TCP17 acts as a key factor in regulating shade-induced hypocotyl rapid growth by directly promoting the transcriptional levels of PIFs and auxin biosynthesis genes.
Abstract
Light quality surrounding a plant is largely determined by the density of its neighboring vegetation. Plants are able to sense shade light signals and initiate a series of adaptation responses, which is known as shade avoidance syndrome (SAS). PHYTOCHROME INTERACTING FACTORS (PIFs) are key factors in the SAS network by regulating the biosynthesis of multiple phytohormones and the expression of cell expansion genes. Although the protein levels of PIFs were found to be acumulated in shade, the transcriptional regulation of PIFs in response to such an environmental signal remains poorly understood. Here we show that TCP17 and its two closely related homologs, TCP5 and TCP13, play an important role in mediating shade-induced hypocotyl elongation by up-regulating auxin biosynthesis via a PIF-dependent and a PIF-independent pathway. In constitutive white light, a tcp5, 13, 17 triple mutant (3tcp) showed a subtle hypocotyl defective phenotype. In shade, however, 3tcp showed a significantly reduced hypocotyl elongation phenotype, indicating a positive role of TCPs in regulating SAS. Our in-depth biochemical and genetic analyses indicated that TCP17 can be significantly accumulated in shade. TCP17 binds to the promoters of PIFs and YUCCAs to indirectly or directly up-regulate auxin levels in shade. These data provide new insights into our better understanding of the regulatory mechanisms of SAS in plants.
Shade-avoidance syndrome (SAS) is a phenomenon widely observed in plant kingdom (Mathews, 2006). Shade light signals regulate various growth and developmental processes such as repressing seed germination, promoting hypocotyl and petiole growth, changing leaf angle, arresting leaf and root development, accelerating flowering, and reducing branching (Cerdán and Chory, 2003; Casal, 2012; González-Grandío et al., 2013). These adaptive responses to shade can increase the fitness of plants in crowded environments.
Shade light signals mainly include a reduced ratio of red light to far red light (low R:FR) and reduced blue light, which can be sensed by PHYB (Halliday et al., 1994; Schepens et al., 2004; Keller et al., 2011) and CRY1 (Yu et al., 2010; Keller et al., 2011; Sellaro et al., 2011; Pedmale et al., 2016), respectively. In response to shade avoidance, the Pr and Pfr forms of PHYB are present in a R:FR ratio-dependent balance. Under white light with a higher R:FR, the photo-equilibrium is displaced toward the active Pfr form, which interacts with a group of PHYTOCHROME INTERACTING FACTORS (PIFs) in the nucleus and mediates their rapid phosphorylation and degradation. Under lower R:FR, on the other hand, the photo-equilibrium is displaced toward the inactive Pr form, leading to the accumulation of PIFs in the nucleus (Cifuentes-Esquivel et al., 2013). The accumulated PIFs subsequently promote the expression of shade responsive genes (Lorrain et al., 2008; Li et al., 2012), including those for the synthesis of multiple phytohormones, to promote rapid growth in shade.
Previous studies indicated that a number of phytohormones including auxin (Tao et al., 2008; Won et al., 2011; Li et al., 2012; Hayes et al., 2014; Hersch et al., 2014), gibberellins (Devlin et al., 2003; Djakovic-Petrovic et al., 2007), and brassinosteroids (Luccioni et al., 2002; Sorin et al., 2009) play key roles in the establishment of SAS. Auxin is thought to be the most important phytohormone regulating shade-induced rapid growth (Casal, 2012). Mutants affecting auxin perception, biosynthesis, or transport show impaired hypocotyl growth under shade (Tao et al., 2008). It was demonstrated that shade can rapidly increase auxin accumulation through promoting transcriptional levels of a number of auxin biosynthesis genes (Tao et al., 2008; Won et al., 2011; Li et al., 2012; Hayes et al., 2014; Hersch et al., 2014). Trp-dependent IAA biosynthesis via indole-3-pyruvic acid (IPA) has been suggested as the most abundant IAA biosynthesis pathway (Mashiguchi et al., 2011). In this pathway, Trp is converted to IPA by TAA1 (Stepanova et al., 2008; Zhou et al., 2011), and IPA is then converted to IAA. The second step is catalyzed by a family of flavin monooxygenases encoded by YUCCA genes (YUCs; Zhao et al., 2001). As YUCs are the rate-limiting enzymes in Trp-dependent auxin biosynthesis pathway, the transcription of YUCs is therefore strictly controlled. PIF transcription factors are critical in regulating the expression of YUCs in shade, and auxin synthesized through PIF-regulated pathway is significant for shade-induced rapid growth (Hornitschek et al., 2012; Li et al., 2012). Although the mechanism that shade light signals increase the stability of PIFs has been elucidated, little is known about the transcriptional network regulating the expression of PIFs in response to shade (Lorrain et al., 2008; Li et al., 2012).
The TEOSINTE BRANCHED1, CYCLOIDEA, and PCF (TCP) family of transcription factors plays important roles in multiple aspects of plant growth and development (Cubas et al., 1999; Martín-Trillo and Cubas, 2010). For example, several TCPs were found to regulate cell elongation, leaf morphogenesis, leaf senescence, and architecture establishment through mediating phytohormone biosynthesis and signal transduction (Aguilar-Martínez et al., 2007; Schommer et al., 2008; Guo et al., 2010; Koyama et al., 2010; Challa et al., 2016). In Arabidopsis, five closely related CIN TCPs including TCP2, TCP3, TCP4, TCP10, and TCP24, whose transcripts can be degraded by miR319, redundantly regulate cell division and elongation during organ morphogenesis. TCP5, TCP13, and TCP17 belong to the CIN subfamily but are not regulated by miR319. Previous studies suggested that they are involved in regulating leaf morphogenesis, petal development, and flowering (Efroni et al., 2008; Huang and Irish, 2015).
Despite the crucial roles of TCP transcription factors in regulating organ morphogenesis have been well studied, detailed mechanisms regarding their functions in integrating environment signals and endogenous gene expressions are not well established. In this study, we provide strong evidence showing that TCP17, TCP5, and TCP13 play critical roles in mediating shade-induced auxin biosynthesis through directly up-regulating the expression of PIFs and YUCs. Our results uncover a new function of TCP transcription factors in linking environmental signals to endogenous gene expression and phytohormone biosynthesis.
RESULTS
TCP17 Acts as a Positive Regulator in Response to Shade
Our previous research indicated that TCP1 can positively regulate brassinosteroid (BR) biosynthesis via directly binding to the promoter of a key BR biosynthetic gene, DWF4 (Guo et al., 2010). To reveal biological functions of other TCP transcription factors in Arabidopsis (Arabidopsis thaliana), we overexpressed all 24 TCPs under the control of a cauliflower mosaic virus 35S promoter in Arabidopsis. Among all the transgenic plants obtained, we found that overexpression of TCP17 (TCP17-OX) resulted in various developmental defects, including narrow leaf blades, abnormal floral organs, and reduced male sterility (Supplemental Fig. S1A, C, and D). Our detailed analyses showed that the hypocotyls of TCP17-OX transgenic seedlings are significantly longer than those of Col-0 in a long-day growing condition (Supplemental Fig. S2, B and C). In dark, TCP17-OX transgenic seedlings, however, showed no obvious phenotypes (Supplemental Fig. S2, D and E). GUS staining analysis of 5-d-old pTCP17::GUS transgenic seedlings grown under a light condition showed that GUS signals can be observed in hypocotyls in addition to cotyledons and mature zones of the roots (Supplemental Fig. S1B). These results suggest that TCP17 plays a crucial role in promoting hypocotyl growth under a light condition.
