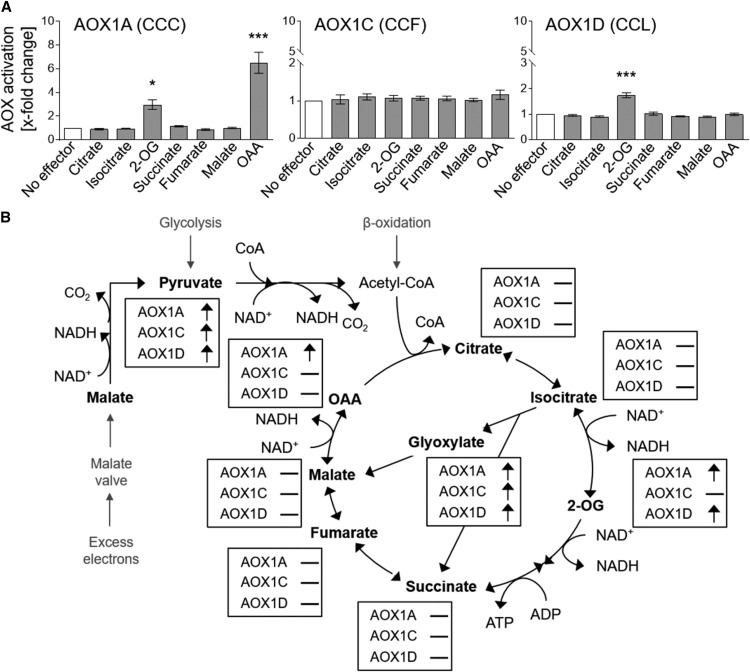
In Arabidopsis, alternative oxidase activation is isoform specific, with AOX1A activated by oxaloacetate and 2-oxoglutarate, AOX1D solely by 2-oxoglutarate, and AOX1C insensitive to both.
Abstract
The cyanide-insensitive alternative oxidase (AOX) is a non-proton-pumping ubiquinol oxidase that catalyzes the reduction of oxygen to water and is posttranslationally regulated by redox mechanisms and 2-oxo acids. Arabidopsis (Arabidopsis thaliana) possesses five AOX isoforms (AOX1A–AOX1D and AOX2). AOX1D expression is increased in aox1a knockout mutants from Arabidopsis (especially after restriction of the cytochrome c pathway) but cannot compensate for the lack of AOX1A, suggesting a difference in the regulation of these isoforms. Therefore, we analyzed the different AOX isoenzymes with the aim to identify differences in their posttranslational regulation. Seven tricarboxylic acid cycle intermediates (citrate, isocitrate, 2-oxoglutarate, succinate, fumarate, malate, and oxaloacetate) were tested for their influence on AOX1A, AOX1C, and AOX1D wild-type protein activity using a refined in vitro system. AOX1C is insensitive to all seven organic acids, AOX1A and AOX1D are both activated by 2-oxoglutarate, but only AOX1A is additionally activated by oxaloacetate. Furthermore, AOX isoforms cannot be transformed to mimic one another by substituting the variable cysteine residues at position III in the protein. In summary, we show that AOX isoforms from Arabidopsis are differentially fine-regulated by tricarboxylic acid cycle metabolites (most likely depending on the amino-terminal region around the highly conserved cysteine residues known to be involved in regulation by the 2-oxo acids pyruvate and glyoxylate) and propose that this is the main reason why they cannot functionally compensate for each other.
Higher plant mitochondria possess two distinct pathways for the transfer of electrons from ubiquinol to molecular oxygen: the cytochrome c oxidase (COX) pathway and the alternative oxidase (AOX) pathway. In the COX pathway, electron transport is coupled to proton translocation and concomitant ATP formation. Electron transport through the AOX pathway occurs without proton translocation and, consequently, is not coupled to ATP synthesis or energy conservation (for review, see Millar et al., 2011). In this case, most of the energy is dissipated as heat (Sluse and Jarmuszkiewicz, 1998; Affourtit et al., 2002). The dimeric AOX mediates the terminal step of the alternative pathway and is localized to the inner mitochondrial membrane, with its catalytic centers oriented toward the matrix (Juszczuk and Rychter, 2003).
In different plant species, the number of nuclear genes encoding AOX isoforms varies between two (e.g. Nelumbo nucifera) and seven (e.g. Arum maculatum). Studies on various transgenic plants indicate that these isoforms are not redundant and cannot compensate for each other under stress or adverse growth conditions (Table I). These include studies on Arabidopsis (Arabidopsis thaliana), which has five nuclear AOX genes: four AOX1 type (A–D) and one AOX2 type (Polidoros et al., 2009). In Arabidopsis, an aox1a knockout cannot be compensated by the expression of other isogenes (Table I; Strodtkötter et al., 2009; Kühn et al., 2015). Although the expression of AOX1D is increased in aox1a mutants, it cannot functionally replace the lack of AOX1A, since, in contrast to the wild type, Ataox1a knockout plants do not survive treatment with antimycin A, an inhibitor of the cytochrome c pathway acting at the site of cytochrome bc1 in complex III (Alexandre and Lehninger, 1984; Campo et al., 1992; Maguire et al., 1992; Xia et al., 1997; Pham et al., 2000; Strodtkötter et al., 2009; Kühn et al., 2015). Moreover, a double mutant impaired in both the COX and AOX pathways (aox1a:rpoTmp) shows more severe growth impairment, even though AOX1D is highly expressed at the transcript and protein levels (Kühn et al., 2015). These results suggest that differences in posttranslational activation of AOX isoforms are more likely to occur than differences in transcriptional regulation in the aox1a:rpoTmp mutants (Strodtkötter et al., 2009; Kühn et al., 2015).
Table I. Overview of studies on aox mutants.
AS, Antisense; ir, RNA interference construct harboring a fragment of a gene in an inverted repeat orientation; OE, overexpression; RNAi, RNA interference; rpoTmp, T3/T7 bacteriophage-type RNA polymerase, mitochondrial and plastidial. So far, in no case could compensation by another isoform be observed.
Besides transcriptional regulation, AOX activity has been shown to be posttranslationally regulated at the two highly conserved Cys residues (CysI and CysII) present in the N-terminal domain of the protein (Millar et al., 1993, 1996; Umbach and Siedow, 1993, 1996; Day and Wiskich, 1995; Day et al., 1995; Rhoads et al., 1998; Siedow and Umbach, 2000; Umbach et al., 2006; Selinski et al., 2016, 2017). Two interrelated mechanisms were identified regulating the activation/inactivation of AOX: (1) oxidation/reduction of the disulfide bridge formed between the two CysI residues in the AOX dime; and (2) further activation of the reduced form via allosteric regulation by 2-oxo acids, also involving the conserved Cys residues (Millar et al., 1993, 1996; Umbach and Siedow, 1993; Rhoads et al., 1998; Umbach et al., 2006; Moore and Albury, 2008; Selinski et al., 2016, 2017).
