A systems biology study reveals that anthocyanin biosynthesis in tomato vegetative tissue is accompanied by changes in the epidermis and architecture of the root.
Abstract
Plants accumulate secondary metabolites to adapt to environmental conditions. These compounds, here exemplified by the purple-colored anthocyanins, are accumulated upon high temperatures, UV-light, drought, and nutrient deficiencies, and may contribute to tolerance to these stresses. Producing compounds is often part of a more broad response of the plant to changes in the environment. Here we investigate how a transcription-factor-mediated program for controlling anthocyanin biosynthesis also has effects on formation of specialized cell structures and changes in the plant root architecture. A systems biology approach was developed in tomato (Solanum lycopersicum) for coordinated induction of biosynthesis of anthocyanins, in a tissue- and development-independent manner. A transcription factor couple from Antirrhinum that is known to control anthocyanin biosynthesis was introduced in tomato under control of a dexamethasone-inducible promoter. By application of dexamethasone, anthocyanin formation was induced within 24 h in vegetative tissues and in undifferentiated cells. Profiles of metabolites and gene expression were analyzed in several tomato tissues. Changes in concentration of anthocyanins and other phenolic compounds were observed in all tested tissues, accompanied by induction of the biosynthetic pathways leading from Glc to anthocyanins. A number of pathways that are not known to be involved in anthocyanin biosynthesis were observed to be regulated. Anthocyanin-producing plants displayed profound physiological and architectural changes, depending on the tissue, including root branching, root epithelial cell morphology, seed germination, and leaf conductance. The inducible anthocyanin-production system reveals a range of phenomena that accompanies anthocyanin biosynthesis in tomato, including adaptions of the plants architecture and physiology.
Anthocyanins are abundant vacuolar pigments derived from the phenylpropanoid pathway and are produced in many different plant species. Depending on the pH and their chemical modifications, anthocyanins can change color from red to purple and blue. Selecting for petal color in ornamental plants has been the subject of extensive research (Sasaki and Nakayama, 2015). This research has revealed many different enzymes involved in chemical modifications such as glycosylation, methylation, and acylation of anthocyanins. Whereas anthocyanins in flowers and fruits are known to function as attractants for pollinators and vectors for seed dispersal, the role of anthocyanin accumulation under stress in vegetative tissues is probably linked to the scavenging of reactive oxygen species (Gould, 2004). In tomato (Solanum lycopersicum), anthocyanins are predominantly found in stem and hypocotyl tissues, as a result of stress conditions (Roldan et al., 2014).
Anthocyanins are powerful antioxidants and, as part of human diet in seeds, fruit, and leaves are proposed to have health-promoting properties (Bassolino et al., 2013; Martin et al., 2011). It has been shown that the consumption of anthocyanins can lower the risk of cancer, diabetes, and cardiovascular diseases (Zafra-Stone et al., 2007; He and Giusti, 2010; Tsuda, 2012; Butelli et al., 2008). To be able to breed for fruits and vegetables that are rich in anthocyanins, it is important to understand both their biosynthesis and functions in plants. By expressing two transcription factor genes, Rosea1 (ROS1) and Delila (DEL), isolated from Antirrhinum majus flowers, under control of the tomato E8 promoter, which is expressed during fruit ripening, tomato plants were engineered that carry purple fruits (Butelli et al., 2008). These purple tomato fruits, that are otherwise isogenic to red fruits, have been essential for defining health claims for anthocyanins (Martin et al., 2011, 2013).
ROS1/DEL tomato fruits are enriched with anthocyanins that predominantly include delphinidin 3,5-diglycosides and are acylated with hydroxycinnamic acids (Butelli et al., 2008). Overexpression of ROS1/DEL transcription factors in tomato led to the induction of expression of a number of genes homologous to known genes from the anthocyanin pathway in Arabidopsis (Arabidopsis thaliana) and petunia (Butelli et al., 2008). In tomato, the role of some of these genes in anthocyanin biosynthesis has been confirmed using mutants disrupted in the FLAVONOID3-HYDROXYLASE (F3H) gene (Maloney et al., 2014) and the DIHYDROFLAVONOL4-REDUCTASE (DFR) gene (Goldsbrough et al., 1994). The tomato anthocyanin-specific O-methyltransferase was identified by a transcriptional analysis of a tomato seedling system, in combination with an interfering RNA strategy (Gomez Roldan et al., 2014). A next step in the understanding of anthocyanin biosynthesis in tomato should focus on the dynamic coordination and gene regulation of the anthocyanin pathways in time, and its integration within plant developmental programs.
Transcription factors (TFs) that regulate anthocyanin biosynthesis have been identified in many plant species (Petroni and Tonelli, 2011). A complex of three TFs, including an R2R3-Myb type TF, a basic helix-loop-helix type TF (bHLH), and a WD repeat TF, was shown to control anthocyanin accumulation (Xu et al., 2015) and in some cases other flavonoids, in many plant species, including Arabidopsis, maize (Zea mays), and petunia (Albert et al., 2014). In tomato, two highly homologous Myb TFs, ANT1 and AN2, have been shown to be involved in the regulation of anthocyanin biosynthesis (Mathews et al., 2003; Zuluaga et al., 2008). R2R3-Myb proteins, such as ROS1 and bHLH proteins such as DEL, serve as transcriptional activators of anthocyanin biosynthesis (Broun, 2005). In contrast, CAPRICE (CPC), a R3-type Myb TF, serves as a negative regulator of anthocyanin biosynthesis in Arabidopsis. CPC inactivates the MBW protein complex by competing with R2R3-MYB binding to a bHLH TF, while being unable to activate transcription (Tominaga et al., 2008). Related Myb and bHLH TF complexes can control other biosynthetic processes, such as glucosinolate biosynthesis (Frerigmann et al., 2014).
Interestingly, specific aspects of cellular differentiation such as root hair and trichome differentiation are also regulated by MBW complexes (for review, see Broun, 2005; Xu et al., 2015; Tominaga-Wada et al., 2013). For example, in Arabidopsis an MBW complex including WEREWOLFE, GLABRA3 (GL3), and Transparent Testa Glabra (TTG) controls the transcription of GLABRA2 (GL2), a TF, which acts on root and trichome developmental programs (Rerie et al., 1994; Bernhardt et al., 2005). Recently it was suggested that MBW complexes controlling secondary metabolism may have evolved from similar MBW complexes that regulate more ancient gene networks for differentiation of cell types (Chezem and Clay, 2016).
Systematic transcriptomics and metabolomics analysis have been employed to obtain a more holistic view of the regulation of the anthocyanin biosynthetic program. Such studies have been done both on tomato seedlings in which anthocyanin formation is induced by nutrient stress (Roldan et al., 2014) and on purple ROS1/DEL fruits (Tohge et al., 2015). In these studies, networks of genes and metabolites were analyzed by combining data from different tissues and cell types. Genes were identified that encode putative anthocyanin-modifying enzymes and transporters. However, to put anthocyanin biosynthesis in a context beyond biosynthetic genes, one needs to make observations on transcription networks and metabolite profiles that are independent of nutrient stress or developmental changes, and with high resolution in time. To achieve this, a uniform and tightly controlled system for steering the anthocyanin biosynthetic program is needed.
Here, we aimed to study the anthocyanin pathway in tomato and its associated cellular and developmental processes using such a tightly regulated transcription system. We developed, for the first time to our knowledge, an inducible dexamethasone-regulated switch that can deliver, on-demand, anthocyanin accumulation in different tissues of the tomato cultivar Micro-tom. We interrogated the transcriptional and metabolic networks associated with anthocyanin biosynthesis in different vegetative tissues of tomato, including undifferentiated totipotent callus cells. This study revealed, to our knowledge, new aspects of transcriptional regulation of anthocyanin accumulation in tomato plants, and linked it to epidermal cell fate, in particular in root tissues. We identified several targets of regulation by ROS1/DEL TFs including genes involved in epidermal cell fate determination, cuticle formation, auxin biosynthesis, and transport as well as several transcription factors. These data can serve as a resource for the identification of genes involved in anthocyanin biosynthesis. As an example, we focused on specific acyl transferases involved in the addition of hydroxycinnamic acids to the glycoside moieties of tomato anthocyanin in vivo. These data provide insight into the processes that may accompany anthocyanin biosynthesis, including physiological and architectural changes in tomato vegetative tissues.
RESULTS
Development of an Inducible System for Anthocyanin Biosynthesis in Tomato cv Micro-tom
Previous studies in tomato fruits have shown that substantial induction of anthocyanin biosynthesis can be achieved by ectopically expressing two transcription factors (ROS1/DEL) from snapdragon (Antirrhinum; Butelli et al., 2008; Tohge et al., 2015). These studies use the fruit-specific promoter E8, which is regulated by ethylene and thereby is specifically activated during the breaker stage of fruit ripening.
To collect comprehensive information about the gene expression program and metabolic changes that specifically accompany anthocyanin biosynthesis in different tissues and at different time points during plant development, a tomato system was engineered in which ROS1 and DEL expression could be experimentally induced by exogenous application of dexamethasone (DEX). This DEX-inducible system was used as described in Aoyama and Chua (1997). The cDNAs of ROS1 and DEL were both inserted behind synthetic promoters containing repeats of the upstream-activating sequence of the yeast gal4 gene. These promoters were regulated by a DEX-regulated chimeric transcription factor GVG, consisting of the DNA-binding domain of the yeast transcription factor GAL4, the transactivation domain of the herpes viral protein VP16, and the receptor domain of the rat glucocorticoid receptor (GR). Expression of the GVG was driven by the constitutive Arabidopsis UBIQUITIN10 (UBQ10) promoter (van Dijken et al., 2004).
