The induction of genes involved in the anaerobic response is repressed if the sugar status of the plant is low.
Abstract
Plants respond to hypoxia, often caused by submergence, by expressing a specific set of genes that contribute to acclimation to this unfavorable environmental condition. Genes induced by low oxygen include those encoding enzymes for carbohydrate metabolism and fermentation, pathways that are required for survival. Sugar availability is therefore of crucial importance for energy production under hypoxia. Here, we show that Arabidopsis (Arabidopsis thaliana) plants require starch for surviving submergence as well as for ensuring the rapid induction of genes encoding enzymes required for anaerobic metabolism. The starchless pgm mutant is highly susceptible to submergence and also fails to induce anaerobic genes at the level of the wild type. Treating wild-type plants under conditions inducing sugar starvation results in a weak induction of alcohol dehydrogenase and other anaerobic genes. Induction of gene expression under hypoxia requires transcription factors belonging to group VII ethylene response factors (ERF-VII) that, together with plant Cys oxidases, act as an oxygen-sensing mechanism. We show that repression of this pathway by sugar starvation occurs downstream of the hypoxia-dependent stabilization of ERF-VII proteins and independently of the energy sensor protein kinases SnRK1.1 (SNF1-related kinase 1.1).
Plants cannot survive for long periods under conditions of limited oxygen availability (hypoxia; Bailey-Serres and Voesenek, 2008). Oxygen is required for ATP production through the mitochondrial electron transport chain and plants thus experience an energy crisis when hypoxic (Perata and Alpi, 1993). Degradation of carbohydrates through glycolysis coupled to the fermentative metabolism only marginally compensates for the drop in ATP production, but it is still considered of great importance for surviving hypoxia (Loreti et al., 2016). Plants that are devoid of genes required for fermentation, such as alcohol dehydrogenase (ADH) and pyruvate decarboxylase (PDC), are much less tolerant to low oxygen conditions (Loreti et al., 2016). Similarly, mutants defective in Suc metabolism, such as those defective in Suc synthases, also are less tolerant to hypoxia (Santaniello et al., 2014). Suc availability in Arabidopsis (Arabidopsis thaliana) influences gene expression, with more sustained expression of ADH and PDC1 and stronger induction of heat shock protein genes (Loreti et al., 2005). In rice (Oryza sativa), starch availability plays a pivotal role for survival to submergence. Rice seeds can germinate even in completely anoxic conditions also thanks to their ability to efficiently degrade starch in the absence of oxygen, while other cereals fail to induce α-amylases under anoxia and cannot therefore germinate in the absence of oxygen (Perata et al., 1992; Guglielminetti et al., 1995b). In rice seeds, sugar signaling is crucial for coordinating anaerobic germination. Rice varieties possessing a functional gene for trehalose 6-phosphate phosphatase (OsTPP7) can degrade starch and germinate more efficiently because removal of trehalose 6-phosphate by OsTPP7 signals the plant for extreme starvation conditions, thus boosting starch degradation through the starvation-induced α-amylase gene RAmy3D (Kretzschmar et al., 2015). Differently from rice germination, rice adult plants that are tolerant to submergence successfully adapt to this condition by reducing starch consumption, so that carbohydrates are retained for the subsequent recovery phase when water recedes (Perata and Voesenek, 2007). Reduction of carbohydrate consumption in rice is also obtained by slowing down growth, a mechanism that requires the Sub1A gene (Xu et al., 2006), which belongs to the group VII of the ethylene response factor (ERF) family (Licausi et al., 2013). The ERF-VII genes were subsequently discovered to play a crucial role in oxygen sensing in plants (Gibbs et al., 2011; Licausi et al., 2011). Members of this family are characterized by their N-terminal Cys oxidation in normoxia, which enables arginylation followed by proteasomal degradation (Bailey-Serres et al., 2012). Under hypoxia, ERF-VII are stabilized and activate transcription of anaerobic genes by binding to the HRPE element present in the promoter of anaerobic genes (Gasch et al., 2016). Anaerobic genes include the fermentative genes PDC1 and ADH, and activation of fermentation at the transcriptional level will lead to successful activation of fermentation that, coupled to glycolysis, can provide cells with a basal level of ATP for housekeeping activities, thus preserving cell integrity and viability (Perata and Alpi, 1993). Obviously, this strategy would be ineffective if Glc, the substrate of glycolysis, is not available. While during the day photosynthesis provides Glc for metabolic activities, transitory starch is required to sustain glycolysis at night (Streb and Zeeman, 2012). In the wild, plants are often submerged in muddy water, severely restricting photosynthesis (Pedersen et al., 2013), thus making starch produced before submergence of importance for maintaining glycolysis active. Differently from rice, little is known about the ability of Arabidopsis to utilize starch under conditions of limited oxygen availability. Given that leaf starch degradation in Arabidopsis requires the activity of glucan water dikinase (GWD), which transfers the β-phosphate of ATP to amylopectin (Ritte et al., 2002; Smith et al., 2005), it is possible that under low-ATP conditions, starch degradation would be impaired. If so, induction of anaerobic genes would be detrimental, since the energy cost for producing the fermentative enzymes would not be compensated for by an effective additional ATP production in the absence of sugars as substrates. Similarly, plants submerged in conditions limiting photosynthesis will need to fine-tune induction of anaerobic genes to the level of available carbohydrates to avoid an excess amount of fermentative activity compromising survival (Paul et al., 2016). It is currently unknown if such a regulatory mechanism exist. Here, we show that survival of submergence requires the presence of transitory starch and that plants that have limited carbohydrate resources drastically down-regulate the induction of anaerobic genes.
RESULTS
Leaf Transitory Starch Is Degraded in Submerged Arabidopsis Plants
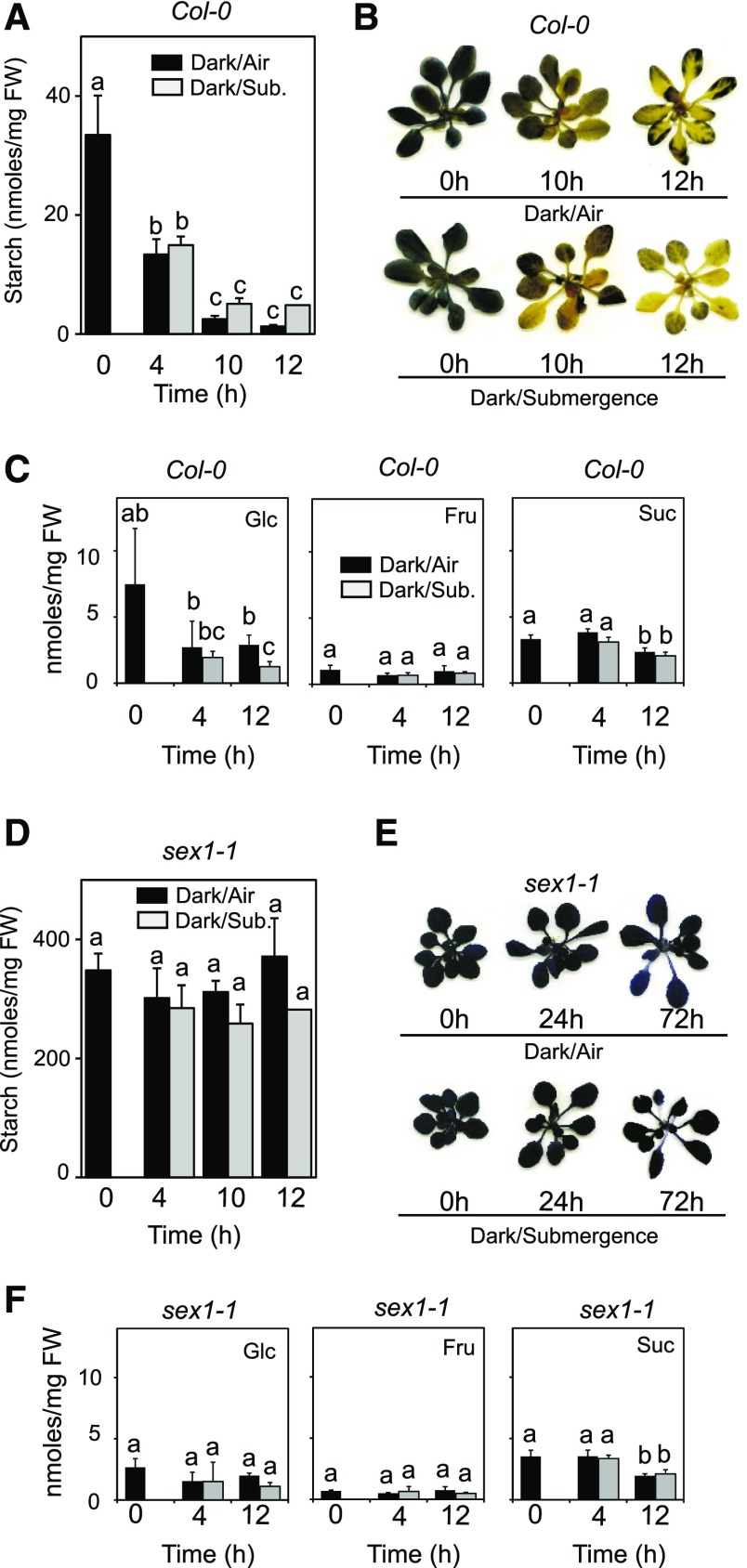
We investigated if starch is degraded in plants that are submerged during the night, when starch degradation normally occurs. The data showed that starch is normally degraded even in plants that are kept under complete submergence for the whole duration of the night (Fig. 1A). A closely similar starch content was measured in control and submerged plants at the end of the night (Fig. 1A), indicating a comparable rate of starch degradation. Iodine staining of the rosette, highlighting the distribution of starch in the leaves, confirmed that starch is uniformly degraded, regardless of oxygen availability (Fig. 1B). Interestingly, starch degradation was also observed in plants that were kept under anoxia (Supplemental Fig. S2). A drop in the sugar content is also observed at the end of the night (Fig. 1C), specifically in Glc and Suc content. However, no statistically differences were detected comparing aerobic versus submerged plants. We explored the possibility that, under submergence, starch degradation could occur through a route that does not require the ATP-dependent phosphorylation of the glucosyl residues of starch by GWD. The sex1 mutant is defective in GWD1 and shows very high starch content that is not degraded during the night. Similarly, starch was not degraded in submerged sex1 plants (Fig. 1D). This was confirmed by iodine staining of sex1 plants, showing high levels of starch even after 72 h of submergence (Fig. 1E). Sugar content was constant under most of the conditions in sex1, with the exception of decreased Suc content after 12h (Fig. 1F).
