The OsbZIP48 gene from rice can complement the hy5 mutant of Arabidopsis but exerts pleiotropic effects and causes semidwarfism when overexpressed in rice, and its mutant/RNAi lines are seedling lethal.
Abstract
Plants have evolved an intricate network of sensory photoreceptors and signaling components to regulate their development. Among the light signaling components identified to date, HY5, a basic leucine zipper (bZIP) transcription factor, has been investigated extensively. However, most of the work on HY5 has been carried out in Arabidopsis (Arabidopsis thaliana), a dicot. In this study, based on homology search and phylogenetic analysis, we identified three homologs of AtHY5 in monocots; however, AtHYH (HY5 homolog) homologs are absent in the monocots analyzed. Out of the three homologs identified in rice (Oryza sativa), we have functionally characterized OsbZIP48. OsbZIP48 was able to complement the Athy5 mutant. OsbZIP48 protein levels are developmentally regulated in rice. Moreover, the OsbZIP48 protein does not degrade in dark-grown rice and Athy5 seedlings complemented with OsbZIP48, which is in striking contrast to AtHY5. In comparison with AtHY5, which does not cause any change in hypocotyl length when overexpressed in Arabidopsis, the overexpression of full-length OsbZIP48 in rice transgenics reduced the plant height considerably. Microarray analysis revealed that OsKO2, which encodes ent-kaurene oxidase 2 of the gibberellin biosynthesis pathway, is down-regulated in OsbZIP48OE and up-regulated in OsbZIP48KD transgenics as compared with the wild type. Electrophoretic mobility shift assay showed that OsbZIP48 binds directly to the OsKO2 promoter. The RNA interference lines and the T-DNA insertional mutant of OsbZIP48 showed seedling-lethal phenotypes despite the fact that roots were more proliferative during early stages of development in the T-DNA insertional mutant. These data provide credible evidence that OsbZIP48 performs more diverse functions in a monocot system like rice in comparison with its Arabidopsis ortholog, HY5.
The light signaling network is one of the most extensively studied networks in plants. The light signal transduction can be divided into sensory photoreceptors, early signaling factors, central integrators, and downstream effectors (Chory, 2010). The photoreceptors, such as phytochromes, cryptochromes, phototropins, UVR8 and zeitlupe, are involved in the perception of light signal. They perceive the light signal and transmit it to early signaling factors like HFR1, FAR1, LAF1, PIFs, and EID1. The central integrators of the COP/DET/FUS class regulate other proteins involved in this pathway by targeting them for degradation (Chory, 2010). Downstream effectors like LONG HYPOCOTYL5 (HY5) regulate the expression of an innumerable number of genes associated with photomorphogenesis. The hy5 mutant was one of the five Arabidopsis (Arabidopsis thaliana) mutants isolated originally by Koornneef et al. (1980) that showed a long-hypocotyl phenotype even when grown in light; the wild type seedlings characteristically developed short hypocotyls. Molecular genetic analysis revealed that HY5 codes for a basic Leu zipper (bZIP) transcription factor that serves as a positive regulator of photomorphogenesis in Arabidopsis (Oyama et al., 1997). Subsequently, it was found that HY5 is constitutively nucleus localized and acts downstream of phytochromes, cryptochromes, and UVR8, indicating that it promotes photomorphogenesis under a broad spectrum of wavelengths, including far-red, red, blue, and UV-B light (Koornneef et al., 1980; Oyama et al., 1997; Osterlund et al., 2000; Ulm et al., 2004; Jiao et al., 2007; Huang et al., 2012; Jiang et al., 2012; Ram and Chattopadhyay, 2013; Zheng et al., 2013). Detailed characterization of the hy5 mutant showed that, apart from elongated hypocotyl, the light-grown hy5 mutant seedlings also develop more lateral roots and exhibit altered gravitropic and touch responses in roots (Oyama et al., 1997). In addition, secondary thickening was reduced and the numbers of lignified xylem vessels and fiber elements were less. The chlorophyll and anthocyanin accumulation in the hy5 mutant seedlings were reduced considerably as compared with the wild type, although the hy5 mutant seedlings had slightly larger cotyledons than the wild type (Oyama et al., 1997; Sibout et al., 2006). ChIP-chip analysis revealed that HY5 binds to the promoters of more than 3,000 genes (Zhang et al., 2011a), indicating that it is a master regulator that binds to an array of genes associated with multiple regulatory circuits and metabolic pathways. Among these genes, 1,173 genes showed HY5-dependent expression, indicating that it probably coregulates many target genes through integrated subprograms; some of those genes were positively regulated and others were negatively regulated by HY5, indicating that it may act as both transcriptional activator and repressor (Zhang et al., 2011a).
Although HY5 has been characterized extensively from Arabidopsis, it has been a subject of investigation in other organisms like Physcomitrella patens, Lotus japonicus, and Pisum sativum (Nishimura et al., 2002; Weller et al., 2009; Yamawaki et al., 2011). But none of the homologs of HY5 in monocots has been characterized so far. In our previous publication on an overall analysis of the bZIP family in rice (Oryza sativa), we had identified three putative homologs in rice, which form part of the same clade of bZIPs as AtHY5 and AtHYH (Nijhawan et al., 2008). In this study, we have functionally characterized OsbZIP48, one of the three AtHY5 homologs in rice, with the aim to find out if it can functionally complement the Athy5 mutant and whether it performs any unique functions in a monocot system like rice.
RESULTS
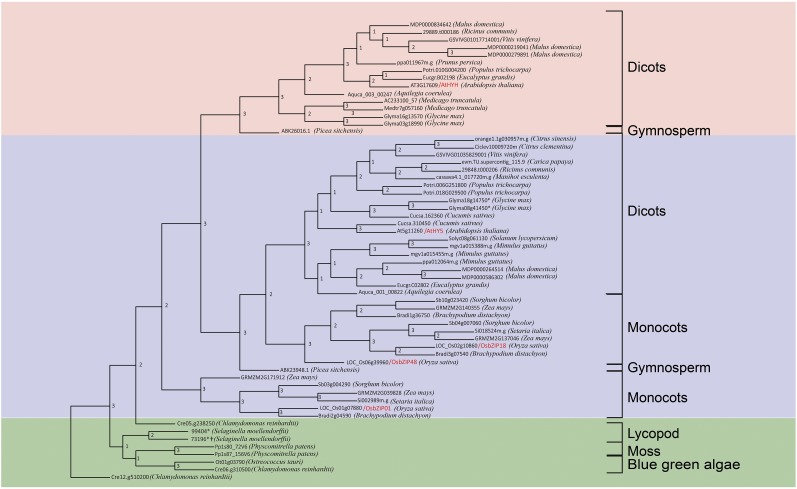
Monocots Have Three Homologs of AtHY5
The phylogenetic and pairwise distance analysis of homologs of AtHY5 showed that there are three homologs of AtHY5 in monocots, while HYH homologs are represented in the genomes of only dicots and gymnosperms (Fig. 1). The HY5/HYH phylogenetic tree in Figure 1 suggests that, out of the three HY5 homologs, the clade of two homologs of HY5 in monocots is closer to the eudicot clade of HY5, while there is no HYH homolog in monocots. However, we were able to retrieve a HYH homolog from Picea sitchensis, a gymnosperm. Moreover, a subclade of monocot HY5 homologs was found to be present as a sister clade of the HY5/HYH homologs found in lower plants along with the HY5/HYH clade. Therefore, pairwise distance analysis of the bZIP domain of these proteins was performed (Supplemental Table S1), which showed that this subclade is closer to AtHY5 than AtHYH. The pairwise distance analysis results indicate that the HY5/HYH homologs in lower organisms like mosses and lycopods are closer to HY5 than HYH. Thus, HYH homologs might be absent in lower plants and monocots but are present in gymnosperms and dicots. Although gymnosperms diverged before the divergence of monocots and dicots, the presence of an AtHYH homolog in gymnosperms but its absence in monocots is intriguing. A detailed evolutionary analysis of AtHYH would be possible only when more genomic data on gymnosperms become available.
Figure 1.
Phylogenetic tree of HY5 and HYH homologous proteins from across species (asterisks represent manually reannotated proteins, and daggers represent incomplete proteins even after manual reannotation but having the bZIP domain). The consensus tree was generated after merging the individual trees generated by phyml, neighbor joining, and the maximum parsimony approach. The numbering at the nodes represents the number of trees (generated by three different methods) that have the same topology as the consensus tree.
The alignment of protein sequences of all HY5 homologs showed that the COP1-interaction motif (as described by Holm et al., 2002) is present in most of the homologs, suggesting that it might be regulated by COP1 in these organisms as well (Supplemental Fig. S1). A consensus casein kinase II phosphorylation site (ESDEE) is conserved in most of the HY5 homologs and in one homolog of Chlamydomonas reinhardtii, indicating that if the COP1-interaction motif is functional in other organisms, HY5 activity and its binding ability to COP1 is likely to be modulated by phosphorylation. The conserved VP pair of this motif is important for the interaction of HY5 with COP1 (Holm et al., 2001). The COP1-interaction motif in green algae lacks this conserved VP pair and has many insertions in the COP1-binding motif. Therefore, interaction studies need to be done in order to find whether HY5 homologs of green algae are able to interact with its COP1 counterpart.
Expression Analysis of OsbZIP48 in Different Tissues of Rice
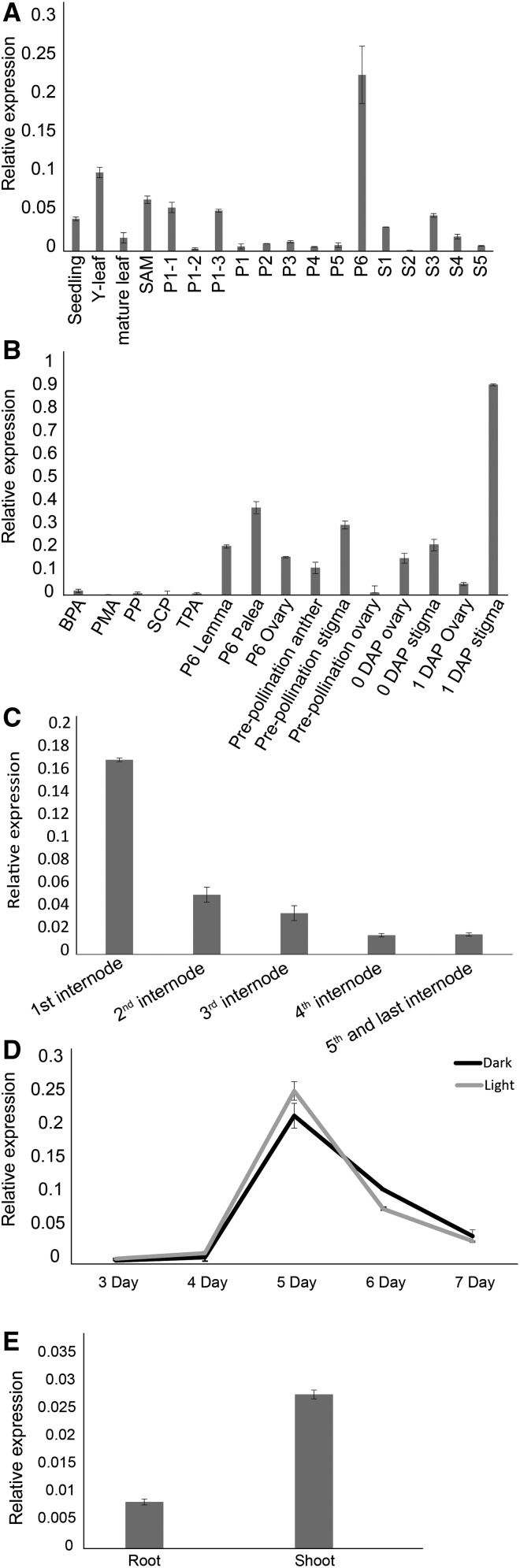
The microarray-based expression of OsbZIP48 was first checked in Affymetrix meta-analysis data and rice atlas data sets (GSE6893 and GSE14298) using the rice oligonucleotide array database; this also includes our own microarray data on whole-genome expression analysis at various stages of panicle and seed development in rice (Sharma et al., 2012). Although OsbZIP48 expression was maximal in stigma followed by ovary, its transcripts could be detected in the plumule, coleoptile, shoot, leaf, flag leaf, spikelet, lemma/palea, and embryo sac of the rice plant. Among different tissues and at the various developmental stages of rice examined, OsbZIP48 expression was found to be significantly high in second leaf and during tiller initiation, P5 panicle, and S1 stage of seed development. Apart from these tissues, OsbZIP48 expression was found to be high in the gynoecium during pollination and fertilization (Supplemental Fig. S2). Real-time PCR analysis was carried out to validate the microarray-based expression data of OsbZIP48 at different stages of development in rice (Fig. 2A). The expression of OsbZIP48 was found to be maximum at the P6 stage of rice panicle development, which was slightly different from the microarray data, which showed maximum expression in the S1 stage as compared with P6; however, these stages are in succession and, thus, closely related. The expression of OsbZIP48 was examined at different stages of pollen development and in P6 ovary, P6 lemma, P6 palea, prepollinated anther, prepollinated ovary, prepollinated stigma, 0-DAP (days after pollination) ovary, 0-DAP stigma, 1-DAP ovary, and 1-DAP stigma. The real-time PCR analysis revealed that OsbZIP48 expression was maximum in 1-DAP stigma followed by P6 palea and prepollinated stigma. Some basal level expression could be detected in 0-DAP ovary, 0-DAP stigma, and P6 lemma. The high level of expression in 1-DAP stigma and prepollinated stigma (Fig. 2B) indicates that it might play some role in pollination and postpollination events of panicle development.
Figure 2.
Expression profile of OsbZIP48 in various tissues and at different stages of development. A, Expression of OsbZIP48 in vegetative (seedling, young [Y] leaf, mature leaf, and shoot apical meristem [SAM]), panicle, and seed stages of development in rice variety IR64 as analyzed by real-time PCR. (Panicle stages are as follows: P1-1, 0.5–2 mm; P1-2, 2–5 mm; P1-3, 5–10 mm; P1, 0–3 cm; P2, 3–5 cm; P3, 5–10 cm; P4, 10–15 cm; P5, 15–22 cm; and P6, 22–30 cm. Seed stages are as follows: S1, 0–2 DAP; S2, 3–4 DAP; S3, 5–10 DAP; S4, 11–20 DAP; and S5, 21–29 DAP.) B, Real-time PCR analysis of OsbZIP48 using different organs of the inflorescence: PMA, premeiotic anther; SCP, single-cell pollen; and TPA, trinucleate pollen anther. C, Real-time PCR analysis to check the expression of OsbZIP48 in different internodes of the mature rice stem. D, Expression analysis of OsbZIP48 using real-time PCR in 3- to 7-d-old light- and dark-grown rice seedlings. E, Expression analysis of OsbZIP48 root and shoot of 5-d-old light-grown seedlings using real-time PCR. Data shown are means ± se. The expression data presented are relative to UBIQUITIN5.
