Genetic disruption of Arabidopsis SEIPINs reveals the relevance of lipid droplets in pollen transmission and in adjusting seed dormancy levels.
Abstract
Lipid droplets (LDs) are ubiquitous organelles in plant cells, but their physiological roles are largely unknown. To gain insight into the function of LDs in plants, we have characterized the Arabidopsis homologs of SEIPIN proteins, which are crucial factors for LD biogenesis in yeast and animals. SEIPIN1 is expressed almost exclusively in embryos, while SEIPIN2 and SEIPIN3 have broader expression profiles with maximal levels in embryos and pollen, where LDs accumulate most abundantly. Genetic analysis demonstrates that all three SEIPINs contribute to proper LD biogenesis in embryos, whereas in pollen, only SEIPIN2 and SEIPIN3 play a significant role. The double seipin2 seipin3 and triple seipin mutants accumulate extremely enlarged LDs in seeds and pollen, which hinders their subsequent mobilization during germination. Interestingly, electron microscopy analysis reveals the presence of nuclear LDs attached to type I nucleoplasmic reticulum in triple seipin mutant embryos, supporting that SEIPINs are essential for maintaining the correct polarity of LD budding at the nuclear envelope, restricting it to the outer membrane. In pollen, the perturbations in LD biogenesis and turnover are coupled to reduced germination in vitro and with lower fertilization efficiency in vivo. In seeds, germination per se is not affected in seipin2 seipin3 and triple seipin mutants, but there is a striking increase in seed dormancy levels. Our findings reveal the relevance of SEIPIN-dependent LD biogenesis in pollen transmission and in adjusting the timing of seed germination, two key adaptive traits of great importance in agriculture.
Lipid droplets (LDs) are cytoplasmic organelles consisting of a central core of neutral lipids, predominantly triacylglycerols (TAGs) and sterol esters, enclosed by a monolayer of phospholipids and proteins (Guo et al., 2009; Wilfling et al., 2014). LDs are found in organisms from all eukaryotic kingdoms, where they play an essential role in cell metabolism, through the dynamic storage and release of their lipid contents on demand. Plants accumulate LDs both in vegetative and reproductive tissues (Penfield et al., 2006; Chapman et al., 2012). LDs are especially abundant in seed tissues, both in the embryo cells and in the endosperm, where they likely function as a store of carbon and energy to be used during germination and seedling establishment (Siloto et al., 2006; Chapman et al., 2012). Pollen grains also contain numerous LDs, which may supply lipids for membrane growth during pollen tube germination (Kim et al., 2002; Ischebeck, 2016). However, the roles of LDs in plant physiology and development remain largely unknown. Moreover, lipids stored in LDs of oleaginous species constitute one of the main agricultural commodities obtained from crops, so understanding the dynamics of LD formation and turnover in plants is also of great economic importance. Genetic characterization of the roles of LDs and their stored lipids in plant physiology and development has been hampered in part by the gametophytic lethality of mutants in TAG biosynthesis enzymes (Zhang et al., 2009; Shockey et al., 2016). Analysis of genes involved in LD formation rather than in lipid synthesis could circumvent this problem.
The prevalent model for LD biogenesis posits that they are formed at specific domains of the endoplasmic reticulum (ER) membrane, through the channeled deposition of lipids between the two membrane leaflets, forming nascent droplets that grow to eventually bud from the ER as mature LDs (Wilfling et al., 2014; Gao and Goodman, 2015; Wang et al., 2016). SEIPIN genes (SEIPINs) encode integral membrane proteins that have been implicated in organizing LD biogenesis at the ER in organism from all eukaryotic kingdoms (Yang et al., 2012; Pol et al., 2014; Wee et al., 2014; Cai et al., 2015). SEIPINs from different organisms show little sequence conservation at the amino acid level, but they share a similar topological organization in all eukaryotes, with two transmembrane domains flanking a large central luminal loop (Lundin et al., 2006; Wee et al., 2014; Cai et al., 2015). SEIPIN was first identified in humans as the protein encoded by the Berardinelli-Seip Congenital Lipodistrophy2 gene, which, when mutated, causes near total absence of adipose tissue and defects in LD morphogenesis (Magré et al., 2001; Szymanski et al., 2007; Boutet et al., 2009). Similar defects in adipogenesis and/or LD formation are observed in yeast, mice, and Drosophila seipin mutants, demonstrating that this protein family has a conserved function in LD biogenesis (Szymanski et al., 2007; Fei et al., 2008; Cui et al., 2011; Tian et al., 2011; Wang et al., 2016). SEIPINs localize to the ER in yeast and animal cells and are enriched at ER-LD contact sites, supporting a direct role in LD formation (Szymanski et al., 2007; Fei et al., 2008; Sim et al., 2012; Wang et al., 2014; Salo et al., 2016; Wang et al., 2016). However, their molecular function is still not well understood. Data from yeast and animals suggest that SEIPINs directly regulate TAG synthesis through physical interaction with GPAT, AGAT2, and Lipin, key enzymes of the TAG biosynthetic pathway (Boutet et al., 2009; Tian et al., 2011; Sim et al., 2012; Han et al., 2015; Talukder et al., 2015; Wolinski et al., 2015; Pagac et al., 2016). However, it has also been proposed that the function of SEIPINs in LD biogenesis is to stabilize ER-LD contacts and hence promote conversion of nascent LDs into mature LDs (Grippa et al., 2015; Salo et al., 2016; Wang et al., 2016).
Genomes from fungi and animals contain single SEIPIN genes, whereas higher plant genomes contain multiple SEIPIN homologs, which can be classified into two separate monophyletic subgroups (Cai et al., 2015). Arabidopsis (Arabidopsis thaliana) contains three SEIPIN homologs: SEIPIN1, belonging to one of the monophyletic groups, and SEIPIN2 and SEIPIN3, which belong to the other group. LD-related phenotypes of a yeast seipin deletion mutant can be partially complemented through heterologous expression of the Arabidopsis SEIPINs (Cai et al., 2015). Moreover, overexpression of Arabidopsis SEIPINs in plants increases the number of LDs and alters their lipid composition (Cai et al., 2015). Interestingly, SEIPIN1 induces accumulation of larger LDs than SEIPIN2 and SEIPIN3, suggesting that their classification in separate phylogenetic groups is corresponded by a certain degree of functional specialization. Although these results are based on gene overexpression, and neomorphic effects cannot be discarded, they do suggest that SEIPINs have conserved a role in LD formation in plants. Based on this premise, we presumed that genetic disruption of the plant SEIPIN gene family could perturb LD biogenesis and hence reveal the physiological relevance of LDs in plants. To gain insight into the functions of Arabidopsis SEIPINs, we have characterized their subcellular localization and expression patterns and analyzed the phenotypes resulting from knocking out the genes individually or in combination. The results reported here unveil a divergence in the expression patterns and the in vivo functions of SEIPINs from the two subfamilies present in plant genomes and reveal the relevance of SEIPIN-mediated LD biogenesis in controlling pollen transmission and seed dormancy.
RESULTS
SEIPIN Overexpression Reshapes the ER into Perinuclear Structures Associated with LDs
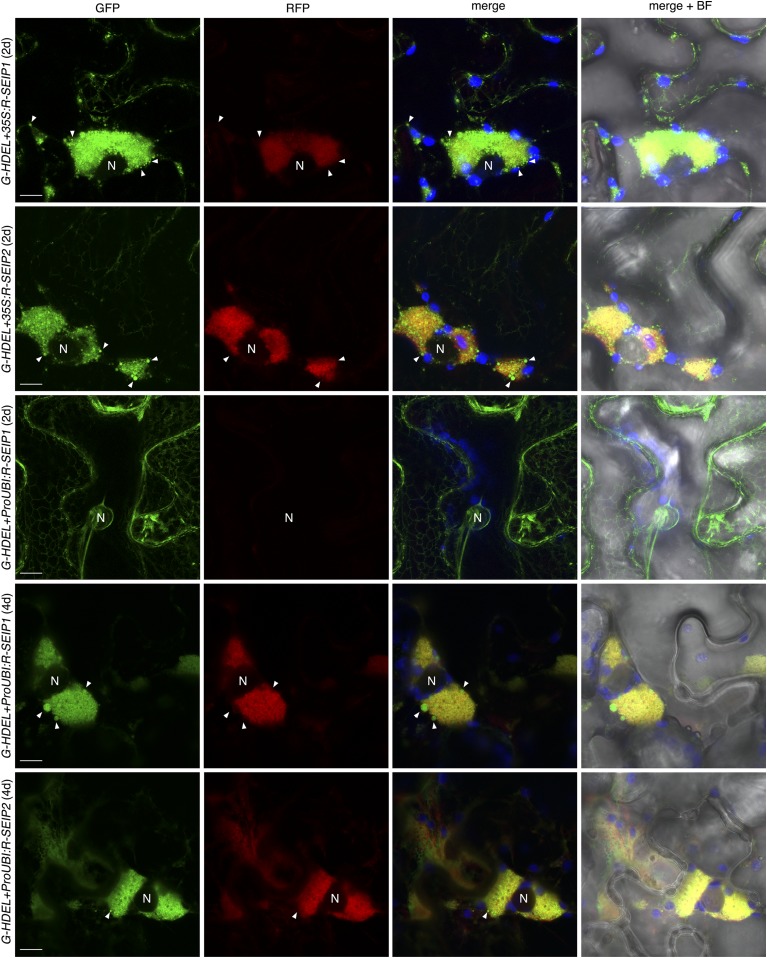
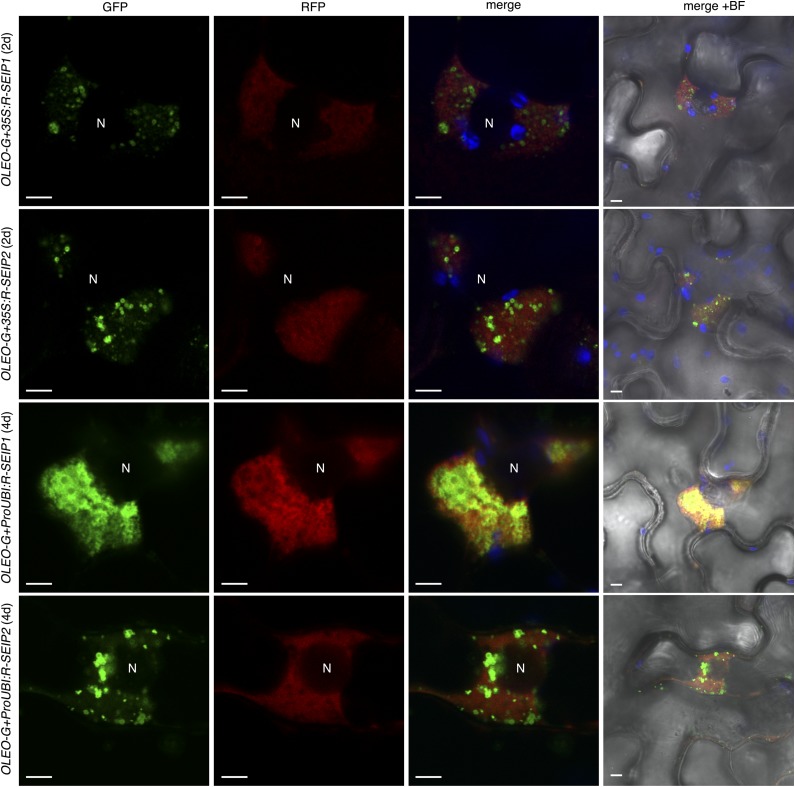
