The transcription factor PIF7 recruits the H3K4me3/H3K36me3-reader protein MRG1/MRG2 to promote histone acetylations in activating genes to promote stem elongation in plant shade response.
Abstract
Shade avoidance syndrome (SAS) allows a plant grown in a densely populated environment to maximize opportunities to access to sunlight. Although it is well established that SAS is accompanied by gene expression changes, the underlying molecular mechanism needs to be elucidated. Here, we identify the H3K4me3/H3K36me3-binding proteins, Morf Related Gene (MRG) group proteins MRG1 and MRG2, as positive regulators of shade-induced hypocotyl elongation in Arabidopsis (Arabidopsis thaliana). MRG2 binds PHYTOCHROME-INTERACTING FACTOR7 (PIF7) and regulates the expression of several common downstream target genes, including YUCCA8 and IAA19 involved in the auxin biosynthesis or response pathway and PRE1 involved in brassinosteroid regulation of cell elongation. In response to shade, PIF7 and MRG2 are enriched at the promoter and gene-body regions and are necessary for increase of histone H4 and H3 acetylation to promote target gene expression. Our study uncovers a mechanism in which the shade-responsive factor PIF7 recruits MRG1/MRG2 that binds H3K4me3/H3K36me3 and brings histone-acetylases to induce histone acetylations to promote expression of shade responsive genes, providing thus a molecular mechanistic link coupling the environmental light to epigenetic modification in regulation of hypocotyl elongation in plant SAS.
Shade avoidance syndrome (SAS) occurs when plants are constrained to the shade of neighbors. A classical SAS phenotype includes elongation of hypocotyls, stems, petioles, or internodes; hyponastic leaves; reduced leaf lamina size; enhanced apical dominance; and/or early flowering (Casal, 2012; Ballaré and Pierik, 2017). Because of cultivable land limitation, crops are planted at a high density in modern agriculture, and if activated, SAS will decrease the crop yield (Carriedo et al., 2016; Ballaré and Pierik, 2017). Thus, understanding the mechanisms underlying SAS has prevalent importance for both natural ecosystem and agricultural breeding interests.
Shade by plant leaves causes reduction of photosynthetically active radiation (radiation with wavelengths between 400 nm and 700 nm) as well as low ratio of Red (R, 660 nm)/Far Red (FR, 730 nm) light and Low Blue light (L; Casal, 2012). Studies in the model plant Arabidopsis (Arabidopsis thaliana) have revealed that the shade-caused reduction of R/FR is perceived by the photoreceptor phytochromes, which physically interact with a subfamily of basic helix-loop-helix (bHLH) proteins, namely PHYTOCHROME INTERACTING FACTORs (PIFs; Leivar and Quail, 2011). In particular, PIF7 plays a major role in shade-induced stem elongation. In response to shade, PIF7 accumulates in its dephosphorylated form, which can subsequently bind and activate its target genes, including PHYTOCHROME RAPIDLY REGULATED1 (PAR1) and PACLOBUTRAZOL RESISTANCE1/BANQUO1 (PRE1/BNQ1) involved in BZR1-mediated brassinosteroid (BR) regulation of cell elongation, and several genes [e.g. INDOLE-3-ACETIC ACID INDUCIBLE19 (IAA19), INDOLE-3-ACETIC ACID-AMIDO SYNTHETASE (GH3.3), FLAVIN MONOOXYGENASE8 (YUCCA8), and YUCCA9] involved in auxin biosynthesis or response, to promote hypocotyl elongation (Li et al., 2012; de Wit et al., 2015; Mizuno et al., 2015). In addition, PIF4 and PIF5 also play regulatory roles in SAS and more recent studies have unraveled their function at downstream of Cryptochromes to mediate Arabidopsis hypocotyl elongation in response to low blue light (Lorrain et al., 2008; de Wit et al., 2016; Pedmale et al., 2016).
Transcription factor binding to DNA and target gene transcription are modulated through chromatin structure (Berr et al., 2011; van Lijsebettens and Grasser, 2014). Lys (K) acetylation neutralizes the positive charge of the histone molecule, thereby decreasing the histone interaction with the negatively charged DNA molecule and transforming the condensed chromatin to a more relaxed structure favorable for gene transcription. Histone acetylation can occur at various K residues and is dynamically regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), two families of enzymes with antagonistic activities in catalyzing acetylation and deacetylation, respectively (Lusser et al., 2001; Liu et al., 2014; Shen et al., 2015). The Arabidopsis HDAC-family member HDA15 has been reported to interact with PIF3 to repress chlorophyll biosynthetic gene expression (Liu et al., 2013) and with PIF1 to regulate seed germination (Gu et al., 2017). A few other histone modifiers have also been described to be involved in light-mediated developmental processes, such as skotomorphogenesis and photomorphogenesis, in Arabidopsis (Barneche et al., 2014). Yet, their roles in SAS remain uncharacterized so far.
In contrast to the negative charge of an acetyl group, a methyl group is neutral and histone methylation may either repress or promote transcription depending on the position of K residues on the histone. In general, methylation on H3K9 or H3K27 is related to gene silencing, whereas methylation on H3K4 or H3K36 is associated with transcription activation (Berr et al., 2011). Interestingly, recent studies demonstrate that the Morf Related Gene (MRG) family proteins MRG1 and MRG2 bind H3K4/H3K36 trimethylation (H3K4me3/H3K36me3) and physically interact with the transcription factor CONSTANS (CO; Bu et al., 2014) as well as with the HAT-family proteins HAM1 and HAM2 (Xu et al., 2014), pointing to a mechanistic connection of CO with histone methylation and acetylation in activation of Arabidopsis FLOWERING LOCUS T (FT) transcription.
In this study, we show that PIF7 physically interacts with MRG2 to promote histone acetylation in transcription activation of shade-responsive genes. More specifically, we found that loss of the Arabidopsis MRG1 and MRG2 impairs shade-induced hypocotyl elongation. PIF7 binds MRG2 and is required for MRG2 recruitment at loci of the shade-responsive genes (YUCCA8, IAA19, and PRE1), which is critical for induction of histone acetylation and activation of transcription. Our study extends previous knowledge and provides, to our knowledge, novel insight about molecular mechanisms that link the transcription factor, the reader of histone methylation, and chromatin structure reprogramming via histone acetylation in shade-induced gene transcription.