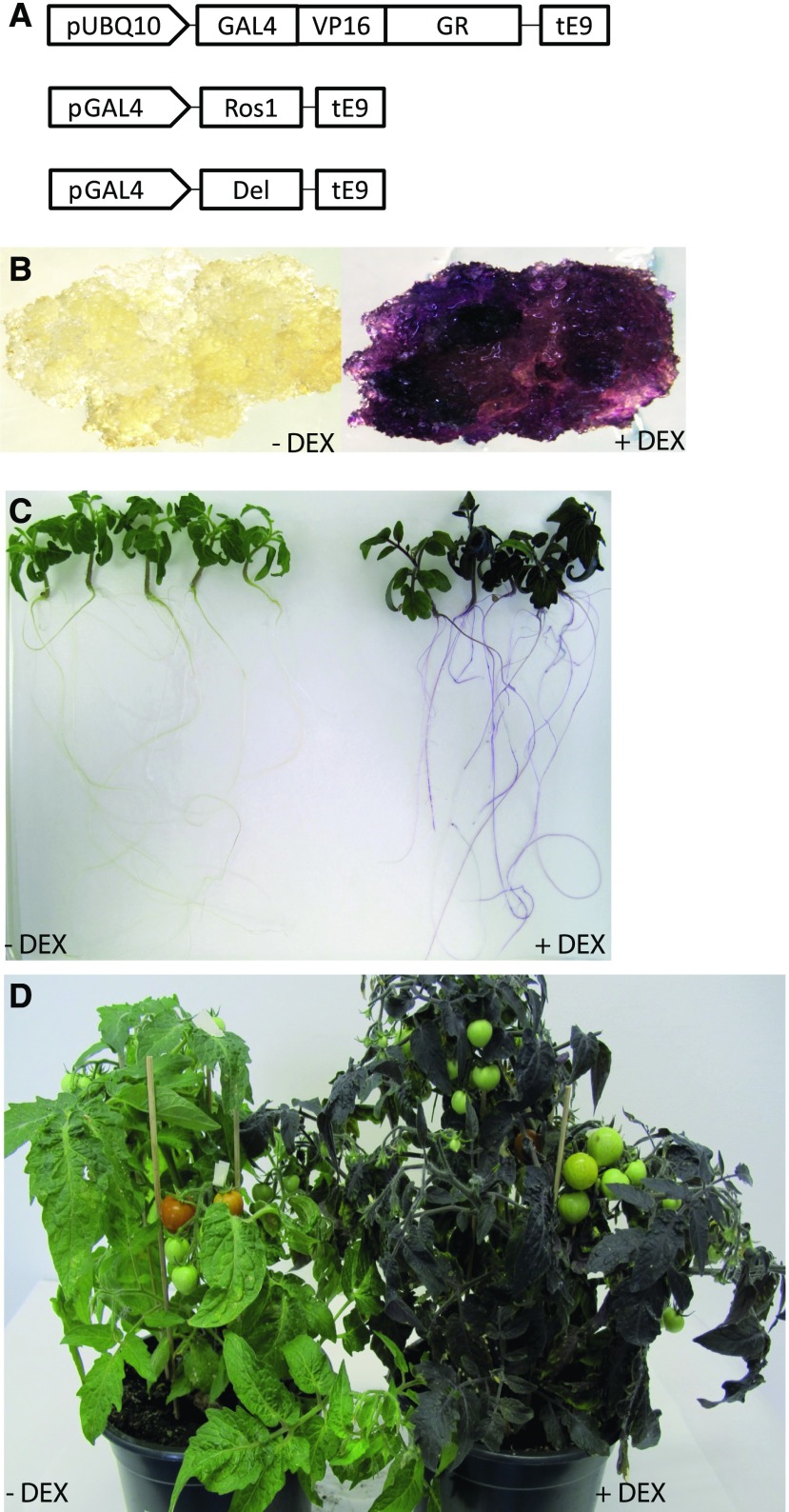
The three cassettes (GAL4-ROS1, GAL4-DEL, and UBQ10-GVG) were combined on a single plasmid as part of the same T-DNA (Fig. 1A). Transformation of this construct to S. lycopersicum Micro-tom yielded a number of transgenic calli, which were analyzed for presence of the transgenes by exposing them to DEX, upon which some of them turn purple (Fig. 1B). Some positive calli were maintained as undefined tissue, whereas others were regenerated into independent transgenic lines, ROS1/DEL lines 4, 8, and 11. Application of DEX to these plants, either by direct contact or by inclusion in the water supply to the soil, resulted in the formation of a purple color in most of the tested tissues (Fig. 1). This included vegetative tissues such as roots, stems, and leaves (Fig. 1, C and D). In contrast, in none of the lines was the formation of purple color observed in flowers or fruits upon application of DEX, even not when DEX was directly applied to these tissues.
Figure 1.
DEX-inducible system for anthocyanin accumulation in tomato. A, Schematic presentation of the constructs used in the study: promoter of the ubiquitin 10 gene from Arabidopsis drives the expression of a chimeric transcription factor consisting of the yeast GAL4 DNA recognition motive-VP16 activation domain and the GR domain. ROS1 and DEL expression is controlled by separate cassettes driven by promoter with gal4 binding sites. B, Representative primary callus before induction and 2 weeks after induction with DEX. C, Representative seedlings of ROS1/DEL line 4, 5 d after DEX induction. D, Plants of ROS1/DEL line 4 in soil two weeks after induction with DEX.
It has been shown before that upon high accumulation of anthocyanins, anthocyanin vacuolar inclusions (AVIs) are formed inside the cell vacuoles in different plant species (Grotewold, 2006; Chanoca et al., 2015). The accumulation of such AVIs were also observed here in tomato leaf epidermal cells when anthocyanin formation was induced by DEX (Supplemental Fig. S1A).
ROS1- and DEL-Induced Metabolites in Tomato Seedlings
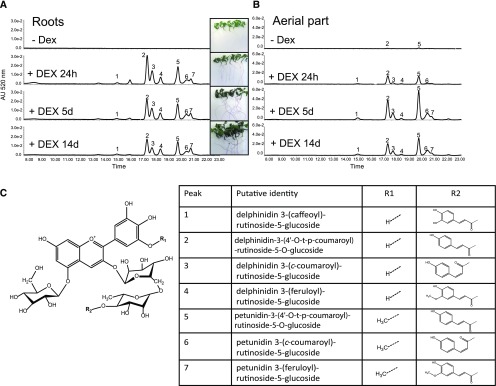
When ROS1/DEL were expressed in the leaves of Nicotiana, a single anthocyanin and a range of nonflavonoid phenolic compounds (e.g. polyamine and nor-nicotine conjugates) was produced (Outchkourov et al., 2014), whereas in tomato fruit, ROS1/DEL expression led to production of a set of complex anthocyanins and flavonoids (Butelli et al., 2008; Tohge et al., 2015). This suggests that the identity of ROS1/DEL-regulated metabolites depends on the species or tissue, but at present there are no data available of the consequence of ROS1/DEL expression in tomato vegetative tissues. To test this, ROS1/DEL were induced in leaf and root tissue from ROS1/DEL line 4, and an untargeted metabolite analysis was performed. Seedlings were incubated with or without the addition of DEX for 24 h, 5 d, and 14 d, then extracted with methanol and analyzed by liquid chromatography-photodiode array-mass spectrometry (LC-PDA-MS; Fig. 2, A to C; Supplemental Fig. S1, B and C). A rapid induction of anthocyanins (absorbing at 520 nm) was observed within less than 24 h after DEX application, in both roots and aerial parts of the seedlings (Fig. 2, A to C; Supplemental Fig. S1, B and C). MS analysis allowed the identification of seven anthocyanins (Fig. 2C; Supplemental Table S1), all of which had been previously identified in tomato hypocotyls and ROS1/DEL fruit (Roldan et al., 2014; Tohge et al., 2015). Both tissues contained the same anthocyanins, but at different ratios and with different kinetics. In roots, maximum induction of anthocyanins was reached within 24 h, whereas in the aerial parts of the seedlings this was reached only after 5 d. In the absence of DEX, roots did not contain detectable anthocyanins, whereas only minor amounts were found in the aerial parts of the seedlings.
Figure 2.
Metabolite analysis of ROS1/DEL-activated tomato plants. Examples of chromatograms from LC-PDA-MS at 520 nm showing seven peaks specific for anthocyanins in roots (A) and shoots (B) of tomato seedling, in the absence of DEX, or induced with DEX and sampled after 24 h, 5 d, and 14 d. C, Core structure of the tomato anthocyanins. Inset table: Identity of the anthocyanins observed in (A).
To identify other metabolites regulated by ROS1/DEL induction, an untargeted analysis of the LC-MS data was performed. In both tissues, the anthocyanins and a number of related flavonoids were found to be the dominant compounds, whereas other compound categories were much less obviously represented. In root tissue, 63 metabolites were found to be more than 2-fold up-regulated by DEX treatment (Supplemental Table S1A). In addition to the seven anthocyanins, seven flavonols were also found to be induced in time by DEX. Furthermore, four dexamethasone-derived metabolites were observed as well, in addition to dexamethasone itself. Thirty metabolites were found to be more than 2-fold down-regulated by dexamethasone treatment. For most of those, no identity could be assigned, but some ferulic acid conjugates (e.g. feruloyl-quinic acid, feruloyl-tyramine, and feruloyl octopamine) were observed. In shoots, 69 metabolites were consistently found to be more than 2-fold up-regulated by dexamethasone treatment. Most identifiable and major compounds (anthocyanins, flavonols, and DEX metabolites) correspond to those observed in roots (Fig. 2; Supplemental Table S1B and Supplemental Fig. S1, D to G). DEX itself was not visible in shoot tissue, suggesting that it can only reach the aerial parts after conjugation. Seventeen metabolites were significantly down-regulated by dexamethasone treatment, none of which could be identified.
A few tissue-specific changes were observed. For example, the root feruloyl conjugates found to be down-regulated by DEX were not detectable in shoot tissue, even in the control samples. In shoots, chlorogenic acid increased upon dexamethasone treatment after 24 h, and had increased further after 5 d and 14 d. In root tissue, an increase was only observed after 24 h, whereas chlorogenic acid levels did not differ significantly from the control after 5 d or 14 d (Supplemental Fig. S1, D to G). Thus, ROS1/DEL expression induced predominantly anthocyanin in tomato leaf and root tissue, in addition to a number of flavonoids, most of which were known to be induced in tomato fruit. In contrast to Nicotiana, no major changes in nonflavonoid metabolites could be observed in tomato root and shoot.
Transcriptome Analysis in Callus and Roots
To obtain a detailed understanding of ROS1/DEL function in regulating secondary metabolism, we identified genes that are controlled by these TFs on a genomewide scale in both undifferentiated and differentiated tissue. Studying callus tissue, consisting of basically uniform, nondifferentiated cells, has the advantage that developmental programs will not influence the transcriptional response to ROS1/DEL expression. On the other hand, root tissue is highly differentiated and can respond rapidly to developmental cues, which will allow us to address the interaction of ROS1/DEL-controlled secondary metabolism with developmental processes. Therefore, transcriptional changes upon ROS1/DEL activation were studied in roots of ROS1/DEL line 4 and of three callus cultures from independent primary transformation events. Time points were selected based on the presence of the anthocyanin biosynthetic proteins, anthocyanin synthase (SlANS), and dihydroflavonol 4-reductase (SlDFR), which have both previously been shown to be induced by ROS1 and DEL (Butelli et al., 2008). Antisera were developed that recognize recombinant SlANS or SlDFR proteins, and these were used to monitor expression of both proteins by western blot. SlANS was detectable as early as 3 h after induction and was increased significantly after 24 h in both callus and roots (Supplemental Fig. S2). The DFR protein was clearly detectable after 24 h. Therefore, samples for transcriptome analysis were taken after 3 h and 24 h.
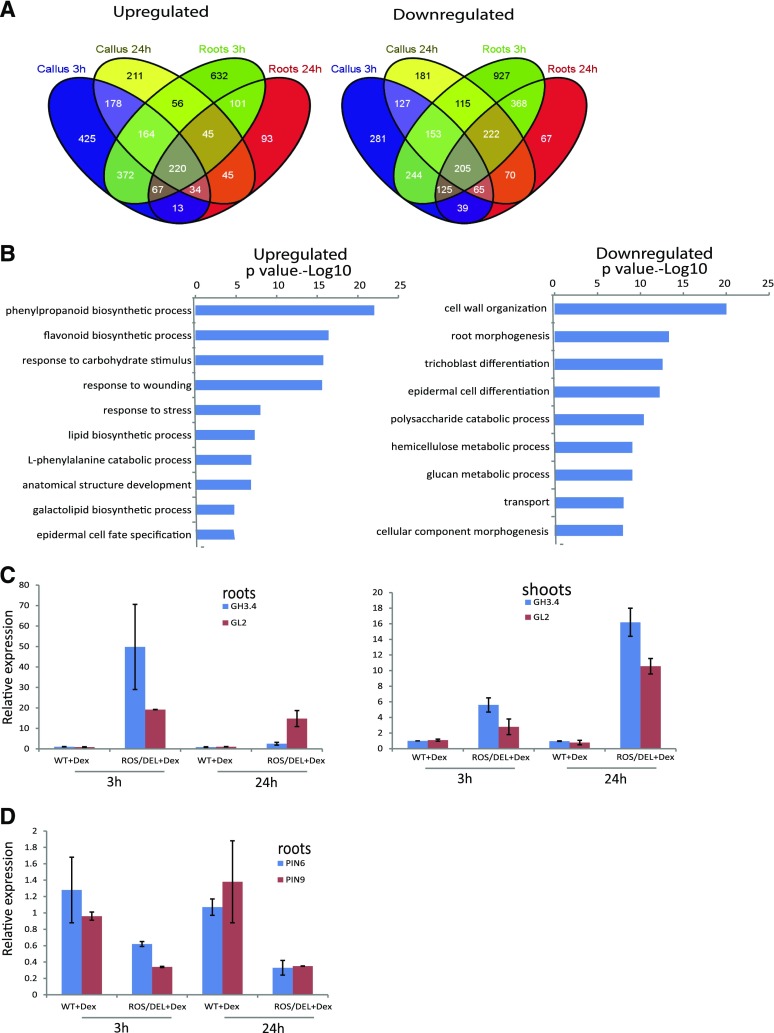
In vitro-grown callus derived from three different primary transformants (T0) and seedlings of line 4 (T2 generation) were transferred to fresh media with and without DEX, and samples were collected at 3 h and 24 h post induction. RNA was extracted, and cDNA was analyzed by Illumina sequencing. Reads were mapped onto tomato gene models to which the ROS1 and DEL cDNA sequences were added, and gene expression data were calculated. In total, expression of 5295 genes significantly changed more than 2-fold (n = 3; FDR < 0.05) for at least one of the time points or tissues, relative to untreated materials. Genes that were affected by ROS1 and DEL induction across all tissues and time points formed only a small subset of these genes, as can be observed in the Venn diagrams in Figure 3A. From a total of 5295 differentially regulated genes, 220 were consistently up-regulated and 205 were consistently down-regulated in both tissue types, i.e. callus and roots, and at both time points. This set of overlapping genes was used as the core of 425 genes affected by ROS1 and DEL (Supplemental Table S2).