To compare the subcellular localization of SEIPINs from the two subfamilies present in plants, we coexpressed Arabidopsis SEIPIN1 and SEIPIN2 fused to different fluorophores in Nicotiana benthamiana leaves under the control of the 35S promoter. We found strict colocalization between the signals of GFP-SEIPIN1 and RFP-SEIPIN2 (Supplemental Fig. S1), indicating that in spite of the large divergence in their sequence they retain the same intracellular targeting information. Interestingly, we found them to colocalize in large perinuclear structures of unknown origin (Supplemental Fig. S1). To determine if the perinuclear structures were derived from the ER, we cotransformed RFP-tagged SEIPIN1 and SEIPIN2 with an ER-targeted GFP, containing a signal peptide and the ER-retention motif HDEL (GFP-HDEL). When expressed alone, GFP-HDEL was found in the typical reticular pattern of the ER (Supplemental Fig. S2), but when coexpressed with RFP-tagged SEIPIN1 or SEIPIN2, GFP-HDEL was additionally found in the large perinuclear structures colocalizing with the RFP-tagged SEIPINs and also in vesicle-like bodies (Fig. 1 and Supplemental Fig. S2). Rearrangement of the ER marker into these perinuclear structures was also observed when coexpressed with untagged SEIPIN1 (Supplemental Fig. S3), excluding that it was due to oligomerization of the GFP or the RFP tag. These results suggest that, when overexpressed under the 35S promoter, SEIPINs reshape the ER. Previously, it was reported that SEIPINs overexpressed in N. benthamiana under the 35S promoter caused the relocalization of an ER-marker into punctate structures (Cai et al., 2015), which may correspond to the vesicle-like bodies found in our experiments. To examine the effect of more moderate levels of overexpression, we expressed RFP-tagged SEIPIN1 and SEIPIN2 under the UBIQUITIN10 promoter, which has at least an order of magnitude lower activity in N. benthamiana than the 35S promoter (Grefen et al., 2010). In accordance with this, RFP-tagged SEIPINs were not detectable until 4 d after infiltration, whereas they were detectable two days after infiltration when using the 35S promoter (Figs. 1 and 2). Importantly, even when expressed at moderate levels, we found SEIPINs localized in the enlarged perinuclear ER structures together with the ER marker (Figs. 1 and 2). To determine if the SEIPINs present in those perinuclear structures were functional in LD biogenesis, we cotransformed RFP-SEIPIN1 and RFP-SEIPIN2 with GFP-tagged Oleosin (De Domenico et al., 2011), a main structural protein of the LD membrane (Kory et al., 2016). We found that Oleosin-GFP was present in the limiting membrane of spherical organelles closely associated with the RFP-labeled perinuclear ER structures (Fig. 2), supporting the functionality in LD biogenesis of the SEIPINS present in those structures.
Figure 1.
SEIPIN overexpression reshapes the ER into perinuclear structures and vesicle-like bodies. Nicotiana benthamiana leaves were cotransformed with 35S:GFP-HDEL (G-HDEL) and 35S:RFP-SEIPIN1 (35S:R-SEIP1), 35S:RFP-SEIPIN2 (35S:R-SEIP2), ProUbiquitin10:RFP-SEIPIN1 (ProUBI:R-SEIP1), or ProUbiquitin10:RFP-SEIPIN2 (ProUBI:R-SEIP2) and imaged 2 d or 4 d after inoculation as indicated. The panels show the GFP signal (green pseudocolor), the RFP signal (red pseudocolor), a merged image of the GFP, the RFP, and the chloroplast autofluorescence (blue pseudocolor), and the same merged image overimposed on a bright field image (right). Scale bar, 10 μm. N, Nucleus. Arrowheads, GFP-positive vesicle-like bodies.
Figure 2.
LDs are associated with perinuclear ER aggregates containing SEIPINs. Nicotiana benthamiana leaves were cotransformed with 35S:Oleosin-GFP (OLEO-G) and 35S:RFP-SEIPIN1 (35S:R-SEIP1), 35S:RFP-SEIPIN2 (35S:R-SEIP2), ProUbiquitin10:RFP-SEIPIN1 (ProUBI:R-SEIP1), or ProUbiquitin10:RFP-SEIPIN2 (ProUBI:R-SEIP2) and imaged 2 d or 4 d after inoculation as indicated. The panels show the GFP signal (green pseudocolor), the RFP signal (red pseudocolor), a merged image of the GFP, the RFP, and the chloroplast autofluorescence (blue pseudocolor), and the same merged image overimposed on a bright field image (right). Scale bar, 5 μm. N, Nucleus.
Arabidopsis SEIPINs Show Distinctive Expression Domains
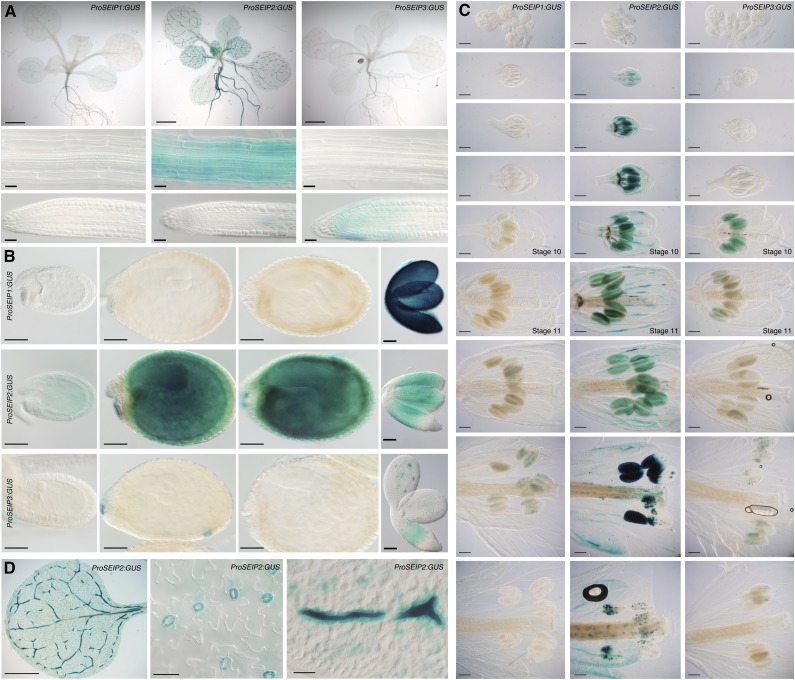
To examine the expression of Arabidopsis SEIPINs with cellular resolution, we analyzed the activity of their promoters. We stably transformed Arabidopsis plants with promoter constructs containing the upstream intergenic regions and the 5′ UTRs up to the start codon of each of the SEIPIN genes driving expression of the GUS reporter (ProSEIP:GUS). We selected 10 independent transgenic lines from each construct, which showed consistent patterns of expression for each of the promoters, as described below. We observed high expression of ProSEIP1:GUS in developing embryos but very weak expression in postgermination stages of plant development (Fig. 3). This embryo-specific SEIPIN1 promoter activity profile is consistent with the pattern published for a slightly shorter SEIPIN1 promoter construct and with mRNA expression data for this gene (Jeong et al., 2014). Contrasting with this restricted expression pattern, we observed high activity of ProSEIP2:GUS, both in developing embryos and in postgermination stages of development. Expression of ProSEIP2:GUS in aerial organs was particularly high in certain cell types, namely guard cells, myrosin cells, and pollen (Fig. 3). Interestingly, guard cells and pollen have been reported to contain abundant LDs (Ischebeck, 2016; McLachlan et al., 2016). The activity of ProSEIP3:GUS was low and restricted to developing embryos, root tips, and pollen. In Arabidopsis, LDs are found in uninucleate pollen (stage 10 flowers, marked in Fig. 3E) and increase greatly in number from pollen mitosis I (stage 11 flowers, marked in Fig. 3E) until pollen maturation (Kuang and Musgrave 1996), which coincides with concurrent expression of both ProSEIP2:GUS and ProSEIP3:GUS at those stages. To compare these promoter activity profiles with the mRNA expression levels of the endogenous genes, we surveyed RNA-seq tissue expression datasets in the TRAVA server (Klepikova et al., 2016). These RNA-seq datasets showed that SEIPIN1 mRNA expression was high in mature seeds and in siliques but was virtually undetectable in all the other vegetative and reproductive tissues of the plant (Supplemental Table S1), matching precisely the activity profile of the ProSEIP1:GUS construct (Fig. 3). In contrast to this, SEIPIN2 and SEIPIN3 mRNA were widely expressed across all the RNA-seq tissue samples. SEIPIN2 mRNA levels were highest in mature seeds, siliques, anthers, and pollen (>7-fold higher than the average expression across all tissues; Supplemental Table S1), in accordance with the activity profile of the SEIPIN2 promoter (Fig. 3). SEIPIN3 mRNA expression levels were more homogeneous across tissues, although they also peaked in mature seeds/siliques and anthers/pollen. The widespread and high levels of expression of SEIPIN3 mRNA in the RNA-seq datasets are not consistent with the low activity of the ProSEIP3:GUS lines generated, suggesting that distant enhancer elements required for full expression are located outside the SEIPIN3 promoter fragment cloned. To confirm these RNA-seq results, we examined the AtGenExpress developmental series ATH1 microarray datasets, which contains probes for SEIPIN1 and SEIPIN2, but not for SEIPIN3. These datasets confirmed that SEIPIN1 is expressed almost exclusively in developing seeds, while SEIPIN2 expression is more broadly distributed in all tissues (Supplemental Table S2). Importantly, the expression of SEIPIN1 and SEIPIN2 was activated in seeds at the transition from late heart to early torpedo stage, reaching maximal levels at the late torpedo/walking stick stage, which coincides with the temporal pattern of activation of TAG biosynthesis during embryogenesis (Baud et al., 2008). We conclude from these analyses that highest expression of SEIPINs is associated with cell types containing abundant LDs, such as guard cells, pollen grain, and embryos, supporting a role of SEIPINs in LD biogenesis in vivo.