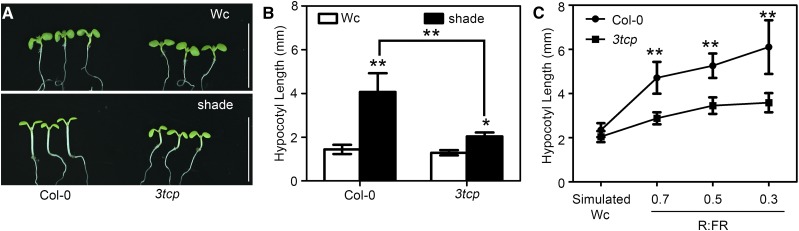
Light quality is an important environmental factor affecting hypocotyl elongation (Casal, 2012). To investigate whether TCPs are involved in regulating hypocotyl growth under a low R:FR, (also known as shade), we analyzed the hypocotyl response of Col-0, tcp17, and TCP17-OX to shade. Hypocotyl measurements showed that TCP17-OX caused longer hypocotyls than wild type under either constitutive white light (Wc) or shade; however, the hypocotyls of tcp17 mutant appeared to be similar to that of wild type (Supplemental Fig. S3). Considering the functional redundancy of TCPs in regulating plant growth and development, we examined the roles of TCP5 and TCP13, the two most closely related homologs of TCP17, in shade avoidance. TCP5-OX and TCP13-OX transgenic plants showed longer hypocotyls than wild type in both Wc and shade conditions (Supplemental Fig. S3). Although tcp17, tcp5, and tcp13 single mutants did not show significant defects in response to shade, a tcp5 tcp17 double mutant showed significantly reduced hypocotyl elongation phenotypes compared to wild-type seedlings. A tcp5 tcp13 tcp17 (3tcp) triple mutant displayed even shorter hypocotyls than the seedlings of wild-type, single, or double mutant in shade (Fig. 1, A and B; Supplemental Fig. S3). We next investigated the sensitivity of Col-0 and 3tcp to lighting conditions with various ratios of R:FR in detail. Treatment in shade with an increasing amount of FR can significantly increase hypocotyl elongation of wild-type seedlings, but the sensitivity of 3tcp in response to shade is greatly reduced (Fig. 1C). Quantitative real-time PCR (qRT-PCR) analysis showed that the expression of many shade response genes, including PIL1, IAA19, IAA29, and GH3.3, is greatly increased in TCP17-OX seedlings but decreased in the 3tcp mutant in shade compared to those of wild type (Supplemental Fig. S4). These results indicated that TCP17, TCP5, and TCP13 are important factors redundantly promoting hypocotyl elongation in response to shade.
Figure 1.
TCP17 positively regulates shade-induced hypocotyl elongation. A and B, Phenotypes and hypocotyl measurements of Col-0 and the triple mutant 3tcp seedlings grown under a Wc or a shade condition. Seedlings are grown under Wc for 3 d and then transferred to shade or stayed in Wc for an additional 5 d before the pictures were taken and the measurements were carried out. Scale bars = 1 cm. C, Hypocotyl elongation in response to the changes of R:FR ratio for Col-0 and 3tcp seedlings. Seedlings were grown under simulated Wc for 3 d before being transferred to simulated Wc or shade at the indicated R:FR for an additional 5 d. Data shown are the average of three independent biological replicates and SE (n ≥ 20 for each experiment). *P < 0.05 and **P < 0.01; based on Student’s t test.
Because TCP17-OX showed a more significant role in promoting hypocotyl growth compared to either TCP5-OX or TCP13-OX (Supplemental Fig. S3), we used TCP17 as a representative of the three TCPs to reveal their detailed molecular mechanisms in regulating hypocotyl elongation in shade.
Shade Causes the Accumulation of TCP17 Protein
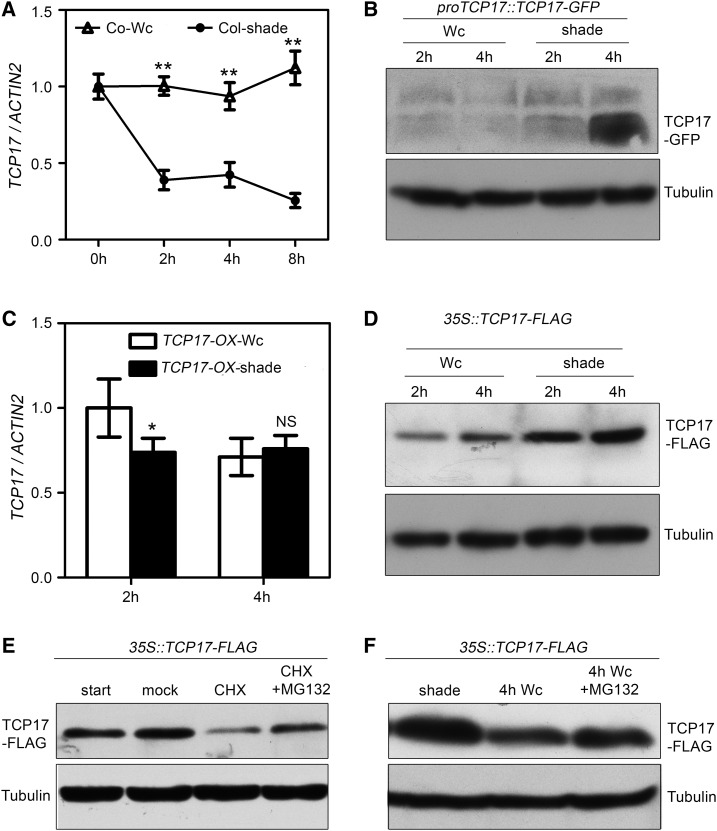
As a positive regulator of SAS, TCP17 was expected to be activated by shade. Our quantitative RT-PCR analysis, however, showed that the transcript abundance of TCP17 was rapidly decreased when Wc grown seedlings were transferred to shade (Fig. 2A). These unexpected results prompted us to investigate the response of TCP17 protein level to shade treatment. Because TCP17 antibody was not available, proTCP17::TCP17-GFP transgenic seedlings were generated for protein accumulation analysis. Our immunoblotting analysis showed that TCP17-GFP was significantly accumulated after transferring from Wc to shade (Fig. 2B; Supplemental Fig. S5A). Further analysis by using 35S::TCP17-FLAG transgenic seedlings, whose transcription was not significantly up-regulated by shade (Fig. 2C), showed the accumulation of TCP17-FLAG is greatly increased in shade as well (Fig. 2D; Supplemental Fig. S5B). To examine whether protein stability plays a role in TCP17 regulation, we examined the in vivo stability of TCP17-FLAG by treating 35S::TCP17-FLAG transgenic seedlings with a protein biosynthesis inhibitor cycloheximide (CHX). We found that TCP17-FLAG was significantly decreased after 1 h treatment with CHX, whereas addition of proteasome inhibitor MG132 largely prevented the TCP17-FLAG from degradation (Fig. 2E; Supplemental Fig. S5C), indicating TCP17 is an unstable protein that may be degraded via a 26S proteasome degradation pathway. Additionally, the accumulation of TCP17-FLAG was greatly decreased after being transferred from shade to Wc. The Wc-induced degradation of TCP17-FLAG can be significantly suppressed by the treatment of MG132 (Fig. 2F; Supplemental Fig. S5D). These results suggest that TCP17 is an unstable protein in Wc, whereas shade can significantly slow its degradation, resulting in protein accumulation.
Figure 2.
The accumulation of TCP17 protein is increased in shade. A, TCP17 transcriptional level is repressed in shade. Seven-day-old Col-0 seedlings grown under Wc were kept in Wc or transferred to shade for the indicated period of time before being harvested for RNA extraction. The error bars represent the SE of three independent biological replicates. *P < 0.05 and **P < 0.01; based on Student’s t test. B, TCP17 protein level is accumulated in shade. Seven-day-old Wc-grown proTCP17::TCP17-GFP transgenic plants were transferred to shade or stayed in Wc for 2 or 4 h. Total protein extracts were separated by SDS-PAGE and analyzed by an immunoblotting approach using an anti-GFP antibody. An immunoblotting assay using antitubulin (TUB) antibody was carried out for a loading control. C and D, TCP17 protein levels were increased, although TCP17 transcription levels were not significantly accumulated, in 35S::TCP17-FLAG transgenic plants after shade treatment. 35S::TCP17-FLAG transgenic plants were treated as shown in B. Total RNA and proteins were extracted for quantitative RT-PCR and immunoblotting assay. Data shown are average of three independent biological replicates and SE. *P < 0.05 and **P < 0.01; NS, not significant (P ≥ 0.05); based on Student’s t test. E, TCP17 is an unstable protein in Wc. Seven-day-old 35S::TCP17-FLAG transgenic seedlings grown in Wc were treated with mock solution, 10 μm CHX, or 10 μm CHX plus 20 μm MG132 for 1 h before the samples were collected for an immunoblotting assay. F, TCP17 is more stable in shade than in Wc. Seven-day-old 35S::TCP17-FLAG transgenic seedlings grown in Wc were transferred to shade for 1 d. The seedlings were kept in shade or exposed to Wc with or without 20 μm MG132 for 4 h. Immunoblotting assays were used to detect protein levels.
