Abstract
TOR signaling regulates plant translation via a specific translation initiation mechanism: reinitiation.
Protein synthesis is an intricate, energy-demanding, and tightly controlled process that plays a fundamental role in cell growth, proliferation, and differentiation. The target of rapamycin (TOR) protein kinase integrates stress-, nutrient-, and energy-related signals to optimize protein synthesis outputs. In mammals, TOR is a main controller of capped mRNA translation; how TOR participates in translation initiation in plants is unclear, but active TOR is required for regulation of translation of mRNA with 5′-untranslated region (5′-UTR) upstream open reading frames (uORFs)—known translation regulatory elements in eukaryotes. Recent data implicates diverse signals such as stress, hormones, and metabolites in regulation of TOR signal transduction pathways and, thus, in response to environmental stress. Here, we review current knowledge of plant TOR complex composition and activation, and its function in translation, compiling data on downstream processes that are under stringent control of TOR in mammals but not yet investigated in plants.
Plants have evolved various adaptation mechanisms to ensure their optimal growth, with plant development and behavior being strongly responsive to various external and internal stimuli. By monitoring their environment, plants trigger signal transduction cascades in order to regulate downstream cellular processes, mainly via protein synthesis—a major energy-consuming process (Buttgereit and Brand, 1995). The TOR protein kinase integrates extracellular signals (hormones, biotic and abiotic stresses, growth factors) together with intracellular nutrient availability and energy status to control protein synthesis and other anabolic processes if conditions are favorable, and represses catabolic processes such as autophagy (Albert and Hall, 2015). The past 10 years has boosted research on plant TOR complex composition, highlighted TOR upstream signals and downstream targets, and revealed an interplay between TOR signaling and hormonal, stress, and other pathways in photosynthetic organisms. We are beginning to understand how TOR is activated, and how it controls many cellular processes, such as transcription, translation, and autophagy. Here, we give an overview of recent results on the role of TOR in plant translation control and draw the reader’s attention to future questions to address. In plants, inactivation of TOR correlates with a decrease in total polysomal levels (Deprost et al., 2007; Schepetilnikov et al., 2011, 2013), strongly suggesting a role for TOR in plant translation. Although a role for TOR in global translation in plants has been documented, the players and mechanisms used to influence different steps of translation initiation—the step most controlled by TOR in mammals—are only starting to emerge. Here, we summarize the current state of knowledge of specific plant translation initiation mechanisms that are controlled by 5′-UTR elements of mRNAs and discuss regulation of these mechanisms by TOR/S6 kinase 1 (S6K1) signaling. Examples where TOR regulates specific mRNAs in other eukaryotic systems are presented. In addition, we discuss current understanding of upstream signaling effectors that link TOR with protein synthesis and plant growth.
PLANT TOR COMPLEX COMPOSITION
TOR belongs to the phosphoinositide 3-kinase-related protein kinase family that exhibits Ser/Thr but no lipid kinase activity (Abraham, 2004). The domain organization of Arabidopsis (Arabidopsis thaliana) TOR is complex and conserved among metazoans, mammals, and plants (Menand et al., 2002; Zoncu et al., 2011). The N-terminal part of TOR contains HEAT (Huntingtin, Elongation Factor 3, protein phosphatase 2A, TOR1) repeat motifs required for substrate recruitment and membrane association, followed by FAT (FRAP, ATM, TRRAP), FRB (FKBP12-Rapamycin Binding), Ser/Thr kinase catalytic, and FATC (FAT Carboxy terminus) domains; the FATC and FAT domains together contribute to kinase activation. TOR is encoded by a single gene in mammals and plants, while two TOR genes, TOR1 and TOR2, are present in yeast (Shimobayashi and Hall, 2014). Mammalian TOR (mTOR) acts in two functionally and structurally distinct complexes—TORC1 and TORC2—that both contain mTOR, mLST8 (mammalian Lethal with Sec13 protein 8) and DEPTOR (DEP-domain-containing mTOR-interacting protein). In addition, mTORC1 harbors the scaffold protein mRaptor (mammalian Regulatory-Associated Protein of mTOR) and PRAS40 (Pro-Rich AKT Substrate 40 kD), while mTORC2 comprises Rictor (Rapamycin-insensitive companion of Introduction mTOR), mSIN1 (mammalian Stress activated protein kinase Interacting Protein 1), and Protor (Protein observed with Rictor; Zoncu et al., 2011). mTORC1 promotes biosynthesis of proteins, lipids, and nucleotides but inhibits autophagy in conditions of substrate sufficiency (Shimobayashi and Hall, 2014), while mTORC2 regulates cell metabolism and cytoskeleton reorganization (Oh and Jacinto, 2011). Furthermore, the TOR pathway is more complex than previously appreciated, and four distinct TOR kinases (TOR1–TOR4) have been identified in the parasite Trypanosoma brucei (Saldivia et al., 2013).
A TOR knockout mutant in Arabidopsis is embryo lethal, and TOR inactivation affects plant growth (Menand et al., 2002). TOR is expressed in all Arabidopsis tissues, particularly in primary and lateral roots, shoot apical meristems, and floral meristems (Menand et al., 2002). Up to now, two core subunits of TORC1 (Raptor and LST8) have been found in Arabidopsis; both are encoded by two genes (RAPTOR1/RAPTOR2 and LST8-1/LST8-2, respectively; Deprost et al., 2007; Moreau et al., 2012). Unlike in yeast and mammals, mutations in LST8 are not lethal but result in developmental defects, whereas RAPTOR1 mutations are conditionally lethal in Arabidopsis (Anderson et al., 2005; Deprost et al., 2007; Moreau et al., 2012). Raptor is an important mTOR accessory protein that facilitates the recruitment and phosphorylation of TOR substrates via targeting both the N-terminal HEAT domain of TOR and a TOR signaling (TOS) motif present in the majority of TOR substrates (Schalm and Blenis, 2002), while mLST8 binds the mTOR kinase domain and likely modulates mTOR complex integrity and activity (Kim et al., 2002; Wullschleger et al., 2005). Accordingly, Raptor1 binding to the TOR HEAT domain and Lst8-1 to the TOR FRB domain was demonstrated in both Arabidopsis and the green alga Chlamydomonas reinhardtii (Anderson et al., 2005; Mahfouz et al., 2006; Deprost et al., 2007; Moreau et al., 2012), suggesting that both proteins are likely to fulfill functions similar to that of mammalian orthologs. Plants may be lacking a TORC2-related complex, since Rictor and SIN1 have not been found in photosynthetic organisms (Dobrenel et al., 2016a). However, we cannot exclude that plants constitute TOR-containing complexes with components that differ from those found in mammals; for example, in neural stem cells, G-protein-coupled receptor kinase-interacting protein 1 nucleates an mTOR complex lacking Raptor and Rictor (Smithson and Gutmann, 2016).