RESULTS
MRG1/2 and PIF7 Positively Regulate Shade-Induced Hypocotyl Elongation
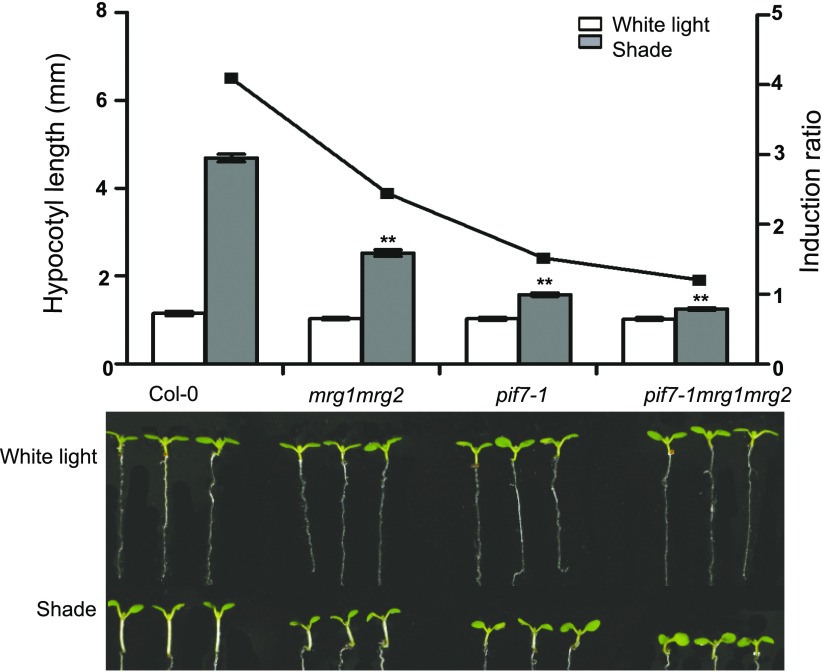
To explore how shade-induced transcription is regulated within the context of chromatin, we analyzed the hypocotyl phenotype of several Arabidopsis mutants defective in deposition or reading of histone methylations (Supplemental Table S1). Interestingly, the mrg1mrg2 double mutant displays a severe defective phenotype in response to shade (Fig. 1, Supplemental Table S1). To investigate whether MRG1 or MRG2 plays any specific role in SAS, we examined the mrg1 and mrg2 single mutants. In contrast to the mrg1mrg2 double mutant, the mrg1 and mrg2 single mutants display a wild-type phenotype, and moreover the transgene expression of MRG2-YFP has fully rescued the defect of the mrg1mrg2 double mutant (Supplemental Fig. S1). These results imply that MRG1 and MRG2 have redundant functions and that their concomitant loss-of-function caused the mrg1mrg2 mutant phenotype.
Figure 1.
Quantification of hypocotyl length and representative seedlings of Col-0, pif7-1, mrg1mrg2, and pif7-1mrg1mrg2. Seedlings were grown under white light for 4 d and maintained in white light or transferred to shade for next 3 d before the measurement of hypocotyl length. The left y axis is the mean of hypocotyl length from three biological replicate measurements presented in Supplemental Table S1, and the right y axis is the ratio of hypocotyl lengths in shade to that in white light. Error bars represent se. Asterisks indicate where the difference between Col-0 and mutant is statistically significant (**P < 0.01).
Hereinafter, we focused on the mrg1mrg2 double mutant for detailed analyses. Under white light or dark growth conditions, mrg1mrg2 shows similar hypocotyl length as compared to the wild-type control Col-0. In contrast, under shade, red light, far-red light, or blue light growth conditions, mrg1mrg2 exhibits significantly shorter hypocotyl as compared to Col-0 (Fig. 1, Supplemental Fig. S2). The shade-response phenotype of mrg1mrg2 prompted us to investigate its relation with pif7-1. Thus, we combined mrg1mrg2 with pif7-1 to generate the triple mutant pif7-1mrg1mrg2. Under shade growth conditions, this triple mutant shows a slightly shorter hypocotyl than mrg1mrg2 and a roughly similar length hypocotyl compared to pif7-1 (Fig. 1), indicating that the function of MRG1/MRG2 on SAS might be mediated by PIF7.
MRG1/2 and PIF7 Regulate Expression of Shade-Responsive Genes
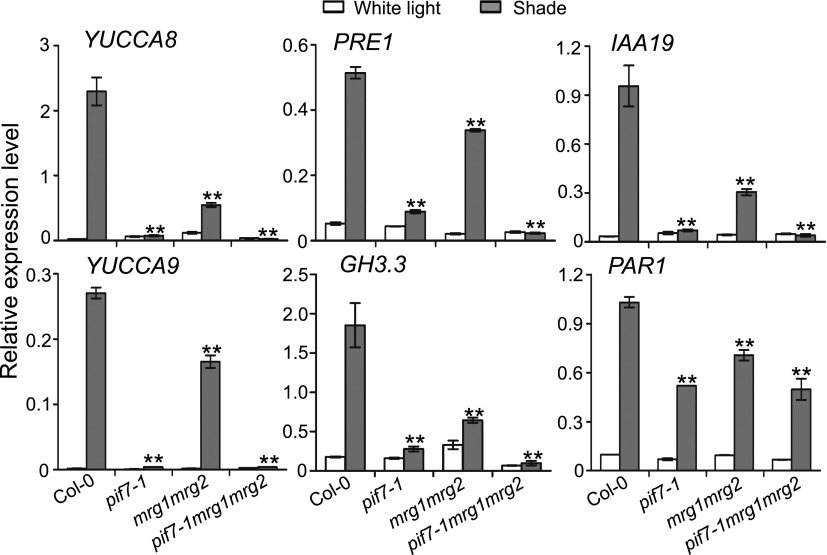
To gain an insight into the mechanism underlying the mutant shade-responsive phenotype, we examined the expression of several shade-induced genes including YUCCA8, PRE1, IAA19, YUCCA9, GH3.3, and PAR1. All these genes were found down-regulated in pif7-1 as well as in mrg1mrg2 when compared to Col-0 under shade growth conditions (Fig. 2). Nevertheless, the reduction is less severe in mrg1mrg2 than in pif7-1. Further analysis revealed that pif7-1 is epistatic to mrg1mrg2 because the pif7-1mrg1mrg2 triple mutant and the pif7-1 mutant display similarly low expression levels of all the analyzed genes (Fig. 2). This is consistent with the hypocotyl phenotypes of these mutants.
Figure 2.
Relative expression levels of YUCCA8, PRE1, IAA19, YUCCA9, GH3.3, and PAR1 in Col-0, pif7-1, mrg1mrg2, and pif7-1mrg1mrg2 under white light or shade treatment conditions. RNA prepared from seedlings grown under white light for 4 d and maintained in white light or transferred to shade for 1 h. The expression levels were normalized to the internal control AT2G39960. Error bars represent se from three independent biological replicates. Asterisks indicate where the difference between Col-0 and mutant under shade is statistically significant (**P < 0.01).
One possible mechanism could be that MRG1/MRG2 modulates the expression of PIF7. However, this assumption is precluded because transcript levels of PIF7 as well as that of PIF1, PIF3 or PIF4 are similar between Col-0 and mrg1mrg2 (Supplemental Fig. S3). Moreover, the expression levels of MRG1 and MRG2 are also not affected in pif7-1 or by shade treatment (Supplemental Fig. S3).