Figure 1.
Starch degradation in submerged Arabidopsis plants. A, Quantitation of starch in rosette leaves of Arabidopsis Col-0 plants that were submerged at the end of the light period. Submergence was carried out in the dark; see Supplemental Figure S1 for oxygen content in the water. Data are mean ± sd of three biological replicates (ANOVA, P < 0.05). B, Iodine staining of starch in plants from the experiment described in A. C, Sugar content in rosette leaves of Arabidopsis plants that were submerged at the end of the light period. Submergence was carried out in the dark. Sugars were quantified at the end of the day (Time 0) and after 4 and 12 h of darkness/submergence. Data are mean ± sd of five biological replicates, n = 3 for sugars (ANOVA, P < 0.05). D, Quantitation of starch in rosette leaves of Arabidopsis sex1-1 mutant. Treatments are as in A. E, Iodine staining of starch in plants from the experiment described in D. F, Sugar content in rosette leaves of Arabidopsis sex1-1 plants (see C for details). Data are mean ± sd of five biological replicates for starch, n = 3 for sugars (ANOVA, P < 0.05).
The pgm Starchless Mutant Is Extremely Susceptible to Submergence
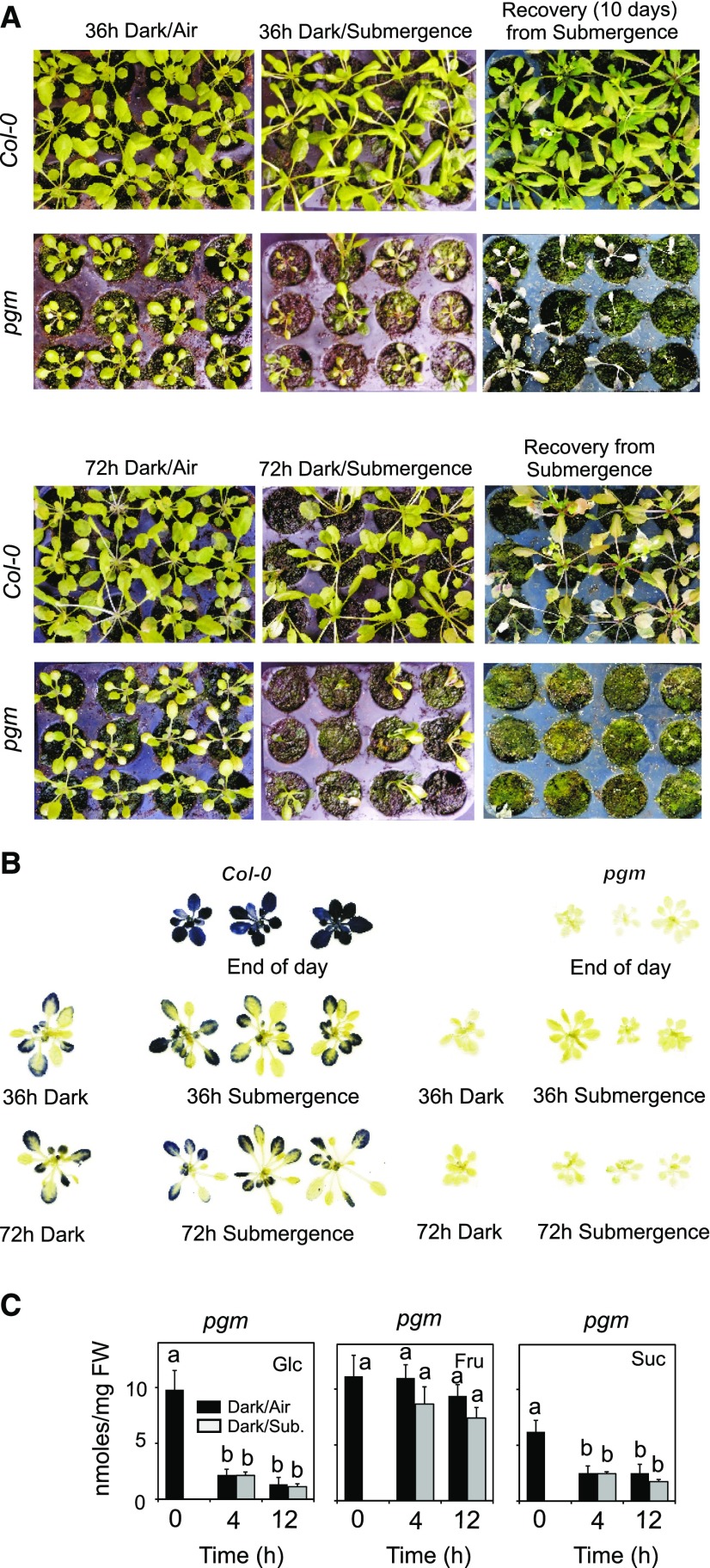
Given that starch is degraded in Arabidopsis plants kept under submergence, we assumed that starch metabolism could be important for plant survival to submergence. To test this hypothesis, we used a starchless mutant that, because of a mutation in the plastidial isoform of phosphoglucomutase (pgm), is unable to synthesize starch during the day. Submergence of pgm plants resulted in a dramatic loss of vitality at the end of the treatment. After 36 h of submergence, most of the pgm plants were dead, while the wild type (Col-0) showed little intolerance symptoms even after 72 h of submergence (Fig. 2A). Recovery from submergence was very poor in pgm (Fig. 2A). Iodine staining of leaves showed that some starch is retained in the Col-0 plants even after 72 h of darkness or submergence (Fig. 2B), suggesting a slower starch degradation after the normal, 12-h-long degradation phase during the night has taken place (Fig. 1A). The pgm plants were completely starchless (Fig. 2B), and their sugar content decreased sharply under submergence, with the exception of the Fru level (Fig. 2C).
Figure 2.
Survival of Arabidopsis plants to submergence. A, Wild-type plants (Col-0) and pgm mutant plants were submerged for 36 and 72 h. The pictures, taken immediately after the submergence treatment or after recovery (10 d), show the consequences of the submergence treatment. B, Iodine staining of starch of plants that were submerged for 36 and 72 h. C, Sugar content in rosette leaves of Arabidopsis pgm plants that were submerged at the end of the light period. Submergence was carried out in the dark. Sugars were quantified at the end of the day (Time 0) and after 4 and 12 h of darkness/submergence. Data are mean ± sd of five biological replicates, n = 3 for sugars (ANOVA, P < 0.05).