Figure 3.
Gene expression profiles after DEX activation of ROS1/DEL. A, Venn diagrams showing the overlap of up- and down-regulated genes (P < 0.05, fold 2) in the different tissues and time points. A core of 220 up- and 205 down-regulated genes were identified as tissue- and time-independent functional targets of ROS1/DEL. B, Functional GO categories of the up-regulated and down-regulated genes. Bars represent P values of GO categories that are significantly overrepresented (left) and underrepresented (right), compared to all GO-annotated genes on the tomato genome. C and D, Quantitative RT-PCR confirmation for GH3.4, GL2 (C), PIN6, and PIN9 (D) gene expression (n = 4 biological replicates; shown are mean values with sds). ROS1/DEL line 4 seedlings (T4 generation) and wild-type seedlings were incubated with DEX for 3 h and 24 h. After that, the roots and the shoots were analyzed separately.
To obtain an overview of the functional implications of transcriptional changes mediated by ROS and DEL induction, gene ontology (GO) annotations for the 425 consistently regulated core genes were analyzed (Fig. 3B). This was done by comparing, for each GO category, its frequency among the 425 core genes to its frequency among all annotated genes in the tomato genome. In this way, GO categories that were overrepresented among the set of up-regulated or among the set of down-regulated genes were obtained. Significantly overrepresented GO categories among the up-regulated genes were involved in the biosynthesis of phenylpropanoids, flavonoids, and anthocyanins, as well as responses to different types of known anthocyanin-related stresses such as carbohydrate stimuli (Das et al., 2012). These functions obviously are relevant for the well-known role of ROS1/DEL in anthocyanin biosynthesis. Interestingly, the up-regulated genes were also enriched for GO categories involved in lipid biosynthesis and epidermal cell specification. Among the genes down-regulated by ROS1/DEL expression, GO terms involved in cell wall organization, root morphogenesis, and the differentiation of trichomes and epidermal cells were overrepresented. It is remarkable that GO categories that are known to function in specific differentiated tissues (e.g. root epidermis, leaf trichomes) were found to be consistently regulated by ROS1/DEL. Apparently, genes with these functions are also regulated by ROS1/DEL in undifferentiated callus, where they have no clear significance to the tissue.
Regulation of Pathways Leading to Anthocyanins by ROS1 and DEL Activation
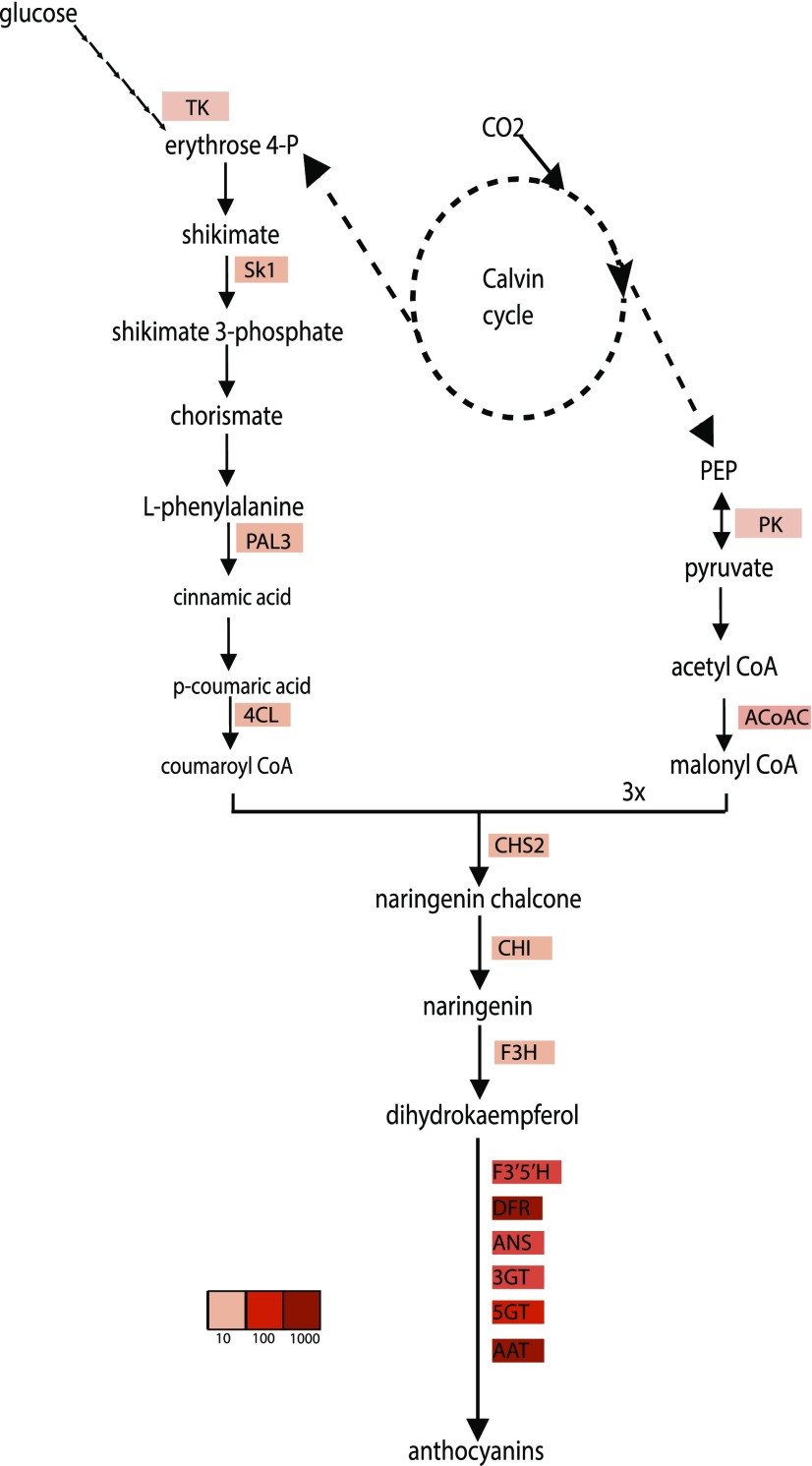
The effect of ROS1 and DEL gene expression on the regulation of individual genes was analyzed. Already after 3 h of induction, ROS1/DEL already strongly activated genes of the anthocyanin pathway, both in callus and in roots (Table I). In view of the observation that no anthocyanins could yet be detected at these stages, this would indicate that these genes are directly regulated by ROS1/DEL, and not by the presence of anthocyanins. Genes involved in converting Phe to polyphenols including anthocyanins mostly overlapped with those identified to be up-regulated in ROS1/DEL fruits, encoding enzymes and transporters from the pathway leading to anthocyanins, flavonoids, and chlorogenic acid (Butelli et al., 2008; Tohge et al., 2015). Interestingly, several genes from the Phe biosynthetic pathways were also activated after 3 h by ROS1/DEL, including shikimate kinase (SK1), a key enzyme of the shikimate pathway toward Phe, and cytosolic pyruvate kinase and acetyl coA carboxylase, involved in malonyl CoA biosynthesis (Table I and Fig. 4). Transketolase, an important enzyme in the pentose phosphate pathway (which converts Glc to supply erythrose 4-phosphate, the starting point of the shikimate pathway), was initially down-regulated in roots, while being up-regulated after 24 h (Table I). Likely this indicates that the pentose phosphate pathway is not under direct regulation of ROS1 and DEL, but is up-regulated in the root when an enhanced supply of carbon into the shikimate pathway is needed.
Table I. Genes regulated upon DEX induction.