The overexpression of the gene encoding AtHY5 lacking the COP1-binding domain is known to cause a reduction in hypocotyl length in Arabidopsis when grown in light. On the other hand, no change in hypocotyl length was observed between the wild type and transgenics overexpressing AtHY5 without the COP1-binding domain when grown in the dark (Ang et al., 1998). The structure of mature rice stem differs from that of Arabidopsis in that it is hollow and consists of nodes and internodes. Therefore, the expression of OsbZIP48 was checked in the internodes of the mature rice plant just after the whole panicle emerged. The expression pattern of OsbZIP48 showed a decline from the bottom to the top internodes, with the first and the bottom-most internode (which does not have shoot-borne roots) having the maximum expression and the second last and top-most internode (the last internode), which bears the panicle, having the least (Fig. 2C). This correlates well with the fact that the bottom-most internode is shortest while the second last and last internodes are the longest in a rice plant.
Since AtHY5 is known to be involved in light signaling, the expression of its ortholog in rice, OsbZIP48, was checked in rice seedlings grown in the dark and the light (75 μmol m−2 s−1) for 3 to 7 d (Fig. 2D). The expression of OsbZIP48 was of basal level in 3- and 4-d-old dark- and light-grown seedlings, reached its peak on day 5 in both dark- and light-grown seedlings, and then began to decline in 6- and 7-d-old seedlings. When expression was checked in 5-d-old shoot and root, OsbZIP48 transcript levels were higher in shoots of the 5-d-old light-grown seedlings than in the roots (Fig. 2E).
OsbZIP48 Protein Levels Are Not Light Regulated
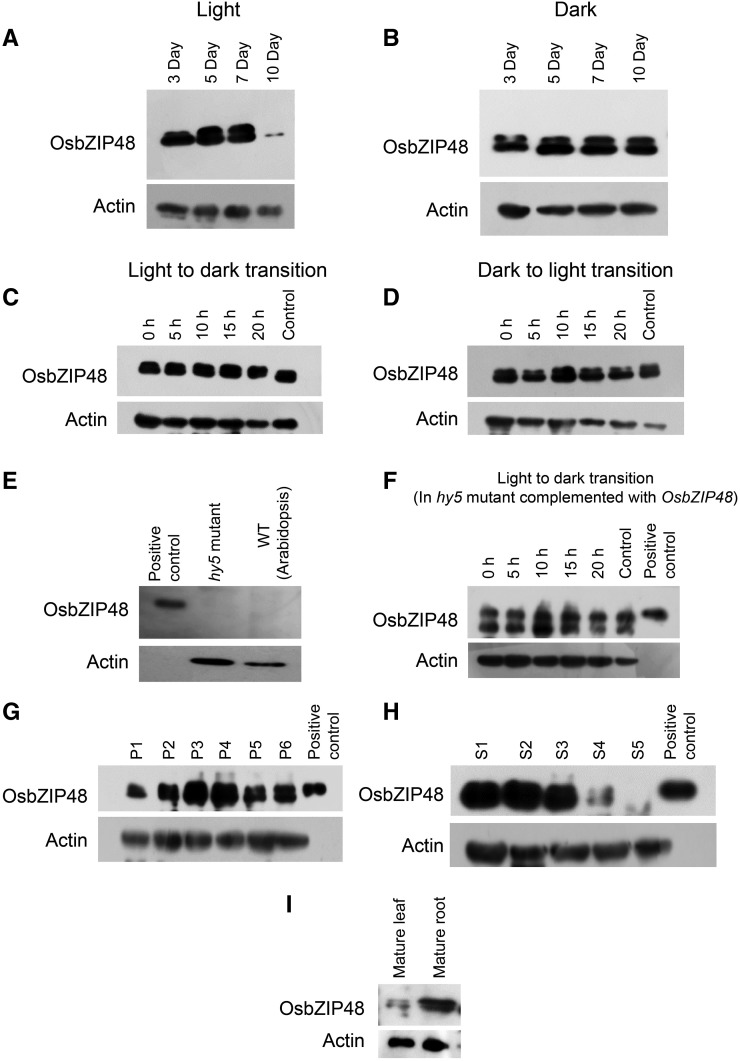
To determine whether OsbZIP48 protein levels are developmentally regulated, the protein extracts from light-grown as well as dark-grown rice seedlings were processed for western analysis. In light-grown seedlings, OsbZIP48 protein levels increased gradually in 3- to 5-d-old seedlings and were maintained in 7-d-old seedlings but decreased in 10-d-old seedlings (Fig. 3A). However, in dark-grown seedlings, the OsbZIP48 protein level was less in 3-d-old seedlings as compared with 5-d-old seedlings but thereafter remained constant until day 10 (Fig. 3B). OsbZIP48 levels also were examined during the light-to-dark transition, and they were found to be constant even after 20 h of light-to-dark and dark-to-light transition, indicating that OsbZIP48 protein levels are not light regulated in rice (Fig. 3, C and D). In order to investigate whether rice OsbZIP48 followed a similar pattern of accumulation when expressed in Arabidopsis, light-to-dark transition experiments were performed. The Athy5 mutant seedlings complemented with OsbZIP48 were used to examine the level of OsbZIP48 in Arabidopsis. Its levels were found to be constant during the light-to-dark transition, suggesting that it is not degraded in Arabidopsis as well (Fig. 3F).
Figure 3.
Western blots showing OsbZIP48 protein expression levels in different tissues of rice and the Arabidopsis hy5 mutant complemented with OsbZIP48. A, OsbZIP48 protein levels in 3-, 5-, 7-, and 10-d-old light-grown rice seedlings (100 μmol m−2 s−1). B, OsbZIP48 protein levels in 3-, 5-, 7-, and 10-d old dark-grown rice seedlings. C, OsbZIP48 protein levels in seedlings grown in continuous light for 4 d and then transferred to dark for 5, 10, 15, and 20 h; the control is 5-d-old seedlings grown in continuous light. D, OsbZIP48 protein levels in seedlings grown in continuous dark for 4 d and then transferred to the light for 5, 10, 15, and 20 h; 5-d-old seedlings grown in continuous dark were used as the control. E, Western blot using OsbZIP48 antibodies shows no cross-reactivity with Athy5 mutant protein extracts. F, OsbZIP48 protein levels in Arabidopsis hy5 mutant seedlings complemented with OsbZIP48, grown in continuous light for 4 d, and then transferred to the dark for 5, 10, 15, and 20 h; control represents 5-d-old seedlings grown in continuous light. G, Changes in OsbZIP48 protein levels during various stages of panicle development in rice. H, OsbZIP48 protein levels during seed development (S1–S5) stages in rice. I, OsbZIP48 protein levels in mature leaf and root in rice. The positive control in E to H is bacterially expressed 6× His-tagged OsbZIP48 protein.
OsbZIP48 protein levels were checked during different stages of panicle and seed development. During panicle development, the protein levels were found to be maximum in P3 and P4 stages, while during seed development, the protein levels were found to be high initially (i.e. during S1 and S2 stages) and then decreased gradually (Fig. 3, G and H). Among the vegetative tissues examined, the level of OsbZIP48 protein was found to be greater in the mature root than in mature leaf (Fig. 3I).
The HY5 protein in Arabidopsis exists in two forms, phosphorylated and unphosphorylated (Hardtke et al., 2000). In this study too, two bands could be recognized by anti-OsbZIP48 antibodies in the western blots, where the higher Mr form may represent a phosphorylated form of OsbZIP48, although this remains to be validated experimentally. It has been shown that the unphosphorylated form of HY5 is more active than the phosphorylated form and is the preferred substrate for COP1-mediated degradation (Hardtke et al., 2000). In the case of OsbZIP48, the phosphorylated form of protein (the upper band in western blots) increased gradually until it attained maximum level in 5- to 7-d-old light-grown seedlings; thereafter, it declined and was virtually undetectable in 10-d-old seedlings (Fig. 3A). It is interesting that, in 5- to 7-d-old light-grown seedlings, the phosphorylated and unphosphorylated forms of protein were almost equal, while in 5- to 10-d-old dark-grown seedlings, the abundance of the phosphorylated form was lower than the unphosphorylated form of OsbZIP48 (Fig. 3, A and B).
OsbZIP48 Is Nucleus Localized and Forms a Homodimer
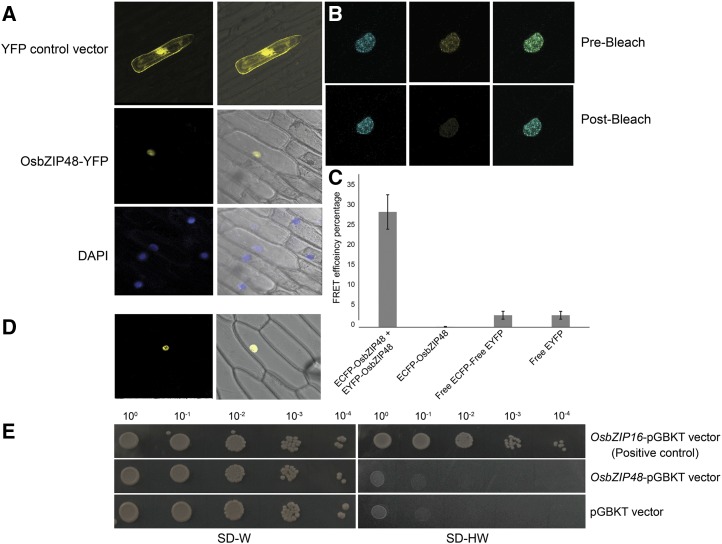
The bZIP proteins are transcription factors; therefore, to elucidate the intracellular localization of OsbZIP48, particle bombardment of onion (Allium cepa) peel cells using gold particles as macrocarriers was done using pSite3CA-OsbZIP48 as the construct. 4′,6-diamidino-2-phenylindole (DAPI) was used as a control for nucleus staining, while the empty pSite3CA was used as a vector control. The YFP-OsbZIP48 fusion protein was found to be localized in the nucleus (Fig. 4A), as predicted by ProtComp 9.0, an online program to predict subcellular localization of plant proteins. The bZIP proteins are known to bind to the DNA as homodimers or heterodimers. This dimerization is mediated by the Leu zipper region of the bZIP domain (Landschulz et al., 1988). Therefore, homodimerization of OsbZIP48 was checked by fluorescence resonance energy transfer (FRET) and bimolecular fluorescence complementation (BiFC) analyses (Fig. 4, B–D). The FRET efficiency of OsbZIP48 was 28%, indicating that it can form a homodimer. BiFC analysis using onion peel cells confirmed that OsbZIP48 homodimerizes exclusively in the nucleus.
Figure 4.
OsbZIP48 is localized in the nucleus, forms a homodimer, and lacks transactivation activity. A, Particle bombardment of the YFP-OsbZIP48 construct in onion cells. The first column shows photographs taken in dark field, and the second column shows merged photographs of dark field and bright field captured using a Leica microscope. The first row (YFP control vector) shows localization of only YFP protein, the second row (OsbZIP48-YFP) shows localization of OsbZIP48 tagged to YFP protein, and the third row (DAPI) shows DAPI-stained nucleus. B, Prebleach and postbleach images showing bleaching of YFP-OsbZIP48 for FRET analysis. C, Histogram showing FRET efficiency of CFP-OsbZIP48 and YFP-OsbZIP48 interaction as compared with the controls. Data shown are means ± se; n = 10. D, BiFC analysis using onion peel cells showing the homodimerization of nEYFPC1-OsbZIP48 and cEYFPC1-OsbZIP48 in the nucleus. E, Transactivation assay of OsbZIP48 in yeast cells. OsbZIP48 lacks transactivation activity, as the yeast cells containing the OsbZIP48-pGBKT construct were unable to grow on SD-HW medium (synthetic defined medium without histidine and tryptophan amino acids).
OsbZIP48 Lacks a Transactivation Domain
Many bZIP transcription factors like OsbZIP16 are known to have transactivation domains through which they can activate the transcription machinery of the cell (Chen et al., 2012). However, there is a debate on whether AtHY5 can act as a transcriptional activator or repressor. AtHY5, in fact, does not show transcription activation in yeast cells (Ang et al., 1998), but ChIP-chip analysis of AtHY5 suggested that it can act both as an activator and a repressor of a rather large number of genes (Zhang et al., 2011a). Therefore, transactivation analysis of OsbZIP48 in yeast was carried out (Fig. 4E). This assay showed that, while the yeast cells containing the positive control grew on SD-HW medium, the cells containing the OsbZIP48-pGBKT construct did not grow on SD-HW medium, indicating that OsbZIP48 does not have a functional transactivation domain.