Exogenous Suc Enhances Arabidopsis Survival during Anoxia
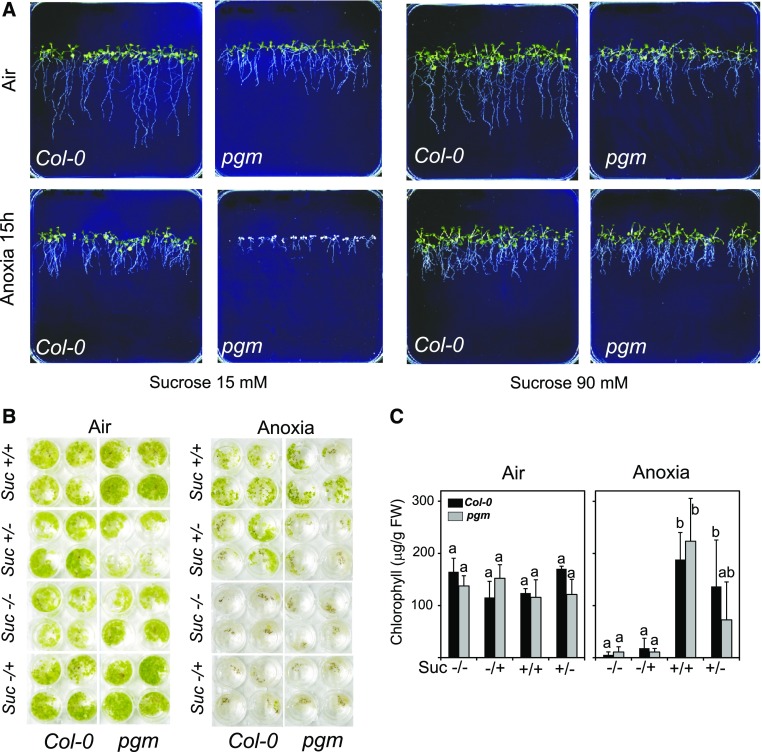
The phenotype of pgm plants after submergence suggested that plants require sugars resulting from starch degradation to withstand the consequence of hypoxia in the darkness. We confirmed that pgm is more sensitive to submergence because of hypoxia together with low sugar availability by observing the intolerance of pgm seedlings grown in vertical plates. When the seedlings were grown in the presence of a low Suc concentration (15 mm), pgm displayed a severe intolerance phenotype after 15 h of anoxia, while the wild type survived without apparently suffering from the treatment (Fig. 3A). Increasing the amount of Suc in the medium to 90 mm resulted in a striking increase in pgm tolerance (Fig. 3A). We set up an experiment to verify if Suc is required during anoxia or recovery from anoxia. Arabidopsis seedlings were grown in a liquid medium that was either deprived of Suc or with exogenous Suc added during the whole duration of the experiment, only during anoxia, or only during recovery from anoxia. Visual inspection of the seedlings showed that the presence of Suc had a positive impact on seedlings survival when fed to the seedling during the whole duration of the experiment (Suc +/+ in Fig. 3B) as well as during the duration of anoxia only (Suc +/− in Fig. 3B). Seedlings that were fed with Suc during the recovery phase only (Suc −/+ in Fig. 3B) were as sensitive to anoxia as those that were kept in a Suc-free medium, while those that were kept in a Suc medium during the duration of the anoxia treatment only recovered well from the stress treatment, as quantitatively demonstrated by their chlorophyll content (Fig. 3C).
Figure 3.
Effects of exogenous Suc on the survival of Arabidopsis seedling. A, Seedlings (1 week old) of Arabidopsis grown on vertical agar plates were treated under anoxia for 15 h. Photographs were taken after 10 d of recovery. Suc was present in the plates at the concentrations shown in the figure. B, Seedlings were treated under anoxia for 22 h in the presence of Suc (30 mm) during anoxia in liquid medium and the subsequent 10 d of recovery in air (Suc +/+), during anoxia only (Suc +/−), during recovery in air (10 d) only (−/+), or in the absence of Suc (Suc −/−). C, Quantitation of chlorophyll in plants from the experiment described in B. Data are mean ± sd of three biological replicates (ANOVA, P < 0.05).
Anaerobic Gene Expression Is Severely Impaired in pgm Plants
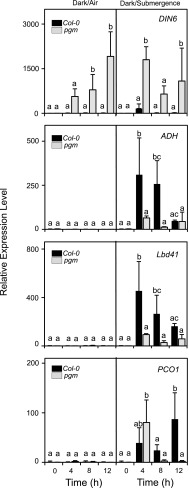
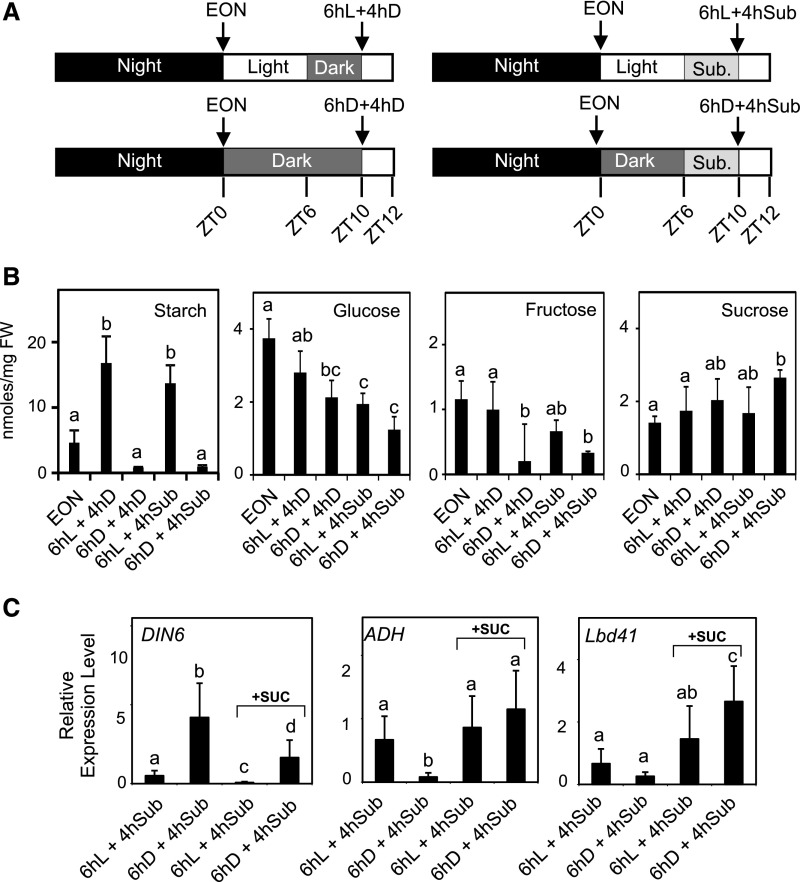
When pgm plants were treated under submergence at the end of the night period, they exhibited a strong expression of the carbon starvation gene marker DIN6 (Fig. 4), confirming that these plants are low in sugars and already entered into a starvation phase. After 4 h of submergence, DIN6 expression peaked in pgm, suggesting that an extra demand of sugars occurred in plants that experience hypoxia. This was also observed in Col-0, although to a much lower extent. Col-0 plants showed a very marked induction of ADH as well as of other anaerobic genes such as LBD41 and PCO1, all belonging to the core anaerobic genes (Mustroph et al., 2010; Fig. 4). The anaerobic response was severely dampened in pgm. The induction of ADH and LBD41 was much lower in pgm than in Col-0. PCO1 expression in pgm was similar to that of Col-0 at 4 h submergence, but declined thereafter (Fig. 4). We decided to check if the reduction of response to hypoxia in terms of anaerobic gene expression that was observed in pgm could be mimicked in the wild type. At ZT0 (end of the night) Col-0 plants were treated under normal light conditions for 6 h (ZT6) to allow starch to be synthesized and then exposed them to 4 h submergence (Fig. 5A). Another set of plants was instead kept in the darkness for six additional hours before being submerged for 4 h (Fig. 5A). The level of starch was very low in plants exposed to the extended night treatment (6hD in Fig. 5B), while those treated in the light for 6 h showed increased starch level compared to the end of night level (Fig. 5B). Submerged plants showed a starch level similar to that of their dark control if pretreated with 6 h in the light, while they were very low in starch if submerged after a 6-h dark treatment (Fig. 5B) The level of Glc and Fru was lower in submerged plants that experienced the ExN treatment (6hD + 4hD and 6hD + 4hSub in Fig. 5), while a higher Suc level was observed (Fig. 5B). At the gene expression level, the extended night (ExN) period induced DIN6 resulted in a lower expression level of ADH and LBD41, thus confirming that in the absence of starch the induction of anaerobic genes is compromised (Fig. 5C). Interestingly, treating plants with exogenous Suc that was sprayed on the plants during the ExN treatment and was also added to the water used to submerge the plants allowed a full recovery of gene expression of ADH and LBD41, while reducing DIN6 expression. The dampening of the anaerobic response observed in plants exposed to an ExN treatment is therefore to be assigned to sugar starvation.