Arabidopsis TOR displays a weak sensitivity to the peptidyl-prolyl cis/trans isomerase (FK506-binding protein 12 [FKBP12])-rapamycin complex (Menand et al., 2002; Sormani et al., 2007), or becomes sensitive when human or yeast FKBP12 is overexpressed in Arabidopsis or high rapamycin concentrations are applied to inhibit growth (Mahfouz et al., 2006; Sormani et al., 2007; Ren et al., 2012; Xiong and Sheen, 2012). Currently, ATP-competitive inhibitors such as Torin-1 and the second-generation inhibitor AZD-8055 are used widely to block TOR in plants (Montané and Menand, 2013; Schepetilnikov et al., 2013, 2017). Nowadays, TOR-deficient, ethanol-inducible, and estradiol-inducible RNA interference lines are widely employed to elucidate TOR functions in planta and avoid the embryo lethality of null tor mutants (Deprost et al., 2007; Caldana et al., 2013; Xiong et al., 2013). mTOR activation status can be monitored by following phosphorylation at Ser-2448, which is likely phosphorylated by S6K1, if TOR is active (Chiang and Abraham, 2005). The corresponding epitope within the C terminus of Arabidopsis TOR is phosphorylated at Ser-2424 in response to TOR activation (Schepetilnikov et al., 2013, 2017).
UPSTREAM CONTROL OF TRANSLATION VIA TOR
Hormone Signaling to Translation
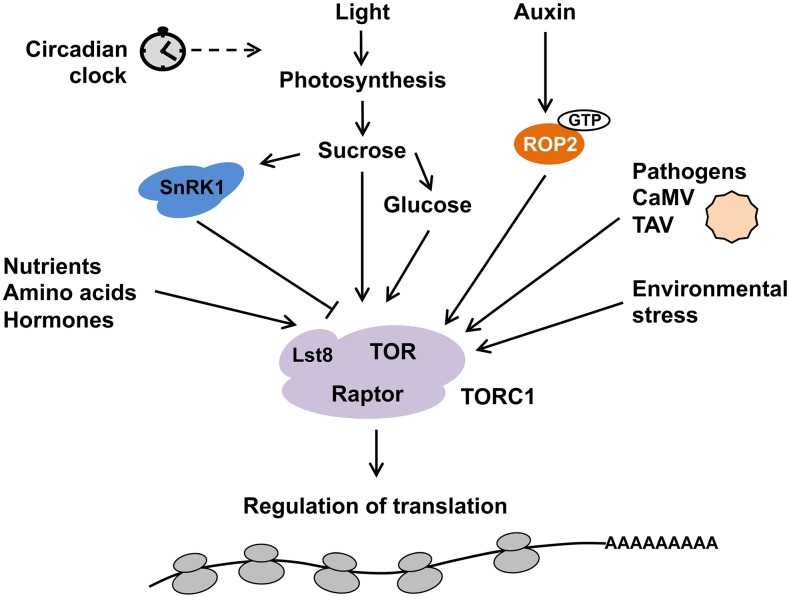
Many experiments strongly suggest that TOR signaling is wired to hormonal control, light, and biotic and abiotic stresses, and recent research has identified some underlying pathways (Fig. 1). Several phytohormones, such as jasmonic acid, abscisic acid, salicylic acid, brassinosteroids, and cytokinin, were implicated in cross talk with the TOR signaling pathway (Dong et al., 2015; Kravchenko et al., 2015; Pfeiffer et al., 2016; Zhang et al., 2016; De Vleesschauwer et al., 2017; Song et al., 2017). However, the most prominent growth hormones, auxin and kinetin, are known to promote S6K phosphorylation in cell suspension cultures, which leads to phosphorylation of ribosomal protein S6 (Turck et al., 2004). A growing body of evidence now demonstrates that TOR acts as an essential factor for auxin signal transduction in Arabidopsis (Schepetilnikov et al., 2013; Deng et al., 2016; Pu et al., 2017). Moreover, auxin was identified as a TOR upstream effector molecule (Schepetilnikov et al., 2013) that acts on TOR via activation of a small GTPase, ROP2 (Schepetilnikov et al., 2017). Indeed, auxin-treated plants, plants with high endogenous auxin accumulation, are characterized by increased levels of active TOR. In addition, plants with high levels of active ROP2, including those expressing constitutively active ROP2 and ROP2-GTP-expressing Arabidopsis plants, display increased phosphorylation of TOR and S6K1 (Schepetilnikov et al., 2013, 2017; Li et al., 2017). Accordingly, TOR inactivation leads to severe gravitropic defects, described as auxin-mediated responses. Currently, the auxin/ROP2/TOR signaling axis represents the most well-studied example of upstream TOR regulation in plants. Auxin-mediated activation of TOR drives translation of specific messages (Schepetilnikov et al., 2013, 2017), inhibits stress-mediated autophagy (Pu et al., 2017), and, at a physiological level, promotes meristem activation (Li et al., 2017). Based on the observed effects of auxin on TOR signaling activation, it was suggested that the auxin-TOR signaling axis regulates translation of the specific class of uORF-mRNAs in the cytoplasm (Schepetilnikov et al., 2013).
Figure 1.
The TOR signaling pathway regulates global translation. The TOR complex integrates plethora of metabolic pathways and environmental signals to drive the global translational rates in photosynthetic organisms.
Auxin-sensing ROP GTPases are conserved molecular switches that function in many signal transduction events (Fehér and Lajkó, 2015). Studies of the mechanism of TOR activation by ROP2 led to the discovery that ROP2 physically binds TOR and promotes its activation, if bound to GTP (Schepetilnikov et al., 2017). In response to ROP2, TOR associates with endosome-like structures, suggesting that plant endosomes serve as a hub for TOR activation upon perception of auxin signal. Currently, ROP2 GTPase represents the only example of plant TOR direct upstream effector that activates TOR in plants.
Light, Energy, and Sugar Signaling in Translation
The most critical environmental input comes from light energy, and inactivation of TOR suppresses light-energy-dependent plant growth (Ren et al., 2012). Light, sugar, and brassinosteroid signaling through TOR maintain the balance between hormone-promoted growth and carbon availability in plants (Xiong et al., 2013; Dong et al., 2015; Zhang et al., 2016). TOR integrates light and metabolic signals for apical shoot meristem activation (Pfeiffer et al., 2016). During photosynthesis, light energy is captured to convert the carbon intake into production of sugars that can be used immediately as material for plant growth or stored as starch for long-term reserves (Zeeman and Ap Rees, 1999). Photosynthesis, the primary source of energy for plants, also appears to be under the control of the TOR pathway. Plants deficient in TOR signaling exhibit abnormalities in thylakoid grana architecture and normal photosynthetic ability (Sun et al., 2016). Interestingly, light promotes translation of mRNAs encoding proteins involved in photosynthesis (Petracek et al., 1997; Tang et al., 2003; Floris et al., 2013). Recent data suggested that sugars act not only as energy source, but also as signaling molecules through TOR, maintaining the balance between hormone-promoted growth and carbon availability (Xiong et al., 2013; Dong et al., 2015; Zhang et al., 2016).
The TOR complex is regulated by AMP protein kinase (AMPK), the main energy sensor in humans (Mihaylova and Shaw, 2011). A similar antagonistic relationship between the AMPK plant ortholog SnRK1 (Suc nonfermenting 1-related Kinase 1) and TOR-related signaling pathways in response to changing nutritional and energy conditions was suggested in Arabidopsis (Robaglia et al., 2012; Broeckx et al., 2016; Baena-González and Hanson, 2017). In mammals, being upstream of TOR, SnRK1 may inhibit TOR activity via direct interaction and phosphorylation of upstream components of TOR signaling and Raptor—the regulatory subunit of the TOR complex (Nukarinen et al., 2016)—possibly leading to complex disassembly (Hughes Hallett et al., 2015). Accordingly, in plants SnRK1 can interact and phosphorylate Raptor (Nietzsche et al., 2016; Nukarinen et al., 2016).