OsbZIP48 Is a Functional Ortholog of AtHY5
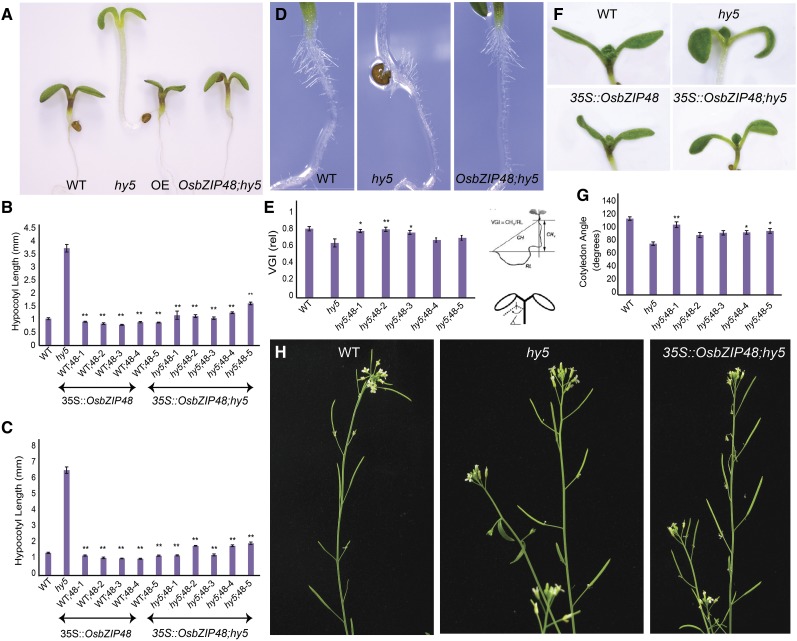
To find out whether OsbZIP48 is functionally similar to Arabidopsis HY5, complementation of the Arabidopsis hy5 mutant was carried out by overexpressing OsbZIP48 in the hy5 mutant. The homozygous transgenic lines were checked for the expression of OsbZIP48 by real-time PCR and for the presence of the hygromycin (hptII) gene by PCR (Supplemental Fig. S3). Since the Arabidopsis hy5 mutant showed an elongated hypocotyl phenotype in light, the homozygous transgenic lines of OsbZIP48/hy5 (mutant background) were checked for hypocotyl length in 3- and 6-d-old white light-grown seedlings. As shown in Figure 5, A and B, the average hypocotyl lengths of a 3-d-old wild-type, hy5 mutant, and OsbZIP48 overexpressed in hy5 mutant background (OsbZIP48;hy5) plants were approximately 1, 3.7, and 1.2 mm, respectively, indicating that OsbZIP48 can compensate for the loss of function of HY5 in the Athy5 mutant. Arabidopsis transgenics overexpressing OsbZIP48 in the Col-0 (wild-type) background (OsbZIP48OE) were generated, and their hypocotyl length was measured under white light. While the average 3-d-old wild-type hypocotyl was approximately 1 mm, the average OsbZIP48OE hypocotyl length was approximately 0.86 mm (Fig. 5B). Since Deng and coworkers had done most of the hypocotyl length measurements in 6-d-old seedlings (Holm et al., 2002), the hypocotyl length of the transgenics also was measured in 6-d-old seedlings (Fig. 5C). While the average hypocotyl lengths of the wild type and the hy5 mutant were approximately 1.4 and 6.6 mm, respectively, the average hypocotyl lengths of OsbZIP48OE and OsbZIP48;hy5 seedlings were approximately 1.2 and 1.6 mm, respectively (Fig. 5C). No significant difference in hypocotyl length was observed in the 3-d-old dark-grown seedlings (data not shown).
Figure 5.
Phenotypic analyses of Arabidopsis hy5 mutant seedlings/plants overexpressing OsbZIP48. A, Phenotypes of 3-d-old white light-grown wild-type (WT), hy5, OsbZIP48OE, and OsbZIP48;hy5 seedlings. B and C, Hypocotyl lengths of 3- and 6-d-old white light (200 μmol m−2 s−1)-grown wild-type, hy5, OsbZIP48OE, and OsbZIP48;hy5 seedlings. D and E, VGI of roots of 3-d-old wild-type, hy5, and OsbZIP48;hy5 seedlings. F and G, Cotyledon opening angle in response to white light. H, Altered gravitropic set angle in siliques of Arabidopsis hy5 mutant plants. Data presented are means ± se, n = 15 plants in each case. Statistically significant differences (*, P < 0.05 and **, P < 0.005) were identified by Dunnett’s test using the wild type as a control for overexpression transgenics and the hy5 mutant as a control for OsbZIP48;hy5 transgenics in B and C and the hy5 mutant as a control in E and G.
OsbZIP48 Is Able to Rescue the Agravitropic Response of the hy5 Mutant
The hy5 mutant seedlings are known to lack a proper gravitropic response (Oyama et al., 1997; Sibout et al., 2006). In order to check whether OsbZIP48 is able to alter the gravitropic response of the hy5 mutant and make it respond in a manner similar to the wild type, the vertical growth index (VGI) was used for the quantitative analysis of root morphology (Fig. 5, D and E). The VGI can be defined as the ratio between a vertical projection of the base-to-tip chord and the root length (Vicente-Agullo et al., 2004). On measuring the VGI of 3-d-old wild-type, hy5 mutant, and OsbZIP48;hy5 seedlings, it was found that, while the average VGI of the wild type was 0.82, the VGI of hy5 and OsbZIP48;hy5 was 0.65 and 0.76, respectively. In fact, some of the transgenics showed VGI similar to the wild type, indicating that OsbZIP48 overexpression is able to rescue the gravitropic response in hy5 mutant roots to the normal wild-type phenotype. Interestingly, the root hairs of the hy5 mutant were found to be agravitropic, and OsbZIP48 was able to rescue this response as well (Fig. 5D). The cotyledon angle of hy5 mutant seedlings also was less as compared with the wild type, and OsbZIP48 was able to complement the loss of AtHY5 in the transgenics in this respect too (Fig. 5, F and G).
In addition to altered root gravitropic response, we found that the gravitropic set angle of siliques in hy5 plants was different from that of the wild type (Fig. 5H), which probably did not come to the notice of previous researchers. In mature plants, the lateral organs generally do not remain parallel to the gravity vector but tend to exist at a particular angle with respect to the vertical growth axis (Wei et al., 2010). The angle between the pedicel and the inflorescence stem influences both the architecture and the yield potential of the plants (Wang and Li, 2008). This stem-pedicel angle of the siliques was larger in the hy5 mutant as compared with the wild type. The stem-pedicel angle of OsbZIP48;hy5 plants was similar to that of the wild type.
OsbZIP48 Complements the hy5 Mutant in Restoring Anthocyanin and Chlorophyll Content
HY5 is known to be involved in anthocyanin and chlorophyll biosynthesis, and hy5 mutant seedlings show reduced greening in the middle and lower parts of the hypocotyl (Oyama et al., 1997; Chattopadhyay et al., 1998). Therefore, anthocyanin and chlorophyll contents of wild-type, hy5 mutant, OsbZIP48OE, and OsbZIP48;hy5 Arabidopsis seedlings were estimated (Supplemental Fig. S4). The anthocyanin contents of 3- and 6-d-old seedlings of OsbZIP48OE were similar to those of the wild type, indicating that overexpression of full-length OsbZIP48 does not cause any increase in anthocyanin content in Arabidopsis seedlings (Supplemental Fig. S4, B and C). The chlorophyll content of 6-d-old OsbZIP48OE transgenics was similar to that of the wild type (Supplemental Fig. S4D). In 3-d-old OsbZIP48;hy5 seedlings, although the anthocyanin content was greater than that in hy5 mutant seedlings, it was comparatively less than that in the wild type (Supplemental Fig. S4B). However, the anthocyanin content of 6-d-old OsbZIP48;hy5 seedlings was almost similar to that of the wild type in three of the five transgenic lines analyzed (Supplemental Fig. S4C). The chlorophyll content of 6-d-old OsbZIP48;hy5 seedlings was more than that of the hy5 mutant but was comparatively less than that of the wild type (Supplemental Fig. S4D).
OsbZIP48OE Rice Transgenics Display a Semidwarf Phenotype
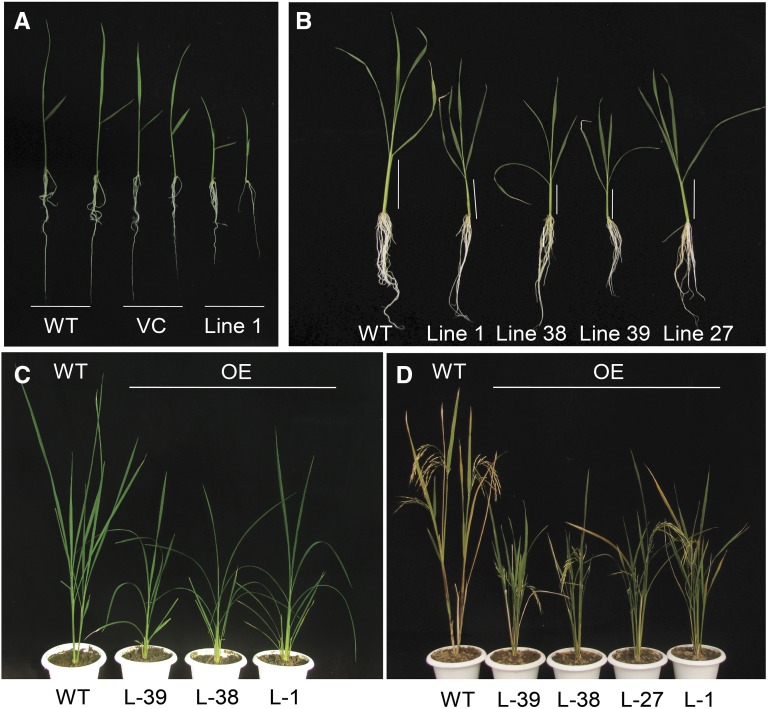
In addition to functional complementation of the hy5 mutant of Arabidopsis by its rice ortholog OsbZIP48, as described above, it was imperative to generate rice transgenics to decipher if OsbZIP48 performs any additional or unique functions in a monocot system like rice. Thus, overexpression transgenics of OsbZIP48 were raised in rice. The level of expression of OsbZIP48 in the leaves of mature plants for overexpression lines was checked and was indeed significantly higher than in the wild type (Supplemental Fig. S5). Southern-blot analysis was performed using the hptII gene as the probe. The genomic DNA of the wild type, vector control, and transgenics was digested by EcoRI restriction enzyme, which is a noncutter of the hygromycin gene. The probe was made using the hptII gene as the template. As a result, the bands on the Southern blot correspond to the hptII gene, which, in turn, indicate the number of inserts present in the transgenic line. As is evident from Supplemental Figure S5, no bands can be seen in the wild type lane of the Southern blot, while three bands are visible in the vector control (pB4NU vector), indicating that there are three inserts in the vector control. Line 1 has one, line 38 has two, and line 39 has three bands, indicating that there may be as many corresponding insertions of the OsbZIP48 coding sequence (CDS) in these three lines. Since no two transgenics have the bands corresponding to the transgene in the same position(s), one can conclude that each transgenic line represents independent event(s) (Supplemental Fig. S5).
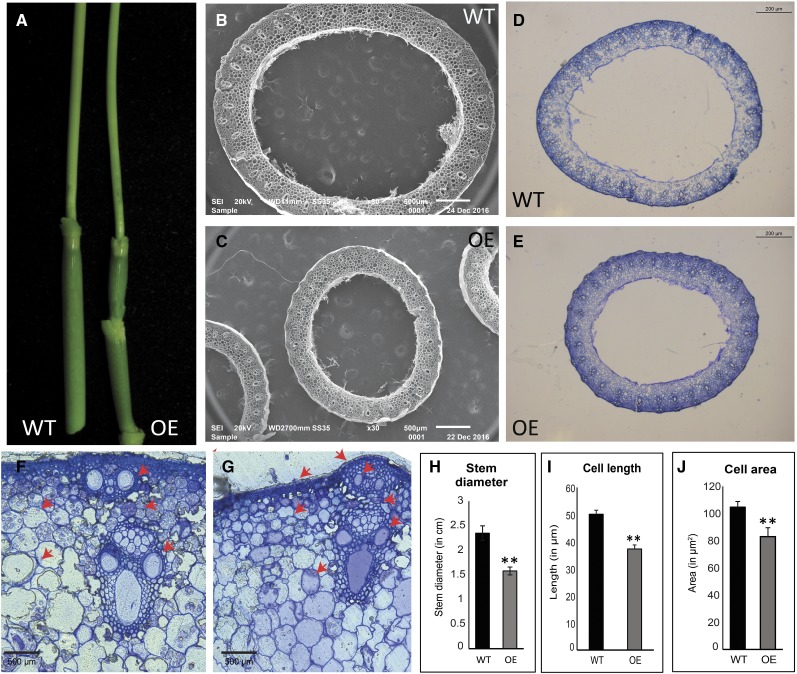
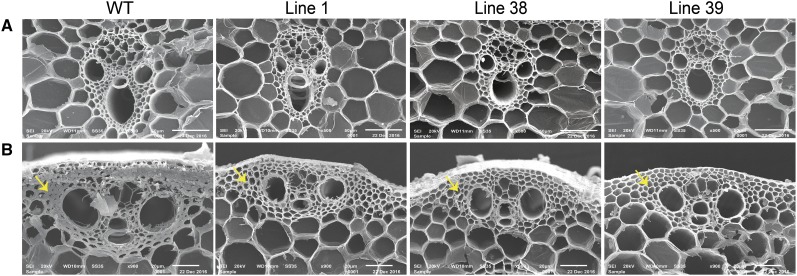
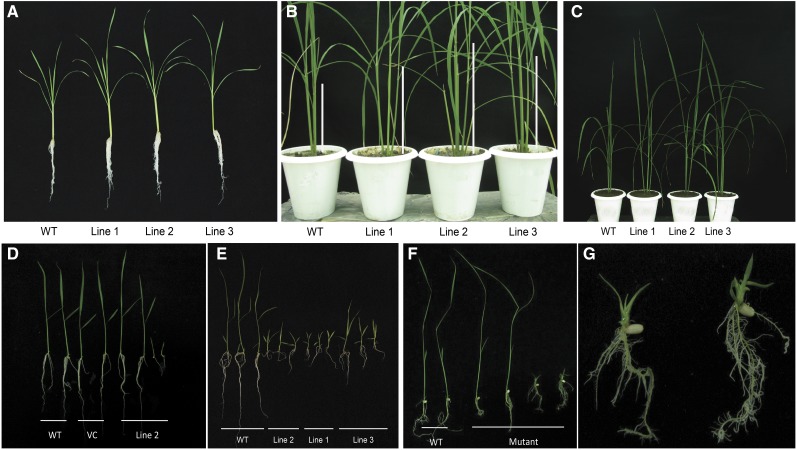
The rice OsbZIP48OE transgenic seedlings displayed a semidwarf phenotype and were distinctly shorter than the wild-type seedlings (Fig. 6, A and B). Even the adult OsbZIP48OE vegetative plants and those bearing panicles were shorter in height as compared with the wild type (Fig. 6, C and D). The overexpression transgenics also were greener for a slightly longer time than the wild type (Fig. 6D). The overexpression of OsbZIP48 resulted in reduction of the internode length as well as the panicle length of the rice plants (Supplemental Fig. S6, B and C). The reduction in plant height was accompanied by reduction in the stem width, as the stem of mature OsbZIP48OE transgenics was thinner than that of the wild type (Fig. 7, A–E). Quantitative analysis of the width of the stem of the wild type and OsbZIP48OE transgenics and of the cell length and cell area showed that there was an overall reduction in the stem width and cell size in overexpression transgenics (Fig. 7, F–J; Supplemental Fig. S7). The transgenics showed smaller vascular bundles and less secondary cell wall thickening as compared with the wild type (Figs. 7, F and G, and 8). Since OsbZIP48OE transgenics showed reductions in internode length and panicle length, a detailed morphometric analysis of these transgenics was performed, and parameters like total plant height, culm length, flag leaf length, panicle length, total number of florets, and total number of fertile florets were considered and measured according to International Rice Research Institute (IRRI) guidelines (Supplemental Figs. S8 and S9). While the average plant height of the wild type was approximately 82 cm, the average plant height of OsbZIP48OE transgenics was approximately 59 cm, about a 32% reduction. Similarly, the culm of OsbZIP48OE showed a 36% reduction as compared with the wild type, with the wild type and OsbZIP48OE having an average culm length of approximately 51 and 32 cm, respectively. There was no change in the flag leaf length of most of the transgenics as compared with the wild type, but the transgenics showed a 19% reduction in the length of their panicle, on average, as compared with the wild type. This indicated that OsbZIP48 overexpression mainly affects the length of the stem and panicle. However, there was close to a 38% reduction in the total number of florets in the panicle, with the average number of florets per panicle in the wild-type plants and transgenics being 73 and 45, respectively. The lines that had much higher expression of OsbZIP48 (lines 38 and 39) had a high degree of sterility. While the fertility percentage of the wild-type plants was 76%, the average fertility percentage in these two transgenics was 47%. However, one transgenic event (line 1 with a single insert), which had only a 2.4-fold increase in OsbZIP48 expression as compared with the wild type, had a fertility percentage nearly similar to that of the wild type.