Figure 4.
Expression level of sugar starvation and anaerobic genes in rosette leaves of Arabidopsis plants that were submerged at the end of the light period. Wild-type plants (Col-0) and plants of the pgm mutant were submerged for 12 h in the darkness or kept in the darkness in air. The mRNA level of genes was measured by RT-qPCR, and data are expressed as relative to the Time 0 of Col-0 (Time 0 = 1). Data are mean ± se of three biological replicates (ANOVA, P < 0.05).
Figure 5.
Effect of submergence and darkness on carbohydrate content and effect of exogenous Suc on the induction of anaerobic genes. A, Schematic representation of the experimental set-up. B, Starch, Suc, Glc, and Fru content in Arabidopsis plants that were submerged after a 6-h treatment in the light or in the darkness. EON, end of night. Data are mean ± sd of three biological replicates (ANOVA, P < 0.05). C, Effect of exogenous Suc on the induction of starvation and anaerobic genes. Suc was applied by spraying (90 mm) the plants during the 6-h-long pretreatment (either in the light or darkness) and Suc was added to the water (45 mm) used to submerge the plants. Data are mean ± sd of three biological replicates (ANOVA, P < 0.05).
Sugar Starvation Dampens the Anaerobic Response Independently of the N-End Rule Pathway
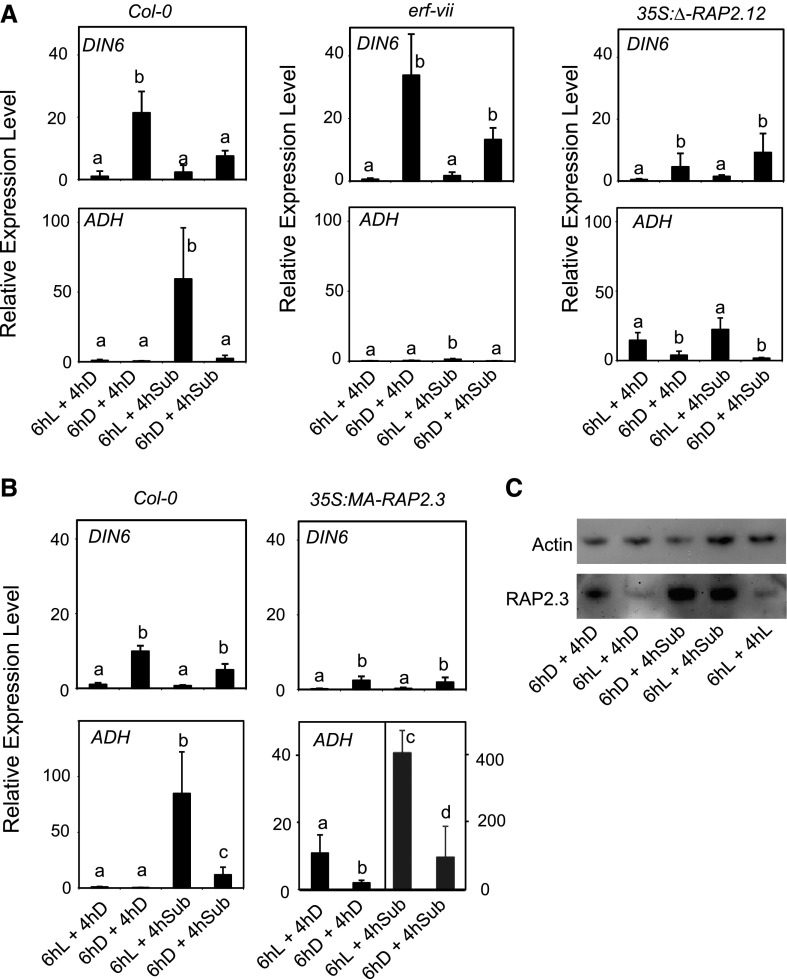
We compared Col-0 plants with a mutant that is defective in all five ERF-VII genes (erf-vii; Marín-de la Rosa et al., 2014) that are known to be responsible for the anaerobic induction of genes, as well as with transgenic lines overexpressing versions of RAP2.12 and RAP2.3 that are constitutively stable even under aerobic conditions (35S:Δ-RAP2.12 [Licausi et al., 2011] and 35S:MA-RAP2.3 [Gibbs et al., 2014]). The starvation response erf-vii was similar to the wild type, but was apparently lower in the 35S:Δ-RAP2.12 and 35S:MA-RAP2.3 lines (Fig. 6, A and B). Under submergence conditions the extended night treatment strongly reduced the induction of ADH, a process that appears to be fully controlled by the three ERF-VII proteins (RAP2.2, RAP2.3, and RAP2.12) that redundantly control the induction of the anaerobic genes (Gasch et al., 2016). The ADH expression was indeed very low in the erf-vii quintuple mutant, but it was already high in the 35S:Δ-RAP2.12 and 35S:MA-RAP2.3 plants even in aerobic conditions, as expected since the Δ-RAP2.12 and MA-RAP2.3 proteins are constitutively stable because they lack the MC motif at the N terminus that is destabilized by oxygen (Fig. 6, A and B). Remarkably, the ExN treatment repressed ADH expression both in aerobic and submerged Δ-RAP2.12 and 35S:MA-RAP2.3 plants, indicating that the signaling mechanism that dampens the anaerobic response occurs downstream of the N-rule-dependent ERF-VII stabilization. The level of the RAP2.3 protein, which is functionally redundant with RAP2.12 and RAP2.2 (Gasch et al., 2016), was increased by submergence, but the darkness pretreatment did not affect the RAP2.3 protein level in submerged plants (Fig. 6C). The light/dark treatment had instead a marked effect on the level of RAP2.3, with light reducing the protein level, an effect previously observed by Abbas et al. (2015). These results indicate that starvation resulting from the ExN treatment does not significantly affect the RAP2.3 protein level.
Figure 6.
Induction of anaerobic genes requires ERF-VII genes but repression by extended night-induced starvation occurs downstream of ERF-VII stabilization by the N-end-rule. A, Wild-type plants (Col-0), the erf-vii mutant, and a transgenic line overexpressing a constitutively stable version of RAP.12 (35S:Δ-RAP2.12) were submerged (4 h) in the darkness or kept in the darkness in air after a pretreatment (6 h) either in the light or darkness. See Figure 5A for the experimental setup. The mRNA level was measured by RT-qPCR and data are expressed as relative to the 6hL+4hD sample of the Col-0 sample. Data are mean ± sd of three biological replicates (ANOVA, P < 0.05). B, same as in A, but using the 35S:MA-RAP2.3 line. C, Immunoblotting using an HA-tag antibody to measure the protein level of RAP2.3-HA. Plants of the 35S:RAP2.3-HA were treated as detailed in the figure. An α-actin antibody was used to verify the loading of protein on the electrophoresis.