TCP17 Is a Key Transcription Factor Regulating Shade-Induced Free IAA Accumulation
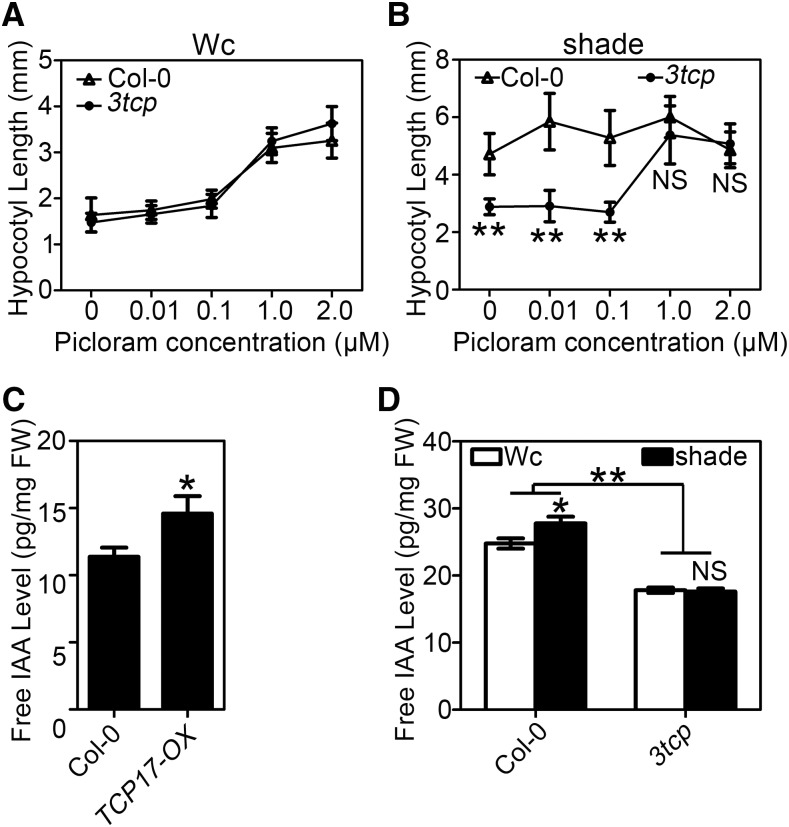
It has been demonstrated that auxin plays a predominant role in response to shade-induced rapid growth (Tao et al., 2008; Casal, 2012). To examine whether TCP17 promotes shade avoidance via regulating an auxin pathway, Col-0 and 3tcp seedlings were grown in media containing various concentrations of picrolam (PIC), an analog of auxin, under Wc for 3 d, and then transferred to shade or remained in Wc for additional 5 d. Hypocotyl measurements showed that in Wc the hypocotyl growth of 3tcp can be stimulated by PIC in a way similar to that of Col-0 (Fig. 3A). In shade, exogenous PIC had no significant effect on hypocotyl elongation of Col-0, whereas it can greatly rescue hypocotyl growth of 3tcp (Fig. 3B). We also treated Col-0 and 3tcp seedlings with two other growth-promoting hormones, GA3 and 24-epiBL, in Wc or shade. Our results showed that the responses of hypocotyls of 3tcp to GA3 and BR treatments in Wc and shade remained unaltered to those of wild type (Supplemental Fig. S6). These results suggested that auxin, instead of other phytohormones, plays an important role in TCP17-mediated hypocotyl growth in shade.
Figure 3.
TCP17 positively regulates IAA biosynthesis. A and B, Responses of Col-0 and 3tcp mutants to an auxin analog picloram (PIC). Col-0 and 3tcp seedlings were grown under Wc with different concentrations (0, 0.01, 0.1, 1.0, 2.0 μm) of PIC for 3 d and then transferred to Wc (A) or shade (B) for an additional 5 d before hypocotyl length was measured. C, TCP17 transgenic plants contain more free IAA than Col-0. Free IAA levels were analyzed in 7-d-old Col-0 and TCP17-OX transgenic seedlings grown under Wc. D, Shade induced free IAA accumulation is significantly impaired in triple mutant 3tcp. Col-0 and 3tcp were grown under Wc for 7 d and transferred to Wc or shade for 2 h before being collected for the measurements. Data shown are the average of three independent biological replicates and SE. *P < 0.05 and **P < 0.01; NS, not significant (P ≥ 0.05); based on Student’s t test.
Previous studies indicated that TCPs are involved in regulating biosynthesis of multiple phytohormones, including jasmonates and brassinosteroids (Schommer et al., 2008; Guo et al., 2010). To investigate whether TCP17 regulates auxin accumulation, we analyzed the expression levels of DR5::GUS reporter in Col-0 and TCP17-OX plants. GUS staining results showed that, compared to that in Col-0, the expression of DR5::GUS is significantly elevated in TCP17-OX (Supplemental Fig. S7). In addition, measurements showed that the accumulation of free IAA is significantly increased in TCP17-OX seedlings while decreased in 3tcp in Wc (Fig. 3, C and D). Moreover, free IAA level elevated in shade is diminished in 3tcp mutant (Fig. 3D). These results demonstrated that TCPs are involved in regulating shade-induced auxin biosynthesis.
TCP17 Promotes Shade Avoidance Partially through PIF Transcription Factors
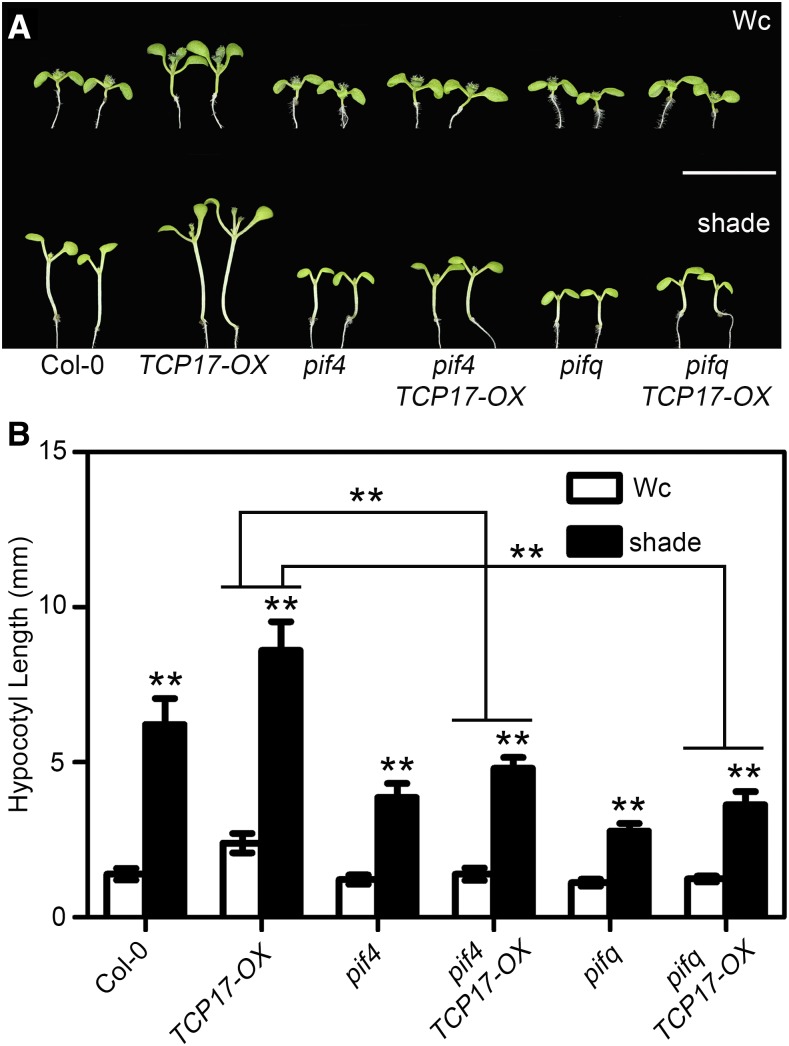
PIF transcription factors are key regulators in integrating environmental signals and endogenous responses (Castillon et al., 2007; Lucyshyn and Wigge, 2009; Wigge, 2013). Previous studies indicated PIF4, PIF5, and PIF7 act as central components in shade-induced rapid growth through promoting free IAA accumulation (Hornitschek et al., 2012; Li et al., 2012). To investigate whether TCP17 promoting hypocotyl elongation depends on PIFs, we overexpressed TCP17 in pif4 single and pif1,3,4,5 (pifq) quadruple mutants. The transgenic lines with transcriptional level of TCP17 similar to that of TCP17-OX seedlings were used for further analyses (Supplemental Fig. S8). Our results showed that in shade, the hypocotyls of pif4 TCP17-OX and pifq TCP17-OX transgenic seedlings are much shorter than that of TCP17-OX seedlings (Fig. 4). Consistently, transgenic plants overexpressing PIF4 in 3tcp appeared a response similar to PIF4-OX plants, displaying a constitutive shade response in white light (Supplemental Fig. S9). These results suggest that TCP17 regulates shade avoidance in a PIF-dependent manner.