Figure 6.
Phenotypes of OsbZIP48OE rice transgenics at different developmental stages. A, Photograph of 10-d-old seedlings of the wild type (WT), pB4NU vector control (VC), and OsbZIP48OE transgenics grown in white light (75 μmol m−2 s−1). B, Photograph of 30-d-old seedlings grown in white light (75 μmol m−2 s−1). C, Photograph of plants at the vegetative phase of life. D, Photograph of plants grown in a greenhouse at the reproductive stage.
Figure 7.
Phenotypic comparison of the stems of wild-type (WT) and OsbZIP48OE rice transgenic plants. A, Photograph showing the difference in the stem diameter of mature green plants. B and C, Scanning electron microscopic images showing differences in the diameter of the stems of wild-type and OsbZIP48OE transgenic plants, respectively. D and E, Methylene Blue-stained transverse sections of wild-type and OsbZIP48OE transgenic stems showing differences in the diameter of the stems taken at 2.5× magnification. F and G, Methylene Blue-stained transverse section of wild-type and OsbZIP48OE transgenic stems, with red arrows showing differences in the size of the vascular bundle, parenchyma cells, and the secondary cell wall thickening of the sclerenchyma cells. H to J, Histograms showing differences in stem diameter, cell length, and cell area of the wild type and overexpression transgenics, respectively. Cortical cells were used to measure cell length and cell area. Data presented are means ± se, n = 10 in each case. Statistically significant differences (*, P < 0.05 and **, P < 0.005) were identified by Student’s t test.
Figure 8.
Scanning electron microscopic images of second last internodes of wild-type (WT) and OsbZIP48OE transgenic plants at different magnifications showing the size of vascular bundles and secondary cell wall thickenings. In B, the yellow arrows show the thickness of secondary cell wall thickenings in wild-type and OsbZIP48OE transgenic plants.
Whether the decrease in height (semidwarfism) of OsbZIP48OE transgenics is because of a uniform reduction of all internodes or some particular internode(s), the length of internodes was measured. Although there was no major change in the number of nodes in most of the transgenics as compared with the wild type, a reduction of 48% was recorded in the length of the second last internode followed by the last internode (30% reduction) in the transgenics as compared with the wild type. The first internode showed the least reduction (Supplemental Fig. S9). There was not much difference in the total number of tillers, as the average tiller number of wild-type plants and of transgenics varied between three and four under the greenhouse conditions maintained (Supplemental Fig. S9). The overexpression transgenics appeared to be greener than the wild type; therefore, their chlorophyll content was estimated. The chlorophyll content in overexpression transgenics was found to be more than that in the wild type (Supplemental Fig. S10). The overexpression of OsbZIP48 also resulted in reduced secondary cell wall thickenings, as revealed by scanning electron microscopy. All three transgenic lines showed reduced secondary cell wall thickenings in sclerenchyma cell walls as compared with the wild type, indicating that cell wall composition might be altered (Fig. 8).
OsbZIP48KD Rice Transgenics Are Lethal
The RNA interference (RNAi) technique was used to create OsbZIP48 knockdown (OsbZIP48KD) lines. The PCR-based confirmation of these transgenics and real-time PCR analysis were carried out to measure the degree of silencing in white light-grown transgenic seedlings (Supplemental Fig. S5). The T1 OsbZIP48KD transgenics showed profuse root growth at the seedling stage, and the plants were slightly taller than the wild type in the contained greenhouse environment (Fig. 9, A–C). However, from the T2 generation onward, two types of seedlings were seen segregating among the populations of RNAi transgenics. One set of seedlings (referred to as Hh) showed similar morphological characters to the wild type but were hygromycin positive. These seedlings showed a growth pattern similar to the wild type when they were shifted to the greenhouse. The other set of seedlings (referred to as hh) showed retarded growth and less chlorophyll content and perished within 1 month on Murashige and Skoog (MS) basal medium (Fig. 9, D and E); these seedlings were hygromycin positive, as they were PCR positive for the hptII gene. The wild-type seedlings died within 5 to 10 d of transfer to hygromycin-containing MS basal medium. Also, the OsbZIP48 transcript levels were reduced drastically in the RNAi transgenic seedlings that perished compared with the ones that were similar to the wild type (Supplemental Fig. S5). Seedlings that resembled wild-type seedlings in the T2 generation, and in the next generation, also showed the similar two-phenotype segregating pattern; even after reaching to the T4 generation, the same segregation pattern was observed. The chlorophyll content in 20-d-old OsbZIP48KD transgenics (showing retarded growth) was considerably less as compared with the wild type (Supplemental Fig. S10).
Figure 9.
Phenotypes of OsbZIP48KD lines and the T-DNA insertion mutant of OsbZIP48. A, T1 OsbZIP48KD lines showing profuse root growth at the seedling stage as compared with the wild type (WT). B and C, T1 OsbZIP48KD plants at the vegetative stage showing increased height at two different magnifications; note the elongated stem of OsbZIP48KD plants in B. D, T2 OsbZIP48KD transgenics showing two types of plants; seedlings of the 10-d-old wild type, pANDA vector control (VC), and T2 OsbZIP48KD transgenics grown in white light (75 μmol m−2 s−1). E, T2 OsbZIP48KD 10-d-old transgenics showing a lethal phenotype in multiple lines. F, Photograph of 15-d-old wild-type and OsbZIP48 mutant lines showing their phenotypes. G, T-DNA insertional mutant plants showing profuse rooting (at higher magnification).
Phenotypic Analysis of the OsbZIP48 T-DNA Insertional Mutant
In order to further evaluate the functional role of OsbZIP48, analysis of a T-DNA insertional mutant obtained from Rice Functional Genomic Express database was carried out. When grown on MS basal medium containing hygromycin, these plants showed a growth pattern similar to the T2 RNAi transgenic lines (OsbZIP48KD). Two types of seedlings were obtained, one that was similar to the wild-type seedlings and one that exhibited a lethal phenotype. It lacked any internode and showed bleached spots on the leaves and profuse root growth (Fig. 9, F and G). However, it was able to grow to a limited extent on MS basal medium containing Suc but was not able to survive at all on medium that lacked Suc (i.e. the rice growth medium used in this study for raising transgenics). The phenotypic analysis of the T-DNA insertional mutant and OsbZIP48KD plants suggests that OsbZIP48 is crucial for proper seedling development and its transition to the adult plant and that its drastic reduction or total absence can be lethal.
Microarray Analysis of OsbZIP48 Overexpression and RNAi Transgenics
Since OsbZIP48 is a transcription factor, its major mode of action would be to alter the expression of downstream genes. In order to get an insight into the mechanism of action of OsbZIP48 through which it regulates such an array of developmental events, whole-genome microarray analysis was performed for the overexpression as well as RNAi transgenics along with respective vector controls (pB4NU for overexpression and pANDA for RNAi). Microarray analysis of both types of RNAi transgenics was performed, and those that resembled wild-type seedlings in phenotype were labeled as Hh and those that showed the lethal phenotype were labeled as hh.
OsbZIP48OE Alters the Expression of Hormone Biosynthesis Pathway Genes
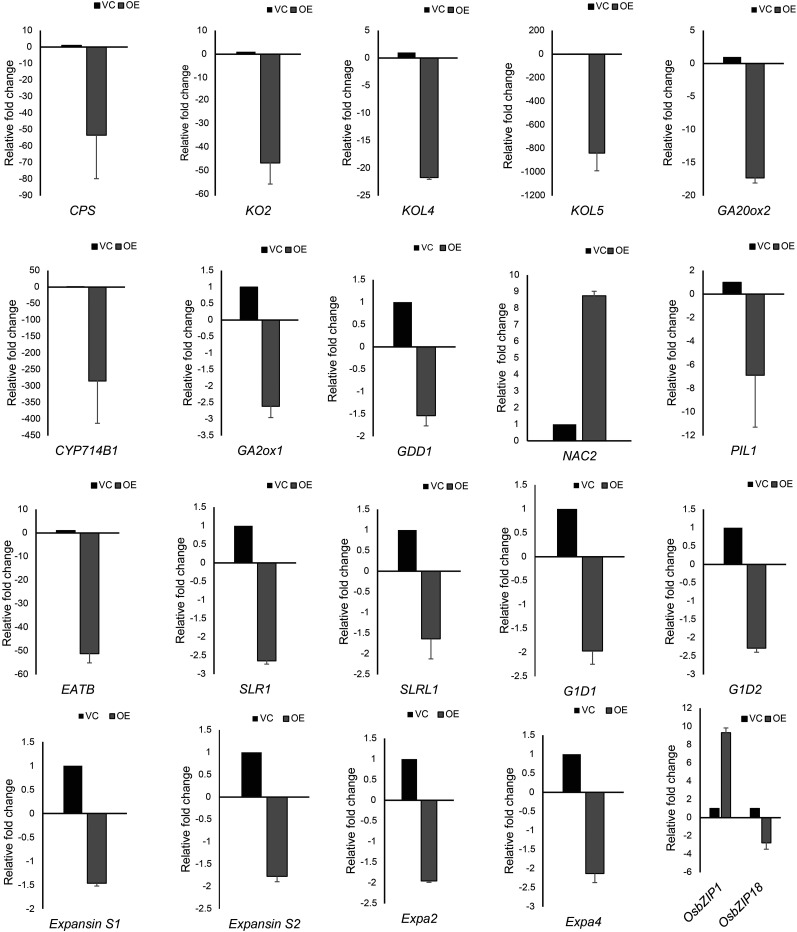
Microarray analysis of 10-d-old white light (75 μmol m−2 s−1)-grown overexpression transgenic seedlings showed that close to 1,741 genes were differentially expressed as compared with the pB4NU vector control, with 251 genes up-regulated and 1,490 genes down-regulated. The overexpression of OsbZIP48 caused the repression of genes associated with GA, jasmonic acid, brassinosteroid, and ethylene biosynthesis pathways (Fig. 10; Supplemental Fig. S11). However, no significant change in the expression of genes involved in abscisic acid (ABA) and auxin biosynthesis/signaling pathways was detected in our analysis. The down-regulation of cytokinin glucosylation also was observed in OsbZIP48OE transgenics as compared with the vector control. Cytokinins can be glucosylated to form O-glucosides and N-glucosides, which are inactive and involved in hormone homeostasis (Hou et al., 2004). This pathway was down-regulated in OsbZIP48OE transgenics and may have resulted in more accumulation of free cytokinins (Supplemental Fig. S12). This might explain the slightly delayed senescence observed in OsbZIP48OE transgenic plants as compared with the wild type, as stated earlier. Strikingly, among two of the homologs of OsbZIP48 in rice, OsbZIP01 (another AtHY5 homolog in rice) was found to be up-regulated by 9-fold in OsbZIP48OE transgenics as compared with the vector control, whereas OsbZIP18 (another AtHY5 homolog in rice) was found to be down-regulated by 2.7-fold (Fig. 11).
Figure 10.
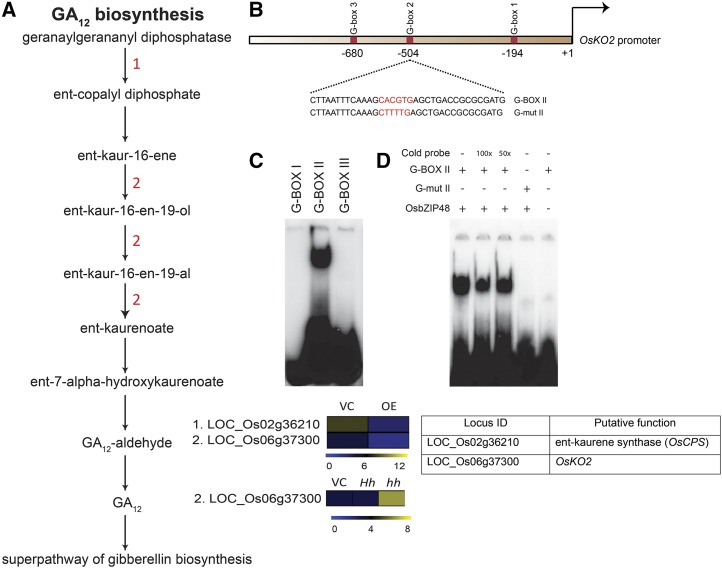
A, Schematic representation of the GA biosynthesis pathway showing altered gene expression in OsbZIP48OE and OsbZIP48KD transgenics. The color bar at the base represents log2 expression values, with blue representing low-level expression, black representing medium-level expression, and yellow signifying high-level expression. The numbers in red in the pathway correspond to the serial number of the locus identifier in the heat map (i.e. the gene with the serial number performs in the step where it is mentioned in the pathway). VC, Vector control. B, Schematic representation of the OsKO2 1-kb promoter showing the location of three G-box motifs. In the diagram, the sequence of the probe is given with the G-box sequence (CACGTG) highlighted in red. The G-mut II probe sequence shows the mutated G-box sequence and is highlighted in red. These probes were labeled with 32P for electrophoretic mobility shift assay (EMSA). C, OsbZIP48 binds in vitro to G-box II in the OsKO2 promoter in EMSA. D, EMSA gel showing OsbZIP48 in vitro binding to G-box II in the OsKO2 promoter with proper controls. For the assay, the radiolabeled probes were incubated with OsbZIP48 protein. Cold (unlabeled) probe (100× or 50×), G-box II probe, and G-mut II probe were used as indicated.
Figure 11.
Real-time PCR-based expression analysis of genes known to be involved in regulating plant height in OsbZIP48OE seedlings. The last histogram shows the transcript levels of OsbZIP1 and OsbZIP18 in the OsbZIP48OE line. Data shown are means ± se. VC, Vector control.