Sugar Starvation Dampens the Anaerobic Response Upstream of Interaction of ERF-VII with the Anaerobic Promoter
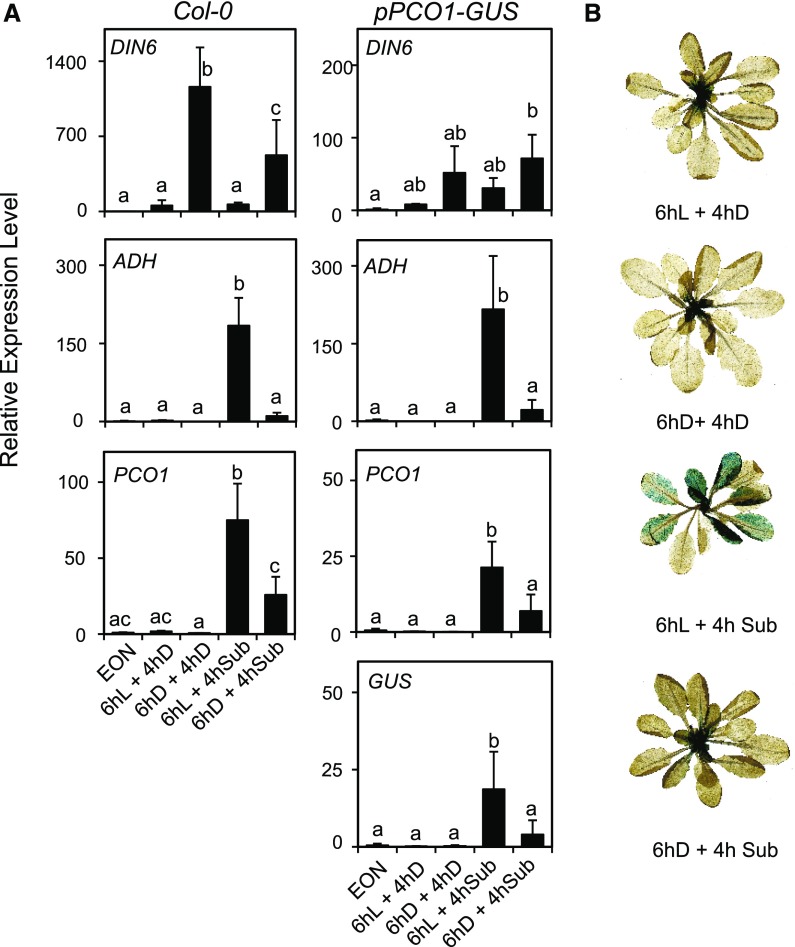
Given that the RAP2.3 protein level was not negatively affected by the ExN treatment, it is tempting to speculate that the reduced ADH expression observed after an ExN treatment might result from either a reduced transcription of anaerobic genes or an effect on ADH mRNA stability. We used a reporter line in which GUS gene expression is driven by the PCO1 promoter, observing that the induction of ADH and PCO1 mRNAs were both repressed in plants that experienced ExN before submergence of the pPCO1:GUS plants (Fig. 7A). The expression of GUS mRNA mirrored that of PCO1 mRNA, indicating that the dampening effect of sugar starvation occurs at the level of gene expression. Staining pPCO1:GUS plants for GUS activity showed a clear GUS staining in the plants that experienced submergence after the 6-h light treatment, while it was restricted to young leaves in the ExN-treated plants, similarly to aerobic plants (Fig. 7B). Given that the ERF-VII proteins bind to the HRPE in the anaerobic promoter (Gasch et al., 2016), this result indicates that their binding to the HRPE is hampered under sugar starvation conditions.
Figure 7.
Darkness-induced sugar starvation interferes with the activation of an anaerobic promoter. A, Wild-type plants (Col-0) and a transgenic lines having the GUS gene under the control of the PCO1 promoter (pPCO1:GUS) were submerged (4 h) in the darkness or kept in the darkness in air after a pretreatment (6 h) either in the light (L) or darkness (D). The mRNA level was measured by RT-qPCR and data are expressed as relative to the EON time point of each genotype. Data are mean ± sd of three biological replicates (ANOVA, P < 0.05). B, GUS staining of the pPCO1:GUS plants at the end of the treatment described in the figure.
Sugar Starvation Dampens the Anaerobic Response Independently of HRA1, the Snrk1 Protein KIN10, and of bZIP63
We investigated if HYPOXIA RESPONSE ATTENUATOR1 (HRA1), a trihelix protein that can counteract the induction of anaerobic genes by RAP2.12 (Giuntoli et al., 2014, 2017a), was possibly involved in the repression triggered by sugar starvation. HRA1 is itself a target of RAP2.12 and, not surprisingly, its expression was repressed by a dark pretreatment (Supplemental Fig. S3), exactly at the same level of ADH, that is always included in independent experiments to provide the reader with a reference gene response. Overexpression of HRA1 reduced the expression of ADH but had little if any effect on the repression of ADH caused by the dark pretreatment (Supplemental Fig. S3), ruling out a possible involvement of this protein in the repression that is observed following an ExN treatment.
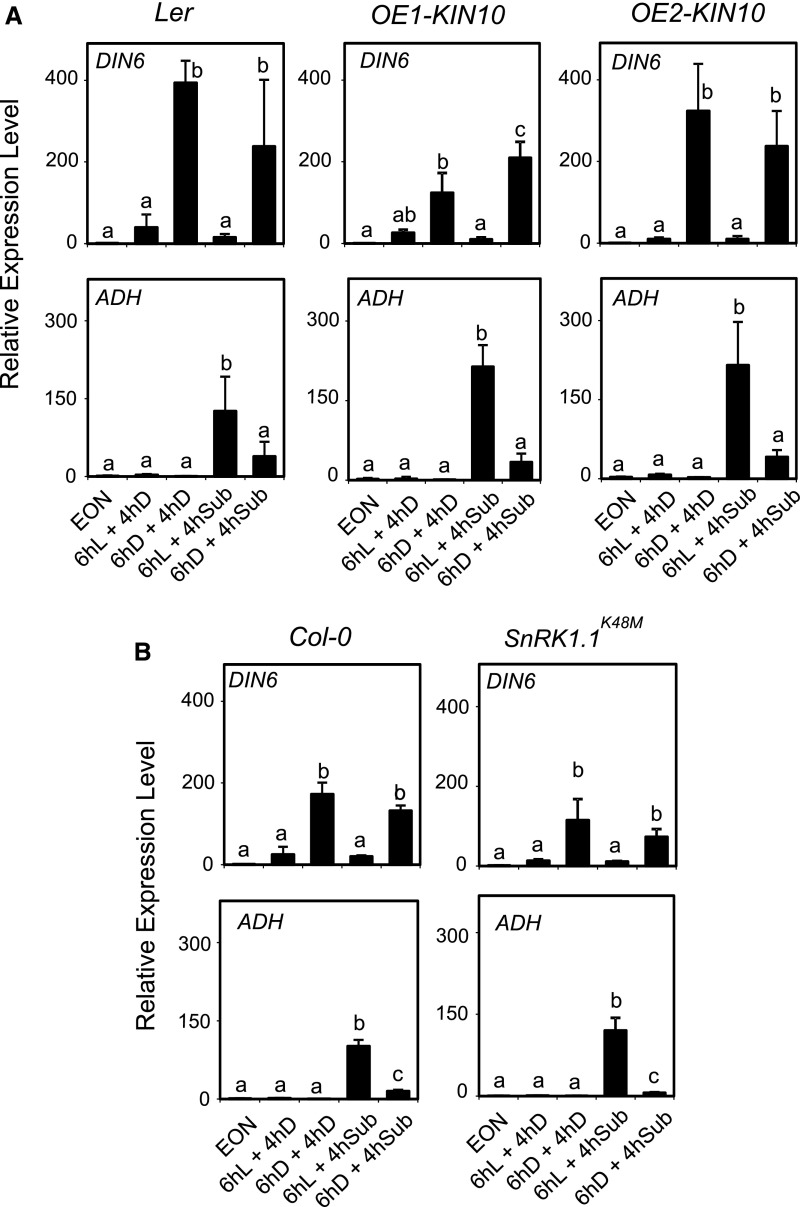
Plants under sugar starvation experience an extensive transcriptional reprogramming that is partly regulated by the energy sensor protein kinases SnRK1 (SNF1-related kinase 1). The SnRK1 subfamily members KIN10 (SnRK1.1) and KIN11 (SnRK1.2) are considered as central regulators of the transcriptome in response to darkness and other stress signals, including hypoxia (Baena-González et al., 2007). We found that two lines overexpressing KIN10 showed higher ADH mRNA level compared to their wild-type background (Ler), but without differences in terms of ExN effects on ADH, since the repression of the ADH mRNA level was evident also in the two independent OE-KIN10 lines (Fig. 8A). We then tested the dominant-negative SnRK1.1K48M line (Cho et al., 2016). The results showed that C starvation repressed ADH expression also in SnRK1.1K48M, suggesting that the starvation-dependent effects of ADH expression are independent of SnrK1.1 (Fig. 8B).
Figure 8.
Darkness-induced sugar starvation dampens the anaerobic response independently of KIN10. A, Wild-type plants (Col-0), the Ler genotype (background of the transgenic lines), and two lines overexpressing KIN10 were submerged (4 h) in the darkness or kept in the darkness in air after a pretreatment (6 h) either in the light (L) or darkness (D). EON, End of night time point. The mRNA levels are expressed as relative to the EON time point of each genotype. Data are mean ± sd (n = 3; ANOVA, P < 0.05). B, Wild-type plants (Col-0) and the dominant-negative SnRK1.1 mutant were treated as described in A. The mRNA levels are expressed as relative to the EON time point of each genotype. Data are mean ± sd (n = 3; ANOVA, P < 0.05).