Figure 3.
Divergent expression patterns of Arabidopsis SEIPINs. A to C, GUS activity of ProSeip1:GUS, ProSeip2:GUS, and ProSeip3:GUS in (A) seedlings (scale bar, 2 mm), mature root, and root tips (scale bar, 25 μm); (B) developing seeds and embryos (scale bar, 100 μm); and (C) developing flowers (scale bar, 200 μm). Equivalent tissues from lines expressing each promoter construct were incubated simultaneously for identical periods of time. Stage 10 and stage 11 flowers (Alvarez-Buylla et al., 2010) are indicated. D, Details of the GUS activity of ProSeip2:GUS in leaves. Scale bars, 2 mm (left), 50 μm (other panels).
SEIPINs Direct LD Formation in Developing Embryos
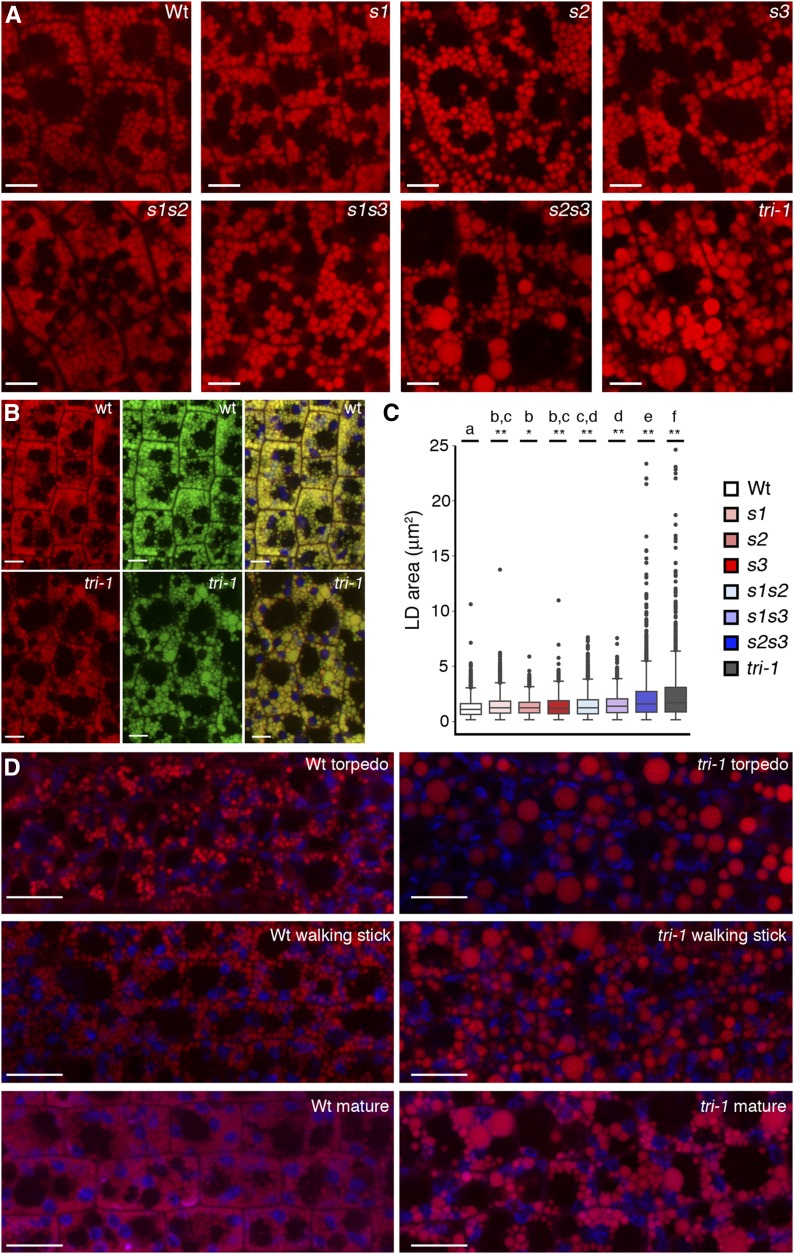
The distinctive expression patterns of Arabidopsis SEIPINs imply a certain degree of functional divergence, but redundancy is also possible wherever two or more isoforms are concomitantly expressed. To determine the biological functions of Arabidopsis, SEIPINs and their degree of divergence and/or redundancy, we characterized T-DNA insertional mutants. We obtained single T-DNA mutant alleles of SEIPIN1 and SEIPIN2, and three T-DNA insertional alleles of SEIPIN3 (Supplemental Fig. S4). The seipin1-1 (s1) and seipin2-1 (s2) alleles have T-DNA insertions in the first exon of the respective genes, which disrupt much of their coding sequence and are thus most likely null alleles. The seipin3-1 (s3), seipin3-2, and seipin3-3 alleles have T-DNA insertions at different positions of the unique exon of SEIPIN3 and show similar phenotypes (see below), suggesting that they are all equivalent null alleles. RT-PCR analysis shows that indeed seeds from the s1, s2, and s3 mutants did not accumulate full-length transcripts of the respective genes (Supplemental Fig. S5). We could not find any obvious developmental or growth phenotype under standard growth conditions in the single, double, or triple seipin mutant combinations, suggesting that the genes are not essential for plant viability or normal development. To determine if LD biogenesis was affected, we first examined embryos, where LDs are particularly abundant. We stained dissected mature embryos with the neutral lipid dye Nile red and examined them by confocal microscopy. Wild-type mature embryos had abundant Nile red-stained bodies of spherical shapes smaller than 1.5 μm in diameter (Fig. 4, A and B). These bodies were also stained with an independent neutral lipid dye, Bodipy 493/503, demonstrating that they were indeed LDs (Fig. 4B). Remarkably, some of the LDs present in double s2s3 and triple s1s2s3 (tri-1) mature embryos were extremely enlarged, more than 30 times larger in volume (up to 5 μm in diameter) than those observed in wild-type embryos (Fig. 4). A similar accumulation of supersized LDs was observed in mature embryos of triple seipin mutant seeds containing the seipin3-2 (tri-2 mutant) or the seipin3-3 (tri-3 mutant) alleles (Supplemental Fig. S6). Interestingly, at earlier stages of embryo development, when LD accumulation begins, differences in LD size between wild-type and tri-1 mutant embryos were even more pronounced, with all the LDs in the mutant being drastically enlarged (Fig. 4D). Severe LD enlargement in the absence of SEIPIN activity in Arabidopsis embryos is consistent with similar phenotypes reported for SEIPIN-deficient mutants from animals and fungi (Fei et al., 2008; Wang et al., 2014; Wang et al., 2016). Albeit to a lower extent than s2s3 and tri-1 mutants, the other seipin mutants also appeared to have alterations in LD size (Fig. 4A). To quantify this effect, we measured areas of LDs in epidermal hypocotyl from mature embryos of the different genotypes. This analysis revealed that wild-type embryos have significantly smaller LDs than all the single mutants, that single mutants have smaller LDs than double mutants and that the largest LDs are found in the triple mutant (Fig. 4C). These results support that all three SEIPIN genes contribute to LD biogenesis in embryos, in accordance with their coordinated expression in those tissues. We hypothesized that reduced packing efficiency of LDs due to their increased size in tri-1 mutants may affect cell growth and ultimately seed size. Indeed, although seed size varies considerably with the growth conditions of the mother plants, we observed that the tri-1 seeds were always significantly larger (around 10% larger in area) than wild-type seeds from the same batch, harvested from plants grown at the same time under identical conditions (Supplemental Fig. S7). These results are consistent with the reduced packing efficiency of larger LDs leading to increased cellular volume and seed size. Intriguingly, RNAi knockdown of SEIPIN1 has been reported to cause reduced seed growth, but LD size was apparently not affected, and the cause of this phenotype is unclear (Cai et al., 2015).
Figure 4.
SEIPINs are required for LD formation in seeds. A, Confocal images (single sections) of hypocotyl cells from mature embryos stained with Nile red. Scale bar, 5 μm. B, Confocal images (single sections) of hypocotyl cells from mature embryos stained with Nile red and Bodipy 493/503. Left, Nile red signal. Middle, Bodipy 493/503 signal. Right, merged image showing chloroplast autofluorescence (in blue) for reference. Scale bar, 5 μm. C, Box plot of the areas of LDs from hypocotyl cells of mature embryos from the different genotypes. In the plots, the box indicates the lower and upper quartile, the horizontal bar the median, the whiskers the largest and lowest data points that fall within 1.5 times the interquartile range from the lower and upper quartile, and the dots the outliers. ANOVA with Tukey’s HSD post-hoc test was used to test for differences in mean LD area among the different genotypes. Genotypes without letters in common are significantly different (P < 0.05). The asterisks indicate the P value for the null hypothesis that the mean LD area in that genotype is not different from the wild-type (Wt) mean. *P < 0.05; **P < 0.01. D, Confocal images (single sections) of hypocotyl cells from embryos at different developmental stages (indicated in the panels) stained with Nile red. Scale bar, 10 μm. Genotypes corresponding to all the images are indicated.