Figure 4.
TCP17 positively regulates shade avoidance partially through PIF transcription factors. The hypocotyl phenotypes and the measurements of Col-0, TCP17-OX, pif4, pif4 TCP17-OX, pifq, and pifq TCP17-OX plants grown in Wc or shade. Three-day-old Wc grown seedlings were transferred to shade or remained in Wc for an additional 5 d before analyses were taken. Scale bars represent 1 cm. Data shown are the average of three independent biological replicates and SE (n ≥ 20 for each experiment). *P < 0.05 and **P < 0.01; based on Student’s t test.
TCP17 Promotes the Expression of PIF4 and PIF5 and Binds to Their Promoters in a Light Quality-Dependent Manner
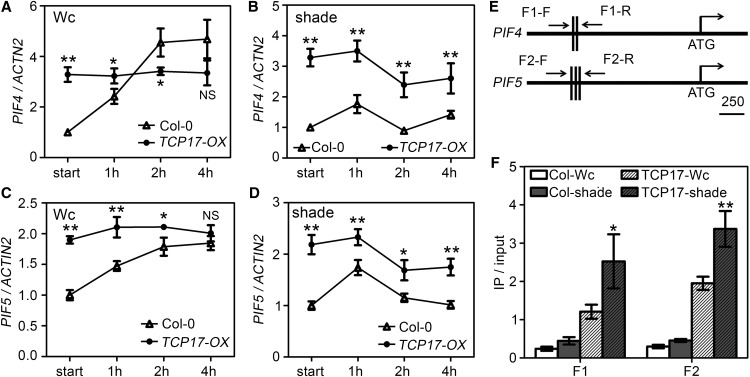
To determine how TCP17 interacts with PIFs in response to shade, we investigated the expression of PIF4 and PIF5 in TCP17-OX transgenic plants after transfer to Wc or shade for various time duration. The transcriptional levels of PIF4 and PIF5 were significantly increased in TCP17-OX seedlings in shade (Fig. 5, A–D). However, the transcriptional levels of PIF4 and PIF5 were not significantly impaired in 3tcp in Wc and shade (Supplemental Fig. S10). These results suggest that TCP17 can positively regulate the expression of PIF4 and PIF5, but the expression of these two transcription factors may also rely on other undetermined cues. We also investigated the expression of TCP17 in pif4 and pifq mutants and found the transcriptional level of TCP17 remained unchanged in both mutants in either Wc or shade (Supplemental Fig. S11). To determine whether TCP17 directly binds to the promoters of PIF4 and PIF5 and regulates their expression, chromatin immuno-precipitation (ChIP) assay with primers flanking TCP binding sites (TBS) in both genes was performed (Fig. 5E). Transgenic plants harboring 35S::TCP17-FLAG were grown in Wc for 2 weeks and transferred to shade or remained in Wc for 2 h before being harvested for analyses. CHIP-qPCR analyses showed that TCP17 binds to the promoters of both PIF4 and PIF5. Interestingly, increased enrichment of PIF4 and PIF5 promoters was observed after shade treatment, suggesting shade can increase the function of TCP17 (Fig. 5F). These results indicated TCP17 can directly associate with the promoters of PIF4 and PIF5 and promotes their expressions in shade.
Figure 5.
TCP17 binds to the promoters of PIF4 and PIF5 and positively regulates their expression in a shade dependent manner. A to D, TCP17 promotes the expression of PIF4 and PIF5 in shade. Col-0 and TCP17-OX plants were grown in Wc for 7 d then transferred to shade or Wc for various times. Total RNA was extracted and the transcriptional levels of PIF4 and PIF5 were analyzed by quantitative RT-PCR. Data shown are average of three independent biological replicates and SE (*P < 0.05 and **P < 0.01; NS, not significant [P ≥ 0.05]; based on Student’s t test). E, Schematic diagrams showing the presence of TBS in the promoters of PIF4 and PIF5. F, Shade improves TCP17 to bind to the promoters of PIF4 and PIF5. Two-week-old Wc grown Col-0 and 35S::TCP17-FLAG plants were transferred to shade or Wc for an additional 2 h before being collected for CHIP-PCR assay using the primers flanking the TBS indicated in C. Data shown are average of three independent biological replicates and SE (*P < 0.05 and **P < 0.01; based on Student’s t test).
TCPs Are Required for Shade-Mediated Up-Regulation of YUCs
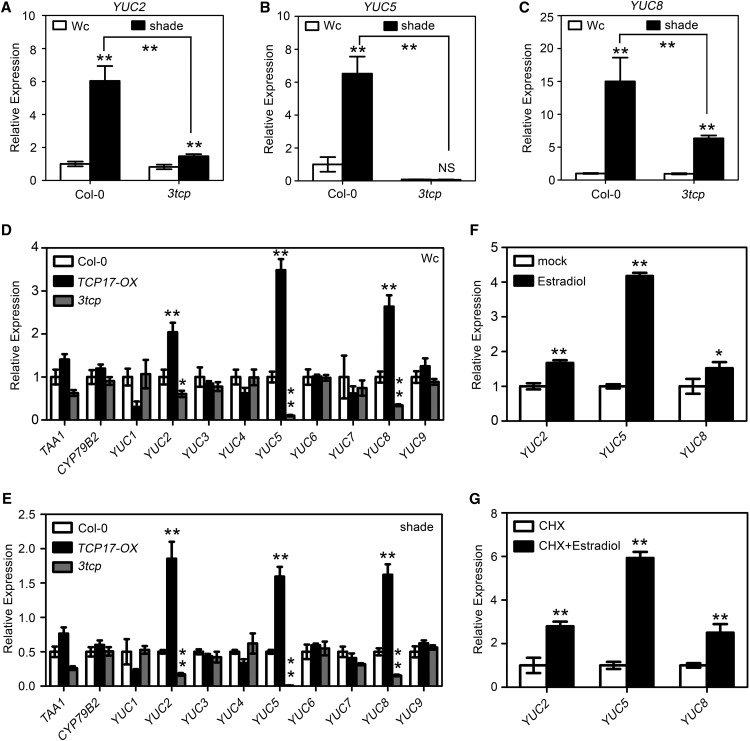
Our results showed that TCP17 promotes shade avoidance through directly regulating the transcript abundance of PIF4 and PIF5 (Fig. 5). However, defects of PIFs cannot completely suppress the hypocotyl elongation response of TCP17-OX transgenic plants in shade (Fig. 4). Additionally, the IAA accumulation induced by shade was significantly impaired in 3tcp (Fig. 3D); however, the transcription levels of both PIF4 and PIF5 were not influenced in 3tcp (Supplemental Fig. S10). These results suggested there is an additional mechanism of TCP17 in promoting shade avoidance. Because the expression of many auxin biosynthesis genes is greatly influenced by shade avoidance, we then assayed the transcriptional levels of auxin biosynthesis genes in 3tcp after treatment with Wc or shade. qRT- PCR analyses showed that YUC2, YUC5, and YUC8 were greatly induced by shade in Col-0 (Fig. 6, A–C). The induced expression levels of these three genes by shade, however, were greatly reduced in 3tcp (Fig. 6, A–C). The responses to shade of other tested auxin biosynthesis genes were not significantly impaired in 3tcp (Supplemental Fig. S12). These data suggested that TCPs are critical for shade-mediated up-regulation of several auxin biosynthesis genes.