MRG2 Physically Interacts with PIF7
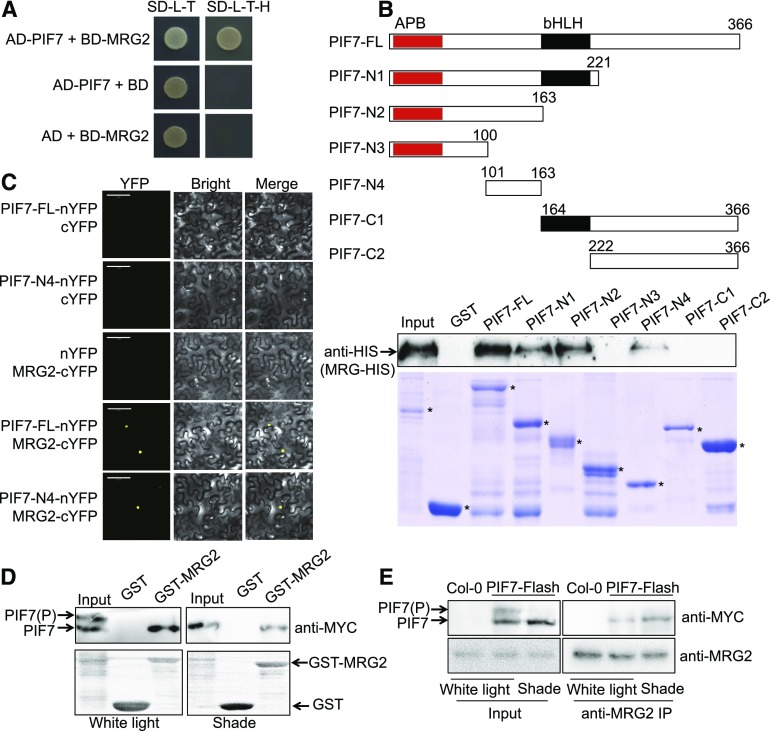
Next, we asked the question whether MRG2 binds PIF7. Indeed, in yeast two-hybrid assays a positive interaction between MRG2 and PIF7 was detected (Fig. 3A). An in vitro GST pull-down experiment further confirmed physical interaction between MRG2 and PIF7. His-MRG2 was precipitated by GST-PIF7 (full-length: 1 to 366 amino acids), GST-PIF7-N1 (N-terminal: 1 to 221 amino acids), GST-PIF7-N2 (1 to 163 amino acids), or GST-PIF7-N4 (101 to 163 amino acids), but not by GST, GST-PIF7-N3 (1 to 100 amino acids), GST-PIF7-C1 (C-terminal: 164 to 366 amino acids), or GST-PIF7-C2 (222 to 366 amino acids; Fig. 3B). The MRG2-binding site is thus located at the 100-to-163 amino acid region between the Active Phytochrome Binding (APB; Khanna et al., 2004) and bHLH domains of PIF7. We further examined the protein interaction in planta by bimolecular fluorescence complementation (BiFC) assay. As shown in Figure 3C, positive signals were detected specifically between MRG2-cYFP and PIF7-nYFP or PIF7-N4-nYFP in transiently transformed Nicotiana benthamiana leaf cells.
Figure 3.
PIF7 interacts with MRG2. A, PIF7 interacts with MRG2 in yeast two-hybrid assay. Each yeast strain containing the pGADT7 or pGADT7-PIF7 together with pGBKT7 or pGBKT7-MRG2 was grown on SD-L-T or SD-L-T-H plates. Yeast growth on SD-L-T-H indicates positive protein-protein interaction. B, PIF7 binds MRG2 in pull-down assay. Top panel, schematic representation of full-length and truncated PIF7 protein. The conserved domains APB and bHLH are indicated as red and black box, respectively. Bottom panel, interaction of purified His-tagged MRG2, and GST-fused different length of PIF7 from Escherichia coli by GST pull-down assays. Western-blot signals detected by the anti-His antibody together with Coomassie Brilliant Blue R250-stained proteins on the SDS-PAGE gel are shown. The positions of GST-PIF7, GST-PIF7-N1, GST-PIF7-N2, GST-PIF7-N3, GST-PIF7-N4, GST-PIF7-C1, GST-PIF7-C2, and GST are indicated by asterisks. C, BiFC analysis of the interaction between PIF7 and MRG2 in tobacco leaf cells. The C-terminal half of YFP was fused to MRG2, and the N-terminal half of YFP was fused to the full length of PIF7 or PIF7-N4. The constructs were cotransformed into tobacco leaf cells, and fluorescence images were obtained by confocal microscopy. White scale bar is 100 μm. D, The interaction between purified GST-fused MRG2 from E. coli and total protein extracts from plants overexpressing PIF7-Flash grown under white light condition or treated by 1-h shade by semi-in vivo GST pull-down assay. (Top panel) The pull-down fractions and inputs were analyzed by western blot using anti-MYC antibody. (Bottom panel) Ponceau S staining is shown. E, Co-IP analysis of the interaction between PIF7 and MRG2. Anti-MRG2 Sepharose beads were used to precipitate PIF7-Flash from PIF7ox plants grown under white light or treated by 1-h shade. Western blot was performed using anti-MRG2 and anti-MYC antibodies, as indicated. AD, pGADT7; AD-PIF7, pGADT7-PIF7; BD, pGBKT7; BD-MRG2, pGBKT7-MRG2; PIF7-FL, full-length PIF7.
It has been known that shade-regulated dephosphorylation of PIF7 is important for PIF7’s transcriptional activity (Li et al., 2012). To test whether the interaction between MRG2 and PIF7 is phosphorylation-dependent, we prepared protein extracts from seedlings overexpressing PIF7-Flash (9xMyc-6xHis-3xFlag, PIF7ox) grown under either white light or shade conditions. GST pull-down assay revealed that only dephosphorylated PIF7 was precipitated by GST-MRG2 (Fig. 3D). A coimmunoprecipitation (Co-IP) experiment further confirmed that the dephosphorylated PIF7 is precipitated with MRG2 (Fig. 3E). These data indicate that shade-dependent dephosphorylation status modulates the interaction between MRG2 and PIF7.
MRG2 and PIF7 Bind Chromatin at Shade-Responsive Genes
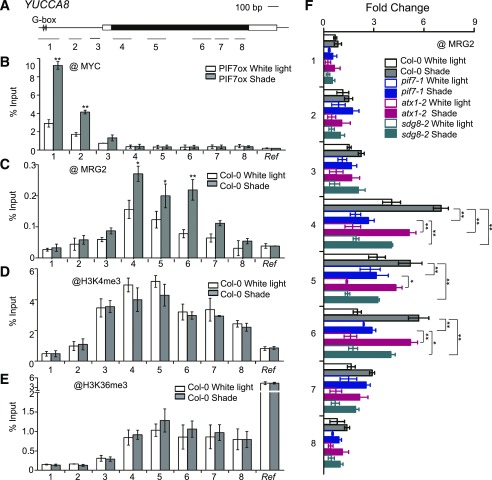
To examine PIF7 and MRG2 binding at target gene loci, we performed chromatin immunoprecipitation (ChIP) analyses. Consistent with previous knowledge that PIFs bind at the G-box (CACGTG) cis-element (Martínez-García et al., 2000; Moon et al., 2008; Hornitschek et al., 2009; Kidokoro et al., 2009), we found that in response to shade, PIF7-Flash is enriched around the G-box site at the promoter region of YUCCA8, IAA19, and PRE1 (Fig. 4, A and B; Supplemental Fig. S4, A B, E, and F). Using the anti-MRG2 antibody (Bu et al., 2014), we found that in response to shade, MRG2 is enriched downstream of the PIF7-binding region, from starting through coding region of YUCCA8 (Fig. 4C). Examination of IAA19 and PRE1 gives similar results, showing MRG2 binding downstream of PIF7 (Supplemental Fig. S4, C and G).
