Modification of GhLac1 expression leads to redirection of phenylpropanoid metabolism and alteration of JA synthesis to confer broad spectrum resistance to both pathogens and pests.
Abstract
Plants are constantly challenged by a multitude of pathogens and pests, which causes massive yield and quality losses annually. A promising approach to reduce such losses is to enhance the immune system of plants through genetic engineering. Previous work has shown that laccases (p-diphenol:dioxygen oxidoreductase, EC 1.10.3.2) function as lignin polymerization enzymes. Here we demonstrate that transgenic manipulation of the expression of the laccase gene GhLac1 in cotton (Gossypium hirsutum) can confer an enhanced defense response to both pathogens and pests. Overexpression of GhLac1 leads to increased lignification, associated with increased tolerance to the fungal pathogen Verticillium dahliae and to the insect pests cotton bollworm (Helicoverpa armigera) and cotton aphid (Aphis gosypii). Suppression of GhLac1 expression leads to a redirection of metabolic flux in the phenylpropanoid pathway, causing the accumulation of JA and secondary metabolites that confer resistance to V. dahliae and cotton bollworm; it also leads to increased susceptibility to cotton aphid. Plant laccases therefore provide a new molecular tool to engineer pest and pathogen resistance in crops.
Microbial pathogens and insect pests are major factors affecting crop yield and quality (Dodds and Rathjen, 2010; Wu and Baldwin, 2010). Agrochemical application is the most common method to mitigate biotic stresses in crop plants, but a more sustainable and environmentally friendly strategy is desirable (Xu et al., 2017). A promising approach to reduce such losses is to enhance the immune system of plants through genetic engineering, and this requires an improved understanding of the molecular mechanisms involved (Lacombe et al., 2010). Plants have evolved two major innate immune responses. One involves pathogen- or herbivore-associated molecular patterns and damage-associated molecular patterns (DAMPs) that triggered immunity. The second is effector-triggered immunity, which predicts that the host plant generates resistance gene products to recognize specifically the effectors that are released from the invader and to confer disease resistance (Howe and Jander, 2008; Dodds and Rathjen, 2010; Tang et al., 2017). These two innate immune strategies trigger downstream responses that are partially overlapping, including the accumulation of phytohormones, reactive oxygen species, and secondary metabolites, including phytoalexins, to activate the corresponding defense systems (La Camera et al., 2004; Pieterse et al., 2009; De Vleesschauwer et al., 2014; Feng and Shan, 2014; Wu et al., 2014).
Phenylpropanoid metabolism is the most important secondary metabolic pathway involved in plant defense against biotic and abiotic stresses (La Camera et al., 2004). Lignin synthesis is one of the branches of the phenylpropanoid pathway, and the polymerization of monolignol to form lignin provides mechanical strength and reinforces cell walls to provide a physical barrier to limit pathogen colonization (Bonello and Blodgett, 2003). Lignin deposition in the infected cells limits the ingress of pathogens and may limit production of pathogen-produced toxins and enzymes, while also preventing the transfer of water and nutrients from plant to pathogen, to restrict pathogen growth. Lignin also makes plant cell walls more difficult for leaf-eating herbivores to digest and piercing-sucking herbivores to penetrate (Naoumkina et al., 2010; Mottiar et al., 2016).
Due to limitations of substrate supply into the phenylpropanoid pathway, the increase of flux into one branch of the pathway will normally decrease flux into another branch (Misra et al., 2010). Flavonoids, synthesized via one branch of the phenylpropanoid metabolism, are involved in myriad biological processes in plants such as the pigmentation of flowers, protection against UV damage, regulation of cotton fiber and pollen development, and defense against microbial pathogens and pests (Harborne and Williams, 2000; Treutter, 2005; Nenaah, 2014). The role of flavonoids in plant-insect interactions is well known, whereby they can serve as feeding deterrents, digestibility reducers, and antinutritional toxins that restrict the development of herbivores (Thoison et al., 2004; Nenaah, 2014; Mechri et al., 2015; Wang et al., 2015).
Jasmonic acid (JA) and its bioactive derivative jasmonoyl-L-Ile (JA-Ile) are fatty acid-derived plant hormones collectively known as “jasmonates” (Sheard et al., 2010). JA alone mediates defense against wounding and herbivores, especially against leaf-eating herbivores that cause wounding or tissue damage to plants (Fürstenberg-Hägg et al., 2013), and activates defense against necrotrophs through cooperation with ethylene. The interaction between the JA and salicylic acid (SA) signaling pathways has been well documented; there are more cases of JA and SA antagonization than cooperation (Derksen et al., 2013). SA is a principal mediator of plant defense against biotrophic or hemibiotrophic pathogens, is involved in systemic acquired resistance, and is known as the most important molecule in the resistance to phloem-feeding aphids and spider mites (Grant and Jones, 2009; Kloth et al., 2016). Although Verticillium dahliae, the causative agent of verticillium wilt, is thought to be a hemibiotrophic pathogen, JA rather than SA plays a dominant role in plant resistance to it (Fradin et al., 2011; Sun et al., 2014). Recent research in Arabidopsis demonstrated that WRKY22 promotes susceptibility to aphids through suppression of the SA signaling pathway, likely mediated by an enhanced JA response soon after aphid infection (Kloth et al., 2016).
Cotton (Gossypium spp.) is one of the most important economic crops and is cultivated globally (Sunilkumar et al., 2006), and pathogens and pests are major limitations to yield and quality. Verticillium wilt is the most notorious disease in cotton production because of its infection strategies and great genetic plasticity. Cotton bollworm (Helicoverpa armigera), once thought to be the most serious insect pest of cotton, has been well controlled by the widely planted Bt cotton (Wu et al., 2008). However, some new challenges have emerged: continuously planted mono-Bt cotton can lead to pest resistance; and in fields planted with Bt cotton, the decreased spraying of pesticides may result in non-Bt-targeted pests becoming new key pests, particularly piercing-sucking insects (Wu et al., 2008; Trapero et al., 2016).
Here, we describe a new strategy for the engineering of cotton resistance to both fungal pathogens and insect pests. We show that the transgenic overexpression of the cotton laccase gene GhLac1 leads to enhanced resistance to V. dahliae, cotton bollworm, and cotton aphid (Aphis gosypii). We demonstrate that the mechanisms underlying the broad-spectrum resistance in transgenic overexpression and RNAi lines are different due to Ghlac1-mediated differential redirection of the metabolic flux in the phenylpropanoid pathway, the biosynthesis of JA, and the balance of JA-SA defense responses, revealing different vulnerabilities of cotton bollworms and cotton aphids. This places GhLac1 as a key regulator in a complex and interacting pathway at the interfaces between phenylpropanoid, JA, and SA metabolism.
RESULTS
Overexpressing or Down-Regulating GhLac1 Enhances Plant Resistance to V. dahliae
The plant cell wall provides not only mechanical strength and rigidity for plants but also acts as a critical barrier against pathogens and pests (Cosgrove, 2000; Santiago et al., 2013). Genes encoding enzymes involved in cell wall construction or modification may represent useful candidates for the manipulation of plant defenses to biotic stresses. Therefore, a suppression subtractive hybridization library was constructed to identify genes expressed during cell wall regeneration in cultured protoplasts of cotton (Yang et al., 2008). Three expressed sequence tags (ESTs; GI: 213047554, GI: 213047555, and GI: 213047556) that are homologous sequences of GaLAC1 (Gossypium arboretum secretory laccase, GI: 40218370) were isolated. Of the three genes, the expression of GI: 213047556 was found to be the most strongly induced by V. dahliae, JA, and H2O2 treatments (Supplemental Fig. S1, A and B). Using a DNA walking strategy, we obtained the full-length cDNA of GI: 213047556 from cotton cultivar YZ1, which consists of 2403 nucleotides and encodes a protein of 566 amino acids; this was named GhLac1. GhLac1 contains a predicted copper binding domain conserved among plant laccase proteins (Supplemental Figs. S1C and S2A) with a distinct phylogenetic relationship with laccases from fungi, insects, and plants (Supplemental Fig. S2B). GhLac1 is predominantly expressed in roots, with some expression in leaves and other tissues (Supplemental Fig. S2C). It was strongly induced in roots by infection with V. dahliae strain V991 and by JA and H2O2 compared to control treatments, but showed no response to SA (Fig. 1A).
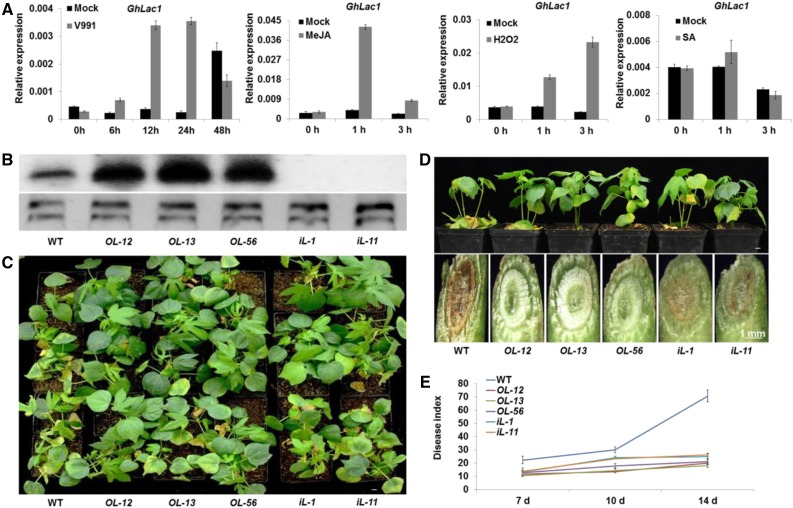
Figure 1.
