proliferation area on Arabidopsis leaves cultivated for 14 d in hormone-free medium. Scale bars: 200 μm. Asterisks: P value < 0.05 by GLM followed by LSD posthoc or χ2 test. AR, adventitious roots.
Auxin signaling is regulated by IAA cofactors acting in combination with AUXIN RESPONSE FACTOR (ARF) transcriptional partners (Li et al., 2016). We reasoned that IAA factors regulating postembryonic (lateral) root formation (Goh et al., 2012) could be involved in de novo organ formation. We assessed the mutants auxin resistant2-1 (axr2-1), solitary-root-1 (slr-1), crane-2, iaa28-1, and the double mutant nonphototrophic hypocotyl4-1 arf19-1 (see “Materials and Methods”); which are, respectively, gain-of-function mutants for the factors IAA7, IAA14, IAA18, IAA28, and a double loss-of-function mutant for ARF7 and ARF19. crane-2 reduced de novo root regeneration, with approximately 40% of petioles not showing any sign of vasculature proliferation or de novo root formation (Fig. 3D). In addition, slr-1 and iaa28-1 showed reduced rooting capacity, although all petioles showed some vasculature proliferation (Fig. 3E). When we quantified vasculature proliferation area of petioles regenerating, we observed a reduction for iaa28-1 and crane-2 but not for slr-1 (Fig. 3F). These results indicate that the auxin signaling module mediated by IAA18 is required for de novo root regeneration at stages of vascular proliferation, that of IAA28 for vascular proliferation and root initiation, whereas IAA14 appears to be required only for de novo root initiation.
We also investigated if these mutants were affected in hormone-induced callus formation. We found that only crane-2 showed reduction in all explants assayed after hormonal incubation, whereas axr2-1 intriguingly showed an increase for callus formed from root explants (Supplemental Fig. S4, A to C). These results indicate that vasculature proliferation during rooting and hormone-induced callus use the auxin signaling pathway mediated by IAA18. Our previous results also showed that cytokinin signaling required for vasculature proliferation during de novo organogenesis was also required for hormone-induced callus formation, suggesting that vasculature proliferation is a type of callus. We investigated ABERRANT LATERAL ROOT FORMATION4 (ALF4) during leaf rooting, as aberrant lateral root formation4 (alf4), in this case, alf4-1 mutants, have been linked to callus formation (Sugimoto et al., 2010) and vascular connection during graft establishment (Melnyk et al., 2015). We observed that during de novo root formation, vasculature proliferation is reduced by 2.5-fold in alf4-1 mutants, which is accompanied by 15-fold decrease in de novo formed root and primordia (Supplemental Fig. S5). Based on these results, we designated the vasculature proliferation developmental stage as the endogenous callus formation.
Auxin Signaling Factors Are Required for De Novo Organ Regeneration in Leaf Blades
In contrast to endogenous callus formation observed in petioles of whole leaves during rooting, limited vasculature proliferation was observed during rooting of leaf blades (Liu et al., 2014). We wondered to what extent auxin and cytokinin signaling factors regulating proliferation at the petiole base would be involved in rooting of leaf blades. When we assessed rooting capacity in the leaf blades of these mutants, we observed that crane-2 and iaa28-1 displayed a reduction in rooting capacity whereas slr-1 presented moderate although nonsignificant reductions (Supplemental Fig. S6A). wol and arr1 arr10 arr12 mutants were similarly affected as slr-1, whereas no change was detected for ahp1 ahp2 ahp3 and ahp2 ahp4 ahp5 mutants (Supplemental Fig. S6B). As IAA18 and IAA28 are required for endogenous callus formation during whole leaf rooting and are shared with leaf blade rooting, it is possible that de novo root regeneration in leaf blades could also involve an endogenous callus developmental program.
Local Auxin Accumulation at the Petiole Base Is Dependent on Polar Auxin Transport
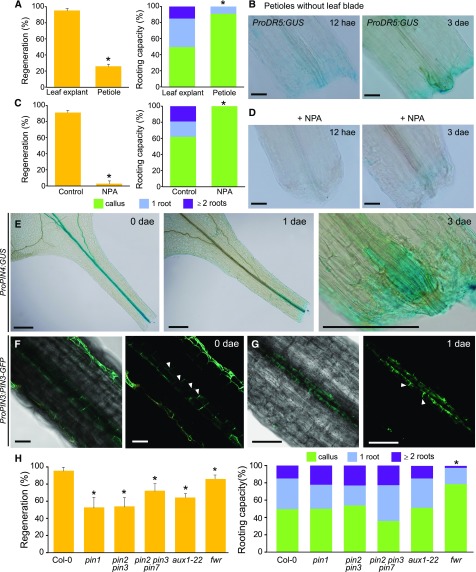
As localized auxin signaling was required for whole leaf rooting, we wondered about the source of auxin. YUCCA-mediated auxin biosynthesis was shown to be ubiquitously enhanced in the mesophyll of leaf blades shortly after wounding (Chen et al., 2016). During rooting of whole leaves, we detected ProYUC9:GUS-enriched expression in leaf mesophyll cells at 12 hae, which progressively decreased at later time points (Supplemental Fig. S7A). ProYUC8:GUS expression was induced in proliferating vascular-associated cells at the petioles’ bases 2 dae (Supplemental Fig. S7B). These results indicate two possible sources of auxin during first stages of regeneration, one from the leaf blade, and the other from the proliferating vasculature itself. To determine if there was differential contribution of these two auxin sources, we removed the leaf blade and found a significant decrease in regeneration and rooting capacity, which in most cases stopped at the endogenous callus stage (Fig. 4A). Local auxin response (ProDR5:GUS expression) in petioles without leaf blade was low or undetectable (Fig. 4B). We also locally inhibited polar auxin transport through application of N-1-naphthylphthalamic acid (NPA) at the blade-petiole junction, resulting in almost complete block of auxin response and subsequent regenerative response (Fig. 4, C and D).
Figure 4.
Polar auxin transport from the leaf blade to the petiole is required for rooting. A and B, Distal blade excision reduces (A) regeneration rate and rooting capacity at 7 dae and (B) ProDR5:GUS expression at 12 hae and 3 dae. C and D, Local treatment at the blade-petiole junction with 1% NPA reduces (C) regeneration rate and rooting capacity at 7 dae, and (D) ProDR5:GUS expression. E to G, Expression of (E) ProPIN4:GUS and (F and G) ProPIN3:PIN3:GFP. Arrowheads point to membrane-localized PIN-GFP. Scale bars: (B, D, F, and G) 0.2 and (E) 0.5 mm. H, Regeneration and rooting capacity in auxin transport mutants at 7 dae. Asterisks: P value < 0.05 by GLM followed by LSD posthoc or χ2 test.
We next characterized PIN-FORMED (PIN) expression. ProPIN4:GUS was ubiquitously expressed in the leaf vasculature at the time of excision; however, at 1 dae it was only expressed at the base of the petiole and in endogenous callus at 3 dae (Fig. 4E). ProPIN3:PIN3:GFP expression in the petiole at the time of excision was polarized toward the base in epidermal cell membranes and both laterally and basally localized in some vascular-associated cell membranes (Fig. 4F, arrowheads). From 12 hae to 1 dae, we found enriched expression of ProPIN3:PIN3:GFP in a subset of vascular-associated cells at the petiole base region (Fig. 4G). Interestingly, PIN3-GFP protein in cells proximal to the excision was oriented toward the apex (upper-left direction in Fig. 4G, arrowheads) whereas its orientation changed to the base in cells at the distal position from the excision.
We also investigated rooting in mutants of genes affected in auxin influx (AUX1) or auxin efflux (PIN1, PIN2, PIN3, and PIN7), which have been described to have low auxin transport rates (Petrásek et al., 2006). Consistent with our previous observations, the regenerative potential of leaves of pin1, pin2 pin3, pin2 pin3 pin7, and aux1 mutants was reduced (Fig. 4H). GNOM loss-of-function mutants have altered polar auxin transport by interfering with PIN internalization (Kleine-Vehn et al., 2009). When we studied the GNOM mutant fewer (fwr; Okumura et al., 2013), we observed significant differences in regeneration and rooting capacity (Fig. 4H). fwr is a weak gnom allele but it is possible that several auxin transporters are simultaneously affected, which could explain why there is also a reduction in rooting capacity whereas no reduction was observed for single auxin transporter mutants.