Although recombinant AOX1A, AOX1C, and AOX1D from Arabidopsis are posttranslationally activated by pyruvate and glyoxylate (Rhoads et al., 1998; Umbach et al., 2002, 2006; Selinski et al., 2016, 2017), the influence of tricarboxylic acid cycle (TCAC) intermediates on AOX activity has not been studied in detail. In soybean (Glycine max) mitochondria and submitochondrial particles, the AOX pathway also is activated by oxaloacetate (OAA) and 2-oxoglutarate (2-OG), but at higher concentrations than pyruvate (Day et al., 1995; Millar et al., 1996). However, these effectors have not been studied in detail, and their effects have not been confirmed directly with AOX protein. Furthermore, these studies used mitochondria isolated from soybean cotyledons, where the only expressed AOX isoform is AOX2a (Finnegan et al., 1997), and tell us nothing about AOX1 isoforms, which are predominant in most other plants. In this work, a detailed comparison of posttranslational activation by TCAC intermediates of the three Arabidopsis isoforms AOX1A, AOX1C, and AOX1D was performed using a sensitive experimental setup with prolonged linear time intervals, based on Escherichia coli membranes enriched in individual AOX isoforms after heterologous expression, as described by Selinski et al. (2016, 2017).
RESULTS
AOX Isoforms Are Differentially Activated by Organic Acids of the TCAC
To analyze the isoform-specific sensitivities of Arabidopsis AOX1A, AOX1C, and AOX1D to TCAC intermediates, each isoform was recombinantly expressed in E. coli BHH8, and membrane vesicles enriched in individual AOX proteins were isolated. A one-letter code for amino acids was used to describe the composition at Cys sites I, II, and III occurring in native and mutant forms combined with a three-letter code, resulting in CCC for the AOX1A wild type (AOX1A-WT), CCF for AOX1C-WT, and CCL for AOX1D-WT, respectively (Selinski et al., 2017). Due to the fact that AOX proteins were heterologously expressed under reducing conditions (these conditions are present in the E. coli cytosol) and that a reductant (DTT) was present during membrane vesicle isolation and activity measurements, AOX proteins are present in their reduced, and therefore activatable, state. Isoform-specific oxygen consumption was measured by linking the NADH dehydrogenases and ubiquinol pool of the E. coli respiratory chain to the heterologously expressed AOX isoform using a Clark-type oxygen electrode (Selinski et al., 2016, 2017).
Based on the observations that monocarboxylic 2-oxo acids such as pyruvate and glyoxylate stimulate AOX1A, AOX1C, and AOX1D activity (Rhoads et al., 1998; Umbach et al., 2002, 2006; Selinski et al., 2016, 2017) and that the dicarboxylic acid succinate activates AOX1A from Arabidopsis after substitution of CysI by Ser or Ala (Djajanegara et al., 1999; Umbach et al., 2002; Selinski et al., 2017), a variety of other organic acids (dicarboxylic acids: fumarate, malate, OAA, 2-OG [and succinate]; tricarboxylic acids: citrate and isocitrate) were tested for their effects on the activity of the three AOX wild-type proteins (Fig. 1A). Neither of the tested tricarboxylic acids (citrate nor isocitrate) influenced the activity of any isoform (Fig. 1A). This also was the case for the dicarboxylic acids fumarate, malate, and succinate. However, OAA and 2-OG (the only TCAC intermediates belonging to the group of 2-oxo acids) significantly stimulated the activity of AOX1A-CCC(WT) (Fig. 1A) but not AOX1A-SCC derivatives (Supplemental Fig. S1). While the AOX1A protein exhibited a 7-fold increase in activity after treatment with OAA compared with its basal activity (no effector), the addition of 2-OG led to a 3-fold increase only. This indicates that AOX1A is more prone to be activated by OAA than by 2-OG. Due to the fact that OAA and 2-OG belong to the group of 2-oxo acids, it is likely that the activation of AOX1A by OAA and 2-OG is based on the same mechanism as is the case for pyruvate and glyoxylate, namely the proposed formation of a thiohemiacetal. AOX1D was activated by 2-OG (2-fold increase) but was insensitive to OAA (Fig. 1A), while the AOX1C-WT protein was insensitive to all tested dicarboxylic and tricarboxylic acids (Fig. 1A). That is, AOX1A responds differentially to metabolites compared with AOX1C and AOX1D, suggesting that isoform-specific functions depend on environmental and/or tissue-specific conditions.
Figure 1.
Influence of organic acids on the activity of AOX wild-type proteins. A, The effect of different organic acids on the activity of AOX1A-, AOX1C-, and AOX1D-WT proteins was analyzed. AOX activity was determined as described by Selinski et al. (2016) using 5 mm citrate, isocitrate, 2-OG, succinate, fumarate, malate, or OAA as effectors. Measurements were carried out as three independent biological replicates. Each biological replicate was measured twice, leading to a total of six values per column. Basal activities (no effector) were 5.7 ± 0.21 nmol oxygen min−1 density units (DU)−1 for AOX1A-WT, 39.28 ± 3.94 nmol oxygen min−1 DU−1 for AOX1C-WT, and 15.26 ± 0.67 nmol oxygen min−1 DU−1 for AOX1D-WT. Asterisks indicate that the differences (*, P < 0.05 and ***, P < 0.001) between the basal activity (no effector) and activities in the presence of the effectors are statistically significant as determined by two-way ANOVA with posthoc Tukey’s honestly significant difference (HSD) test. B, Schematic overview of AOX activation by TCAC intermediates.
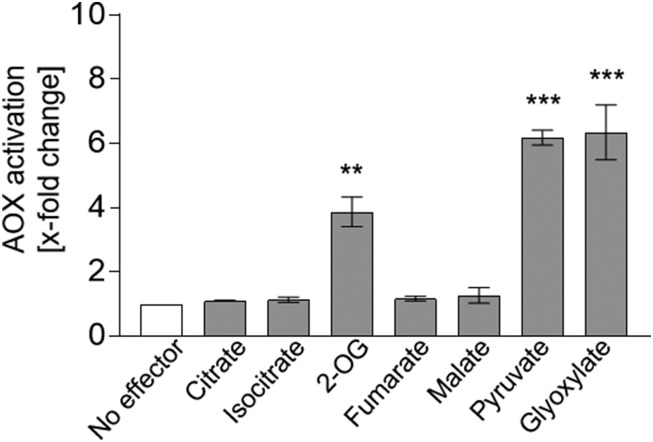
The influence of metabolites on AOX activity also was investigated in isolated mitochondria from Arabidopsis (Fig. 2). The AOX pathway in these mitochondria was stimulated by the 2-oxo acids 2-OG, pyruvate, and glyoxylate in a similar manner to that in isolated E. coli membrane vesicles (Fig. 2). Likewise, the AOX pathway was insensitive to the addition of citrate, isocitrate, fumarate, and malate compared with its activity under reducing conditions solely (no effector; Fig. 2). However, isolated mitochondria pose some problems when testing different metabolites. (1) Isoform-specific responses to the tested metabolites cannot be observed because the isoforms cannot be analyzed separately and, in any case, AOX1A is the predominant form. (2) The effect of OAA on the activity of the AOX pathway cannot be analyzed due to the presence of free malate dehydrogenase (MDH) in isolated mitochondria. Although the intactness of mitochondria was determined to be 90% to 91%, the few broken mitochondria release MDH that then will catalyze NADH oxidation directly in competition with the electron transport chain. (3) The influence of succinate on AOX activity cannot be analyzed accurately because it is an electron transport substrate (Jacoby et al., 2015).
Figure 2.