| Gene Number | Gene Name | Putative Function | Callus 3 h | Roots 3 h | Callus 24 h | Roots 24 h | References |
|---|---|---|---|---|---|---|---|
| Transcription Factors Putatively Involved in the Epidermal Cell Fate | |||||||
| Solyc09g065100 | SlbHLH150 | — | 63 | 199 | 1259 | 2512 | (Sun et al., 2015) |
| Rosea 1 | Rosea 1 | Anthocyanin biosynthesis | 234 | 316 | 63 | 100 | (Goodrich et al., 1992) |
| Delila | Delila | Anthocyanin biosynthesis | 63 | 398 | 40 | 199 | (Schwinn et al., 2006) |
| Solyc07g052490 | MYB-CPC-like | Trichome and root hair regulation | 20 | 316 | 79 | 158 | — |
| Solyc03g120620 | GL2 | Cuticle regulation | 6 | 39 | 5 | 31 | (Lashbrooke et al., 2015) |
| Solyc01g109120 | WD40 repeat | — | 12 | 16 | 25 | 6 | — |
| Solyc10g055410 | SlTHM27 | Anthocyanin biosynthesis | 31 | 6 | 79 | 2 | (Matus et al., 2009) |
| Solyc04g072890 | WD40 repeat | — | 16 | 10 | 8 | 4 | — |
| Solyc03g098200 | HDG11-like/GL2- like | — | 2.5 | 6 | 2.5 | 10 | (Yu et al., 2008) |
| Solyc02g088190 | MIXTA-like | Epidermis and cuticle development | 2 | 3 | 4 | 8 | (Lashbrooke et al., 2015) |
| Solyc03g097340 | TTG1-like | Trichome and root hair regulation | 5 | 4 | 2.5 | 3 | (Galway et al., 1994) |
| Solyc10g081320 | SlMYB69 | — | −4 | −5 | −2 | −2 | (Stracke et al., 2001) |
| Solyc09g057710 | SlbHLH057 | — | 4 | 5 | 2 | 2 | (Sun et al., 2015) |
| Solyc04g074170 | SlMYB39 | — | −3 | −2.5 | −3 | −5 | — |
| Solyc05g051550 | SlMYB | — | −5 | −2.5 | −5 | −6 | — |
| Solyc09g018500 | SlbHLH | — | −20 | −10 | −20 | −32 | — |
| Cutin Pathway | |||||||
| Solyc00g278110 | CER1-like | Cutin synthesis | 29 | 252 | 17 | 199 | — |
| Solyc01g079240 | SlLACS1 | Cutin synthesis | 100 | 422 | 4.3 | 17 | (Girard et al., 2012) |
| Flavonoid Pathway | |||||||
| Solyc09g091510 | CHS2 | Chalcone synthase | 4 | 398 | 125 | 5011 | (Tohge et al., 2015) |
| Solyc05g052240 | CHIL | Chalcone isomerase | 20 | 316 | 100 | 631 | (Tohge et al., 2015) |
| Solyc05g010320 | CHI | Chalcone isomerase | 2.5 | 3.1 | 7.9 | 7.9 | (Tohge et al., 2015) |
| Solyc02g083860 | F3H | Flavanone 3-hydroxylase | 12 | 158 | 16 | 251 | (Tohge et al., 2015) |
| Solyc11g066580 | F3′5’H | Flavonoid-3′-monooxygenase | 501 | 1585 | 25,118 | 31,623 | (Tohge et al., 2015) |
| Solyc02g085020 | DFR | Dihydroflavonol 4-reductase | 15,849 | 5012 | 19,953 | 5012 | (Butelli et al., 2008) |
| Solyc08g080040 | ANS | Leucoanthocyanidin dioxygenase | 126 | 251 | 251 | 251 | (Tohge et al., 2015) |
| Solyc09g082660 | AnthOMT | Anthocyanin-O-methyltransferase | 501 | 100 | 5012 | 1995 | (Gomez Roldan et al., 2014) |
| Solyc10g083440 | A3GT | Anthocyanin-3-O-Glucosyltransferase | 501 | 630 | 398 | 501 | (Butelli et al., 2008) |
| Solyc09g059170 | A3G2”GT | Anthocyanin-3-O-gluc-2″ GT | 148 | 631 | 631 | 1000 | (Tohge et al., 2015) |
| Solyc12g098590 | A5GT | Anthocyanin-5-O- Glucosyltransferase | 1585 | 630 | 25,119 | 10,000 | (Tohge et al., 2015) |
| Solyc12g088170 | SlFdAT1 | Anthocyanin acyltransferase | 794 | 3162 | 1585 | 10,000 | (Tohge et al., 2015) |
| Solyc08g068700 | THT7-1 | Acyltransferase | 4 | 2.5 | 10 | 2.5 | (Von Roepenack et al., 2003) |
| Solyc03g097500 | HCT-like | Acyltransferase | −2 | −2.5 | −2.5 | −3 | — |
| Solyc08g078030 | HCT-like | Acyltransferase | −12 | −3 | −12 | −5 | — |
| Solyc10g078240 | C3H | p-Coumarate 3-hydroxylase | 5 | 4 | 4 | 2.5 | (Butelli et al., 2008) |
| Solyc02g081340 | GST | Glutathione-S-transferase | 1585 | 10,000 | 2511 | 10,000 | (Butelli et al., 2008) |
| Solyc10g084960 | GST | Glutathione-S-transferase | 10 | 10 | 6 | 5 | — |
| Solyc07g056510 | GST | Glutathione-S-transferase | 6 | 8 | 3 | 3 | — |
| Solyc06g009020 | GST | Glutathione-S-transferase | 4 | 5 | 3 | 2.5 | — |
| Solyc03g025190 | FFT | Flower flavonoid transporter/putative anthocyanin permease | 316 | 10,000 | 2000 | 50,119 | (Mathews et al., 2003) |
| Pentose Phosphate Pathway | |||||||
| Solyc10g018300 | Tansketolase | Transketolase | −2.5 | −4 | 2 | 2.5 | — |
| Solyc09g008840 | Pyruvate kinase | Pyruvate kinase | 3.4 | 2.5 | 4 | 2.6 | — |
| Solyc12g056940 | Acetyl-CoA carboxylase | Acetyl-CoA carboxylase | 17 | 12 | 12 | 9 | — |
| Shikimate Pathway | |||||||
| Solyc04g051860 | SK1 | Shikimate kinase | 4 | 4 | 2.5 | 2 | (Schmid et al., 1992) |
| Phenylpropanoid Pathway | |||||||
| Solyc02g086770 | CCR | Cinnamoyl CoA reductase | 40 | 32 | 20 | 10 | (Tohge et al., 2015) |
| Solyc08g005120 | CCR | Cinnamoyl CoA reductase | 4 | 8 | 5 | 5 | — |
| Solyc03g036480 | PAL | Phe ammonia lyase | 6 | 8 | 5 | 5 | — |
| Solyc03g036470 | PAL | Phe ammonia lyase | 6 | 8 | 6 | 4 | — |
| Solyc05g056170 | PAL6 | Phe ammonia lyase | 5 | 3 | 10 | 3 | (Tohge et al., 2015) |
| Solyc09g007910 | PAL3 | Phe ammonia lyase | 10 | 2 | 5 | 2 | (Tohge et al., 2015) |
| Solyc03g042560 | PAL | Phe ammonia lyase | 5 | 6 | 6 | 3 | (Butelli et al., 2008) |
| Solyc01g079240 | 4CL6 | 4-Coumaroyl CoA ligase | 16 | 4 | 398 | 16 | (Tohge et al., 2015) |
Shown are accession numbers (1st column), gene abbreviation (2nd column), putative function (3rd column), fold-change induction upon DEX application in callus after 3 h (4th column), in roots after 3 h (5th column), in callus after 24 h (6th column), in roots after 24 h (7th column), and a reference for the function of the gene (8th column). Only genes for which a t test indicates that expression upon DEX induction is significantly different from noninducing conditions (t test n = 3; P ≤ 0.05) are shown. More genes can be found in Supplemental Table S2.
Figure 4.
Genes regulated by ROS1/DEL in the anthocyanin pathway. Schematic overview of the anthocyanin biosynthetic pathway and the genes changing upon induction with DEX. More intense red color indicates stronger up-regulation.
Biosynthetic genes not known to participate in anthocyanin biosynthesis were also found to be regulated by ROS1/DEL expression, including, for instance, genes important for auxin homeostasis. A number of auxin transporter genes (PINs and LAXs) were already found to be down-regulated 3 h after DEX induction (Supplemental Table S2). Gene homologous to Auxin-regulated Indole-3-acetic acid-amido synthetase (GH3.4; Liao et al., 2015) was up-regulated upon DEX addition (Supplemental Table S2). GH3 genes in plants have been shown to regulate auxin homeostasis levels by conjugating the excess of active indole acetic acid to an inactive form.
Regulation of a Transcriptional Network Involved in Epidermal Differentiation
Upon up-regulation of ROS1 and DEL, a considerable number of other transcription factors were observed to be activated in both callus and roots (Table I and Supplemental Table S2). From the core of 425 genes a total of 27 genes were transcription factors, most of which (22 genes) were up-regulated. This indicated the possibility that ROS1/DEL not only directly activate biosynthetic genes in the anthocyanin pathway but also regulate, or at least influence, a more complex transcriptional network. Interestingly, among the ROS1/DEL-regulated TF genes, were several that are annotated with the GO-category epidermal cell fate. One of these genes is the tomato MIXTA-like TF, which is a key regulator of epidermal cell patterning and cuticle assembly in tomato fruit (Lashbrooke et al., 2015). Also, a homolog of Glabra2 (GL2), known to be involved in root and trichome developmental programs in Arabidopsis (Rerie et al., 1994; Bernhardt et al., 2005), was 6-fold up-regulated in callus and 30-folds in roots after DEX induction. Surprisingly, one of the most strongly up-regulated TFs (125-fold in roots) only 3 h after DEX induction is homologous to the Arabidopsis root-hair regulator CPC.
Confirmation of the Activation of Root Morphology and Auxin-Related Genes Using Quantitative Real Time PCR
Transcriptome analysis was based on a comparison of tissues from induced and noninduced plants from the same genotype, to avoid noise from genotype-related differences in the data. Gene expression changes relevant for root morphology and for auxin regulation were validated to confirm their dependence on the expression of ROS1/DEL, and not to result from the application of DEX per se. Real-time quantitative PCR (qPCR) analyses were performed for a subset of genes, comparing ROS1/DEL plants (T4 generation) to wild-type seedlings, both of which were treated with DEX. The qPCR results confirmed the up-regulation of GH3.4 and GL2 and the down-regulation of PIN6 and PIN9, observed from the transcriptomics results in roots treated with DEX for 3 h and 24 h (Fig. 3, C and D). Furthermore, we observed up-regulation of GH3.4 and GL2 in the aerial parts of seedlings incubated with DEX for 3 h and 24 h (Fig. 3C).
Activation of the Tomato GL2 Promoter by ROS1/DEL in Nicotiana
To confirm that the tomato GL2 homolog is a possible direct target of ROS1/DEL, a reporter transactivation assay was used. The tomato GL2 promoter was fused to a luciferase reporter gene (pGL2-LUC), which enables the visualization of GL2 promoter activation. N. benthamiana leaves were agro-infiltrated with pGL2-LUC alone, or in combination with 35S:ROS1 and 35S:DEL constructs (Supplemental Fig. S3, A and B). After 3 d postinfiltration, the leaves were sprayed with luciferin and incubated for one more day, after which luciferase activity was measured. The pGL2-LUC construct alone resulted in very low luminescence (Supplemental Fig. S3, A and B), whereas the luciferase activity was strongly induced by coinfiltration of pGL2-LUC with ROS1/DEL, confirming ROS1/DEL control GL2 expression also in leaf tissue.
Tissue-Specific Gene Activation by ROS1/DEL
Besides the genes found to be regulated in both callus and root tissues and at both time points, a number of GO categories was found to be overrepresented among genes regulated differentially in only one of the tissues (Supplemental Fig. S4A). This analysis was performed in the same way as described above for the set of 425 genes, but this time separately per tissue. Specifically for genes differentially regulated in callus, a set of genes involved in transcriptional regulation was observed. These genes consisted of a set of 18 TFs, from diverse TF families. These TFs are different from those identified to be regulated in all tissues (the core set of genes). Also, the GO term “oxidoreductase activity” was found exclusively in callus; this category included genes involved in ethylene biosynthesis, such as five 1-aminocyclopropane-1-carboxylate oxidases (ACOs), two of which (including ACO2) were up-regulated by ROS1/DEL in callus, whereas three were down-regulated.
In roots, compared to callus, a larger set of GO categories was found to be overrepresented in genes that were differentially regulated by DEX-induced ROS1/DEL expression (Supplemental Fig. S4 and Supplemental Table S3). A number of these categories relate to lipid metabolism, for instance categories lipid transport and lipid localization and lipid metabolic processes. The regulated genes in these categories encode enzymes involved in cuticle polymerization (e.g. GDSL1/cutin deficient1, which was 94-fold up-regulated in roots; Girard et al., 2012; Yeats et al., 2014) and several lipid transfer proteins. Interestingly, two of the highly up-regulated genes identified in both callus and roots correspond to long chain acyl-CoA synthase1 (LACS1) and the tomato homolog of CER1, both of which have been predicted to be involved in cutin monomer synthesis (Lü et al., 2009; Girard et al., 2012; Table I). Thus, the transcriptomics data suggest a positive link between anthocyanins’ biosynthesis and cuticular wax biosynthesis. Another GO category, antioxidant activity, comprised five peroxidases, four of which were strongly down-regulated upon ROS1/DEL expression in roots. In the GO category, response to biotic stimuli, we found three highly up-regulated genes (from 87-fold to 160-fold), which encode homologs of the birch pollen allergen Bet v 1 (Muñoz et al., 2010), whereas other genes from the same protein family were down-regulated by ROS1/DEL.
Functional Analysis of the ROS1- and DEL-Activated Genes
For functional analysis of genes regulated by ROS1 and DEL, a virus-induced gene silencing (VIGS) system was used in combination with DEX induction of ROS1/DEL in Micro-tom seedlings. The efficiency of VIGS was tested by silencing the PHYTOENE DESATURASE (PDS) gene in parallel to the genes of interest. Silencing of PDS resulted in photobleaching (Supplemental Fig. S5A), causing white patches and white leaves, and is therefore easily detected visually (Velásquez et al., 2009; Zheng et al., 2010). After 11 d, white patches and white leaves were observed, mainly in the newly formed leaves. After induction, by transferring seedlings to DEX-containing medium, the white patches became bright purple (Supplemental Fig. S5A).