Postembryonic Root Founder Cells Establish on Endogenous Callus Prior Primordia Formation
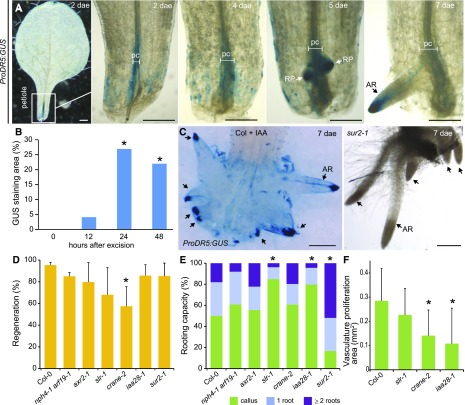
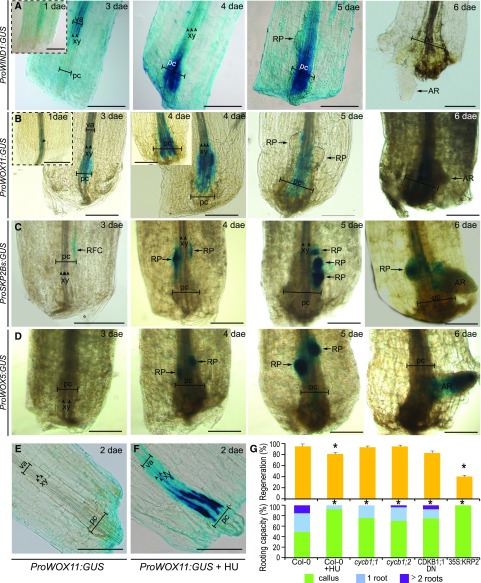
WOUND INDUCED DEDIFFERENTIATION1 (WIND1) is rapidly induced at the wound site to promote callus formation through the ARR-dependent signaling pathway (Iwase et al., 2011). From 1 to 4 dae, we found ProWIND1:GUS expression in vascular cells at the petiole near the excision site, whereas expression was downregulated in new root primordia and no expression was detected at the time of root emergence by 6 dae (Fig. 5A). Postembryonic development involves specification of organ founder cells (Chandler, 2011). Thus, we hypothesized that root founder cells (RFCs) could be specified within the endogenous callus to de novo form a root. It has been proposed that RFCs during de novo root formation in leaf blades could be marked by ProWOX11:GUS expression (Liu et al., 2014; Hu and Xu, 2016). We observed discrete ProWOX11:GUS signals early detected, at 1 dae, in a few xylem-associated cells at the petiole base (Fig. 5B). From 1 dae onwards, during callus formation, high ProWOX11:GUS expression was observed in many cells within this domain, but not simultaneously in all proliferating cells. Later on, ProWOX11:GUS expression was observed near the central zone of the endogenous callus but excluded from the dome-shape root primordia. WOX11 expression thus appears to associate with endogenous callus formation, although not all marked cells resulted in primordia initiation. We wondered whether cell division of vascular-associated cells was downstream of the WOX11 signal. ProWOX11:GUS expression was increased in vasculature of petioles of excised leaves incubated with the G1/S cell cycle inhibitor hydroxyurea, even when little vasculature proliferation was observed (Fig. 5, E and F).
Figure 5.
Root founder cells establish on endogenous callus prior de novo primordium formation. A to F, Expression of (A) ProWIND1:GUS, (B, E, and F) ProWOX11:GUS, (C) ProSKP2Bs:GUS, and (D) ProWOX5:GUS in leaf petioles at indicated dae. E, Control conditions or (F) upon 5 mm HU treatment. GUS staining time in (E) was set for no expression. Scale bars: 200 μm. G, Regeneration percentage and rooting capacity at 7 dae of mutants impaired in cell cycle progression and HU-treated Col-0 leaves. Asterisks: P value < 0.05 by GLM followed by LSD posthoc or χ2 test. DN, de novo-formed root; pc, proliferating cells; RFC, root founder cell; RP, root primordium; va, vasculature; xy, xylem.
ProSKPB2s:GUS expression has been shown to mark RFCs and their progeny during early stages of lateral root formation (Manzano et al., 2012). We detected ProSKPB2s:GUS expression at 3 dae restricted to few cells within the endogenous callus (Fig. 5C). From 4 to 5 dae, we observed marker expression in developing root primordia whereas its expression disappeared from functional primordia during the emerging process, remaining in some cells of the endogenous callus (Fig. 5C). Interestingly, RFCs did not appear to be specified simultaneously, supporting our initial observations about asynchrony in the regeneration process. To follow primordia formation, we used ProWOX5:GUS. WOX5 is expressed early during lateral root primordia formation (Tian et al., 2014; Goh et al., 2016), and also after root primordia initiation during de novo organogenesis (Liu et al., 2014; Hu and Xu, 2016). Expression of ProWOX5:GUS was first detected at 4 dae in clusters of cells within the endogenous callus (Fig. 5D), similarly to ProSKPB2s:GUS after root primordia initiation. At 6 dae, ProWOX5:GUS expression was enriched at root meristem tip, coinciding with meristem activation and growth prior emergence. Interestingly, we did not detect ProWOX5:GUS in the endogenous callus (Fig. 5D), although WOX5 expression has been associated to hormone-induced callus (Sugimoto et al., 2010), suggesting specific regulation.
Cell division is required for primordia initiation and formation, and petioles of leaves treated with the cell cycle inhibitor HU, which started to form an endogenous callus, remained almost blocked at this stage (Fig. 5G). We wondered if general regulators of cell cycle progression would be involved in de novo primordia formation. Our results indicate that rooting capacity is compromised in cycb1;1, cycb1;2, and CDKB1;1 DN161 mutants. Accordingly, overexpression of the cell cycle inhibitor KRP2 blocked progression to root initiation and formation, along with reduction in callus formation by 60% previously shown in Figure 1Q. Taken together, our results indicate that WIND1 and WOX11 expression associates with vasculature proliferation leading to endogenous callus formation at the petiole base, whereas ProSKPB2s:GUS is restricted to a few RFCs within the callus that quickly acquire WOX5 expression associated to cell division, in turn mediated by CYCB1;1, CYCB1;2, CDKB1;1, and KRP2, to initiate de novo root primordia regeneration (Fig. 5G).
SHORT-ROOT, PLETHORA1, and PLETHORA2 Are Required for De Novo Initiation of Roots
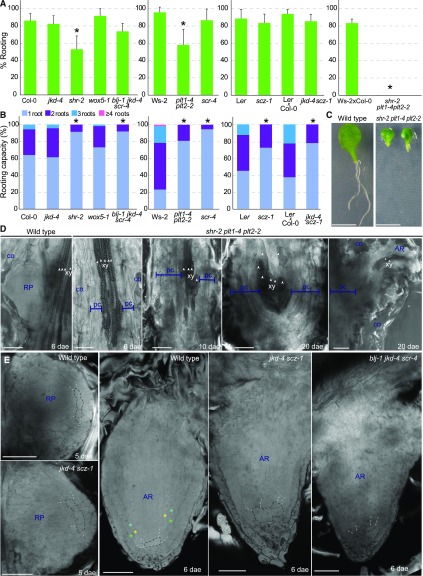
We investigated whether factors specifying stem cell fate or their activity, such as PLETHORA (PLT), JACKDAW (JKD), BLUEJAY (BLJ), SCARECROW (SCR), SHORT-ROOT (SHR), SCHIZORIZA (SCZ), and WOX5 were required during de novo root regeneration. In addition to single loss-of-function mutants, double plt1-4 plt2-2 and triple blj-1 jkd-4 scr-4, we generated and tested the double mutant jkd-4 scz-1. We found significant differences in percentage of leaves rooting and in rooting capacity for shr-2 and plt1-4 plt2-2 (Fig. 6, A and B). In addition, scr-4, blj-1 jkd-4 scr-4, scz-1, and jkd-4 scz-1 were impaired in rooting capacity. These phenotypes suggested impairment in de novo root initiation or primordia formation. As in shr-2 and plt1-4 plt2-2, many leaves failed to regenerate any root. We generated shr-2 plt1-4 plt2-2. Notably, leaves of shr-2 plt1-4 plt2-2 were not capable of rooting (Fig. 6, A and C).
Figure 6.