SEIPIN Are Involved in Directional Biogenesis of LDs at the Nuclear Envelope
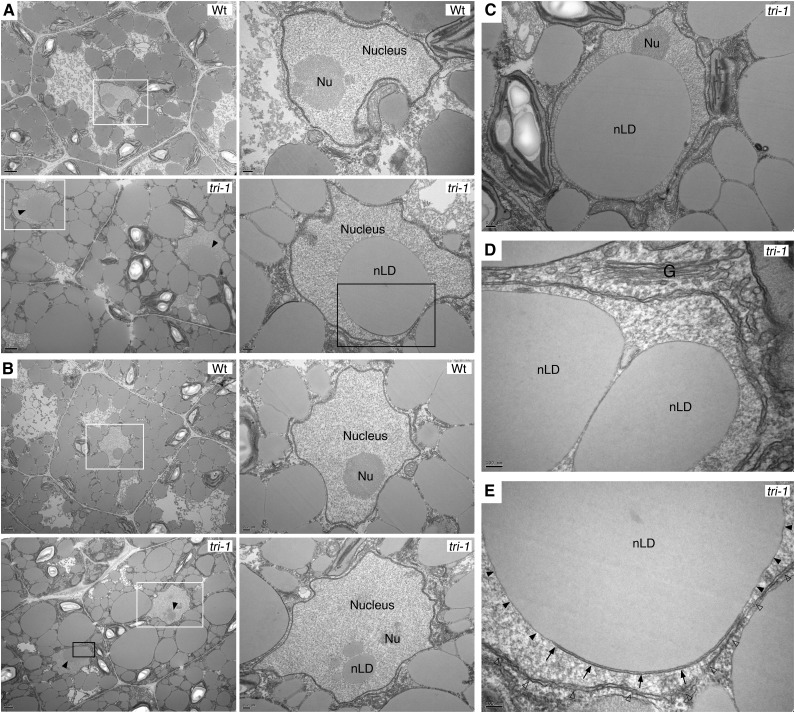
To investigate potential subcellular phenotypes caused by SEIPIN deficiency, we examined mature wild-type and tri-1 mutant embryos by transmission electron microscopy. Consistent with the confocal analysis of Nile red-stained samples, tri-1 mutants accumulated greatly enlarged LDs in the cytosol of both cotyledon and hypocotyl cells (Fig. 5, A and B). In contrast, the morphology of the Golgi apparatus, chloroplasts, mitochondria, ER, and nuclear envelope appeared unaltered in the mutants (Fig. 5). Remarkably, tri-1 mutant cells, but not wild-type cells, accumulated nuclear LDs (nLDs) (Fig. 5), a phenotype that is also observed in a yeast SEIPIN deletion mutant (Cartwright et al., 2015; Wolinski et al., 2015). Some of the nLDs in the tri-1 mutant had very large size, but the integrity of the nuclear membrane and the appearance of subnuclear structures like the nucleolus was not affected (Fig. 5, C and D). Interestingly, we could also detect nLDs still attached to nucleoplasmic reticulum. In Figure 5E, an nLD is seen budding from one of the membranes of double membrane type I nucleoplasmic reticulum (Malhas et al., 2011). nLDs have been recently described in yeast and animal cells (Farese and Walther, 2016), but they were yet to be reported in plants. Likewise, extensive infoldings of the nuclear envelope and type II nuclear invaginations of the cytosol had been reported in onion cells and in Podocarpus microspores, respectively (Collings et al., 2000; Aldrich and Vasil, 1970), but evidence for the presence of type I nucleoplasmic reticulum in plants was not available. These results support that, as proposed for their yeast counterparts (Cartwright et al., 2015; Wolinski et al., 2015), plant SEIPINs are required for maintaining the correct polarity of LD biogenesis and budding at the nuclear envelope, restricting it to the outer membrane. Moreover, this function is consistent with the predominant localization of the overexpressed SEIPINs in perinuclear membrane structures (Figs. 1 and 2).
Figure 5.
Nuclear LDs accumulate in SEIPIN-deficient mutants. A and B, Transmission electron microscopy images of (A) cotyledon cells and (B) hypocotyl cells from developing embryos of the indicated genotypes. Solid arrowheads, nLDs. Right, magnifications of the areas boxed in white. C, A tri-1 cotyledon cell with a supersized nLD, but with apparently intact nuclear membrane and nucleolus. D, Magnification of the area boxed in black in B showing a detail of the Golgi apparatus and the nuclear membrane. E, Magnification of the area boxed in black in A showing an nLD budding from one of the membranes (solid arrowheads) of a double membrane type I nucleoplasmic reticulum (arrows) and surrounded by the nuclear envelope membrane (empty arrowheads). Scale bar sizes are indicated in the panels. G, Golgi apparatus; Nu, nucleolus; nLD, nuclear LD; Wt, wild type.
LD Enlargement Interferes with TAG Mobilization
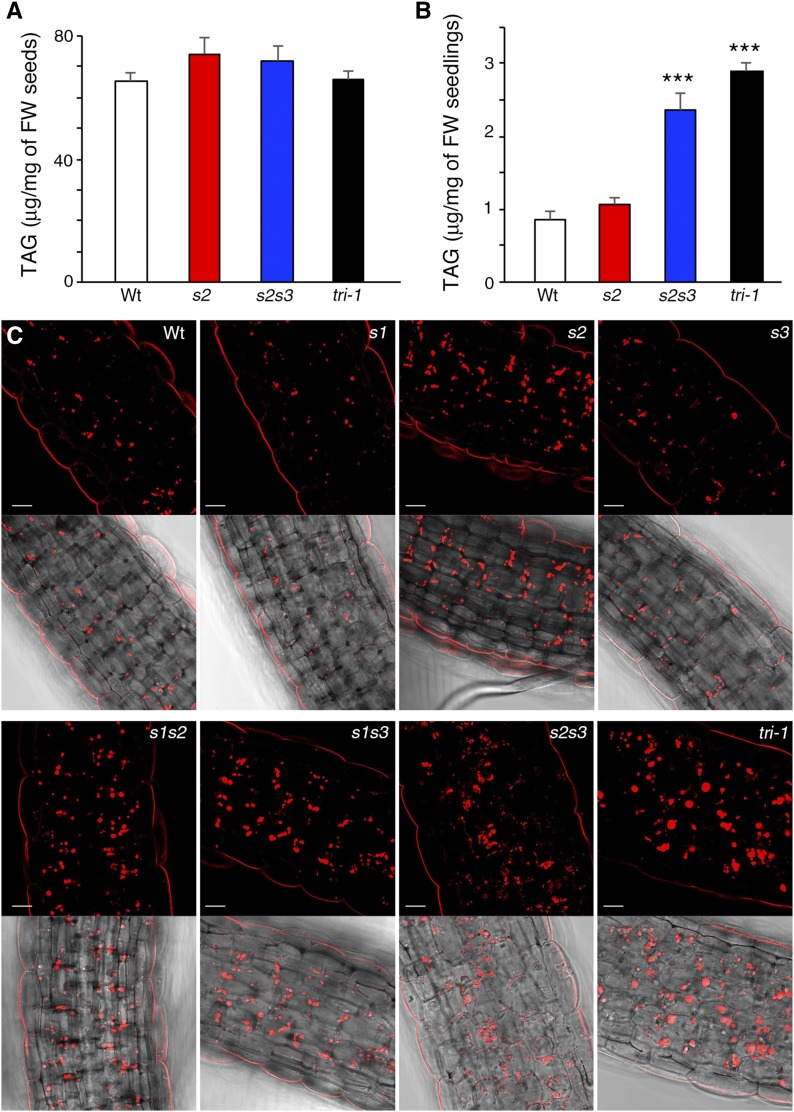
LD enlargement reduces the surface-to-volume ratio of the organelle and thus limits the access of lipases to the TAG core. Hence, TAG mobilization from LDs should be altered in the s2s3 and triple seipin mutants that accumulate supersized LDs. To test this, we measured TAG levels in mature seeds and in germinating seedlings from wild-type, s2, s2s3, and tri-1 mutant plants. In spite of the alterations in LD morphogenesis, the concentration of TAGs in mature s2s3 and tri-1 seeds was unchanged in comparison to wild-type and s2 seeds (Fig. 6A). This is similar to what occurs in SEIPIN-depleted Drosophila S2 cells (Wang et al., 2016) and supports the view that the role of SEIPINs in LD formation is to stabilize ER-LD connections rather than to directly promote TAG synthesis. Importantly, although the initial TAG concentration in mature s2s3 and triple tri-1 seeds was similar to wild-type seeds, in germinating seedlings the TAG levels remained significantly higher in the mutants than in wild-type plants (Fig. 6B). Interestingly, microscopy analysis of germinating seedlings showed that tri-1 plants retained many apparently intact supersized LDs, whereas the s2s3 seedlings accumulated many small amorphous LDs (Fig. 6C), supporting that LD turnover is perturbed but to a larger extent in the tri-1 mutant. These results are consistent with LD enlargement impairing efficient TAG mobilization, although we cannot exclude the possibility of increased TAG synthesis in the germinating s2s3 and triple tri-1 seedlings. In the sdp1sdp1l mutant, which is defective for the two main TAG lipases in seedlings (Eastmond, 2006; Kelly et al., 2011), hypocotyl growth of etiolated plants and seedling establishment is severely compromised when grown in the absence of Suc (Kelly et al., 2011), suggesting that TAG hydrolysis is important in those conditions. In tri-1 mutants, there were no significant changes in seedling establishment or in etiolated hypocotyl growth (Supplemental Fig. S8) relative to wild-type plants, neither in the presence nor in the absence of Suc in the germination media, possibly because LD mobilization is perturbed but not blocked.