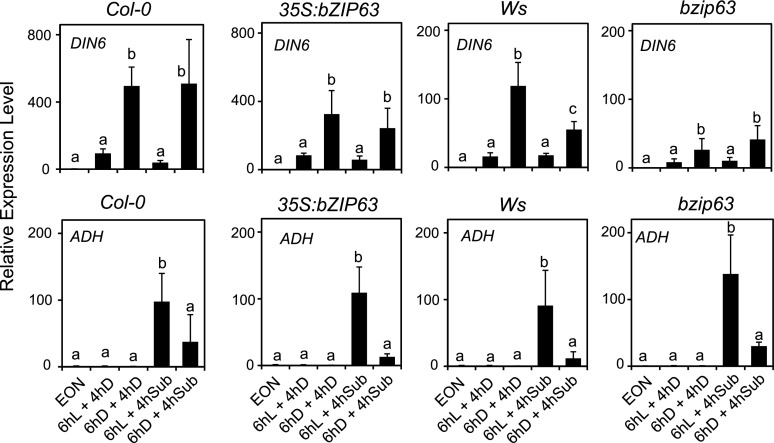
Given that DIN6 expression always correlated with the dampening of the anaerobic response, we also explored the possibility that the Snrk1-regulated bZIP63 transcription factor, which regulates DIN6 expression, could be involved. The bzip63 mutant displayed lower DIN6 expression, but comparable ADH mRNA level as well as repression induced by ExN (Fig. 9). Similarly, an overexpression line (35S:bZIP63) did not show any effect of ADH repression by ExN (Fig. 9). We can therefore conclude that the sugar starvation-dependent effects of ADH expression are largely independent of SnRK1.1.
Figure 9.
Darkness-induced sugar starvation dampens the anaerobic response independently of bZIP63. Wild-type plants (Col-0), the bzip63 mutant, and a transgenic line overexpressing bZIP63 were treated as described in Figure 8. The mRNA levels are expressed as relative to the EON time point of each genotype. Data are mean ± sd (n = 3; ANOVA, P < 0.05).
DISCUSSION
Plants under hypoxia need to reconfigure several aspects of their metabolism, and this is accomplished by a drastic modulation of gene expression (Mustroph et al., 2010). Transcription factors belonging to the ERF-VII group were recently identified as master regulators of this transcriptional reconfiguration (Gibbs et al., 2011; Licausi et al., 2011). While the discovery of the mechanism of oxygen sensing modulating ERF-VII stability shed light on the events upstream of anaerobic gene regulation (Bailey-Serres et al., 2012), little is known about how this pathway is fine-tuned to allow the best possible fitness to plants under hypoxia. A subset of genes induced by hypoxia encodes enzymes required for an efficient anaerobic metabolism of carbohydrates (e.g. Suc synthase, alcohol dehydrogenase, and pyruvate decarboxylase). Mutants defective in these enzymatic activities are less tolerant to hypoxia, pointing out their importance in the survival mechanism enabling plants to tolerate submergence and other low-oxygen conditions (Loreti et al., 2016). Soluble carbohydrates are not present in plants at levels allowing an efficient sugar metabolism for prolonged periods of time. Starch can instead represent an adequate source of carbohydrates for several hours or even days. In the case of leaf transitory starch, the amount of this polysaccharide that is accumulated during the day can provide the plant with a more than adequate carbon supply for the full duration of the night (Smith et al., 2005).
Our experimental evidence showed that starch degradation is possible in plants that experience darkness under complete submergence, with a pattern mirroring that of aerobic plants. Given that transitory starch degradation is under circadian control (Graf et al., 2010), these results suggests that starch degradation in submerged plants is preserved both at the level of regulation and enzymatic efficiency, although we cannot exclude that differences exist in the mechanism operating under submergence. Remarkably, the energy-requiring step catalyzed by GWD was required for starch degradation under submergence (Fig. 1D). This result was not necessarily expected. In cereals, starch degradation in seeds under anoxia is thought to occur only in rice, a species well adapted to submergence (Loreti et al., 2016). Arabidopsis is not tolerant to submergence and starch degradation in leaves is an energy-requiring process, but its degradation occurs normally even in submerged plants (Fig. 1; Supplemental Fig. S2). It is possible that the requirement of ATP for phosphorylation of starch by GWD is very low, an aspect that deserves further investigation. Starch utilization in submerged Arabidopsis is essential for survival. Starchless mutants such as pgm (Fig. 2), but also mutants unable to degrade starch such as sex1 (Supplemental Fig. S4), are highly intolerant to low-oxygen conditions. Feeding exogenous Suc rescued pgm plants from the consequences of anoxia (Fig. 3A) and we showed that Suc is required mostly during the low-oxygen treatment rather than during the recovery phase (Fig. 3B). Exogenous Suc treatment had a remarkably positive effect of submergence tolerance in rice varieties that do not possess the SUB1A gene (Kudahettige et al., 2011). Interestingly, inhibition of growth in submerged FR13A rice plants, a SUB1A variety, was abolished if plants were submerged in a Suc-containing solution, suggesting that Suc availability can circumvent the inhibitory role of SUB1A, an ERF-VII gene (Xu et al., 2006). Suc exerts a very positive effect on Arabidopsis survival (Banti et al., 2008). Experiments using the 35S:Δ-RAP2.12 line demonstrated that constitutive expression of Δ-RAP2.12 results in stronger fermentative metabolism leading to lower carbohydrate availability and decreased tolerance to hypoxia (Paul et al., 2016). Tolerance was restored by feeding exogenous Suc, indicating that low levels of carbon reserves were the most likely reason for reduced anoxic resistance in 35S:Δ13RAP2.12 plants (Paul et al., 2016). The strong intolerance to submergence of pgm demonstrates that starch availability plays an important role for Arabidopsis tolerance in vivo. The major consequences of the lack of carbon resources in terms of tolerance to hypoxia is likely at the metabolic level, but we cannot rule out the possibility that sugars also play a regulatory role on the signaling pathway regulating the anaerobic response at the transcriptional level.
Unexpectedly, we indeed found that pgm plants drastically reduce their responses to submergence at the mRNA level of anaerobic response genes (Fig. 4). This suggested that an adequate level of sugars is required to ensure a full response to hypoxia in terms of expression of anaerobic genes, also given that transcription and translation are cost-intensive processes. Also, wild-type plants displayed a dampened anaerobic response if they were left starving of carbohydrates by exposing them to a 6-h-long extended night (Fig. 5). Interestingly, submerging the plants in a solution containing Suc abolished the darkness-depended dampening of ADH expression (Fig. 5C), thus confirming that it is not darkness itself, but carbon starvation arising from the extended night period the signal leading to repression of the anaerobic response. However, the level of sugars itself did not correlate with the observed effects on ADH expression. The level of sugars in plants experiencing submergence after a light or dark period is similar, while the starch level is drastically different (Fig. 5). The expression of DIN6, however, indicates that starvation is occurring at the cellular level. Further work is required to elucidate the sugar sensing and signaling pathway that senses starvation.
Carbon starvation is regulated by the energy sensor protein kinases SnRK1. An Arabidopsis SnRK1.1 dominant-negative mutant showed lower tolerance to submergence (Cho et al., 2016). However, overexpression of the SnRK1 subfamily members KIN10 as well as the dominant-negative mutation in SnRK1.1 had no consequences on the starvation-dependent dampening of ADH expression (Fig. 8), suggesting that the sugar starvation pathway that negatively affects ADH expression is independent of SnRK1, although we cannot exclude an involvement of KIN11 (SnRK1.2). The expression of the starvation-induced DIN6 gene always correlated with repression of ADH following an extended night treatment. Expression of DIN6 is under control of bZIP63 (Mair et al., 2015); however, neither the bzip63 mutant nor a bZIP63 overexpression line displayed any alteration of the expression of ADH (Fig. 9), thus ruling out this possible link. Besides bZIP63, other bZIP genes such as bZIP1 and bZIP53 (Hartmann et al., 2015) could be involved in the regulation of anaerobic genes, a hypothesis requiring further work. The lower expression of DIN6, coding for an Asn synthase, observed in Δ-RAP2.12 and 35S:MA-RAP2.3 plants (Fig. 6) may reflect the altered amino acid metabolism in plants expressing a stable version of ERF-VII proteins (Paul et al., 2016).