SHORT-ROOT, PLETHORA1, PLETHORA2, JACKDAW, BLUEJAY, SCARECROW, and SCHIZORIZA are required for de novo root primordia initiation and formation. A and B, Rooting percentage and rooting capacity, respectively, at 10 dae for loss-of-function mutants in stem cell regulators. Asterisks: P value < 0.05 by GLM followed by LSD posthoc or χ2 test. C, Triple shr-2 plt1-4 plt2-2 mutant does not regenerate roots at 10 dae. D, Confocal images of control and shr-2 plt1-4 plt2-2 during root regeneration. Malformed primordia (3%, n = 90) in shr-2 plt1-4 plt2-2 do not emerge through petiole tissues. E, Confocal images of de novo root primordia of wild type, jkd4-4 scz-1, and blj-1 jkd-4 scr-4 at 5 and 6 dae. Black dashed line corresponds to cells in the position of quiescent center; white dashed line cells correspond to the position of the ground tissue in contact to quiescent center. Green asterisk, cortex; turquoise asterisk, middle cortex; yellow asterisk, endodermis. Scale bars: (C) 5 mm and (D to I) 50 μm. AR, adventitious root; co, collenchyma; pc, proliferative cells; RP, root primordium; xy, xylem.
Petioles of shr-2 plt1-4 plt2-2 were observed through confocal microscopy at 6, 10, and 20 dae. We did not observe any primordia at 6 dae in shr-2 plt1-4 plt2-2, whereas most control leaves had one or more primordia (Fig. 6D). Inspection at later days indicated that most leaves (97%, n = 90) of shr-2 plt1-4 plt2-2 did not form any primordia up to 20 dae. The few primordia found (3%, n = 90) remained blocked or developed aberrant shapes with presence of mature xylem indicating premature differentiation. These primordia did not emerge through petiole tissues. We observed endogenous callus formation, which maintained growth over time up to 20 dae in all observed petioles of shr-2 plt1-4 plt2-2. These results indicate that the combined activity of SHR and PLT1 and PLT2 is required to de novo initiate root primordia. In addition, these factors maintain proliferative activity in the forming primordia; when these factors are removed, primordia differentiate.
JACKDAW, BLUEJAY, SCARECROW, and SCHIZORIZA Are Required for De Novo Formation of Root Primordia
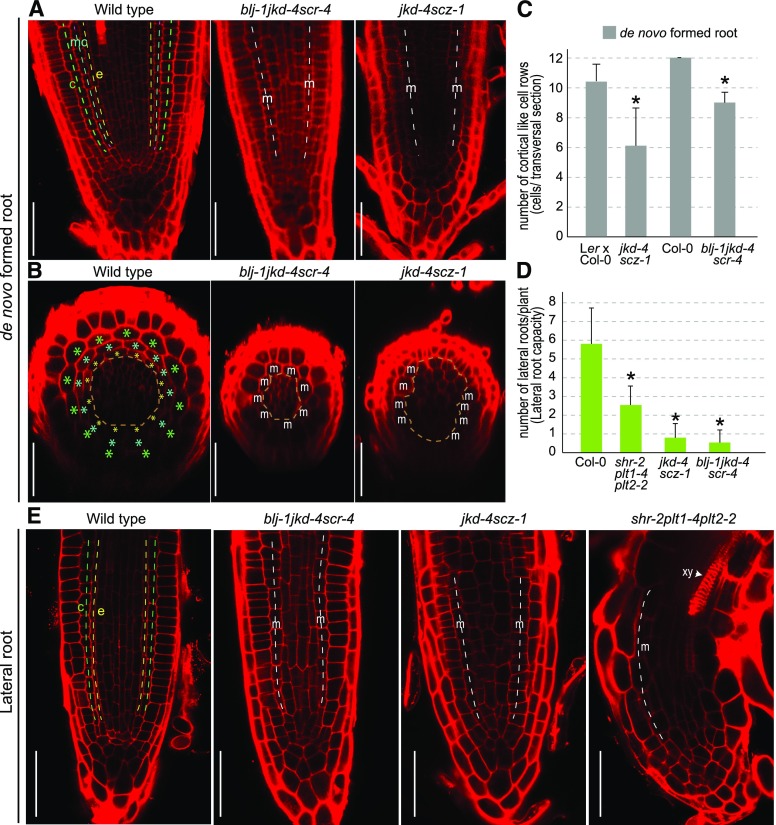
We observed formation of root primordia in petioles of jkd-4 scz-1 and blj-1 jkd-4 scr-4 through confocal microscopy. We observed abnormal formative divisions in jkd-4 scz-1 primordia at 5 dae, which did not organize properly in layers or rows as compared to control roots (Fig. 6E). At 6 dae, we observed reduced and disorganized number of cell rows in jkd-4 scz-1 and blj-1 jkd-4 scr-4 primordia. Although endodermis, cortex, and middle cortex could be identified in control roots at this developmental stage based on position, corresponding rows in jkd-4 scz-1 and blj-1 jkd-4 scr-4 could not be identified (Fig. 6E). These results suggest that cell lineages or positional identity could not be correctly established in these mutants during de novo root formation.
JACKDAW, BLUEJAY, SCARECROW, SHORT-ROOT, and SCHIZORIZA Regulate Patterning of De Novo Formed Roots and of Lateral Roots
Primordia formation occurred incorrectly in blj-1 jkd-4 scr-4 and jkd-4 scz-1, and therefore patterning of these de novo formed roots could be compromised after emergence. Longitudinal- and cross sections of de novo root meristems were examined through confocal microscopy after emergence at 10 dae (encompassing roots at 1 d to 3 d postemergence). The de novo wild-type roots showed ground tissue with middle cortex formation, a layer located between endodermis and cortex and associated to postembryonic development (Paquette and Benfey, 2005), whereas de novo roots of shr-2, scr-4, blj-1 jkd-4 scr-4, and jkd-4 scz-1 had a single layer of ground tissue that we denoted the “mutant layer” (Fig. 7A; Fig. S8, A and B). In addition, the number of cell rows making the ground tissue, which indicates the number of tissues initializing, was reduced in de novo emerged roots of blj-1 jkd-4 scr-4 and jkd-4 scz-1 as compared to wild-type roots (Fig. 7E; Supplemental Fig. S8C). As a result, the stele region in these mutants was not delimited by a closed ring of ground tissue as in wild-type roots, as shown in cross sections (Fig. 7B). In addition, shr-2, scr-4, blj-1 jkd-4 scr-4, and jkd4 scz-1 mutants also had a smaller stele region (Fig. 7, B and C; Supplemental Fig. 8, D and E).
Figure 7.
JACKDAW, BLUEJAY, SCARECROW, SHORT-ROOT, and SCHIZORIZA regulate patterning of postembryonic roots. A and B, Confocal (A) longitudinal and (B) transversal sections of de novo-formed roots at 10 dae (1 to 3 d post emergence). Transversal sections were taken at the end of the lateral root cap. Dashed lines and asterisks: green, cortex; turquoise, middle cortex; yellow, endodermis; brown, stele; white, mutant undivided ground tissue. C, Number of cortical-like cell or undivided mutant cell rows in control accessions, jkd-4 scz-1 and blj-1 jkd-4 scr-4. D, Lateral root capacity in Col-0, shr-2 plt1-4 plt2-2, jkd-4 scz-1, and blj-1 jkd-4 scr-4 and at 4 d after seed imbibition. E, Confocal longitudinal sections of emerged lateral roots of Col-0, shr-2 plt1-4 plt2-2, jkd-4 scz-1, and blj-1 jkd-4 scr-4 at 7 d after seed imbibition. Scale bars: 50 μm. Asterisks: P value < 0.05 by GLM and LSD posthoc test. m, mutant; xy, xylem.
Lateral roots are organs formed postembryonically the same as de novo-formed roots. We decided to investigate if stem cell regulators regulating de novo root formation could also regulate lateral root formation. We quantified lateral root capacity. In this assay, the root tip is cut to induce growth of lateral roots as described previously (van Norman et al., 2014). We confirmed, under the microscope, there were no unemerged primordia. We observed reduced lateral root capacity in shr-2 plt1-4 plt2-2, blj-1 jkd-4 scr-4, and jkd-4 scz-1 mutant combinations indicating defects in founder cell specification or lateral root initiation (Fig. 7D), similarly to defects observed in these mutants during de novo root formation. We also observed patterning defects in emerged lateral roots of these mutants, which showed a single mutant layer of ground tissue and reduced number of cell rows as compared to wild-type lateral roots (Fig. 7E). In addition, we observed signs of premature differentiation in shr-2 plt1-4 plt2-2, with presence of mature xylem in the meristem similarly to de novo formed roots.