Response of GhLac1 to JA, H2O2, SA, and V. dahliae, and the overexpressing or down-regulating GhLac1 each confers enhanced tolerance to V. dahliae. A, qRT-PCR analysis of V. dahliae-induced, JA-induced, H2O2-induced, and SA-induced GhLac1 expression patterns at different time points. Total RNA for V. dahliae-induced, JA-induced, H2O2-induced, and SA-induced GhLac1 expression patterns was isolated from the roots of the wild-type cotton line YZ1. The values are the means ± sd; n = 3. The expression levels of GhLac1 were normalized to those of GhUB7. B, Expression of GhLac1 in wild-type and transgenic lines was detected by northern blotting for roots from three-leaf-stage plants that were inoculated with V991 for 24 h. OL-12, OL-13 and OL-56: overexpression lines; iL-1 and iL-11: RNAi lines. C, Disease symptoms in wild type, overexpression lines, and RNAi plants after inoculation with V. dahliae strain V991. Three-leaf-stage seedlings of wild-type and transgenic lines were inoculated with V. dahliae and replanted in soil, with at least 25 plants for each line, and photographed 10 d after inoculation. D, V991 hyphal growth in wild-type and transgenic plants after inoculation for 14 d. E, Disease indices of wild-type and transgenic plants were determined after inoculation with V991 for 7 d, 10 d, and 14 d. The values are the means ± sd; n = 3. Statistical analyses were performed using Student’s t test. All of the experiments were repeated at least three times with similar results. Bars = 1 cm.
To investigate the function of GhLac1 in cotton, we selected three independent transgenic GhLac1 overexpression (OL12, OL13, and OL56) and two independent RNAi (iL-1 and iL-11) lines based on the expression levels among all the generated transformants (Fig. 1B and Supplemental Fig. S3, A to D). Protoplasts were isolated from GhLac1 overexpression line OL-12 or YZ1 embryogenic callus to investigate the cell wall regeneration. Results showed that overexpression of GhLac1 accelerates cell wall regeneration, which confirms that GhLac1 participates in cell wall synthesis (Supplemental Fig. S3E). No obvious architectural phenotypic differences were observed between the wild-type and transgenic lines. Seedlings of wild-type and transgenic lines were inoculated at the three leaf-stage with V. dahliae strain V991. Typical disease symptoms, including chlorosis, wilt, and dark brown streaks in the stem, were much more severe in the wild type than in either the overexpression or RNAi lines (Fig. 1, C and D). A disease index analysis also supported these results (Fig. 1E). Therefore both overexpressing and down-regulating GhLac1 in cotton confer enhanced tolerance to V. dahliae, although the V. dahliae tolerance scales are different.
Overexpressing or Down-Regulating GhLac1 Confers Feeding Deterrence to Cotton Bollworm
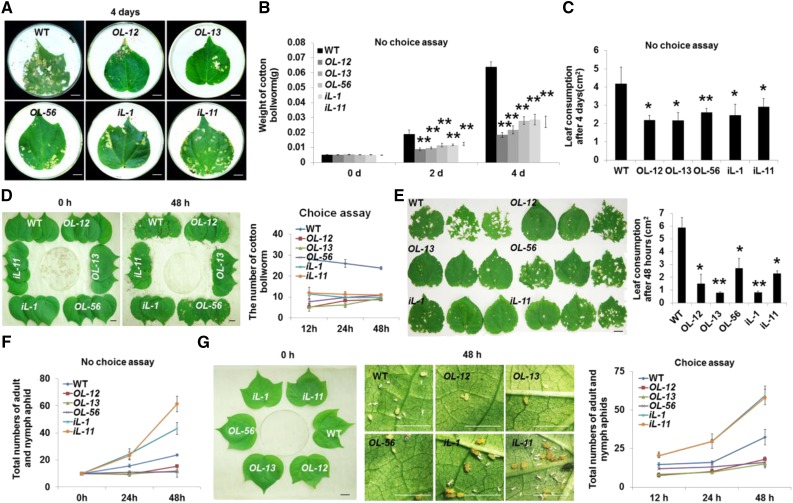
To determine any effect of GhLac1 in tolerance to insect pests, we chose one leaf-eating insect, cotton bollworm, and one piercing-sucking insect, cotton aphid, for no-choice and choice feeding assays. For the no-choice assay with cotton bollworm, prestarved second-instar larvae were placed on the leaves of wild-type and transgenic plants and allowed to feed for up to 4 d (Fig. 2A). The larvae feeding on wild-type leaves were significantly heavier, and the leaves were more severely damaged, compared with feeding on GhLac1 transgenic leaves (Fig. 2, B and C). In a further experiment, cotton bollworms were allowed to feed for 7 d. The larvae fed with the overexpression and RNAi lines were small in size with arrested development, whereas larvae fed with wild-type cotton were well developed (Supplemental Fig. S4A). A choice assay, in which second-instar larvae were provided with wild type, overexpression, and RNAi leaves, showed that the population size of cotton bollworm and the leaf consumption by bollworms on either overexpression and RNAi lines were lower than those on wild-type leaves at 48 h, suggesting that overexpressing or down-regulating GhLac1 confers a feeding deterrent effect on cotton bollworm (Fig. 2, D and E).
Figure 2.
GhLac1 effect on tolerance to insect pests. A, No-choice feeding assay for cotton bollworms. The second-instar cotton bollworms were prestarved for 6 h, and photographed at 4 d after cotton bollworm feeding on leaves from transgenic and wild-type plants. B, The weights of cotton bollworms fed with the leaves from each line. The values are the means ± sd; n = 10. C, Leaf consumption in the no-choice assay for cotton bollworms determined using the software ImageJ (National Institutes of Health). Values are means ± sd; n = 10. D, Choice feeding assay of cotton bollworms. The second-instar cotton bollworms were prestarved for 6 h, and photographed at 0 h and 48 h. The number of cotton bollworms on the leaves from each line was recorded at 12 h, 24 h, and 48 h. Values are the means ± sd; n = 4. E, Leaf consumption in the choice feeding assay for cotton bollworms using ImageJ. Values are the means ± sd; n = 10, and photographed 48 h after cotton bollworm treatment. F, No-choice feeding assay for cotton aphids. The number of adult and nymph aphids on the leaves from each line in the no-choice assay was recorded at 24 h and 48 h. G, Choice feeding assay for cotton aphids. Photos were taken at 48 h after introduction of aphids using a microscope (DM2500, Leica). The number of cotton aphids on the leaves from each plant line was recorded at 12 h, 24 h, and 48 h. Values are the means ± sd; n = 4. Statistical analyses were performed using Student’s t test. *, P < 0.05; **, P < 0.01. All of the experiments were repeated at least three times with similar results. Bars = 1 cm.
GhLac1 Overexpression and Suppression Show Opposite Effects Against Cotton Aphid
No-choice and choice feeding assays for cotton aphids fed with excised leaves showed that aphid number was reduced in the overexpression lines, but increased in the RNAi lines compared to wild type (Fig. 2, F and G; Supplemental Fig. S4B). An outdoor choice feeding assay carried out in the field also demonstrated that fewer aphids were found on overexpression lines and more were found on the RNAi lines compared to wild type (Supplemental Fig. S5). These data show that overexpression of GhLac1 in transgenic cotton acts as a feeding deterrent to cotton aphids as well as cotton bollworm, but the different response of bollworms and aphids to the GhLac1 RNAi lines suggests species-specific effects of this gene.
Redirection of Metabolic Flux in Phenylpropanoid Pathway in GhLac1 Transgenic Lines
To investigate the mechanism(s) by which laccase deters insect feeding and V. dahlia infection, we carried out a metabolic study. Consistent with previous reports that laccase is involved in lignin polymerization in stem vascular tissue (Berthet et al., 2011), phloroglucinol staining of the stem cross sections showed that the overexpression lines possessed a higher lignin content than did wild type, and the RNAi lines had less lignin (Supplemental Fig. S6). The lignin content of roots and young leaves (first and second leaves from top) from plants used in the fungal infection and insect feeding assays also corresponded to the detected lignin accumulation in stems (Fig. 3A). The expression levels of most of the monolignol biosynthesis enzyme-encoding genes were not significantly different between GhLac1 overexpression lines and wild type in roots and young leaves, but showed up-regulation for all of these genes in RNAi lines (Supplemental Fig. S7A). This suggests a negative feedback of low lignin content on transcript levels of the biosynthetic enzyme genes. This was also reflected in an increase in Phe ammonia lyase (PAL, EC. 4.3.1.5) enzyme activity (Supplemental Fig. S7B).
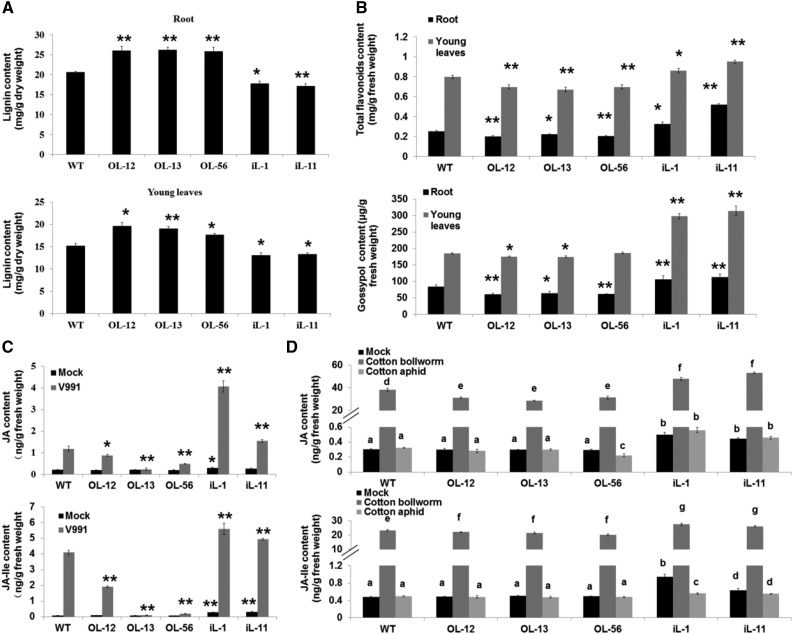
Figure 3.