Different light regimes affect general translation; the massive increase in translation during photomorphogenesis (Liu et al., 2012, 2013), and changes in polysomal loading of specific pull of mRNAs after reillumination (Juntawong and Bailey-Serres, 2012) and high light intensity stress (Floris et al., 2013), indirectly indicate the involvement of TOR signaling in response to light energy. Oxygen singlets, which are a consequence of excess light, repress translation and phosphorylation of 40S ribosomal protein S6 (Khandal et al., 2009), suggesting that TOR orchestration of translation and growth is light dependent and that, in turn, TOR controls photosynthesis at the level of gene expression.
Circadian Clock and TOR
An endogenous biological timekeeper that produces rhythmic outputs with roughly 24-h periodicity—the circadian clock—is based on an autoregulatory transcriptional feedback loop that schedules physiological processes to occur at specific and appropriate times of day (Nohales and Kay, 2016). Several studies in animals proposed that TORC1 activity is controlled by the circadian clock, providing some rhythmicity of TOR signaling (Cornu et al., 2014; Khapre et al., 2014; Lipton et al., 2015). In higher plants, translation of the core component of the circadian clock LHY occurs in a light-sensitive manner (Kim et al., 2003). More generally, overall translation efficiency in plants correlates with diurnal changes in energy status mediated by light perception and carbon metabolism (Piques et al., 2009; Pal et al., 2013). Accordingly, rhythmical oscillations of mRNA loading on polysomes support a connection between circadian clock and translational regulation (Missra et al., 2015). However, whether circadian oscillations regulate TOR signaling in plants remains to be elucidated.
TOR Signaling under Biotic Stress
Viral and bacterial infection are significant stresses that affect many signaling pathways in plants, including TOR signaling—a significant target during viral infection in both mammals and plants (Walsh et al., 2013). TOR-deficient plants appear to be more resistant to both viral and bacterial infection (Schepetilnikov et al., 2011; Ouibrahim et al., 2015; Popa et al., 2016; Zvereva et al., 2016; Meteignier et al., 2017). There are several explanations for this phenomenon. First, TOR signaling is required for translation of specific polycistronic viral mRNAs; for example, the plant pararetrovirus Cauliflower mosaic virus (CaMV) protein transactivator/viroplasmin (TAV) directly binds and mediates TOR activation—a characteristic unique among plant and mammalian viruses (Schepetilnikov et al., 2011). Second, recent translatome analysis proposes a role for TOR-mediated translational control in immune response by regulation of defense gene expression (Meteignier et al., 2017). Finally, beside translational input, TOR-mediated stress induction of autophagy and innate immunity can eliminate pathogens, thus protecting plants from infection. The pathogenicity effectors can trigger TOR activity, thus modulating the timing, or extent, of autophagy induction (Popa et al., 2016; Zvereva et al., 2016).
SIGNALING OF PLANT TORC1 COMPLEX TOWARD TRANSLATION
S6K Activation by TORC1
The main downstream targets of TORC1 in mammals are found within the cell translation apparatus, including ribosomal protein S6Ks and eIF4E-binding proteins (4E-BPs; Albert and Hall, 2015; Rexin et al., 2015). In mammals, activation of the S6Ks p70S6K1 and p85S6K1 critically depends on direct phosphorylation by mTORC1 at the C-terminal hydrophobic motif residue, and the phosphoinositide dependent kinase 1 that contributes to full activation of S6K1 by phosphorylating a motif within the activation loop (Thr-389 and Thr-229 in S6K1, respectively; Fenton and Gout, 2011; Liko and Hall, 2015). According to the crystal structure of S6K1, phosphorylation of the hydrophobic motif residue occurs independently and likely precedes phosphorylation of the activation loop (Wang et al., 2013). Phosphorylation of S6K1 by TOR is aided by Raptor, which mediates interaction between TOR and the S6K1 TOS motif (FDIDL in S6K1; Kim et al., 2002; Schalm and Blenis, 2002).
Arabidopsis harbors two closely related homologs of mammalian S6Ks: AtS6K1 and AtS6K2 (Zhang et al., 1994; Mizoguchi et al., 1995). Mutational analysis suggests that Arabidopsis S6K single mutants show different expression patterns and stress sensitivity (Henriques et al., 2010). Accumulating data suggest that Arabidopsis AtS6K1 most likely substitutes for mammalian p70S6K1 (Mahfouz et al., 2006; Schepetilnikov et al., 2011; Xiong and Sheen, 2012), while AtS6K2, which is likely localized in the nucleus, may be functionally equivalent to p85S6K (Mahfouz et al., 2006). As in mammals, Arabidopsis TOR likely cooperates with Raptor in phosphorylation of direct targets, since interaction between Raptor and both TOR and S6K1 has also been found in Arabidopsis (Mahfouz et al., 2006). The TOS motif recently uncovered within the N terminus of Arabidopsis S6K1 displays a low level of conservation with canonical consensus motifs found in mTOR kinase substrates, indicating plant specificity in Raptor-substrate interactions (Son et al., 2016, 2017; Sun et al., 2016).
4E-BPs in Plants
In eukaryotes, cap-dependent translation initiation requires mRNA binding to the eIF4F complex, which contains cap-binding eIF4E, an ATP-dependent helicase eIF4A and their interacting scaffold protein eIF4G (Browning and Bailey-Serres, 2015; Hinnebusch et al., 2016). mTOR, if active, maintains eIF4F integrity via phosphorylation of a family of translational initiation repressors, eIF4E-binding proteins (4E-BPs), in mammals and Drosophila (Jackson et al., 2010). In the generally accepted scenario, 4E-BPs, when not phosphorylated by TOR, efficiently compete with eIF4G for eIF4E binding, inhibiting eIF4F complex formation and cap-dependent initiation of translation. TOR phosphorylates 4E-BPs in response to particular physiological conditions that trigger their release from cap-bound eIF4E and restoration of the eIF4F complex. Notably, 4E-BPs are the least abundant factors among eukaryotic translation initiation factors (eIFs), and misregulation of their expression leads to various diseases and cancers in animals (Furic et al., 2010). Additionally, 4E-BPs preferentially repress so-called eIF4E-sensitive mRNAs, translation initiation of which is strictly dependent on eIF4E binding to the cap structure (Koromilas et al., 1992). In plants, despite the growing number of eIF4E-binding proteins that harbor the so-called canonical eIF4E-binding motif, none is a known TOR target. However, we cannot exclude that the small size and low abundance of 4E-BP-like proteins might limit identification of their orthologs in plants. Thus, to date, there is no experimental evidence that TOR is involved in control of the main mechanism of translation initiation mediated by the mRNA 5′ cap structure, opening a door to future research.
eIF3 and Polysomes Are Platforms for TOR Phosphorylation Events
In higher eukaryotes, including plants, eIF3 is a 700- to 800-kD multisubunit complex comprising 13 different subunits (eIF3a–eIF3m), with only five subunits—eIF3a, eIF3b, eIF3c, eIF3g, and eIF3i—that function in Saccharomyces cerevisiae (Hinnebusch, 2006). eIF3 was implicated at every step of translation initiation, including 43S and 48S preinitiation complex formation assembly, AUG start codon recognition, and reinitiation of translation, when ribosomes terminating translation of the leader uORF resume scanning and reinitiate at a further downstream ORF on the same mRNA (Park et al., 2001; Kim et al., 2004; Pöyry et al., 2007; Schepetilnikov et al., 2013).