Figure 4.
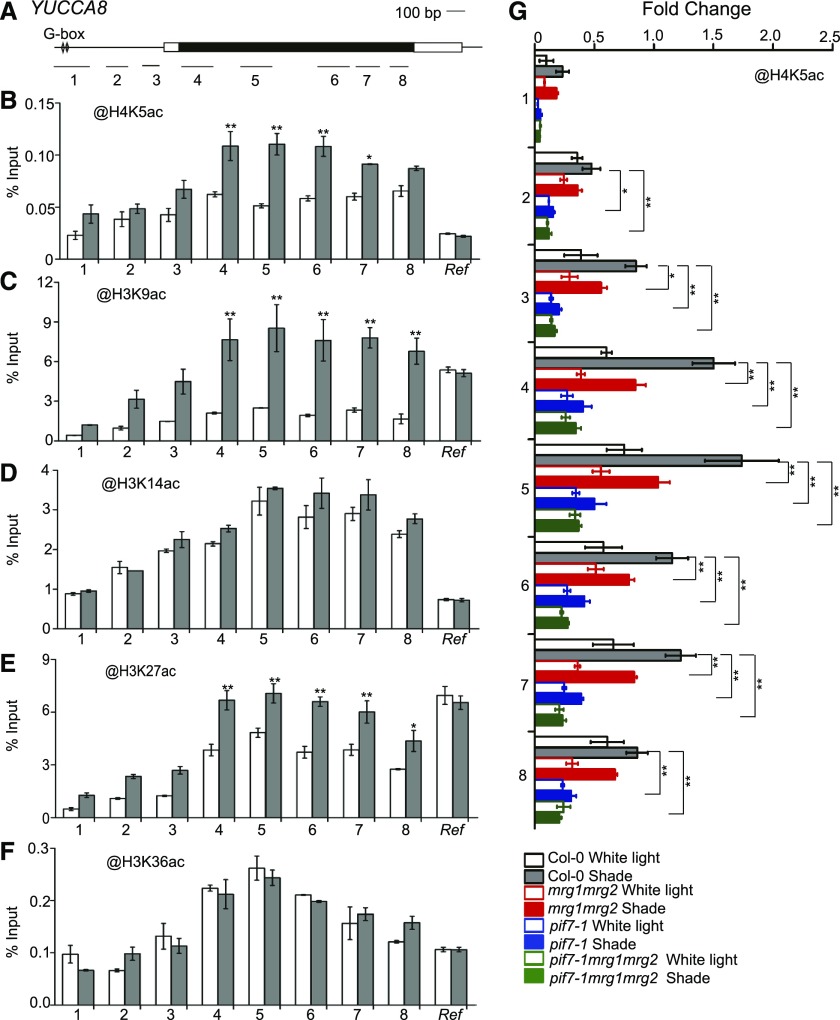
MRG2 and PIF7 bind chromatin at shade-responsive genes. A, Schematic of YUCCA8 gene structure. Black box represents the coding region, white boxes represent untranslated regions, and lines represent the promoter. The G-box elements within gene promoter are indicated. Bars underlabeled with numbers represent regions amplified by PCR. B, ChIP-PCR analyses using anti-MYC antibody at various chromatin regions of YUCCA8 in PIF7ox seedlings grown under white light and shade conditions. C, ChIP-PCR analyses using anti-MRG2 antibody at YUCCA8 chromatin in Col-0 seedlings grown under white light and shade conditions. d and E, ChIP-PCR analyses of H3K4me3 (D) and H3K36me3 (E) at YUCCA8 chromatin using Col-0 grown under white light and shade. F, ChIP-PCR analyses using anti-MRG2 antibody at YUCCA8 chromatin in the pif7-1, atx1-2, and sdg8-2 mutants as compared to Col-0 grown under white light and shade conditions. Seedlings were grown under white light for 4 d and maintained in white light or transferred to shade for 1 h ChIP and quantitative real-time PCR were performed using specific antibodies and gene primers, as indicated. A region of AT2G39960 served as an internal control and was used in normalization. Error bars represent se from three independent biological replicates. Asterisks in (B), (C), (D), and (E) indicate where the differences between white light and shade are statistically significant, and those in (F) indicate that the differences between two genotypes under shade are statistically significant (*P < 0.05, **P < 0.01). Ref, internal control.
Similar to the MRG2-binding pattern, the H3K4me3 and H3K36me3 distribution is generally found at high levels at the 5′-end of the gene body (Zhang et al., 2009; Li et al., 2015). To investigate the histone methylation pattern in response to shade, we analyzed H3K4me3 and H3K36me3 levels at YUCCA8. Both H3K4me3 and H3K36me3 were found enriched at 5′-end and along the gene body of YUCCA8 (Fig. 4, D and E), which overlaps MRG2 binding regions (Fig. 4C) and thereby is in agreement with MRG1/MRG2 acting as readers of H3K4me3 and H3K36me3 (Bu et al., 2014; Xu et al., 2014). Strikingly, significant difference was undetected at either H3K4me3 levels (Fig. 4D) or H3K36me3 levels (Fig. 4E) between white-light and 1-h-shade-treated Col-0 seedlings, implying that shade-induced MRG2 enrichment is not mediated via H3K4me3 or H3K36me3 increase.
We further asked the question whether MRG2 recruitment is dependent on H3K4me3/H3K36me3 and/or PIF7. The H3K4-methyltransferase mutant atx1-2 (Pien et al., 2008) and the H3K36-methyltransferase mutant sdg8-2 (Zhao et al., 2005; Xu et al., 2008) showed a weak shade-responsive hypocotyl phenotype (Supplemental Table S1) and slightly reduced level of YUCCA8, IAA19, and PRE1 expression (Supplemental Fig. S5). Comparison of MRG2 binding in Col-0 and atx1-2 or sdg8-2 revealed that shade-induced MRG2 enrichment is slightly but significantly reduced at YUCCA8 in the atx1-2 or sdg8-2 mutants (Fig. 4F), implying that albeit not induced by shade, H3K4me3/H3K36me3 at basal level still play a promoting role in MRG2 recruitment. Interestingly, in pif7-1 shade-induced MRG2 enrichment at YUCCA8 is severely attenuated (Fig. 4F), indicating strong dependency of MRG2 recruitment on PIF7. These essential conclusions drawn from analyses at YUCCA8 also hold true from analyses at PRE1 and IAA19 (Supplemental Fig. S4, D and H).