GhLac1 transgenic lines show an altered phenylpropanoid pathway and phytohormone homeostasis. A, Lignin content in the roots and young leaves. Values are the means ± sd; n = 6. B, Total flavonoid content and gossypol equivalent content in the roots and young leaves. Values are the means ± sd; n = 6. C, JA and JA-Ile contents in the roots between Mock and V. dahliae-inoculated plants. Roots were collected after inoculation with V. dahliae spores or sterile distilled water for 6 h. Values are the means ± sd; n = 9. D, JA and JA-Ile contents in the leaves between Mock, cotton bollworm-, and cotton aphid-infected plants. Statistical analyses were performed using Student’s t test. *, P < 0.05; **, P < 0.01. Values are the means ± sd; n = 9. All of the experiments were repeated at least three times with similar results.
We speculated that the altered expression level of GhLac1 might lead to a redirection of the metabolic flux in the phenylpropanoid pathway. Thus, we determined the total flavonoid and gossypol equivalents content in roots and young leaves. The results revealed a significant increase of flavonoids and gossypol in the GhLac1 RNAi lines compared to wild type, with the overexpressing lines showing small reductions (Fig. 3B). The expression levels of those genes encoding enzymes involved in flavonoid and gossypol biosynthesis were consistent with the observed redirection of metabolic flux, which exhibited significant up-regulation in the RNAi lines but slight down-regulation in the overexpression lines, especially in leaf tissues (Supplemental Fig. S8). Therefore, the alterations in lignin, flavonoid, and gossypol accumulation in laccase transgenic lines are associated with changes in the transcriptional activity of genes encoding relevant biosynthetic enzymes, resulting in more lignin in the laccase overexpressers and more flavonoids and gossypol in the laccase RNAi lines.
Lignin Modification Affects Phytohormone Homeostasis in Transgenic Plants
It is reported that antisense down-regulation of the enzyme hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase (HCT) in alfalfa (Medicago sativa L.) led to decreased lignin content but increased concentrations of SA, JA, and abscisic acid (ABA), coupled with enhanced resistance to abiotic stress and pathogens (Gallego-Giraldo et al., 2011). This illustrates the interconnectedness of lignin and other defense pathways. To investigate possible links between laccase expression level and defense signaling, we measured JA and JA-Ile concentrations in roots and young leaves in GhLac1 transgenic lines and wild type. The prediction was that the low lignin RNAi lines would contain more JA.
In untreated roots and young leaves, the RNAi lines showed significantly higher levels of JA and JA-Ile than wild-type, but the overexpression lines showed no significant difference compared with wild type (Fig. 3, C and D). After inoculation with V. dahliae for 6 h or cotton bollworm for 3 h, the JA and JA-Ile content sharply increased in all of the lines compared to mock treatments, with the highest levels in RNAi lines and lowest in overexpression lines (Fig. 3, C and D). The expression levels of marker genes involved in the JA signaling pathway were also in agreement with the dosage of JA and JA-Ile (Supplemental Figs. S9 and S10), which showed significant up-regulation in the RNAi lines under normal conditions (Supplemental Fig. S9A). Moreover, after treatment with V. dahliae or cotton bollworm, the JA defense pathway was activated further in the RNAi lines and slightly suppressed in overexpressing lines (Supplemental Figs. S9B and S10).
Aphid attack produced a different response. After inoculation of wild-type and transgenic plants with cotton aphids for 3 h, the JA and JA-Ile content showed no significant difference compared with the mock treatment in most lines, except for iL-1 (which had a lower JA-Ile content), but retained the highest levels in RNAi lines and lowest levels in overexpression lines (Fig. 3, C and D). The results suggest that JA synthesis was constitutively activated in the RNAi lines.
The Altered Cell Wall as a Stress Generator Affects Levels of Jasmonates
It was reported that, in cinnamoyl CoA reductase-deficient tobacco lines and HCT-deficient alfalfa lines, the stress response was enhanced (Dauwe et al., 2007). One possibility is that the altered cell walls of these plants mimics wounding when under penetration by fungal pathogens or chewing by herbivores, as such pests secrete cell-wall-hydrolyzing enzymes. Consequently, the altered cell wall releases bioactive oligosaccharides acting as DAMPs that are perceived by the pattern recognition receptors to initiate downstream immune responses (Boller and Felix, 2009).
To investigate this possibility, we extracted the cell wall pectic polysaccharides of wild-type and GhLac1 transgenic lines (Supplemental Fig. S11A). The cell wall fractions from RNAi lines released much more cold-water-soluble pectic material than did wild type, whereas the overexpression lines showed no significant difference compared with wild type (Supplemental Fig. S11A). After treatment with V991 or cotton bollworm for 48 h, all the plants accumulated cold water-soluble polysaccharide material to different degrees, indicating the possible existence of a response caused by wounding or a mechanism by which polysaccharide hydrolase is released by the pathogen or herbivores (Supplemental Fig. S11A). Moreover, the cold water-soluble pectic levels of overexpression lines and RNAi lines were the lowest and highest, respectively, compared with wild type (Supplemental Fig. S11A).
Cotton suspension-cultured cells were collected after being treated with the crude cold water-soluble polysaccharide material from each line for 10 h, and the JA and JA-Ile content was determined. The results show that the cold water-soluble polysaccharide material could induce the JA and JA-Ile accumulation, and the JA and JA-Ile content was positively correlated with the concentration of cold-water-soluble pectic material (Supplemental Fig. S11B).
Altered Metabolites in GhLac1 RNAi Lines Are Associated with Tolerance to Verticillium Wilt and Cotton Bollworm
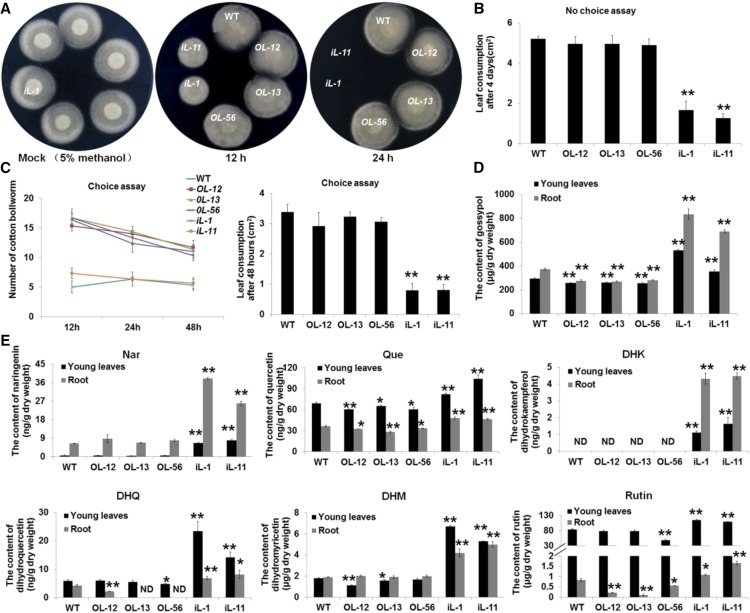
The effects of flavonoids and gossypol on pathogens and insects are well documented (Nenaah, 2014; Iranshahi et al., 2015; Mechri et al., 2015). To test the biological activities of metabolites in extracts from the GhLac1 transgenic plants, methanolic extracts from each line were prepared. Spores of V. dahliae strain V991 were treated with the methanolic extracts for 12 h or 24 h and then the mixture was spot-inoculated with an equal volume on potato-dextrose agar (PDA) medium and cultured for 3 d. The results show that the extracts from RNAi lines could inhibit the growth of V. dahliae. Compared with the spores treated with methanolic extracts from overexpression lines and wild type, some spores treated with the methanolic extracts from RNAi lines for 12 h could germinate and grew into a small circle on the PDA medium, whereas the spores treated with the RNAi methanolic extracts for 24 h were unable to germinate at all, and there was no difference found between the overexpression lines and wild type (Fig. 4A).
Figure 4.
Bioassay of the methanolic extracts and determination of endogenous phenolic materials. A, Evaluation of the effects of methanolic extracts from transgenic lines and wild type on the growth of V. dahliae. Photos were taken 3 d after the mixtures of methanolic extracts and V991 strain spores (mixed and placed at room temperature for 12 h or 24 h) were dropped onto potato-dextrose agar media. The mixture of methanol extraction buffer (5% methanol) and V991 strain spores (placed at room temperature for 24 h) served as the Mock. B, No-choice feeding assay to determine the effects of methanolic extracts from each line on cotton bollworm. Wild-type leaves were smeared with the methanolic extracts from each line, and recorded 4 d after cotton bollworm treatment. Values are the means ± sd; n = 4. C, Choice feeding assay to determine the effects of methanolic extracts from each plant line for cotton bollworm. The population size and leaf consumption of each leaf was recorded at 12 h, 24 h, and 48 h after cotton bollworm treatment. Values are the means ± sd; n = 4. All of the leaves used in the no-choice and choice assays were from wild type and were smeared with the methanolic extracts from each line. D, Gossypol content in roots and young leaves. Values are the means ± sd; n = 9. E, Nar, Que, DHK, DHQ, DHM, and rutin content in roots and young leaves. Values are the means ± sd; n = 9. Statistical analyses were performed using Student’s t test. *, P < 0.05; **, P < 0.01. All of the experiments were repeated at least three times with similar results. Bars = 1 cm.