The experiments carried out using the erf-vii mutant and lines overexpressing stable versions of RAP2.12 highlighted that the process being repressed by sugar starvation depends on the ERF-VII group of transcription factors but is independent of the stabilization of these transcription factors by the N-end rule pathway (Fig. 6). The 35S:Δ-RAP2.12 line and the 35S:MA-RAP2.3 express versions of RAP2.12 and RAP2.3 that are constitutively stable and, as a consequence, induce the expression of anaerobic genes also in air (Fig. 6). Repression of ADH is also observed in air in the 35S:Δ-RAP2.12 and 35S:MA-RAP2.3 lines and, given that the Δ-RAP2.12 and MA-RAP2.3 proteins are stable and insensitive to the oxygen-dependent destabilization of RAP2.12, we conclude that sugar starvation does not reduce ADH induction by enhancing destabilization of ERF-VII proteins via the N-end rule. We actually observed that the protein level of one of these ERF-VII proteins, namely, RAP2.3, was not reduced by the extended night treatment when plants were submerged (Fig. 6C), thus indicating that repression occurs downstream of RAP2.3 stabilization by hypoxia. Interestingly, light reduced significantly the RAP2.3 protein level, as previously observed by Abbas et al. (2015). Expression of RAP2.2 was reported to be induced by darkness (Hinz et al., 2010). The fact that darkness stabilized RAP2.3 (Fig. 6C) with negative consequences in terms of ADH gene expression if submergence follows (Fig. 6B) is intriguing and counterintuitive. We would have expected that a higher level of RAP2.3 in aerobic, dark-treated plants, in which RAP proteins are protected by binding to the ACBP proteins (Licausi et al., 2011), would have resulted in a stronger anaerobic response if submergence follows, releasing RAP2.3 from ACBPs. The fact that a light treatment reduces the aerobic level of RAP2.3 without any consequence on the subsequent induction of anaerobic genes if the plant is submerged suggests that most of the anaerobic response is the consequence of de novo ERF-VII synthesis during submergence and does not relies much on stored ERF-VII proteins. ERF-VII binding to ACBP proteins could be required during the very initial phases of the anaerobic response, but not to guarantee sustained expression of the anaerobic genes. The fact that darkness actually leads to accumulation of RAP2.3 in air suggests that RAP2.3 could be bound to darkness-induced proteins, different from ACBPs, that sequester the ERF-VII proteins, protecting them from degradation, but making them unable to interact to the promoter of anaerobic genes. This may also occur under submergence thus making the ERF-VII proteins unavailable for binding to the HRPE element in the anaerobic promoters. We indeed observed that the extended darkness treatment reduced the activity of the PCO1 promoter in submerged plants (Fig. 7), thus indicating that repression occurs at the level of interaction or availability of ERF-VII proteins for transactivation of the promoter.
Here, we demonstrate that the ERF-VII-based anaerobic response is fine-tuned so that it does not exceed the actual needs of the anaerobic metabolism that are estimated on the level of available carbohydrates. Sugar starvation triggers repression of the anaerobic response downstream of the hypoxic stabilization of ERF-VII proteins but upstream of their interaction with the promoter of anaerobic genes. Further work is required to identify the molecular mechanism by which sugar starvation interferes with the anaerobic response of plants.
MATERIALS AND METHODS
Plant Material
Genotypes of Arabidopsis (Arabidopsis thaliana) used included Col-0 ecotype, Ler ecotype, Wassilewskija ecotype, the starchless pgm mutant, and the sex1-1 mutant. The quintuple erfVII mutant (Marín-de la Rosa et al., 2014) is a null mutant for RAP2.2, RAP2.3, RAP2.12, HRE1, HRE2, and a knockdown for RAP2.2 (Giuntoli et al., 2017b); a line constitutively expressing a version of the RAP2.12 protein that is also stable in air (35S:Δ-RAP2.12; Licausi et al., 2011); a line expressing GUS under the control of the PCO1 promoter (pPCO1:GUS; Weits et al., 2014); and a line overexpressing HRA1 (Giuntoli et al., 2014). The bzip63 mutant and the bZIP63 overexpressor (35S:bZIP63) (Mair et al., 2015), two independent KIN10 overexpressor lines (OE1 KIN10 and OE2 KIN10) (Baena-González et al., 2007); the Arabidopsis SnRK1.1 dominant-negative mutant (SnRK1.1 K48M; Cho et al., 2016); a line expressing RAP2.3 with an HA tag (35S:RAP2.3-HA; Gibbs et al., 2014).
Plant Growth Conditions
Plants were grown in pots for 3 to 4 weeks at 23°C with a 12/12-h photoperiod at 120 μmol photons m–2 s–1 before being utilized for experiments. Plants were submerged in tanks with a water level of 10 cm above leaf level. The submergence treatment was carried out in the dark. For experiments with 12-d-old seedlings (Fig. 3B), sterilized seeds were sown in liquid half-strength Murashige and Skoog medium, in the presence or absence of Suc (30 mm). Seeds were kept for 72 h in the dark at 4°C and then transferred at 23°C with a 12-h light photoperiod (100 mE m2 s2 intensity) with shaking. To obtain plants grown on vertical agar plates (Fig. 3A), seeds were germinated on agar (0.9% [w/v]) medium supplemented with Suc using the same growing conditions as reported above. Anoxia treatment assays were carried out using an enclosed anaerobic workstation (Anaerobic System model 1025; Forma Scientific) to provide an oxygen-free environment (<10 μg mL−1 oxygen) in the dark.
Total RNA Extraction, qPCR, and RNA Gel Blots
Total RNA was extracted as previously described (Perata et al., 1997) with a minor modification (omission of aurintricarboxylic acid) to make the protocol compatible with the subsequent PCR procedures. Electrophoresis using a 1% agarose gel was performed for all RNA samples to check for RNA integrity, followed by spectrophotometric quantification. Contaminated DNA was removed using a TURBO DNA-free kit (Ambion). RNA was then reverse-transcribed using an iScript TM cDNA synthesis kit (Bio-Rad Laboratories). Expression analysis was performed by real-time PCR using an ABI Prism 7300 sequence detection system (Applied Biosystems). Quantitative PCR was performed using 30 ng cDNA and iQTM SYBR Green Supermix (Bio-Rad Laboratories), according to the manufacturer’s instructions. Expression of Ubiquitin10 (At4g05320) was used as an endogenous control. Relative expression levels were calculated using GeNorm (https://genorm.cmgg.be/). For a list of the primers used and designed by QuantPrime Tool (http://quantprime.mpimp-golm.mpg.de/; Arvidsson et al., 2008), see Supplemental Table S1.
GUS Staining
Histochemical GUS staining was carried out according to Jefferson et al. (1987). Briefly, plant material was fixed immediately after sampling in ice-cold 90% acetone for 1 h, rinsed several times in 100 mm phosphate buffer (pH 7.2), and then stained in a freshly prepared reaction solution (0.2% Triton X-100, 2 mm potassium ferrocyanide, 2 mm potassium ferricyanide, and 2 mm X-Gluc [5-bromo-4-chloro-3-indolyl β-d-glucuronide, sodium salt dissolved in DMSO] in 100 mm phosphate buffer, pH 7.2). Plants were stained for 2 to 4 h (seedlings) or overnight (adult plants). Chlorophyll was eliminated from green tissues by washing them with absolute ethanol.
Extraction and Analysis of Carbohydrates
Four-week-old rosettes with a fresh weight of 300 to 500 mg were harvested rapidly into liquid N2. Frozen samples were homogenized and then extracted in 80% ethanol. Glc, Fru, and Suc were measured enzymatically (soluble sugars) (Guglielminetti et al., 1995a), and starch was assayed in the insoluble pellet (starch) (Critchley et al., 2001).
Qualitative Analysis of Leaf Starch Content by Lugol Staining
Rosettes of individual plants were harvested at the end of the light period and boiled in 50 mL 80% (v/v) ethanol. Decolored plants were stained with a fresh iodine solution (I2/KI [5 g KI and 0.5 g I2 in 500 mL distilled water]) for 5 min, destained in water for 1 to 2 h, and photographed immediately.
Analysis of Chlorophyll
The quantitation of the chlorophyll content in leaves was carried our spectrophotometrically using the method described by Lichtenthaler (1987).
Oxygen Measurement
Oxygen concentration was measured in the tanks used for submerging the plants using a FireStingO2 high-precision, PC-controlled fiber-optic oxygen meter produced by Pyro Science. The oxygen probe used was OXROB10. Water temperature was contemporarily measured using the TDIP15 temperature probe.