PIF7/MRG2-Mediated Shade-Induction of Gene Expression Is Associated with Histone Hyperacetylation
MRG2 interacts with HAM1/HAM2, which has been proposed to bridge H3K36me3 to H4K5 acetylation (H4K5ac) in transcription activation of FT in flowering time control (Xu et al., 2014). To examine whether a similar mechanism exists in shade response, we analyzed histone acetylation levels at YUCCA8. Indeed, increased levels of H4K5ac were detected at several regions of YUCCA8 after 1-h shade treatment as compared to white light control in Col-0 (Fig. 5, A and B). Beside HAM1/HAM2-mediated H4K5ac (Earley et al., 2007; Xiao et al., 2013), several other K-residues of histone H3 are also known as being subject to acetylation modification. We further extend our analysis to examine H3K9, H3K14, H3K27, and H3K36 acetylation in shade response. Interestingly, levels of H3K9ac and H3K27ac but not H3K14ac nor H3K36ac were found induced at YUCCA8 after 1-h shade treatment as compared to white light control in Col-0 (Fig. 5, C to F).
Figure 5.
Histone acetylation modification at YUCCA8 chromatin. A, Schematic of YUCCA8 gene structure. B to F, ChIP-PCR analyses of H4K5ac (B), H3K9ac (C), H3K14ac (D), H3K27ac (E), and H3K36ac (F) at various chromatin regions of YUCCA8 in Col-0 grown under white light and shade conditions. Asterisks indicate where the difference between white light and shade is statistically significant (*P < 0.05, **P < 0.01). G, ChIP-PCR analyses of H4K5ac at YUCCA8 chromatin in Col-0, mrg1mrg2, pif7-1, and pif7-1mrg1mrg2 grown under white light and shade. Asterisks indicate where the difference between wild type and mutant under shade is statistically significant (*P < 0.05, **P < 0.01). Seedlings were grown under white light for 4 d and maintained in white light or transferred to shade for 1 h ChIP and quantitative real-time PCR were performed using specific antibodies and gene primers, as indicated. A region of AT2G39960 served as an internal control and used in normalization. Error bars represent se from three independent biological replicates. Ref, internal control.
To test whether the shade-induced H4K5ac accumulation is dependent on MRG1/MRG2 and PIF7, we compared H4K5ac levels at YUCCA8 in Col-0 and the mrg1mrg2, pif7-1, and pif7-1mrg1mrg2 mutants. As shown in Figure 5G, the shade-induced H4K5ac elevation is drastically reduced in mrg1mrg2 and is completely lost in pif7-1 and pif7-1mrg1mrg2, indicating a partial MRG1/MRG2 but full PIF7 dependency. Examination of H3K9ac at YUCCA8 revealed that its shade-induced elevation is independent of MRG1/MRG2 but partly dependent on PIF7 (Supplemental Fig. S6).
Taken together, our results indicate that PIF7 recruits MRG2-HATs, most likely including HAM1/HAM2 in H4K5ac deposition, and other HATs in H3K9ac and H3K27ac deposition, to induce transcription of shade-responsive genes.
DISCUSSION
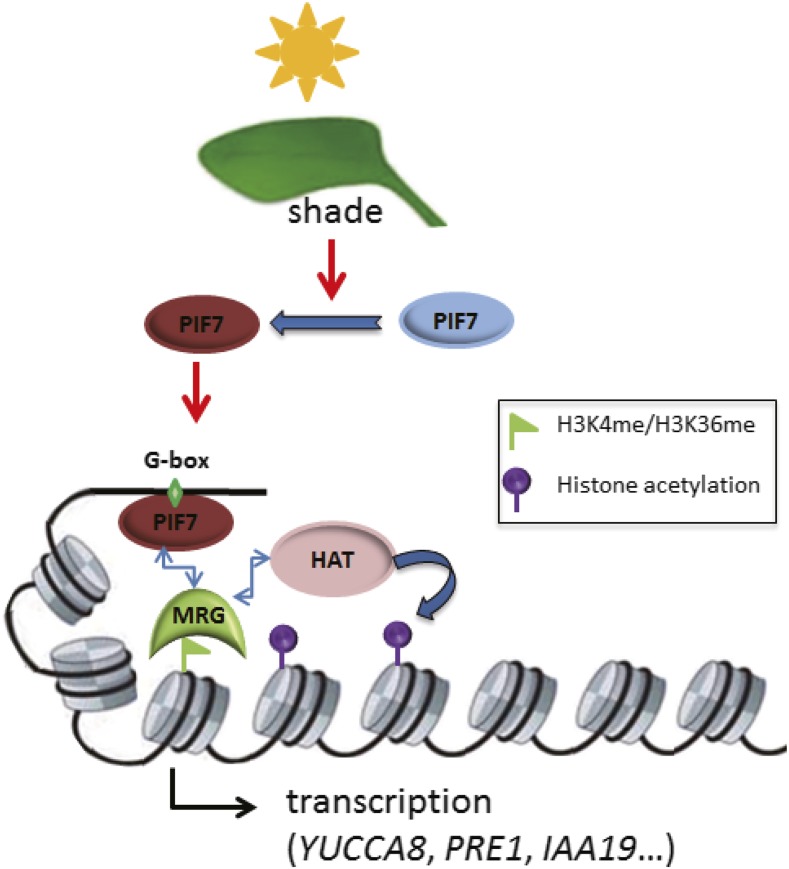
Although much had been learned from both the input pathway perceiving shade and multiple output pathways involving phytohormone production/signaling in SAS (Casal, 2012; Carriedo et al., 2016; Ballaré and Pierik, 2017), how transcription of multiple genes is coordinately induced within the chromatin context in response to shade had been largely obscure. Our current study demonstrates that PIF7 recruits the H3K4me3/H3K36me3-reader MRG2 (possibly also MRG1 because of their high sequence homology) to promote histone acetylations in transcription induction, providing a crucial mechanistic link to couple shade perception to epigenetic regulation of gene transcription in plant SAS (Fig. 6).
Figure 6.
Model for PIF7 and MRGs in regulating the shade-induced hypocotyl elongation. Under shade, the transcription factor PIF7 is getting dephosphorylated and subsequently binds to the cis-element G-box of a target gene (e.g. YUCCA8, PRE1, or IAA19). Physical interactions between PIF7 and the H3K4me/H3K36me-reader MRG as well as between MRG and a histone-acetyltransferase HAT lead to formation of a multiprotein complex to acetylate nucleosomal histones to promote the target gene transcription within chromatin.
This proposed mechanistic model is based on numerous points of evidence: (1) PIF7 and MRG2 physically interact in vitro and in vivo; (2) PIF7 is dephosphorylated (Li et al., 2012) when plants are exposed to shade, and it is this shade-induced PIF7 form that binds MRG2; (3) both pif7-1 and mrg1mrg2 mutants display impaired hypocotyl elongation and reduced expression levels of a common set of shade-responsive genes under shade treatment; (4) pif7-1 is epistatic to mrg1mrg2 in both hypocotyl elongation and gene expression regulation; (5) PIF7 recognizes the G-box in the promoter (Li et al., 2012) and binds chromatin around it, whereas MRG2 binds H3K4me3/H3K36me3 (Bu et al., 2014; Xu et al., 2014) and is enriched together with H3K4me3/H3K36me3 at the 5′-end of gene body; (6) shade-induced binding of MRG2 at chromatin is PIF7-dependent and is enhanced by basal level of H3K4me3/H3K36me3; (7) H4K5ac/H3K9ac/H3K27ac are induced under shade treatment, although H3K4me3/H3K36me3 levels remain unchanged; and (8) MRG2 physically interacts with the H4K5-acetyltransferase HAM1/HAM2 (Xu et al., 2014), and a consistently shade-induced H4K5ac elevation is dependent on PIF7 and MRG1/MRG2.