Influence of organic acids on the activity of the AOX pathway in plant mitochondria. The effect of different organic acids on the activity of the AOX pathway in isolated mitochondria from Arabidopsis was analyzed. AOX activity was determined as described by Jacoby et al. (2015) using 5 mm citrate, isocitrate, 2-OG, fumarate, malate, pyruvate, or glyoxylate as effectors. Measurements were carried out as three independent biological replicates. Each biological replicate was measured twice, leading to a total of six values per column. The basal activity (no effector) of the AOX pathway was 3.96 ± 0.6 nmol oxygen min−1 mg−1 protein. Asterisks indicate that the differences (**, P < 0.01 and ***, P < 0.001) between the basal activity (no effector) and activities in the presence of the effectors are statistically significant as determined by two-way ANOVA with posthoc Tukey’s HSD test.
AOX Isoforms Cannot Be Transformed into One Another by Amino Acid Substitutions at Position CysIII
Given that the AOX1A-WT protein is differentially regulated by metabolites compared with AOX1C and AOX1D (Fig. 1) and that AOX1A possesses a third Cys residue (CysIII) near the diiron center, while AOX1C and AOX1D contain a Phe or Leu residue at this position (Selinski et al., 2017), derivatives containing Phe or Leu at the position of CysIII in AOX1A were generated to mimic AOX1C or AOX1D and vice versa.
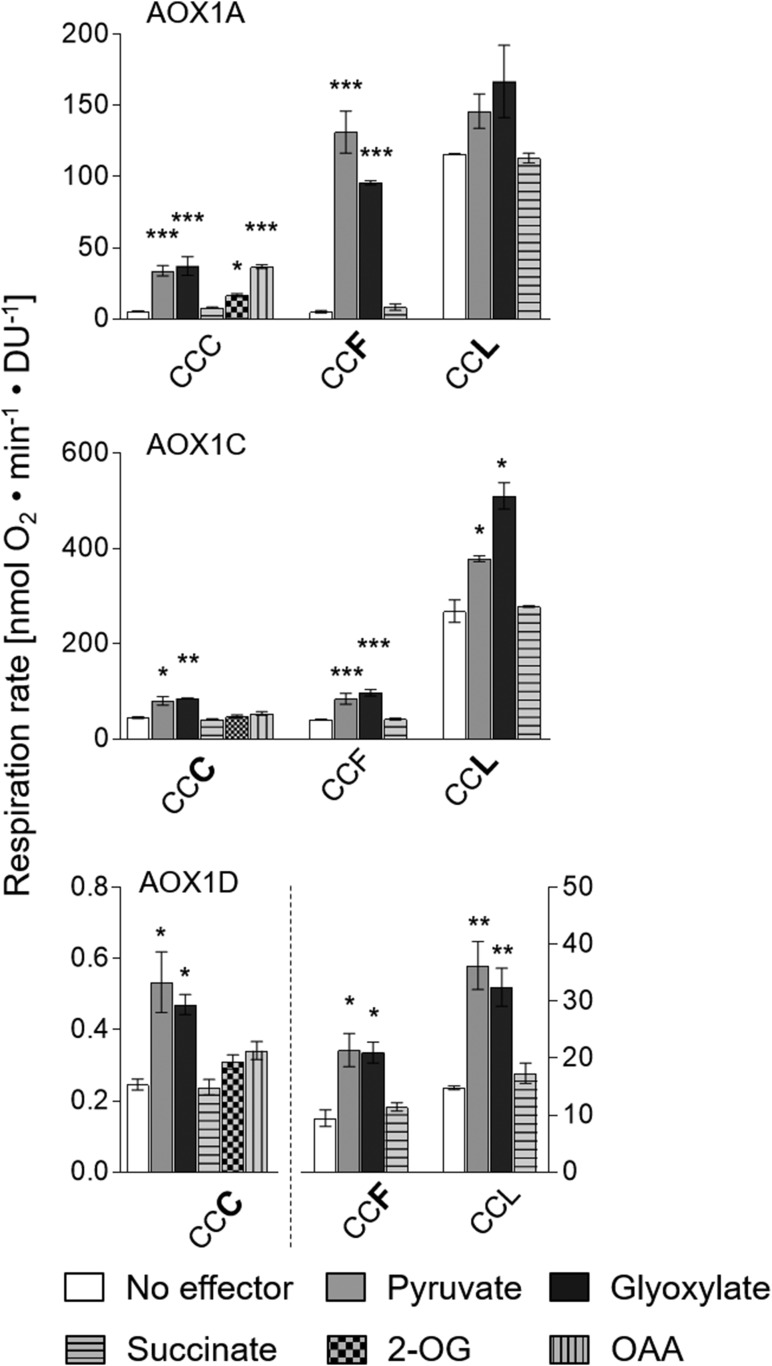
Conversion of AOX1A into the CCF form (i.e. like AOX1C-WT) did not change its activation by 2-oxo acids or its basal activity (Fig. 3, top); in this respect, AOX1A-CCF resembled AOX1C. However, AOX1A-CCF was much more sensitive to pyruvate and glyoxylate (25- to 18-fold increase) compared with AOX1A-CCC#(WT) (7-fold increase) and AOX1C (Fig. 3).
Figure 3.
Convertibility of AOX isoforms into one another by substitutions of amino acid residues at position CysIII. Oxygen consumption measurements and calculations of specific respiration rates (nmol oxygen min−1 density units [DU]−1) were performed as described by Selinski et al. (2016). Measurements were carried out as three independent biological replicates. Each biological replicate was measured twice, leading to a total of six values per column. Basal activities (no effector) were 5.71 ± 0.15 nmol oxygen min−1 DU−1 for AOX1A-WT (CCC), 40.89 ± 0.87 nmol oxygen min−1 DU−1 for AOX1C-WT (CCF), and 14.81 ± 0.34 nmol oxygen min−1 DU−1 for AOX1D-WT (CCL). Asterisks indicate that the differences (*, P < 0.05; **, P < 0.01; and ***, P < 0.001) between the basal activity (no effector) and activities in the presence of the effectors are statistically significant as determined by two-way ANOVA with posthoc Tukey’s HSD test. Wild types are as follows: AOX1A, CCC; AOX1C, CCF; and AOX1D, CCL. Substitutions are presented in the one-letter code for amino acids in enlarged boldface letters. Note the difference in scale for AOX1D proteins: the left y axis belongs to AOX1D-CCC, and the right y axis belongs to AOX1D-CCF and AOX1D-CCL.
Conversion of AOX1C to the CCC form (like AOX1A-WT), on the other hand, had little effect on its response to organic acids: it remained stimulated by pyruvate and glyoxylate and insensitive to the other, larger 2-oxo acids tested (Figs. 1 and 3, middle). Obviously, the transformation of AOX1A into AOX1C and vice versa is not possible simply by substituting CysIII in AOX1A by Phe or PheIII in AOX1C by Cys. Likewise, neither isoform can be transformed into AOX1D simply by substitution of CysIII in AOX1A or PheIII in AOX1C by Leu. In both cases, the basal activity was increased dramatically (20-fold for AOX1A-CCL and 7-fold for AOX1C-CCL) over the activity of the WT proteins. In addition, AOX1A-CCL activity was insensitive to 2-oxo acids (Fig. 3), in contrast to the AOX1D-WT protein, which is stimulated by 2-OG (Fig. 1).