Figure 6.
LD turnover is hindered in SEIPIN-deficient mutants. A, TAG quantification carried out by TLC in samples from mature dried seeds. No significant differences were found between the mean TAG concentration in wild-type and in seipin mutant seeds; one-way ANOVA with Dunnett’s post-hoc hest. FW, fresh weight. B, TAG quantification in germinating seedlings 3 d after sowing. Asterisks indicate significant difference with the mean TAG concentration in wild-type (Wt) seedlings, ANOVA with Dunnett’s post-hoc test (P < 0.001). FW, fresh weight. C, Confocal sections (single images) of hypocotyl cells from Nile red-stained seedlings 3 d after sowing. Scale bar, 20 μm.
SEIPIN Deficiency Imposes Seed Dormancy
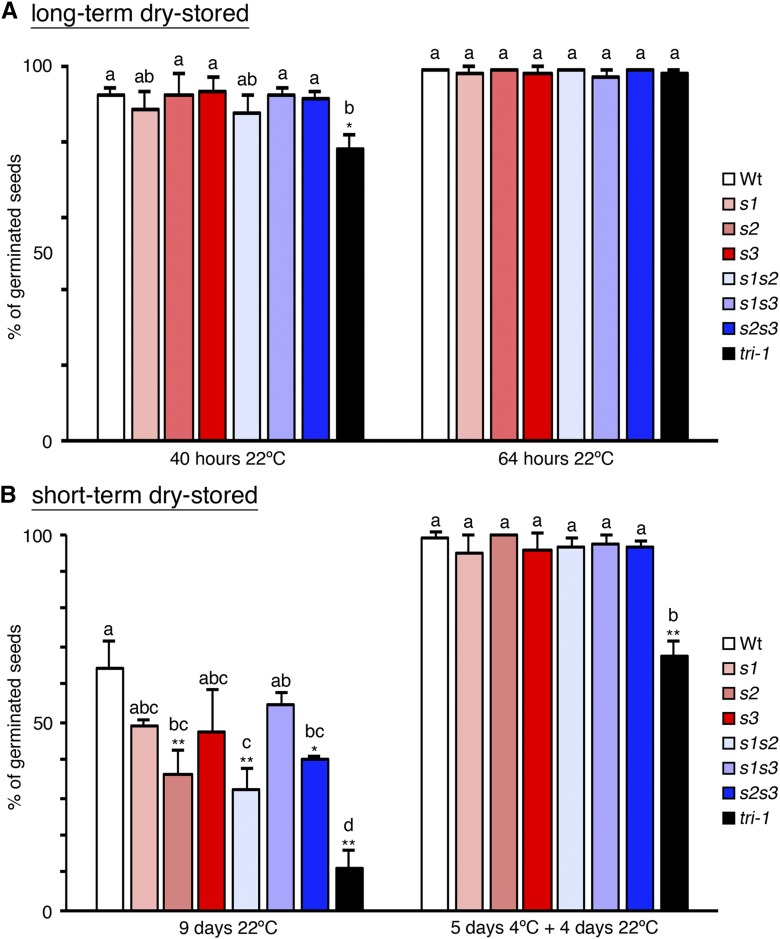
We next analyzed if the defects in LD biogenesis and mobilization in seeds of the seipin mutants were associated with lower seed germination rates. We could only detect a small reduction of germination rate in the tri-1 mutant shortly after imbibition (40 h) but not 24 h later, when all genotypes presented close to 100% germination, similar to wild-type seeds (Fig. 7A). This weak effect agrees with the slight delay in germination reported for the oleosin1 mutant, which also accumulates enlarged LDs that are not efficiently mobilized (Siloto et al., 2006; Shimada et al., 2008; Miquel et al., 2014), and for the double sdp1sdp1l mutant (Kelly et al., 2011). Our results support the view that lipid mobilization is not essential for seed germination (Kelly et al., 2011), but they also suggest that alterations in LDs can have an effect on the timing of germination. In this regard, the main process controlling the timing of germination is seed dormancy, which arrests germination of viable seeds to ensure that it occurs at ecologically favorable conditions (van Der Schaar et al., 1997; Alonso-Blanco et al., 2003). Dormancy is established as reserves accumulate during seed maturation (Bentsink and Koornneef, 2008) and may thus be regulated by the LD storage status of the embryo. The initial germination assays (Fig. 7A) were done with long-term dry-stored seeds (more than 3 months of storage), which is much longer than the period required to release dormancy in the Col-0 background used in this study (van Der Schaar et al., 1997; Alonso-Blanco et al., 2003). Hence, we measured germination of seeds shortly after harvesting (short-term dry-stored for 2 weeks), when wild-type (Col-0) seeds may still retain some dormancy (van Der Schaar et al., 1997). Indeed, short-term dry-stored wild-type seeds showed reduced germination rates relative to long-term dry-stored wild-type seeds (Fig. 7, A and B). Remarkably, short-term dry-stored seeds from all the seipin mutant genotypes showed even stronger inhibition of germination than wild-type plants, indicating that dormancy levels were increased. In particular, in all the genotypes that included the seipin2-1 mutant allele (s2, s1s2, s2s3, tri-1, tri-2, and tri-3) germination rates were significantly decreased relative to wild-type (Fig. 7B and Supplemental Fig. S9). Germination was particularly compromised in the triple seipin mutant genotypes, which had germination rates significantly lower from wild type but also from all the other seipin mutant genotypes, supporting that all three SEIPIN genes contribute to reducing dormancy levels in seeds. The germination rates of freshly harvested seed were not increased in media containing 1% Suc, which is consistent with dormancy not being released by an external carbon source. In contrast, germination of dormant seeds can be recovered by cold stratification (Shu et al., 2016), so we tested the effect of this treatment. Importantly, 5 d of cold stratification fully rescued full germination of s1, s2, s3, s1s2, s1s3, and s2s3 short-term dry-stored seeds and greatly increased the germination rate of triple seipin mutant seeds (Fig. 7B and Supplemental Fig. S9). Moreover, those same batches of seeds were analyzed 3 months later, and full germination of all genotypes was recovered (over 97% germination rate without prior cold-stratification), confirming that the freshly harvested mutant seeds were viable but dormant. These findings uncover SEIPINs as key factors determining the timing of seed germination in Arabidopsis and suggest that perturbations in LD biogenesis during embryo development impose increased seed dormancy.
Figure 7.
SEIPIN deficiency imposes seed dormancy. A, Bar plot of the mean germination (as % of total seeds) from three experiments with independent batches of long-term dry-stored seeds (over 3 months of dry storage) measured at 40 h and 64 h after imbibition. Error bars indicate the sd. B, Bar plots of the mean germination rate (as % of the total seeds) from three experiments with independent batches of short-term dry-stored seeds (2 weeks of dry storage) measured after 9 d in the growth chamber (9 d at 22°C) or after stratification for 5 d at 4°C followed by 4 additional d in the growth chamber (5 d 4°C + 4 d 22°C). Error bars indicate the sd. ANOVA with Tukey’s HSD post-hoc test was used to test for differences among the means of the different genotypes in each experiment. Means without letters in common are significantly different (P < 0.05). The asterisks indicate the P value for the null hypothesis that the mean germination rate in that genotype is not different from the wild-type (Wt) mean in that particular experiment. *P < 0.05, **P < 0.01.