A new connection between the TOR/S6K1 signaling axis and translation initiation was discovered (Holz et al., 2005), with eIF3 identified as a TOR-interacting partner serving as a platform for TOR phosphorylation events in humans. Later results have shown that polysomes serve as a docking site for plant TORC1 (Schepetilnikov et al., 2011, 2013) and yeast TORC2 (Oh et al., 2010). A simplified model states that, in the absence of extracellular stimuli such as auxin, for example, S6K1 associates with either eIF3-containing complexes or polysomes, while inactive TOR does not. Upon TOR stimulation, TOR is recruited to eIF3 and, to a lesser extent, polysomes, where it phosphorylates S6K1, triggering S6K1 concomitant release from both platforms. The patterns of association with TORC1 are opposite to those of S6K1. Indeed, in TOR-deficient mutants, characterized by nondetectable S6K1 activity, S6K1 is constitutively bound to polysomes (Schepetilnikov et al., 2011).
Ribosomal Protein S6 Is a Major Target of S6Ks within the Cell Translation Machinery
S6Ks phosphorylate numerous targets to control various cellular functions (Ma and Blenis, 2009), with translation factors, ribosomal protein S6 (eS6; Fenton and Gout, 2011; Liko and Hall, 2015), and eukaryotic elongation factor-2 kinase (Wang et al., 2001) being the most famous S6K targets.
Initial observations that eS6 is a major TOR/S6K signaling downstream target prompted much subsequent research into the role in translation played by eS6 when phosphorylated in response to TOR activation. The crystal structures of the 80S ribosome and 40S ribosomal subunit have established that eS6 is located at the right foot of the 40S ribosomal subunit (Ben-Shem et al., 2011). The C-terminal α-helix of eS6, which harbors multiple TOR/S6K1 phosphorylation sites, protrudes out of 40S, thus providing an alternative bridge to 60S or external protein binding (Ben-Shem et al., 2011). Despite these advances, the role of eS6 phosphorylation in initiation or elongation steps of mRNA translation remains a matter of debate (Meyuhas, 2015).
In plants, eS6 is encoded by two well-conserved genes encoding proteins having equivalent and interchangeable functions (Creff et al., 2010). Activation and phosphorylation of S6Ks strongly correlates with eS6 phosphorylation (Turck et al., 1998; Mahfouz et al., 2006). Comparative studies of eS6 posttranslational modifications occurring in response to hormones and various abiotic signals documented phosphorylation of several sites within the C-terminal α-helix of eS6 (for review, see Browning and Bailey-Serres, 2015). Recently, phosphoproteomic analysis and western blot with an antibody specifically recognizing phosphorylated Ser-240 has suggested that phosphorylation of Ser-240 is TOR/S6K1 responsive, and, accordingly, TOR inactivation decreases eS6 phosphorylation at this site (Dobrenel et al., 2016b).
Recent work by Nukarinen et al. (2016) has indicated that activation of SnRK1 is implicated in repression of protein synthesis (as main energy consumer) and implicated SnRK1 in regulation of photosynthesis (as main energy source); Weckwerth and colleagues found that ribosomal protein S6 was highly phosphorylated in Arabidopsis SnRK-deficient plants at Ser-240, Ser-237, and Ser-231, indicating that these phosphorylation sites are responsive to TOR activation (Nukarinen et al., 2016).
ROLE OF TOR IN REGULATION OF TRANSLATION OF SPECIFIC MRNAS
mRNAs Harboring uORFs within Their Leader Regions
More than 30% of eukaryotic mRNAs are loaded with uORFs within their 5′-UTRs, and there is mounting evidence that uORFs are major repressors of translation and thus play a key role in growth and development (Tran et al., 2008; Calvo et al., 2009). uORF-mRNAs, which are common in Arabidopsis (Hanada et al., 2007; Hayden and Jorgensen, 2007; Kim et al., 2007), encode many regulatory proteins, including transcriptional factors (e.g. auxin response factors [ARFs]), protein kinases, and other potent proteins, expression levels of which are limited by posttranscriptional mechanism such as ribosomal reinitiation. Where the 5′-UTR of mRNA contains a short uORF (typically < 30–50 codons), a fraction of ribosomes (usually < 50%) can resume scanning after translation of this uORF and reinitiate at a further downstream ORF (Kozak, 2001). According to the current model, the short translation elongation event leaves eIFs that have been recruited during the cap-dependent initiation event bound to the 80S ribosome, making the ribosome competent for reinitiation (Kozak, 1987; Mohammad et al., 2017). The temporary retention of these reinitiation-promoting factors (RPFs) can assist the terminating ribosome to resume scanning, rapidly reacquire the ternary complex, eIF2-GTP-Met-tRNA, and the 60S ribosomal subunit de novo. This model helps explain why reinitiation is precluded after a long elongation event, when all factors gradually dissociate from the translating ribosome before the termination step. eIF3 has been suggested as a ribosome-associated RPF in yeast (Cuchalová et al., 2010; Munzarová et al., 2011) and plants (Park et al., 2001; Schepetilnikov et al., 2013), while eIF4F, elongation factor 2, and DENR-MCT-1 promote reinitiation in mammals (Pöyry et al., 2004; Skabkin et al., 2013; Schleich et al., 2014).
Identification of noncanonical RPFs promoting efficient translation of uORF-mRNAs was pioneered in plants, implicating the 60S ribosomal protein L24B (Nishimura et al., 2005; Zhou et al., 2010), eIF3 subunit h (Kim et al., 2004; Zhou et al., 2010; Roy et al., 2010), and TOR (Schepetilnikov et al., 2013, 2017). Two critical Arabidopsis mutants, eif3h-1, which expresses C-terminal truncation alleles of eIF3h, and rpl24b (short valve 1), have contributed greatly to our understanding of reinitiation phenomena (Nishimura et al., 2005; Zhou et al., 2010). Both mutants result in strongly reduced translation of uORF-mRNAs that encode ARF transcription factors. Although the mechanisms of eIF3h and eL24 in reinitiation are not yet known, and likely differ (Tiruneh et al., 2013), both mutants display defects in translation reinitiation and auxin-mediated organogenesis (Nishimura et al., 2005; Zhou et al., 2010). Despite its critical role in translation reinitiation, there was no significant effect of eIF3h C-terminal truncation on cap-dependent initiation events (Zhou et al., 2010; Roy et al., 2010; Schepetilnikov et al., 2013). Moreover, eIF3h was identified as a new TOR/S6K1 target, and its phosphorylation site was mapped to Ser-178 (Schepetilnikov et al., 2013). Accordingly, auxin, which mediates activation of the TOR/S6K1 signaling axes, promotes phosphorylation of eIF3h in Arabidopsis extracts, in contrast to the TOR inhibitor Torin-1, which nearly abolishes its phosphorylation. Interestingly, the human ortholog of eIF3h is highly expressed in many cancer cells, and its high levels trigger dysregulation of protein synthesis. This oncogenic potential of eIF3h is strongly enhanced by phosphorylation at the conserved residue Ser-183 (Zhang et al., 2008). Recent research revealed that translation regulation via uORFs contributes to plant development, playing important roles in organogenesis. Indeed, translation of many meristematic mRNAs, including CLV1 and AS1, appears to be under uORF control; their suppression leads to enlarged shoot apical meristem and defects in organ polarity (Zhou et al., 2014). Despite these findings, the mechanism by which eIF3h and its phosphorylation by TOR up-regulates the reinitiation capacity of ribosomes is unclear. The eIF3h C-terminal domain is critical to eIF3h integration within eIF3 (Roy et al., 2010), suggesting that the reinitiation function depends on the complete eIF3 complex. Interestingly, the eIF3 noncore subunit h was implicated in assembly of subunits e, d, k, and l into the functional eIF3 complex; however, there are no data on this complex formation in plants (des Georges et al., 2015; Smith et al., 2016).