The effects of methanolic extracts on cotton bollworm were also examined. All the tested leaves were collected from the wild type, but smeared with the methanolic extracts from different transgenic lines and wild type. In both no-choice and choice feeding assays with cotton bollworm, the methanolic extracts from RNAi lines significantly deterred feeding, but little difference was observed for extracts from overexpression lines and wild type (Fig. 4, B and C; Supplemental Fig. S12). Leaf consumption and population size data in no-choice and choice assays with cotton bollworm also supported the results of the methanolic extract analysis (Fig. 4, B and C).
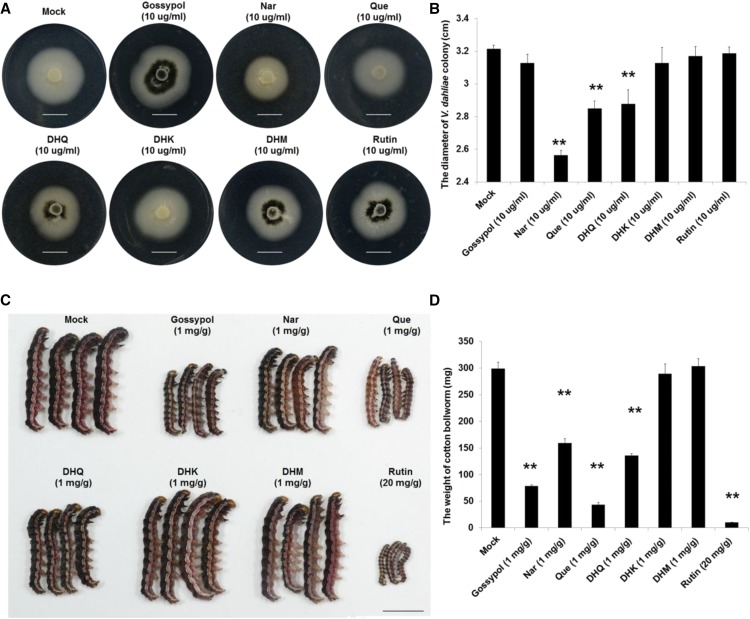
To identify the possible chemical basis of the toxic effects, phenolic metabolism was analyzed in GhLac1 transgenic lines and wild type. In accordance with expectations, the contents of gossypol and several flavonoids such as naringenin (Nar), quercetin (Que), dihydrokaempferol (DHK), dihydroquercetin (DHQ), dihydromyricetin (DHM), and rutin were significantly accumulated in the RNAi lines (Fig. 4, D and E). We tested the toxicity of these secondary metabolites to V. dahliae strain V991 and cotton bollworm (Fig. 5), and the concentration of each chemical used was based on previous reports (König et al., 2014; Jadhav et al., 2012; Mellon et al., 2014; Krempl et al., 2016). The results showed that Nar, Que, and DHQ significantly inhibited the growth of V. dahliae mycelium, but gossypol, DHK, DHM, and rutin induced the generation of microsclerotia at the concentrations indicated in Fig. 5, A and B. Moreover, all the secondary metabolites except DHK and DHM were found to inhibit the growth and development of cotton bollworm at the concentrations indicated in Fig. 5, C and D. These results show that secondary metabolites accumulated after GhLac1 suppression likely contribute to the resistance effects to V. dahliae and cotton bollworm.
Figure 5.
Influence of gossypol and flavonoids on V. dahliae and cotton bollworm growth. A and B, The fungus was grown for 5 d on the PDA plates with the indicated concentration for each chemical and the colony diameter was determined. C and D, The second-instar cotton bollworm was fed with the artificial diet with the indicated concentration for each chemical for 7 d, and the weight was determined. Statistical analyses were performed using Student’s t test. *, P < 0.05; **, P < 0.01. All of the experiments were repeated at least three times with similar results for DHK, DHM, DHQ, Nar, and Que. Bars = 1 cm.
The Suppressed SA Signaling Pathway in RNAi Lines Accounts for the Susceptibility to Cotton Aphid
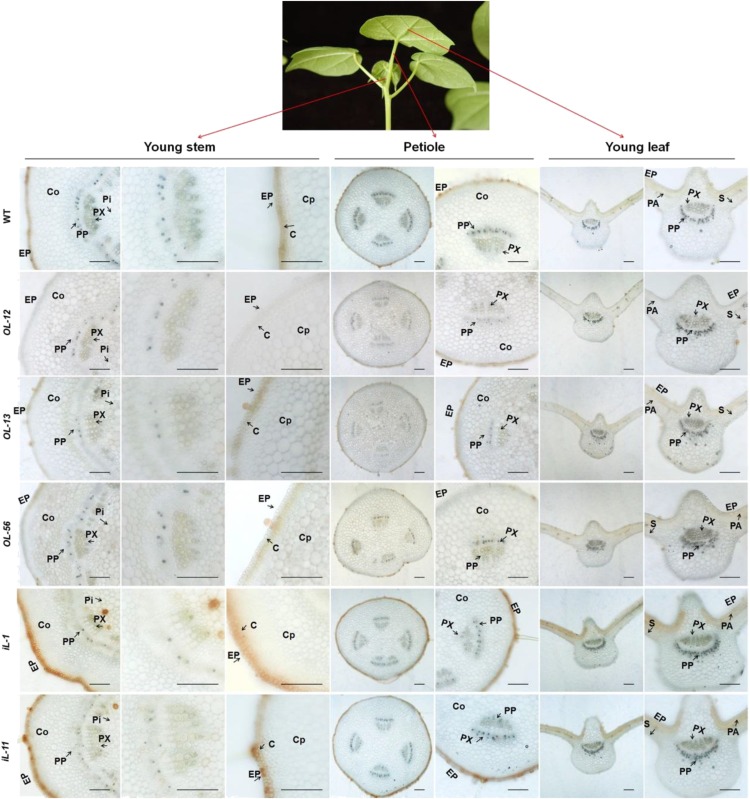
Cotton aphid, a sap-sucking insect, uses a stylet to pierce plant tissue and navigate between plant cells to locate the phloem for access to plant sap. We wanted to determine whether the toxic secondary compounds accumulate at the aphid feeding sites. Histochemical localization suggested that flavonoids indeed accumulate in the RNAi lines, and specifically in the epidermis, collenchyma, and pith cells in the young stem; the epidermis and collenchyma cells in the petiole; and the upper epidermis and palisade cells in the leaf. Interestingly the flavonoid content was not significantly different between the wild-type and GhLac1 transgenic lines in vascular tissues, including the primary phloem (Fig. 6). This suggests that aphid feeding by stylet may avoid the ingestion of potentially toxic compounds in the RNAi lines. This leads us to conclude that the increased content of the secondary metabolites in the RNAi lines probably do not contribute to the observed aphid resistance.
Figure 6.
The histochemical localization of flavonoids in the aphid feeding sites. The histochemical localization of flavonoids in young stem, petiole, and young leaf. Brown color indicates the presence of flavonoids. Compared with wild type, the RNAi lines of GhLac1 showed enhanced accumulation of flavonoids in the epidermis, collenchyma, and pith, and the overexpression lines of GhLac1 showed decreased content of flavonoids in those tissues. In addition, the content of the flavonoids showed no significant difference between the wild-type and GhLac1 transgenic lines in vascular tissue, especially in primary phloem. C, collenchyma; Co, cortex; Cp, cortex parenchyma; EP, epidermis, PA, palisade; Pi, pith; PP, primary phloem; PX, primary xylem; S, spongy.
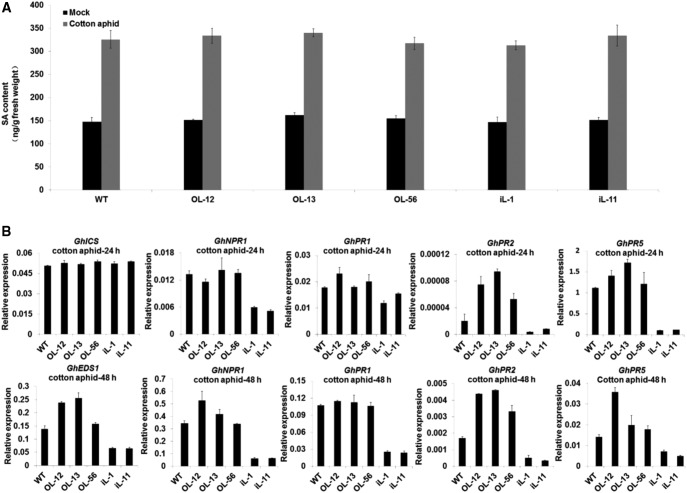
Given the importance of SA in plant defense to aphid infection (Pieterse et al., 2009; Derksen et al., 2013), the known antagonistic relationship between JA and SA (Derksen et al., 2013), and to explain the increased aphid feeding on RNAi leaves (Fig. 2), we measured the SA content in transgenic lines and wild type. As shown in Figure 7A, SA content showed no significant difference between wild-type and transgenic lines in young leaves under noninfected condition, but the SA-responsive genes were significantly suppressed in the RNAi lines (Supplemental Fig. S13). However, SA levels significantly increased in each line after infection with cotton aphids for 3 h, although there was no difference between wild-type and transgenic lines. After infection with cotton aphids, the expression levels of all the selected SA-responsive genes were induced in each line to different degrees, but showed highest induction in the overexpression lines, and lowest in the RNAi lines (Supplemental Fig. S13; Fig. 7B). These results suggest a suppressed SA-mediated defense capability in the laccase. RNAi lines were associated with enhanced JA signaling, consistent with a preference of cotton aphids for the RNAi lines in the choice assay.
Figure 7.