Altered Expression of Genes Involved in GA Biosynthesis and Signaling in OsbZIP48OE Transgenics
Microarray analysis and real-time PCR showed that OsCPS (LOC_Os02g36210) and OsKO2 (LOC_Os06g37300) were down-regulated in OsbZIP48OE transgenics (Figs. 10 and 11). A literature search was carried out to identify rice genes that are known to be involved in controlling plant height in rice. The list of genes involved in controlling rice plant height and their expression in overexpression transgenics is provided in Table I. The expression of some of them, including OsNAC2, OsGDD1, OsPIL1, and OsEATB, was checked (Li et al., 2011a; Qi et al., 2011; Todaka et al., 2012; Chen et al., 2015). The expression of OsNAC2 was high, but OsPIL1, OsGDD1, and OsEATB were down-regulated in these transgenics (Fig. 11).
Table I. List of genes known to be involved in regulating plant height in rice.
| Gene Name | Pathway Involved | Function | Expression Level in OsbZIP48OE | Literature |
|---|---|---|---|---|
| CPS | GA biosynthesis | Take part in the early steps of GA biosynthesis | Down-regulated | Yamaguchi (2008) |
| KO2 | GA biosynthesis | Take part in the early steps of GA biosynthesis | Down-regulated | Yamaguchi (2008) |
| OsKOL4 | GA biosynthesis | Take part in the early steps of GA biosynthesis | Down-regulated | Yamaguchi (2008) |
| OsKOL5 | GA biosynthesis | Take part in the early steps of GA biosynthesis | Down-regulated | Yamaguchi (2008) |
| OsGA20ox2 | GA biosynthesis | Convert GA12 into bioactive GA1 and GA4 | Down-regulated | Yamaguchi (2008) |
| OsGA2ox1 | GA biosynthesis | Convert bioactive GA1 into nonbioactive GAs | Down-regulated | Yamaguchi (2008) |
| OsGA13ox | GA biosynthesis | Convert GA12 to GA53, which is then converted to GA1 by OsGA20ox | Down-regulated | Yamaguchi (2008) |
| (OsCYP714B1)OsGA2ox1 | GA biosynthesis | Convert bioactive GA1 and GA4 in bioinactive GAs | Down-regulated | Yamaguchi (2008) |
| OsNAC2 | Transcription factor | Negative regulator of plant height; binds to the promoter of OsKO2 and represses its expression | Up-regulated | Chen et al. (2015) |
| OsGDD1 | Kinesin-like protein | Positive regulator of plant height; binds to the promoter of OsKO2 and promotes its expression | Down-regulated | Li et al. (2011a) |
| OsPIL1 | Phytochrome-interacting factor-like protein | Positive regulator of plant height | Down-regulated | Todaka et al. (2012) |
| OsEATB | Rice ethylene-response AP2/ERF factor | Restrict the ethylene-induced enhancement of GA responsiveness by down-regulating the GA biosynthetic gene, ent-kaurene synthase A | Down-regulated | Qi et al. (2011) |
| OsEXPANSIN S1 | Cell wall-related genes | Regulate cell elongation by cell wall expansion, which is caused by acid-induced cell wall relaxation | Down-regulated | Todaka et al. (2012) |
| OsEXPANSIN S2 | Cell wall-related genes | Regulate cell elongation by cell wall expansion, which is caused by acid-induced cell wall relaxation | Down-regulated | Todaka et al. (2012) |
| OsEXPA2 | Cell wall-related genes | Regulate cell elongation by cell wall expansion, which is caused by acid-induced cell wall relaxation | Down-regulated | Todaka et al. (2012) |
| OsEXPA4 | Cell wall-related genes | Regulate cell elongation by cell wall expansion, which is caused by acid-induced cell wall relaxation | Down-regulated | Todaka et al. (2012) |
| OsSLR1 | Involved in GA signaling | Central repressor of GA signaling | Down-regulated | Hauvermale et al. (2012) |
| OsSLRL | Involved in GA signaling | Central repressor of GA signaling | Down-regulated | Hauvermale et al. (2012) |
| OsGID1 | Involved in GA signaling | Positive regulator of GA signaling | Down-regulated | Hauvermale et al. (2012) |
| OsGID2 | Involved in GA signaling | Positive regulator of GA signaling | Down-regulated | Hauvermale et al. (2012) |
The real-time PCR analysis revealed that the expression of OsGA20ox2, OsGA2ox1, and the KO-like genes OsKOL4 and OsKOL5 in OsbZIP48OE transgenics was down-regulated (Fig. 11). OsCYP714B1, which encodes GA 13-oxidase, also was significantly down-regulated. The expression of genes that are involved in GA signaling, like OsSLR1, OsSLRL, OsGID1, and OsGID2, also was checked. OsSLR1 and OsSLRL1 are central repressors of GA signaling, while OsGID1 and OSGID2 are positive regulators of GA signaling (Hauvermale et al., 2012). The expression of OsSLR1, OsSLRL1, OsGID1, and OsGID2 was down-regulated. An essentially similar trend also was observed with regard to the expression of OsEXPANSIN S1, OsEXPANSIN S2, OsEXPA2, and OsEXPA4 genes (Fig. 11).
Changes in Gene Expression in RNAi Transgenics
Microarray analysis of OsbZIP48 RNAi transgenics (OsbZIP48KD) showed altered expression of 1,857 genes, with 856 and 1,001 genes showing up-regulation and down-regulation, respectively. The heat map of 10-d-old hh, Hh, and pANDA vector control seedlings grown in white light (75 μmol m−2 s−1) showed that the degree of differential expression of these genes was greater in hh seedlings as compared with Hh seedlings (Supplemental Fig. S13). This also corresponds to the phenotype that these two seedlings show: hh seedlings correspond to the RNAi seedlings, which show a lethal phenotype and, therefore, exhibit a greater degree of differential expression; Hh seedlings are phenotypically similar to the wild type and, therefore, do not show any drastic difference in their expression pattern as compared with the wild type.
In OsbZIP48KD lines, the activation of GA, jasmonic acid, and indole-3-acetic acid (IAA) biosynthesis pathways must have occurred, as genes encoding for enzymes involved in these pathways showed higher expression as compared with the vector control (Fig. 10; Supplemental Fig. S14). In relation to GA12 biosynthesis, while two genes (LOC_Os02g36210 and LOC_Os06g37300) were down-regulated in OsbZIP48OE transgenics, only one of them (LOC_Os06g37300 or OsKO2) was up-regulated in OsbZIP48KD transgenics. With respect to jasmonic acid biosynthesis, four genes encoding for the enzymes involved in this pathway showed altered expression in comparison with the vector control. Only one gene (LOC_Os08g39850) is common in OsbZIP48OE and OsbZIP48KD transgenics, and it showed decreased expression in OsbZIP48OE transgenics and its expression was high in OsbZIP48KD transgenics. For the brassinosteroid biosynthesis pathway, one gene (LOC_Os01g01650) showed reduced expression in OsbZIP48KD transgenics as compared with the vector control, and this gene is different from the genes altered in OsbZIP48OE transgenics but catalyzes the same step. IAA biosynthesis pathway genes were not altered in OsbZIP48OE transgenics but were up-regulated in OsbZIP48KD transgenics. Thus, down-regulation of OsbZIP48 does indeed alter the homeostasis of major hormonal pathways. Real-time PCR confirmation of the changes in expression of genes involved in hormonal biosynthesis pathways and showing differential expression in overexpression and RNAi transgenics by microarray also was done, and these data are presented in Supplemental Figures S15 and S16. The expression of two other homologs of OsbZIP48 (i.e. OsbZIP01 and OsbZIP18) was not altered significantly in OsbZIP48KD transgenics as compared with the vector control (Supplemental Fig. S16).
OsbZIP48 Binds Directly to the Promoter of OsKO2 through a G-Box Element
Since OsKO2 was found to be down-regulated in OsbZIP48OE and up-regulated in OsbZIP48KD (Fig. 10A), promoter analysis of OsKO2 was performed manually, as it could be a direct target of OsbZIP48. The analysis revealed that there are three G-box-binding elements (CACGTG) within the 1-kb region upstream of the transcription start site of OsKO2 gene (Fig. 10B). EMSA showed that OsbZIP48 binds to the G-box at −504 bp (G-box II) from the transcription start site (Fig. 10C) and not to the other two G-box elements. In the same assay, we were able to demonstrate that OsbZIP48 binds to the labeled probe containing this G-box but not the mutated version (G-mut II) where the G-box had been disrupted (Fig. 10D). This indicates that OsbZIP48 binds directly to the promoter of OsKO2 and may regulate its expression.
DISCUSSION
AtHY5 Has Three Homologs in Monocots
Phylogenetic tree and pairwise distance analyses of AtHY5 and AtHYH homologs across different plant species show that, while there are three AtHY5 homologs in monocots, AtHYH homologs are present only in gymnosperms and dicots. Also, the homologs in mosses and lycopods seem to be closer to AtHY5 than AtHYH, indicating that HYH might have evolved more recently as compared with HY5. OsbZIP48 represents one of the three homologs of AtHY5 in rice and has been functionally characterized in this study. The other two homologs of OsbZIP48 are OsbZIP1 and OsbZIP18 (Nijhawan et al., 2008), which have different expression profiles in the vegetative and reproductive tissues examined and may have undergone neofunctionalization; their functional validation will throw more light on the specific and/or overlapping functions they perform vis-à-vis OsbZIP48.
OsbZIP48 Has an Expression Profile and Molecular Characteristics Essentially Similar to AtHY5
OsbZIP48 was found to have maximum expression in 1-DAP stigma, although it is expressed in prepollination stigma, P6 panicle stage, and the first internode of rice plants. In Arabidopsis too, the maximum expression of AtHY5 has been reported in the floral organs and stem of the plant (Oyama et al., 1997), indicating that, in terms of expression pattern, OsbZIP48 resembles AtHY5 to a large extent. GAs are known to be required for seed development and pollen tube growth in Arabidopsis (Singh et al., 2002), and HY5 is known to regulate GA levels (Weller et al., 2009). In our analysis, we have found that there is high level of expression of OsbZIP48 in the stigma. The genes involved in GA biosynthesis are down-regulated in OsbZIP48OE transgenics. This indicates that OsbZIP48 might play a role in pollen tube elongation and fertilization by modulating GA levels. The expression of OsbZIP48 was found to be high in 5-d-old dark- and light-grown seedlings, indicating that the expression of OsbZIP48 is developmentally regulated instead of being regulated by light. OsbZIP48 is nucleus localized and lacks transactivation activity as assayed using the yeast system; this is akin to the observation made for AtHY5 (Ang et al., 1998; Chattopadhyay et al., 1998; Stracke et al., 2010).
OsbZIP48 Protein Accumulation Is Developmentally Regulated But It Is Not Light Labile
Western-blot analysis of total protein extracts from light- and dark-grown rice seedlings as well as from different developmental stages of rice showed that anti-OsbZIP48 antibody recognizes a protein band running at an apparent molecular mass of 30 kD, which is similar to what was observed in the case of the HY5 protein in Arabidopsis (Osterlund et al., 2000). OsbZIP48 was expressed in the bacterial system, and the protein purified was used as a positive control in order to confirm its migration in SDS-PAGE.
AtHY5 protein levels were found to be maximum 2 to 3 d after seed germination, and after that they declined gradually (Hardtke et al., 2000). In contrast, OsbZIP48 protein levels in rice were found to be maximum 5 to 7 d after seed germination and, like AtHY5, declined gradually thereafter. In 3- to 10-d-old dark-grown rice seedlings, levels of OsbZIP48 were rather high. It is worth mentioning here that, as compared with photomorphogenesis in dicots like Arabidopsis, monocots undergo partial photomorphogenesis in the dark. In monocots like rice, first and second leaf emergence takes place in the dark (Zhang et al., 2006). It is possible that the accumulation of OsbZIP48 might play a role in partial photomorphogenesis in the dark during rice seedling development, as its levels increase in 5-d-old seedlings as compared with 3-d-old seedlings grown in the dark.
HY5 protein levels change within 5 h of light-to-dark and dark-to-light transitions in Arabidopsis, with its levels decreasing drastically within 20 h of light-to-dark transition and, similarly, increasing within 20 h of dark-to-light transition, which shows that HY5 levels are light regulated (Osterlund et al., 2000). However, unlike AtHY5, light-to-dark and dark-to-light transition experiments wherein 4-d-old rice seedlings grown in continuous light or dark conditions were transferred to opposite light/dark conditions for 5, 10, 15, and 20 h showed no drastic change in OsbZIP48 protein levels; the OsbZIP48 protein levels were found to be unaffected by the presence or absence of light. OsbZIP48 protein levels did not change even when hy5 mutant seedlings complemented with OsbZIP48 were subjected to light-to-dark transition, implying that OsbZIP48 is not degraded in the dark by AtCOP1 as well. This might be due to the slightly different amino acid composition of the conserved COP1-binding domain of OsbZIP48 as compared with that of AtHY5. Despite being an ortholog of AtHY5 and having a COP1-binding domain, it is intriguing that OsbZIP48 is not degraded in the dark. COP1 plays a major role in the light regulation of HY5 in Arabidopsis. In the dark, COP1 localizes in the nucleus and binds to HY5, thus causing proteasome-mediated degradation of HY5 in the nucleus (Oyama et al., 1997; Ang et al., 1998; Osterlund et al., 2000), whereas in the light, it localizes to the cytoplasm (von Arnim and Deng, 1994). It is likely that the mechanism of regulation of the HY5 ortholog in rice might be different from that in Arabidopsis.
OsbZIP48 protein levels also were checked during different developmental stages of rice, and it was found to be present in tissues representing all the stages of panicle development, from P1 to P6. The protein levels were high in P3 and P4 stages, while real-time PCR analysis showed its transcript levels to be maximum in P6 stage. This indicates that OsbZIP48 levels are regulated at both transcriptional and translational levels during panicle development; however, the significance of these changes in expression of OsbZIP48 during various stages of panicle development remains to be elucidated. Interestingly, the protein levels were found to vary during seed development, being high in the initial stages of seed development (S1 and S2), declining gradually in later stages (S3 and S4), and were hardly detectable in the last stage of seed development (S5). This shows that OsbZIP48 accumulation starts after seed germination and peaks during early seedling development, after which it declines gradually, and its accumulation resumes during panicle development and continues until early stages of seed development. This correlates with the transcript profile of OsbZIP48 checked by real-time PCR. To a large extent, it appears that the protein profile of OsbZIP48 is similar to that of AtHY5, which also is known to accumulate in the inflorescences of Arabidopsis and is hardly detectable in the leaves, shoots, and siliques (Hardtke et al., 2000).