SEIPIN2 and SEIPIN3 Are Required for Efficient Pollen Germination and Ovule Fertilization
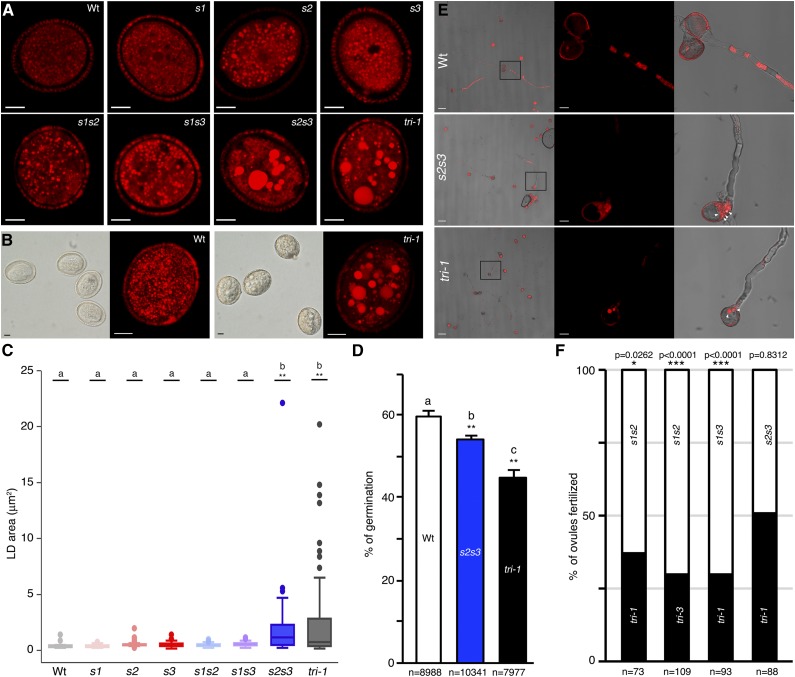
To analyze other in planta functions of SEIPINs we next focused on pollen grains, which accumulate abundant LDs and express high levels of SEIPIN2 and SEIPIN3 mRNA (Fig. 3; Supplemental Table S1). Nile red staining of mature pollen grains showed alterations in LD morphogenesis in some of the mutant combinations (Fig. 8, A and B). Quantification of LD area showed that the size of LDs was not significantly different in pollen from wild type, single mutants, and double s1s2 and s1s3 mutants (Fig. 8C). In contrast, in the s2s3 double and the tri-1 triple mutant, the number of LDs was reduced (Fig. 8, A and B) and their size dramatically increased (Fig. 8C). Likewise, tri-2 and tri-3 triple mutants contained a reduced number of enlarged LDs in pollen grains (Supplemental Fig. S10). We conclude from these results that SEIPIN2 and SEIPIN3 are the main isoforms involved in LD formation in pollen, while the contribution of SEIPIN1 is marginal, consistent with the low expression of that isoform (Fig. 1; Supplemental Tables S1 and S2). It is assumed, but not experimentally tested, that LDs in pollen grains function as a reservoir of energy and lipids to be used during germination and for de novo membrane synthesis for pollen tube growth (Ischebeck, 2016). In fact, no defects in pollen transmission have been reported for mutants affected in LD formation or turnover (Eastmond, 2006; Siloto et al., 2006; Shimada et al., 2008; Kelly et al., 2011; Gidda et al., 2013; López-Ribera et al., 2014). Interestingly, we observed a significant reduction in germination rate of s2s3 and tri-1 pollen relative to wild-type pollen in an in vitro germination assay (Fig. 8D), supporting that perturbed LD morphogenesis in the mutant pollen impairs germination. We also observed an impaired entrance of LDs into the pollen tube in s2s3 and tri-1 pollen (Fig. 8E), suggesting that LD mobilization was perturbed. However, pollen tube growth was not significantly affected in the in vitro assay conditions. To test if alterations in LD biogenesis affected fertilization capacity in vivo, we set up a pollen competition assay. We pollinized stigma from wild-type plants with pollen from plants that were homozygous mutants for two of the SEIPIN genes and heterozygous for the third one and thus segregate 50% double and 50% triple seipin mutant pollen. We then genotyped the progeny from these crosses to measure the transmission efficiency of the competing pollen. This analysis showed that the triple seipin mutant pollen, which has supersized LDs, had greatly reduced fertilization efficiency when competing with s1s2 or s1s3 double mutant pollen, which has normal-sized LDs (Fig. 8F, first three columns). In contrast, when the triple seipin mutant pollen were set to compete with s2s3 double mutant pollen, which also contains supersized LDs, both genotypes were equally transmitted (Fig. 8F, fourth column). We conclude from this that the aberrant LD morphogenesis in the s2s3 and triple seipin mutant pollen causes lower transmission efficiency, but this is only evident when pollen from these mutants competes with pollen having normal-sized LDs. In accordance with this, seed set in the self-fertilized homozygous s2s3 and triple seipin mutants was unaffected under the laboratory growth conditions optimized for Arabidopsis growth. All together, these results support that LD enlargement in mutants deficient for SEIPIN2 and SEIPIN3 mutant hinders pollen germination and transmission efficiency.
Figure 8.
SEIPIN2 and SEIPIN3 promote pollen germination and fertilization capacity. A, Confocal images (single sections) of Nile red-stained pollen grains from the indicated genotypes. Scale bar, 5 μm. B, Nomarski images (left) and maximal intensity projection images (from 14 serial confocal sections) of Nile red-stained pollen. Scale bar, 5 μm. C, Box plot of the LD areas in pollen grains of the different genotypes. The box plot features and statistical analysis are as explained in Figure 4C. D, Bar plot of the mean percentage of pollen grains germinated after 15 h in solid germination media in three independent experiments. The total number of pollen grains counted in the three experiments for each genotype is indicated below the graphs. Error bars, sd. ANOVA with Tukey’s HSD post-hoc test was used to test for differences in mean germination rate among the different genotypes. Genotypes without letters in common are significantly different (P < 0.05). The asterisks indicate the P value for the null hypothesis that the mean germination rate in that genotype is not different from the wild-type (Wt) mean. **P < 0.01. E, Confocal and bright field images of Nile red-stained pollen grains from the indicated genotypes after 15 h in liquid “hanging drop” germination media. Scale bar, 50 μm left panels, 10 μm other panels. Solid arrowheads, large LDs retained in the pollen grain. F, Graph showing the percentage of ovules fertilized by the competing pollen genotypes (indicated inside the bars) in each of the crosses. The P value for the null hypothesis that the observed transmission ratios do not deviate from the 1:1 ratio for equal transmission (two-tailed χ2 test) is shown above the graphs. The total number of plants genotyped for each cross is indicated below the graphs.
DISCUSSION
On the Nuclear Role of SEIPINs
In yeast, Pah1, an essential enzyme for TAG synthesis, is targeted to the nuclear ER membrane (Barbosa et al., 2015). Likewise, the yeast SEIPIN protein localizes primarily to subdomains of the nuclear envelope (Szymanski et al., 2007; Wang et al., 2014) and the corresponding mutant accumulates nLDs (Cartwright et al., 2015; Wolinski et al., 2015). Moreover, mutations or treatments perturbing TAG synthesis and LD biogenesis cause the abnormal proliferation of nuclear ER membranes closely associated with clustered LDs (Wolinski et al., 2011, 2015; Cartwright et al., 2015; Barbosa et al., 2015). Based on this evidence, it has been proposed that yeast cells have a specific “LD assembly domain” at the nuclear envelope for localized neutral lipid synthesis and LD biogenesis (Wolinski et al., 2015). The fact that tri-1 embryos accumulate nLDs and that overexpressed SEIPINs localize in perinuclear ER membranes associated with LD clusters suggests that plants may also have an LD assembly domain at the nuclear membrane. Why would this specific localization be selected during evolution? LD formation at the ER most likely interferes with budding of coat protein complex II (COP II)-coated vesicles and/or fusion of coat protein complex I (COP I)-coated vesicles, so a possible explanation is that directing LD biogenesis to a nuclear ER subdomain minimizes the interference with trafficking in the secretory pathway, which is essential for cell viability. A prediction from this model is that organisms should have developed mechanisms to cope with situations imposing high demands for LD biogenesis and for vesicular trafficking. In that respect, Arabidopsis embryos would be an interesting model to study how this is achieved, because during a short developmental window, cells have to coordinate extensive production of LDs and highly active vesicular trafficking of vacuolar storage proteins (Zouhar and Rojo, 2009). Another important question to resolve would be how SEIPINs in yeast and plants may be targeted to nuclear ER subdomains. In this regard, comparative sequence analysis of Arabidopsis SEIPIN1 and SEIPIN2, which strictly colocalize but show significant sequence divergence, may reveal conserved motifs involved in their specific targeting to nuclear ER membrane subdomains.
On SEIPINs and Seed Dormancy
Much has been learned in recent years about the genetic and hormonal control of seed dormancy (Nonogaki, 2014; Shu et al., 2016). In contrast, little is known about how environmental or developmental conditions regulate this trait. In particular, it is unclear how the metabolic status of the seeds impacts seed dormancy. Seed metabolism is largely directed toward the accumulation of reserves that provide energy for germination and seedling establishment and should then be a key factor in the decision of when to germinate. However, no evidence had been reported for a relation between storage status and the timing of germination. Now, our results show that perturbations in LD biogenesis in SEIPIN-deficient mutants markedly increase seed dormancy, suggesting that the LD status controls the degree of dormancy. Interestingly, mobilization of TAGs from LDs starts prior to germination (Penfield et al., 2006) at late stages of seed maturation, when dormancy is established (Bentsink and Koornneef, 2008). Hence, the perturbations in LD turnover rather than in LD biogenesis could underlie the higher dormancy observed in seipin mutants. Seeds might then sense energy availability from released TAGs to adjust dormancy. Interestingly, alterations in this pathway may have been selected to regulate dormancy in nature. Analysis of natural allelic variation for seed dormancy among Arabidopsis accessions has uncovered several delay of germination loci regulating this trait (Alonso-Blanco et al., 2003; Bentsink et al., 2010). The DELAY OF GERMINATION3 locus maps to a chromosomal region containing SEIPIN2, making it a candidate for this yet-unidentified locus. This opens the possibility that SEIPIN2 has been selected in nature to modulate LD-dependent dormancy and ensure proper timing of germination in different environments.
SEIPIN-Dependent LD Biogenesis Contributes to Pollen Fitness
Very few data were previously available on the function of TAGs and LDs in pollen, but given their abundance, it was generally thought that they played relevant roles in pollen germination and pollen tube growth (Ischebeck, 2016). However, prior studies on mutants in genes involved in LD formation or in their mobilization did not report defects in pollen germination or in transmission of the mutant alleles, even when the major pollen TAG-lipases SDP1 and SDP1L were mutated (Eastmond, 2006; Kelly et al., 2011), thus questioning the relevance of LDs for pollen fertilization efficiency. Now, through in vitro germination and in vivo pollen competition assays, we have provided solid evidence supporting that altered biogenesis of LDs in the s2s3 and triple seipin mutant pollen lowers germination rates in vitro and reduces the transmission efficiency in vivo. Although much less-studied than in seeds, pollen dormancy has been reported in Arabidopsis, and it may involve regulatory elements in common with seed dormancy (Šírová et al., 2011). It will be interesting to determine if the reduced germination rate observed in triple seipin mutant pollen is also related with increased dormancy as observed in seeds. In laboratory conditions optimized for Arabidopsis development, we only detected defects in pollen transmission when pollen from the triple seipin mutant was set to compete with pollen having normal-sized LDs. It is thus possible that defects in pollen transmission of spd1, spd1L, and other LD-related mutants may have gone undetected because only self-fertilization was analyzed. In fact, transmission and seed-set defects in self-fertilized triple seipin mutant plants may also become evident under growth conditions that are less favorable for pollen germination and fertilization. Considering the low levels of outcrossing in Arabidopsis, a negative effect on self-fertilization under suboptimal conditions would provide the selective pressure that maintains SEIPIN-dependent LD biogenesis in pollen.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The T-DNA insertion lines in the Col-0 background seipin1-1 (Salk_Seq0958), seipin2-1 (GabiGK_183F09), seipin3-1 (Salk_019429c), seipin3-2 (Sail_771_G12), and seipin3-3 (Sail_1280_H09) were obtained from the Arabidopsis Stock Center and genotyped using specific primers (Supplemental Table S3). Plants were grown on soil in the greenhouse under natural light, supplemented with Osram HQL 400w sodium lamps when illuminance fell below 5,000 l×, and a 16-h light/8-h dark cycle at a temperature range between 22°C maximum/18°C minimum. For in vitro culture, plants were grown at 22°C under 6,000 lux of illuminance in a 16-h light/8-h dark cycle.