In contrast to stable PIF7, most other PIFs including PIF1, PIF3, PIF4, and PIF5 are subject to light-induced phosphorylation and degradation via the ubiquitin-proteasome system (Leivar and Quail, 2011). Albeit responding to the shade signal, the quartet mutant pifq (deprived of PIF1, PIF3, PIF4, and PIF5) showed shorter hypocotyls than the wild-type control under all tested conditions including white light and dark (Leivar et al., 2012a, 2012b). This differs from pif7-1 and mrg1mrg2, which display shorter hypocotyls than Col-0 under shade but not under white light or dark growth conditions. In general, PIF1 is primarily involved in seed germination regulation; only PIF3/PIF4/PIF5 play some redundant roles in shade-response of hypocotyl elongation (Lorrain et al., 2008; Leivar et al., 2012a, 2012b), likely by regulating downstream target genes independently in an additive manner from the more prominent PIF7 function (Li et al., 2012; de Wit et al., 2015; Mizuno et al., 2015). Nevertheless, PIF7 can form not only homodimers but also heterodimers with PIF4, and also possibly with other PIFs (Kidokoro et al., 2009). Thus, this proposed model, based primarily on PIF7, will likely be extendable to include additional PIFs.
MRG1/MRG2 also can form homodimers and heterodimers (Liu et al., 2016). The tethered PIF-MRG multiprotein complex may contribute to function together for the PIF-binding and MRG-binding sites that are separated with variable distances among different target genes. In line with this idea, for all three examined genes, PIF7 was found enriched at the chromatin region around the G-box within the promoter and MRG2 was found at the 5′-end of the gene body, two regions that are located closely in the case of IAA19 but much more distantly in the case of YUCCA8 and PRE1. YUCCA8 and IAA19 are important regulators involved in auxin biosynthesis and response (Zhao, 2014; Leyser, 2017), and PRE1 is implicated in BR signaling (Clouse, 2011)—two major types of hormones combined to act in SAS (Keuskamp et al., 2011). Thus, the PIF7-MRG2 module constitutes a crucial step linking shade response to output of hormonal regulation of cell growth.
In agreement with MRG1/MRG2 acting as readers of H3K4me3 and H3K36me3 (Bu et al., 2014; Xu et al., 2014), the MRG2-bound but not the PIF7-bound chromatin regions at shade-responsive genes are covered by high levels of H3K4me3 and H3K36me3. The H3K4me3/H3K36me3 levels at YUCCA8 are unchanged upon shade treatment. Shade-induced MRG2 enrichment at YUCCA8 is primarily mediated by PIF7. Nevertheless, the atx1-2 and sdg8-2 mutants, in which H3K4me3 and H3K36me3 deposition is respectively impaired (Zhao et al., 2005; Pien et al., 2008; Xu et al., 2008), exhibit reduction of MRG2 recruitment and hypocotyl elongation, supporting a role of basal level of H3K4me3/H3K36me3 in shade response. Basal levels of H3K4me3/H3K36me3 likely build a poised chromatin state advantageous for target genes to rapidly respond to shade. Both atx1-2 and sdg8-2 are hypostatic to mrg1mrg2 (Supplemental Table S1), implying that MRG1/MRG2 read H3K4me3/H3K36me3 to effect downstream transcription events in response to shade.
Histone acetylation is generally well known to be associated with active transcription (Lusser et al., 2001). In response to shade, levels of H4K5ac, H3K9ac, and H3K27ac but not H3K14ac nor H3K36ac were found increased at YUCCA8. The H4K5ac increase is positively coregulated by PIF7 and MRG1/MRG2. Because MRG1/MRG2 physically interact with HAM1/HAM2 (Xu et al., 2014), two homologous HATs specifically catalyzing H4K5 acetylation (Earley et al., 2007), our findings strongly support that the transcription factor PIF7, the H3K4me3/H3K36me3-reader MRG1/MRG2 and the H4K5ac-effector HAM1/HAM2, together form a mechanistic module in induction of shade-responsive genes. Similarly, the transcription factors CO, MRG1/MRG2, and HAM1/HAM2 likely also form a module in promoting FT transcription and plant flowering (Bu et al., 2014; Xu et al., 2014), albeit it is remaining to be verified whether CO is required for H4K5ac deposition. Because the ham1ham2 mutant is lethal (Latrasse et al., 2008), direct examination of function of H4K5ac deposition by HAM1/HAM2 in either the PIF7-MRG1/MRG2 or CO-MRG1/MRG2 module has not been performed so far. Differing from H4K5ac, H3K9ac responded to shade induction in only a PIF7-dependent but not a MRG1/MRG2-dependent manner. H3K9 acetylation as well as H3K27 acetylation had already been reported to be associated with expression of light-responsive genes in previous studies (Barneche et al., 2014). Several HATs, including the most extensively studied one, GENERAL CONTROL NONDEREPRESSIBLE5 (Benhamed et al., 2006; Barneche et al., 2014), and the broad-specificity ones, such as HAC1/HAC5/HAC12 (Earley et al., 2007), are capable of catalyzing H3K9/H3K27 acetylation. Future work will be required to identify precisely which HAT(s) is (are) involved and through which mechanism PIF7 recruits it (them), independently of MRG1/MRG2, to shade-responsive genes in H3K9/H3K27 acetylation.
Lastly, it is worth pointing out that PIF7 has a repressive role in transcription of DRE-Binding1 and C-repeat Binding Factor genes (Kidokoro et al., 2009; Lee and Thomashow, 2012). Evidence from yeast and animals shows that MRG proteins interact with both HAT and HDAC (Eisen et al., 2001; Pardo et al., 2002; Carrozza et al., 2005), and Arabidopsis PIF3 associates with HDA15 in repression of chlorophyll biosynthesis and photosynthesis genes (Liu et al., 2013) whereas PIF1 associates with HDA15 in repression of seed germination genes (Gu et al., 2017). Together, it is emerging that PIF transcription factors could recruit diverse chromatin modulators for transcriptional activation or repression, likely in a gene-context-dependent manner, to enable plant effectively responding to environmental shade or light.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants used in this study were of the Columbia-0 ecotype. The mutants used in this study have been described previously: pif7-1 (Li et al., 2012), mrg1mrg2, MRG2-YFP/mrg1mrg2, and pMRG2::MRG2-GUS (Bu et al., 2014), atx1-2 (Pien et al., 2008), sdg8-2 (Zhao et al., 2005; Xu et al., 2008), and PIF7-Flash (PIF7ox; Li et al., 2012). For the phenotypic analysis, seeds were germinated on 1/2 Murashige & Skoog medium (Duchefa Biochemie) plates with 1% (w/v)agar (Sangon) and without Suc. After stratification, the plates were incubated in growth chambers under continuous LED white light for 4 d, then the plates were either left in white light (R: approximately 25 μE × m−2 × s1, B: approximately 27 μE × m−2 × s−1, FR: approximately 5 μE × m−2 × s−1) or transferred to simulated shade (R: approximately 25 μE × m−2 × s−1, B: approximately 27 μE × m−2 × s−1, FR: approximately 35 μE × m−2 × s−1) for 3 d before hypocotyl measurements were made. For single light treatment, the plates were either left in Dark or Red (approximately 10 μE × m−2 × s−1), Far-Red (approximately 2 μE × m−2 × s−1), and Blue light (approximately 40 μE × m−2 × s−1) for 4 d.