Figure 6.
TCP17 positively regulates the transcriptional levels of YUCs in response to shade. A to C, The transcription levels of YUC2 (A), YUC5 (B), and YUC8 (C) in Col-0 and triple mutant 3tcp in response to Wc and shade. Seven-day-old Wc grown seedlings were transferred to Wc or shade for an additional 2-h treatment before being harvested for total RNA extraction. D and E, Quantitative PCR results showing transcript levels of auxin biosynthesis genes in Col-0, 3tcp, and TCP17-OX transgenic seedlings grown in Wc (D) or shade (E). Seven-day-old seedlings grown under Wc were transferred to Wc or shade for an addition l-day treatment before being collected for measurements. F and G, The transcriptional levels of YUC2, YUC5, and YUC8 can be induced by TCP17 directly. Seven-day-old 35S::TCP17-ER transgenic seedlings grown under Wc were treated with mock solution (1/10 000 ethanol), 10 μm estradiol, 10 μm CHX, or a combination of 10 μm CHX and 10 μm estradiol for 4 h before being harvested for measurements. All data shown are the average of three independent biological replicates and SE (*P < 0.05 and **P < 0.01; NS, based on Student’s t test.).
Our in-depth analysis showed that the transcription levels of YUC2, YUC5, and YUC8 are significantly elevated in TCP17-OX and decreased in 3tcp under a Wc or a shade condition (Fig. 6, D and E). To determine whether these three YUCs are the early targets of TCP17, we fused TCP17 with a hormone-binding domain of a human estrogen receptor (ER) at its C terminus. Transgenic plants overexpressing TCP17-ER under the control of a 35S promoter were generated. Treated 35S::TCP17-ER transgenic plants with estradiol induced phenotypes mimicking those of TCP17-OX plants (Supplemental Fig. S13). RT-PCR analyses indicated that the transcriptional levels of YUC2, YUC5, and YUC8 were increased significantly after 4-h treatment with estradiol (Fig. 6F). To exclude the possibility that TCP17 induces the expression of these YUCs through other transcription factors, we pretreated 35S::TCP17-ER seedlings with CHX for 2 h before estradiol was applied. Consistently, the expression levels of these three YUCs were significantly enhanced after a combination of CHX and estradiol treatment (Fig. 6G). These results suggested that the transcriptional levels of YUC2, YUC5, and YUC8 can be rapidly and directly induced by TCP17.
TCP17 Directly Binds to the Promoter Regions of YUCs and Promotes Their Expression
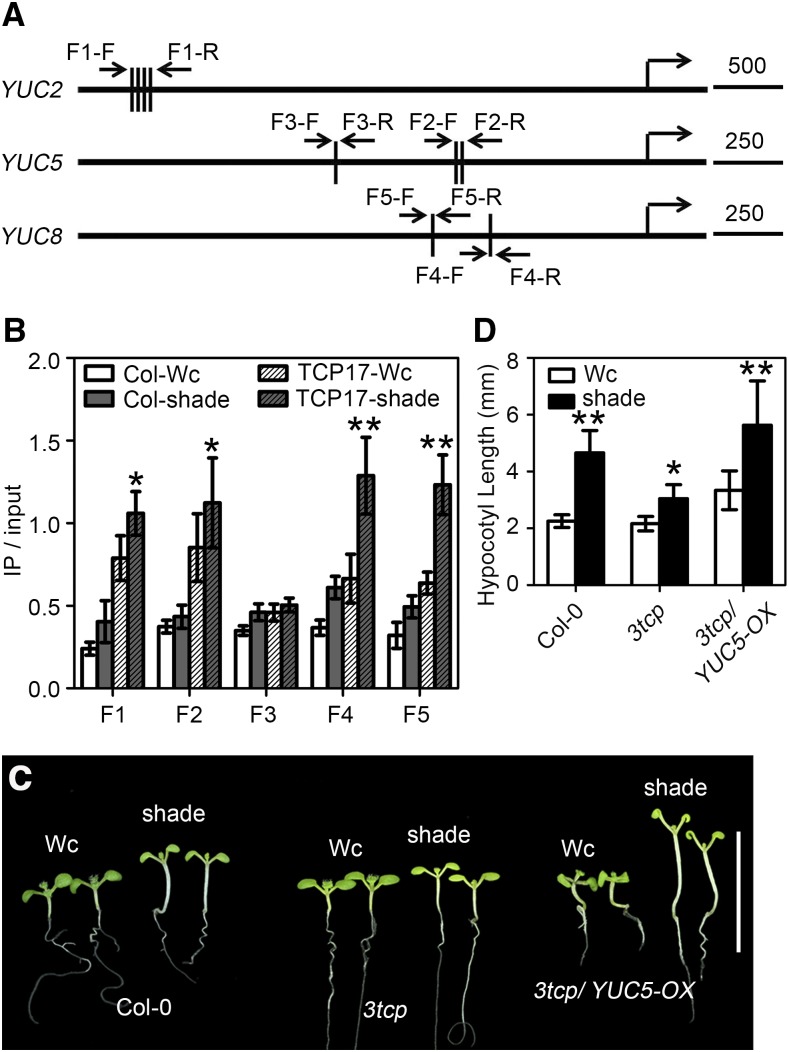
Previous studies demonstrated the DNA motifs that class-I and class-II TCPs prefer to bind to are GGNCCCAC and GTGGNCCC, respectively. GGNCCC is the core sequence of the TBS; mutation in this motif can reduce the interaction between TCPs and TBS (Kosugi and Ohashi, 2002). Because the binding specificity of each TCP is determined by the certain residues in the DNA binding motif (Kosugi and Ohashi, 2002; Viola et al., 2011, 2012), there are slight differences between sequences preferred by each TCP member. The sequences CIN members prefer to bind to are GGACCA and its complement sequence, TGGTCC (Schommer et al., 2008). To further confirm that TCP17 can directly regulate the expression of YUC2, YUC5, and YUC8, we first searched for proposed TBS (GGACCA and TGGTCC) in the promoter regions of these three genes (1.5 kb to 10 bp upstream of the transcription starting codon ATG). We identified three classical TBSs in the promoter of YUC5 and two in YUC8 (Fig. 7A). Unfortunately, no binding motif was identified within the 1.5-kb promoter region of YUC2. However, there are two GGACC and two GGTCC sequences in the promoter region of YUC2 at about 4.0 kb upstream of the initial codon ATG (Fig. 7A). To test whether TCP17 can associate with the promoter regions of these YUCs, we performed ChIP assays using 35S::TCP17-FLAG transgenic plants. Quantitative RT-PCR analysis showed that TCP17-FLAG can specifically enrich the TBS of these YUCs, and shade can significantly increase the function of TCP17 in associating with the promoters of YUCs (Fig. 7B). These results suggested that TCP17 can directly interact with the promoters of these YUCs.
Figure 7.