Extraction of Proteins, SDS-PAGE, and Immunoblots
Plant material was extracted by grinding in liquid nitrogen precooled samples with a pestle to a fine powder. The extraction buffer (50 mm Tris-HCl, pH 7, and 1% SDS with Sigma P9599 protease inhibitor cocktail) was added, vortexed vigorously, and then centrifuged for 30 min at 14,000 rpm to obtain a supernatant. Protein content in the supernatant was quantified with Bio-Rad DC reagent (Lowry method). Samples were dissolved in 5× Laemmli buffer, treated at 95°C for 10 min, and loaded (30 mg) to Invitrogen NuPAGE gels (10% Bis-Tris Midi Gels). After electrophoresis, proteins were transferred to a PVDF membrane using the Bio-Rad Trans-Blot turbo transfer pack. An anti-HA antibody (Sigma-Aldrich; SAB4300603), an antiactin antibody (Agrisera AS13 2640), and secondary goat anti-rabbit IgG HRP conjugated antibody (Agrisera AS09 602) were used. Detection was performed using the LiteAblot Turbo Chemiluminescence substrate (Euroclone).
Statistics
Values that significantly differ from each other are indicated by different letters in figures (according to two-way ANOVA test, Bonferroni posthoc test, P < 0.05).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Oxygen concentrations in the water in which plants were submerged (experiment described in Fig. 1A).
Supplemental Figure S2. Iodine staining of starch in Col-0 plants that were kept in the dark under aerobic or anoxic conditions for the time indicated.
Supplemental Figure S3. Darkness-induced sugar starvation dampens the anaerobic response independently of HRA1.
Supplemental Figure S4. Effects of anoxia on the survival of the Arabidopsis sex1-1 seedling.
Supplemental Table S1. Primers used for gene expression analysis using real-time quantitative RT-PCR.
Acknowledgments
We thank Professor Michael J. Holdsworth (University of Nottingham) for providing the pentuple erfVII mutant and the 35S:RAP2.3-HA and 35S:MA-RAP2.3 seeds, Dr. Tinne Boeckx (University of Nottingham) for useful information for performing the immunoblotting analysis, and Professor Markus Teige (University of Vienna) for providing the bZIP63 seeds.
Footnotes
Articles can be viewed without a subscription.
This work was supported by the Italian Ministry of Education, Research and Universities, Special Program PRIN 2015.
These authors contributed equally to this work.
References
- Abbas M, Berckhan S, Rooney DJ, Gibbs DJ, Vicente Conde J, Sousa Correia C, Bassel GW, Marín-de la Rosa N, León J, Alabadí D, Blázquez MA, Holdsworth MJ (2015) Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr Biol 25: 1483–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B (2008) QuantPrime--a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Voesenek LACJ (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Fukao T, Gibbs DJ, Holdsworth MJ, Lee SC, Licausi F, Perata P, Voesenek LACJ, van Dongen JT (2012) Making sense of low oxygen sensing. Trends Plant Sci 17: 129–138 [DOI] [PubMed] [Google Scholar]
- Banti V, Loreti E, Novi G, Santaniello A, Alpi A, Perata P (2008) Heat acclimation and cross-tolerance against anoxia in Arabidopsis. Plant Cell Environ 31: 1029–1037 [DOI] [PubMed] [Google Scholar]
- Cho HY, Wen TN, Wang YT, Shih MC (2016) Quantitative phosphoproteomics of protein kinase SnRK1 regulated protein phosphorylation in Arabidopsis under submergence. J Exp Bot 67: 2745–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley JH, Zeeman SC, Takaha T, Smith AM, Smith SM (2001) A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J 26: 89–100 [DOI] [PubMed] [Google Scholar]
- Gasch P, Fundinger M, Müller JT, Lee T, Bailey-Serres J, Mustroph A (2016) Redundant ERF-VII transcription factors bind an evolutionarily-conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28: 160–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Md Isa N, Movahedi M, Lozano-Juste J, Mendiondo GM, Berckhan S, Marín-de la Rosa N, Vicente Conde J, Sousa Correia C, Pearce SP, et al. (2014) Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol Cell 53: 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, Holdsworth MJ (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuntoli B, Lee SC, Licausi F, Kosmacz M, Oosumi T, van Dongen JT, Bailey-Serres J, Perata P (2014) A trihelix DNA binding protein counterbalances hypoxia-responsive transcriptional activation in Arabidopsis. PLoS Biol 12: e1001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuntoli B, Licausi F, van Veen H, Perata P (2017a) Functional balancing of the hypoxia regulators RAP2. 12 and HRA1 takes place in vivo in Arabidopsis thaliana plants. Front Plant Sci 8: 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuntoli B, Shukla V, Maggiorelli F, Giorgi FM, Lombardi L, Perata P, Licausi F (2017b) Age-dependent regulation of ERF-VII transcription factor activity in Arabidopsis thaliana. Plant Cell Environ 40: 2333–2346 [DOI] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM (2010) Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA 107: 9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielminetti L, Perata P, Alpi A (1995a) Effect of anoxia on carbohydrate metabolism in rice seedlings. Plant Physiol 108: 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielminetti L, Yamaguchi J, Perata P, Alpi A (1995b) Amylolytic activities in cereal seeds under aerobic and anaerobic conditions. Plant Physiol 109: 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann L, Pedrotti L, Weiste C, Fekete A, Schierstaedt J, Göttler J, Kempa S, Krischke M, Dietrich K, Mueller MJ, et al. (2015) Crosstalk between two bZIP signaling pathways orchestrates salt-induced metabolic reprogramming in Arabidopsis roots. Plant Cell 27: 2244–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R (2010) Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol 153: 757–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar T, Pelayo MAF, Trijatmiko KR, Gabunada LFM, Alam R, Jimenez R, Mendioro MS, Slamet-Loedin IH, Sreenivasulu N, Bailey-Serres J, et al. (2015) A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat Plants 1: 15124. [DOI] [PubMed] [Google Scholar]
- Kudahettige NP, Pucciariello C, Parlanti S, Alpi A, Perata P (2011) Regulatory interplay of the Sub1A and CIPK15 pathways in the regulation of α-amylase production in flooded rice plants. Plant Biol (Stuttg) 13: 611–619 [DOI] [PubMed] [Google Scholar]
- Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LACJ, Perata P, van Dongen JT (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479: 419–422 [DOI] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199: 639–649 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic membranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, van Veen H, Perata P (2016) Plant responses to flooding stress. Curr Opin Plant Biol 33: 64–71 [DOI] [PubMed] [Google Scholar]
- Mair A, Pedrotti L, Wurzinger B, Anrather D, Simeunovic A, Weiste C, Valerio C, Dietrich K, Kirchler T, Nägele T, et al. (2015) SnRK1-triggered switch of bZIP63 dimerization mediates the low-energy response in plants. eLife 4: 05828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-de la Rosa N, Sotillo B, Miskolczi P, Gibbs DJ, Vicente J, Carbonero P, Oñate-Sánchez L, Holdsworth MJ, Bhalerao R, Alabadí D, Blázquez MA (2014) Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol 166: 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, Yaghoubi-Masihi A, Fukao T, Bailey-Serres J (2010) Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MV, Iyer S, Amerhauser C, Lehmann M, van Dongen JT, Geigenberger P (2016) Oxygen sensing via the ethylene response transcription factor RAP2.12 affects plant metabolism and performance under both normoxia and hypoxia. Plant Physiol 172: 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen O, Colmer TD, Sand-Jensen K (2013) Underwater photosynthesis of submerged plants - recent advances and methods. Front Plant Sci 4: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perata P, Alpi A (1993) Plant responses to anaerobiosis. Plant Sci 93: 1–17 [Google Scholar]
- Perata P, Matsukura C, Vernieri P, Yamaguchi J (1997) Sugar repression of a gibberellin-dependent signaling pathway in barley embryos. Plant Cell 9: 2197–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perata P, Pozueta-Romero J, Akazawa T, Yamaguchi J (1992) Effect of anoxia on starch breakdown in rice and wheat seeds. Planta 188: 611–618 [DOI] [PubMed] [Google Scholar]
- Perata P, Voesenek LA (2007) Submergence tolerance in rice requires Sub1A, an ethylene-response-factor-like gene. Trends Plant Sci 12: 43–46 [DOI] [PubMed] [Google Scholar]
- Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M (2002) The starch-related R1 protein is an alpha-glucan, water dikinase. Proc Natl Acad Sci USA 99: 7166–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaniello A, Loreti E, Gonzali S, Novi G, Perata P (2014) A reassessment of the role of sucrose synthase in the hypoxic sucrose-ethanol transition in Arabidopsis. Plant Cell Environ 37: 2294–2302 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Smith SM (2005) Starch degradation. Annu Rev Plant Biol 56: 73–98 [DOI] [PubMed] [Google Scholar]
- Streb S, Zeeman SC (2012) Starch metabolism in Arabidopsis. Arabidopsis Book 10: e0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weits DA, Giuntoli B, Kosmacz M, Parlanti S, Hubberten HM, Riegler H, Hoefgen R, Perata P, van Dongen JT, Licausi F (2014) Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat Commun 5: 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]