The suppressed SA-mediated defense response accounts for the aphid susceptibility in GhLac1 RNAi lines. A, SA content of roots and young leaves of transgenic and wild-type plants under uninfected and cotton aphid-infected conditions. Values are the means ± sd; n = 9. B, qRT-PCR analysis of the expression levels of selected genes involved in the SA synthesis and signaling pathway in transgenic and wild-type plants after infection with cotton aphid. The values are normalized to GhUB7 and expressed as the means ± sd; n = 3. Statistical analyses were performed using Student’s t test. All experiments were repeated at least three times with similar results.
DISCUSSION
Pleiotropic Functions of Ghlac1 in Plant Defense against Pathogens and Insects
Plant laccases, copper-containing glycoproteins related to blue copper oxidase, are involved in lignin polymerization in many plant species and are specifically expressed in lignifying cells (Sterjiades et al., 1992; Bao et al., 1993; Berthet et al., 2011). They can catalyze the oxidation of a broad range of phenolic compounds (Wang et al., 2004; Iqbal et al., 2009). A laccase GaLAC1, from Gossypium arboreum, has been reported to be a secretory laccase. Overexpression of GaLAC1 in Arabidopsis is able to enhance plant resistance to several phenolic allelochemicals and 2,4,6-trichlorophenol (Wang et al., 2004). Interestingly, when the rice laccase OsLAC is overexpressed, the grain size, main panicle length, number of primary and secondary branches, and number of grains per main panicle decreases (Zhang et al., 2013), suggesting that plant laccases participate in developmental processes other than lignification.
Here we show that the overexpression of GhLac1 leads to enhanced tolerance of V. dahliae, cotton bollworm, and cotton aphids, and suppression of GhLac1 by RNAi leads to enhanced tolerance to V. dahliae and cotton bollworm, but increased susceptibility to cotton aphid (Figs. 1 and 2). The differential response of bollworms and aphids to the down-regulation of GhLac1 suggests the metabolic effects in the plant affect different insect pests in different ways. Up-regulation of GhLac1 led to an increased lignin content (Fig. 3, A to C) and unchanged JA signaling pathway (Supplemental Fig. S9A), then led to the restriction of initial infection and colonization by V. dahliae and to the decreased feeding preference of cotton bollworm and cotton aphids (Figs. 1 and 2). Down-regulation of GhLac1 led to reduced lignin content and tolerance to V. dahlia and cotton bollworm, but not cotton aphids. Interestingly, decreased lignin content in HCT antisense alfalfa resulted in elevated levels of SA, JA, and ABA, associated with a massive up-regulation of pathogenesis- and abiotic stress-related genes and enhanced tolerance to fungal infection and drought (Gallego-Giraldo et al., 2011). We therefore conclude that down-regulation of laccase leads to enhanced tolerance to V. dahliae and cotton bollworm through an increase in JA and JA-Ile levels (Fig. 3, C and D), associated with an up-regulation of genes involved in the JA signaling pathway (Supplemental Figs. S9 and S10).
Secondary Metabolites Play a Complex Role in Plant-Invader Interactions
Our metabolic data further suggest that the suppressed lignin polymerization in the laccase RNAi lines leads to a redirection of metabolic flux in the phenylpropanoid pathway to the formation of flavonoids and phenolics (Figs. 3B and 4E), and the increased contents of flavonoids and gossypol likely account for the observed inhibitory effects on V. dahliae mycelial growth and cotton bollworm feeding (Figs. 4, A to D and 5; Supplemental Fig. S12). This interpretation is supported by the observation that silencing GbCAD1, a key enzyme involved in gossypol biosynthesis, compromises cotton resistance to V. dahliae (Gao et al., 2013); and overexpression of the Arabidopsis transcription factor AtMYB12 in tobacco (Nicotiana tabacum) results in the enhanced expression of genes involved in the phenylpropanoid pathway, leading to a severalfold higher accumulation of flavonoids and to the development of resistance against the leaf-eating insect pests Spodoptera litura and H. armigera (Misra et al., 2010).
Unlike cotton bollworm, aphids are piercing-sucking insects; they insert a stylet into the phloem to suck out the sap (Howe and Jander, 2008). Most studies of plant-aphid interactions show that SA is the dominant molecule in plant defense to aphid infestation; an increased SA level and an induced SA defense pathway are prerequisites for plant defense against aphids (Avila et al., 2012; Spoel et al., 2003; Kloth et al., 2016). In the GhLac1 overexpression lines, the observed enhanced tolerance to aphids may derive from the increased lignin deposition and the activated SA-mediated defense responses (Figs. 3A and 4F). The down-regulation of GhLac1 led to a greater susceptibility to cotton aphids. The reduced lignin content in these plants might facilitate the insertion of aphid stylets (Fig. 3A). However, the observed reduced activity of the SA signaling pathway in the laccase RNAi lines suggests this accounts for plant susceptibility (Fig. 7).
The different mode of feeding by cotton bollworm and cotton aphid may explain why RNAi plants accumulated many more flavonoids, but led to a distinct effect on aphid feeding (Fig. 4, A to D). Feeding by stylet may avoid aphid ingestion of potentially toxic phenolic from the phloem. Although numerous studies show that the aphid infection can induce flavonoid synthesis and accumulation in plants, and some flavonoids possess inhibitory effects on aphid growth and reproduction rate, details about the location of toxic compounds have not previously been determined (Smith and Chuang, 2014; Morkunas et al., 2016; Woźniak et al., 2017). Flavonoids are likely synthesized in a cytoplasmic metabolic environment, and the flavonoid biosynthetic genes are regulated by light, but the long distance movement of flavonoids is largely unknown (Buer et al., 2007). A recent study in Arabidopsis showed that overexpression of AtMYB75 results in accumulated flavonoids and enhanced plant resistance to caterpillars, but no effects on aphid (Onkokesung et al., 2014), and this is due to the differential kaempferol-3,7-dirhamnoside accumulation after infection by either caterpillars or aphids. This suggests that the precise transport and localization of flavonoids may be an important component of the interaction between plants and specific insect herbivores, and may determine tolerance levels by the plant.
Although up- or down-regulation of GhLac1 had no significant effect on cotton plant growth and development, the cotton fiber development showed a dramatic response. The overexpression lines had a poor fiber quality, with high micronaire, and the RNAi lines had a shorter fiber length (data not shown). Using a more restricted expression promoter to replace the 35S promotor may be a better approach for breeding pest and pathogen resistance in cotton, and for a reduced effect on cotton fiber quality.
Altered Cell Wall Polysaccharides in GhLac1 Transgenic Lines Act as a Stress Generator
The lignin synthesis deficiency in the RNAi lines of GhLac1, associated with enhanced resistance to V. dahliae and cotton bollworm, was associated also with an activated JA signaling pathway, so it compensates for defects in structural resistance otherwise provided by lignin. Our evidence supports a model in which GhLac1 acts as a stress generator to modulate JA levels. The altered dosage of GhLac1 in the transgenic lines leads to cell wall modifications, resulting in altered DAMPs (oligogalacturonides, OGAs) signal in the transgenic lines. We suggest that the cell wall fragments could induce JA synthesis (Supplemental Fig. S11B). Although the elicitor activity of OGAs is well documented, the mechanism for the perception of OGAs remains obscure (D’Ovidio et al., 2004). The Arabidopsis wall-associated kinase 1 (WAK1) has the ability to bind these oligosaccharides in vitro, suggesting WAK1 or its homologs might be the receptor for OGAs (Brutus et al., 2010). It has been reported that during the early physiological responses to DAMPs, ion flux exchange, including Ca2+ and H+, was the most notable characteristic (Boller and Felix, 2009). Whether altered JA synthesis and the related gene expression changes depend on a Ca2+ signal or on pattern recognition receptor activity requires further research.
CONCLUSION
Plants live in complex environments in which they continuously encounter a broad range of pathogens and insects with various invasion strategies. The coevolutionary arms race between plants and invaders provides plants with a highly sophisticated defense system to recognize the invaders and activate response systems to protect themselves. We have found that the modulation of GhLac1 expression leads to a redirection of metabolic flux in the phenylpropanoid pathway, associated with a rebalance of phytohormones that confer broad tolerance to pests and pathogens, and this opens the opportunity for new strategies for breeding biotic resistance in crops.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Gossypium hirsutum cv YZ1 cotton plants and transgenic lines of GhLac1 derived from YZ1 were either cultivated in Wuhan, China, under normal farming practices or grown in the greenhouse during the winter. Indoor experiments were maintained at a constant temperature of 25°C to 28°C under long-day conditions with an 8-h/16-h dark/light photoperiod and a relative humidity of 60%. Experiments with excised leaves were performed in 29-cm × 44-cm plastic boxes or 9-cm round glass dishes supplied with Hoagland’s solution. All biological samples were stored at −80°C until further analysis.
GhLac1 Cloning, Sequence Analysis, Vector Construction, and Plant Transformation
ESTs were isolated from a suppression subtractive hybridization library of cell wall regeneration during protoplast culture in cotton (Yang et al., 2008). The full-length GhLac1 gene was obtained through DNA walking using the Genome Walker Universal Kit (Clontech) with YZ1 DNA as the PCR template. The open reading frame (ORF) of GhLac1 was predicted by ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and further confirmed by sequencing the cDNA. The amino acid sequence alignments and phylogenetic relationship were generated using ClustalX (http://www.clustal.org/) and MEGA5 (http://www.megasoftware.net/), respectively.
Full-length GhLac1 was cloned into the Gateway vector pK2GW7.0 (Ghent University) by attB with attP site (BP) recombination and attL with attR site recombination (Invitrogen) to generate the overexpression vector. The conserved region of GhLac1 was inserted into pHellsgate4 through BP recombination reactions to generate the GhLac1 RNAi vector. The GhLac1 overexpression and RNAi vectors were used to transform YZ1 by Agrobacterium tumefaciens (strain GV3101)-mediated transformation according to previous methods (Jin et al., 2006). The primers used in this study are listed in Supplemental Table S2.