Among the genes that were highly up-regulated by ROS1/DEL were putative anthocyanin acyltransferases (AAT), which could potentially contribute to modification of the anthocyanins. Two AAT genes, Solyc08g068710, here referred to as AAT-1 and AAT-2 (Solyc12g088170), were tested for their effect on anthocyanin biosynthesis. Although AAT-1 silencing did not result in any significant changes in anthocyanin content or composition, the effects of AAT-2 were clearly visible in an LC-MS analysis (Supplemental Fig. S5B). Major tomato anthocyanins, such as petunidin 3-(transp-coumaroyl)-rutinoside-5-glucoside and delphinidin 3-(transp-coumaroyl)-rutinoside-5-glucoside, were down-regulated by silencing AAT-2, whereas delphinidin 3-rutinoside, lacking an acyl group, was strongly up-regulated. Similarly, some less-abundant quercetin acyl conjugates were strongly down-regulated upon silencing of AAT-2 (Supplemental Fig. S5B). These metabolic changes support a role for this gene in acylation of anthocyanins as well as other flavonoids in tomato. The role of AAT-2 in acylation of anthocyanins was already supported by in vitro enzyme activity experiments, and overexpression of the gene in tobacco flowers (Tohge et al., 2015).
Physiological and Developmental Effects of ROS1/DEL Induction in Different Tomato Tissues
As observed from the RNA sequencing data and the qRT-PCR data, ROS1/DEL overexpression up-regulates the expression of the tomato GL2 homolog (Fig. 3C and Table I). In Arabidopsis, GL2 has been reported to promote trichome development and to inhibit both root-hair formation (Ohashi et al., 2002; Masucci et al., 1996) and stomata formation (Lin and Aoyama, 2012). We therefore hypothesized that ROS1/DEL overexpression, through up-regulation of a homolog of GL2, would have similar effects on tomato. Therefore, several tissues were inspected for morphological perturbation.
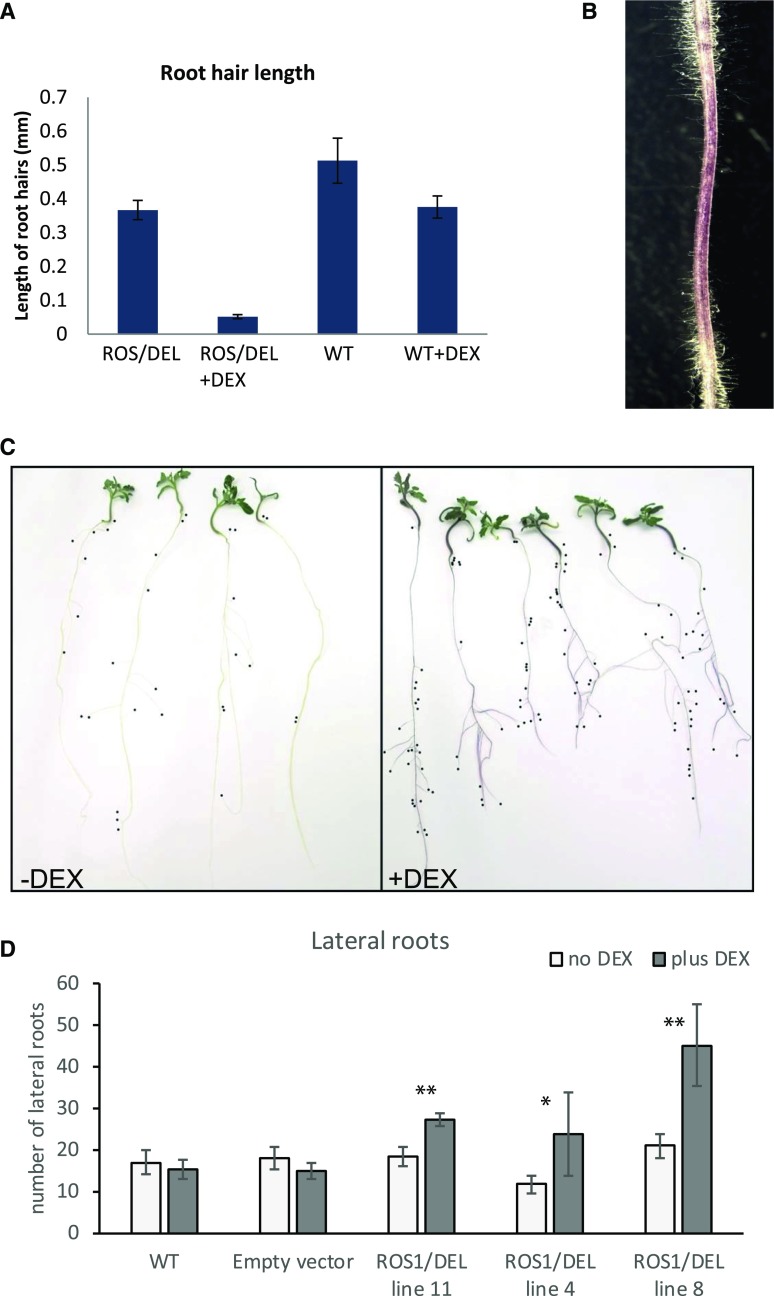
To address the effect of ROS1/DEL overexpression on root morphology, 5-d-old seedlings of ROS1/DEL lines 4, 8, and 11, wild-type, and EV (control) plants were transferred to tilted MS-agar plates with or without DEX. After 4 d of growth, the newly formed parts of the root were studied for root-hair length. The ROS1/DEL plants showed purple roots with much shorter root hairs than plants grown on plates without DEX (Supplemental Fig. S6A). For ROS1/DEL line 4 and wild-type seedlings, root hair length was quantified (n = 12), and was observed to be 10-fold reduced upon DEX induction (Fig. 5). Within one root, the intensity of the purple color of the root appeared to correlate with a reduction in root hairs (Fig. 5B). No significant change in root-hair length or density occurred when wild-type tomato plants were placed on DEX (Supplemental Fig. S6B). Also, the number of lateral roots was quantified after 12 d on plates with and without DEX. Whereas EV and wild-type plants did not display a significant difference in the number of lateral roots upon DEX induction, seedlings from ROS1/DEL lines 4, 8, and 11 developed significantly more (up to 2-fold) lateral roots on DEX, in comparison to plants transferred to medium without DEX (Fig. 5, C and D).
Figure 5.
Phenotypic effects of ROS1/DEL activation on root morphology. A, Root-hair length of ROS1/DEL line 4 plants and wild-type plants in the presence and absence of DEX. Shown are mean values (n = 12) and sd. B, Sectors with less purple coloration have longer root hairs. C, Influence of ROS1/DEL activation on the number of lateral roots in ROS1/DEL line 4. Plants were transferred to media with or without DEX 5 d after germination and scored for lateral roots after 12 d of DEX induction. Black dots are placed at the end of each lateral root. D, Number of lateral roots in the absence and presence of DEX in wild-type Micro-tom, EV control and ROS1/DEL lines 4, 8, and 11. Shown are mean number of lateral roots (n = 7): asterisks indicate significant differences between induced and noninduced seedlings from the same line after 12 d of growth (Student’s t test: *: P < 0.05; **: P < 0.01). Error bars represent the sd.
In leaf epidermis tissue, effects of ROS1/DEL induction on the morphology could not be observed, apart from the purple-colored AVIs (see above). Upon prolonged exposure of 6-week-old wild-type and ROS1/DEL plants to DEX for 5 d to 10 d, no changes in the number of trichomes or number of stomata on newly formed leaves could be observed using microscopy (Supplemental Fig. S7, A and B). Also, no changes in chlorophyll levels were observed by spectral analysis (Supplemental Fig. S7D). Notably, leaf conductance in the DEX-induced ROS1/DEL plants was reduced by ± 25% (Supplemental Fig. S7C). However, no consistent differences in stomatal opening could be observed using a binocular microscope, indicating that transpiration rates are potentially lowered in these plants by other mechanisms.
Seed germination was strongly affected by ROS1/DEL overexpression. Although DEX itself hardly affects germination of control seeds, germination of ROS1/DEL seeds of lines 4 and 11 was strongly delayed on DEX medium, compared to non-DEX medium (Supplemental Fig. S6C). When seedlings were germinated on noninducing medium, and then transferred to DEX containing medium after 5 d, no growth retardation upon ROS1 and DEL activation was observed.
Combined, these results strongly indicate that regulation of anthocyanin signaling is linked to various developmental programs, some of which are executed in a tissue-specific manner.
DISCUSSION
In this work, we have engineered tomato plants with a DEX-inducible system for anthocyanin biosynthesis, to study the systems biology of a secondary metabolic pathway in tomato. This is the first time, to our knowledge, that such a system has been used in a crop species such as tomato. The use of DEX-mediated promoter control allows for the precise characterization of responses to regulatory genes, such as transcription factors, and is here used to drive expression of a well-known transcription-factor pair, ROS1 and DEL. In Arabidopsis, the DEX promoter system has mainly been deployed to study gene expression programs associated with organ development, e.g. by overexpression of TFs regulating flower formation (Kaufmann et al., 2010) or trichome patterning (Lloyd et al., 1994), and recently, processes such as secondary cell wall formation (Li et al., 2016). Myb transcription factors such as ROS1 have hardly been studied before with such tight DEX-inducible systems (Morohashi and Grotewold, 2009), probably because these TFs often do not tolerate the presence of C-terminal tags such as the glucocorticoid binding domain. An indirect induction system, using an artificial TF and artificial promoters, such as used here, avoids this limitation and allows studying the activity of the native TFs. One could exploit such an inducible system to monitor specific responses of plants and plant organs to secondary metabolic pathways.
Expression of ROS1 and DEL in tomato is known to lead to the production of anthocyanins in fruits (Butelli et al., 2008), but was never addressed in root or callus tissues. In particular, purple coloration of tomato root is not known to occur in wild-type tomato plants. The DEX-inducible system as it was deployed here in tomato led to controlled anthocyanin production in leaves, stems, roots, and undifferentiated callus within 24 h of induction. It can be used in tissue culture, in whole plants that are grown in vitro or in soil. This allows us to monitor gene expression programs and metabolite profiles that are directly controlled by the ROS1/DEL TF pair, which will take place in all these tissues and conditions. In addition, one can observe tissue-specific responses at the transcriptional level that provide insights into the interactions between the ROS1/DEL-controlled processes, and the local, organ-specific physiological conditions and developmental programs. These interactions define the role of anthocyanins and their master regulators in the physiology and development of the tomato plant.
Notably, the DEX-inducible system used in this study did not result in anthocyanin formation in tomato flower and fruit tissues. Clearly this provides a limitation to the applications of this system in tomato. Likely this observation relates to poor expression of the GVG TF from the UBI10 promoter in these tissues, because direct application of DEX to fruits and flowers also did not result in appreciable coloration. On the other hand, in the absence of DEX, no anthocyanins could be detected in roots, indicating that the regulation of the UBI10-GVG system is sufficiently tight to provide a no-expression condition, which is very useful for systems biology approaches.
ROS1/DEL Control Anthocyanin-Biosynthetic Genes
At the metabolite level, the response of tomato tissues to ROS1/DEL expression is visibly dominated by anthocyanins and related polyphenolic compounds. Although other metabolites were also consistently induced in all analyzed tissues, their numbers are limited and less prominent in the metabolic profiles. Similarly, in tomato fruit, metabolite changes controlled by these TFs were found to be confined to anthocyanins and flavonols (Butelli et al., 2008; Tohge et al., 2015). Accordingly, the ROS1/DEL-induced genes that encode biosynthetic enzymes for anthocyanins and their precursors largely correspond to those observed in these earlier transgenic studies.