OsbZIP48 Can Functionally Complement the hy5 Mutant
When overexpressed in the hy5 mutant of Arabidopsis, OsbZIP48 was able to complement the hy5 mutant with respect to hypocotyl elongation growth in the light, indicating that it is a functional ortholog of AtHY5. However, overexpression of OsbZIP48 in the Arabidopsis Col-0 wild type showed no significant effect on hypocotyl elongation growth or anthocyanin and chlorophyll content in comparison with the wild type. Morphometric analysis of Arabidopsis hy5 mutant seedlings showed that AtHY5 might be involved in root hair gravitropic response and silique gravitropic set angle (observation made in this study) in addition to the agraviotropic root response that has already been reported previously (Oyama et al., 1997; Sibout et al., 2006). It is surprising that alteration in the positioning of root hairs and silique angle in hy5 mutant plants has escaped notice in previous studies. OsbZIP48 overexpression could functionally complement these traits as well. Arabidopsis hy5 mutant roots are known to show reduced basipetal auxin transport, which is consistent with their agravitropic response (Sibout et al., 2006). However, the genes involved in the agraviotrpic response of the hy5 mutant and the mode of action through which AtHY5 controls the gravitropic response have not been completely elucidated. Auxin is known to play an important role in regulating root gravitropism, and the involvement of polar auxin transport is well documented in this response. Polar auxin transport is mediated mainly by membrane-localized auxin influx and efflux carriers like AUX1/LAX proteins, PIN-formed proteins, and the multidrug resistance/p-glycoprotein class of ATP-binding cassette auxin transporters (Petrásek et al., 2006; Bandyopadhyay et al., 2007; Swarup et al., 2008; Zhang et al., 2011b). Among the components of polar auxin transport, AUX1, PIN2, PIN3, PIP5K (phosphatidylinositol monophosphate 5-kinase), and PINOID (PID) proteins have been found to be involved in root gravitropic responses (Bennett et al., 1996; Müller et al., 1998; Friml et al., 2002; Sukumar et al., 2009; Mei et al., 2012). ChIP-chip analysis showed that AtHY5 binds to the promoters of AUX1, PIN3, PIP5K, and PID (Lee et al., 2007). Therefore, it is quite possible that AtHY5 might regulate the root gravitropic response by altering the expression of these genes, which, in turn, might alter the polar auxin transport machinery of the plant.
OsbZIP48 Overexpression Does Not Alter Anthocyanin and Chlorophyll Content in Arabidopsis
In an earlier study, although overexpression of the full-length AtHY5 did not cause any increase in anthocyanin content in Arabidopsis, the partial CDS of AtHY5 (HY5-ΔN77) that lacked the COP1-binding domain caused increased anthocyanin accumulation in the upper region of the hypocotyl (Ang et al., 1998). HY5-ΔN77 transgenics showed more chlorophyll accumulation and precocious chloroplast development (Ang et al., 1998). In this study, the chlorophyll and anthocyanin content in Arabidopsis transgenics overexpressing OsbZIP48 were not affected significantly as compared with the wild type, although it could complement the hy5 mutant in practically all aspects of photomorphogenesis studied.
OsbZIP48 Controls Plant Height in Rice
The overexpression of OsbZIP48 in rice caused a decrease in plant height resulting in a semidwarf phenotype of the transgenics. This was caused due to the reduction in cell size, and maximum reduction was visible in the second last and last internodes. In wild-type rice plants, the expression of OsbZIP48 is least in the second last internode followed by the last internode and is maximum in the first internode. This is in accordance with the observation that, in wild-type plants, the last internode (which bears the panicle) is longest, followed by the second last internode, while the first internode is shortest in length. Therefore, the ectopic expression of OsbZIP48 caused maximum compression in the last and second last internodes, as the natural expression of OsbZIP48 is normally less in these two internodes. This observation is quite different from what is known for AtHY5, as overexpression of full-length AtHY5 does not show any significant difference in hypocotyl length of light-grown Arabidopsis seedlings as compared with the wild type, whereas the overexpression of truncated AtHY5, which lacks the COP1-binding domain, can shorten the hypocotyl (Ang et al., 1998). It is intriguing that only a partial HY5 CDS lacking the COP1-binding domain can cause hypocotyl reduction and increase in chlorophyll and anthocyanin accumulation in Arabidopsis, while in rice, the full-length CDS of its ortholog, OsbZIP48, can cause reduction in height and increase in chlorophyll accumulation. Moreover, both the rice T-DNA insertional mutant and RNAi lines of OsbZIP48 resulted in a lethal phenotype, although root proliferation in early stages of rice seedling development was profuse, a phenotype essentially similar to that of hy5 mutant seedlings. AtHY5 orthologs have been characterized in P. patens, L. japonicus, and P. sativum, and its role has been defined by analyzing the mutants defective in this gene. However, the overexpression transgenics in wild-type Arabidopsis have been raised only for PpHY5a lacking the COP1-binding domain and, like truncated AtHY5, the hypocotyl length was shorter than in the wild type. In fact, besides Arabidopsis, the overexpression transgenics of the full-length AtHY5 orthologs in the same organism have not been raised, and that makes it difficult for us to conclude whether full-length HY5 overexpression causes semidwarfism only in rice (this study) or in other organisms too.
Several Plant Hormone Pathways Are Altered in OsbZIP48OE and OsbZIP48KD Transgenics of Rice
AtHY5 is known to be involved in auxin, GA, cytokinin, ABA, ethylene, and strigolactone signaling; while it represses ethylene, GA, and auxin signaling, it promotes ABA signaling (Cluis et al., 2004; Vandenbussche et al., 2007; Alabadí et al., 2008; Chen and Xiong, 2008; Li et al., 2011b; Yu et al., 2013). In this study too, the expression of genes associated with the biosynthesis of GA, jasmonic acid, ethylene, and brassinosteroid as well as cytokinin glucosylation was altered in OsbZIP48OE transgenics. The overexpression of OsbZIP48 caused the repression of GA and ethylene biosynthesis-related genes. The HY5 of P. sativum was shown to bind to the promoter of GA2ox2 and increase its expression (Weller et al., 2009). In Arabidopsis also, chromatin immunoprecipitation-sequencing (ChIP-seq) analysis showed that AtHY5 binds to the promoter of AtGA2ox2 (Lee et al., 2007). GA2ox2 converts bioactive GA1 into bioinactive GA8. Thus, evidently, HY5 reduces the amount of active GA1, and consequently, that may cause a decrease in elongation of the hypocotyl. AtHY5 has been shown to bind to the promoter of AtERF11 and induce its expression, which, in turn, represses the ethylene biosynthesis pathway (Li et al., 2011b). In the microarray analysis carried out in this study, we found that the transcript levels of rice homologs of these two Arabidopsis genes were elevated in OsbZIP48OE transgenics, but their fold change was less than the 2-fold cutoff used in our analysis. We found that genes involved in GA12 biosynthesis were repressed by more than 2-fold. GA12 is the precursor for the synthesis of active GAs in rice. In addition to the repression of genes involved in GA biosynthesis, genes involved in GA signaling, like OsSLR1 and OsSLRL1, also were repressed. The expression of OsPIL1 was reduced significantly in OsbZIP48OE transgenics. OsPIL1 is known to be a positive regulator of internode elongation and causes internode elongation by increasing the cell size (Todaka et al., 2012). OsPIL1 does not alter GA levels but up-regulates the expression of cell wall-related genes like OsEXPANSIN S1, OsEXPANSIN S2, OsEXPA2, and OsEXPA4. In OsbZIP48OE transgenics too, the expression of OsEXPANSIN S1, OsEXPANSIN S2, OsEXPA2, and OsEXPA4 was reduced. Thus, OsbZIP48 might regulate plant height in two ways. First, it decreases the biosynthesis of active GA by inhibiting the expression of GA biosynthesis genes or stimulating genes involved in its catabolism; and second, it down-regulates the expression of OsPIL1, which, in turn, decreases the expression of cell wall-related genes coding for expansins, which then leads to a decrease in cell elongation. In addition, the expression of OsbZIP01 and OsbZIP18, which are the other two AtHY5 homologs in rice, was found to be up-regulated and down-regulated, respectively, in OsbZIP48OE transgenics. The significance of this observation will be realized only when these two genes are functionally characterized.
OsbZIP48 overexpression results in the down-regulation of OsGDD1. The gdd1 mutant also shows a phenotype similar to OsbZIP48OE transgenics. The gdd1 mutant is defective in GA biosynthesis and has reduced secondary cell wall thickenings (Zhang et al., 2010; Li et al., 2011a). Therefore, the reduction in secondary cell wall thickening in OsbZIP48OE might be because of the reduction of OsGDD1 expression.
In ethylene biosynthesis, the major role of OsbZIP48 could be to reduce the expression of genes encoding Tyr transaminase enzyme, as the expression of OsERF11 was increased only slightly as compared with the vector control. However, no significant change in the expression of genes associated with the ABA and auxin signaling pathways was detected in our analysis, although genes associated with the jasmonic acid and brassinosteroid biosynthesis pathways were repressed. In Arabidopsis, hy5 mutant seedlings are known to be hypersensitive to brassinosteroid, and it is known that AtHY5 plays a role in brassinosteroid signaling by altering the expression of Arabidopsis MEMBRANE STEROID BINDING PROTEIN1 (AtMSBP1) and by interacting with AtBZR1 (Shi et al., 2011; Li and He, 2016). AtHY5 has been shown to bind to the promoter of Arabidopsis MSBP1 and trigger its expression. AtMSBP1 is a negative regulator of brassinosteroid signaling, and its expression is less in hy5 mutant seedlings, causing the hypersensitive response of hy5 mutant seedlings to brassinosteroid (Shi et al., 2011). We were not able to find any putative homolog of AtMSBP1 in rice through The Institute for Genomic Research (TIGR) ortholog finder. The genes that code for enzymes like 3-β-hydroxysteroid dehydrogenase/isomerase, leucoanthocyanidin reductase, and NAD-dependent epimerase/dehydratase, which are involved in brassinosteroid biosynthesis, showed more than 2-fold repression in OsbZIP48OE transgenics, which could possibly result in the repression of brassinosteroid biosynthesis. Brassinosteroids are known to affect cell expansion and division, reproductive development, tissue differentiation, and stress resistance (Wang and Irving, 2011). Therefore, reduction in brassinosteroid biosynthesis also may be involved in the semidwarf phenotype of OsbZIP48OE transgenics raised in this study.
AtHY5 is known to work in a synergistic manner with AtHY1 to control jasmonic acid responsiveness in Arabidopsis (Prasad et al., 2012). However, the role of AtHY5 in jasmonic acid biosynthesis has yet to be elucidated. In rice, jasmonic acid is known to play diverse roles in various growth and developmental stages and is involved in defense and environmental responses. It positively regulates spikelet development, senescence, photomorphogenesis, and defense against microbes and nematodes but negatively regulates germination, shoot and root growth, and gravitropism (Liu et al., 2015). We found that OsbZIP48 down-regulates genes encoding lipoxygenase enzyme involved in jasmonic acid biosynthesis. Jasmonic acid is known to be involved in plant defense, and insufficient jasmonic acid results in sterile floral organs. It plays a central role in the biosynthesis of secondary metabolites that protect plants from biotic and abiotic stresses (Wang and Irving, 2011). Therefore, it will be interesting to study the role of OsbZIP48 in biotic and abiotic stresses.
The genes associated with cytokinin glucosylation were down-regulated in OsbZIP48OE transgenics as compared with the vector control. Cytokinins can be glucosylated to form O-glucosides and N-glucosides, which are inactive and are involved in hormone homeostasis (Hou et al., 2004). The down-regulation of this pathway in OsbZIP48OE transgenics could result in greater accumulation of free cytokinins. Cytokinins are involved in plastid development and are known to delay senescence. Thus, the accumulation of elevated levels of free (active) cytokinins could account for prolonged greening in the OsbZIP48OE plants.
The knockdown of OsbZIP48 could have resulted in the activation of GA, jasmonic acid, and IAA biosynthesis pathways, as genes encoding for enzymes involved in these pathways showed higher expression as compared with the vector control. The IAA biosynthesis pathway was not altered in OsbZIP48OE transgenics but was up-regulated in OsbZIP48KD transgenics. In Arabidopsis also, the hy5 mutant has higher auxin levels than the wild type. AtHY5 binds to the promoters of AXR2 and SLR/IAA14 genes and increases their expression, which, in turn, inhibits auxin signaling (Cluis et al., 2004); these genes encode for Aux/IAA proteins that act as negative regulators of auxin signaling. We were not able to find any putative homologs of AXR2/IAA7 and SLR/IAA14 through TIGR ortholog finder. Therefore, the mode of action of OsbZIP48 might be slightly different from AtHY5 or else it may stimulate the expression of another set of Aux/IAA genes. Ethylene biosynthesis pathway genes, however, were not altered in their expression in OsbZIP48KD transgenics as compared with the vector control.
OsbZIP48 May Regulate OsKO2 Expression by Binding Directly to Its Promoter
We found that OsbZIP48 binds directly to the G-box II (at −504 bp) present in the promoter upstream region of OsKO2. ChIP-seq analysis of AtHY5, however, did not show the promoter of AtKO2 to be a binding site of AtHY5 (Lee et al., 2007). OsNAC2 is known to repress the expression of OsKO2 by binding directly to its promoter, as shown by ChIP-seq analysis (Chen et al., 2015). In OsNAC2 overexpression transgenics, the expression of OsbZIP48 was shown to increase by 4-fold (Chen et al., 2015). We also found that the expression of OsNAC2 is enhanced in OsbZIP48OE transgenics. Therefore, it will be interesting to see whether OsbZIP48 binds directly to the promoter of OsKO2 as a homodimer or as a heterodimer with OsNAC2 or if they act independently.