DISCUSSION
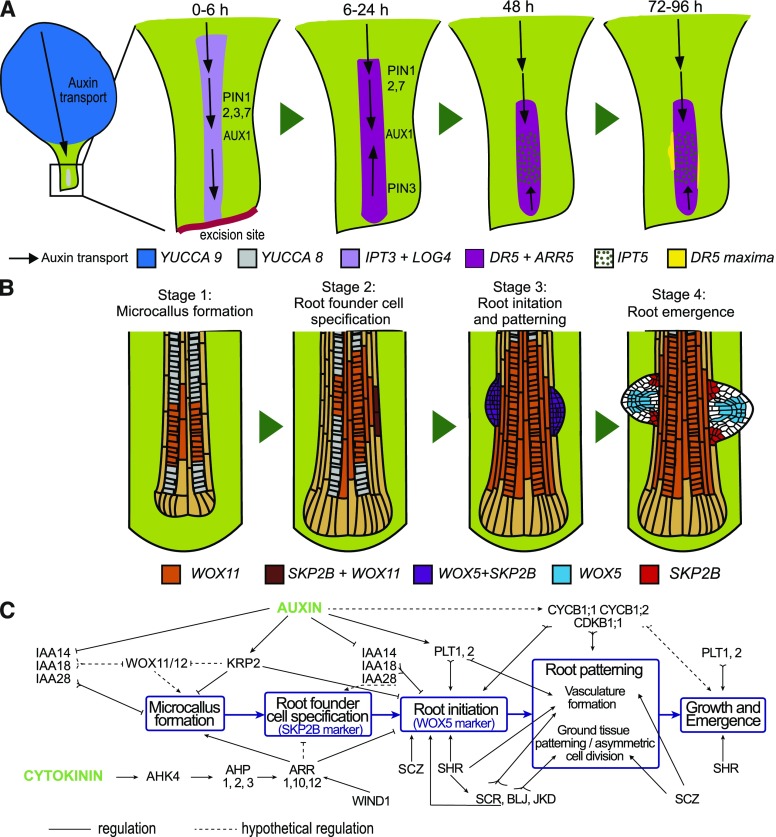
Plants have the remarkable ability to regenerate a new entire individual, specific organs, or their tissues from explants or even a few cells (Birnbaum and Sánchez Alvarado, 2008; Della Rovere et al., 2016). Plant regeneration through de novo organogenesis can be achieved through hormonal induction, directly or indirectly (Ikeuchi et al., 2013), but also in hormone free medium. The molecular pathways involved (Ikeuchi et al., 2016; Kareem et al., 2016) and the relationship between hormonal-induced and endogenous programs are not well understood. Moreover, callus formation, which is a prerequisite for hormonal-induced regeneration, does not appear to occur during endogenous organogenesis, although both processes share regulation (Sugimoto et al., 2010; Liu et al., 2014; Perianez-Rodriguez et al., 2014; Ramirez-Parra et al., 2017). Using a simple method to study de novo root organogenesis without hormone supplementation (Chen et al., 2014; Liu et al., 2014) but applied to whole leaves (Fig. 8A), we found that formation of an endogenous callus is a required step for de novo root organogenesis, and thus we established a direct connection between both regenerative processes. Our research has also identified the distinctive and required developmental stages leading to novo root organogenesis (Fig. 8B): 1) vasculature proliferation and endogenous callus formation, 2) root founder cell specification, 3) root primordia initiation and patterning, and 4) root meristem activation and emergence. Furthermore, we have genetically dissected the primary developmental pathways involved in its regulation and identified some of the key regulators involved (Fig. 8C).
Figure 8.
Model of de novo root regeneration in Arabidopsis leaves. A, Auxin is transported to the petiole base and acts in combination with locally produced cytokinin to induce endogenous callus formation: LOG4, IPT3, and IPT5 for cytokinin biosynthesis; ARR5 for cytokinin response; YUCCA8/9 for auxin synthesis; PIN1/2/3/7 and AUX1 for auxin transport; and DR5 for auxin response. B, Stages of de novo formation of roots based on morphological changes and expression of markers. C, Regulation of distinctive stages of de novo organogenesis based on mutant phenotypes. Regulation of functions that could not be directly assigned are indicated as hypothetical.
Vasculature Proliferation and Endogenous Callus Formation
Vasculature division is the first morphological change we detect, and by chemically or genetically inhibiting vasculature proliferation, we affected de novo root organogenesis. We also observed that root primordia originated from vasculature. Pericycle-like cells, particularly those expressing the J0121 marker, have been associated to regenerative and morphogenic processes as the source of reprogrammable cells (Sugimoto et al., 2010; Chen et al., 2014). When we assessed the J0661 and J0121 pericycle markers, we observed that vasculature proliferation associated to J0661 identity, whereas some proliferating cells were devoid of J0121 expression. Intriguingly, J0661 marks cells around xylem and procambium in petioles whereas J0121 only marks cells in contact to xylem. Proliferation competence, therefore, appears to involve different cell types. Closer examination showed that all primordia expressed a J0121 marker, which indicates that regeneration competence associates with J0121 identity and shows parallelisms with callus and lateral root formation. Cell lineage tracing using clonal markers and live imaging could dissect the exact source of reprogrammable cells during de novo root regeneration from whole leaves.
We also found that proliferating vascular cells express WIND1, a positive regulator of wound-induced callus formation (Iwase et al., 2011), suggesting together with J0121 analysis that these proliferating tissues could be a type of callus. Callus formation requires confluence of auxin and cytokinin responses in the same set of cells (Gordon et al., 2007). In agreement with this idea, our results show specific expression of auxin and cytokinin signaling reporters ProDR5:GUS and ProARR5:GUS, respectively, in the vasculature. These results also indicate that specific regulation was required to induce auxin and cytokinin signaling at the petiole base. In contrast to early notions that cytokinins are produced only in roots, it is now recognized that they are synthesized throughout the plant (Zürcher and Müller, 2016). Our results are consistent with ISOPENTENYLTRANSFERASE3 (IPT3), IPT5, and LONELY GUY4 locally mediating cytokinin biosynthesis at the petiole base to contribute to vascular proliferation during root regeneration. Thus, cytokinin signaling mutants displayed reduced regeneration potential as well as defective hormone-induced callus formation. Interestingly, the triple cytokinin signaling mutant ahp2 ahp4 ahp5 is affected in hormone-induced callus formation but not in vasculature proliferation during rooting, indicating the existence of specific genetic differences between hormone-induced callus formation and de novo root formation.
ProDR5:GUS expression in the proximal petiole vasculature could be indicative of auxin accumulation. A study in leaf blades has shown that YUCCA1 (YUC1) and YUC4 appear to mediate synthesis of auxin in mesophyll cells (Chen et al., 2016). If this occurred in our system, auxin would also need to be transported to cells near the wound to induce de novo root organogenesis. We observed that YUC9 expression was induced in the leaf blade mesophyll but not in the petiole (Fig. 8A), suggesting a predominant function of the leaf blade mesophyll as source of auxin for regeneration. We confirmed this by removing leaf blades, which resulted in inhibition of regeneration. YUC9 expression has been shown to respond to methyl-jasmonate (MeJA) treatment in a COI1-dependent manner (Hentrich et al., 2013). As excision of whole leaves or leaf blades involves wounding and therefore MeJA production, MeJA might activate YUC9 expression to rapid increase auxin levels in leaf blades, likely in combination with YUC1 and YUC4 activity. We also found that YUC8 was specifically upregulated in the vascular region of the petiole associated with proliferation, and thus, it is possible that YUC8 might have a specific role in maintaining auxin levels during vasculature proliferation or at later regenerative steps.