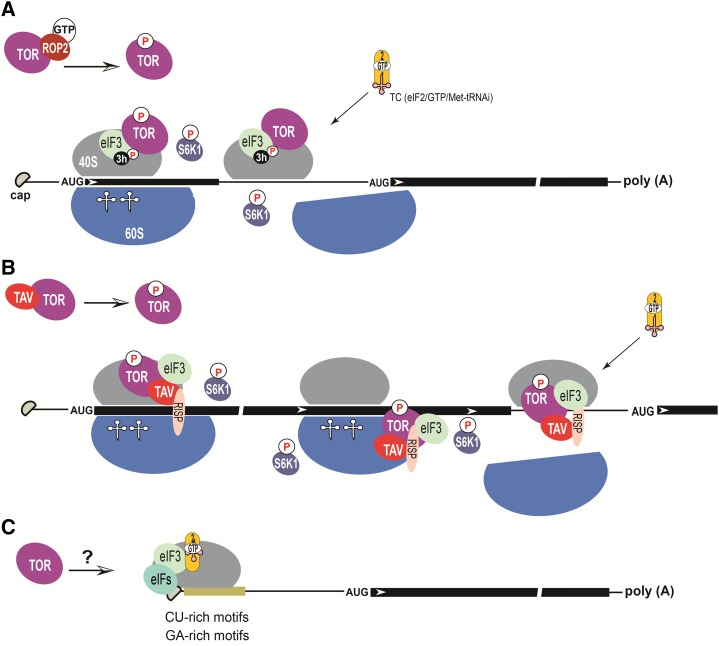
In plants, TOR, when active, promotes translation of uORF-containing mRNAs, likely via the posttranscriptional regulation of the active phosphorylation state of some RPFs (Schepetilnikov et al., 2011, 2013). According to the current model (Fig. 2A), during the short elongation event, eIF3/other RPFs might still be bound to the translating ribosome, and TOR is required to maintain the active phosphorylation status of eIF3h and possibly other RPFs in polysomes. When active, TOR is recruited to polysomes, leading to phosphorylation of bound and inactive S6K1 at Thr-449 and its dissociation. Notably, association of TOR and eIF3h with polysomes is a hallmark of efficient polysomal loading of uORF-containing mRNAs and their translation (Schepetilnikov et al., 2013). Indeed, active TOR promotes polysomal loading of many uORF-containing mRNAs, including auxin-related ARFs and auxin-unrelated bZIP11.
Figure 2.
TOR activates translation reinitiation mechanisms. A, The pattern of reinitiation after short uORF translation. TOR is activated in response to various stimuli. For example, TOR can be activated by auxin, which acts on TOR through GTP-bound ROP2. TOR-P binds polysomes to maintain the high phosphorylation status of S6K1 and thus the eIF3 subunit h (3h), which promotes the reinitiation event. eIF3, together with other eIFs, remains associated with the translating ribosome during the short elongation event to form reinitiation-competent ribosomal complexes capable of resuming scanning, as well as recruiting ternary complex (TC) and the 60S ribosomal subunit. B, The pattern of virus-activated reinitiation after long ORF translation. TAV binds and activates TOR (TOR-P). TAV retains eIF3/RISP on the translating ribosome during the long elongation event, likely by transferring to the rear side of the 60S subunit through association with eL18 or eL24 in the 60S subunit (Park et al., 2001; Thiébeauld et al., 2009). TOR-P associates with polysomes, where it maintains high RISP phosphorylation status. During translation termination and release of 60S, eIF3/TAV/RISP-P might be transported back to 40S and assist in 60S/TC recruitment. C, Sequence features of mRNA 5′-UTRs that might influence translation initiation.
Several studies have highlighted the role of auxin in TOR signaling activation (Schepetilnikov et al., 2013, 2017). Translation reinitiation control by TOR appears to be an important component of the auxin response pathway, as TOR-deficient plants and eif3h-1 mutants are characterized by altered root gravitropism (Zhou et al., 2010; Schepetilnikov et al., 2013). Accordingly, TOR inactive plants expressing yeast FKBP12 display auxin-defective phenotypes if treated by rapamycin (Deng et al., 2016).
Virus-Activated Reinitiation after Long ORF Translation
An extreme case of reinitiation—reinitiation after long ORF translation—operates on the polycistronic 35S RNA from CaMV and artificial bicistronic RNAs under the control of a CaMV reinitiation factor, TAV (Bonneville et al., 1989; Ryabova et al., 2006). TAV can interact with the cell translation machinery via the 60S ribosomal subunit, where it targets at least two ribosomal proteins, eL18 and eL24 (Park et al., 2001). Previously, it was shown that TAV is involved in the complex formation with eIF3 and a reinitiation supporting protein, RISP, that lead to both protein accumulation in polysomes of TAV transgenic plants, indicating that TAV can prevent a loss of eIF3/RISP by the translating ribosome during the long elongation event, such that a terminating ribosome is competent to reinitiate translation of the downstream ORF (Thiébeauld et al., 2009). Relatively recent research has uncovered TOR as a critical TAV partner in reinitiation of translation, demonstrating that TAV directly binds to, and activates, TOR and thus promotes its association with polysomes (Schepetilnikov et al., 2011). Moreover, the same study identified RISP as a downstream target of the TOR/S6K1 signaling axis and showed that phosphorylated RISP promotes TAV-activated reinitiation of translation (Schepetilnikov et al., 2011). According to the current model, TOR, if active, associates with polysomes, where it can maintain the high phosphorylation status of RISP and possibly other RPFs bound to polysomes in the presence of TAV during the long elongation event. Finally, after termination of translation, 40S ribosomal subunits equipped with necessary RPFs, including eIF3 and phosphorylated RISP (RISP-P), are capable to resume scanning, rebind the ternary complex, and reinitiate (Fig. 2B). Although many details of RISP function in reinitiation are now known, the mechanism remains to be established.
5′-UTR Motifs That Are Controlled by TOR in Animals
5′-leader motifs and secondary structures can significantly modulate ribosome loading on mRNA, movement toward the start codon, and thus translation of the downstream ORF. In animals, mRNAs with a 5′-terminal oligopyrimidine (TOP) motif (CCUUUCU) at the 5′-end, encoding mainly ribosomal proteins and translation factors, are subject to TOR control, regulating protein synthesis and thus cell growth (Jefferies et al., 1994; Meyuhas and Kahan, 2015). It was first thought that the S6K/eS6 signaling axis was required for TOP mRNA translation regulation (Jefferies et al., 1994); however, this hypothesis was discarded following experiments showing that TOP mRNA translation was not disturbed in S6K-deficient mice or in phosphorylation site knockout eS6 (Tang et al., 2001; Barth-Baus et al., 2002; Ruvinsky and Meyuhas, 2006). Relatively recently, global high-resolution transcriptome-scale ribosome profiling revealed that another TOR downstream target, 4E-BPs, might mediate mTOR effects in translation of TOP-mRNAs (Thoreen et al., 2012). Several other potential regulatory factors, including the RNA-binding protein LARP1 (La-related protein), have been demonstrated to function under TOR control in TOP mRNA regulation (Tcherkezian et al., 2014). In plants, TOP-like motifs are abundant within mRNA 5′-UTRs of ribosomal proteins (Dobrenel et al., 2016b), but whether these cis-elements are involved in translation regulation in plants requires further research. Interestingly, several recent publications revealed the existence of so-called R-motifs containing GA, G(A)3, G(A)6, etc., repeats (Dobrenel et al., 2016; Xu et al., 2017), and provided evidence that the R-motif regulates translation of its mRNA via interaction with poly(A)-binding proteins during immune regulation in plants (Xu et al., 2017). Whether R-motifs are targets of TOR signaling in plants remains a very interesting subject of study (Fig. 2C).