AOX1D-CCL#(WT) and AOX1D-CCF (like AOX1C) exhibited nearly identical basal activities and were activated by pyruvate and glyoxylate (Fig. 3, bottom). The substitution of LeuIII in AOX1D by Cys (as in the AOX1A-WT protein), on the other hand, resulted in a very low overall activity, but this form was nonetheless stimulated by the 2-oxo acids pyruvate and glyoxylate. However, as found with AOX1C-CCC, AOX1D-CCC was insensitive to the 2-oxo acids OAA and 2-OG (Fig. 3), indicating that substitution of LeuIII by Cys or Phe does not, in itself, confer similar responsiveness to 2-OG to that seen with AOX1A-WT or AOX1C-WT.
Overall, these results demonstrate that the differential regulation by metabolites is not dependent solely on the amino acid present at position CysIII in the AOX protein.
DISCUSSION AND CONCLUSION
About 25 years ago, the posttranslational regulation of plant AOX activity by redox mechanisms and 2-oxo acids like pyruvate and glyoxylate was discovered (Millar et al., 1993; Umbach and Siedow, 1993). Succinate also was shown to activate specific AOX isoforms that contain a Ser residue instead of a Cys residue at position I (Djajanegara et al., 1999; Holtzapffel et al., 2003). It also has been shown that AOX1A, AOX1C, and AOX1D are activated by pyruvate and glyoxylate, although AOX1A is activated to a greater extent (Selinski et al., 2017). Consequently, we used isolated membrane vesicles of E. coli BHH8 containing heterologously expressed AOX protein and have confirmed that AOX-WT proteins are insensitive to succinate but are all activated by pyruvate and glyoxylate, consistent with the previous studies. It has been shown that AOX2 in soybean is activated by OAA and 2-OG (Day et al., 1995; Millar et al., 1996), but these studies were carried out with mitochondria that contain multiple isoforms of AOX; thus, isoform-specific activation could not be determined. Using a bacterial expression system expressing individual AOX isoforms (Selinski et al., 2016, 2017), differential activation by metabolites is possible to detect. In this study, we used this system to demonstrate several important differences between the different isoforms in their responsiveness to the 2-oxo acids OAA and 2-OG. AOX1A was stimulated by both OAA and 2-OG, AOX1D was activated by 2-OG solely, while AOX1C was insensitive to all tested TCAC intermediates (Fig. 1).
Citrate does not directly influence AOX activity, but its accumulation, triggered by the inhibition of aconitase, was shown to induce AOX gene expression, especially AOX1A and AOX1D, and therefore leads to an increased capacity of the AOX pathway under stress conditions in plants (Vanlerberghe and McIntosh, 1997; Gray et al., 2004; Gupta et al., 2012; Konert et al., 2015). Malate itself also does not influence AOX activity directly (Fig. 1). However, under various stress conditions, malate [e.g. generated in the chloroplast via NAD(P)-dependent MDH] can be transported into mitochondria, where its conversion to pyruvate (via malic enzyme) or OAA (via MDH) occurs (Fig. 1B). The 2-oxo acids pyruvate and OAA both stimulate AOX activity (the latter only AOX1A) and, therefore, could prevent an overreduction of the mitochondrial electron transport chain under various stress conditions when excess reducing equivalents are exported indirectly from the chloroplast via the malate valve (Scheibe, 2004). This is likely to be particularly important under photoinhibitory conditions in leaf cells. The accumulation of OAA also could occur when citrate levels increase and inhibit citrate synthase (Wiegand and Remington, 1986). In isolated mitochondria, OAA accumulates substantially during malate oxidation under conditions where malic enzyme is not activated and the cytochrome c pathway is restricted by adenylates, curtailing NADH oxidation and causing a severe inhibition of electron transport (Tobin et al., 1980; Day et al., 1984). This is a consequence of the equilibrium conditions of the MDH reaction. If this occurs in vivo, for example under conditions where respiration is limited by the energy status of the cell, then OAA activation of AOX would help to alleviate this problem by providing an additional avenue for NADH oxidation.
While pyruvate and OAA possibly represent the most important regulatory metabolites for AOX(1A) activity under stress conditions, 2-OG and glyoxylate also can accumulate in mitochondria, especially during photorespiration (Bari et al., 2004). Glyoxylate, 2-OG, and pyruvate also are substrates of the γ-aminobutyric acid (GABA) shunt, which is functionally linked to the TCAC (Narayan and Nair, 1990; Bouché and Fromm, 2004; Studart-Guimarães et al., 2007). The interconversion of Glu to GABA takes place in the cytosol. Subsequently, GABA is transported into mitochondria, where it is converted to succinic-semialdehyde catalyzed by GABA-transaminase (GABA-T). Depending on the enzyme’s substrate specificity, the GABA-T enzyme can be divided into two types: the 2-OG-dependent GABA-T (GABA-TK) and the pyruvate-dependent GABA-T (GABA-TP). While GABA-TK uses 2-OG to generate Glu, the bispecific GABA-TP uses pyruvate to generate Ala or glyoxylate to generate Gly (Clark et al., 2009a, 2009b; Shimajiri et al., 2013; Trobacher et al., 2013). Succinic-semialdehyde generated by GABA-T is further converted to succinate, which can be used in the TCAC and as an electron donor of the mitochondrial electron transport chain. In both cases, increased activity of AOX will be important to avoid an overreduction of the mitochondrial electron transport chain. Studies in vivo with a variety of plant species show that AOX activities are correlated with GABA and 2-OG (Florez-Sarasa et al., 2016), and the correlation of increases in AOX activity with 2-OG is consistent with the stimulatory effect of 2-OG observed in the study outlined here.
In Arabidopsis, AOX1A and AOX1D are highly stress responsive at the transcriptional level, in contrast to the other AOX genes, which do not respond to various stress conditions (Clifton et al., 2006). It is interesting, therefore, that these two isoforms are activated by 2-OG (both), OAA (only AOX1A), while AOX1C is insensitive to all tested TCAC intermediates. This may ensure that electron flux through the AOX pathway is optimized, helping to minimize oxidative stress under various environmental conditions.
Differential posttranslational activation of AOX1 isoforms also may explain why these proteins are unable to functionally substitute for each other in planta. While this is clearly the case for plants under stress conditions, cyanide-resistant respiration has been reported in vivo in aox1a T-DNA lines under nonstress conditions but was proposed to be due to a variety of other oxidases, as it was inhibited by salicylhydroxamic acid, an inhibitor of AOX (Watanabe et al., 2008, 2010). In our hands in two different laboratories, the Ataox1a T-DNA lines do not show expression of any of the other AOX genes under normal conditions, as measured by real-time quantitative reverse transcription-PCR, and mitochondria isolated from these lines do not contain detectable AOX protein or activity (J. Selinski, D.A. Day, and J. Whelan, unpublished data).