Quantitative RT-PCR Analysis
RNA was extracted from Arabidopsis seedlings as described in Oñate-Sánchez and Vicente-Carbajosa (2008). Reverse transcription was performed using Superscript III reverse transcriptase (Invitrogen). Relative levels of cDNA were quantified with SYBR-Green I master mix in the LightCycler 480-2 according to the manufacturer's instructions (Roche). All primer sequences are listed in Supplemental Table S2. cDNA levels were normalized to internal reference genes AT2G39960 that are transcriptionally stable after shade treatment (Tao et al., 2008).
Yeast Two-Hybrid Assay
For yeast two-hybrid assay, full length of the PIF7-coding sequence was cloned into pGADT7 (AD-PIF7) and that of MRG2 was cloned into pGBKT7 (BD-MRG2). Positive protein-protein interaction was tested by growth on SD medium without Leu, Trp, and His (SD-l-T-H), and transformation was verified by growth on SD-l-T, using the yeast strain AH109 according to the manufacturer’s manual (Clontech).
GST Pull-Down Assays
Full-length PIF7 cDNA, truncated PIF7-N1 (1 to 221 amino acids), PIF7-N2 (1 to 163 amino acids), PIF7-N3 (1 to 100 amino acids), PIF7-N4 (101 to 163 amino acids), PIF7-C1 (164 to 366 amino acids), and PIF7-C2 (222 to 366 amino acids) were cloned into BamHI and EcoRI sites of pGEX-6P-1. The pET-28a-MRG2 construct have been previously described (Bu et al., 2014). His-MRG2 protein was incubated with pretreated GST-PIF7 bead for 2 h. GST was used as a negative control. Beads were resuspended with SDS-PAGE loading buffer and analyzed by SDS-PAGE and immunoblotting using anti-His antibody.
In a semi-in vivo pull-down assay, total protein was extracted from PIF7ox plants grown in duplicate under white light for 4 d and then one of the duplicates was treated with shade for 1 h and the other of duplicate was maintained under white light for an additional hour. MRG2 cDNA were cloned into BamHI and EcoRI sites of pGEX-6P-1 expression vector. Plant materials were ground with liquid N and resuspended in extraction buffer [100 mm Tris-HCl (pH 7.5), 300 mm NaCl, 2 mm EDTA, 1% Trion X-100, 10% glycerol, and protease inhibitor cocktail]. Protein extracts were centrifuged at 20,000g for 10 min, and the resulting supernatant was incubated with pretreated GST-MRG2 beads for 2 h. GST was used as a negative control. Beads were resuspended with SDS-PAGE loading buffer and analyzed by SDS-PAGE and immunoblotting using anti-Myc antibody (9E10; Covance).
Co-IP Assay
Total protein extracts were prepared from Col-0 and PIF7ox seedlings grown in white light for 4 d and then transferred to shade for 1 h or kept in white light. A Co-IP assay was performed as described in Molitor et al., (2014). IP was performed using anti-MRG2 agarose beads. Input and IP-resulted fractions were analyzed on western blot using anti-MYC and anti-MRG2 antibodies. The polyclonal antibody against MRG2 was produced by Abmart (Bu et al., 2014).
ChIP-PCR
ChIP-PCR was performed as in Li et al. (2012) and Bu et al. (2014). Whole seedlings were grown in duplicate under white light for 4 d, then one of the duplicates was treated with shade for 1 h and the other duplicate was maintained under white light for an additional hour. They were harvested and cross linked for 15 min under vacuum in a cross-linking buffer (extraction buffer 1 with 1% formaldehyde). Cross linking was quenched in 125 mm Gly, pH 8.0, under vacuum for 5 min, and then seedlings were washed three times in double-distilled water and rapidly frozen. Bioruptor was used at high power with 30-s-on/30-s-off cycles for fifteen times until the average chromatin size was approximately 300 bp. Antibodies used in this study were anti-MYC affinity gel (E6654; Sigma-Aldrich), antitrimethyl-H3K4 (07-473; Millipore), antitrimethyl-H3K36 (ab9050; Abcam), antitrimethyl-H3K27 (07-449; Millipore), antiacetylation-H3K9 (ab4441; Abcam), antiacetylation-H3K14 (ab8201; Abcam), antiacetylation-H3K27 (ab4729; Abcam), antiacetylation-H3K36 (39379; Active Motif), and antiacetylation-H4K5 (39699; Active Motif). Quantitative real-time PCR was performed with a kit from Takara to determine the enrichment of DNA immunoprecipitated in the ChIP experiments, using the gene-specific primers listed in Supplemental Table S2. To facilitate comparisons, fold-change values were obtained from the enrichment data, expressed as % Input, by using AT2G39960 as an internal reference gene (Tao et al., 2008; Li et al., 2012).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers PIF7, AT5G61270; MRG1, AT4G37280; MRG2, AT1G02740; SDG8, AT1G77300; ATX1, AT1G66240; YUCCA8, AT4G28720; YUCCA9, AT1G04180; PRE1, AT5G39860; IAA19, AT3G15540; GH3.3, AT2G23170; PAR1, AT2G42870.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Quantification of hypocotyl length and representative seedlings of Col-0, mrg1, mrg2, mrg1mrg2, and MRG2-YFP/mrg1mrg2.
Supplemental Figure S2. Hypocotyl lengths of Col-0 and mrg1mrg2 seedlings grown in Dark, Red, Far-Red or Blue light.
Supplemental Figure S3. Relative gene expression of PIFs in mrg1mrg2 and the expression of MRG1/2 in pif7-1 under white and shade.
Supplemental Figure S4. PIF7 and MRG2 binding at IAA19 and PRE1 chromatin.
Supplemental Figure S5. Relative expression levels of YUCCA8, PRE1, and IAA19 in Col-0, mrg1mrg2, atx1-2, and sdg8-2 under white light or shade treatment conditions.
Supplemental Figure S6. Comparison of H3K9ac levels at YUCCA8 in Col-0, mrg1mrg2, pif7-1, and pif7-1mrg1mrg2 grown under white light and shade.