TCP17 directly binds to the promoters of YUCs and regulates their expression. A, Schematic diagram showing the TCP binding motifs in the promoters of YUC2, YUC5, and YUC8. B, CHIP assay showed that shade can increase the capacity of TCP17 to bind to the promoters of YUCs. Col-0 and 35S::TCP17-FLAG plants were treated and analyzed similar to those shown in Figure 5 (F), except primers used for qPCR analyses were different. Data shown are average of three independent biological replicates and SE (*P < 0.05 and **P < 0.01; based on Student’s t test). C and D, YUC5 can rescue the insensitive phenotype of 3tcp in response to shade. Col-0, 3tcp, and 3tcp/YUC5-OX transgenic seedlings were grown in Wc for 3 d and then transferred to Wc or shade for an additional 5 d before the measurements were taken. Scale bar = 1 cm. Data shown are the average of three independent biological replicates and SE (n ≥ 20 for each experiment). *P < 0.05 and **P < 0.01; based on Student’s t test.
In addition, we used a transient expression assay in Nicotiana benthamiana leaves to verify the role of TCP17 on the expression of YUC5. We fused wild-type YUC5 promoter (pYUC5-WT), and a mutated promoter (pYUC5-m) with a firefly luciferase (LUC) gene. Co-infiltration of pYUC5-WT::LUC with 35S::TCP17-FLAG led to an obvious induction of luminescence intensity. However, the activation effect of TCP17 on pYUC5-m::LUC, in which the TCP binding motifs were altered from GGNCC to GTNAC, was largely abolished (Supplemental Fig. S14). This result suggested that TCP17 can activate the expression of YUC5, and TCP binding motif is critical for TCP17 to activate its target genes.
Since TCP17 elevates endogenous auxin accumulation in response to shade through directly up-regulating the expression of YUCs, we tested whether ectopic expression of these YUCs could rescue the hypocotyl elongation of 3tcp under a shade condition. Because YUC5 is the gene whose expression is most significantly influenced by TCP17, we generated transgenic plants overexpressing YUC5 in 3tcp background. Transgenic seedlings with elevated expression levels of YUC5 can completely rescue the reduced response of hypocotyl growth of 3tcp to shade (Fig. 7, C and D). This result proved that TCP17 genetically regulates hypocotyl growth in response to shade through promoting the transcriptions of auxin biosynthesis genes.
DISCUSSION
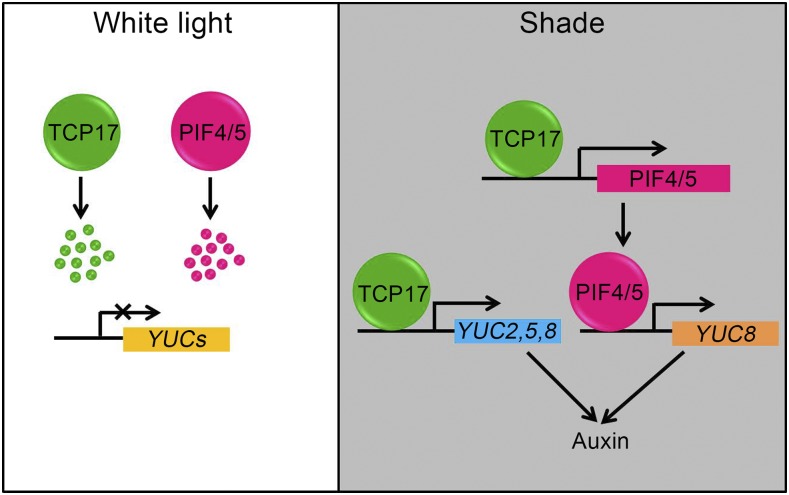
Light quality, as a signal, regulates many aspects of plant growth and development (Lau and Deng, 2010). SAS alters the function and architecture of a plant in response to shade light signals (Casal, 2012). SAS is a unique developmental process plants evolved to avoid detrimental consequences caused by shade. Rapid growth in response to shade light signals is beneficial for a plant to survive in a crowed environment. In this report, we illustrate a molecular framework that TCP17 integrates shade light signals and hypocotyl growth regulation in Arabidopsis (Fig. 8). Our detailed analyses demonstrated that shade light increases the stability and function of TCP17, allowing it to bind to the promoters of PIF4, PIF5, and auxin biosynthesis genes, YUCs, to up-regulate their expression. The increased free IAA accumulation regulated by TCP17 is important for rapid hypocotyl growth in shade.
Figure 8.
A hypothetical model of TCP17 in regulating hypocotyl growth in shade. Under a white light condition, TCP17 and PIFs are unstable and degraded, resulting in decreased expression levels of YUCs. Under a shade condition, on the other hand, TCP17 protein is stable and accumulated, resulting in elevated expressions of PIF4, PIF5, and YUCs. As a result, auxin level is increased.
TCP17 belongs to the CIN subfamily, in which many members, including TCP2, TCP3, TCP4, TCP10, TCP24, are regulated by miR319 at a posttranscriptional level (Palatnik et al., 2003). The important roles of TCPs in regulating cell proliferation and expansion have been well documented (Palatnik et al., 2003; Efroni et al., 2008; Schommer et al., 2008; Nag et al., 2009; Tao et al., 2013; Huang and Irish, 2015). Their roles in response to various environmental signals, however, are largely unknown. In this study we demonstrated a novel function of TCP17 in promoting hypocotyl elongation in response to fluctuating light quality in environment. Overexpression of TCP17 results in constitutive shade response in white light, whereas a 3tcp triple mutant shows a reduced response to shade-induced hypocotyl elongation (Fig. 1; Supplemental Fig. S3). Although significantly impaired, the attenuated responses of 3tcp hypocotyls to shade treatment can still be observed, implying that additional factors other than TCP5, TCP13, and TCP17 may be required for a full response of a plant to shade. Previous studies demonstrated that JAW-D mutants, with elevated expression of miR319, showed shorter hypocotyls than wild type, suggesting important roles of miR319-targeted TCPs in promoting hypocotyl elongation (Palatnik et al., 2003; Challa et al., 2016). These results raise the possibility that in addition to the three TCPs we studied, many other CIN transcription factors may also be involved in regulating cell elongation in response to changing light quality.
As key regulators in response to environmental signals, PIF transcription factors were found to regulate multiple phytohormone biosynthetic or signaling pathways via mediating the expression of their target genes (Castillon et al., 2007). For example, DELLA proteins, the key repressors in GA signaling pathway, can directly interact with PIF3 and PIF4 to repress their transcriptional activities (de Lucas et al., 2008). BZR1, BZR2/BES1, and ARFs, key transcription factors in BR and auxin signaling pathways, can interact with PIFs to regulate the expression of cell elongation related genes (Oh et al., 2014). Previous studies also demonstrated that PIFs can directly associate with the G-box motifs in the promoters of auxin and BR biosynthetic genes to regulate endogenous levels of auxin and BRs, respectively (Hornitschek et al., 2012; Wei et al., 2017). Accumulated evidence indicated that regulation of the functions of PIFs is an important strategy for a plant to increase their fitness to environments. Many environmental cues were found to regulate the function of PIFs. For instance, high temperature increases the transcriptional level and binding activity of PIF4 (Koini et al., 2009; Franklin et al., 2011). Red light sensed by photo-receptor phyB promotes the degradation of PIF3, PIF4, and PIF5 through a 26S proteasome-dependent pathway (Leivar and Quail, 2011). Shade light with low R:FR increases the stability of PIF4, PIF5, and PIF7, allowing them to be accumulated in the nucleus and promote the expression of shade response genes (Lorrain et al., 2008; Li et al., 2012). Despite the fact that the mechanism of PIF degradation has been well studied, however, the transcription factors regulating the expression of PIFs are poorly understood. In this study, we demonstrated that TCP17 can bind to the promoters of PIF4 and PIF5 and increase their expression in a shade-dependent manner (Fig. 5). Our results uncover a molecular mechanism by which shade light signals regulate the transcriptional levels of PIF transcription factors.