CONCLUSIONS AND PERSPECTIVES
Gene expression is intensively controlled, particularly at the level of transcription and translation, but control of mRNA translation has to date been much less studied than mechanisms of transcription. In plants, many strategies of protein synthesis and the role of TOR in regulation of mRNA translation remain to be discovered. The past 10 years of TOR research provided clear evidence that TOR is a critical regulator of cell growth and proliferation in photosynthetic organisms that acts, at least in part, via promoting protein synthesis in favorable environmental conditions. Several reports have uncovered the role of TOR and translation initiation factors in both short- and long-term reprogramming of the translational landscape. Today, the mechanisms by which TOR regulates translation of specific plant mRNAs, for example, to control reinitiation events used to modulate production of critical effector proteins, are under intensive investigation. Further work will be required to understand the mechanisms and function of TOR and its downstream targets in plant translation. An important question is to understand whether, and if so how, TOR regulates cap-dependent translation initiation in plants. Several growth hormones have been implicated in cross talk with the TOR signaling pathway, and our focus now is to study the components of these pathways and how hormones are wired into TOR translation control.
Acknowledgments
The authors apologize for limited bibliography not directly related to TOR in translation due to manuscript length limitations.
Footnotes
This work was supported by the French Agence Nationale de la Recherche (BLAN-2011_BSV6 010 03 and ANR-14-CE19-0007 to L.A.R.).
Articles can be viewed without a subscription.
References
- Abraham RT. (2004) mTOR as a positive regulator of tumor cell responses to hypoxia. Curr Top Microbiol Immunol 279: 299–319 [DOI] [PubMed] [Google Scholar]
- Albert V, Hall MN (2015) mTOR signaling in cellular and organismal energetics. Curr Opin Cell Biol 33: 55–66 [DOI] [PubMed] [Google Scholar]
- Anderson GH, Veit B, Hanson MR (2005) The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Hanson J (2017) Shaping plant development through the SnRK1-TOR metabolic regulators. Curr Opin Plant Biol 35: 152–157 [DOI] [PubMed] [Google Scholar]
- Barth-Baus D, Stratton CA, Parrott L, Myerson H, Meyuhas O, Templeton DJ, Landreth GE, Hensold JO (2002) S6 phosphorylation-independent pathways regulate translation of 5′-terminal oligopyrimidine tract-containing mRNAs in differentiating hematopoietic cells. Nucleic Acids Res 30: 1919–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M (2011) The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529 [DOI] [PubMed] [Google Scholar]
- Bonneville JM, Sanfaçon H, Fütterer J, Hohn T (1989) Posttranscriptional trans-activation in cauliflower mosaic virus. Cell 59: 1135–1143 [DOI] [PubMed] [Google Scholar]
- Broeckx T, Hulsmans S, Rolland F (2016) The plant energy sensor: evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J Exp Bot 67: 6215–6252 [DOI] [PubMed] [Google Scholar]
- Browning KS, Bailey-Serres J (2015) Mechanism of cytoplasmic mRNA translation. Arabidopsis Book 13: e0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit F, Brand MD (1995) A hierarchy of ATP-consuming processes in mammalian cells. Biochem J 312: 163–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Li Y, Leisse A, Zhang Y, Bartholomaeus L, Fernie AR, Willmitzer L, Giavalisco P (2013) Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J 73: 897–909 [DOI] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK (2009) Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA 106: 7507–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang GG, Abraham RT (2005) Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem 280: 25485–25490 [DOI] [PubMed] [Google Scholar]
- Cornu M, Oppliger W, Albert V, Robitaille AM, Trapani F, Quagliata L, Fuhrer T, Sauer U, Terracciano L, Hall MN (2014) Hepatic mTORC1 controls locomotor activity, body temperature, and lipid metabolism through FGF21. Proc Natl Acad Sci USA 111: 11592–11599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creff A, Sormani R, Desnos T (2010) The two Arabidopsis RPS6 genes, encoding for cytoplasmic ribosomal proteins S6, are functionally equivalent. Plant Mol Biol 73: 533–546 [DOI] [PubMed] [Google Scholar]
- Cuchalová L, Kouba T, Herrmannová A, Dányi I, Chiu W-L, Valásek L (2010) The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol Cell Biol 30: 4671–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Filipe O, Hoffman G, Seifi HS, Haeck A, Canlas P, Van Bockhaven J, De Waele E, Demeestere K, Ronald P, et al. (2017) Target of rapamycin signaling orchestrates growth-defense trade-offs in plants. New Phytol [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J, Hashem Y (2015) Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525: 491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng K, Yu L, Zheng X, Zhang K, Wang W, Dong P, Zhang J, Ren M (2016) Target of rapamycin is a key player for auxin signaling transduction in Arabidopsis. Front Plant Sci 7: 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C (2007) The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 8: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C (2016a) TOR signaling and nutrient sensing. Annu Rev Plant Biol 67: 261–285 [DOI] [PubMed] [Google Scholar]
- Dobrenel T, Mancera-Martínez E, Forzani C, Azzopardi M, Davanture M, Moreau M, Schepetilnikov M, Chicher J, Langella O, Zivy M, et al. (2016b) The Arabidopsis TOR kinase specifically regulates the expression of nuclear genes coding for plastidic ribosomal proteins and the phosphorylation of the cytosolic ribosomal protein S6. Front Plant Sci 7: 1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P, Xiong F, Que Y, Wang K, Yu L, Li Z, Ren M (2015) Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Front Plant Sci 6: 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér A, Lajkó DB (2015) Signals fly when kinases meet Rho-of-plants (ROP) small G-proteins. Plant Sci 237: 93–107 [DOI] [PubMed] [Google Scholar]
- Fenton TR, Gout IT (2011) Functions and regulation of the 70kDa ribosomal S6 kinases. Int J Biochem Cell Biol 43: 47–59 [DOI] [PubMed] [Google Scholar]
- Floris M, Bassi R, Robaglia C, Alboresi A, Lanet E (2013) Post-transcriptional control of light-harvesting genes expression under light stress. Plant Mol Biol 82: 147–154 [DOI] [PubMed] [Google Scholar]
- Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, et al. (2010) eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci USA 107: 14134–14139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Zhang X, Borevitz JO, Li W-H, Shiu SH (2007) A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res 17: 632–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden CA, Jorgensen RA (2007) Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol 5: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Magyar Z, Monardes A, Khan S, Zalejski C, Orellana J, Szabados L, de la Torre C, Koncz C, Bögre L (2010) Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J 29: 2979–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. (2006) eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 31: 553–562 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N (2016) Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352: 1413–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J (2005) mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123: 569–580 [DOI] [PubMed] [Google Scholar]