Visualization of Cell Wall Regeneration
To determine the role of GhLac1 in plant cell wall regeneration, protoplasts were isolated from YZ1 or the GhLac1 overexpression lines embryogenic calli according to previous methods (Min et al., 2015) and transferred to a cell wall regeneration medium described by Yang et al. (2007). The visualization of cell wall regeneration was performed as described by Yang et al. (2008).
Treatments with Methyl Jasmonate, H2O2, SA, and Verticillium dahliae
Cotton seedlings were treated with 200 μm methyl jasmonate (MeJA; Sigma-Aldrich), 2 mm SA (Sigma-Aldrich), and 1 mm H2O2 (Sigma-Aldrich) according to the experiment. Four YZ1 seedlings were cultivated in a pot in a growth chamber until reaching the three-leaf stage and then treated with MeJA or H2O2 by soil drench application. Control plants were treated with distilled water in the same way. The roots of four individual seedlings were collected for each treatment at each time point.
For treatment with V. dahliae, three-leaf-stage YZ1 seedlings were inoculated with V. dahliae strain V991 spore solutions as described previously (Xu et al., 2011). Control plants were treated with distilled water in the same way. The roots of the control and treated plants were harvested at each time point. Experiments were maintained at a constant temperature of 25°C under long-day conditions with an 8-h/16-h dark/light photoperiod and a relative humidity of 60%.
Southern and Northern Blotting
To determine the T-DNA insertion copy number of transgenic lines of GhLac1, genomic DNA was extracted using the plant genomic DNA kit DP305 (Tiangen Biotech). A quantity of 20 μg of genomic DNA was digested with HindIII (New England Biolabs) for 60 h, and Southern blotting was performed using the DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche) according to the manufacturer’s instructions. A fragment of the NPTII gene was used as a probe to detect the copy number.
For northern blotting, total RNA was isolated from the leaves using a minor modified guanidine thiocyanate method as described previously (Deng et al., 2012). A quantity of 25 μg of total RNA was separated in 1.2% agarose formaldehyde gels for 4 h and then transferred onto a nylon membrane. A specific frame of GhLac1 was amplified using the primer pair GhLac1-northern-F/R and used as the probe, and blot hybridization was performed using the DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche) according to the manufacturer’s instructions. The primers used in our study are listed in Supplemental Table S2.
RT-PCR and qRT-PCR
To determine the expression levels of the genes involved in the plant defense response, RT-PCR and qRT-PCR analyses were performed. First-strand cDNA was generated from 3 μg of total RNA using SuperScript III reverse transcriptase (Invitrogen). qRT-PCR was performed in 15 μL reactions using the ABI 7500 Real-Time PCR System (Applied Biosystems), with at least three technical replicates and three independent biological replicates. For each independent biological replicate, we took material samples from at least 5 to 10 plants of each line or treatment. GhUB7 was used as an internal control. The primers used in our study are listed in Supplemental Table S2.
Pathogen Infection and Disease Assay
To determine the performance of wild-type and transgenic lines in response to fungal pathogens, three-leaf-stage seedlings of wild-type and transgenic lines were inoculated with V. dahlia strain V991 as described above. The disease index was scored using at least 25 plants per treatment and repeated at least three times. The plant disease index was calculated according to Xu et al. (2011); a higher disease index indicates more serious symptoms and increased susceptibility to V. dahliae. Experiments were maintained at a constant temperature of 25°C to 28°C under long-day conditions with an 8-h/16-h dark/light photoperiod and a relative humidity of 60%.
Choice and No-Choice Assays of Cotton Bollworm and Cotton Aphid
Choice and no-choice assays were performed using cotton bollworms and cotton aphids on leaves of wild-type and transgenic lines of GhLac1 in 29 × 44 cm plastic boxes or 9-cm round glass dishes as described previously, with slight modification (Zhang et al., 2014). Briefly, for the choice assay using aphids, 80 apterous aphids were placed in the center of a culture dish, and the leaves of wild-type and transgenic plants were placed symmetrically around the dish. The total numbers of adult and nymph aphids per leaf were recorded at 12 h, 24 h, and 48 h. For the no-choice assay of aphids, 10 apterous adult aphids were released onto a wild-type or transgenic leaf. The total numbers of adult and nymph aphids per leaf were counted at 12 h, 24 h, and 48 h. The design of the choice assay using cotton bollworms was the same as the choice assay of aphids. For the cotton bollworm no-choice assay, one second-instar cotton bollworm was released onto one wild-type or transgenic leaf in a 9-cm round glass dish, and leaf consumption and larval weight were recorded at 0 d, 2 d, and 4 d. Experiments were performed in an incubator and maintained at a constant temperature of 25°C under long-day conditions with an 8-h/16-h dark/light photoperiod and a relative humidity of 60%.
Histochemical Staining and Determination of the Total Lignin Content
Histochemical staining of hand-cut cross sections of cotton stem tissue was used to visualize lignin deposition. Sections were cut at the same position of inoculated and mock-treated stems. Lignin was visualized using Wiesner reagent (Xu et al., 2011). Briefly, sections were dipped in a phloroglucinol solution (2% v/v in 95% ethanol) for 10 min, followed by 18% HCl for 5 min, and immediately photographed under a fluorescence microscope (DM2500; Leica) with bright field illumination.
The lignin content of roots and young leaves was determined for at least three biological replicates from protein-free cell wall fractions using the lignin-thioglycolic acid reaction according to a previous study (Bubna et al., 2011). The lignin content was expressed as the weight percentage of dry weight.
Measurement of PAL Enzyme Activity
Root and leaf samples were collected from three-leaf-stage seedlings of wild-type and transgenic lines of GhLac1, and PAL activity was measured according to the method of Dubery and Smit (1994) with slight modifications. The samples were frozen in liquid nitrogen and quickly ground into a fine powder. Samples of 100 mg were homogenized in extraction buffer [0.05 m borate buffer, pH 8.8, with 5 mm 2-mercaptoethanol and 2% (w/v) polyvinylpyrrolidone]. The lysate was centrifuged at 13,000 rpm for 20 min at 4°C, and the supernatant contained crude enzyme extract. The supernatant was incubated for 1 h at 37°C with 0.025 m borate buffer, pH 8.8, and 5 mm L-Phe. The reaction was stopped by adding 6 m HCl. The mixture was measured spectrophotometrically at 290 nm. The PAL enzyme activity was expressed as micrograms of cinnamic acid formed per gram of fresh weight.
Determination of the Total Flavonoid and Gossypol Equivalent Contents
The total flavonoid content was measured according to a previously reported spectrophotometric method with minor modifications (Dewanto et al., 2002). Briefly, samples of leaves or roots (approximately 100 mg, the precise weight was recorded) were frozen in liquid nitrogen and quickly ground. 1 mL of 80% (v/v) methanol was added. After centrifugation at 13,000 rpm, the supernatant was collected, and the residual pellet was reextracted with 1 mL of 80% (v/v) methanol. The supernatants were combined and clarified by centrifugation in a microfuge at 2000 rpm for 30 min. 0.1 mL of the extract or rutin standard solution was mixed with 0.5 mL distilled water in a test tube, to which was added 30 μL of 5% NaNO2 (w/v) solution. After 6 min, 60 μL of 10% (w/v) AlCl3 solution was added to the test tube. After 5 min of standing time, 0.2 mL of 1 m NaOH solution was added. The mixture was brought to 1 mL with distilled water and mixed well. The absorbance was measured immediately against the blank at 420 nm using a Multimode Plate Reader (PerkinElmer).
The gossypol equivalents were extracted and quantitated following a previously reported phloroglucinol/HCl method (Gao et al., 2013). A standard curve was made using gossypol (Sigma-Aldrich).
Measurement of Phytohormones
To measure the endogenous concentrations of JA, JA-Ile, OPDA, and SA, leaf, or root samples of approximately 100 mg were homogenized twice with 80% (v/v) cold methanol and shaken at 4°C overnight in the dark. The dissolution, filtration, storage, and quantification of the combined extract were performed as described in Sun et al. (2014). To each sample were added 7.5 ng (±) of 9-, 10-dihydro-JA (OlChemim), and 75 ng of 1-naphthaleneacetic acid (Sigma-Aldrich) as internal standards for the JA, JA-Ile, OPDA, and SA content assays.
Effects of Methanolic Extracts on Pathogens and Herbivores
To determine the effects of methanolic extracts on pathogens and herbivores, whole three-leaf-stage wild-type and transgenic seedlings were separately frozen in liquid nitrogen and quickly ground. A 10 g sample of each line was extracted twice with 10 mL of 80% (v/v) methanol and shaken at 4°C overnight. The combined extract was evaporated to the aqueous phase with N2, and dissolved in 10 mL of 5% (v/v) methanol. To detect the effects of methanolic extracts on pathogens, the equivalent volume of methanolic extracts and V991 strain spores (prepared as described in Xu et al., 2011, and adjusted to 105 spores/mL with sterile distilled water) were mixed and placed at room temperature for 12 h or 24 h. 10 μL of the mixture was dropped on PDA medium and cultured for 3 d to detect the growth of V. dahliae spores. To detect the effects of methanolic extracts on herbivores, no-choice and choice assays of cotton bollworms and cotton aphids were performed with wild-type leaves that had been daubed with methanolic extracts.