Among the identified biosynthetic genes affected were two AAT-encoding genes that could putatively mediate the acylation of anthocyanins. The role of these genes was further explored using a VIGS approach in combination with DEX-induced anthocyanin biosynthesis. Tomato anthocyanins are derived from delphinidin 3-rutinoside, which is modified by methylation on the 3′ and 4′ position, glucosylated at the 5 position and acylated on the rhamnose (Butelli et al., 2008; Gomez Roldan et al., 2014). Silencing the expression of one of the putative AATs leads to the accumulation of delphinidin 3-rutinoside in tomato leaves, demonstrating the role of this gene in anthocyanin acylation in tomato. Modifications such as methylation and 5-glucosylation apparently depend on rhamnose acylation, because no anthocyanins lacking acyl groups but carrying methyl groups were observed. Also, only small amounts of delphinidin 3-rutinoside-5-glucoside could be observed upon silencing of AAT-2. Recently, it was shown that the SlFdAT1 gene, which corresponds to AAT-2, mediates acylation of cyanidin rutinoside in Nicotiana tabacum flowers (Tohge et al., 2015). The observed dependence of anthocyanin methylation and, to some extent, 5-glucosylation, on acylation is similar to the situation in Petunia, where the gf mutant, which localizes in an acyltransferase gene, produces anthocyanins that lack acyl-groups, 5-glucosides, and methyl groups (Jonsson et al., 1984). Thus, the AAT-2/SlFdAT1 is not only required for acylation of anthocyanins, but it is also necessary for their 5-glucosylation and O-methylation.
Interestingly, several tissue specific changes in gene expression were observed that can be related to anthocyanin biosynthesis and function. For example, a number of genes of the Bet v 1 family were observed to be highly regulated by ROS1/DEL overexpression only in roots. The Bet v 1 protein family encodes small lipocalin-like proteins with a hydrophobic core, which may contain polyphenolic compounds such as quercetin (Roth-Walter et al., 2014). A function of these proteins in anthocyanin accumulation was previously suggested from work in strawberry, where down-regulation of Fra a1, a BetV1 homolog, led to colorless fruits (Muñoz et al., 2010).
A set of genes encoding peroxidases was found to be strongly down-regulated only in roots, but not in callus. The role of these peroxidases is likely in controlling damage by scavenging reactive oxygen species (Davletova et al., 2005). Overexpression of ROS1/DEL in tomato fruit leads to a higher antioxidant capacity and control of the oxygen burst during fruit ripening (Zhang et al., 2013). Possibly the scavenging of reactive oxygen species by accumulating anthocyanins makes expression of peroxidases redundant, thus leading to their down-regulation. However, it remains unclear why such down-regulation is only observed in roots, and not in callus.
Regulation of Epidermal Programs
The DEX-controlled system for expression of ROS1/DEL apparently controls processes beyond anthocyanin biosynthesis. In fruits, effects of ROS1/DEL on ripening and glycoalkaloid accumulation have been observed (Tohge et al., 2015; Zhang et al., 2013). In this work, substantial morphological changes were observed in roots, with regard to architecture and epidermal morphology, and in stomatal conductance. The transcriptomics analysis provides leads for mechanistic explanations of these phenomena. For instance, genes involved in auxin homeostasis and auxin flux, such as GH3.4, PINs, and LAX (Liao et al., 2015), were already found to be strongly regulated 3 h after DEX induction in root, callus, and leaf tissue. Auxin is known to control formation of lateral roots and root-hair elongation (Overvoorde et al., 2010; Maloney et al., 2014). In addition, genes involved in epidermal cell fate regulation are strongly regulated by ROS1/DEL expression. Among those is a MIXTA-like TF gene, which was shown to regulate the epidermal cell patterning and cuticle assembly in tomato fruit (Lashbrooke et al., 2015). Genes known to act downstream of MIXTA-like were also observed to be strongly regulated by ROS1/DEL, for example genes involved in cuticular wax biosynthesis, such as CER1 and GDSL1-cutin deficient 1, and GL2. GL2 is known in Arabidopsis to play a role in root-hair patterning (Girard et al., 2012; Yeats et al., 2014), but its role in tomato has never been addressed. Now that these genes have been identified as putative actors on architecture and epithelial morphology in roots, more detailed functional studies are needed to elucidate their exact role in those processes.
Our results suggest that ROS1/DEL orchestrates anthocyanin biosynthesis by integrating and regulating a network of transcription factors, metabolic enzymes, and transporters, and growth, patterning, and hormonal pathways in tomato. Moreover, the presented data indicate that anthocyanin-regulating TFs like ROS1 and DEL can be linked to a broader set of stress responses than solely anthocyanin accumulation. As with anthocyanins, the architectural changes in roots can also be related to stress due to nutrient availability and drought (López-Bucio et al., 2003; Kovinich et al., 2015). Also, effects on cuticle-related gene expression and stomatal conductance could similarly be linked to protection against stress. One could interpret these results to hypothesize that MYB/bHLH transcription factor complexes share a number of programs that allow the adaptation of the plant to environmental changes. Thus, regulation of secondary metabolites such as anthocyanins appears as an integral part of the plant’s adaptive repertoire, which also includes developmental and physiological programs. By using an inducible system for expression of such TF complexes, one can study these programs and their phenotypic consequences in different tissues, and provide, to our knowledge, novel leads for mechanisms that can be recruited by a plant for its survival and adaptation.
CONCLUSIONS
Anthocyanins are important in plants for protecting them against stress, and they are also important antioxidants in the human diet. Here a fully inducible system to make anthocyanins on demand is presented. With this we show that anthocyanin biosynthesis is integrated with changes in the architecture of the root.
MATERIALS AND METHODS
Plant Growth Conditions
Tomato plants (Solanum lycopersicum) cultivars Micro-tom and transgenic ROS1/DEL plants were grown in the greenhouse at ambient temperatures (>20°C) under natural light supplemented with artificial sodium lights, following a 16-h-light/8-h-dark cycle. Activation of ROS1/DEL seedlings by dexamethasone was done by placing seedlings on agar containing 10 mg/L dexamethasone. Dexamethasone was dissolved in ethanol at 10 mg/mL and diluted 1000× in the agar. As a negative control, the same volume of ethanol was used. To increase ROS1/DEL activation, 3 mg/mL DEX was dripped with 10 μL drops on top of the agar near the plants. Induction experiments were done in round (base diameter, 9 cm; top, 10.5 cm × 14 cm) sterile plastic plant containers with a breathing strip. For the VIGS experiments, 33-d-old plants were used with a maximum of three plants per pot supplemented with 200 mg/L cefotaxime and 50 mg/L vancomycin in the medium. Root development studies were done on large 24.5 cm × 24.5 cm plates with 0.6 cm of 1.3% Duchefa Daisin agar and 0.5× Duchefa MS medium. 5 d after germination, seedlings were transferred to plates with or without DEX. Only seedlings with intact roots were used. Plates were tilted between 45° and 60° and grown at 25°C.
Generation of Inducible ROS1/DEL Transgenic Plants
The DEX-inducible system pTA7002 (van Dijken et al., 2004) was modified by placing the genes ROSEA1 and DELILA from Antirrhinum majus under the control of a DEX-activated fusion protein constitutively expressed by the UBIQUITIN10 promoter. Coding sequences of ROS1 and DEL from Outchkourov et al. (2014) were amplified using the primers: XhoI_ROS, ROS_SpeI and XhoI_Del, and Del_Spe (Table S4) and ligated into the pTA7002 (UBQ10; van Dijken et al., 2004) vector digested with XhoI and SpeI restriction enzymes to generate pTA7002(UBQ10)-ROS1 and pTA7002(UBQ10)-DEL constructs. Next, the pTA7002(UBQ10)-DEL vector was modified to contain an extra multiple cloning site (pTA7002(UBQ10)DEL-MSC). Briefly, pTA7002(UBQ10)-DEL was amplified by PCR using oligonucleotides AsiSI_FW and MauBI_Rev (Table S4). The obtained PCR fragment was ligated into pTA7002(UBQ10)-DEL predigested with the same enzymes to generate pTA7002(UBQ10)DEL-MSC. A new PCR was conducted using the oligonucleotides MauBI_B_FW and ApaI_FW using pTA7002(UBQ10)-ROS1 as a template and the obtained PCR fragment was ligated in pTA7002(UBQ10)DEL-MSC digested with ApaI and MauBI to generate pTA7002(UBQ10) ROS1/DEL. The resulting vectors (pTA7002(UBQ10) ROS1/DEL and pTA7002) were transformed into Agrobacterium tumefaciens strain AGL0 using electroporation and transformed to Micro-tom wild-type plants as described in Karlova et al. (2011) and Bemer et al. (2012). Primary (transgenic) callus was obtained during the plant transformation procedure and was maintained on MS medium supplemented with 1.0 mg/L 2,4-dichlorophenoxyacetic acid and 0.1 mg/l kinetin. ROS1/DEL positive callus was selected by transferring part of the callus to DEX-containing medium and screening for formation of purple color after 48 h. Seed from T2- to T4-generation, self-pollinated transgenic plants were used for all experiments, including wild-type segregants from the same population. Seeds were sterilized in 1% bleach for 20 min and washed 2 times for 5 min in sterile water, dried on sterile filter paper for 10 min, and sowed on 0.8% Duchefa Daisin agar containing 2.2 g/L Duchefa MS with vitamins. A quantity of 25 seeds was used per sterile plastic rectangular container (Duchefa; base, 12 cm × 6.5 cm; top, 8 cm × 14 cm; height, 7 cm) with a breathing strip in the lid. Containers with seeds were placed at 4°C overnight and grown for 5 d to 9 d at 25°C at 16-h light/8-h dark.
Generation of the Constructs
Construction of pTRV2 Plasmids for VIGS
Fragments for VIGS were obtained from a frozen sample of purple tomato fruit (Butelli et al., 2008), which was used as a source of RNA. mRNA extraction was performed using the RNeasy kit (Qiagen). cDNA was synthesized with the Iscript cDNA synthesis kit (BioRad). VIGS fragments were designed by using the VIGS tool from https://www.solgenomics.net/ PCR fragments were obtained with Phusion DNA polymerase (Thermo Fisher Scientific). AAT VIGS fragments were amplified using primers AAT-2VIGSFw (300-bp fragment) and AAT-2VIGSRev, AAT-1Fw, and AAT-1Rev. The PDS-VIGS fragment was described in Romero et al. (2011). PCR products sizes were confirmed on 1% agarose and excised from gel. The PCR products and pTRV2 vector were digested with EcoRI and XhoI (New England Biolabs). Digested PCR products were purified from the gel. pTRV2 and VIGS fragments were ligated with T4 ligase for 3 h at room temperature. The full-length open-reading frame (ORF) of AAT-2 was obtained from genomic DNA with the primers AAT-2ORFFw and AAT-2ORFRev. PCR products were gel purified and TOPO cloned into the pCR8/GW/TOPO-TA vector (Invitrogen). After sequence verification, the ATT fragment was transferred by GATEWAY recombination to pK7WG2 to create p35S-AAT. The plasmids obtained were then introduced into Agrobacterium tumefaciens AGL0 as described in Outchkourov et al. (2014). All the constructs were verified by sequencing (EZ-seq; Macrogen). AGL0 harboring an empty pBINPLUS plasmid (van Engelen et al., 1995) was used as a negative control.
Constructs Used for Transactivation Assay
The GL2 promoter (pGL2, 1938bp) was amplified from tomato genomic DNA using the primers Gl2promoterFw and Gl2promoterRev (Supplemental Table S4). The fragment was A-tailed and inserted into the pCR8/GW/TOPO-TA vector (Invitrogen). The sequence was verified and pGL2 was recombined into pGKGWG (containing the GFP reporter gene) and pGreen-LUC vectors (containing the luciferase reporter gene, kindly provided by Dr. Franziska Turck; Adrian et al., 2010). Plasmids were transformed into A. tumefaciens strain AGL0 for plant infiltration.