- Hughes Hallett JE, Luo X, Capaldi AP (2015) Snf1/AMPK promotes the formation of Kog1/Raptor-bodies to increase the activation threshold of TORC1 in budding yeast. eLife 4: 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CUT, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Reinhard C, Kozma SC, Thomas G (1994) Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci USA 91: 4441–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Bailey-Serres J (2012) Dynamic light regulation of translation status in Arabidopsis thaliana. Front Plant Sci 3: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandal D, Samol I, Buhr F, Pollmann S, Schmidt H, Clemens S, Reinbothe S, Reinbothe C (2009) Singlet oxygen-dependent translational control in the tigrina-d.12 mutant of barley. Proc Natl Acad Sci USA 106: 13112–13117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khapre RV, Kondratova AA, Patel S, Dubrovsky Y, Wrobel M, Antoch MP, Kondratov RV (2014) BMAL1-dependent regulation of the mTOR signaling pathway delays aging. Aging (Albany NY) 6: 48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B-H, Cai X, Vaughn JN, von Arnim AG (2007) On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol 8: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110: 163–175 [DOI] [PubMed] [Google Scholar]
- Kim J-Y, Song H-R, Taylor BL, Carré IA (2003) Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J 22: 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Kim BH, Yahalom A, Chamovitz DA, von Arnim AG (2004) Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell 16: 3341–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromilas AE, Lazaris-Karatzas A, Sonenberg N (1992) mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J 11: 4153–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1987) At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol 196: 947–950 [DOI] [PubMed] [Google Scholar]
- Kozak M. (2001) Constraints on reinitiation of translation in mammals. Nucleic Acids Res 29: 5226–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchenko A, Citerne S, Jéhanno I, Bersimbaev RI, Veit B, Meyer C, Leprince A-S (2015) Mutations in the Arabidopsis Lst8 and Raptor genes encoding partners of the TOR complex, or inhibition of TOR activity decrease abscisic acid (ABA) synthesis. Biochem Biophys Res Commun 467: 992–997 [DOI] [PubMed] [Google Scholar]
- Li X, Cai W, Liu Y, Li H, Fu L, Liu Z, Xu L, Liu H, Xu T, Xiong Y (2017) Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc Natl Acad Sci USA 114: 2765–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liko D, Hall MN (2015) mTOR in health and in sickness. J Mol Med (Berl) 93: 1061–1073 [DOI] [PubMed] [Google Scholar]
- Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Güttler T, Davis F, Asara JM, Sahin M (2015) The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell 161: 1138–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M-J, Wu S-H, Chen H-M, Wu S-H (2012) Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol Syst Biol 8: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M-J, Wu S-H, Wu J-F, Lin W-D, Wu Y-C, Tsai T-Y, Tsai H-L, Wu S-H (2013) Translational landscape of photomorphogenic Arabidopsis. Plant Cell 25: 3699–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J (2009) Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318 [DOI] [PubMed] [Google Scholar]
- Mahfouz MM, Kim S, Delauney AJ, Verma DP (2006) Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meteignier L-V, El Oirdi M, Cohen M, Barff T, Matteau D, Lucier J-F, Rodrigue S, Jacques P-E, Yoshioka K, Moffett P (2017) Translatome analysis of an NB-LRR immune response identifies important contributors to plant immunity in Arabidopsis. J Exp Bot 68: 2333–2344 [DOI] [PubMed] [Google Scholar]
- Meyuhas O. (2015) Ribosomal protein S6 phosphorylation: four decades of research. Int Rev Cell Mol Biol 320: 41–73 [DOI] [PubMed] [Google Scholar]
- Meyuhas O, Kahan T (2015) The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta 1849: 801–811 [DOI] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ (2011) The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol 13: 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missra A, Ernest B, Lohoff T, Jia Q, Satterlee J, Ke K, von Arnim AG (2015) The circadian clock modulates global daily cycles of mRNA ribosome loading. Plant Cell 27: 2582–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Hayashida N, Yamaguchi-Shinozaki K, Kamada H, Shinozaki K (1995) Two genes that encode ribosomal-protein S6 kinase homologs are induced by cold or salinity stress in Arabidopsis thaliana. FEBS Lett 358: 199–204 [DOI] [PubMed] [Google Scholar]
- Mohammad MP, Munzarová Pondelícková V, Zeman J, Gunišová S, Valášek LS (2017) In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res 45: 2658–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montané MH, Menand B (2013) ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J Exp Bot 64: 4361–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M, Azzopardi M, Clément G, Dobrenel T, Marchive C, Renne C, Martin-Magniette M-L, Taconnat L, Renou J-P, Robaglia C, et al. (2012) Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24: 463–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzarová V, Pánek J, Gunišová S, Dányi I, Szamecz B, Valášek LS (2011) Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-acting mRNA elements preceding short uORFs. PLoS Genet 7: e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nietzsche M, Landgraf R, Tohge T, Bornke F (2016) A protein-protein interaction network linking the energy-sensor kinase SnRK1 to multiple signaling pathways in Arabidopsis thaliana. Current Plant Biology 5: 36–44 [Google Scholar]
- Nishimura T, Wada T, Yamamoto KT, Okada K (2005) The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17: 2940–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohales MA, Kay SA (2016) Molecular mechanisms at the core of the plant circadian oscillator. Nat Struct Mol Biol 23: 1061–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukarinen E, Nägele T, Pedrotti L, Wurzinger B, Mair A, Landgraf R, Börnke F, Hanson J, Teige M, Baena-Gonzalez E, et al. (2016) Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci Rep 6: 31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Jacinto E (2011) mTOR complex 2 signaling and functions. Cell Cycle 10: 2305–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E (2010) mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J 29: 3939–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouibrahim L, Rubio AG, Moretti A, Montané MH, Menand B, Meyer C, Robaglia C, Caranta C (2015) Potyviruses differ in their requirement for TOR signalling. J Gen Virol 96: 2898–2903 [DOI] [PubMed] [Google Scholar]
- Pal SK, Liput M, Piques M, Ishihara H, Obata T, Martins MCM, Sulpice R, van Dongen JT, Fernie AR, Yadav UP, et al. (2013) Diurnal changes of polysome loading track sucrose content in the rosette of wild-type arabidopsis and the starchless pgm mutant. Plant Physiol 162: 1246–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Himmelbach A, Browning KS, Hohn T, Ryabova LA (2001) A plant viral “reinitiation” factor interacts with the host translational machinery. Cell 106: 723–733 [DOI] [PubMed] [Google Scholar]
- Petracek ME, Dickey LF, Huber SC, Thompson WF (1997) Light-regulated changes in abundance and polyribosome association of ferredoxin mRNA are dependent on photosynthesis. Plant Cell 9: 2291–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Janocha D, Dong Y, Medzihradszky A, Schöne S, Daum G, Suzaki T, Forner J, Langenecker T, Rempel E, et al. (2016) Integration of light and metabolic signals for stem cell activation at the shoot apical meristem. eLife 5: e17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Höhne M, Usadel B, Gibon Y, Rohwer J, Stitt M (2009) Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol Syst Biol 5: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa C, Li L, Gil S, Tatjer L, Hashii K, Tabuchi M, Coll NS, Ariño J, Valls M (2016) The effector AWR5 from the plant pathogen Ralstonia solanacearum is an inhibitor of the TOR signalling pathway. Sci Rep 6: 27058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Connell EJ, Fraser CS, Jackson RJ (2007) The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev 21: 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Jackson RJ (2004) What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev 18: 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y, Luo X, Bassham DC (2017) TOR-dependent and -independent pathways regulate autophagy in Arabidopsis thaliana. Front Plant Sci 8: 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Venglat P, Qiu S, Feng L, Cao Y, Wang E, Xiang D, Wang J, Alexander D, Chalivendra S, et al. (2012) Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 24: 4850–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexin D, Meyer C, Robaglia C, Veit B (2015) TOR signalling in plants. Biochem J 470: 1–14 [DOI] [PubMed] [Google Scholar]