To conclude, the results obtained in this study provide evidence that, in monocot systems like rice, there are at least three homologs of AtHY5, the bZIP transcription factor that plays a central role in regulating photomorphogenesis in plants. Although only one of them (OsbZIP48) has been functionally characterized in this study, their differential expression profile (A.B., N.B., and J.P.K, unpublished data) indicates that they may have evolved to regulate at least some unique functions. It is striking that the knockdown of OsbZIP48, either by insertional mutagenesis or RNAi, results in arresting growth at an early stage of development, leading to lethality; however, like the Arabidopsis hy5 mutant, these OsbZIP48KD lines did display excessive root proliferation. Its ectopic expression caused semidwarfism in rice, but, like AtHY5, it had no significant effect in altering seedling height when expressed ectopically in wild-type Arabidopsis. This raises the question of why both rice OsbZIP48 and Arabidopsis HY5 are ineffective in reducing seedling height when overexpressed in wild-type Arabidopsis, whereas the hy5 mutant develops an elongated hypocotyl when grown in the light; moreover, it can be complemented functionally by both AtHY5 and OsbZIP48, reducing its height that is fairly comparable to wild-type seedlings. OsbZIP48 from rice could complement the Arabidopsis hy5 mutant with respect to agravitropic responses displayed by root and root hairs and gravitropic set angle of the silique. That the semidwarf plants of rice overexpressing OsbZIP48 are compromised in its secondary cell wall thickenings is unusual; rather than providing strength to the semidwarf plant, these transgenics are fragile. OsbZIP48 appears to have a role in regulating panicle development and seed set, since its ectopic expression caused partial sterility. Although this study has unraveled some novel functions that the AtHY5 ortholog OsbZIP48 performs in rice, more work on HY5 homologs in both monocots and dicots is required to elucidate the specific and redundant functions they perform in regulating plant development. Whether the three AtHY5 homologs present in rice (and other monocots) have undergone neofunctionalization is another aspect that is under investigation in our laboratory.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Rice (Oryza sativa indica) seeds (varieties PB1 and IR64) were obtained from the Indian Agricultural Research Institute (IARI). These seeds were surface sterilized by treating with 0.1% HgCl2 (v/v) for 10 min, washed repeatedly with autoclaved Milli-Q water, and then kept at 28°C ± 1°C for 16 h for imbibition. Seeds were allowed to germinate, and seedlings were grown hydroponically for 7 d on Yoshida medium in a culture room maintained at 28°C ± 1°C for white light-grown seedlings and in incubators maintained at 28°C ± 1°C in a dark room for dark-grown seedlings. Light was provided by fluorescent lamps with the fluence rate of 200 μmol m−2 s−1 for Arabidopsis (Arabidopsis thaliana) and 75 μmol m−2 s−1 for rice. For RNA isolation, the tissues were harvested every day from day 3 until day 7, frozen in liquid nitrogen, and kept at −70°C for long-term storage and used when required. White light was supplied from a bank of Cool Daylight fluorescent lamps (Philips; TL 5800°K). For harvesting of tissues belonging to different developmental stages, plants were grown in the rice fields of IARI, and the tissue was harvested as per the requirement. Tissue samples were frozen in liquid nitrogen and kept at −80°C until use.
Real-Time PCR Analysis
Rice tissues of different developmental stages, harvested in liquid N2 and stored at −80°C, were used for RNA extraction. Total RNA was isolated using TRIzol Reagent (Invitrogen) as per the manufacturer’s instructions. The primers for real-time PCR were designed by using Primer Express 2.0 (Applied Biosystems). cDNA synthesis was carried out with random hexamer primers as per the manufacturer’s instructions using the High Capacity cDNA Archive Kit (Applied Biosystems). Each sample with two biological replicates and three technical replicates was used for real-time PCR analysis in the LightCycler 480II Real Time system (Roche) as per the manufacturer’s instructions. The relative mRNA levels of OsbZIP48 in different samples were computed with respect to the internal standard UBIQUITIN5 for rice (Jain et al., 2006). The conditions used for real-time PCR cycles were as follows: preincubation for 10 min at 95°C; followed by 45 cycles of 10 s at 95°C, 20 s at 60°C, and 10 s at 72°C; then cooling at 40°C for 30 s. The relative mRNA levels corresponding to different genes in different RNA samples were measured by the ΔΔCt method (relative mRNA level = 2−(ΔΔCt)). The heat map representing the expression profile of OsbZIP48 was generated using the Rice Oligonucleotide Array Database (Cao et al., 2012).
Western-Blot Analysis
Rice seedlings were grown hydroponically in rice growth medium at 28°C. For light treatment, seedlings were grown under white light at an intensity of 100 µmol m−2 s−1 in a culture room (16 h of light/8 h of dark). For dark treatment, seedlings were grown in complete darkness. Seedlings were harvested 3, 5, 7, and 10 d after germination. For light-shift experiments, the rice seedlings were grown for 4 d in continuous light or darkness under conditions as described above, and on day 5, they were transferred to the opposite light conditions for the designated duration. In the case of light-to-dark transition, 5-d-old seedlings grown in continuous light were used as a control, and similarly, in the case of dark-to-light transition, 5-d-old seedlings grown in continuous darkness were used as a control. In the case of Arabidopsis, the transgenic line overexpressing OsbZIP48 in the hy5 mutant background was used. The transgenic seeds were surface sterilized and plated on one-half-strength MS basal medium followed by 72 h of stratification at 4°C and then transferred to continuous light conditions (100 µmol m−2 s−1) at 22°C for 4 d. These 4-d-old seedlings were then transferred to dark for the desired duration, harvested at different time points, and processed for western analysis.
Panicles and seeds were harvested at different stages of development from rice plants grown under field conditions. For panicles, the stages were as follows: P1, 0–3 cm; P2, 3–5 cm; P3, 5–10 cm; P4, 10–15 cm; P5, 15–22 cm; and P6, 22–30 cm. For seeds, the stages were as follows: S1, 0 to 2 DAP; S2, 3 to 4 DAP; S3, 5 to 10 DAP; S4, 11 to 20 DAP; and S5, 21 to 29 DAP (Jain et al., 2007). Frozen seedlings and different stages of panicle and seed development were ground in a buffer (200 mm Tris, pH 8, 100 mm NaCl, 10 mm EDTA, 10 mm DTT, 5% glycerol, 0.05% Tween 20, and protease inhibitor cocktail [Sigma]), and the extracts were centrifuged at 13,000 rpm and 4°C. The supernatant was centrifuged again at 13,000 rpm and 4°C, and the concentration of proteins was estimated by the Bradford assay (Bradford, 1976). Equal concentrations of proteins were loaded on SDS-PAGE gels, and the protein extracts were subjected to western blotting (Sharma et al., 2014). For detection of OsbZIP48 protein, peptide antibody synthesis was done. Actin was used as a loading control, and its levels were detected by using ThermoFisher Scientific actin monoclonal antibody (catalog no. MA1-744).
Phylogenetic Analysis
Homologs of AtHY5 were identified by using the reverse best hit approach (using BLASTP and an e value cutoff of 10−5). For Cyanidioschyzon merolae and Ostreococcus tauri, the Kyoto Encyclopedia of Genes and Genomes database (http://www.genome.jp/kegg/kegg1.html) was used, while for other genomes, the Phytozome database version 9 (http://www.phytozome.net/) was used (Kanehisa and Goto, 2000; Goodstein et al., 2012; Kanehisa et al., 2014). To retrieve gymnosperm sequences, the National Center for Biotechnology Information BLAST tool was used. For reciprocal hits in the Arabidopsis genome, the wu-BLAST tool of TAIR was used (Huala et al., 2001). Since there is a large variation in the degree and quality of annotation of genomic information available across species, reverse best hit (RBH) was performed manually, and if no homolog was identified, a tBLASTN approach was used to manually annotate the genes in those species using FGENESH+ (http://www.softberry.com/berry.phtml?topic=fgenes_plus&group=programs&subgroup=gfs), a gene-predicting software, which uses similar protein support. Effort was made to reannotate the incomplete proteins using this software. The sequences of the reannotated proteins are available on request. The genes retrieved were then checked for the presence of the requisite domains using InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/; Quevillon et al., 2005; Jones et al., 2014; Mitchell et al., 2015). EST evidence also was checked using tBLASTN in the National Center for Biotechnology Information database for the particular species in which no ortholog was identified by the above method. ESTs were considered only if they had the requisite domain. The protein phylogenetic tree was made using the conserved bZIP domain. Three phylogenetic trees were prepared using phyml, maximum parsimony, and the neighbor-joining method using the Phylip package, and then a consensus tree was generated. The pairwise distances between protein sequence were calculated using MEGA6 (Tamura et al., 2013).
Amplification of OsbZIP48
The OsbZIP48 CDS is predicted to be of 552 bp, but the KOME clone of OsbZIP48 (AK241558) shows a truncated protein. Therefore, we amplified a 552-bp amplicon as predicted by TIGR from the cDNA derived from P6 stage panicle tissue and cloned it in a primary vector.
Generation of Rice and Arabidopsis Transgenics
Arabidopsis transgenics were raised using the floral dip method (Clough and Bent, 1998), while rice transgenics were raised using the protocol described by Toki et al. (2006) with some modifications. Seeds of indica rice variety PB1 were grown under light on NB medium (Himedia labs, cat no. PT107) at 32°C. The 7-d-old calli were cocultivated with the EHA105 strain of Agrobacterium tumefaciens. The calli were washed on day 3 after cocultivation and kept in selection medium with appropriate antibiotics. The positive calli were then transferred to regeneration medium with appropriate antibiotics until they formed plantlets. They were transferred to rooting medium and then rice growth medium. The Arabidopsis hy5 mutant (SALK_096651C) was obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/), while the rice OsbZIP48 mutant (PFG_3A-07378.R) was procured from the Rice Functional Genomic Express database (http://signal.salk.edu/cgi-bin/RiceGE). Genotyping of the T-DNA insertional mutant was done as described by Jung et al. (2008).
Southern Analysis
Southern analysis was carried out as described by Sharma et al. (2014) with some modifications. A 10-µg aliquot of genomic DNA from each transgenic line, vector control, and the wild-type plant was restriction digested using EcoRI (Roche Molecular Bio Labs) and probed with the hptII gene cassette (850 bp). The PCR-amplified and gel-eluted hptII gene was used as a positive control.
Particle Bombardment and Transactivation Assay
In silico prediction for intracellular localization was done using ProtComp 9.0 (http://www.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc). Particle bombardment for BiFC, FRET, and intracellular localization in onion (Allium cepa) epidermal peel cells was carried out using the Biolistic PDS-1000/He particle delivery system (Bio-Rad) according to the protocol described earlier (Thakur et al., 2005; Giri et al., 2011). The spring onion peels were kept on MS basal plates, and particle bombardment was carried out using the following parameters: 27 mm Hg vacuum, 1,100 p.s.i. He pressure, and target distance of 6 cm. The plates were then kept at 28°C in the dark for 16 h. A confocal microscope (Leica TCS, SP5) was used to observe the onion peels for BiFC, FRET, and intracellular localization. For FRET and BiFC, the experiment was repeated three times.
For the transactivation assay, the full-length OsbZIP48 CDS was cloned in pDEST-GBKTT7 (CD3-764) obtained from TAIR (https://www.arabidopsis.org) and transformed in the AH109 yeast strain (Clontech). The cloned construct was transformed into yeast as described in the Yeastmaker Yeast Transformation System 2 User Manual (Clontech). Serially diluted transformed colonies were dropped on –Trp/-His synthetic dropout selection medium. The sealed plates were incubated at 30°C for 3 to 6 d.
Arabidopsis Morphometric Analyses
Hypocotyl Length Measurement
The Arabidopsis seeds were surface sterilized, washed with sterile autoclaved reverse osmosis (RO) water, and plated on Murashige and Skoog (1962) medium supplemented with 0.8% agar and 1% Suc. The plates with seeds were then kept at 4°C for 3 to 4 d (for stratification) and transferred to light chambers maintained at 22°C with 200 μmol m−2 s−1 white light for 3 or 6 d, and the hypocotyl length was measured using ImageJ1.41 software (National Institutes of Health).
Chlorophyll and Anthocyanin Estimation
Chlorophyll a/b was estimated by overnight incubation of 20 Arabidopsis seedlings in 500 μL of dimethyl sulfoxide at 65°C in the dark as described by Hiscox and Israelstam (1979) with some modifications. Absorbance was recorded at 645 and 663 nm in a DUTM 640B (Beckman Instruments) spectrophotometer. Chlorophyll a/b contents were calculated according to the following formulae:
where V = volume of dimethyl sulfoxide in mL, Y = path length of 1 cm, and n = number of seedlings.
Anthocyanin was estimated by overnight incubation of 20 Arabidopsis seedlings in 150 μL of 1% HCl in methanol at room temperature in the dark. After the addition of 100 μL of MQ water, an equal volume (i.e. 250 μL) of chloroform was added to remove chlorophyll. The quantity of anthocyanins was determined by spectrophotometric measurements of the aqueous phase (A530–A657; Neff and Chory, 1998).
Cotyledon Angle Measurement
Cotyledon angle represented the angle between a straight line drawn between the tip of the cotyledon and the cotyledon blade/cotyledon petiole junction and a straight line drawn through the cotyledon petiole. An angle of 180° reflected a fully folded cotyledon (Christians et al., 2012). The cotyledon angles were measured using ImageJ1.41 software (National Institutes of Health).
Root Gravitropic Response
Root gravitropic response was measured as described by Vicente-Agullo et al. (2004) and Grabov et al. (2005). To analyze root geometry, plates with seedlings were photographed with a Leica S8AP0 stereomicroscope. The obtained images were analyzed using ImageJ 1.41 software (National Institutes of Health). The VGI was calculated using the following formula: VGI = CHα/RL, where CHα is a projection of the base-to-tip chord CH on the vertical axis and RL is total root length.
Rice Morphometric Analysis
The morphometric analysis of rice transgenics was carried out according to the IRRI guidelines as described in Descriptors for Rice by the IBPGR-IRRI Rice Advisory Committee (http://books.irri.org/getpdf.htm?book=971104000X). The internode closest to the roots was considered as the first internode, while the panicle-bearing internode was considered to be the last internode.
Microarray
Microarray analysis was done with transgenic plants expressing transgene(s) constitutively and showing the desired phenotype. Both the vector control and transgenic lines were grown on one-half-strength MS medium with antibiotics for 10 d under white light (100 µmol m−2 s−1). Seedlings were gently pulled out of the medium and immediately frozen in liquid nitrogen. Total RNA was isolated (Trizol method) as described by Chomczynski and Sacchi (1987). The isolated RNA (500 ng) was used for microarray experiments, and subsequent analysis was carried out as per the manufacturer’s protocol (Affymetrix; Jain et al., 2007). The pathway analysis was carried out using Plant MetGenMAP (http://bioinfo.bti.cornell.edu/cgi-bin/MetGenMAP/home.cgi; Joung et al., 2009). The metabolic pathways were reconstructed using Adobe Illustrator software. The microarray data have been submitted to the Gene Expression Omnibus with accession number GSE90472.