Our results indicate that a long distance basipetal transport system concentrates auxin generated in the leaf blade mesophyll toward defined vascular cells at the petiole base. We showed that genetic and localized chemical inhibition of auxin transport significantly affected regeneration. Despite known redundancy among auxin transporters (Blilou et al., 2005; Péret et al., 2012), we detected phenotypes in single mutants, suggesting spatial compartmentalization. Supporting this idea, PIN-FORMED3 (PIN3) was expressed in the petiole vasculature whereas PIN4 was later restricted to the proliferating vascular region. In contrast, more delocalized auxin transport is involved in rooting of leaf blades (Liu et al., 2014; Chen et al., 2016). As we observed predominant expression of ProDR5:GUS in the proximal petiole vasculature, it is possible that auxin would need to be retained in this area. Our data suggest a model in which subcellular PIN3 localization shifts from basal to apical membranes in vascular cells near the wound to redirect auxin flow backward and thus maintaining high auxin levels in the proximal petiole vasculature. Interestingly, an auxin-dependent switch in PIN3 polarization contributing to auxin-flow reversal is involved in the shoot gravitropic response (Rakusová et al., 2016), where basal-to-apical shift in PIN localization has been described to depend on phosphorylation (Dai et al., 2012). It is thus tempting to speculate that auxin-dependent phosphorylation of PIN3 would be involved in maintaining high auxin levels in the petiole base vasculature during root regeneration.
The alf4 mutant (DiDonato et al., 2004) has been linked to hormone-induced callus formation (Sugimoto et al., 2010), but not to wound-induced callus formation during graft establishment (Melnyk et al., 2015). We observed reduced vasculature proliferation along with great reduction in de novo root formation from whole leaves in alf4-1 mutants. As vasculature proliferation during de novo root formation associates to J0121- and WIND1-marked cells, requires auxin and cytokinin signaling and involves ALF4, we propose it is a type of callus, and therefore we refer to it as “endogenous callus” to differentiate it from callus obtained by exogenous hormone supplementation. In our model, time-dependent auxin accumulation in a subset of vascular cells activates proliferation, whereas cytokinins might regulate the expression of genes that are directly involved in callus formation (such as WIND1 or ALF4) or that are downstream targets of the auxin signal involved in callus formation (LATERAL ORGAN BOUNDARIES DOMAIN factors; Schaller et al., 2015). Particularly, we found that IAA18 and IAA28 are both involved in endogenous callus formation, although only mutations in IAA18 affect whole regeneration response, whereas ARABIDOPSIS HIS PHOSPHOTRANSFER PROTEIN1 (AHP1) to AHP3 and ARABIDOPSIS RESPONSE REGULATOR1 (ARR1), ARR10, and ARR12 were involved in vasculature proliferation and regeneration response.
Specification of Root Founder Cells
Hormone-induced callus is organized in layers showing root tissue identities that resemble a root meristem and therefore a new organ could be theoretically initiated through a differentiation process (Sugimoto et al., 2010). WUSCHEL-RELATED HOMEOBOX11 (WOX11) expression has been associated to first cell-fate transition from regeneration-competent cells to root founder cells during leaf blade rooting (Liu et al., 2014; Hu and Xu, 2016). WOX11 activates WOX5 during root formation in leaf blades; however, we did not find WOX5 expression in endogenous callus during de novo regeneration from whole leaves, suggesting that WOX11 could be involved in an earlier step in the reprograming process. On the other hand, specific expression associated to root founder cell specification (SKP2B) revealed the establishment of a cell lineage capable of forming a new root. These results indicate that additional reprogramming processes are required.
PLT1 and PLT2 have been shown to be required to establish pluripotency during de novo shoot regeneration (Kareem et al., 2015). Our results show that during de novo root formation, PLT1 and PLT2, in combination with SHR, could also be involved in specification of root founder cells, which are pluripotent. In addition, persistent expression of PLT1, PLT2, and SHR appears to be necessary during subsequent formative stages to maintain primordia growth, as the very primordia found in shr-2 plt1-4 plt2-2 quickly differentiate. In contrast, in the de novo shoot regeneration system, transient induction of PLT2 has been shown to be sufficient to specify shoot progenitors, whereas subsequent expression of other regulators is required to accomplish de novo shoot formation from these progenitors (Kareem et al., 2015).
Root Primordia Initiation and Patterning
Multiple INDOLE-3-ACETIC ACID INDUCIBLE (IAA)-ARF modules cooperatively regulate lateral root formation (Goh et al., 2012). We observed that factors regulating auxin signaling, such as SOLITARY ROOT (IAA14), could be also involved in de novo root initiation; we detected decreased root capacity for slr-1, which could be indicative of fewer primordia initiation. In addition, the IAA28 module, which is upstream of lateral root founder cell specification (De Rybel et al., 2010), also regulates de novo root founder cell specification or initiation, although further experiments could dissect more precisely at which stage IAA28 is involved. Our results show that factors primarily involved in formation of lateral roots are also affected in rooting of leaves, suggesting the existence of partially overlapping auxin signaling modules during postembryonic root development. Conversely, cytokinin mutants (ahp1 ahp2 ahp3 and arr1 arr10 arr12) showed increased rooting capacity and thus, a repressor role in de novo root initiation can be assigned for these factors, likely in an analog manner as their role during lateral root initiation (Lavenus et al., 2013; Chang et al., 2015). In agreement with this model, we restored regeneration potential of cytokinin signaling mutants by a moderate increase in auxin levels.
PLT1 and PLT2 expression is considered to be a slow readout of auxin response and prolonged auxin treatment results in PLT1 and PLT2 activation and the de novo specification of WOX5-marked stem cells (Mähönen et al., 2014). We found severe impairment in de novo primordia initiation in shr-2 plt1-4 plt2-2, and WOX5 expression during de novo primordia formation requires auxin input through an unknown pathway (Hu and Xu, 2016). Therefore, it is possible that PLT1 and PLT2 are required for specification of WOX5-marked cells downstream of auxin during de novo organ initiation, which could occur in combination with activity of WOX5 transcription factor itself and/or SHR, in turn acting in an auxin-independent pathway. Further experiments will be required to dissect the molecular pathway including PLTs, SHR, and WOX5.
We have also identified specific factors involved in formation of de novo root primordia. We have found that the stem cell regulators BLJ, JKD, SCR, SHR, and SCZ regulate ground tissue patterning and vasculature formation prior emergence at the step of dome-shape primordia. Subsequently, more developed primordia are not properly organized in cell layers or rows and by the time of emergence, these defects persist and aggravate. Our results indicate that ground tissue patterning appears to be regulated in newly formed roots at two levels: First, we observed impairment in asymmetric divisions specifying cortex, middle cortex, and endodermis in mutants of SHR and SCR, although a few asymmetric divisions were still observed. In agreement, shr mutants have been shown to form endodermis in anchor roots (Lucas et al., 2011), which are a type of adventitious root. Furthermore, we observed that ground tissue asymmetric divisions were absent in mutant combinations of scr-4 with bjl-1 and jkd-4 and in double mutants jkd-4 scz-1. Interestingly, when we studied if these mutant combinations had defects in lateral roots, which are also organs formed postembryonically, we also observed absence of ground tissue asymmetric divisions suggesting a conserved developmental program for endodermis and cortex specification. Second, we observed that SCR, JKD, BLJ, and SCZ could function as ground tissue lineage determinants during de novo root organogenesis. The combined action of BLJ, JKD, and SCR is required to maintain the ground tissue lineage postembryonically, and lacking these three factors results in missing ground tissue initializations (hence fewer ground tissue cell rows are observed in cross sections: Moreno-Risueno et al., 2015). We observed that de novo formed roots in blj-1 jkd-4 scr-4 and jkd-4 scz-1 mutants were missing ground tissue cell rows shortly after emergence, which indicates incorrect specification of ground tissue initials during primordia formation or later on in the course of development. Thus, it is possible that SCZ, SHR, PLT1, and PLT2 function as lineage or cell fate determinants during postembryonic development, and particularly so during de novo organogenesis.
MATERIALS AND METHODS
Growth Conditions
Seeds were surfaced-sterilized in 10% (m/v) NaClO and rinsed with sterile water before being transferred to 120 × 120 × 10 mm petri dishes containing 65 mL of one-half-Murashige & Skoog (MS) medium with 1% Suc and 10 g/L Plant Agar (Duchefa). After 2 d of stratification at 4°C in darkness, plates were transferred to an MLR-352-PE growth chamber (Panasonic) at 22°C, 16/8 photoperiod or continuous light (50 µmol·m−2·s−1). Twelve d after germination, the first pair of leaves was excised across the junction of the petiole with the stem and transferred to Gamborg B5 medium with 2.5% Suc, 10 g/L Difco Agar (Becton Dickinson), or 3 g/L Gelrite (Sigma-Aldrich) and Gamborg B5 vitamin mixture (Duchefa). Leaves after excision were grown in darkness at 22°C, routinely for 10 d, or for number of days indicated in corresponding experiment.