The differential activation by metabolites may be an example of a neofunctionalization of AOX isoforms, where, due to activation by different metabolites, various isoforms are only active under certain conditions. This proposal is consistent with the variation in the number of AOX genes that are observed in plants, which is not linked to their phylogenetic history. Expression of a number of isoproteins that are differentially activated by metabolites may allow fine-tuning of their activity in different tissues and under different circumstances. Differential activation is likely to occur at the N-terminal end of the AOX protein that displays more variation in sequence identity (amino acid identity of 19%–43%, calculated for the first 50 amino acids in the N-terminal region of the mature proteins) compared with the C-terminal end (amino acid identity of 82%–94%, calculated for the last 50 amino acids in the C-terminal region). The fact that the stimulation of AOX isoforms does not depend just on the size of the binding pockets (otherwise, OAA also would stimulate AOX1D, because this molecule is smaller than 2-OG, which is activating; Fig. 1) further supports the importance of the N-terminal region. Especially the position of the additional negative charge in the 2-oxo acids OAA and 2-OG in relation to the 2-oxo group appears to be essential for their interaction with the amino acid residues in close proximity to the conserved Cys residues.
In this study, we used a concentration of 5 mm of added metabolites to ensure maximal activation, as the mitochondrial concentration of these metabolites in vivo is unknown. The concentration of OAA in vivo has not been measured even in whole cells, presumably due to rapid conversion to citrate or the spontaneous decarboxylation to pyruvate. While it is recognized that the TCAC exists in plants in many situations, it does not always operate as a complete cycle, with intermediates used in various biosynthetic reactions. For example, 2-OG is used in the assimilation of nitrogen and GABA synthesis (Sweetlove et al., 2010). Again, the extent of these pathways differs between species and tissues, and in mature illuminated leaves, noncyclic flux is proposed (Sweetlove et al., 2010). Added onto this is the occurrence of metabolic channeling of citrate and fumarate, but not 2-OG, in plant mitochondria (Zhang et al., 2017), and the fact that most steps in the TCAC can be bypassed by similar nonmitochondrial activities in other cellular locations means that the matrix subcompartment concentrations are unknown. Finally, the existence of alternative respirasomes, where AOX isoforms are associated with alternative NAD(P)H dehydrogenases, suggests subcompartmentalizations of respiration and that local concentrations of substrates may differ significantly from the concentrations in a given compartment (Senkler et al., 2017).
While our results offer a possible explanation for the finding that different AOX isoforms cannot completely substitute for each other, the situation in planta needs to be considered under normal and stress conditions. Inactivation of AOX1 in Arabidopsis and tobacco (Nicotiana tabacum) has no apparent physiological or growth consequences under normal, optimal growth conditions. This can be interpreted as either that AOX is not required for growth under normal conditions or that other isoforms can compensate for each other. However, an alternative explanation is that, under normal conditions, changes in the underlying transcriptional program compensate for a lack of AOX. In Arabidopsis, it has been shown that the transcriptome of aox1a plants is greatly altered compared with that in wild-type plants under conditions where no changes in growth or physiology are apparent (Giraud et al., 2008). In these and other studies, no other AOX isoforms are expressed or detected in aox1a T-DNA lines (Giraud et al., 2008; Watanabe et al., 2008, 2010). This differs from antisense lines, where significant, albeit reduced, AOX can be detected (Guy and Vanlerberghe, 2005; Florez-Sarasa et al., 2011), and in Arabidopsis, the remaining AOX capacity in antisense lines appears to be fully engaged (Florez-Sarasa et al., 2011). Thus, under normal growth conditions, while the other isoforms are not expressed, changes in the complete underlying transcriptional program in T-DNA lines suggest that compensation occurs via other mechanisms.
In contrast, under stress conditions, other isoforms are induced in Arabidopsis, in particular AOX1D (Table I). Despite this induction, the altered phenotypes of aox1a mutants indicate that they are not able to compensate for the absence of AOX1a. This may be due to the fact that there is not sufficient AOX1D protein, but in several studies, it took several days for the altered phenotypes to emerge and significant AOX1D transcripts and protein could be detected (Giraud et al., 2008; Strodtkötter et al., 2009; Kühn et al., 2015). This suggests that the biochemical properties of isoforms may differ, as shown in this study. Supportive for this suggestion is that, in investigations of the in vivo activities in different plant species under high light, the activity of AOX was independent of AOX protein abundance, and differential posttranslational regulation of AOX was proposed (Florez-Sarasa et al., 2016). Our findings are consistent with this in vivo study highlighting that differences may occur between species.
Some further questions remain to be answered in future studies. (1) Homodimerization/heterodimerization of AOX isoforms. AOX has been proposed to be present in plant mitochondria as covalently and noncovalently linked homodimers (Umbach and Siedow, 1993). However, it is not possible to distinguish AOX homodimers and heterodimers based on SDS-PAGE, because monomers of different isoforms are of nearly identical size (e.g. for Arabidopsis, molecular masses of mature monomeric proteins vary between 32 kD [AOX1B and AOX1D] and 33 kD [AOX1A and AOX1C]). AOX2 is larger (38 kD) but is poorly expressed in Arabidopsis. It is possible that heterodimers possess different properties from homodimers, which would allow further fine-tuning of their activities. (2) Amount of total AOX protein and capacity in plant cells. Although the expression of AOX1D is increased when AOX1A is lacking (Strodtkötter et al., 2009), the total amount of AOX protein is lower compared with wild-type plants (data not shown). Therefore, the total AOX protein present in aox1a mutants might not be sufficient to compensate for the lack of AOX1A, depending on the AOX pathway capacity. As shown by Selinski et al. (2017), fully activated AOX1A and AOX1D (after the addition of pyruvate or glyoxylate) exhibit similar activities, indicating that AOX1D should be able to compensate for the lack of AOX1A in aox1a knockout mutants. Since this is not the case, other regulatory mechanisms must play a role in this context. (3) Cell-specific expression patterns of AOX isoforms. It is also conceivable that AOX isoforms cannot compensate for each other because of differences in their cell-specific localizations. This has to be investigated further.
MATERIALS AND METHODS
Plasmids Used in This Study
Plasmids p536 (AOX1A in pET-22b), p537 (AOX1D in pET-22b), and p583 (AOX1C in pET-22b) were obtained from former studies by Selinski et al. (2016, 2017). For mutagenesis, plasmids p536, p537, and p583 were amplified via PCR using PfuUltra II Fusion HS DNA-Polymerase (Agilent Technologies) and specific mutagenesis primers for amplification (Supplemental Table S1). Following PCR, products were treated with DpnI for 1 h at 37°C to eliminate the maternal DNA template. Constructs were verified by sequencing.
Isolation and Oxygen Consumption Measurements of Plant Mitochondria
Intact plant mitochondria were isolated from Arabidopsis (Arabidopsis thaliana Columbia-0) water cultures, and subsequent respiratory measurements were carried out as described by Murcha and Whelan (2015) with some modifications. To analyze the effect of metabolites on the activity of the AOX pathway, citrate, isocitrate, 2-OG, fumarate, malate, pyruvate, or glyoxylate was added to the reaction chamber. The final concentration of each effector used in this study was 5 mm solubilized in respiration medium (for details, see Jacoby et al., 2015) with a pH adjusted to 7.
Activity Measurements of Recombinantly Expressed AOXs
Cell growth, protein expression, Escherichia coli membrane vesicle isolation, and AOX activity measurements with concomitant immunoblot analysis and calculations were carried out according to Selinski et al. (2016). The final concentration of each effector (citrate, isocitrate, 2-OG, succinate, fumarate, malate, or OAA) used in this study was 5 mm. The pH of all effectors was adjusted to 7 using NaOH before use.