Our results showed that the function of TCP17 was greatly impaired in pif4 and pifq mutants, indicating TCP17 promotes hypocotyl elongation partially through PIFs (Fig. 4). We observed that TCP17 significantly up-regulates the transcriptional level of PIF4 and PIF5 in shade (Fig. 5, A–D). On the other hand, the expression of PIF4 and PIF5 was not significantly altered in 3tcp compared to wild type in both Wc and shade (Supplemental Fig. S10). These results suggest the transcription levels of PIFs can also be regulated by other unknown factors yet to be elucidated. In addition, our results raised the possibility that TCP transcription factors form a heterodimer with PIFs and act as transcription activators to improve the activities of PIFs.
Auxin can be rapidly accumulated in shade and regulate shade-induced cell elongation (Tao et al., 2008). The observation that exogenous PIC can rescue the defective phenotype of hypocotyl growth of 3tcp in shade suggests a close relationship between TCP17 and auxin biosynthesis (Fig. 3, A and B). Our in-depth analysis showed that TCP17 is necessary for shade-induced auxin accumulation (Fig. 3, C and D). The balance of endogenous auxin is important for plant growth and development. As rate-limited enzymes in catalyzing auxin biosynthesis, the spatio-temporal transcriptional regulation of YUCs is critical (Cheng et al., 2006, 2007; Chen et al., 2014). As an important transcription factor regulating shade avoidance, TCP17 controls free IAA accumulation in shade through at least two independent pathways. One is the TCP17-PIF-auxin pathway, in which TCP17 increases the IAA level via up-regulating the expression of PIFs. Recently, PIFs have been reported to regulate the transcriptional levels of several YUCs in response to shade (Hornitschek et al., 2012; Li et al., 2012). PIF7 directly regulates the expression of YUC5, YUC8, and YUC9; PIF4 can bind to the G box regions of YUC8. The other is the TCP17-YUCs pathway, in which TCP17 can directly bind to the promoters of YUCs and promotes their expression. In our study, TCP17 associates with the promoters of YUC2, YUC5, and YUC8 and up-regulates their expressions (Figs. 6 and 7). These results suggest that there are not only overlaps but also differences between TCPs and PIFs in regulating auxin biosynthesis. The complex transcriptional networks regulating the expression of YUCs under developmental and environmental changes are yet to be clarified. Nevertheless, our study demonstrated a flexible mechanism enabling plants to adapt to the fluctuating environments.
MATERIALS AND METHODS
Plant Materials and Growth Condition
Wild-type and mutant plants used in this study are all in Col-0 accessions. 3tcp triple mutant was obtained from Yuval Eshed’s lab and was previously described (Efroni et al., 2008). All plants were grown at 22°C. The light condition used in this study is Wc (50 μE·m−2·s−1) or simulated shade (LED light, red: 13 μE·m−2·s−1, blue: 1.23 μE·m−2·s−1, far-red light: 20.2 μE·m−2·s−1, R:FR = 0.7) as previously described (Tao et al., 2008; Li et al., 2012), if not specified. For hypocotyl measurements, surface-sterilized seedlings were grown on half-strength Murashige and Skoog (MS) media containing 1% Suc and 0.8% agar under Wc for 3 d and then were left in Wc or transferred to simulated shade for 5 d before hypocotyl measurements were carried out. For analyzing responses to different R:FR ratios, seedlings were grown in simulated white light (LED light, red: 13 μE·m−2·s−1, blue: 1.23 μE·m−2·s−1) for 3 d and then were kept in simulated Wc or transferred to various shade conditions (simulated Wc supplemented with increasing amounts of FR) for 5 d before measurements were taken. For PIC treatment, seeds were planted on half-strength MS medium containing various concentrations of PIC. Image J software was used to quantify hypocotyl lengths. At least 20 seedlings were measured for each independent experiment.
Plant Transformation
DNA fragments including the full-length ORF of TCP17, TCP5, TCP13, YUC5, and PIF4 were amplified with primers listed in Supplementary Table S1 and then were cloned into entry vector pDONR (Invitrogen) by BP reaction. To create 35S::TCP17-FLAG and 35S::TCP17-GFP transgenic plants, pDONR-TCP17 was inserted into the binary vectors pBIB-BASTA-35S::GWR-FLAG and pBIB-HYG-35S::GWR-GFP by LR reaction (Invitrogen). 35S::HA-TCP5 and 35S::HA-TCP13 transgenic plants were generated by LR recombination of pDONR-TCP5 and pDONR-TCP13 to pEarleyGate201 vector. To generate estradiol induction transgenic seedlings, pDONR-TCP17 was inserted into pBIB-BASTA-35S-GWR-ER vector. pDONR-YUC5 was inserted into pBIB-HYG -35S–GWR-GFP vector. To generate proTCP17::TCP17-GFP transgenic plants, DNA fragment including the promoter and full-length ORF of TCP17 was amplified from genomic DNA, followed by BP and LR reactions to insert into the pBIB-BASTA-GWR-GFP binary vector. These binary constructs were introduced into the pGV3101 strain of Agrobacterium and transformed into wild-type Col-0, 3tcp, pif4, pifq, or DR5:GUS transgenic plants using the floral dip transformation method. Transformants were selected on BASTA or HYG-containing medium. Homozygous lines were selected for followed experiments.
RNA Extraction and Reverse Transcriptase Quantitative PCR
Seven-day-old seedlings were grown under Wc and then treated with Wc or shade for 2 h before being collected for further analyses, if not specified. RNA was extracted from whole seedlings using a Tiangen Plant Total RNA kit. Then 1 μg total RNA was used for the first-strand cDNA synthesis using an Invitrogen reverse transcriptase kit. PCR was performed with SYBR-Green PCR Mastermix (Takara) and amplification was detected on an Applied Biosystems One Step Plus Real-time PCR system. Expression was normalized against ACTIN2. At least three biological replicates were performed, with three technical replicates for each. The mean and SE from three biological replicates are shown.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation assays were performed as described previously (Ni et al., 2009). For analyzing the binding activity of TCP17 to the promoters of PIF4, PIF5, and YUCs, 35S::TCP17-FLAG transgenic seedlings were used for CHIP assays and Col-0 was used as negative control. Col-0 and 35S:TCP17-FLAG grown under Wc condition for 2 weeks and then transferred to shade or Wc for 2 h before harvested. Two grams of seedlings and 50 μL of the anti-FLAG Affinity Matrix (SIGMA) were used for chromatin immunoprecipitation. Precipitated DNA was dissolved in 50 μL of ddH2O, and 1 μL was used for PCR amplification.
Measurement of Free IAA
For quantification of free IAA in Col-0 and 3tcp in response to shade, Col-0 and 3tcp were grown in Wc for 7 d and then treated with shade light or Wc for 2 h. Whole seedlings were harvested and approximately 200 mg of fresh tissues were used for IAA extraction and measurement as previously described (Wang et al., 2015). Three biological replicates were performed.
Immunoblotting
Twenty seedlings for each treatment were collected. Plant tissues were ground to a fine powder in liquid nitrogen. Total proteins were extracted with extraction buffer (100 mm Tris-HCl, pH 7.8, 4 m urea, 5% SDS, 15% glycerol, 10 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 1 mm cocktail). Protein extracts were separated on 12% SDS-PAGE gels and transferred to nitrocellulose membrane. The FLAG and GFP tags were detected by western blotting assay using an anti-FLAG and anti-GFP antibody (Abmart), respectively. An antitubulin antibody (sigma) was used for probing tubulin as a loading control. Signal was detected using the western Lightning Plus-ECL (Perkin Elmer) kit. The experiments were repeated three times and similar results were obtained. Here shows one of the representative results.
GUS Staining
The promoter of TCP17 (from -1,500 to -1 bp) was amplified by PCR from Arabidopsis (Arabidopsis thaliana) genomic DNA and cloned into the pBIB-BASTA-GWR-GUS binary vector. pTCP17::GUS transgenic seedlings were used for GUS staining as described (Hornitschek et al., 2012; Fukazawa et al., 2014).
Transient Expression in N. benthamiana
Agrobacterium (pGV3101 strain) harboring each plasmid of interest were incubated in Luria-Bertani broth containing 10 mm MES (pH 5.7) and 20 mm acetosyringone at 28°C overnight with shaking. The pellets were collected by centrifugation and resuspended in MS media with 10 mm MES (pH 5.7), 10 mm MgCl2, and 150 mm acetosyringone to an OD600 of 0.6. For cotransfections, an equal volume of appropriate agrobacteria was mixed and the mixtures were incubated at room temperature for 3 h before injection. After 48 h of infiltration, the leaves were used for further analysis.