VIGS of In Vitro Tomato Seedlings
VIGS was based on the method described in Gomez Roldan et al. (2014). Seedlings (15 per container) were raised in a container on 0.5 × MS agar. The pTRV1, pTRV2, pTRV2-AAT1, and pTRV2-AAT2 vectors in A. tumefaciens were grown at 28°C with shaking in LB medium containing 50 μg/mL kanamycin and 25 μg/mL rifampicin. After 24 h, culture aliquots were transferred to 1.6 mL YEB medium (0.5% beef extract, 0.1% yeast extract, 0.5% peptone, 0.5% Suc, 2 mm MgSO4, 20 μm acetosyringone, and 10 mm MES, pH 5.6) plus antibiotics, and grown for 3 h. After this, the cells were washed twice in 10 mm MgCl2, 100 µM acetosyringone, and resuspended in cocultivation medium (0.25× MS with vitamins pH 6, 0.1% Suc, 100 μM acetosyringone, 0.005% Silwet L-77), and painted onto leaves of 9-d-old seedlings (T2 generation). After inoculation with A. tumefaciens, seedlings were kept at 21°C and allowed to recover and grow for 24 d before DEX induction. Activation of ROS1/DEL by dexamethasone was done by placing seedlings on agar containing 10 mg/L dexamethasone for 3 d, after which the seedlings were harvested for analysis.
Transactivation Assays
Agrobacterium clones were grown for 24 h at 28°C in LB medium (10 g/L tryptone, 5 g/L Yeast extract, 10 g/L NaCl) with antibiotics (kanamycin 50 μg/mL or spectinomycin 100 μg/mL and rifampicin 25 μg/mL). The optical density (OD) of the cultures was measured at 600 nm and the bacteria were resuspended in infiltration media (10 mM MES buffer, 10 mM MgCL2, 100 μm acetosyringone) to an OD of 0.5. After 3 h incubation with rotation, leaves of 4-week-old to 5-week-old Nicotiana benthamiana plants were infiltrated as described in Outchkourov et al. (2014). After 3 d, agro-infiltrated leaves were sprayed with luciferin (1 mM) to inactivate accumulated luciferase. Next day, the leaves were again sprayed with luciferin, and 5 min after the treatment the leaves were cut from the plant and measured with a cooled CCD camera. Leaves were placed on a plastic tray in a box coated with aluminum foil to reduce noise in the pictures caused by cosmic rays. Measurements were made for 5 min or 10 min. Emission of luminescence has a maximum at 560 nm, therefore a filter in the camera was used to block most other wavelengths. Analysis of the pictures was performed with the software ImageJ (National Institutes of Health). The measured intensity is proportional to the amount of luciferase produced. The intensity of selected areas was measured, and values were processed using the software SPSS Statistics 22 (IBM).
Generation of Anti-ANS Antibody
ORFs of tomato DFR and ANS genes were PCR-amplified from a total cDNA isolated from purple tomato fruits (Butelli et al., 2008) using the oligonucleotides tDFR_Fw, tDFR_Rev, and tANS_Fw, tANS_Rev. The obtained PCR fragments were gel-purified, digested with EcorI-BglII (for DFR) and EcoRI-SalI (for ANS) restriction enzymes, and ligated into the pACYCDuet-1 vector (Novagen). The newly prepared constructs were sequence-verified and immobilized into Escherichia coli BL21DE3 cells. Bacterial cultures at an OD600 nm of 0.8 were induced with 0.1 mm isopropyl β-d-1-thiogalactopyranoside and incubated overnight at 18°C. Soluble protein was extracted and His-tagged proteins were purified on a Ni-NTA column (Qiagen). Subsequently, fractions containing tDFR and tANS4 protein were further purified over a Superdex 75 10/30 column (GE Healthcare Life Sciences) in 100 mm Tris-HCL pH8 buffer. Protein purity of >95% as visible using SDS-PAGE was used for rabbit immunization at Eurogentec. Western-blot analysis was done as described in Outchkourov et al. (2014). Primary antibodies were diluted 1:1000, and secondary anti-Rabbit IgG (whole molecule)-Alkaline Phosphatase antibody produced in goat (A3687; Sigma-Aldrich) was diluted 1:6000. After washing, signals from the blots were developed using substrate tablet BCIP/NBT from Sigma-Aldrich.
LC-PDA-MS Analysis
Samples of three biological replicates were used for analysis. Semipolar compounds were extracted and analyzed as described in Moco et al. (2006). Tissues were snap-frozen, and subsequently ground to a fine powder using mortar and pestle. The powder derived from the tomato tissues was weighed exactly (90 mg to 110 mg dry weight), and was extracted using 10 volumes of 70% methanol solution acidified with 1% v/v formic acid. Samples were sonicated and filtered through a 0.45-μm filter before LC-MS analysis.
Separation was achieved using two Luna C18 (2) precolumns (2.0 mm × 4 mm and an analytical column (2.0 mm × 150 mm, 100 Å, particle size 3 μm), both from Phenomenex. Samples (5 μL) were injected and eluted using formic acid/water (1:1000 v/v; eluent A) and formic acid/acetonitrile (1:1000 v/v; eluent B) as elution solvents. The flow rate was set at 0.190 mL min1 with the following linear gradient elution program: 5% B to 35% B over 45 min, with washing for 15 min to equilibrate before the next injection. The column temperature was maintained at 40°C.
UV absorbance analysis was performed with a 2996 photodiode array detector (range from 240 nm to 600 nm; Waters) and metabolite masses were detected using a LTQ Orbitrap XL hybrid MS system (Waters) operating in positive electrospray ionization mode heated at 300°C with a source voltage of 4.5 kV for full-scan LC-MS in the m/z range 100 to 1500.
Acquisition and visualization of the LC-FTMS data were performed using the software Xcalibur (Thermo Fisher Scientific). The MetAlign software package (www.metAlign.nl; Lommen, 2009) was used for baseline correction, noise estimation, and spectral alignment. Aligned masses were directly used for further analysis. Comparison and visualization of the main features of the LC-MS data were performed by loading the data matrix into the software GeneMaths XT 1.6 (www.applied-maths.com). Metabolite intensities were normalized using log2 transformation and standardized using range scaling (autoscaling normalization).
RNA Sequencing and Data Analysis
Primary callus from three independent transformation events (T0 generation) was grown in vitro in MS medium supplemented with 1.0 mg/L 2,4-dichlorophenoxyacetic acid (2,4-D) and 0.1 mg/L kinetin. From line ROS1/DEL-04 (T2 generation), 4-week-old seedlings were grown in 1/2 MS medium, both with 8% agar. Both calli and seedlings were placed on new medium with or without 10 mg/L dexamethasone. Dexamethasone was dissolved in ethanol at 10 mg/mL and diluted 1000× in the agar. As a negative control, the same volume of ethanol was used. Triplicates of each induction timepoint, –DEX, 3 h +DEX, and 24 h +DEX of both tissues, were taken for analysis giving in total 18 samples. Total RNA was extracted from 50 mg ground tissues using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. Purified poly(A) RNA was used to produce libraries using a TrueSeq RNA library Prep Kit (Illumina) following the manufacturer’s instructions. Pooled libraries were sequenced on a HiSEquation 2000 (Illumina; WUR-Applied Bioinformatics). Differential expression was analyzed by the software CLCBIO (Qiagen Bioinformatics), using tomato ITAG2.4 gene models (Tomato Genome, 2012).
GO term analysis was performed using the tool available at http://bioinfo.bti.cornell.edu/cgi-bin/MetGenMAP/home.cgi (Boyle et al., 2004). This determines whether any GO terms annotated to a specified list of genes occur at a frequency greater than that which would be expected by chance. It calculates a P value using the hypergeometric distribution followed by Benjamin-Hochberg multiple testing correction, applying a false discovery rate cutoff of 0.1.
Gene Expression Analysis by Quantitative PCR
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). cDNA synthesis and qPCR analyses were performed as described in Karlova et al. (2013). The primers used for qPCR are listed in Supplemental Table S4.
Microscopy
Micro-tom wild-type and ROS1/DEL 7-d-old seedlings were transferred to plates (base, 11 cm; height, 11 cm) with or without DEX. Seedlings were placed ±2 cm from the top of the plate to allow the roots to grow downward, and root length was marked. Plates were placed at an angle of 60° (16 h light, 25°C). Microscopy was performed 3 d after transferring the seedlings to the plates. Two pictures were made of five roots per treatment. Close-up root pictures to show root-hair development were also made by placing the camera on a Stemi SV11 binocular microscope (Zeiss) by means of an adapter. From each picture the length of 10 root hairs was measured using the software ImageJ. In total, 20 root hairs per root and 100 root hairs per treatment were measured.
Scanning Electron Microscopy
Small pieces of Micro-tom wild-type and ROS1/DEL leaves were attached to a brass sample holder (Leica) with carbon glue (Leit-C; Neubauer Chemikalien). The holder was fixed onto the cryo-sample loading system (VCT 100; Leica) and simultaneously frozen in liquid nitrogen. The frozen holder was transferred to the cryo-preparation system (MED 020/VCT 100; Leica) onto the sample stage at −92°C. For removal of frost contamination on the sample surface, the samples were freeze-dried for 5 min at −92°C and 1.3 × 10−6 mbar. After sputter-coating with a layer of 20 nm tungsten at the same temperature the sample holder was transferred into the field-emission scanning electron microscope (SEM; Magellan 400; FEI) onto the sample stage at −120°C. The analysis was performed with SEM detection at 2 kV and 6.3 pA. SEM pictures were taken at a magnitude of 250×, and trichomes, hairs, and stomata were counted on squares of 500 μm × 500 μm.
Measurement of Chlorophyll Content with Pigment Analyzer
For nondestructive measurements of total chlorophyll levels, a CP Pigment Analyzer PA1101 (Control in Applied Physiology) was used according to the manufacturer’s instructions.
Measurement of Stomatal Conductance
Stomatal conductance (mmol H2O m−2 s−1) was measured on the abaxial side of the leaf, using an SC-1 leaf porometer (Decagon Devices).
Pictures
Photos were made with a Powershot G12 (Canon).
Statistical Analyses
Statistical analyses were performed using the software SPSS Statistics 23 (IBM). For root-hair analysis, an ANOVA was performed with genotype and DEX treatment in a model. The natural logarithm of the root-hair length was calculated to obtain a normal distribution and values were used in the ANOVA (ref) and LSD test. For the luciferase assay, background values were subtracted, and log values were calculated from remaining luminescence values to achieve a normal distribution. Values were used for an lsd test.
Accession Numbers
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE109772 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE109772).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Metabolite analysis of ROS1/DEL-activated tomato plants.
Supplemental Figure S2. Induction of ANS and DFR proteins upon DEX induction.
Supplemental Figure S3. Direct activation of GL2 by ROS1/DEL and pleiotropic effects of anthocyanin induction.
Supplemental Figure S4. Functional categories of genes significantly regulated by ROS1/DEL in a tissue-specific manner.
Supplemental Figure S5. VIGS of anthocyanin acyltransferase.
Supplemental Figure S6. Phenotypic effects of anthocyanin induction in root tissue.
Supplemental Figure S7. Phenotypic effects of anthocyanin induction in leaf tissue.
Supplemental Table S1. Metabolite analysis of roots and shoots of ROS1/DEL-activated tomato plants.
Supplemental Table S2. Gene expression profiles after DEX activation of ROS1/DEL TFs.
Supplemental Table S3. GO categories only in roots. Functional categories of the up- and down-regulated tissue specific genes by ROS1/DEL.
Supplemental Table S4. Oligonucleotides used in this study.
Acknowledgments
The authors have no conflict of interest. The authors acknowledge Bert Schipper for LC-MS measurements. The authors thank Prof. Cathie Martin for E8:ROS1/DEL tomato seeds.