Plasmid Construction
Promoter fragments containing the upstream intergenic regions and the 5′UTRs up to the start codon of each of the SEIPIN genes were amplified using the primers shown in Supplemental Table S3, cloned using Gateway technology in the destination vector pGWB3, sequence verified and transformed into Arabidopsis (Arabidopsis thaliana). At least 10 independent lines with a single insertion of the transgene for each of the three SEIPIN promoter constructs were obtained, and they showed essentially identical patterns of expression, as described in the figures. The coding sequences of SEIPIN1 and SEIPIN2 were amplified by PCR using the primers shown in Supplemental Table S3, cloned into pGEM-T Easy vector (Promega) and sequence verified. SEIPIN1 and SEIPIN2 coding sequences were then amplified with specific att-B modified primers (Supplemental Table S3), cloned into the pDONOR207 entry vector and recombined into the pH7WGR2 and pUBN-RFP-Dest (for RFP fusions), pK7WGF2 (for GFP fusions), or pGWB2 (for untagged expression) destination vectors (Grefen et al., 2010; Karimi et al., 2002; Nakagawa et al., 2007).
Tissue Staining and Light Microscopy Analyses
Embryos of Arabidopsis were dissected from seeds, stained with 5 mg/L Nile red in 50 mm PIPES buffer (1,4-Piperazinediethanesulfonic acid/NaOH, pH 7.0) for 20 min and mounted in 50 mm PIPES buffer for imaging. Pollen grains were directly mounted in 50 mm PIPES buffer, 18% Suc, 15 mg/L Nile red for imaging. Confocal microscopy was performed on an inverted Leica SP5II laser-scanning microscope using a dry (20× 0.75 numerical aperture) objective or a water immersion (63× 1.2 numerical aperture) objective. Fluorophores were excited (ex) and emission (em) was detected in sequential mode to prevent signal bleed-through: GFP (ex/em, 488 nm/498–555 nm), RFP (ex/em, 561 nm/598–658 nm), Nile red (ex/em, 561 nm/571–627 nm), Bodipy 493/503 (ex/em, 488 nm/498–552 nm), Chlorophyll (ex/em, 633 nm/653–695 nm). Pinholes were adjusted to 1 Airy unit and photomultiplier tube settings were chosen to avoid amplifier saturation. For quantification of LD size in embryos, the area of the LDs present in 30 μm × 30 μm fields from single-section confocal images of epidermal hypocotyl cells was automatically measured using a script in the Image J package. For quantification of LD size in pollen, the area of the LDs present in single-section confocal images of mature pollen grains was manually measured using the ImageJ package. For GUS staining, samples were incubated at 37°C in the dark in 100 mm sodium phosphate buffer (pH 7.4), 10 mm EDTA, 0.1% Triton X-100, 10 mm K3Fe(CN)6, 10 mm K4Fe(CN)6, and 1 mg/mL 5-bromo-4-chloro-3-indolyl-beta-D-glucuronide. Then the samples were cleared in 70% ethanol and mounted in chloral hydrate for imaging in a Zeiss Axioskop microscope with differential interference contrast optics.
Electron Microscopy
Flowers were marked at the time of anthesis and siliques collected 11 d after. The embryos were dissected from the ovules and fixed for 2 h at room temperature in 4% formaldehyde/2.5% glutaraldehyde in 50 mm phosphate-buffered saline, followed by an overnight incubation at 4°C in fresh fixing solution. After washing with water, the embryos were postfixed in 1% osmium tetroxide for 1 h at 4°C, washed with water, incubated in uranyl acetate 2% for 1 h at 4°C, washed with water, dehydrated in an acetone series and included in TAAB 812 resin (Taab Laboratories Equipment). Ultrathin (60 nm) sections were stained with uranyl acetate and lead citrate by standard procedures and analyzed in a JEOL JEM 1011 electron microscope operating with a Gatan Erlangshen ES1000W camera.
Seed and Pollen Germination Assays
For germination assays, seeds were imbibed in water for 1 h and sown on water-soaked filter paper inside petri dishes, placed in moisture trays to maintain humidity, and grown in in vitro chambers at 22°C under 6,000 lux of illuminance in a 16-h light/8-h dark cycle. For cold stratification, water-imbibed seeds were maintained at 4°C in the dark. Seedlings were considered germinated when the radicle had emerged from the seed coats. For all experiments, we measured germination of three independent seed batches obtained at different times. Each seed batch was collected from plants of the set of genotypes analyzed grown together under the same conditions. The time after imbibition when germination was scored and the period of after-ripening dry storage of the seeds are indicated for each assay in the figure legends. For pollen germination, pollen grains from freshly opened anther‐dehisced flowers were set to germinate in pollen-germination media (1 mm CaCl2, 1 mm Ca(NO3)2, 1 mm MgSO4, 0.01% boric acid, 18% Suc, set at pH 7.0 and supplemented with 0.5% bacto-agar). Pollen germination was scored as grains showing a distinguishable pollen tube after incubation for 15 h in the dark at 24°C in a humid chamber. For hypocotyl growth measurements, sterilized seeds from three independent batches were sown in 1% agar plates containing Murashige and Skoog media, with or without 1% Suc. Seeds in plates were stratified for 3 d at 4°C in the dark, set horizontally under light in the growth chamber for 5 h, covered with tinfoil, and set vertically in the growth chamber. Five days later, pictures of the seedlings were taken to measure hypocotyl length.
Lipid Extraction and thin layer chromatography (TLC)
Total lipids from mature seeds (5 mg) and seedlings (30 mg) were extracted using chloroform:methanol as described (Zhou et al., 2014). The lipid phase was washed once with methanol:water (1:1) and recovered after centrifugation. The solvent was then evaporated, and the sample used for TLC analyses. TLC silica gel 60 plates (20 × 20 cm in size) (Merck, Darmstadt, Germany) were activated at 110°C for at least 30 min before lipid loading. Neutral lipids were resolved using hexane:diethyl ether:acetic acid (70:30:1, v/v/v). Plates were sprayed uniformly with 10% cupric sulfate in 8% aqueous phosphoric acid, allowed to dry 10 min at room temperature, and then placed into an oven at 165°C for 10–15 min. Glyceryl trioleate (Sigma-Aldrich) was used as external standard for TAGs. Spots were quantified by densitometric analysis using ImageJ program, and the amount of TAG was calculated from the standard curves obtained.
Data Analysis and Bioinformatics
ANOVA, χ2 or Student’s t test were used to determine statistical significance. Differences were considered statistically significant for P values < 0.05. In all the graphs, *P < 0.05; **P < 0.01; ***P < 0.001.
Accession Numbers
SEIPIN1 (At5g16460); SEIPIN2 (At1g29760); SEIPIN3 (At2g34380).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. SEIPIN1 and SEIPIN2 colocalize in perinuclear structures.
Supplemental Figure S2. Overexpressed SEIPIN1 colocalizes with an ER marker in perinuclear structures.
Supplemental Figure S3. Overexpression of untagged SEIPIN1 reshapes the ER into perinuclear structures and vesicle-like bodies.
Supplemental Figure S4. Graphical representation of T-DNA insertional alleles.
Supplemental Figure S5. RT-PCR analysis of SEIPIN transcript accumulation
Supplemental Figure S6. Increased LD size in tri-2 and tri-3 embryos.
Supplemental Figure S7. The tri-1 mutant plants produces larger seeds.
Supplemental Figure S8. Hypocotyl growth in etiolated seedlings.
Supplemental Figure S9. Increased dormancy in tri-2 and tri-3 seeds.
Supplemental Figure S10. LD distribution in tri-2 and tri-3 mutant pollen.
Supplemental Table S1. RNA-seq expression profiles of SEIPIN1, SEIPIN2, and SEIPIN3.
Supplemental Table S2. Microarray expression profiles of SEIPIN1 and SEIPIN2.
Supplemental Table S3. Sequences of primers used in this work.
Acknowledgments
The authors thank C. Alonso-Blanco for critical reading and helpful comments on the manuscript and the CNB electron microscopy facility and Y. Fernandez for excellent technical assistance.
Footnotes
This work was supported by the Spanish Ministry of Economy and Competitivity and FEDER funds (BIO2015-69582-P MINECO/FEDER to E.R., J.J.S.-S., and M.T.).
These authors contributed equally to the article.