- Robaglia C, Thomas M, Meyer C (2012) Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr Opin Plant Biol 15: 301–307 [DOI] [PubMed] [Google Scholar]
- Roy B, Vaughn JN, Kim B-H, Zhou F, Gilchrist MA, Von Arnim AG (2010) The h subunit of eIF3 promotes reinitiation competence during translation of mRNAs harboring upstream open reading frames. RNA 16: 748–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Meyuhas O (2006) Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci 31: 342–348 [DOI] [PubMed] [Google Scholar]
- Ryabova LA, Pooggin MM, Hohn T (2006) Translation reinitiation and leaky scanning in plant viruses. Virus Res 119: 52–62 [DOI] [PubMed] [Google Scholar]
- Saldivia M, Barquilla A, Bart JM, Diaz-González R, Hall MN, Navarro M (2013) Target of rapamycin (TOR) kinase in Trypanosoma brucei: an extended family. Biochem Soc Trans 41: 934–938 [DOI] [PubMed] [Google Scholar]
- Schalm SS, Blenis J (2002) Identification of a conserved motif required for mTOR signaling. Curr Biol 12: 632–639 [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA (2013) TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J 32: 1087–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Kobayashi K, Geldreich A, Caranta C, Robaglia C, Keller M, Ryabova LA (2011) Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO J 30: 1343–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Makarian J, Srour O, Geldreich A, Yang Z, Chicher J, Hammann P, Ryabova LA (2017) GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. EMBO J 36: 886–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich S, Strassburger K, Janiesch PC, Koledachkina T, Miller KK, Haneke K, Cheng Y-S, Kuechler K, Stoecklin G, Duncan KE, et al. (2014) DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature 512: 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimobayashi M, Hall MN (2014) Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol 15: 155–162 [DOI] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Hellen CUT, Pestova TV (2013) Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol Cell 51: 249–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Arake-Tacca L, Nitido A, Montabana E, Park A, Cate JH (2016) Assembly of eIF3 mediated by mutually dependent subunit insertion. Structure 24: 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithson LJ, Gutmann DH (2016) Proteomic analysis reveals GIT1 as a novel mTOR complex component critical for mediating astrocyte survival. Genes Dev 30: 1383–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son O, Kim S, Hur Y-S, Cheon C-I (2016) Identification of the Raptor-binding motif on Arabidopsis S6 kinase and its use as a TOR signaling suppressor. Biochem Biophys Res Commun 472: 83–87 [DOI] [PubMed] [Google Scholar]
- Son O, Kim S, Hur Y-S, Cheon C-I (2017) Molecular details of the Raptor-binding motif on Arabidopsis S6 kinase. Biochem Biophys Res Commun 486: 137–142 [DOI] [PubMed] [Google Scholar]
- Song Y, Zhao G, Zhang X, Li L, Xiong F, Zhuo F, Zhang C, Yang Z, Datla R, Ren M, et al. (2017) The crosstalk between Target of Rapamycin (TOR) and Jasmonic Acid (JA) signaling existing in Arabidopsis and cotton. Sci Rep 7: 45830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormani R, Yao L, Menand B, Ennar N, Lecampion C, Meyer C, Robaglia C (2007) Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol 7: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Yu Y, Hu W, Min Q, Kang H, Li Y, Hong Y, Wang X, Hong Y (2016) Ribosomal protein S6 kinase1 coordinates with TOR-Raptor2 to regulate thylakoid membrane biosynthesis in rice. Biochim Biophys Acta 1861: 639–649 [DOI] [PubMed] [Google Scholar]
- Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D, Avruch J, Meyuhas O (2001) Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol Cell Biol 21: 8671–8683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Bhat S, Petracek ME (2003) Light control of nuclear gene mRNA abundance and translation in tobacco. Plant Physiol 133: 1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP (2014) Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev 28: 357–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiébeauld O, Schepetilnikov M, Park H-S, Geldreich A, Kobayashi K, Keller M, Hohn T, Ryabova LA (2009) A new plant protein interacts with eIF3 and 60S to enhance virus-activated translation re-initiation. EMBO J 28: 3171–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruneh BS, Kim B-H, Gallie DR, Roy B, von Arnim AG (2013) The global translation profile in a ribosomal protein mutant resembles that of an eIF3 mutant. BMC Biol 11: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran MK, Schultz CJ, Baumann U (2008) Conserved upstream open reading frames in higher plants. BMC Genomics 9: 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Kozma SC, Thomas G, Nagy F (1998) A heat-sensitive Arabidopsis thaliana kinase substitutes for human p70s6k function in vivo. Mol Cell Biol 18: 2038–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Zilbermann F, Kozma SC, Thomas G, Nagy F (2004) Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol 134: 1527–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mathews MB, Mohr I (2013) Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harb Perspect Biol 5: a012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG (2001) Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J 20: 4370–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhong C, Wang F, Qu F, Ding J (2013) Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif. Biochem J 454: 39–47 [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Oppliger W, Hall MN (2005) Molecular organization of target of rapamycin complex 2. J Biol Chem 280: 30697–30704 [DOI] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J (2013) Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J (2012) Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem 287: 2836–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Greene GH, Yoo H, Liu L, Marqués J, Motley J, Dong X (2017) Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 545: 487–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Ap Rees T (1999) Changes in carbohydrate metabolism and assimilate export in starch‐excess mutants of Arabidopsis. Plant Cell Environ 22: 1445–1453 [Google Scholar]
- Zhang L, Smit-McBride Z, Pan X, Rheinhardt J, Hershey JWB (2008) An oncogenic role for the phosphorylated h-subunit of human translation initiation factor eIF3. J Biol Chem 283: 24047–24060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SH, Broome MA, Lawton MA, Hunter T, Lamb CJ (1994) atpk1, a novel ribosomal protein kinase gene from Arabidopsis. II. Functional and biochemical analysis of the encoded protein. J Biol Chem 269: 17593–17599 [PubMed] [Google Scholar]
- Zhang Z, Zhu J-Y, Roh J, Marchive C, Kim S-K, Meyer C, Sun Y, Wang W, Wang Z-Y (2016) TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr Biol 26: 1854–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Roy B, Dunlap JR, Enganti R, von Arnim AG (2014) Translational control of Arabidopsis meristem stability and organogenesis by the eukaryotic translation factor eIF3h. PLoS One 9: e95396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Roy B, von Arnim AG (2010) Translation reinitiation and development are compromised in similar ways by mutations in translation initiation factor eIF3h and the ribosomal protein RPL24. BMC Plant Biol 10: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvereva AS, Golyaev V, Turco S, Gubaeva EG, Rajeswaran R, Schepetilnikov MV, Srour O, Ryabova LA, Boller T, Pooggin MM (2016) Viral protein suppresses oxidative burst and salicylic acid-dependent autophagy and facilitates bacterial growth on virus-infected plants. New Phytol 211: 1020–1034 [DOI] [PubMed] [Google Scholar]