Footnotes
N.S.O., R.D.H., and J.B. were supported by the “Platform Green Synthetic Biology” Program 3, funded by the Netherlands Genomics Initiative. J.B. and R.D.H. acknowledge support by the EU 7th Framework ATHENA project (FP7-KBBE-2009-3-245121-ATHENA).
Equal authorship.
Articles can be viewed without a subscription.
References
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F (2010) cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26: 962–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Bassolino L, Zhang Y, Schoonbeek HJ, Kiferle C, Perata P, Martin C (2013) Accumulation of anthocyanins in tomato skin extends shelf life. New Phytol 200: 650–655 [DOI] [PubMed] [Google Scholar]
- Bemer M, Karlova R, Ballester AR, Tikunov YM, Bovy AG, Wolters-Arts M, Rossetto PdeB, Angenent GC, de Maagd RA (2012) The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24: 4437–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J (2005) The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298 [DOI] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G (2004) GO::TermFinder-open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P. (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol 8: 272–279 [DOI] [PubMed] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EG, Hall RD, Bovy AG, Luo J, Martin C (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Chanoca A, Kovinich N, Burkel B, Stecha S, Bohorquez-Restrepo A, Ueda T, Eliceiri KW, Grotewold E, Otegui MS (2015) Anthocyanin vacuolar inclusions form by a microautophagy mechanism. Plant Cell 27: 2545–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chezem WR, Clay NK (2016) Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs. Phytochemistry 131: 26–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PK, Shin DH, Choi SB, Park YI (2012) Sugar-hormone cross-talk in anthocyanin biosynthesis. Mol Cells 34: 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerigmann H, Berger B, Gigolashvili T (2014) bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol 166: 349–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW (1994) The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol 166: 740–754 [DOI] [PubMed] [Google Scholar]
- Girard AL, Mounet F, Lemaire-Chamley M, Gaillard C, Elmorjani K, Vivancos J, Runavot JL, Quemener B, Petit J, Germain V, Rothan C, Marion D, et al. (2012) Tomato GDSL1 is required for cutin deposition in the fruit cuticle. Plant Cell 24: 3119–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsbrough A, Belzile F, Yoder JI (1994) Complementation of the Tomato anthocyanin without (aw) mutant using the dihydroflavonol 4-Reductase gene. Plant Physiol 105: 491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Roldan MV, Outchkourov N, van Houwelingen A, Lammers M, Romero de la Fuente I, Ziklo N, Aharoni A, Hall RD, Beekwilder J (2014) An O-methyltransferase modifies accumulation of methylated anthocyanins in seedlings of tomato. Plant J 80: 695–708 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Carpenter R, Coen ES (1992) A common gene regulates pigmentation pattern in diverse plant species. Cell 68: 955–964 [DOI] [PubMed] [Google Scholar]
- Gould KS. (2004) Nature’s Swiss Army Knife: the diverse protective roles of anthocyanins in leaves. J Biomed Biotechnol 2004: 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57: 761–780 [DOI] [PubMed] [Google Scholar]
- He J, Giusti MM (2010) Anthocyanins: natural colorants with health-promoting properties. Annu Rev Food Sci Technol 1: 163–187 [DOI] [PubMed] [Google Scholar]
- Jonsson LM, Aarsman ME, Poulton JE, Schram AW (1984) Properties and genetic control of four methyltransferases involved in methylation of anthocyanins in flowers of Petunia hybrida. Planta 160: 174–179 [DOI] [PubMed] [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23: 923–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R, van Haarst JC, Maliepaard C, van de Geest H, Bovy AG, Lammers M, Angenent GC, de Maagd RA (2013) Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J Exp Bot 64: 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueño F, Krajewski P, Meyerowitz EM, Angenent GC, Riechmann JL (2010) Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Kovinich N, Kayanja G, Chanoca A, Otegui MS, Grotewold E (2015) Abiotic stresses induce different localizations of anthocyanins in Arabidopsis. Plant Signal Behav 10: e1027850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrooke J, Adato A, Lotan O, Alkan N, Tsimbalist T, Rechav K, Fernandez-Moreno JP, Widemann E, Grausem B, Pinot F, Granell A, Costa F, et al. (2015) The Tomato MIXTA-like transcription factor coordinates fruit epidermis conical cell development and cuticular lipid biosynthesis and assembly. Plant Physiol 169: 2553–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Omranian N, Neumetzler L, Wang T, Herter T, Usadel B, Demura T, Giavalisco P, Nikoloski Z, Persson S (2016) A transcriptional and metabolic framework for secondary wall formation in Arabidopsis. Plant Physiol 172: 1334–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Chen X, Chen A, Wang H, Liu J, Liu J, Gu M, Sun S, Xu G (2015) The characterization of six auxin-induced tomato GH3 genes uncovers a member, SlGH3.4, strongly responsive to arbuscular mycorrhizal symbiosis. Plant Cell Physiol 56: 674–687 [DOI] [PubMed] [Google Scholar]
- Lin Q, Aoyama T (2012) Pathways for epidermal cell differentiation via the homeobox gene GLABRA2: update on the roles of the classic regulator. J Integr Plant Biol 54: 729–737 [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW (1994) Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science 266: 436–439 [DOI] [PubMed] [Google Scholar]
- Lommen A. (2009) MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal Chem 81: 3079–3086 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6: 280–287 [DOI] [PubMed] [Google Scholar]
- Lü S, Song T, Kosma DK, Parsons EP, Rowland O, Jenks MA (2009) Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J 59: 553–564 [DOI] [PubMed] [Google Scholar]
- Maloney GS, DiNapoli KT, Muday GK (2014) The anthocyanin reduced tomato mutant demonstrates the role of flavonols in tomato lateral root and root hair development. Plant Physiol 166: 614–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Butelli E, Petroni K, Tonelli C (2011) How can research on plants contribute to promoting human health? Plant Cell 23: 1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y, Tonelli C, Petroni K (2013) Plants, diet, and health. Annu Rev Plant Biol 64: 19–46 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW (1996) The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, Wagner DR (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15: 1689–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus JT, Loyola R, Vega A, Pena-Neira A, Bordeu E, Arce-Johnson P, Alcalde JA (2009) Post-veraison sunlight exposure induces MYB-mediated transcriptional regulation of anthocyanin and flavonol synthesis in berry skins of Vitis vinifera. J Exp Bot 60: 853–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moco S, Bino RJ, Vorst O, Verhoeven HA, de Groot J, van Beek TA, Vervoort J, de Vos CHR (2006) A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol 141: 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Grotewold E (2009) A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz C, Hoffmann T, Escobar NM, Ludemann F, Botella MA, Valpuesta V, Schwab W (2010) The strawberry fruit Fra A allergen functions in flavonoid biosynthesis. Mol Plant 3: 113–124 [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Oka A, Ruberti I, Morelli G, Aoyama T (2002) Entopically additive expression of GLABRA2 alters the frequency and spacing of trichome initiation. Plant J 29: 359–369 [DOI] [PubMed] [Google Scholar]
- Outchkourov NS, Carollo CA, Gomez-Roldan V, de Vos RC, Bosch D, Hall RD, Beekwilder J (2014) Control of anthocyanin and non-flavonoid compounds by anthocyanin-regulating MYB and bHLH transcription factors in Nicotiana benthamiana leaves. Front Plant Sci 5: 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P, Fukaki H, Beeckman T (2010) Auxin control of root development. Cold Spring Harb Perspect Biol 2: a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K, Tonelli C (2011) Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci 181: 219–229 [DOI] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Roldan MVG, Engel B, de Vos RCH, Vereijken P, Astola L, Groenenboom M, van de Geest H, Bovy A, Molenaar J, van Eeuwijk F, Hall RD (2014) Metabolomics reveals organ-specific metabolic rearrangements during early tomato seedling development. Metabolomics 10: 958–974 [Google Scholar]
- Romero I, Tikunov Y, Bovy A (2011) Virus-induced gene silencing in detached tomatoes and biochemical effects of phytoene desaturase gene silencing. J Plant Physiol 168: 1129–1135 [DOI] [PubMed] [Google Scholar]
- Roth-Walter F, Gomez-Casado C, Pacios LF, Mothes-Luksch N, Roth GA, Singer J, Diaz-Perales A, Jensen-Jarolim E (2014) Bet v 1 from birch pollen is a lipocalin-like protein acting as allergen only when devoid of iron by promoting Th2 lymphocytes. J Biol Chem 289: 23329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki N, Nakayama T (2015) Achievements and perspectives in biochemistry concerning anthocyanin modification for blue flower coloration. Plant Cell Physiol 56: 28–40 [DOI] [PubMed] [Google Scholar]
- Schmid J, Schaller A, Leibinger U, Boll W, Amrhein N (1992) The in-vitro synthesized tomato shikimate kinase precursor is enzymatically active and is imported and processed to the mature enzyme by chloroplasts. Plant J 2: 375–383 [PubMed] [Google Scholar]
- Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18: 831–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Sun H, Fan HJ, Ling HQ (2015) Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics 16: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Zhang Y, Peterek S, Matros A, Rallapalli G, Tandrón YA, Butelli E, Kallam K, Hertkorn N, Mock HP, Martin C, Fernie AR (2015) Ectopic expression of snapdragon transcription factors facilitates the identification of genes encoding enzymes of anthocyanin decoration in tomato. Plant J 83: 686–704 [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga R, Iwata M, Sano R, Inoue K, Okada K, Wada T (2008) Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135: 1335–1345 [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R, Nukumizu Y, Sato S, Wada T (2013) Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS One 8: e54019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda T. (2012) Dietary anthocyanin-rich plants: biochemical basis and recent progress in health benefits studies. Mol Nutr Food Res 56: 159–170 [DOI] [PubMed] [Google Scholar]
- van Dijken AJ, Schluepmann H, Smeekens SC (2004) Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol 135: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 4: 288–290 [DOI] [PubMed] [Google Scholar]
- Velásquez AC, Chakravarthy S, Martin GB (2009) Virus-induced gene silencing (VIGS) in Nicotiana benthamiana and tomato. J Vis Exp 28: 1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Roepenack-Lahaye E, Newman MA, Schornack S, Hammond-Kosack KE, Lahaye T, Jones JD, Daniels MJ, Dow JM (2003) p-Coumaroylnoradrenaline, a novel plant metabolite implicated in tomato defense against pathogens. J Biol Chem 278: 43373–43383 [DOI] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20: 176–185 [DOI] [PubMed] [Google Scholar]
- Yeats TH, Huang W, Chatterjee S, Viart HMF, Clausen MH, Stark RE, Rose JKC (2014) Tomato Cutin Deficient 1 (CD1) and putative orthologs comprise an ancient family of cutin synthase-like (CUS) proteins that are conserved among land plants. Plant J 77: 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB (2008) Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafra-Stone S, Yasmin T, Bagchi M, Chatterjee A, Vinson JA, Bagchi D (2007) Berry anthocyanins as novel antioxidants in human health and disease prevention. Mol Nutr Food Res 51: 675–683 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, De Stefano R, Schoonbeek HJ, Magusin A, Pagliarani C, Wellner N, Hill L, Orzaez D, Granell A, Jones JD, Martin C (2013) Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr Biol 23: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SJ, Snoeren TA, Hogewoning SW, van Loon JJ, Dicke M (2010) Disruption of plant carotenoid biosynthesis through virus-induced gene silencing affects oviposition behaviour of the butterfly Pieris rapae. New Phytol 186: 733–745 [DOI] [PubMed] [Google Scholar]
- Zuluaga DL, Gonzali S, Loreti E, Pucciariello C, Degl’Innocenti E, Guidi L, Alpi A, Perata P (2008) Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct Plant Biol 35: 606–618 [DOI] [PubMed] [Google Scholar]