Supplemental Table S1. Analysis of hypocotyl length of wild-type (Col-0) and different mutants under white light or shade growth conditions.
Supplemental Table S2. Primers used in this study.
Acknowledgments
This research was conducted within the context of the International Associated Laboratory Plant Epigenome Research (LIA PER).
Footnotes
This work was supported by National Key R&D Program of China grant no. 2017YFA0503800 National Natural Science Foundation of China (grant nos. 31300263, 31371304, 31470374, 31500973, and 31571319), the French Agence Nationale de la Recherche (no. ANR-12-BSV2-0013-02), and the Centre National de la Recherche Scientifique (CNRS, LIA PER).
Articles can be viewed without a subscription.
References
- Ballaré CL, Pierik R (2017) The shade-avoidance syndrome: multiple signals and ecological consequences. Plant Cell Environ 40: 2530–2543 [DOI] [PubMed] [Google Scholar]
- Barneche F, Malapeira J, Mas P (2014) The impact of chromatin dynamics on plant light responses and circadian clock function. J Exp Bot 65: 2895–2913 [DOI] [PubMed] [Google Scholar]
- Benhamed M, Bertrand C, Servet C, Zhou DX (2006) Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18: 2893–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, Shafiq S, Shen WH (2011) Histone modifications in transcriptional activation during plant development. Biochim Biophys Acta 1809: 567–576 [DOI] [PubMed] [Google Scholar]
- Bu Z, Yu Y, Li Z, Liu Y, Jiang W, Huang Y, Dong AW (2014) Regulation of Arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genet 10: e1004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriedo LG, Maloof JN, Brady SM (2016) Molecular control of crop shade avoidance. Curr Opin Plant Biol 30: 151–158 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Casal JJ. (2012) Shade avoidance. Arabidopsis Book 10: e0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD. (2011) Brassinosteroids. The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD: 10.1199/tab.0151 [Google Scholar]
- de Wit M, Keuskamp DH, Bongers FJ, Hornitschek P, Gommers CMM, Reinen E, Martínez-Cerón C, Fankhauser C, Pierik R (2016) Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Curr Biol 26: 3320–3326 [DOI] [PubMed] [Google Scholar]
- de Wit M, Ljung K, Fankhauser C (2015) Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol 208: 198–209 [DOI] [PubMed] [Google Scholar]
- Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS (2007) In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J 52: 615–626 [DOI] [PubMed] [Google Scholar]
- Eisen A, Utley RT, Nourani A, Allard S, Schmidt P, Lane WS, Lucchesi JC, Cote J (2001) The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J Biol Chem 276: 3484–3491 [DOI] [PubMed] [Google Scholar]
- Gu D, Chen CY, Zhao M, Zhao L, Duan X, Duan J, Wu K, Liu X (2017) Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic Acids Res 45: 7137–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C (2009) Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp DH, Sasidharan R, Vos I, Peeters AJ, Voesenek LA, Pierik R (2011) Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J 67: 208–217 [DOI] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH (2004) A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro S, Maruyama K, Nakashima K, Imura Y, Narusaka Y, Shinwari ZK, Osakabe Y, Fujita Y, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2009) The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol 151: 2046–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D, Benhamed M, Henry Y, Domenichini S, Kim W, Zhou DX, Delarue M (2008) The MYST histone acetyltransferases are essential for gametophyte development in Arabidopsis. BMC Plant Biol 8: 121.10.1186/1471-2229-8-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Thomashow MF (2012) Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc Natl Acad Sci USA 109: 15054–15059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Cohn MM, Quail PH (2012a) Phytochrome signaling in green Arabidopsis seedlings: impact assessment of a mutually negative phyB-PIF feedback loop. Mol Plant 5: 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, Quail PH (2012b) Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. (2017) Auxin signaling. Plant Physiol 10.1104/pp.17.0076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, Ecker JR, Kay SA, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Mukherjee I, Thum KE, Tanurdzic M, Katari MS, Obertello M, Edwards MB, McCombie WR, Martienssen RA, Coruzzi GM (2015) The histone methyltransferase SDG8 mediates the epigenetic modification of light and carbon responsive genes in plants. Genome Biol 16: 79 10.1186/s13059-015-0640-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen CY, Wang KC, Luo M, Tai R, Yuan L, Zhao M, Yang S, Tian G, Cui Y, Hsieh HL, Wu K (2013) PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25: 1258–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang S, Zhao M, Luo M, Yu CW, Chen CY, Tai R, Wu K (2014) Transcriptional repression by histone deacetylases in plants. Mol Plant 7: 764–772 [DOI] [PubMed] [Google Scholar]
- Liu Y, Wu H, Yu Y, Huang Y (2016) Structural studies on MRG701 chromodomain reveal a novel dimerization interface of MRG proteins in green plants. Protein Cell 7: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Lusser A, Kölle D, Loidl P (2001) Histone acetylation: lessons from the plant kingdom. Trends Plant Sci 6: 59–65 [DOI] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Mizuno T, Oka H, Yoshimura F, Ishida K, Yamashino T (2015) Insight into the mechanism of end-of-day far-red light (EODFR)-induced shade avoidance responses in Arabidopsis thaliana. Biosci Biotechnol Biochem 79: 1987–1994 [DOI] [PubMed] [Google Scholar]
- Molitor AM, Bu Z, Yu Y, Shen WH (2014) Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet 10: e1004091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Zhu L, Shen H, Huq E (2008) PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci USA 105: 9433–9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J (2008) DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes 1: 93 10.1186/1756-0500-1-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo PS, Leung JK, Lucchesi JC, Pereira-Smith OM (2002) MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation. J Biol Chem 277: 50860–50866 [DOI] [PubMed] [Google Scholar]
- Pedmale UV, Huang SC, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PAB, Sridevi P, Nito K, Nery JR, Ecker JR, Chory J (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S, Fleury D, Mylne JS, Crevillen P, Inzé D, Avramova Z, Dean C, Grossniklaus U (2008) ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20: 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Wei W, Zhou DX (2015) Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci 20: 614–621 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, Cheng Y, Lim J, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lijsebettens M, Grasser KD (2014) Transcript elongation factors: shaping transcriptomes after transcript initiation. Trends Plant Sci 19: 717–726 [DOI] [PubMed] [Google Scholar]
- Xiao J, Zhang H, Xing L, Xu S, Liu H, Chong K, Xu Y (2013) Requirement of histone acetyltransferases HAM1 and HAM2 for epigenetic modification of FLC in regulating flowering in Arabidopsis. J Plant Physiol 170: 444–451 [DOI] [PubMed] [Google Scholar]
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH (2008) Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol 28: 1348–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Gan ES, Zhou J, Wee WY, Zhang X, Ito T (2014) Arabidopsis MRG domain proteins bridge two histone modifications to elevate expression of flowering genes. Nucleic Acids Res 42: 10960–10974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE (2009) Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol 10: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2014) Auxin biosynthesis. In Arabidopsis Book 12: e0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Yu Y, Meyer D, Wu C, Shen WH (2005) Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 7: 1256–1260 [DOI] [PubMed] [Google Scholar]