References
- Aldrich HC, Vasil IK (1970) Ultrastructure of the postmeiotic nuclear envelope in microspores of Podocarpus macrophyllus. J Ultrastruct Res 32: 307–315 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M (2003) Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164: 711–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Benítez M, Corvera-Poiré A, Chaos Cador A, de Folter S, Gamboa de Buen A, Garay-Arroyo A, García-Ponce B, Jaimes-Miranda F, Pérez-Ruiz RV, et al. (2010) Flower development. Arabidopsis Book 8: e0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AD, Sembongi H, Su WM, Abreu S, Reggiori F, Carman GM, Siniossoglou S (2015) Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol Biol Cell 26: 3641–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Dubreucq B, Miquel M, Rochat C, Lepiniec L (2008) Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. Arabidopsis Book 6: e0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Hanson J, Hanhart CJ, Blankestijn-de Vries H, Coltrane C, Keizer P, El-Lithy M, Alonso-Blanco C, de Andrés MT, Reymond M, et al. (2010) Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc Natl Acad Sci USA 107: 4264–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Koornneef M (2008) Seed dormancy and germination. Arabidopsis Book 6: e0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet E, El Mourabit H, Prot M, Nemani M, Khallouf E, Colard O, Maurice M, Durand-Schneider AM, Chrétien Y, Grès S, et al. (2009) Seipin deficiency alters fatty acid Delta9 desaturation and lipid droplet formation in Berardinelli-Seip congenital lipodystrophy. Biochimie 91: 796–803 [DOI] [PubMed] [Google Scholar]
- Cai Y, Goodman JM, Pyc M, Mullen RT, Dyer JM, Chapman KD (2015) Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell 27: 2616–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright BR, Binns DD, Hilton CL, Han S, Gao Q, Goodman JM (2015) Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol Biol Cell 26: 726–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Dyer JM, Mullen RT (2012) Biogenesis and functions of lipid droplets in plants: Thematic review series: Lipid droplet synthesis and metabolism: From yeast to man. J Lipid Res 53: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings DA, Carter CN, Rink JC, Scott AC, Wyatt SE, Allen NS (2000) Plant nuclei can contain extensive grooves and invaginations. Plant Cell 12: 2425–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Wang Y, Tang Y, Liu Y, Zhao L, Deng J, Xu G, Peng X, Ju S, Liu G, et al. (2011) Seipin ablation in mice results in severe generalized lipodystrophy. Hum Mol Genet 20: 3022–3030 [DOI] [PubMed] [Google Scholar]
- De Domenico S, Bonsegna S, Lenucci MS, Poltronieri P, Di Sansebastiano GP, Santino A (2011) Localization of seed oil body proteins in tobacco protoplasts reveals specific mechanisms of protein targeting to leaf lipid droplets. J Integr Plant Biol 53: 858–868 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ. (2006) SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18: 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese RV Jr., Walther TC (2016) Lipid droplets go nuclear. J Cell Biol 212: 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, Wenk MR, Parton RG, Yang H (2008) Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol 180: 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Goodman JM (2015) The lipid droplet-a well-connected organelle. Front Cell Dev Biol 3: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda SK, Watt S, Collins-Silva J, Kilaru A, Arondel V, Yurchenko O, Horn PJ, James CN, Shintani D, Ohlrogge JB, et al. (2013) Lipid droplet-associated proteins (LDAPs) are involved in the compartmentalization of lipophilic compounds in plant cells. Plant Signal Behav 8: e27141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Grippa A, Buxó L, Mora G, Funaya C, Idrissi FZ, Mancuso F, Gomez R, Muntanyà J, Sabidó E, Carvalho P (2015) The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J Cell Biol 211: 829–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Cordes KR, Farese RV Jr., Walther TC (2009) Lipid droplets at a glance. J Cell Sci 122: 749–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Binns DD, Chang YF, Goodman JM (2015) Dissecting seipin function: The localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1p(ΔNterm) only in combination with Ldb16p. BMC Cell Biol 16: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T. (2016) Lipids in pollen—They are different. Biochim Biophys Acta 1861: 1315–1328 [DOI] [PubMed] [Google Scholar]
- Jeong HJ, Choi JY, Shin HY, Bae JM, Shin JS (2014) Seed-specific expression of seven Arabidopsis promoters. Gene 553: 17–23 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kelly AA, Quettier AL, Shaw E, Eastmond PJ (2011) Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol 157: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Hsieh K, Ratnayake C, Huang AH (2002) A novel group of oleosins is present inside the pollen of Arabidopsis. J Biol Chem 277: 22677–22684 [DOI] [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA (2016) A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J 88: 1058–1070 [DOI] [PubMed] [Google Scholar]
- Kory N, Farese RV Jr., Walther TC (2016) Targeting fat: Mechanisms of protein localization to lipid droplets. Trends Cell Biol 26: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang A, Musgrave ME (1996) Dynamics of vegetative cytoplasm during generative cell formation and pollen maturation in Arabidopsis thaliana. Protoplasma 194: 81–90 [DOI] [PubMed] [Google Scholar]
- López-Ribera I, La Paz JL, Repiso C, García N, Miquel M, Hernández ML, Martínez-Rivas JM, Vicient CM (2014) The evolutionary conserved oil body associated protein OBAP1 participates in the regulation of oil body size. Plant Physiol 164: 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C, Nordström R, Wagner K, Windpassinger C, Andersson H, von Heijne G, Nilsson I (2006) Membrane topology of the human seipin protein. FEBS Lett 580: 2281–2284 [DOI] [PubMed] [Google Scholar]
- Magré J, Delépine M, Khallouf E, Gedde-Dahl T Jr., Van Maldergem L, Sobel E, Papp J, Meier M, Mégarbané A, Bachy A, et al. ; BSCL Working Group (2001) Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat Genet 28: 365–370 [DOI] [PubMed] [Google Scholar]
- Malhas A, Goulbourne C, Vaux DJ (2011) The nucleoplasmic reticulum: Form and function. Trends Cell Biol 21: 362–373 [DOI] [PubMed] [Google Scholar]
- McLachlan DH, Lan J, Geilfus CM, Dodd AN, Larson T, Baker A, Hõrak H, Kollist H, He Z, Graham I, et al. (2016) The breakdown of stored triacylglycerols is required during light-induced stomatal opening. Curr Biol 26: 707–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M, Trigui G, d’Andréa S, Kelemen Z, Baud S, Berger A, Deruyffelaere C, Trubuil A, Lepiniec L, Dubreucq B (2014) Specialization of oleosins in oil body dynamics during seed development in Arabidopsis seeds. Plant Physiol 164: 1866–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nonogaki H. (2014) Seed dormancy and germination-emerging mechanisms and new hypotheses. Front Plant Sci 5: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagac M, Cooper DE, Qi Y, Lukmantara IE, Mak HY, Wu Z, Tian Y, Liu Z, Lei M, Du X, et al. (2016) SEIPIN regulates lipid droplet expansion and adipocyte development by modulating the activity of glycerol-3-phosphate acyltransferase. Cell Reports 17: 1546–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Pinfield-Wells HM, Graham IA (2006). Storage reserve mobilisation and seedling establishment in Arabidopsis. Arabidopsis Book 4: e0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A, Gross SP, Parton RG (2014) Review: biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. J Cell Biol 204: 635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo VT, Belevich I, Li S, Karhinen L, Vihinen H, Vigouroux C, Magré J, Thiele C, Hölttä-Vuori M, Jokitalo E, et al. (2016) Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J 35: 2699–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J 55: 798–809 [DOI] [PubMed] [Google Scholar]
- Shockey J, Regmi A, Cotton K, Adhikari N, Browse J, Bates PD (2016) Identification of Arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiol 170: 163–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH (2016) Two faces of one seed: Hormonal regulation of dormancy and germination. Mol Plant 9: 34–45 [DOI] [PubMed] [Google Scholar]
- Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim MF, Dennis RJ, Aubry EM, Ramanathan N, Sembongi H, Saudek V, Ito D, O’Rahilly S, Siniossoglou S, Rochford JJ (2012) The human lipodystrophy protein seipin is an ER membrane adaptor for the adipogenic PA phosphatase lipin 1. Mol Metab 2: 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šírová J, Sedlářová M, Piterková J, Luhová L, Petřivalský M (2011) The role of nitric oxide in the germination of plant seeds and pollen. Plant Sci 181: 560–572 [DOI] [PubMed] [Google Scholar]
- Szymanski KM, Binns D, Bartz R, Grishin NV, Li W-P, Agarwal AK, Garg A, Anderson RG, Goodman JM (2007) The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci USA 104: 20890–20895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder MM, Sim MF, O’Rahilly S, Edwardson JM, Rochford JJ (2015) Seipin oligomers can interact directly with AGPAT2 and lipin 1, physically scaffolding critical regulators of adipogenesis. Mol Metab 4: 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Bi J, Shui G, Liu Z, Xiang Y, Liu Y, Wenk MR, Yang H, Huang X (2011) Tissue-autonomous function of Drosophila seipin in preventing ectopic lipid droplet formation. PLoS Genet 7: e1001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Der Schaar W, Alonso-Blanco C, Léon-Kloosterziel KM, Jansen RC, van Ooijen JW, Koornneef M (1997) QTL analysis of seed dormancy in Arabidopsis using recombinant inbred lines and MQM mapping. Heredity (Edinb) 79: 190–200 [DOI] [PubMed] [Google Scholar]
- Wang H, Becuwe M, Housden BE, Chitraju C, Porras AJ, Graham MM, Liu XN, Thiam AR, Savage DB, Agarwal AK, et al. (2016) Seipin is required for converting nascent to mature lipid droplets. eLife 5: e16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Miao YH, Chang YS (2014) Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J Cell Sci 127: 1214–1228 [DOI] [PubMed] [Google Scholar]
- Wee K, Yang W, Sugii S, Han W (2014) Towards a mechanistic understanding of lipodystrophy and seipin functions. Biosci Rep 34: e00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Haas JT, Walther TC, Farese RV Jr (2014) Lipid droplet biogenesis. Curr Opin Cell Biol 29: 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinski H, Hofbauer HF, Hellauer K, Cristobal-Sarramian A, Kolb D, Radulovic M, Knittelfelder OL, Rechberger GN, Kohlwein SD (2015) Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim Biophys Acta 1851: 1450–1464 [DOI] [PubMed] [Google Scholar]
- Wolinski H, Kolb D, Hermann S, Koning RI, Kohlwein SD (2011) A role for seipin in lipid droplet dynamics and inheritance in yeast. J Cell Sci 124: 3894–3904 [DOI] [PubMed] [Google Scholar]
- Yang H, Galea A, Sytnyk V, Crossley M (2012) Controlling the size of lipid droplets: Lipid and protein factors. Curr Opin Cell Biol 24: 509–516 [DOI] [PubMed] [Google Scholar]
- Zhang M, Fan J, Taylor DC, Ohlrogge JB (2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XR, Callahan DL, Shrestha P, Liu Q, Petrie JR, Singh SP (2014) Lipidomic analysis of Arabidopsis seed genetically engineered to contain DHA. Front Plant Sci 5: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhar J, Rojo E (2009) Plant vacuoles: Where did they come from and where are they heading? Curr Opin Plant Biol 12: 677–684 [DOI] [PubMed] [Google Scholar]