Hormonal and Inhibition Treatments
For exogenous hormone treatment, filter-sterilized indole-3-acetic acid, 6-BAP, or thidiazuron (TDZ) stock solutions were added to warm growth medium before pouring into plates to provide a final concentration of 1 μm indole-3-acetic acid, 5 μm 6-BAP, or 4 μm TDZ, respectively. NPA was applied locally by preparing a lanoline solution containing 1% w/w NPA. For cell cycle arrest, leaf explants were incubated with growth medium supplemented with 0.5, 2.5, or 5 mm hydroxyurea (Sigma-Aldrich).
Plant Material
Columbia-0 (Col-0), Landsberg erecta (Ler), and Wassilewskija-2 (Ws-2) accessions were used as a genetic background as corresponding. The reporter lines ProIPT3:GUS, ProIPT5:GUS (Miyawaki et al., 2004), ProLOG4:GUS s (Kuroha et al., 2009) obtained from RIKEN, and ProARR5:GUS (D’Agostino et al., 2000) were used for tracing cytokinin biosynthesis and signaling during rooting. ProYUC8:GUS, ProYUC9:GUS (Hentrich et al., 2013) and ProDR5:GUS (Ulmasov et al., 1997) were used to investigate auxin biosynthesis and signaling; ProPIN3:PIN3:GFP (Xu et al., 2006) and ProPIN4:GUS (Friml et al., 2004) were used for auxin transport. To trace the molecular mechanisms during de novo root formation we used: ProWIND1:GUS (Iwase et al., 2011), ProWOX11:GUS (Liu et al., 2014), ProWOX5:GUS (Sarkar et al., 2007), and ProSKP2Bs:GUS (Manzano et al., 2012), which corresponds to a promoter deletion containing 0.5 Kb upstream from the ATG. The J0121 and J0661 lines (Laplaze et al., 2005) were used to locate pericycle-like cells during rooting. The following mutant lines were used: wol-1 (Scheres et al., 1995), ahp1 ahp2 ahp3 (Hutchison et al., 2006), ahp2 ahp4 ahp5, arr1 arr10 arr12 (Mason et al., 2005), aux1-22 (Bennett et al., 1996), axr2-1 (Timpte et al., 1994), slr-1 (Fukaki et al., 2005), crane-2 (Uehara et al., 2008), sur2-1 (Delarue et al., 1998), and iaa28-1 (Rogg et al., 2001), which were obtained from NASC; and pin1, pin2 pin3, pin2 pin3 pin7 (Blilou et al., 2005), fwr (Okumura et al., 2013), cyclinb1;1 (cycb1;1) and cycb1;2 (Nowack et al., 2012), and CDKB1;1 DN161 and Pro35S:KRP2 (Boudolf et al., 2004; Verkest et al., 2005). We also used the following stem cell niche mutants: blj-1 jkd-4 scr-4 (Moreno-Risueno et al., 2015), scr-4 (Fukaki et al., 1996), shr-2 ProSHR:SHR:GR (Levesque et al., 2006), plt1-4 plt2-2 (Aida et al., 2004), jkd-4 (Welch et al., 2007), scz-1 (Mylona et al., 2002), and wox5-1 (Sarkar et al., 2007), and jkd-4 scz-1 and shr-2 plt1-4 plt2-2 ProSHR:SHR:GR (generated in this study).
Genotyping
The triple mutant shr-2 plt1-4 plt2-2 ProSHR:SHR:GR was generated by crossing shr-2 ProSHR:SHR:GR and plt1-4 plt2-2. F2 seedlings were grown and preselected on one-half-MS medium with 10 µM dexamethasone followed by DNA extraction and genotyping by PCR to finally obtain homozygous lines. The following oligonucleotides were used for genotyping shr-2 (while discriminating ProSHR:SHR:GR): F 5′-CCAATACCATCCCGCCAC-3′ and R 5′-TGAACCGGTCATGCGGTTG-3′; for plt1-4: F 5′-AGACGGCCACGCCAAGAC-3′ and R 5′CTAGTATCACGACATTATTTGC-3′; and for plt-2-2: F 5′-ACCTACAGTCGTCACTTGTGC-3′ and R 5′-ACTCTTGTCTCGTCATGTTTTTC-3′. T-DNA insertions of PLT1 and PLT2 were amplified using LB 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′ and respective reverse primer. Double mutant jkd-4 scz-1 was generated by crossing jkd-4 and scz-1. DNA extraction from F2 seedlings was used for genotyping by PCR. The following oligonucleotides were used for genotyping jkd-4: F 5′-GGATGAAAGCAATGCAAAACA-3′ and R 5′-AATGTCGGGATGATGAACTCC-3′. The T-DNA insertion in the jkd-4 line was genotyped using respective forward primer and RB 5′-TCAAACAGGATTTTCGCCTGCT-3′. For scz-1 genotyping, a SCZ fragment was amplified using F 5′-CGAAGGTCAAGGCAAAGCTG-3′ and R 5′-GAGCAACAGGCTTGACATGG-3′primers, followed by digestion with NlaIII restriction enzyme. Upon digestion, an SCZ fragment from scz-1 renders a band of 900 bp, while the wild-type fragment is cut by the enzyme, resulting in two fragments of 530 bp and 360 bp.
Cell Death, GUS Staining, and Microscopy Analysis
In leaf tissues, dying cells were visualized by Lactophenol-Trypan Blue staining as described in Pavet et al. (2005). For GUS staining, leaf explants were incubated at 37°C for variable times (4 h to 24 h) in multiwell plates in the presence of the GUS staining solution as described in Pérez-Pérez et al. (2010) or Manzano et al. (2012). To bleach chlorophyll, two different methods were used indistinctly, with similar results. For the first method, samples were dehydrated after GUS staining using increasing ethanol concentrations (15, 50, 70, 96, and 100%) for 15 min in each one, and kept overnight in 100% ethanol; then samples were rehydrated following the same ethanol concentration series to 15% ethanol and mounted in 15% glycerol. For the second method, samples were fixed in 96% ethanol for 48 h and washed with 0.1 m P buffer (pH 6.8) before being transferred to a clearing solution (80 g chloral hydrate and 30 mL distilled water). Leaf explants were mounted on slides using a mixture of 80 g chloral hydrate, 20 mL distilled water, and 10 mL glycerol. After GUS staining, pictures were taken in a DM 2000 with a DCF300 camera (Leica), or in a bright-field AX70 microscope (Olympus) equipped with a PM-C35DX microphotography system (Olympus). The area of proliferating tissues (callus or vasculature) was manually drawn from microscopic images using a Bamboo tablet (Wacon) and areas or diameters of their best-fitting ellipses were measured with the software ImageJ (v1.50; National Institutes of Health).
Microscopy Analysis
Leaf petioles (for studying developmental stages during rooting) and mature embryos were stained using an Aniline Blue staining method as described in Bougourd et al. (2000). Processed samples were observed with a TCS SP8 laser scanning confocal microscope (Leica) with the settings described in Bougourd et al. (2000). A J0121 fluorescent reporter line was processed before imaging as indicated in Kurihara et al. (2015). Cross sections (10 µm of thickness) of J0121 petioles were obtained using a Vibratome 1000 Plus (Ted Pella) when indicated. Other fluorescent reporter lines were incubated in a methanol/acetone solution for 20 min at −20°C, and immediately transferred to a 0.1-m P buffer (pH 6.8). Imaging was performed by confocal microscopy using a Digital Eclipse C1 (Nikon) equipped with EZ-C1 control software (Nikon Instruments) or a TCS SP8 laser scanning microscope (Leica). GFP was excited with an Argon laser at 488 nm and emission was collected at 505 nm to 530 nm, whereas YFP was excited at 514 nm and emission collected at 535 nm to 560 nm. To exclude autofluorescence contamination, sample emission was collected at 605 nm to 675 nm before excitation with an He-Ne laser at 543 nm, and used as a background reference. Root samples for radial and longitudinal analyses in patterning studies were fluorescently stained with 10 mm propidium iodide (Sigma-Aldrich), and imaged using standard settings on a TCS SP8 confocal microscope (Leica).