Statistical Analysis
Statistical evaluations were conducted by means of two-way ANOVA with posthoc Tukey’s HSD test integrated in GraphPad Prism 7 (GraphPad Software). Differences with P < 0.05, P < 0.01, and P < 0.001 were considered as significant and indicated as *, **, and ***, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Influence of organic acids on the activity of AOX derivatives after substitution of CysI by Ser.
Supplemental Table S1. Primers used in this study.
Footnotes
This work was supported by the Collaborative Research Center 944, Physiology and Dynamics of Cellular Microcompartments (R.S. and G.D.-H.), the government of Lower Saxonia (Lichtenberg Fellowship to J.S.), the Frauenförderpool of Osnabrück University (J.S.), La Trobe University School of Life Science Publication Booster Award, and the Alexander von Humboldt Foundation (Feodor Lynen Research Fellowship to J.S.). J.W. was supported by the Australian Research Council Centre of Excellence Program in Plant Energy Biology (CE140100008).
Articles can be viewed without a subscription.
References
- Affourtit C, Albury MS, Crichton PG, Moore AL (2002) Exploring the molecular nature of alternative oxidase regulation and catalysis. FEBS Lett 510: 121–126 [DOI] [PubMed] [Google Scholar]
- Alexandre A, Lehninger AL (1984) Bypasses of the antimycin A block of mitochondrial electron transport in relation to ubisemiquinone function. Biochim Biophys Acta 767: 120–129 [DOI] [PubMed] [Google Scholar]
- Bari R, Kebeish R, Kalamajka R, Rademacher T, Peterhänsel C (2004) A glycolate dehydrogenase in the mitochondria of Arabidopsis thaliana. J Exp Bot 55: 623–630 [DOI] [PubMed] [Google Scholar]
- Bouché N, Fromm H (2004) GABA in plants: just a metabolite? Trends Plant Sci 9: 110–115 [DOI] [PubMed] [Google Scholar]
- Campo ML, Kinnally KW, Tedeschi H (1992) The effect of antimycin A on mouse liver inner mitochondrial membrane channel activity. J Biol Chem 267: 8123–8127 [PubMed] [Google Scholar]
- Chai TT, Simmonds D, Day DA, Colmer TD, Finnegan PM (2010) Photosynthetic performance and fertility are repressed in GmAOX2b antisense soybean. Plant Physiol 152: 1638–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SM, Di Leo R, Dhanoa PK, Van Cauwenberghe OR, Mullen RT, Shelp BJ (2009a) Biochemical characterization, mitochondrial localization, expression, and potential functions for an Arabidopsis gamma-aminobutyrate transaminase that utilizes both pyruvate and glyoxylate. J Exp Bot 60: 1743–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SM, Di Leo R, Van Cauwenberghe OR, Mullen RT, Shelp BJ (2009b) Subcellular localization and expression of multiple tomato gamma-aminobutyrate transaminases that utilize both pyruvate and glyoxylate. J Exp Bot 60: 3255–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton R, Millar AH, Whelan J (2006) Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim Biophys Acta 1757: 730–741 [DOI] [PubMed] [Google Scholar]
- Cvetkovska M, Dahal K, Alber NA, Jin C, Cheung M, Vanlerberghe GC (2014) Knockdown of mitochondrial alternative oxidase induces the ‘stress state’ of signaling molecule pools in Nicotiana tabacum, with implications for stomatal function. New Phytol 203: 449–461 [DOI] [PubMed] [Google Scholar]
- Cvetkovska M, Vanlerberghe GC (2012) Alternative oxidase modulates leaf mitochondrial concentrations of superoxide and nitric oxide. New Phytol 195: 32–39 [DOI] [PubMed] [Google Scholar]
- Cvetkovska M, Vanlerberghe GC (2013) Alternative oxidase impacts the plant response to biotic stress by influencing the mitochondrial generation of reactive oxygen species. Plant Cell Environ 36: 721–732 [DOI] [PubMed] [Google Scholar]
- Dahal K, Martyn GD, Alber NA, Vanlerberghe GC (2017) Coordinated regulation of photosynthetic and respiratory components is necessary to maintain chloroplast energy balance in varied growth conditions. J Exp Bot 68: 657–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahal K, Vanlerberghe GC (2017) Alternative oxidase respiration maintains both mitochondrial and chloroplast function during drought. New Phytol 213: 560–571 [DOI] [PubMed] [Google Scholar]
- Dahal K, Wang J, Martyn GD, Rahimy F, Vanlerberghe GC (2014) Mitochondrial alternative oxidase maintains respiration and preserves photosynthetic capacity during moderate drought in Nicotiana tabacum. Plant Physiol 166: 1560–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DA, Neuburger M, Douce R (1984) Activation of NAD-linked malic enzyme in intact plant mitochondria by exogenous coenzyme A. Arch Biochem Biophys 231: 233–242 [DOI] [PubMed] [Google Scholar]
- Day DA, Whelan J, Millar AH, Siedow JN, Wiskich JT (1995) Regulation of the alternative oxidase in plants and fungi. Aust J Plant Physiol 22: 497–509 [Google Scholar]
- Day DA, Wiskich JT (1995) Regulation of alternative oxidase activity in higher plants. J Bioenerg Biomembr 27: 379–385 [DOI] [PubMed] [Google Scholar]
- Djajanegara I, Holtzapffel R, Finnegan PM, Hoefnagel MHN, Berthold DA, Wiskich JT, Day DA (1999) A single amino acid change in the plant alternative oxidase alters the specificity of organic acid activation. FEBS Lett 454: 220–224 [DOI] [PubMed] [Google Scholar]
- Finnegan PM, Whelan J, Millar AH, Zhang Q, Smith MK, Wiskich JT, Day DA (1997) Differential expression of the multigene family encoding the soybean mitochondrial alternative oxidase. Plant Physiol 114: 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorani F, Umbach AL, Siedow JN (2005) The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature: a study of Arabidopsis AOX1a transgenic plants. Plant Physiol 139: 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez-Sarasa I, Flexas J, Rasmusson AG, Umbach AL, Siedow JN, Ribas-Carbo M (2011) In vivo cytochrome and alternative pathway respiration in leaves of Arabidopsis thaliana plants with altered alternative oxidase under different light conditions. Plant Cell Environ 34: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Florez-Sarasa I, Ribas-Carbo M, Del-Saz NF, Schwahn K, Nikoloski Z, Fernie AR, Flexas J (2016) Unravelling the in vivo regulation and metabolic role of the alternative oxidase pathway in C3 species under photoinhibitory conditions. New Phytol 212: 66–79 [DOI] [PubMed] [Google Scholar]
- Gandin A, Denysyuk M, Cousins AB (2014) Disruption of the mitochondrial alternative oxidase (AOX) and uncoupling protein (UCP) alters rates of foliar nitrate and carbon assimilation in Arabidopsis thaliana. J Exp Bot 65: 3133–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin A, Duffes C, Day DA, Cousins AB (2012) The absence of alternative oxidase AOX1A results in altered response of photosynthetic carbon assimilation to increasing CO2 in Arabidopsis thaliana. Plant Cell Physiol 53: 1627–1637 [DOI] [PubMed] [Google Scholar]