Luciferase Imaging
The transient expression assays were performed in N. benthamiana leaves as previously described (Walley et al., 2007). The wild-type and mutant YUC5 promoter was inserted into binary vector pGWB235 to generate the reporter constructs pYUC5-WT::LUC and pYUC5-m::LUC. 35S:TCP17-FLAG was used as an effector. The experiments were repeated three times with similar results. Luciferase activities were imaged using a Lumazone CA 1300B camera.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers TCP5 (AT5G60970), TCP13 (AT3G02150), TCP17 (AT5G08070), PIF4 (AT2G43010), PIF5 (AT3G59060), YUC2 (AT4G13260), YUC5 (AT5G43890), and YUC8 (AT4G28720).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. List of primers used in this study.
Supplemental Figure S1. TCP17 regulates many aspects of Arabidopsis growth and development.
Supplemental Figure S2. TCP17 plays a critical role in regulating hypocotyl elongation.
Supplemental Figure S3. TCP17 acts redundantly with TCP5 and TCP13 in promoting hypocotyl elongation in shade.
Supplemental Figure S4. Expression of shade-regulated genes.
Supplemental Figure S5. Quantification of TCP17 protein shown in Figure 3 normalized to Tubulin using image J.
Supplemental Figure S6. The response of triple mutant 3tcp to BR and GA3 treatment.
Supplemental Figure S7. TCP17 overexpression increased the GUS activity of DR5:GUS reporter.
Supplemental Figure S8. The transcriptional level of TCP17 in Col-0, TCP17-OX, TCP17-OX/pif4, and TCP17-OX/pifq plants.
Supplemental Figure S9. PIF4 rescued the defect of 3tcp in promoting hypocotyl elongation in shade.
Supplemental Figure S10. The transcription levels of PIF4 and PIF5 in 3tcp.
Supplemental Figure S11. The expression of TCP17 was not impaired in pif4 and pifq mutants.
Supplemental Figure S12. Transcript levels of auxin biosynthesis genes in Col-0 or 3tcp under Wc or shade condition.
Supplemental Figure S13. The phenotype of 35S::TCP17-ER transgenic plants.
Supplemental Figure S14. TCP17 increased the promoter activity of YUC5 in a transient assay.
Acknowledgments
We thank Dr. Yuval Eshed for kindly providing the 3tcp mutant. This study is supported by the National Natural Science Foundation of China (31470380, 31530005, and 91317311 to J. Li), and by the Program of Science and Technology of Gansu Province (17ZDZNA015-06 and 17ZDZNA016-05 to J. Li).
Footnotes
This study is supported by the National Natural Science Foundation of China (31470380, 31530005, and 91317311 to J.L.), and by the Program of Science and Technology of Gansu Province (17ZDZNA015-06 and 17ZDZNA016-05 to J.L.).
References
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. (2012) Shade avoidance. Arabidopsis Book 10: e0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E (2007) Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Cerdán PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Challa KR, Aggarwal P, Nath U (2016) Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28: 2117–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Dai X, De-Paoli H, Cheng Y, Takebayashi Y, Kasahara H, Kamiya Y, Zhao Y (2014) Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol 55: 1072–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Esquivel N, Bou-Torrent J, Galstyan A, Gallemí M, Sessa G, Salla Martret M, Roig-Villanova I, Ruberti I, Martínez-García JF (2013) The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J 75: 989–1002 [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière J-M, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA (2003) A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol 133: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic-Petrovic T, de Wit M, Voesenek LA, Pierik R (2007) DELLA protein function in growth responses to canopy signals. Plant J 51: 117–126 [DOI] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Teramura H, Murakoshi S, Nasuno K, Nishida N, Ito T, Yoshida M, Kamiya Y, Yamaguchi S, Takahashi Y (2014) DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis. Plant Cell 26: 2920–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Grandío E, Poza-Carrión C, Sorzano COS, Cubas P (2013) BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 25: 834–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Fujioka S, Blancaflor EB, Miao S, Gou X, Li J (2010) TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Koornneef M, Whitelam GC (1994) Phytochrome B and at least one other phytochrome mediate the accelerated flowering response of Arabidopsis thaliana to low red/far-red ratio. Plant Physiol 104: 1311–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Velanis CN, Jenkins GI, Franklin KA (2014) UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc Natl Acad Sci USA 111: 11894–11899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch M, Lorrain S, de Wit M, Trevisan M, Ljung K, Bergmann S, Fankhauser C (2014) Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc Natl Acad Sci USA 111: 6515–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Huang T, Irish VF (2015) Temporal control of plant organ growth by TCP transcription factors. Curr Biol 25: 1765–1770 [DOI] [PubMed] [Google Scholar]
- Keller MM, Jaillais Y, Pedmale UV, Moreno JE, Chory J, Ballaré CL (2011) Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J 67: 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Luccioni LG, Oliverio KA, Yanovsky MJ, Boccalandro HE, Casal JJ (2002) Brassinosteroid mutants uncover fine tuning of phytochrome signaling. Plant Physiol 128: 173–181 [PMC free article] [PubMed] [Google Scholar]
- Lucyshyn D, Wigge PA (2009) Plant development: PIF4 integrates diverse environmental signals. Curr Biol 19: R265–R266 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S. (2006) Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol Ecol 15: 3483–3503 [DOI] [PubMed] [Google Scholar]
- Nag A, King S, Jack T (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Kim E-D, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ (2009) Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu J-Y, Bai M-Y, Arenhart RA, Sun Y, Wang Z-Y (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Pedmale UV, Huang SC, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PAB, Sridevi P, Nito K, Nery JR, et al. (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepens I, Duek P, Fankhauser C (2004) Phytochrome-mediated light signalling in Arabidopsis. Curr Opin Plant Biol 7: 564–569 [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellaro R, Yanovsky MJ, Casal JJ (2011) Repression of shade-avoidance reactions by sunfleck induction of HY5 expression in Arabidopsis. Plant J 68: 919–928 [DOI] [PubMed] [Google Scholar]
- Sorin C, Salla-Martret M, Bou-Torrent J, Roig-Villanova I, Martínez-García JF (2009) ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J 59: 266–277 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie D-Y, Doležal K, Schlereth A, Jürgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Tao Q, Guo D, Wei B, Zhang F, Pang C, Jiang H, Zhang J, Wei T, Gu H, Qu L-J, et al. (2013) The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer J-L, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola IL, Reinheimer R, Ripoll R, Manassero NG, Gonzalez DH (2012) Determinants of the DNA binding specificity of class I and class II TCP transcription factors. J Biol Chem 287: 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola IL, Uberti Manassero NG, Ripoll R, Gonzalez DH (2011) The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem J 435: 143–155 [DOI] [PubMed] [Google Scholar]
- Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, Harmer SL, Dehesh K (2007) Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet 3: 1800–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Chu J, Yu T, Xu Q, Sun X, Yuan J, Xiong G, Wang G, Wang Y, Li J (2015) Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc Natl Acad Sci USA 112: 4821–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Yuan T, Tarkowská D, Kim J, Nam HG, Novák O, He K, Gou X, Li J (2017) Brassinosteroid biosynthesis is modulated via a transcription factor cascade of COG1, PIF4, and PIF5. Plant Physiol 174: 1260–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA. (2013) Ambient temperature signalling in plants. Curr Opin Plant Biol 16: 661–666 [DOI] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Liu H, Klejnot J, Lin C (2010) The cryptochrome blue light receptors. Arabidopsis Book 8: e0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
- Zhou Z-Y, Zhang C-G, Wu L, Zhang C-G, Chai J, Wang M, Jha A, Jia P-F, Cui S-J, Yang M, et al. (2011) Functional characterization of the CKRC1/TAA1 gene and dissection of hormonal actions in the Arabidopsis root. Plant J 66: 516–527 [DOI] [PubMed] [Google Scholar]