Quantification and Statistical Analysis
Regeneration percentage was scored as the number of leaf explants that showed swelling of petioles, proliferation of vascular-associated cells, and/or outgrowth of roots at the proximal region of the petiole, at 7 dae or as indicated in corresponding experiments. Rooting percentage was scored as the number of leaf explants showing roots at 10 dae or as indicated. Proliferation of vascular-associated cells and number of de novo formed roots were scored on individual leaf samples and used to estimate rooting capacity categories at indicated days in corresponding experiments. Ten-dae de novo-formed roots were used for establishing differences in tissue patterning using number of cortical cells. The number of cortical cells was quantified using confocal cross sections taken at the site of lateral root cap ending. To determine lateral root capacity, root meristems of 4 d postimbibition seedlings were removed and the number of lateral roots quantified 3 d later. Roots were observed at the microscope to confirm there were not unemerged primordia. Data values referred to % rooting, % regeneration, number of cortical cells, lateral root capacity, and vasculature or callus area were statistically analyzed by a univariate general linear model (GLM) and ANOVA with a least significant difference (LSD) posthoc test, using SPSS Statistics 21 software (IBM). For rooting capacity, χ2 test was performed to assay if there were differences in distribution frequency between lines, analyzed two-by-two, using the software Centurion XVI.I (STATGRAPHICS). Significant differences were collected with 5% level of significance (P value < 0.05).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Rooting of whole leaves and leaf blades of Arabidopsis wild-type accessions.
Supplemental Figure S2. Vascular proliferation and callus formation during rooting of whole leaves.
Supplemental Figure S3. AHK4, AHP1, AHP2, AHP3, AHP4, and AHP5, and ARR1, ARR10, and ARR12 cytokinin signaling factors regulate hormone-induced callus formation.
Supplemental Figure S4. IAA7/AXR2 and IAA18 auxin signaling factors regulate hormone-induced callus formation.
Supplemental Figure S5. ALF4 is required for vasculature proliferation and de novo root formation.
Supplemental Figure S6. IAA7/AXR2, CRANE/IAA18, and IAA28 signaling factors regulate de novo root formation in leaf blades.
Supplemental Figure S7. The YUC8 and YUC9 auxin biosynthesis genes are induced in excised leaves.
Supplemental Figure S8. SCARECROW and SHORT-ROOT regulate ground tissue patterning of de novo formed roots.
Acknowledgments
We are especially indebted to M.A. Fernández-López for her expert technical assistance. We thank Dr. M. Pernas for providing the scz-1 mutant and advice about its genotyping, and two anonymous reviewers for their useful suggestions.
Footnotes
This work was supported by grants from Ministerio de Economía y Competitividad (MINECO) of Spain, the European Regional Development Fund (ERDF) and FP7 Funds of the European Commission, BFU2013-41160-P, BFU2016-80315-P, and PCIG11-GA-2012-322082 to M.A.M.-R., AGL2012-33610 and BIO2015-64255-R to J.M.P.-P., and BIO2014-52091-R to J.C.P. M.A.M.-R. was supported by a Ramon y Cajal contract from MICINN.
Articles can be viewed without a subscription.
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Atta R, Laurens L, Boucheron-Dubuisson E, Guivarc’h A, Carnero E, Giraudat-Pautot V, Rech P, Chriqui D (2009) Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J 57: 626–644 [DOI] [PubMed] [Google Scholar]
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C (2000) The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci USA 97: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini C, Pacurar DI, Perrone I (2014) Adventitious roots and lateral roots: similarities and differences. Annu Rev Plant Biol 65: 639–666 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Birnbaum KD, Sánchez Alvarado A (2008) Slicing across kingdoms: regeneration in plants and animals. Cell 132: 697–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GT, Magyar Z, Torres Acosta JA, Maes S, van der Schueren E, Inzé D, De Veylder L (2004) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougourd S, Marrison J, Haseloff J (2000) Technical advance: an aniline blue staining procedure for confocal microscopy and 3D imaging of normal and perturbed cellular phenotypes in mature Arabidopsis embryos. Plant J 24: 543–550 [DOI] [PubMed] [Google Scholar]
- Chandler JW. (2011) Founder cell specification. Trends Plant Sci 16: 607–613 [DOI] [PubMed] [Google Scholar]
- Chandler JW, Werr W (2015) Cytokinin-auxin crosstalk in cell type specification. Trends Plant Sci 20: 291–300 [DOI] [PubMed] [Google Scholar]
- Chang L, Ramireddy E, Schmülling T (2015) Cytokinin as a positional cue regulating lateral root spacing in Arabidopsis. J Exp Bot 66: 4759–4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Tong J, Xiao L, Ruan Y, Liu J, Zeng M, Huang H, Wang JW, Xu L (2016) YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J Exp Bot 67: 4273–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Qu Y, Sheng L, Liu J, Huang H, Xu L (2014) A simple method suitable to study de novo root organogenesis. Front Plant Sci 5: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Zhang C, Kania U, Chen F, Xue Q, McCray T, Li G, Qin G, Wakeley M, Terzaghi W, Wan J, Zhao Y, et al. (2012) A PP6-type phosphatase holoenzyme directly regulates PIN phosphorylation and auxin efflux in Arabidopsis. Plant Cell 24: 2497–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Prinsen E, Onckelen HV, Caboche M, Bellini C (1998) Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J 14: 603–611 [DOI] [PubMed] [Google Scholar]
- Della Rovere FD, Fattorini L, D’Angeli S, Veloccia A, Falasca G, Altamura MM (2013) Auxin and cytokinin control formation of the quiescent centre in the adventitious root apex of Arabidopsis. Ann Bot 112: 1395–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Rovere FD, Fattorini L, Ronzan M, Falasca G, Altamura MM (2016) The quiescent center and the stem cell niche in the adventitious roots of Arabidopsis thaliana. Plant Signal Behav 11: e1176660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, van Campenhout J, Overvoorde P, Jansen L, Vanneste S, Möller B, Wilson M, et al. (2010) A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol 20: 1697–1706 [DOI] [PubMed] [Google Scholar]
- DiDonato RJ, Arbuckle E, Buker S, Sheets J, Tobar J, Totong R, Grisafi P, Fink GR, Celenza JL (2004) Arabidopsis ALF4 encodes a nuclear-localized protein required for lateral root formation. Plant J 37: 340–353 [DOI] [PubMed] [Google Scholar]
- Drisch RC, Stahl Y (2015) Function and regulation of transcription factors involved in root apical meristem and stem cell maintenance. Front Plant Sci 6: 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Gambetta GA, Hernández-Barrera A, Shishkova S, González I (2006) Lateral root initiation in Arabidopsis: developmental window, spatial patterning, density and predictability. Ann Bot 97: 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Mello A, Nawy T, Ip PL, Rahni R, DelRose N, Powers A, Satija R, Birnbaum KD (2016) Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165: 1721–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Showk S, Ruonala R, Helariutta Y (2013) Crossing paths: cytokinin signalling and crosstalk. Development 140: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Fan M, Xu C, Xu K, Hu Y (2012) LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res 22: 1169–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forzani C, Aichinger E, Sornay E, Willemsen V, Laux T, Dewitte W, Murray JA (2014) WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr Biol 24: 1939–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, Hooykaas PJ, Palme K, et al. (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Fujisawa H, Tasaka M (1996) SGR1, SGR2, SGR3: novel genetic loci involved in shoot gravitropism in Arabidopsis thaliana. Plant Physiol 110: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M (2005) Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J 44: 382–395 [DOI] [PubMed] [Google Scholar]
- Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H (2012) Multiple AUX/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos Trans R Soc Lond B Biol Sci 367: 1461–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Toyokura K, Wells DM, Swarup K, Yamamoto M, Mimura T, Weijers D, Fukaki H, Laplaze L, Bennett MJ, Guyomarc’h S (2016) Quiescent center initiation in the Arabidopsis lateral root primordia is dependent on the SCARECROW transcription factor. Development 143: 3363–3371 [DOI] [PubMed] [Google Scholar]
- Gordon SP, Heisler MG, Reddy GV, Ohno C, Das P, Meyerowitz EM (2007) Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134: 3539–3548 [DOI] [PubMed] [Google Scholar]
- Hartmann HT, Kester DE, Davies FT, Geneve R (2010) Hartmann and Kester’s Plant Propagation: Principles and Practices, 8th Ed Prentice-Hall, Englewood Cliffs, NJ [Google Scholar]