- Giraud E, Ho LH, Clifton R, Carroll A, Estavillo G, Tan YF, Howell KA, Ivanova A, Pogson BJ, Millar AH, et al. (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray GR, Maxwell DP, Villarimo AR, McIntosh L (2004) Mitochondria/nuclear signaling of alternative oxidase gene expression occurs through distinct pathways involving organic acids and reactive oxygen species. Plant Cell Rep 23: 497–503 [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Shah JK, Brotman Y, Jahnke K, Willmitzer L, Kaiser WM, Bauwe H, Igamberdiev AU (2012) Inhibition of aconitase by nitric oxide leads to induction of the alternative oxidase and to a shift of metabolism towards biosynthesis of amino acids. J Exp Bot 63: 1773–1784 [DOI] [PubMed] [Google Scholar]
- Guy RD, Vanlerberghe GC (2005) Partitioning of respiratory electrons in the dark in leaves of transgenic tobacco with modified levels of alternative oxidase. Physiol Plant 125: 171–180 [Google Scholar]
- Holtzapffel RC, Castelli J, Finnegan PM, Millar AH, Whelan J, Day DA (2003) A tomato alternative oxidase protein with altered regulatory properties. Biochim Biophys Acta 1606: 153–162 [DOI] [PubMed] [Google Scholar]
- Jacoby RP, Millar AH, Taylor NL (2015) Assessment of respiration in isolated plant mitochondria using Clark-type electrodes. In Whelan J, Murcha MW, eds, Plant Mitochondria: Methods and Protocols. Humana Press, New York, pp 165–185 [DOI] [PubMed] [Google Scholar]
- Juszczuk IM, Rychter AM (2003) Alternative oxidase in higher plants. Acta Biochim Pol 50: 1257–1271 [PubMed] [Google Scholar]
- Keunen E, Schellingen K, Van Der Straeten D, Remans T, Colpaert J, Vangronsveld J, Cuypers A (2015) ALTERNATIVE OXIDASE1a modulates the oxidative challenge during moderate Cd exposure in Arabidopsis thaliana leaves. J Exp Bot 66: 2967–2977 [DOI] [PubMed] [Google Scholar]
- Konert G, Trotta A, Kouvonen P, Rahikainen M, Durian G, Blokhina O, Fagerstedt K, Muth D, Corthals GL, Kangasjärvi S (2015) Protein phosphatase 2A (PP2A) regulatory subunit B′γ interacts with cytoplasmic ACONITASE 3 and modulates the abundance of AOX1A and AOX1D in Arabidopsis thaliana. New Phytol 205: 1250–1263 [DOI] [PubMed] [Google Scholar]
- Kühn K, Yin G, Duncan O, Law SR, Kubiszewski-Jakubiak S, Kaur P, Meyer E, Wang Y, Small CC, Giraud E, et al. (2015) Decreasing electron flux through the cytochrome and/or alternative respiratory pathways triggers common and distinct cellular responses dependent on growth conditions. Plant Physiol 167: 228–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire JJ, Kagan VE, Packer L (1992) Electron transport between cytochrome c and alpha tocopherol. Biochem Biophys Res Commun 188: 190–197 [DOI] [PubMed] [Google Scholar]
- Mathy G, Cardol P, Dinant M, Blomme A, Gérin S, Cloes M, Ghysels B, DePauw E, Leprince P, Remacle C, et al. (2010) Proteomic and functional characterization of a Chlamydomonas reinhardtii mutant lacking the mitochondrial alternative oxidase 1. J Proteome Res 9: 2825–2838 [DOI] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Hoefnagel M, Day DA, Wiskich JT (1996) Specificity of the organic acid activation of alternative oxidase in plant mitochondria. Plant Physiol 111: 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Soole KL, Day DA (2011) Organization and regulation of mitochondrial respiration in plants. Annu Rev Plant Biol 62: 79–104 [DOI] [PubMed] [Google Scholar]
- Millar AH, Wiskich JT, Whelan J, Day DA (1993) Organic acid activation of the alternative oxidase of plant mitochondria. FEBS Lett 329: 259–262 [DOI] [PubMed] [Google Scholar]
- Moore AL, Albury MS (2008) Further insights into the structure of the alternative oxidase: from plants to parasites. Biochem Soc Trans 36: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Murcha MW, Whelan J (2015) Isolation of intact mitochondria from the model plant species Arabidopsis thaliana and Oryza sativa. In Whelan J, Murcha MW, eds, Plant Mitochondria: Methods and Protocols. Humana Press, New York, pp 1–12 [DOI] [PubMed] [Google Scholar]
- Narayan VS, Nair PM (1990) Metabolism, enzymology and possible roles of 4-aminobutyrate in higher plants. Phytochemistry 29: 367–375 [Google Scholar]
- Parsons HL, Yip JY, Vanlerberghe GC (1999) Increased respiratory restriction during phosphate-limited growth in transgenic tobacco cells lacking alternative oxidase. Plant Physiol 121: 1309–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham NA, Robinson BH, Hedley DW (2000) Simultaneous detection of mitochondrial respiratory chain activity and reactive oxygen in digitonin-permeabilized cells using flow cytometry. Cytometry 41: 245–251 [DOI] [PubMed] [Google Scholar]
- Polidoros AN, Mylona PV, Arnholdt-Schmitt B (2009) AOX gene structure, transcript variation and expression in plants. Physiol Plant 137: 342–353 [DOI] [PubMed] [Google Scholar]
- Rhoads DM, Umbach AL, Sweet CR, Lennon AM, Rauch GS, Siedow JN (1998) Regulation of the cyanide-resistant alternative oxidase of plant mitochondria: identification of the cysteine residue involved in alpha-keto acid stimulation and intersubunit disulfide bond formation. J Biol Chem 273: 30750–30756 [DOI] [PubMed] [Google Scholar]
- Robson CA, Vanlerberghe GC (2002) Transgenic plant cells lacking mitochondrial alternative oxidase have increased susceptibility to mitochondria-dependent and -independent pathways of programmed cell death. Plant Physiol 129: 1908–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe R. (2004) Malate valves to balance cellular energy supply. Physiol Plant 120: 21–26 [DOI] [PubMed] [Google Scholar]
- Selinski J, Hartmann A, Höfler S, Deckers-Hebestreit G, Scheibe R (2016) Refined method to study the posttranslational regulation of alternative oxidases from Arabidopsis thaliana in vitro. Physiol Plant 157: 264–279 [DOI] [PubMed] [Google Scholar]
- Selinski J, Hartmann A, Kordes A, Deckers-Hebestreit G, Whelan J, Scheibe R (2017) Analysis of posttranslational activation of alternative oxidase isoforms. Plant Physiol 174: 2113–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkler J, Senkler M, Eubel H, Hildebrandt T, Lengwenus C, Schertl P, Schwarzländer M, Wagner S, Wittig I, Braun HP (2017) The mitochondrial complexome of Arabidopsis thaliana. Plant J 89: 1079–1092 [DOI] [PubMed] [Google Scholar]
- Shimajiri Y, Ozaki K, Kainou K, Akama K (2013) Differential subcellular localization, enzymatic properties and expression patterns of γ-aminobutyric acid transaminases (GABA-Ts) in rice (Oryza sativa). J Plant Physiol 170: 196–201 [DOI] [PubMed] [Google Scholar]
- Siedow JN, Umbach AL (2000) The mitochondrial cyanide-resistant oxidase: structural conservation amid regulatory diversity. Biochim Biophys Acta 1459: 432–439 [DOI] [PubMed] [Google Scholar]
- Sieger SM, Kristensen BK, Robson CA, Amirsadeghi S, Eng EW, Abdel-Mesih A, Møller IM, Vanlerberghe GC (2005) The role of alternative oxidase in modulating carbon use efficiency and growth during macronutrient stress in tobacco cells. J Exp Bot 56: 1499–1515 [DOI] [PubMed] [Google Scholar]