EMSA
The G-box element (CACGTG) was searched manually in a 1-kb promoter of the OsKO2 gene. The probe for the G-box element was designed, and the sequence is given in Supplemental Table S2. For EMSA, OsbZIP48 cDNA was cloned in pET28A vector, and 6× His-tagged OsbZIP48 was induced using 0.5 mm isopropylthio-β-galactoside and overexpressed in Escherichia coli. The overexpressed 6× His-OsbZIP48 was affinity purified according to the manufacturer's protocol (Qiagen). All DNA-binding reactions were carried out in 15 mm HEPES-KOH, pH 7.5, 35 mm KCl, 1 mm EDTA, pH 8, 6% glycerol, 1 mm DTT, 1 mm MgCl2, and 1 µg µL−1 poly(dI-dC). Gel-shift assays were performed as described by Jain et al. (2009).
Sectioning and Scanning Electron Microscopy Analysis
The second last internode of the wild type and overexpression transgenics was fixed in fixative (100 mm PIPES, pH 7.2, 10% formaldehyde [37%], and 80% reverse osmosis [RO] water) by vacuum infiltration and kept overnight at 4°C. The tissue was then dehydrated in a graded ethanol series (70%, 80%, 90%, and 100%) followed by a tertiary butanol series (25%, 50%, and 100%) before placing in Paraplast plus (Sigma-Aldrich). Paraplast-embedded internodes were sectioned by using a Leica RM2245 rotary microtome producing 50- to 100-μm-thick sections that were placed on poly-l-Lys-coated slides (Polysciences). Quantitative analysis of cell size was done using ImageJ 1.41 software (National Institutes of Health). Cell area is cell base area, which is similar to the base area of a cylinder and was calculated using ImageJ 1.41 software. For scanning electron microscopy analysis, the second last internode of overexpression transgenics and the wild type was harvested in Trumps 4F:1G fixative, vacuum infiltrated, and sent for fixation and gold coating to the University Science Instrumentation Centre, Delhi University. Scanning of coated samples was carried out by using a JEOL scanning electron microscope (JSM 6610LV).
Accession Number
The Gene Expression Omnibus accession number for the microarray data is GSE90472.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Protein alignment of HY5 orthologs showing the presence of the COP1-interaction motif in most of the homologs.
Supplemental Figure S2. Heat maps showing the expression of OsbZIP48 in different anatomical and developmental stages of rice.
Supplemental Figure S3. PCR confirmation and real-time PCR analysis of Arabidopsis transgenics.
Supplemental Figure S4. Anthocyanin and chlorophyll estimation of the Arabidopsis transgenics.
Supplemental Figure S5. Confirmation of rice transgenics through PCR, real-time PCR, and southern analysis.
Supplemental Figure S6. OsbZIP48 overexpression reduces panicle and internodal length in rice.
Supplemental Figure S7. Longitudinal sections of wild-type and OsbZIP48OE transgenic stems showing the difference in cell length.
Supplemental Figure S8. Morphometric analysis of the wild type and OsbZIP48OE transgenics.
Supplemental Figure S9. Morphometric analysis of the wild type and OsbZIP48OE transgenics.
Supplemental Figure S10. Chlorophyll content of rice transgenic seedlings.
Supplemental Figure S11. Schematic representation of different hormone pathways showing altered gene expression in OsbZIP48OE transgenics.
Supplemental Figure S12. Schematic representation of cytokinin glucosylation pathways showing altered gene expression in OsbZIP48OE transgenics.
Supplemental Figure S13. Heat map showing differentially expressed genes in RNAi transgenics (Hh and hh) and the pANDA vector control.
Supplemental Figure S14. Schematic representation of different hormone pathways showing altered gene expression in OsbZIP48KD transgenics in rice.
Supplemental Figure S15. Real-time PCR of genes of different hormonal biosynthetic pathways that were shown to be altered by microarray in OsbZIP48OE.
Supplemental Figure S16. Real-time PCR of genes of different hormonal biosynthetic pathways that were shown to be altered by microarray in OsbZIP48KD (RNAi).
Supplemental Table S1. Pairwise distance result of protein sequences of HY5/HYH homologs.
Supplemental Table S2. Primers used for quantitative reverse transcription-PCR analysis.
Acknowledgments
The Research Fellowship provided by the Council of Scientific and Industrial Research, New Delhi, to N.B. and A.B. is gratefully acknowledged. The National Post-Doctoral Fellowship provided by SERB to N.B. is also acknowledged. We gratefully acknowledge the support extended by Dr. Ashok Kumar. Singh, Division of Genetics, IARI, during the course of this study. We also acknowledge the infrastructural support provided by the Department of Science and Technology, Government of India, and the University Grants Commission, New Delhi. J.P.K. also thanks SERB for financial support in the form of a JC Bose Fellowship award.
Glossary
- DAP
days after pollination
- FRET
fluorescence resonance energy transfer
- BiFC
bimolecular fluorescence complementation
- VGI
vertical growth index
- CDS
coding sequence
- RNAi
RNA interference
- MS
Murashige and Skoog
- ABA
abscisic acid
- EMSA
electrophoretic mobility shift assay
- TIGR
The Institute for Genomic Research
- IARI
Indian Agricultural Research Institute
- DAPI
4′,6-diamino-phenylindole
Footnotes
This research was funded by the Department of Biotechnology, Government of India (grant no. BT/AGIII/CARI/01/2012).
Articles can be viewed without a subscription.
References
- Alabadí D, Gallego-Bartolomé J, Orlando L, García-Cárcel L, Rubio V, Martínez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, et al. (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53: 324–335 [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Blakeslee JJ, Lee OR, Mravec J, Sauer M, Titapiwatanakun B, Makam SN, Bouchard R, Geisler M, Martinoia E, et al. (2007) Interactions of PIN and PGP auxin transport mechanisms. Biochem Soc Trans 35: 137–141 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cao P, Jung KH, Choi D, Hwang D, Zhu J, Ronald PC (2012) The Rice Oligonucleotide Array Database: an atlas of rice gene expression. Rice (N Y) 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chen W, Zhou J, He H, Chen L, Chen H, Deng XW (2012) Basic leucine zipper transcription factor OsbZIP16 positively regulates drought resistance in rice. Plant Sci 193-194: 8–17 [DOI] [PubMed] [Google Scholar]
- Chen H, Xiong L (2008) Role of HY5 in abscisic acid response in seeds and seedlings. Plant Signal Behav 3: 986–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lu S, Wang Y, Zhang X, Lv B, Luo L, Xi D, Shen J, Ma H, Ming F (2015) OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J 82: 302–314 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Chory J. (2010) Light signal transduction: an infinite spectrum of possibilities. Plant J 61: 982–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians MJ, Gingerich DJ, Hua Z, Lauer TD, Vierstra RD (2012) The light-response BTB1 and BTB2 proteins assemble nuclear ubiquitin ligases that modify phytochrome B and D signaling in Arabidopsis. Plant Physiol 160: 118–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cluis CP, Mouchel CF, Hardtke CS (2004) The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J 38: 332–347 [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K (2002) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Giri J, Vij S, Dansana PK, Tyagi AK (2011) Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol 191: 721–732 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A, Ashley MK, Rigas S, Hatzopoulos P, Dolan L, Vicente-Agullo F (2005) Morphometric analysis of root shape. New Phytol 165: 641–651 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauvermale AL, Ariizumi T, Steber CM (2012) Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol 160: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox JD, Israelstam GF (1979) A method for the extraction of chlorophyll from leaf tissue without maceration. Can J Bot 57: 1332–1334 [Google Scholar]
- Holm M, Hardtke CS, Gaudet R, Deng XW (2001) Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J 20: 118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou B, Lim EK, Higgins GS, Bowles DJ (2004) N-Glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J Biol Chem 279: 47822–47832 [DOI] [PubMed] [Google Scholar]
- Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, et al. (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 29: 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Yang P, Lau OS, Li G, Li J, Chen H, Deng XW (2012) Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 24: 4590–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Roy N, Chattopadhyay D (2009) CaZF, a plant transcription factor functions through and parallel to HOG and calcineurin pathways in Saccharomyces cerevisiae to provide osmotolerance. PLoS ONE 4: e5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice: genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143: 1467–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345: 646–651 [DOI] [PubMed] [Google Scholar]
- Jiang L, Wang Y, Li QF, Björn LO, He JX, Li SS (2012) Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res 22: 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JG, Corbett AM, Fellman SM, Tieman DM, Klee HJ, Giovannoni JJ, Fei Z (2009) Plant MetGenMAP: an integrative analysis system for plant systems biology. Plant Physiol 151: 1758–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Lee J, Dardick C, Seo YS, Cao P, Canlas P, Phetsom J, Xu X, Ouyang S, An K, et al. (2008) Identification and functional analysis of light-responsive unique genes and gene family members in rice. PLoS Genet 4: e1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42: D199–D205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP (1980) Genetic control of lightinhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol 100: 147–160 [Google Scholar]
- Landschulz WH, Johnson PF, McKnight SL (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240: 1759–1764 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jiang J, Qian Q, Xu Y, Zhang C, Xiao J, Du C, Luo W, Zou G, Chen M, et al. (2011a) Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell 23: 628–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, Wang Deng X (2011b) Phytochrome signaling mechanisms. The Arabidopsis Book 9: e0148, doi/10.1199/tab.0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QF, He JX (2016) BZR1 interacts with HY5 to mediate brassinosteroid- and light-regulated cotyledon opening in Arabidopsis in darkness. Mol Plant 9: 113–125 [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhang S, Sun N, Liu H, Zhao Y, Liang Y, Zhang L, Han Y (2015) Functional diversity of jasmonates in rice. Rice (N Y) 8: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Jia WJ, Chu YJ, Xue HW (2012) Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Res 22: 581–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, et al. (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43: D213–D221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura R, Ohmori M, Fujita H, Kawaguchi M (2002) A Lotus basic leucine zipper protein with a RING-finger motif negatively regulates the developmental program of nodulation. Proc Natl Acad Sci USA 99: 15206–15210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wisniewska J, Tadele Z, Kubes M, Covanová M, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Prasad BR, Kumar SV, Nandi A, Chattopadhyay S (2012) Functional interconnections of HY1 with MYC2 and HY5 in Arabidopsis seedling development. BMC Plant Biol 12: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Sun F, Wang Q, Chen M, Huang Y, Feng YQ, Luo X, Yang J (2011) Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol 157: 216–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33: W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram H, Chattopadhyay S (2013) Molecular interaction of bZIP domains of GBF1, HY5 and HYH in Arabidopsis seedling development. Plant Signal Behav 8: e22703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Chatterjee M, Burman N, Khurana JP (2014) Cryptochrome 1 regulates growth and development in Brassica through alteration in the expression of genes involved in light, phytohormone and stress signalling. Plant Cell Environ 37: 961–977 [DOI] [PubMed] [Google Scholar]
- Sharma R, Agarwal P, Ray S, Deveshwar P, Sharma P, Sharma N, Nijhawan A, Jain M, Singh AK, Singh VP, et al. (2012) Expression dynamics of metabolic and regulatory components across stages of panicle and seed development in indica rice. Funct Integr Genomics 12: 229–248 [DOI] [PubMed] [Google Scholar]
- Shi QM, Yang X, Song L, Xue HW (2011) Arabidopsis MSBP1 is activated by HY5 and HYH and is involved in photomorphogenesis and brassinosteroid sensitivity regulation. Mol Plant 4: 1092–1104 [DOI] [PubMed] [Google Scholar]
- Sibout R, Sukumar P, Hettiarachchi C, Holm M, Muday GK, Hardtke CS (2006) Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet 2: e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DP, Jermakow AM, Swain SM (2002) Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell 14: 3133–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R (2010) The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ 33: 88–103 [DOI] [PubMed] [Google Scholar]
- Sukumar P, Edwards KS, Rahman A, Delong A, Muday GK (2009) PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiol 150: 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur JK, Jain M, Tyagi AK, Khurana JP (2005) Exogenous auxin enhances the degradation of a light down-regulated and nuclear-localized OsiIAA1, an Aux/IAA protein from rice, via proteasome. Biochim Biophys Acta 1730: 196–205 [DOI] [PubMed] [Google Scholar]
- Todaka D, Nakashima K, Maruyama K, Kidokoro S, Osakabe Y, Ito Y, Matsukura S, Fujita Y, Yoshiwara K, Ohme-Takagi M, et al. (2012) Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc Natl Acad Sci USA 109: 15947–15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Máté Z, Adám E, Oakeley EJ, Schäfer E, Nagy F (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Habricot Y, Condiff AS, Maldiney R, Van der Straeten D, Ahmad M (2007) HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J 49: 428–441 [DOI] [PubMed] [Google Scholar]
- Vicente-Agullo F, Rigas S, Desbrosses G, Dolan L, Hatzopoulos P, Grabov A (2004) Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant J 40: 523–535 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J (2008) Molecular basis of plant architecture. Annu Rev Plant Biol 59: 253–279 [DOI] [PubMed] [Google Scholar]
- Wang YH, Irving HR (2011) Developing a model of plant hormone interactions. Plant Signal Behav 6: 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Tan C, Qi B, Zhang Y, Xu G, Zheng H (2010) Changes in gravitational forces induce the modification of Arabidopsis thaliana silique pedicel positioning. J Exp Bot 61: 3875–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Hecht V, Vander Schoor JK, Davidson SE, Ross JJ (2009) Light regulation of gibberellin biosynthesis in pea is mediated through the COP1/HY5 pathway. Plant Cell 21: 800–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yamawaki S, Yamashino T, Nakanishi H, Mizuno T (2011) Functional characterization of HY5 homolog genes involved in early light-signaling in Physcomitrella patens. Biosci Biotechnol Biochem 75: 1533–1539 [DOI] [PubMed] [Google Scholar]
- Yu Y, Wang J, Zhang Z, Quan R, Zhang H, Deng XW, Ma L, Huang R (2013) Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLoS Genet 9: e1004025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW (2011a) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65: 346–358 [DOI] [PubMed] [Google Scholar]
- Zhang J, Vanneste S, Brewer PB, Michniewicz M, Grones P, Kleine-Vehn J, Löfke C, Teichmann T, Bielach A, Cannoot B, et al. (2011b) Inositol trisphosphate-induced Ca2+ signaling modulates auxin transport and PIN polarity. Dev Cell 20: 855–866 [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhang B, Qian Q, Yu Y, Li R, Zhang J, Liu X, Zeng D, Li J, Zhou Y (2010) Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J 63: 312–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YC, Gong SF, Li QH, Sang Y, Yang HQ (2006) Functional and signaling mechanism analysis of rice CRYPTOCHROME 1. Plant J 46: 971–983 [DOI] [PubMed] [Google Scholar]
- Zheng X, Wu S, Zhai H, Zhou P, Song M, Su L, Xi Y, Li Z, Cai Y, Meng F, et al. (2013) Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell 25: 115–133 [DOI] [PMC free article] [PubMed] [Google Scholar]