- Hayashi K, Hashimoto K, Kusaka N, Yamazoe A, Fukaki H, Tasaka M, Nozaki H (2006) Caged gene-inducer spatially and temporally controls gene expression and plant development in transgenic Arabidopsis plant. Bioorg Med Chem Lett 16: 2470–2474 [DOI] [PubMed] [Google Scholar]
- Hentrich M, Böttcher C, Düchting P, Cheng Y, Zhao Y, Berkowitz O, Masle J, Medina J, Pollmann S (2013) The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J 74: 626–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J, Cools T, Vandenbussche F, Heyndrickx KS, van Leene J, Vercauteren I, Vanderauwera S, Vandepoele K, De Jaeger G, van der Straeten D, De Veylder L (2013) ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342: 860–863 [DOI] [PubMed] [Google Scholar]
- Hu X, Xu L (2016) Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol 172: 2363–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Ogawa Y, Iwase A, Sugimoto K (2016) Plant regeneration: cellular origins and molecular mechanisms. Development 143: 1442–1451 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Sugimoto K, Iwase A (2013) Plant callus: mechanisms of induction and repression. Plant Cell 25: 3159–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, Ohme-Takagi M (2011) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol 21: 508–514 [DOI] [PubMed] [Google Scholar]
- Kareem A, Durgaprasad K, Sugimoto K, Du Y, Pulianmackal AJ, Trivedi ZB, Abhayadev PV, Pinon V, Meyerowitz EM, Scheres B, Prasad K (2015) PLETHORA genes control regeneration by a two-step mechanism. Curr Biol 25: 1017–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareem A, Radhakrishnan D, Sondhi Y, Aiyaz M, Roy MV, Sugimoto K, Prasad K (2016) De novo assembly of plant body plan: a step ahead of Deadpool. Regeneration (Oxf) 3: 182–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Huang F, Naramoto S, Zhang J, Michniewicz M, Offringa R, Friml J (2009) PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara D, Mizuta Y, Sato Y, Higashiyama T (2015) ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142: 4168–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H (2009) Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21: 3152–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha T, Ueguchi C, Sakakibara H, Satoh S (2006) Cytokinin receptors are required for normal development of auxin-transporting vascular tissues in the hypocotyl but not in adventitious roots. Plant Cell Physiol 47: 234–243 [DOI] [PubMed] [Google Scholar]
- Laplaze L, Parizot B, Baker A, Ricaud L, Martinière A, Auguy F, Franche C, Nussaume L, Bogusz D, Haseloff J (2005) GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J Exp Bot 56: 2433–2442 [DOI] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L (2013) Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 18: 450–458 [DOI] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, Scheres B, Benfey PN (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SB, Xie ZZ, Hu CG, Zhang JZ (2016) A review of auxin response factors (ARFs) in plants. Front Plant Sci 7: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, Xu L (2014) WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26: 1081–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Swarup R, Paponov IA, Swarup K, Casimiro I, Lake D, Peret B, Zappala S, Mairhofer S, Whitworth M, Wang J, Ljung K, et al. (2011) Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol 155: 384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B (2014) PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano C, Ramirez-Parra E, Casimiro I, Otero S, Desvoyes B, De Rybel B, Beeckman T, Casero P, Gutierrez C, C Del Pozo J (2012) Auxin and epigenetic regulation of SKP2B, an F-box that represses lateral root formation. Plant Physiol 160: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk CW, Schuster C, Leyser O, Meyerowitz EM (2015) A developmental framework for graft formation and vascular reconnection in Arabidopsis thaliana. Curr Biol 25: 1306–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37: 128–138 [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Sozzani R, Yardımcı GG, Petricka JJ, Vernoux T, Blilou I, Alonso J, Winter CM, Ohler U, Scheres B, Benfey PN (2015) Transcriptional control of tissue formation throughout root development. Science 350: 426–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Salvi E, Giustini L, Terpstra I, Heidstra R, Costantino P, Sabatini S (2016) A SCARECROW-based regulatory circuit controls Arabidopsis thaliana meristem size from the root endodermis. Planta 243: 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona P, Linstead P, Martienssen R, Dolan L (2002) SCHIZORIZA controls an asymmetric cell division and restricts epidermal identity in the Arabidopsis root. Development 129: 4327–4334 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack MK, Harashima H, Dissmeyer N, Zhao X, Bouyer D, Weimer AK, De Winter F, Yang F, Schnittger A (2012) Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev Cell 22: 1030–1040 [DOI] [PubMed] [Google Scholar]
- Okumura K, Goh T, Toyokura K, Kasahara H, Takebayashi Y, Mimura T, Kamiya Y, Fukaki H (2013) GNOM/FEWER ROOTS is required for the establishment of an auxin response maximum for Arabidopsis lateral root initiation. Plant Cell Physiol 54: 406–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette AJ, Benfey PN (2005) Maturation of the ground tissue of the root is regulated by gibberellin and SCARECROW and requires SHORT-ROOT. Plant Physiol 138: 636–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavet V, Olmos E, Kiddle G, Mowla S, Kumar S, Antoniw J, Alvarez ME, Foyer CH (2005) Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol 139: 1291–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Swarup K, Ferguson A, Seth M, Yang Y, Dhondt S, James N, Casimiro I, Perry P, Syed A, Yang H, Reemmer J, et al. (2012) AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez JM, Candela H, Robles P, López-Torrejón G, del Pozo JC, Micol JL (2010) A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol 51: 1661–1673 [DOI] [PubMed] [Google Scholar]
- Perianez-Rodriguez J, Manzano C, Moreno-Risueno MA (2014) Post-embryonic organogenesis and plant regeneration from tissues: two sides of the same coin? Front Plant Sci 5: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wisniewska J, Tadele Z, Kubes M, Covanová M, Dhonukshe P, Skupa P, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T (2015) Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev Cell 33: 576–588 [DOI] [PubMed] [Google Scholar]
- Rakusová H, Abbas M, Han H, Song S, Robert HS, Friml J (2016) Termination of shoot gravitropic responses by auxin feedback on PIN3 polarity. Curr Biol 26: 3026–3032 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Perianez-Rodriguez J, Navarro-Neila S, Gude I, Moreno-Risueno MA, Del Pozo JC (2017) The transcription factor OBP4 controls root growth and promotes callus formation. New Phytol 213: 1787–1801 [DOI] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B (2001) A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13: 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ (2015) The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 27: 44–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Di Laurenzio L, Willemsen V, Hauser MT, Janmaat K, Weisbeek P, Benfey PN (1995) Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121: 53–62 [Google Scholar]
- Skoog F, Miller CO (1957) Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol 11: 118–130 [PubMed] [Google Scholar]
- Sugimoto K, Jiao Y, Meyerowitz EM (2010) Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell 18: 463–471 [DOI] [PubMed] [Google Scholar]
- Tian H, Jia Y, Niu T, Yu Q, Ding Z (2014) The key players of the primary root growth and development also function in lateral roots in Arabidopsis. Plant Cell Rep 33: 745–753 [DOI] [PubMed] [Google Scholar]
- Timpte C, Wilson AK, Estelle M (1994) The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H (2008) Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol 49: 1025–1038 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B (1997) Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390: 287–289 [DOI] [PubMed] [Google Scholar]
- van Norman JM, Zhang J, Cazzonelli CI, Pogson BJ, Harrison PJ, Bugg TD, Chan KX, Thompson AJ, Benfey PN (2014) Periodic root branching in Arabidopsis requires synthesis of an uncharacterized carotenoid derivative. Proc Natl Acad Sci USA 111: E1300–E1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Manes CL, Vercruysse S, Maes S, van der Schueren E, Beeckman T, Genschik P, Kuiper M, Inzé D, De Veylder L (2005) The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17: 1723–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B (2007) Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev 21: 2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B (2006) A molecular framework for plant regeneration. Science 311: 385–388 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jiao Y, Liu Z, Zhu YX (2015) ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nat Commun 6: 6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zürcher E, Müller B (2016) Cytokinin synthesis, signaling, and function—advances and new insights. Int Rev Cell Mol Biol 324: 1–38 [DOI] [PubMed] [Google Scholar]