To test the toxicity of those secondary metabolites to V. dahliae strain V991 and cotton bollworm, 2 mg/mL gossypol (Aladdin), Nar (Sigma-Aldrich), Que (Sigma-Aldrich), DHK (Sigma-Aldrich), DHQ (Sigma-Aldrich), DHM (Aladdin), and rutin (Aladdin) were prepared with 80% (v/v) methanol. Referring to the previous studies, the final concentration of those materials in PDA medium was 10 μg/mL, and the PDA medium with equal volume of 80% (v/v) methanol was used as Mock (König et al., 2014; Jadhav et al., 2012; Mellon et al., 2014; Krempl et al., 2016). The artificial diet for cotton bollworm was prepared as described in Chen et al. (2017). The final concentration of gossypol, Nar, Que, DHK, DHQ, DHM, and rutin were 1 mg/g, 1 mg/g, 1 mg/g, 1 mg/g, 1 mg/g, 1 mg/g, and 20 mg/g, respectively. Second-instar cotton bollworms were fed with those artificial diets for 7 d, and then photographed and weighted. Experiments were performed in an incubator and maintained at a constant temperature of 25°C under long-day conditions with an 8-h/16-h dark/light photoperiod and a relative humidity of 60%.
Determination and Histochemical Staining of the Flavonoid and Gossypol Contents
To determinate the endogenous concentrations of gossypol, Nar, Que, DHK, DHQ, DHM, and rutin, leaf, or root samples were dried in a lyophilizer and ground to a fine powder. The powder (40 mg) was extracted with 2 mL 80% (v/v) methanol or solvent containing acetonitrile/water/phosphoric acid (80:20:0.1, v/v/v) by ultrasonification for 20 min for the detection of flavonoids or gossypol, respectively. The sample was centrifuged for 10 min at 13,000 rpm. The extract was analyzed on LC-MS or HPLC equipment for flavonoids or gossypol detection according to previously studies (Sunilkumar et al., 2006; Tan et al., 2013).
Histochemical staining of hand-cut cross sections of cotton tissue was used to visualize flavonoid localization. Sections were cut at the same position and visualized using 5% (w/v) NaOH. Briefly, sections were dipped in 5% (w/v) NaOH for 10 min, and immediately photographed under a fluorescence microscope (DM2500; Leica) with bright field illumination, the brown color indicating the presence of flavonoids.
Isolation of Cell Wall Water-Soluble Polysaccharides
The content of cell wall polysaccharides was measured according to a previously reported method with minor modifications (Rosli et al., 2004; Gallego-Giraldo et al., 2011). Briefly, samples (approximately 5 g) of leaves or roots were frozen in liquid nitrogen and quickly ground. The powder was homogenized with 10 mL 80% (v/v) methanol, and incubated overnight at 4°C. The homogenate was centrifuged at 13,000 rpm for 10 min. The methanol-insoluble cell wall residue was washed twice with 10 mL 80% (v/v) ethanol and followed with 10 mL absolute ethanol. After centrifugation at 13,000 rpm, the insoluble cell wall residue was dried with N2 and weighted.
Elicitation of Cell Cultures
The cell suspensions used in this experiment was derived from YZ1 embryogenic callus after two weeks subculture in dark (Sun et al., 2004). The 5-mL cell suspension cultures were treated with 5 mL of the water soluble fraction from wild type or the transgenic lines in 50 mL Erlenmeyer flasks, using the equal volume of distilled water or PGA (final concentration 0.5 mg/mL) as negative or positive control, respectively. The cell suspension cultures were harvested from each group of elicited samples at 10 h post elicitation, and frozen at 80°C for further analysis (Gallego-Giraldo et al., 2011).
Accession Numbers
Sequence data from this article can be found in the GenBank databases under the following accession numbers: GhLac1, KX822020.1, GhUB7, DQ116441.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of three homologous ESTs of GaLac1.
Supplemental Figure S2. Alignment analysis, phylogenetic analysis of the laccase family, and the tissue expression pattern of GhLac1.
Supplemental Figure S3. Molecular characterization of transgenic lines of GhLac1 and wild type.
Supplemental Figure S4. The indoor no-choice feeding assay for cotton bollworm and indoor choice assay for cotton aphid.
Supplemental Figure S5. The outdoor choice feeding assay for cotton aphid.
Supplemental Figure S6. Histochemical analysis of lignin deposition in stem hand-cut cross sections of Mock and V. dahliae-inoculated plants.
Supplemental Figure S7. Expression of lignin synthesis-related genes and assay of PAL enzyme activity in wild-type and transgenic plants.
Supplemental Figure S8. Expression of flavonoid and gossypol synthesis-related genes.
Supplemental Figure S9. Expression of JA signaling pathway genes after infection by V. dahliae.
Supplemental Figure S10. Expression of JA signaling pathway genes after infection by cotton bollworm.
Supplemental Figure S11. Measurement of cell wall polysaccharides and the elicitation of cell cultures.
Supplemental Figure S12. Efficacy of methanolic extracts from GhLac1 transgenic plants in conferring tolerance to cotton bollworm.
Supplemental Figure S13. Expression of SA-responsive genes under noninfected conditions.
Supplemental Table S1. The sequences used for phylogenetic analysis.
Supplemental Table S2. Oligonucleotides used in this study.
Acknowledgments
We are indebted to Professor Changying Niu (Huazhong Agricultural University, China) for providing cotton bollworms, and Huazhi Song (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, China) for assistance with the laser scanning confocal microscope.
Footnotes
This work was supported by the International Science and Technology Cooperation Program of China (grant no. 2015DFA30860), the Program of Introducing Talents of Discipline to Universities in China (grant no. B14032), and the National Science and Technology Major Project (2014ZX0800503B).
Articles can be viewed without a subscription.
References
- Avila CA, Arévalo-Soliz LM, Jia L, Navarre DA, Chen Z, Howe GA, Meng QW, Smith JE, Goggin FL (2012) Loss of function of FATTY ACID DESATURASE7 in tomato enhances basal aphid resistance in a salicylate-dependent manner. Plant Physiol 158: 2028–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, O’Malley DM, Whetten R, Sederoff RR (1993) A laccase associated with lignification in loblolly pine xylem. Science 260: 672–674 [DOI] [PubMed] [Google Scholar]
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, Lapierre C, Jouanin L (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bonello P, Blodgett JT (2003) Pinus nigra-Sphaeropsis sapinea as a model pathosystem to investigate local and systemic effects of fungal infection of pines. Physiol Mol Plant Pathol 63: 249–261 [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubna GA, Lima RB, Zanardo DY, Dos Santos WD, Ferrarese MdeL, Ferrarese-Filho O (2011) Exogenous caffeic acid inhibits the growth and enhances the lignification of the roots of soybean (Glycine max). J Plant Physiol 168: 1627–1633 [DOI] [PubMed] [Google Scholar]
- Buer CS, Muday GK, Djordjevic MA (2007) Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiol 145: 478–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Shi X, Desneux N, Han P, Gao X (2017) Elevated carboxylesterase activity contributes to the lambda-cyhalothrin insensitivity in quercetin fed Helicoverpa armigera (Hübner). PLoS One 12: e0183111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2000) Loosening of plant cell walls by expansins. Nature 407: 321–326 [DOI] [PubMed] [Google Scholar]
- Dauwe R, Morreel K, Goeminne G, Gielen B, Rohde A, van Beeumen J, Ralph J, Boudet AM, Kopka J, Rochange SF, Halpin C, Messens E, et al. (2007) Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J 52: 263–285 [DOI] [PubMed] [Google Scholar]
- Deng F, Tu L, Tan J, Li Y, Nie Y, Zhang X (2012) GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol 158: 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen H, Rampitsch C, Daayf F (2013) Signaling cross-talk in plant disease resistance. Plant Sci 207: 79–87 [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D, Xu J, Höfte M (2014) Making sense of hormone-mediated defense networking: from rice to Arabidopsis. Front Plant Sci 5: 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewanto V, Wu X, Adom KK, Liu RH (2002) Thermal processing enhances the nutritional value of tomatoes by increasing total antioxidant activity. J Agric Food Chem 50: 3010–3014 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- D’Ovidio R, Mattei B, Roberti S, Bellincampi D (2004) Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant-pathogen interactions. Biochim Biophys Acta 1696: 237–244 [DOI] [PubMed] [Google Scholar]
- Dubery IA, Smit F (1994) Phenylalanine ammonia-lyase from cotton (Gossypium hirsutum) hypocotyls: properties of the enzyme induced by a Verticillium dahliae phytotoxin. Biochim Biophys Acta 1207: 24–30 [DOI] [PubMed] [Google Scholar]
- Feng B, Shan L (2014) ROS open roads to roundworm infection. Sci Signal 7: pe10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GC, Joosten MH, Thomma BP (2011) Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol 156: 2255–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürstenberg-Hägg J, Zagrobelny M, Bak S (2013) Plant defense against insect herbivores. Int J Mol Sci 14: 10242–10297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA (2011) Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol 190: 627–639 [DOI] [PubMed] [Google Scholar]
- Gao W, Long L, Zhu LF, Xu L, Gao WH, Sun LQ, Liu LL, Zhang XL (2013) Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol Cell Proteomics 12: 3690–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MR, Jones JD (2009) Hormone (dis)harmony moulds plant health and disease. Science 324: 750–752 [DOI] [PubMed] [Google Scholar]
- Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55: 481–504 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Iqbal MJ, Ahsan R, Afzal AJ, Jamai A, Meksem K, El-Shemy HA, Lightfoot DA (2009) Multigenic QTL: the laccase encoded within the soybean Rfs2/rhg1 locus inferred to underlie part of the dual resistance to cyst nematode and sudden death syndrome. Curr Issues Mol Biol 11(Suppl 1): i11–i19 [PubMed] [Google Scholar]
- Iranshahi M, Rezaee R, Parhiz H, Roohbakhsh A, Soltani F (2015) Protective effects of flavonoids against microbes and toxins: the cases of hesperidin and hesperetin. Life Sci 137: 125–132 [DOI] [PubMed] [Google Scholar]
- Jadhav D, Mallikarjuna N, Rathore A, Pokle D (2012) Effect of some flavonoids on survival and development of Helicoverpa armigera (Hübner) and Spodoptera litura (Fab) (Lepidoptera: Noctuidae). Asian J Agric Sci 4: 298–307 [Google Scholar]
- Jin S, Zhang X, Nie Y, Guo X, Liang S, Zhu H (2006) Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol Plant 50: 519–524 [Google Scholar]
- Kloth KJ, Wiegers GL, Busscher-Lange J, van Haarst JC, Kruijer W, Bouwmeester HJ, Dicke M, Jongsma MA (2016) AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signalling. J Exp Bot 67: 3383–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König S, Feussner K, Kaever A, Landesfeind M, Thurow C, Karlovsky P, Gatz C, Polle A, Feussner I (2014) Soluble phenylpropanoids are involved in the defense response of Arabidopsis against Verticillium longisporum. New Phytol 202: 823–837 [DOI] [PubMed] [Google Scholar]
- Krempl C, Heidel-Fischer HM, Jiménez-Alemán GH, Reichelt M, Menezes RC, Boland W, Vogel H, Heckel DG, Joußen N (2016) Gossypol toxicity and detoxification in Helicoverpa armigera and Heliothis virescens. Insect Biochem Mol Biol 78: 69–77 [DOI] [PubMed] [Google Scholar]
- La Camera S, Gouzerh G, Dhondt S, Hoffmann L, Fritig B, Legrand M, Heitz T (2004) Metabolic reprogramming in plant innate immunity: the contributions of phenylpropanoid and oxylipin pathways. Immunol Rev 198: 267–284 [DOI] [PubMed] [Google Scholar]
- Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BP, Staskawicz B, Jones JD, Zipfel C (2010) Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol 28: 365–369 [DOI] [PubMed] [Google Scholar]
- Mechri B, Tekaya M, Cheheb H, Attia F, Hammami M (2015) Accumulation of flavonoids and phenolic compounds in olive tree roots in response to mycorrhizal colonization: a possible mechanism for regulation of defense molecules. J Plant Physiol 185: 40–43 [DOI] [PubMed] [Google Scholar]
- Mellon JE, Dowd MK, Beltz SB, Moore GG (2014) Growth inhibitory effects of gossypol and related compounds on fungal cotton root pathogens. Lett Appl Microbiol 59: 161–168 [DOI] [PubMed] [Google Scholar]
- Min L, Hu Q, Li Y, Xu J, Ma Y, Zhu L, Yang X, Zhang X (2015) LEAFY COTYLEDON1-CASEIN KINASE I-TCP15-PHYTOCHROME INTERACTING FACTOR4 network regulates somatic embryogenesis by regulating auxin homeostasis. Plant Physiol 169: 2805–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra P, Pandey A, Tiwari M, Chandrashekar K, Sidhu OP, Asif MH, Chakrabarty D, Singh PK, Trivedi PK, Nath P, Tuli R (2010) Modulation of transcriptome and metabolome of tobacco by Arabidopsis transcription factor, AtMYB12, leads to insect resistance. Plant Physiol 152: 2258–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morkunas I, Woźniak A, Formela M, Mai VC, Marczak Ł, Narożna D, Borowiak-Sobkowiak B, Kühn C, Grimm B (2016) Pea aphid infestation induces changes in flavonoids, antioxidative defence, soluble sugars and sugar transporter expression in leaves of pea seedlings. Protoplasma 253: 1063–1079 [DOI] [PubMed] [Google Scholar]
- Mottiar Y, Vanholme R, Boerjan W, Ralph J, Mansfield SD (2016) Designer lignins: harnessing the plasticity of lignification. Curr Opin Biotechnol 37: 190–200 [DOI] [PubMed] [Google Scholar]
- Naoumkina MA, Zhao Q, Gallego-Giraldo L, Dai X, Zhao PX, Dixon RA (2010) Genome-wide analysis of phenylpropanoid defence pathways. Mol Plant Pathol 11: 829–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenaah GE. (2014) Toxic and antifeedant activities of prenylated flavonoids isolated from Tephrosia apollinea L. against three major coleopteran pests of stored grains with reference to their structure-activity relationship. Nat Prod Res 28: 2245–2252 [DOI] [PubMed] [Google Scholar]
- Onkokesung N, Reichelt M, van Doorn A, Schuurink RC, van Loon JJ, Dicke M (2014) Modulation of flavonoid metabolites in Arabidopsis thaliana through overexpression of the MYB75 transcription factor: role of kaempferol-3,7-dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae. J Exp Bot 65: 2203–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Leon-Reyes A, van der Ent S, van Wees SC (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Rosli HG, Civello PM, Martínez GA (2004) Changes in cell wall composition of three Fragaria x ananassa cultivars with different softening rate during ripening. Plant Physiol Biochem 42: 823–831 [DOI] [PubMed] [Google Scholar]
- Santiago R, Barros-Rios J, Malvar RA (2013) Impact of cell wall composition on maize resistance to pests and diseases. Int J Mol Sci 14: 6960–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, He SY, Rizo J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Chuang WP (2014) Plant resistance to aphid feeding: behavioral, physiological, genetic and molecular cues regulate aphid host selection and feeding. Pest Manag Sci 70: 528–540 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SM, Korzelius JP, van Pelt JA, Mueller MJ, Buchala AJ, Métraux JP, Brown R, Kazan K, van Loon LC, Dong X, et al. (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterjiades R, Dean JF, Eriksson KE (1992) Laccase from Sycamore Maple (Acer pseudoplatanus) polymerizes monolignols. Plant Physiol 99: 1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhu L, Xu L, Yuan D, Min L, Zhang X (2014) Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat Commun 5: 5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhang X, Nie Y, Guo X, Jin S, Liang S (2004) Production and characterization of somatic hybrids between upland cotton (Gossypium hirsutum) and wild cotton (G. klotzschianum Anderss) via electrofusion. Theor Appl Genet 109: 472–479 [DOI] [PubMed] [Google Scholar]
- Sunilkumar G, Campbell LM, Puckhaber L, Stipanovic RD, Rathore KS (2006) Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc Natl Acad Sci USA 103: 18054–18059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Tu L, Deng F, Hu H, Nie Y, Zhang X (2013) A genetic and metabolic analysis revealed that cotton fiber cell development was retarded by flavonoid naringenin. Plant Physiol 162: 86–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou JM (2017) Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29: 618–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoison O, Sévenet T, Niemeyer HM, Russell GB (2004) Insect antifeedant compounds from Nothofagus dombeyi and N. pumilio. Phytochemistry 65: 2173–2176 [DOI] [PubMed] [Google Scholar]
- Trapero C, Wilson IW, Stiller WN, Wilson LJ (2016) Enhancing integrated pest management in GM cotton systems using host plant resistance. Front Plant Sci 7: 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutter D. (2005) Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol (Stuttg) 7: 581–591 [DOI] [PubMed] [Google Scholar]
- Wang GD, Li QJ, Luo B, Chen XY (2004) Ex planta phytoremediation of trichlorophenol and phenolic allelochemicals via an engineered secretory laccase. Nat Biotechnol 22: 893–897 [DOI] [PubMed] [Google Scholar]
- Wang Q, Eneji AE, Kong X, Wang K, Dong H (2015) Salt stress effects on secondary metabolites of cotton in relation to gene expression responsible for aphid development. PLoS One 10: e0129541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woźniak A, Drzewiecka K, Kęsy J, Marczak Ł, Narożna D, Grobela M, Motała R, Bocianowski J, Morkunas I (2017) The Influence of lead on generation of signalling molecules and accumulation of flavonoids in pea seedlings in response to pea aphid infestation. Molecules 22: 9 10.3390/molecules22091404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Baldwin IT (2010) New insights into plant responses to the attack from insect herbivores. Annu Rev Genet 44: 1–24 [DOI] [PubMed] [Google Scholar]
- Wu KM, Lu YH, Feng HQ, Jiang YY, Zhao JZ (2008) Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science 321: 1676–1678 [DOI] [PubMed] [Google Scholar]
- Wu S, Shan L, He P (2014) Microbial signature-triggered plant defense responses and early signaling mechanisms. Plant Sci 228: 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Yuan M, Ai C, Liu L, Zhuang E, Karapetyan S, Wang S, Dong X (2017) uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545: 491–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhu L, Tu L, Liu L, Yuan D, Jin L, Long L, Zhang X (2011) Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J Exp Bot 62: 5607–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Tu L, Zhu L, Fu L, Min L, Zhang X (2008) Expression profile analysis of genes involved in cell wall regeneration during protoplast culture in cotton by suppression subtractive hybridization and macroarray. J Exp Bot 59: 3661–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XY, Zhang XL, Jin SX, Fu LL, Wang LG (2007) Production and characterization of asymmetric hybrids between upland cotton Coker 201 (Gossypium hirsutum) and wild cotton (G. klozschianum Anderss). Plant Cell Tissue Organ Cult 89: 225–235 [Google Scholar]
- Zhang JM, Huang GQ, Li Y, Zheng Y, Li XB (2014) Cotton photosynthesis-related PSAK1 protein is involved in plant response to aphid attack. Mol Biol Rep 41: 3191–3200 [DOI] [PubMed] [Google Scholar]
- Zhang YC, Yu Y, Wang CY, Li ZY, Liu Q, Xu J, Liao JY, Wang XJ, Qu LH, Chen F, Xin P, Yan C, et al. (2013) Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol 31: 848–852 [DOI] [PubMed] [Google Scholar]