- Sluse FE, Jarmuszkiewicz W (1998) Alternative oxidase in the branched mitochondrial respiratory network: an overview on structure, function, regulation, and role. Braz J Med Biol Res 31: 733–747 [DOI] [PubMed] [Google Scholar]
- Strodtkötter I, Padmasree K, Dinakar C, Speth B, Niazi PS, Wojtera J, Voss I, Do PT, Nunes-Nesi A, Fernie AR, et al. (2009) Induction of the AOX1D isoform of alternative oxidase in A. thaliana T-DNA insertion lines lacking isoform AOX1A is insufficient to optimize photosynthesis when treated with antimycin A. Mol Plant 2: 284–297 [DOI] [PubMed] [Google Scholar]
- Studart-Guimarães C, Fait A, Nunes-Nesi A, Carrari F, Usadel B, Fernie AR (2007) Reduced expression of succinyl-coenzyme A ligase can be compensated for by up-regulation of the gamma-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol 145: 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Beard KF, Nunes-Nesi A, Fernie AR, Ratcliffe RG (2010) Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci 15: 462–470 [DOI] [PubMed] [Google Scholar]
- Tobin A, Djerdjour B, Journet E, Neuburger M, Douce R (1980) Effect of NAD on malate oxidation in intact plant mitochondria. Plant Physiol 66: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobacher CP, Clark SM, Bozzo GG, Mullen RT, DeEll JR, Shelp BJ (2013) Catabolism of GABA in apple fruit: subcellular localization and biochemical characterization of two γ-aminobutyrate transaminases. Postharvest Biol Technol 75: 106–113 [Google Scholar]
- Umbach AL, Fiorani F, Siedow JN (2005) Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol 139: 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach AL, Gonzàlez-Meler MA, Sweet CR, Siedow JN (2002) Activation of the plant mitochondrial alternative oxidase: insights from site-directed mutagenesis. Biochim Biophys Acta 1554: 118–128 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Ng VS, Siedow JN (2006) Regulation of plant alternative oxidase activity: a tale of two cysteines. Biochim Biophys Acta 1757: 135–142 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN (1993) Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol 103: 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN (1996) The reaction of the soybean cotyledon mitochondrial cyanide-resistant oxidase with sulfhydryl reagents suggests that alpha-keto acid activation involves the formation of a thiohemiacetal. J Biol Chem 271: 25019–25026 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L (1997) Alternative oxidase: from gene to function. Annu Rev Plant Physiol Plant Mol Biol 48: 703–734 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, Vanlerberghe AE, McIntosh L (1997) Molecular genetic evidence of the ability of alternative oxidase to support respiratory carbon metabolism. Plant Physiol 113: 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma A, Bashyam L, Senthilkumaran B, Scheibe R, Padmasree K (2014) Physiological role of AOX1a in photosynthesis and maintenance of cellular redox homeostasis under high light in Arabidopsis thaliana. Plant Physiol Biochem 81: 44–53 [DOI] [PubMed] [Google Scholar]
- Vishwakarma A, Tetali SD, Selinski J, Scheibe R, Padmasree K (2015) Importance of the alternative oxidase (AOX) pathway in regulating cellular redox and ROS homeostasis to optimize photosynthesis during restriction of the cytochrome oxidase pathway in Arabidopsis thaliana. Ann Bot 116: 555–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rajakulendran N, Amirsadeghi S, Vanlerberghe GC (2011) Impact of mitochondrial alternative oxidase expression on the response of Nicotiana tabacum to cold temperature. Physiol Plant 142: 339–351 [DOI] [PubMed] [Google Scholar]
- Wang J, Vanlerberghe GC (2013) A lack of mitochondrial alternative oxidase compromises capacity to recover from severe drought stress. Physiol Plant 149: 461–473 [DOI] [PubMed] [Google Scholar]
- Watanabe CK, Hachiya T, Takahara K, Kawai-Yamada M, Uchimiya H, Uesono Y, Terashima I, Noguchi K (2010) Effects of AOX1a deficiency on plant growth, gene expression of respiratory components and metabolic profile under low-nitrogen stress in Arabidopsis thaliana. Plant Cell Physiol 51: 810–822 [DOI] [PubMed] [Google Scholar]
- Watanabe CK, Hachiya T, Terashima I, Noguchi K (2008) The lack of alternative oxidase at low temperature leads to a disruption of the balance in carbon and nitrogen metabolism, and to an up-regulation of antioxidant defence systems in Arabidopsis thaliana leaves. Plant Cell Environ 31: 1190–1202 [DOI] [PubMed] [Google Scholar]
- Wiegand G, Remington SJ (1986) Citrate synthase: structure, control, and mechanism. Annu Rev Biophys Biophys Chem 15: 97–117 [DOI] [PubMed] [Google Scholar]
- Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J (1997) Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science 277: 60–66 [DOI] [PubMed] [Google Scholar]
- Xu F, Yuan S, Zhang DW, Lv X, Lin HH (2012) The role of alternative oxidase in tomato fruit ripening and its regulatory interaction with ethylene. J Exp Bot 63: 5705–5716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Shibata M, Terashima I, Noguchi K (2010) Simultaneous determination of in vivo plastoquinone and ubiquinone redox states by HPLC-based analysis. Plant Cell Physiol 51: 836–841 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Terashima I, Noguchi K (2011a) How and why does mitochondrial respiratory chain respond to light? Plant Signal Behav 6: 864–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Watanabe CK, Hachiya T, Tholen D, Shibata M, Terashima I, Noguchi K (2011b) Distinct responses of the mitochondrial respiratory chain to long- and short-term high-light environments in Arabidopsis thaliana. Plant Cell Environ 34: 618–628 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Watanabe CK, Terashima I, Noguchi K (2011c) Physiological impact of mitochondrial alternative oxidase on photosynthesis and growth in Arabidopsis thaliana. Plant Cell Environ 34: 1890–1899 [DOI] [PubMed] [Google Scholar]
- Zhang L, Oh Y, Li H, Baldwin IT, Galis I (2012) Alternative oxidase in resistance to biotic stresses: Nicotiana attenuata AOX contributes to resistance to a pathogen and a piercing-sucking insect but not Manduca sexta larvae. Plant Physiol 160: 1453–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Beard KFM, Swart C, Bergmann S, Krahnert I, Nikoloski Z, Graf A, Ratcliffe RG, Sweetlove LJ, Fernie AR, et al. (2017) Protein-protein interactions and metabolite channelling in the plant tricarboxylic acid cycle. Nat Commun 8: 15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xi D, Wang J, Zhu D, Guo X (2009) Functional analysis reveals effects of tobacco alternative oxidase gene (NtAOX1a) on regulation of defence responses against abiotic and biotic stresses. Biosci Rep 29: 375–383 [DOI] [PubMed] [Google Scholar]